Abstract
The tumor suppressor genes EAF2 and p53 are frequently dysregulated in prostate cancers. Recently, we reported that concurrent p53 nuclear staining and EAF2 downregulation were associated with high Gleason score. Combined loss of EAF2 and p53 in a murine model induced prostate tumors, and concurrent knockdown of EAF2 and p53 in prostate cancer cells enhanced proliferation and migration, further suggesting that EAF2 and p53 could functionally interact in the suppression of prostate tumorigenesis. Here, RNA-seq analyses identified differentially regulated genes in response to concurrent knockdown of p53 and EAF2. Several of these genes were associated with the STAT3 signaling pathway, and this was verified by significantly increased p-STAT3 immunostaining in the Eaf2−/−p53−/− mouse prostate. STAT3 knockdown abrogated the stimulation of C4-2 cell proliferation by concurrent knockdown of EAF2 and p53. Furthermore, immunostaining of p-STAT3 was increased in human prostate cancer specimens with EAF2 downregulation and/or p53 nuclear staining. Our findings suggest that simultaneous inactivation of EAF2 and p53 can act to activate STAT3 and drive prostate tumorigenesis.
Introduction
The transcription elongation factor ELL-associated factor 2 (EAF2) was recently shown to functionally interact with p53 in the repression of prostate carcinogenesis [1]. EAF2 is an androgen-responsive gene that is expressed by luminal epithelial cells in benign prostate tissues and significantly decreased in high–Gleason score prostate cancer specimens [2], [3]. Overexpression of EAF2 in prostate cancer cell lines induced apoptosis and inhibited colony formation and xenograft tumor growth [2], [4]. Conventional deletion of Eaf2 in the murine model induced murine prostatic intraepithelial neoplasia (mPIN) lesions in several strains [5], [6], further suggesting that EAF2 can act as a tumor suppressor in the prostate. Previously, EAF2 was shown to colocalize and co-immunoprecipitate with the tumor suppressor p53 [4], which is frequently mutated or overexpressed in advanced prostate cancer but infrequently mutated in localized tumors [7], [8], [9], [10], [11]. In prostate cancer cell lines, EAF2 was shown to interact with p53 to alleviate the repression of TSP-1 expression by p53, suggesting that EAF2 and p53 could functionally interact [4]. In a recent report, we showed that combined conventional deletion of Eaf2 and p53 in a murine model induced prostate carcinogenesis, and concurrent knockdown of EAF2 and p53 increased prostate cancer cell proliferation and migration [1]. Endogenous p53 and EAF2 interaction in prostate cancer cells was mediated through the C-terminus of EAF2 and the DBD of p53 [1], which frequently harbors mutations [12]. EAF2 downregulation and p53 nuclear staining in human prostate cancer specimens were correlated with high Gleason score, suggesting that simultaneous inactivation of EAF2 and p53 is associated with prostate cancer progression.
The p53 tumor suppressor controls DNA damage response, cell cycle regulation, and apoptosis. In the prostate, tumors with inactive p53 are more resistant to anticancer treatment [13], [14]. Wild-type but not mutant p53 has been reported to inhibit the phosphorylation of STAT3 at tyrosine residue 705 (Tyr705) and STAT3 DNA binding in prostate cancer cells [15]. The Janus kinase-signal transducer and activator of transcription (JAK/STAT) signaling pathway is activated by interferons and can be triggered by chronic inflammation, immune response, and cancer (reviewed in [16], [17]). STAT3, which is activated by interferon-gamma, plays a role in promoting cell survival and proliferation [18] and has been classified as an oncogene [19]. STAT3 activation is mediated by phosphorylation of cytoplasmic STAT3 on tyrine residue 705 and serine residue 727 leading to dimerization and nuclear translocation. STAT3 can transcriptionally repress p53 expression, and blocking STAT3 can activate p53 expression in cancer cells [20]. Transfection of wild-type p53 into prostate cancer cell line DU145, which expresses a mutant p53 and constitutively activated STAT3 [21], dramatically reduced expression of p-STAT3, suggesting that wild-type p53 could regulate activation of STAT3 [15]. Recently, Pencik et al. showed STAT3 transcriptionally regulated ARF, which is upstream of p53 [22]. Further elucidating the mechanisms of STAT3 activation and regulation in prostate carcinogenesis could provide new insights for developing more effective prostate cancer treatment strategies.
In the current study, we explored molecular changes associated with combined loss of EAF2 and p53 in prostate cancer cell lines, the murine prostate and human prostate cancer specimens. RNA-seq analysis was utilized to identify the genes altered in response to concurrent knockdown of p53 and EAF2 in order to identify pathways targeted by functional interaction between these two tumor suppressors in prostate cancer. We identified the activation of the STAT3 signaling pathway in C4-2 prostate cancer cells with concurrent knockdown of EAF2 and p53 and verified increased expression of p-STAT3 (Tyr705) in the p53−/−Eaf2−/− mouse prostate. STAT3 knockdown abrogated the stimulation of C4-2 cell proliferation by concurrent knockdown of EAF2 and p53. Immunostaining of p-STAT3 (Tyr705) was increased in human prostate cancer specimens with EAF2 downregulation and/or p53 nuclear staining. These findings suggest that combined loss of EAF2 and p53 contributes to the activation of the p-STAT3 pathway and promotes prostate tumorigenesis.
Materials and Methods
Cell Culture, Transfection, and RNA Interference
The human prostate cancer cell line C4-2 was a gift from Dr. Leland W. K. Chung and maintained in RPMI-1640 medium. The human prostate cancer cell line LNCaP was obtained from American Type Culture Collection (Manassas, VA) and maintained in RPMI-1640 medium. Medium was supplemented with 10% heat-inactivated fetal bovine serum. The C4-2 and LNCaP cell lines were authenticated in 2016 using DNA fingerprinting by examining microsatellite loci in a multiplex PCR (AmpFlSTR Identifiler PCR Amplification Kit, Applied Biosystems, Foster City, CA) by the University of Pittsburgh Cell Culture and Cytogenetics Facility. For knockdown experiments, cells were transfected with 100 pmol nontargeted siRNA or siRNA against p53, EAF2, and/or STAT3 using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) in a six-well plate. Targeted siRNA was complemented with nontargeted siRNA in the single-knockdown group such that the total amount of siRNAs in each well was identical. At 24 hours posttransfection, cells were treated with 2 nM R1881 for an additional 48 hours in order to induce higher levels of androgen-responsive EAF2 [1], [2] and then used for further experiments. Nontargeted control siRNA (sc-37007, Santa Cruz Biotechnology, Dallas, TX, USA) as well as targeted siRNA against EAF2 and p53 [Integrated DNA Technologies (IDT), Coralville, IA] were utilized. Pooled siRNA against p53 was purchased from Santa Cruz (sc-29435); siRNA against EAF2 was designed using the IDT RNAi Design Tool [23] as follows: AAACAGUUACUGGUGGAGUUGAACCUU (IDT); and siSTAT3: AAGUUUACAUUCUUGGGAUUGUUGGUC (IDT). Additional siRNA sequences for validation experiments were obtained from IDT and included sip53-2: 5-AGUGUUUCUGUCAUCCAAAUACUCCAC, siEAF2-2: 5-CUGUUCACCUUCACCAACCUCAAGGUA, siSTAT3-2: 5-UGAAGUACACAUUGGAAUUUGAAUGCA and siSTAT3-3: AUACUUUCCGAAUGCCUCCUCCUUGGG.
RNA-seq Analysis
For RNA-seq analysis, C4-2 cells were subjected to RNA interference for 24 hours using Lipofectamine 2000 (Invitrogen), followed by treatment with 2 nM R1881 for an additional 48 hours. The amount of each siRNA was 1200 pmol in each 10-mm dish. Samples included nontargeted siRNA control, siEAF2, and/or sip53. Knockdown of EAF2 and/or p53 by siRNA was verified by Western blotting of a small aliquot of the cells. The remainder of cells were used for total RNA isolation using TRIzol (Invitrogen). Sequencing was carried out by the Beijing Genomics Institute (Hong Kong, China). Briefly, total RNA samples were treated with DNaseI, and then mRNA was enriched using oligo(dT) magnetic beads followed by fragmentation. Double-strand cDNA was synthesized and then purified. End reparation and 3’-end single nucleotide A addition were performed, and finally, sequencing adaptors were ligated to the fragments which were then enriched by PCR amplification. The sample library was qualified and quantified using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) and ABI StepOnePlus Real-Time PCR system (Applied Biosystems, Foster, CA). The library products were sequenced via Illumina HiSeq 2000 (Illumina, San Diego, CA). In the current study, transcript quantification was done using Salmon (0.7.2) [24], and the differential expression analysis was done using the Bioconductor package edgeR (v3.18.1) using R [25], [26]. The final gene list considered was obtained from filtering to a final gene list of ratios greater than two-fold. Functional and ontology enrichment analysis was performed using Ingenuity Pathways Analysis (IPA) 5.0 (Ingenuity Systems, Redwood City, CA) as described in Haram et al. [27]. Interferon-regulated genes were identified using Interferome v2.01 (Monash University, Melbourne, Australia).
Quantitative Real-Time RT-PCR Validation
Validation of RNA-seq results were performed with a separate experimental group of C4-2 cells treated as described for the RNA-seq analysis experiment above. The RNA reverse transcription was carried out using the first-strand cDNA synthesis kit (Promega, Madison, WI). Quantitative real-time RT-PCR (qPCR) was performed using SYBR green mix (Thermo Scientific, Rockford, IL). The expression of indicated genes was normalized with respect to the GAPDH mRNA level. The sequences of primers used were listed in Supplemental Table S1.
Western Blotting
Cell samples were lysed in RIPA buffer [50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% (v/v) NP-40, 0.1% SDS, 0.25% sodium deoxycholate, 1mM sodium orthovanadate, 1 mM PMSF, 1:100 dilution of protease inhibitor cocktail P8340 (Sigma-Aldrich, St. Louis, MO)], and protein concentration was determined by BCA Protein Assay (Thermo Scientific). Tissue lysates were boiled in SDS sample buffer, separated on a NEXT GEL 10% gel (Amresco, Solon, OH) under reducing conditions, and transferred onto a nitrocellulose membrane. Blotted proteins were probed with primary antibodies (Supplemental Table S2) followed by horseradish peroxidase–labeled secondary antibody (Santa Cruz Biotechnology). Signals were visualized using chemiluminescence (ECL Western Blotting Detection Reagents, GE Healthcare, Pittsburgh, PA) and exposed to X-ray film (Fujifilm, Tokyo, Japan). All RNA-seq data have been deposited in NCBI's Gene Expression Omnibus [28] and are accessible through GEO Series accession number GSE104729.
Generation of Male EAF2 and p53 Gene Deletion Mice
Preparation of mice with specific deletion of the Eaf2 or p53 genes has been described previously [4], [5], [29]. Heterozygous Eaf2 mice on a C57BL6/J background were crossed with heterozygous p53 mice (#002101, B6.129S2-Trp53tm1Tyj/J, Jackson Laboratory, Bar Harbor, ME), and these mice were again intercrossed to generate various male cohorts with either p53−/−, p53+/−, or p53+/+ background [1] Genotyping was performed using PCR analysis of mouse tail genomic DNA at age 21 days and after euthanization [5], [29]. All mice were maintained identically under approval by the Institutional Animal Care and Use Committee of the University of Pittsburgh.
Immunohistochemical Staining
The methods of tissue collection and immunostaining have been published previously [5]. Briefly, tissues were fixed in 10% phosphate-buffered formalin at 4°C overnight. Samples were then embedded in paraffin, sectioned at 5 μm, and stained with hematoxylin and eosin. Immunostaining was performed with primary antibodies (Supplemental Table S2) using the ImmunoCruz rabbit ABC staining System (SantaCruz Biotechnology) followed by Vector NovaRED substrate (Vector Laboratories, Burlingame, CA). Slides were then counterstained in hematoxylin and coverslipped. Immunostained sections were imaged with a Leica DM LB microscope (Leica Microsystems Inc, Bannockburn, IL) equipped with an Imaging Source NII 770 camera (The Imaging Source Europe GmbH, Bremen, Germany) and NIS-Elements Documentation v 4.6 software (Nikon Instruments, Inc., Mellville, NY). All tissues were examined by a board-certified veterinary pathologist (L.H.R.) or a board-certified genitourinary pathologist (A.V.P.) using light microscopy.
Immunostaining Image Analysis
Slides stained with EAF2, p53, and p-STAT3 were evaluated semiquantitatively. The percentage of prostate epithelial cells in each core that expressed the antigen was estimated at a final magnification of 40×. Protein expression was assessed as a function of staining intensity and percentage of cells exhibiting each level of intensity. The intensity of the reaction product was based on a 4-point scale: none, faint/equivocal, moderate, and intense. An H-score was calculated for each immunostain by cell type using the following formula: H-score = 0(% no stain) + 1(% faint/equivocal) + 2(% moderate) + 3(% intense).
BrdU Assay
C4-2 cells seeded on coverslips were treated with 2 nM R1881 for 48 hours following RNA interference. Cells were subsequently cultured in the presence of 10 μM BrdU for 2 hours and then fixed with Carnoy fixative (3:1 volume/volume methanol and glacial acetic acid) for 20 minutes at −20°C. After treatment with 2 M HCl and 0.1 M boric acid, cells were incubated with 3% hydrogen peroxide (H2O2) for 10 minutes at room temperature, followed by blocking with 10% goat serum for 1 hour. Cells were then incubated with anti-BrdU antibody (Supplemental Table S2) overnight at 4°C and CY3-labeled goat anti-mouse secondary antibody (A10521, Life Technologies) for 1 hour at 37°C. The nucleus was stained with SYTOX Green (S7020, Life Technologies). Images were acquired using a fluorescence microscope (Nikon TE2000-U). Cells were counted using the Photoshop CS5 counting tool (Adobe, San Jose, CA) or Image-Pro Plus 6.0 (Media Cybernetics, Rockville, MD), and the number of BrdU-positive cells was assessed as percent of the total cell number by counting the total number of BrdU-positive cells in at least five nonoverlapping fields for each condition at 40× magnification.
Human Prostate Tissue Specimens
Human prostate tissue specimens without any previous chemo-, radio-, or hormone therapy were obtained from the surgical pathology archives of the University of Pittsburgh Prostate Tumor Bank under approval by the University of Pittsburgh Institutional Review Board from deidentified tumor specimens consented for research at time of treatment (Table 1). Specimens included two prostate tissue microarray slides (TMA). TMA specimens scored included 104 normal prostate specimens adjacent to malignant glands plus 16 normal donor prostate (normal) specimens, 64 high-grade prostatic intraepithelial neoplasia (PIN) specimens, and 216 acinar type specimens of prostatic adenocarcinoma.
Table 1.
Demographics of Human Prostate TMA Specimens
| Tissue Type | Mean Age (Years) | Gleason Score | Number of Specimens |
|---|---|---|---|
| Adenocarcinoma | 62.9 | ≤6 | 32 |
| 7 | 100 | ||
| 8 | 34 | ||
| 9 | 52 | ||
| High-grade PIN | 61 | 64 | |
| Normal adjacent to tumor | 62.9 | 104 | |
| Donor | 30.9 | 16 |
Statistics
Comparison between groups was calculated using the unpaired Student’s t test and nonparametric Mann-Whitney test. A Grubb’s test (using 5% significance level critical values) was used to detect outliers for genes validated by qPCR. Pearson correlation and Spearman coefficient were calculated for immunostaining intensity of EAF2 and CD34-positive microvessel density in human prostate tissue specimens and categorized as follows: no or weak correlation (0.00-0.30), moderate correlation (0.31-0.79), or strong correlation (0.80-0.99). A P value < .05 was considered significant. GraphPad Prism version 4 was used for graphics (GraphPad Software, San Diego, CA).
Results
EAF2 and p53 Target Genes in C4-2 Cells
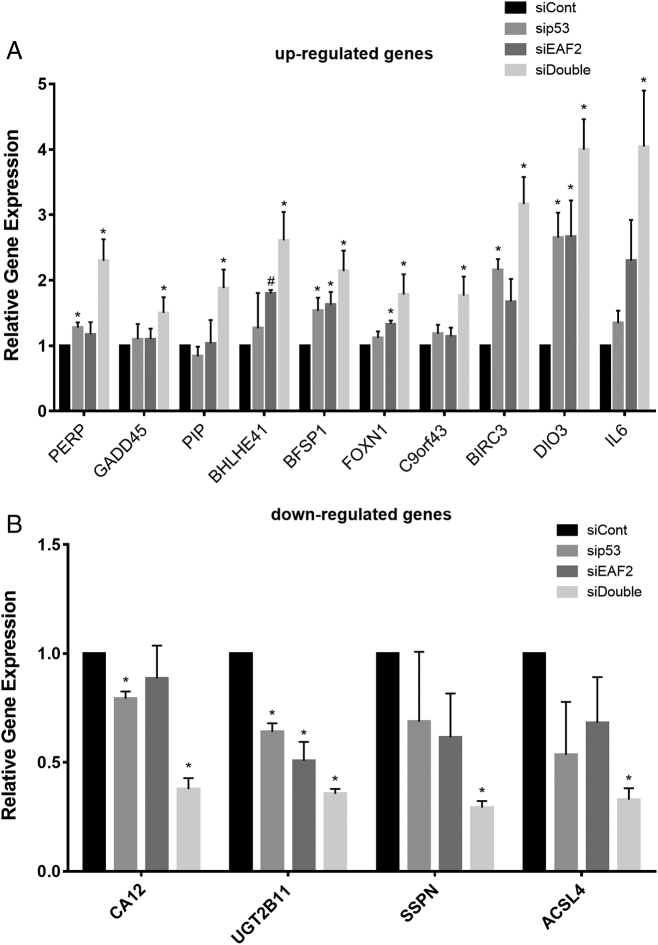
We recently showed evidence that EAF2 and p53 could functionally interact in prostate tumor suppression [1]. Here, RNA interference mediated knockdown of EAF2 and/or p53 followed by RNA-seq analysis was used to determine target genes for these two tumor suppressors in C4-2 prostate cancer cells. Several genes were altered in response to combined knockdown of p53 and EAF2. The most differentially regulated genes are listed in Supplemental Table S3. qPCR analysis verified the additive up-regulation of PERP, GADD45, PIP, BHLHE41, BFSP1, FOXN1, C9orf43, BIRC3, DIO3, and IL6 and the additive down-regulation of CA12, UGT2B11, SSPN, and ACSL4 in knockdown of p53, EAF2, and combined p53 and EAF2 (Figure 1).
Figure 1.
(A) qPCR verification of several genes identified by RNA-seq as upregulated in C4-2 p53 and EAF2 double-knockdown cells. C4-2 cells were treated with siRNA control (siCont), sip53, siEAF2, or concurrent siEAF2 and sip53 (siDouble). #, one outlier excluded using Grubb’s test. (B) qPCR verification of several genes identified by RNA-seq as downregulated in C4-2 p53 and EAF2 double-knockdown cells. Results for A and B are expressed as mean ± S.D. relative to siCont (* P < .05).
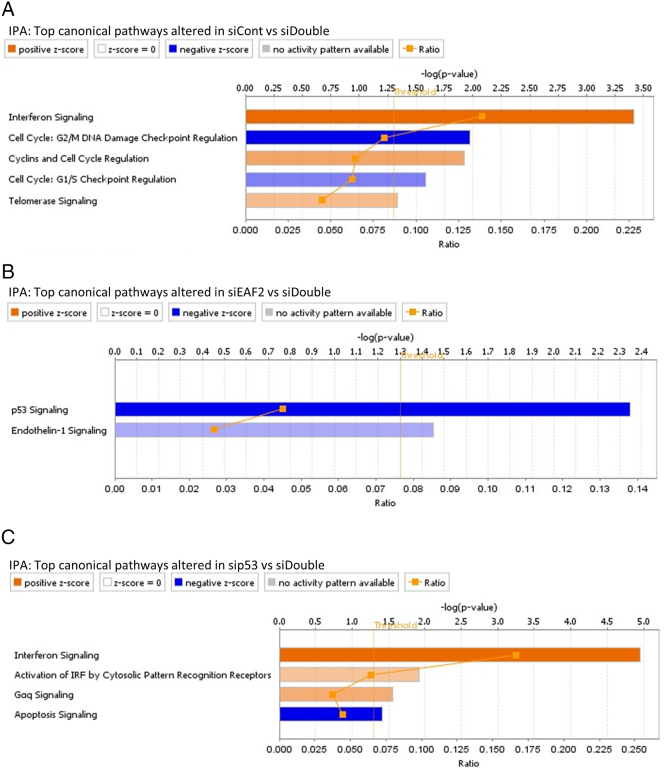
IPA identified the top canonical pathways altered in response to knockdown of combined knockdown of EAF2 and p53 (siDouble) compared to nontargeted control (siCont) (Supplemental Figure S1). Interferon signaling, cyclins, and cell cycle regulation and telomerase signaling were activated, while cell cycle: G2/M DNA damage checkpoint regulation and cell cycle: G1/S checkpoint regulation were inhibited (Supplemental Figure S1A). Compared to knockdown of EAF2 alone, combined knockdown of EAF2 and p53 resulted in the inhibition of p53 signaling and endothelin-1 signaling (Supplemental Figure S1B). Compared to knockdown of p53 alone, combined knockdown of EAF2 and p53 resulted in the activation of interferon signaling, activation of IRF by cytosolic pattern recognition receptors, Gaq signaling, and the inhibition of apoptosis signaling compared to knockdown of p53 in the presence of EAF2 (Supplemental Figure S1C).
Supplemental Figure S1.
(A) IPA of RNA-seq data identified top canonical pathways altered in C4-2 cells treated with siRNA control (siCont) compared to concurrent siEAF2 and sip53 (siDouble). (B) IPA top canonical pathways altered in C4-2 cells treated with siEAF2 alone compared to cells treated with concurrent siEAF2 and sip53 (siDouble). (C) IPA top canonical pathways altered in C4-2 cells treated with sip53 alone compared to cells treated with concurrent siEAF2 and sip53 (siDouble).
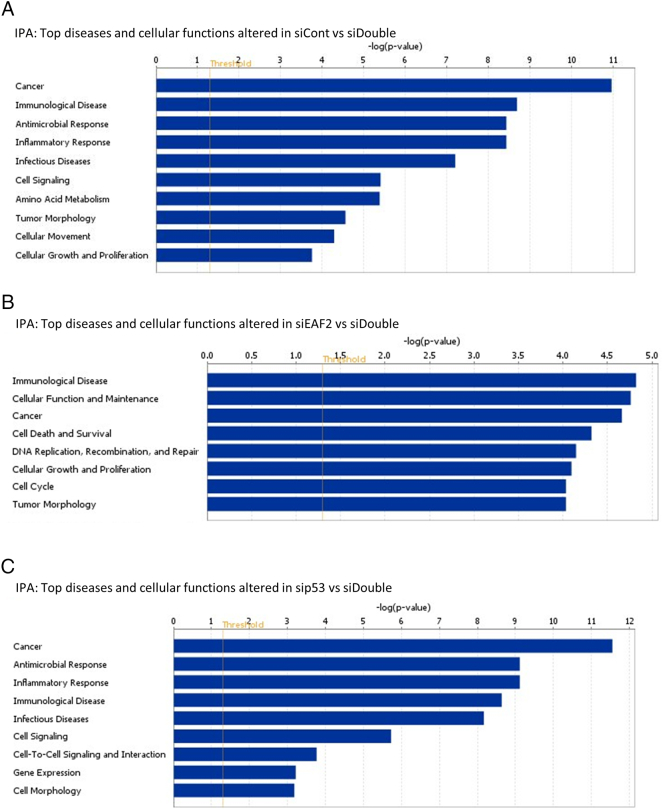
IPA downstream effects analysis identified pathways altered in response to combined knockdown of EAF2 and p53 (Supplemental Figure S2). Concurrent knockdown of EAF2 and p53 induced alteration in several pathways associated with carcinogenesis, including cancer, inflammatory response, cell signaling, tumor morphology, cellular movement, and cellular growth and proliferation (Supplemental Figure S2A). Carcinogenesis associated pathways altered in concurrent knockdown of EAF2 and p53 compared to EAF2 knockdown alone included cellular function and maintenance, cancer, cell death and survival, DNA replication, recombination and repair, cellular growth and proliferation, cell cycle, and tumor morphology (Supplemental Figure S2B). Concurrent knockdown compared to p53 knockdown included carcinogenesis-associated pathways such as cancer, inflammatory response, cell signaling, cell-to-cell signaling and interaction, gene expression, and cell morphology (Supplemental Figure S2C).
Supplemental Figure S2.
(A) IPA of RNA-seq data identified top diseases and cellular functions altered in C4-2 cells treated with siRNA control (siCont) compared to concurrent siEAF2 and sip53 (siDouble). (B) IPA top diseases and cellular functions altered in C4-2 cells treated with siEAF2 alone compared to cells treated with concurrent siEAF2 and sip53 (siDouble). (C) IPA top diseases and cellular functions altered in C4-2 cells treated with sip53 alone compared to cells treated with concurrent siEAF2 and sip53 (siDouble).
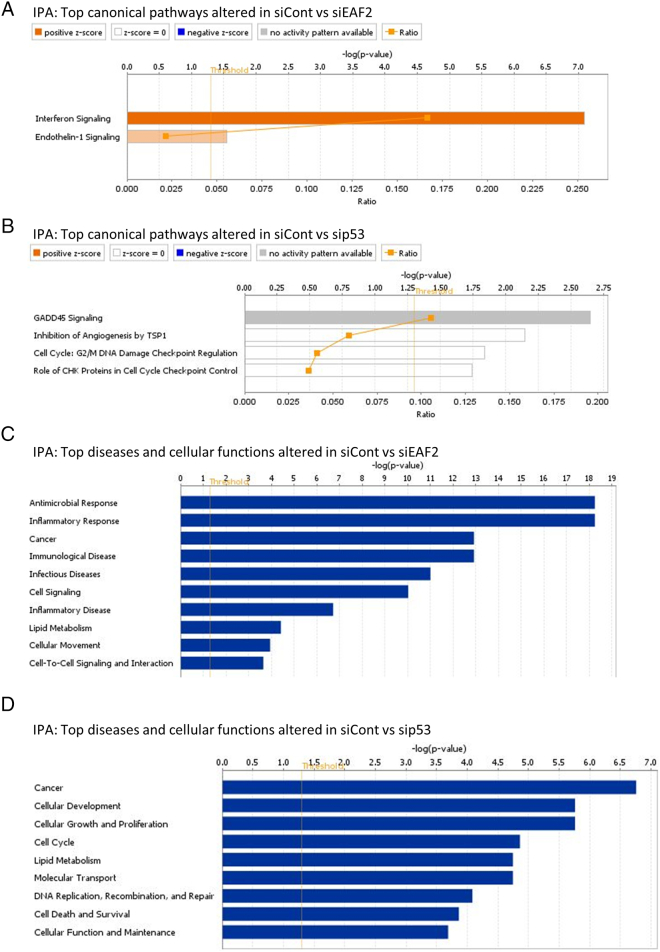
Knockdown of EAF2 alone resulted in the activation of interferon signaling and the activation of endothelin-1 signaling (Supplemental Figure S3A). Endothelin-1 has been reported to inhibit apoptosis in prostate cancer [30] and is a transcriptional target of p53 [31]. As reported by others, knockdown of p53 alone inhibited GADD45 signaling cell cycle: G2/M DNA damage checkpoint regulation [32] and inhibition of angiogenesis by TSP1 (Supplemental Figure S3B). Down-regulation of GADD45A has been shown to increase STAT3 phosphorylation and inhibit angiogenesis [33]. The pathways inflammatory response, cancer, cell signaling, cellular movement, lipid metabolism, and cell-to-cell signaling and interaction were altered in EAF2 knockdown cells compared to control siRNA (Supplemental Figure S3C). The pathways cancer, cellular development, cellular growth and proliferation, cell cycle, lipid metabolism, molecular transport, DNA replication, recombination and repair, cell death and survival, and cellular function and maintenance were altered in p53 knockdown compared to controls (Supplemental Figure S3D).
Supplemental Figure S3.
(A) IPA of RNA-seq data identified top canonical pathways altered in C4-2 cells treated with siRNA control (siCont) compared to siEAF2 alone. (B) IPA top canonical pathways altered in C4-2 cells treated with siCont compared to sip53. (C) IPA top diseases and cellular functions altered in C4-2 cells treated with siCont compared to siEAF2. (D) IPA top diseases and cellular functions altered in C4-2 cells treated with siCont compared to sip53.
STAT3 Activation in Response to Concurrent Loss of EAF2 and p53 in C4-2 and LNCaP Cells
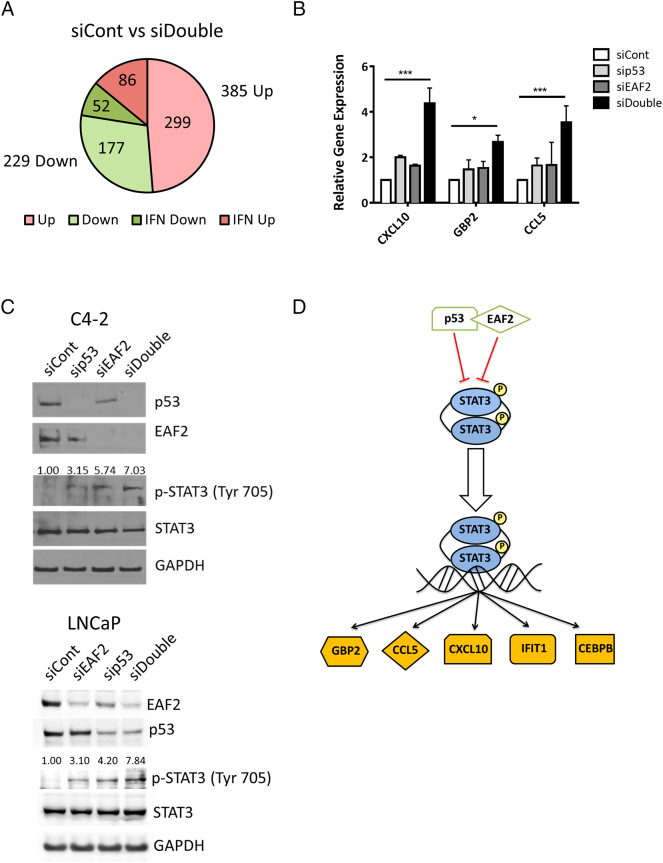
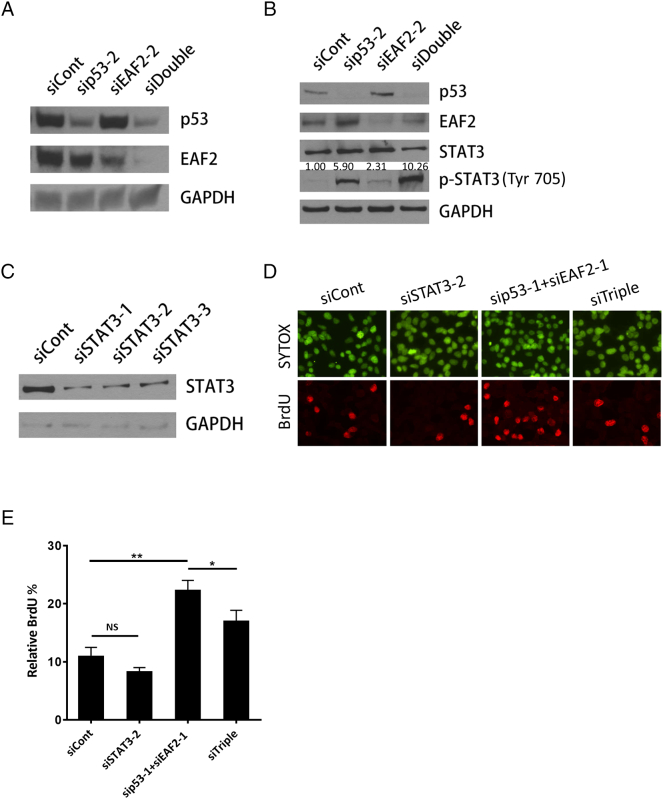
Since interferon-signaling was identified as the top canonical pathway altered in response to combined knockdown of EAF2 and p53 (see Supplemental Figure S1A), the list of genes differentially regulated in response to combined knockdown of EAF2 and p53 was also analyzed by Interferome v2.01 [34]. A total of 138 genes were identified as interferon-stimulated (Supplemental Table S4, Figure 2A), of which 24 genes were identified by IPA analysis as STAT3-regulated genes (Table 2). The upregulation of STAT3 target genes CXCL10, GBP2, and CCL5 in response to combined knockdown of p53 and EAF2 was verified by qPCR (Figure 2B). STAT3 is frequently phosphorylated at Tyr705 in prostate cancer [35] but not in LNCaP [21], [36] or C4-2 cells [37]. STAT3 protein level was not altered in response to individual knockdown of p53 and/or EAF2; however, p-STAT3 (Tyr705) level in double-knockdown cells was elevated significantly in both C4-2 and LNCaP prostate cancer cells (Figure 2C). These results suggest that EAF2 and p53 may cooperate in suppressing the STAT3 pathway (Figure 2D).
Figure 2.
(A) RNA-seq identification of 614 altered genes in C4-2 cells treated with siRNA control (siCont) compared to concurrent siEAF2 and sip53 (siDouble). Of the 385 upregulated genes (Up), 86 were identified as interferon-regulated (IFN) genes. Of the 229 downregulated genes (Down), 52 were identified as IFN-regulated genes. (B) qPCR verification of several STAT3 target genes identified by RNA-seq as upregulated in C4-2 p53 and EAF2 double-knockdown cells expressed as mean ± S.D. relative to control (siCont). (C) Western immunoblotting of STAT3 phosphorylation (Tyr 705) in C4-2 cells (top panel) and LNCaP cells (bottom panel) with knockdown of p53 and/or EAF2. GAPDH served as loading control. (D). Graphical depiction of potential EAF2 and p53 interaction in the STAT3 pathway. Gold molecules represent genes identified by RNA-Seq as upregulated in siDouble C4-2 cells.
Table 2.
STAT3-Regulated Genes Altered in Response to Combined EAF2 and p53 Knockdown in C4-2 Prostate Cancer Cells Identified by IPA
| Gene | HUGO* Gene Name | Fold-Change (siCont:siDouble) |
|---|---|---|
| ARG2 | arginase 2 | 0.430074586 |
| BST2 | bone marrow stromal cell antigen 2 | 2.741465156 |
| CCL5 | C-C motif chemokine ligand 5 | 12.95052 |
| CDKN1A | cyclin dependent kinase inhibitor 1A | 0.299638 |
| CEBPB | CCAAT/enhancer binding protein beta | 2.110388 |
| CXCL10 | C-X-C motif chemokine ligand 10 | 46.08148 |
| DDIT3 | DNA-damage inducible transcript 3 | 2.10529 |
| GBP2 | guanylate binding protein 2 | 5.109627 |
| IFI27 | interferon alpha inducible protein 27 | 4.871708 |
| IFI44 | interferon induced protein 44 | 3.399659 |
| IFI6 | interferon alpha inducible protein 6 | 7.756538586 |
| IFIT1 | interferon induced protein with tetratricopeptide repeats 1 | 20.14326215 |
| IFIT2 | interferon induced protein with tetratricopeptide repeats 2 | 24.6528095 |
| IFIT3 | interferon induced protein with tetratricopeptide repeats 3 | 30.08867001 |
| IFITM1 | interferon induced transmembrane protein 1 | 20.05318114 |
| IL4R | interleukin 4 receptor | 0.448043593 |
| OAS1 | 2'-5'-oligoadenylate synthetase 1 | 4.805611452 |
| OAS2 | 2'-5'-oligoadenylate synthetase 2 | 44.93395535 |
| OAS3 | 2'-5'-oligoadenylate synthetase 3 | 2.159541374 |
| OASL | 2'-5'-oligoadenylate synthetase like | 73.02252524 |
| PIM2 | Pim-2 proto-oncogene, serine/threonine kinase | 0.479701624 |
| RSAD2 | radical S-adenosyl methionine domain containing 2 | 9.931982035 |
| SP110 | SP110 nuclear body protein | 2.157420835 |
| TP53 | Tumor protein 53 | 0.393462413 |
pSTAT3 (Tyr 705) Levels in Eaf2−/−p53−/− Mouse Prostate
We recently generated mice with combined conventional deletion of Eaf2 and p53 [1]. Eaf2−/−p53−/− mice developed mPIN and prostate cancer lesions. Lesions were identified as mPIN and prostate cancer according to criteria commonly used in scoring prostate lesions in transgenic mouse models [38]. Briefly, mPINs were characterized by dysplasia appearing mainly as cribriform structures along with occasional stratification of cells, papilliferous structures, and tufts of cells [38]. mPIN lesions do not invade the basement membrane but may fill and expand the glandular lumen. Prostate cancer lesions were characterized by a loss of basal cells [39], unencapsulated, often poorly circumscribed, and composed of haphazard acini and lobules of pleomorphic cells with limited or no fibrovascular stroma [38]. Necrosis, vascular invasion, and/or local invasion of the tumor beyond the basement membrane into surrounding stromal tissues may also be observed [38]. Prostate cancer lesions in the Eaf2−/−p53−/− mice were characterized by increased vascularity, increased proliferation, and loss of p63-positive basal epithelial cells [1]. Basal cells in the prostate specifically express p63, and loss of p63-positive immunostaining is frequently observed in human prostate tumors [40], [41], [42].
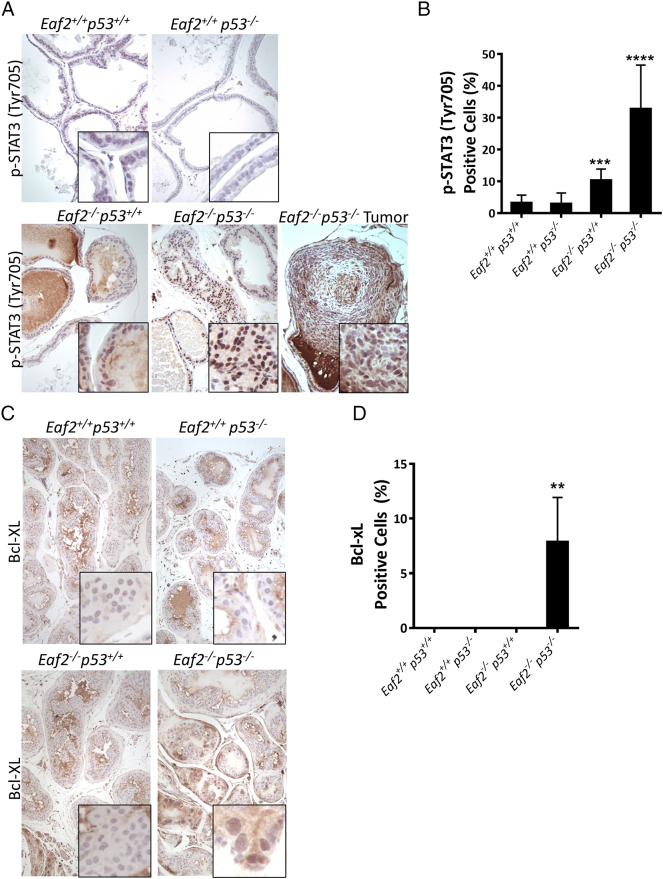
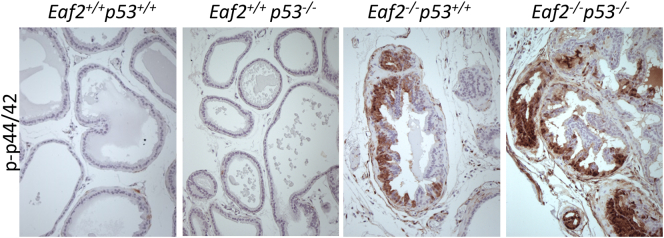
Immunostaining analysis of p-STAT3 (Tyr 705) in Eaf2−/−p53−/− mouse prostate demonstrated a dramatic increase in p-STAT3–expressing cells (Figure 3, A and B). The prostates of Eaf2+/+p53+/+ and Eaf2+/+p53−/− mice did not express p-STAT3 protein, suggesting that p53 knockout alone does not activate the STAT3 pathway. In the Eaf2−/− p53+/+ mouse prostate, p-STAT3 was focally expressed in regions displaying epithelial hyperplasia. In the Eaf2−/−p53−/− mouse prostate, p-STAT3 expression was widespread and enhanced compared to loss of Eaf2 alone, with large sections of prostatic tissues exhibiting intense staining. STAT3 can also positively regulate the antiapoptotic factor Bcl-XL [43]. Bcl-XL immunostaining was not observed in the prostates of Eaf2+/+p53+/+, Eaf2+/+p53−/−, or Eaf2−/− p53+/+ mice; however, Bcl-XL immunostaining was increased in the prostates of Eaf2−/−p53−/− mice (Figure 3C). Eaf2−/− mice have previously been reported to display increased ERK phosphorylation [3], [44]. As in these previous studies, the prostates of Eaf2−/− mice had increased p-p44/42 as evidenced by immunostaining of mPIN lesions in mice at 4 to 6 months of age (Figure 3, D and E). The prostate tumors in the Eaf2−/−p53−/− mouse also displayed a significant up-regulation of p-p44/42, while the prostates of Eaf2+/+p53+/+ and Eaf2+/+p53−/− mice did not express p-p44/42 (Supplemental Figure S4). The widespread increased immunostaining of p-STAT3 and Bcl-XL in Eaf2−/−p53−/− mice suggests that activation of the STAT3 pathway is a significant response mediated by loss of both EAF2 and p53.
Figure 3.
(A) Immunostaining of p-STAT3 expression in mouse ventral prostate. Representative images showing the upregulation of p-STAT3 (Tyr 705) in mice with combined loss of p53 and Eaf2 (Eaf2−/−p53−/−) prostate and tumor compared with wild-type (Eaf2+/+p53+/+) and Eaf2+/+p53−/− or Eaf2−/−p53+/+ mice prostate. Images are representative of three mice. (B) Quantitative analysis of p-STAT3–positive epithelial cells of the prostate from different genotype mice. For each genotype, three mice were used. (***P < .001, ****P < .0001) (C) Immunostaining of Bcl-XL in mouse ventral prostate. Representative images showing the upregulation of Bcl-XL in mice with combined loss of p53 and Eaf2 (Eaf2−/−p53−/−) prostate and tumor compared with wild-type (Eaf2+/+p53+/+) and Eaf2+/+p53−/− or Eaf2−/−p53+/+ mice prostate. Images are representative of three mice. (D) Quantitative analysis of Bcl-xL-positive epithelial cells of the prostate from different genotype mice. For each genotype, three mice were used (**P < .01) Magnification is 20×.
Supplemental Figure S4.
Immunostaining of p-p44/42 in mouse ventral prostate. Representative images showing the upregulation of p-p44/42 in mice with combined loss of p53 and Eaf2 (Eaf2−/−p53−/−) prostate and tumor compared with wild-type (Eaf2+/+p53+/+) and Eaf2+/+p53−/− or Eaf2−/−p53+/+ mice prostate. Magnification is 20×.
STAT3 Requirement for C4-2 Cell Proliferation Upon Concurrent EAF2 and p53 Knockdown
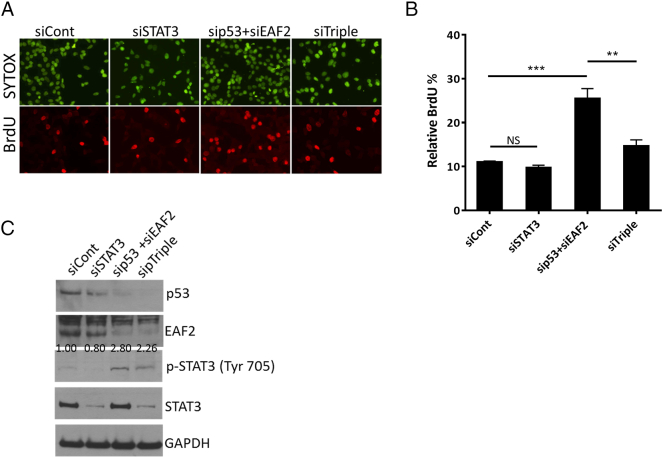
To determine if activation of STAT3 might contribute to increased proliferation, thereby contributing to the prostate carcinogenesis seen in the murine model, knockdown of STAT3 was performed in C4-2 cells treated with p53 and EAF2 siRNAs. Knockdown of STAT3 alone did not affect proliferation in C4-2 cells; however, knockdown of STAT3 abrogated the enhanced proliferation induced by concurrent EAF2 and p53 knockdown (Figure 4). Similar effects were achieved when EAF2, p53, and STAT3 were targeted with different siRNA sequences (siTriple), excluding potential nonspecific siRNA effects (Supplemental Figure S5). These results suggest that EAF2 and p53 functionally interact to repress STAT3 activation and that the proliferation in prostate cancer cells induced by combined loss of EAF2 and p53 was largely mediated through activation of STAT3 in the C4-2 model.
Figure 4.
(A) BrdU incorporation in C4-2 cells transfected with nontargeted control (siCont) siRNA; targeted to p53 (sip53), EAF2 (siEAF2), and EAF2 and p53 (sip53+siEAF2); and EAF2, p53, and STAT3 (siTriple) knockdown. Bottom panel shows BrdU-positive nuclei (red), and top panel shows nuclear staining with SYTOX green (green). (B) Quantification of BrdU incorporation shown as mean percentage ± S.D. of BrdU-positive cells relative to the total number of cells. NS, nonsignificant. (**P < .01, ***P < .001). (C) Western immunoblotting analysis of endogenous protein expression of p53, EAF2, STAT3, and p-STAT3 (Tyr705) in C4-2 cells following treatment with siRNA knockdown. GAPDH served as loading control.
Supplemental Figure S5.
(A) Western blot analysis of C4-2 cells transfected with different siRNA (siRNA-2) nontargeted control (siCont) siRNA, sip53-2, siEAF2-2, or concurrent EAF2 and p53 (siDouble) to verify knockdown efficiency for RNA-seq analysis. (B) Western blot analysis of STAT3 phosphorylation using siRNA-2 siRNA targeting nontargeted control (siCont) siRNA, targeted to p53 or EAF2 siRNA, or concurrent EAF2 and p53 (siDouble) knockdown. (C) Western blot analysis of three different siRNAs targeting STAT3 in C4-2 cells. Endogenous protein level of STAT3 was detected by Western blot using anti-STAT3. GAPDH protein was detected as a loading control. (D) BrdU incorporation following the additional siRNA knockdown of p53, EAF2, and p53 + EAF2 and triple knockdown (siTriple) of p53, EAF2, and STAT3 in C4-2 cells. Top panel shows nuclear staining with SYTOX green (green); bottom panel shows BrdU staining (red). Images shown are representative pictures of different treatment groups. (E) Quantification of BrdU incorporation (D) shown as percentage of BrdU-positive cells relative to the total number of cells.
Expression of EAF2, p53, and p-STAT3 in Human Prostate Tissue Specimens
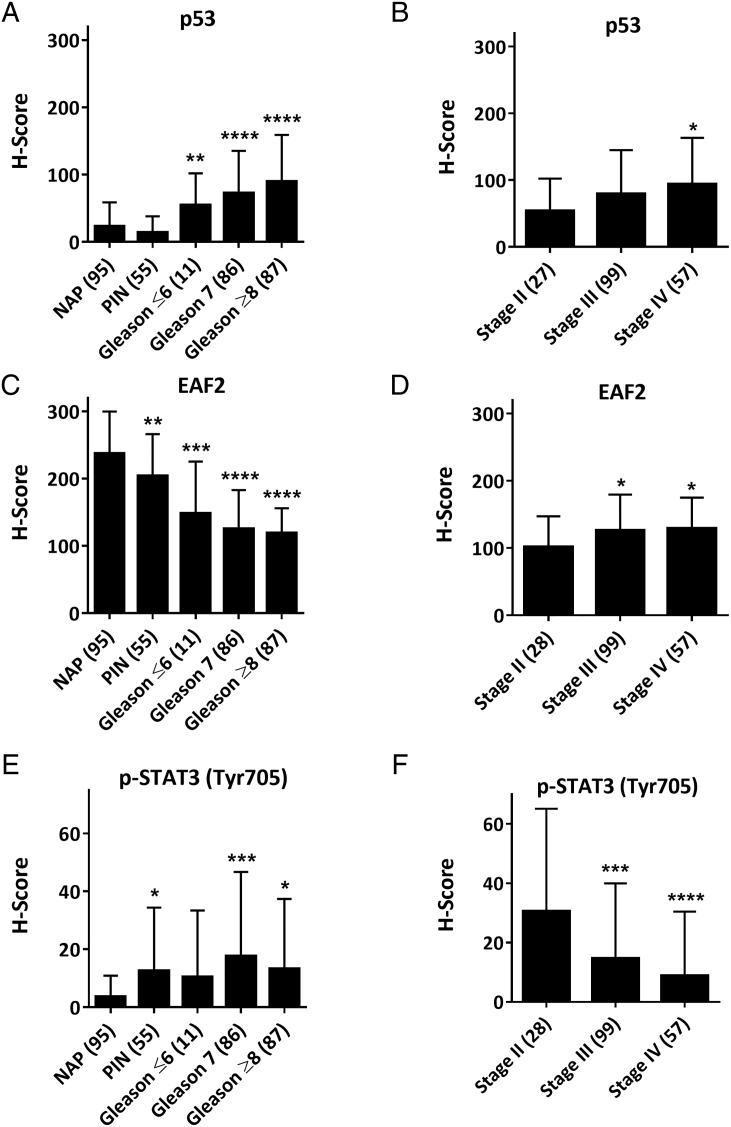
EAF2 is down-regulated in advanced human prostate tumor tissue specimens [2], [3], [6], and p53 mutation/loss is frequent in advanced and castration-resistant prostate cancers [7], [45]. EAF2 immunostaining was significantly decreased in PIN lesions and in prostate cancer tissues, suggesting that EAF2 downregulation is associated with early carcinogenesis, while p53 immunostaining was increased in Gleason ≥7 prostate cancer specimens [1]. We previously reported that the frequency of prostate cancer specimens with combined alteration in EAF2 and p53 was significantly higher in specimens with Gleason score ≥8 [1]. Here, the expression of p53, EAF2, and pSTAT3 (Tyr705) was examined in human prostate cancer tissue specimens using immunostaining of human prostate tissue specimens assembled in two TMAs (Table 1, Table 3). Tissue specimens in the TMA that did not contain prostate epithelial cells or that were washed away during the staining process were not scored. Similar to our previous findings, p53 immunostaining was significantly increased in prostate cancer compared to normal adjacent and donor prostate specimens (NAP) or PIN and increased in a stepwise fashion in Gleason ≤6, Gleason 7, and Gleason ≥8 (Figure 5A). p53 immunostaining also was increased in stage IV tumors compared to stage II (Figure 5B). EAF2 immunostaining was decreased in both PIN lesions as well as prostate cancer tissues compared to NAP, and was increased in stage III and stage IV tumors compared to stage II (Figure 5, C and D). The H-score of p-STAT3 (Tyr705) was increased in PIN lesions and prostate specimens with Gleason score 7 and Gleason score ≥8 compared to NAP (Figure 5E). Interestingly, p-STAT3 (Tyr705) H-score was decreased in stage III and stage IV specimens compared to stage II (Figure 5F). p-STAT3 (Tyr705) H-score was not correlated with Gleason score (data not shown).
Table 3.
Statistics of H-Score Data for EAF2, p53, and p-STAT3 (Tyr705) Immunostaining Study
| p53 |
|||||
|---|---|---|---|---|---|
| NAP (95) | PIN (55) | Gleason ≤6 (11) | Gleason 7 (86) | Gleason ≥8 (87) | |
| Minimum | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 25% Percentile | 0.0 | 0.0 | 10.00 | 8.750 | 40.00 |
| Median | 0.0 | 0.0 | 70.00 | 67.50 | 80.00 |
| 75% Percentile | 30.00 | 20.00 | 100.0 | 120.0 | 140.0 |
| Maximum | 190.0 | 105.0 | 125.0 | 240.0 | 300.0 |
| Mean | 22.85 | 13.64 | 54.55 | 72.62 | 89.83 |
| Std. Deviation | 35.87 | 24.41 | 47.56 | 62.75 | 69.22 |
| Std. Error of Mean | 3.681 | 3.291 | 14.34 | 6.767 | 7.421 |
| Lower 95% CI of mean | 15.54 | 7.038 | 22.59 | 59.16 | 75.07 |
| Upper 95% CI of mean | 30.16 | 20.24 | 86.50 | 86.07 | 104.6 |
|
EAF2 |
|||||
| NAP (95) |
PIN (55) |
Gleason ≤6 (11) |
Gleason 7 (86) |
Gleason ≥8 (87) |
|
| Minimum | 50.00 | 80.00 | 50.00 | 0.0 | 20.00 |
| 25% Percentile | 190.0 | 160.0 | 60.00 | 90.00 | 100.0 |
| Median | 260.0 | 200.0 | 150.0 | 120.0 | 120.0 |
| 75% Percentile | 290.0 | 270.0 | 200.0 | 162.5 | 140.0 |
| Maximum | 300.0 | 300.0 | 280.0 | 300.0 | 200.0 |
| Mean | 237.6 | 204.0 | 148.2 | 125.2 | 119.0 |
| Std. Deviation | 62.25 | 62.20 | 77.18 | 57.86 | 37.10 |
| Std. Error of Mean | 6.387 | 8.387 | 23.27 | 6.239 | 3.977 |
| Lower 95% CI of mean | 224.9 | 187.2 | 96.33 | 112.8 | 111.1 |
| Upper 95% CI of mean | 250.3 | 220.8 | 200.0 | 137.6 | 126.9 |
|
p-STAT3 (Tyr705) |
|||||
| NAP (95) | PIN (55) | Gleason ≤6 (11) | Gleason 7 (86) | Gleason ≥8 (87) | |
| Minimum | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 25% Percentile | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Median | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 75% Percentile | 3.000 | 15.00 | 15.00 | 26.25 | 15.00 |
| Maximum | 31.00 | 80.00 | 75.00 | 115.0 | 110.0 |
| Mean | 3.568 | 12.53 | 10.45 | 17.57 | 13.30 |
| Std. Deviation | 7.344 | 21.86 | 22.96 | 29.13 | 24.12 |
| Std. Error of Mean | 0.7534 | 2.947 | 6.923 | 3.141 | 2.586 |
| Lower 95% CI of mean | 2.072 | 6.618 | -4.972 | 11.32 | 8.158 |
| Upper 95% CI of mean | 5.064 | 18.44 | 25.88 | 23.82 | 18.44 |
Figure 5.
(A and B) Quantification of mean p53 H-score in prostate TMA of normal adjacent and donor (NAP), PIN, and prostate cancer specimens stratified by Gleason score or Tumor stage. (C and D) Quantification of mean EAF2 H-score in prostate TMA. (E and F) Quantification of mean p-STAT3 (Tyr705) H-score.
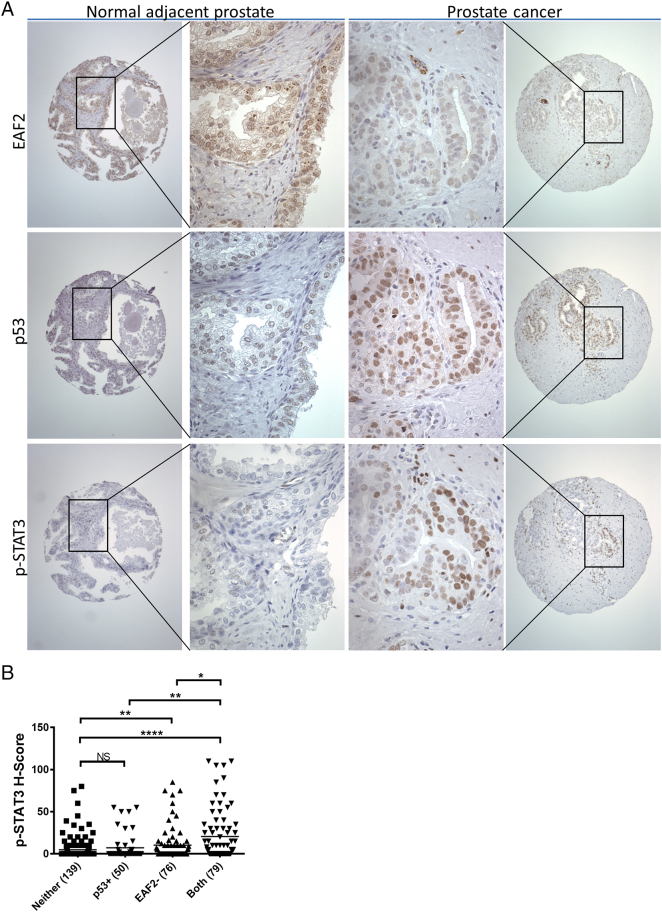
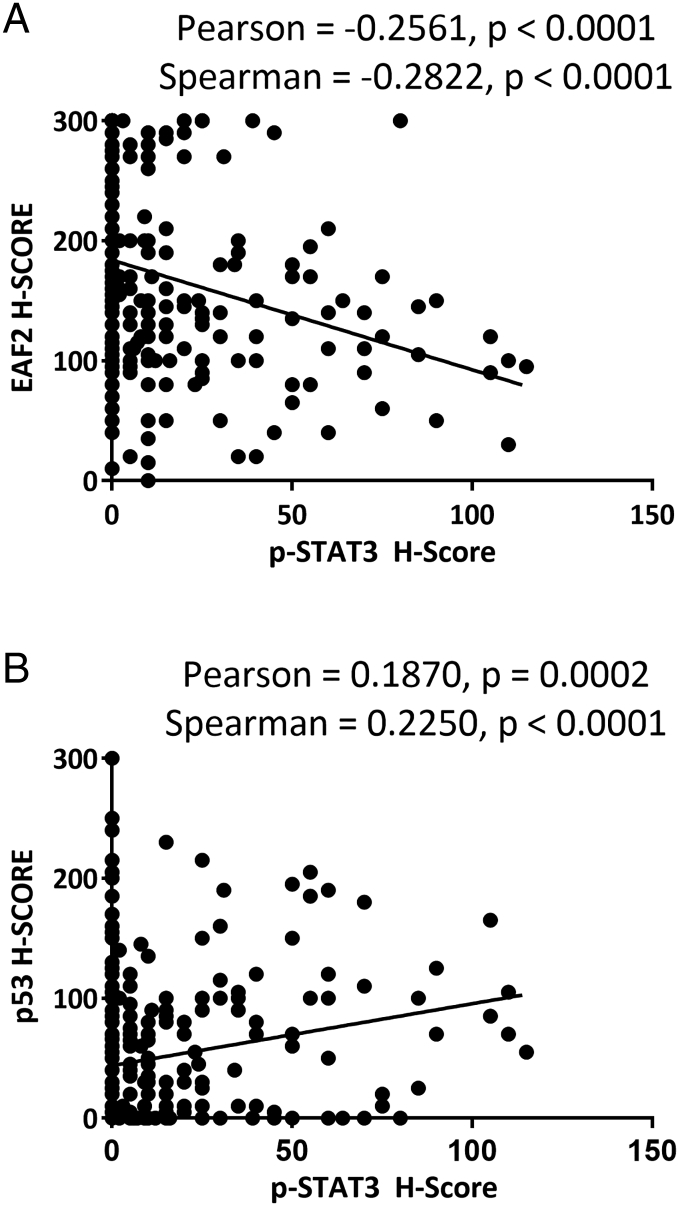
To determine the association between combined alterations in p53 and EAF2 expression and p-STAT3 (Tyr705) immunostaining, the nuclear accumulation of p-STAT3 was also determined for the TMA specimens (Figure 6). As others have reported [35], p-STAT3 (Tyr705) immunostaining was detected primarily in the nuclei of epithelial tumor cells in tumor specimens and was rare or absent in normal adjacent and donor prostate specimens (Figure 6A). Aberrant expression was defined as an H-score ≥55 for p53 [which corresponded to the mean H-score for p53 immunostaining in Gleason ≤6 (see Figure 5A)], and an H-Score of <150 for EAF2 [which corresponded to the mean H-Score for EAF2 immunostaining in Gleason ≤6 (see Figure 5C)]. Specimens were categorized as having concurrent p53 and EAF2 alteration (both), p53 only (p53+), EAF2 only (EAF2−), or alteration in neither p53 nor EAF2 (neither). p-STAT3 (Tyr705) staining was significantly increased in specimens with decreased EAF2 (P < .01) compared to normal adjacent and donor prostate specimens (NAP) but was not increased in specimens with increased p53 (P = .31) (Figure 6B). Specimens with alterations in both p53 and EAF2 had significantly higher p-STAT3 staining (P < .0001) compared to NAP specimens and compared to p53 or EAF2 alteration alone. Interestingly, p-STAT3 (Tyr705) staining was significantly higher in specimens with increased p53, or p-STAT3 was significantly higher in specimens with alterations in both p53 and EAF2 compared to EAF2 alteration alone (P < .01). p-STAT3 immunostaining was negatively correlated with EAF2 H-score and positively correlated with p53 H-score (Supplemental Figure S6). The Pearson correlation was −0.2561 (P < .0001) and Spearman coefficient was −0.2822 (P < .0001), indicating a weak negative correlation between EAF2 and p-STAT3 (Supplemental Figure S6A). The Pearson correlation was 0.1870 (P = .0002) and Spearman coefficient was 0.2250 (P < .0001), indicating a weak positive correlation between p53 and p-STAT3 (Supplemental Figure S6B). These results suggest that combined aberrant expression of EAF2 and p53 is associated with increased STAT3 signaling. These results are in agreement with the murine model which showed an increase in p-STAT3 staining in Eaf2−/− p53+/+ mice but not Eaf2+/+p53−/−mice, as well as a significant increase in p-STAT3 staining in Eaf2−/−p53−/− mice.
Figure 6.
(A) p-STAT3 (Tyr 705) immunostaining in TMA prostate tissue specimens. (B) Quantification of mean p-STAT3 (Tyr 705) nuclear staining intensity H-score in prostate tissue microarray. Specimens are categorized as no alteration in EAF2 or p53 expression (Neither), upregulated p53 (p53+), downregulated EAF2 (EAF2−), or concurrent upregulation of p53 and downregulation of EAF2 (Both). Scoring was quantified for patients with p-STAT3, EAF2, and p53 immunostaining scores; specimens missing either p-STAT3, EAF2, or p53 data due to tissue loss during the immunostaining process were not included (number of patients in parentheses, *P < .05, **P < .01, ****P < .0001).
Supplemental Figure S6.
(A) Correlation of EAF2 downregulation and p-STAT3 (Tyr705) upregulation in human prostate cancer tissue specimens. Scatter plot of EAF2 and p-STAT3 immunostaining intensity in human prostate tissue specimens. Statistical Pearson correlation was −0.2561 (P < .0001) and Spearman coefficient was −0.2822 (P < .0001). EAF2 immunostaining intensity was negatively correlated with p-STAT3 immunostaining intensity in human prostate tissue specimens. (B) Correlation of p53 and p-STAT3 upregulation in human prostate cancer tissue specimens. Scatter plot of p53 and p-STAT3 immunostaining intensity in human prostate tissue specimens. Statistical Pearson correlation was 0.1870 (P = .0002), and Spearman coefficient was 0.2250 (P < .0001). p53 immunostaining intensity was positively correlated with p-STAT3 immunostaining intensity in human prostate tissue specimens.
Discussion
Our findings here identified the STAT3 signaling pathway as an important target highly activated by concurrent inactivation of p53 and EAF2 in both cultured C4-2 cells and in the mouse prostate. STAT3 knockdown abrogated the effect of concurrent knockdown of EAF2 and p53 on the proliferation of C4-2 cells. Furthermore, immunostaining of p-STAT3 (Tyr705) correlated with combined EAF2 downregulation and increased p53 nuclear staining in human prostate cancer specimens. These findings suggest that simultaneous inactivation of EAF2 and p53 can act to activate STAT3 signaling and drive the progression of prostate cancer.
STAT3 phosphorylation by p53 knockdown in C4-2 cells is consistent with a previous finding that p53 transfection inhibited STAT3 phosphorylation at Tyr705 in prostate cancer cells [15]. However, p53 knockout in the mouse prostate did not affect STAT3 (Tyr705) phosphorylation (see Figure 3). This suggests that although p53 knockdown induced STAT3 (Tyr705) phosphorylation in prostate cancer cells (see Figure 2C), p53 deletion alone in nonmalignant cells was insufficient to activate STAT3 signaling. EAF2 knockdown in prostate cancer cells and knockout in normal mouse prostate induced STAT3 (Tyr705) phosphorylation, suggesting that EAF2 is a repressor of STAT3 signaling and that p53 is insufficient to fully repress the phosphorylation of STAT3 in the absence of EAF2. The more profound effect of EAF2 deletion on STAT3 phosphorylation than p53 deletion is also consistent with the development of mPIN in EAF2 knockout mice but not in p53 knockout prostate. Additionally, previous studies have demonstrated that EAF2 knockout in C57BL/6J mice induced an increase in inflammation in the prostate [6], [46]. Knockout of p53 in addition to EAF2 in the murine model further increased p-STAT3 (Tyr705) staining and increased Bcl-XL staining, indicating the importance of p53 in STAT3 signaling regulation when EAF2 is absent (see Figure 3). STAT3 knockdown inhibited C4-2 cell proliferation induced by the knockdown of both p53 and EAF2, suggesting that activation of STAT3 is oncogenic when EAF2 and p53 are both inactivated in prostatic cells. Furthermore, p-STAT3 (Tyr705) immunostaining was significantly higher in human prostate tissue specimens with alterations in both p53 and EAF2 compared to p53 or EAF2 alteration alone (see Figure 6B). These findings provided evidence for p53 and EAF2 cooperation in the regulation of STAT3 signaling in the prostate and suggested an important role of STAT3 signaling in EAF2 and p53 suppression of prostate carcinogenesis.
The potential importance of STAT3 signaling in prostate carcinogenesis has been demonstrated in multiple studies [21], [35], [47]. However, the role of STAT3 signaling in prostate carcinogenesis appears to be controversial. Most published studies have suggested an oncogenic role for STAT3 signaling in prostate carcinogenesis. Immunostaining of tyrosine 705-phosphorylated STAT3 in prostate cancer specimens was associated with poor survival [48] and high Gleason score [35], and STAT3 inhibition or knockdown decreased proliferation of prostate cancer cells [35]. However, in a recently published study, inhibition of STAT3 signaling promoted prostate tumor growth and progression, and loss of STAT3 staining correlated with increased risk of disease recurrence and metastatic prostate cancer [22]. Since STAT3 level may not reflect the p-STAT3 level, the STAT3 staining pattern in prostate cancer specimens may not be comparable to the p-STAT3 staining. The potential functional differences of STAT3 signaling in prostate cancer observed in different studies may reflect differences in experimental models. According to Pencik and colleagues, blocking STAT3 signaling can bypass senescence through disrupting the ARF-MDM2-p53 tumor suppressor axis in a PTEN-knockout prostate cancer model [22]. Thus, the presence of intact ARF-MDM2-p53 axis may be required for STAT3 to act as a tumor suppressor in prostate cancer cells. When p53 is mutated or deleted, STAT3 may no longer function as a tumor suppressor. Instead, activation of STAT3 signaling may become oncogenic in prostate cancer cells with defects in the ARF-MDM2-p53 axis. Future investigation will be needed to address the reasons for STAT3 to act as a tumor suppressor or an oncogene in different circumstances.
Suppression of STAT3 phosphorylation by p53 overexpression in prostate cancer cells has been previously reported [15], which is consistent with the increased STAT3 phosphorylation by p53 knockdown in this study. These observations suggested that p53 or a p53 downstream gene product may directly or indirectly inhibit STAT3 phosphorylation or upstream activators of STAT3 such as JAK2. More studies will be required to clarify the mechanisms of p53 regulation of STAT3 activation. Similarly, the mechanisms regulating STAT3 phosphorylation by EAF2 are also not clear. One potential mechanism for EAF2 knockout–induced STAT3 phosphorylation may involve the activation of RAS/ERK pathway, which was activated in the EAF2 knockout prostate [44]. Under physiological conditions, the phosphorylation of STAT3 seems to be modulated by stromal-epithelial interactions. The prostate tumors of EAF2 and p53 double-knockout mice exhibited more intense p-STAT3 immunostaining in the tumor cells of nearby stromal areas, which is consistent with a previous report showing more intense p-STAT3 staining in prostate tumor cells close to the stroma [49]. These findings suggest that the regulation of STAT3 phosphorylation by EAF2 and/or p53 involves multiple mechanisms.
Strong nuclear p53 immunostaining was closely associated with point mutations of p53 and rapid biochemical recurrence in an analysis of a tissue microarray containing 11,152 prostate cancer samples [11]. Although the correlation of p-STAT3 staining in clinical prostate cancer specimens with EAF2 downregulation and/or nuclear p53 staining was not strong, it is consistent with our hypothesis that defects in EAF2 and/or p53 signaling can enhance STAT3 phosphorylation and promote prostate carcinogenesis.
In summary, these studies demonstrate physical and functional interactions between two important tumor suppressors, p53 and EAF2, in prostate carcinogenesis and an important role for STAT3 signaling in prostate cancer cells lacking both p53 and EAF2. Since the proliferation of prostate cancer cells induced by concurrent knockdown of EAF2 and p53 could be blocked by STAT3 knockdown, therapeutic agents targeting STAT3 signaling may be effective for a subset of prostate cancer exhibiting EAF2 downregulation, p53 nuclear staining, and elevated phosphorylated STAT3.
The following are the supplementary data related to this article.
Primers for qPCR
Antibodies Used for Western Blot (WB), Immunofluorescence (IF), and Immunohistochemical Staining (IHC)
Differentially expressed genes identified in RNA-seq analysis of C4-2 cells treated with siRNA control (siCont), siEAF2, sip53, or concurrent siEAF2 and sip53 (siDouble)
Interferon-stimulated genes identified using Interferome v2.01 analysis of RNA-seq data from C4-2 cells treated with concurrent siEAF2 and sip53
Acknowledgements
We thank Anthony Green, Robin Frederick, Megan Lambert, Jianhua Zhou, and Aiyuan Zhang for technical support. This work was funded in part by National Institutes of Health Grants R01 CA186780 (Z.W.), P50 CA180995 (Z.W.), R50 CA211242 (L.E.P.), and T32 DK007774 (Z.W.) as well as by the Tippins Foundation (L.E.P.), Mellam Family Foundation (L.E.P. and D.W.), and AUA Foundation (D.W.). This project used the UPCI Animal Facility, the Cancer Bioinformatics Services and Cancer Genomics Facility, and the Tissue and Research Pathology Services and was supported in part by award P30 CA047904.
Footnotes
Disclosure Summary: The authors have nothing to disclose.
References
- 1.Wang Y, Pascal LE, Zhong M, Ai J, Wang D, Jing Y, Pilch J, Song Q, Rigatti LH, Graham LE. Combined loss of EAF2 and p53 induces prostate carcinogenesis in male mice. Endocrinology. 2017;158:4189–4205. doi: 10.1210/en.2017-00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao W, Zhang Q, Jiang F, Pins M, Kozlowski JM, Wang Z. Suppression of prostate tumor growth by U19, a novel testosterone-regulated apoptosis inducer. Cancer Res. 2003;63:4698–4704. [PubMed] [Google Scholar]
- 3.Ai J, Pascal LE, O'Malley KJ, Dar JA, Isharwal S, Qiao Z, Ren B, Rigatti LH, Dhir R, Xiao W. Concomitant loss of EAF2/U19 and Pten synergistically promotes prostate carcinogenesis in the mouse model. Oncogene. 2014;33:2286–2294. doi: 10.1038/onc.2013.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su F, Pascal LE, Xiao W, Wang Z. Tumor suppressor U19/EAF2 regulates thrombospondin-1 expression via p53. Oncogene. 2010;29:421–431. doi: 10.1038/onc.2009.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao W, Zhang Q, Habermacher G, Yang X, Zhang AY, Cai X, Hahn J, Liu J, Pins M, Doglio L. U19/Eaf2 knockout causes lung adenocarcinoma, B-cell lymphoma, hepatocellular carcinoma and prostatic intraepithelial neoplasia. Oncogene. 2008;27:1536–1544. doi: 10.1038/sj.onc.1210786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pascal LE, Ai J, Masoodi KZ, Wang Y, Wang D, Eisermann K, Rigatti LH, O'Malley KJ, Ma HM, Wang X. Development of a reactive stroma associated with prostatic intraepithelial neoplasia in EAF2 deficient mice. PLoS One. 2013;8:e79542. doi: 10.1371/journal.pone.0079542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heidenberg HB, Sesterhenn IA, Gaddipati JP, Weghorst CM, Buzard GS, Moul JW, Srivastava S. Alteration of the tumor suppressor gene p53 in a high fraction of hormone refractory prostate cancer. J Urol. 1995;154:414–421. doi: 10.1097/00005392-199508000-00024. [DOI] [PubMed] [Google Scholar]
- 8.Moul JW. Angiogenesis, p53, bcl-2 and Ki-67 in the progression of prostate cancer after radical prostatectomy. Eur Urol. 1999;35:399–407. doi: 10.1159/000019916. [DOI] [PubMed] [Google Scholar]
- 9.Schlomm T, Iwers L, Kirstein P, Jessen B, Kollermann J, Minner S, Passow-Drolet A, Mirlacher M, Milde-Langosch K, Graefen M. Clinical significance of p53 alterations in surgically treated prostate cancers. Mod Pathol. 2008;21:1371–1378. doi: 10.1038/modpathol.2008.104. [DOI] [PubMed] [Google Scholar]
- 10.Ecke TH, Schlechte HH, Schiemenz K, Sachs MD, Lenk SV, Rudolph BD, Loening SA. TP53 gene mutations in prostate cancer progression. Anticancer Res. 2010;30:1579–1586. [PubMed] [Google Scholar]
- 11.Kluth M, Harasimowicz S, Burkhardt L, Grupp K, Krohn A, Prien K, Gjoni J, Hass T, Galal R, Graefen M. Clinical significance of different types of p53 gene alteration in surgically treated prostate cancer. Int J Cancer. 2014;135:1369–1380. doi: 10.1002/ijc.28784. [DOI] [PubMed] [Google Scholar]
- 12.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 13.Chappell WH, Lehmann BD, Terrian DM, Abrams SL, Steelman LS, McCubrey JA. p53 expression controls prostate cancer sensitivity to chemotherapy and the MDM2 inhibitor Nutlin-3. Cell Cycle. 2012;11:4579–4588. doi: 10.4161/cc.22852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehmann BD, McCubrey JA, Jefferson HS, Paine MS, Chappell WH, Terrian DM. A dominant role for p53-dependent cellular senescence in radiosensitization of human prostate cancer cells. Cell Cycle. 2007;6:595–605. doi: 10.4161/cc.6.5.3901. [DOI] [PubMed] [Google Scholar]
- 15.Lin J, Tang H, Jin X, Jia G, Hsieh JT. p53 regulates Stat3 phosphorylation and DNA binding activity in human prostate cancer cells expressing constitutively active Stat3. Oncogene. 2002;21:3082–3088. doi: 10.1038/sj.onc.1205426. [DOI] [PubMed] [Google Scholar]
- 16.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 17.Biron CA. Interferons alpha and beta as immune regulators—a new look. Immunity. 2001;14:661–664. doi: 10.1016/s1074-7613(01)00154-6. [DOI] [PubMed] [Google Scholar]
- 18.Tanabe Y, Nishibori T, Su L, Arduini RM, Baker DP, David M. Cutting edge: role of STAT1, STAT3, and STAT5 in IFN-alpha beta responses in T lymphocytes. J Immunol. 2005;174:609–613. doi: 10.4049/jimmunol.174.2.609. [DOI] [PubMed] [Google Scholar]
- 19.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 20.Niu G, Wright KL, Ma Y, Wright GM, Huang M, Irby R, Briggs J, Karras J, Cress WD, Pardoll D. Role of Stat3 in regulating p53 expression and function. Mol Cell Biol. 2005;25:7432–7440. doi: 10.1128/MCB.25.17.7432-7440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ni Z, Lou W, Leman ES, Gao AC. Inhibition of constitutively activated Stat3 signaling pathway suppresses growth of prostate cancer cells. Cancer Res. 2000;60:1225–1228. [PubMed] [Google Scholar]
- 22.Pencik J, Schlederer M, Gruber W, Unger C, Walker SM, Chalaris A, Marie IJ, Hassler MR, Javaheri T, Aksoy O. STAT3 regulated ARF expression suppresses prostate cancer metastasis. Nat Commun. 2015;6:7736. doi: 10.1038/ncomms8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Owczarzy R, Tataurov AV, Wu Y, Manthey JA, McQuisten KA, Almabrazi HG, Pedersen KF, Lin Y, Garretson J, McEntaggart NO. IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008;36:W163–169. doi: 10.1093/nar/gkn198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haram KM, Peltier HJ, Lu B, Bhasin M, Otu HH, Choy B, Regan M, Libermann TA, Latham GJ, Sanda MG. Gene expression profile of mouse prostate tumors reveals dysregulations in major biological processes and identifies potential murine targets for preclinical development of human prostate cancer therapy. Prostate. 2008;68:1517–1530. doi: 10.1002/pros.20803. [DOI] [PubMed] [Google Scholar]
- 28.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strasser A, Harris AW, Jacks T, Cory S. DNA damage can induce apoptosis in proliferating lymphoid cells via p53-independent mechanisms inhibitable by Bcl-2. Cell. 1994;79:329–339. doi: 10.1016/0092-8674(94)90201-1. [DOI] [PubMed] [Google Scholar]
- 30.Nelson JB, Udan MS, Guruli G, Pflug BR. Endothelin-1 inhibits apoptosis in prostate cancer. Neoplasia. 2005;7:631–637. doi: 10.1593/neo.04787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hyter S, Coleman DJ, Ganguli-Indra G, Merrill GF, Ma S, Yanagisawa M, Indra AK. Endothelin-1 is a transcriptional target of p53 in epidermal keratinocytes and regulates ultraviolet-induced melanocyte homeostasis. Pigment Cell Melanoma Res. 2013;26:247–258. doi: 10.1111/pcmr.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naidu KA, Fang Q, Naidu KA, Cheng JQ, Nicosia SV, Coppola D. P53 enhances ascorbyl stearate-induced G2/M arrest of human ovarian cancer cells. Anticancer Res. 2007;27:3927–3934. [PubMed] [Google Scholar]
- 33.Yang F, Zhang W, Li D, Zhan Q. Gadd45a suppresses tumor angiogenesis via inhibition of the mTOR/STAT3 protein pathway. J Biol Chem. 2013;288:6552–6560. doi: 10.1074/jbc.M112.418335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, Chapman R, Hertzog PJ. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41:D1040–1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mora LB, Buettner R, Seigne J, Diaz J, Ahmad N, Garcia R, Bowman T, Falcone R, Fairclough R, Cantor A. Constitutive activation of Stat3 in human prostate tumors and cell lines: direct inhibition of Stat3 signaling induces apoptosis of prostate cancer cells. Cancer Research. 2002;62:6659–6666. [PubMed] [Google Scholar]
- 36.Jacobberger JW, Sramkoski RM, Zhang D, Zumstein LA, Doerksen LD, Merritt JA, Wright SA, Shults KE. Bivariate analysis of the p53 pathway to evaluate Ad-p53 gene therapy efficacy. Cytometry. 1999;38:201–213. doi: 10.1002/(sici)1097-0320(19991015)38:5<201::aid-cyto2>3.3.co;2-6. [DOI] [PubMed] [Google Scholar]
- 37.Delk NA, Farach-Carson MC. Interleukin-6: a bone marrow stromal cell paracrine signal that induces neuroendocrine differentiation and modulates autophagy in bone metastatic PCa cells. Autophagy. 2012;8:650–663. doi: 10.4161/auto.19226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shappell SB, Thomas GV, Roberts RL, Herbert R, Ittmann MM, Rubin MA, Humphrey PA, Sundberg JP, Rozengurt N, Barrios R. Prostate pathology of genetically engineered mice: definitions and classification. The consensus report from the Bar Harbor meeting of the Mouse Models of Human Cancer Consortium Prostate Pathology Committee. Cancer Res. 2004;64:2270–2305. doi: 10.1158/0008-5472.can-03-0946. [DOI] [PubMed] [Google Scholar]
- 39.Brawer MK, Peehl DM, Stamey TA, Bostwick DG. Keratin immunoreactivity in the benign and neoplastic human prostate. Cancer Res. 1985;45:3663–3667. [PubMed] [Google Scholar]
- 40.Parsons JK, Gage WR, Nelson WG, De Marzo AM. p63 protein expression is rare in prostate adenocarcinoma: implications for cancer diagnosis and carcinogenesis. Urology. 2001;58:619–624. doi: 10.1016/s0090-4295(01)01311-5. [DOI] [PubMed] [Google Scholar]
- 41.Parsons JK, Saria EA, Nakayama M, Vessella RL, Sawyers CL, Isaacs WB, Faith DA, Bova GS, Samathanam CA, Mitchell R. Comprehensive mutational analysis and mRNA isoform quantification of TP63 in normal and neoplastic human prostate cells. Prostate. 2009;69:559–569. doi: 10.1002/pros.20904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Signoretti S, Waltregny D, Dilks J, Isaac B, Lin D, Garraway L, Yang A, Montironi R, McKeon F, Loda M. p63 is a prostate basal cell marker and is required for prostate development. Am J Pathol. 2000;157:1769–1775. doi: 10.1016/S0002-9440(10)64814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong A, Yang Z, Shen Y, Zhou J, Shen Q. Transcription factor STAT3 as a novel molecular target for cancer prevention. Cancers (Basel) 2014;6:926–957. doi: 10.3390/cancers6020926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su F, Correa BR, Luo J, Vencio RZ, Pascal LE, Wang Z. Gene expression profiling reveals regulation of ERK Phosphorylation by androgen-induced tumor suppressor U19/EAF2 in the mouse prostate. Cancer Microenviron. 2013;6:247–261. doi: 10.1007/s12307-013-0132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bookstein R, MacGrogan D, Hilsenbeck SG, Sharkey F, Allred DC. p53 is mutated in a subset of advanced-stage prostate cancers. Cancer Res. 1993;53:3369–3373. [PubMed] [Google Scholar]
- 46.Pascal LE, Ai J, Rigatti LH, Lipton AK, Xiao W, Gnarra JR, Wang Z. EAF2 loss enhances angiogenic effects of Von Hippel-Lindau heterozygosity on the murine liver and prostate. Angiogenesis. 2011;14:331–343. doi: 10.1007/s10456-011-9217-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spiotto MT, Chung TD. STAT3 mediates IL-6-induced growth inhibition in the human prostate cancer cell line LNCaP. Prostate. 2000;42:88–98. doi: 10.1002/(sici)1097-0045(20000201)42:2<88::aid-pros2>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 48.Campbell CL, Jiang Z, Savarese DM, Savarese TM. Increased expression of the interleukin-11 receptor and evidence of STAT3 activation in prostate carcinoma. Am J Pathol. 2001;158:25–32. doi: 10.1016/S0002-9440(10)63940-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nowak DG, Cho H, Herzka T, Watrud K, DeMarco DV, Wang VM, Senturk S, Fellmann C, Ding D, Beinortas T. MYC Drives Pten/Trp53-deficient proliferation and metastasis due to IL6 secretion and AKT suppression via PHLPP2. Cancer discov. 2015;5:636–651. doi: 10.1158/2159-8290.CD-14-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers for qPCR
Antibodies Used for Western Blot (WB), Immunofluorescence (IF), and Immunohistochemical Staining (IHC)
Differentially expressed genes identified in RNA-seq analysis of C4-2 cells treated with siRNA control (siCont), siEAF2, sip53, or concurrent siEAF2 and sip53 (siDouble)
Interferon-stimulated genes identified using Interferome v2.01 analysis of RNA-seq data from C4-2 cells treated with concurrent siEAF2 and sip53