Abstract
Whether the antimutagenic DNA repair protein MGMT works solo in human cells and if it has other cellular functions is not known. Here, we show that human MGMT associates with PCNA and in turn, with the cell cycle inhibitor, p21cip1 in glioblastoma and other cancer cell lines. MGMT protein was shown to harbor a nearly perfect PCNA-Interacting Protein (PIP box) motif. Isogenic p53-null H1299 cells were engineered to express the p21 protein by two different procedures. Reciprocal immunoprecipitation/western blotting, Far-western blotting, and confocal microscopy confirmed the specific association of MGMT with PCNA and the ability of p21 to strongly disrupt the MGMT-PCNA complexes in tumor cells. Alkylation DNA damage resulted in a greater colocalization of MGMT and PCNA proteins, particularly in HCT116 cells deficient in p21 expression. p21 expression in isogenic cell lines directly correlated with markedly higher levels of MGMT mRNA, protein, activity and greater resistance to alkylating agents. In other experiments, four glioblastoma cell lines synchronized at the G1/S phase using either double thymidine or thymidine-mimosine blocks and subsequent cycling consistently showed a loss of MGMT protein at mid- to late S-phase, irrespective of the cell line, suggesting such a downregulation is fundamental to cell cycle control. MGMT protein was also specifically degraded in extracts from S-phase cells and evidence strongly suggested the involvement of PCNA-dependent CRL4Cdt2 ubiquitin-ligase in the reaction. Overall, these data provide the first evidence for non-repair functions of MGMT in cell cycle and highlight the involvement of PCNA in MGMT downregulation, with p21 attenuating the process.
Abbreviations: MGMT, O6-methylguanine DNA methyltransferase; PCNA, proliferating nuclear antigen; His Tag, Hexahistidine tag; DAPI, 4,6-Diamidino-2-phenylindole; rMGMT, recombinant MGMT protein; tet, tetracycline; IB, immunoblot; WB, western blot; LV, lenti-viral transfection; AS, asynchronous cells; CS0, G1/S synchronized cells before release for cycling; CS2, CS4 and CS8, synchronized cells 2, 4 and 8 hours after release into cycling; TMZ, temozolomide; BCNU, 1,3-bis (2-chloroethyl)-1-nitrosourea; Conc, concentration; CRL4Cdt2, PCNA-dependent ubiquitin ligase; Ub, ubiquitin; IP, immunoprecipitation; PIP-box, PCNA interacting protein motif.
Introduction
O6-Methylguanine DNA methyltransferase (MGMT) is a DNA repair protein that stoichiometrically removes the mutagenic alkyl adducts introduced at the O6-position of guanine and O4-of thymine by various exogenous and endogenous agents [1]. This unique and conserved direct reversal repair prevents the GC to AT transitions and maintains the stability of human genome [2]. Unlike the other DNA repair pathways, human MGMT works as a single protein to remove the guanine O6-bound alkyl groups to cysteine145 at its active site [3]. Because the alkyl groups are covalently linked to Cys145, MGMT is self-inactivated after each reaction and the inactive protein is degraded through the ubiquitin/proteasome pathway [[4], [5]]. The alkylation DNA damage introduced by the clinically used anticancer alkylating agents is also targeted and scavenged by MGMT. These drugs bearing a single (temozolomide) or two (chloroethylnitrosoureas) electrophilic groups generate mutagenic lesions and cytotoxic G-C interstrand crosslinks respectively to exert the antitumor effects. Paradoxically, however, the human cancers express higher levels of MGMT, and the resulting repair of alkyl lesions confers a high level of tumor drug resistance and failure of chemotherapy [6]. Diverse groups of MGMT inhibitors have emerged, and O6-benzylguanine, a pseudo-substrate inhibitor has been extensively studied [[7], [8]]. Therefore, understanding the molecular aspects of MGMT function is highly important from several angles ranging from the mechanism of DNA repair, carcinogenesis, and cancer therapy.
The stoichiometric reaction mechanism of MGMT raises many puzzling questions such as why human cells synthesize a protein for a single repair event at a significant cost, why does it undergo several posttranslational modifications [[9], [10], [11]], whether MGMT serves other functions in the cell, and whether MGMT accomplishes the de-alkylation all by itself in vivo or does it associate with accessory /replication proteins? Although answers to these are unknown, evidence from our laboratory and others suggests that MGMT specifically interacts with many cellular proteins and may have other functions [[12], [13]]. For example, using MGMT-Sepharose affinity chromatography and tandem mass spectrometry, we showed the specific interaction of MGMT with a diverse group of proteins involved in DNA replication (ORC1, MCM helicases, PCNA, DNA pol δ) and cell cycle progression (CDKs, p21cip1, and ubiquitin pathway components) [12]. A similar study by another group confirmed the specific binding of MGMT with various regulatory proteins in human glioma cells [13]. Recently, we showed a specific association and fine interplay between human MGMT and estrogen receptor-α proteins and their co-degradation after tumor cell treatments with either O6-benzylguanine or fulvestrant, their respective inhibitors [14]. A previous study implied that the alkylated (inactivated) human MGMT is a negative regulator of ER-mediated transcription following DNA alkylation damage [15].
Previously, we reported briefly on MGMT binding with PCNA [16] and the presence of MGMT in p21cip1-PCNA complexes [17]. Human PCNA is a homotrimeric protein that encircles the duplex DNA forming a ring-shaped clamp and functions as a processivity factor by tethering replicative DNA polymerases [18]. PCNA also provides a molecular platform that facilitates the myriad protein-protein and protein–DNA interactions that occur at the replication fork. Numerous PCNA-associated proteins such as the FEN1 nuclease, DNA cytosine methyltransferase, and topoisomerase II compete for binding to a common surface on PCNA [19]. Many of these partners contain a highly conserved PCNA-binding motif, QXXhXXaa (where ‘h’ is a hydrophobic, ‘aa’ are aromatic and X is any amino acid), referred to as a PCNA interacting protein (PIP) box [[18], [19]]. Besides being an essential part of DNA replication machinery, PCNA also plays important roles in cell cycle regulation (primarily in S-phase), translesion synthesis, long-patch base excision repair and recombination [20].
All DNA repair pathways including nucleotide excision repair (XP-A, XP-G), mismatch repair (MSH2, MSH6), and PARP-1 are associated with PCNA in some way or another [20]. While PCNA exists in free and chromatin-bound states, the abundance of cellular PCNA is strictly controlled by the cyclin-dependent kinase (CDK) inhibitor p21CDKN1A, which is an avid binder and sequestrator of the former [21]. p21cip1, which is activated by the p53 tumor suppressor, plays essential roles in the DNA damage response by inducing cell cycle arrest, direct inhibition of DNA replication and apoptotic regulation. p21 interferes with PCNA-dependent DNA polymerase activity, thereby inhibiting DNA replication, however, DNA repair processes dependent on PCNA appear to be largely unaffected [[22], [23]]. Additionally, PCNA interfaces with the cell cycle by forming PCNA-p21/CDK-cyclin quaternary complexes with both positive and negative signaling roles in cell cycle progression. Furthermore, PCNA is an integral component of the regulated and timely destruction of proteins during the S-phase and facilitates the formation of pre-replication complexes for the next cell cycle [[24], [25]]. In this process, PCNA assists with the recognition of target proteins by CRL4Cdt2 ub-ligase [[26], [27]]. No information is available on the involvement of either PCNA or other cell cycle proteins in the regulation of the one-step direct reversal reaction performed by MGMT. Here we describe a detailed analysis of MGMT interaction with PCNA and p21cip1 in isogenic cancer cell lines and demonstrate a PCNA-dependent downregulation of MGMT protein in S-phase.
Materials and Methods
Cell Culture and Reagents
All cell lines were obtained within 6 months of time and were authenticated by the original sources and investigators. The human glioblastoma cell lines, SF188 (Univ. of California Brain Tumor Bank, San Francisco, CA), GBM10 (Dr. Jann Sarkaria, Mayo Clinic, Rochester, MN), and T98G (ATCC). Other cell lines, HT29 colon cancer, H1299 lung cancer All cell lines were from ATCC. The human medulloblastoma cell line UW228 was provided by Dr. Francis Ali-Osman (Duke University, Durham, NC). All tumor cells were grown in Dulbecco's Modified Eagle's Medium with 10% fetal bovine serum and antibiotics at 37°C in 95% humidified air, and 5% CO2. Thymidine, mimosine and all other chemicals were obtained from Sigma-Aldrich. Monoclonal antibodies to MGMT were purchased from EMD Millipore. Other antibodies were purchased as follows: actin, PCNA, p21 (Cell Signaling), Ubiquitin (Santa Cruz Biotechnology).
Isogenic Cell Lines
Three different isogenic cell line pairs, with one member in each group differing in p21 expression level, were used in this study. The first model was a p21-inducible cell line generated using the tetracycline (Tet)-regulated vector system (H1299-p21) that repressed p21 expression in the presence of Tet [28]. The plasmid constructs for this conditional gene expression were from Clonetech Laboratories (Mountain View, CA). Full-length cDNA for human p21cip1 was cloned into the HindIII-SpeI sites of the pTET-SPLICE plasmid. The p53-null H1299 cells were first transfected with the ‘regulatory plasmid’, pUDH15-1 and the clones selected in the presence of 450 μg/ml of G418. These resulting clones were next transfected with the pTET-p21 ‘response plasmid’ and selected against zeocin. The resistant clones were screened for p21 expression and maintained in a medium containing Tet (1 μg/ml). The second isogenic cell line was developed by stable transfection of H1299 cells with p21 lentiviral particles (H1299-LV/p21) obtained from the Vigene Biosciences (Rockville, MD). Blasticidin was used for selecting the transfected cells and clones were created. The third cell line model was the HCT116 colon cancer cells proficient or deficient in p21 expression. HCT116 p21++ and HCT116 p21—cells [29] were kindly provided by Dr. Bert Vogelstein (Johns Hopkins University, Baltimore, MD).
Immunoprecipitation and Western Blotting
For IP, whole cell lysates were prepared in 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% glycerol, 1 mM EDTA, 1 X protease inhibitors, 0.5% NP-40 (IP buffer). The extracts were pre-cleaned with 10 μl of protein A/G -Sepharose beads. Antibodies or normal IgG (isotype control) were added at 1 μg and the samples mixed overnight at 4oC. Protein A/G-Sepharose beads were (20 μl) added, and samples incubated for another 2-3 h in a rotator. The immune complexes were pelleted by centrifugation, washed three times with IP buffer and re-suspended in reducing sample buffers. The samples were boiled for 2 min before loading on 10% SDS-polyacrylamide gels. For direct western blots, the cell pellets were sonicated in Pellets were then sonicated in a lysis buffer containing 50mM Tris-HCl (pH 8.0),1% glycerol, 0.5 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 2 mM benzamidine and 20 μM spermidine. Proteins were then electrophoretically transferred to PVDF membranes, blocked with 5% non-fat dry milk in TBS containing 0.1% Tween 20 for 2 h. The membranes were then incubated with appropriate primary antibodies followed by washing and exposure to Peroxidase-labeled secondary antibodies. The signals on the western blots were visualized by enhanced chemiluminescence and the band densities quantitated using a Bio-Rad Versadoc imager.
Expression and Purification of Recombinant MGMT Protein (rMGMT)
Hexa-histidine tagged human MGMT was expressed in E. coli (JM109 strain) and the native protein was purified using Talon affinity-chromatography as described by us previously [9].
Pull-Down Assays
For pull-down assays, the rMGMT or bovine serum albumin (as control) (2 μg) was first bound with the Talon metal-affinity resin (~15 μl) in a binding buffer (25 mM sodium phosphate pH 8.3, 250 mM NaCl, 1 mM PMSF, 2 mM benzamidine, 5% glycerol, 0.2 mM EDTA, 0.1% Nonidet P-40), and washed with the same buffer. Cell lysates were prepared in the above binding buffer + 0.2 mM mercaptoethanol. Next, the clarified cell lysates were added to the his-rMGMT–bound Talon resin and mixed on a rotator for 3 h. After pelleting the resin and washing three times, the proteins bound to the resin were eluted with 10 mM imidazole, and the supernatants were mixed with SDS-sample buffer followed by western blotting using the anti-His tag or anti-PCNA antibodies.
MGMT DNA Repair Activity Assay
Transfer of 3H-labeled methyl groups from the O6-position of guanine in the DNA to the MGMT protein was measured by an established procedure in our laboratory [9].
RNA Isolation and Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Total RNA from HCT116 (p21++, p21--) cells was isolated using the TRIzol Reagent (Invitrogen). Equal amounts (0.1 μg) of RNA were taken reverse-transcribed using Qiagen One-Step RT-PCR kit. The primers used were: MGMT, forward 5’ TGGAGCTGTCTGGTTGTGAG 3’ and reverse 5’ TGGAAAACATGCCGTTATCA 3’; GAPDH, forward 5’ GAAGGTGAAGGTCGGAGTC 3’ and reverse 5’ GAAGATGGTGATGGGATTTC 3’. The PCR products were analyzed by electrophoresis on 1% agarose gels and visualized by ethidium bromide staining.
Measurement of Interstrand DNA Crosslinking in Tumor Cells
BCNU-induced DNA crosslinking was quantitated by ethidium bromide fluorescence assay as described previously [30]. Briefly, cells were exposed to BCNU (100 μM) for 1 hour and then incubated in drug-free media for up to 48 hours to allow DNA crosslinking and its disappearance. DNA was isolated after SDS lysis, RNase and proteinase K (1 mg/ml) treatments followed by ethanol precipitation. To Equal amounts (2 μg) of DNA from control and drug-treated cells, ethidium bromide (1μg/ml) was added in 20 mM potassium phosphate and 2 mM EDTA, (pH 11.8) in the buffer. One set of DNA samples were boiled for 5 min and cooled to room temperature. Fluorescence of the native and denatured DNA was measured (305 nm excitation and 585 nm emission) using a Perkin Elmer LS-50 spectrofluorometer. Crosslink index (CLI) was calculated from the fluorescence readings by following a published method [30].
MTT Cytotoxicity Assays
For in vitro cytotoxicity assays, cells were seeded and cultured overnight (7000 per well in 96-well microtiter plates) followed by drug treatments. Then 10 μl of MTT [1-(4,5-Dimethylthiazol-2-yl)-3,5-diphenylformazan;5 mg/ml] solution was added to each well and incubation continued for 3 hours to allow the formation of insoluble formazan. Supernatants were removed, dimethyl sulfoxide was used to solubilize the formazan crystals and colorimetric absorbance was measured using a plate reader at 570 nm. GraphPad Prism 7 software was used for cytotoxicity analysis.
Cell Cycle Synchronization and Flow Cytometry
Three different cell synchronization methods were used, namely double thymidine block (G1/S phase arrest), thymidine-mimosine block (G1 phase arrest) and thymidine-nocodazole block (G2/M phase arrest) using previously described procedures [31]. For double thymidine block, cells were first treated with 2 mM thymidine in culture medium for 18 h followed by a release period of 10 h in thymidine-free media. Cells were then treated with 2 mM thymidine for another 17 h to enrich the G1/S phase cells. The arrested cells were then washed with DMEM and allowed to release from the G1 phase in fresh medium. Thymidine treatment in excess induces imbalance in dNTP pools, which results in feedback-inhibition of ribonucleotide reductase. In contrast to thymidine-induced early S-phase arrest, mimosine is a more effective cell synchronization reagent that arrests >90% of cells at a tight G1-S phase border by inhibiting the transition of the pre-replication complex to the preinitiation complex [32]. The thymidine-mimosine block was obtained by the following treatment sequence: thymidine (2 mM, 12-14 h), release (10-12 h), mimosine (400 μM, 12-14 h), and subsequent release in drug-free medium. An orderly treatment with thymidine (24 h) and nocodazole (100 ng/ml, 12 h) was also used to synchronize cells at G2/M phase. For flow cytometry, cells were trypsinized and the suspensions fixed in ethanol: PBS (70:30 v/v). DNA labeling was performed with propidium iodide (50 μg/ml) preceded by incubation with RNase A for 45 min at 37oC. Stained cells were sorted and analyzed using a BD FACSVerse. FlowJo (version 10) was used to analyze the histograms.
Chromatin Fractionation
Chromatin fractionation for analyzing the DNA-bound proteins was performed by a previous method [33]. After synchronizing into different cell cycle phases, cell pellets were collected and washed with PBS. Briefly, cell pellets were lysed with the cytoskeleton buffer (10 mM PIPES pH 7.0, 300 mM sucrose, 100 mM NaCl, 1.5 mM MgCl2, protease inhibitors, 0.5% Triton X-100) for 30 min and a small fraction of this lysate was saved as a whole cell extract. The remaining extract was centrifuged at 2000g and supernatant was collected (soluble fraction). To obtain the chromatin fraction, pellets were again resuspended in 200 mM HCl and incubated at 4oC for 2h. The resulting supernatant (chromatin-bound fraction) was neutralized with Tris-HCl buffer (pH 7.5) before SDS-PAGE.
Far-Western Blotting
Purified recombinant MGMT and PCNA proteins (700 ng) were electrophoresed on 10% SDS-polyacrylamide gels and transferred to nitrocellulose membranes using 150 mM glycine-Tris buffer containing 5% methanol without SDS. To facilitate a native folding of the immobilized proteins, the membranes were sequentially treated with guanidine hydrochloride (6 M, 3 M, 1 M, 0.1 M and 0 M in a buffer [100 mM NaCl, 20 mM Tris (pH 7.6), 0.5 mM EDTA, 10% glycerol, 0.1% Tween-20, 2% non-fat dry milk and 0.5 mM DTT] as described previously [34]. Next, the washed membranes were incubated with recombinant MGMT protein (100 μg/ml) in the same buffer (without guanidine hydrochloride) for 3 h at room temperature. After washing, the membranes were western blotted using MGMT antibodies.
Immunofluorescence Procedure
Cells were cultured on sterile coverslips and treated with chemotherapy drugs as indicated. After 24 h, they were washed with PBS and fixed in 4% paraformaldehyde. Permeabilization was performed with ice-cold methanol followed by blocking in 5% normal goat serum containing 0.3% Triton X-100 for 2 hours. Incubation with primary (overnight) and fluorochrome-conjugated secondary (1-2 h at room temperature in dark) antibodies were performed in the presence of 1% BSA. Next, cells were washed in PBS and stained with DAPI for 10 min, finally, the coverslips were mounted on glass slides using Prolong Gold antifade reagent (Life Technology/Molecular Probes). Images were visualized using a Nikon A1 MP+ multiphoton confocal microscope. The extent of protein colocalization was quantitated by determining Pearson's correlation coefficient using the ImageJ program.
Assay for MGMT Ubiquitination In Vitro
Cells were lysed in a buffer containing 50 mM Tris-HCl (pH 8.0), 1 mM DTT, a proteasome inhibitor (PS-341/Velcade, 10μM), 4 mM MgCl2, centrifuged for extract preparation. Reaction mixtures containing His-tagged recombinant MGMT (0.5 μg), ubiquitin (0.5 μg) or in some cases, biotinylated ubiquitin (0.25 μg), MgCl2 (2.5 mM), ATP (2 mM) and cell extracts (75 μg protein, as a source of endogenous ubiquitinating enzymes). After incubation at 37oC for 30 min, the samples were boiled and subjected to SDS-PAGE followed by western blotting with antibodies to MGMT or ubiquitin or hexa-histidine or simply, the streptavidin-HRP to detect the biotinylated polyubiquitin residues.
Statistical Analysis
All experiments were performed in triplicates and data presented as mean±SD. Statistical comparisons were done in GraphPad Prism using methods as appropriate (Anova / Student’s t-test). P values are reported using a star system as follows: *, P < 0.05; **, P < 0.005; and ***, P < 0.0005.
Results
Presence of a Near Typical PCNA-Interaction Motif (PIP-box) in Human MGMT Protein
Since our previous study indicated that PCNA may be an interactive partner for human MGMT [16], efforts were made to check for a PCNA binding domain in the MGMT protein sequence. Most PCNA-binding proteins like p21, Cdt1, DNA polymerases-β and δ interact with the interdomain connector loop of PCNA through a motif of 8 amino acids called a PIP box, with the consensus sequence Q-xx-(h)-x-x-(a)-(a), where “h” represents moderately hydrophobic amino acid residues (e.g. leucine, isoleucine, methionine, alanine), “a” represents residues with highly hydrophobic, aromatic side chains (e.g. phenylalanine, tyrosine) and “x” is any amino acid (Figure 1A). In human MGMT protein (GenBank, AAH00824.1), we identified a region from amino acids 61 to 70 containing all essential residues confirming to a PIP box domain (QCTAWLNAYF) (Figure 1B). While this sequence strictly adheres to a canonical PIP-box, it is two residues longer with additional aspargine67 and alanine68 residues flanked by two aromatic amino acids, tyrosine, and phenylalanine respectively. Therefore, this sequence confirms to a consensus but flexible PIP-box structure of Qxxh(xx)naa [[18], [19]]. Further, a bioinformatic Clustal Omega analysis revealed that this PIP motif in MGMT amino acid sequence is evolutionary conserved across the phyla ranging from yeast, Drosophila, amphibians to humans (Figure 1B). MGMT crystal structure shows presence this region in interface helices, well accessible for protein-protein interactions [35].
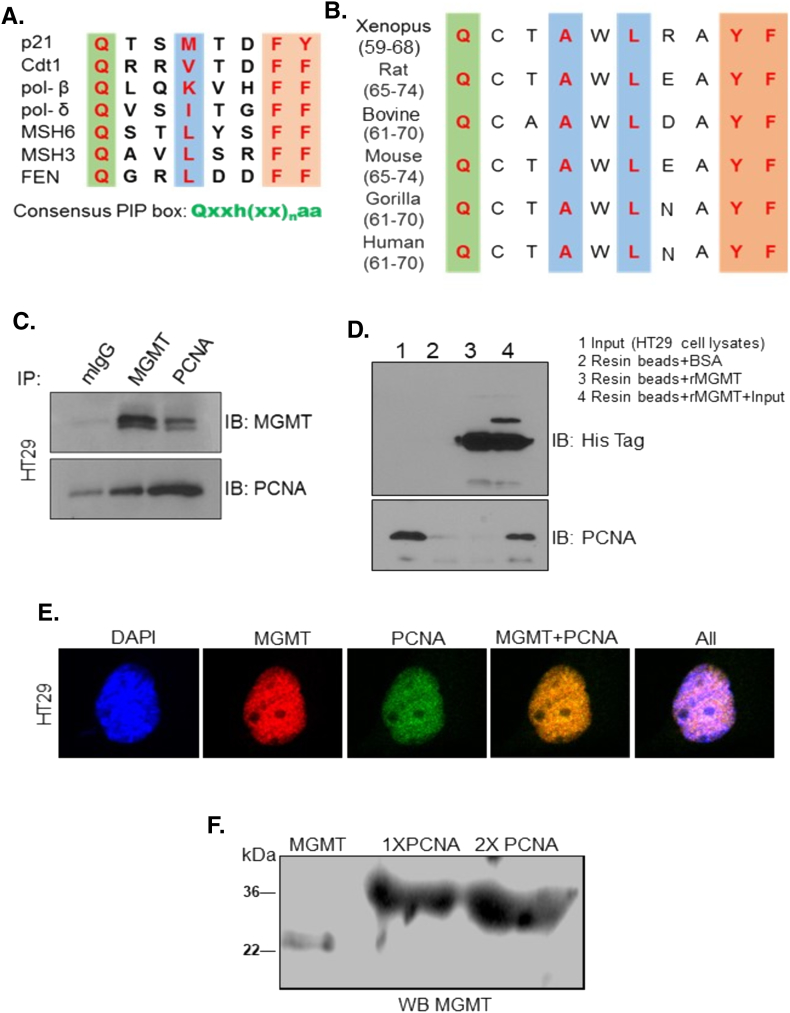
Figure 1.
The PIP-box in MGMT protein and evidence for MGMT-PCNA interaction in human tumor cells. (A) Established PIP-box sequence in different proteins and the consensus motif, where h -hydrophobic, a- aromatic and x – any amino acids are shown. (B) Presence of a conserved PIP-motif in human MGMT and MGMT from other species. (C) HT29 cell extracts were immunoprecipitated with antibodies to MGMT or PCNA followed by immunoblotting with reciprocal antibodies to show the protein-protein associations. Mouse IgG (mIgG) was used as the isotype control. (D) Pull-down assays: Histidine-tagged recombinant MGMT (rMGMT) protein was incubated with HT29 cell lysates, followed by addition of Talon metal affinity resin-beads, washing and immunoblotting with antibodies to hexahistidine tag or PCNA; lanes, 1- 10% HT29 lysate input, 2-resin beads incubated with BSA, 3 -resin beads incubated with rMGMT, and 4- rMGMT and cell extracts mixed together. (E) Confocal microscopy showing a co-localization of endogenous MGMT with PCNA in HT29 cells. Permeabilized HT29 cells were treated with mouse monoclonal Ab to MGMT or polyclonal Abs to PCNA either alone or together and stained with anti-mouse IgG-Alexa 594 or anti-rabbit IgG-Alexa 488 respectively. Merged yellow fluorescence shows the colocalization. (F) Far-western analysis showing in vitro interaction of purified MGMT and PCNA proteins. rMGMT and rPCNA were electrophoresed, blotted, renatured and the blot was incubated with rMGMT protein followed by western blotting for MGMT.
Evidence for Specific Interaction Between MGMT and PCNA Proteins In Vitro and in Tumor Cells
To determine if MGMT physically interacts with PCNA, HT29 cell extracts were subjected to reciprocal co-immunoprecipitation using anti-MGMT or anti-PCNA antibodies along with an isotype control. As shown in, Figure 1C, western blotting with the opposing antibodies revealed a significant association of MGMT with PCNA and vice versa. The data imply that MGMT interacts with PCNA through the canonical PIP-box domain. To validate the protein-protein interaction between MGMT and PCNA, we used pull-down assays using histidine-tagged recombinant MGMT protein. The cobalt affinity resin (Talon) beads were coated with His-tagged MGMT or BSA (as control) followed by their incubation with HT29 cell lysates containing PCNA. As shown in Figure 1D, the rMGMT protein was able to pull-down a significant amount of PCNA from the cell lysates, suggesting a strong physical and specific association of PCNA with MGMT.
Next, we demonstrated the association between MGMT and PCNA in HT29 cells through confocal microscopic imaging (Figure 1E). For this, the PCNA and MGMT proteins in permeabilized tumor cells were stained with Alexa flour 594 (red) and Alexa 488 (green) respectively, and the overlaid images are shown in last two panels. The images show that these two proteins localized closely in the vicinity of each other as evident from the strong merged yellow signals in the immunofluorescence photomicrographs (micrograph 4 inFigure 1E).
As an additional evidence to prove the direct interaction of these protein partners, far-western blotting was performed. In this procedure, purified PCNA was electrophoresed by SDS-PAGE at 0.7 and 1.4 μg levels and the protein immobilized on the blots were denatured and renatured to its native state. The blots were then incubated with the rMGMT protein as the bait and western blotted using MGMT antibodies. The far western blot (Figure 1F) shows that MGMT antibodies recognized the dose-dependent complex formation of PCNA with MGMT (at 36 kDa) on the nitrocellulose membrane. Note that MGMT electrophoresed on the left lane migrated at 22 kDa, as expected. Collectively, the results in Figure 1E-F demonstrate a clear physical interaction of human MGMT with PCNA both in vitro and in vivo.
Dynamic Interplay Between MGMT, PCNA, and p21cip1 Proteins in Human Cancer Cells
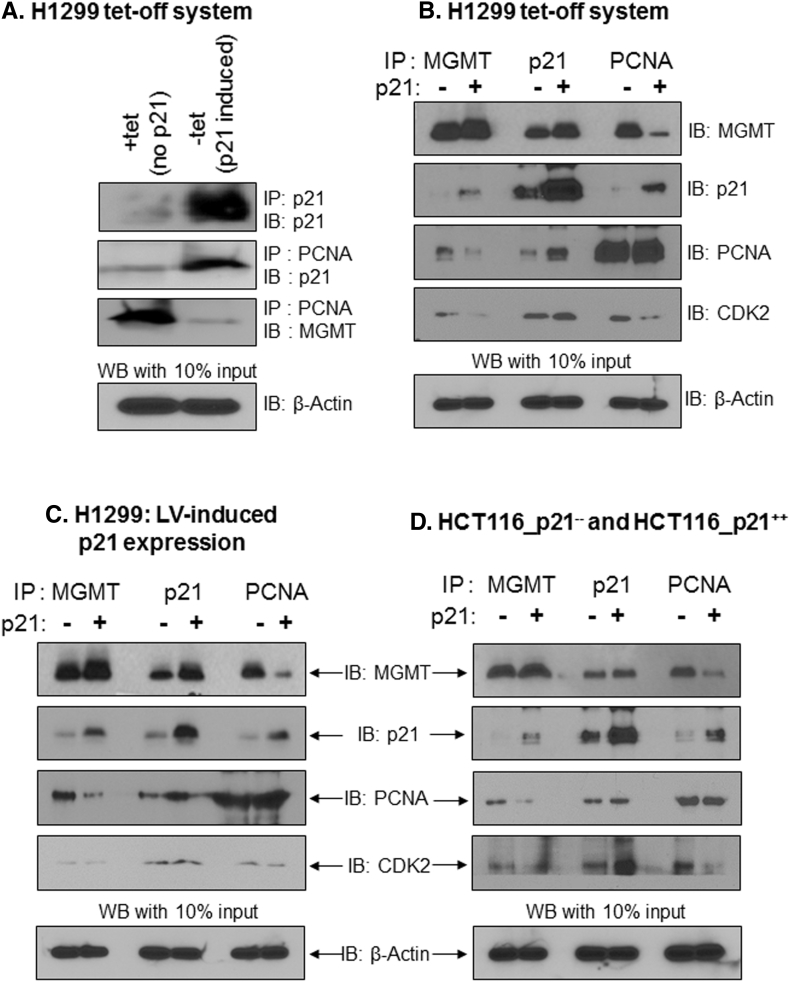
Distinct spatiotemporal interactions of the DNA repair, replication and cell cycle proteins with PCNA and p21 have been noted frequently in the literature [[23], [36]]. In many of these cases, the CDK inhibitor p21 plays a significant role in modulating the DNA repair and short- and long-patch repair synthesis that follow the damage processing [37]. The physical interaction of MGMT with PCNA prompted us to investigate a possible interplay of these regulators with p21, with which PCNA is known to bind avidly. For analyzing these relationships, we used three different isogenic cell lines with or without heightened p21 expression. The first model was a conditional gene expression model in which the p21 was cloned in a tetracycline (Tet)-regulated vector system (H1299-p21) that repressed p21 expression in the presence of Tet. H1299 Cells are p53-null and this choice enabled us to study MGMT- PCNA-p21 independent of the p53 influence on p21. This stably transfected cell line was maintained in the presence of Tet (1μg/ml) during early log phase and then cultured in the absence of Tet for successive 3 days to induce p21 expression, another group cultured in Tet-containing media (null or low p21) served as the control. The second isogenic pair was engineered to overexpress the p21 protein stably through lentiviral infection of a p21 cDNA construct (H1299-LV/p21). The third model was the normal p21-proficient HCT116 cells (HCT116.p21++) and its –deficient counterpart (HCT116.p21--). Reciprocal co-immunoprecipitation assays were performed to determine the dynamic interactions between the MGMT, PCNA, and p21 proteins in the three isogenic experimental models described above. Specifically, the cell lysates from the isogenic cell line pairs were immunoprecipitated using antibodies to MGMT or p21 or PCNA, the immune-complexes were western blotted and probed with antibodies to the self and the partner proteins (Figure 2). Figure 2A shows a huge induction of p21 protein after tet withdrawal in the conditional expression model; as expected, the p21 induction was followed by sequestration of PCNA and a great decrease in PCNA-associated MGMT. These results demonstrate that p21can bind PCNA away from MGMT and can regulate the amount of the DNA repair protein bound with the sliding clamp at a given time. Figure 2B represents a second experiment in the same conditional gene expression system (performed months later) and shows an extended but essentially a similar result. A slightly higher p21 expression seen in Figure 2B in the presence of tet (compared to 2A) indicates a slightly leaky gene expression, which is common after passing and freezing these cells. The recombinant p21 and MGMT proteins failed to form a complex (data not shown), suggesting a direct interaction between these two proteins is unlikely. Therefore, MGMT appears to interact with p21 through PCNA and the presence of ternary complexes containing the MGMT-PCNA-p21 proteins explains the pattern of proteins pulled down in the immune complexes in Figure 2. The overall MGMT content associated with p21 was significantly higher in p21-overexpressing isogenic cells (Figure 2B and C). The most important results from Figure 2A-C is that elevated p21 levels displace PCNA from MGMT, and perhaps from other partners through a higher affinity to PCNA. Analysis of these protein partners in the p21-deficient and –proficient HCT116 cells (Figure 2D) revealed similar results. Interestingly, p21-proficient HCT116 cells had higher MGMT protein levels. p21-deficient cells reproducibly showed lower MGMT protein levels. Again, PCNA-associated MGMT was much less in p21++ cells. p21 overexpressing cell lines showed a slightly increased expression of MGMT in all three isogenic models. Most importantly, a dramatic change was observed in MGMT-PCNA interaction upon p21 overexpression. In the first model, p21 overexpression caused around 70% reduction (quantified by densitometry) in PCNA-associated MGMT (Figure 2A). Second (80%) and third model (68%) also showed a similar pattern. Another significant finding was the presence of cell cycle kinases in MGMT immunoprecipitates, suggesting an association and a possible modulation of cell cycle progression by MGMT; this was shown to occur in all three isogenic cell line models used. Collectively, these data point to a dynamic interaction, with p21 strongly interfering with PCNA- MGMT association and dissociating this partnership.
Figure 2.
The spatiotemporal interplay between the MGMT, PCNA and p21cip1 proteins in three isogenic cell line pairs. (A) The first model was a H1299 cell line with Tet-off inducible gene expression system for p21. The H1299-p21 cells were maintained in the presence or absence of tet (1μg/ml) for 3 days, and the cell lysates were immunoprecipitated with antibodies to p21 or PCNA. Equal portions of immune-complexes were western blotted and probed with antibodies to p21 or MGMT. A strong reduction in PCNA-associated MGMT is evident after p21 expression. (B) In a second experiment, lysates from the tet-off p21 expression system (described above) were immunoprecipitated with antibodies to the triad of proteins and immunoblotted to show the protein associations. These cells had a slightly leaky p21 expression in the presence of tet but showed similar results. (C) The second isogenic cell line was the parent H1299 and the same cell line with stable expression of p21 developed after lentiviral transduction. Immunoprecipitation followed by immunoblotting for the partner proteins were performed as indicated. (D) The third pair, HCT116 cells proficient and deficient for p21 were similarly subjected to IP and immunoblotting to show the interplay between these proteins. Note the presence of CDK2 too in the complexes. Similar results were obtained in three independent experiments. In all cases, 10% of the cell lysates used for immunoprecipitations were western blotted for β-actin protein to verify the use of equivalent protein levels for IP.
Regulatory Role of p21 in MGMT expression, DNA Repair Activity and Decreased Drug-Induced DNA Interstrand Crosslinking
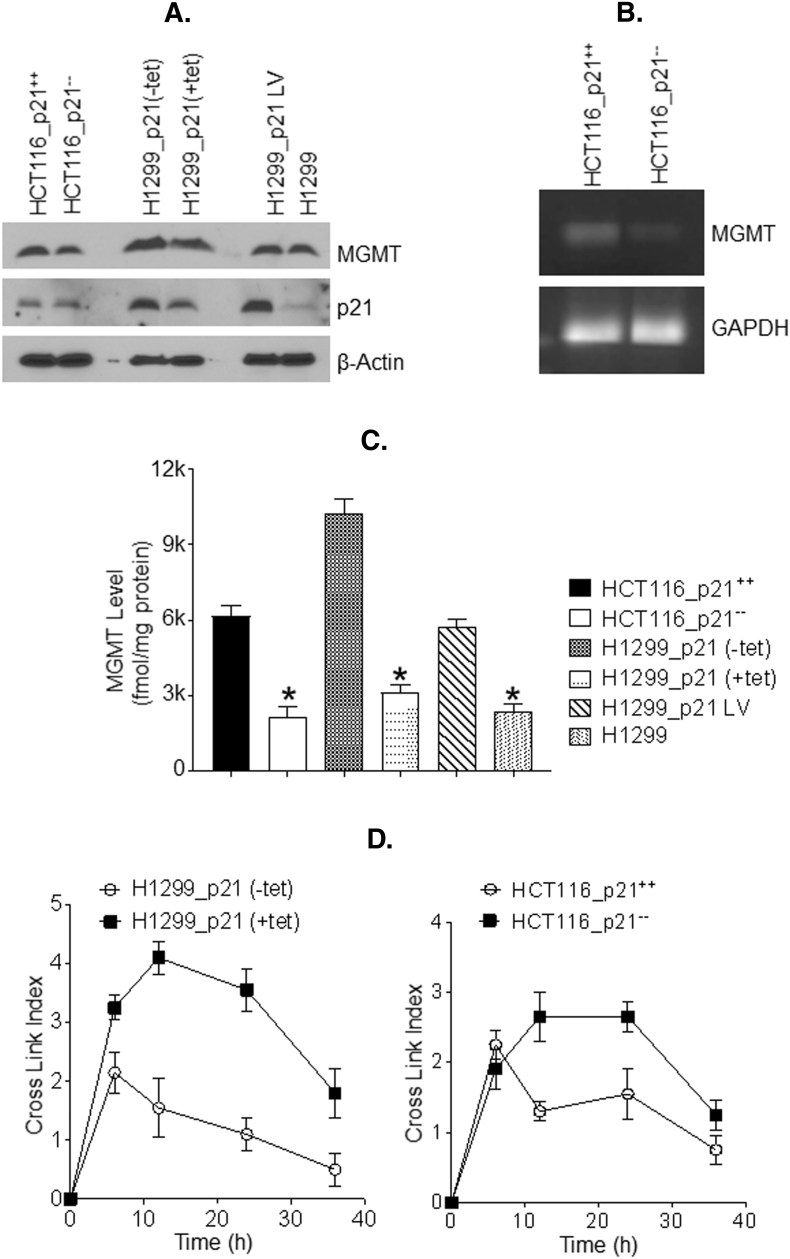
Because our co-immunoprecipitation (Figure 2A-D) and immunofluorescence experiments showed an upregulation of MGMT in p21 overexpressing cells, we validated this finding at both the transcriptional and translational levels and performed RT-PCR and western blotting. MGMT expression correlated positively with that of p21 in all three isogenic models; approximately 30%-80% higher levels of MGMT protein was present when p21 was induced or expressed endogenously in tumor cells (Figure 3A). RT-PCR performed with HCT116 cells with or without p21 expression (HCT116.p21-- and HCT116.p21++) showed higher levels of MGMT mRNA in HCT116.p21++ cells, suggesting an unexpected involvement of p21 in the transcriptional regulation of MGMT (Figure 3B).
Figure 3.
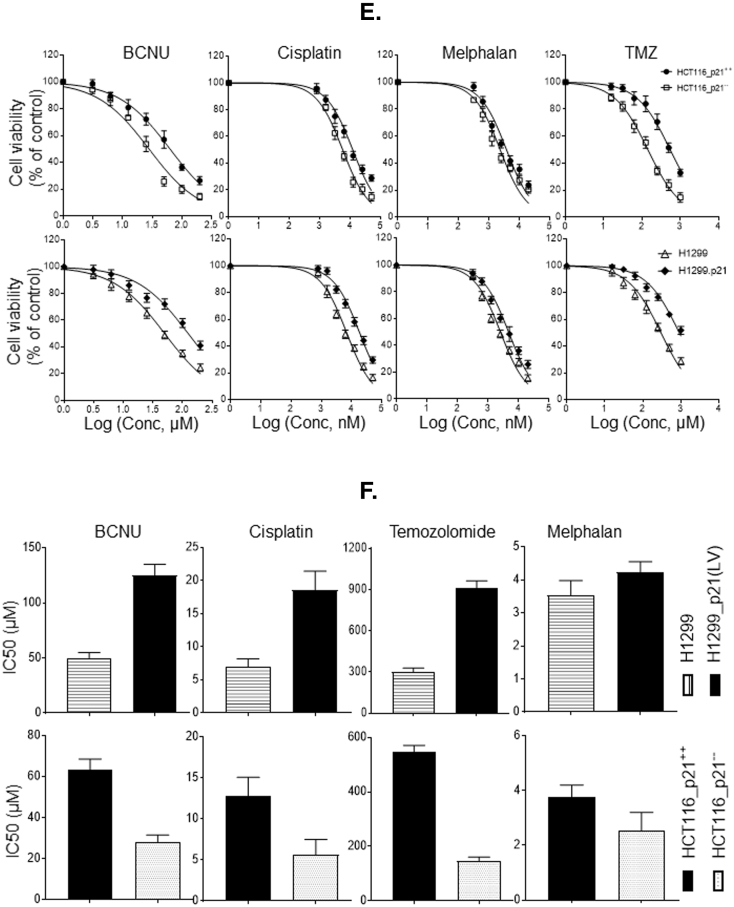
Regulatory role of p21 in MGMT expression, DNA repair activity, and drug resistance. (A) Levels of MGMT and p21 proteins in isogenic cells with p21-deficient or enforced p21 expression were analyzed by western blotting. (B) RT-PCR analysis of MGMT mRNA in HCT116.p21-- and HCT116.p21++ cell lines. (C) Quantitation of MGMT activity in three pairs of isogenic cell lines with or without increased p21 expression. Data are the means of three independent experiments; bars, SD. * p, significant at P <0.05. (D) Kinetics and extent of interstrand DNA cross-link induction by BCNU in isogenic cell lines with or without increased p21 expression. Cells were exposed to BCNU (100 μM) for 1h and then incubated in drug-free media for hours specified. DNA was isolated and the extent of interstrand crosslinking was determined by the ethidium bromide fluorescence assay as described in Methods. (E) p21-proficient cancer cells display significant resistance to MGMT-targeted anticancer alkylating agents. p21-deficient and –proficient HCT116 cells and H1299 cells with and without stable expression of p21 were exposed to BCNU, cisplatin, Melphalan and TMZ for 72 h followed by MTT assays. (F) IC50 values for cell viability from the above data are shown as bar graphs.
To verify whether MGMT upregulation also correlates with its DNA repair activity, the catalytic transfer of O6-methylguanine in DNA to the MGMT protein was measured in the tumor cell extracts. Cell lines with enforced p21 expression consistently showed higher enzymatic activity (fold increase) of MGMT. A consistent and marked increase (2.4 to 3.3 -fold) in MGMT activity was observed when cells expressed p21 compared with their non-expressing counterparts (Figure 3C). A higher MGMT content in tumor cells is expected to diminish the levels of alkylation DNA damage and hence the interstrand cross-linking of DNA induced by bifunctional alkylating agents. This postulate was tested by treating HCT116 (p21 -proficient and -deficient) cells and H1299 cells with conditional p21 expression with BCNU, an anticancer alkylating agent well known to induce G-C interstrand crosslinks in DNA [38]. An ethidium bromide fluorescence assay, well established in our laboratory [30], was used to determine the levels of cellular DNA cross-linking after BCNU exposure, up to 48 h. p21-proficient cells in this setting showed a range of about 1.7 to 4-fold reduction in the net DNA interstrand cross-links relative to their isogenic p21-deficient counterparts (Figure 3D) thereby confirming a positive regulation of MGMT DNA repair by p21 in human cancers.
p21 Expression Confers Decreased Cytotoxicity and to Resistance to Alkylating Agents
To determine if p21-induced enhancement of DNA repair by MGMT translates into increased survival of cancer cells after alkylation damage, we performed the MTT cell survival assays. For this, the isogenic cell lines described in previous sections were treated with three different alkylating agents (BCNU, Melphalan, and TMZ) and cisplatin. Cisplatin was tested in this setting because higher MGMT expression has been previously shown to result in resistance to the platinating agent [39]. Results from these assays revealed remarkable differences in sensitivity towards cytotoxic drugs (Figure 3E). Higher levels of p21 in tumor cells conferred significant resistance to BCNU and TMZ. Comparison of IC50 values showed around 2-4 fold increase in drug’s concentration required 50% cell killing (bar graphs are shown inFigure 3F). Most significant differences were observed in TMZ-induced cytotoxicity (3.8 fold increased IC50 in cells with enforced p21 expression). Interestingly, p21 overexpressing cells also showed to some extent (1.6-1.8 fold) less sensitivity towards Melphalan that predominantly generates N7–guanine alkylations [40], which are not removed by MGMT. Similarly, the p21 expression bestowed significant resistance to cisplatin. These data suggest that p21 reduces the cytotoxicity to MGMT-targeted alkylating agents. However, the CDK inhibitor, through its involvement in the regulatory network of numerous other DNA repair systems can also impact the cell killing by different classes of anticancer drugs [37].
Co-Localization of MGMT with PCNA in p21-proficient and –Deficient Cells Following Alkylation Damage
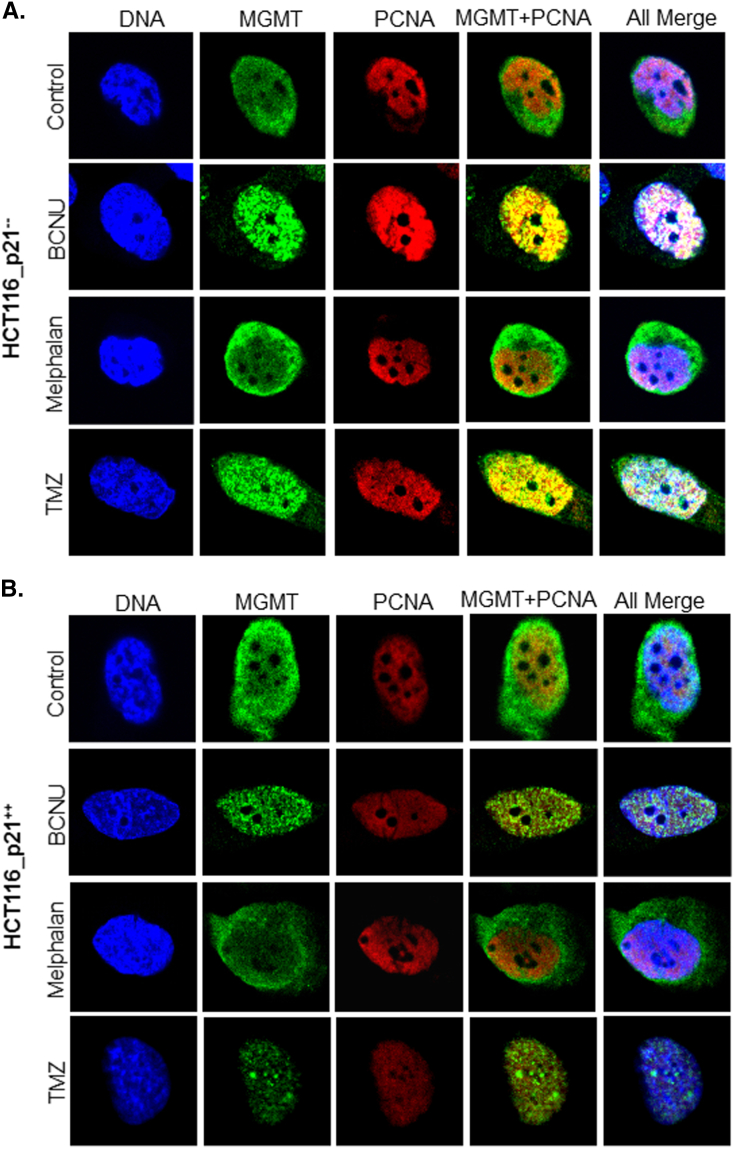
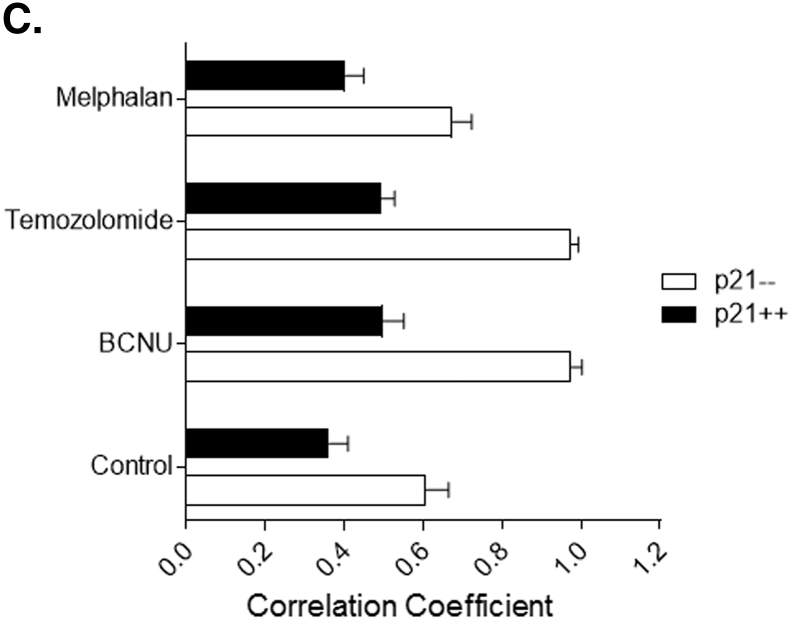
To explore whether PCNA and/ or p21 have roles in subcellular distribution or localizing the MGMT to the sites of alkylation DNA damage, the HCT116.p21-- and HCT116.p21++ cells were treated with the alkylating agents (BCNU, TMZ, and Melphalan) followed by immunostaining for MGMT and PCNA proteins and multiphoton confocal microscopy. On treatment with BCNU (30μM) or TMZ (250μM) at near IC50 doses for 24 hours, a significant change in the subcellular localization of MGMT was evident. In the control groups (both HCT116.p21-- and HCT116.p21++ cells), MGMT was found to be distributed both in nucleus and cytoplasm (Figure 4A and B, top panel). In contrast, BCNU or TMZ-treated groups showed more MGMT staining in the nuclear area (Figure 4A and B, 2ndand 4thpanels). This change in MGMT distribution was observed both in HCT116.p21-- and HCT116.p21++ cells, indicating the null effect of p21 on DNA damage-induced translocation of MGMT. However, MGMT localization was not altered in Melphalan-treated cells irrespective of p21 expression (Figure 4A and B, 3rdpanel), which was expected as the N7- alkyl lesions inserted by this DNA damaging agent are not subjected to MGMT-mediated repair. Another significant feature noted was the spotted or punctate appearance of MGMT in damaged cells, implying a selective distribution within the nuclei. A remarkable change in MGMT-PCNA co-localization was also observed upon treatment with BCNU and TMZ, but only in p21-deficient cells (HCT116.p21--). In HCT116.p21++ cells, decreased levels of MGMT protein co-localized with PCNA relative to a greater merging in p21-deficient cells. The extent of colocalization in these experiments was quantitated and compared (Pearson’s correlation coefficient) using the ImageJ analysis (Figure 4C). At least five different images were used to obtain an average correlation coefficient values. Careful analysis of these co-localization data revealed that though p21 showed no effect on nuclear translocation of MGMT upon induction of DNA damage, however, it certainly interfered with the MGMT-PCNA interaction, because the immunocytochemical association of MGMT-PCNA was markedly less in p21- proficient HCT116 cells. Taken together, the results suggest that following DNA damage, PCNA binding with MGMT may have unspecified functions and that p21 association is likely to downregulate such an interplay. As discussed in sections to follow, increased PCNA association in the absence of p21 may target the MGMT protein for elimination by CRL4Cdt2, and this may explain the decreased MGMT content in p21-deficient cells as well.
Figure 4.
Alkylation DNA damage-induced distribution of MGMT protein and its co-localization with PCNA in p21-proficient and –deficient HCT116 cells. (A) p21-deficient cells, (B) p21-proficient cells. Cells were treated with DNA-damaging agents (30 μM BCNU or 2 μM Melphalan or 250 μM TMZ for 24 h followed by fixation and permeabilization. Immunostaining was performed for both MGMT (green) and PCNA (red) using the murine and polyclonal antibodies respectively and the Alexa–conjugated secondary antibodies followed by confocal microscopy as described in the legend for Figure 1E. DNA was stained with DAPI to distinguish nuclear and cytoplasmic locations and co-localizations were depicted as merged signals (yellow fluorescence). Single-cell images are shown for better visualization of MGMT, PCNA localizations, and their physical interactions. (C) Pearson’s correlation coefficients (r) for the co-localization of MGMT and PCNA proteins in the above experiments were done by using ImageJ software. The correlation is based on the average of 5–10 independent cells, with standard deviations shown. The correlation coefficient value (r) ranged from -1 (a perfect negative correlation) to 1 (a perfect positive correlation). MGMT molecules co-localized with PCNA are likely to be targeted for degradation by CRLCdt2 as described in this report.
Specific Elimination of MGMT Protein During S-Phase of the Cell Cycle in Human Glioblastoma Cells
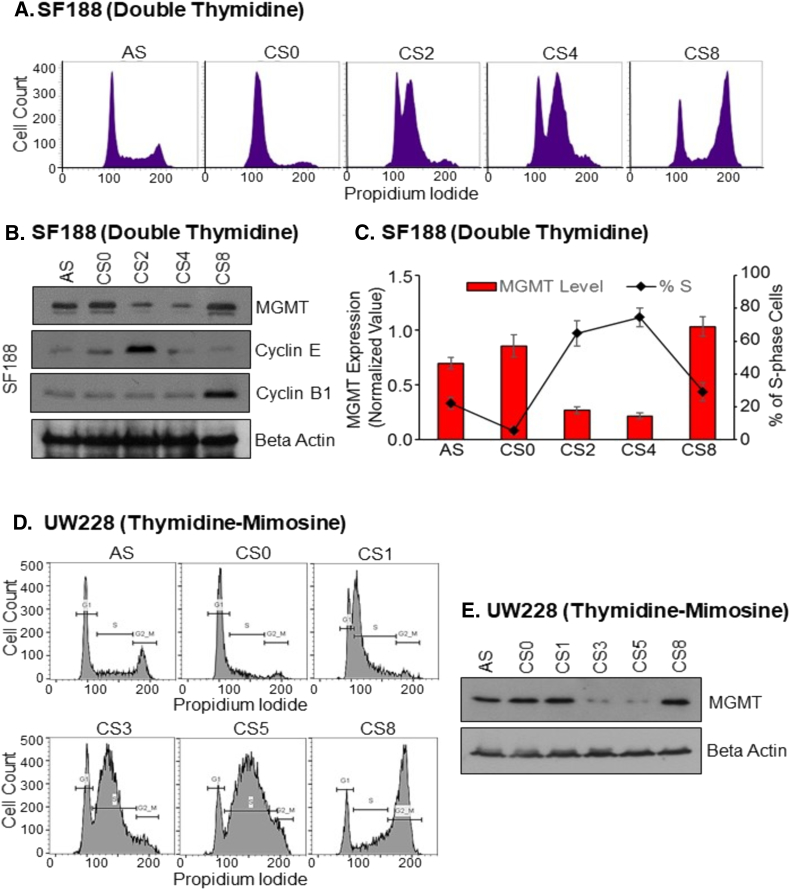
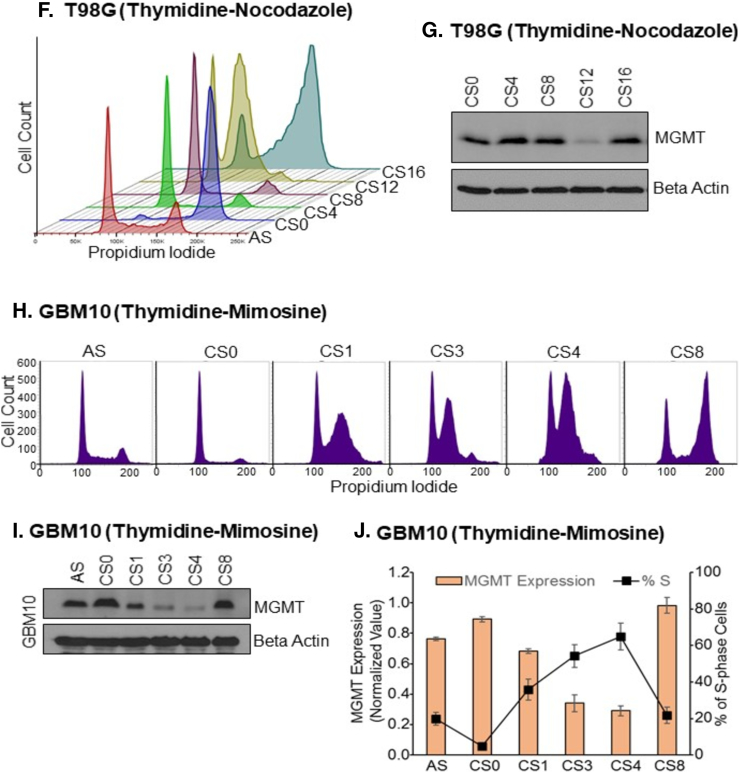
Timely removal of DNA damage is highly integrated with accurate DNA replication and cell cycle progression. Therefore, the repair proteins often show cell cycle-specific expression patterns reflecting their role in repairing different types of DNA damage [[41], [42]]. MGMT transcription appears to be uniform during cell cycle progression [41]. However, in view of association with p21 described here, we hypothesized that MGMT may exhibit periodic oscillations during the cell cycle. To quantitate MGMT levels during cell cycle progression, we synchronized glioblastoma cell lines (SF188, GBM10, and T98G) and the UW228 medulloblastoma cells, by using three different methods, namely, double thymidine block, thymidine/mimosine block, and thymidine/nocodazole block. Double thymidine and thymidine/mimosine blocks arrested the cells at an early S or G1/S, whereas thymidine/nocodazole block induced a G2/M phase arrest. The synchronized cells were then suspended in fresh media and allowed pass through the rest of cell cycle in unison. The cells were collected at different times (4-16 h) during their recycling to reflect the early, late S-phase and G2/M phase populations. The cell lines used in these experiments showed slight differences in progression rates and accumulation in various cell cycle phases as revealed by flow cytometry; therefore, optimization was required for collection times. In case of SF188 with double thymidine arrest (Figure 5A), cells released for 0, 2, 4 and 8 h represented the G1/S, early S, late S and G2/M phases respectively. After confirming the time points, we prepared cell lysates to analyze MGMT expression level at different cell cycle phases by western blotting (Figure 5B). The relative levels of MGMT were quantified by densitometry and compared with the respective percent of the S-phase population (Figure 5C). Known markers of cell cycle phases (cyclin E and cyclin B1) were used to validate the synchronization protocols where cyclin E appeared with the initiation of S-phase and remained high until transition into G2/M phase. G2/M phase population mostly expressed cyclin B1. A similar and more pronounced downregulation of MGMT during the S-phase also occurred in UW228 cells after synchronization by the thymidine-mimosine procedure (Figure 5D and E). Also evident is the quick reappearance of MGMT at the G2/M phase (Figure 5B, E, G). A dramatic loss of MGMT protein corresponding to a huge S-phase cell population was also observed after arresting theT98 glioblastoma cells (T98G) at G2/M arrest using thymidine–nocodazole combination (Figure 5F and G). Confirmation of the S-phase dependent elimination of MGMT protein was also made in GBM10, another human glioblastoma cell line. Cell synchronization and release profiles of the histograms following the thymidine-mimosine block are shown in Figure 5H. A downregulation of MGMT protein in the S-phase (labeled CS3 and CS4) is again evident in Figure 5I. The bar diagram in Figure 5J shows a 3-fold depletion of the DNA repair in S-phase cells occurred in comparison with the asynchronous GBM-10 control cells. Collectively, the experiments performed in four brain tumor cell lines (3 glioblastomas and 1medulloblastoma) using three different synchronization procedures, all unequivocally and reproducibly demonstrated a downregulation of MGMT as an S-phase-specific event, suggesting a crucial function performed by this repair protein during the cell cycle.
Figure 5.
S-phase-specific degradation of MGMT protein in three human glioblastoma cell lines. In these experiments, GBM cells were first synchronized in G1/S by double thymidine block or thymidine-mimosine block or G2/M phase by thymidine-nocodazole block as described in Methods. The arrested population was released into cell cycle in a uniform manner. “AS” represents an asynchronous population whereas CS0 is the arrested cells. CS2, CS4, and CS8 represent cells at 0h, 2h, 4h, and 8h after release into the cycle respectively. CS2-early S phase, CS4-late S phase, and CS8-G2/M phase. In the thymidine-nocodazole block, the release times were extended to CS12 and CS16 hours. FACS analysis was performed at times indicated to analyze the cell cycle phases. Cells from the AS, CS0 and at different release times were analyzed for expression of MGMT and cell cycle phase markers by western blotting. (A) SF188 GBM cells were synchronized by double thymidine-block and subjected to FACS. (B) Protein levels of MGMT, cyclin E (S-phase marker) and cyclin B (G2/M marker) in synchronized and released SF188 cells. (C) Comparison of normalized levels of MGMT protein and respective percentages of S-phase cell population at the indicated time points after double thymidine block in three independent experiments. (D) Cell cycle histograms of SF188 cells synchronized by thymidine-mimosine block and release. (E) Western blot analysis of MGMT in SF-188 cells synchronized by thymidine-mimosine. (F) Stacked histograms showing the synchronized progression of SF188 cells after release from the thymidine-nocodazole block at different time points indicated in the Figure. At 0h release (CS0), most cells were in G2/M phase and from there cells progressed into subsequent phases; CS4-G1 phase, CS8-G1/S boundary, CS12-S phase, and CS16-again G2/M phase. (G) MGMT protein levels in cells synchronized by thymidine-nocodazole block and released. (H) FACS analysis of GBM10 cells synchronized by thymidine-mimosine block and release. (I) MGMT protein levels in synchronized and released GBM10 cells. (J) Relative MGMT protein levels and S-phase cell population in thymidine-mimosine blocked and released GBM10 cells. (K) Cell cycle histograms of T98G cells synchronized at different cell cycle phases by double thymidine block. (L) MGMT protein levels in T98G cells arrested by double thymidine block and released.
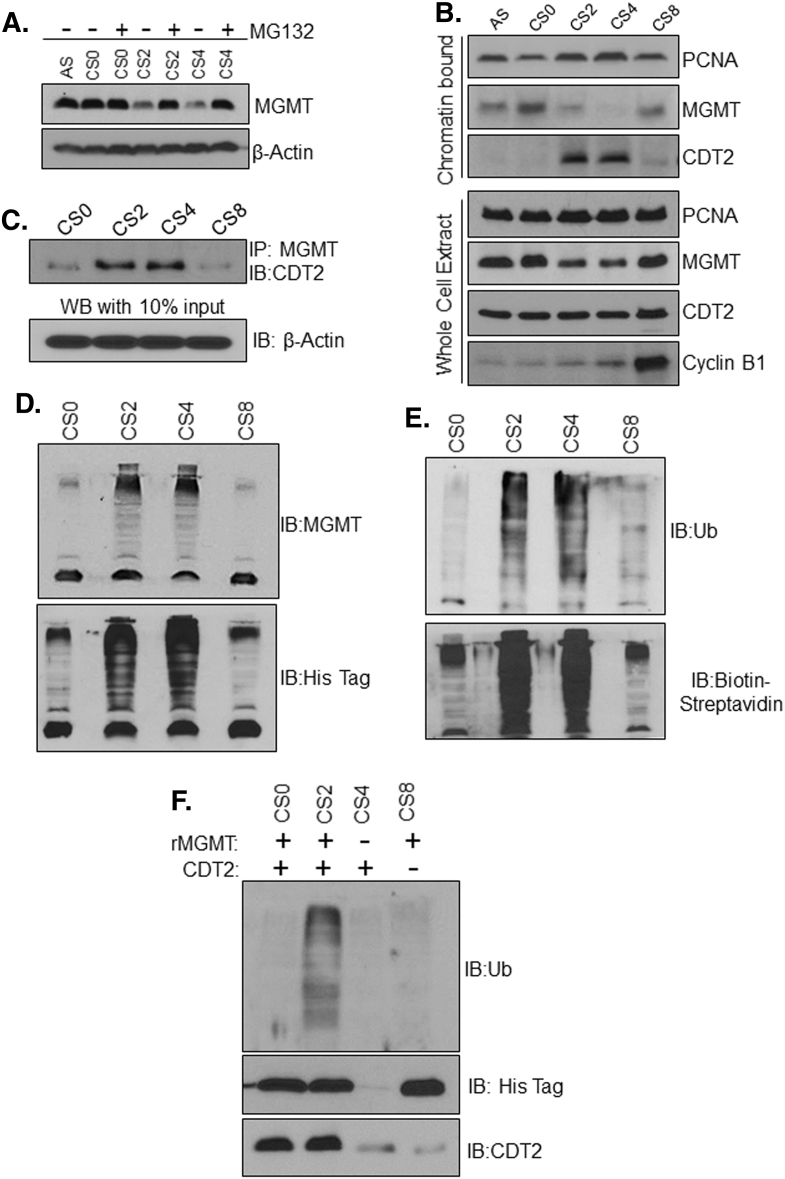
To elucidate the mechanisms of MGMT turnover during the S-phase, the G1/S synchronized cells were exposed to the proteasomal inhibitor MG132; we observed a significant stabilization of MGMT in cells released into S-phase (Figure 6A). The data suggested a role for ubiquitination-dependent proteolysis in S-phase-specific elimination of MGMT. To obtain further insight into how the MGMT downregulation occurs, we explored the putative ubiquitin ligase enzyme responsible for MGMT ubiquitination. It has been reported that the sliding clamp processivity factor, PCNA cooperates with the E3 ubiquitin ligase enzyme CRL4Cdt2 in the replication-coupled destruction of critical cell cycle proteins during the S-phase [43]. In human cells, several S-phase regulatory proteins such as Cdt1, PR-SET7/SET8, p21, and DNA polymerase δ subunit p12 have been identified as CRL4Cdt2 substrates [[26], [44], [45]]. The CRL4cdt2 specifically adds ubiquitin moieties to the substrate proteins which are chromatin-bound (not free in the cytosol) and complexed with PCNA [43]. The PIP-box dependent association of PCNA with their substrates in their DNA-bound state is a prime requirement for ubiquitination by CRL4cdt2 ubiquitin-ligase that regulates the S-phase exit and maintenance of genomic integrity [35]. Presence of a typical PIP box domain in the MGMT protein and the strong evidence MGMT-PCNA interaction uncovered in this study strongly suggested the possible involvement of PCNA and CRL4Cdt2 ubiquitin ligase in the proteasomal digestion of MGMT. To address this possibility, we performed cell synchronization assays to characterize the cell cycle specific expression pattern of MGMT, Cdt2, and PCNA proteins. SF188 cells were synchronized at G1/S phase boundary through thymidine-mimosine block followed by the release of cells and collection of cell lysate at subsequent phases. Both the chromatin-bound fractions and whole cell lysates were immunoblotted to determine the abundance of these proteins and chromatin occupancy. As shown in Figure 6B, chromatin localization of MGMT was lowest in mid to late S phase whereas PCNA and Cdt2 showed highest chromatin binding during the entire S- phase. In contrast, no significant differences were observed in the levels of PCNA and Cdt2 in the whole cell lysates, demonstrating their equivalent levels in the cytosol; however, the MGMT protein, as shown in previous experiments (Figure 5B, E, G, I), was downregulated during mid to late S- phase (Figure 6Blower panel). These results clearly indicate the involvement of CRL4Cdt2 in the replication-coupled destruction of MGMT protein.
Figure 6.
Evidence for the involvement of CRLCdt2, a PCNA-dependent ub-ligase, and proteasome in S-phase downregulation of MGMT protein. (A) Proteasome inhibition curtails MGMT degradation in S-phase cells. SF188 cells were synchronized by thymidine-mimosine block and released into the cell cycle as described in the legend to Figure 6. AS, asynchronous, CS0, 0 h release and CS2 (2 h release, early S) CS4 (4 h, late S), CS8 (8h, G2/M) phases. The cells were treated with proteasome inhibitor MG132 during release from the G1/S block. (B) Inverse correlation of MGMT with PCNA and CRLCdt2 proteins in the chromatin-bound fractions. PCNA is increasingly loaded on to chromatin during the S-phase and PCNA-bound proteins in chromatin are essential substrates for CRLCdt2. Chromatin-bound proteins from the G1/S synchronized and released SF188 cells were isolated as described in Methods and western blotted along with the whole cell extracts for comparison. A decrease in MGMT correlated with increased PCNA and Cdt2 occupancy in the S-phase chromatin. (C) Evidence for the Interaction of MGMT with CDT2 during the S-phase. SF188 cells were synchronized by thymidine-mimosine block and released into cycling in the time frame described above. Cell lysates from the times specified were immunoprecipitated with anti-MGMT antibody followed by SDS-PAGE and immunoblotting with the anti-CDT2 antibody. Ten percent of the protein input used for IP were subjected to western blotting for actin to verify the use of equivalent protein levels for IP. (D) Selective ubiquitination of MGMT during the S-phase. In vitro ubiquitination assays using the histidine-tagged recombinant MGMT and unlabeled ubiquitin were performed as described in Methods. Extracts from the G1/S -arrested and -released SF188 cells served as the sources of ubiquitin-conjugating enzymes. The reaction samples were electrophoresed and western blotted using antibodies to MGMT or hexahistidine sequence. (E) In vitro ubiquitination assays similar to the ones described in Figure 6D was performed by using biotinylated ubiquitin instead of the unlabeled ubiquitin. The resulting blots were probed with anti-ubiquitin antibodies or Streptavidin-linked HRP to obtain intense polyubiquitinated bands. (F) Association of MGMT with CDT2 plays an essential role in S-phase ubiquitination of MGMT protein. Extracts from SF188 cells arrested at G1/S and 4 h release (CS4) were prepared and CDT2 was immunodepleted as indicated. Extracts with or without CDT2 immunodepletion were used as the enzyme source and rMGMT as the substrate for in vitro ubiquitination assays. The resulting blots were western blotted with antibodies to ubiquitin, histidine-tag or CDT2. The reactions in lane 4 had the CDT2 immunodepleted extract. N=no, Y=yes.
As a further proof, we probed the physical interaction between MGMT and Cdt2 proteins during different cell cycle phases. For this, the G1/S synchronized SF188 cells (labeled CS0 in Figure 6C) and cells released into subsequent cycling were lysed and subjected to immunoprecipitation with MGMT antibody followed by western blotting for Cdt2. As shown in Figure 6C, increased levels of Cdt2 were found to be associated with MGMT specifically at 2 and 4 h release time points corresponding to the mid and late S-phases. This evidence demonstrates that MGMT is indeed a substrate for the CRL4Cdt2 ubiquitin ligase, and the enzyme binds with MGMT only during the S phase. To visualize the S-phase specific polyubiquitination of MGMT, we performed both in vivo and in vitro ubiquitination assays using cell lysates collected from different cell cycle phases. For the in vitro assay, the histidine-tagged recombinant MGMT protein was used as the substrate and the cell lysates from different cell cycle phases served as the enzyme source. The reaction samples were split into two and western blotted separately, and probed with antibodies to MGMT or hexa-histidine tag. The resulting immunoblots shown in Figure 6D reveal that the MGMT underwent a high level of ubiquitination (as represented higher MW protein bands and smear) in CS2 and CS4 extracts which originated from cells enriched in the S-phase. Recognition of the same protein bands by the anti-hexa histidine antibodies confirms the higher MW forms of MGMT protein were ubiquitinylated (Figure 6Dlower panel).
Another in vitro experiment was done for the same purpose using the biotinylated ubiquitin as one of the substrates with the endogenous MGMT present in the cell extracts (from synchronized and released cells) as the reactant. The reaction samples were again electrophoresed and the resulting protein blots were probed with streptavidin-linked HRP to detect the biotin-ub linkages or the ubiquitin antibody. Here again, the extracts from cells enriched in S-phase had the highest capacity to MGMT ubiquitination compared to cells in G1/S or G2/M phases (Figure 6E). The data support our conclusion that S-phase ubiquitination and subsequent proteasomal digestion targets the MGMT protein.
To further verify the involvement of Cdt2 in MGMT polyubiquitination, we immunodepleted the Cdt2 protein from the S phase lysates (CS4) and performed the in vitro ubiquitination assays. As shown in Figure 6F, immunodepletion of Cdt2 from cell lysates led to a diminished polyubiquitination of MGMT (lane 4) relative to the intense ubiquitin-conjugation observed with the Cdt2-proficient (control, undepleted) S-phase lysates (lane 2). These results validate and confirm that CRL4Cdt2 is the ubiquitin ligase responsible for MGMT-modification mid to late S-phase.
Discussion
As is common with other DNA metabolic proteins to bind with PCNA, this study discovered a physical and functional interaction between MGMT and PCNA for the first time. We also found the association of the CDK inhibitor p21cip1 in this triad, because p21 is an inseparable companion of PCNA [[21], [46]]. First, we identified the presence of a canonical PIP box in the MGMT protein sequence (amino acids 61-70), which was conserved in the DNA repair protein across the higher order species. We confirmed a direct association between MGMT and PCNA by using multiple lines of approaches including pulldown assays, reciprocal co-immunoprecipitations, far-western blot analysis, and confocal immunofluorescence imaging. Three isogenic cell line pairs, with one member in each group, differing in p21 expression levels were used to elucidate the MGMT-PCNA interaction.
The cell cycle inhibitor p21 functions as an essential hub for integrating accurate DNA replication with precise cell cycle progression. With its ability to inhibit all CDKs, the p21 engineers a G1 or G2-phase block to prevent the replication of damaged genomes and enable DNA repair, if possible [[29], [47]]. In this process, p21 competitively binds with PCNA and blocks the interaction of replication system with PCNA and thus stalling of replication forks [47]. Results for our reciprocal co-immunoprecipitation assays conducted in three isogenic cell line pairs conclusively showed that p21 interferes with binding of MGMT with PCNA, wherein increased p21 expression significantly attenuated the level (70 to 90%) of PCNA-associated MGMT. As expected, enforced expression of p21 also resulted in the increased PCNA-p21 association. While a direct binding of p21 with the MGMT was not detected, all indications were that p21 interacts with MGMT via the PCNA in ternary complexes. Binding of p21 to PCNA occurs at the interdomain connector loop, which is also the region involved in the binding of several PCNA-interacting proteins [23]. It is possible that the PIP box sequence of MGMT, being longer by 2 residues, may bind PCNA with a decreased affinity (Figure 1A and B), and, therefore, p21 may efficiently displace PCNA from MGMT. Such a functional dislodgement of PARP-1, DNA cytosine methyltransferase and other partners from PCNA by p21 is well known [[22], [48]].
Our studies did not address the structural aspects of PCNA binding with MGMT, however, looked at some functional aspects dependent and independent of p21cip1. Unlike other DNA repair (BER, NER, MMR) pathways, removal of O6-alkylguanines by MGMT does not involve a repair synthesis. In our experiments, the addition of recombinant PCNA protein (0.1-1 μg) to the MGMT assays did not affect the extent of DNA repair (not shown). Following alkylation DNA damage, there was an increased co-localization of MGMT and PCNA proteins, particularly in p21-deficient HCT116 cells (Figure 4A). For proteins involved in mismatch repair, NER, and homologous recombination, all requiring a repair synthesis, such an association has been interpreted to mean that PCNA recruits the repair proteins to the damage sites and facilitate a repair process [49]. While an effective halting of DNA replication is known to occur by PCNA sequestration by p21, many studies have confirmed that p21 does not interfere with post-repair DNA synthesis steps where PCNA is involved in the short patch repair of gaps [[37], [49]]. Although the mechanistic consequences of PCNA binding on substrate loading and catalytic rate for MGMT remains to be solved, many of our key observations clearly point to a negative regulatory role for PCNA in controlling MGMT function(s). First, higher levels of p21 that reduce MGMT-PCNA association correlated directly with significantly greater levels of mRNA, protein, and activity for MGMT in all three isogenic cell lines tested (Figure 2, Figure 3). Second, there was an increased co-localization of PCNA and MGMT in p21-deficient cells that had lower levels of MGMT protein and activity (Figure 4A). Third, as discussed in the sections to follow, PCNA-bound MGMT in chromatin was subjected to degradation via CRL4cdt2, an ubiquitin-ligase active during the S-phase.
The mechanism of transcriptional upregulation of MGMT by p21 (Figure 3B) is currently unclear, however, may involve the interactions with E2F transcription factors, Rb-protein and p300 acetyltransferase [50]. In line with a higher MGMT content, the p21-expressing cells were markedly resistant to the O6-alkylguanine-generating alkylating agents and exhibited lower levels of interstrand DNA crosslinks when challenged with BCNU (Figure 3D, E, F). Consistent with this observation was the marked sensitivity of p21-deficient tumor cells to the anticancer alkylating agents (Figure 3E, F). Such an increased sensitivity of p21-deficient HCT116 cells to many other anticancer drugs has been reported previously [51]. These findings imply that p21-orchestrated cell cycle arrest is likely to promote the repair of alkylation DNA damage, consistent with the functions ascribed to G1 or G2 phase blockades.
The second part of this study, a logical extension, looking into the cell cycle distribution of MGMT, which is largely considered to have a repair function through all phases, revealed interesting features in the cell cycle. Our studies provided the first and conclusive evidence for an S-phase specific downregulation of the MGMT protein. Using three different cell synchronization procedures, in four human brain cancer cell lines, we observed a highly reproducible downregulation of MGMT protein in mid to late S-phase.
A timed destruction of cyclins that mediate the cell cycle phase transitions drives an orderly and invariant cell cycle progression. Two families of E3 ubiquitin ligases, the Skp/cullin/F-box-containing, and anaphase-promoting complex/cyclosome complexes, respectively are largely responsible for G1 to S and G2 to M phase transitions [25]. In contrast, the S-phase progression is driven by the CRL4Cdt2, a cullin 4-based E3 ubiquitin ligase (CRL4) that associates with a substrate recognition factor called the Cdt2, a WD40 repeat domain protein [[43], [52]]. The mechanism of CRL4Cdt2 substrate recognition is unique in that the substrates must first interact with DNA-loaded PCNA; the PCNA is loaded on DNA during both S phase and DNA repair [52]. PCNA exists in the cell both as a free/detergent-soluble protein, and a chromatin-bound /detergent-insoluble form which encircling the DNA is directly involved in DNA synthesis and other transactions. The target substrates of the CRL4Cdt2 E3 ubiquitin ligase complex are known to have a PIP degron comprising a consensus PCNA-interacting protein motif (PIP box) with additional sequence requirements [[43], [52]]. The substrates of CRL4Cdt2 include the DNA replication initiation factor Cdt1, the cell cycle inhibitor p21, the histone H4 Lys-20 methyltransferase Set8, Cdt2 itself and p12, a subunit of DNA polymerase δ [[24], [26], [43], [44]]. The destruction of the above regulators in S phase is crucial to ensure precise and efficient genome duplication [25]. Thus, CDT1 is required in G1 for rendering DNA replication origins competent for initiation in S phase otherwise called replication licensing [53]. SET8 is the only enzyme responsible for histone H4 lysine 20 monomethylation (H4K20me1) and, like CDT1, is also required for origin licensing [44]. Degradation of p21 in early S phase stimulates CDK2 activity, which, in turn, triggers key S phase events, including DNA replication initiation and inhibition of origin licensing [[24], [26]]. Failure to degrade these regulators during S-phase results in multiple rounds of origin firing, leading to DNA re-replication and genome instability [25].
Our studies unequivocally demonstrated the S-phase elimination of MGMT protein. Compared to G1, the mid to late S-phase cells in four glioma cell lines had 10 to 25% of MGMT content (Figure 6). Like CDT1 and p21, the downregulation of MGMT was also mediated by proteasomal degradation preceded by polyubiquitination. We also identified CRL4Cdt2 as the probable ubiquitin ligase that preferentially promotes ubiquitylation of chromatin- and PCNA-bound MGMT (Figure 6B-F). The Involvement of CRL4Cdt2 in MGMT ubiquitination is consistent with MGMT protein harboring a PIP-box, co-immunoprecipitation of CRL4Cdt2, immunodepletion of CRL4Cdt2 leading to loss of MGMT ubiquitination (Figure 6).
Most CRL4Cdt2 substrates are toxic for S phase progression [[43], [52]]. In analogy with the other regulatory proteins that are eliminated before the onset of G2 phase by CRL4Cdt2 to maintain genomic stability, the removal of MGMT at the same time supports the notion that the presence of MGMT may harm the termination of genomic replication. The results can also be interpreted to mean that MGMT performs an unknown, perhaps an important function, in the cell cycle until its downregulation in the S-phase. Noteworthy in this context is the targeting of thymine DNA glycosylase by CRL4Cdt2 during S-phase to avoid competition with mismatch repair and prevent mutations [27]. Further studies are required to delineate the exact reason(s) for removal of MGMT protein during the S-phase.
Our findings on MGMT are likely to have significant consequences for cancer progression, therapy, and prevention. However, further studies are required to unravel the details. For example, MGMT overexpression in a large number of human cancers may indicate an oncogenic role facilitating an increased proliferation. In this context, whether the brain cancers with MGMT promoter methylation are less invasive and aggressive is an important question to explore. Any role of MGMT in modulating the CDK activities, DNA synthesis, and cell cycle maintenance needs to be deciphered as well. Since MGMT inhibitors are readily available, such an understanding may help to create replication stress in gliomas and other cancers. On the therapy front, such a replication distress could be potentially exploited for improved antitumor efficacy by combining the MGMT inhibitors with antimetabolites and other drugs. Therefore, MGMT can be considered as a target not only for alkylating agents but also other anticancer drugs. Furthermore, we have shown that MGMT is inducible to some extent by certain cysteine prodrugs and phytochemicals [[54], [55]]; whether our current findings bear any significance for MGMT-driven cancer prevention strategies also merits attention.
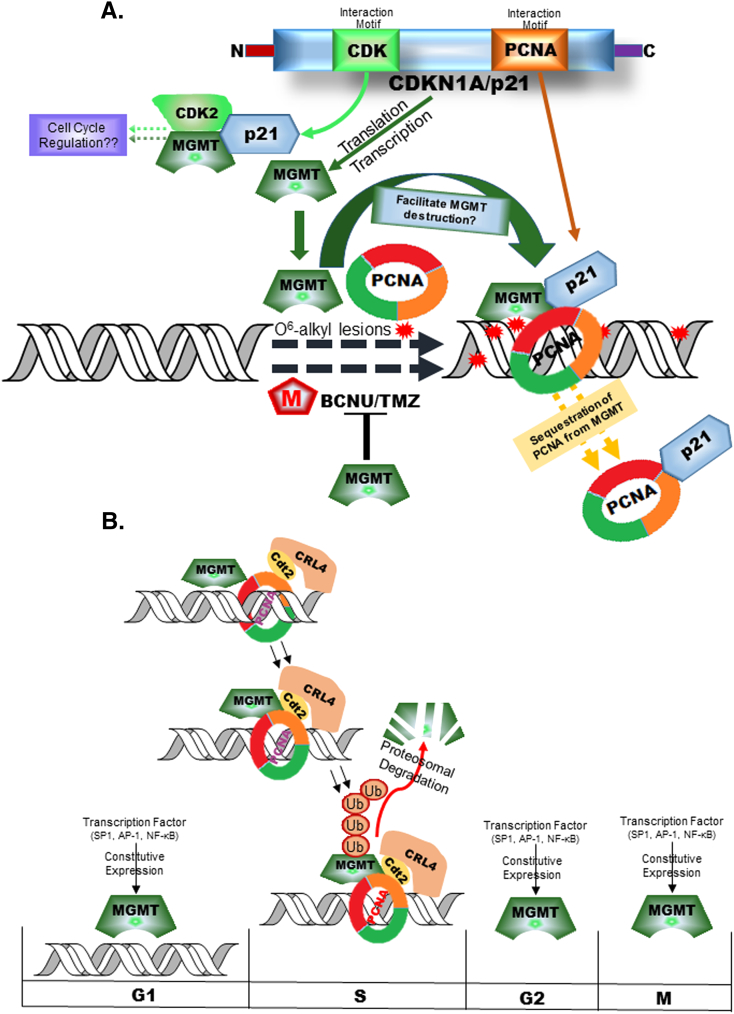
The scheme presented in Figure 7 summarizes our salient findings. p21cip1 is a critical protein with binding domains for the CDKs and PCNA. Besides inhibiting the CDKs, p21 also functions to sequestrate PCNA during cell cycle arrest and thereby restrict the DNA synthesis. Our results for the first time indicate that p21 has multiple effects on MGMT, all working to enhance its DNA repair activity to remove the mutagenic damage; while being a positive regulator of MGMT expression, it reduces the PCNA-binding with MGMT through a competition. Such interference may attenuate the destruction of MGMT by preventing its ubiquitination by CRL4Cdt2, an enzyme that couples ubiquitination to DNA-bound PCNA. Through these mechanisms, an elevated p21 expression, particularly, in response to DNA damage and during cell cycle blockade, while facilitating the removal of alkylation damage, appears to confer resistance to anticancer alkylating agents. Because MGMT associates with p21 and the CDKs (Figure 2), it may have additional roles in the cell cycle. Understanding these fundamental regulatory networks that hugely influence the biochemistry of MGMT is critical to its role in both oncogenesis and drug resistance. Such knowledge is likely to provide new MGMT-targeted therapeutic strategies for treatment of gliomas and other cancers.
Figure 7.
Schematic of the regulatory role of p21cip1 on MGMT expression, MGMT-PCNA interaction and tumor drug resistance. MGMT is a constitutively expressed repair protein thought to be active throughout the cell cycle. The transcription factors known to be involved in MGMT expression are shown in panel B. Our findings showed that p21 (which harbors binding domains for both CDKs and PCNA) is a positive regulator of MGMT expression enhancing its transcription and protein levels, and in turn, the glioma resistance to O6-alkylguanine-generating anticancer drugs (Panel A). MGMT was also found to associate with PCNA in a process that led to its elimination by the CRL4Cdt2, an S-phase-specific ub-ligase. The ability to bind PCNA away from MGMT by p21 may contribute to an increased stability and content of MGMT protein in tumor cells, as observed in this study.
Acknowledgements
This study was supported by grants RP130266 and RP170207 from the Cancer Prevention and Research Institute of Texas (CPRIT) and funding from the Carson Leslie Research Grants for Pediatric Brain Cancers. We are grateful to Drs. Francis Ali-Osman (Duke University) and Bert Vogelstein (John’s Hopkins University) for providing the tumor cell lines.
References
- 1.Mitra S, Kaina B. Regulation of repair of alkylation damage in mammalian genomes. Prog Nucleic Acid Res Mol Biol. 1993;44:109–142. doi: 10.1016/s0079-6603(08)60218-4. [DOI] [PubMed] [Google Scholar]
- 2.Pegg AE. Repair of O(6)-alkylguanine by alkyltransferases. Mutat Res. 2000;462:83–100. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- 3.Pegg AE. Multifaceted roles of alkyltransferase and related proteins in DNA repair, DNA damage, resistance to chemotherapy, and research tools. Chem Res Toxicol. 2011;24:618–639. doi: 10.1021/tx200031q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srivenugopal KS, Yuan XH, Friedman HS, Ali-Osman F. Ubiquitination-dependent proteolysis of O6-methylguanine-DNA methyltransferase in human and murine tumor cells following inactivation with O6-benzylguanine or 1,3-bis(2-chloroethyl)-1-nitrosourea. Biochemistry. 1996;35:1328–1334. doi: 10.1021/bi9518205. [DOI] [PubMed] [Google Scholar]
- 5.Xu-Welliver M, Pegg AE. Degradation of the alkylated form of the DNA repair protein, O(6)-alkylguanine-DNA alkyltransferase. Carcinogenesis. 2002;23:823–830. doi: 10.1093/carcin/23.5.823. [DOI] [PubMed] [Google Scholar]
- 6.Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4:296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- 7.Paranjpe A, Zhang R, Ali-Osman F, Bobustuc GC, Srivenugopal KS. Disulfiram is a direct and potent inhibitor of human O6-methylguanine-DNA methyltransferase (MGMT) in brain tumor cells and mouse brain and markedly increases the alkylating DNA damage. Carcinogenesis. 2014;35:692–702. doi: 10.1093/carcin/bgt366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolan ME, Pegg AE. O6-benzylguanine and its role in chemotherapy. Clin Cancer Res. 1997;3:837–847. [PubMed] [Google Scholar]
- 9.Srivenugopal KS, Mullapudi SR, Shou J, Hazra TK, Ali-Osman F. Protein phosphorylation is a regulatory mechanism for O6-alkylguanine-DNA alkyltransferase in human brain tumor cells. Cancer Res. 2000;60:282–287. [PubMed] [Google Scholar]
- 10.Velu CS, Niture SK, Bailey N, Srivenugopal KS. Posttranslational regulation of human MGMT by sumoylation in brain tumor cells. Proc Amer Assoc Cancer Res. 2004;45:137. [Google Scholar]
- 11.Srivenugopal KS, Rawat A, Niture SK, Paranjpe A, Velu C, Venugopal SN, Madala HR, Basak D, Punganuru SR. Posttranslational regulation of O(6)-methylguanine-DNA methyltransferase (MGMT) and new opportunities for treatment of brain cancers. Mini Rev Med Chem. 2016;16:455–464. doi: 10.2174/1389557515666150722101046. [DOI] [PubMed] [Google Scholar]
- 12.Niture SK, Doneanu CE, Velu CS, Bailey NI, Srivenugopal KS. Proteomic analysis of human O6-methylguanine-DNA methyltransferase by affinity chromatography and tandem mass spectrometry. Biochem Biophys Res Commun. 2005;337:1176–1184. doi: 10.1016/j.bbrc.2005.09.177. [DOI] [PubMed] [Google Scholar]
- 13.Candib A. Thesis submitted to McGill University. 2016. Characterization and validation of MGMT-binding partners using a proteomic-based approach in glioblastoma; pp. 1–96. [ http://digitool.library.mcgill.ca/webclient/StreamGate?folder_id=0&dvs=1510014967660~570] [Google Scholar]
- 14.Paranjpe A, Bailey NI, Konduri S, Bobustuc GC, Ali-Osman F, Yusuf MA, Punganuru SR, Madala HR, Basak D, Mostofa A. New insights into estrogenic regulation of O6-methylguanine DNA-methyltransferase (MGMT) in human breast cancer cells: Co-degradation of ER-alpha and MGMT proteins by fulvestrant or O6-benzylguanine indicates fresh avenues for therapy. J Biomed Res. 2016;30:393–410. doi: 10.7555/JBR.30.20160040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teo AK, Oh HK, Ali RB, Li BF. The modified human DNA repair enzyme O(6)-methylguanine-DNA methyltransferase is a negative regulator of estrogen receptor-mediated transcription upon alkylation DNA damage. Mol Cell Biol. 2001;21:7105–7114. doi: 10.1128/MCB.21.20.7105-7114.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srivenugopal KS, Niture SK, Velu CS, Bailey N, Doneanu C. Interaction of human MGMT with the sliding clamp PCNA and DNA polymerase delta: new regulatory modes for repair of alkylation DNA damage? Proc Amer Assoc Cancer Res. 2007;48:954. [Google Scholar]
- 17.Srivenugopal KS, Bailey N, Velu CS, Niture SK. The CDK inhibitor p21waf1/cip1 regulates human MGMT repair protein by two distinct and complementary mechanisms to increase tumor resistance to alkylating agents. Proc Amer Assoc Cancer Res. 2004;45:185. [Google Scholar]
- 18.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J Cell Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 19.Boehm EM, Gildenberg MS, Washington MT. The Many Roles of PCNA in Eukaryotic DNA Replication. Enzymes. 2016;39:231–254. doi: 10.1016/bs.enz.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gary R, Ludwig DL, Cornelius HL, MacInnes MA, Park MS. The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA-binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21. J Biol Chem. 1997;272:24522–24529. doi: 10.1074/jbc.272.39.24522. [DOI] [PubMed] [Google Scholar]
- 21.Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. Structure of the c-terminal region of p21waf1/cip1 complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 22.Frouin I, Maga G, Denegri M, Riva F, Savio M, Spadari S, Prosperi E, Scovassi AI. Human proliferating cell nuclear antigen, poly(ADP-ribose) polymerase-1, and p21waf1/cip1. A dynamic exchange of partners. J Biol Chem. 2003;278:39265–39268. doi: 10.1074/jbc.C300098200. [DOI] [PubMed] [Google Scholar]
- 23.Perucca P, Cazzalini O, Mortusewicz O, Necchi D, Savio M, Nardo T, Stivala LA, Leonhardt H, Cardoso MC, Prosperi E. Spatiotemporal dynamics of p21CDKN1A protein recruitment to DNA-damage sites and interaction with proliferating cell nuclear antigen. J Cell Sci. 2006;119:1517–1527. doi: 10.1242/jcs.02868. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi A, Suenaga N, Shiomi Y, Nishitani H. PCNA-dependent ubiquitination of Cdt1 and p21 in mammalian cells. Methods Mol Biol. 2014;1170:367–382. doi: 10.1007/978-1-4939-0888-2_19. [DOI] [PubMed] [Google Scholar]
- 25.Coleman KE, Grant GD, Haggerty RA, Brantley K, Shibata E, Workman BD, Dutta A, Varma D, Purvis JE, Cook JG. Sequential replication-coupled destruction at G1/S ensures genome stability. Genes Dev. 2015;29:1734–1746. doi: 10.1101/gad.263731.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibata E, Dar A, Dutta A. CRL4Cdt2 E3 ubiquitin ligase and proliferating cell nuclear antigen (PCNA) cooperate to degrade thymine DNA glycosylase in S phase. J Biol Chem. 2014;289:23056–23064. doi: 10.1074/jbc.M114.574210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Todd MC, Langan TA, Sclafani RA. Doxycycline-regulated p16MTS1 expression suppresses the anchorage-independence and tumorigenicity of breast cancer cell lines that lack endogenous p16. J Cancer. 2017;8:190–199. doi: 10.7150/jca.15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 30.Ali-Osman F, Rairkar A, Young P. Formation and repair of 1,3-bis-(2-chloroethyl)-1-nitrosourea and cisplatin-induced total genomic DNA interstrand crosslinks in human glioma cells. Cancer Biochem Biophys. 1995;14:231–241. [PubMed] [Google Scholar]
- 31.Harper JV. Synchronization of cell populations in G1/S and G2/M phases of the cell cycle. Methods Mol Biol. 2005;296:157–166. doi: 10.1385/1-59259-857-9:157. [DOI] [PubMed] [Google Scholar]
- 32.Kubota S, Fukumoto Y, Ishibashi K, Soeda S, Kubota S, Yuki R, Nakayama Y, Aoyama K, Yamaguchi N, Yamaguchi N. Activation of the pre-replication complex is blocked by mimosine through reactive oxygen species-activated ATM without DNA damage. J Biol Chem. 2014;289:5730–5746. doi: 10.1074/jbc.M113.546655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rizzardi LF, Coleman KE, Varma D, Matson JP, Oh S, Cook JG. CDK1-dependent inhibition of the E3 ubiquitin ligase CRL4CDT2 ensures robust transition from S Phase to Mitosis. J Biol Chem. 2015;290:556–567. doi: 10.1074/jbc.M114.614701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y, Li Q, Chen XZ. Detecting protein-protein interactions by Far western blotting. Nat Protoc. 2007;2:3278–3284. doi: 10.1038/nprot.2007.459. [DOI] [PubMed] [Google Scholar]
- 35.Daniels DS, Tainer JA. Conserved structural motifs governing the stoichiometric repair of alkylated DNA by O(6)-alkylguanine-DNA alkyltransferase. Mutat Res. 2000;460:151–163. doi: 10.1016/s0921-8777(00)00024-0. [DOI] [PubMed] [Google Scholar]
- 36.Cazzalini O, Perucca P, Savio M, Necchi D, Bianchi L, Stivala LA, Ducommun B, Scovassi AI, Prosperi E. Interaction of p21(CDKN1A) with PCNA regulates the histone acetyltransferase activity of p300 in nucleotide excision repair. Nucleic Acids Res. 2008;36:1713–1722. doi: 10.1093/nar/gkn014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cazzalini O, Scovassi AI, Savio M, Stivala LA, Prosperi E. Multiple roles of the cell cycle inhibitor p21(CDKN1A) in the DNA damage response. Mutat Res. 2010;704:12–20. doi: 10.1016/j.mrrev.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 38.Wilds CJ, Xu F, Noronha AM. Synthesis and characterization of DNA containing an N1-2'-deoxyinosine-ethyl-N3-thymidine interstrand cross-link: a structural mimic of the cross-link formed by 1,3-bis-(2-chloroethyl)-1-nitrosourea. Chem Res Toxicol. 2008;21:686–695. doi: 10.1021/tx700422h. [DOI] [PubMed] [Google Scholar]
- 39.D'Atri S, Graziani G, Lacal PM, Nisticò V, Gilberti S, Faraoni I, Watson AJ, Bonmassar E, Margison GP. Attenuation of O(6)-methylguanine-DNA methyltransferase activity and mRNA levels by cisplatin and temozolomide in Jurkat cells. J Pharmacol Exp Ther. 2000;294:664–671. [PubMed] [Google Scholar]
- 40.Tilby MJ, Lawley PD, Farmer PB. Alkylation of DNA by melphalan in relation to immunoassay of melphalan–DNA adducts: characterization of mono-alkylated and cross-linked products from reaction of melphalan with dGMP and GMP. Chem Biol Interact. 1990;73:183–194. doi: 10.1016/0009-2797(90)90002-5. [DOI] [PubMed] [Google Scholar]
- 41.Mjelle R, Hegre SA, Aas PA, Slupphaug G, Drablos F, Saetrom P, Krokan HE. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair (Amst) 2015;30:53–67. doi: 10.1016/j.dnarep.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 42.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 43.Abbas T, Dutta A. CRL4Cdt2: master coordinator of cell cycle progression and genome stability. Cell Cycle. 2011;10:241–249. doi: 10.4161/cc.10.2.14530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jorgensen S, Eskildsen M, Fugger K, Hansen L, Larsen MS, Kousholt AN, Syljuasen RG, Trelle MB, Jensen ON, Helin K. SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation. J Cell Biol. 2011;192:43–54. doi: 10.1083/jcb.201009076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Senga T, Sivaprasad U, Zhu W, Park JH, Arias EE, Walter JC, Dutta A. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J Biol Chem. 2006;281:6246–6252. doi: 10.1074/jbc.M512705200. [DOI] [PubMed] [Google Scholar]
- 46.Prives C, Gottifredi V. The p21 and PCNA partnership: a new twist for an old plot. Cell Cycle. 2008;7:3840–3846. doi: 10.4161/cc.7.24.7243. [DOI] [PubMed] [Google Scholar]
- 47.Waga S, Hannon GJ, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 48.Chuang L.S., Ian H.I., Koh T.W., Ng H.H., Xu G., Li B.F. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 49.Matsumoto Y. Molecular mechanism of PCNA-dependent base excision repair. Prog Nucleic Acid Res Mol Biol. 2001;68:129–138. doi: 10.1016/s0079-6603(01)68095-4. [DOI] [PubMed] [Google Scholar]
- 50.Bhakat KK, Mitra S. Regulation of the human O(6)-methylguanine-DNA methyltransferase gene by transcriptional coactivators cAMP response element-binding protein-binding protein and p300. J Biol Chem. 2000;275:34197–34204. doi: 10.1074/jbc.M005447200. [DOI] [PubMed] [Google Scholar]
- 51.Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, Williams J, Lengauer C, Kinzler KW, Vogelstein B. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Havens CG, Walter JC. Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev. 2011;25:1568–1582. doi: 10.1101/gad.2068611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pozo PN, Cook JG. Regulation and function of Cdt1; A key factor in cell proliferation and genome stability. Genes (Basel) 2016;8(1) doi: 10.3390/genes8010002. [pii:E2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niture SK, Velu CS, Smith QR, Bhat GJ, Srivenugopal KS. Increased expression of the MGMT repair protein mediated by cysteine prodrugs and chemopreventative natural products in human lymphocytes and tumor cell lines. Carcinogenesis. 2007;28:378–389. doi: 10.1093/carcin/bgl155. [DOI] [PubMed] [Google Scholar]
- 55.Niture SK, Rao US, Srivenugopal KS. Chemopreventative strategies targeting the MGMT repair protein: augmented expression in human lymphocytes and tumor cells by ethanolic and aqueous extracts of several Indian medicinal plants. Int J Oncol. 2006;29:1269–1278. [PubMed] [Google Scholar]