Abstract
TARS and TARS2 encode cytoplasmic and mitochondrial threonyl-tRNA synthetases (ThrRSs) in mammals, respectively. Interestingly, in higher eukaryotes, a third gene, TARSL2, encodes a ThrRS-like protein (ThrRS-L), which is highly homologous to cytoplasmic ThrRS but with a different N-terminal extension (N-extension). Whether ThrRS-L has canonical functions is unknown. In this work, we studied the organ expression pattern, cellular localization, canonical aminoacylation and editing activities of mouse ThrRS-L (mThrRS-L). Tarsl2 is ubiquitously but unevenly expressed in mouse tissues. Different from mouse cytoplasmic ThrRS (mThrRS), mThrRS-L is located in both the cytoplasm and nucleus; the nuclear distribution is mediated via a nuclear localization sequence at its C-terminus. Native mThrRS-L enriched from HEK293T cells was active in aminoacylation and editing. To investigate the in vitro catalytic properties of mThrRS-L accurately, we replaced the N-extension of mThrRS-L with that of mThrRS. The chimeric protein (mThrRS-L-NT) has amino acid activation, aminoacylation and editing activities. We compared the activities and cross-species tRNA recognition between mThrRS-L-NT and mThrRS. Despite having a similar aminoacylation activity, mThrRS-L-NT and mThrRS exhibit differences in tRNA recognition and editing capacity. Our results provided the first analysis of the aminoacylation and editing activities of ThrRS-L, and improved our understanding of Tarsl2.
INTRODUCTION
As one of the essential components for translation of the genetic code, aminoacyl-tRNA synthetases (aaRSs) catalyze the aminoacylation of their cognate tRNAs (1–3). Mammalian cells contain two separate translation machineries in the cytoplasm and mitochondria. In the human nuclear genome, there are 37 genes encoding all the aaRSs for the two machineries (4,5). Cytoplasmic and mitochondrial aaRSs are usually encoded by two different nuclear genes, except for lysyl- and glycyl-tRNA synthetases (LysRS and GlyRS), which functioning in both subcellular areas and are encoded by KARS and GARS, respectively (5).
We have revealed the existence of two human cytoplasmic ThrRSs, hThrRS and hThrRS-like (hThrRS-L), which are encoded by genes TARS and TARSL2, respectively (6). In mouse, the corresponding genes are Tars (ID: 110960) and Tarsl2 (ID: 272396). They are separate genes and are located on different chromosomes. In our previous report, phylogenetic analysis revealed that the similarity between ThrRS-L and ThrRS is higher in different mammalian cells compared with lower eukaryotes, bacteria and mitochondria, indicating that Tarsl2 is likely derived from Tars gene duplication, rather than from horizontal gene transfer of a bacterial ThrRS gene or a mitochondrial ThrRS (mtThrRS) gene (6). Detailed primary sequence alignment of the three mouse ThrRSs is shown in Supplementary Figure S1 (6).
Aminoacylation usually proceeds in two steps, amino acid activation and transfer of the amino acid to tRNA (1). Aminoacylation requires remarkable accuracy for translational fidelity. However, accurate aminoacylation is difficult to maintain for some aaRSs because of their inability to accurately select and recognize cognate amino acids, as opposed to their non-cognate analogs. A proofreading mechanism (editing) has evolved in some aaRSs to ensure translation quality (7). Editing steps occur on the formation of misaminoacyl-AMP (in pre-transfer editing) and/or misaminoacyl-tRNA (in post-transfer editing) (7). Pre-transfer editing can be classified into tRNA-independent and tRNA-dependent pre-transfer editing, according to whether tRNA is required for hydrolysis of aminoacyl-AMP (7). The post-transfer editing reaction is usually catalyzed by a particular domain, such as the N2 domain of ThrRS, or the CP1 domain of leucyl-tRNA synthetase (LeuRS) (8–13). Aminoacylation and editing of aaRSs are essential capacities for translation quality control and maintaining normal cell metabolism.
Bacterial ThrRSs, such as Escherichia coli ThrRS (EcThrRS), comprise an N1 domain (regulating the editing function), an N2 domain (editing), an aminoacylation domain and a C-terminal tRNA binding domain (CTD) (14). However, eukaryotic ThrRSs have evolved an extra ThrRS-specific N-extension (Figure 1). Mammalian ThrRS and ThrRS-L show high sequence consistency in their N1, N2, aminoacylation and CTD domains (76.4% identity between mouse ThrRS and ThrRS-L), but differ in their N-extensions. The N-extension of mouse cytoplasmic ThrRS (mThrRS, containing 722 amino acid residues) consists of 82 amino acid residues, while that of mouse ThrRS-L (mThrRS-L, containing 790 amino acid residues) is a longer peptide with 150 amino acid residues (Figure 1 and Supplementary Figure S1). Recently, human ThrRS-L (hThrRS-L) has been reported to be a component of the multiple-synthetase complex in human embryonic kidney 293T (HEK293T) cells, according to affinity purification-mass spectrometry (MS) analysis (15,16). However, the tissue expression pattern, level and cellular distribution of ThrRS-L in cells remain unexplored. Furthermore, based on the observation that ThrRS-L shares conserved aminoacylation and editing domains with ThrRS, it is an open question whether ThrRS-L has tRNA aminoacylation and editing activities.
Figure 1.
Domain arrangement of bacterial/eukaryotic ThrRSs. Domain arrangement of EcThrRS, human cytoplasmic ThrRS (hThrRS), human ThrRS-L (hThrRS-L), mouse cytoplasmic ThrRS (mThrRS) and mouse ThrRS-L (mThrRS-L). Various domains of ThrRSs or ThrRS-Ls (including N1, N2 editing, aminoacylation and CTD domains) are presented in white or diagonal boxes, while the N-extension of ThrRSs or ThrRS-Ls is colored gray or black, respectively. The domain arrangement of mThrRS-L-NT is also shown and the lengths of enzymes and N-extensions are indicated. The identity of the main body or different domains between hThrRS and hThrRS-L or mThrRS and mThrRS-L is shown.
In the present study, we studied the expression of Tarsl2, including the levels of mRNA and protein, the subcellular localization and the catalytic properties of ThrRS-L. Our results showed that ThrRS-L could catalyze aminoacylation of tRNA like ThrRS. We compared the aminoacylation and editing activities of ThrRS-L and ThrRS from mouse and reported the kinetic properties of mammalian ThrRS-L in vitro for the first time.
MATERIALS AND METHODS
Materials
L-Thr, L-Ser, dithiothreitol (DTT), ribonucleoside triphosphate (NTP), guanosine monophosphate (GMP), tetrasodium pyrophosphate, pyrophosphatase (PPiase), Tris-base, MgCl2, NaCl, activated charcoal, horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Sigma (St Louis, MO, USA). [14C]Thr, [14C]Ser and [α-32P]ATP were obtained from Perkin Elmer Inc. (Waltham, MA, USA). KOD-plus mutagenesis kits, ReverTra Ace quantitative polymerase chain reaction (qPCR) RT Kit and SYBR Green Realtime PCR Master Mix were obtained from TOYOBO (Osaka, Japan). Dynabeads protein G, Lipofectamine 2000 transfection reagent, and the protein synthesis kit (1-Step Human Coupled IVT Kit) were obtained from Thermo Scientific (Waltham, MA, USA). Qproteome nuclear protein kits and Ni2+-NTA Superflow resin were purchased from Qiagen Inc. (Hilden, Germany). Polyethyleneimine cellulose plates were purchased from Merck (Darmstadt, Germany).
Antibodies
The N-extension of mThrRS-L (Met1-Val149) was used as an epitope to raise rabbit polyclonal antibodies against mThrRS-L (Abclonal, China). The anti-FLAG (M20008L) antibody was purchased from Abmart (Shanghai, China). Anti-His6-tag (SAB4301134) and anti-GlyRS (HPA019097) antibodies were from Sigma. Anti-Histone 3.1 (9715) and anti-α-Tubulin (3873S) antibodies were obtained from Cell Signaling Technology (Danvers, MA, USA).
Cloning, expression, protein purification, tRNA transcription and expression
Genes encoding mThrRS and mThrRS-L were amplified from cDNA, obtained by reverse-transcription PCR from total RNA extracted from NIH/3T3 cells. Supplementary Table S2 summarizes all gene constructs. Gene mutagenesis was performed according to the protocol provided with the KOD-plus mutagenesis kit. Proteins were purified using a previously reported method (17). In vitro mouse tRNAThrs (mtRNAThrs), including mtRNAThr(AGU), -(CGU) and -(UGU) transcripts were obtained using the T7 RNA polymerase run-off procedure, as previously described (18). The gene encoding EctRNAThr(UGU) was inserted into pTrc99b and the construct was transformed into E. coli MT102 for tRNA gene overexpression, as described previously (19).
Cell culture and transfection
NIH/3T3 cells were cultured in Dulbecco’s modified Eagle’s medium (high glucose) supplemented with 10% fetal bovine serum in a 37°C incubator with 5% CO2. Dishes (100 mm) of NIH/3T3 cells were transfected with 30 μg of plasmid using the Lipofectamine 2000 transfection reagent at a ratio of 1:2.5, according to the manufacturer’s protocol. Twenty-four hours after transfection, the cells were washed with 5 ml of ice-cold phosphate-buffered saline (PBS) twice, and lysed with 1 ml of ice-cold lysis buffer [50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 5 mM ethylenediaminetetraacetic acid, 1% Triton X-100] supplemented with a protease inhibitor cocktail. The supernatant was collected using centrifugation at 12 000 × g for 30 min. All procedures were performed on ice.
Immunoprecipitation
Whole cell lysate was incubated with the anti-FLAG antibody with agitation overnight, and then the mixture was incubated with Dynabeads protein G for 3 h. Recovered immune complexes were washed three times with ice-cold PBS plus 0.05% Tween-20 (PBST) buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 and 0.5‰ Triton X-100). All procedures are performed at 4°C. Proteins were eluted from the beads in 2 × protein loading buffer (100 mM Tris–HCl, 4% sodium dodecyl sulphate, 0.2% bromophenol blue, 20% glycerol, 200 mM DTT).
Nuclear fractionation
After the cells were collected, the cytoplasmic and nuclear fractions were separated and extracted using the Qproteome nuclear protein kit (Qiagen) according to the manufacturer's protocol. Briefly, the cells were swelled in a hypotonic buffer and detergent was added to rupture the plasma membrane. The cell lysate was centrifuged to separate the cytosolic fraction. A hypertonic buffer was then used to extract the nucleic-acid-binding proteins, and the benzonase nuclease was used to digest genomic DNA to obtain the insoluble nuclear proteins. The nucleic-acid-binding proteins and insoluble nuclear proteins were combined together as the nuclear fraction.
Western blotting
Proteins in the whole cell lysate, or different cell fraction extracts, were separated by 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene fluoride (PVDF) membranes. The PVDF membrane was then cropped to blot different proteins in the same lane. After blocking with 5% (w/v) non-fat dried milk, the membranes with targeted proteins were incubated with the corresponding primary antibodies overnight at 4°C. Membranes were then washed three times with PBST, and incubated with HRP-conjugated secondary antibody at room temperature for 30 min. After washing three times with PBST, the membranes were treated with the chemiluminescent substrate, and imaging was performed using the LAS-4000 system (GE, CA, USA). The quantitative analysis was carried out by measuring the gray value of the main blot band in the MultiGauge software platform (Fujifilm, Tokyo, Japan).
Epifluorescence
HEK293T cells were transfected with specific plasmids. After 8 h, cells were collected in 1 ml of PBS, washed twice by PBS, and pelleted by centrifugation at 400 × g at 4°C. After being fixed in 4% paraformaldehyde at room temperature for 30 min, the cells were permeated in 0.2% Triton X-100 for 5 min in ice and subsequently stained in 0.1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI). The cells were plated on glass slides and the epifluorescence of GFP was observed by super-resolution structured illumination microscopy (Zeiss, Oberkochen, Germany).
RNA extraction, reverse transcription (RT)-PCR and quantitative real-time PCR (qPCR)
Total RNAs from mouse tissues and cultured cells were extracted using the Trizol Reagent (Thermo Fisher) according to the user's manual. cDNAs were reverse transcribed using ReverTra Ace qPCR RT Kit (TOYOBO) and then applied for qPCR analysis. Gapdh mRNA was examined as an internal control for normalization. The relative expression of each examined gene was determined with triplicate experiments. qPCR analysis was carried on with the SYBR Green Realtime PCR Master Mix (TOYOBO). The sequences of the primers for Gapdh, Tars and Tarsl2 are listed in Supplementary Table S3.
32P-labeling of tRNA
32P-labeling of EctRNAThr was performed at 37°C in a mixture containing 60 mM Tris–HCl (pH 8.0), 12 mM MgCl2, 15 μM tRNA, 0.5 mM DTT, 15 μM adenosine triphosphate (ATP), 50 μM tetrasodium pyrophosphate, 0.666 μM [α-32P]ATP and 10 μM E. coli CCA-adding enzyme (CCase) for 5 min, as described previously (20).
ATP-PPi exchange
The amino acid activation activity of aaRSs was assayed by an ATP-PPi exchange reaction (21). The kinetics of mThrRS or mThrRS-L-NT (the design of mThrRS-L-NT is described in detail in the fourth part of the RESULTS) in ATP-PPi exchange were determined at 37°C in a reaction containing 60 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 5 mM DTT, 0.1 mg/ml bovine serum albumin (BSA), 2.5 mM ATP, 2 mM tetrasodium [32P]pyrophosphate and 100 nM enzyme with 0–15 mM Thr (for mThrRS), 0–100 mM Thr (for mThrRS-L-NT) or 0–1000 mM Ser.
Aminoacylation
According to a previously described procedure (6), time course curves of aminoacylation by mThrRS, mThrRS-L-NT or human mitochondrial ThrRS (hmtThrRS) were determined at 37°C in a reaction mixture (30 μl) containing 60 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 5 mM DTT, 2.5 mM ATP, 114 μM [14C]Thr and 200 nM enzyme with 5 μM of various tRNAs. The time course curve for aminoacylation by EcThrRS for various tRNAs was determined in similar conditions, except with 20 mM MgCl2 and 30 mM KCl added. The samples were processed as described previously (6).
As signal from [32P]AMP or aminoacyl-[32P]AMP is sensitive and easy to detect, we used [32P]EctRNAThr(UGU) to determine the kinetics, as follows. The kinetics of mThrRS or mThrRS-L-NT for Thr were determined in a reaction mixture containing 60 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 5 mM DTT, 2.5 mM ATP, 0.5 μM [32P]EctRNAThr(UGU) (66000 cpm/μl), 10 μM unlabeled EctRNAThr(UGU), 46.875–1500 μM Thr and 75 nM enzyme. For ATP, 1.5 mM Thr and 0.104–7.5 mM ATP were used. For EctRNAThr(UGU), 1.5 mM Thr, 2.5 mM ATP and 0.5–15.5 μM unlabeled EctRNAThr were used. After various incubation intervals at 37°C, 9-μl aliquots were taken for ethanol precipitation with NaAc (pH 5.2) at −20°C overnight. The samples were processed as described previously (20). The amount of aminoacyl-[32P]AMP produced was calculated by multiplying the total amount of tRNAThr (including [32P]EctRNAThr(UGU) and unlabeled tRNAThr) by the relative level of charged tRNAThr in the aliquots: [aminoacyl-[32P]AMP/(aminoacyl-[32P]AMP + [32P]AMP)].
Post-transfer editing
Mischarged mouse tRNAThr(AGU) ([14C]Ser-tRNAThr) was prepared by editing-deficient EcThrRS-H73A-H77A (8). Post-transfer editing by mThrRS or mThrRS-L-NT, or their mutants, was carried out in a reaction mixture containing 60 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 5 mM DTT, 0.1 mg/ml BSA, 1 μM [14C]Ser-tRNAThr and 0.1 μM enzyme at 37°C and measured as previously described (22).
AMP formation assay
The Adenosine monophosphate (AMP) formation assay was carried out by thin layer chromatography (TLC) at 37°C in a reaction mixture containing 60 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 10 U/ml PPiase, 40 mM Ser, 3 mM [α-32P]ATP, and 2 μM mThrRS, mThrRS-L-NT or variants, in the presence or absence of mouse tRNAThr(AGU) as described previously (20). The rate of non-enzymatic hydrolysis of adenylate was measured in pulse-chase experiments (6,23). In detail, a solution of 2 μM mThrRS or mThrRS-L-NT was first incubated with 60 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 10 U/ml PPiase, 40 mM Ser, 100 μM ATP and 0.25 μM [α-32P]ATP for 10 min at 37°C to prepare Ser-[32P]AMP. Subsequently, unlabeled ATP (50 mM, equal to a 500-fold molar excess of initial unlabeled ATP) was added and the hydrolysis activity was quenched at various time points.
Enrichment of wild-type and mutant mThrRS-Ls from their overexpressing HEK293T cells and determination of their aminoacylation and editing activities
HEK293T cells in two 150 mm dishes were transfected with each plasmid encoding the wild-type or mutant mThrRS-Ls by the calcium phosphate transfection method. Cells were lysed after 24 h and the supernatant was incubated with 50 μl of slurry beads covalently attached with the anti-FLAG antibody (ANTI-FLAG M2 Affinity Gel, Sigma) for 3 h. After washing three times, the packed beads were resuspended in 30 μl working buffer [60 mM Tris–HCl (pH 7.5), 10 mM MgCl2] for aminoacylation activity determination. The amounts of the enriched enzyme on beads were normalized based on the western blotting (WB) assay result.
Animal work
Mice from the Institute of Cancer Research were used in this study. All experimental animal procedures were approved by the Institutional Animal Care and Research Advisory Committee at the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. All methods were performed in accordance with the relevant guidelines and regulations of the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences.
RESULTS
Tarsl2 is ubiquitously expressed
hThrRS-L has been detected in HEK293T cells by MS (15). To observe mouse tissue-specific expression of Tarsl2, we prepared a polyclonal antibody (anti-mThrRS-L) by immunizing rabbits with its N-extension (Met1-Val149). Anti-mThrRS-L only reacts with mThrRS-L, but not mThrRS (Supplementary Figure S2). Male mice were killed and total proteins and RNAs from various tissues including the lung, spleen, fat, stomach, heart, muscle, testis, brain, kidney and liver were extracted. The result of WB assays showed that mThrRS-L was present in all tested mouse tissues with different levels (Figure 2A and B). Large amounts of mThrRS-L were observed in the muscle and heart and lower amounts in the lung and fat, hinting that mThrRS-L is possibly involved in muscle development and/or energy metabolism. It is possible that high levels of protein synthesis in these organs require high amounts of functional ThrRSs.
Figure 2.
Protein expression levels of mThrRS-L in various mouse tissues. (A) A typical image showing the protein level of mThrRS-L in various tissues using α-Tubulin as a loading control. (B) Quantification of relative protein levels of mThrRS-L, based on α-Tubulin. The ratio of mThrRS-L to α-Tubulin (mThrRS-L/α-Tubulin) in the lung was designated as 1; the value of the ratio in other tissues relative to that in lung is indicated on the top of each bar (values represent average levels from three independent WB analyses with SD indicated).
We quantified the relative amount of mRNA transcribed from Tars and Tarsl2 by quantitative real-time PCR (qPCR), using Gapdh as a control. The data showed that the mRNA level of Tarsl2 differed among various tissues, and the expression level was much higher in the testis or spleen (Supplementary Table S1). Clearly, the levels of protein and mRNA are not coincident, possibly because translation is differentially regulated at the post-transcription level in various tissues. Furthermore, by calculating Ct difference values (2−ΔΔCt) (24), we showed that the mRNA level of Tarsl2 was significantly lower than that of Tars in various tissues of mouse, with the ratio differing from 3.4 to 100. Similar results were also observed in different mouse cell lines, with ∼125-fold more mRNA of Tars than that of Tarsl2 (Supplementary Table S1).
Overall, our data showed that Tarsl2 was expressed in mouse tissues and its mRNA expression level was notably lower than that of Tars.
ThrRS-L is located in both the cytoplasm and nucleus
To determine the cellular localization of mThrRS-L in NIH/3T3 and GC2 cells, we separated the cytoplasmic and nuclear fractions of NIH/3T3 cells. WB analysis showed that mThrRS-L is mainly located in the cytoplasm and a minor fraction is located in the nucleus in both mouse somatic cells (NIH/3T3, ∼7.3%) and germ cells (GC2, ∼6.7%) (Figure 3A). By contrast, as a cytoplasmic protein control, mouse GlyRS (mGlyRS) was exclusively detected in the cytoplasmic fraction using an anti-mGlyRS antibody (anti-mThrRS antibodies were not successfully obtained) (Figure 3A).
Figure 3.
Subcellular localization of native or overexpressed mThrRS and mThrRS-L. Cytoplasmic (C) and nuclear (N) fractions were separated from NIH/3T3 cells or NIH/3T3 cells overexpressing mThrRS-L-FLAG or mThrRS-FLAG. The localization of native mThrRS-L in NIH/3T3 or GC2 (A), overexpressed mThrRS-L-FLAG, or mThrRS-FLAG in NIH/3T3 (B) was detected using anti-mThrRS-L or anti-FLAG antibodies. Note that a qualified anti-mThrRS antibody was not obtained, thus mGlyRS was detected using anti-mGlyRS in (A) as a comparison. α-Tubulin and Histone 3.1 were used as indicators of the cytoplasmic and nuclear fractions, respectively. The distribution pattern of α-Tubulin and Histone 3.1 between NIH/3T3 cells overexpressing mThrRS-L-FLAG or mThrRS-FLAG is the same; thus, only the localization of α-Tubulin and Histone 3.1 from NIH/3T3 cells overexpressing mThrRS-L-FLAG is shown in (B). The gene encoding mThrRS-L-EGFP, mThrRS-EGFP, mThrRS-L-Δ149-EGFP or mThrRS-L-A4-EGFP was overexpressed in HEK293T cells and the localization of each protein was observed. The nucleus was stained by DAPI (C).
Furthermore, we introduced a gene encoding C-terminal FLAG-tagged mThrRS (mThrRS-FLAG) or mThrRS-L (mThrRS-L-FLAG) into NIH/3T3 cells, and separated the cytoplasmic and nuclear fractions. A small amount of overexpressed mThrRS-L-FLAG (∼25.0%) was located in the nucleus, while overexpressed mThrRS-FLAG was exclusively located in the cytoplasm (Figure 3B). In addition, we constructed two genes encoding proteins fusing the C-terminus of mThrRS-L and mThrRS with EGFP (mThrRS-L-EGFP and mThrRS-EGFP) and then expressed them in HEK293T cells, separately. Epifluorescence assays showed that mThrRS-L-EGFP was distributed in both the cytoplasm and nucleus, while mThrRS-EGFP was only present in the cytoplasm (Figure 3C).
The above data revealed that ThrRS-L has the potential to function in both the cytoplasm and the nucleus.
ThrRS-L has a nuclear localization sequence (NLS) at its C-terminus
On account of the nuclear localization of mThrRS-L-FLAG but not mThrRS-FLAG, we hypothesized that the specific N-extension might mediate its nuclear localization. Based on the sequence alignment of various ThrRSs, the N-extension of mThrRS-L includes the terminal 149 amino acid residues (Supplementary Figure S1, Val149 of mThrRS-L indicated). To identify the NLS, we firstly deleted the full-length N-extension or nearly half of the N-extension of mThrRS-L to obtain truncated mutants mThrRS-L-Δ149 and mThrRS-L-Δ82, respectively (Supplementary Figure S1, Glu82 of mThrRS-L indicated). Their genes were transfected and expressed in NIH/3T3 cells, and fractions of the transfected cells were assayed. Data from the WB assay showed that mThrRS-L-Δ149 was absent from the nuclear fraction, while mThrRS-L-Δ82 was still present in the nuclear fraction (Figure 4A). In addition, epifluorescence analysis apparently showed that overexpressed mThrRS-L-Δ149-EGFP was only located in the cytoplasm of the host cells (Figure 3C). To identify a possible NLS with clusters of basic residues, we mutated all Lys in the probable string of Lys from Ala83 to Val149 of mThrRS-L; however, because all mutants (including mThrRS-L-143AA144 and mThrRS-L-101AAAALA106) still entered the nucleus, we concluded that the NLS is not in this region (data not shown). In addition, through sequence alignment, we noticed a string of basic residues that is only present in the C-terminus of ThrRS-Ls, but not ThrRSs, separate from the high homology in their N1, N2, aminoacylation and CTD domains. The four Lys residues (K774, K776, K779 and K780) in the sequence 774KLKNLKK780 of mThrRS-L were homologous with the four basic residues (K786, K788, R791 and K792) in 786KLKNLRK792 of hThrRS-L, and the four basic residues are conserved in all known ThrRS-Ls (Figure 4B). Therefore, when the four basic residues were simultaneously substituted with Ala, the mutant (mThrRS-L-A4) nearly disappeared from the nucleus as compared with wild-type mThrRS-L (Figure 4C). Correspondingly, mThrRS-L-A4-EGFP was only observed in the cytoplasm of HEK293T cells (Figure 3C). The above data indicated that the NLS of mThrRS-L is located its C-terminal region. Based on the crystal structures of both EcThrRS (PDB 1QF6) (14) and hThrRS (PDB 4TTV) (25), this peptide is located in the last α-helix of the CTD domain and at the domain surface, suggesting its suitability as an NLS. Meanwhile, the fact that mThrRS-L-Δ149 (lacking the entire N-extension) failed to enter the nucleus, suggested that the N-extension of ThrRS-L probably mediates a crucial protein-protein interaction during nuclear entry.
Figure 4.
Identification of the NLS of mThrRS-L. (A) Subcellular localization analysis of wild-type mThrRS-L, partial N-extension deletion mutant (mThrRS-L-Δ82) or nearly full-length N-extension deletion mutant (mThrRS-L-Δ149). (B) Sequence alignment of the C-terminus of ThrRSs and ThrRS-Ls from various species, highlighting the Lys-rich stretch in ThrRS-Ls. Nl, Nomascus leucogenys; Cf, Canis familiaris; Ss, Sus scrofa; Bt, Bos taurus; Mm, Macaca mulatta. (C) Subcellular localization analysis of wild-type mThrRS-L or mThrRS-L-A4-FLAG. α-Tubulin and Histone 3.1 were used as indicators of the cytoplasmic and nuclear fractions, respectively.
Native mThrRS-L is active in aminoacylation and editing
To understand whether native mThrRS-L is able to catalyze aminoacylation and editing reactions as a bone fide ThrRS, we first overexpressed the gene encoding FLAG-tagged mThrRS-L in HEK293T cells. Immunoprecipitation was performed to enrich the FLAG-tagged mThrRS-L (Figure 5A). The results showed that the immunoprecipitated FLAG-tagged mThrRS-L was indeed able to synthesize Thr-tRNAThr (Figure 5B). To confirm that the reaction was catalyzed by the mThrRS-L, two absolutely conserved residues in the active site of ThrRSs, according to the crystal structure of E. coli ThrRS (PDB 1QF6), Cys480 (EcThrRS Cys334 counterpart, forming a zinc chelation pocket for Thr side-chain binding) and Arg509 (EcThrRS Arg363 counterpart, binding the α-phosphate of Thr-AMP) (14), were mutated to Ala, respectively (mThrRS-L-C480A or mThrRS-L-R509A). The aminoacylation activity of both the mutants was obviously decreased (Figure 5B). The remaining activity of the sample containing mThrRS-L-C480A or mThrRS-L-R509A likely resulted from the co-precipitated hThrRS and/or hThrRS-L, which could form heterodimers or homodimers with mThrRS-L (second lane from left, Figure 5A), because the dimer interface of ThrRS locates in the aminoacylation domains and is conserved in ThrRS and ThrRS-L (26). We also found that the enriched FLAG-tagged mThrRS-L was able to hydrolyze pre-formed Ser-tRNAThr (Figure 5C), synthesized by editing-defective EcThrRS-H73A-H77A (10). After mutating the two His counterparts (His222 and His226) in the potential active site of mThrRS-L, the editing activity of mThrRS-L-H222A-H226A decreased obviously (Figure 5C), but with nearly intact aminoacylation activity (Figure 5B). Again, the remaining editing activity was likely derived from the co-immunoprecipitated hThrRS and/or hThrRS-L. The above data clearly showed that native mThrRS-L is able to catalyze both aminoacylation and editing reactions with similar mechanism to canonical ThrRSs. It is difficult to accurately determine the concentration of mThrRS-L or its mutants in the immunoprecipitated products, which also contain other co-precipitated proteins (Figure 5A), the data only revealed the qualitative abilities of mThrRS-L in aminoacylation and editing.
Figure 5.
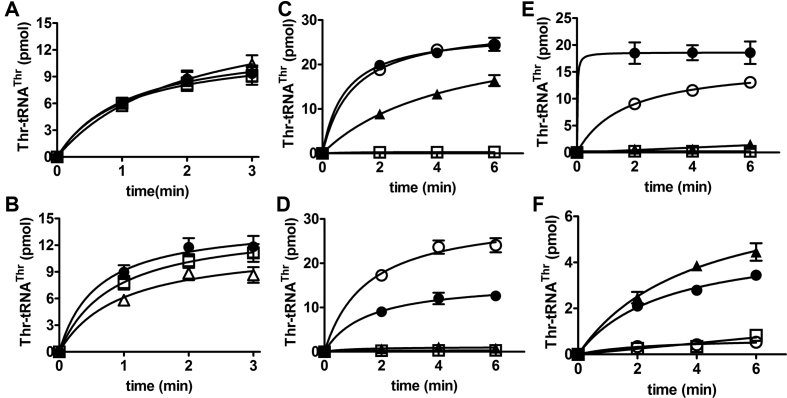
Native mThrRS-L enriched from HEK293T cells has aminoacylation and editing activities. The empty vector pCMV-3Tag-3A was transfected as a negative control (no enzyme). (A) WB assay of immunoprecipitated proteins. hThrRS and hThrRS-L were co-immunoprecipitated; α-Tubulin was immunoblotted as a negative control. (B) The aminoacylation activities of FLAG-tagged mThrRS-L (black square), mThrRS-L-C480A (black diamond), mThrRS-L-R509A (black triangle), mThrRS-L-H222A-H226A (white triangle) and no enzyme (white circle). (C) The post-transfer editing activities of FLAG-tagged mThrRS-L (black square), mThrRS-L-H222A-H226A (white triangle), mThrRS-L-NT (black inverted triangle), mThrRS-L-NT-H222A-H226A (white diamond) and no enzyme (black circle). (D) The aminoacylation activities of FLAG-tagged mThrRS-L-NT (black inverted triangle), mThrRS-L-NT-C480A (white square), mThrRS-L-NT-R509A (white inverted triangle), mThrRS-L-NT-H222A-H226A (white diamond) and no enzyme (white circle). Data represent averages of three independent experiments and the corresponding standard errors. Some error bars are hidden by symbols.
Obtaining a soluble chimeric ThrRS-L containing active aminoacylation and editing domains
To compare the kinetic parameters in aminoacylation and editing reactions of mThrRS-L with those of mThrRS in vitro requires purified mThrRS-L and mThrRS. The gene encoding mThrRS was successfully overexpressed in E. coli transformants, and mThrRS was purified to homogeneity (Figure 6A). However, mThrRS-L could not be obtained because inclusion bodies formed during gene expression. Moreover, we were unable to obtain soluble mThrRS-L-Δ149 or mThrRS-L-Δ82 via gene expression in E. coli. Additionally, we also tried to obtain active mThrRS-L using a baculovirus expression system or an in vitro protein synthesis kit; however, we could not acquire mThrRS-L with sufficient purity. The main sequences of mThrRS and mThrRS-L are mostly conserved, including the N1 domain, N2 editing domain, aminoacylation domain and CTD (Figure 1). To obtain a soluble protein with intact aminoacylation and editing domains of mThrRS-L, we replaced the N-extension from Met1 to Val149 of mThrRS-L with that of mThrRS (from Met1 to Pro81) (Supplementary Figure S1, Pro81 of mThrRS indicated) to form chimeric mThrRS-L-NT, which comprised the N-extension of mThrRS and the other domains of mThrRS-L (Figure 1). The gene encoding mThrRS-L-NT was successfully expressed in E. coli transformants, and soluble mThrRS-L-NT was purified to homogeneity (Figure 6A). Despite containing a different N-extension compared with wild-type mThrRS-L, mThrRS-L-NT retained the domains potentially catalyzing the aminoacylation and editing reactions. We hypothesized that the replacement would influence activities of mThrRS-L quantitatively but not qualitatively; thus, it is reasonable to use mThrRS-L-NT with its native aminoacylation and editing active sites to reveal the relevant enzymatic properties of mThrRS-L. We determined the activities of mThrRS-L-NT and compared them with those of mThrRS in subsequent experiments. Furthermore, compared with the mThrRS-L immunoprecipitated from HEK393T cells, the purified mThrRS-L-NT from E. coli transformants could be used to assay the aminoacylation and editing activities of the enzymes.
Figure 6.
SDS-PAGE analysis of purified mThrRS and mThrRS-L-NT from Escherichia coli, editing and aminoacylation activities of ThrRSs. (A) SDS-PAGE analysis of purified mThrRS and mThrRS-L-NT. Samples of 2 μg were loaded onto a 10% SDS-PAGE gel and stained with Coomassie Brilliant Blue. (B) Preformed Ser-tRNAThr was hydrolyzed by mThrRS (black square), mThrRS-H154A-H158A (white square), mThrRS-L-NT (black inverted triangle) or mThrRS-L-NT-H222A-H226A (white square). Spontaneous hydrolysis was measured as a negative control, by adding no enzyme (black circle). Aminoacylation of mThrRS (black square) and mThrRS-H154A-H158A (white triangle) (C) or mThrRS-L-NT (black inverted triangle) and mThrRS-L-NT-H222A-H226A (white diamond) (D) was compared. Data represent averages of three independent experiments and the corresponding standard errors. Some error bars are hidden by symbols.
The aminoacylation domain of mThrRS-L-NT has amino acid activation and aminoacylation activities
We measured the amino acid activation activities of mThrRS and mThrRS-L-NT using an ATP-PPi exchange reaction (Table 1). mThrRS-L-NT catalyzed Thr activation with a nearly 11-fold higher KM (3.26 ± 0.29 mM) compared with that of mThrRS (0.30 ± 0.05 mM); however, its kcat value (25.61 ± 2.01 s−1) was approximately double that of mThrRS (11.16 ± 0.85 s−1); the catalytic efficiency (kcat/KM) of mThrRS (37.20 mM−1 s−1) was nearly five times greater than that of mThrRS-L-NT (7.86 mM−1 s−1) (Table 1). We used the transcribed mouse tRNAThr(AGU) as a substrate to compare their aminoacylation activities. In the aminoacylation reaction, the KM value of mThrRS-L-NT for the tRNA (1.31 ± 0.07 μM) was approximately double that of mThrRS (0.74 ± 0.05 μM) and the kcat values of the two enzymes were similar; their catalytic efficiency differed by 1.4-fold (391.89 mM−1 s−1 for mThrRS versus 282.44 mM−1 s−1 for mThrRS-L-NT) (Table 2). Despite their different efficiencies in amino acid activation, mThrRS and mThrRS-L-NT aminoacylated tRNAThr with a similar reaction rate in vitro. However, their relative aminoacylation activities may differ in vivo because the mature tRNAThrs are well modified in cells.
Table 1. Kinetic parameters of mThrRS or mThrRS-L-NT for Thr or Ser in amino acid activationa.
| Enzyme | Amino acid | k cat (s−1) | K M (mM) | k cat/KM (mM−1 s−1) | Discriminator factor (D)b |
|---|---|---|---|---|---|
| mThrRS | Thr | 11.16 ± 0.85 | 0.30 ± 0.05 | 37.20 | 1 |
| Ser | 2.73 ± 0.32 | 76.64 ± 6.38 | 0.036 | 1033 | |
| mThrRS-L-NT | Thr | 25.61 ± 2.01 | 3.26 ± 0.29 | 7.86 | 1 |
| Ser | 1.10 ± 0.08 | 152.12 ± 10.21 | 0.0072 | 1092 |
aResults are the average of three independent repeats, with standard deviations indicated.
b D was calculated using the formula D = (kcat/KM)Thr/(kcat/KM)Ser.
Table 2. Aminoacylation kinetic parameters of mThrRS and mThrRS-L-NT for mouse tRNAThr(AGU)a.
| Enzyme | K M (μM) | kcat (s−1) | k cat/KM (mM−1 s−1) |
|---|---|---|---|
| mThrRS | 0.74 ± 0.05 | 0.29 ± 0.04 | 391.89 |
| mThrRS-L-NT | 1.31 ± 0.07 | 0.37 ± 0.05 | 282.44 |
aResults are the average of three independent repeats, with standard deviations indicated.
We further compared the time-course curves of aminoacylation or post-transfer editing reactions of immunoprecipitated FLAG-tagged mThrRS-L and mThrRS-L-NT and found that they synthesized Thr-tRNAThr or hydrolyzed Ser-tRNAThr with a similar catalytic rate (Figure 5B–D), indicating that changing the N-extension of mThrRS-L had little effect on aminoacylation or post-transfer editing. Again, in the context of mThrRS-L-NT, the aminoacylation activities of the two mutants at the active site residues, mThrRS-L-NT-C480A and mThrRS-L-NT-R509A (native mThrRS-L numbering), decreased significantly, suggesting that the aminoacylation activity was indeed derived from mThrRS-L-NT (Figure 5D).
Both mThrRS and mThrRS-L-NT misactivate non-cognate Ser
The requirement for the editing function of ThrRS is based on insufficient discrimination of cognate Thr and non-cognate Ser during amino acid activation. We measured and compared Ser misactivation by mThrRS and mThrRS-L-NT using an ATP-PPi exchange reaction. Both of them obviously misactivated Ser (Table 1). mThrRS-L-NT had higher KM and lower kcat values than mThrRS in the activation of Ser. The catalytic efficiency of mThrRS-L-NT was ∼5-fold lower than that of mThrRS in Ser activation, which was similar to the Thr activation. The discrimination factor D can be calculated using the formula D = (kcat/KM)Thr/(kcat/KM)Ser. The discrimination factors of mThrRS and mThrRS-L-NT were 1/1033 and 1/1092, respectively, indicating that both ThrRSs require editing, based on a misincorporation threshold of 1/3300 for protein synthesis (27).
The editing domain of ThrRS-L-NT has proofreading activity
The above misactivation analysis suggested that both ThrRS and ThrRS-L-NT need an editing activity to ensure accuracy in tRNA aminoacylation. Editing activity has been confirmed in bacterial EcThrRS (8), hmtThrRS (28), archaeal ThrRSs (29,30) and Saccharomyces cerevisiae cytoplasmic and mitochondrial ThrRSs (ScThrRS and ScmtThrRS) (6,22). However, editing by mammalian cytoplasmic ThrRS has never been reported.
We found that the pre-formed Ser-tRNAThr, prepared by the editing-deficient EcThrRS-H73A-H77A (8), could be hydrolyzed by both mThrRS and mThrRS-L-NT, indicating that they both have a post-transfer editing activity (Figure 6B), as revealed by the native mThrRS-L overexpressed in HEK293T cells. To confirm that editing was catalyzed by the N2 domain, we simultaneously mutated His154 and His158 within the domain of mThrRS to Ala residues to obtain the double variant mThrRS-H154A-H158A. According to sequence alignment, the two residues are homologous to His73 and His77 of ThrRS from E. coli (8) or His151 and His155 of that from S. cerevisiae cytoplasm (6), which are crucial to the post-transfer editing activity of the ThrRSs. mThrRS-L-NT-H222A-H226A (numbering based on native mThrRS-L) was also purified from E. coli. The post-transfer editing of mThrRS-H154A-H158A and mThrRS-L-NT-H222A-H226A was abolished (Figure 6B). However, the replacements did not affect their aminoacylation activity (Figure 6C and D).
We also enriched FLAG-tagged mThrRS-L-NT-H222A-H226A from HEK293T cells. Post-transfer editing by the immunoprecipitated mThrRS-L-NT-H222A-H226A was obviously decreased when compared with immunoprecipitated mThrRS-L-NT, but with some activity remaining (Figure 5C), which was different from the abolished editing activity of bacteria-derived mThrRS-L-NT-H222A-H226A (Figure 6B). This was also likely caused by the co-precipitated hThrRS and/or hThrRS-L (Figure 5A).
Collectively, the above results showed that both mThrRS and mThrRS-L use their post-transfer editing activity, catalyzed by the N2 domain, to remove mischarged Ser-tRNAThr, similar to ThrRSs from E. coli and S. cerevisiae.
mThrRS and mThrRS-L-NT use similar editing pathways, but exhibit different editing activities
The net effect of the proofreading reaction is the consumption of ATP. Therefore, editing can be measured by AMP formation in a TLC-based AMP formation assay (7). tRNA-independent pre-transfer editing is determined by AMP formation in the absence of tRNA. In the presence of tRNA, total editing (including tRNA-independent, tRNA-dependent pre-transfer editing and post-transfer editing) is measured (7).
In the presence of non-cognate Ser, we assayed AMP formation catalyzed by mThrRS with or without tRNAThr. From the amounts of AMP observed in TLC assays (Figure 7A), the observed catalytic constants (kobs) of mThrRS, calculated as AMP formation, were (8.4 ± 1.0) × 10−2 (with tRNAThr) and (3.4 ± 0.6) × 10−2 s−1 (without tRNAThr) (Figure 7B). To determine the type of the tRNA-dependent editing, we assayed AMP formation by mThrRS-H154A-H158A with Ser in the presence and absence of tRNA, because mThrRS-H154A-H158A does not have the post-transfer editing activity. The kobs values of mThrRS-H154A-H158A with and without tRNA were (4.6 ± 0.7) × 10−2 and (4.3 ± 0.5) × 10−2 s−1, respectively. Therefore, after abolishment of post-transfer editing, mThrRS only has the tRNA-independent pre-transfer editing activity, and has a negligible tRNA-dependent pre-transfer editing activity (Figure 7A and B).
Figure 7.
Editing properties of mThrRS. (A) A representative TLC image showing the generation of [32P]AMP or Ser-[32P]AMP in the absence (−tRNA) or presence (+tRNA) of tRNAThr by mThrRS and mThrRS-H154A-H158A. A 2-fold dilution of [α-32P]ATP (initial concentration 3 mM) was included for quantification. The standard point of 3 mM [α-32P]ATP and the loading origins were over-exposed to make the AMP and Ser-AMP signals more obvious. The remaining six points of diluted [α-32P]ATP are enough and the loading origins were not quantified; therefore the over-exposure has no effect on quantification and data interpretation. Quantification of [32P]AMP (B) or Ser-[32P]AMP (C) formation by mThrRS with (white circle) or without black circle) tRNA, or by H154A-H158A with (white square) or without (black inverted triangle) tRNA. Data represent averages of three independent experiments and the corresponding standard errors.
The kobs values for mThrRS in Ser-AMP formation, without or with tRNAThr, were (1.2 ± 0. 2) × 10−2 and (0.8 ± 0.2) × 10−2 s−1, respectively (Figure 7C). In the absence of tRNA, the activated Ser could not be transferred to tRNA; therefore, Ser-AMP accumulated. Ser-AMP is probably released from the active site of the enzyme and then hydrolyzed in solution (selective release) to produce AMP, because the concentration of accumulated Ser-AMP exceeded the enzyme concentration (2 μM) in the reaction (31). To investigate this selective release, pulse-chase experiments were performed to measure the rate of non-enzymatic hydrolysis of Ser-AMP. We prepared Ser-[32P]AMP by incubating mThrRS with Ser and [α-32P]ATP. The reaction was quenched by adding a 500-fold molar excess of unlabeled ATP. The kobs value for the conversion of Ser-[32P]AMP to [32P]AMP was (0.89 ± 0.12) × 10−2 s−1, indicating a negligible rate of non-enzymatic hydrolysis after Ser-AMP release into solution (Supplementary Figure S3). Thus, editing by mThrRS in the absence of tRNA was almost all tRNA-independent pre-transfer editing.
We investigated the editing pathways of mThrRS-L-NT using similar methods. The kobs values of mThrRS-L-NT in the absence or presence of tRNAThr were (0.9 ± 0.1) × 10−2 and (1.7 ± 0.2) × 10−2 s−1, calculated from AMP formation, respectively, which were lower than those for mThrRS ((3.4 ± 0.6) × 10−2 and (8.4 ± 1.0) × 10−2 s−1, respectively) (Figure 8A and B). The kobs values of mThrRS-L-NT for Ser-AMP formation were (0.42 ± 0.07) × 10−2 and (0.36 ± 0.5) × 10−2 s−1, with and without tRNA, respectively, which were lower than those for mThrRS ((0.8 ± 0.2) × 10−2 and (1.2 ± 0.2) × 10−2 s−1, respectively) (Figure 8A and C). Similarly, selective release of mThrRS-L-NT [kobs of (0.037 ± 0.004) × 10−2 s−1] made a negligible contribution to the total editing by the enzyme (Supplementary Figure S3A and C). The kobs values of mThrRS-L-NT-H222A-H226A in AMP formation were very similar without ((1.0 ± 0. 2) × 10−2 s−1) or with tRNA ((1.2 ± 0.3) × 10−2 s−1) (Figure 8A and B). Therefore, like mThrRS, mThrRS-L has negligible tRNA-dependent pre-transfer editing.
Figure 8.
Editing properties of mThrRS-L-NT. (A) A representative TLC image showing generation of [32P]AMP or Ser-[32P]AMP in the absence (−tRNA) or presence (+tRNA) of tRNAThr by mThrRS-L-NT and mThrRS-L-NT-H222A-H226A. A two-fold dilution of [α-32P]ATP (initial concentration 3 mM) was included for quantification. The standard point of 3 mM [α-32P]ATP and the loading origins were over-exposed to make the AMP and Ser-AMP signals more obvious. The remaining six points of diluted [α-32P]ATP are enough and the loading origins were not quantified; therefore, the over-exposure has no effect on quantification and data interpretation at all. Quantification of [32P]AMP (B) or Ser-[32P]AMP (C) formation by mThrRS-L-NT with (white circle) or without (black circle) tRNA or by mThrRS-L-NT-H222A-H226A with (white square) or without (black inverted triangle) tRNA. Data represent averages of three independent experiments and the corresponding standard errors.
Thus, mThrRS and mThrRS-L-NT have both tRNA-independent pre-transfer and post-transfer editing pathways, but lack tRNA-dependent pre-transfer editing. However, the editing activity of mThrRS in various pathways is significantly higher than that of mThrRS-L-NT. Notably, mThrRS and mThrRS-L-NT have similar aminoacylation activities.
mThrRS and mThrRS-L-NT recognize a full set of cytoplasmic tRNAThrs but display different recognition of tRNAThrs from E. coli and mitochondria
Mouse expresses three cytoplasmic tRNAThr isoacceptors: tRNAThr(AGU), -(CGU) and -(UGU). To reveal whether the tRNA recognition capacities of mThrRS and mThrRS-L-NT are same, we assayed their threonylation activities for these three tRNAThr isoacceptors and found that the two ThrRSs have comparable activities (Figure 9A and B), suggesting that they equally threonylate the full set of tRNAThr isoacceptors. We used one of the three isoacceptors, tRNAThr(AGU), as a representative and measured the kinetic parameters of the two enzymes for the tRNAThr substrate in the threonylation reaction. mThrRS and mThrRS-L-NT had similar Km and kcat values (Table 2).
Figure 9.
Aminoacylation of various tRNAThrs by various ThrRSs. Aminoacylation of mouse tRNAThr(AGU) (white triangle), tRNAThr(CGU) (white square) or tRNAThr(UGU) (black circle) by either mThrRS (A) or mThrRS-L-NT (B). (C–F) Aminoacylation of EctRNAThr(UGU) (black circle), mouse tRNAThr(UGU) (white circle) or hmt-tRNAThr(UGU) (black triangle) by mThrRS (C), mThrRS-L-NT (D), EcThrRS (E) or hmtThrRS (F). Negative controls were performed by not adding tRNA substrate (white square). Data represent averages of three independent experiments and the corresponding standard errors. Some error bars are hidden by symbols.
We tested the cross-recognition of various tRNAThrs, including mouse cytoplasmic tRNAThr(UGU) (mtRNAThr), E. coli tRNAThr(UGU) (EctRNAThr) and human mitochondrial tRNAThr(UGU) (hmt-tRNAThr) by mThrRS, mThrRS-L-NT, hmtThrRS and EcThrRS, respectively. As expected, each of the four ThrRSs could threonylate its cognate tRNA. mThrRS threonylated EctRNAThr as well as mtRNAThr; however, it aminoacylated hmt-tRNAThr at a lower rate (Figure 9C). mThrRS-L-NT threonylated EctRNAThr more slowly compared with mtRNAThr; however, it was unable to charge hmt-tRNAThr completely (Figure 9D). Although EcThrRS could aminoacylate mtRNAThr, it failed to charge hmt-tRNAThr (Figure 9E). hmtThrRS could threonylate EctRNAThr, but failed to recognize mtRNAThr (Figure 9F).
Thus, mThrRS and mThrRS-L-NT recognized the full set of cytoplasmic tRNAThrs, but exhibited different recognition of bacterial or mitochondrial tRNAThr, indicating that the two enzymes recognize tRNAThrs in different ways. Mitochondrial ThrRS could charge both bacterial and mitochondrial tRNAThr but not mtRNAThr; however, hmt-tRNAThr is not a substrate of the bacterial ThrRS. Notably, mtRNAThr, EctRNAThr and hmt-tRNAThr contain the same G35 and U36, the well-defined identity elements for ThrRSs (32), suggesting that other elements also function as determinants and/or anti-determinants for recognition by various ThrRSs.
We also compared the affinities between mThrRS and mThrRS-L-NT for small molecular substrates, please see Supplementary Result.
DISCUSSION
Alongside two genes encoding cytoplasmic and mitochondrial ThrRSs (TARS and TARS2), the third gene (TARSL2) encoding a ThrRS-like protein exists uniquely in higher eukaryotes. In mammals, the existence of two aaRSs responsible for one given amino acid in a single cellular compartment is rare. Genes encoding tRNA synthetases for Arg and Glu are the only two reported instances. Through alternative translational initiation, RARS encodes two forms of arginyl-tRNA synthetase (ArgRS), and the only distinction between them is with or without the N-extension (33). Both ArgRS isoforms exhibit similar tRNAArg charging activities (34). The long form ArgRS synthesizes Arg-tRNAArg for protein biosynthesis on the ribosomes, while the short form is thought to participate in post-translational modification of proteins for ubiquitin-dependent protein degradation (35). Co-existence of two ArgRSs is non-essential in Chinese hamster ovary (CHO) cells, suggesting that the short form ArgRS participates in translation (34). EPRS encodes bi-functional glutamyl-prolyl-tRNA synthetase (GluProRS); in addition, a truncated GluProRS (containing intact GluRS) is also found in the cytoplasm to maintain a translational trickle of inflammatory gene expression in myeloid cells (36). Whether the truncated GluProRS has aminoacylation activity and participates in cytoplasmic translation remains unknown. In addition, human aaRS genes produce many natural catalytic nulls (CNs), and these CNs usually lack an intact catalytic domain. For example, the C8 isoform of human alanyl-tRNA synthetase lacks part of the aminoacylation domain (37). Considering other eukaryotes, a seryl-tRNA synthetase (SerRS) duplicated gene, SLIMP, is present in insect mitochondria, encoding a SerRS-like protein (SLIMP). Even though SLIMP binds tRNASer, it is unable to catalyze tRNASer aminoacylation (38). Thus, insect mitochondrial SerRS/SLIMP is different from mammalian ThrRS/ThrRS-L, based on tRNA aminoacylation activity. For ThrRS systems, duplicated ThrRS genes have recently been reported in the cyanobacterial genus Anabaena. Both ThrRSs (T1 and T2) have tRNAThr aminoacylation activity; however, only the constitutively expressed T1 has an editing function; the conditionally expressed T2 is editing-deficient and present only in conditions of zinc deprivation (39). Here, both mThrRS and mThrRS-L have editing activities.
Our study on mThrRS-L clearly showed that, in vivo, native mouse Tarsl2 is ubiquitously expressed in mouse tissues and cell lines, and its organ distribution varies. The majority of native or overexpressed mThrRS-L is located in the cytoplasm, possibly binding and/or charging tRNAThr, based on the in vitro data. A small amount of native or overexpressed mThrRS-L is also located in the nucleus. TARS is conserved across sampled bacteria and eukaryotes, thus mThrRS is likely to be responsible for translation on the ribosomes. TARSL2 is only present in higher eukaryotes and its nuclear localization suggests that it might play some different roles with mThrRS. Thus, we propose that ThrRS-L is not essential for translation on the ribosomes and is likely to manufacture Thr-tRNAThr under particular conditions, for example, tRNAThr threonylation for nuclear tRNA transport, or tRNA quality control (40,41). We are currently studying the in vivo function of ThrRS-L.
mThrRS-L has a distinct N-extension from mThrRS, and both native and overexpressed mThrRS-L are located in the nucleus. We identified an NLS at the C-terminus of mThrRS-L; however, overexpressed mThrRS-L-Δ149 (without the N-extension) is only located in the cytoplasm, suggesting that the N-extension of mThrRS-L possibly mediates a crucial protein–protein interaction during nuclear entry.
Our data showed that the enriched mThrRS-L from HEK293T cells is active in aminoacylation of tRNA and deacylation of mis-charged tRNA. To the best of our knowledge, this is the first report describing the kinetic parameters of mammalian ThrRS-L in vitro. mThrRS-L-NT and mThrRS aminoacylate tRNAThr with similar efficiency. However, they exhibit different recognition of bacterial and mitochondrial tRNAThrs. Recent bioinformatics studies have revealed that hundreds of mitochondrial tRNA-lookalike genes (including tRNAThr-lookalikes), expressing tRNAs identical or highly similar to mitochondrial tRNAs, are distributed throughout the nuclear genome (42). Whether ThrRS and ThrRS-L have distinct roles in charging these tRNAThr-lookalikes (if they are expressed) is an open question. Both ThrRS and ThrRS-L have retained efficient post-transfer editing, suggesting that aminoacylation of tRNAThr by either enzyme requires a high level of accuracy.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mr Xian Xia for purifying EctRNAThr from E. coli.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program of China [2017YFA0504000]; Natural Science Foundation of China [31670801, 91440204, 31500644]; Strategic Priority Research Program of the Chinese Academy of Sciences [XDB19010203]; Committee of Science and Technology in Shanghai [12JC1409700]; Shanghai Rising-Star Program [16QA1404400]; Youth Innovation Promotion Association (Chinese Academy of Sciences) [Y119S41291 to X.L.Z.]; China Postdoctoral Science Foundation [2014T70438 to Q.H.]. Funding for open access charge: National Key Research and Development Program of China [2017YFA0504000].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ibba M., Söll D.. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000; 69:617–650. [DOI] [PubMed] [Google Scholar]
- 2. Cusack S. Aminoacyl-tRNA synthetases. Curr. Opin. Struct. Biol. 1997; 7:881–889. [DOI] [PubMed] [Google Scholar]
- 3. Schimmel P. Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu. Rev. Biochem. 1987; 56:125–158. [DOI] [PubMed] [Google Scholar]
- 4. Antonellis A., Green E.D.. The role of aminoacyl-tRNA synthetases in genetic diseases. Annu. Rev. Genomics Hum. Genet. 2008; 9:87–107. [DOI] [PubMed] [Google Scholar]
- 5. Bonnefond L., Fender A., Rudinger-Thirion J., Giege R., Florentz C., Sissler M.. Toward the full set of human mitochondrial aminoacyl-tRNA synthetases: characterization of AspRS and TyrRS. Biochemistry. 2005; 44:4805–4816. [DOI] [PubMed] [Google Scholar]
- 6. Zhou X.L., Ruan Z.R., Huang Q., Tan M., Wang E.D.. Translational fidelity maintenance preventing Ser mis-incorporation at Thr codon in protein from eukaryote. Nucleic Acids Res. 2013; 41:302–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou X.L., Wang E.D.. Transfer RNA: a dancer between charging and mis-charging for protein biosynthesis. Sci. China Life Sci. 2013; 56:921–932. [DOI] [PubMed] [Google Scholar]
- 8. Dock-Bregeon A.C., Rees B., Torres-Larios A., Bey G., Caillet J., Moras D.. Achieving error-free translation; the mechanism of proofreading of threonyl-tRNA synthetase at atomic resolution. Mol. Cell. 2004; 16:375–386. [DOI] [PubMed] [Google Scholar]
- 9. Chen J.F., Guo N.N., Li T., Wang E.D., Wang Y.L.. CP1 domain in Escherichia coli leucyl-tRNA synthetase is crucial for its editing function. Biochemistry. 2000; 39:6726–6731. [DOI] [PubMed] [Google Scholar]
- 10. Zhou X.L., Yao P., Ruan L.L., Zhu B., Luo J., Qu L.H., Wang E.D.. A unique insertion in the CP1 domain of Giardia lamblia leucyl-tRNA synthetase. Biochemistry. 2009; 48:1340–1347. [DOI] [PubMed] [Google Scholar]
- 11. Zhou X.L., Wang E.D.. Two tyrosine residues outside the editing active site in Giardia lamblia leucyl-tRNA synthetase are essential for the post-transfer editing. Biochem. Biophys. Res. Commun. 2009; 386:510–515. [DOI] [PubMed] [Google Scholar]
- 12. Xu M.G., Li J., Du X., Wang E.D.. Groups on the side chain of T252 in Escherichia coli leucyl-tRNA synthetase are important for discrimination of amino acids and cell viability. Biochem. Biophys. Res. Commun. 2004; 318:11–16. [DOI] [PubMed] [Google Scholar]
- 13. Zhou X.L., Chen Y., Fang Z.P., Ruan Z.R., Wang Y., Liu R.J., Xue M.Q., Wang E.D.. Translational quality control by bacterial threonyl-tRNA synthetases. J. Biol. Chem. 2016; 291:21208–21221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sankaranarayanan R., Dock-Bregeon A.C., Romby P., Caillet J., Springer M., Rees B., Ehresmann C., Ehresmann B., Moras D.. The structure of threonyl-tRNA synthetase-tRNA(Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell. 1999; 97:371–381. [DOI] [PubMed] [Google Scholar]
- 15. Kim K., Park S.J., Na S., Kim J.S., Choi H., Kim Y.K., Paek E., Lee C.. Reinvestigation of aminoacyl-tRNA synthetase core complex by affinity purification-mass spectrometry reveals TARSL2 as a potential member of the complex. PLoS One. 2013; 8:e81734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park S.J., Ahn H.S., Kim J.S., Lee C.. Evaluation of multi-tRNA synthetase complex by multiple reaction monitoring mass spectrometry coupled with size exclusion chromatography. PLoS One. 2015; 10:e0142253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou X.L., Zhu B., Wang E.D.. The CP2 domain of leucyl-tRNA synthetase is crucial for amino acid activation and post-transfer editing. J. Biol. Chem. 2008; 283:36608–36616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Y., Chen J.F., Wang E.D., Wang Y.L.. T7 RNA polymerase transcription of Escherichia coli isoacceptors tRNA(Leu). Sci. China C. Life Sci. 1999; 42:185–190. [DOI] [PubMed] [Google Scholar]
- 19. Li Y., Wang E.D., Wang Y.L.. Overproduction and purification of Escherichia coli tRNA(Leu). Sci. China C. Life Sci. 1998; 41:225–231. [DOI] [PubMed] [Google Scholar]
- 20. Zhou X.L., Fang Z.P., Ruan Z.R., Wang M., Liu R.J., Tan M., Anella F.M., Wang E.D.. Aminoacylation and translational quality control strategy employed by leucyl-tRNA synthetase from a human pathogen with genetic code ambiguity. Nucleic Acids Res. 2013; 41:9825–9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fang Z.P., Wang M., Ruan Z.R., Tan M., Liu R.J., Zhou M., Zhou X.L., Wang E.D.. Coexistence of bacterial leucyl-tRNA synthetases with archaeal tRNA binding domains that distinguish tRNA(Leu) in the archaeal mode. Nucleic Acids Res. 2014; 42:5109–5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou X.L., Ruan Z.R., Wang M., Fang Z.P., Wang Y., Chen Y., Liu R.J., Eriani G., Wang E.D.. A minimalist mitochondrial threonyl-tRNA synthetase exhibits tRNA-isoacceptor specificity during proofreading. Nucleic Acids Res. 2014; 42:13873–13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gruic-Sovulj I., Uter N., Bullock T., Perona J.J.. tRNA-dependent aminoacyl-adenylate hydrolysis by a nonediting class I aminoacyl-tRNA synthetase. J. Biol. Chem. 2005; 280:23978–23986. [DOI] [PubMed] [Google Scholar]
- 24. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 25. Mirando A.C., Fang P., Williams T.F., Baldor L.C., Howe A.K., Ebert A.M., Wilkinson B., Lounsbury K.M., Guo M., Francklyn C.S.. Aminoacyl-tRNA synthetase dependent angiogenesis revealed by a bioengineered macrolide inhibitor. Sci. Rep. 2015; 5:13160–13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eriani G., Delarue M., Poch O., Gangloff J., Moras D.. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990; 347:203–206. [DOI] [PubMed] [Google Scholar]
- 27. Loftfield R.B., Vanderjagt D.. The frequency of errors in protein biosynthesis. Biochem. J. 1972; 128:1353–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Y., Zhou X.L., Ruan Z.R., Liu R.J., Eriani G., Wang E.D.. A human disease-causing point mutation in mitochondrial threonyl-tRNA synthetase induces both structural and functional defects. J. Biol. Chem. 2016; 291:6507–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hussain T., Kruparani S.P., Pal B., Dock-Bregeon A.C., Dwivedi S., Shekar M.R., Sureshbabu K., Sankaranarayanan R.. Post-transfer editing mechanism of a D-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea. EMBO J. 2006; 25:4152–4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Korencic D., Ahel I., Schelert J., Sacher M., Ruan B., Stathopoulos C., Blum P., Ibba M., Soll D.. A freestanding proofreading domain is required for protein synthesis quality control in Archaea. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:10260–10265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hati S., Ziervogel B., Sternjohn J., Wong F.C., Nagan M.C., Rosen A.E., Siliciano P.G., Chihade J.W., Musier-Forsyth K.. Pre-transfer editing by class II prolyl-tRNA synthetase: role of aminoacylation active site in “selective release” of noncognate amino acids. J. Biol. Chem. 2006; 281:27862–27872. [DOI] [PubMed] [Google Scholar]
- 32. Dock-Bregeon A., Sankaranarayanan R., Romby P., Caillet J., Springer M., Rees B., Francklyn C.S., Ehresmann C., Moras D.. Transfer RNA-mediated editing in threonyl-tRNA synthetase. The class II solution to the double discrimination problem. Cell. 2000; 103:877–884. [DOI] [PubMed] [Google Scholar]
- 33. Zheng Y.G., Wei H., Ling C., Xu M.G., Wang E.D.. Two forms of human cytoplasmic arginyl-tRNA synthetase produced from two translation initiations by a single mRNA. Biochemistry. 2006; 45:1338–1344. [DOI] [PubMed] [Google Scholar]
- 34. Kyriacou S.V., Deutscher M.P.. An important role for the multienzyme aminoacyl-tRNA synthetase complex in mammalian translation and cell growth. Mol. Cell. 2008; 29:419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ferber S., Ciechanover A.. Role of arginine-tRNA in protein degradation by the ubiquitin pathway. Nature. 1987; 326:808–811. [DOI] [PubMed] [Google Scholar]
- 36. Yao P., Potdar A.A., Arif A., Ray P.S., Mukhopadhyay R., Willard B., Xu Y., Yan J., Saidel G.M., Fox P.L.. Coding region polyadenylation generates a truncated tRNA synthetase that counters translation repression. Cell. 2012; 149:88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lo W.S., Gardiner E., Xu Z., Lau C.F., Wang F., Zhou J.J., Mendlein J.D., Nangle L.A., Chiang K.P., Yang X.L. et al. Human tRNA synthetase catalytic nulls with diverse functions. Science. 2014; 345:328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guitart T., Leon Bernardo T., Sagales J., Stratmann T., Bernues J., Ribas de Pouplana L.. New aminoacyl-tRNA synthetase-like protein in insecta with an essential mitochondrial function. J. Biol. Chem. 2010; 285:38157–38166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rubio M.A., Napolitano M., Ochoa de Alda J.A., Santamaria-Gomez J., Patterson C.J., Foster A.W., Bru-Martinez R., Robinson N.J., Luque I.. Trans-oligomerization of duplicated aminoacyl-tRNA synthetases maintains genetic code fidelity under stress. Nucleic Acids Res. 2015; 43:9905–9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Azad A.K., Stanford D.R., Sarkar S., Hopper A.K.. Role of nuclear pools of aminoacyl-tRNA synthetases in tRNA nuclear export. Mol. Biol. Cell. 2001; 12:1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Steiner-Mosonyi M., Mangroo D.. The nuclear tRNA aminoacylation-dependent pathway may be the principal route used to export tRNA from the nucleus in Saccharomyces cerevisiae. Biochem. J. 2004; 378:809–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Telonis A.G., Loher P., Kirino Y., Rigoutsos I.. Nuclear and mitochondrial tRNA-lookalikes in the human genome. Front. Genet. 2014; 5:344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.