Abstract
Tomato is a high value crop and the primary model for fleshy fruit development and ripening. Breeding priorities include increased fruit quality, shelf life and tolerance to stresses. To contribute towards this goal, we re-sequenced the genomes of Corbarino (COR) and Lucariello (LUC) landraces, which both possess the traits of plant adaptation to water deficit, prolonged fruit shelf-life and good fruit quality. Through the newly developed pipeline Reconstructor, we generated the genome sequences of COR and LUC using datasets of 65.8 M and 56.4 M of 30–150 bp paired-end reads, respectively. New contigs including reads that could not be mapped to the tomato reference genome were assembled, and a total of 43, 054 and 44, 579 gene loci were annotated in COR and LUC. Both genomes showed novel regions with similarity to Solanum pimpinellifolium and Solanum pennellii. In addition to small deletions and insertions, 2, 000 and 1, 700 single nucleotide polymorphisms (SNPs) could exert potentially disruptive effects on 1, 371 and 1, 201 genes in COR and LUC, respectively. A detailed survey of the SNPs occurring in fruit quality, shelf life and stress tolerance related-genes identified several candidates of potential relevance. Variations in ethylene response components may concur in determining peculiar phenotypes of COR and LUC.
Keywords: traditional varieties, SNPs, next generation sequencing, genomic platform
1. Introduction
Cultivated tomato (Solanum lycopersicum L.) belongs to the large Solanaceae family and, together with 12 wild species, is classified in the Solanum section Lycopersicon.1 Following the first introduction in Europe from South America in the XVI century, tomato has been the subject of continuous selection that resulted in reduced genetic diversity for this crop.2 As an effect of this process, regions of the cultivated tomato genomes are nearly fixed, causing linkage drag and hindering further breeding efforts.3 Nevertheless, thanks to mutations and introgressions from wild species, progress has been made for several traits, including fruit colour, shape, size and environment adaptation.4 Recent sequencing efforts contributed to promoting tomato as a model crop to understand the molecular and biochemical processes underlying climacteric fleshy fruit evolution.5,6 In addition, hundreds of genomes from landraces, cultivars, and wild relatives have been re-sequenced, laying the foundation to better exploit genome variation.3,7,8 Spain and Italy are considered secondary centres of tomato diversification.9–11In these countries, farmers’ selection and adaptation to local climates and low-input agricultural practices resulted in a plethora of landraces, differing in growth habit and fruit shape and size, and frequently carrying abiotic and biotic stress resistances as well as peculiar organoleptic characteristics and high nutritional quality.11–14 This biodiversity has been already used to undertake comparative genomics studies for the identification of fruit quality traits, stress tolerance mechanisms as well as fleshy fruit development and composition under harsh environments.15–17
Increased shelf-life is among the most sought-after targets for tomato,18 though the molecular mechanisms and genetic determinants underlying this trait have not been fully elucidated yet. However, a large number of ripening-related mutants have been identified, including ripening-inhibitor (rin), non-ripening (nor), Never-ripe (Nr), Colorless non-ripening (Cnr) and Green-ripe (Gr), some of which represent a useful resource for extending tomato shelf-life.19,20 For instance, the rin mutation has been widely used by breeders, and nowadays hybrids (rin/Rin) constitute the basis for most slow ripening, long shelf-life (LSL), fresh market tomatoes. Landraces contributed with additional mutations to the identification of alleles associated to the LSL trait. Previous studies on Spanish tomatoes provided evidence that delayed fruit deterioration was associated with the presence of the alcobaça (alc) mutation in the coding sequence of the NOR gene.21–23 An additional mutant in NOR, delayed fruit deterioration (dfd), also causes attenuation of over-ripening processes, contributing to increased fruit shelf-life.24 Studying tomato fruit development, Zhong and collaborators25 demonstrated that epigenetic changes, such as variations in methylation profiles, are also involved in the ripening process and are associated with tomato shelf-life. The growing evidence connecting plant epigenomes to the interactions with the environment suggests that adaptation to local conditions may as well be accomplished by genotype specific changes in epi mark profiles.26,27
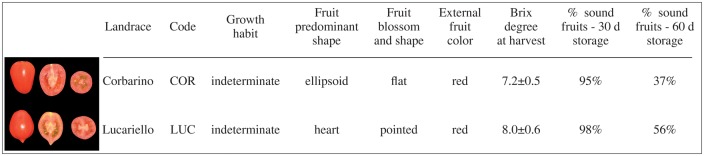
In this paper, we describe the resequencing of the two tomato landraces Corbarino (COR) and Lucariello (LUC), the latter belonging to the Vesuviano type. They are traditionally grown in the Campania region (Southern Italy) and are highly prized by consumers for the superior quality of their fruits.28,29 They share some common traits, along with distinctive peculiarities (Fig. 1). COR produces obovoid fruits with an intense red colour and high levels of soluble and total solids,29 and LUC produces hearth-shaped fruit with a pronounced pointed apex of a less intense red and with a thick cuticle.30 Both are grown with very low water inputs that mostly rely on the infrequent rains of the spring-summer season in Southern Italy. These cultivation practices contribute to the distinctive trait of both landraces, which are the high fruit quality and extended shelf-life (Fig. 1). Fruits are traditionally stored in unrefrigerated, well-aerated, semi-shaded premises, thus the name ‘da serbo,’ i.e. preservable. Notably, LUC fruits are stored in bunches (named ‘piennoli’), which can be kept for 5–10 months. These peculiar features of COR and LUC offer a unique opportunity to identify, on a genome-wide scale, variants putatively associated with traits of great economic importance. Therefore, polymorphism identification was largely focused on classes of genes related to prolonged shelf-life, fruit quality and stress tolerance.
Figure 1.
Main morpho-physiological characteristics of the tested genotypes, Corbarino (COR) and Lucariello (LUC). Brix degrees values represent average ± SD (n = 6). % of sound fruits is defined as the percentage of fruits which do not present signs of decay (e.g. pathogen attacks or injuries) or wrinkling after 30 or 60 days of storage (n = 100).
2. Materials and methods
2.1. Plant material, DNA isolation and sequencing
Seeds of the Italian landraces Corbarino (COR, selection Crovarese, catalogue n. TM326) and Lucariello (LUC) were kindly provided by Semiorto Sementi s.r.l., Lavorate di Sarno (Salerno, Italy) and Farm Casa Barone, Massa di Somma (Naples, Italy; http://www.casabarone.it (October 2017, date last accessed)), respectively. Seeds of Heinz 1706 (accession LA4345) were obtained from the Tomato Genetics Resource Center, USA. Plants of COR, LUC and Heinz 1706 were grown in greenhouse at the Institute of Biosciences and Bioresources (IBBR-CNR) in Portici (Italy). DNA was extracted from five plants per each accession. DNA was isolated using the DNeasy Plant Mini kit (QIAGEN, Valencia, CA) according to manufacturer’s recommendations. DNA quality and concentration were evaluated on agarose gel and using the Nanodrop spectrophotometer (Thermo Fisher Scientific, Wilmington, USA) and the Qubit 2.0 Fluorometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). High quality DNA of COR and LUC was sequenced by Genomix4Life S.r.l. (Baronissi, Italy). Indexed libraries were prepared from 1 µg purified DNA with the TruSeq DNA Sample Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Libraries were quantified using the Agilent 2100 Bioanalyzer (Agilent Technologies) and pooled such that each index-tagged sample was present in equimolar amounts, with 2 nM final concentration. Pooled samples were subject to cluster generation and sequencing using a NextSeq 500 System (Illumina, San Diego, CA, USA) in a 2 × 150 paired-end format at a final concentration of 1.8 pmol. The DNA for InDel and SNP validation was freshly isolated from the same source plants according to the same reported protocol.
2.2. Reference-guided assembly
Raw Illumina reads were processed with Trimmomatic (v. 0.33) to remove adapter/primer sequences and trim 5′ and 3′-end bases (minimum quality 35, minimum length 35 bp). Quality of trimmed sequences was checked using FastQC (v0.11.3; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (October 2017, date last accessed)). Reference-guided assembly was performed using Reconstructor (v1.0) combined with the Heinz 1706 reference genome. A two-step strategy was used: iterative read mapping and de novo assembly of unmapped reads. In the first step, high-quality reads were aligned along the reference genome using SUPER-W (10.1093/molbev/msv152). Five iterations were performed in order to identify sequence variations such as SNPs, deletion and insertion polymorphisms (DIPs), and structural variations (SVs). At each iteration, only the variants with a genotype quality (GQ) higher than 30, with a coverage higher than 6× score and an allele frequency higher than 0.75 were kept. The identified polymorphisms were used to edit the tomato reference genome using VCF tools (v. 0.1.12 b, 10.1093/bioinformatics/btr330), thus obtaining cultivar specific sequences. The second step based on the de novo assembly of the reads that did not map on the reference genome was performed by using SOAPdenovo (v2.04, 10.1186/2047-217X-1-18) setting two k-mer sizes: 50 and 120 nt. Resulting contigs were filtered for size (> 400 bp) and subjected to BLASTN (v2.2.30+) search (e-value ≤ 0.01) against the NCBI non-redundant nucleotide database section ‘Viridiplantae’ in order to remove possible contaminants. Using paired-read and split read approaches, Reconstructor was employed to combine the genotype-specific reconstructed genomes with de novo assembled contigs in order to obtain final private genome sequences. At least four paired-end reads covering the junction between the reference chromosomes and the de novo assembled contigs were required to generate new insertions. Low-complexity sequences, repetitive sequences and interspersed repeats within the reconstructed genomes were identified and classified using RepeatMasker (v4.0.5).31 The regions highlighted from the assemblies of COR and LUC as deletions or insertions were validated by bioinformatic and experimental approaches. For bioinformatic validation, the deleted regions in COR and LUC were localized in the Heinz 1706 genome (v. 2.50) and classified as belonging to repetitive, intergenic or genic regions. The hypothetical insertions in COR and LUC were re-checked versus the Heinz 1706 genome (v. 2.50) and the 2, 764 unassembled BAC sequences available at https://www.solgenomics.net/ (October 2017, date last accessed), using BLASTn. The Heinz 1706 regions matching the putative COR/LUC inserted regions were further analysed to check if they belong to repetitive, intergenic or genic regions. In this latter case, we selected the regions that presented mismatches between Heinz 1706 and COR/LUC in the coding sequence for further validation.
For experimental validation, all the InDels and 12 randomly selected SNPs-containing genes found in COR and LUC were analysed through PCR and Sanger sequencing. Primer pairs (Supplementary Table S1) were designed on Heinz 1706 using the Primer3 software32 to amplify fragments from 0.2 to 0.9 kbp in length with optimum primer length of 23 bp and Tm of 62 °C. Amplification products were cycle-sequenced from both directions with a BigDye version 3.1 terminator kit and visualized on an ABI PRISM 3130 Genetic Analyser (Applied Biosystems, CA, USA).
2.3. Genome annotation
Before the annotation process, COR and LUC transcripts (including splicing isoforms) were assembled from custom RNA-seq data (papers in preparation). Reads from LUC fruits and COR leaves were subjected to normalization using Trinity (v2.0.6, 10.1038/nbt.1883). Then, normalized reads were aligned on the corresponding reconstructed genome sequences with STAR (v. 2.4.2a, 10.1093/bioinformatics/bts635). The resulting BAM files were filtered to remove duplicates with Picard MarkDuplicates (v. 1.31, http://broadinstitute.github.io/picard/ (October 2017, date last accessed)) and then used as input for Trinity (v2.0.6) to perform genome-guided RNA-Seq assembly. In order to remove redundancy, transcripts were post-processed using the CD-HIT-EST (v4.6, 10.1093/bioinformatics/btl158) tool, which clusters similar transcripts based on a similarity threshold (99% of identity). Trinity assembled transcripts and the official ITAG2.4 gene models were merged and fed into the PASA genome annotation pipeline (v.2.0.2, 10.1093/nar/gkg770). The annotation was fully post-processed through different filtering steps in order to reduce false positive most likely due to assembly errors. Firstly, a transcript-level quantification was carried out using eXpress (v1.5.1, 10.1093/bioinformatics/btt034). Those splicing variants with an expression level < 15% compared to the most expressed isoform were removed due to their high probability of being assembly artifacts. In addition, single exon sequences overlapping multi-exon transcripts on the same strand were filtered out as they were also considered mis-assemblies. To classify the predicted transcripts, a lncRNA prediction analysis was carried out following the approach described in by Paytuví-Gallart and collaborators.33 Only the coding transcripts were further analysed using TransDecoder (v2.0.1, 10.1038/nprot.2013.084) in order to identify candidate coding regions (CDS) and untranslated regions (UTRs). CDS were functionally annotated using InterProScan (v. 5, 10.1093/bioinformatics/btu031). In addition, BLASTP (v2.2.30+) searches were performed against the Arabidopsis thaliana (TAIR10) protein complement. Only those hits with an e-value ≤ 0.001 and a percentage match length > 20% were taken into account. In order to assess whether the COR and LUC loci could be mapped back to the Heinz genome, the gene body sequences were extracted with bedtools and then a BLASTn was performed against a database of Heinz 2.50 reference chromosomes and unplaced scaffolds obtained from the NCBI Nucleotide database (keywords: unplaced scaffolds SL2.50). Only the alignments with an e-value less than 0.01 and covering at least 75% of the gene length were kept.
2.4. Public genome browser
A genome browser was set-up to collect genomic sequences and their annotations as described by Kent and collaborators.34 The platform works as a web-based application running on the Net Framework 4.0. The embedded databases are organized in a relational model and implemented in MySQL (v. 5.6.14 InnoDB engine). All key fields and query dependent tuples were indexed using the BTree indexing algorithm.35 The genome browser includes the 12 pseudomolecules and the chromosome 0 (unplaced regions) of Heinz 1706 (v. 2.50), as well as the associated reference gene annotation (iTAG 2.4, downloaded from the SOLgenomics website), and the COR and LUC genomes. Gene annotations of COR, LUC, and Heinz, were included in species-specific partition in the Gbrowse database. A download data section allows sequences and data to be accessed upon request. Association of all the genes among the three genomes from each annotation (Heinz 1706, COR and LUC) was carried out by GenomeThreader36 using transcript to genome mapping of each landrace to the two alternative ones, with 95% and 85% of coverage and identity, respectively. This permitted to create crosslinks between each genome annotation to the corresponding similar loci in the other two genomes.
2.5. Annotation and functional analysis of polymorphic regions
SNPs identified in COR and LUC during the iterative variant calling were functionally annotated respect to the ITAG2.40 genome annotation with SNPEff (http://dx.doi.org/10.4161/fly.19695 (October 2017, date last accessed)). Gene Ontology Enrichment Analysis (GOEA) was performed on the genes showing missense mutations as well as on genes with polymorphisms altering CDS length (i.e. disruptive inframe deletions, disruptive in-frame insertions, frameshift variants, stop coding gain/loss and start codon loss). GOEA was performed with in-house scripts and was based on hypergeometric test comparing the proportion of genes in each GO category in the genome and in the groups of mutated genes; an FDR < =0.05 was considered significant.
3. Results and discussion
3.1. Reference-guided assembly through Reconstructor
To develop genomic tools for molecular breeding and explore genetic variability for LSL and environmental adaptation traits, we performed genome resequencing of two LSL Italian tomato landraces, namely Corbarino (COR), and Lucariello (LUC). The Reconstructor pipeline (available at http://www.sequentiabiotech.com/omicstools/pipeline/ (October 2017, date last accessed)),37 which combines iterative variant calling and de novo assembly, was used to generate COR and LUC genome sequences. A similar approach is also implemented in the IMR-DENOM pipeline, which has been used for the genome reconstruction of 18 Arabidopsis accessions (Gan et al 2011). The final size of the reconstructed genomes of COR and LUC were 823, 643, 567 bp and 823, 719, 762 bp, respectively. Both genomes were slightly shorter than the tomato annotated Heinz 1706 reference (823, 944, 041 bp). The dataset used for reconstruction included 65.8 M and 56.4 M of 30–150 bp paired-end reads with an average insert size of 380 bp (± 52 bp) and 364 bp (± 49 bp) for COR and LUC, respectively. With the first iteration of variant calling, the average coverage was estimated to be 15.34X (± 81X) for COR and 13.42X (± 67X) for LUC. After five iterations, 189, 797 variants were identified in COR and 173, 764 in LUC (Table 1). On average, 95% of the variable sites could be already observed after the first iteration of the variant calling workflow (Supplementary Fig. S1). Conversely, the IMR-DENOM pipeline needed five iterations to reach convergence with Arabidopsis data,38 possibly due to the larger reads size of our re-sequencing effort (150 bp vs 32–52 bp of the IMR-DENOM dataset). The two landraces showed a similar ratio of SNPs, small deletions and insertions. In fact, 68.8% of SNPs, 8.9% of small deletions and 22.1% of insertions were observed in COR, whereas 69.9%, 8.6% and 21.3% of the polymorphic sites were SNPs, deletions and insertions in LUC. Twelve randomly selected SNPs and DIPs were experimentally validated. Sanger sequencing following PCR amplifications confirmed all the mutations, and thus the high quality of the reconstructed genomes. Since the position of SNPs and DIPs may influence the functionality of the encoded protein and is important for the developing of molecular markers, we evaluated the chromosomal distribution of genomic variants. In both landraces these variants were evenly distributed between the 12 chromosomes on a percent length basis. A slightly higher number of variants was found on Chr01, Chr04, Chr09 and Chr11, the latter mainly for LUC (Supplementary Fig. S2). It has been previously reported that Chr04, Chr09 and Chr11 in the Heinz 1706 reference genome display large introgressions from S. pimpinellifolium.5 Other resequencing studies on cultivated tomatoes found that these chromosomes carry a higher number of SNPs with respect to Heinz 1706.7,39 Higher SNP density reported for Chr09 in ‘San Marzano’ was associated to resistance loci.17
Table 1.
Resequencing data from Corbarino (COR) and Lucariello (LUC) were used to perform an iterative variant calling analysis to identify polymorphisms with respect to the reference tomato genome Heinz 1706
| Variant calling | COR | LUC | Common |
|---|---|---|---|
| SNPs | 129,426 | 120,112 | 85,578 |
| Deletions | 16,847 | 14,894 | 10,958 |
| Insertions | 44,152 | 39,289 | 27,651 |
| Total | 190,425 | 174,295 | 124,187 |
Here the number of high quality SNPs and DIPs across COR and LUC are showed.
3.2. Structural variants
The fifth iteration of variant calling allowed the identification of 269 and 261 structural variants (SVs) in COR and LUC, respectively. In COR, 265 SVs were deletions and four were duplications, whereas 255 deletions and six duplications were identified in LUC. Other resequencing studies in tomato landraces and heirloom varieties evidenced, on average, a higher number of SVs.8,17 It should be pointed out that low genetic diversity is a common finding in cultivated tomatoes.40,41 However, despite the limited number of identified polymorphic variants, a clear discrimination of the two investigated landraces was possible.
A high proportion of the deleted genomic regions (67.9% in COR and 67.8% in LUC) was characterized by the presence of stretches of unidentified nucleotides (Ns). They are usually introduced during the scaffolding process and were thus considered non informative (Table 2A). Regions including < 87% of Ns were classified on the basis of their genomic context (i.e. genic, intergenic and highly repetitive regions; Table 2A). Sequences with a N content that exceeded the 87% threshold could not be adequately processed by Blast similarity searches. The COR and/or LUC deletions occurring in genic regions concerned 14 Heinz 1706 gene loci (Supplementary Table S2A), that interrogation of the NextGenEx-Tom database42 indicated to be expressed in different tissues, including fruits. Therefore, the absence of these genes may have a functional relevance for the ‘da serbo’ phenotype of COR and LUC.
Table 2.
Validation of the putative structural variants through reciprocal alignments of Corbarino (COR) and Lucariello (LUC) genomes to the assembled and unassembled (BAC sequences) Heinz 1706 reference genome and verification by PCR
| A. Deletions | COR | LUC | Common |
|---|---|---|---|
| Size range, bp | 256–19,538 | 229–18,654 | |
| Sequences including > 87% of Ns | 180 | 173 | |
| Sequences including < 87% of Ns | |||
| in genic regions | 12 | 7 | 2 |
| in intergenic regions | 13 | 16 | 3 |
| in highly repetitive regions | 60 | 59 | 15 |
| Total | 265 | 255 | |
| B. Insertions | COR | LUC | Common |
| Size range, bp | 446–1012 | 441–1032 | |
| Unconfirmed insertions | 3 | 3 | 1 |
| Confirmed total length insertionsa | 6 | 5 | 0 |
| Confirmed fragmented insertions | |||
| in genic regions | 4 | 4 | 2 |
| in intergenic regions | 10 | 13 | 6 |
| in highly repetitive regions | 6 | 11 | 3 |
| Total | 29 | 36 | |
The size range of the variants are indicated. A: % of unspecified nucleotides (Ns) and genomic context of the COR and LUC deletions. B: number of unconfirmed and confirmed insertions and genomic context of the scattered insertions.
aVerified also by PCR.
By performing de novo assembly of unmapped reads, 29 and 36 novel contigs were identified in COR and LUC, respectively, and successfully placed within pseudo-chromosomes (Table 2B). To confirm that these putative novel genotype-specific genomic regions were absent in the tomato reference genome, they were aligned versus all the genomic sequences available for Heinz 1706, including the unassembled BAC sequences. As a result, five (two COR-specific, two LUC-specific and one common) sequences were found to be total length insertion (Table 2B) while six novel fragments from COR and five from LUC did not find matches in Heinz 1706 and were thus confirmed as inserted regions. Interestingly, most of these novel sequences displayed highly significant matches with Solanum pimpinellifolium (https://solgenomics.net (October 2017, date last accessed); Supplementary Table S2B). Moreover, among all Solanaceae species included in the NBCI database (March 6, 2017), all of them showed highly significant matches with S. pennellii (Supplementary Table S2B). Most of these matches corresponded to annotated gene loci in S. pennellii and could therefore be of significance for the characteristic phenotypes of COR and LUC. These findings suggest that the presence of a stretch of nucleotide base pairs into COR and LUC DNA sequences (i.e. full length insertions) may be the result of introgressions from these wild relatives followed by breeding efforts. Fragmented insertions, i.e. COR/LUC de novo regions only partially similar to Heinz, were identified for the remaining 20 and 28 sequences from COR and LUC, respectively, and were grouped in the three above-mentioned categories (Table 2B).
3.3. Genome annotation
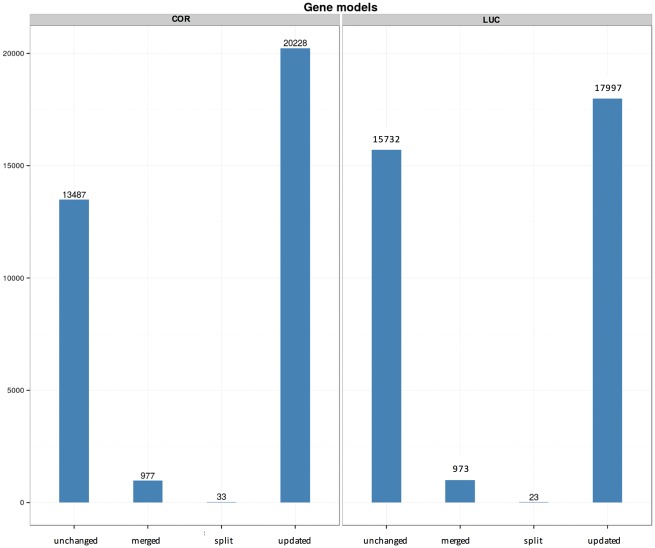
RNA-seq data from COR (leaves; 164 M of 35–100 bp single reads) and LUC (fruits at red ripe stage; 421 M of 35–100 bp single reads) were firstly assembled in reference-guided mode, obtaining ∼108, 000 transcripts in COR and ∼94, 000 transcripts in LUC. Then, both sequence datasets were individually combined with transcripts derived from the iTAG annotation (version 2.40) and used to identify gene loci as well as derive transcript structural annotations within the reconstructed genomes. After removing redundant transcripts (i.e. duplicated and/or fragmented sequences), 43, 054 gene loci (corresponding to 46, 065 protein coding transcripts) were annotated in the COR genome, whereas 44, 579 genes (corresponding to 45, 815 protein coding transcripts) were detected in LUC (Table 3). These figures include about 10, 000 genes not annotated in the reference genome by iTAG and are similar to those reported for other Solanaceae such as Solanum tuberosum, S. pennellii and Solanum commersonii, which harbour 39, 028,43 44, 96644 and 37, 66245 genes, respectively. The higher number of annotated genes in COR and LUC respect to Heinz could be explained either by an actual higher gene content of the two landraces or by a lack of gene annotations in the reference genome. In order to confirm either hypothesis, the sequences of all the COR and LUC gene bodies were mapped on the Heinz genome and on Heinz unplaced scaffolds. All COR and LUC loci could be mapped back to the Heinz genome with a similarity higher than 90% and a coverage higher than 75%. The only exception was the LUC gene SolycLC11g018560, which shows a high similarity with a S. pennellii transcript with unknown function (LOC107005075), which could not be mapped. These results show that the iTAG2.40 annotation of the Heinz genome is still to be completed. By our hybrid liftover/de novo approach, new genes (i.e. either genotype-specific or missing in the official annotation) and novel splicing isoforms were identified. In addition, ∼20, 000 gene models were refined and corrected. Moreover, RNA-seq data allowed to revise the structure of ∼1, 000 mis-annotated gene loci. Most of the mis-annotated genes had small changes with respect to the reference genome. In most cases these changes affected the start and/or the end coordinates of the genes (Fig. 2). We also found evidences supporting gene merges (separated genes in Heinz 1706 were actually part of the same locus, see Supplementary Fig. S3). This occurred for 977 Heinz 1706 genes that were merged in 330 loci in COR and for 973 Heinz 1706 genes that were merged in 552 loci in LUC (Fig. 2 and Supplementary Fig. S3). Based on prediction analysis (Table 3), 26% and 29% of transcripts were classified as lncRNAs in COR and LUC, respectively. Notably, the recent release of version 3.1 of the tomato annotation by iTAG, occurred during the writing of this manuscript, also fixed some errors in gene models that were incorrectly fused or split. The exploitation of both leaf and fruit RNA-seq data, for COR and LUC, respectively, allowed robust discovery and annotation of alternative transcripts, providing an insight into the alternative splicing landscape in these two landraces. The official tomato genome annotation by iTAG did not deliberately provide an annotation of transcript variants; as a consequence, the ∼60, 000 transcripts we annotated in COR and LUC using combined genome and transcriptome assembly RNA-seq data represent a valuable, publicly available, resource for studying tomato transcriptome plasticity for functional studies. The ratio of variants per gene, 1.44 in COR and 1.45 in LUC, is very similar to the 1.54 ratio observed in the latest A. thaliana genome annotation (Araport11),46 which was produced through a similar pipeline. A recent effort on tomato fruit transcriptome assembly identified more than 31, 000 splicing variants in about 11, 000 fruit-expressed genes,47 suggesting that alternative splicing may occur more frequently in fruits than in other tissues. It must be pointed out that the number of annotated genes and transcripts can vary significantly using different bioinformatic methods. For instance, by combining ab initio and genome-guided transcriptome assembly methods, about 33, 000 genes and 41, 000 transcripts were identified in A. thaliana Col-038versus ∼27, 600 loci and 48, 000 transcripts described in Araport11. The remaining transcripts were translated into proteins, and Gene Ontology terms were assigned to 22, 198 transcripts both in COR and LUC.
Table 3.
Number and classification of genes annotated in Corbarino (COR) and Lucariello (LUC)
| Annotation | COR | LUC |
|---|---|---|
| Genes | 43,054 | 44,579 |
| Transcripts | 62,369 | 65,047 |
| Protein coding transcripts | 46,065 | 45,815 |
| High confidence lncRNAsa | 14,091 | 16,838 |
| Low confidence lncRNAsa | 2,213 | 2,394 |
aAccording to the criteria listed in reference 33.
Figure 2.
Barplot showing the changes of Heinz 1706 gene annotations in Corbarino (COR) and Lucariello (LUC). Loci are described as (1) ‘unchanged’ if they are unchanged between Heinz 1706 and the two ecotypes; (2) ‘merged’ if they are originated from the fusion of two or more Heinz 1706 genes; (3) ‘split’ if they derive from a Heinz 1706 gene that was split in two and (4) ‘updated’ if the locus in COR and/or LUC showed a different exon/UTR structure.
3.4. Public genome browser
A dedicated genome browser-based platform, accessible at http://cab.unina.it/genotom (October 2017, date last accessed), allows the genomic sequences of Heinz 1706, COR and LUC to be accessed and investigated. In particular, data can be queried by ‘gene ID’ and ‘keywords.’ The iTAG annotation (version 2.4) as well as the novel annotations from COR and LUC were reciprocally mapped on the three reference genomes. This allows to query using a specific gene ID and to receive information on where a locus similar to the one corresponding to that ID is detected on the alternative genomes. In Table 4, we report the number of transcripts per genome and the total corresponding number of hits found in the alternative genomes.
Table 4.
Mapping of one genome transcripts versus the other genome considering multiple hits
| Mapping | Starting number of transcripts | Number of mapped transcripts | Number of identified loci |
|---|---|---|---|
| COR-Trans_vs_Heinz250 | 62,369 | 62,206 | 77,220 |
| COR-Trans_vs_LUC250 | 62,369 | 62,200 | 77,227 |
| Heinz-Trans_vs_COR250 | 34,725 | 34,484 | 42,337 |
| Heinz-Trans_vs_LUC250 | 34,725 | 34,489 | 42,352 |
| LUC-Trans_vs_COR250 | 65,094 | 64,820 | 79,259 |
| LUC-Trans_vs_Heinz250 | 65,094 | 64,813 | 79,277 |
3.5. Functional classification of the genomic variants
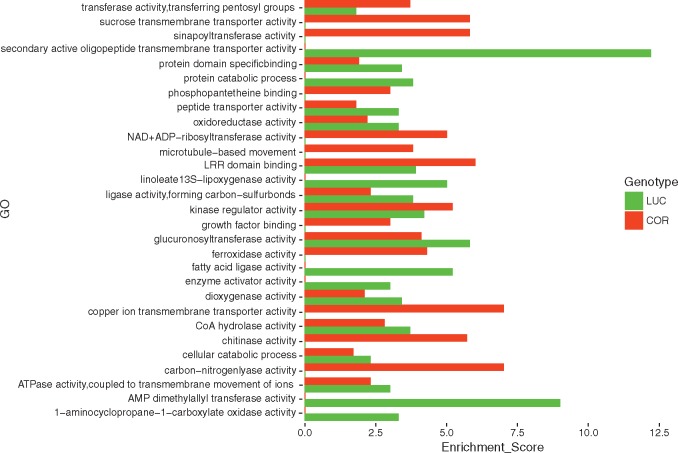
COR and LUC genes showing sequence polymorphisms and whose structure was very similar to that of Heinz 1706 were analysed to get an interpretation of the effects of sequence changes. We focused on 30, 830 COR and 29, 935 LUC genes, respectively, corresponding to 86% and 88% of the genes annotated in iTAG 2.40. Interestingly, 3, 360 genes in COR and 2, 908 in LUC included more than one variation. In addition, approximately 2, 000 and 1, 700 SNPs with potential disruptive effects on 1371 COR and 1201 LUC genes were identified (Table 5 and Supplementary Tables S2 and S3). Gene Ontology Enrichment Analysis (GOEA) was performed on genes harbouring missense mutations as well as on genes that included polymorphisms altering CDS length (i.e. disruptive in-frame deletions, disruptive in-frame insertions, frame-shift variants, stop coding gain/loss and start codon loss). No significant enrichment (FDR ≤ 0.05) was found either in COR or in LUC. The most abundant GO categories are reported in Fig. 3. Genes carrying SNPs possibly associated with the peculiar LSL and stress-resistant phenotype of COR and LUC were further surveyed in detail. These include genes associated with fruit ripening and quality, epigenetic regulation of gene expression, and abiotic stress adaptation.
Table 5.
Types and number of SNPs with potential disruptive effect on gene function in Corbarino (COR) and Lucariello (LUC)
| Type | Subtype | COR | Number of genes | LUC | Number of genes |
|---|---|---|---|---|---|
| Mutations creating reading frame shifts | Disruptive inframe deletions | 24 | 24 | 18 | 18 |
| Disruptive inframe insertions | 13 | 12 | 9 | 9 | |
| Frameshift variants | 146 | 140 | 100 | 96 | |
| Inframe deletions | 9 | 9 | 11 | 11 | |
| Inframe insertions | 15 | 14 | 14 | 14 | |
| Mutations affecting ORF length | Start codon loss | 2 | 2 | 5 | 5 |
| Stop codon gain | 29 | 27 | 28 | 27 | |
| Stop codon loss | 1 | 1 | 1 | 1 | |
| Mutations affecting protein sequence | Missense variants | 1,749 | 1,186 | 1,477 | 1,055 |
| Mutations affecting splicing sites | Stop retained variants | 1 | 1 | 2 | 2 |
| Complex mutations creating reading frame shifts and affecting ORF length | Frameshift variants and start loss | 13 | 13 | 11 | 11 |
| Frameshift variants and stop gain | 3 | 3 | 2 | 2 | |
| Frameshift variants and stop loss | 1 | 1 | 1 | 1 | |
| Complex mutations affecting splicing sites and ORF length | Missense variants and splice region variants | 1 | 1 | 1 | 1 |
| Splice region variants and stop retained variants | 2 | 2 | 1 | 1 | |
| Stop loss and splice region variant | 6 | 6 | 6 | 6 |
Figure 3.
A gene ontology enrichment analysis (GOEA) was performed to identify enriched functions among the LUC and COR polymorphic genes. The barplot shows the mostly enriched GO categories (molecular function and biological process domains) of the genes affected by missense and non-sense mutations.
3.6. Ripening-, shelf-life- and quality-related genes
Two hundred and four and 207 sequence variants were observed in 122 COR and 140 LUC genes putatively related to the process of fruit ripening, which also affects fruit shelf-life and quality (Supplementary Tables S3 and S4). The two genotypes shared 136 missense variations in 83 genes. All these sequence changes were predicted to have moderate impact on protein function (Supplementary Table S3). In addition to common missense variations in six ethylene-responsive transcription factors (TFs) (e.g. Solyc05g050010, Solyc08g078170, Solyc10g076370), COR and LUC shared a splicing-site mutation in ERF13, a member of the ethylene-responsive transcription factor (ERF) family. This includes several TFs that trigger and orchestrate ripening. Their position in the regulatory network, however, is not fully understood.48 Some mutations affecting ethylene-related loci, previously identified also in ‘San Marzano’ and ‘Vesuviano’ landraces,17 included a 1-aminocyclopropane-1-carboxylate synthase (Solyc05g050010—T82A) and a transmembrane protein 222 (Solyc08g065320—V157L). This latter locus showed high identity with the tomato Green-ripe like1. Dominant mutations in this locus have been reported to induce a reduction in ethylene responsiveness in fruit tissues and may provide useful tools for breeding, although the impact of the identified mutation on protein function remains to be verified.49
In LUC, a disruptive in-frame deletion of six nucleotides in exon 7 and an I5N polymorphism were identified in the coding sequence of the TAGL1 TF. TAGL1 is a key component of the regulatory network controlling ripening, being involved in cuticle development and fruit softening.50,51 The mutations identified in TAGL1 might putatively contribute to the LUC shelf- life phenotype.
Common missense variations were found in several genes coding for cell wall degradation enzymes including polygalacturonases (Solyc12g019130, Solyc09g098270, Solyc02g067650), pectinacetylesterases (Solyc08g074950, Solyc08g075030) and rhamnogalacturonate lyases (e.g. Solyc04g014430, Solyc04g014450) that play a major role in fruit softening.52,53 This class of genes, together with ripening-related and polyamines genes, have been widely used for transgenic approaches to extend shelf life.54–57 Indeed, the first genetically engineered food brought to market was Flavr-savr® tomato, displaying decreased softening of ripe fruits due to reduced activity of a polygalacturonase.58 However, the expression modulation of some of the abovementioned genes has led so far to modest improvements in shelf-life.59,60 Nevertheless, the polymorphisms here identified might collectively have an impact on fruit preservation. In addition, LUC-specific missense variants were identified in the genes coding for an expansin (Solyc03g115310—E146G) and a fasciclin-like arabinogalactan protein 13 (Solyc01g091530—S201Y), possibly involved in fruit texture and firmness.61 Notably, the same sequence variants were also identified by Ercolano and collaborators.17 In particular, the variant (E146G) observed in the expansin was the same found in the other LSL landrace ‘Vesuviano,’ whereas the one (S201Y) detected in the gene coding for the fasciclin-like arabinogalactan protein 13 was identified in both ‘Vesuviano’ and ‘San Marzano’ landraces.
A missense variant was also found in COR and LUC in Solyc11g072030, which encodes for a non-specific lipid transfer protein (nsLTP). nsLTPs are well known as involved in the transfer of lipids through the extracellular matrix for the formation and deposition of the surface cuticle layer,62 which provides structural support and limits water loss, thus influencing fruit softening and storage longevity.24
Fruit shelf-life can be extended also through alleviation of oxidative tissue damaging, which is the most likely cause of the delay in over-ripening and softening observed in purple and orange tomatoes.57 In COR and LUC, common missense mutations were identified in genes mapping to antioxidant pathways. Among these genes an aldo/keto reductase (Solyc09g097960) might control ascorbate synthesis and accumulation.63 Also, a LUC-specific missense variation in an ascorbate peroxidase gene (Solyc01g006310) may affect the cell redox state and level of hydrogen peroxide.64 Finally, polymorphisms were identified in genes involved in the biosynthesis and accumulation of phenolic compounds (Solyc09g007910, Solyc03g005090, Solyc10g009520) as well as in glutathione S-transferases genes that control their conjugation and compartmentalization.65 Besides missense variants, a COR-specific in-frame deletion K91del was found in a beta-glucosidase gene (Solyc01g081170) involved in the biosynthesis of the polyphenol coumarin (via 2-coumarate), whereas a COR-specific nonsense mutation (Y234*) was detected in a polyphenol oxidase gene (Solyc08g074640). These sequence variants might contribute to the fruit antioxidant activity of COR and LUC and to their LSL phenotype.
Surprisingly, neither in COR nor in LUC we detected mutations in ripening and extended shelf-life associated TFs, such as ripening inhibitor (rin), non-ripening (nor), alcobaça (alç), or delayed fruit deterioration (dfd) (Conesa et al. 2014). This finding suggests that the COR and LUC long shelf-life has a different genetic basis compared to the Spanish LSL varieties ‘de penjar’ and ‘Tomàtiga de Ramellet’. In these varieties, the LSL phenotype was mostly associated to the presence of the alç mutation in NOR,22,23 a TF controlling the expression of several ripening-related genes and acting upstream of ethylene-dependent regulation of this process.66
Besides extended shelf-life, COR and LUC fruits have other desirable traits, including high soluble solids content.29 Compared to the Heinz 1706 reference genome, COR and LUC showed missense variations in genes putatively involved in carbohydrate metabolism, transport and partitioning. For instance, the sucrose transporter SUT4, having a function in phloem loading, may control sugar translocation to sink tissues, enhancing the fruit soluble solids content.67,68Additional contribution to the fruit soluble solids content may come from shared mutations in the gene sequence of a 1, 4-alpha-glucan branching enzyme, involved in the immobilization of reducing sugars into starch.
The plethora of sequence variations identified in COR and LUC in genes involved in ripening as well as in fruit storability and quality suggest that these traditional landraces could represent a pool of unique candidate genes never so far exploited in breeding for fruit LSL and improved quality.
3.7. Epigenetic regulation of gene expression
Given the crucial role of epigenetic control in fruit development and ripening,20 variations in epiregulator-encoding genes may also contribute to the peculiar traits of COR and LUC. Several SNPs within genes encoding for epiregulators involved in histone modifications and RNA-directed DNA methylation (RdDM) were found. Overall, 26 genes were affected by 58 non-synonymous (57 missense and 1 nonsense) mutations which were in common between COR and LUC for twelve genes, while were COR- and LUC-specific for 8 and 6 genes, respectively (Supplementary Table S4). Approximately half of the mutations occurred in highly conserved portions of the proteins (data not shown). Common genes included SWI/SNF chromatin remodelling enzymes (Solyc08g077580, Solyc09g042480), a histone acetyltransferase of the HAG superfamily (Solyc08g068770), and histone demethylases (HDMs) of the Jumonji family (Solyc04g074490, Solyc08g081000, Solyc09g065690). The knowledge paucity about histone architecture in fruit genomes does not allow attributing significance to the identified SNPs relating to these loci. However, the evidence that Solyc04g074490 is preferentially expressed in fruit69 (https://bar.utoronto.ca/eplant/ (October 2017, date last accessed)) could be suggestive of a ripening function. Among common genes we also identified a SU(VAR)3-9-related histone methyltransferase (HMTs) (Solyc10g074370) showing 41% sequence similarity with the Arabidopsis SU(VAR)3-9 homolog 5 (SUVH5). This gene is required for CROMOMETHYLASE 3 dependent CHG methylation.70 It harbours a premature stop codon in COR, resulting in a 9-amino acid shorter protein product. Chen and collaborators71 reported that CMT3 affects the fruit DNA methylation status and the ripening phenotype in tomato. COR and LUC also shared polymorphisms in genes (Solyc01g012550 and Solyc06g051170) participating in the biogenesis of natural antisense siRNAs and in transitive silencing of transgene transcripts. Among genotype-specific variants, those affecting LUC were found in genes involved in small RNA-directed silencing mechanism, such as Argonaute slicer proteins (AGO1 and AGO10) and RNA-directed DNA methylation (RDM1). Similarly, a COR gene was found to code for a small RNA methyltransferase (HEN1) (Supplementary Table S4). RdDM is essential to maintain DNA methylation in CHH context and, strikingly, methylation at CHH is two times higher in fruit than in leaf, being enriched at promoter regions of the most highly expressed genes in the pericarp.25 Further studies are needed to understand whether the identified polymorphisms in RdDM-related genes could have effects on epigenetics-mediated fruit development, ripening and storability. COR-specific missense mutations were also identified in the DDB1- and CUL4-associated factor homolog 1 (DCAF1) gene (Solyc08g074370). In tomato, DDB1 affects the fruit DNA methylation status72 that is remarkably important for the developmental transition from unripe to ripe fruit.25 Equally interesting is a COR specific in frame disruptive duplication found in the ENHANCED DOWNY MILDEW 2 gene (EDM2, Solyc10g006090) whose Arabidopsis homolog regulates DNA methylation.73,74 The expression of this gene is preferential in fruit and increases during ripening (https://bar.utoronto.ca/eplant/ (October 2017, date last accessed)).
3.8. Stress-related genes
Since COR and LUC traditionally grow and produce in non-irrigated conditions, the genomic variations identified are expected to have functional importance on adaptation to low water input conditions. We identified 122 putative stress-related genes carrying at least one non-synonymous polymorphism in COR and/or LUC (Supplementary Table S4). Several SNPs were common to both landraces, e.g. those present in two Cation/H+ antiporters (Solyc03g032240, Solyc09g010530) and in several heat shock and heat shock-like proteins (e.g. Solyc05g055200, Solyc08g078720, Solyc09g011710). A stop gain variant shared between COR and LUC, leading to the formation of a truncated protein, is present in Solyc02g066890, encoding an AFP3-like Ninja-family protein. AFPs (ABI Five Binding Protein) were identified as a small family of uncharacterized proteins that may have a role in the attenuation of the stress signal through interaction with bZIPs responsible for ABA-dependent transcription.75 Similarly to Ninja, they also acted as co-repressors of Jasmonate responses.76
COR-specific sequence variants included two amino acid substitutions (L878Q; V186I) found in a Tudor/nuclease-containing protein (Solyc03g118020), which is similar to the TSN1/TSN2 RNA binding proteins from Arabidopsis, essential for stress adaptation and RNA stability.77 An additional COR-specific polymorphism (C115W) affected Solyc00g154980, a putative ethylene insensitive class 3 transcription factor. The tryptophan found in COR is also present in several other Solanaceae, including S. pennellii and S. tuberosum. Moreover, Solyc12g094700, which codes for a Cathepsin B-like cysteine proteinase, shows a stop loss mutation in COR. Its Arabidopsis homolog is involved in tracheary element formation leading to functional water-transporting xylem vessels.78
A putative heat stress transcription factor A3 (Solyc03g097120) showed a W469L substitution only in LUC. Few substitutions predicted to cause premature stop codons or disruptive in-frame deletions were also identified (Supplementary Tables S3 and S4). Two LUC-specific stop gain variants were predicted to have a high impact on protein function and affect a putative plasma membrane aquaporin (SlPIP2.10, Solyc09g007760) and a putative Homeobox leucine zipper protein (Solyc01g096320). The premature stop in this LUC transcription factor results in the genesis of a truncated protein of 71 aa in length compared to 239 aa in Heinz 1706. The closest orthologues of Solyc01g096320 in Arabidopsis (AtHB7 and HB12) have been associated to primary drought stress responses via ABA-dependent mechanisms.79,80 This LUC-specific stop gain mutation, together with the one identified in COR and LUC in Solyc02g066890, likely lead to a different or abolished activity of these proteins, resulting in a modified response to water deficit of the two landraces.
Since fruit shelf life and water availability during the vegetative season were shown to be negatively correlated in ‘Tomàtiga de Ramellet’ tomatoes,23 it is possible that individual mutations may affect both traits. This could be the case for the splicing-site mutation in ERF13, since ethylene has a predominant role not only in fruit ripening but also in plant stress responses, and several ERFs are induced in response to stress.81
In this work, we provided high quality genome sequences of two noteworthy long shelf-life tomato landraces, while contributing to quality improvement of the reference genome. A dedicated web-based platform allows browsing the distinct genomes, including information on the pseudomolecules representing Heinz 1706, COR and LUC chromosomes and their respective gene annotations. The possibility to query by ‘gene ID’ and ‘keywords’ to investigate genome tracks available in the platform offers an immediate access to integrated information from the three genomics resources. We believe that our findings highlight the wealth of diversity present in tomato landraces and may be of importance in exploiting DNA variation associated to traits of great economic importance. Future functional validation experiments and studies to examine the identified sequence polymorphisms in different landraces will shed new light on the genetics and molecular basis of fruit metabolism and storability in tomato, which is the model for climacteric fleshy fruits as well as on abiotic stress response mechanisms. Our data can be also employed to develop genetic markers and chip arrays for gene mapping and functional studies.
4. Availability
Raw sequencing reads are available at NCBI SRA (accession number SRP076324).
The Reconstructor pipeline is available at t http://www.sequentiabiotech.com/omicstools/pipeline/ (October 2017, date last accessed).
The annotated COR and LUC genomes are available at the public Genotom platform (http://cab.unina.it/genotom (October 2017, date last accessed)).
Supplementary Material
Acknowledgements
The authors thank all lab members for helpful discussions and Gaetano Guarino for technical assistance. This work was supported by the Italian Ministry of University and Research, project GenoPOM-PRO (PON02_00395_3082360).
Conflict of interest
None declared.
Accession number
SRP076324
Supplementary data
Supplementary data are available at DNARES online.
References
- 1. Peralta I.E., Knapp S., Spooner D.M.. 2006, Nomenclature for wild and cultivated tomatoes, Tomato Genetics Cooperative Rep., 56, 6–12. [Google Scholar]
- 2. Jenkins J.A. 1948, The origin of the cultivated tomato, Econ. Bot., 2, 379–92. [Google Scholar]
- 3. Lin T., Zhu G., Zhang J., et al. 2014, Genomic analyses provide insights into the history of tomato breeding, Nat. Genet., 46, 1220–6. [DOI] [PubMed] [Google Scholar]
- 4. Labate J. A., Grandillo S., Fulton T., et al. 2007, Tomato In: Kole C., ed. Genome Mapping and Molecular Breeding in Plants, Vol. 5, Vegetables, pp. 11–135. Springer Verlag, Berlin, Heidelberg. [Google Scholar]
- 5. The Tomato Genome Consortium, 2012, The tomato genome sequence provides insights into fleshy fruit evolution, Nature, 485, 635–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gapper N.E., McQuinn R.P., Giovannoni J.J.. 2013, Molecular and genetic regulation of fruit ripening, Plant Mol. Biol., 82, 575–91. [DOI] [PubMed] [Google Scholar]
- 7. Causse M., Desplat N., Pascual L., et al. 2013, Whole genome resequencing in tomato reveals variation associated with introgression and breeding events, BMC Genom, 14, 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aflitos S., Schijlen E., de Jong H., The 100 Tomato Genome Sequencing Consortium., et al. 2014, Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing, Plant J., 80, 136–48. [DOI] [PubMed] [Google Scholar]
- 9. Bauchet G., Causse M.. 2012, Genetic diversity in tomato (Solanum lycopersicum) and its Wild Relatives, In: Caliskan M. (ed.) Genetic Diversity in Plants, pp. 133–162. InTech, Rijeka. [Google Scholar]
- 10. Garcia-Martinez S., Andreani L., Garcia-Gusano M., et al. 2006, Evaluation od amplified fragment length polymorphism and simple sequence repeats for tomato germplasm fingerprinting: utility for grouping closely related traditional cultivars, Genome, 49, 648–56. [DOI] [PubMed] [Google Scholar]
- 11. Mazzucato A., Papa R., Bitocchi E., et al. 2008, Genetic diversity, structure and marker-trait associations in a collection of Italian tomato (Solanum lycopersicum L.) landraces, Theor. Appl. Genet. 116, 657–69. [DOI] [PubMed] [Google Scholar]
- 12. Mazzucato A., Ficcadenti N., Caioni M., et al. 2010, Genetic diversity and distinctiveness in tomato (Solanum lycopersicum L.) landraces; The Italian case study of ‘A pera Abruzzese’, Sci. Hort., 125, 55–62. [Google Scholar]
- 13. Galmés J., Conesa M. A., Ochogavía J. M., et al. 2011, Physiological and morphological adaptations in relation to water use efficiency in Mediterranean accessions of Solanum lycopersicum, Plant Cell Environ., 34, 245–60. [DOI] [PubMed] [Google Scholar]
- 14. Siracusa L., Patanè C., Avola G., et al. 2012, Polyphenols as chemotaxonomic markers in Italian “long-storage” tomato genotypes, J. Agric. Food Chem., 60, 309–14. [DOI] [PubMed] [Google Scholar]
- 15. Moyle L.C. 2008, Ecological and evolutionary genomics in the wild tomatoes (Solanum Sect. Lycopersicon), Evolution, 62, 2995–3013. [DOI] [PubMed] [Google Scholar]
- 16. Moyle L.C., Muir C.D.. 2010, Reciprocal insights into adaptation from agricultural and evolutionary studies in tomato, Evol. Appl., 3, 409–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ercolano M.R., Sacco A., Ferriello F., et al. 2014, Patchwork sequencing of tomato San Marzano and Vesuviano varieties highlights genome-wide variations, BMC Genomics, 15, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bai Y., Lindhout P.. 2007, Domestication and breeding of tomatoes: what have we Gained and what can we gain in the future?, Ann. Bot ., 100, 1085–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Giovannoni J.J. 2007, Fruit ripening mutants yield insights into ripening control, Curr. Opin. Plant Biol., 10, 283–9. [DOI] [PubMed] [Google Scholar]
- 20. Giovannoni J.J., Nguyen C., Ampofo B., Zhong S., Fei Z.. 2017, The epigenome and transcriptional dynamics of fruit ripening, Annu. Rev. Plant Biol., 68, 61–84. [DOI] [PubMed] [Google Scholar]
- 21. Bota J., Conesa M.A., Ochogavia J.M., et al. 2014, Characterization of a landrace collection for Tomàtiga de Ramellet (Solanum lycopersicum L.) from the Balearic Islands, Genet. Resour. Crop Evol., 61, 1131–46. [Google Scholar]
- 22. Casals J., Pascual L., Canizares J., et al. 2012, Genetic basis of long shelf life and variability into Penjar tomato, Genet. Resour. Crop Evol., 59, 219–29. [Google Scholar]
- 23. Conesa M.A., Galmes J., Ochogavia J.M., et al. 2014, The postharvest tomato fruit quality of long shelf-life Mediterranean landraces is substantially influenced by irrigation regimes, Postharvest Biol. Technol., 93, 114–21. [Google Scholar]
- 24. Saladié M., Matas A.J., Isaacson T., et al. 2007, A re-evaluation of the key factors that influence tomato fruit softening and integrity, Plant Physiol., 144, 1012–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhong S., Fei Z., Chen Y.R., et al. 2013, Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening, Nat. Biotechnol., 31, 154–9. [DOI] [PubMed] [Google Scholar]
- 26. Bressan R.A., Zhu J.-K., Van Oosten M.J., Maggio A., Bohnert H. J., Chinnusamy V.. 2014, Epigenetics connects the genome to its environment, In: Janick J.,ed. Plant Breeding Reviews, Vol. 38, pp. 69–142. John Wiley & Sons, Inc., Hoboken. [Google Scholar]
- 27. Van Oosten M.J., Bressan R.A., Zhu J.-K., Bohnert H.J., Chinnusamy V.. 2014, The role of the rpigenome in gene expression control and the epimark changes in Response to the Environment, CRC Crit. Rev. Plant Sci., 33, 64–87. [Google Scholar]
- 28. Scalfi L.V., Fogliano A., Pentangelo G., et al. 2000, Antioxidant activity and general fruit characteristics in different ecotypes of corbarino small tomatoes, J. Agric. Food Chem., 48, 1363–6. [DOI] [PubMed] [Google Scholar]
- 29. Andreakis N., Giordano I., Pentangelo A., et al. 2004, DNA fingerprinting and quality traits of Corbarino cherry-like tomato landraces, J. Agri. Food Chem., 52, 3366–71. [DOI] [PubMed] [Google Scholar]
- 30. Caiazzo R., Ricci S., Cantarella C., et al. 2014, Combining transcriptomics and metabolomics to investigate ripening and post-harvest fruit withering in a cherry-like tomato landrace. In: 11th Solanaceae Conference (SOL 2014), Arraial d´Ajuda, Brazil, November 2-6. Book of abstracts, p. 114, abs. N. 1.
- 31. Smit A.F.A., Hubley R., Green P.. 2015, RepeatMasker Open-4.0. 2013–2015. http://www.repeatmasker.org.
- 32. Rozen S., Skaletsky H.. 2000, Primer3 on the WWW for general users and for biologist programmers, Methods Mol. Biol., 132, 365–86. [DOI] [PubMed] [Google Scholar]
- 33. Paytuví-Gallart A., Hermoso A., Anzar I., Sanseverino W., Aiese Cigliano R.. 2016, GREENC: a Wiki-based database of plant lncRNAs, Nucleic Acids Res., 44, 1161–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kent W.J., Sugnet C.W., Furey T.S., et al. 2002, The human genome browser at UCSC, Genome Res., 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bayer R. 1997, The universal B-tree for multidimensional indexing: general concepts, In: International Conference on Worldwide Computing and Its Applications, pp. 198–209. Springer, Berlin, Heidelberg. [Google Scholar]
- 36. Gremme G., Brendel V., Sparks M.E., Kurt S.. 2005, Engineering a software tool for gene structure prediction in higher organisms, Inf. Softw. Technol., 47, 965–78. [Google Scholar]
- 37. Scala V., Grottoli A., Aiese Cigliano R., et al. 2017, Careful with that Axe, Gene, Genome perturbation after a PEG-mediated protoplast transformation in Fusarium verticillioides, Toxins, 9, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gan X., Stegle O., Behr J., et al. 2011, Multiple reference genomes and transcriptomes for Arabidopsis thaliana, Nature, 477, 419–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shirasawa K., Fukuoka H., Matsunaga H., et al. 2013, Genome-wide association studies using single nucleotide polymorphism markers developed by re-sequencing of the genomes of cultivated tomato, DNA Res., 20, 593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miller J.C., Tanksley S. D.. 1990, RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon, Theor. Appl. Genet., 80, 437–48. [DOI] [PubMed] [Google Scholar]
- 41. Zhou R., Wu Z., Cao X., Jiang F.L.. 2015, Genetic diversity of cultivated and wild tomatoes revealed by morphological traits and SSR markers, Genet. Mol. Res., 14, 13868–79. [DOI] [PubMed] [Google Scholar]
- 42. Bostan H., Chiusano M.L.. 2015, NexGenEx-Tom: a gene expression platform to investigate the functionalities of the tomato genome, BMC Plant Biol., 15, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sharma S.K., Bolser D., de Boer J., et al. 2013, Construction of Reference Chromosome-scale pseudomolecules for potato: integrating the potato genome with genetic and physical maps, 2013, G3: Genes, Genomes, Genetics, 3, 2031–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bolger A., Scossa F., Bolger M.E., et al. 2014, The genome of the stress-tolerant wild tomato species Solanum pennellii, Nat. Genet., 46, 1034–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aversano R., Contaldi F., Ercolano M.R., et al. 2015, The Solanum commersonii genome sequence provides insights into adaptation to stress conditions and genome evolution of wild potato relatives, Plant Cell, 27, 954–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cheng C.-Y., Krishnakumar V., Chan A.P., Thibaud-Nissen F., Schobel S., Town C.D.. 2017, Araport11: a complete reannotation of the Arabidopsis thaliana reference genome, Plant J., 89, 789–804. [DOI] [PubMed] [Google Scholar]
- 47. Scarano D., Rao R., Corrado G.. 2017, In Silico identification and annotation of non-coding RNAs by RNA-seq and De Novo assembly of the transcriptome of tomato fruits, PLoS One, 12, e0171504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu M., Gomes B.L., Mila I., et al. 2016, Comprehensive profiling of ethylene response factor expression identifies ripening-associated ERF genes and their link to key regulators of fruit ripening in tomato, Plant Physiol., 170, 1732–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Barry C.S., Giovannoni J.J.. 2006, Ripening in the tomato Green-ripe mutant is inhibited by ectopic expression of a protein that disrupts ethylene signalling, Proc. Natl. Acad. Sci., 103, 7923–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vrebalov J., Pan I.L., Arroyo A.J.M., et al. 2009, Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1, Plant Cell, 21, 3041–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gimenez E., Dominguez E., Pineda B., et al. 2015, Transcriptional activity of the MADS box ARLEQUIN/TOMATO AGAMOUS-LIKE1 gene is required for cuticle development of tomato fruit, Plant Physiol., 168, 1036–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Molina-Hidalgo F.J., Franco A.R., Villatoro C., et al. 2013, The strawberry (Fragaria×ananassa) fruit-specific rhamnogalacturonate lyase 1 (FaRGLyase1) gene encodes an enzyme involved in the degradation of cell-wall middle lamellae, J. Exp. Bot., 64, 1471–83. [DOI] [PubMed] [Google Scholar]
- 53. Uluisik S., Chapman N.H., Smith R., et al. 2016, Genetic improvement of tomato by targeted control of fruit softening, Nat. Biotechnol., 34, 950–2. [DOI] [PubMed] [Google Scholar]
- 54. Xie Y., Zhu B., Yang X., et al. 2006, Delay of postharvest ripening and senescence of tomato fruit through virus-induced LeACS2 gene silencing, Postharvest Biol. Technol., 42, 8–15. [Google Scholar]
- 55. Centeno D.C., Osorio S., Nunes-Nesi A., et al. 2011, Malate plays a crucial role in starch metabolism, ripening, and soluble solid content of tomato fruit and affects postharvest softening, Plant Cell, 23, 162–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nambeesan S., Datsenka T., Ferruzzi M. G., Malladi A., Mattoo A. K., Handa A.K.. 2010, Overexpression of yeast spermidine synthase impacts ripening, senescence and decay symptoms in tomato, Plant J., 63, 836–47. [DOI] [PubMed] [Google Scholar]
- 57. Zhang Y., Butelli E., De Stefano R., et al. 2013, Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold, Curr. Biol., 23, 1094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kramer M.G., Redenbaugh K.. 1994, Commercialization of a tomato with an antisense polygalacturonase gene: The FLAVR SAVRTM tomato story, Euphytica, 79, 293–7. [Google Scholar]
- 59. Smith C.J.S., Watson C.F., Ray J., et al. 1988, Antisense RNA inhibition of polygalacturonase gene expression in transgenic tomatoes, Nature, 334, 724–6. [Google Scholar]
- 60. Hall L.N., Tucker G.A., Smith C.J.S., et al. 1993, Antisense inhibition of pectinesterase gene expression in transgenic tomatoes, Plant J., 3, 121–9. [Google Scholar]
- 61. Brummell D.A., Harpster M.H., Civello P.M., Palys J.M., Bennett A.B., Dunsmuir P.. 1999, Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening, Plant Cell, 11, 2203–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu F., Zhang X., Lu C., et al. 2015, Non-specific lipid transfer proteins in plants: presenting new advances and an integrated functional analysis, Exbotj., 66, 5663–81. [DOI] [PubMed] [Google Scholar]
- 63. Di Matteo A., Sacco A., Anacleria M., et al. 2010, The ascorbic acid content of tomato fruits is associated with the expression of genes involved in pectin degradation, BMC Plant Biol., 10, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Duan M., Feng H.L., Wang L.Y., et al. 2012, Overexpression of thylakoidal ascorbate peroxidase shows enhanced resistance to chilling stress in tomato, J. Plant Physiol., 169, 867–77. [DOI] [PubMed] [Google Scholar]
- 65. Di Matteo A., Ruggieri V., Sacco A., et al. 2013, Identification of candidate genes for phenolics accumulation in tomato fruit, Plant Sci., 205–206, 87–96. [DOI] [PubMed] [Google Scholar]
- 66. Ito Y. 2016, Regulation of tomato fruit ripening by MADS-Box transcription factors, Jarq, 50, 33–8. [Google Scholar]
- 67. Weise A., Barker L., Kühn C., et al. 2000, A new subfamily of sucrose transporters SUT4 with low affinity/high capacity localized in enucleate sieve elements of plants, Plant Cell, 12, 1345–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Reuscher S., Akiyama M., Yasuda T., et al. 2014, The sugar transporter inventory of tomato: genome-wide identification and expression analysis, Plant Cell Physiol., 55, 1123–41. [DOI] [PubMed] [Google Scholar]
- 69. Aiese Cigliano R., Sanseverino W., Cremona G., Ercolano M.R., Conicella C., Consiglio F.M.. 2013, Genome-wide analysis of histone modifiers in tomato: gaining an insight into their developmental roles, BMC Genomics, 14, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ebbs M.L., Bender J.. 2006, Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase, Plant Cell, 18, 1166–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen W., Kong J., Qin C., et al. 2015, Requirement of CHROMOMETHYLASE3 for somatic inheritance of the spontaneous tomato epimutation Colourless non-ripening, Sci. Rep., 5, 9192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liu J., Tang X., Gao L., et al. 2012, A role of tomato UV-damaged DNA Binding Protein 1 (DDB1) in organ size control via an epigenetic manner, PLoS One, 7, e42621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lei M., La H., Lu K., et al. 2014, Arabidopsis EDM2 promotes IBM1 distal polyadenylation and regulates genome DNA methylation patterns, Proc. Natl. Acad. Sci. USA., 111, 527–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tsuchiya T., Eulgem T.. 2013, Mutations in EDM2 selectively affect silencing states of transposons and induce plant developmental plasticity, Sci. Rep., 3, 1701–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Garcia M.E., Lynch T., Peeters J., Snowden C., Finkelstein R.. 2008, A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings, Plant Mol. Biol., 67, 643–58. [DOI] [PubMed] [Google Scholar]
- 76. Pauwels L., Barbero G.F., Geerinck J., et al. 2010, NINJA connects the co-repressor TOPLESS to jasmonate signalling, Nature, 464, 788–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. dit Frey N.F., Muller P., Jammes F., et al. 2010, The RNA binding protein Tudor-SN is essential for stress tolerance and stabilizes levels of stress-responsive mRNAs encoding secreted proteins in Arabidopsis, Plant Cell, 22, 1575–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Avci U., Earl Petzold H., Ismail I.O., Beers E.P., Haigler C.H.. 2008, Cysteine proteases XCP1 and XCP2 aid micro-autolysis within the intact central vacuole during xylogenesis in Arabidopsis roots, Plant J., 56, 303–15. [DOI] [PubMed] [Google Scholar]
- 79. Ré D.A., Capella M., Bonaventure G., Chan R. L.. 2014, Arabidopsis AtHB7 and AtHB12 evolved divergently to fine tune processes associated with growth and responses to water stress, BMC Plant Biol., 14, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Valdés A.E., Overnäs E., Johansson H., Rada-Iglesias A., Engström P.. 2012, The homeodomain-leucine zipper (HD-Zip) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities, Plant Mol. Biol., 80, 405–18. [DOI] [PubMed] [Google Scholar]
- 81. Day S., Vlot C.. 2015, Ethylene responsive factors in the orchestration of stress responses in monocotyledonous plants, Front. Plant Sci., 28, 640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.