Abstract
Simple sequence repeats (SSRs) are found throughout the genome, and under some conditions can change in length over time. Germline and somatic expansions of trinucleotide repeats are associated with a series of severely disabling illnesses, including Huntington's disease. The underlying mechanisms that effect SSR expansions and contractions have been experimentally elusive, but models suggesting a role for DNA repair have been proposed, in particular the involvement of transcription-coupled nucleotide excision repair (TCNER) that removes transcription-blocking DNA damage from the transcribed strand of actively expressed genes. If the formation of secondary DNA structures that are associated with SSRs were to block RNA polymerase progression, TCNER could be activated, resulting in the removal of the aberrant structure and a concomitant change in the region's length. To test this, TCNER activity in primary human fibroblasts was assessed on defined DNA substrates containing extrahelical DNA loops that lack discernible internal base pairs or DNA stem–loops that contain base pairs within the stem. The results show that both structures impede transcription elongation, but there is no corresponding evidence that nucleotide excision repair (NER) or TCNER operates to remove them.
INTRODUCTION
Repeated DNA sequences are ubiquitous in eukaryotic genomes (reviewed in (1)). Among them are DNA elements called microsatellites, which are often called short sequence repeats or simple sequence repeats (SSRs). SSRs consist of short segments of DNA usually between one and nine nucleotides in length that are in turn arranged head-to-tail in tandem (2). The number of tandemly repeated units in a microsatellite region typically ranges from 1 to ∼60 repeat units (3), but can exceed 200 repeat units (for example, an ACC repeat tract in the human genome contains 210 repeat units) (4). (It is worth noting that the defined length of the individual repeats within SSRs and the total number of tandemly arranged repeats that constitutes a microsatellite are not uniformly agreed upon; hence, there is significant variation in the literature.)
SSRs are abundant in eukaryotes, with estimates ranging from 103 to 106 per genome, and they are found in both intergenic and intragenic regions (1,5). SSRs within genes can be located in regulatory sequences and transcription units, and they are even observed in open reading frames where they typically exist as tri- and hexanucleotide repeats (4,6). Among the trinucleotide repeats is the well-known CAG/CTG repeat that is found in several human genetic loci whose gene products are associated with disorders such as Huntington's disease (HD) (7), spinobulbar muscular atrophy (8) and spinocerebellar ataxias (9).
SSRs generally exhibit instability, which leads to expansions and contractions in these regions (10). This instability may well play a role in the CAG/CTG expansions observed in the HD gene that leads to the onset of the disease (11). While the cellular traits that contribute to SSR instability are not clear, slippage during DNA replication and aberrant DNA repair have been proposed as possible events involved in the observed expansions and contractions (12). Some hypotheses related to the underlying cause of genetic instability in repeat regions invoke a common theme—the formation of structures such as DNA loops that lack discernible internal base pairs or DNA stem–loops that contain base-pairing within the stem. Indeed, slippage events in repeat regions of DNA produce DNA heteroduplexes (13) that can consist of DNA loops and DNA stem–loops. Both DNA loops and DNA stem–loops have been suggested to be putative substrates for DNA repair (14,15), but in the absence of actual damage or nicks in the region, the removal or expansion of such structures would require the extruded DNA itself to be recognized as damage. To date, several DNA repair pathways have been implicated in genetic instability in general and in somatic instability of trinucleotide repeats in particular: These include nucleotide excision repair (NER) (16), transcription-coupled nucleotide excision repair (TCNER) (17), base excision repair (BER) (18) and mismatch repair (MMR) (19,20).
During NER, DNA damage recognition requires the presence of the XPC-RAD23B heterodimer that binds to distorted regions of DNA. This step is more than likely followed by the assembly of additional repair factors that include XPA and TFIIH. TFIIH is a multi-protein complex that contains the helicases XPB and XPD, which operate in tandem to unwind the duplex. Current evidence suggests that XPD stalls at DNA damage, acting to verify the presence of a lesion and in turn allowing XPG and XPF-ERCC1 to incise the damaged DNA strand on either side of the lesion (21,22). This results in the release of a DNA oligomer ∼24–32 nucleotides in length that contains the damaged base (23). A DNA polymerase catalyses the synthesis of nascent DNA to fill the gap and DNA ligase seals the nick (23).
Cells harbor another pathway, TCNER, that overlaps with NER and removes damage from the transcribed strand of actively expressed genes. TCNER does not use XPC-HR23B to identify distortions in DNA, relying instead on RNA polymerase that stalls at sites of DNA damage as a means for identifying a lesion. The stalled RNA polymerase then triggers downstream events that repair the DNA (24), which involves the interaction of additional proteins unique to TCNER, including CSA and CSB, and some of those described earlier for NER, including TFIIH. Indeed, TFIIH serves the same role during TCNER as in NER—to unwind the duplex and confirm the presence of DNA damage.
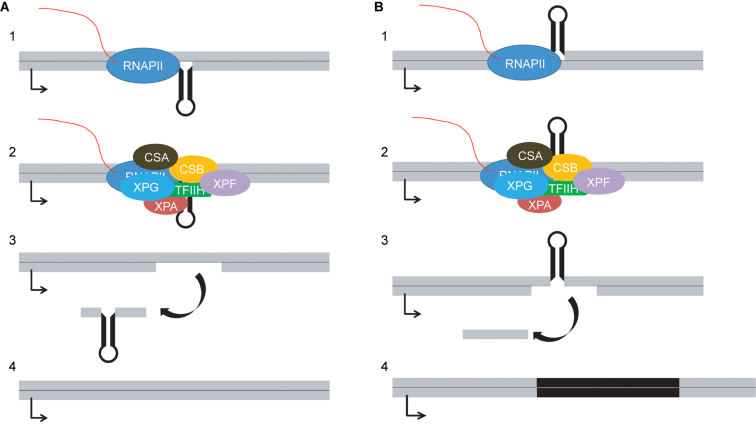
Several models have proposed that extrahelical unpaired DNA loops and DNA stem–loops can be recognized as DNA damage, despite the lack of modified bases, due to their propensity to stall an elongating RNA polymerase during transcription (25–28). If DNA loops that lack internal base-pairing, which will be referred to as unpaired DNA loops, or DNA stem–loops that contain base-pairing in the stem were to block transcription, the stalled RNA polymerase complex could induce TCNER. TCNER would treat the aberrant structure as DNA damage, resulting in the excision of a patch of DNA from the transcribed strand. Re-synthesis of DNA would result in a deletion or expansion of the DNA sequence, depending on which strand the extruded DNA had been located. A DNA loop or DNA stem–loop on the transcribed strand would be removed, resulting in a contraction; a DNA loop or DNA stem–loop on the non-transcribed strand could result in an event in which an expansion would occur (Figure 1). However, such models disregard the potential role of TFIIH in confirming the presence of DNA damage, which would not necessarily be present in the DNA loops or stem-loops that block transcription elongation. In fact, this suggests that a model invoking NER or TCNER as players in the instability of SSRs would not accurately describe the currently understood mechanism underlying the events leading to DNA repair.
Figure 1.
Model for DNA contractions and expansions caused by DNA secondary structure and TCNER. (A) 1. A DNA stem–loop on the transcribed strand blocks transcription elongation, triggering association of the TCNER proteins CSA and CSB. 2. NER factors assemble in response to the stalled transcription complex; specific displacement or retention of the RNA polymerase machinery is unknown. 3. A patch of DNA containing the stem–loop is excised from the transcribed strand. 4. DNA polymerase synthesizes nascent DNA in the resulting gap, resulting in a contraction. (B) 1. A DNA stem–loop on the non-transcribed strand blocks transcription elongation, triggering association of the TCNER proteins CSA and CSB. 2. NER factors assemble in response to the stalled transcription complex; the specific displacement or retention of the RNA polymerase machinery is unknown. 3. A patch of DNA opposite the stem–loop is removed from the transcribed strand. 4. DNA polymerase fills the gap, causing an expansion of the stem–loop sequence which is replicated during DNA synthesis.
To test if transcriptional stalling at a DNA loop or DNA stem–loop can trigger a repair event through TCNER in human cells, DNA templates were assembled in which extrahelical DNA in the form of DNA stem–loops or unpaired DNA loops were situated on the transcribed or non-transcribed strand of a transcription unit. For these structures, all unpaired DNA loops and DNA stem–loops impeded transcription elongation in vitro, but to varying extents. DNA stem–loops appeared to be stable in human cells, but a small quantity of expansions and contractions were detected. Unpaired DNA loops appeared to undergo preferential deletion in human cells. Importantly, DNA stem–loops and unpaired DNA loops were subject to equivalent processing in fibroblasts proficient in DNA repair, fibroblasts defective in NER and fibroblasts defective in TCNER, indicating that neither NER nor TCNER is responsible for removal of undamaged DNA that forms unpaired DNA loops or DNA stem–loops in cells.
MATERIALS AND METHODS
Construction of templates containing extrahelical DNA loops or DNA stem–loops for in vitro transcription
Construction of vectors with extrahelical DNA positioned within a transcription unit was as follows. DNA oligomers and primers were procured from Integrated DNA Technologies (Coralville, IA). The 5′-ends of oligomers (Supplemental Table S1) were phosphorylated in the presence of T4 polynucleotide kinase (PNK) (New England BioLabs Inc., Ipswich, MA, USA) and ATP at 37°C for 30 min. Equimolar quantities of complementary oligomers were mixed in Tris–EDTA buffer (TEB) (10 mM Tris–HCl (pH 7.8), 50 mM NaCl, 1 mM EDTA) and annealed in a MyCycler™ Thermal Cycler (Bio-Rad Laboratories Inc., Hercules, CA) by heating to 90°C and cooling the mixture to 4°C, decrementing the temperature by 0.5°C during each 30 s interval. Annealed DNA oligomers were ligated into the BbsI site of vector pCI-neo-G-less-T7 (29) at a 3-fold molar excess of insert to vector using T4 DNA ligase (New England BioLabs Inc., Ipswich, MA) at 16°C for 18 h. The resulting vectors were propagated in Escherichia coli (DH5α) and recovered using the commercially available GeneJet Miniprep Kit (Thermo Fisher Scientific, Waltham, MA). DNA sequences were verified by Sanger sequencing using primer 5′-AACGCAGTCAGTGCTTCTGACA-3′ (Genewiz, Inc., South Plainfield, NJ, USA). Following sequencing, each vector was propagated in E. coli at a scale sufficient to produce at least 1 mg of product that was ultimately purified with the Plasmid Plus Maxi Kit (QIAGEN, Germantown, MD, USA).
Escherichia coli strain MV1190 (American Type Culture Collection, Manassas, VA) was transformed with pCI-neo-G-Less-T7-C (29) by electroporation and grown on Luria broth (LB) agar plates containing 100 μg/ml ampicillin (amp). A single, isolated colony was expanded in 5 ml LB-amp as an overnight culture. The overnight culture was further expanded in 500 ml double-strength, yeast-tryptone extract medium (2 × YT) supplemented with 100 μg/ml amp, and the culture was superinfected with helper bacteriophage VCSM13 (Agilent Technologies, Santa Clara, CA, USA) to a level greater than 100 phage per bacterium. Phage were recovered by polyethylene glycol precipitation, and single-strand pCI-neo-G-Less-T7-C was purified from the phage particles.
Control templates and templates containing extrahelical DNA for in vitro transcription were formed by linearizing the appropriate double-strand vector with BamHI, mixing it with single-strand, closed-circular DNA derived from pCI-neo-G-Less-T7-C, and denaturing and annealing using the oscillating phenol reassociation technique (OsPERT) (30). This resulted in the transcribed strand from the linear double-strand DNA annealing to the closed-circular, single-strand DNA. A typical OsPERT reaction contained 10 μg of double-strand vector cut with BamHI and 60 μg of closed-circular, single-strand DNA in a 600 μl volume reaction. Note that the closed-circular, single-strand DNA was essentially present in 12 fold-molar excess over the double-strand DNA.
In OsPERT reactions, the mixture of double-strand, linear DNA and single-strand, circular DNA was denatured by the addition of NaOH to a 0.3 M final concentration and held at room temperature for 15 min. MOPS free acid was added to a final concentration of 0.4 M to bring the solution to near neutral pH as confirmed with pH paper. NaCl and Na2EDTA were added to final concentrations of 0.75 M and 1 mM, respectively. Phenol that was buffered with 10 mM Tris-HCl (pH 8.0), 1 mM EDTA (Sigma Aldrich, St. Louis, MO, USA) was added to a final concentration of 10% of the volume of the annealing mixture. The solution was mixed with a micropipette and divided into 100 μl aliquots. Each aliquot was heated to 65°C for 15 s and chilled to 4°C for 15 s, repeating the cycles for 1 h. The mixtures were extracted once with an equal volume of chloroform-isoamyl alcohol (v:v 24:1). The aqueous layer was recovered, 3 M sodium acetate was added to a final concentration of 0.3 M, and an equal volume of isopropanol was added to the mixture. After 10 min at room temperature, the samples were centrifuged for 15 min at 12 500 × g. The DNA pellet was washed once with 700 μl 70% ethanol, air dried and re-suspended in 100 μl Tris-EDTA (TE) buffer (10 mM Tris–HCl, 1 mM Na2EDTA (pH 7.8)). The appearance of double-strand, nicked-circular DNA product was assessed by electrophoresis at 135 V for 45 min using 0.65% agarose containing 0.5 μg/ml ethidium bromide.
Annealed DNA was digested with the restriction enzyme AlwNI for 1 h at 37°C in a 60 μl volume. The 5061 base-pair fragment representing heteroduplex DNA was separated by gel electrophoresis using a 0.8% agarose gel and was isolated by excising the band and recovering the DNA using the QIAGEN gel extraction kit. Product was quantified by absorbance at 260 nm (NanoDrop 1000, NanoDrop, Wilmington, DE, USA). A diagram of the procedure can be found in Burns, J.A. (2012), Figure 3.2 (31).
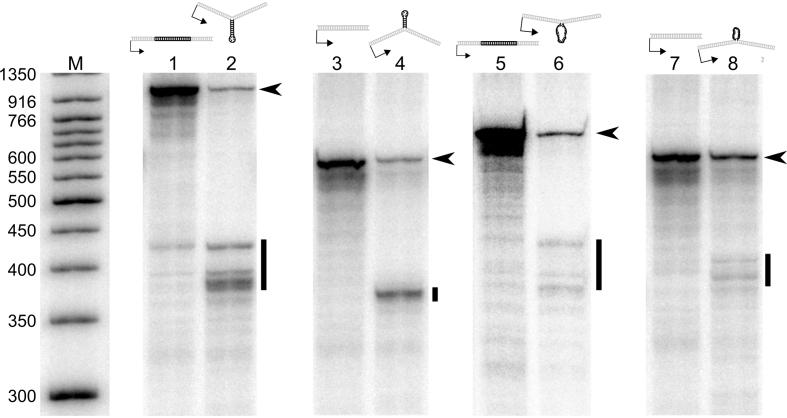
Figure 3.
DNA stem–loops are strong blocks to elongating hRNAPII during in vitro transcription reactions with HeLa nuclear extract. The diagram above each pair of lanes represents the template used in the reactions containing extruded DNA secondary structure for the respective experiment. The black arrowhead to the right of each gel indicates the position of full length transcripts for that experiment. The black bar to the right of each gel indicates the position of truncated transcripts in the respective experimental lanes. Lane 1: Control for a stem–loop on the transcribed strand. Lane 2: stem–loop on the transcribed strand. Lane 3: Control for a stem–loop on the non-transcribed strand. Lane 4: stem–loop on the non-transcribed strand. Lane 5. Control for a loop on the transcribed strand. Lane 6: Loop on the transcribed strand Lane 7: Control for a loop on the non-transcribed strand. Lane 8: Loop on the non-transcribed strand. Gels were cropped and ordered for clarity, maintaining their relationship to the marker. Brightness and contrast were adjusted as needed to allow visualization of RNA bands.
Templates for in vitro transcription were attached to a solid support by phosphorylating 120 pmol of the DNA oligomer 5′-GGTTACTCGAGATTCGTCGTGGGCCCTACGTTCTTCCG-3′ in an 80 μl reaction containing 20 units T4 PNK and 1 mM ATP for 30 min at 37°C. The phosphorylated oligomer was mixed with an equimolar ratio of its 5′-biotinylated complement 5′-biotin-TTTTTTTTTCGGAAGAACGTAGGGCCCACGACGAATCTCGAGTAACCTGG-3′ and annealed by heating to 90°C and cooling to 4°C, decrementing the temperature by 0.5°C during each 30 s cycle in a MyCycler™ Thermal Cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA). A ten-fold molar excess of the annealed, biotinylated DNA fragment was added to the gel-purified heteroduplex DNA and ligated overnight at 16°C using T4 DNA ligase. Streptavidin-coated magnetic particles (Magnesphere, Promega Corporation, Madison, WI) were washed three times with 0.5 SSC, mixed with 50 μg denatured salmon sperm DNA and washed three additional times with 500 μl of 0.5 × SSC. (SSC is 15 mM sodium citrate (pH 7.0) and 150 mM NaCl.) Paramagnetic particles were added to the biotinylated DNA mixture to a final ratio of 20 μg particles/pmol DNA, and incubated with gentle mixing for 1 h at room temperature. The beads were captured with a magnet (MagneSphere® Technology Magnetic Separation Stand, Promega Corporation, Madison, WI, USA), washed with NEBuffer 4 (New England BioLabs Inc., Ipswich, MA) and resuspended in 50 μl NEBuffer 4. SmaI (20 units) was added, and the mixture was incubated at room temperature for 1 h. The beads were captured, and the supernatant was retained for analysis. The beads were washed three times with 10 mM Tris buffer (pH 7.8) and resuspended in 100 μl of the same buffer. An aliquot of the beads, typically 8.5 μl, was used in a 10 μl digest in NEBuffer 3 (New England BioLabs Inc., Ipswich, MA) with 5 units BglII for 1 h at 37°C, which cuts upstream of the CMV promoter. The beads were captured, and the supernatant was analyzed on a 0.8% agarose gel to verify the presence of DNA templates. A diagram of the procedure can be found in Burns, J.A. (2012), Figure 3.4 (31).
In vitro transcription
Human RNA polymerase II (hRNAPII) in vitro transcription was performed as published (29,32). Briefly, 100 ng of DNA template attached to paramagnetic particles was incubated with transcription-competent HeLa nuclear extract (HeLaScribe®, Promega Corporation, Madison, WI, USA) in a 25 μl reaction containing 400 μM ATP, 400 μM GTP, 400 μM UTP and 16 μM [α-32P]CTP (25 Ci/mmol) for 1 h at 30°C. The reaction was quenched with 175 μl transcription stop solution (300 mM Tris–HCl (pH 7.4), 300 mM sodium acetate, 0.5% SDS, 2 mM EDTA, 3 μg/ml tRNA). The RNA was recovered by phenol-chloroform extraction and ethanol precipitation (HeLaScribe® Manual, Promega Corporation, Madison, WI, USA) and resolved by denaturing polyacrylamide gel electrophoresis (29). Experiments were performed in triplicate.
To test whether truncated transcripts remained associated with the DNA template and whether stalled hRNAPII complexes were present, in vitro transcription was allowed to proceed as noted above for 1 h at 30°C using 200 ng DNA template in 50 μl reactions. DNA templates were captured in a magnetic field, the buffer was removed, and the pellet was washed with 500 μl Sarkosyl buffer (200 mM Tris–HCl (pH 8.0), 1 mM DTT, 0.2 mM EDTA, 20% glycerol (v/v), 1% sarkosyl (w/v), 1 M KCl) to remove protein that was not tightly bound to the DNA template. The captured templates were washed again with buffer containing 200 mM Tris–HCl (pH 8.0), 1 mM DTT, 0.2 mM EDTA, 20% glycerol (v/v) and 100 mM KCl. Captured templates were re-suspended in 10 mM Tris–HCl buffer (pH 7.8). Transcripts tightly associated with the captured DNA templates were released by the addition of transcription stop solution, purified by phenol-chloroform extraction and ethanol precipitation, and analyzed by denaturing PAGE as described above.
Potential backtracking of hRNAPII was examined by isolating transcripts associated with DNA templates as described above, but they were not released from the DNA templates following re-suspension. HeLa nuclear extract was incubated in transcription buffer with 100 ug/ml heparin for 20 min at 30°C and then added to the re-suspended templates in the presence of NTPs (400 μM each) or with no additional NTPs added. The samples were incubated for 30 min at 30°C. RNA was released by the addition of transcription stop solution, purified by phenol-chloroform extraction and ethanol precipitation, and then analyzed by denaturing PAGE as described above.
Production of heteroduplex DNA vectors with extrahelical DNA regions
The vector pW-GR_control was made by modification of plasmid pWHE467 (33) using standard protocols. The modifications to pWHE467 were as follows. The F1 origin of replication was amplified by PCR from vector pCI-neo (Promega Corporation, Madison, WI) using primers 5′-TAACATGGATGACGTCTGATAGCCTGAATGGCGAATGGAC-3′ and 5′-ACTACAGTGATGACGTCACACGGAGAAAATACCGCATCAGGAA-3′, and inserted into the AatII restriction site of pWHE467. The puromycin resistance gene of plasmid pWHE467 was replaced with ZsGreen1 between restriction sites BamHI and SacII in pWHE467. The bidirectional promoter pBI was replaced with the unidirectional promoter TRE-tight derived from vector pTRE-tight (Clontech Laboratories, Inc., Mountain View, CA, USA). The red fluorescent protein (RFP) coding sequence DsRed-express, which was derived from plasmid pDsRed-express (Clontech Laboratories, Inc., Mountain View, CA), was inserted downstream of the TRE-tight promoter, forming vector pW-GR_control. The RFP gene in vector pW-GR was further modified by PCR and insertion of DNA oligomers using standard cloning techniques to make vectors pW-GR_stem_loop_insertion; pW-GR_stem_loop_deletion; pW-GR_loop_insertion; pW-GR_loop_deletion; pW-GR_Esp3I_insert. The series of vectors was used to make vectors containing extrahelical DNA.
Two methods were used to obtain the final experimental vectors containing an extrahelical region. The first involved annealing M13 phage-derived, single-strand circular vector (the non-transcribed strand of pW-GR_control with respect to promoter TRE-tight) to linear, double-strand vector (pW-GR_stem_loop_insertion, pW-GR_stem_loop_deletion, pW-GR_loop_insertion, or pW-GR_loop_deletion cut with XhoI and SphI) by OsPERT (30,34). The resulting double-strand, circular vector with an extrahelical region in the RFP gene also contained a gap between the XhoI and SphI restriction sites that was filled by annealing and ligating a complementary DNA oligomer (5′- TCGAGCCCCAGCTGGTTCTTTCCGCCTCAGAAGCCATAGAGCCCACCGCATCCCCAGCATG-3′) into the gap, thus forming the covalently, closed-circular vector. A diagram of the procedure for an analogous vector can be found in Appendix C, Figure C-11 of Burns, J.A. (2012) (31). The second followed a protocol outlined in (35). A vector that contained a cleavable insert in place of a 44 bp gap in the RFP gene (pW-GR_Esp3I_insert) was cut with Esp3I. Following restriction digestion, the linearized vector was purified from the insert by anion exchange chromatography. To insert an extrahelical DNA loop or DNA stem–loop into the linearized vector, DNA oligomers of two different lengths were annealed to form a DNA fragment with an extrahelical region and sticky ends complementary to the ends of the linearized pW-GR_Esp3I_insert. The annealed DNA oligomers were ligated into the 44 bp gap of Esp3I digested vector pW-GR_Esp3I_insert, resulting in covalent, closed-circular DNA. For both methods, the final products were purified through a CsCl gradient (36). Resolution of restriction fragments by gel electrophoresis was used to verify the presence of extrahelical DNA and assess final purity of the experimental vectors (Supplementary Figure S1).
Transfection of primary human fibroblasts
Primary human fibroblasts were obtained from the Coriell Cell Repository (Camden, NJ). They included normal human fibroblasts proficient in DNA repair (GM03651), XPA(−/−) fibroblasts deficient in NER and TCNER (GM05509) and CSB(−/−) fibroblasts deficient in TCNER (GM01629). Cells were maintained in culture up to passage 18 counting from the passage recorded upon receipt. For flow cytometry experiments, 75,000 cells were plated in a twelve-well cell-culture dish (3.8 cm2 growth area) with 500 μl minimal essential medium (MEM) (Mediatech, Inc., Manassas, VA) supplemented with 15% fetal bovine serum (FBS) that was certified tetracycline free (Atlanta Biologicals, Lawrenceville, GA), penicillin (100 units/ml) and streptomycin (100 μg/ml). Transfection complexes were made by mixing TransIT®-2020 Transfection Reagent (Mirus Bio LLC, Madison, WI) and DNA at a 2:1 ratio in 100 μl Opti-MEM I® Reduced Serum Medium (Life Technologies Corporation, Grand Island, NY) for 30 min at room temperature. Complexes were added to cells in MEM containing FBS and antibiotics, and the cells were incubated at 37°C under 5% CO2. The medium was removed 24 h following transfection, and cells were washed once with phosphate buffered saline (PBS) (10mM phosphate (pH 7.4), 2.7 mM potassium chloride, 137 mM sodium chloride). Transcription of the transfected RFP gene was induced by adding medium containing 1 μg/ml doxycycline. Uninduced cells received growth medium without doxycycline. Cells were harvested for flow cytometry 24 h after transcription induction by removing the medium, washing the cells with PBS and incubating with an appropriate volume of 0.5 × trypsin-EDTA (Life Technologies Corporation, Grand Island, NY) for approximately 5 min. Following cell detachment, MEM supplemented with FBS and antibiotics was added. The cells were collected and centrifuged at 100 × g for 10 min. The supernatant was removed, and the cell pellet was suspended in 300 μl PBS for fluorescence activated cell sorting (FACS).
Transcription in cells
Primary human fibroblasts were plated on 48-well plates (Corning Inc., Corning, NY, USA) and transfected with TransIT®-2020 Transfection Reagent as described. Transcription was induced 24 h after transfection by the addition of fresh medium containing 1 μg/ml doxycycline. (Note that the point immediately preceding addition of antibiotic was considered time 0.) Medium was removed at various times following induction, and the cells were lysed for RNA analysis using the RNeasy® Mini Kit (QIAGEN, Germantown, MD).
RFP and GFP mRNA levels were quantified by real-time quantitative reverse transcriptase PCR q(RT)2-PCR. Reverse transcription of the RNA utilized an anchored, tailed poly(dT) primer, 5′ ATGTTGACGCAGCCAGTGAC(T)20VN-3′, where V is A, C or G. This strategy ensured that the cDNA produced contained a unique sequence at the 3′-end that could be used for further amplification without interference caused by amplifying residual vector DNA. Reverse transcription reactions were carried out with AffinityScript™ Multiple Temperature Reverse Transcriptase (Agilent Technologies, Inc., Santa Clara, CA) following the manufacturer's protocol.
qPCR reactions were carried out using GoTaq Colorless Hot Start Master Mix (Promega Corporation, Madison, WI, USA) in a MyIQ2 Thermocycler (Bio-Rad Laboratories, Hercules, CA, USA) with an optical module. qPCR amplicons utilized the gene specific forward primers (5′-TAGTTGCCAGCCATCTGTTG-3′ for GFP cDNA and 5′-GAGGCTAACTGAAACACGGA-3′ for RFP cDNA) and a common reverse primer complementary to the tail added to each cDNA during the RT reaction (5′- ATGTTGACGCAGCCAGTGAC-3′). The quenched, 5′-nuclease probes for the fluorescent qPCR signal were complimentary to regions located between the primers (5′- AAGGTGCCACTCCCACTGTCCTTT-3′ for GFP cDNA and 5′-AGGAGACAATACCGGAAGGAACCC-3′ for RFP cDNA). Efficiency correction, mRNA quantification, and statistics were performed as in Nadkarni and Burns et al. (34).
Flow cytometry to quantify RFP and GFP fluorescence
GFP and RFP fluorescence was measured in a FACSaria II cell sorter (BD Biosciences, San Jose, CA, USA) using a 488 nm laser for excitation. Photomultiplier tube voltages were chosen such that fluorescent cells remained within the dynamic range of the instrument without accumulating in the last channel. The cytometer settings were 290 V for the RFP photomultiplier tube and 355 V for the GFP photomultiplier tube. Significantly fluorescent cells were gated based on RFP or GFP fluorescence in RFP versus allophycocyanin (APC) and GFP versus APC. To acquire data for fluorescence analysis, gates were drawn by hand in the program Cyflogic (CyFlo Ltd., Turku, Finland). Data for fluorescent cells were analyzed with custom scripts written in R (37), making use of the flow cytometry tools in Bioconductor (38) and graphics in the ggplot2 package (39). Experiments were performed at least in triplicate.
Nuclear fractionation and DNA purification
Fluorescent cells were sorted into PBS using the BD FACSaria II, and they were chilled on ice. Nuclei were immediately isolated from the cells (40). Briefly, cells were pelleted by centrifugation for 10 min at 100 × g at 4°C. Cells were suspended in cell lysis buffer (60 mM Tris-HCl (pH8.0), 0.3 M sucrose, 60 mM KCl, 15 mM NaCl, 0.5 mM spermidine, 0.15 mM spermine, 2 mM EDTA and 0.5% NP-40) and placed on ice for 5 min. Nuclei were isolated by centrifugation for 5 min at 1000 × g and washed once in cell lysis buffer without detergent, centrifuged for 5 min at 1000 × g and frozen at −80°C. Vector DNA was recovered from nuclei by small scale alkaline lysis (41). DNA recovered from nuclei was digested with NheI and XhoI, and quantified by qPCR against a standard curve of known quantities of vector.
Strand-specific primer extension and PCR
Extension primers were designed with a region complementary to either the transcribed or non-transcribed strand of the RFP gene, and each had a non-complementary 5′-tail (transcribed strand extension primer: 5′-TCAAGGCGTGGTGCTCAGTGAACCGTCAGATCGCCTGGAG-3′; non-transcribed strand extension primer: 5′-TCAAGGCGTGGTGCTCAGCTTGGTCACCTTCAGCTTGG-3′). The region complementary to the vector allowed for annealing and extension where the extrahelical DNA was placed. The non-complementary tail was used to amplify extended products without amplifying the vector. This arrangement permitted probing the sequence of a single strand within the parent vector's DNA. Extension reactions were carried out using Phusion® High-Fidelity DNA Polymerase (New England BioLabs Inc., Ipswich, MA, USA) in 10 μl reactions using a high concentration of primers (10 μM) with 1 cycle at 95°C for 2 min denaturation, 54°C for 10 s annealing and 72°C for 60 s extension. The extension reaction contained between 104 and 106 molecules of template DNA. Excess primers were removed using Ampure XP magnetic particles (Beckman Coulter, Brea, CA) which precipitated the template DNA and long extension products, while the short, single-strand primers were washed away. Following purification, extension products and total plasmid were amplified by PCR using a primer complementary to the tail on the primer extension products, and a primer complementary to the vector downstream from the extension primer. Cycling conditions were one cycle at 95°C for 2 min, then 60 cycles of 95°C for 10 s followed by 60°C for 30 s. The products from PCR were resolved by native PAGE in Tris/borate/EDTA (TBE) buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA, pH 8.2–8.4). The gels were stained with SYBR Green, and images were captured using a Typhoon FLA-9000 Laser Scanner (GE Healthcare Life Sciences, Pittsburgh, PA, USA). Densitometry measurements of bands in the gel were performed with FIJI (42). Experiments were performed in triplicate unless noted otherwise.
RESULTS
DNA loops and DNA stem–loops situated on the transcribed strand or non-transcribed strand of a transcription unit impede RNA synthesis in vitro
DNA loops and DNA stem–loops impede transcription
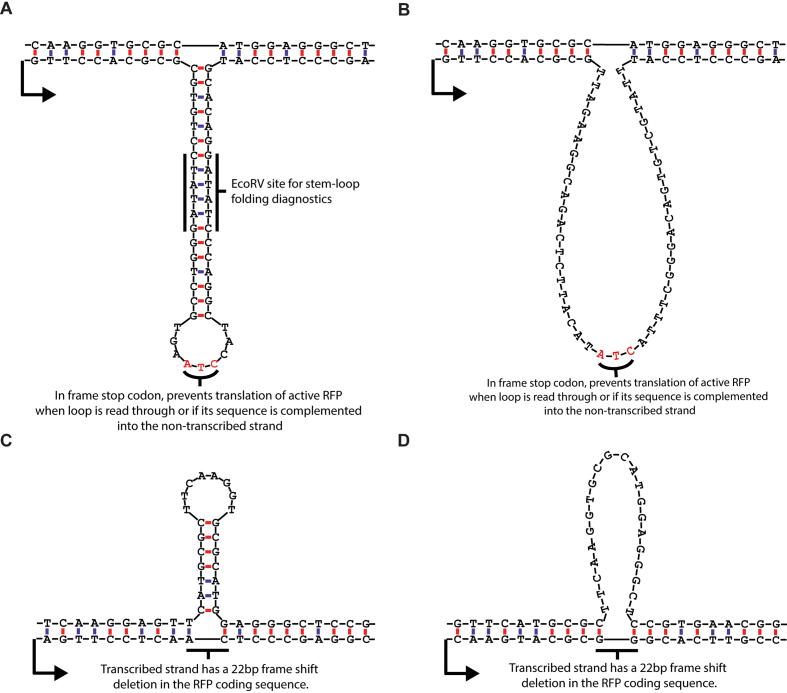
To test the hypothesis that extrahelical loops of DNA can block or impede transcription, DNA templates that contained either a DNA stem–loop or an unpaired DNA loop were assembled. Each template contained a cytomegalovirus (CMV) promoter to support hRNAPII transcription. In addition, a DNA stem–loop or an unpaired DNA loop was positioned on either the transcribed strand or non-transcribed strand. The DNA stem–loops were designed such that the structure contained an 18 base-pair stem and a 9-nucleotide loop when positioned on the transcribed strand (Figure 2A) or a 7 base-pair stem and 8-nucleotide loop when positioned on the non-transcribed strand (Figure 2C). The unpaired DNA loops, which were designed to exhibit no predictable Watson-Crick base pairs, were 45 nucleotides in length when positioned on the transcribed strand (Figure 2B) and 22 nucleotides in length when positioned on the non-transcribed strand (Figure 2D).
Figure 2.
DNA loop and stem–loop sequences and structures. (A) stem–loop, transcribed strand. The sequence of the extrahelical DNA on the transcribed strand was designed such that the stem–loop contained a restriction site (EcoRV) in the stem with a sufficient length of dsDNA (6 base pairs) on either side of the restriction site for recognition and cutting. This was done for diagnostic purposes to confirm that the stem loop folded as expected (Supplementary Figure S2). (B) Unpaired loop, transcribed strand. The unpaired loop on the transcribed strand was matched in length to the stem–loop structure. Both sequences contained in-frame stop codons for cell culture experiments, where removal of the loop or stem–loop from the transcribed strand would lead to RFP activation, but retention or expansion of the sequence would produce a truncated, non-functional RFP. (C) Stem–loop, non-transcribed strand. The sequences of the extrahelical DNA on the non-transcribed strand were limited by the native RFP coding sequence. The length of 22 nucleotides was chosen because there was a sequence in the RFP coding region that would form a stem loop with a 7 base-pair stem and a 9 nucleotide loop if the corresponding sequence on the transcribed strand were missing. The 22 nucleotide deletion on the transcribed strand also caused a frame-shift mutation that produced a non-functional protein product in cell culture experiments. (D) Unpaired loop, non-transcribed strand. Similar to the stem–loop on the non-transcribed strand, this is a native sequence in the RFP coding sequence that was not predicted to form significant internal base-pairing. The transcribed strand also has a 22 nucleotide frame-shift deletion in the RFP coding sequence that knocks out fluorescence.
In vitro transcription reactions were conducted using HeLa nuclear extract as the source of the transcription machinery. Nascent transcripts were radiolabeled with [32P] phosphate, and the resulting RNA was resolved on denaturing polyacrylamide gels (43). Extrahelical DNA present on the transcribed strand of the DNA templates should produce truncated RNA approximately 380 nucleotides in length if such structures were to disrupt transcription; similarly, extrahelical DNA present on the non-transcribed strand of the DNA templates should produce truncated RNA approximately 365 nucleotides in length if these structures were to disrupt transcription. In all cases in vitro, extrahelical DNA disrupted transcription regardless of whether or not the structures were present on the transcribed or non-transcribed strand, but each interfered with RNA synthesis to a very different extent as shown in Figure 3 and Table 1.
Table 1. Summary of in vitro transcription results showing transcriptional blockage at extrahelical DNA. ND = no data.
| Extrahelical DNA | Position | % Block | Position of extrahelical DNA with respect to the transcribed strand. | Approximate position of discrete bands indicating stalled hRNAPII | Characterization | hRNAPII Backtracking? | hRNAPII Re-extension? |
|---|---|---|---|---|---|---|---|
| DNA stem Loop | Transcribed Strand | 92% ± 3% | (+)384 to +429 | 380; 400; 430 | Strong Block | Yes | Yes |
| Nontranscribed Strand | 75% ± 2% | (+)365 | 365 | Strong Block | Yes | Yes | |
| Unpaired DNA Loop | Transcribed Strand | 56% ± 13% | (+)384 to +429 | 380; 395; 430 | Moderate Block | Yes | Yes |
| Nontranscribed Strand | 28% ± 10% | (+)372 | 370 | Weak block | ND | ND |
hRNAPII stalled at unpaired DNA loops or DNA stem–loops can remain associated with the DNA template as a competent transcription complex
The presence of a DNA stem–loop or unpaired DNA loop on either the transcribed or non-transcribed strand of a transcription unit clearly interferes with the progression of hRNAPII during transcription elongation. This observation led to a fundamental question: Does the hRNAPII complex remain associated with the DNA template at the sites of extrahelical DNA, possibly poised to continue transcription elongation, or does it dissociate and release truncated RNA?
Following transcription initiation and elongation in the presence of [α-32P]CTP, DNA templates containing a biotin moiety at the 5′-end of the non-transcribed strand were mixed with paramagnetic beads that contained streptavidin. The resulting biotin–streptavidin complexes were subsequently captured with a magnetic field and washed with a buffer containing 1% sarkosyl to remove proteins and RNAs that were not stably associated with the DNA template (44). Stable transcription complexes that remained associated with the beads were released by phenol-chloroform extraction.
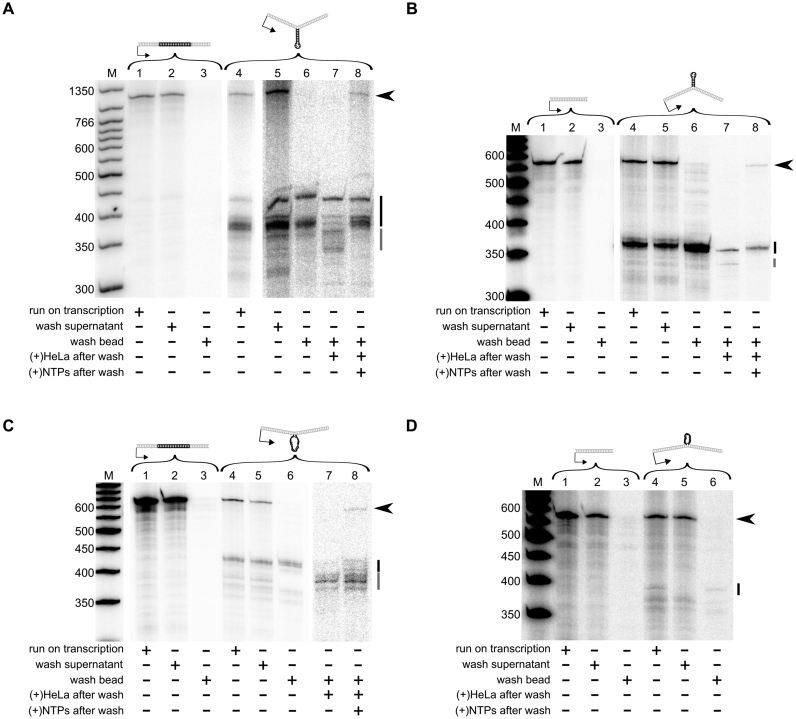
Control DNA templates with no extrahelical DNA exhibited no evidence of stalled transcription complexes (Figure 4, panels A–D, lanes 1–3). In contrast, templates with extrahelical DNA structures produced truncated transcripts that precipitated with the DNA during the capture protocol. Furthermore, the size of these truncated transcripts was equivalent to the size of the truncated transcripts observed during run-on transcription reactions. These results indicate that the hRNAPII complex remained stably associated with the template DNA in the vicinity of extrahelical DNA structures or that stable DNA–RNA hybrids were formed (Figure 4, panels A–D, lanes 4–6). This is analogous to the situation when RNA polymerase stalls at sites of DNA damage, a scenario that can lead to backtracking of stalled transcription complexes. Backtracking results from endonucleolytic cleavage of the stalled transcript, yielding a product that is several bases shorter than those found at the immediate site of stalling (44,45), which might provide a means for the stalled transcription complex to re-enter elongation in order to bypass the block and yield a transcript.
Figure 4.
DNA loops and stem–loops induce stably stalled hRNAPII during transcription elongation. In all cases, the following samples from in vitro transcription reactions were resolved by gel electrophoresis. Each gel (A–D) represents one DNA loop or stem–loop template and its paired control template. Reaction conditions are indicated by the table below each gel. The black arrowhead to the right of each gel indicates the position of full length transcripts for that experiment. The black bar to the right of each gel indicates the truncated transcripts. The gray bar indicates truncated transcripts after hRNAPII backtracking. Lanes 1–3 = control template; Lanes 4–8 = experimental template. (A) Stem–loop on the transcribed strand. (B) Stem–loop on the non-transcribed strand. ( C) Loop on the transcribed strand (D). Loop on the non-transcribed strand. For D, there were not sufficient stalled transcripts to obtain signal in add back experiments so lanes 7 and 8 are not present. Gels were cropped and ordered for clarity, maintaining their relationship to the marker. Brightness and contrast were adjusted as needed to allow visualization of RNA bands.
To test if hRNAPII exhibits backtracking when stalled at extrahelical DNA, HeLa nuclear extract mixed with heparin was added to stalled complexes. Heparin prevents free hRNAPII from initiating transcription, but it does not interfere with factors that could cause a stalled hRNAPII complex to stimulate endonucleolytic cleavage of RNA by factors like TFIIS (46), which is involved in recovery from backtracking (47). To test whether backtracked complexes were elongation competent, NTPs were added, allowing backtracked complexes to elongate back to the site of the extrahelical DNA, and in some cases through the block to the end of the template (Figure 4, panels A–D, lanes 7 and 8). This was true for all cases in which stalling occurred, suggesting that hRNAPII stalled at extrahelical DNA behaves in a manner similar to hRNAPII stalled at chemically modified DNA.
Unpaired DNA loops and DNA stem–loops on the transcribed strand of a gene block RNA polymerase progression in human cells, but extrahelical DNA on the non-transcribed strand of an expressed gene poses no observable impediment to RNA synthesis
Transcription time courses measured the quantity of RFP mRNA produced in cells from vectors containing stem–loops within the RFP transcription unit
In vitro transcription reactions indicated that extrahelical DNA impedes transcription elongation by hRNAPII in a manner similar to hRNAPII stalled at chemically modified DNA. Hence, to assess the effect of DNA stem–loops or unpaired DNA loops on transcription elongation in vivo, cells were transfected with vectors (Figure 5A) containing extrahelical DNA positioned in the RFP gene's transcription unit (Figure 5B–E), and mRNA levels were measured. The vectors and experiments were analogous to those published earlier (34), in which the contributions of NER and TCNER were determined for bulky DNA lesions. The sequences and structures of the extrahelical regions for each vector used in cellular transfection experiments were identical to those used for in vitro transcription reactions (Figure 2).
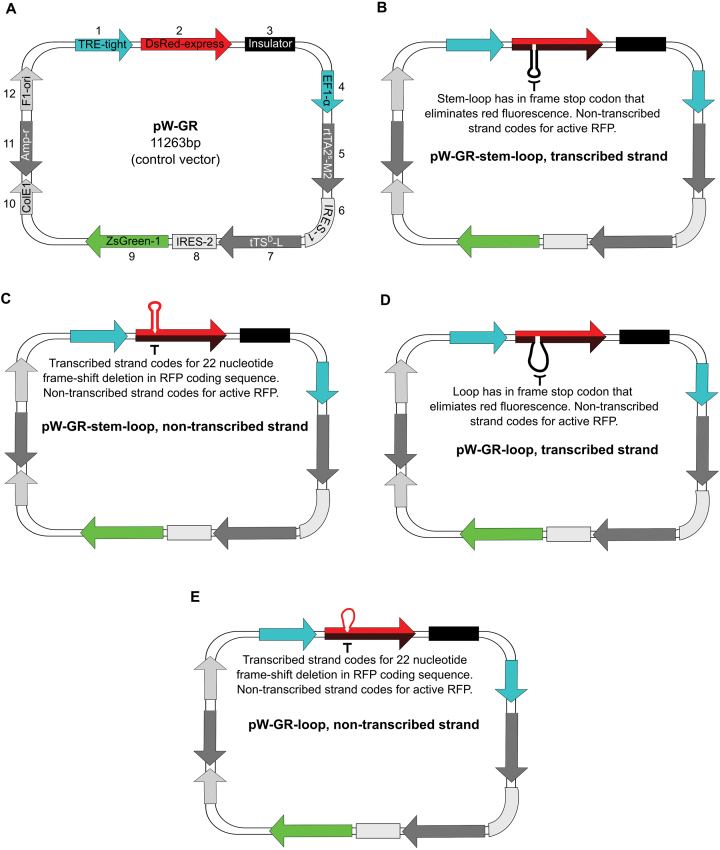
Figure 5.
Map of the control vector, pW-GR, and of experimental vectors indicating the position of extrahelical DNA within the RFP transcription unit. (A) Vector pW-GR map. The vector contains red fluorescent protein (RFP), DsRed-express under the control of inducible promoter TRE-tight. It also contains an insulator element to separate the RFP transcription unit from the inducer elements transcription unit. The inducer elements, rtTA (the tet-activator protein) and tTS (a tet-responsive repressor), and a green fluorescent protein (GFP), ZsGreen1, are under the control of the constitutive elongation factor 1-α (EF1-α) promoter. The vector also contains the F1-origin of replication (F1ori) for single strand phage packaging, and bacterial propagation components, ColE1 (E. coli origin of replication) and Ampr (ampicillin resistance gene). (B) Vector pW-GR-stem–loop, transcribed strand, has an extrahelical stem–loop on the transcribed strand of the RFP transcription unit. The non-transcribed strand (red) codes for RFP, while the transcribed strand with the stem loop codes for a truncated, non-fluorescent protein. ( C) pW-GR-stem–loop, non-transcribed strand. (D) pW-GR-loop, transcribed strand. (E) pW-GR-loop, non-transcribed strand.
In human fibroblasts proficient in DNA as well as XPA(−/−) human fibroblasts deficient in NER, a DNA stem–loop present on the transcribed strand of the RFP gene significantly reduced the RFP mRNA levels compared to a control vector that lacked extrahelical DNA (Figure 6, Panels A and B). Furthermore, the reduction in RFP mRNA was observed at each time point during the experiment, and the transcript levels did not reach those of the comparable control vector. In contrast, a DNA stem–loop present on the non-transcribed strand of the RFP gene had no significant effect on RFP mRNA synthesis compared to an unmodified control vector (Figure 6A and B). These results suggest that the behavior of hRNAPII at DNA stem–loops in cells differs from that observed in vitro, where a DNA stem–loop on either strand of a transcription unit posed a block to elongation. This phenomenon could be due to removal of the extrahelical DNA stem–loop from the non-transcribed strand or from fundamental differences between in vitro reactions and cell-based assays. The similar curves obtained from fibroblasts proficient in DNA repair and XPA(−/−) cells suggest that the presence of NER does not modulate the effect of extrahelical DNA on transcription elongation.
Figure 6.
DNA stem–loops on the transcribed strand interfere with transcription elongation in normal and NER deficient cells. (A) Time course of RFP mRNA accumulation in repair proficient cells from a control vector, a vector containing a stem–loop on the transcribed strand, or a vector containing a stem–loop on the non-transcribed strand. (B) Time course of RFP mRNA accumulation in XPA cells. Error bars indicate the standard deviation of each measurement. (C) mRNA quantity at 24 hours post transcription induction in cells with different repair backgrounds transfected with vectors containing loops and stem–loops. Error bars indicate the standard deviation. *, Statistically significant difference for that sample compared to the within cell type control (P < 0.05). †, statistically significant difference for comparison between repair deficient and repair normal cells (P < 0.05). Statistics were calculated using the standardized mean of a contrast variable as in Nadkarni and Burns et al. (34).
RFP mRNA synthesis from vectors containing DNA stem–loops or unpaired DNA loops within the RFP transcription unit was measured in human fibroblasts proficient and deficient in DNA repair
The effect of DNA stem–loops or unpaired DNA loops on transcription was measured in primary human cells, specifically fibroblasts proficient in DNA repair, XPA(−/−) fibroblasts deficient in NER and TCNER and CSB(−/−) fibroblasts deficient in TCNER. The results show that a DNA stem–loop positioned on the transcribed strand of the RFP gene decreased the amount of accumulated, full-length RFP mRNA relative to a control vector. This trend was seen in fibroblasts proficient in DNA repair as well as cells that are defective in NER or TCNER (Figure 6C, stem–loop transcribed strand). In contrast, a DNA stem–loop present on the non-transcribed strand of the RFP gene exerted no significant effect on transcription in any of the cell types examined (Figure 6C, stem–loop, non-transcribed strand).
An unpaired DNA loop present on the transcribed strand of the RFP gene significantly reduced the amount of RFP mRNA in all cells examined (Figure 6C, loop, transcribed strand). However, an unpaired DNA loop located on the non-transcribed strand of the RFP gene exerted no observable effect on transcription in normal and XPA(−/−) cells, but did inhibit transcription in CSB(−/−) cells (Figure 6C, loop, non-transcribed strand).
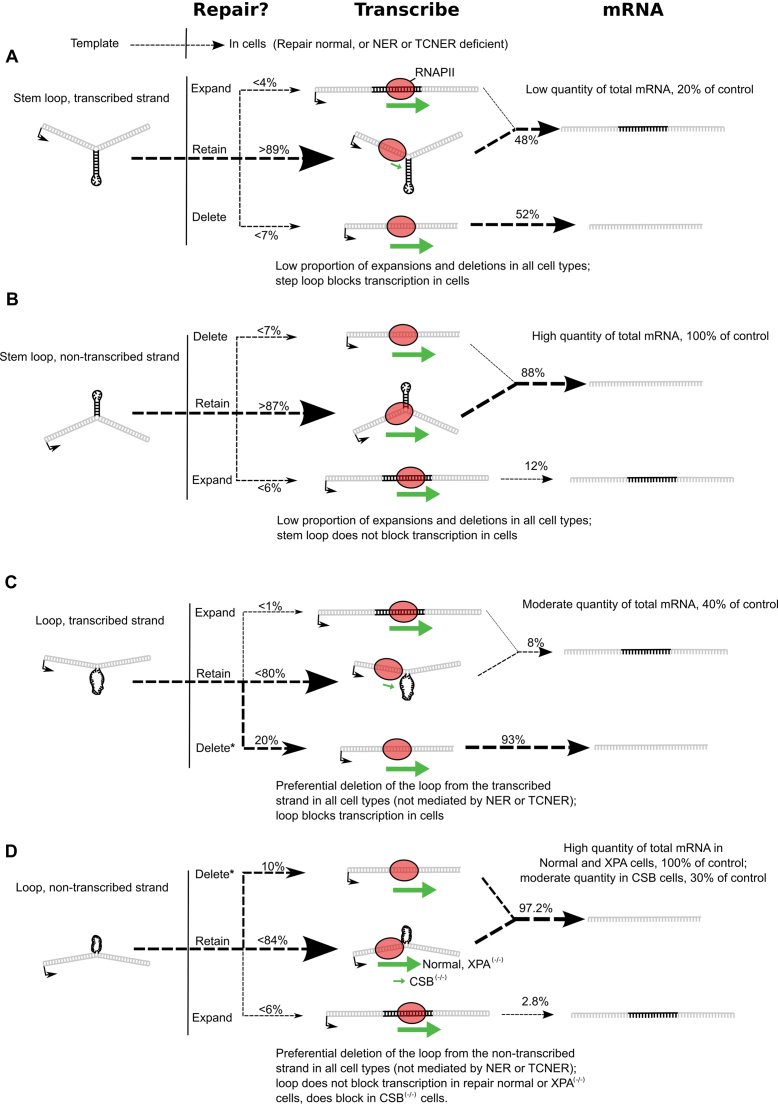
These results show that extrahelical DNA in the form of DNA stem–loops or unpaired DNA loops located on the transcribed strand of a gene hinder transcription elongation in human fibroblasts that are proficient in DNA repair. Comparable results are obtained in cells deficient in both NER and TCNER, while similar structures located on the non-transcribed strand do not tend to affect transcription elongation in terms of the quantity of mRNA produced in a given period of time. However, an unpaired DNA loop on the non-transcribed strand does hinder transcription in CSB cells, suggesting a role for the CSB protein in assisting hRNAPII to move through an unpaired DNA loop present on the non-transcribed strand.
RFP activity was observed in normal human fibroblasts, and human fibroblasts deficient in NER or TCNER following transfection with vectors containing extrahelical DNA
RFP fluorescence was measured to quantify reactivation of the RFP gene
The transcript abundance data show that RFP mRNA is made in cells transiently transfected with vectors containing extrahelical DNA within the RFP transcription unit, albeit to a very low extent when the DNA stem–loop or unpaired DNA loop is positioned on the transcribed strand. But does the resulting RFP mRNA encode functional RFP? Recall that vectors containing unpaired DNA loops or DNA stem–loops positioned within the RFP gene were assembled such that the sequence on the non-transcribed strand reflected the actual coding sequence for functional RFP mRNA. Vectors with extruded DNA on the transcribed strand would result in active RFP only if the extrahelical DNA were removed. In contrast, extruded DNA positioned on the non-transcribed strand would result in an active RFP if the extrahelical DNA were retained during TCNER and expanded into the transcribed strand during DNA synthesis.
When normal human fibroblasts were transfected with a vector containing a DNA stem–loop on the transcribed strand, a red-fluorescent signal was observed. This result suggests that the extrahelical DNA stem–loop was removed and replaced with a sequence that encoded active RFP in some of the vectors (Figure 7A). Note that the RFP signal was lower than that found for the control vector. Furthermore, when CSB(−/−) cells or XPA(−/−) cells were transfected with a vector containing a DNA stem–loop on the transcribed strand, an RFP signal equivalent to that seen in normal human fibroblasts was observed (Figure 7A).
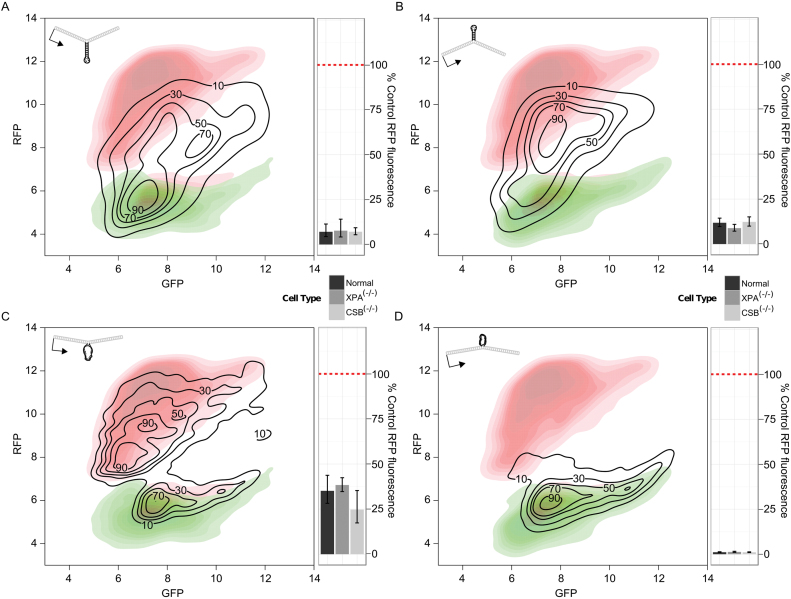
Figure 7.
Flow cytometry plots indicate recovery of red fluorescence from vectors containing extruded stem–loops, but not from a vector containing an unpaired loop on the non-transcribed strand. (A–D) Black contour lines show the distribution of red and green fluorescence in cells transfected with the respective experimental vector containing an extrahelical DNA loop or stem–loop. The red contoured heatmap in each plot shows the red and green fluorescence from control vector pW-GR, representing the maximal red fluorescence (y-axis) attainable through deletion or expansion of a stem loop. The green contoured heatmap in each plot shows the expected distribution if only GFP is made without any RFP. Associated bar-plots show average RFP levels, normalized to RFP fluorescence from control vector pW-GR, in different cell types. (A) Stem–loop, transcribed strand. (B) Stem–loop, non-transcribed strand. (C) Loop, transcribed strand. (D) Loop, non-transcribed strand.
Normal human fibroblasts transfected with a vector containing a DNA stem–loop on the non-transcribed strand, which did not appear to block transcription in human cells, displayed RFP recovery similar to a vector containing a DNA stem–loop on the transcribed strand (Figure 7B). Furthermore, RFP fluorescence was equivalent to that found for the same vector transfected into in CSB(−/−) or XPA(−/−) fibroblasts (Figure 7B).
Fibroblasts transfected with a vector containing an unpaired DNA loop on the transcribed strand exhibited significantly more recovery of RFP fluorescence compared to cells transfected with a vector containing a DNA stem–loop (Figure 7C). This suggested that a loop of DNA without internal base pairing is processed more efficiently than a DNA stem–loop. However, the increased signal was seen in XPA(−/−) and CSB(−/−) human fibroblasts. (Figure 7C).
In sharp contrast to results observed in cells transfected with the unpaired DNA loops on the transcribed strand and DNA stem–loops on either strand, cells transfected with a vector containing an unpaired DNA loop on the non-transcribed strand exhibited very little red fluorescence (Figure 7D). This suggests that such a structure escapes processing or is preferentially deleted. Additionally, equivalent red fluorescence signal was observed in CSB(−/−) or XPA(−/−) fibroblasts (Figure 7D).
The results reported here indicate that DNA stem–loops and unpaired DNA loops are resulting in expansions, deletions, or both in cells. However, the results also indicate that such processing occurs in cells proficient in DNA repair as well as those deficient in NER or TCNER. Hence, these results also show that neither of these DNA repair pathways is involved in the expansion or contraction of such structures.
Unpaired DNA loops are preferentially deleted and DNA stem–loops are predominantly retained in primary human fibroblasts
To characterize further the expansions or deletions of extrahelical DNA, the vectors used in these experiments were recovered from human cells, and changes to the length of DNA in the vicinity of the extrahelical sequences were assessed using primer extension, amplifying the products, and resolving them using native PAGE (see Supplementary Figure S2A for a schematic of the method and expected outcomes). In parallel to these studies, the length of mRNA produced from each vector following transfection into human fibroblasts was determined using RT-PCR and resolving the products by native PAGE. The proportion of the different mRNA species present was expected to agree with the observed proportion of expansions or deletions of extrahelical DNA in the vector templates (see Supplementary Figure S2B for a schematic of the method and expected outcomes).
PCR analysis of a vector containing a DNA stem–loop positioned on the transcribed strand showed no expansions or deletions of the region above background (Figure 8, panel 1). The result was the same in cells proficient in DNA repair as well as XPA(−/−) cells and CSB(−/−) cells (Supplementary Figure S3). However, RT-PCR analysis indicated that short mRNA was indeed produced and represented 51 ± 1.4% of the total RFP mRNA in repair proficient cells, suggesting that the DNA stem–loop was removed (Figure 9, panel 1). The result was not significantly different in XPA(−/−) or CSB(−/−) cells (Table 2) and might not be surprising. The DNA stem–loop itself blocks transcription, thwarting the formation of longer, full-length mRNA; hence, shorter mRNA made from a small fraction of the templates from which the structure was deleted may well be over-represented in the population.
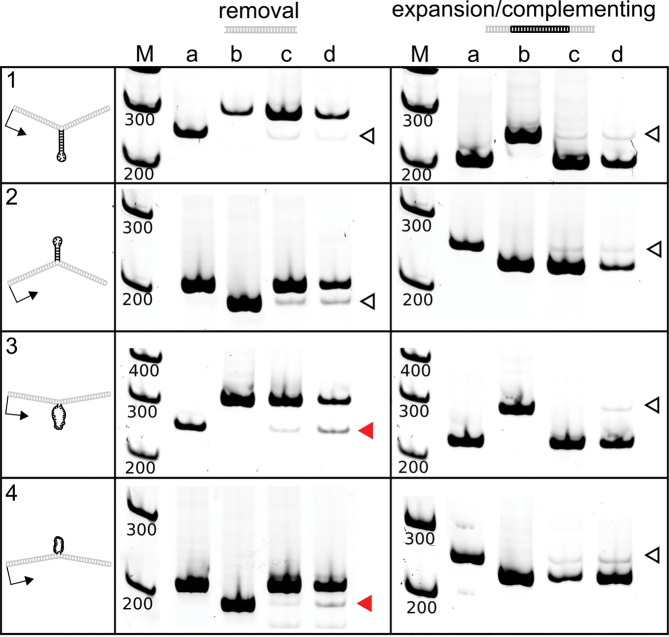
Figure 8.
Human fibroblasts delete unpaired DNA loops, but do not cause measurable expansions or contractions of DNA stem–loops. These gels show the length of DNA bands after strand specific PCR of vector controls and DNA recovered from normal primary fibroblasts. Lane (a) is strand specific PCR of control vector pW-GR. Lane (b) is strand specific PCR the respective expanded or deleted control vector. Lane (c) is strand specific PCR of the experimental vector prior to introduction to cells. Although only one band should be present in this lane, there was a faint second band limiting the sensitivity of the assay due to difficulty removing 100% of the linear extension primer prior to PCR. Leftover extension primer caused some amplification of the opposite strand in heteroduplex vectors. Significant expansions or deletions were measured as signal above this background. Lane (d) is strand specific PCR of the experimental vector after recovery from normal primary fibroblasts. Open arrowheads indicate bands with an equal representation in control and experimental samples. Filled red arrowheads indicate bands where there was a significant difference (P< 0.05) in the experimental samples. Statistical tests were performed by one-way analysis of variance with contrasts to delineate specific comparisons. Gels were cropped and ordered for clarity, maintaining their relationship to the marker. Brightness and contrast were adjusted as needed to allow visualization of DNA bands.
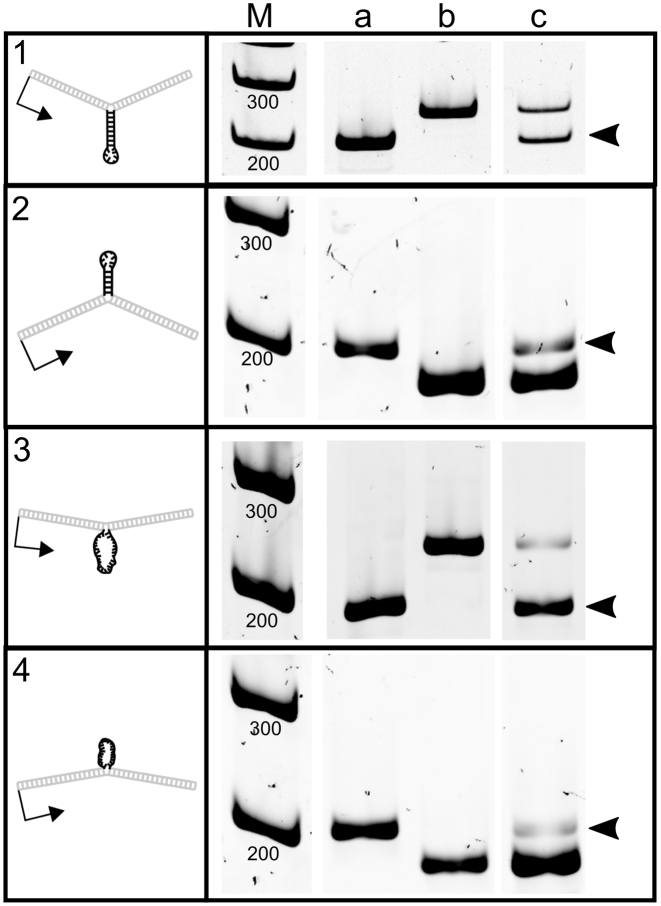
Figure 9.
cDNA length indicates extrahelical stem–loop and loop deletions and expansions. These gels show the length of cDNA amplicons containing sequences transcribed from the heteroduplex region in the template DNA in human cells. Lane (a) is cDNA recovered from cells transfected with control vector pW-GR. Lane (b) is cDNA recovered from cells transfected with the respective expanded or deleted control vector. Lane (c) is cDNA recovered from cells transfected with heteroduplex vectors containing extrahelical DNA. Black arrowheads indicate the 180 bp sequence that was not present on the transcribed strand of heteroduplex templates prior to introduction into cells. Gels were cropped and ordered for clarity, maintaining their relationship to the marker. Brightness and contrast were adjusted as needed to allow visualization of cDNA bands.
Table 2. Summary of vector repair and mRNA quantification shows low levels of expansions/contractions at DNA stem loops, and significant deletions of unpaired loops.
| Cell line | Extrahelical DNA | Location in vector | Extrahelical DNA deleted | mRNA indicating deletion1 | Extrahelical DNA expanded | mRNA indicating expansion2 |
|---|---|---|---|---|---|---|
| Normal human fibroblasts | DNA stem–loop | Transcribed strand | <7% | 51 ± 14% | <4% | Not applicable |
| Nontranscribed strand | <7% | Not applicable | <6% | 12 ± 6% | ||
| Unpaired DNA loop | Transcribed strand | 19 ± 5%* | 93 ± 1% | <1% | Not applicable | |
| Nontranscribed strand | 10 ± 5%* | Not applicable | <6% | 3 ± 2% | ||
| XPA(−/−) human fibroblasts | DNA stem–loop | Transcribed strand | <7% | 52 ± 3% | <4% | Not applicable |
| Nontranscribed strand† | 9%† | Not applicable | 6%† | 9 ± 1% | ||
| Unpaired DNA loop | Transcribed strand | 20 ± 5%* | 91 ± 4% | <1% | Not applicable | |
| Nontranscribed strand | 10 ± 2%* | Not applicable | <6% | 6 ± 1% | ||
| CSB(−/−) human fibroblasts | DNA stem–loop | Transcribed strand | <7% | 55 ± 7% | <4% | Not applicable |
| Nontranscribed strand | <7% | Not applicable | < 6% | 8 ± 0.4% | ||
| Unpaired DNA loop | Transcribed strand | 23 ± 4%* | 85 ± 2% | < 1% | Not applicable | |
| Nontranscribed strand | 8 ± 5%* | Not applicable | < 6% | 6 ± 6% |
*P < 0.05 for deletions or expansions in DNA repair assay.†No replicate data. Percent repair is reported observed repair over background. 1For extrahelical structures on the non-transcribed strand, mRNA bands would not detect deletions as the mRNA length would be the same in the case of deletion and retention of the structure. 2For extrahelical structures on the transcribed strand, mRNA bands would not detect expansions as the mRNA length would be the same in the case of expansion and retention of the structure. Statistical significance was estimated using ANOVA followed by Tukey's HSD to get individual P-values, testing for significance over background.
PCR analysis of a vector containing a DNA stem–loop positioned on the non-transcribed strand also showed no expansions or deletions of the region above background (Figure 8, panel 2), regardless of the DNA repair phenotype investigated (Supplementary Figure S3). Although the repair assay did not detect DNA expansions above background, long mRNA was observed, suggesting that some processing of the DNA stem–loop occurred in which the transcribed strand was expanded. The proportion of mRNA corresponding to an expansion of the stem–loop was 12% ± 5.8% in cells proficient in DNA repair (Figure 9, panel 2), and was not different in cells lacking NER or TCNER (Table 2).
In contrast to DNA stem–loops, processing of unpaired DNA loops could be directly detected at levels above background using the PCR assay described (Figure 8, panel 3). An unpaired DNA loop present on the transcribed strand was removed from 18.6% ± 4.7% (P = 1.35 × 10−7) of the vector DNA recovered from cells proficient in DNA repair. A similar result was observed in cells deficient in NER or TCNER (Table 2). The proportion of mRNA corresponding to a deletion of the unpaired DNA loop from the transcribed strand was 93% ± 1.3% (Figure 9, panel 3), and this value was not significantly different in cells deficient in DNA repair (Table 2). While approximately 20% of the vectors exhibited removal of the unpaired DNA loop from the transcribed strand, the remaining 80% of vectors retained extrahelical loops that posed blocks to transcription, resulting in overrepresentation of the mRNA from the repaired vectors (see Figure 10C).
Figure 10.
Schematic summary of results. (A) Following transfection into primary human fibroblasts, a stem–loop on the transcribed strand appears to be stable, with more than 89% of the vectors retaining an extrahelical stem–loop. The stem–loop blocks transcription elongation, while transcription from a small percentage of repaired or replicated vectors proceeds at a normal rate. Transcripts from the small percentage of repaired or replicated vectors are overrepresented in the final mRNA product. (B) A stem–loop on the non-transcribed strand appears to be stable, with more than 87% of the vectors retaining an extrahelical stem–loop. The stem–loop on the non-transcribed strand does not block transcription elongation so transcription proceeds at a normal rate. Transcription from a small percentage of repaired or replicated vectors also proceeds at a normal rate. Transcripts from the small percentage of repaired or replicated vectors are overrepresented in the final mRNA product. (C) A loop on the transcribed strand is preferentially deleted, with about 20% of recovered vectors indicating deletion of the loop. Deletion occurs in repair normal and NER and TCNER deficient cell types. The loop in vectors from which the loop had not been deleted blocks transcription elongation, while transcription from the repaired vectors proceeds at a normal rate. Transcripts from the repaired vectors indicating deletion of the DNA loop are overrepresented in the final mRNA product. (D) A loop on the non-transcribed strand is preferentially deleted, with about 10% of recovered vectors indicating deletion of the loop. Deletion occurs in repair normal and NER and TCNER deficient cell types. The loop in vectors from which the loop had not been deleted blocks transcription elongation in TCNER deficient CSB cells, but not in repair normal or NER deficient XPA cells. Transcription from the repaired vectors proceeds at a normal rate. Transcripts in the final mRNA product indicate bypass of the loop on the non-transcribed strand or overrepresentation of mRNA from vectors where the loop was deleted (potentially the case in CSB cells). *Statistically significant experimental evidence for deletion of unpaired loops.
Extrahelical DNA in the form of an unpaired DNA loop on the non-transcribed strand was also deleted (Figure 8, panel 4; Table 2). Note that these deletions resulted in the removal of any information that could encode active RFP from the vector. This resulted in little to no reactivation of RFP fluorescence in cells transfected with a vector containing an unpaired DNA loop on the non-transcribed strand, and the absence of RFP reactivation was true regardless of the DNA repair phenotype (Figure 7). The proportion of mRNA indicating an expansion of the loop into the transcribed strand, which would cause re-activation of RFP, was very low at 2.8 ± 2.0% (Figure 9, panel 4) and not significantly different in cells deficient in DNA repair (Table 2). This correlates well with the repair data and RFP fluorescence, suggesting that the unpaired DNA loops were deleted from the non-transcribed strand (see Figure 10D).
DISCUSSION
The work presented here tested if extrahelical DNA—specifically DNA stem–loops or unpaired DNA loops—positioned within a transcription unit would interfere with hRNAPII elongation in vitro and in vivo. The biochemical results clearly indicate that extrahelical DNA on the transcribed strand or non-transcribed strand of a gene impedes the progression of hRNAPII, with the order of inhibition as follows: DNA stem–loop on the transcribed strand > DNA stem–loop on the non-transcribed strand > an unpaired DNA loop on the transcribed strand > an unpaired DNA loop on the non-transcribed strand. While the cellular assays also suggest that DNA stem–loops and unpaired DNA loops on the transcribed strand of a gene interfere with transcription, similar structures exert minimal if any influence on transcription elongation when they are positioned on the non-transcribed strand of a transcription unit in cells.
The ability of extrahelical DNA to interfere with hRNAPII elongation, especially when the structure is a DNA stem–loop, is analogous to the effect on transcription of certain types of DNA damage. In most cases, chemically modified bases in DNA that result from exposure to genotoxins impede or block hRNAPII elongation when the damage is located on the transcribed strand (48). The stalled hRNAPII complex then induces the stable association of DNA repair factors such as CSA, CSB and TFIIH that initiate TCNER (49). The studies reported here show that extrahelical DNA extruded from the transcribed strand of a gene also interferes with hRNAPII transcription elongation. However, in contrast to damaged DNA, these structures are not substrates for TCNER or NER, casting into doubt models in which either of these repair pathways effect somatic expansions or contractions of DNA.
Stalling of hRNAPII at DNA loops and stem–loops during in vitro transcription has been observed and reported (25). The data provided here serve as additional evidence that hRNAPII complexes stalled at DNA stem–loops or unpaired DNA loops remain associated with the DNA template, are able to backtrack and are elongation competent. This behavior is reminiscent of hRNAPII stalled at a cyclobutane pyrimidine dimer (45); however, unlike DNA damage, DNA stem–loops and unpaired DNA loops are not recognized as aberrant DNA that is subject to TCNER.
When vectors containing an unpaired DNA loop or DNA stem–loop positioned on the transcribed strand within an RFP reporter gene were introduced into cells, the structures inhibited transcription elongation, resulting in a sharp reduction in the number of transcripts relative to control templates that lacked extrahelical DNA structures. This reduction was observed in cells proficient in DNA repair as well as cells deficient in NER or TCNER. In contrast, a DNA stem–loop or unpaired DNA loop present as extrahelical DNA on the non-transcribed strand of a gene did not inhibit transcription in cells, results that differ from the biochemical data.
For DNA stem–loops, the detection of RFP fluorescence by flow cytometry suggests that a percentage of this DNA underwent expansions or contractions, with an expansion occurring in the opposite strand due to the addition of DNA complementary to the stem–loop, or a deletion occurring when the DNA stem–loop was removed. However, the observation that RFP fluorescence was identical in cells proficient DNA repair-proficient, XPA, and CSB cells indicates that neither NER nor TCNER is involved in the process. Furthermore, when vectors were recovered from cells, the proportion of DNA stem–loops that underwent expansions or contractions was below the detection limit of our assay. The apparent resistance to processing stem–loop DNA through expansions or deletions of the stem loop suggests that stem loops, even transcription blocking ones, can exist stably in human cells.
With the system employed in this work, replication of the vector could act as a potential source of RFP fluorescence recovery, but this is unlikely for several reasons. First, the vectors used have no human origins of replication. Second, extensive vector replication, which would complement extrahelical DNA in one daughter molecule and remove it from the other, should lead to recovery of transcription, which was not observed for a DNA loop or stem-loop on the transcribed strand. Third, asymmetrical processing of DNA loops by preferential deletion of the extrahelical region would not be an expected outcome of vector replication as was observed in this work: replication would result in equal expansions and deletions.
Considered in toto, the data presented here do not support a model in which NER or TCNER removes extrahelical DNA from the transcription units of genes. While the in vitro data show that hRNAPII does indeed stall in an elongation-competent conformation at unpaired DNA loops and stem–loops, the observed transcriptional blockage is not sufficient to trigger DNA repair in cells. This suggests that TCNER may well require transcriptional stalling as necessary to trigger repair, but that stalling alone is not sufficient. Importantly, this same system detected the relative contributions of NER and TCNER in removing bulky DNA lesions (34), showing that the system would detect NER and TCNER at extrahelical DNA, were it occurring.
Coupling of transcriptional blockage at DNA damage to subsequent DNA repair requires the CSA and CSB proteins (50,51); but the mechanistic details as to how the coupling is accomplished remain unclear (52). Following RNAPII stalling and association of CSB, NER factors must gain access to the site of the lesion, verify the damage and carry out repair (24,53–55). Recent studies point to the importance of DNA damage verification during NER. The XPC protein complex that recognizes DNA damage can bind to undamaged DNA as well, but the event does not lead to nicking because the cell fails the subsequent mechanistic steps that verify the actual presence of DNA damage (22,56–58). The data reported here indicate that extrahelical DNA can stall the progress of elongating hRNAPII, which is the DNA damage recognition step in TCNER, but the extrahelical DNA is not removed, possibly because it fails the DNA damage verification test as well.
The results reported here might seem somewhat surprising in light of other findings concerning SSR instability. For example, a model that describes CAG expansions and contractions proposes that TCNER is involved (17) since downregulation of the CSB protein product by RNAi significantly decreased the number of transcription-associated CAG contraction events, thus implicating TCNER in the contraction process (17,26). Our data do not contradict this work, but rather suggest that the mechanism of the observed contractions is not likely to proceed through the formation of extrahelical structures alone since processing of extrahelical DNA is not impacted by the CSB or XPA proteins. Additional key DNA repair proteins have since been associated with significantly reduced or increased CAG contractions in human cells, including MSH2, MSH3, PARP1, ERCC1-XPF and FEN1, among others (59–65). Some of these proteins and their respective repair pathways are candidate effectors for the sparse processing of stem loop DNA and significant deletions of unpaired loops observed here. For example, Rad1 protein, which is the yeast homolog of the XPF protein, acts in concert with the mismatch repair protein MSH2 to implement the removal of large DNA loops during meiosis in yeast (66). Importantly, the results reported here do not rule out the involvement of proteins other than XPA and CSB in the deletion of unpaired DNA loops or DNA stem loops. Moreover, this work focused on four model DNA structures, but the space of folded DNA is large. Sequence context and different forms of extrahelical DNA, including stem–loops with unpaired bubble regions, and four-way DNA junctions (Holliday junctions) might behave differently than the model structures investigated here.
The results reported here clearly show that DNA stem–loops and unpaired DNA loops impede transcription in vitro and in vivo (see Figure 10 for a summary of the findings). Additionally, NER does not affect the expansion or contraction of these regions, and while DNA stem–loops block transcription in cells, the event is not sufficient to trigger repair by TCNER. Hence, these experiments rule out a general model of TCNER operating on undamaged DNA that blocks transcription, indicating that this repair pathway does not affect the expansion or contraction of SSRs, quite possibly including the physiologically important CAG repeats. Both CAG and the complementary GTC extrahelical structures are known to form stable DNA stem–loops (65), so these results predict that both structures would be stably maintained in human cells and would not be affected by NER and TCNER. Therefore, the previously observed contributions of NER and TCNER to CAG/CTG repeat instability are not likely due to the formation of extrahelical DNA structures alone.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Professor Nadrian Seeman (New York University) for helpful discussions on forming extrahelical DNA structures. We also thank Professor Suse Broyde (New York University), Dr Aine McCabe (NYU Abu Dhabi) and David Cano for valuable suggestions and comments on an earlier version of this article.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [ES-010581 to D.A.S.]; New York University Abu Dhabi. Funding for open access charge: New York University Abu Dhabi.
Conflict of interest statement. None declared.
REFERENCES
- 1. Richard G.F., Kerrest A., Dujon B.. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol. Mol. Biol. Rev.: MMBR. 2008; 72:686–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W. et al. Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921. [DOI] [PubMed] [Google Scholar]
- 3. Lai Y., Sun F.. The relationship between microsatellite slippage mutation rate and the number of repeat units. Mol. Biol. Evol. 2003; 20:2123–2131. [DOI] [PubMed] [Google Scholar]
- 4. Kozlowski P., de Mezer M., Krzyzosiak W.J.. Trinucleotide repeats in human genome and exome. Nucleic Acids Res. 2010; 38:4027–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tóth G., Gáspári Z., Jurka J.. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 2000; 10:967–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J.. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuroendocrinology. 2011; 72:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. MacDonald M.E., Ambrose C.M., Duyao M.P., Myers R.H., Lin C., Srinidhi L., Barnes G., Taylor S.A., James M., Groot N.. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993; 72:971–983. [DOI] [PubMed] [Google Scholar]
- 8. La Spada A.R., Roling D.B., Harding A.E., Warner C.L., Spiegel R., Hausmanowa-Petrusewicz I., Yee W.-C., Fischbeck K.H.. Meiotic stability and genotype–phenotype correlation of the trinucleotide repeat in X–linked spinal and bulbar muscular atrophy. Nat. Genet. 1992; 2:301–304. [DOI] [PubMed] [Google Scholar]
- 9. Jodice C., Malaspina P., Persichetti F., Novelletto A., Spadaro M., Giunti P., Morocutti C., Terrenato L., Harding A.E., Frontali M.. Effect of trinucleotide repeat length and parental sex on phenotypic variation in spinocerebellar ataxia I. Am. J. Hum. Genet. 1994; 54:959. [PMC free article] [PubMed] [Google Scholar]
- 10. Gemayel R., Vinces M.D., Legendre M., Verstrepen K.J.. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 2010; 44:445–477. [DOI] [PubMed] [Google Scholar]
- 11. Swami M., Hendricks A.E., Gillis T., Massood T., Mysore J., Myers R.H., Wheeler V.C.. Somatic expansion of the Huntington's disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum. Mol. Genet. 2009; 18:3039–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pearson C.E., Nichol Edamura K., Cleary J.D.. Repeat instability: mechanisms of dynamic mutations. Nat. rev. Gen. 2005; 6:729–742. [DOI] [PubMed] [Google Scholar]
- 13. Viterbo D., Michoud G., Mosbach V., Dujon B., Richard G.-F.. Replication stalling and heteroduplex formation within CAG/CTG trinucleotide repeats by mismatch repair. DNA Repair. 2016; 42:94–106. [DOI] [PubMed] [Google Scholar]
- 14. Parsa J.-Y., Ramachandran S., Zaheen A., Nepal R.M., Kapelnikov A., Belcheva A., Berru M., Ronai D., Martin A.. Negative supercoiling creates single-stranded patches of DNA that are substrates for AID–mediated mutagenesis. PLoS Genet. 2012; 8:e1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Saini N., Zhang Y., Usdin K., Lobachev K.S.. When secondary comes first—the importance of non-canonical DNA structures. Biochimie. 2012; 95:117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin Y., Wilson J.H.. Nucleotide excision repair, mismatch repair, and R-loops modulate convergent transcription-induced cell death and repeat instability. PLoS One. 2012; 7:e46807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin Y., Wilson J.H.. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol. Cell. Biol. 2007; 27:6209–6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kovtun I.V., Liu Y., Bjoras M., Klungland A., Wilson S.H., McMurray C.T.. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007; 447:447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin Y., Wilson J.H.. Diverse effects of individual mismatch repair components on transcription-induced CAG repeat instability in human cells. DNA Repair. 2009; 8:878–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tome S., Manley K., Simard J.P., Clark G.W., Slean M.M., Swami M., Shelbourne P.F., Tillier E.R., Monckton D.G., Messer A. et al. MSH3 polymorphisms and protein levels affect CAG repeat instability in Huntington's disease mice. PLoS Genet. 2013; 9:e1003280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fuss J.O., Tainer J.A.. XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase. DNA Repair. 2011; 10:697–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mathieu N., Kaczmarek N., Naegeli H.. Strand-and site-specific DNA lesion demarcation by the xeroderma pigmentosum group D helicase. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:17545–17550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Laat W.L., Jaspers N.G., Hoeijmakers J.H.. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999; 13:768–785. [DOI] [PubMed] [Google Scholar]
- 24. Svejstrup J.Q. Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell Biol. 2002; 3:21–29. [DOI] [PubMed] [Google Scholar]
- 25. Salinas-Rios V., Belotserkovskii B.P., Hanawalt P.C.. DNA slip-outs cause RNA polymerase II arrest in vitro: potential implications for genetic instability. Nucleic Acids Res. 2011; 39:7444–7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin Y., Dion V., Wilson J.H.. Transcription promotes contraction of CAG repeat tracts in human cells. Nat. Struct. Mol. Biol. 2006; 13:179–180. [DOI] [PubMed] [Google Scholar]
- 27. Wang G., Vasquez K.M.. Non-B DNA structure-induced genetic instability. Mutat. Res. 2006; 598:103–119. [DOI] [PubMed] [Google Scholar]
- 28. Kim N., Jinks-Robertson S.. Transcription as a source of genome instability. Nat. Rev. Genet. 2012; 13:204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dimitri A., Burns J.A., Broyde S., Scicchitano D.A.. Transcription elongation past O6-methylguanine by human RNA polymerase II and bacteriophage T7 RNA polymerase. Nucleic Acids Res. 2008; 36:6459–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bruzel A., Cheung V.G.. DNA reassociation using oscillating phenol emulsions. Genomics. 2006; 87:286–289. [DOI] [PubMed] [Google Scholar]
- 31. Burns J.A. RNA Polymerase II Transcription: Effects of DNA Damage and DNA Secondary Structure on Elongation and Fidelity. 2012; NY: New York University; Doctoral dissertation. [Google Scholar]
- 32. Perlow R.A., Schinecker T.M., Kim S.J., Geacintov N.E., Scicchitano D.A.. Construction and purification of site-specifically modified DNA templates for transcription assays. Nucleic Acids Res. 2003; 31:e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krueger C., Danke C., Pfleiderer K., Schuh W., Jack H.M., Lochner S., Gmeiner P., Hillen W., Berens C.. A gene regulation system with four distinct expression levels. J. Gene Med. 2006; 8:1037–1047. [DOI] [PubMed] [Google Scholar]
- 34. Nadkarni A., Burns J.A., Gandolfi A., Chowdhury M.A., Cartularo L., Berens C., Geacintov N.E., Scicchitano D.A.. Nucleotide excision repair and transcription-coupled DNA repair abrogate the impact of DNA damage on transcription. J. Biol. Chem. 2016; 291:848–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Enoiu M., Long D.T., Walter J.C., Schärer O.D.. DNA Repair Protocols. 2012; Springer; 203–219. [Google Scholar]
- 36. Sambrook J., Russell D.W.. Purification of closed circular DNA by equilibrium centrifugation in CsCl-ethidium bromide gradients: continuous gradients. Cold Spring Harbor Protocols. 2006; 2006, doi:10.1101/pdb.prot3927. [DOI] [PubMed] [Google Scholar]
- 37. R Core Team R: A Language and Environment for Statistical Computing. 2016; Vienna: R Foundation for Statistical Computing. [Google Scholar]
- 38. Ellis B., Haaland P., Hahne F., Le Meur N., Gopalakrishnan N.. flowCore: basic structures for flow cytometry data. 2009; R package version 1.
- 39. Wickham H. ggplot2: Elegant Graphics for Data Analysis. 2009; Springer Publishing Company; Incorporated. [Google Scholar]
- 40. Lee H.-W., Lee H.-J., Hong C.-M., Baker D.J., Bhatia R., O’Connor T.R.. Monitoring repair of DNA damage in cell lines and human peripheral blood mononuclear cells. Anal. Biochem. 2007; 365:246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stary A., Menck C.F., Sarasin A.. Description of a new amplifiable shuttle vector for mutagenesis studies in human cells: application to N-methyls-N′-nitro-N-nitrosoguanidine-induced mutation spectrum. Mutat. Res./Environ. Mutagen. Related Subj. 1992; 272:101–110. [DOI] [PubMed] [Google Scholar]
- 42. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Perlow R.A., Kolbanovskii A., Hingerty B.E., Geacintov N.E., Broyde S., Scicchitano D.A.. DNA adducts from a tumorigenic metabolite of benzo[a]pyrene block human RNA polymerase II elongation in a sequence- and stereochemistry-dependent manner. J. Mol. Biol. 2002; 321:29–47. [DOI] [PubMed] [Google Scholar]
- 44. Jung Y., Lippard S.J.. RNA polymerase II blockage by cisplatin-damaged DNA Stability and polyubiquitylation of stalled polymerase. J. Biol. Chem. 2006; 281:1361–1370. [DOI] [PubMed] [Google Scholar]
- 45. Tornaletti S., Reines D., Hanawalt P.C.. Structural characterization of RNA polymerase II complexes arrested by a cyclobutane pyrimidine dimer in the transcribed strand of template DNA. J. Biol. Chem. 1999; 274:24124–24130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reinberg D., Roeder R.. Factors involved in specific transcription by mammalian RNA polymerase II. Transcription factor IIS stimulates elongation of RNA chains. J. Biol. Chem. 1987; 262:3331–3337. [PubMed] [Google Scholar]
- 47. Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012; 149:1438–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tornaletti S. Transcription arrest at DNA damage sites. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2005; 577:131–145. [DOI] [PubMed] [Google Scholar]
- 49. Sarker A.H., Tsutakawa S.E., Kostek S., Ng C., Shin D.S., Peris M., Campeau E., Tainer J.A., Nogales E., Cooper P.K.. Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: insights for transcription-coupled repair and Cockayne Syndrome. Mol. Cell. 2005; 20:187–198. [DOI] [PubMed] [Google Scholar]
- 50. Orren D.K., Dianov G.L., Bohr V.A.. The human CSB (ERCC6) gene corrects the transcription-coupled repair defect in the CHO cell mutant UV61. Nucleic Acids Res. 1996; 24:3317–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tu Y., Bates S., Pfeifer G.P.. The transcription-repair coupling factor CSA is required for efficient repair only during the elongation stages of RNA polymerase II transcription. Mutat. Res./Fundam. Mol. Mech. Mutagen. 1998; 400:143–151. [DOI] [PubMed] [Google Scholar]
- 52. Marteijn J.A., Lans H., Vermeulen W., Hoeijmakers J.H.. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 2014; 15:465–481. [DOI] [PubMed] [Google Scholar]
- 53. Walmacq C., Cheung A., Kireeva M.L., Lubkowska L., Ye C., Gotte D., Strathern J.N., Carell T., Cramer P., Kashlev M.. Mechanism of translesion transcription by RNA polymerase II and its role in cellular resistance to DNA damage. Mol. Cell. 2012; 46:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Laine J.P., Egly J.M.. Initiation of DNA repair mediated by a stalled RNA polymerase IIO. EMBO J. 2006; 25:387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tremeau-Bravard A., Riedl T., Egly J.M., Dahmus M.E.. Fate of RNA polymerase II stalled at a cisplatin lesion. J. Biol. Chem. 2004; 279:7751–7759. [DOI] [PubMed] [Google Scholar]
- 56. Luijsterburg M.S., von Bornstaedt G., Gourdin A.M., Politi A.Z., Moné M.J., Warmerdam D.O., Goedhart J., Vermeulen W., van Driel R., Höfer T.. Stochastic and reversible assembly of a multiprotein DNA repair complex ensures accurate target site recognition and efficient repair. J. Cell Biol. 2010; 189:445–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Naegeli H., Sugasawa K.. The xeroderma pigmentosum pathway: decision tree analysis of DNA quality. DNA Repair. 2011; 10:673–683. [DOI] [PubMed] [Google Scholar]
- 58. Mathieu N., Kaczmarek N., Rüthemann P., Luch A., Naegeli H.. DNA quality control by a lesion sensor pocket of the xeroderma pigmentosum group d helicase subunit of TFIIH. Curr. Biol. 2013; 23:204–212. [DOI] [PubMed] [Google Scholar]
- 59. Dion V., Lin Y., Hubert L. Jr, Waterland R.A., Wilson J.H.. Dnmt1 deficiency promotes CAG repeat expansion in the mouse germline. Hum. Mol. Genet. 2008; 17:1306–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hubert L. Jr, Lin Y., Dion V., Wilson J.H.. Xpa deficiency reduces CAG trinucleotide repeat instability in neuronal tissues in a mouse model of SCA1. Hum. Mol. Genet. 2011; 20:4822–4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hubert L. Jr, Lin Y., Dion V., Wilson J.H.. Topoisomerase 1 and single-strand break repair modulate transcription-induced CAG repeat contraction in human cells. Mol. Cell. Biol. 2011; 31:3105–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lin Y., Dent S.Y., Wilson J.H., Wells R.D., Napierala M.. R loops stimulate genetic instability of CTG.CAG repeats. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:692–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mittelman D., Sykoudis K., Hersh M., Lin Y., Wilson J.H.. Hsp90 modulates CAG repeat instability in human cells. Cell Stress Chaperones. 2010; 15:753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hou C., Chan N.L., Gu L., Li G.-M.. Incision-dependent and error-free repair of (CAG) n/(CTG) n hairpins in human cell extracts. Nat. Struct. Mol. Biol. 2009; 16:869–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lai Y., Budworth H., Beaver J.M., Chan N.L., Zhang Z., McMurray C.T., Liu Y.. Crosstalk between MSH2–MSH3 and polβ promotes trinucleotide repeat expansion during base excision repair. Nat. Commun. 2016; 7:12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kirkpatrick D.T., Petes T.D.. Repair of DNA loops involves DNA-mismatch and nucleotide-excision repair proteins. Nature. 1997; 387:929–931. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.