Abstract
The diploid annual legume Medicago truncatula has been developed as a tractable genetic system for studying biological questions that are unique to, or well suited for study in legume species. An efficient mutagenesis protocol using ethyl-methyl sulfonate and a polymorphic ecotype with properties appropriate for use as a mapping parent are described. Isolation and characterization of three developmental mutants are described. The mtapetala mutation results in homeotic conversions of floral organ whorls 2 and 3 into sepals and carpelloid structures, respectively, similar to mutations in the apetala3/pistillata genes of Arabidopsis. The palmyra mutation primarily affects seedling shoot meristem initiation, and thus phenocopies meristem function mutations identified in Arabidopsis such as the zwille locus. The phenotype of the palmyra and mtapetala double mutant is additive, with seedling shoot meristems and floral organs indistinguishable from those of the single palmyra and mtapetala mutants, respectively. These results are consistent with a lack of genetic interaction between these loci. A third mutant, speckle, is characterized by spontaneous necrotic lesion formation on leaves, root, and stems, similar to necrosis mutants identified in other plant species. In addition to documenting the efficient mutagenesis of M. truncatula, the availability of developmental mutants that phenocopy characterized Arabidopsis mutants will provide a basis for establishing orthologous gene function between M. truncatula and Arabidopsis, once the genes responsible are cloned. Moreover, the male-sterile, female-fertile nature of the mtapetala mutant provides a convenient tool for genetic analyses in M. truncatula.
The molecular dissection of plant growth and development in model plants such as Arabidopsis has largely relied on genetic approaches involving the isolation and subsequent characterization of mutants affected in specific biological processes. The utility of this approach now encompasses virtually every facet of plant biology, beginning from plant embryogenesis to flowering and senescence, and plant-pathogen interactions.
Despite the general validity of Arabidopsis as a model plant species, several biological processes do not occur in Arabidopsis and are better studied in other species. Examples of such species-processes combinations include tomato for climacteric fruit ripening, maize for regulation of seed storage protein biosynthesis, and legumes for study of plant symbiotic interactions with nitrogen-fixing bacteria (collectively termed rhizobia) and fungal symbionts (collectively termed mycorrhizae).
In contrast to the large impact plant mutants have had on a molecular dissection of plant development and physiology in maize and Arabidopsis, legume mutants have had a relatively minor impact on our understanding of the legume-rhizobial symbiosis. Although numerous nodulation mutants are available in the economically important legumes such as pea, soybean, and alfalfa (for review, see Caetano-Anolles and Gresshoff, 1991), molecular genetic analyses in these species are difficult due to inefficient transformation and regeneration (pea, soybean), large genome size (pea, soybean), and polyploidy (soybean, alfalfa). Diploid autogamous legumes with small genomes that can be efficiently transformed and regenerated, such as Lotus japonicus and Medicago truncatula have thus emerged as model legumes (Cook et al., 1997, 1999; Schauser et al., 1998, 1999). In particular, M. truncatula is amenable to highly efficient vacuum infiltration genetic transformation techniques (Trieu et al., 2000).
Mutagenesis efforts in these genetically tractable legumes will be an important means in identifying host genes required for symbiotic function. In M. truncatula mutagenesis efforts have led to the isolation of several symbiotic mutants including a single ethyl-methyl sulfonate (EMS)-induced mutant (Benaben et al., 1995), and a set of 18 γ-ray-induced mutants (Sagan et al., 1995), whereas in Lotus japonicus, a set of 20 mutants representing 14 complementation groups with altered symbiotic phenotypes has been recently described (Szczyglowski et al., 1998). Complementation tests with pea nodulation mutants have identified over 30 loci with nodulation phenotypes (Weeden et al., 1990; Sagan et al., 1995). By analogy to pea, mutagenesis efforts in M. truncatula are likely to have produced mutants in only a subset of the nodulation loci, implying that more loci and a range of mutant alleles at each locus remain to be identified.
In an effort to develop an efficient mutagenesis method for M. truncatula and to isolate additional nodulation mutants, we undertook a large-scale mutagenesis program. After considering ease of use and the diversity of mutations possible, EMS, a base alkylating agent that generates point mutations (most commonly, G to A transitions), was selected as the mutagen. Point mutagens such as EMS typically produce a broader range of mutant alleles including loss-of-function, gain-of-function, altered function, and novel function mutations when compared with deletion and insertional mutagens that provide primarily loss-of-function mutants.
In this report we describe the results of our mutagenesis, and as evidence of the efficacy of the mutagenesis, we describe the isolation and characterization of three developmental mutants that affect diverse developmental processes.
RESULTS
Development of the Mutagenized Population: Optimizing EMS Dose
Our initial goal was to generate a highly mutagenized population of M. truncatula as a resource for identification of plant nodulation mutants. Based on EMS dosages commonly used for mutagenesis in plants, we selected EMS concentrations ranging from 0.025% to 2.025% and quantified putative somatic and germinal effects; the objective of this analysis was to maximize EMS dose while retaining a high level of gametic transmission to the subsequent (M2) generation. Thus, we assayed seedling survival, frequency of fertile individuals, and seed production in treated seed (M1) as a measure reproductive capacity, and we assayed the frequency of arrested embryos in the M2 population as a measure of mutagenicity. The analysis was conducted on six lots of 500 seeds each, treated in parallel with different EMS concentrations. As shown in Table I, fertility of treated individuals became a limiting factor at 0.225% EMS despite a seedling survival rate of near 40%. Lower doses of EMS had a relatively small effect on seedling survival and the frequency of fertile individuals, although fecundity (seed per pod) decreased significantly as EMS dose increased from 0.025% to 0.150%. The frequency of arrested embryos was also correlated with EMS dose, increasing roughly 6-fold from 0.025% to 0.15% EMS. EMS dose did not significantly affect seed size or weight.
Table I.
Somatic and germinal effects of EMS dose in M. truncatula A17
| EMS | Seedling Survivala | Fertile Plants | Seed/Podb | Seed Weightb | Arrested Embryosc |
|---|---|---|---|---|---|
| % | mg | ||||
| 0 | 69 ± 5.9 | 100 | 9.7 ± 0.07 | 3.8 ± 0.07 | 0 |
| 0.025 | 68 ± 11.9 | 100 | 8.6 ± 0.38 | 3.5 ± 0.12 | 1.1 ± 0.2 |
| (6.7 ± 2.8) | |||||
| 0.075 | 64 ± 5.7 | >95 | 4.6 ± 0.47 | 4.1 ± 0.03 | 8.4 ± 1.8 |
| (27 ± 10.2) | |||||
| 0.150 | 71 ± 13.9 | >95 | 2.3 ± 0.15 | 3.7 ± 0.20 | 21.0 ± 2.2 |
| (42.0 ± 2.6) | |||||
| 0.225 | 40 ± 6.4 | <2 | ndd | nd | nd |
| 0.675 | 0 | 0 | nd | nd | nd |
| 2.025 | 0 | 0 | nd | nd | nd |
Seedling survival was scored as the emergence of the first trifoliate.
Seed/pod and seed wt were scored individually on >120 randomly selected pod for each EMS treatment.
The frequency of arrested embryos was assessed in four groups of =30 seed pod each. Data are expressed both as the percentage of seed pod containing at least one arrested embryo and as the percentage of total seed analyzed (in parentheses).
nd, Values not determined due the absence of fertile individuals.
Based on results of the preceding analysis we selected EMS concentrations of 0.1% and 0.15% for large scale seed treatment. Typically 5,000 to 10,000 seeds were treated simultaneously and subdivided into lots of 500 seeds each for growth and seed collection. Seeds were planted in bulk rather than as individuals to circumvent the need to handle such a large number of individuals. Seed bulks derived from individual lots represented unique mutant populations and comprised the basic unit used in subsequent mutant screens. To assess the efficacy of EMS treatment on large seed lots, we analyzed seedling survival, plant fecundity, and the frequency of arrested embryos in a single M2 seed bulk (bulk C, 0.15% EMS treatment) derived from 354 M1 individuals. The values obtained for seedling survival, seed weight, seed per pod, and fertility were consistent with our previous dose-response data presented in Table I. In particular, the frequency of arrested embryos (a measure of mutagen efficacy) was over 20% of M2 assayed and was the highest of all EMS concentrations tested without significantly diminished fecundity (Table I). As shown in Table II, altered pigmentation phenotypes were observed at high frequencies for seeds (17.5 ± 3.2) and seedlings (5.8 ± 3.1) in the bulk C population. Taken together these data indicate that, under these treatment conditions, 0.15% EMS is probably close to the maximum effective dose of EMS on M. truncatula.
Table II.
Frequency of selected seed and seedling phenotypes in M2 of 0.15% EMS-treated seed
| Seed Phenotypes
|
Seedling Pigmentationc
|
||||
|---|---|---|---|---|---|
| Tan seed coata | Stay greenb | Albino | Chlorotic | Pale green | Total seedling pigmentation |
| 17.5 ± 3.2 | 0.20 ± 0.03 | 0.4 ± 0.7 | 3.5 ± 1.8 | 2.0 ± 1.7 | 5.8 ± 3.1 |
Tan seed coat phenotypes were scored on seed obtained from individual pods representing four groups of >30 pod each group. Data are expressed as the percentage of seed pod containing at least one scorable phenotype.
Stay green phenotypes were scored on three samples of =4,000 seed each.
Pigmentation phenotypes were scored on 7-d-old aeroponically grown seedlings. A total of 17 seed pools were analyzed, each derived from 40 randomly collected seed pods, with an average of 70 ± 9.4 germinated individuals/pool.
Plant embryos typically contain two to three cells that serve as germline precursors (Li and Redei, 1969; Carroll et al., 1988); therefore, each M1 individual can potentially give rise to two to three unique cell lineages, each carrying a distinct set of mutant alleles. As most of our 0.15% EMS-treated seed bulks were derived from an average of 280 surviving M1 individuals, the corresponding M2 populations should contain on average 840 unique lineages. The level of confidence for recovery of a given recessive phenotype from a bulk comprised of 840 unique M1 lineages is 76% if 4,200 M2 individuals are screened, 94% if 8,400 M2 individuals are screened, and >99% if 16,800 M2 individuals are screened. In subsequent screens of 0.15% EMS-treated seed bulks, we examined approximately 3,000 to 6,000 individuals per bulk for specific phenotypic classes.
Identification of a Male-Sterile Floral Homeotic Mutant
Although cross hybridization in M. truncatula is relatively simple, we sought to obtain male-sterile, female-fertile lines for situations where unambiguous cross pollination was desirable; for example, when crossing phenotypically similar individuals that may represent an allelic series. Three thousand seeds of the 0.15% EMS-treated bulk C were scarified for germination and distributed among six flats. As the resulting plants matured, those plants that set seeds were removed from the flat and discarded. Through this process of elimination, 24 putative sterile mutants were identified. As shown in Figure 1, a and b, four of these individuals shared a common altered floral morphology, with sepaloid structures in place of petals and presumed carpeloid structures in place of anthers. Despite these homeotic organ transformations, organ numbers in each whorl (5 in whorl 1, 5 in whorl 2, 10 in whorl 3, and 1 in whorl 4) in the mutant were identical to those in wild type (data not shown). These four individuals (presumably siblings) were male-sterile, but readily developed viable seeds when outcrossed. Molecular proof of cross pollination was obtained by crossing mutant flowers (derived from genotype A17) with the polymorphic M. truncatula ecotype A48. As shown in Figure 1c, six F1 progeny that were randomly selected were all heterozygous for random-amplified polymorphic DNA (RAPD) markers that distinguished the two parental genotypes. Similar results have been obtained with other polymorphic ecotypes of M. truncatula, including ecotypes A20 and A68 (data not shown), and in all cases F1 plants were fertile and produced viable and fertile F2 progeny. In backcross experiments to the wild-type parent A17, F1 progeny produced fertile flowers with normal morphology, whereas the F2 progeny segregated for the floral homeotic phenotype as expected for a single recessive allele (Table III). We have named the corresponding gene tap, for M. truncatula apetala.
Figure 1.
Floral phenotype of the homeotic mutant mtapetala and molecular evidence of artificial hybridization. a and b, Flowers of a similar developmental stage from genotypes mtapetala (a) and A17 (b). Numbers refer to floral whorl number beginning with the outermost whorl that form in wild-type flowers: 1, sepals; 2, petals; 3, stamens; and 4, carpels. c, RAPD analysis of progeny of a cross into mtapetala confirms cross-hybridization. Lane M, Mr marker; 1, parental ecotype A17 (parent 1); 2, ecotype A48 (parent 2); 3 to 8, six independent F1 progeny. Primer 1, “gaacggactc”; primer 2, “tggaccggtg.” Bar = 1 mm.
Table III.
Genetic analysis of M. truncatula developmental mutants and seedling albinism
| Cross | Total Progeny Analyzed | Observed
|
χ2 Value | P Value | No. of Genes and Inferred Nature of Dominance | |
|---|---|---|---|---|---|---|
| Wild type | Mutant | |||||
| tap × A17 | 134 | 102 | 32 | 0.090 | >0.750 | 1 Gene, recessive |
| plm × A17 | 187 | 145 | 42 | 0.644 | >0.400 | 1 Gene, recessive |
| spk × A17 | 141 | 114 | 27 | 2.577 | >0.100 | 1 Gene, recessive |
| albino × A20a | 495 | 458 | 37 | 1.267 | >0.250 | 2 Genes, recessivea |
Tested with F2 ratios of 15:1, wild type:mutant expected for duplicate dominant epistasis.
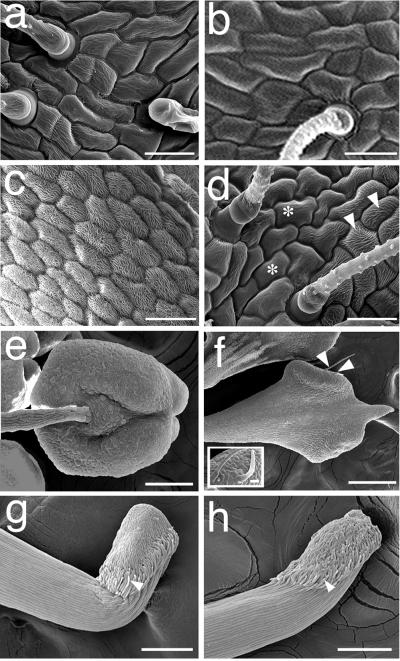
Similar floral homeotic mutants have been obtained and the responsible genes have been cloned in species such as Arabidopsis, Antirrhinum majus, and petunia (for review, see Theisen and Saedler, 1999). For these cases transformed organs typically show a corresponding conversion of epidermal cell morphology, with the extent of conversion varying according to the strength of the mutant alleles. To determine if tap was also characterized by conversion of epidermal cell morphology, we analyzed transformed and wild-type organs by scanning electron microscopy. The results shown in Figure 2, a, b, g, and h indicate that cell morphology of whorls 1 and 4 in tap were identical to those found in wild type, consistent with the absence of macroscopic transformations in these organs. As shown in Figure 1a, whorl 2 in tap has macroscopic features typical of wild-type whorl 1 sepals, including the replacement of yellow pigmentation by green chloroplast bearing cells and the presence of trichomes that are characteristic of wild-type sepals, but not wild-type petals (Fig. 2, a–c). By contrast, the epidermal cell morphology of whorl 2 in tap is a mosaic of sectors of whorl 1 and whorl 2 cell identities. Transition of cell morphologies across sectors is gradual as evidenced by cells of varying levels of intermediate morphology (compare Fig. 2, c and d with Fig. 2, a and b). Furthermore, unlike the uniform organ shape of whorl 1 in both tap and wild-type flowers, organs in whorl 2 of tap are varied in shape, similar to the distinct petal shapes that are characteristic of flowers of the subfamily Papilionoideae, including M. truncatula (data not shown). In wild-type flowers whorl 3 organs are composed of orbicular anthers that hinge on the subtending stamen filaments, with distinct cell morphologies characterizing the stamen, filament, and the stamen/filament junction (Fig. 2e). In contrast whorl 3 organs in tap are characterized by filaments that radiate gradually into club-shaped structures and are devoid of the sharp transitions in cell morphology from the filament to the terminal region (Fig. 2f). Moreover, whorl 3 organs in tap, but not in wild type, bear hair-like structures (arrow, Fig. 2f, and inset), which are similar to stigmatic papillae that occur at the stigma/style junction in whorl 4 of wild type and tap (Fig. 2, g and h, respectively). Taken together these data suggest that the tap mutation results in a moderate transformation of whorls 2 and 3, whereas whorls 1 and 4 are unaffected.
Figure 2.
Morphology of floral organs in wild-type A17 and in floral homeotic mutant mtapetala. Scanning electron micrographs of whorl 1/sepal (a and b); whorl 2/petal (c and d); whorl 3/stamen (e and f); and whorl 4/carpel (g and h). Whorl 2 in the tap mutant is a partial transformation of whorl 2 identity and consists of cellular morphologies that are diagnostic of whorl 1/sepals (asterisk), and whorl 2/petals (arrowheads) of wild-type flowers. Whorl 3 in tap is a partial transformation of organ shape into a club shaped structure (Fig. 2f), and these organs are devoid of the sharp transition in cell morphologies found in wild-type whorl 3 (Fig. 2e). Moreover, whorl 3 in tap contains stigmatic papillae (arrowheads, Fig. 3f, and inset) that occur at the stigma/style junction in whorl 4 of wild type and tap (arrowheads, Fig. 2, g and h, respectively). Bars in a through d = 25 μm; bars in e through h = 100 μm; bar in f inset = 10 μm.
Identification of a Shoot Meristem Initiation Mutant
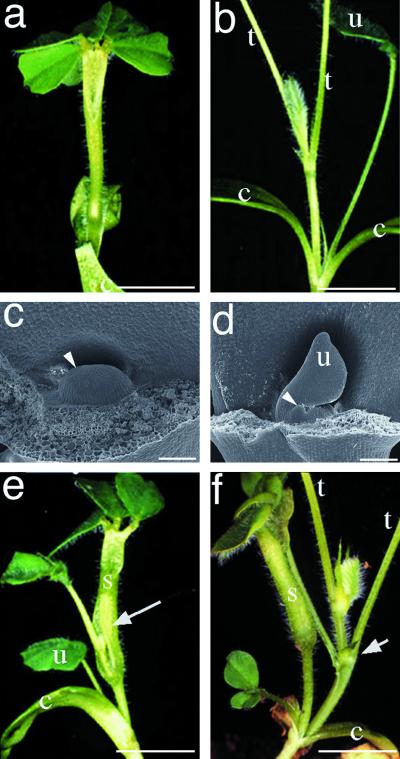
Another mutant that exhibited an aberrant shoot phenotype was selected for further study. This mutant was designated palmyra for its superficial resemblance to a palm tree-like shoot architecture (Fig. 3a). Specifically, new shoots are not produced by the seedling-derived apical meristem and normal shoot phyllotaxy is absent from palmyra seedlings. Instead the young palmyra shoot is crowned by a cluster of leaflets (Fig. 3a) and lacks both unifoliate and trifoliate leaves. Following a prolonged lag phase of 2 to 3 weeks, an adventitious shoot ruptures through the basal portion of the stem (Fig. 3e) or from the base of the cotyledons (Fig. 3f). Continued development from the adventitious meristem produces nearly normal shoot architecture and the development of apparently normal inflorescences, flowers, and fruit pods. Infrequently, vegetative shoot meristems in palmyra terminate growth, similar to the seedling apical meristem, with subsequent growth re-initiating from a subterminal leaf axillary meristem. Despite the gross effect on seedling shoot morphology, development of the cotyledons and the root system in palmyra are indistinguishable from wild type. To determine if the seedling shoot growth phenotype of palmyra might be correlated with an abnormal seedling apical meristem, we examined the morphology of apical meristems in mature embryos of palmyra and wild-type plants. The apical meristem of wild-type embryos has features typical of a normally functioning shoot apical meristem (Fig. 3d), including a smooth apical dome, flanking shoot primordia, and a unifoliate leaf. In contrast the apical meristem of the palymra mature embryo (Fig. 3c) is characterized by a smooth apical dome with an absence of lateral leaf primordia, and no unifoliate leaf (arrowhead, Fig. 3c). Thus the effects of the palmyra mutation are largely restricted to early vegetative shoot meristem function, but not post-embryonic development.
Figure 3.
Phenotypic analysis of shoot meristem formation in mutant palmyra. a and b, Shoot systems of 25-d-old seedlings of genotypes palmyra (a) and wild type (b). c and d, Morphological analysis of shoot meristem of mature embryos from palmyra (c) and wild type (d). The shoot meristem is undifferentiated in palmyra with no organ primordia evident (arrowhead, c). In comparison, wild-type shoot meristem is differentiated and contains leaf primordia (arrowhead, d). e and f, Adventitious shoot formation in palmyra from flank of stalk-like structure (e) and base of cotyledons (f) is marked by an arrow. c, Cotyledon; u, unifoliate leaf; t, trifoliate leaf petiole; s, stem-like shoot. Bars in a, b, e, and f = 5 mm; bars in c and d = 100 μm.
To determine the mode of inheritance of this mutation, pollen from palmyra plants was used to cross into the homozygous male sterile crossing line tap. F1 progeny derived from this cross were wild type in seedling shoot morphology, and the F2 progeny segregated 3:1 for wild type: palmyra phenotypes as expected for a monogenic, recessive mutant allele (Table III). We have designated the corresponding locus Plm and the recessive mutant allele plm. F2 palmyra homozygotes obtained from the above cross were grown to maturity to determine possible interactive effects of the palmyra mutation with the floral homeotic mutant mtapetala. Among the population of palmyra homozygotes, two floral phenotypes were observed. In the majority (approximately 75%) of the F2 progeny, flowers were identical to wild-type flowers. In the residual fraction (approximately 25%) of the F2 plants, flowers were indistinguishable from the parental tap homozygotes, corresponding to the plm/tap double mutant genotype. The recovery of only wild type and the parental mutant phenotypes in the F2 population is consistent with the absence of genetic interaction between these two developmental mutations.
Identification of a Spontaneous Pigmentation Mutant
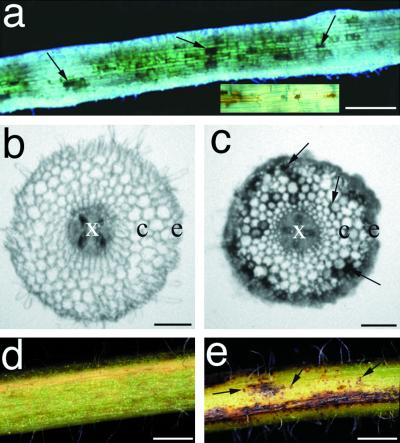
As a by-product of a visual screen for altered early root phenotypes (R.V. Penmetsa and D.R. Cook, unpublished data), we identified mutants exhibiting normal nodule morphogenesis, but were altered in root morphology and/or development. One such mutant was initially selected for further study based on brown pigmentation on the roots. Light microscopic examination of M2 and M3 individuals revealed that the macroscopically brown appearance of the root system was due to patches of dark brown pigmented cells. Pigmented cells were distributed sporadically along the entire length of the root system (Fig. 4a) and apparently in random manner in cortical and epidermal root layers (Fig. 4c). Furthermore, this spontaneous pigment formation also extended to the root-hypocotyl junction (crown), leaves, and stem (Fig. 4, e and f). All M3 progeny obtained from selfing the original mutant displayed a similar pigmentation phenotype, suggesting that the original mutant was homozygous for the mutant allele. To determine the genetic nature of this mutation, pollen from homozygotes M3 individuals were used for crosses into homozygous tap. The resulting F1 seedlings were wild type in root and shoot morphology, and F2 seedlings segregated 3:1 (wild type:mutant) for both root and shoot pigmentation phenotypes, as expected for a monogenic recessive mutation (Table III). We have designated this mutant speckle, and the corresponding gene spk.
Figure 4.
Phenotypic analysis of pigmentation mutant speckle. a, Light micrograph of aeroponically grown speckle roots. b and c, Transverse sections through roots of wild type (c) and speckle (d) prior to inoculation with Rhizobium. d and e, Light micrographs of stems from wild type (d) and speckle (e). Pigmented cells are sporadically distributed in the epidermis and cortex of speckle roots (arrows, Fig. 5, a and c) and stems (arrows, Fig. 5e). x, Xylem; e, epidermis; c, cortex. Bars in a = 250 μm; bars in c and d = 50 μm; bars in e and f = 1 mm.
Identification of a Polymorphic Ecotype for Use as Mapping Parent
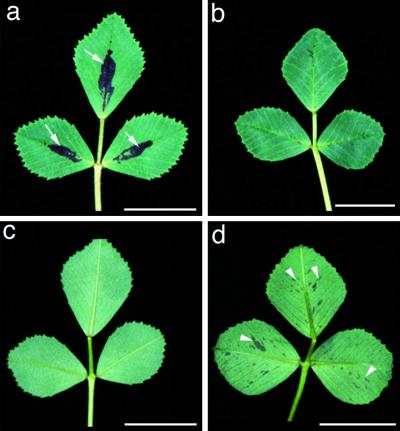
For genetic mapping and subsequent map based cloning of genes, we screened ecotypes of M. truncatula to identify genotypes that were both highly polymorphic and exhibited similar nodulation properties as ecotype A17, which was used as the parent in our mutagenesis experiments (data not shown). Based primarily on its nodulation kinetics with Sinorhizobium meliloti strains, ABS7 M, 1021, and 2011 (as compared with genotype A17), ecotype A20 was selected for further analysis. It is interesting that morphological traits that are characteristic of A20 such as a clockwise direction of pod coiling (versus anticlockwise in A17) and leaf spot pattern (Fig. 5) were inherited in a dominant manner in F1 individuals. The results of χ2 tests are consistent with separate, single genes conditioning leaf spot phenotypes in the two ecotypes, whereas the A17 and A20 loci behave as recessive and dominant genetic characters, respectively (P values > 0.85 and > 0.20, respectively). It is also interesting that the dominant nature of A20 leaf spot provides a simple means to distinguish hybrid F1 from selfed progeny when A20 is used as a male parent in crosses to A17.
Figure 5.
Pigmentation patterns on both the adaxial and abaxial leaf surfaces distinguish ecotypes A17 and A20. Typical leaf pigmentation found on ecotype A17 (a) and (c), and ecotype A20 (b) and (d). Adaxial leaf surfaces (a) and (b); abaxial leaf surfaces (c) and (d). Arrowheads denote the adaxial leaf spot of A17 (a), and the abaxial freckles of A20 (d). Bar = 1 cm.
Isolation of a M. truncatula ap3/pi-Like MADS-Domain-Containing Sequence
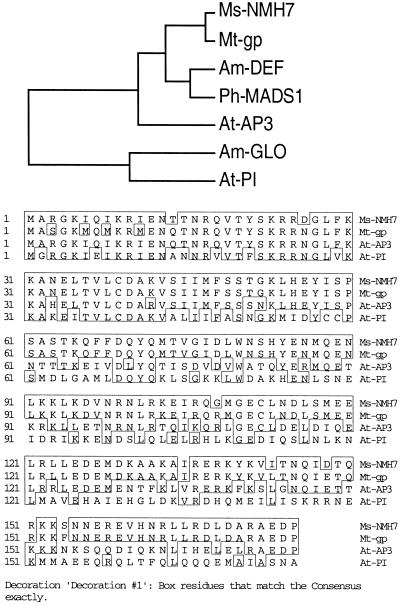
Developmental mutants in M. truncatula that strongly phenocopy mutant phenotypes where the responsible genes have been cloned in other plant species are prospects for a candidate gene approach to map-based cloning. The mtapetala locus phenotype of M. truncatula closely resembles class B function mutations that have been cloned and characterized in Arabidopsis, Antirrhinum majus, and petunia, including apetala3/deficiens/greenpetals and pistillata/globosa/fbp1 (Theisen and Saedler, 1999). To determine whether the tap locus might be orthologous to apetala3/deficiens in a cosegregation test, we isolated a cDNA clone (designated mtgp) from an immature flower cDNA library using degenerate primers designed against the apetala3-subfamily of MADS-box transcriptional activators. As shown in Figure 6a, mtgp is most similar to NMH7, a MADS-box cDNA of Medicago sativa (Heard and Dunn, 1995), with greater than 90% nucleotide and deduced-amino acid sequence identity to NMH7. Phylogenetic analysis of the mtgp with class B floral homeotic MADS-box genes places mtgp within the ap3/def subfamily, and distinct from the pi/glo subfamily (Fig. 6b), reflecting the higher (62%–74%) sequence similarity of mtgp to ap3/def, versus a lower (36%–37%) similarity to pi/glo. The high sequence similarity and presence of this transcript in developing flowers is consistent with the inference that mtgp is orthologous to ap3/def.
Figure 6.
Analysis of mtgp, a MADS-domain containing gene expressed in developing M. truncatula flowers. a, Phylogenetic analysis of mtgp with functionally characterized class B floral homeotic genes, places mtgp within the ap3/def/gp subgroup, and distinct from the pi/glo subgroup of class B floral homeotic MADS-box genes. b, Alignment of deduced amino acid sequences of mtgp with class B MADS-domain containing genes nmh7 of M. sativa, and ap3 and pi of Arabidopsis. For clarity, only sequences corresponding to the partial mtgp cDNA are shown.
To map mtgp, the cDNA clone was used as a probe to identify two overlapping clones from the M. truncatula bacteria artificial chromosome (BAC) library (Nam et al., 1999). Sequences from subclones of these BACs were used to design primers for PCR amplification and sequence characterization of the corresponding regions in the mapping parent A20. Sequence polymorphisms were converted into codominant PCR markers for mapping mtgp cDNA. In a F2 population segregating for the mtapetala mutant phenotype, the mtgp genetic marker did not cosegregate with the tap locus (data not shown).
DISCUSSION
The diploid small genome legume M. truncatula has emerged as a model plant species for the study of biological processes unique to or best studied in legumes (Cook et al., 1997, 1999). In M. truncatula in particular, several tools to facilitate molecular genetic analyses have been developed. Such tools include efficient transformation and regeneration protocols (Trieu and Harrison, 1996; Trieu et al., 2000), naturally occurring ecotypes (Bonnin et al., 1996), a large-insert BAC library (Nam et al., 1999), and a molecular marker genetic map (T. Huguet, personal communication; D.J. Kim and D.R. Cook, unpublished data).
Although the genome size of M. truncatula (450–500 million bp) is roughly 3 to 4 times that of Arabidopsis (100–150 million bp/1 C), it is unlikely that M. truncatula contains a correspondingly larger number of genes. Rather, a higher abundance of non-coding DNA in M. truncatula, relative to that in Arabidopsis, would seem more likely to account for this difference in DNA content. Consequently, at optimal mutagen dosage for each species, the frequencies of visible mutant phenotypes might be expected to be similar. To compare the efficacy of our mutagenesis with optimally mutagenized Arabidopsis we compared mutant frequencies for phenotypes measured in both species.
At the optimal EMS concentration of 0.15%, we observed embryonic lethality (21% ± 2.2%) and chlorophyll variants (5.8% ± 3.1%; Table II) at frequencies similar to those observed with optimally mutagenized Arabidopsis populations (Koornneef et al., 1982). This concordance of mutant frequencies for common phenotypes between these two species implies that our dose-response study has indeed identified a near optimal EMS dose for M. truncatula. Further corroborative evidence of the efficacy of our mutagenesis comes from the observation that screens of these bulks have yielded mutants with altered metal homeostasis phenotypes (M. Grusak, personal communication), mutants altered in leaf calcium oxalate content (P. Nakata, personal communication; M.A. Webb, personal communication), and many mutants with altered nodulation properties (R.V. Penmetsa and D.R. Cook, manuscript in preparation; J. Denarie, personal communication).
Identification of a Polymorphic Ecotype for Use as Mapping Parent
To facilitate genetic mapping and subsequent map-based cloning of genes defined by their mutant phenotypes, we sought to identify ecotypes that would be useful as mapping parents. Since the ultimate focus of our mutagenesis efforts was to identify genes required for nodulation, it was essential to identify ecotypes that met two criteria. First, a desirable mapping parent had to exhibit a high level of polymorphism when compared with our standard (wild-type) ecotype, A17. Second, the polymorphic ecotype had to possess nodulation characteristics similar to, and compatible with, genotype A17. Selection for this second criterion would minimize confounding effects of pre-existing natural variation in nodulation genes, and thereby permit the unambiguous assignment of phenotypes in populations segregating for the EMS-induced mutations. Ecotype A20 satisfied both criteria described above (data not shown), while the dominant leaf spot pattern provides a simple means to identify F1 progeny from crosses where A20 serves as the male parent.
Mtapetala as a Tool for Genetic Analysis
Although artificial hybridization by hand emasculation and pollination is relatively simple in M. truncatula (Pathipanawat et al., 1994), male-sterile, female-fertile lines would be useful tools for crossing in certain situations, for example when crossing phenotypically similar individuals that may represent an allelic series, or in test cross analyses. The phenotype of mtapetala, including homeotic conversion of petals to sepaloid organs and stamens to carpelloid organs, most closely resembles that of Arabidopsis mutants with mutations in the class B organ identity genes apetala3 and pistillata (Bowman et al., 1989). Our current data do not allow us to distinguish whether the moderate degree of homeotic transformation that is observed in tap is the result of a weak mutant allele caused by the point mutagen used to generate the mutant, or the phenotype of a complete loss-of-function of the tap gene. Nonetheless, the effects of the mtapetala mutation are restricted to floral organs. This is of particular importance, since this allows the use of mtapetala as a male-sterile line to facilitate genetic analysis of mutants affected in other processes, including nodulation. We have experimentally verified that nodulation in mtapetala is indistinguishable from that of wild type using morphological, cytological, and molecular assays (data not shown). We anticipate that the availability of mtapetala will provide a useful tool for genetic analyses in M. truncatula.
Palmyra
Mutant palmyra was initially selected for further study based on its aberrant shoot meristem phenotype (Fig. 3). This recessive mutation specifically affects embryonic and early post-embryonic shoot meristem morphology, but not other plant parts such as cotyledons, roots, and flowers. Genetic analysis indicates that palmyra is a monogenic recessive mutation (Table III), whereas double mutant analysis with the floral homeotic mutant mtapetala is consistent with the absence of genetic interaction between these two developmental mutations. Embryogenesis, shoot meristem formation, and organ formation at the vegetative meristem are most extensively characterized in Arabidopsis (for review, see Clark 1997; Kerstetter and Hake, 1997; Laux and Jurgens, 1997). Of three Arabidopsis genes known to play a role in shoot meristem initiation, the palmyra mutant most closely phenocopies mutations in the pinhead (McConnell and Barton, 1995), a locus independently described as zwille (zll; Endrizzi et al., 1996; Moussian et al., 1998). Specifically, the shoot meristem in mature embryos in both zll and plm is reduced to a flat surface, lacking leaf primordial bulges (Fig. 3, c and d). Furthermore, in plm seedlings, initial growth at the shoot meristem is variable resulting in the production of a stalk-like stem, solitary leaves, or adventitious meristems (Fig. 3, a, e, and f), similar to a range of structures observed in zll mutants (Moussian et al., 1998). In addition, postembryonically in plm, vegetative growth is largely similar to wild type. Infrequently, shoot meristems in plm terminate in modified structures, and subsequent growth occurs from a subterminal leaf axillary meristem. Although inflorescence meristem formation in plm has not been studied in detail, no obvious differences were observed between the plm and wild-type inflorescences. Moreover, floral organ development in plm homozygotes and plm tap double homozygotes was indistinguishable from the parental phenotype. Taken together these observations suggest that the spatial and temporal domain of the plm mutation are restricted to early postembryonic shoot meristem initiation with minor effects on subsequent shoot meristem maintenance and without an effect on floral organ identity in a manner strongly similar to the Arabidopsis zll mutants.
Speckle
The recessive, monogenic mutant speckle is characterized by spontaneous pigmentation pattern throughout the plant (Fig. 4); however, nodule morphogenesis appears unaffected although total numbers of rhizobial infections may be reduced (Prabhu, 1998). Superficially the spontaneous lesion formation on roots in speckle is similar to the phenotype of mutations at the root necrosis (rn) locus of soybean (Kosslak et al., 1997). However, a closer examination of the phenotypes indicates that these mutations are unlikely to be homologous. For example, in contrast to rn of soybean, speckle affects aerial plant parts in addition to roots. Furthermore, in speckle the sites of root lesions are also distinct; specifically, in the speckle mutant lesions are localized to the epidermis and/or outer cortex, whereas in the rn mutants lesions initiate in the inner cortex and subsequently spread toward the epidermis (Kosslak et al., 1997).
Mapping mtgp
Based on the availability of the complete genome sequence and a growing data set on plant function in Arabidopsis we anticipate an increase in candidate gene approaches to cloning phenotypically similar mutant loci in species other than Arabidopsis. For example several floral homoetic mutants have been described in pea (Ferrandiz et al., 1999) and we have more recently identified additional floral homeotic mutants in M. truncatula (R.V. Penmetsa and D.R. Cook, unpublished data). Because pea is refractory for molecular genetic analysis, M. truncatula mutants that phenocopy pea mutants could facilitate molecular genetic analysis of the underlying developmental pathways in legumes, underscoring the utility of M. truncatula as a “bridge” species.
Here we used a candidate gene approach to test for genetic correspondence between a class B function mutant of M. truncatula and the putative M. truncatula ortholog of ap3/def. Based on our analysis it appears unlikely that tap is orthologous to the apetala3 locus of Arabidopsis. Instead tap may be an ortholog of pistillata, or an as yet unidentified class B function gene. The MADS-box ap3 subfamily contains several genes in Arabidopsis, and ap3-like genes are also likely to exist as a paralogous gene family in legumes. Thus despite its isolation from a floral cDNA library, mtgp may not in fact the ap3 ortholog.
Thorough genetic dissection of plant processes is facilitated by the isolation of a large collection of mutants that define an allelic series at given loci. To achieve this goal an efficient mutagenesis protocol wherein mutants occur at high frequencies is highly desirable. The mutagenesis procedure, identification of a polymorphic ecotype, as well as the isolation of the male sterile mutant mtapetala that are described in this report are likely to represent important tools for efficient molecular genetic studies in the model legume M. truncatula. Experiments to test whether tap and plm define the M. truncatula orthologs of cloned Arabidopsis genes pi and zll are being initiated.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Medicago truncatula cv Jemalong genotype A17 was used as wild-type control and as the genetic background for mutagenesis. The plant growth conditions employed have been previously described (Cook et al., 1995).
Mutagenesis
For germination, seeds were scarified and surface sterilized by immersion in concentrated sulfuric acid for 5 to 10 min, rinsed in distilled water five times, treated with commercial bleach (approximately 5% [w/v] NaOCl) for 3 min, and rinsed six to eight times in sterile deionized water. Scarified seeds were soaked for 15 h in deionized water containing selected concentrations of EMS on a rotary shaker set at 30 rpm. Treated seeds were rinsed extensively (12 times for 30 min each) in sterile deionized water to remove residual EMS. Rinsed seeds were suspended in 0.1% (w/v) agar (to aid even distribution) and aliquoted on to water-soaked soil in flats of size 35 × 50 × 10 cm at a rate of 500 seeds/flat, and covered with a very thin layer of soil. Flats were covered with plastic wrap and placed at 4°C for 36 h before transfer to the greenhouse. Once seedlings had emerged from the soil, the plastic wrap was punctured to allow humidity to decline gradually; the plastic wrap was eventually removed completely after 3 d. Flats were fertilized with granular slow release fertilizer (Osmicote, Marysville, OH). Pods from each flat (typically 500 treated seeds) were harvested as M2 bulks.
Crossing M. truncatula
Artificial hybridization was performed essentially as described by Pathipanawat et al. (1994) except that flowers were not placed in polystyrene tubes after artificial pollination. Briefly, the size and appearance of floral buds prior to tripping and pollen release (full anthesis) were recorded by daily monitoring of tagged buds. Crosses were performed into emasculated buds corresponding to 1, 2, and 3 d prior to full anthesis (d.p.a.). We determined that buds 2 d.p.a. were most amenable to cross pollination, givin a high degree of cross pollination and relatively low (< 20%) self pollination. By comparison, seed set in buds 3 d.p.a. was inefficient (<25%), although self pollination was never observed. Buds 1 d.p.a. often had mature anthers and consequently gave a high degree of self pollination, despite emasculation. Visual identification of F1 hybrids in the progeny of A17 (female) × A20 (male) crosses was facilitated by scoring for the dominant A20 leaf pigmentation phenotypes in the F1 seedlings. Prior to hybridization flowers were suction emasculated using vacuum applied through a 1-mL micropipette tip. A 2.5× binocular headset with a 10-inch working distance was used to facilitate crossing. Pollen was obtained from freshly tripped flowers and applied to the stigma of emasculated flowers using extra fine forceps. Crossing to the male sterile, floral homeotic mutant (mtapetala) was conducted without emasculation and flowers were pollinated on the day of anthesis up to 2 d later. In all cases the resulting pods were wrapped in surgical gauze and tied to the stem at 2 to 3 weeks after crossing to prevent mixing of pods upon their abscission from the stem.
DNA Extraction and PCR Analysis
For PCR analysis, three trifoliate leaves (approximately 0.15 g) were harvested from each F1 individual and ground in liquid nitrogen. Ground tissue was resuspended in 1 mL of DNA extraction buffer (100 mm Tris [Tris(hydroxymethyl)aminomethane], pH 8.0, 50 mm Na2 EDTA, pH 8.0, 500 mm NaCl, and 1.25% [w/v] SDS), followed by addition of 0.3 mL of 5 m KOAc. Particulates were removed from the sample by centrifugation and filtration through Miracloth. DNA was precipitated by the addition of 0.6 volume of isopropanol, pelleted by centrifugation, and resuspended in 300 μL of TE buffer (10 mm Tris, pH 8.0, and 1 mm EDTA, pH 8.0). The resulting solution was clarified by centrifugation for 20 min at 18.5K rpm in a microcentrifuge. The DNA-containing supernatant was treated for 15 min at 21°C with 1 μL (10 mg/mL) of DNA-free RNAse A, and reprecipated by addition of 0.1 volume of 3 m NaOAc, pH 5.2, and 2.5 volumes of ethanol. DNA was recovered by centrifugation, and the resulting pellet was washed in 70% (w/v) ethanol before resuspension in 25 μL of TE buffer. PCR was conducted on a thermal cycler (Perkin Elmer, Foster City, CA; 30 cycles of 94°C for 1 min, 40°C for 1 min, and 72°C for 2 min) using 0.1 μL of DNA solution for a 50-μL PCR reaction (0.2 mm dNTPs, 1.25 units of Taq polymerase, 1× Taq polymerase buffer [Promega, Madison, WI], 2 mm MgCl2, and 20 pm oligonucleotide primers). RAPD oligonucleotide primers were “gatgaccgcc,” “gaacggactc,” “gtcccgacga,” “tggaccggtg,” and “ctcaccgtcc.” PCR products were resolved by electrophoresis on a 0.8% (w/v) agarose gel and visualized by staining with ethidium bromide.
Isolation of M. truncatula ap3-Like Sequence
A developing flower cDNA library was prepared from a pool of buds at stages 2 to 7 d.p.a. using the SMART cDNA kit (CLONTECH, Palo Alto, CA), according to the manufacturer's instructions. Degenerate primers were designed against the conserved regions MARGKIQIKRIENTQ (Ap3-C, 5′-atggcamgtggtaaraticaratiaarmgiatigaraaycarac-3′), and EDPH/YY/FGLVDD/E (Ap3-E, 5′-gargaycciyaytwyggiytigtiga-3′), of the ap3/def subfamily and used for amplification from cDNA library. Amplified PCR products were cloned into bluescript vector using pCR Script kit (Stratagene, La Jolla, CA) and clones were sequenced according to manufacturer's instructions.
Genetic Mapping of ap3-Like Sequence
BAC clones 5J18 and 42C6 from the M. truncatula BAC library containing the ap3-like sequence, mtgp, were identified and characterized according to previously published procedures (Nam et al., 1999). BamHI, EcoRI, and HindIII restriction enzyme digests were cloned into the bluescript vector using pCR Script kit (Stratagene) and random subclones were sequenced to obtain sequences, including J-11/12. For genetic mapping, primer J-11/12 A2 (5′-gagggcattcttttctgtcttac-3′) and primer J-11/12 B2 (5′-ccggtagtagaaaatttagaggaa-3′) were used in 10-μL PCR reactions (30 ng of genomic DNA, 0.25 m dNTPs, 2.5 mm MgCl2, 1 unit of Taq polymerase, and 1 pmol of each primer), using a F2 mapping population segregating for the mtapetala mutant phenotype. Following PCR amplification (96°C for 3 min, 96°C for 10 s, 55°C for 10 s, and 72°C for 1 min, 40 cycles), reactions were digested to completion with 1 unit of NlaIV restriction enzyme (New England Biolabs, Beverly, MA) in a 20-μL reaction. The codominant cleaved-amplified polymorphic sequence marker was scored on restriction digests of PCR reactions following electrophoresis on a 2% (w/v) agarose gel.
Light and Scanning Electron Microscopy
Light micrographs of specimens were obtained using a dissection microscope (Olympus, Tokyo). Specimens were either freshly harvested or fixed in glutaraldehyde. Specimens were fixed in PIPES (1,4-piperazinediethanesulfonic acid) buffer (0.1 m; pH 7.2) containing 2.5% (w/v) glutaraldehyde by drawing a vacuum (three times for 30 s each), followed by immersion at atmospheric pressure for 1 h. Fixed samples were rinsed three times for 1 h each in the buffer, embedded in 3% (w/v) agarose, and dissected with a micro-slicer (DTK-1000, Ted Pella, Redding, CA). For electron microscopy, samples were fixed in an aqueous solution of 1% (w/v) formaldehyde and 2% (w/v) glutaraledehyde for 1 h, and rinsed three times for 1 h each in sterile water. Following fixation, samples were dehydrated in a graded ethanol series, critical-point dried in carbon dioxide, coated with gold, and examined under a scanning electron microscope (JEOL-JSM-6400, JEOL USA, Dallas).
ACKNOWLEDGMENTS
We wish to thank Thomas Stephens for help with the scanning electron microscopy. A portion of results described in this report include work undertaken in partial fulfillment of requirements for the doctoral dissertation (by R.V.P.) submitted to Texas A&M University.
Footnotes
This work was supported by the Samuel Roberts Noble Foundation, by the National Science Foundation (grant no. IBN 9507535), and by a Tom Slick Graduate Fellowship from the College of Agriculture and Life Sciences, Texas A&M University.
LITERATURE CITED
- Benaben V, Duc G, Lefebvre V, Huguet T. TE7, an inefficient symbiotic mutant of Medicago truncatula (Gaertn.) cv Jemalong. Plant Physiol. 1995;107:53–62. doi: 10.1104/pp.107.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnin I, Huguet T, Gherardi M, Prosperi JM, Olivieri I. High level of polymorphism and spatial structure in a selfing plant species, Medicago truncatula (Leguminosae), shown using RAPD markers. Am J Bot. 1996;83:843–855. [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano-Anolles G, Gresshoff PM. Plant genetic control of nodulation. Annu Rev Microbiol. 1991;45:345–382. doi: 10.1146/annurev.mi.45.100191.002021. [DOI] [PubMed] [Google Scholar]
- Carroll BJ, Gresshoff PM, Delves AC. Inheritance of supernodulation in soybean and estimation of the genetically effective cell number. Theor Appl Genet. 1988;76:54–58. doi: 10.1007/BF00288831. [DOI] [PubMed] [Google Scholar]
- Clark S. Organ formation at the vegetative shoot meristem. Plant Cell. 1997;9:1067–1076. doi: 10.1105/tpc.9.7.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D, Dreyer D, Bonnet D, Howell M, Nony E, VandenBosch K. Transient induction of a peroxidase gene in Medicago truncatula precedes infection by Rhizobium meliloti. Plant Cell. 1995;7:43–55. doi: 10.1105/tpc.7.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DR. Medicago truncatula: a model in the making. Curr Opin Plant Biol. 1999;2:301–304. doi: 10.1016/s1369-5266(99)80053-3. [DOI] [PubMed] [Google Scholar]
- Cook DR, VandenBosch K, De Bruijn FJ, Huguet T. Model legumes get the nod. Plant Cell. 1997;9:275–281. [Google Scholar]
- Endrizzi K, Moussian B, Haecker A, Levin JZ, Laux T. The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 1996;6:967–979. doi: 10.1046/j.1365-313x.1996.10060967.x. [DOI] [PubMed] [Google Scholar]
- Ferrandiz C, Navarro C, Gomez MD, Canas LA, Beltran JP. Flower development in Pisum sativum: from the war of the whorls to the battle of the common primordia. Dev Genet. 1999;25:280–290. doi: 10.1002/(SICI)1520-6408(1999)25:3<280::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Heard J, Dunn K. Symbiotic induction of a MADS-box gene during development of alfalfa root nodules. Proc Natl Acad Sci USA. 1995;92:5273–5277. doi: 10.1073/pnas.92.12.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter RA, Hake S. Shoot meristem formation in vegetative development. Plant Cell. 1997;9:1001–1010. doi: 10.1105/tpc.9.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Dellaert LWM, Veen JH. EMS and radiation induced mutation frequencies at individual loci in Arabidopsis thaliana (L.) Heynh. mouse ear cress. Mutat Res. 1982;93:109–123. doi: 10.1016/0027-5107(82)90129-4. [DOI] [PubMed] [Google Scholar]
- Kosslak RM, Chamberlin MA, Palmer RG, Bowen BA. Programmed cell death in the root cortex of soybean root necrosis mutants. Plant J. 1997;11:729–745. doi: 10.1046/j.1365-313x.1997.11040729.x. [DOI] [PubMed] [Google Scholar]
- Laux T, Jurgens G. Embryogenesis: a new start in life. Plant Cell. 1997;9:989–1000. doi: 10.1105/tpc.9.7.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SL, Redei GP. Estimation of mutation rate in autogamous diploids. Radiat Bot. 1969;9:125–131. [Google Scholar]
- McConnell JR, Barton MK. Effects of mutations in the PINHEAD gene of Arabidopsis on the formation of shoot apical meristems. Dev Genet. 1995;16:358–366. [Google Scholar]
- Moussian B, Schoof H, Haecker A, Jurgens G, Laux T. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 1998;17:1799–1809. doi: 10.1093/emboj/17.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam YW, Penmetsa RV, Endre G, Uribe P, Kim DJ, Cook DR. Construction of a bacterial artificial chromosome library of Medicago truncatula and identification of clones containing ethylene-response genes. Theor Appl Genet. 1999;98:638–646. [Google Scholar]
- Pathipanawat W, Jones RAC, Sivasithamparam K. An improved method for artificial hybridization in annual Medicago species. Aust J Agric Res. 1994;45:1329–1335. [Google Scholar]
- Prabhu R. Characterization of Medicago truncatula mutants defective in infection persistence and defense response during Rhizobium-legume symbiosis. MS thesis. TX: Texas A&M University, College Station; 1998. [Google Scholar]
- Sagan M, Morandi D, Tarenghi E, Duc G. Selection of nodulation and mycorrhizal mutants in the model legume Medicago truncatula (Gaertn.) after gamma-ray mutagenesis. Plant Sci. 1995;111:63–71. [Google Scholar]
- Schauser L, Handberg K, Sandal N, Stiller J, Thykjaer T, Pajuelo E, Nielsen A, Stougaard J. Symbiotic mutants deficient in nodule establishment identified after T-DNA transformation of Lotus japonicus. Mol Gen Genet. 1998;259:414–423. doi: 10.1007/s004380050831. [DOI] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402:191–195. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- Szczyglowski K, Shaw RS, Wopereis J, Copeland S, Hamburger D, Kasiborski B, Dazzo FB, deBruin FJ. Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus. Mol Plant-Microbe Interact. 1998;11:684–697. [Google Scholar]
- Theisen G, Saedler H. The golden decade of molecular floral development (1990–1999): a cheerful obituary. Dev Genet. 1999;25:181–193. doi: 10.1002/(SICI)1520-6408(1999)25:3<181::AID-DVG1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Trieu AT, Burleigh SH, Kardailsky IV, Maldonado-Mendoza IE, Versaw WK, Blaylock LA, Shin H, Chiou TJ, Katagi H, Dewbre GR, Weigel D, Harrison MJ. Transformation of Medicago truncatula via infiltration of seedlings or flowering plants with Agrobacterium. Plant J. 2000;22:531–541. doi: 10.1046/j.1365-313x.2000.00757.x. [DOI] [PubMed] [Google Scholar]
- Trieu AT, Harrison MJ. Rapid transformation of Medicago truncatula: regeneration via shoot organogenesis. Plant Cell Reports. 1996;16:6–11. doi: 10.1007/BF01275439. [DOI] [PubMed] [Google Scholar]
- Weeden NF, Kneen BE, LaRue TA. Genetic analysis of sym genes and other nodule-related genes in Pisum sativum. In: Gresshoff PM, Roth LE, Stacey G, Newton WE, editors. Nitrogen Fixation: Achievements and Objectives. New York: Chapman & Hall; 1990. pp. 323–330. [Google Scholar]