Abstract
Paraspeckles are nuclear bodies that regulate multiple aspects of gene expression. The long non-coding RNA (lncRNA) NEAT1 is essential for paraspeckle formation. NEAT1 has a highly ordered spatial organization within the paraspeckle, such that its 5′ and 3′ ends localize on the periphery of paraspeckle, while central sequences of NEAT1 are found within the paraspeckle core. As such, the structure of NEAT1 RNA may be important as a scaffold for the paraspeckle. In this study, we used SHAPE probing and computational analyses to investigate the secondary structure of human and mouse NEAT1. We propose a secondary structural model of the shorter (3,735 nt) isoform hNEAT1_S, in which the RNA folds into four separate domains. The secondary structures of mouse and human NEAT1 are largely different, with the exception of several short regions that have high structural similarity. Long-range base-pairing interactions between the 5′ and 3′ ends of the long isoform NEAT1 (NEAT1_L) were predicted computationally and verified using an in vitro RNA–RNA interaction assay. These results suggest that the conserved role of NEAT1 as a paraspeckle scaffold does not require extensively conserved RNA secondary structure and that long-range interactions among NEAT1 transcripts may have an important architectural function in paraspeckle formation.
INTRODUCTION
Long non-coding RNAs (lncRNAs) are defined as non-protein coding RNAs that are longer than 200 nucleotides. In the human genome, more than thirteen thousand lncRNAs have been annotated (1), making up a large proportion of human genes. lncRNAs are involved in gene regulatory functions through diverse mechanisms including chromatin binding (Xist) (2), regulating gene transcription in cis (ANRIL) (3), and scaffolding of nuclear bodies (NEAT1). Intriguingly, although many lncRNA have important conserved functions, they usually have relatively low sequence conservation (1). This is counterintuitive, as sequence conservation is often assumed to be required for genes with important functions (4). One possible explanation is that lncRNA preserve higher order conservation, such as conservation of secondary structure (base pairing interactions) or tertiary structure (three dimensional shape of folded RNA).
Large RNAs fold into secondary structures, which then influence their 3D tertiary structures. Resolving the secondary structures of lncRNAs in vivo is a difficult task due to their large size and low abundance in cells. High-throughput in vivo structure probing using reverse transcription truncation (-seq) methods requires extreme sequence depth for low abundance lncRNAs. Till now, there is only one human lncRNA, Xist, whose structure has been probed in vivo (5). Furthermore, lncRNAs are expressed in alternative isoforms and bound by a variety of RNA binding proteins in vivo, both of which can obscure interpretation of chemical modification patterns. In vitro structure probing interrogates an RNA’s inherent folding potential without interference by bound proteins or alternative transcript isoforms. Although this simplifies the task, the large size of lncRNA still poses a significant challenge, and only a few lncRNA structures have been experimentally characterized in vitro (6) (HOTAIR (7), Xist (8,9) and ncSRA (10) RepA (11) and lincRNAp21 (12)).
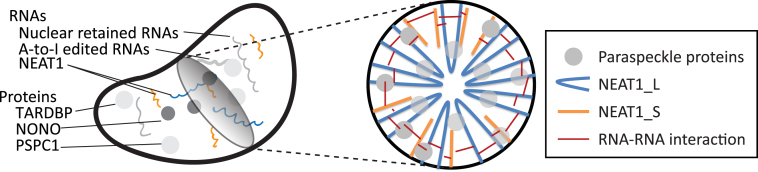
NEAT1 is an especially interesting lncRNA for structural study. It is a key structural component of paraspeckles and is essential for paraspeckle formation. Paraspeckles are nuclear bodies located in the nucleus interchromatin space. Though paraspeckle functions and regulatory mechanisms are not completely understood, recent studies showed they are involved in multiple gene regulatory processes, such as mRNA retention, mRNA cleavage, A-to-I editing (13) and protein sequestration (14). These regulatory functions are responsible for several cellular responses and shown to be associated with the pathology of multiple cancers and neurodegenerative diseases (15–17). Deletion of NEAT1 in mice disrupts development of female reproductive tissues, underscoring the biological importance of this lncRNA (18,19).
NEAT1 has two isoforms that share the same transcription start site, but have different termination sites. In humans, the short isoform NEAT1_S is 3735 nt long with a polyA tail. The long isoform, which is essential for paraspeckle formation, is 22 741 nt in length and has a non-polyadenylated 3′ end produced by RNase P cleavage (20,21). The expression level of NEAT1_S is estimated to be at least five-fold higher than NEAT1_L, and even higher in many tissues and cell types (22,23). Though less abundant, NEAT1_L is considered to be the key isoform for paraspeckle formation. Targeted knock down of NEAT1_L leads to loss of paraspeckles, while de novo paraspeckle formation can be rescued by transient expression of NEAT1_L (20,24). Intriguingly, NEAT1_S can be found outside of the paraspeckle in tissue culture cells, suggesting it may have independent biological functions (25). The two isoform gene structure and the function of NEAT1 in paraspeckle formation were observed in both humans and mice. However, the sequence of NEAT1 is not well conserved between human and mouse. This suggests higher-order conservation of NEAT1 RNAs, such as secondary structural conservation or conserved RNA-protein interactions.
Interestingly, evidence has emerged indicating that the specific structural conformation of NEAT1 might be important for paraspeckle architecture. EM-ISH (electron microscopy-in situ hybridization) studies using DNA probes to the 5′ and 3′ ends of NEAT1_L RNA showed that NEAT1_L has a highly ordered spatial organization within the paraspeckle (15). The 5′ and 3′ ends of NEAT1_L were localized to the paraspeckle periphery, while the central region of NEAT1_L was found within the paraspeckle core. Since the 5′ end of NEAT1_L is identical to NEAT1_S, the short isoform NEAT1_S should also localize to the periphery of paraspeckle. Based on these observations, an ultrastructural paraspeckle model was proposed with two salient features. First, NEAT1_L folds end-to-end. Secondly, multiple folded NEAT1_L and NEAT1_S molecules are regularly organized in the cross sections of paraspeckle, forming a circular skeleton. However, the actual secondary structure of NEAT1 has not yet been characterized. The nature of the spatial organization of NEAT1 and its contribution to paraspeckle architecture is yet to be understood.
Here, we combined high throughput RNA structure probing (Mod-seq) (26) with computational analyses to investigate the structural features of NEAT1. Mapping and comparing the structures of human and mouse NEAT1_S revealed two short regions of similar SHAPE reactivity, and phylogenetic comparisons found relatively little evidence for conservation of RNA secondary structure. Computational analysis identified putative long-range RNA–RNA base paring interactions between NEAT1_L’s 5′ and 3′ ends, which are common in mammals. We propose that the NEAT1 lncRNA has maintained its function as a paraspeckle scaffold with little structural conservation, and identify a strong propensity for long-range intramolecular base-pairing that may contribute to scaffolding the paraspeckle.
MATERIALS AND METHODS
In vitro transcription
hNEAT1_S and mNEAT1_S plasmids were generously provided by Dr Gérard Pierron (27) and Dr Lingling Chen (28), respectively. PCR primers were designed for both full length NEAT1 RNA and short segments, and the SP6 promoter sequence was included in the forward primers. The DNA template for in vitro transcription was amplified from the plasmids using Phusion high-fidelity polymerase and purified by agarose gel extraction. The RNA was in vitro transcribed using Promega RiboMAX large scale RNA production systems (SP6), as described in the manufacturer's instructions. Briefly, 200–500 ng cDNA template, 4 μl 5X SP6 buffer, 4 μl 25 mM rNTPs and 2 μl SP6 enzyme mix were mixed in a 20 μl reaction and incubated at 37°C for 3.5 hours. 0.5 μl RQ1 RNase-Free DNase (1u/μl) were added to each reaction and incubated at 37°C for 15 min to destroy DNA template. 0.5 μl proteinase K (20 mg/ml) was then added to reaction and incubated at 37°C for 1 h to destroy SP6 transcriptase and RQ1 DNase.
Non-denaturing purification of RNA
A non-denaturing purification was adapted from Somarowthu et al. (7) to maintain the co-transcriptionally folded structure for SHAPE probing experiments. Briefly, after proteinase K treatment, the RNA was diluted with 200 μl 1× SHAPE buffer (111 mM NaCl, 111 mM HEPES, 6.67 mM MgCl2), transferred to Amicon Ultra 100K column and centrifuged at 14 000 g for 10 min to concentrate the RNA sample to approximately 30 μl. This dilution/concentration step was repeated for a total of two rounds. The purified RNA was then collected by centrifuging the column upside down 2 min at 1000g. The RNAs were verified on a TapeStation. The RNAs were kept on ice and were immediately used for SHAPE probing.
1M7 synthesis procedure
We synthesized 1M7 using a novel procedure. In brief, 2-amino-4-nitrobenzoic acid was converted to 2-((ethoxycarbonyl)amino)-4-nitrobenzoic acid through the addition of ethyl chloroformate by reflux for 1 h. This product was converted to 7-nitro-1H-benzo[d][1,3]oxazine-2,4-dione by heating at 65°C in the presence of thionylchloride for 30 min, cooled to room temperature and washed with chloroform. The 7-nitro-1H-benzo[d][1,3]oxazine-2,4-dione dissolved in DMF was then treated with potassium carbonate and iodomethane, similar to published methods (29), yielding an orange precipitate containing both 1M7 and a hydrolyzed contaminant (as determined by NMR). Pure 1M7 (light yellow in color) hydrolizes to 2-(methylamino)-4-nitrobenzoic acid (orange in color). Published synthesis methods describe an orange product that is likely contaminated with the hydrolysis product. We purified 1M7 by fractional crystallization from ethyl acetate/hexane where the contaminant crystallized first to yield (40%) of orange crystals, mp 256–258°C. 1M7 crystallized second to yield (50%) of light yellow crystals, mp 206–208°C. 1M7 was resuspended in DMSO at 65 mM and stored at –80°C. The solution retained a light yellow color that turned bright orange when mixed with the RNA sample in SHAPE buffer.
In vitro SHAPE probing with 1M7
RNA secondary structure probing was performed using 1M7 as the SHAPE reagent, as described in Mortimer et al. (29). 2 pmol RNA product were diluted in 13.3 μl 1× SHAPE buffer, incubated at 37°C for 5 min. 1.7 μl 1M7 (65 mM, in DMSO) were then added into each reaction, continue incubation at 37°C for 70 s. The control samples were incubated with same volume of DMSO instead of 1M7. 1M7 probed RNA was then purified using ethanol precipitation method.
Mod-seq library preparation and data processing by mod-seeker pipeline
Probed RNA samples were pooled together for Mod-seq library preparation. At least two replicates were sequenced for 1M7 treated samples and negative control samples (Supplementary Table S1). Mod-seq libraries were generated as previously described (30) and sequenced with an Illumina Miseq sequencer. Sequencing reads were aligned to hNEAT1 or mNEAT1 sequences and replicates were combined for further analysis after checking for correlations. The SHAPE reactivity score is calculated using the equation: SHAPE reactivity = normalized count(treated) – α × normalized count(Ctrl), as described in Spitale et al.(31). Parameter α was set to 0.35 by using in vitro transcribed and probed tetrahymena P4P6 domain (32) (Supplementary Figure S1) as a positive control.
RNA secondary structure modeling
RNA secondary structure models with or without SHAPE probing constraints were generated using RNAstructure software (Linux text interface 64 bit, version 5.8.1; default parameters) (33). SHAPE reactivity scores were used as constraints for RNA secondary structure predictions. To generate RNA secondary structures models of NEAT1 segments, partition functions (34) were first calculated with the ‘partition’ command in RNAstructure; the ‘max expect’ structures (35) were used as RNA structure models, which was calculated using the ‘MaxExpect’ command. For full length hNEAT1_S and mNEAT1_S structure modeling, partition function predictions are computationally intense, so minimum free energy structures were instead calculated with the ‘Fold’ command in RNAstructure. Structure models were stored in ct files and visualized with VARNA (v3.92) (36).
Comparing structures of full length NEAT1 and 3S shotgun segments
To compare structures of full length NEAT1 and segments, we calculated Pearson's correlations of their SHAPE reactivity scores between segments and the corresponding regions in full length NEAT1_S. A similar correlation analysis was done in sliding windows with a window size of 60 nt and a step size of 1 nt.
Infernal alignment and covariation analysis
To identify conserved secondary structure in NEAT1_S, we first used Infernal (default parameters) (37) to generate improved multiple alignments of regions in NEAT1_S as described in (7). Multiple alignments of 99 vertebrates were downloaded from UCSC genome browser database (38), where 64 sequences have alignments to human NEAT1_S region. Covariation models were built using Infernal cmbuild on eight sequences including hNEAT1_S and mNEAT1_S, and then calibrated with cmcalibrate. Improved multiple alignments across 64 species were then generated using cmsearch and cmalign. Finally, covariant base pairs were identified with both R2R (39) using a 15% threshold (7,10) and R-scape using default parameters (6). To compare R-scape results from NEAT1 to those of well-characterized structured RNAs, we subsampled sequence alignments to have similar numbers of sequences in each alignment (∼50) and pairwise sequence identity (average: ∼68%). For covariation score analysis, R-scape's default scoring metric (APC G-test statistics) was used. With Infernal improved alignments of hNEAT1_S and mNEAT1_S, we calculated Pearson's correlation coefficients of SHAPE reactivity scores in each region after aligning SHAPE scores to their sequence alignment.
Generating synthetic NEAT1 alignments with random mutations
For each Infernal aligned region, the hNEAT1_S sequence was used as an ancestor sequence to build random synthetic alignments. In each round of sequence generation, two child sequences were generated from their parent sequence, where point mutations were introduced at random for each nucleotide position with a fixed mutation rate (probability). After seven rounds, 128 sequences were generated. Fifty out of 128 sequences were randomly selected to build each synthetic alignment. This simulation was repeated 100 times each with mutation rates ranging from 0.5% to 5% to generate random null alignment models with average pairwise identity ranging from 60% to 95%. These null alignments were used directly for R2R analyses, or realigned with Infernal before R2R analyses (Supplementary Figure S4).
RNA–RNA interaction prediction
Prediction of long range interactions in NEAT1 was done with RNAduplex (40,41). The sequence of NEAT1_S and the rest of NEAT1_L sequence (after trimming off NEAT1_S sequence) were used as input. In sliding window analyses, NEAT1_L sequence was separated into 120 nt long windows with a step size of 40 nt. The pairwise minimum free energy of each duplex was then predicted using RNA duplex using default parameters.
In vitro gel shift assay
NEAT1 segment templates were generated by PCR from genomic DNA (HEK genomic DNA for hNEAT1 and mouse kidney genomic DNA for mNEAT1). After in vitro transcription with SP6, the predicted interacting NEAT1 segments were treated with RQ DNase and purified with phenol–chloroform extraction and ethanol precipitation as described in RiboMax SP6 kit (Promega). An RNA gel shift experiment was adapted from Gavazzi et al. (42). Briefly, 2 pmol of each RNA segment were mixed in 8 μl H2O, incubated at 90°C for 2 min and then chilled on ice. 4 μl 3× pairing buffer (50 mM Sodium Cacodylate, 40 mM KCl, 0.5/2/6 mM MgCl2) and 0.25U SUPERase-in was added into each reaction and incubated at 37°C for 30 min. RNA duplexes were then assayed by agarose electrophoresis. The duplexes were electrophoresed through a 3% agarose gel in TBM buffer (45 mM Tris, 43 mM borate, 2 mM MgCl2, pH 8.3) for 1 h at 4°C.
eCLIP data analysis
eCLIP RNA binding protein binding site data was downloaded from ENCODE (43) in narrowPeak format. Protein binding sites on NEAT1 were filtered using bedtools intersect. To map the binding sites of TARDBP on NEAT1_S structure, each nucleotide in NEAT1_S was assigned an eCLIP score that equals to the highest signal value among all peaks covering that nucleotide. Nucleotide that has no crosslinking has score of zero. hNEAT1_S structure model was then visualized by VARNA and colored by eCLIP scores. For hierarchy clustering analysis, eCLIP score on each nucleotide was filtered such that it has enough signal enrichment (signal value: >3), and is statistically significant (P-value: <1e–5), and has significant binding sites in both replicates. The mean scores of the two replicates were then used in clustering analysis, where correlation was used as distance matrix with average-link clustering algorithm.
RESULTS
In vitro secondary structure probing of human NEAT1_S
We first used Mod-seq (26) (Figure 1) to probe the in vitro structure of the 3,735 nt human NEAT1 short isoform (hNEAT1_S). Large RNAs often adopt multiple structural folds after heat denaturation and refolding in vitro. To avoid this, we purified in vitro transcribed NEAT1_S under non-denaturing conditions designed to preserve its co-transcriptionally folded structure (7). hNEAT1_S RNA were probed with 1M7 (29), and modification sites were identified using Mod-seq. SHAPE reactivity scores for each nucleotide were then calculated as previously described (31), where higher scores suggest structural flexibility (Supplementary Figure S2). Although modeling long RNA structures with Mod-seq has not been validated, Mod-seq measures SHAPE reactivity accurately (Supplementary Figure S1) and SHAPE reactivity data have been used to model many long RNA secondary structures (6–12,44,45).
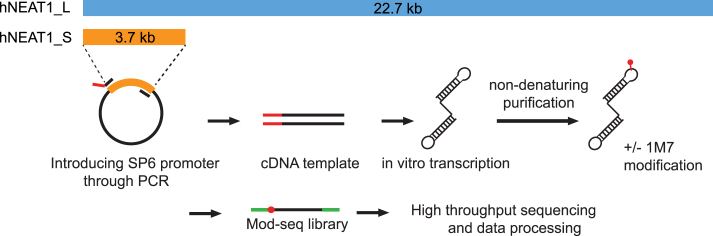
Figure 1.
Overview of NEAT1 secondary structure probing. cDNA templates of NEAT1 regions were generated by PCR using primers that incorporated the SP6 promoter sequence. NEAT1 RNA was then generated by SP6 in vitro transcription. After non-denaturing RNA purification, RNA were probed with the SHAPE reagent, 1M7. The negative controls were treated with DMSO only. Mod-seq libraries were then made and sequenced to an average combined depth of ∼100 reverse transcriptase stops per nucleotide. SHAPE reactivity was calculated by comparing reverse transcriptase stops from 1M7 treated and untreated control samples.
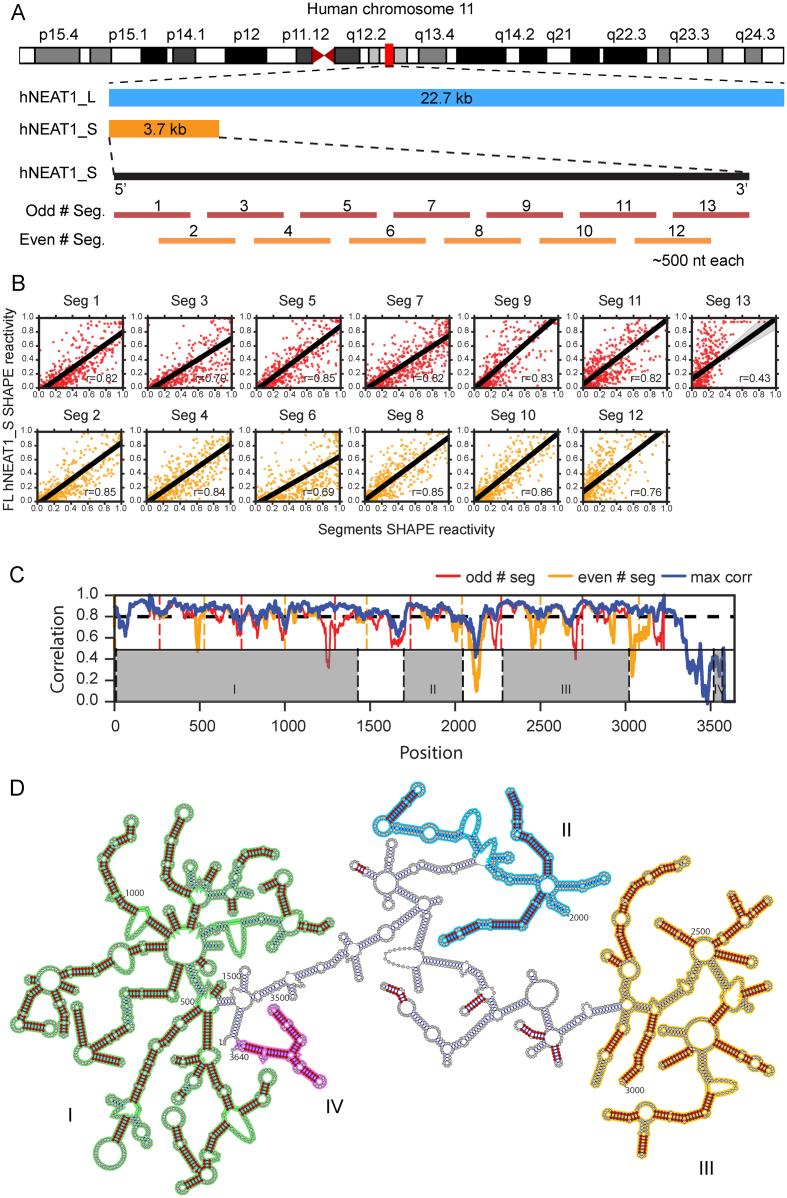
We investigated the domain structure of NEAT1_S using an approach similar to the 3S shotgun method (46). In this approach, full length NEAT1_S was divided into 13 overlapping ∼500 nt segments (Figure 2A and Supplementary Table S2). Each segment was in vitro transcribed and SHAPE probed individually using the same non-denaturing method that we used in full length NEAT1_S probing. If nucleotides within a segment exhibit similar SHAPE reactivity to that seen in the context of full length RNA, they likely form base-pairs within a sub-domain with relatively independent and stable local structure. The similarity of SHAPE scores between each segment and full length NEAT1_S was measured by Pearson's correlation (Figure 2B), finding that most regions appear to have stable local structures. To identify boundaries between local structures, we also evaluated Pearson's correlations in 60-nucleotide sliding windows across NEAT1_S (Figure 2C). These results indicate that hNEAT1_S has primarily local base-pairing interactions when prepared under non-denaturing conditions.
Figure 2.
Identification of local stable structures in hNEAT1_S. (A) Illustration of gene locus of hNEAT1_S and hNEAT1_L. The secondary structure of full length hNEAT1_S and 13 ∼500 nucleotide sub-segments were probed in vitro. (B) Scatter plots showing the correlation of SHAPE reactivity scores in each segment with the corresponding region in full length hNEAT1_S. (C) Pearson's correlations of SHAPE reactivity scores between full length hNEAT1_S and each segment were calculated using a 60-nucleotide sliding window with 1 nucleotide step size. The correlations of hNEAT1_S and even number segments are shown in red, while the correlations of hNEAT1_S and odd number segments are shown in orange. The blue line indicates the larger correlation of the two (odd versus even segments). Odd and even segment boundaries are marked as upper dashed lines. The lower dashed lines indicate boundaries of identified structural domains. (D) Secondary structure models in hNEAT1_S. Shared base-pairs between full length hNEAT1 and the 500 nucleotide sub-segments are marked in red. The four structural domains are highlighted with colors.
To identify stable local subdomains of hNEAT1_S, we compared the secondary structure models of each segment with the 100 lowest free energy structures of full length hNEAT1_S and searched for shared base-pairs (Figure 2D). Six hundred ninety-six shared base-pairs were identified in total, accounting for 57.7% of all base pairs in the full length hNEAT1_S structure. By manually clustering adjacent shared base-pairs, we demarcated four domains in hNEAT1_S that have relatively stable local structures, as highlighted by colors (Figure 2D). Domain I encompasses most of the 5′ end of NEAT1_S, while domains II, III and IV are more separated. Domain IV marks a folded 3′ end. The separation of domains is also observed in the sliding window correlation analysis (Figure 2C), where the correlation of SHAPE reactivity scores is higher within each domain, but drops in junction regions between domains. These results support a model in which NEAT1 folds into a modular multi-domain RNA.
Phylogenetic analyses of NEAT1 secondary structure conservation
We used phylogenetic analyses to investigate the conservation of the NEAT1_S structure. We first used Infernal (37) to generate improved mammalian multiple alignments of NEAT1_S using our SHAPE constrained structure model. As it is possible that only small subdomains of NEAT1_S have conserved structure, we applied Infernal to compact helical regions from the domains defined using the 3S shotgun procedure (see methods; Supplementary Table S3). For 12 of 14 subdomains, Infernal identified at least 40 out of 64 mammalian species with significant alignment to human NEAT1_S. Two regions in domain III (nt 2470–2609 and nt 3199–3316) had only 12 and 25 alignments, respectively, and the former one only had alignments within primates.
We used R2R (39) and R-scape (6) to evaluate the conservation of NEAT1_S secondary structure. R2R classifies base-pairs as covarying if at least one compensatory mutation is present in an alignment, given there are less non-canonical base pairs than a user-defined threshold. R-scape uses a background null distribution to identify statistical significant covariant base-pairs, but performance depends on the number of alignments used and their average pairwise identity. Some lncRNAs have covariant base-pairs identified by R2R (7,11) but many failed the statistical tests in R-scape (6). Similarly, R2R identified many more covariant base pairs than R-scape on NEAT1_S (Supplementary Figure S3 A and B). However, R2R may be too liberal and / or R-scape too conservative for analysis of NEAT1_S structural conservation. Further analyses suggest R2R is prone to false-positive covariation calls on NEAT1_S (Supplementary materials; Supplementary Figures S4D and E), and that R-scape has reasonably strong performance on well-structured RNAs (tRNA, riboswitches, TERC, etc.) after matching alignment number and pairwise identity to that of NEAT1_S (Supplementary Figure S5). NEAT1_S alignments had higher R-scape co-variation scores than random null alignments (Supplementary Figure S4F and G), however NEAT1_S had relatively few significant covariant base pairs (E value < 0.05; Supplementary Figure S5). These results suggest that NEAT1_S is under less selective pressure for specific RNA structures than well-known highly-structured RNAs.
SHAPE probing of mouse NEAT1_S identifies several structurally similar regions
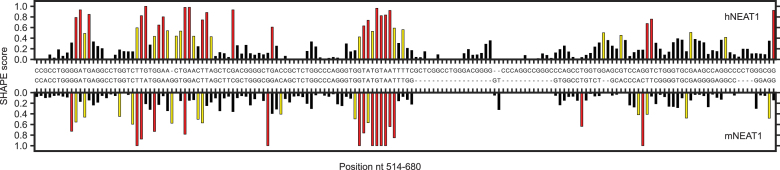
Since most human lncRNAs only exist in mammals and are much younger than structured small non-coding RNAs, the R-scape E-value significance threshold of 0.05 may be too stringent for lncRNAs. In addition, it's possible that lncRNAs like NEAT1 have conserved single-stranded regions that would be undetectable using R-scape. To experimentally evaluate the conservation of NEAT1 structure, we compared the in vitro structures of human NEAT1_S and mouse NEAT1_S. A secondary structural model of mNEAT1_S was determined using the same pipeline for hNEAT1_S (Supplementary Figure S6). Both full-length mNEAT1_S and 12 overlapping segments (Supplementary Table S2) were in vitro transcribed and probed with 1M7, and their SHAPE reactivity profiles were assayed by Mod-seq. We compared the SHAPE reactivity profiles of hNEAT1_S and mNEAT1_S using the Infernal derived mammalian NEAT1_S sequence alignment to align their SHAPE scores. Out of 10 regions with well-defined sequence alignments, 5 had significantly positive correlations (nt 514–680, nt 901–1036, nt 1037–1268, nt 1269–1467, nt 1710–1833) (Supplementary Table S3). The nt 514–680 region had the highest correlation (R = 0.43; Figure 3), suggesting higher structural similarity, even though R-scape identified no covariant base pairs in this region. These results show NEAT1 has several small regions with evidence for structural similarity, while other regions have much lower structural conservation.
Figure 3.
Comparison of SHAPE reactivity profiles for the region of hNEAT1_S found to have the highest correlation with the corresponding region in mNEAT1_S (nts 514–680). SHAPE scores (see methods) are plotted for hNEAT1 (upper) and mNEAT1 (lower).
Long range RNA–RNA interactions in NEAT1
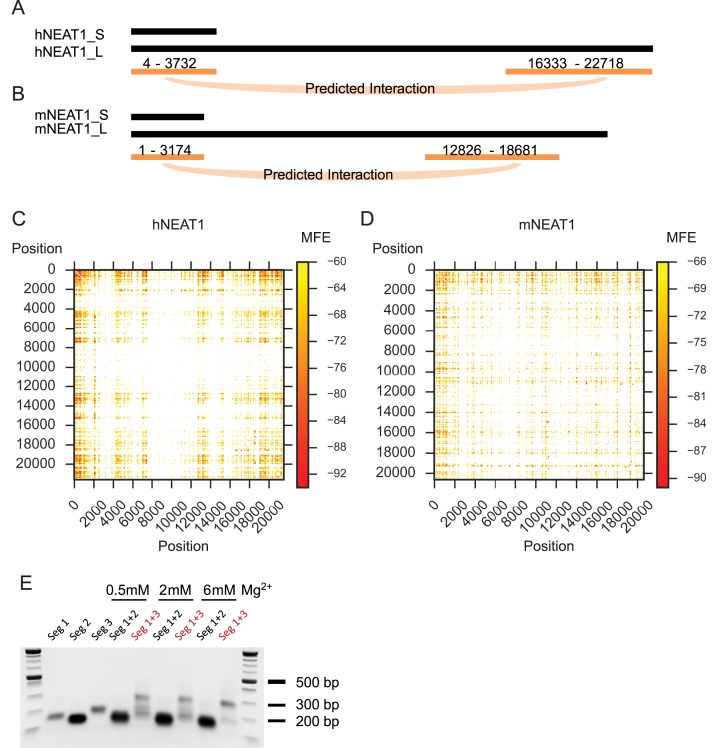
Previous studies have reported that the 5′ and 3′ ends of NEAT1 are co-localized in the paraspeckle periphery, and speculated that this is a consequence of interactions among RNA-binding proteins (27), We investigated the possibility that long range RNA–RNA interactions might contribute to colocalization. We used RNAduplex, a software package for predicting structure upon hybridization of two RNA, with hNEAT1_S sequence and the remaining 19,006 nt sequence of hNEAT1_L to identify potential long range interactions. Surprisingly, RNAduplex predicted a large interaction of almost the entire short hNEAT1 with the 3′ end of long hNEAT1. The prediction is similar in mouse NEAT1, with mNEAT1_S predicted to form a duplex with the 3′ end sequence of mNEAT1_L (Figure 4A and B). To further investigate the potential for long range interactions, we separated human and mouse NEAT1_L sequences into 120 nt windows and calculated the minimum free energy of each pair of windows (Figure 4C and D). Both in human and mouse, duplex minimum free energy heat maps show darker colors at the edges and corners. These long range interaction regions in hNEAT1_L and mNEAT1_L have significantly lower minimum free energy (z-scores < –3) than random pairs of NEAT1_L sequences (Supplementary Figure S7A and B). This pattern is consistent across mammals (Supplementary Figure S7B). These results show that NEAT1 has a conserved inherent capacity to form long-range interactions between its 5′ and 3′ ends.
Figure 4.
Putative long-range base-pairing interaction in mammalian NEAT1 RNAs. (A and B) RNAduplex analyses of NEAT1_S and NEAT1_L predict NEAT1_S is likely to interact with the 3′ end of NEAT1_L, in both human and mouse. (C and D) RNAduplex analysis of pair-wise 120 nt window regions of NEAT1_L. The heatmaps are colored by the predicted minimum free energy of each RNA duplex. These predicted interactions are significantly stronger than expected by chance along NEAT1 RNAs in mammals (Supplementary Figure S6). (E) In vitro gel shift assay shows the predicted interacting RNA segments (seg 1 and seg 3) form a duplex in vitro. The duplex product is visible as a band that migrates similar to the 300 nt DNA ladder on the native agarose gel.
Based on our windowed analysis of base-pairing potential, we predicted RNA segments most likely to form long-range interactions by searching for the best candidate segment pairs (Supplementary Table S4). Selected RNA–RNA interactions of predicted regions were tested using an in vitro RNA–RNA gel shift assay (Figure 4E and Supplementary Figure S8). As predicted, hNEAT1 segment 1 (nt 282–546) and hNEAT1 segment 2 (nt 600–840) formed a stable duplex structure with segment 3 (nt 20761–21120). In mNEAT1, the predicted regions also show RNA–RNA interaction ability, though the interaction seems to be weaker than the tested hNEAT1 segments (Supplementary Figure S8). These results show that sequences in the 5′ and 3′ ends of NEAT1 can form base-pairing interactions under physiological Mg2+ concentration.
Mapping RBP binding sites on the NEAT1_S secondary structure model
A recent study by West et al. (47) investigated the localization of proteins within the paraspeckle. TARDBP was identified as a shell component that co-localizes with the NEAT1_L 3′ and 5′ ends, while other paraspeckle proteins such as SFPQ, NONO, FUS and PSPC1 were identified as core components expected to associate the with middle region of NEAT1_L. Public eCLIP data generated by the ENCORE project shows four significant clusters of TARDBP binding sites on NEAT1. Two sites are located within NEAT1_S, while one is in 3′ end of NEAT1_L (Supplementary Figures S9 and S10). Strikingly, our predicted long-range interacting region in each of the 5′ end and 3′ end is adjacent to a TARDBP associated region (∼40 nt apart). Thus RNA–RNA interactions and NEAT1–TARDBP interactions could act cooperatively to stabilize a NEAT1 circular scaffold within the paraspeckle (Figure 5).
Figure 5.
Model of NEAT1’s architectural function in scaffolding the paraspeckle. NEAT1_L RNA folds end to end. RNA–RNA interactions between the 5′ end and 3′ end of NEAT1_L, or between NEAT1_S and NEAT1_L 3′end help form a circular skeleton for the paraspeckle.
We also examined the binding sites of all 160 proteins with available ENCODE eCLIP data. After stringent filtering, 50 out of 160 proteins have significant binding sites on NEAT1_L. Hierarchical clustering analyses of these binding sites are shown in (Supplementary Figure S11). Two other paraspeckle proteins, SFPQ and NONO, are clustered together. These two proteins are known to form dimers and localize to the core region of the paraspeckle, consistent with their eCLIP binding sites.
DISCUSSION
It has been an intriguing mystery that lncRNA often have very little sequence conservation even when they appear to have conserved biological functions. One hypothesis is that secondary structures, rather than primary sequences, are more likely to be conserved in lncRNA. In this study, we compared the structure of human and mouse NEAT1, the lncRNA component of paraspeckles. Our phylogenetic analyses and Mod-seq structure probing results suggest that most of the NEAT1 secondary structure is undergoing evolutionary drift, leaving only a few short regions of structural similarity and very few specific base pairs with significant covariation. Thus, secondary structure conservation alone is not sufficient to explain NEAT1’s functional conservation. Other molecular interactions are likely important for scaffolding the paraspeckle.
Previous studies on the organization of NEAT1 within paraspeckles reported that the 5′ and 3′ ends are co-localized to the paraspeckle periphery. However, the nature of co-localization is not well understood. Our computational analyses and in vitro gel shift experiments suggest that the 5′ and 3′ ends of NEAT1 could form long-range base-pairing interactions. In the 5′ end of NEAT1, the regions most likely to form such interactions (nt 282–546 and nt 600–840) flank a region of highly conserved SHAPE probing (nt 514–680). It's possible that local structures in the interacting segments may be required for long-range interactions with the 3′ end of NEAT1_L. Future studies, including targeted mutation around this region, would help evaluate its role in paraspeckle formation. Since NEAT1_S and NEAT1_L share the same transcription start site, the NEAT1_S sequence is identical to the NEAT1_L 5′ end sequence. Thus, our predicted intramolecular interaction between the 5′ and 3′ ends of NEAT1_L could also occur between separate molecules of NEAT1_S and NEAT1_L. Such interactions could form a network of RNA–RNA basepairs that help shape the architecture of the paraspeckle (Figure 5).
Recently, several groups reported high-throughput analysis of RNA–RNA interactions mapped by in vivo psoralen crosslinking of RNA helices (PARIS (48), LIGR-Seq (49) and SPLASH (50) methods). Notably, 435 out of 1206 base-pairs (36.1%) in our in vitro hNEAT1_S structure model are supported by PARIS data (48), (Supplementary Figure S12). However, only 59 out of 298 PARIS RNA–RNA interactions were also observed in our structure model. This discord likely stems from the fact that PARIS samples a population of alternative or intermediate structures, while SHAPE probing of in vitro transcribed NEAT1 assays a homogenous, single RNA transcript. Interestingly, the PARIS data include seven crosslink reads consistent with a long-range base-pairing interaction between the 5′ and 3′ ends of NEAT1_L (nt 3172–3190 and nt 21219–21264, Supplementary Figure S10). The fact that this is a very small fraction of the total mapped interactions suggests that each NEAT1 molecule may have only few intramolecular interactions in the paraspeckle. Alternatively, as NEAT1_S is expressed 5–8 fold more than NEAT1_L and can be localized as single-transcript ‘microspeckles’ outside of the paraspeckle (25), the PARIS data may reflect mostly intermolecular interactions among separate NEAT1_S transcripts. Finally, the AMT psoralen used in PARIS is biased towards crosslinking U residues in adjacent AU pairs (51), such that long-range interactions involving GC pairs would be difficult to identify with PARIS. In addition, some RNA–RNA interactions supported by PARIS may require protein binding in the in vivo environment.
Previous work suggested that two other lncRNAs, SRA and HOTAIR, have conserved secondary structure supported by co-varying nucleotides in genomic sequence alignments (7,10). A more recent computational analysis using R-scape (6) reported that the apparently conserved base pairing seen in these lncRNAs was no more common than expected by chance. However, R-scape may have suffered from a lack of power due to having too few alignments of lncRNA genes. Our analyses suggest that R-Scape has the power to identify conserved base pairs in highly structured RNAs, even when applied to a smaller number of alignments with mutation rates similar to those of lncRNAs. Furthermore, our simulations illustrate that using R2R can result in random mutations being interpreted as evidence of co-varying base pairs on NEAT1_S.
As more and more genomes are sequenced, the power to identify significant covariation with tools like R-scape will increase. However, it may be wrong to assume that lncRNA structural conservation is comparable to that of deeply conserved, ancient structured RNAs like tRNA, rRNA, and RNase P RNA. Because lncRNA are relatively young (in evolutionary terms), they may not have yet evolved as many constraints on their secondary and tertiary structure. For example, tRNA must be recognized by multiple processing enzymes and synthetases, in addition to their interactions with the translation machinery, all in the space of ∼70 nucleotides. In comparison, lncRNAs are much longer and may have fewer sequence and structural-specific interactions. This would explain the observation that these RNAs have generally less conserved structure (6).
Our comparative structural analysis on NEAT1 serves as a case study of lncRNA structural evolution. With the exception of a few short regions, the secondary structure of NEAT1 has changed extensively over evolutionary time. Thus the conserved function of NEAT1 cannot be explained solely by conserved secondary structure. It is possible that maintaining certain small regions of NEAT1 in single-stranded conformation, is a conserved structural feature. This is consistent with the regions of correlated SHAPE signal we observed in human and mouse NEAT1_S. In addition, there may be non-canonical RNA–RNA interactions in NEAT1 (e.g. pseudoknots) that are not accommodated by most structure modeling software. We propose a model in which a small number of short regions in the NEAT1 RNA have important specific base-pairs, while the rest remains structurally heterogeneous, allowing multiple intermolecular interactions among RNA binding proteins and separate molecules of NEAT1 RNA.
DATA AVAILABILITY
Mod-seq data have been deposited to the NCBI Sequence Read Archive, under accession number SRP128926.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Ling-Ling Chen and Dr Gérard Pierron for sharing plasmids encoding mouse and human NEAT1 lncRNA, Dr Andrea Berman for sharing plasmids encoding the Tetrahymena ribozyme. We thank Howard Chang and Zhipeng Lu for correspondence regarding PARIS data interpretation. We also thank members of the McManus lab for helpful comments on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Kaufman Foundation (to C.J.M.); David Scaife Family Charitable Foundation (to M.P.B.). Funding for open access charge: Laboratory start-up funds (to C.J.M.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012; 22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Simon M.D., Pinter S.F., Fang R., Sarma K., Rutenberg-Schoenberg M., Bowman S.K., Kesner B.A., Maier V.K., Kingston R.E., Lee J.T.. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013; 504:465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Congrains A., Kamide K., Ohishi M., Rakugi H.. ANRIL: molecular mechanisms and implications in human health. Int. J. Mol. Sci. 2013; 14:1278–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Graur D., Zheng Y., Price N., Azevedo R.B.R., Zufall R.A., Elhaik E.. On the immortality of television sets: ‘Function’ in the human genome according to the evolution-free gospel of encode. Genome Biol. Evol. 2013; 5:578–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smola M.J., Christy T.W., Inoue K., Nicholson C.O., Friedersdorf M., Keene J.D., Lee D.M., Calabrese J.M., Weeks K.M.. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:10322–10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rivas E., Clements J., Eddy S.R.. A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat. Methods. 2016; 14:45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Somarowthu S., Legiewicz M., Chillón I., Marcia M., Liu F., Pyle A.M.. HOTAIR forms an intricate and modular secondary structure. Mol. Cell. 2015; 58:353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maenner S., Blaud M., Fouillen L., Savoye A., Marchand V., Dubois A., Sanglier-Cianférani S., Van Dorsselaer A., Clerc P., Avner P. et al. 2-D structure of the a region of Xist RNA and its implication for PRC2 association. PLoS Biol. 2010; 8:e1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fang R., Moss W.N., Rutenberg-Schoenberg M., Simon M.D.. Probing Xist RNA structure in cells using targeted structure-seq. PLoS Genet. 2015; 11:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Novikova I.V., Hennelly S.P., Sanbonmatsu K.Y.. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012; 40:5034–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu F., Somarowthu S., Marie Pyle A.. Visualizing the secondary and tertiary architectural domains of lncRNA RepA. Nat. Chem. Biol. 2017; 13:282–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chillón I., Pyle A.M.. Inverted repeat Alu elements in the human lincRNA-p21 adopt a conserved secondary structure that regulates RNA function. Nucleic Acids Res. 2016; 44:9462–9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bond C.S., Fox A.H.. Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 2009; 186:637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hirose T., Virnicchi G., Tanigawa A., Naganuma T., Li R., Kimura H., Yokoi T., Nakagawa S., Bénard M., Fox A.H. et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell. 2014; 25:169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu X., Li Z., Zheng H., Chan M.T.V., Wu W.K.K.. NEAT1: A novel cancer-related long non-coding RNA. Cell Prolif. 2017; 50:e12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nishimoto Y., Nakagawa S., Hirose T., Okano H.J., Takao M., Shibata S., Suyama S., Kuwako K.-I., Imai T., Murayama S. et al. The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain. 2013; 6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sunwoo J.-S., Lee S.-T., Im W., Lee M., Byun J.-I., Jung K.-H., Park K.-I., Jung K.-Y., Lee S.K., Chu K. et al. Altered expression of the long noncoding RNA NEAT1 in Huntington's disease. Mol. Neurobiol. 2017; 54:1577–1586. [DOI] [PubMed] [Google Scholar]
- 18. Nakagawa S., Shimada M., Yanaka K., Mito M., Arai T., Takahashi E., Fujita Y., Fujimori T., Standaert L., Marine J.-C. et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development. 2014; 141:4618–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Standaert L., Adriaens C., Radaelli E., Van Keymeulen A., Blanpain C., Hirose T., Nakagawa S., Marine J.. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA. 2014; 20:1844–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Naganuma T., Nakagawa S., Tanigawa A., Sasaki Y.F., Goshima N., Hirose T.. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012; 31:4020–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sunwoo H., Dinger M.E., Wilusz J.E., Amaral P.P., Mattick J.S., Spector D.L.. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009; 19:347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sasaki Y.T.F., Ideue T., Sano M., Mituyama T., Hirose T.. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:2525–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakagawa S., Naganuma T., Shioi G., Hirose T.. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 2011; 193:31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mao Y.S., Sunwoo H., Zhang B., Spector D.L.. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 2011; 13:95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li R., Harvey A.R., Hodgetts S.I., Fox A.H.. Functional dissection of NEAT1 using genome editing reveals substantial localisation of the NEAT1_1 isoform outside paraspeckles. RNA. 2017; 23:872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Talkish J., May G., Lin Y., Woolford J.L., McManus C.J.. Mod-seq: high-throughput sequencing for chemical probing of RNA structure. RNA. 2014; 20:713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Souquere S., Beauclair G., Harper F., Fox A., Pierron G.. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol. Biol. Cell. 2010; 21:4020–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu S., Xiang J., Li X., Xu Y., Xue W., Huang M., Wong C.C., Sagum A., Bedford M.T., Yang L. et al. Protein arginine methyltransferase CARM1 attenuates the paraspeckle- mediated nuclear retention of mRNAs containing IR Alus. Genes Dev. 2015; 29:630–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mortimer S.A., Weeks K.M.. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007; 129:4144–4145. [DOI] [PubMed] [Google Scholar]
- 30. Lin Y., May G.E., McManus C.J.. Mod-seq: A High-Throughput Method for Probing RNA Secondary Structure. 2015; 1st ednElsevier Inc. [DOI] [PubMed] [Google Scholar]
- 31. Spitale R.C., Flynn R. a., Zhang Q.C., Crisalli P., Lee B., Jung J.-W., Kuchelmeister H.Y., Batista P.J., Torre E. a., Kool E.T. et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015; 519:486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guo F., Gooding A.R., Cech T.R.. Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol. Cell. 2004; 16:351–362. [DOI] [PubMed] [Google Scholar]
- 33. Reuter J.S., Mathews D.H.. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010; 11:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCaskill J.S. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990; 29:1105–1119. [DOI] [PubMed] [Google Scholar]
- 35. Lu Z.J., Gloor J.W., Mathews D.H.. Improved RNA secondary structure prediction by maximizing expected pair accuracy. RNA. 2009; 15:1805–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Darty K., Denise A., Ponty Y.. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009; 25:1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nawrocki E.P., Kolbe D.L., Eddy S.R.. Infernal 1.0: Inference of RNA alignments. Bioinformatics. 2009; 25:1335–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D.. The human genome browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weinberg Z., Breaker R.R.. R2R–software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics. 2011; 12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lorenz R., Bernhart S.H., Höner zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L., Thirumalai D., Lee N., Woodson S. et al. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hofacker I.L., Fekete M., Stadler P.F.. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 2002; 319:1059–1066. [DOI] [PubMed] [Google Scholar]
- 42. Gavazzi C., Isel C., Fournier E., Moules V., Cavalier A., Thomas D., Lina B., Marquet R.. An in vitro network of intermolecular interactions between viral RNA segments of an avian H5N2 influenza A virus: Comparison with a human H3N2 virus. Nucleic Acids Res. 2013; 41:1241–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Van Nostrand E.L., Pratt G.A., Shishkin A.A., Gelboin-Burkhart C., Fang M.Y., Sundararaman B., Blue S.M., Nguyen T.B., Surka C., Elkins K. et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods. 2016; 13:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Watts J.M., Dang K.K., Gorelick R.J., Leonard C.W., Bess Jr J.W., Swanstrom R., Burch C.L., Weeks K.M.. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009; 460:711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pollom E., Dang K.K., Potter E.L., Gorelick R.J., Burch C.L., Weeks K.M., Swanstrom R.. Comparison of SIV and HIV-1 genomic RNA structures reveals impact of sequence evolution on conserved and non-conserved structural motifs. PLoS Pathog. 2013; 9:e1003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Novikova I. V, Dharap A., Hennelly S.P., Sanbonmatsu K.Y.. 3S: shotgun secondary structure determination of long non-coding RNAs. Methods. 2013; 63:170–177. [DOI] [PubMed] [Google Scholar]
- 47. West J.A., Mito M., Kurosaka S., Takumi T., Tanegashima C., Chujo T., Yanaka K., Kingston R.E., Hirose T., Bond C. et al. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J. Cell Biol. 2016; 214:817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lu Z., Zhang Q.C., Lee B., Flynn R.A., Smith M.A., Robinson J.T., Davidovich C., Gooding A.R., Goodrich K.J., Mattick J.S. et al. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell. 2016; 165:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sharma E., Sterne-Weiler T., O’Hanlon D., Blencowe B.J.. Global mapping of human RNA–RNA interactions. Mol. Cell. 2016; 62:1–9. [DOI] [PubMed] [Google Scholar]
- 50. Aw J.G.A., Shen Y., Wilm A., Sun M., Lim X.N., Boon K.-L., Tapsin S., Chan Y.-S., Tan C.-P., Sim A.Y.L. et al. In vivo mapping of eukaryotic RNA interactomes reveals principles of higher-order organization and regulation. Mol. Cell. 2016; 62:1–15. [DOI] [PubMed] [Google Scholar]
- 51. Cimino G.D., Gamper H.B., Isaacs S.T., Hearst J.E.. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu. Rev. Biochem. 1985; 54:1151–1193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mod-seq data have been deposited to the NCBI Sequence Read Archive, under accession number SRP128926.