Abstract
EZH2 is a subunit of polycomb repressive complex 2 (PRC2) that silences gene transcription via H3K27me3 and was shown to be essential for mammalian liver circadian regulation and hematopoiesis through gene silencing. Much less, however, is known about how Ezh2 acts in live zebrafish. Here, we show that zebrafish ezh2 is regulated directly by the circadian clock via both E-box and RORE motif, while core circadian clock genes per1a, per1b, cry1aa and cry1ab are down-regulated in ezh2 null mutant and ezh2 morphant zebrafish, and either knockdown or overexpression of ezh2 alters locomotor rhythms, indicating that Ezh2 is required for zebrafish circadian regulation. In contrast to its canonical silencing function, zebrafish Ezh2 up-regulates these key circadian clock genes independent of histone methyltransferase activity by directly binding to key circadian clock proteins. Similarly, Ezh2 contributes to hematopoiesis by enhancing expression of hematopoietic genes such as cmyb and lck. Together, our findings demonstrate for the first time that Ezh2 acts in both circadian regulation and hematopoiesis independent of silencing PRC2.
INTRODUCTION
Circadian rhythms are endogenous oscillations generated by interlocking negative transcriptional and translational feedback loops, allowing for organisms to coordinate physiological and behavioral rhythms with a period ∼24 h for anticipating environmental changes (1). In mammals, molecular oscillators operate in the master clock, the suprachiasmatic nucleus (SCN) in the hypothalamus of the brain (2) and also in peripheral organs or tissues (3,4). The bHLH-PAS proteins CLOCK and BMAL1 form a heterodimer to drive transcription of Period genes (Per1, Per2 and Per3) and Cryptochrome genes (Cry1 and Cry2) through E-boxes (5). PER and CRY proteins then are translocated into the nucleus, and interact with the CLOCK–BMAL1 complex to repress their own expression (6). CLOCK and BMAL1 also drive the expression of the orphan nuclear receptor Rev-erbα in the SCN and liver (7). Bmal1 contains Rev-Erbα/ROR response elements (ROREs) in its promoter, and its expression is activated by RORα but repressed by Rev-Erbα (7).
Roles of histone modifiers in circadian regulation have recently been investigated (8). In mice, transcriptional activation of Per1 and Per2 correlates with rhythmic acetylation of histone H3 at their promoter regions (9,10). In addition to histone acetylation, histone methylation also has been suggested to be important for circadian regulation. WDR5, a member of a histone methyltransferase complex, mediates rhythmic methylation of H3K4 and H3K9 at the promoters of PER1-regulated genes (11), and methylation of H3K4 and H3K9 are in phase with transcriptional activity of Dbp (albumin D-site binding protein) (12). JMJD5 and JMJ30, two Jumonji C domain-containing histone demethylase proteins, function in both the plant and human circadian systems (13,14).
Polycomb group proteins (PcG) are involved in heritable epigenetic regulation of gene expression during development (15). To date, two principal polycomb repressive complexes (PRCs), PRC1 and PRC2, have been identified (16). Mammalian PRC1 contains CBX, MPH, RING, BMI1 and MEL185, whereas PRC2 contains EZH, EED and SU(Z)12 (16). EZH2, enhancer of zeste homolog 2, silences gene transcription through trimethylation of histone H3 lysine 27 (H3K27me3) via its SET domain (17) and is essential for early development and tumor progression in most model organisms (18–20). However, EZH2 also functions positively independent of PRC2 in various cancers (21–24). For instance, EZH2 oncogenic activity in castration-resistant prostate cancer cells is independent of H3K27me3 (24,25). Interestingly, EZH2 was shown to be required for the mouse liver clock through binding to the clock complex and mediating methylation of H3K27 at the promoters of Per1 and Per2 (26). BMAL1 inhibits tumorigenesis and increases paclitaxel sensitivity in tongue squamous cell carcinoma through interaction with EZH2 (27).
In addition to its involvement in circadian regulation, EZH2 also was shown to play important roles for normal and malignant hematopoiesis (28,29). EZH2 was shown to be critical for B-cell development (30), and overexpression of mutated EZH2 was observed in B- and T-cell lymphoproliferative disorders and myeloid malignancy (28). During early megakaryopoiesis, Notch1 intracellular domain increases cytoplasmic EZH2 levels (31) and GATA-1 utilizes IKAROS and EZH2 to promote erythropoiesis through suppressing HES1 (32). EZH2 is up-regulated with differentiation of bone marrow cells (33) and down-regulated during differentiation of HL-60 cells to mature granulocytes (34). Ectopic expression of Ezh2 in the hematopoietic system increases the repopulation potential of hematopoietic stem cells (HSCs) and serial transplantation of these Ezh2-overexpressing HSCs resulted in mice developing myeloproliferative neoplasm (35).
Because Ezh2 knockout mice cease development at early gastrulation (19), circadian roles of EZH2 in live animals, and its functions during hematopoiesis are far from certain to date. Zebrafish have become an attractive vertebrate model organism for studying circadian rhythms (36–39) and hematopoiesis (40). Here, we found that zebrafish ezh2 is regulated directly by the circadian clock. Loss of Ezh2 disrupts rhythmic expression of circadian clock genes and either knockdown or overexpression of ezh2 alters locomotor rhythms of zebrafish larvae. Loss of Ezh2 also results in defects in zebrafish primitive and definitive hematopoiesis. In contrast to its predominant gene silencing, Ezh2 enhances circadian regulation and hematopoiesis independent of silencing PRC2 in zebrafish.
MATERIALS AND METHODS
Experimental animals
Wild-type zebrafish (Danio rerio), ezh2 heterozygous, per1b–/– mutant (41), rev-erbα–/– mutant (42), Tg(gata1:DsRED) (43), Tg(mpx:EGFP) (44), Tg(lyz:EGFP) (45), Tg(cmyb:EGFP) (46) and Tg(coro1a:EGFP) (47) zebrafish lines were maintained on a 14-h light/10-h dark cycle and fed three times daily at the Soochow University Zebrafish Facility. Embryos were obtained by natural crosses and fertilized eggs were raised at 28.5°C. PTU (2-phenylthiourea) (0.003%, wt/vol) was added to embryo medium to inhibit pigment formation for whole mount in situ hybridization experiments. All experiments were conducted in accordance with guidelines approved by the Soochow University Committee on the Use and Care of Animals.
Plasmid construction
cDNAs of zebrafish ezh2 and mice Ezh2 were PCR amplified with primer pairs ezh2F3 and ezh2R3, mEzh2F and mEzh2R, and cloned into pCDNA3.1/Myc-His(–) with XhoI and BamHI sites, respectively. Zebrafish eed cDNA was PCR amplified with primers eedF and eedR, and cloned into pCDNA3.1/Myc-His(–) with XhoI and EcoRI sites. Zebrafish ezh2 cDNA was PCR amplified with primer pair ezh2F4 and ezh2R4 and cloned into pDNA3.1-HA with BamHI and XhoI sites. Two amino acids, Phe681 and His703, critical for zabrafish Ezh2 H3K27 methyltransferase activity (25,48,49) were mutated to Ile and Ala to generate the mutated form of zebrafish ezh2 by two rounds of mutagenesis using primer pairs ezh2F681F and ezh2F681R, ezh2H703AF and ezh2H703AR, respectively. Similarly, the mutated form of mouse Ezh2 with mutations at F667I and H689A was generated with primer pairs mEzh2F667IF and mEzh2F667IR, mEzh2H689AF and mEzh2H689AR.
An ezh2 RNA probe for whole mount in situ hybridization was prepared as described previously (50). The fragment containing ezh2 E-box was amplified with primers ezh2-E-luc-F and ezh2-E-luc-R, while the fragment containing ezh2 RORE was amplified with primers ezh2-R-luc-F and ezh2-R-luc-R, and then both fragments were inserted into the pGL3.17-promoter vector with SacI and XhoI sites, respectively, generating ezh2-E-luc and ezh2-R-luc. Then, the E-box sequence, CACGTG, was mutated to CCCGTC, with primers ezh2-Emu-F and ezh2-Emu-R, and the RORE sequence, TTGGGTC, also was mutated to TTCCCTC, with primers ezh2-Rmu-F and ezh2-Rmu-R. The cmyb and lck promoter fragments containing E-boxes were amplified with primer pairs cmyb-E-luc-F and cmyb-E-lucR, lck-E-luc-F and lck-E-luc-R, respectively, and then both fragments were inserted into the pGL3.17-promoter vector with KpnI and XhoI sites, respectively, generating cmyb-luc and lck-luc. Plasmids for bmal1b, clock1a and cry1aa were constructed elsewhere (41). All primers are listed in Supplementary Table S1.
Generation of ezh2–/– mutant zebrafish
The ENU-induced ezh2–/– mutant zebrafish (sa1199) was recovered from the Zebrafish Mutation Project (51). A point mutation (C to T) results in a premature stop codon that leads to a truncated protein of only 17 amino acids (aa). The fragment containing the mutation was PCR amplified with primers ezh2F1 and ezh2R1 (Supplementary Table S1), and confirmed by DNA sequencing. ezh2 homozygous zebrafish was obtained by incrossing ezh2 heterozygotes.
Generation of ezh2 heat shock-inducible transgenic zebrafish
Transgenic zebrafish were generated using the Tol2-basse Multisite Gateway system (52). Briefly, zebrafish ezh2 or mutated ezh2 was cloned into the pME-MCS vector with XhoI and BamHI sites, using primers ezh2pMEF and ezh2pMER (Supplementary Table S1). The resulting plasmids, pME-ezh2 and pME-ezh2m, were mixed with p5E-hsp70l, p3E-polyA and pDestTol2CG2 (EGFP driven by the cmlc2 promoter) and performed LR reaction, respectively, resulting in pDestTol2-hsp70l:ezh2;CG2 and pDestTol2-hsp70l:ezh2m;CG2. To generate transgenic fish, 50 pg of pDestTol2-hsp70l:ezh2;CG2 or pDestTol2-hsp70l:ezh2m;CG2 with 25 pg of Tol2 transposase mRNA was injected into one-cell stage zebrafish embryos, respectively. Then injected embryos were cultured in Petri dishes with embryo medium at 28.5°C for 3 days. The larvae with GFP fluorescence in the heart were selected and raised to adulthood. Tg(hsp70l:ezh2;CG) and Tg(hsp70l:ezh2m;CG) were identified by EGFP fluorescence in the heart and confirmed by PCR using primers ezh2-F5 and ezh2-R5 (Supplementary Table S1). Transgenic larvae were heat shocked at 37°C for one hour at ZT0 or ZT12 and used for locomotor assays or qRT-PCR analyses.
Cell culture, transfection and luciferase assay
Human embryonic kidney (HEK) 293T cells purchased from American Type Culture Collection (Manassas, VA) were cultured according to ATCC’s instructions. Transfection and luciferase assays were performed as described previously (53). Briefly, 0.05 × 106 HEK293T cells were seeded into 24-well plate. After growing to 90% confluency, cells were transfected with 50 ng of per1b-luc or other reporter vectors, 100 ng each of bmal1b and clock1a with or without 600 ng of ezh2. pRL-TK was co-transfected to normalize transfection efficiency. Twenty four hours after transfection, cells were harvested to perform luciferase assays.
Co-immunoprecipitation and Western Blotting
HEK293T cells were seeded in 10-cm Petri dishes. After reaching 90% confluency, cells were transfected with 6 ug each of bmal1b-HA or bmal1b-His, clock1a-Flag and ezh2-His or ezh2-HA, and 2 ug of cry1aa-His. Forty eight hours after transfection, cells were lysed in RIPA buffer with protease inhibitor (Sigma) and subjected to co-immunoprecipitation (Co-IP) according to manufacturer's instructions (Sigma). Briefly, cell lysates were released with protein G–Sepharose beads conjugated with anti-HA antibodies. After washing five times, the precipitates were re-suspended in SDS–PAGE sample buffer, boiled for 5 min and run on 12% SDS–PAGE gel, and then western blotting (WB) was conducted with mouse monoclonal anti-His (Cell Signaling Technology), anti-HA (Sigma) or anti-Flag (Cell Signaling Technology) antibody or rabbit polyclonal anti-Tubulin antibody. To detect the protein levels in zebrafish embryos, 48-hpf wild-type and ezh2–/– mutant embryos were collected and lysated with RIPA buffer, respectively. Equal amount of wild-type and ezh2 embryo extracts were loaded on SDS-PAGE, and then WB was performed using antibodies against EZH2 (Millipore), Eed (Millipore), H3K27m3 (Millipore), H3K27m2 (Millipore), H3K27m1 (Millipore), H3 (Cell Signaling Technology).
Whole mount in situ hybridization (WISH) and quantitative real-time PCR analysis
Whole mount in situ hybridization (WISH) was conducted as previously described (54), zebrafish embryos were fixed with PFA at 26 hpf, 28 hpf or 96 hpf and the fixed embryos were dehydrated with methanol and stored at –20°C. Embryos were rehydrated with sequential rehydration solution and then incubated in 50% formamide hybridization buffer with DIG-labeled probes at 65°C for overnight. Nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Roche) were used for colorimetric detection. For each WISH experiment, 25–30 larvae were used. At least three independent WISH experiments were conducted for each probe.
For quantitative real-Time PCR (qRT-PCR) analysis, total RNAs were extracted using TRIzol Reagent (Invitrogen) according to the manufacturer's instruction. After DNase treatment, RNA (1 μg) was reverse-transcribed with oligo(dT)18 primers and M-MLV reverse transcriptase. qRT-PCR was carried out in an Step-One real-time PCR detection system (ABI) using SYBR® Premix Ex Taq™ (Takara Bio Inc., Dalian, Liaoning). Primers for qRT-PCR are listed in Supplementary Table S2 and some were previously reported (41). At least three independent samples were examined each with triplicate for qRT-PCR. The mRNA level of interested genes were calculated using 2−ΔΔCT method (55) and presented as relative (-fold) values of the control group after normalized by β-actin mRNA levels.
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed as described previously (41). Briefly, two groups of 200 wild-type larvae at 120 or 132 hpf were collected, cross-linked in 2% formaldehyde at room temperature for 30 min, and the crosslinking was terminated by adding 1/10 V of 1.12 M glycine, followed by PBS washing (3 times, each for 10 min). The following procedures were performed according to the manufacturer's protocol (Millipore's ChIP assay kit). The purified rabbit or mouse IgG (Invitrogen) was used as negative controls. ChIP PCR reactions were performed using primers flanking the E-box, D-box, and RORE sites as well as primers not flanking the these sites in the 5′ promoter regions of genes as controls, respectively. Primers for the ChIP-qPCR are listed in Supplementary Table S1.
Morpholino and mRNA microinjection
ezh2 translation blocking Morpholinos (ezh2-MO: 5′-CCGATTTCCTCCCGGTCAATCCCAT-3′) and a standard control MO (cMO: 5′-CCTCTTACCTCAGTTACAATTTATA-3′) (Gene Tools) were re-suspended in water as a 4 mM stock solution and diluted to the appropriate concentrations for microinjections in 0.2% PhenolRed and 0.1 M KCl. Different amounts of ezh2-MO and control MO were injected into zebrafish embryos at one-two cell stage. The efficacy of ezh2-MO was determined by GFP reporter assays (56).
ezh2 mRNA was synthesized using a commercial kit and linearized plasmid DNA as a template (mMESSAGE mMACHINE kit; Ambion). 1 nl (200 pg) ezh2 mRNA was microinjected into one-cell stage zebrafish embryos. After microinjection, embryos were raised in Petri dishes with embryo medium at 28.5°C. Embryos were collected for RNA extraction or fixed with PFA for in situ hybridization at 26 hpf and 28 hpf, respectively, and kept in methanol at -20°C until use.
Zebrafish behavioral assays
Zebrafish larvae behavioral assays were performed under both DD and LD conditions, respectively, as previously reported (41). Briefly, a single zebrafish larva was placed in each wells of 48-well plate at 4 dpf (24 wild-type, 24 ezh2 MO-injected or ezh2-overexpressing transgenic fish). For ezh2-overexpressing transgenic fish, the fish was heat shocked at 37°C for one hour at ZT12. The 48-well plate was placed inside the Zebrabox (Videotrack; ViewPoint Life Sciences, Montreal, France) where continuous infrared light and white light were illuminated from 9:00 A.M. to 11:00 P.M. (LD) and constant 10% dim light (30 lx) was illuminated throughout the procedure (DD) (57). Instruments were placed in a chamber to maintain a constant temperature of 28.5°C. The Videotrack quantization parameters were set as described previously (58). Locomotor activities of larvae were monitored for five consecutive days using an automated video-tracking system (ViewPoint Life Sciences), and the movement of each larva was recorded using Zebralab3.10 software (ViewPoint Life Sciences).
Deep sequencing-based transcriptome analysis
Total RNAs from ezh2–/– mutant and wild-type zebrafish at 28 hpf were extracted and purified. A total amount of 3 μg RNA per sample was used for the RNA sample preparations. Sequencing libraries were generated using NEBNext®Ultra™ RNA Library Prep Kit (NEB, USA) following the manufacturer's instructions. Clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit (Illumia, PE-401–3001). After clustering, the library preparations were sequenced on an Illumina Hiseq 2500 platform. Perl scripts were used for removing the adapter and duplication sequences, calculating the Q20, Q30, GC-content, and then generating the raw reads. Transcriptome assembly was performed according to the protocol described previously (59). Assembled sequences were batch blasted against the latest version of the annotated zebrafish genome assembly (GRCz10) and to determine their gene names and Ensembl IDs. Gene expression levels were estimated using FPKM values (fragments per kilobase of exon per million fragments mapped) by the Cufflinks software (60). Gene abundance differences between those samples were calculated based on the ratio of the FPKM values. For the Gene Ontology (GO) enrichment analysis, the differentially expressed genes (DEGs) was implemented by the GOseq R packages based Wallonia noncentral hyper-geometric distribution (61). For the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, we used KOBAS software to test the statistical enrichment of DEGs in KEGG pathways to predict and classify functions of the DEGs (62).
Statistical analysis
Values are presented as means ± standard deviation (S.D.). Expression levels of genes were analyzed by the JTK cycle method (63), where P < 0.05 was considered as rhythmic expression. Differences between two groups were analyzed by unpaired Student's t-test and statistical significance was accepted at P < 0.05 or smaller.
RESULTS
Zebrafish ezh2 is a circadian clock-controlled gene
While Ezh2 is known to be regulated by various oncogenic transcription factors, miRNAs and non-coding RNAs (18,64,65), little is known about its circadian regulation. We first examined expression of ezh2 mRNA in two consecutive days with qRT-PCR. Results show that zebrafish ezh2 mRNA is rhythmically expressed under both DD (Figure 1A) and LD conditions (Figure 1B). Because Per1b inhibits clock gene expression through E-box (41), and Rev-erbα suppresses gene expression through RORE (42), we examined ezh2 expression in per1b–/– and rev-erbα–/– mutant zebrafish. Results show that ezh2 mRNA is up-regulated in the per1b–/– mutant (Figure 1C and D) and rev-erbα–/– mutant zebrafish (Figure 1E and F) under both DD (Figure 1C and E) and LD conditions (Figure 1E and F). These results suggest that zebrafish ezh2 is controlled by the circadian clock.
Figure 1.
Zebrafish ezh2 is a circadian clock-controlled gene. (A, B) ezh2 is expressed rhythmically in developing zebrafish embryos under DD (A) and LD (B) conditions. (C–F) Up-regulation of ezh2 in per1b–/– (C, D) and rev-erbα–/– (E, F) mutant zebrafish under DD (C, E) and LD (D, F) conditions. For (A) to (F), at least three independent experiments were performed. Levels of mRNA expression were analyzed with the JTK-CYCLE method. ADJ.P for adjusted minimal P-values (P < 0.05). Student's t-test was conducted. *P < 0.05; **P < 0.01; ***P < 0.001. (G) Schematic of the E-box in the ezh2 promoter and the RORE in its first intron. The fragments containing the E-box and RORE were cloned into the pGL3.17-promoter vector, generating the ezh2-E-luc and ezh2-R-luc constructs, respectively. (H, I) Luciferase reporter assays show that ezh2-E-luc is activated by Clock1a and Bmal1b but inhibited by Cry1aa (H) and ezh2-R-luc is activated by Rarαa but inhibited by Rev-erbα (I). ezh2-Em-luc wherein the E-box was mutated; ezh2-Rm-luc wherein the RORE was mutated. Student's t-test was performed. **P < 0.01. (J, K) ChIP-qPCR assays show that Bmal1b binds to the E-box region in the ezh2 promoter (J) and Roaα binds to the RORE in the first intron (K). Student's t-test was performed. **P < 0.01.
Bioinformatic analysis revealed one E-box in the zebrafish ezh2 promoter and one RORE in its first intron (Figure 1G), implying that zebrafish ezh2 expression may be mediated by these cis-regulatory elements. We isolated the DNA fragments harboring the E-box and the RORE and cloned them into the pGL3.17-promoter vector, respectively. Luciferase reporter assays show that ezh2 E-box-mediated luciferase activities are activated by the Clock1a-Bmal1b complex (Figure 1H) but repressed by Cry1aa, a negative element in the circadian clock; and these E-box-mediated luciferase activities were completely abolished when the E-box sequence, CACGTG, was mutated into CCCGTC (Figure 1H). Further, ezh2 RORE-mediated luciferase activities are activated by Rorαa but suppressed by Rev-erbα, and these activities were diminished when RORE was mutated (Figure 1I). We also performed ChIP-qPCR assays with zebrafish embryos. Results show that Bmal1b rhythmically binds to the E-box in the ezh2 promoter (Figure 1J and Supplemental Figure S1A), while Rorαa rhythmically binds to the RORE in its first intron (Figure 1K and Supplemental Figure S1B). Together, these results indicate that zebrafish ezh2 is regulated directly by the circadian clock via both E-box and RORE.
Generation and characterization of an ezh2 null mutant zebrafish
To investigate the functions of ezh2 in vivo, we recovered an ENU-induced ezh2 mutant line (sa1199) from the Zebrafish Mutation Project (ZMP) (51). A point mutation (C to T) (Figure 2A) in Exon 2 (Figure 2B) results in a premature stop codon that leads to a truncated peptide with only 17 amino acids (aa), missing its major functional domains (Figure 2C). The ezh2 heterozygous fish appear normal outwardly and can survive until adulthood, while ezh2 homozygous embryos display a little curved tail and thin yolk sac extension ∼28 hpf (hours postfertilization) (Figure 2D), short body length and small eyes after 48 hpf (Figure 2D), and die after 7 dpf (days postfertilization). We further confirmed this phenotype by knocking it down with an ezh2 Morpholino (ezh2-MO). We conducted EGFP reporter assays to determine the efficacy of the ezh2-MO. As shown in Supplemental Figure S2, 4 ng ezh2-MO effectively inhibits ezh2 expression. Results showed that ezh2 morphants phenocopy the ezh2–/– mutant (Supplemental Figure S3A and B).
Figure 2.
Generation and characterization of a zebrafish ezh2 null mutant. (A) The C to T point mutation in the ezh2 coding region was confirmed by DNA sequencing. WT, wild-type. (B) Gene structure of ezh2 and the point mutation occurs in Exon 2. (C) The C to T nonsense mutation results in a truncated peptide and losses of its major functional domains. SANT, SWI3, ADA2, N-CoR and TFIIIB'' DNA-binding domains; CXC, Tesmin/TSO1-like CXC domain; SET, Su(var)3–9, Enhancer-of-zeste, Trithorax domain. (D) Images of WT and ezh2–/– mutant zebrafish from 24 hpf to 72 hpf. Note that the ezh2–/– mutant zebrafish display curved tail and thin yolk sac extension around 28 hpf and short body length and small eyes after 48 hpf. Scale bar, 0.5 mm. (E) Western Blotting shows that expression of Ezh2 is abolished and that of H3K27 mono-, di- and trimethylation are reduced in ezh2–/– mutant fish. Proteins of zebrafish larvae were extracted with RIPA buffer at 48 hpf. Western Bloting was performed with indicated antibodies. (F) Quantification of western bloting images shown in (E) with ImageJ. Student's t-test was conducted. ***P <0.001. (G, H) qRT-PCR analysis of Ezh2-targeted gene dab2ipa (G) and tmem48 (H) in 28 hpf WT and ezh2–/– mutant zebrafish. Three independent experiments were conducted. Statistical analysis was performed using student's t-test. *P < 0.05.
To examine whether this ezh2 mutation is a null allele, we first performed western blotting analysis. Results show that the expression of Ezh2 protein is completely abolished, and mono-, di- and trimethylation of H3K27 are largely reduced in the mutant fish (Figure 2E and F), indicating that Ezh2 plays critical roles in regulating all methylations of H3K27 in zebrafish, which was reconfirmed with ezh2 morphant embryos at 48 hpf (Supplemental Figure S3C). It has been reported that DAB2IP (DOC-2/DAB2 interactive protein) is repressed by EZH2 (66), while tmem48 (NDC1 transmembrane nucleoporin) is activated by EZH2 (25). We examined expression of dab2ipa and tmem48 in ezh2–/– mutant zebrafish with qRT-PCR. Results show that dab2ipa is up-regulated (Figure 2G), while tmem48 is down-regulated (Figure 2H) in ezh2–/– mutant zebrafish, consistent with previous results (25,66). These results suggest that the ezh2–/– mutant zebrafish is a null allele.
Ezh2 plays important roles in the zebrafish circadian clock
EZH2 is shown to play an important role in the mouse liver circadian clock in vitro (26). To investigate the role of Ezh2 in zebrafish circadian regulation, we examined expression of core circadian clock genes in ezh2–/– mutant fish by qRT-PCR. Results show down-regulation of per1a, per1b, cry1aa, and cry1ab in the ezh2–/– mutant zebrafish under both DD and LD conditions (Figure 3A–H and Supplemental Figure S4A–H). Down-regulation of these genes is largely resulted from loss of ezh2, rather than zebrafish embryonic lethality, because we also observed that a zebrafish developmental gene, her9 (67,68) and its target gene dab2ipa are up-regulated (Figure 2G and Supplementary Figure S5) during early development. Moreover, knocking down ezh2 by a Morpholino resulted in down-regulation of these circadian clock genes (Figure 3I–L). We then generated an ezh2 heat shock-inducible transgenic zebrafish, Tg(hsp70l:ezh2;CG2) (Supplemental Figure S6). In contrast, qRT-PCR results showed that overexpression of ezh2 by either heat shock-induction or capped mRNA microinjection up-regulates these clock genes (Figure 3M–P and Supplemental Figure S7). These results suggest that Ezh2 contributes to zebrafish circadian regulation.
Figure 3.
Ezh2 is required for the zebrafish circadian clock. (A–H) qRT-PCR shows down-regulation of key circadian clock genes per1a (A, E), per1b (B, F), cry1aa (C, G), and cry1ab (D, H) in ezh2–/– mutants under DD (A–D) and LD (E–H) conditions. Three independent experiments were conducted. mRNA expression levels were analyzed as in Figure 1. *P < 0.05; **P < 0.01; ***P < 0.001. (I–L) Down-regulation of per1a (I), per1b (J), cry1aa (K) and cry1ab (L) in ezh2 morphants. After injection, total RNAs were extracted at 26 hpf (ZT2) and 38 hpf (ZT14),respectively. Student's t-test was conducted. *P < 0.05. (M–P) Overexpression of ezh2 driven by a heat shock promoter results in up-regulation of per1a (M), per1b (N), cry1aa (O) and cry1ab (P). Heat shock was conducted at ZT0 (96 hpf) and ZT12 (108 hpf) for 1 h, respectively, and samples were collected at ZT2 and ZT14. Three independent experiments were performed. Statistical analysis was performed using Student's t-test. *P < 0.05; **P < 0.01.
The ezh2–/– mutant zebrafish lose movement after 3 dpf. In order to examine the effects of Ezh2 on zebrafish circadian regulation, we used ezh2 morphants and Tg(hsp70l:ezh2;CG2) fish to perform locomotor activity assays. Results showed that ezh2 morphants display 0.2-h lengthened period (Figure 4A and B), reduced locomotor amplitude (Figure 4A and C) and 0.25-h delayed phase (Figure 4A and D) under DD condition, and enhanced locomotion activity under LD condition (Figure 4E–H); whereas overexpression of ezh2 by heat shocking inducible promoter hsp70l led to 0.8-h shortened period (Figure 4I and J), elevated amplitude (Figure 4G and K) and 1.4-h advanced phase (Figure 4I and L) under DD condition, and elevated locomotor activities under LD condition (Figure 4M–P). It is intriguing that both the morphant larvae and heat shock-induced overexpressing larvae display elevated locomotor activities under LD condition, but the underlying mechanisms need further investigation. Together, these results indicate that Ezh2 plays important roles in the zebrafish circadian clock.
Figure 4.
Altered locomotor rhythms in ezh2 morphants and ezh2-overexpressing zebrafish larvae. (A–H) Locomotor activities were monitored and analyzed in ezh2 morphants and control zebrafish larvae under DD (A–D) or LD (E–H) condition. The period (B), amplitude (C) and phase (D) under DD condition and total average moving distances (F), at day (G) and night (H) under LD conditions were shown. Student's t-test was conducted. *P < 0.05; **P < 0.01; ***P < 0.001. (I–P) Locomotor assays were conducted in ezh2-overexpressing and control zebrafish larvae under LD (I–L) or LD (N–P) condition. The period (J), amplitude (K) and phase (L) under DD condition and total average moving distances (N), at day (O) and at night (P) under LD conditions were shown. Overexpression of ezh2 was done with heat shock at 108 hpf (ZT12) for 1 h, and then locomotor assays were performed. Student's t-test was conducted. *P < 0.05, **P < 0.01; ***P < 0.001.
Zebrafish Ezh2 enhances clock gene expression independent of its histone methyltransferase activity
To delineate how zebrafish Ezh2 regulates the circadian clock, we performed Co-IP assays to examine whether zebrafish Ezh2 can interact with clock proteins. Results show that zebrafish Ezh2 binds to the Clock1a–Bmal1b–Cry1aa complex (Figure 5A and B). We also performed luciferase reporter assays with E-boxes-containing per1b-luc. Results show that per1b promoter activities are activated by the Clock1a–Bmal1b complex, and Ezh2 enhances Clock1a-Bmal1b-mediated activities in a dosage-dependent manner, while Cry1aa represses the activities (Figure 5C). However, EZH2 was shown to bind to the clock complex in the mouse liver, and particularly to enhance transcriptional repression mediated by mouse CRY proteins (26). Indeed, we reconfirmed the results that mouse EZH2 inhibits CLOCK–BMAL1 transcriptional activities on Per1-luc (Supplementary Figure S8A), implicating different mechanisms underlying roles of EZH2/Ezh2 in mice and zebrafish circadian regulation.
Figure 5.
Zebrafish Ezh2 enhances expression of clock genes through binding to core clock components independent of its H3K27 methyltrasnsferase activity. (A, B) Co-IP assays show that zebrafish Ezh2 complexes with Bmal1b, Clock1a, and Cry1aa. (C) Luciferase reporter assays show that zebrafish Ezh2 enhances transcription activities of Clock1a and Bmal1b in a dose-dependent manner. Co-transfection was performed as indicated, and luciferase activities were assayed 24 h after transfection. Experiments were performed in three independent experiments. Transactivation activities are expressed as fold increase over the control group. Values are expressed as means ± S.D., n = 3. Student's t-test was conducted. *P < 0.05; **P < 0.01; ***P < 0.001. (D) Luciferase reporter assays show that Ezh2 enhances expression of circadian clock genes independent of its H3K27 methyltrasnsferase activity. Student's t-test was conducted. **P < 0.01; ***P < 0.001. (E) Co-IP assays show that Clock1a and Bmal1b bind to Ezh2 but not to Eed. (F, G) per1b-luc expression was up-regulated in ezh2-overexpressing zebrafish embryos (F) and down-regulated in ezh2–/– mutant zebrafish embryos (G). 50 pg of per1b-luc plasmid was injected into wild-type or ezh2–/– mutant zebrafish embryos with or without 100 pg ezh2 mRNA. Embryos were homogenized at 28 hpf and luciferase assays were performed. Student's t-test was conducted. **P < 0.01. (H) Co-IP assays show that Bmal1b binds to Ezh2 in zebrafish. (I, J) ChIP-qPCR assays show that Bmal1b (I) and Ezh2 (J) bind to the E-box of the zebrafish per1b promoter region. Student's t-test was conducted. *P < 0.05; **P < 0.01; ***P < 0.001.
While EZH2 was shown to primarily mediate gene silencing via recruitment of the PRC2 (17), EZH2 also functions to enhance target genes in human cancers independent of PRC2 (24,25). We hypothesized that mouse EZH2 and zebrafish Ezh2 contribute to circadian regulation with different mechanisms. To examine this hypothesis, the two amino acid residuals, F681 and H703 in zebrafish and F667 and H689 in mice, critical for the Ezh2/EZH2 H3H27 methyltransferase activity were mutated (Supplementary Figure S9) (25,48,49), respectively. Luciferase reporter assays showed that zebrafish Ezh2 enhances Clock1a-Bmal1b transcriptional activity regardless of mutations of the two critical residuals (Figure 5D), and one of the PRC2 members, Eed, has no such activity (Figure 5D). Intriguingly, mouse EZH2 represses CLOCK-BMAL transcriptional activity (Supplementary Figure S8A). However, the mutated EZH2 loses this repressive activity and surprisingly elevates CLOCK-BMAL transcriptional activity (Supplementary Figure S8B). Moreover, Co-IP assays showed that Eed binds to Ezh2 but not Clock1a and Bmal1b in zebrafish (Figure 5E), suggesting that just Ezh2 alone, rather than PRC2, interacts with clock proteins. These results indicate that Ezh2 enhances Bmal1b-Clock1a transcription activity independent of its histone methyltransferase activity in zebrafish.
We also examine the functions of Ezh2 in vivo by injecting per1b-luc and ezh2 mRNA into wild-type or ezh2–/– mutant zebrafish. Results showed that overexpressing ezh2 up-regulates per1b-luc promoter activities in zebrafish embryos (Figure 5F), while per1b-luc activities are down-regulated in the ezh2–/– mutant zebrfish embryos (Figure 5G). In line with in vitro result, in vivo Co-IP assays showed that Bmal1b binds to Ezh2 but not to Eed in zebrafish embryos (Figure 5H). ChIP-qPCR assays showed that both Bmal1b and Ezh2 rhythmically bind to the E-box in the per1b promoter (Figure 5I, J and Supplementary Figure S1C) and these bindings are largely reduced in ezh2–/– mutant (Figure 5I and J). Moreover, zebrafish larvae overexpressing ezh2 with two mutated residues by heat shock display 0.5-h shortened period, elevated amplitude and 0.8-h advanced phase of locomotor activities under DD condition (Supplementary Figure S10A–D), elevated locomotor activities under LD condition (Supplementary Figure S10E–H), and up-regulates per1a, per1b, cry1aa and cry1ab (Supplementary Figure S10I–L), similar to the results obtained from heat shock induction of wild-type Ezh2, further indicating that Ezh2 functions in zebrafish circadian regulation independent of PRC2.
Taken together, these results suggest that Ezh2 plays positive roles in circadian regulation by directly binding to key circadian clock proteins occupying the E-box containing promoter region, and Ezh2 enhances clock function independent of its histone methyltransferase activity.
Hematopoietic defects in ezh2 –/– mutant zebrafish
EZH2 is known to play important roles in normal and malignant hematopoiesis in vitro (16,29). However, the in vivo role of Ezh2 in hematopoiesis is still unclear. Whole mount in situ hybridization (WISH) shows that ezh2 is expressed in the ICM (intermediate cell mass) at 24, 28 and 33 hpf and PBI (posterior blood island) at 33 hpf (Supplementary Figure S11), consistent with a previous observation (50). We also observed that ezh2–/– mutant embryos display reduced blood cells at 32 hpf (Supplemental Movie S1). To search for Ezh2-affected genes involved in hematopoiesis, we performed high-throughput RNA sequencing of ezh2–/– mutant and wild-type embryos at 28 hpf. The transcriptome analysis reveals 893 up-regulated genes and 425 down-regulated genes in the ezh2–/– mutant zebrafish (Figure 6A, and Supplementary Table S2) (The data was deposited into NCBI Gene Expression Omnibus, GEO accession number GSE103913). Further, both GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) analyses of these DEGs show that genes involved in developmental processes, transcriptional activities, MAPK and calcium signaling pathways, and vascular smooth muscle contraction are markedly altered in the ezh2–/– mutant (Supplemental Figure S12). In particular, genes related to hematopoiesis, such as hbaa1, cmyb, gata1a, scl, alas2, hbbe1, hbbe3, hbbe3, hbae3 and hbae1, are down-regulated (Figure 6B), which was reconfirmed with qRT-PCR (Figure 6C). These results implicate that Ezh2 plays a pivotal role in hematopoiesis in vivo.
Figure 6.
Disrupted expression of hematopoietic genes in ezh2–/– mutant zebrafish revealed by transcriptome analysis. (A) Numbers of differentially expressed genes (DEGs) in ezh2–/– mutant zebrafish at 28 hpf, revealed by transcriptome analysis. (B) Histogram of 10 hematopoietic DEGs in ezh2–/– mutant zebrafish. Red and green colors represent up-regulation and down-regulation, respectively. (C) Nine out of 10 down-regulated hematopoietic genes revealed by transcriptome analysis are reconfirmed by independent qRT-PCR analysis. Three independent experiments were performed. Statistical analysis was performed using Student's t-test. *P < 0.05; **P < 0.01.
Ezh2 is essential for primitive hematopoiesis in zebrafish
To further investigate roles of Ezh2 in hematopoiesis, we performed blood cell staining and WISH. O-dianisidine staining shows that red blood cells are dramatically reduced in the ezh2–/– mutant embryos at 33 hpf (Figure 7A and B). To determine whether the primitive hematopoiesis is affected in ezh2–/– mutant fish, we performed whole mount in situ hybridization using gata1 (a marker for erythroid progenitors) (69–71), pu.1 (a marker for myeloid progenitors) (72), hbbe1 (a marker for erythroid cells) (73), mpx (a marker for granulocytes) (74), and l-plastin (a marker for macrophages) (75). Results show that gata1 (Figure 7C and D) and pu.1 (Figure 7E and F) are markedly reduced in the ezh2–/– mutant ICM at 26 hpf, while hbbe1 (Figure 7G and H), mpx (Figure 7I and J), and l-plastin (Figure 7K and L) are down-regulated in the ezh2–/– mutant PBI at 28 hpf. In addition, primitive marker genes gata1 (Supplementary Figure S13A and B) and pu.1 (Supplementary Figure S13C and D) are also down-regulated in ezh2 morphant embryos, consistent with the results observed in the ezh2–/– mutant embryos.
Figure 7.
Primitive hematopoiesis is disrupted in ezh2–/– mutant zebrafish. (A, B) O-dianisidine (O-d) staining of wild-type sibling (A) and ezh2–/– mutant fish (B) at 33 hpf. Black arrowhead indicates hemoglobin-staining, which is significantly reduced in ezh2–/– mutant zebrafish. (C–L) In situ hybridization of wild-type (C, E, G, I and K) and ezh2–/– mutant embryos (D, F, H, J and L) using probes of gata1 (C, D), pu.1 (E, F), hbbe1 (G, H), mpx (I, J), l-plastin (K, L) at 26 hpf (C-F) and 28 hpf (G-L), respectively. White arrowheads in (C) and (G) indicate ICM (Intermediate cell mass) and black arrowheads in (E, I and K) PBI (Posterior blood island). Scale bar, 0.25 mm. (M–P) Images of gata1:dsRed cells in WT (M, O) and ezh2–/– mutant (N, P) fish at 24 hpf (M, N) and 28 hpf (O, P). (Q–T) Images of mpx:eGFP cells in wild-type (Q, S) and ezh2–/– mutant (R, T) fish at 28 hpf (Q, R) and 48 hpf (S, T). (U, V) Images of lyz:eGFP cells in wild-type (U) and ezh2–/– mutant (V) fish at 28 hpf. White arrowheads in (M–R) indicate ICM and red arrowheads in (S–V) PBI. Scale bar in (M–V), 0.25 mm. All images shown are lateral view, anterior to left.
To further visualize primitive hematopoiesis defects in ezh2–/– mutant zebrafish, we crossed ezh2–/– mutant zebrafish with transgenic zebrafish lines of several hematopoietic markers. Living imaging results show that loss of Ezh2 leads to reduced gata1:dsRed cells in the ICM at 24 hpf (Figure 7M and N) and 28 hpf (Figure 7O and P), reduced mpx:eGFP cells (Figure 7Q to T) and reduced lyz:eGFP cells (Figure 7U and V) in the trunk region at 28 hpf (Figure 7Q, R, U and V) and 48 hpf (Figure 7S and T). Together, these results suggest that Ezh2 is required for primitive hematopoiesis.
Ezh2 is required for definitive hematopoiesis and specification of HSCs in zebrafish
To investigate the role of Ezh2 in definitive hematopoiesis, we examined expression of runx1 (76), c-myb (77), and ikaros (78), markers for HSCs in the ventral wall of the dorsal aorta in ezh2–/– mutant zebrafish by WISH. Results show that runx1, c-myb, and ikaros are dramatically reduced in the ventral wall of the dorsal aorta in ezh2–/– mutant embryos at 28 hpf (Figure 8A to F), suggesting that the specification of HSCs is affected in ezh2–/– mutant zebrafish. We also examined expression of markers for lymphoid cells in ezh2–/– mutant zebrafish. WISH results show that ikaros, lck (79), and rag-1 (80) all lose their expression domains in ezh2–/– mutant zebrafish at 96 hpf (Figure 7G–L), implicating the lymphoid defects in ezh2–/– mutant zebrafish. Further, definitive hematopoiesis markers runx1 (Supplementary Figure S13E and F) and cmyb (Supplementary Figure S13G and H) are all down-regulated in ezh2 morphant embryos. In vivo living image results show that cmyb:eGFP cells (Figure 8M to P) and coro1a:eGFP cells (Figure 8Q–T) are reduced in the ICM (Figure 8M, N, Q and R) and the thymus (Figure 8O, P, S and T) in ezh2–/– mutant zebrafish. Together, these results suggest that Ezh2 plays a critical role in definitive hematopoiesis and specification of HSCs in zebrafish.
Figure 8.
Definitive hematopoiesis is disrupted in ezh2–/– mutant zebrafish. (A–L) In situ hybridization of WT sibling (A, C, E, G, I and K) and ezh2–/– mutant embryos or larvae (B, D, F, G, J and L) using probes of runx1 (A, B), cmyb (C, D), ikaros (E, H), lck (I, J), rag-1 (K, L) at 28 hpf (A, F) and 96 hpf (G, L), respectively. White arrowheads indicate ICM in A, C and E and black arrowheads the thymus in G, I and K. Scale bar, 0.25 mm. (M–P) Images of cmyb:eGFP cells in wild-type (M, O) and ezh2–/– mutant (N, P) fish at 32 hpf (M, N) and 72 hpf (O, P). (Q–T) Images of coro1a:eGFP cells in wild-type (Q, S) and ezh2–/– mutant (R, T) fish at 28 hpf (Q, R) and 72 hpf (S, T). White arrowheads indicate ICM in M, N, Q and R and dashed circle the thymus in O, P, S and T. Scale bar in (M–T), 0.25 mm. All images shown are lateral view, anterior to left.
Specification and development of hemangioblasts and vascular endothelial cells are not affected in ezh2–/– mutant zebrafish during early development
Because HSCs and endothelial cells (ECs) are both derived from hemangioblast (40,81), we next examined expression of hemangioblast markers. WISH shows that expression of fli1a, a marker for hemangioblas (77) at zebrafish early development, is not altered in ezh2–/– mutant zebrafish in comparison with that of its wild-type siblings at 80% epiboly (Supplementary Figure S14A and B). scl, a marker for hemangioblasts (82), HSCs, and angioblasts, has the same expression pattern in ezh2–/– mutant zebrafish as in its wide-type siblings at 80% epiboly (Supplementary Figure S14C and D). We then examined endothelial development using markers fli1a and flk1. Results show that the vasculature develops normally in ezh2–/– mutant zebrafish at 24 hpf (Supplementary Figure S14E to H). These results indicate that specification and development of hemangioblasts and ECs are not affected during early development of ezh2–/– mutant embryos in spite of its hematopoietic defects.
Hematopoietic genes are regulated by the circadian clock and Ezh2 complex
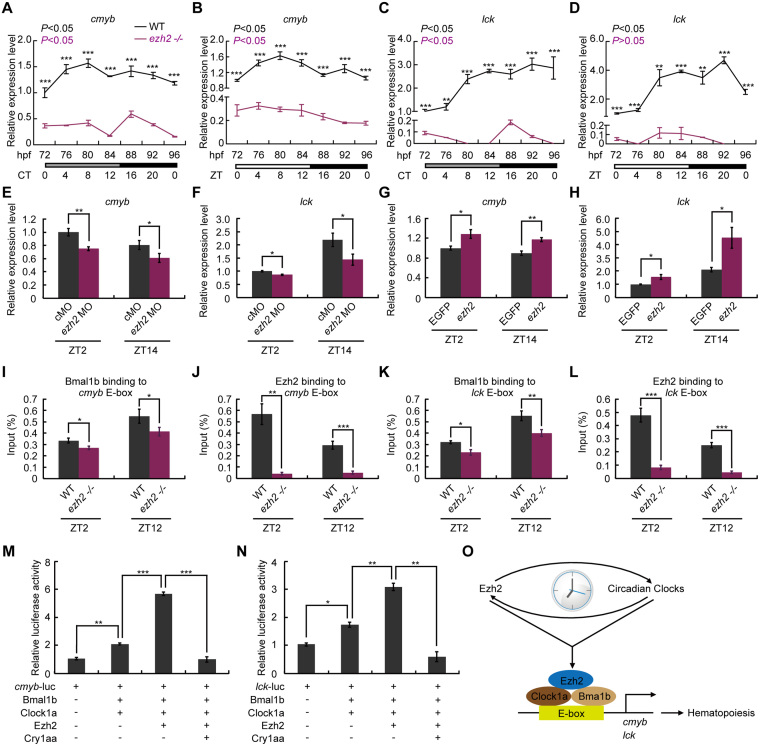
The results that ezh2 is controlled by the circadian clock and hematopoietic genes are down-regulated in ezh2–/– mutant fish led us to hypothesize that most, if not all, hematopoietic genes, are rhythmically expressed. To examine this hypothesis, we first analyzed approximately 2.5-kb 5′ promoter regions of hematopoietic genes and found that there are E-boxes in the cmyb and lck contain promoter regions. qRT-PCR results show that cymb (Figure 9A and B) and lck (Figure 9C and D) are rhythmically expressed in zebrafish larvae under both DD and LD conditions but are down-regulated in ezh2–/– mutant zebrafish, which were also observed in ezh2 morphants (Figure 9E and F). In contrast, overexpressing ezh2 by mRNA injection up-regulates these genes (Figure 9G and H). ChIP-qPCR results showed that Bmal1b and Ezh2 rhythmically bind to the E-boxes of cmyb (Figure 9I, J and Supplemental S1D) and lck (Figure 9K, L and Supplemental S1E). Furthermore, luciferase reporter assays showed that Bmal1b-Clock1a activates cmyb-luc (Figure 9M) and lck-luc (Figure 9N), which is enhanced by Ezh2 (Figure 9M and N). Taken together, these results suggest that both the circadian clock and Ezh2 play regulatory roles in zebrafish hematopoiesis.
Figure 9.
Ezh2 and the circadian clock regulate zebrafish hematopoietic genes. (A–D) qRT-PCR analysis of cmyb (A, B) and lck (C, D) in WT and ezh2–/– mutant under DD (A, C) and LD (B, D) conditions. Three independent experiments were conducted. Rhythmic mRNA expression was analyzed with the JTK-CYCLE method. ADJ.P for adjusted minimal P-values (P < 0.05). Two-way ANOVA with Tukey's post hoc test was conducted, **P < 0.01; ***P < 0.005. (E, F) Down-regulation of cmyb (E) and lck (J) in ezh2 morphants. Student's t-test was conducted. *P < 0.05; **P < 0.01. (G, H) Up-regulation of cmyb (G) and lck (H) in ezh2- overexpressing zebrafish. Student's t-test was conducted. *P < 0.05; **P < 0.01. (I–L) ChIP-qPCR results show that Bmal1b and Ezh2 bind to E-boxes in the promoters of cmyb (I, J) and lck (K, L). Student's t-test was conducted. *P < 0.05; **P < 0.01; ***P < 0.001. (M, N) Luciferase reporter assays show that cmyb-luc (M) and lck-luc (N) are activated by Bmal1b-Clock1a, respectively, and Ezh2 enhances Bmal1b-Clock1a transcription activities. Three independent experiments were performed. Statistical analysis was performed using Student's t-test. *P < 0.05; **P < 0.01; ***P < 0.005. (O) A model for regulation of hematopoiesis by Ezh2 and the circadian clock in zebrafish. Zebrafish ezh2 is controlled directly by the circadian clock and contributes to circadian regulation by binding to the clock complex. The Ezh2 and clock complex controls hematopoiesis through directly regulating hematopoietic genes, cmyb and lck, mediated by E-boxes, respectively.
DISCUSSION
Regulation of EZH2 expression has been studied extensively in different types of human cancers (18). For instance, E2Fs activate EZH2 expression via binding to its promoter (83). EWS-FLI1 and NF-κB2 activate EZH2, while SNF5 represses it (64,84,85). It was also reported that EZH2 is transcriptionally induced by Estradiol, Bisphenol-A, and Diethylstilbestrol through Estrogen-response elements (EREs) (65). But how it is regulated in normal physiology in animals still remains unclear. In animals, ∼43% of transcripts show circadian expression (86,87). However, whether EZH2 is regulated by the circadian clock is unknown to date. Here we show that zebrafish ezh2 is rhythmically expressed but up-regulated in per1b–/– and rev-erbα–/– mutants under both DD and LD condition. Luciferase and ChIP-qPCR reveal that ezh2 is regulated directly by the circadian clock through E-box and RORE. However, statistical analysis of qRT-PCR data shows that ezh2 still remains rhythmic with reduced amplitudes in per1b–/– mutant fish under LD condition and in rev-erbα–/– mutant under DD condition, indicating that both Per1b and Rev-erbα play large roles in the amplitude of ezh2 expression. In addition, the expression level of ezh2 appears gradually decreased during development (Figure 1A and B), suggesting that ezh2 is also affected by unknown developmental cues. A previous study showed that transcription factor CLOCK protein, an essential component of the mammalian circadian system, is a histone acetyltransferase (88). This and our results reveal that the circadian clock could regulate histone remodeling at both transcriptional and translational level.
Posttranslational modifications of histones are essential for transcriptional regulation of gene expression. Histone modification also has found to be evolved as an important mechanism at the core circadian machinery (9). EZH2 was shown to be required for the mouse liver clock in vitro (26). However, because loss of Ezh2 results in lethality during mouse early development (19), effects of EZH2 on clock gene expression and locomotor rhythmicity in live animals are unknown. Here we recovered an ENU-induced ezh2–/– mutant zebrafish that can live up to seven days. Key circadian clock genes are down-regulated in the ezh2–/– mutant zebrafish. Knockdown or overexpression of ezh2 alters zebrafish larval locomotor rhythms. Moreover, consistent with mice EZH2 (26), zebrafish Ezh2 contributes to circadian regulation through binding to the core clock components and regulates clock genes by E-box. Interestingly, we found that zebrafish Ezh2 enhances clock function independent of polycomb repressive complex 2, differing from mice EZH2 that augments the repressive action of CRY1 (26). Our data suggest that zebrafish Ezh2 and mice EZH2 contribute to circadian function through different mechanisms.
EZH2 was shown to play pivotal roles in stem cell maintenance and have a dual role as either oncogene or tumor-suppressor gene, depending on gene dosages and cell context progression of several types of tumors, including breast and prostate cancers (18,20,89). EZH2 also is important for normal and malignant hematopoiesis (28,29). However, due to embryonic lethality of Ezh2–/– mutant mice, whether EZH2 regulates hematopoiesis in early development remains unclear. We found that both primitive and definitive hematopoiesis is affected in ezh2–/– mutant zebrafish, as evidenced that loss of Ezh2 results in reduced numbers of erythroid cells, granulocytes, macrophages, T cells, and HSCs in live zebrafish. These are not likely due to developmental delay because the mutant fish embryos appear normal outwardly before 28 hpf. Our results are consistent with a previous study that knocking out Ezh2 specifically in the mouse liver impaired expansion of HSCs and reduced the number of megakaryocyte-erythroid progenitors, but moderately reduced common myeloid progenitors and granulocyte-macrophage progenitors (90).
Circadian rhythms are highly conserved among organisms ranging from prokaryotes to humans, to maintain coordination with the daily changes of light and temperature. However, little is known about circadian control of hematopoiesis. It has been reported that hematopoietic stem cell release is regulated by circadian oscillations (91). Circadian clock gene Bmal1 regulates diurnal oscillations of Ly6Chi inflammatory monocytes (92). EZH2 was found to be overexpressed in natural killer/T-cell lymphoma (NKTL), and directly activates CCND1 in NKTL without histone methyltransferase activity (93). We found that cmyb and lck involved in hematopoiesis display rhythmic expression, which are down-regulated in ezh2–/– mutant zebrafish; importantly, zebrafish Bmal1b and Ezh2 regulate cmyb and lck through directly binding to the E-box in their promoters, indicating that Ezh2 promotes normal hematopoiesis via directly enhancing key hematopoietic marker genes in zebrafish. However, even though we observed rhythmic expression of other important hematopoietic genes, for instance, gata1a, pu.1, mpx and ikaros, exactly how Ezh2 and the circadian clock regulate them would need to be investigated in the future. Nonetheless, our results reveal that the circadian clock modulates hematopoiesis in an intricate manner, i.e. the circadian clock regulates ezh2 directly and the Ezh2-circadian clock complex in turn regulates hematopoiesis (Figure 9O).
In summary, zebrafish ezh2 is regulated directly by the circadian clock. Ezh2 promotes clock function and hematopoiesis independent of its histone methyltransferase activity.
DATA AVAILABILITY
The data was deposited into NCBI Gene Expression Omnibus, GEO accession number GSE103913.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Feng Liu and Wenqing Zhang for sharing plasmids of pu.1, hbbe1, mpx, plastin, c-myb, runx1, ikaros, lck and rag-1; Leonard Zon, Philip Crosie, Zilong Wen, Shuo Lin and Stephen Renshaw for sharing transgenic lines of Tg(cmyb:EGFP), Tg(lyz:EGFP),Tg(coro1a:EGFP),Tg(gata1:DsRED) and Tg(mpx:EGFP); and Mishra Kumar for carefully reading the manuscript.
Author contributions: Y.Z. and H.W. designed the study; Y.Z., Q.Y., C.C. and M.W. performed experiments; Y.Z. and H.W. analyzed data and wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Basic Research Program of China (973 Program) [2012CB947600]; National Natural Science Foundation of China (NSFC) [31200877, 31030062, 81570171]; Jiangsu Planned Projects for Postdoctoral Research Funds [1401013A]; Jiangsu Distinguished Professorship Program [SR13400111]; Natural Science Foundation of Jiangsu Province [BK2012052]; Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions [YX13400214]; High-Level Innovative Team of Jiangsu Province, and the ‘333’ project of Jiangsu Province [BRA2015328]. Funding for open access charge: National Basic Research Program of China (973 Program) [2012CB947600]; National Natural Science Foundation of China (NSFC) [31200877, 31030062, 81570171]; Jiangsu Planned Projects for Postdoctoral Research Funds [1401013A]; Jiangsu Distinguished Professorship Program [SR13400111]; Natural Science Foundation of Jiangsu Province [BK2012052]; Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions [YX13400214]; High-Level Innovative Team of Jiangsu Province, and the ‘333’ project of Jiangsu Province [BRA2015328].
Conflict of interest statement. None declared.
REFERENCES
- 1. Takahashi J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017; 18:164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Klein D.C., Moore R.Y., Reppert S.M.. Suprachiasmatic Nucleus: The Mind's Clock. 1991; NY: Oxford University Press. [Google Scholar]
- 3. Yamazaki S., Numano R., Abe M., Hida A., Takahashi R., Ueda M., Block G.D., Sakaki Y., Menaker M., Tei H.. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000; 288:682–685. [DOI] [PubMed] [Google Scholar]
- 4. Lee C., Etchegaray J.P., Cagampang F.R., Loudon A.S., Reppert S.M.. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001; 107:855–867. [DOI] [PubMed] [Google Scholar]
- 5. Gekakis N., Staknis D., Nguyen H.B., Davis F.C., Wilsbacher L.D., King D.P., Takahashi J.S., Weitz C.J.. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998; 280:1564–1569. [DOI] [PubMed] [Google Scholar]
- 6. Kume K., Zylka M.J., Sriram S., Shearman L.P., Weaver D.R., Jin X., Maywood E.S., Hastings M.H., Reppert S.M.. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999; 98:193–205. [DOI] [PubMed] [Google Scholar]
- 7. Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., Schibler U.. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002; 110:251–260. [DOI] [PubMed] [Google Scholar]
- 8. Ripperger J.A., Merrow M.. Perfect timing: epigenetic regulation of the circadian clock. FEBS Lett. 2011; 585:1406–1411. [DOI] [PubMed] [Google Scholar]
- 9. Etchegaray J.P., Lee C., Wade P.A., Reppert S.M.. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003; 421:177–182. [DOI] [PubMed] [Google Scholar]
- 10. Naruse Y., Oh-hashi K., Iijima N., Naruse M., Yoshioka H., Tanaka M.. Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol. Cell. Biol. 2004; 24:6278–6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brown S.A., Ripperger J., Kadener S., Fleury-Olela F., Vilbois F., Rosbash M., Schibler U.. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science. 2005; 308:693–696. [DOI] [PubMed] [Google Scholar]
- 12. Ripperger J.A., Schibler U.. Rhythmic CLOCK–BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 2006; 38:369–374. [DOI] [PubMed] [Google Scholar]
- 13. Jones M.A., Covington M.F., DiTacchio L., Vollmers C., Panda S., Harmer S.L.. Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:21623–21628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu S.X., Knowles S.M., Webb C.J., Celaya R.B., Cha C., Siu J.P., Tobin E.M.. The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 2011; 155:906–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Paro R., Harte P.J.. Russo VEA, Martienssen RA, Riggs AD. The role of Polycomb group and trithorax group chromatin complexes in the maintenance of determined cell states. Epigenetic Mechanisms of Gene Regulation. 1996; NY: Cold Spring Harbor Laboratory Press; 507–528. [Google Scholar]
- 16. Lund A.H., van Lohuizen M.. Polycomb complexes and silencing mechanisms. Curr. Opin. Cell Biol. 2004; 16:239–246. [DOI] [PubMed] [Google Scholar]
- 17. Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y.. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002; 298:1039–1043. [DOI] [PubMed] [Google Scholar]
- 18. Chang C.J., Hung M.C.. The role of EZH2 in tumour progression. Br. J. Cancer. 2012; 106:243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. O’Carroll D., Erhardt S., Pagani M., Barton S.C., Surani M.A., Jenuwein T.. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 2001; 21:4330–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim K.H., Roberts C.W.. Targeting EZH2 in cancer. Nat. Med. 2016; 22:128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee S.T., Li Z., Wu Z., Aau M., Guan P., Karuturi R.K., Liou Y.C., Yu Q.. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol. Cell. 2011; 43:798–810. [DOI] [PubMed] [Google Scholar]
- 22. LaJeunesse D., Shearn A.. E(z): a polycomb group gene or a trithorax group gene. Development. 1996; 122:2189–2197. [DOI] [PubMed] [Google Scholar]
- 23. Strutt H., Cavalli G., Paro R.. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 1997; 16:3621–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cavalli G. Molecular biology. EZH2 goes solo. Science. 2012; 338:1430–1431. [DOI] [PubMed] [Google Scholar]
- 25. Xu K., Wu Z.J., Groner A.C., He H.H., Cai C., Lis R.T., Wu X., Stack E.C., Loda M., Liu T. et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012; 338:1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Etchegaray J.P., Yang X., DeBruyne J.P., Peters A.H., Weaver D.R., Jenuwein T., Reppert S.M.. The polycomb group protein EZH2 is required for mammalian circadian clock function. J. Biol. Chem. 2006; 281:21209–21215. [DOI] [PubMed] [Google Scholar]
- 27. Tang Q., Cheng B., Xie M., Chen Y., Zhao J., Zhou X., Chen L.. Circadian clock gene Bmal1 inhibits tumorigenesis and increases paclitaxel sensitivity in tongue squamous cell carcinoma. Cancer Res. 2017; 77:532–544. [DOI] [PubMed] [Google Scholar]
- 28. Lund K., Adams P.D., Copland M.. EZH2 in normal and malignant hematopoiesis. Leukemia. 2014; 28:44–49. [DOI] [PubMed] [Google Scholar]
- 29. Herviou L., Cavalli G., Cartron G., Klein B., Moreaux J.. EZH2 in normal hematopoiesis and hematological malignancies. Oncotarget. 2016; 7:2284–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Su I.H., Basavaraj A., Krutchinsky A.N., Hobert O., Ullrich A., Chait B.T., Tarakhovsky A.. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 2003; 4:124–131. [DOI] [PubMed] [Google Scholar]
- 31. Roy A., Basak N.P., Banerjee S.. Notch1 intracellular domain increases cytoplasmic EZH2 levels during early megakaryopoiesis. Cell Death Dis. 2012; 3:e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ross J., Mavoungou L., Bresnick E.H., Milot E.. GATA-1 utilizes Ikaros and polycomb repressive complex 2 to suppress Hes1 and to promote erythropoiesis. Mol. Cell. Biol. 2012; 32:3624–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lessard J., Baban S., Sauvageau G.. Stage-specific expression of polycomb group genes in human bone marrow cells. Blood. 1998; 91:1216–1224. [PubMed] [Google Scholar]
- 34. Fukuyama T., Otsuka T., Shigematsu H., Uchida N., Arima F., Ohno Y., Iwasaki H., Fukuda T., Niho Y.. Proliferative involvement of ENX-1, a putative human polycomb group gene, in haematopoietic cells. Br. J. Haematol. 2000; 108:842–847. [DOI] [PubMed] [Google Scholar]
- 35. Herrera-Merchan A., Arranz L., Ligos J.M., de Molina A., Dominguez O., Gonzalez S.. Ectopic expression of the histone methyltransferase Ezh2 in haematopoietic stem cells causes myeloproliferative disease. Nat. Commun. 2012; 3:623. [DOI] [PubMed] [Google Scholar]
- 36. Vatine G., Vallone D., Gothilf Y., Foulkes N.S.. It's time to swim! Zebrafish and the circadian clock. FEBS Lett. 2011; 585:1485–1494. [DOI] [PubMed] [Google Scholar]
- 37. Vallone D., Lahiri K., Dickmeis T., Foulkes N.S.. Start the clock! Circadian rhythms and development. Dev. Dyn. 2007; 236:142–155. [DOI] [PubMed] [Google Scholar]
- 38. Wang M.Y., Huang G.D., Wang H.. [Advances in the zebrafish circadian clock mechanisms]. Yi chuan = Hereditas. 2012; 34:1133–1143. [PubMed] [Google Scholar]
- 39. Zhong Z., Wang M., Huang G., Zhang S., Wang H.. Kumar V. Molecular genetic and genomic analyses of zebrafish circadian rhythmicity. Biological Timekeeping: Clocks, Rhythms and Behaviour. 2017; New Delhi: Springer (India) Pvt. Ltd; 193–209. [Google Scholar]
- 40. de Jong J.L., Zon L.I.. Use of the zebrafish system to study primitive and definitive hematopoiesis. Annu. Rev. Genet. 2005; 39:481–501. [DOI] [PubMed] [Google Scholar]
- 41. Huang J., Zhong Z., Wang M., Chen X., Tan Y., Zhang S., He W., He X., Huang G., Lu H. et al. Circadian modulation of dopamine levels and dopaminergic neuron development contributes to attention deficiency and hyperactive behavior. J. Neurosci. 2015; 35:2572–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang G., Zhang F., Ye Q., Wang H.. The circadian clock regulates autophagy directly through the nuclear hormone receptor Nr1d1/Rev-erbalpha and indirectly via Cebpb/(C/ebpbeta) in zebrafish. Autophagy. 2016; 12:1292–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Traver D., Paw B.H., Poss K.D., Penberthy W.T., Lin S., Zon L.I.. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat. Immunol. 2003; 4:1238–1246. [DOI] [PubMed] [Google Scholar]
- 44. Renshaw S.A., Loynes C.A., Trushell D.M., Elworthy S., Ingham P.W., Whyte M.K.. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006; 108:3976–3978. [DOI] [PubMed] [Google Scholar]
- 45. Hall C., Flores M.V., Storm T., Crosier K., Crosier P.. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev. Biol. 2007; 7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. North T.E., Goessling W., Walkley C.R., Lengerke C., Kopani K.R., Lord A.M., Weber G.J., Bowman T.V., Jang I.H., Grosser T. et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007; 447:1007–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li L., Yan B., Shi Y.Q., Zhang W.Q., Wen Z.L.. Live imaging reveals differing roles of macrophages and neutrophils during zebrafish tail fin regeneration. J. Biol. Chem. 2012; 287:25353–25360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., Reinberg D.. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002; 16:2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Joshi P., Carrington E.A., Wang L., Ketel C.S., Miller E.L., Jones R.S., Simon J.A.. Dominant alleles identify SET domain residues required for histone methyltransferase of Polycomb repressive complex 2. J. Biol. Chem. 2008; 283:27757–27766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sun X.J., Xu P.F., Zhou T., Hu M., Fu C.T., Zhang Y., Jin Y., Chen Y., Chen S.J., Huang Q.H. et al. Genome-wide survey and developmental expression mapping of zebrafish SET domain-containing genes. PLoS One. 2008; 3:e1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kettleborough R.N., Busch-Nentwich E.M., Harvey S.A., Dooley C.M., de Bruijn E., van Eeden F., Sealy I., White R.J., Herd C., Nijman I.J. et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013; 496:494–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kwan K.M., Fujimoto E., Grabher C., Mangum B.D., Hardy M.E., Campbell D.S., Parant J.M., Yost H.J., Kanki J.P., Chien C.B.. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 2007; 236:3088–3099. [DOI] [PubMed] [Google Scholar]
- 53. Zhong Y., Lu L., Zhou J., Li Y., Liu Y., Clemmons D.R., Duan C.. IGF binding protein 3 exerts its ligand-independent action by antagonizing BMP in zebrafish embryos. J. Cell Sci. 2011; 124:1925–1935. [DOI] [PubMed] [Google Scholar]
- 54. Wang H., Zhou Q., Kesinger J.W., Norris C., Valdez C.. Heme regulates exocrine peptidase precursor genes in zebrafish. Exp. Biol. Med. 2007; 232:1170–1180. [DOI] [PubMed] [Google Scholar]
- 55. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 56. Li Y., Xiang J., Duan C.. Insulin-like growth factor-binding protein-3 plays an important role in regulating pharyngeal skeleton and inner ear formation and differentiation. J. Biol. Chem. 2005; 280:3613–3620. [DOI] [PubMed] [Google Scholar]
- 57. Appelbaum L., Wang G., Yokogawa T., Skariah G.M., Smith S.J., Mourrain P., Mignot E.. Circadian and homeostatic regulation of structural synaptic plasticity in hypocretin neurons. Neuron. 2010; 68:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Prober D.A., Rihel J., Onah A.A., Sung R.J., Schier A.F.. Hypocretin/orexin overexpression induces an insomnia-like phenotype in zebrafish. J. Neurosci. 2006; 26:13400–13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011; 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L.. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Young M.D., Wakefield M.J., Smyth G.K., Oshlack A.. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010; 11:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T. et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008; 36:D480–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hughes M.E., Hogenesch J.B., Kornacker K.. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms. 2010; 25:372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. De Donatis G.M., Pape E.L., Pierron A., Cheli Y., Hofman V., Hofman P., Allegra M., Zahaf K., Bahadoran P., Rocchi S. et al. NF-kB2 induces senescence bypass in melanoma via a direct transcriptional activation of EZH2. Oncogene. 2016; 35:2735–2745. [DOI] [PubMed] [Google Scholar]
- 65. Bhan A., Hussain I., Ansari K.I., Bobzean S.A., Perrotti L.I., Mandal S.S.. Histone methyltransferase EZH2 is transcriptionally induced by estradiol as well as estrogenic endocrine disruptors bisphenol-A and diethylstilbestrol. J. Mol. Biol. 2014; 426:3426–3441. [DOI] [PubMed] [Google Scholar]
- 66. Chen H., Tu S.W., Hsieh J.T.. Down-regulation of human DAB2IP gene expression mediated by polycomb Ezh2 complex and histone deacetylase in prostate cancer. J. Biol. Chem. 2005; 280:22437–22444. [DOI] [PubMed] [Google Scholar]
- 67. Latimer A.J., Shin J., Appel B.. her9 promotes floor plate development in zebrafish. Dev. Dyn. 2005; 232:1098–1104. [DOI] [PubMed] [Google Scholar]
- 68. Bae Y.K., Shimizu T., Hibi M.. Patterning of proneuronal and inter-proneuronal domains by hairy- and enhancer of split-related genes in zebrafish neuroectoderm. Development. 2005; 132:1375–1385. [DOI] [PubMed] [Google Scholar]
- 69. Martin D.I., Zon L.I., Mutter G., Orkin S.H.. Expression of an erythroid transcription factor in megakaryocytic and mast cell lineages. Nature. 1990; 344:444–447. [DOI] [PubMed] [Google Scholar]
- 70. Romeo P.H., Prandini M.H., Joulin V., Mignotte V., Prenant M., Vainchenker W., Marguerie G., Uzan G.. Megakaryocytic and erythrocytic lineages share specific transcription factors. Nature. 1990; 344:447–449. [DOI] [PubMed] [Google Scholar]
- 71. Long Q., Meng A., Wang H., Jessen J.R., Farrell M.J., Lin S.. GATA-1 expression pattern can be recapitulated in living transgenic zebrafish using GFP reporter gene. Development. 1997; 124:4105–4111. [DOI] [PubMed] [Google Scholar]
- 72. Bennett C.M., Kanki J.P., Rhodes J., Liu T.X., Paw B.H., Kieran M.W., Langenau D.M., Delahaye-Brown A., Zon L.I., Fleming M.D. et al. Myelopoiesis in the zebrafish, Danio rerio. Blood. 2001; 98:643–651. [DOI] [PubMed] [Google Scholar]
- 73. Quinkertz A., Campos-Ortega J.A.. A new beta-globin gene from the zebrafish, betaE1, and its pattern of transcription during embryogenesis. Dev. Genes Evol. 1999; 209:126–131. [DOI] [PubMed] [Google Scholar]
- 74. Lieschke G.J., Oates A.C., Crowhurst M.O., Ward A.C., Layton J.E.. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood. 2001; 98:3087–3096. [DOI] [PubMed] [Google Scholar]
- 75. Herbomel P., Thisse B., Thisse C.. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development. 1999; 126:3735–3745. [DOI] [PubMed] [Google Scholar]
- 76. Kataoka H., Ochi M., Enomoto K., Yamaguchi A.. Cloning and embryonic expression patterns of the zebrafish Runt domain genes, runxa and runxb. Mech. Dev. 2000; 98:139–143. [DOI] [PubMed] [Google Scholar]
- 77. Thompson M.A., Ransom D.G., Pratt S.J., MacLennan H., Kieran M.W., Detrich H.W. 3rd, Vail B., Huber T.L., Paw B., Brownlie A.J. et al. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev. Biol. 1998; 197:248–269. [DOI] [PubMed] [Google Scholar]
- 78. Willett C.E., Kawasaki H., Amemiya C.T., Lin S., Steiner L.A.. Ikaros expression as a marker for lymphoid progenitors during zebrafish development. Dev. Dyn. 2001; 222:694–698. [DOI] [PubMed] [Google Scholar]
- 79. Langenau D.M., Ferrando A.A., Traver D., Kutok J.L., Hezel J.P., Kanki J.P., Zon L.I., Look A.T., Trede N.S.. In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:7369–7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Willett C.E., Zapata A.G., Hopkins N., Steiner L.A.. Expression of zebrafish rag genes during early development identifies the thymus. Dev. Biol. 1997; 182:331–341. [DOI] [PubMed] [Google Scholar]
- 81. Choi K., Kennedy M., Kazarov A., Papadimitriou J.C., Keller G.. A common precursor for hematopoietic and endothelial cells. Development. 1998; 125:725–732. [DOI] [PubMed] [Google Scholar]
- 82. Liao E.C., Paw B.H., Oates A.C., Pratt S.J., Postlethwait J.H., Zon L.I.. SCL/Tal-1 transcription factor acts downstream of cloche to specify hematopoietic and vascular progenitors in zebrafish. Genes Dev. 1998; 12:621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bracken A.P., Pasini D., Capra M., Prosperini E., Colli E., Helin K.. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003; 22:5323–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Richter G.H., Plehm S., Fasan A., Rossler S., Unland R., Bennani-Baiti I.M., Hotfilder M., Lowel D., von Luettichau I., Mossbrugger I. et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:5324–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wilson B.G., Wang X., Shen X., McKenna E.S., Lemieux M.E., Cho Y.J., Koellhoffer E.C., Pomeroy S.L., Orkin S.H., Roberts C.W.. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010; 18:316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Doherty C.J., Kay S.A.. Circadian control of global gene expression patterns. Annu. Rev. Genet. 2010; 44:419–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhang R., Lahens N.F., Ballance H.I., Hughes M.E., Hogenesch J.B.. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:16219–16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Doi M., Hirayama J., Sassone-Corsi P.. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006; 125:497–508. [DOI] [PubMed] [Google Scholar]
- 89. Tan J.Z., Yan Y., Wang X.X., Jiang Y., Xu H.E.. EZH2: biology, disease, and structure-based drug discovery. Acta Pharmacol. Sinica. 2014; 35:161–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Mochizuki-Kashio M., Mishima Y., Miyagi S., Negishi M., Saraya A., Konuma T., Shinga J., Koseki H., Iwama A.. Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood. 2011; 118:6553–6561. [DOI] [PubMed] [Google Scholar]
- 91. Mendez-Ferrer S., Lucas D., Battista M., Frenette P.S.. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008; 452:442–447. [DOI] [PubMed] [Google Scholar]
- 92. Nguyen K.D., Fentress S.J., Qiu Y., Yun K., Cox J.S., Chawla A.. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science. 2013; 341:1483–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yan J., Ng S.B., Tay J.L., Lin B., Koh T.L., Tan J., Selvarajan V., Liu S.C., Bi C., Wang S. et al. EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood. 2013; 121:4512–4520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data was deposited into NCBI Gene Expression Omnibus, GEO accession number GSE103913.