Abstract
Many bacterial genomes exclusively display an N4-methyl cytosine base (m4C), whose physiological significance is not yet clear. Helicobacter pylori is a carcinogenic bacterium and the leading cause of gastric cancer in humans. Helicobacter pylori strain 26695 harbors a single m4C cytosine methyltransferase, M2.HpyAII which recognizes 5′ TCTTC 3′ sequence and methylates the first cytosine residue. To understand the role of m4C modification, M2.hpyAII deletion strain was constructed. Deletion strain displayed lower adherence to host AGS cells and reduced potential to induce inflammation and apoptosis. M2.hpyAII gene deletion strain exhibited reduced capacity for natural transformation, which was rescued in the complemented strain carrying an active copy of M2.hpyAII gene in the genome. Genome-wide gene expression and proteomic analysis were carried out to discern the possible reasons behind the altered phenotype of the M2.hpyAII gene deletion strain. Upon the loss of m4C modification a total of 102 genes belonging to virulence, ribosome assembly and cellular components were differentially expressed. The present study adds a functional role for the presence of m4C modification in H. pylori and provides the first evidence that m4C signal acts as a global epigenetic regulator in H. pylori.
INTRODUCTION
In prokaryotes, many DNA methyltransferases (MTases) are associated with restriction enzymes (REases) to form a restriction-modification system (R-M system). R-M systems act as primitive immune systems against bacteriophages in prokaryotes (1). In addition to bacterial defense, MTases of R-M system or solitary MTases play a significant role in bacterial evolution and epigenetic gene regulation (2). DNA methylation in the bacterial genome is of three types: N6-adenine methylation (m6A), C5-cytosine methylation (m5C) and N4-cytosine methylation (m4C). Methylation of adenine (m6A) and cytosine (m5C and m4C) leads to protrusion of methyl-group from the major groove of the DNA double helix (2). As a result of this modification the interaction between DNA binding proteins and their recognition sites could be altered. Epigenetic gene regulation is common in mammals and plants where m5C is used as a primary epigenetic signal (3,4). Unlike eukaryotes, bacteria utilize m6A as a main signal for epigenetic regulation. It regulates the virulence of several different human pathogens such as Escherichia coli, Mycobacterium tuberculosis, Salmonella enterica, Haemophilus influenza and Helicobacter pylori (2).
Helicobacter pylori is a Gram-negative, spiral-shaped, microaerophilic bacterium which causes various gastric diseases such as peptic and duodenal ulcers, gastritis and gastric cancer (5). H. pylori exhibit a high level of allelic variation and genetic diversity which likely reflects its adaptability in the host (6). Methylome analyses of H. pylori strains reveal that R-M systems are abundant in the genome, and each strain displays a strain specific DNA methylation pattern (7,8). Helicobacter pylori 26695 strain contains a phase variable type IIS HpyAII R-M system which recognizes the sequence 5′ GAAGA 3′/ 5′ TCTTC 3′. This system is composed of N6-adenine (M1.HpyAII), N4-cytosine (M2.HpyAII) MTases and a novel phase variable type IIS endonuclease, R.HpyAII (9). M1.HpyAII methylates the underlined adenine residue of the 5′ GAAGA 3′ sequence and M2.HpyAII methylates the underlined cytosine in the sequence 5′ TCTTC 3′ at N4- position (8) in the complimentary strand. H. pylori 26695 strain has numerous N6-methyl adenine and C5-methyl cytosine MTases. However, M2.HpyAII is the only N4-methyl cytosine MTase present in the genome (8). We had previously shown that H. pylori utilizes m5C modification signal for the regulation of gene expression, and differential methylation plays a central role in motility, adhesion, and virulence (10). In another study, lack of m5C in E. coli led to overexpression of stress-responsive sigma factor RpoS (11). However, no physiological role of m4C has been found in any bacteria except for protection against restriction endonucleases (12). Recently m4C modification was observed in the type I R-M systems which are known to have exclusive m6A modification (13). Suggesting that type I R-M systems can also use m4C modification for host protection in addition to m6A modification.
In this study, we used H. pylori 26695 strain to answer whether m4C modification plays an epigenetic role in H. pylori pathogenesis and gene expression. The full genome sequence (14), transcriptome data (15), and global methylome data (8) of H. pylori 26695 strain are available making it an ideal candidate for in-depth analysis. To answer whether m4C modification plays an epigenetic role in H. pylori physiology and pathogenesis, deletion strain of M2.HpyAII MTases was prepared followed by genome-wide gene expression, proteomics analysis and coculture studies with AGS cell line.
MATERIALS AND METHODS
Strains and growth conditions
Helicobacter pylori strains were grown on Brain-heart infusion (BHI) agar (Difco) plate supplemented with 8% horse serum (Invitrogen), 0.2% Iso-vitaleX (BD), antibiotics vancomycin (6 μg ml−1), trimethoprim (8 μg ml−1), and amphotericin B (8 μg ml−1) under microaerobic conditions in an incubator equilibrated with 10% CO2 and 5% O2. Selective plates were prepared by additionally supplementing the bacterial agar media with 8.0 μg ml−1 chloramphenicol, 10 μg ml−1 streptomycin or 5 μg ml−1 gentamicin or kanamycin 20 μg ml−1.
H. pylori growth analysis
Helicobacter pylori strains were grown in Brain-heart infusion (BHI) broth supplemented with 8% horse serum (Invitrogen), 0.2% Iso-vitaleX (BD) and antibiotics vancomycin (6 μg ml−1), trimethoprim (8 μg ml−1), and amphotericin B (8 μg ml−1) under microaerobic conditions with constant shaking at 130 rpm. Growth was measured by the addition of 24 h old exponentially growing H. pylori cells at a final O.D.600 nm of 0.02 in 30 ml of BHI broth. Bacterial growth was monitored by measuring O.D.600 nm at 2 h time intervals. Growth in acidic pH medium was carried out by adjusting the pH of BHI medium to 4.5 by HCl. The generation time of H. pylori strains was estimated as described in (16).
PCR-based screening of HpyAII R-M system
Fourty nine clinical H. pylori isolates from patients diagnosed with gastric cancer, duodenal ulcer, or non-ulcer dyspepsia and twenty-nine isolates from healthy adult volunteers (HVs) of both sexes who had no symptoms of gastritis or dyspeptic syndromes like abdominal pain were analyzed. PCR was carried out to detect R.hpyAII, M1.hpyAII and M2.hpyAII genes using primers 1 to 6 (Supplementary Table S1). PCR-based screening was done in the presence of positive and negative controls for each gene. Distribution of R.hpyAII, M1.hpyAII and M2.hpyAII genes in completely sequenced H. pylori strains was analyzed using NCBI BLAST tool. Nucleotide sequences were submitted as a query in NCBI BLAST tool, and H. pylori (taxid: 210) was selected as the database for the analysis.
In silico genome analysis for GAAGA/TCTTC sites distribution
The complete genome sequence of the 26695 strain of H. pylori, annotated by (14) was used in this study. Intergenic regions of the genome were identified based on the given annotations. Intergenic region in the vicinity of the transcription start sites (50 nucleotides upstream) were screened for distribution of 5′ TCTTC 3′. To check the distribution of 5′ TCTTC 3′ sites in the gene body and the possible promoter region spanning 200 bp upstream of start codon till the stop codon of the gene was screened using Python scripts. Trends in the frequency and spatial distribution of 5′ TCTTC 3′ repeats were also analyzed.
Control intergenic sequences was generated by randomly shuffling 10 000 existing intergenic sequences from the whole genome, and analyzing the frequency and spatial distribution of 5′ TCTTC 3′ sites. As expected, the frequency distribution of randomly generated 5′ TCTTC 3′ sites was observed to follow a Gaussian distribution. To determine if test and control distributions were significantly different, Gaussian distributions analysis was carried out. The P-value described in this study represents the probability of our test data occurring at or below its given location in the Gaussian function by pure chance.
Frequency differences of GAAGA/TCTTC sites between intragenic and intergenic regions
To determine whether the frequencies of GAAGA/TCTTC differ between intragenic and intergenic regions, their frequencies per region in 26695 genomes was calculated. For each individual intragenic/intergenic region, the total number of GAAGA/TCTTC sites was counted. Most regions possessed no GAAGA/TCTTC sites. A frequency list of regions possessing ≥1 GAAGA/TCTTC site(s) was prepared. For example, if two regions possessed three GAAGA/TCTTC sites each, and three regions possessed four GAAGA/TCTTC sites each, this frequency list would be generated: 3, 3, 4, 4, 4. By randomly shuffling the sequence of individual intragenic/intergenic regions (10 times per region), control frequency lists were prepared. The test and control frequency lists were plotted as length and log-10 normalized frequency histograms to evaluate frequency differences. For determining statistical significance, the Welch Two Sample t-test directly on unshuffled and shuffled frequency lists was performed.
Construction of HpyAII R-M system mutant strains
HpyAII R-M system was deleted in 26695 strain in two steps by using PCR-based gene deletion method described previously (17) with modifications. First, R.hpyAII gene deletion cassette was prepared by overlap extension PCR method using primers 7–10 (Supplementary Table S1). R.hpyAII gene deletion was constructed by disrupting the ORF with cam resistance cassette by allelic replacement through natural transformation. Helicobacter pylori R.hpyAII deletion in 26695 strain was used as a template to delete the M2.hpyAII gene by the integration of gentamicin resistance cassette in the ORF. To generate HpyAII R-M system deletion, R.hpyAII deletion strain was used as a template. Gentamicin resistance cassette was PCR amplified to make deletion construct which has 5′ region homologous to M2.hpyAII and 3′ region homologous to M1.hpyAII genes. Allelic replacement through natural transformation was performed to inactivate the two MTases genes. Deletion strains lacking the M2.hpyAII gene were selected on BHIA plate containing both gentamicin and chloramphenicol Deletion strains were screened on BHIA plate containing 8.0 μg ml−1 chloramphenicol or 5 μg ml−1 gentamicin or both. Mutants were scored by using combination of gene specific and gentamicin or chloramphenoicol cassette primers (Supplementary Table S1).
Complementation of M2.hpyAII gene in H. pylori R.hpyAII-M2.hpyAII deletion strain
A region containing M2.hpyAII coding region with 5′ flanking region (250 bp) was PCR amplified (1126 bp) from H. pylori 26695 genome using primer Sm1 and Sm2 (Supplementary Table S1). Purified insert was cloned in P1175 vector (modified pJet 1.2 vector with ampicillin resistance). P1175 vector has rdxA gene which is disrupted by kanamycin resistance cassette (kind gift from Dr Pablo Radicella's laboratory). M2.hpyAII was cloned at 5′ end of kanamycin resistance cassette with flanking rdxA gene using overlapping primers Sm3 and Sm4. Clones were confirmed by sequencing and PCR. Purified clone was used to transform H. pylori 26695 R.hpyAII-M2.hpyAII deletion strain through natural transformation. Complementation strain was screened on BHIA plate containing 20 μg ml−1 kanamycin. Complementation and deletion strains were confirmed by PCR using primers Rdx F (–100) and Rdx R (–100) (Supplementary Table S1), in vitro digestion with purified HpyAII endonuclease, DNA dot-blot assay and by methylome analysis by SMRT PacBio sequencing.
DNA dot-blot assay
Purified genomic DNA was diluted to 500 ng μl−1 stock and 200 and 500 ng was spotted per spot on the dry nitrocellulose membrane. DNA spots were allowed to air dry at 37°C followed by UV-crosslinking in UVC 500 Crosslinker (Amersham Biosciences) at 2400 × 100 μJ cm2 in energy mode. Membrane was allowed to air dry for 10 min and blocked with 5% skimmed milk in 1× TBST (Tris-buffered saline with 0.1% Tween-20) for 2 h at 4°C. Primary antibody (anti-m6A or anti-m4C) was added to the membrane at 1:1000 dilutions and membrane was probed for 4 h at 4°C with gentle shaking. After incubation with primary antibody, membrane was washed three times (10 min each) with 1× TBST solution. Secondary antibody conjugated with HRP was added at 1:20 000 dilution and membrane was incubated at room temperature for 1 h with gentle shaking. Three washes were given and membrane was developed with ECL kit. We thank NEB (USA) for providing us the monoclonal anitibodies against m4C and m6A residues.
Methylome analysis by SMRT PacBio sequencing
Genomic DNA from various deletions and the complemented strains was prepared by CTAB reagent followed by QIAGEN genomic-tip 100/G. SMRTbell template libraries were prepared from total H. pylori genomic DNA following the 10 kb Template Preparation Procedure (Pacific Biosciences, Menlo Park, CA, USA). One SMRT cell was run on an RSII sequencer using P6/C4 chemistry, producing between 800 and 1100 Mb of raw sequence (two SMRT cells for the dR_dM1_dM2 strain). The genome sequence was assembled for each strain using Pacific Biosciences SMRT Analysis 2.3 program ‘RS_HGAP_Assembly.3′, with coverage between 411X and 513X, and the assembly was closed to a single circular genome manually. SMRT sequencing and methylation detection was performed as described earlier in (8,18). PacBio motif files and the sequencing data was submitted to NCBI database (Table 1).
Table 1. Methylation status at GAAGA/TCTTC sites in H. pylori deletion strains. SMRT sequencing was performed to determine the loss of m4C methylation in the genome.
| Strain | Number of GAAGA sites | Total GAAGA sites methylated | Number of TCTTC sites | Total TCTTC sites methylated | NCBI accession number |
|---|---|---|---|---|---|
| 26695 ΔR.hpyAII | 4816 | 4816 (100%) | 4816 | 4813 (99.94%) | CP026326 |
| 26695 ΔR.hpyAII-M2.hpyAII | 4818 | 4813 (99.9%) | 4816 | 0 | CP026324 |
| 26695 ΔR.hpyAII-M2.hpyAII:: M2.hpyAII:: gen | 4828 | 4828 (100%) | 4828 | 4826 (99.96%) | CP026515 |
| 26695 ΔR.hpyAII-M1.hpyAII | 4827 | 4826 (99.99%) | 4827 | 4826 (99.99%) | CP026325 |
| 26695 ΔR.hpyAII-M1.hpyAII-M2.hpyAII | 4817 | 4750 (98.61%) | 4817 | 0 | CP026323 |
Estimation of natural transformation capacity using point-mutated and larger fragment of donor DNA
H. pylori recombination assays were performed as described in (19). Streptomycin resistance genomic DNA (A128G mutation in rpsL gene) of H. pylori 26695 strain lacking GAAGA/TCTTC site methylation was used as the donor DNA. Exponentially growing H. pylori cells (6 × 106 CFU) were naturally transformed by incubating it with 200 ng of donor DNA under room temperature on BHIA plates. Cells were recovered for 24 h at 37°C under microaerobic conditions, H. pylori cells were serially diluted and plated on BHIA plates with or without streptomycin (10 μg ml−1). CFU were counted after incubation for 3–5 days at 37°C. The recombination rates were calculated as the number of streptomycin resistance colonies per recipient CFU. P values were calculated using the Mann-Whitney U test. Experiments were repeated at least six times for each sample.
To determine whether loss of m4C methylation regulates uptake of larger DNA fragments, recombination assays using 1478 bp DNA fragment were performed. This fragment has kanamycin resistance cassette with flanking region of rdxA gene at both the ends (317 and 385 bp). Successful integration of 1478 bp fragment in the rdxA locus will disrupt the rdxA gene and provides kanamycin resistance to the transformed cells. The experiment was carried out in a similar manner as done for point-mutated streptomycin DNA. Transformation frequencies of the wild-type and the deletion strains were measured by selecting the bacteria on kanamycin plate.
Cell culture and co-culture
AGS cells (ATCC CRL 1739, a human gastric adenocarcinoma cell line) were grown in a complete medium consisting of RPMI 1640 containing 10% fetal bovine serum (FBS), 2 mM l-glutamine and 1× antibiotic and antimycotic mixture (10 000 units ml−1 of penicillin, 10 000 μg ml−1 of streptomycin, and 25 μg ml−1 of Amphotericin B) at 37°C in a humidified incubator equilibrated with 5% CO2 and 95% air. For coculture experiments, confluent cells were trypsinized and seeded in 6- or 24- or 96-well culture plates for 24 h and then replaced with an antibiotic-free RPMI 1640 medium containing 2% FBS. AGS cells were then cocultured in the presence or absence of H. pylori strains under microaerobic conditions.
IL-8 proinflammatory cytokine expression analysis in AGS cells
IL-8 secretion from H. pylori -infected AGS cells was determined as described in (10). Briefly, exponentially growing 24 h old H. pylori cells were resuspended in phosphate-buffered saline (PBS). Bacterial cells were harvested by centrifugation for 5 min at 2000 g. The supernatant was discarded, and the pellet was resuspended in RPMI 1640 containing 2% FBS to obtain 5 × 107 CFU. This bacterial suspension was cocultured with AGS cells at an MOI of 100:1 for 8 h at 37°C. A negative control (RPMI alone) was taken. All samples were tested at least in duplicates. The medium was removed and centrifuged at 13 000 g for 10 min in order to remove the bacteria and the cell fragments. The supernatant was frozen in –80°C prior to IL-8 measurement by Sandwich ELISA. The IL-8 measurement was performed using the specific ELISA kit provided by Puregene (Human IL-8/CXCL8 ELISA kit) according to the manufacturer's instructions.
AGS cells apoptosis assay
Apoptosis analysis of AGS cells (3 × 106) co-cultured with H. pylori strains at an MOI of 1:100 for 24 h was detected by the appearance of a typical sub-Go/G1 fraction of fragmented nuclei. Apoptosis analysis was carried out as described in (20). Briefly, cells were scraped and resuspended in PBS followed by centrifugation at 200 × g. The cells were washed with PBS and fixed by the drop wise addition of 70% chilled ethanol. Fixed cells were stored in 4°C overnight before analysis. The fixed cells were washed thrice with PBS and stained with propidium iodide (50 μg ml−1 in PBS) solution containing, 1 mg ml−1 RNase and 0.05% Triton X-100. A minimum of 10 000 events per sample was measured. Acquisition and analysis were conducted using FACS Diva software (Becton Dickinson, USA).
Lipopolysaccharide isolation
LPS was isolated from exponentially growing wild-type or H. pylori deletion strains (4.5 × 107 CFU) by proteinase K method as described previously (21). Briefly H. pylori cells were collected and diluted to 0.6 at O.D.600 nm. Bacterial cells from 1.5 ml suspension were harvested and resuspended in the lysis buffer (0.125% Tris–Cl pH 6.8, 4.6% SDS, 5% 2-mercaptoethanol 0.004% bromophenol blue, 20% glycerol). Mixture was incubated at 95°C for 15 min and cooled to room temperature for the addition of 180 μg proteinase K. Mixture was then incubated for 1 h at 60°C and purified LPS was analyzed on SDS-PAGE (15% polyacrylamide) followed by detection by silver staining.
Adhesion of H. pylori to AGS cells
In vitro H. pylori adhesion was quantified by alamarBlue assay as described previously (22). Briefly, AGS cells (2 × 104) were cocultured (under microaerobic conditions at 37°C) for 1 h with exponentially growing H. pylori wild-type or deletion strains in RPMI media containing 2% FBS. The bacterial suspension was prepared in RPMI media containing 2% FBS, and different dilutions were prepared to obtain MOI of 50:1, 100:1 and 200:1 with a constant number of AGS cells. Adherence measurements were performed by monitoring the development of fluorescent pink color from blue. Fluorescent measurements were performed at an excitation wavelength of 544 nm and emission at 590 nm. As a control, fluorescent measurements from AGS cells, which were not incubated with H. pylori but treated, similarly were used as blank. Experiments were performed twice with three technical replicates.
RNA isolation and transcriptomic analysis
Total RNA was isolated from exponentially growing H. pylori culture (O.D. 600 nm ∼ 0.8) using TRIzol reagent. Two biological replicates were used for both wild-type and R.hpyAII-M2.hpyAII gene deletion strain. Total RNA was treated with DNaseI as per the manufacturer's protocol to remove contaminating DNA. Integrity of isolated RNA was checked using Qubit measurements and RNA with RIN value greater than 8.6 were used for further analysis. Ribosomal RNAs were removed by Ribo-Zero kit from Illumina, Inc. as per the manufacture's protocol.
RNA sequencing and data analysis
Total 3 μg of RNA was used to prepare RNA seq library. To prepare library mRNA molecules were purified using magnetic beads. Following purification, the mRNA was fragmented into small pieces using divalent cations under elevated temperature. The cleaved RNA fragments were used to synthesize first strand cDNA using reverse transcriptase and random primers. This was followed by second strand cDNA synthesis using DNA polymerase I and RNase H. cDNA fragments generated were used to generate RNA libraries. Paired end run was performed on Illumina HiSeq 2500 platform to obtain 2 × 100 bp reads. Based on the quality of sequence reads sequences were trimmed to retain the high quality sequence for further analysis. Low-quality sequence reads were excluded from the analysis. The contamination removal step had been performed using bowtie2 (version 2.2.2), in-house Perl scripts and picard tools (version 1.119). The pre-processed reads were aligned to the Helicobacter pylori 26695 reference genome and gene model downloaded from ENSEMBL (ftp://ftp.ensemblgenomes.org/pub/bacteria/release32/gtf/bacteria_0_collection/helicobacter_pylori_26695/Helicobacter_pylori_26695.ASM852v1.32.gtf.gz).The alignment was performed using Tophat program (version 2.1.1) with default parameters. After aligning the reads with reference gene model, the aligned reads were used for estimating expression of the genes and transcripts using cufflinks program (version 2.2.1). Raw data are publicly available at NCBI GEO accession number GSE94268. Genes with a fold change value of log2fold >1.0 and a P-value of <0.05 were considered to be differentially expressed, and represented by CiVi circle diagram (23).
Sample preparation for two-dimensional polyacrylamide gel electrophoresis
H. pylori protein lysates were prepared by sonicating the bacterial pellet in sonication buffer (100 mM Tris–Cl pH 8.0, 25 mM NaCl and 2 mM DTT). Samples were then centrifuged at 14 000 g for 15 min to remove cell debris. Supernatants (∼500 μg proteins) were then acetone precipitated by adding 9 volumes of chilled acetone to remove excess salt from the sample. Samples were stored overnight at –80°C followed by the collection of protein precipitate by centrifugation at 14 000 g for 20 min at 4°C. The precipitate was then air dried followed by solubilization in 125 μl buffer containing 7 M urea, 2 M thiourea, 0.5% pH 3–10 carrier ampholytes (GE Healthcare), 2% 3-([3-cholamidopropyl] dimethylammonio)-1-propanesulfonate (CHAPS) and 2 mM DTT. Solubilized protein samples were then used to rehydrate the immobilized pH-gradient 7 cm linear pH 3–10 IPG strip. Rehydration was done at room temperature for 8 h. The first dimension electrophoresis was performed at 20°C with the following conditions: 100 V for 1 h, 300 V for 1.45 h, 500 V for 1.30 h, 1000 V for 1.30 h, 3000 V for 1.45 h, 5000 V for 2 h. Following isoelectric focusing, IPG strips were equilibrated with the buffer containing 130 mM Tris–Cl pH 8.5, 10 mM DTT, 135 mM Iodoacetamide, 2% SDS and 10% glycerol for 10 min. The second dimension electrophoresis was performed to separate proteins according to their molecular weight using 0.1% SDS–10% polyacrylamide gel. Proteins were visualized by Coomassie Brilliant Blue or Silver staining.
Mass spectrometry and protein identification
In gel trypsin of protein bands were performed as described (24). Digested peptides were reconstituted in 15 μl of the 0.1% formic acid and 1 μl of the same was injected on column. Digested peptides were subjected to 70 min RPLC gradient, followed by acquisition of the data on LTQ-Orbitrap-MS. Generated data was searched for the identity on MASCOT as search engine and using Swiss-Prot, TrEMBL and H. pylori databases. Proteins with >20% coverage were used for further analysis.
RESULTS
Distribution of HpyAII R-M system in H. pylori clinical isolates
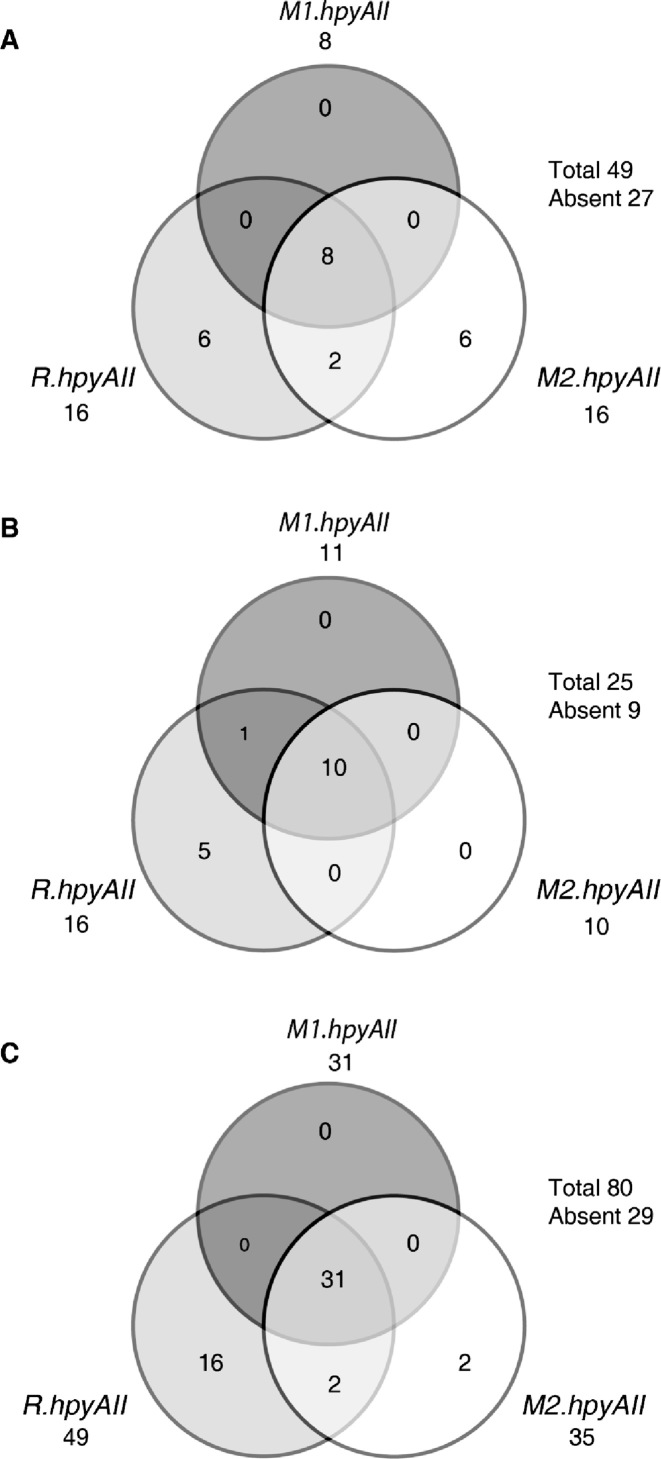
HpyAII R-M system is a type IIS R-M system present in 26695 strain of H. pylori coded by three open reading frames R.hpyAII, M1.hpyAII and M2.hpyAII (8,14). We analyzed the presence of HpyAII R-M system in 74 Indian clinical isolates of H. pylori. PCR-based screening revealed that 16% (8/49) of the H. pylori strains taken from symptomatic patients harbors a complete HpyAII R-M system (Figure 1A). However, 12% (6/49) of symptomatic strains were positive for R. hpyAII gene alone. Upon analysis of strains from asymptomatic people, it was found that 40% (10/25) of the strains contain a complete HpyAII R-M system, and 20% (5/25) were positive only for R. hpyAII gene (Figure 1B). Thus, HpyAII R-M system belongs to the strain-specific repertoire of genes. This was further validated, by evaluating completely sequenced strains of H. pylori for distribution of HpyAII R-M system. In silico analysis, which corroborated with our PCR results revealed HpyAII R-M is present in 39% (31/80) of sequenced strains (Figure 1C). Also, 20% (16/80) of sequenced strains carry R. hpyAII gene alone without any cognate MTases (Figure 1C). Multiple sequence alignment of the R. hpyAII gene in the sequenced strains lacking cognate mtases genes revealed frame-shift mutations and truncations in the open-reading frame compared to 26695 strain (Supplementary Figure S1), suggesting that an inactive copy of the R. hpyAII is present in these strains.
Figure 1.
Distribution of HpyAII R-M system in H. pylori clinical strains. Venn-diagram for the distribution of HpyAII R-M system in H. pylori clinical isolates. (A) H. pylori Indian isolates from symptomatic patients (n = 49). Eight out of forty-nine strains screened, harbor complete HpyAII R-M system, singletons: six strains carry R.hpyAII and M2.HpyAII genes. (B) H. pylori Indian isolates from healthy volunteers (n = 25). Ten out of twenty-five strains screened contain complete HpyAII R-M system, singletons: five strains positive for R.hpyAII. (C) H. pylori completely sequenced strains (n = 80). Thirty one out of eighty strains screened for HpyAII R-M system carry complete HpyAII R-M system, Singletons: sixteen strains positive for R.hpyAII and 2 strains for M2.HpyAII gene.
To test the activity of M1.HpyAII and M2.HpyAII MTases in the isolates, in vitro digestion assay was carried out using purified genomic DNA from 14 strains (11 strains positive and 3 negative for HpyAII R-M system) and HpyAII endonuclease. If both the MTases are active in a strain they will methylate GAAGA/TCTTC sites in the genome. Methylation will protect the genomic DNA from the action of cognate HpyAII endonuclease. However, in those strains where the MTases are inactive the GAAGA/TCTTC sites will remain unmethylated and will therefore, be susceptible to HpyAII digestion in vitro (Supplementary Figure S2A). It was observed that all 11 strains screened positive for HpyAII R-M system were refractory to in vitro HpyAII digestion (Supplementary Figure S2B). However, the three strains which were negative for HpyAII R-M system were found to be sensitive to HpyAII. These results suggest that both the MTases are functional in the 11 strains screened.
Screening of 5′ TCTTC 3′ sites in the intergenic region of H. pylori 26695 strain
To evaluate whether the methylation of 5′ TCTTC 3′ sites by M2.HpyAII could play role in bacterial physiology, the frequency of 5′ TCTTC 3′ sites was examined near (-50 nt) the known transcription start sites (TSS) of H. pylori strain 26695 (15). It was observed that a total 103 sites were present near the TSS of 99 ORFs respectively. Genes involved in metabolism, natural transformation, transcription, DNA replication, ion-transport and virulence carry sites near TSS region (Supplementary Figure S3). A null hypothesis was developed stating that if there is no selection pressure on the distribution of 5′ TCTTC 3′ sites upstream TSS, then the occurrence of such sites will be random. To test this hypothesis, the observed number of 5′ TCTTC 3′ sites in the complete intergenic region (143 sites) was compared with the control data obtained by randomly shuffling (n = 10,000) the intergenic region. Results obtained from such randomized data set show that 5′ TCTTC 3′ site occurred 272 times on average with the maximum being 338 and minimum 217. The difference in values obtained after shuffling the intergenic regions from those present in the actual genome is statistically significant (P- value: 4.796424e−16). We then compared the site distribution within individual intergenic regions. Each intergenic region was individually shuffled ten times and after each round of shuffling the number of TCTTC sites was calculated. It was observed that GAAGA/TCTTC sites were under-represented in the intergenic region of the genome (Supplementary Figure S4A) (Welch's two sample t test P-value: 3.018 e−12). These results provide evidence contrary to the null hypothesis and indicate the presence of selection pressure on 5′ TCTTC 3′ sites. On further investigation into the spatial distribution of these sites in the intergenic region it was observed that the sites were evenly distributed across the intergenic region of any two genes (Supplementary Figure S4C). We also checked the distribution of 5′ TCTTC 3′ sites in the coding region or intragenic region of H. pylori 26695 strain (Supplementary Figure S4B). Interestingly we observed that 5′ TCTTC 3′ sites were over-represented in the coding region or intragenic region of the genome (Welch two sample t-test, P-value: 3.582 e−11). These results suggest that distribution of 5′ TCTTC 3′ sites varies between the intergenic and intragenic region of the genome. The sites are present more in coding region compared to intergenic region. Similar observations were seen for the complementary sequence 5′ GAAGA 3′ (data not shown).
Stepwise inactivation of R.hpyAII and M2.hpyAII genes in H. pylori 26695 strain
To determine the cellular function of m4C methylation, deletion strains with an inactive copy of R.hpyAII, and M2.hpyAII genes were made. Type II R-M systems are selfish in nature and thus, any mutation which makes MTases nonfunctional will be lethal unless the cognate endonuclease is inactive (25). First, R. hpyAII gene was disrupted by inserting chloramphenicol resistance cassette in the open reading frame, followed by the integration of gentamicin resistance cassette in M2.hpyAII open reading frame M2.hpyAII gene deletion strain thus have two different antibiotic resistance compared to parent wild-type 26695. Gene deletions were confirmed by PCR (Supplementary Figure S5A and B) using gene-specific primers (Supplementary Table S1). Loss of DNA methylation in the deletion strains was confirmed by subjecting the isolated genomic DNA to in vitro HpyAII digestion. Strains with inactive R.hpyAII- M2.hpyAII genes will be unmethylated at 5′ TCTTC 3′ sites (Supplementary Figure S2A and S5C). It was observed that wild-type strain was refractory to cleavage. However, deletion of M2.hpyAII gene leads to loss of 5′ TCTTC 3′ site DNA methylation in the genome making the sequence hemi-methylated. Hemi-methylated DNA at GAAGA/TCTTC site was found to be sensitive to HpyAII cleavage in vitro (Supplementary Figure S5C). It was found that inactivation of R.hpyAII alone does not affect the activity M2.HpyAII MTase as the genomic DNA of R.hpyAII deletion strain was refractory to HpyAII cleavage (Supplementary Figure S5C) suggesting the presence of GAAGA/TCTTC methylation on the genome. Complemented strain carrying an active copy of M2.hpyAII mtase was found to be resistant to HpyAII digestion in vitro (Supplementary Figure S5C).
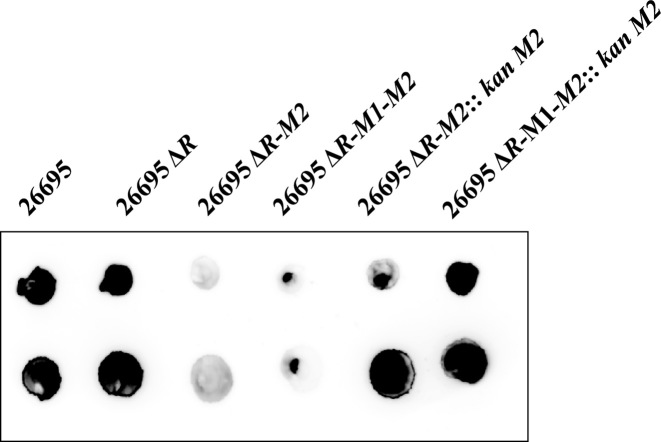
Inactivation of M2.hpyAII was further confirmed by performing DNA-dot blot assay using monoclonal antibodies against m4C modification. In this assay chromosomal signal of m4C modification was probed using anti-m4C antibodies. Since M2.HpyAII is the only N4-cytosine MTase in H. pylori 26695 strain its activity can be directly monitored by examining the genomic DNA for the presence of m4C modification. Both R.hpyAII-M2.hpyAII and R.hpyAII-M1.hpyAII.M2.hpyAII deletion strains were found to be negative for the presence of m4C modification (Figure 2). However, when R.hpyAII-M2.hpyAII strain was complemented with an active copy of M2.hpyAII with its own promoter, m4C signal was restored on the genomic DNA confirming the expression of cis copy of M2.HpyAII in the complemented strain (Figure 2).
Figure 2.
Confirmation of loss of m4C modification using DNA Dot-blot assay. Presence of m4C modification on H. pylori genome was detected using monoclonal antibodies against m4C. Purified genomic DNA was spotted on the nitrocellulose membrane and probed with monoclonal antibodies (1:1000 dilution) for the presence of m4C. Blot to detect m4C modification in wild-type and deletion strains.
Detection of m4C methylation at TCTTC site by SMRT sequencing
SMRT sequencing provides complete methylome profile and the genome sequence and thus can be used to compare different deletion and complemented strains prepared in our study. The R.hpyAII, R.hpyAII-M2.hpyAII and the R.hpyAII-M2.hpyAII::M2.hpyAII kan strains were sequenced by using PacBio RS sequencing platform. The R.hpyAII gene was disrupted by a large insertion in all 5 strains studied. The native genome copy of the M1.hpyAII and M2.hpyAII genes was intact in the R.hpyAII gene deletion strain (Table 1) and complete methylation at 5′ GAAGA 3′ and 5′ TCTTC 3′ sites was seen in the genome. SMRT sequencing of R.hpyAII-M2.hpyAII deletion strain confirmed the loss of m4C methylation at 5′ TCTTC 3′ sites in the genome. Inactivation of M2.hpyAII gene had no effect on the activity of M1.hpyAII gene since m6A methylation at complementary 5′ GAAGA 3′ sites was observed in R.hpyAII-M2.hpyAII deletion strain (Table 1). The native genomic locus copy of the M2.hpyAII gene was disrupted by an insertion in all 4 MTase deletion strains, with an additional 156 bp deletion in the R.hpyAII-M2.hpyAII and R.hpyAII-M2.hpyAII::M2.hpyAII kan strains. M2.HpyAII modification in the R.hpyAII-M1.hpyAII and R.hpyAII-M2.hpyAII::M2.hpyAII kan was obtained from a wild-type copy of the M2.hpyAII gene located at a rdxA locus in the H. pylori chromosome. Complete restoration of m4C methylation at 5′ TCTTC 3′ sites was seen in the complemented strain carrying active copy of M2.hpyAII gene at the rdxA locus (Table 1). The results of SMRT sequencing validate the results observed with DNA-dot blot and in vitro digestion assay.
SMRT sequencing revealed that M1.hpyAII deletion was not successful in the R.hpyAII-M1.hpyAII and R.hpyAII-M1.hpyAII-M2.hpyAII deletion strains. The M1.hpyAII gene in the R.hpyAII-M1.hpyAII and R.hpyAII-M1.hpyAII-M2.hpyAII deletion strains encoded a full length M1.HpyAII MTase having 9 individual amino acid mutations, all located near the C-terminal portion of the protein. The reason for the acquired point-mutations in M1.hpyAII gene is not well understood. The R.hpyAII-M1.hpyAII gene strain was thus has a point-mutated copy of M1.hpyAII gene which retained the methylation activity. From this, the results can be reinterpreted to reflect the fact that the m4C deletions and reinstatement were complete and as planned, however the m6A methylation was always present (ie, the m6A methylation was not deleted as planned, making the R.hpyAII and R.hpyAII-M1.hpyAII strains identical (along with strain R.hpyAII-M2.hpyAII::M2.hpyAII kan); and the R.hpyAII-M2.hpyAII and R.hpyAII-M1.hpyAII-M2.hpyAII strains identical, in terms of methylation at the GAAGA/TCTTC site being investigated (Table 1).
SMRT sequencing DNA modification
Genome-wide detection of modified bases, and analysis of DNA motifs for those modified bases, was performed using SMRT Analysis 2.3 program ‘RS_Modification_and_Motif_Analysis.1’. Coverage for the motifs identified was high, ranging from 195X to 240X per DNA strand. Modification of the 3-prime adenine in the 5′-GAAGA-3′ motif was observed in all five strains, with nearly 100% of the sites called by the program as modified, while m4C modification of the complement strand motif, 5′-TCTTC-3′, was called at nearly 100% of the sites in the three strains containing the M2.HpyAII MTase (R.hpyAII, dR_M1 and R.hpyAII-M2.hpyAII::M2.hpyAII kan), but was completely absent in the two M2.HpyAII deletion strains (R.hpyAII-M2.hpyAII and R.hpyAII-M1.hpyAII-M2.hpyAII) having a deletion of the M2.HpyAII MTase (Table 1). All other methylated motifs were consistent across all five of the various deletion strains. Slightly lower average IPD ratio and percent of sites called as modified was observed in the R.hpyAII-M1.hpyAII-M2.hpyAII deletion strain which carried the M1.HpyAII protein with nine mutated amino acids and lacking the M2.HpyAII protein, suggesting the mutant form of the M1.HpyAII MTase was slightly less active than the wild type M1.HpyAII, particularly in the absence of its partner M2.HpyAII MTase.
m4C DNA methylation does not regulate growth of H. pylori 26695 strain
To determine whether 5′ TCTTC 3′ site methylation plays regulatory role in H. pylori growth, growth comparison was carried out between different deletion strains in broth medium. It was observed that deletion of R.hpyAII, or M2.hpyAII genes had no effect on the growth of H. pylori in neutral pH media (Supplementary Figure S6A). To answer whether presence of 5′ TCTTC 3′ methylation provides growth benefit under stress condition like low pH, the growth profile was compared in acidic medium (pH 4.5). However, under acidic growth medium no growth defect was observed in any of the deletion strains compared to the wild type (Supplementary Figure S6B). In addition, loss of m4C methylation does not contribute to change in the generation time as evident from the overlapping exponential phase of the deletion and parent strain in both neutral as well as in acidic medium (Supplementary Figure S6C and D).
DNA m4C modification epigenetically regulates natural transformation capacity in H. pylori 26695 strain
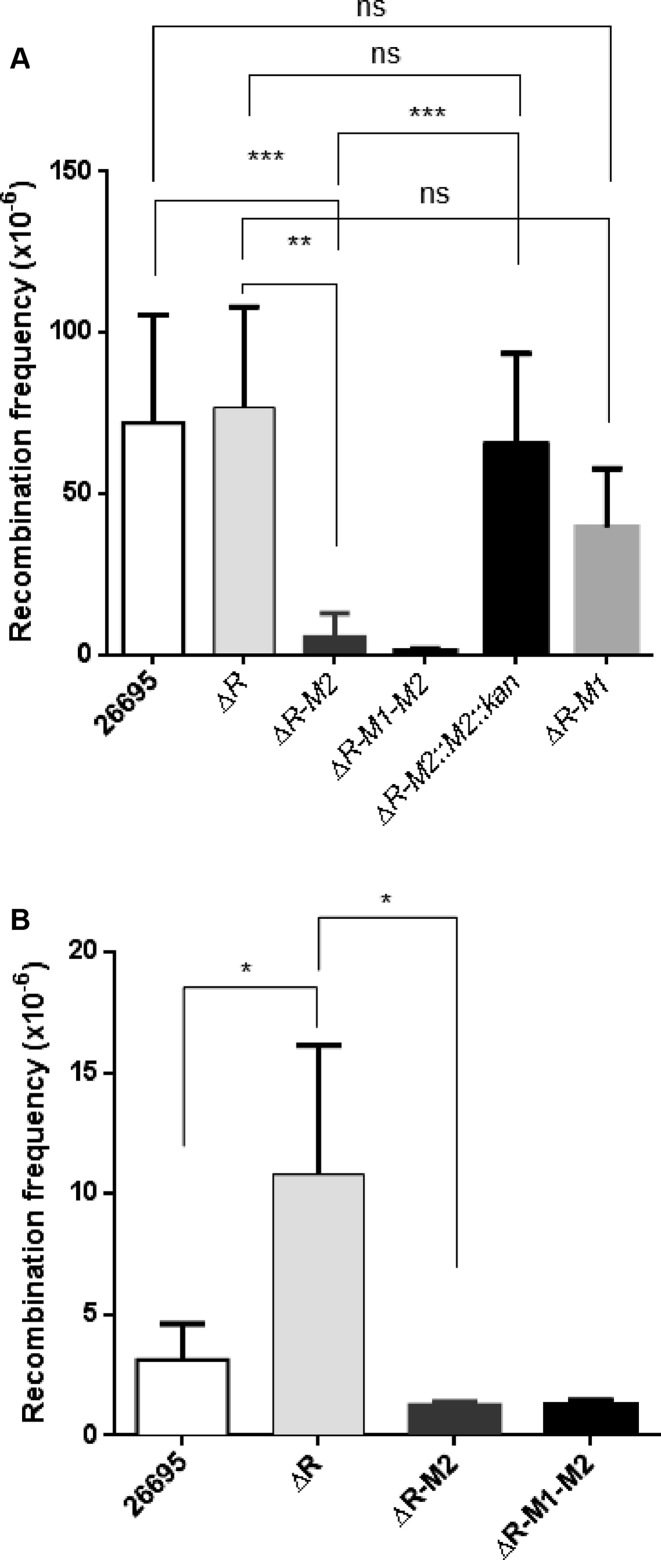
To understand the role of m4C modification in H. pylori natural transformation, recombination assays were performed using point-mutated and a fragment carrying antibiotic resistance gene. To monitor uptake of point-mutated DNA and recombination into the genome, isogenic chromosomal DNA carrying the A128G point mutation in the rpsL gene and lacking methylation at 5′ TCTTC 3′ sites was used as a donor. Transformation frequencies of wild type and the deletion strains were measured by selecting the bacteria on streptomycin plate. Deletion of R.hpyAII-M2.hpyAII leads to a 16-fold (Mann-Witney test, P-value: 0.0043) reduction in transformation frequency compared to R.hpyAII deletion strain and wild-type strain (Figure 3A). These results provide the first evidence that natural transformation in H. pylori can be regulated by m4C modification. No significant difference in the transformation frequency was seen between R.hpyAII-M1.hpyAII and R.hpyAII deletion strains (Figure 3A). Similarly, no difference was observed for R.hpyAII-M2.hpyAII and R.hpyAII-M1.hpyAII-M2.hpyAII in terms of natural competence (Figure 3). These results suggest that mutant copy of M1.HpyAII MTase retained the activity and had no detectable effect in natural competence.
Figure 3.
Effect of N4-cytosine DNA methylation on H. pylori natural transformation. Streptomycin resistant genomic DNA (200 ng) or 1487 bp long linear donor DNA (harboring kanamycin resistance cassette) was used as a donor DNA to transform the H. pylori 26695 (wild-type), R.hpyAII (ΔR), R.hpyAII-M2.hpyAII (ΔR-M2) and R.hpyAII-M1.hpyAII-M2.hpyAII (ΔR-M1-M2) deletion strains. Recombination frequency was expressed as, number of streptomycin or kanamycin resistant colonies per recipient colony-forming unit. (A) Donor DNA carrying point-mutation in rpsL gene (A128G), (B) 1487 bp linear DNA fragment carrying rdxA gene disrupted with kanamycin resistance cassette. Data was represented as mean ± SD values of pooled experiments. P-values were calculated using the Mann–Whitney U test.
To understand the role of m4C on uptake of larger DNA fragment similar experiments were carried out using 1487 bp long donor DNA. Successful recombination of this fragment will disrupt rdxA, a non-essential gene in the genome by integration of kanamycin resistance cassette. Recombination frequency of wild-type and deletion strains was compared by selection on kanamycin plate. It was observed that upon deletion of R.hpyAII gene recombination frequency increased by 3.5 fold (Mann–Whitney test, P-value: 0.0159), suggesting R.HpyAII limits uptake of large DNA fragment (Figure 3B). However, upon loss of m4C modification natural transformation capacity of R.hpyAII-M2.hpyAII deletion strain was reduced by 8 fold compared to the R.hpyAII deletion strain (Mann–Whitney test, P-value: 0.0357). These results confirm that m4C modification regulates uptake of both point-mutated and larger DNA fragment in H. pylori.
To further confirm that the reduction in natural transformation capacity is due to loss of m4C modification and not due to acquired secondary mutations. Complementation experiments were performed where the active copy of M2.hpyAII gene with its natural promoter was introduced in the R.hpyAII-M2.hpyAII deletion strain under kanamycin selection marker at rdxA locus. Natural transformation capacity of complemented was compared with the R.hpyAII-M2.hpyAII deletion strain using streptomycin resistance donor DNA. It was observed that reintroduction of M2.hpyAII gene enhances recombination frequency by 11.3-fold (Mann–Whitney test, P-value: 0.0003) compared to the R-hpyAII-M2.hpyAII deletion strain (Figure 3A). Comparable rate of recombination was observed for R.hpyAII deletion and the M2.hpyAII complemented strain (Mann–Whitney test, P-value: 0.7290). The rescue in the natural transformation capacity further strengthens the observation that presence of the m4C modification at 5′ TCTTC 3′ sites in the genome enhances natural transformation capacity of H. pylori.
DNA m4C modification controls H. pylori-induced inflammation in AGS cells
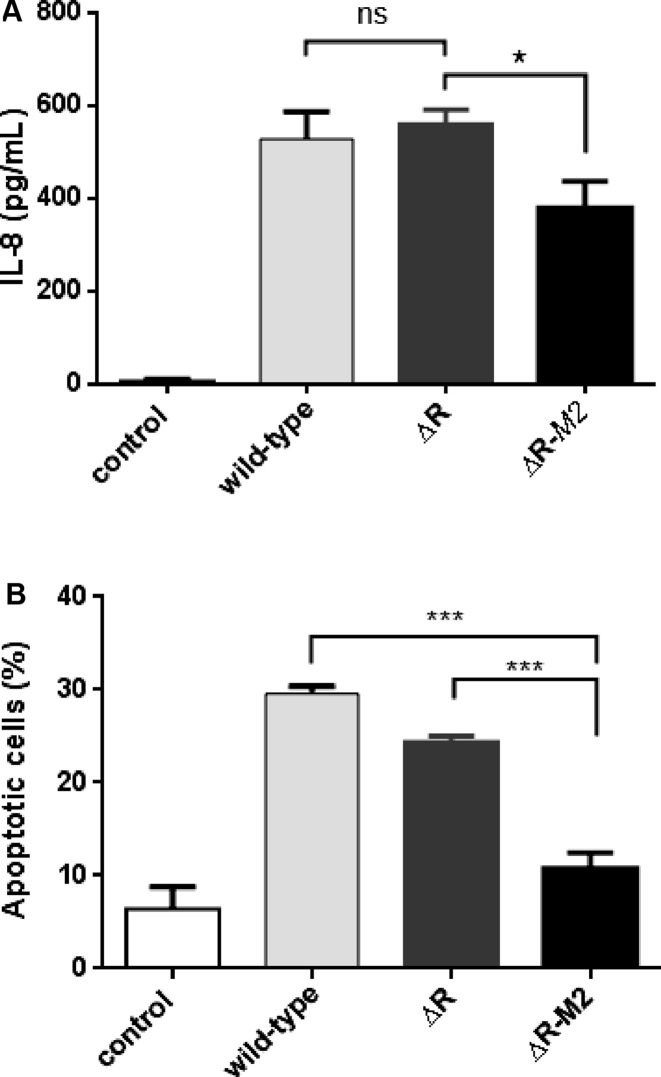
To determine the effect of m4C modification on H. pylori-induced inflammation coculture experiments with AGS cells were performed. The ability of H. pylori 26695 strain, R.hpyAII and R.hpyAII-M2.hpyAII deletion strains to elicit proinflammatory cytokine (IL-8) induction in AGS cells was assessed. Deletion of R.hpyAII resulted in the similar level of IL-8 induction from the host cells compared to wild-type strain (Figure 4A). Conversely, R.hpyAII-M2.hpyAII deletion lowered the IL-8 release by 30% compared to R.hpyAII deletion strain (P-value: 0.0165, unpaired t-test) (Figure 4A).
Figure 4.
Effect of N4-cytosine DNA methylation on H. pylori-induced IL-8 production and apoptosis in AGS cells. (A) H. pylori induced induction of IL-8 cytokine, AGS cells (5 × 105) were cultured in the absence (control) or presence of H. pylori (5 × 107 CFU) 26695 and the respective deletion strains, at MOI of 100 in RPMI 1640 medium containing 2% FBS, 2 mM l-glutamine. Supernatants were collected after 8 h for the quantification of IL-8 by sandwich ELISA. Experiments were performed in duplicates in two independent infections (*P <0.05). (B) H. pylori-induced apoptosis was assayed by coculturing AGS cells (3 × 106) in the absence (control) or presence of H. pylori (3 × 108 CFU) 26695 strain and its respective deletion strains for 24 h (MOI 100) in RPMI 1640 medium containing 2% FBS, 2 mM l-glutamine. Flow cytometry analysis was performed using propidium iodide staining. Apoptosis was analysed by plotting Sub-G0 population versus the strains used for coculture. Experiments were done in duplicates in three independent infections. Percentage apoptotic cells were expressed as mean ± SD. *P <0.05 as compared between groups. P values were calculated using unpaired t test.
DNA m4C modification regulates H. pylori-induced apoptosis in AGS cells
Twenty four hours coculture of H. pylori 26695 strain and deletion strains with AGS cells were carried out to monitor the effect of m4C on H. pylori-induced AGS cell apoptosis. Cell cycle progression of AGS cells was monitored by flow cytometry analysis (FACS) after propidium iodide staining. Proportion of cells in sub-G0 phase was used as a measure for apoptosis (Figure 4B and Supplementary Figure S7A). About 30% ± 0.8% (P-value: 0.0002) of apoptotic AGS cells were observed during coculture with wild type strain compared to 6.4 ± 2.3% seen in non-treated cells (Figure 4B, Supplementary Figure S7A and B).Inactivation of the R.hpyAII had only a modest effect on the induction of apoptosis compared to the wild-type strain (Figure 4B and Supplementary Figure S7B and C). The R.hpyAII-M2.hpyAII inactivation drastically lowered the bacteria-induced apoptosis to 10.8 ± 1.7% (P-value: 0.004) compared to 24.4 ± 0.6% observed in R.hpyAII deletion (Figure 4B and Supplementary Figure S7C and D). These results show that m4C modification in H. pylori epigenetically modulates the induced apoptosis of the AGS cells.
Investigating the role of m4C modification on Lipopolysaccharide profile, and adhesion of H. pylori to AGS cells
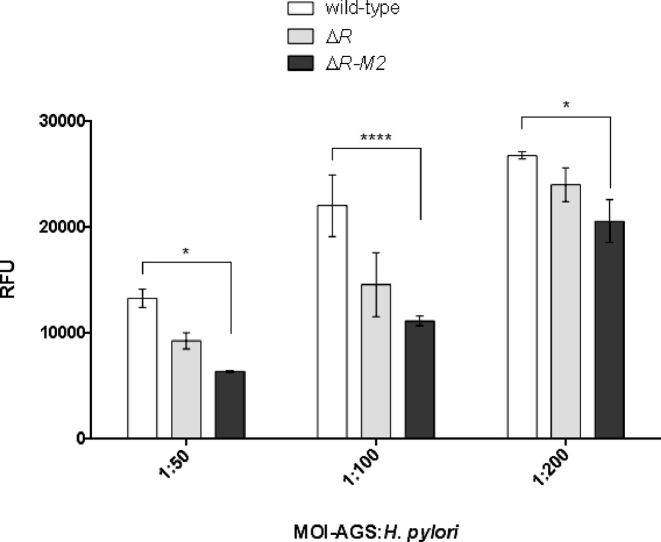
Lipopolysaccharide (LPS) of H. pylori comprises O-antigen, which acts as a vital determinant of H. pylori adhesion and pathogenicity (26,27). LPS from wild-type and deletion strains was isolated to monitor possible alteration in LPS profile. It was observed that the deletion of R.hpyAII or R.hpyAII-M2.hpyAII resulted in decrease in core LPS (Supplementary Figure S8) compared to wild-type strain. Furthermore, the implication of the altered LPS profile was assessed by performing in vitro H. pylori adhesion assay on host AGS cells using alamarBlue indicator dye. Various dilutions of wild-type and deletion strains of H. pylori were used to quantitate the cell adhesion on AGS cells. It was observed that the deletion of R.hpyAII-M2.hpyAII genes resulted in reduction in adherence (50% at 1:50 and 1:100 dilution and 23% reduction at 1:200 dilution) compared to wild-type strain (Figure 5) (P-value: 0.018, MOI: 50, P-value: <0.0001, MOI: 100 and P-value: 0.0014, MOI: 200). Non-significant reduction in adherence was seen when R.hpyAII gene alone was deleted. These results indicate that bacterial m4C modification can regulate H. pylori adherence to host cells, the first step in H. pylori pathogenesis.
Figure 5.
Comparison of in-vitro adherence of H. pylori 26695, R.hpyAII, R.hpyAII-M2.hpyAII gene deletion strains on AGS cells. Adhesion of H. pylori 26695 and deletion strains to AGS cells was quantified by alamarBlue assay. Varying bacterial dilutions were added to constant number of AGS cells per well. The P-values were calculated using two-way ANOVA followed by Bonferroni's post-test. *P< 0.01, **P< 0.001, ****P< 0.0001 compared between groups. RFU indicates relative fluorescence units.
Gene expression analysis using RNA-seq
In silico genome analysis revealed that occurrence of 5′ TCTTC 3′ sites is under natural selection and not random. It is possible that loss of methylation at 5′ TCTTC 3′ sites can influence the transcription of genes resulting in a different phenotype. RNA-seq profile of the wild-type strain was compared with R.hpyAII-M2.hpyAII deletion strain. It was observed that 102 genes were differentially expressed in the deletion strain (52 upregulated and 50 downregulated) P-value ≤0.05 (Figure 6 and Supplementary Figure S9). The major pathways which were affected in the deletion strain belonged to DNA recombination, membrane components, virulence, purine metabolism and ribosome assembly (Figure 6). To determine the relation between differentially expressed genes and with the occurrence of 5′ TCTTC 3′ sites, region encompassing 200 bp upstream of the gene and gene body was screened for 5′ TCTTC 3′ sites. It was observed that 22 downregulated genes out of 50 had at least one recognition site in the screened region. In total 49 sites were found to be present in 22 downregulated genes. Analysis of the coding region of downregulated genes showed the distribution of 5′ TCTTC 3′ sites follows a trend where majority of sites are located in the 60–90% spatial region of the gene (coding region of the gene and 200 bp upstream). In case of upregulated genes a total 124 sites were found to be present in 34 genes out of 52 genes. Genes such as cag15, ruvC, omp19 which had known roles in pathogenesis were also found to be downregulated in the R.hpyAII-M2.hpyAII deletion strain (Figure 6 and Supplementary Figure S9). This supports the less virulent phenotype of the deletion strain compared to wild-type. RNA-seq analysis results showed that the loss of genome wide m4C modification affected gene expression in H. pylori. However, no statistical correlation with the distribution of 5′ TCTTC 3′ sites was seen in the upregulated genes (Fisher's t test, P-value: 0.3049). A total of nine genes involved in ribosome assembly were found to be upregulated in the R.hpyAII-M2.hpyAII deletion strain. It has been shown previously that m5C can regulate ribosome biogenesis and loss of m5C results in upregulation of 45 genes in E. coli (11,28).
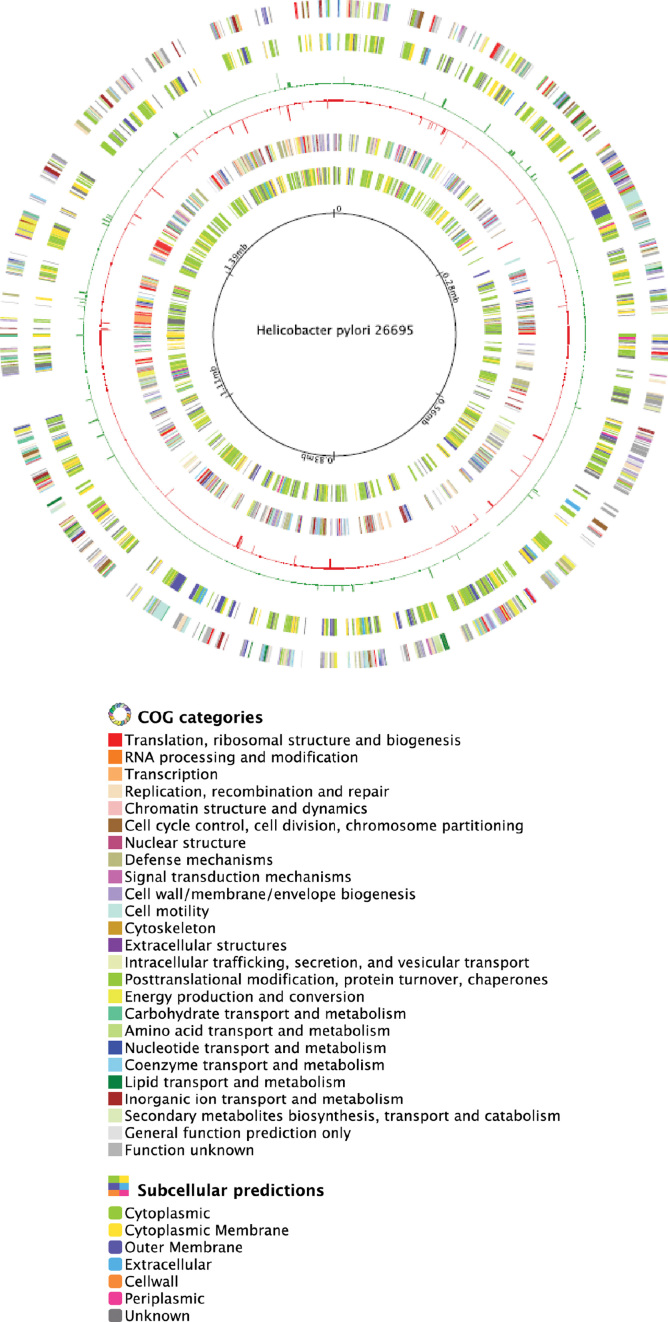
Figure 6.
Circular genome map of H. pylori strain 26695. First ring indicates ORFs on the plus strand classified according to COG category, second ring indicates subcellular location of the ORFs present in the plus strand, third ring indicates upregulated genes in R.hpyAII-M2.hpyAII deletion strain, fourth ring indicates downregulated in R.hpyAII-M2.hpyAII deletion strain, fifth ring indicates the ORFs present in the negative strand classified according to COG category, sixth ring indicates the subcellular localization of ORFs present in the negative strand, seventh ring indicates the genomic coordinates of H. pylori strain 26695.
Understanding the impact of m4C modification on H. pylori proteome
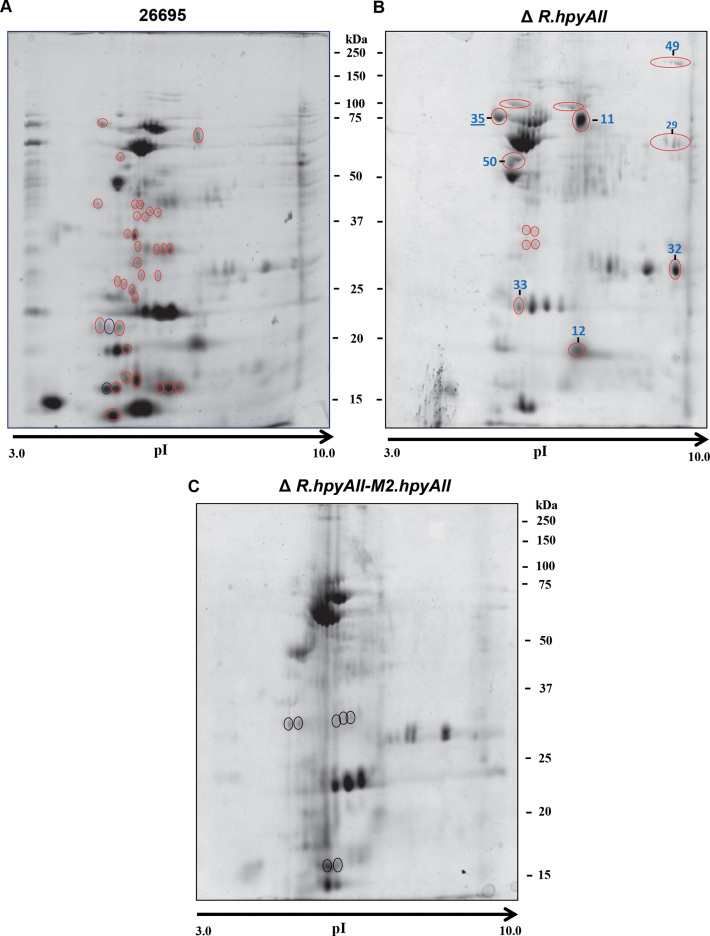
Changes in the mRNA levels are not always found at the protein level (29) and thus to gain insight into the effect of m4C modification on proteome, global proteome profiles of H. pylori 26695 wild-type strain and deletions were compared by running 2D gel electrophoresis using linear pH 3–10 gradient IPG strips. The R.hpyAII-M2.hpyAII genes deletion resulted in altered global protein profile compared to wild-type and the deletion strains (Figure 7A–C). These results confirm our hypothesis that the m4C modification alters the physiology of H. pylori by globally affecting the gene expression. The protein spots which were found to be absent in R.hpyAII-M2.hpyAII gene deletion strain but present in other strains were marked with open red circles (Figure 7). Protein spots which are present exclusively in R.hpyAII-M2.hpyAII gene deletion strain were marked with open black circles (Figure 7C). A total 12 protein spots (marked with numerical) which are absent in R.hpyAII-M2.hpyAII gene deletion strain but present in either wild-type or R.hpyAII gene deletion strain were excised from the gel and identified by mass spectrometry (Supplementary Table S2, Figure 7B and C). Protein identification revealed that the many spots were comprised of a mixture of proteins, and a total of 55 different proteins (coverage ≥ 20%) were identified from 12 different spots (Supplementary Table S2). Identified proteins were found to be part of various metabolic pathways and virulence factors (Supplementary Table S2). Interestingly, the previously known virulence factors which directly affect the pathogenesis of H. pylori were found to be absent or present in a very low amount in the R.hpyAII-M2.hpyAII gene deletion strain (Figure 7C and Supplementary Table S2). The loss of m4C modification in R.hpyAII-M2.hpyAII gene deletion strain resulted in the low expression of about 16 crucial virulence factors including CagA, CagZ, GroES, KatA and others (Supplementary Table S2) suggesting that their expression is epigenetically regulated by m4C genome modification.
Figure 7.
Global effect of N4-cytosine methylation on the proteome profile of H. pylori 26695 strain. Two-dimensional gel electrophoresis was performed for comparison of global proteomic variations between the wild-type and deletion strains. (A) 26695, (B) R.hpyAII, (C) R.hpyAII-M2.hpyAII. Red open circle indicate the differentially expressed proteins, which are absent in R.hpyAII-M2.hpyAII deletion strain but present in other strains. Black open circles represent protein spots present exclusively in R.hpyAII-M2.hpyAII deletion strain. Spots labeled were used for identification by MS–MS analysis.
DISCUSSION
H. pylori is a genetically diverse bacteria and to date 80 different clinical isolates have been sequenced (NCBI database). Methylome analysis of H. pylori isolates revealed strain-specific methylation patterns on the genome (7). Interestingly, H. pylori codes for a large number of N6-adenine MTases (8) and many of these have been shown to be involved in the epigenetic regulation of gene expression (30,31). Unlike eukaryotic cells, bacterial epigenetic regulation is centered on the modification of adenine residues at the N6-position. We have previously shown that the deletion of a solitary C5-cytosine MTase in H. pylori leads to altered gene expression affecting the transcription of genes involved in motility, adherence, and virulence (10). Prior to our study the existence of C5-cytosine methylation in the bacterial genome was mainly associated with restriction protection. Bacterial genomes exclusively contain N4-cytosine methylation (m4C) which is not present in other prokaryotes or eukaryotes. The very presence of this type of modification was thought to be an evolutionary strategy in order to overcome the hypermutability associated with m5C signal (32). Whether the transition in the methylation position of cytosine from m5C to m4C is also associated with possible epigenetic regulation is not known. Recent study
In the present study, we address the possible association of m4C with gene expression regulation in H. pylori 26695 strain. The recognition site of M2.HpyAII is a non-palindromic sequence of 5 bp (5′ TCTTC 3′), which can be found at an average frequency of one in 512 (45/2) base pairs, respectively. The longer fragments of DNA (>1 kb) tend to have many sites for M2. HpyAII. In silico genome analysis of H. pylori strain 26695 reveals that the occurrence of 5′ TCTTC 3′ sites are not random in the intergenic or intragenic region of the genome and the frequency of occurrence is tightly regulated in the genome. It is likely that the distribution of 5′ TCTTC 3′ sites could play a regulatory role. The proposed mechanism of methylation mediated epigenetic gene regulation is DNA methylation at promoter elements which results in altered DNA-protein interaction leading to differential gene expression (2). We observed that coding region of H. pylori 26695 strain over-represents the 5′ TCTTC 3′ sites. However, a recent study has also shown that methylation within the coding region can also affect gene expression (11). In eukaryotes C5-cytosine DNA methylation in the coding region facilitates transcription while it blocks transcription of CpG islands in the promoters (33).
An indirect phenomenon of gene regulation has been observed for phase variable type IIG MTase in Campylobacter jejuni where there is no correlation between the differentially expressed genes and the distribution of recognition sites (34). In the present study RNA-seq analysis of the strain lacking the m4C modification revealed that 102 genes were differentially expressed. Seqeunce analysis revealed that out of 102 genes, 56 genes were positive for 5′ TCTTC 3′ sites and total 173 sites are present in the region -200 bp upstream of the start codon till the end of the other gene. How the loss of m4C methylation affected expression of other 46 differentially expressed genes which are negative for M2.HpyAII recognition site is not known. It could be possible that the differential expression of ribosome assembly components had an indirect role in the regulation of these 46 genes. We also observe that number of expression of housing keeping genes is altered upon the loss of m4C modification. This might suggets indirect effects on cell physiology which in turn impair viruelnce. Finally, expression of mfd homologue (hp1541) which is involved in antibiotics sensitivity and recombination (35) was upregulated along with HcpA and HcpD genes in the R.hpyAII-M2.hpyAII deletion strain. We hypothesize that m4C methylation could also indirectly regulate gene expression as seen in Dam methylase mutants of E. coli and Salmonella (2,36,37), where Dam mutants overexpress SOS pathway genes leading to altered gene expression. Similary in Salmonella enterica, Dam mediated DNA methylation indirectly control expression of key virulence factors through transcriptional control of other unrelated genes (37). Dam mediated phenotype changes are proposed to be indirect, direct, secondary or due to indirect regulation of transcriptional machinery (36). In Salmonella regulation of finP gene occurs due to altered nucleoid topology due to loss of m6A modification at GATC sites (38). DNA methylation can alter DNA curvature and thus lower the thermodynamical stability of the double helix (39). We propose that M2.HpyAII controls gene expression in a similar way and loss of TCTTC site methylation might result in altered nucleoid toplogy with concomitant effects in gene expression. Altogether we observe that m4C modification in H. pylori genome acts as a epigenetic regulator of gene expression.
Previous studies have shown that DNA adenine methylation controls cell cycle progression and regulation of DNA replication initiation in Gamma-proteobacteria (40,41). Here we report that m4C modification at TCTTC motif has no effect in 26695 strain growth under normal or stress condition (acidic medium). This is in contrast with the pleiotropic effects as seen in dam metylase mutants where loss of GATC site methylation lead to asynchronic cell division and virulence defects (41). Similarly, loss of CcrM methylase in Caulobacter was found to be essential in rich medium (42).
Natural transformation in H. pylori plays a major role in genomic diversity, evolutionary fitness and virulence regulation (43,44) and studies have shown that R-M systems can regulate DNA uptake in the organism (45–48). It was observed that natural transformation capacity was reduced when m4C modification was erased from the genome. Uptake of both point-mutated DNA and larger DNA fragment was reduced upon loss of m4C modification. Our results also confirm the previous observation that type II endonucleases do not play a role in uptake of point-mutated DNA but regulate uptake of larger DNA fragement in H. pylori (48). We propose that the presence of m4C can regulate the natural transformation pathway through epigenetic control of gene expression in H. pylori. An alternative explanation for the reduction of natural transformation in R.hpyAII-M2.hpyAII deletion could be the several fold preference of M2.HpyAII for ssDNA over dsDNA substrate (unpublished data, Kumar et al.). It was reported that the incoming DNA enters the cell in double-stranded form and gets converted to single-stranded form in the cytoplasm (49). This conversion along with the methylation of ssDNA by M2.HpyAII might play a crucial role in deciding the success of natural transformation process.
IL-8 cytokine is produced by epithelial cells during inflammatory response and is a hallmark of H. pylori pathogenesis (50). It was observed that deletion of M2.hpyAII in H. pylori affected IL-8 induction in the host cells, suggesting the association of m4C with H. pylori-induced inflammation. It is known that H. pylori induces apoptosis of epithelial cells by inhibiting the cell cycle progression at G1 to S phase and altering the host gene expression (51). Significant reduction of AGS cell apotosis by R.hpyAII-M2.hpyAII deletion strain indicates the role of m4C modification in the bacterial genome. LPS is one of the central component of H. pylori pathogenesis. However, a notable phenotypic variation of LPS occurs in unrelated clinical strains (52). It is known that decreased expression of O-antigen is associated with low adherence of H. pylori in the mouse model (53). There is evidence that H. pylori can reversibly regulate the synthesis of O-antigen during colonization, but the mechanisms remain unclear (27). Here, we report that the level of O-antigen can be regulated epigenetically by m4C modification. It was also observed that R.hpyAII-M2.hpyAII deletion strain exhibited 50% less adherence to AGS cells compared to wild-type strain. This is the first study suggesting the role of m4C in the LPS assembly and in H. pylori adherence to the host cells.
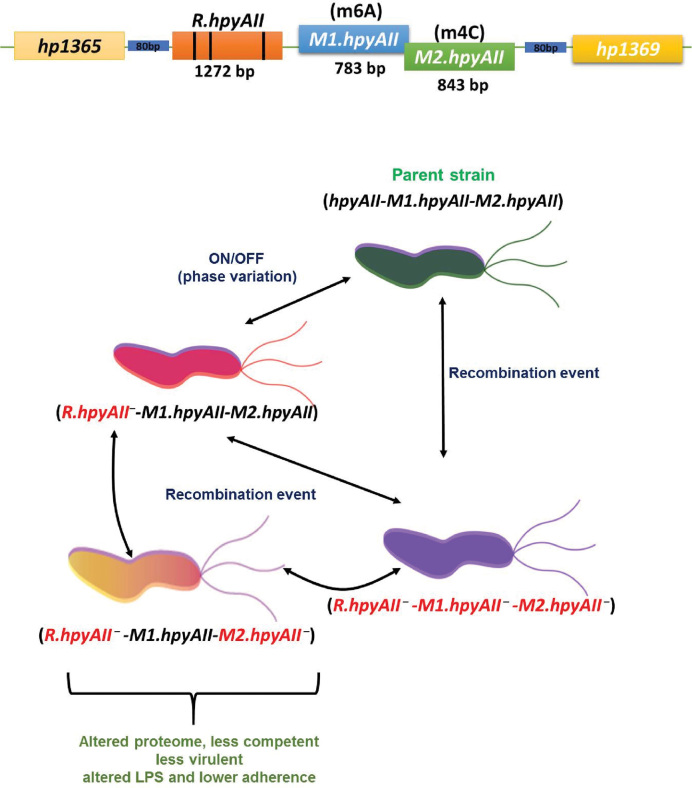
Proteomics analysis revealed more information about the differentially expressed genes in R.hpyAII-M2.hpyAII deletion strain. Identification of differentially expressed proteins spots revealed that the expression of many virulence factors were affected due to the loss of m4C modification. The low expression of virulence factors such as CagA, GroES, CagZ, catalase (KatA) and superoxide dismutase (SOD) in the R.hpyAII-M2.hpyAII gene deletion strain highlights the epigenetic regulation mediated by m4C modification. It was observed that factors such as thiol peroxidase, ferritin non-heme binding protein, Ni/Fe hydrogenase and HslV which were previously shown to be essential for colonization (54–57) were found to be downregulated. The absence of multiple virulence factors which directly affects the colonization, H. pylori-induced inflammation and pathogenesis further supports the less virulent phenotype of R.hpyAII-M2.hpyAII gene deletion strain. Previously it was reported that the deletion of N6-adenine M.HpyAIV methyltransferase in H. pylori strain 26695 resulted in down-regulation of katA gene suggesting the epigenetic regulation. In this study we provide the first evidence that H. pylori gene expression and pathogenesis can be regulated through the modification of cytosine at N4-position. Earlier studies had shown that HpyAII R-M system undergoes spontaneous deletion and horizontal re-acquisition in H. pylori genome (45,58) leading to a generation of a mixed population. However, the functional significance of this novel observation is not known. Our study explains the phenotypic effect of loss of m4C modification which could result from spontateneous deletion of HpyAII R-M system. We propose that, in a given niche of H. pylori, phenotypically diverse populations can occur as a consequence of harbouring different variants of HpyAII R-M system.The spontaneous event leading to deletion or restoration of the entire R-M system will result in the sub-population of the bacteria which lack the GAAGA/TCTTC methylation signature from the genome. The subsequent loss of m4C modification in the sub-population will attenuate natural transformation and virulence, by epigenetic gene regulation (Figure 8). Considering the harsh environment which persists in the niche of H. pylori, it is but apparent that the organism would constantly adapt to acquire a fitter or more robust phenotype. This persistent struggle for existence is essential for the H. pylori to survive in its only host, evade the host immune system and fine tune its pathogenicity. The m4C modification plays a novel epigenetic role in enabling the above mentioned fundamental necessities for the long-term survival of the bacteria in the host. Our present work suggests that H. pylori have evolved and acquired a novel way of gene regulation by exploiting the extensive m4C modification in the genome. This study opens up a new avenue for an alternate way of gene regulation by m4C modification in bacteria.
Figure 8.
Proposed functional role of N4-cytosine methylation in the pathogenesis and gene regulation of H. pylori. Diagrammatic illustration explaining the differential occurrence of phenotypically diverse population of H. pylori based on HpyAII R-M system variations. Representation of HpyAII R-M system, phase variable region of R.hpyAII gene (poly-adenine residues) are shown with solid black vertical lines.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Utpal Tatu and his lab members for their help in 2D gel electrophoresis. Members of D.N.R. laboratory are acknowledged for their support and valuable suggestions for the study. We thank Dr Ritesh Kumar, Dr Prashant Damke, Dr Gajendra Dwiwedi, Semanti Ray, Yedu Prasad and Sushant Bangru for crucial discussions and critical reading of the manuscript. Dr Avijit Sarkar and Dr Raghavan are acknowledged for their support during the study in NICED. We thank the FACS facility at IISc and NICED. D.N.R. acknowledges DST for J.C. Bose Fellowship. We thank Dr Pablo Radicella for providing plasmid P1175 and for critical suggestions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Biotechnology-Indian Institute of Science (DBT-IISc) Partnership program and the Indo-French Centre for Promotion of Advanced Research (CEFIPRA) [5203-5]; IISc Integrated-Ph.D. (to S.K.); Council of Scientific and Industrial Research (CSIR) [Ref. No. 37(1640)/14/EMR-II in part]; Indian Council of Medical Research, New Delhi. Funding for open access charge: J.C. Bose Fellowship, Department of Science and Technology, India.
Conflict of interest statement. None declared.
REFERENCES
- 1. Vasu K., Nagaraja V.. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev.: MMBR. 2013; 77:53–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sanchez-Romero M.A., Cota I., Casadesus J.. DNA methylation in bacteria: from the methyl group to the methylome. Curr. Opin. Microbiol. 2015; 25:9–16. [DOI] [PubMed] [Google Scholar]
- 3. Kim J.K., Samaranayake M., Pradhan S.. Epigenetic mechanisms in mammals. Cell. Mol. Life Sci.: CMLS. 2009; 66:596–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vanyushin B.F., Ashapkin V.V.. DNA methylation in higher plants: past, present and future. Biochim. Biophys. Acta. 2011; 1809:360–368. [DOI] [PubMed] [Google Scholar]
- 5. Backert S., Neddermann M., Maubach G., Naumann M.. Pathogenesis of Helicobacter pylori infection. Helicobacter. 2016; 21(Suppl. 1):19–25. [DOI] [PubMed] [Google Scholar]
- 6. Suerbaum S., Smith J.M., Bapumia K., Morelli G., Smith N.H., Kunstmann E., Dyrek I., Achtman M.. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:12619–12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Furuta Y., Namba-Fukuyo H., Shibata T.F., Nishiyama T., Shigenobu S., Suzuki Y., Sugano S., Hasebe M., Kobayashi I.. Methylome diversification through changes in DNA methyltransferase sequence specificity. PLoS Genet. 2014; 10:e1004272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krebes J., Morgan R.D., Bunk B., Sproer C., Luong K., Parusel R., Anton B.P., Konig C., Josenhans C., Overmann J. et al. The complex methylome of the human gastric pathogen Helicobacter pylori. Nucleic Acids Res. 2014; 42:2415–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin L.F., Posfai J., Roberts R.J., Kong H.. Comparative genomics of the restriction-modification systems in Helicobacter pylori. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:2740–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kumar R., Mukhopadhyay A.K., Ghosh P., Rao D.N.. Comparative transcriptomics of H. pylori strains AM5, SS1 and their hpyAVIBM deletion mutants: possible roles of cytosine methylation. PLoS One. 2012; 7:e42303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kahramanoglou C., Prieto A.I., Khedkar S., Haase B., Gupta A., Benes V., Fraser G.M., Luscombe N.M., Seshasayee A.S.. Genomics of DNA cytosine methylation in Escherichia coli reveals its role in stationary phase transcription. Nat. Commun. 2012; 3:886. [DOI] [PubMed] [Google Scholar]
- 12. Chung D., Farkas J., Huddleston J.R., Olivar E., Westpheling J.. Methylation by a unique alpha-class N4-cytosine methyltransferase is required for DNA transformation of Caldicellulosiruptor bescii DSM6725. PLoS One. 2012; 7:e43844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morgan R.D., Luyten Y.A., Johnson S.A., Clough E.M., Clark T.A., Roberts R.J.. Novel m4C modification in type I restriction-modification systems. Nucleic Acids Res. 2016; 44:9413–9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tomb J.F., White O., Kerlavage A.R., Clayton R.A., Sutton G.G., Fleischmann R.D., Ketchum K.A., Klenk H.P., Gill S., Dougherty B.A. et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997; 388:539–547. [DOI] [PubMed] [Google Scholar]
- 15. Sharma C.M., Hoffmann S., Darfeuille F., Reignier J., Findeiss S., Sittka A., Chabas S., Reiche K., Hackermuller J., Reinhardt R. et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010; 464:250–255. [DOI] [PubMed] [Google Scholar]
- 16. Sharma A., Kamran M., Verma V., Dasgupta S., Dhar S.K.. Intracellular locations of replication proteins and the origin of replication during chromosome duplication in the slowly growing human pathogen Helicobacter pylori. J. Bacteriol. 2014; 196:999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zeng X., He L.H., Yin Y., Zhang M.J., Zhang J.Z.. Deletion of cagA gene of Helicobacter pylori by PCR products. World J. Gastroenterol.: WJG. 2005; 11:3255–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Clark T.A., Murray I.A., Morgan R.D., Kislyuk A.O., Spittle K.E., Boitano M., Fomenkov A., Roberts R.J., Korlach J.. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 2012; 40:e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Orillard E., Radicella J.P., Marsin S.. Biochemical and cellular characterization of Helicobacter pylori RecA, a protein with high-level constitutive expression. J. Bacteriol. 2011; 193:6490–6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuck D., Kolmerer B., Iking-Konert C., Krammer P.H., Stremmel W., Rudi J.. Vacuolating cytotoxin of Helicobacter pylori induces apoptosis in the human gastric epithelial cell line AGS. Infect. Immun. 2001; 69:5080–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Preston M.A., Penner J.L.. Structural and antigenic properties of lipopolysaccharides from serotype reference strains of Campylobacter jejuni. Infect. Immun. 1987; 55:1806–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Skindersoe M.E., Rasmussen L., Andersen L.P., Krogfelt K.A.. A Novel Assay for Easy and Rapid Quantification of Helicobacter pylori Adhesion. Helicobacter. 2015; 20:199–205. [DOI] [PubMed] [Google Scholar]
- 23. Overmars L., van Hijum S.A., Siezen R.J., Francke C.. CiVi: circular genome visualization with unique features to analyze sequence elements. Bioinformatics. 2015; 31:2867–2869. [DOI] [PubMed] [Google Scholar]
- 24. Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M.. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006; 1:2856–2860. [DOI] [PubMed] [Google Scholar]
- 25. Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001; 29:3742–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Edwards N.J., Monteiro M.A., Faller G., Walsh E.J., Moran A.P., Roberts I.S., High N.J.. Lewis X structures in the O antigen side-chain promote adhesion of Helicobacter pylori to the gastric epithelium. Mol. Microbiol. 2000; 35:1530–1539. [DOI] [PubMed] [Google Scholar]
- 27. Li H., Liao T., Debowski A.W., Tang H., Nilsson H.O., Stubbs K.A., Marshall B.J., Benghezal M.. Lipopolysaccharide structure and biosynthesis in Helicobacter pylori. Helicobacter. 2016; 21:445–461. [DOI] [PubMed] [Google Scholar]
- 28. Militello K.T., Simon R.D., Qureshi M., Maines R., VanHorne M.L., Hennick S.M., Jayakar S.K., Pounder S.. Conservation of Dcm-mediated cytosine DNA methylation in Escherichia coli. FEMS Microbiol. Lett. 2012; 328:78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vogel C., Marcotte E.M.. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012; 13:227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Skoglund A., Bjorkholm B., Nilsson C., Andersson A.F., Jernberg C., Schirwitz K., Enroth C., Krabbe M., Engstrand L.. Functional analysis of the M.HpyAIV DNA methyltransferase of Helicobacter pylori. J. Bacteriol. 2007; 189:8914–8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Srikhanta Y.N., Gorrell R.J., Steen J.A., Gawthorne J.A., Kwok T., Grimmond S.M., Robins-Browne R.M., Jennings M.P.. Phasevarion mediated epigenetic gene regulation in Helicobacter pylori. PLoS One. 2011; 6:e27569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ehrlich M., Gama-Sosa M.A., Carreira L.H., Ljungdahl L.G., Kuo K.C., Gehrke C.W.. DNA methylation in thermophilic bacteria: N4-methylcytosine, 5-methylcytosine, and N6-methyladenine. Nucleic acids research. 1985; 13:1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rishi V., Bhattacharya P., Chatterjee R., Rozenberg J., Zhao J., Glass K., Fitzgerald P., Vinson C.. CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:20311–20316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anjum A., Brathwaite K.J., Aidley J., Connerton P.L., Cummings N.J., Parkhill J., Connerton I., Bayliss C.D.. Phase variation of a Type IIG restriction-modification enzyme alters site-specific methylation patterns and gene expression in Campylobacter jejuni strain NCTC11168. Nucleic Acids Res. 2016; 44:4581–4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dorer M.S., Sessler T.H., Salama N.R.. Recombination and DNA repair in Helicobacter pylori. Annu. Rev. Microbiol. 2011; 65:329–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heusipp G., Falker S., Schmidt M.A.. DNA adenine methylation and bacterial pathogenesis. Int. J. Med. Microbiol.: IJMM. 2007; 297:1–7. [DOI] [PubMed] [Google Scholar]
- 37. Marinus M.G., Casadesus J.. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 2009; 33:488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Camacho E.M., Serna A., Madrid C., Marques S., Fernandez R., de la Cruz F., Juarez A., Casadesus J.. Regulation of finP transcription by DNA adenine methylation in the virulence plasmid of Salmonella enterica. J. Bacteriol. 2005; 187:5691–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Diekmann S. DNA methylation can enhance or induce DNA curvature. EMBO J. 1987; 6:4213–4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reisenauer A., Kahng L.S., McCollum S., Shapiro L.. Bacterial DNA methylation: a cell cycle regulator. J. Bacteriol. 1999; 181:5135–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wion D., Casadesus J.. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 2006; 4:183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gonzalez D., Collier J.. DNA methylation by CcrM activates the transcription of two genes required for the division of Caulobacter crescentus. Mol. Microbiol. 2013; 88:203–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baltrus D.A., Guillemin K., Phillips P.C.. Natural transformation increases the rate of adaptation in the human pathogen helicobacter pylori. Evolution. 2008; 62:39–49. [DOI] [PubMed] [Google Scholar]
- 44. Dorer M.S., Cohen I.E., Sessler T.H., Fero J., Salama N.R.. Natural competence promotes Helicobacter pylori chronic infection. Infect. Immun. 2013; 81:209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aras R.A., Small A.J., Ando T., Blaser M.J.. Helicobacter pylori interstrain restriction-modification diversity prevents genome subversion by chromosomal DNA from competing strains. Nucleic Acids Res. 2002; 30:5391–5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Humbert O., Dorer M.S., Salama N.R.. Characterization of Helicobacter pylori factors that control transformation frequency and integration length during inter-strain DNA recombination. Mol. Microbiol. 2011; 79:387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Humbert O., Salama N.R.. The Helicobacter pylori HpyAXII restriction-modification system limits exogenous DNA uptake by targeting GTAC sites but shows asymmetric conservation of the DNA methyltransferase and restriction endonuclease components. Nucleic Acids Res. 2008; 36:6893–6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang X.S., Blaser M.J.. Natural transformation of an engineered Helicobacter pylori strain deficient in type II restriction endonucleases. J. Bacteriol. 2012; 194:3407–3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stingl K., Muller S., Scheidgen-Kleyboldt G., Clausen M., Maier B.. Composite system mediates two-step DNA uptake into Helicobacter pylori. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:1184–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee K.E., Khoi P.N., Xia Y., Park J.S., Joo Y.E., Kim K.K., Choi S.Y., Jung Y.D.. Helicobacter pylori and interleukin-8 in gastric cancer. World J. Gastroenterol.: WJG. 2013; 19:8192–8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shirin H., Sordillo E.M., Oh S.H., Yamamoto H., Delohery T., Weinstein I.B., Moss S.F.. Helicobacter pylori inhibits the G1 to S transition in AGS gastric epithelial cells. Cancer Res. 1999; 59:2277–2281. [PubMed] [Google Scholar]
- 52. Gibson J.R., Chart H., Owen R.J.. Intra-strain variation in expression of lipopolysaccharide by Helicobacter pylori. Lett. Appl. Microbiol. 1998; 26:399–403. [DOI] [PubMed] [Google Scholar]
- 53. Moran A.P., Sturegard E., Sjunnesson H., Wadstrom T., Hynes S.O.. The relationship between O-chain expression and colonisation ability of Helicobacter pylori in a mouse model. FEMS Immunol. Med. Microbiol. 2000; 29:263–270. [DOI] [PubMed] [Google Scholar]
- 54. Wang G., Olczak A.A., Walton J.P., Maier R.J.. Contribution of the Helicobacter pylori thiol peroxidase bacterioferritin comigratory protein to oxidative stress resistance and host colonization. Infect. Immun. 2005; 73:378–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Waidner B., Greiner S., Odenbreit S., Kavermann H., Velayudhan J., Stahler F., Guhl J., Bisse E., van Vliet A.H.M., Andrews S.C. et al. Essential Role of Ferritin Pfr in Helicobacter pylori Iron Metabolism and Gastric Colonization. Infect. Immun. 2002; 70:3923–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kusters J.G., van Vliet A.H., Kuipers E.J.. Pathogenesis of Helicobacter pylori infection. Clin. Microbiol. Rev. 2006; 19:449–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Du R.J., Ho B.. Surface localized Heat Shock Protein 20 (HslV) of Helicobacter pylori. Helicobacter. 2003; 8:257–267. [DOI] [PubMed] [Google Scholar]
- 58. Aras R.A., Takata T., Ando T., van der Ende A., Blaser M.J.. Regulation of the HpyII restriction-modification system of Helicobacter pylori by gene deletion and horizontal reconstitution. Mol. Microbiol. 2001; 42:369–382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.