Abstract
PURPOSE: Bile-containing gastroesophageal reflux may promote cancer at extraesophageal sites. Acidic bile can accelerate NF-κB activation and molecular events, linked to premalignant changes in murine hypopharyngeal mucosa (HM). We hypothesize that short-term in vivo topical application of NF-κB inhibitor BAY 11-7082 can prevent acidic bile–induced early preneoplastic molecular events, suggesting its potential role in disease prevention. EXPERIMENTAL DESIGN: We topically exposed HM (C57Bl/6j wild-type) to a mixture of bile acids at pH 3.0 with and without BAY 11-7082 3 times/day for 7 days. We used immunofluorescence, Western blotting, immunohistochemistry, quantitative polymerase chain reaction, and polymerase chain reaction microarrays to identify NF-κB activation and its associated oncogenic mRNA and miRNA phenotypes, in murine hypopharyngeal cells in vitro and in murine HM in vivo. RESULTS: Short-term exposure of HM to acidic bile is a potent stimulus accelerating the expression of NF-κB signaling (70 out of 84 genes) and oncogenic molecules. Topical application of BAY 11-7082 sufficiently blocks the effect of acidic bile. BAY 11-7082 eliminates NF-κB activation in regenerating basal cells of acidic bile–treated HM and prevents overexpression of molecules central to head and neck cancer, including bcl-2, STAT3, EGFR, TNF-α, and WNT5A. NF-κB inhibitor reverses the upregulated “oncomirs” miR-155 and miR-192 and the downregulated “tumor suppressors” miR-451a and miR-375 phenotypes in HM affected by acidic bile. CONCLUSION: There is novel evidence that acidic bile–induced NF-κB–related oncogenic mRNA and miRNA phenotypes are generated after short-term 7-day mucosal exposure and that topical mucosal application of BAY 11-7082 can prevent the acidic bile–induced molecular alterations associated with unregulated cell growth and proliferation of hypopharyngeal cells.
Abbreviations: HM, hypopharyngeal mucosa; MHPC, murine hypopharyngeal primary cell
Introduction
Gastroesophageal reflux disease (GERD) is known to promote extraesophageal inflammatory and neoplastic diseases [1], [2]. In fact, a recent well-powered epidemiologic study now confirms that GERD is an independent risk factor in laryngopharyngeal cancer [3]. In 60% percent of patients with GERD, both gastric and duodenal (bile) fluids flow retrograde through the esophagus, reaching the upper aerodigestive tract [1], [4]. The combination of bile refluxate and acid seems to be a critical factor in damaging hypopharyngeal mucosa (HM) [4], [5], [6]. We previously showed in vitro that a mixture of bile salts, in concentration and composition similar to what have been previously identified in GERD refluxate [1], [6], [7], [8], could induce NF-κB activation and transcriptional activation of genes with oncogenic function in treated normal hypopharyngeal cells [9], whereas acidic pepsin appeared oncologically inactive [10]. Constitutive activation of NF-κB has been observed in various cancers, linking inflammation to neoplastic transformation of epithelium [11], [12], [13], [14], [15]. Lee et al. suggest that the constitutive activation of NF-κB provides an alternative mechanism for head and neck carcinogenesis [16]. We also showed, using an in vivo animal model, that a 45-day intermittent exposure to acidic bile (pH 3.0) induced histopathologic changes and early preneoplastic events including cancer-related mRNA and miRNA phenotypes in treated HM [17], [18]. We further demonstrated in vitro that BAY 11-7082, a specific pharmacologic inhibitor of NF-κB, could prevent the acidic bile–induced NF-κB signaling pathway and its cancer-related mRNA phenotype in normal human hypopharyngeal cells [19].
The canonical pathway of NF-κB activation includes phosphorylation of IKB-α, leading to translocation of NF-κB complexes p50/Rela(p65) or p50/cRel from cytoplasm to the nucleus and consequent binding to the promoters of target genes, regulating their expression [20], [21]. NF-κB activation induces a signaling pathway by accelerating the expression of several receptors, ligands, and transcriptional factors that regulate NF-κB activation and immune response through NF-κB, while inhibition of NF-κB negatively affects the gene expression profiling of the NF-κB signaling pathway and has been considered promising for improving anticancer therapies [21], [22]. Pierce et al. have shown that the inhibition of NF-κB through BAY 11-7082 inhibits IκB-α phosphorylation, blocking proteosomal degradation of IκB-α and allowing NF-κB to sequester in the cytoplasm in an inactivated state [23], [24], [25], [26], [27].
We hypothesize that short-term intermittent topical exposure of murine HM to BAY 11-7082 can suppress acidic bile–induced cancer-related mRNA and miRNA phenotypes. To explore our hypothesis, we describe an experimental paradigm employing a short-term topical application of BAY 11-7082 to acidic bile–treated murine HM using wild-type mice C57Bl6J as previously described. In so doing, we will explore potential use of NF-κB inhibitors, such as BAY 11-7082, in the reduction of NF-κB–related early preneoplastic molecular events.
Materials and Methods
In Vitro Model
Cell Culture
We cultured murine hypopharyngeal primary cells (MHPC) from Celprogen Inc., Torrance, CA, as described previously by Vageli et al. [17], [19].
Briefly, “Bile” and a “Bile + BAY 11-7082” treatment procedures were performed in parallel. MHPC (second passage) underwent repeated exposure for 10 to 15 minutes three times per day for 5 days to a mixture of conjugated bile salts (400 μM) (Glycocholic acid:taurocholic acid:glycochenodeoxycholic acid:taurodeoxycholic acid:glycodeoxycholic acid:taurodeoxycholic acid at molar concentration 20:3:15:3:6:1) (Sigma, St. Louis, MO; Calbiochem, San Diego, CA), as previously described [8], [9], [17], [28], with and without BAY 11-7082 (10 μM) [29], a pharmacologic inhibitor of NF-κB (Calbiochem 2016 EMD Millipore Corporation; Germany), and corresponding controls. Experimental groups included 1) acidic bile mixture at pH 4.0, 2) acidic bile mixture with BAY 11-7082 at pH 4.0, 3) neutral bile mixture at pH 7.0, and 4) neutral bile with BAY 11-7082 at pH 7.0. Control groups included identical media used for experimental groups, at pH 4.0 and 7.0, without the addition of bile mixture, as follows: 1) acid alone at pH 4, 2) acid alone with BAY 11-7082 at pH 4.0, 3) neutral control at pH 7.0, and 4) neutral control with BAY 11-7082 at pH 7.0. We also included an untreated control and a reference control for the NF-κB inhibitor vehicle (DMSO).
In Vivo Model
We used Mus Musculus, mouse strain C57BL/6J (Jax mice, Jackson Laboratory USA) [16 males and 16 females; 8 mice (4 males+4 females) per group). We performed repeated topical exposure of murine HM, three times per day for 7 days (20-21 applications), to a mixture of bile salts (10 mmol/l in buffered saline) (~5 μmol per day) at molar concentrations previously described and considered to be close to “physiologic” [8], [28], at pH 3.0, with and without 0.25 μmol of BAY 11-7082 (~0.75 μmol per day) (Calbiochem; EDM Millipore Corp.) [29]. Acidic pH (3.0) was selected, as previously described by Vageli D et al. to induce a marked NF-κB activation and overexpression of a cancer mRNA and miRNA phenotype in murine laryngopharyngeal mucosa in vivo [17], [18]. The experimental and control groups included 1) acidic bile at pH 3.0, 2) acidic bile plus BAY 11-7082 at pH 3.0, 3) a saline-treated control group at pH 7.0 (reference control for the mechanical effect of the feeding tube on HM and the NF-κB inhibitor vehicle), and 4) an untreated control group (negative control). Procedures followed the approved protocol 11039 by IACUC (Yale University).
At the end of the procedures, experimental and control animals were euthanatized (IACUC euthanasia policy and guidelines). The euthanized animals were kept on ice for dissection of HM tissue fragments. The HM from four animals (two males and two females) of each group and corresponding tissue fragments from the mucosa of the tip of the tongue (TM), to represent an internal control, were placed immediately into 10% neutral buffered formalin (Thermo Fisher Scientific, Middletown, VA) to be submitted for embedding in paraffin blocks (Yale Pathology Facilities), while the remaining tissue fragments from each experimental and control groups were immersed in RNA stabilization solution (RNAlater, Life Technologies, Grand Island, NY) and kept in −80°C for RNA isolation.
Western Blotting
We performed Western blot analysis, as described previously [9], [10], [19], to determine the nuclear and cytoplasmic protein expression levels of p-NF-κB (p65 S536) and phospho-inhibitor kappaB-α (p-IκB-α), respectively, on treated MHPC with and without NF-κB inhibitor BAY 11-7082.
Immunofluorescence Assay
We performed immunofluorescence assay in treated human hypopharyngeal primary cells, as described previously [19], to determine acidic bile–induced DNA damage using the primary rabbit polyclonal anti-gamma H2A.X antibody (1:500) (pS139, ab11174, Abcam, Cambridge, UK).
Tissue Examination Ki67 Staining
Histologic Staining
We examined hematoxylin and eosin–stained 3- to 4-μm–thick tissue sections of formalin-fixed and paraffin-embedded HM by light microscopy. All tissue specimens were examined to exclude histological signs of local treatment toxicity, such as hemorrhagic lesions, ulceration, or inflammation.
Immunohistochemical (IHC) Analysis
We performed IHC analysis, using immunoperoxidase (DAB peroxidase substrate), for Ki67 cell proliferation markers in hypopharyngeal tissue sections from all experimental and control specimens to explore the effect of short-term exposure to acidic bile in increasing regenerative activity of mucosal basal/parabasal layers and consequently the effect of NF-κB inhibitor in reducing that effect.
We used 1:200 dilutions of anti-Ki67 (rabbit mAb, SP6, Thermo Scientific Lab Vision, UK) to detect protein in nuclei of basal/suprabasal cells of HM. We analyzed the slides using a Leica light microscope and captured images using Aperio CS2. The images were analyzed by Image Scope software (Leica Microsystems, Buffalo Grove, IL) that generated algorithm(s) illustrating the mucosal and cellular compartments by Ki67 staining. Nuclear Ki67 protein levels in treated HM with/without NF-κB inhibitor, saline-treated HM, and untreated HM were expressed as positive nuclei to total number (defined as positivity) derived from two independent images per tissue section (at least four tissue sections per group) (mean±SD by multiple t test).
IHC Analysis for NF-κB (p65 Phosphorylated at Ser536)
We performed IHC analysis to detect acidic bile–induced p-NF-κB (p65 S536) activation in murine HM, in vivo, with and without BAY 11-7082 and in corresponding controls. We performed chromogenic IHC using immunoperoxidase (DAB peroxidase substrate) for p-NF-κB on hypopharyngeal tissue sections from all experimental and control specimens as previously described [17], [18]. Nuclear p-p65 (S536) protein levels in treated HM with/without NF-κB inhibitor, saline-treated HM, and untreated HM were expressed as ratios of positive nuclei to total number (defined as positivity) derived from two independent images per tissue section (at least four tissue sections per group) (mean ± SD by multiple t test).
Quantitative Real-Time Polymerase Chain Reaction
Real-time quantitative polymerase chain reaction (qPCR) analysis was performed in MHPC and HM (two animals per group) exposed to bile with or without BAY 11-7082 and in corresponding controls to evaluate the transcriptional levels of RELA (p65), bcl-2, TNF-α, EGFR, STAT3, WNT5A, IL-1β, and IL6 as previously described [17], [19] (Supplementary Table S1). Relative mRNA expression levels were estimated for each target gene relative to the reference gene (GAPDH) (ΔΔCt).
miRNA Analysis
We performed miRNA analysis to determine the expression levels of “oncomirs” and ‘tumor suppressor” miRNA specific markers in MHPC and murine HM exposed to bile and corresponding controls, with and without BAY 11-7082, as described in our in vivo model [18] (Supplementary Table S2). We explored the expression of “oncomirs” miR-21, miR-155, and miR-192, as well as “tumor suppressors” miR-34a, miR-375, and miR-451a previously characterized in laryngopharyngeal cancer and/or gastroduodenal reflux [13], [14], [15], [18], [26], [30], [31], [32] and that we recently found to be inhibited by BAY 11-7082, in normal human hypopharyngeal cells in vitro (unpublished data).
Statistical Analysis
We used GraphPad Prism 6 software and one-way analysis of variance (ANOVA) (Kruskal-Wallis and Dunn’s multiple-analysis test; P values <.05) as well as t test analysis (multiple comparisons by Holm-Sidak) to reveal statistically significant reduction in protein, mRNA, or miRNA expression levels among the studied groups. We performed Pearson correlation to estimate the correlation coefficient between expression levels of the analyzed genes and miRNA markers in the studied groups (P values <.05).
PCR Array for NF-κB Signaling Pathway
We performed a PCR microarray analysis of the NF-κB signaling pathway in acidic bile–treated groups with and without BAY 11-7082 to identify the effect of NF-κB inhibitor on the acidic bile–induced gene expression profiling of the NF-κB signaling pathway. Specifically, we used a transcriptome of murine HM and a PCR array kit for murine NF-κB signaling pathway (RT2-Profiler PCR array, PAMM-025ZD; SABiosciences, Qiagen), following the manufacturer’s instructions. The PCR array profiled the expression of 84 key genes related to NFκB-mediated signal transduction, including genes that encode members of the Rel, NFκB, and IκB families; NFκB-responsive genes; extracellular ligands and receptors that activate the pathway; and kinases and transcription factors that propagate the signal. The data were analyzed online by RT2-Profiler PCR Array Data Analysis version 3.5 software, and differential expression of more than two-fold change of gene expression (up- and downregulation) was estimated between the untreated HM (control group) and HM exposed to acidic bile–treated group (group 1) or acidic bile–treated with NF-κB inhibitor (BAY 11-7082) group (group 2).
Results
BAY 11-7082 Inhibits NF-κB Activation in Acidic Bile–Treated MHPC In Vitro
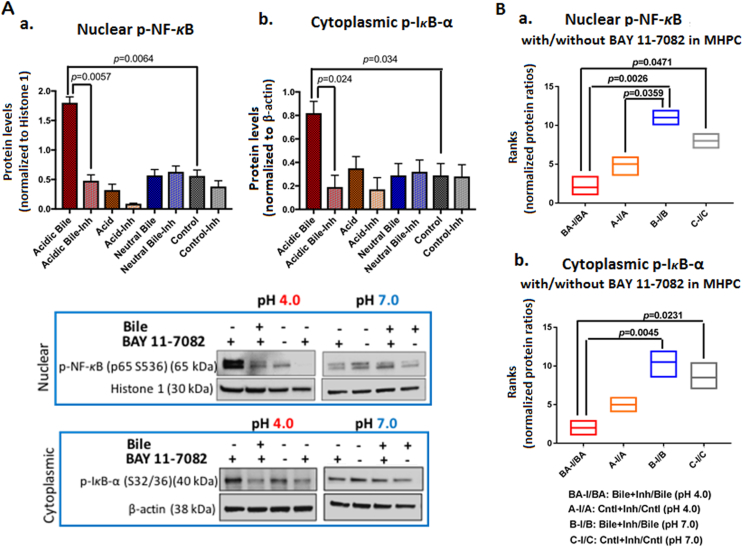
We first used a previously described in vitro model [9] to explore the role of BAY 11-7082 in preventing acidic bile–induced NF-κB activation and mRNA phenotype in MHPC. Our data from Western blot analysis (Figure 1) showed that BAY 11-7082 effectively suppressed NF-κB activation. We observed that the effect of NF-κB inhibition resulted in a significant reduction in acidic bile–induced nuclear p-NF-κB (p65 S536) and cytoplasmic phospho-IκB-α levels (Figure 1B). Specifically, the effect of BAY 11-7082 was significantly more intense in the presence of acidic bile relative to neutral bile, acid alone, or neutral control (P<.05), resulting in reduced nuclear p-NF-κB (p65) and cytoplasmic p-IκB-α protein levels (Supplementary Figure 2). NF-κB inhibitor suppressed acidic bile–induced NF-κB activation and decreased its expression levels in treated MHPC similar to what we recently demonstrated in human hypopharyngeal primary cells in vitro [19].
Figure 1.
BAY 11-7082 inhibits the acidic bile–induced NF-κB activation in MHPC. (A) Western blot analysis was performed in nuclear and cytoplasmic protein extracts of treated MHPC with and without BAY 11-7082, demonstrating that NF-κB inhibitor induced a significant reduction of activated NF-κB. Graphs depict significantly lower (a) nuclear p-NF-κB (p65 S536) and (b) cytoplasmic p-IκB-α (Ser32/36) levels in acidic bile–treated groups (pH 4.0) (P values by t test; multiple comparisons by Holm-Sidak; GraphPad Prism 6.0) (Nuclear p-NF-κB protein levels were normalized to Histone 1; cytoplasmic p-IκB-α and bcl-2 protein levels were normalized to β-actin). (B) Graphs created by Graph Pad Prism 6.0 software reveal ranks of BAY 11-7082–induced (a) nuclear p-NF-κB(p65 S536) and (b) cytoplasmic p-IκB-α (Ser32/36) protein ratios (with/without NF-κB inhibitor) between experimental and control treated groups. Acidic bile (pH 4.0)–treated MHPC demonstrates the most significant reduction of activated NF-κB and p-IκB-α protein levels in the presence of ΒΑΥ 11-7082 (P<.05; by one-way ANOVA; Kruskal-Wallis; GraphPad Prism 6.0) (Data are from three independent experiments).
BAY 11-7082 Prevents the Acidic Bile–Induced mRNA Phenotype in MHPC
We performed qPCR to characterize the effect of BAY 11-7082 in preventing overexpression of NF-κB and related oncogenic genes in acidic bile–treated MHPC (Supplementary results; Supplementary Figure S1). RELA(p65), TNF-α, STAT3, EGFR, bcl-2, and WNT5A were selected because they had been previously found to be overexpressed in the acidic bile–treated murine HM [17]. Specifically, we found that BAY 11-7082 induced a significant reduction in acidic bile–induced transcriptional levels of RELA(p65), STAT3, TNF-α, bcl-2, and EGFR (P<.05; multiple t test). STAT3, RELA(p65), TNF-α, and bcl-2 appeared to be the most profoundly affected by NF-κB inhibition (Supplementary Figure S1, A-a). We observed that the acidic bile–treated group was most affected by the NF-κB inhibitor, resulting in lower mRNA gene ratios with/without inhibitor relative to other experimental and control groups (Supplementary Figure S1, A-b). We found that RELA(p65), TNF-α, and STAT3 showed a more intense reduction of their mRNA levels by BAY 11-7082 in acidic bile relative to neutral control and neutral bile–treated MHPC (P=.0022 and P=.0022; P=.0415 and P=.0415; P=.0415 and P=.0022).
We used Pearson analysis to identify correlations of BAY 11-7082–induced transcriptional levels of the analyzed genes and found a significant linear correlation between BAY 11-7082–induced mRNA ratios (with/without inhibitor) of RELA(p65) and TNF-α (r=0.972473, P value=.0276) in MHPC-treated groups.
All the above support the observation that NF-κB inhibition significantly prevented the acidic bile–induced overexpression of NF-κB and key oncogenic molecules in MHPC similar to our prior observations in human hypopharyngeal primary cells [19].
Acidic Bile May Induce DNA Damage
Phosphorylation of H2AX at Ser139 is commonly used as a marker for general DNA damage [33] and is generally accepted as consistent marker of DNA double-strand breaks (DSBs), applicable even under conditions where only a few DSBs are present [34].
Immunofluorescence assay revealed that acidic bile and not neutral bile, acid alone, or neutral control treatment upregulated γH2AX (pS139) in treated human hypopharyngeal primary cells (Supplementary Figure 1B), indicating that acidic bile may induce DNA damage causing DSBs.
BAY 11-7082 Reverses the Acidic Bile–Induced miRNA Phenotype in MHPC
We performed miRNA analysis to identify the role of BAY 11-7082 in preventing alterations in small regulatory molecules, such as miRNAs, under the stimulation by acidic bile (Supplementary Results; Supplementary Figure S2). We analyzed the expression of “oncomirs” miR-21, miR-192, and miR-155 and “tumor suppressors” miR-451a, miR-34a, and miR-375, previously linked to acidic bile–induced early neoplastic events in treated laryngopharyngeal mucosa [18].
We found that application of BAY 11-7082 reversed the acidic bile–induced miRNA phenotype in treated MHPC (Supplementary Figure S2). Specifically, we observed significantly lower levels of the analyzed “oncomirs” and higher levels of the analyzed “tumor suppressor” miRNAs in MHPC exposed to acidic bile with inhibitor compared to MHPC exposed to acidic bile without inhibitor (P<.05, t test; means±SD; multiple comparisons by Holm-Sidak) (Supplementary Results and Supplementary Figure S2A).
The effect of NF-κB inhibition was found to be particularly intense in the acidic bile group, demonstrating significantly higher miR-34a, miR-375, and miR-451a and lower miR-21, miR-155, and miR-192 expression ratios (with/without inhibitor) relative to other experimental and control groups (P<.05, by Kruskal-Wallis) (Supplementary Figure S2B).
Finally, we found that the calculated miR-21/miR-375 ratio with and without inhibitor was significantly lower in acidic bile compared to neutral bile and neutral control–treated MHPC (P<.05; ANOVA) (Supplementary Figure S3).
Pearson analysis revealed correlations between expression levels of the analyzed miRNA markers. We identified a positive but not significant correlation between BAY 11-7082–induced expression changes of “tumor suppressor” miR-375 and miR-451a (r=0.733097). We observed an inverse but not significant correlation between BAY 11-7082–induced expression changes of miR-21 and miR-34a (r=−0.718682), miR-21 and miR-375 (r=−0. 853456), miR-155 and miR-451a (r=−414677), and miR-192 and miR-451a (r=−0.629922) in treated MHPC.
Correlations between BAY 11-7082–Induced Changes of Oncogenic miRNA Markers and NF-κB–Related Genes in Treated MHPC
Pearson analysis revealed a strong linear correlation between BAY 11-7082–induced expression changes (with/without NF-κB inhibitor) of miRNA markers and NF-κB–related genes in treated MHPC. We identified a strongly positive correlation between BAY 11-7082–induced expression changes of “oncomiR” miR-21 and RELA(p65) (r=0.994943, P=.0051) or TNF-α (r=0.954683, P=.0454). We also found a strongly inverted correlation of the expression changes between BAY 11-7082–induced “tumor suppressor” miR-375 and TNF-α (r=−0.954396, P=.0046) and between miR-451a and STAT3 (r=−0.957702, P=.0043). There was an inverted but not statistically significant correlation between BAY 11-7082–induced expression changes of miR-375 and RELA(p65) (r=−0.897911), STAT3 (r=−0.7919789), or TNF-α (r=−0.870275), as well as between miR-34a and RELA(p65) (r=−0.645360), and miR-451a and RELA(p65) (r=−0.5820112), or bcl-2 (r=−0.405298).
In Vivo BAY 11-7082 Prevents the Acidic Bile–Induced NF-κB Activation in Murine HM
Short-term, repeated topical application of BAY 11-7082 on murine HM was performed to characterize its effect in preventing acidic bile–induced NF-κB activation and early transcriptionally activated molecular alterations.
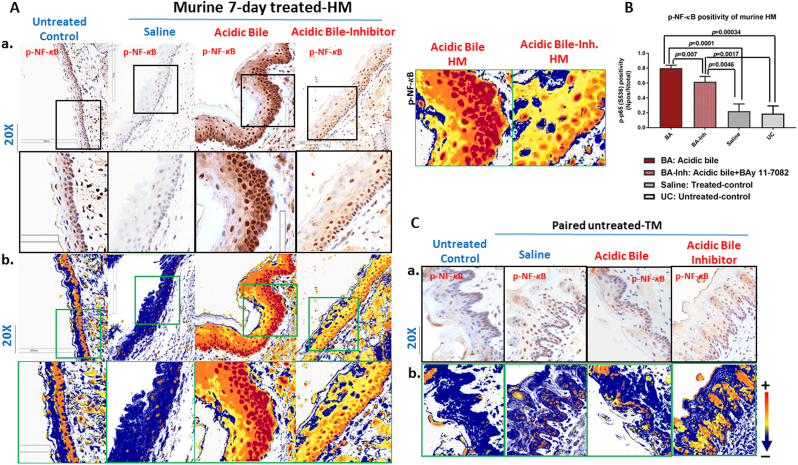
IHC analysis revealed that short-term exposure of HM to acidic bile without NF-κB inhibitor enhanced NF-κB activation (Figure 2). We found an intense p-NF-κB (p65 S536) nuclear staining in basal/parabasal and suprabasal layers of acidic bile–treated HM (Figure 2A). However, the in vivo exposure of HM to NF-κB inhibitor BAY 11-7082 prevented the acidic bile–induced NF-κB activation. BAY 11-7082 induced a less intense p-NF-κB (p65 SS536) nuclear staining in basal and suprabasal layers compared to acidic bile alone (Figure 2A). Saline-treated HM, used as a control, demonstrated low levels of NF-κB activation in a few sporadic cells and was considered negative for NF-κB activation (Figure 2A), its minimal NF-κB positivity possibly reflecting the mechanical stress of saline application. The untreated HM did not show NF-κB activation, representing negative nuclear p-NF-κB staining (Figure 2A).
Figure 2.
Topical application of BAY 11-7082 inhibits the acidic bile–induced NF-κB activation in short-term 7-day–exposed murine HM. (A-a) IHC analysis for p-NF-κB (p65 S536) (from left to right): control untreated HM, revealing cytoplasmic staining; saline-treated HM, revealing sporadic and weak cytoplasmic or nuclear staining in a few basal cells; acidic bile–treated HM, producing intense nuclear and cytoplasmic staining of basal and parabasal/suprabasal layers; and acidic bile+inhibitor (BAY 11 7082)–treated HM, demonstrating nuclear or cytoplasmic staining mainly of cells of basal layer and weak cytoplasmic staining of suprabasal layers. (b) Image analysis algorithm(s) (red: positive nuclear staining of p-NF-κB; orange: intense positive cytoplasmic staining of p-NF-κB; yellow: weak cytoplasmic staining of p-NF-κB; blue: negative p-NF-κB staining) [Images were captured using Aperio CS2 and analyzed by Image Scope software (Leica Microsystems, Buffalo Grove, IL) that generated algorithm(s) illustrating mucosal and cellular compartments demonstrating p-NF-κB staining]. (B) Acidic bile (pH 3.0) induces significantly higher positive nuclear p-NF-κB (p65 S536) levels compared to saline-treated HM or untreated control. Topical application of BAY 11-7082 significantly decreases nuclear p-NF-κB levels in acidic bile (pH 3.0) HM (P values by t test; multiple comparisons by Holm-Sidak; GraphPad Prism 6.0) (positivity=nuclear-positive/total-positive p-p65 staining). (C-a) IHC analysis for p-NF-κB (p65 S536) and (b) corresponding image analysis algorithms in murine tongue mucosa (used as internal control for analyzed HM in each group) (from left to right): control untreated TM, revealing weak cytoplasmic staining; saline-treated TM, acidic-bile-treated TM and acidic-bile+inhibitor (BAY 11 7082)–treated TM, producing sporadic weak cytoplasmic or nuclear staining in scattered basal cells (red: positive nuclear staining of p-NF-κB; orange: intense positive cytoplasmic staining of p-NF-κB; yellow: weak cytoplasmic staining of p-NF-κB; blue: negative p-NF-κB staining).
Scoring of nuclear p-NF-κB (p65 S536) by ImageScope software revealed that the short-term exposure of HM to acidic bile induced significantly higher levels of activated NF-κB compared to saline and untreated controls (Figure 2B) (P<.05, t test; means±SD; multiple comparisons by Holm-Sidak). In contrast, the effect of NF-κB inhibitor resulted in significantly lower NF-κB activated levels compared to acidic bile without inhibitor (Figure 2B).
Mucosa from TM used as internal control for each animal and considered to be negative for NF-κB activation revealed only a few sporadic cells positive for p-NF-κB nuclear staining, allowing its use as internal control for molecular analysis (Figure 2C).
Short-Term Topical In Vivo Exposure of Murine HM to Acidic Bile, Activating Cell Proliferation, Is Prevented in the Presence of BAY 11-7082
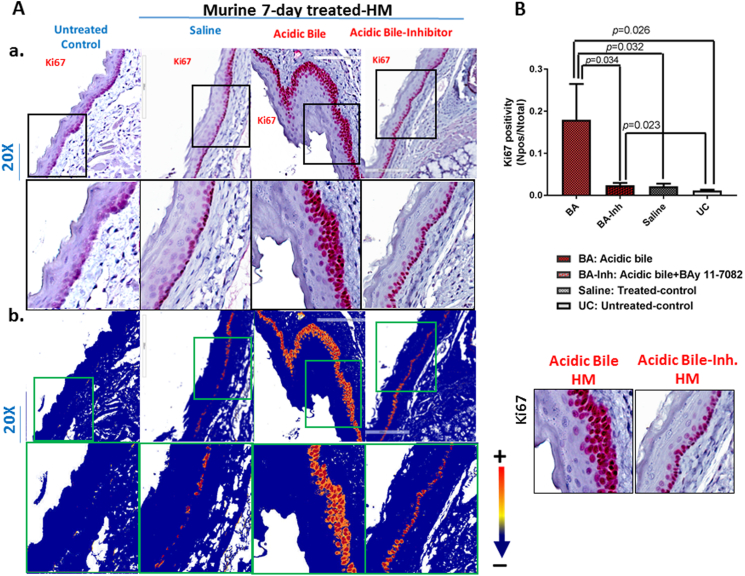
The short-term effect of acidic bile (pH 3.0) on murine HM resulted in increased proliferation of regenerating cells under acidic bile stimuli. The elevated cell proliferation was documented by intense staining of cell proliferation marker Ki67 in basal and parabasal layers of acidic bile–treated HM (Figure 3A). Scoring of nuclear Ki67 by ImageScope software showed that the short-term exposure of HM to acidic bile induced significantly higher levels of nuclear Ki67 compared to saline and untreated controls (Figure 3B) (P<.05, t test; means±SD; multiple comparisons by Holm-Sidak).
Figure 3.
Topical application of BAY 11-7082 prevents acidic bile–induced cell proliferation in short-term exposed murine HM. (A-a) IHC analysis for Ki67 (from left to right): control untreated HM, revealing negative nuclear or cytoplasmic staining; saline-treated HM, demonstrating sporadic and weak cytoplasmic or nuclear staining in few basal cells; acidic bile–treated HM, revealing intense nuclear and cytoplasmic staining of cells of basal and parabasal layers; and acidic-bile+inhibitor (BAY 11 7082)–treated HM, producing nuclear or cytoplasmic staining mainly in cells of the basal layer. (b) Image analysis algorithm(s) (red: nuclear positive staining of Ki67; orange: intense positive cytoplasmic staining of Ki67; yellow: weak cytoplasmic staining of Ki67; blue: negative Ki67 staining) [Images were captured using Aperio CS2 and analyzed by Image Scope software (Leica Microsystems, Buffalo Grove, IL) that generated algorithm(s) illustrating the mucosal and cellular Ki67 staining compartments]. (B) Graphs depict short-term exposure of HM to acidic bile (pH 3.0) inducing significantly higher positive nuclear Ki67 levels compared to saline-treated HM or untreated control. Topical application of BAY 11-7082 significantly decreases nuclear Ki67 levels in acidic bile (pH 3.0) HM (P values by t test; multiple comparisons by Holm-Sidak; GraphPad Prism 6.0).(positivity=nuclear-positive/total-positive Ki67 staining).
BAY 11-7082 prevented the acidic bile–induced regeneration of the basal layer. Microscopic examination of HM exposed to BAY 11-7082 revealed that Ki67 staining was limited to the single basal layer of treated MH (Figure 3A), demonstrating significantly lower levels of positivity compared to acidic bile–treated HM (Figure 3B). Histologic examination of treated HM revealed that there were no histological signs of local treatment toxicity.
Short-Term Topical In Vivo Exposure of Murine HM to Acidic Bile, Upregulating the NF-κB–Related Oncogenic mRNA Phenotype, Is Prevented in the Presence of BAY 11-7082
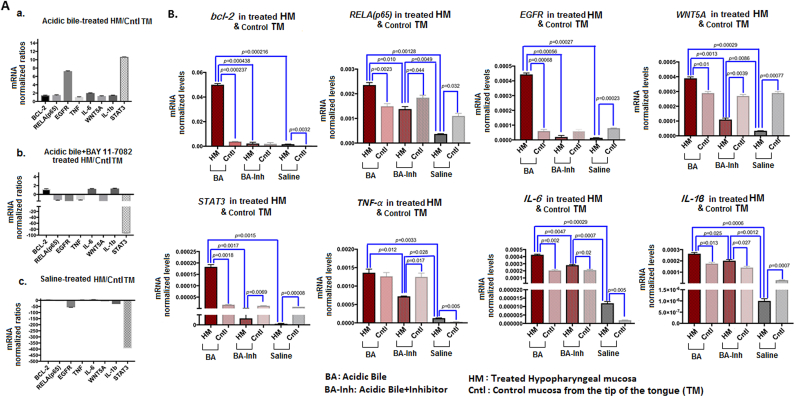
We performed qPCR analysis to identify the effect of short-term topical application of BAY 11-7082 on murine HM. We found that short-term exposure of HM to acidic bile induced a significant upregulation of RELA(p65), STAT3, EGFR, bcl-2, WNT5Α, IL-6, and IL-1β compared to its corresponding TM (P<.05; multiple t test) (Figure 4). Short-term exposure of HM to acidic bile therefore upregulated the expression of NF-κB and related genes with oncogenic function (Figure 4, A-a), previously identified in acidic bile–induced premalignant HM [19]. Topical application of BAY 11-7082 prevented the acidic bile–induced upregulation of the analyzed genes linked to a cancer-related mRNA phenotype. Specifically, HM exposed to acidic bile with NF-κB inhibitor produced a reversed mRNA phenotype compared to HM exposed to acidic bile alone (Figure 4, A-b). BAY 11-7082–induced mRNA phenotype was similar to that induced by saline (Figure 4, A-c).
Figure 4.
In vivo short-term exposure to acidic bile, inducing transcriptional activation of NF-κB related genes with antiapoptotic, cell proliferation, or oncogenic function, is prevented by topical application of BAY 11-7082. (A) Columns of graphs created by Graph Pad Prism software 6.0 depict mRNA phenotype in short-term exposed HM to (a) acidic bile (pH 3.0), (b) acidic bile (pH 3.0) + BAY 11-7082, and (c) saline (pH 7.0). mRNA phenotypes correspond to transcriptional expression ratios of NF-κB–related genes bcl-2, RELA(p65), EGFR, WNT5A, STAT3, TNF-α, IL-6, and IL-1β comparing treated HM to corresponding mucosa from TM, used as internal control, by real-time qPCR analysis (mRNA levels of each target gene were normalized to GAPDH). We observe an upregulation of mRNA expression of the analyzed genes in acidic bile–treated HM. BAY 11-7082 reverses the acidic bile–induced mRNA phenotype, demonstrating a phenotype similar to saline-induced mRNA. (B) Graphs, created by Graph Pad Prism 6 software, reveal transcriptional levels (normalized to GAPDH) for the analyzed genes comparing HM exposed to acidic bile, acidic bile+BAY 11-7082, and saline to corresponding TM by real-time qPCR analysis (P value <0.05; by t test; multiple comparisons by Holm-Sidak; data obtained from four analyzed samples).
Acidic bile–treated HM demonstrated significantly higher mRNA levels of RELA(p65), bcl-2, STAT3, EGFR, TNF-α, WNT5A, IL-6, and IL-1β compared to saline-treated HM (P<.00; multiple t test) (Figure 4B). In contrast, we observed that the in vivo topical application of BAY 11-7082 on HM inhibited the acidic bile–induced overexpression of all the analyzed genes (Figure 4B). Specifically, we found significantly lower transcriptional levels of RELA(p65), bcl-2, STAT3, EGFR, TNF-α, WNT5A, IL-6, and IL-1β in HM exposed to acidic bile with BAY 11-7082 compared to HM exposed to acidic bile without BAY 11-7082 (Figure 4B). Interestingly, the NF-κB inhibitor restored the transcriptional levels of crucial oncogenic factors, such as bc-2, EGFR, and STAT3, to levels expressed in saline-treated HM.
Short-Term Topical In Vivo Exposure of Murine HM to Acidic Bile, Resulting in Deregulation of Cancer-Related miRNA Phenotype, Is Prevented by BAY 11-7082
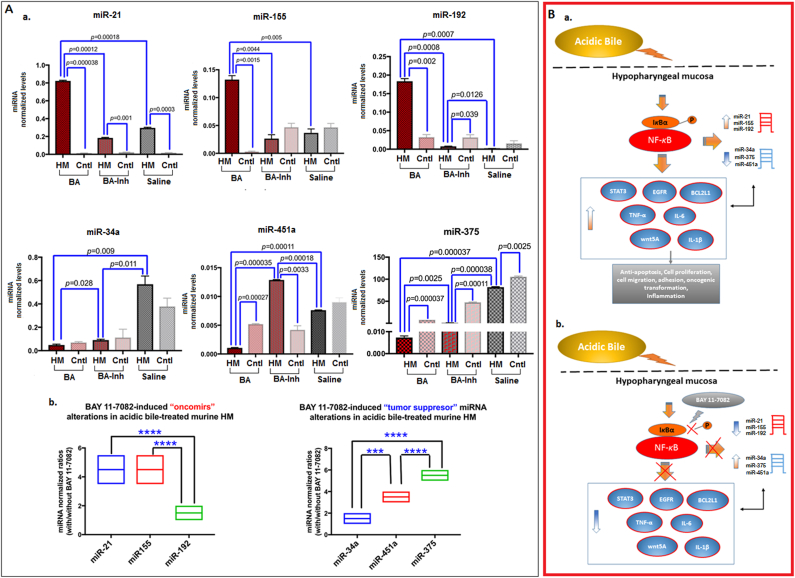
We performed miRNA analysis to demonstrate 1) the short-term in vivo effect of topical acidic bile on murine HM with regard to the expression of “oncomirs” and “tumor suppressor” specific miRNA markers, previously linked to acidic bile–induced early preneoplastic events of the laryngopharyngeal mucosa, and 2) their possible inversion when exposed to topically applied BAY 11-7082 (Figure 5).
Figure 5.
(A) In vivo short-term exposure of murine HM to acidic bile, resulting in deregulation of cancer-related miRNA phenotype, is prevented by topical application of BAY 11-7082. (a) Upregulation of “oncomirs” miR-21, miR-155, and miR-192 and downregulation of tumor suppressors miR-375, -34a, and -451a in short-term exposed murine HM to acidic bile (pH 3.0) compared to saline control and to mucosa from TM, used as internal control, by real-time qPCR analysis. Topical application of BAY 11-7082 prevents these effects (miRNA levels normalized to RNU6) (columns of graphs created by Graph Pad Prism software 6.0; P value <.05; by t test; multiple comparisons by Holm-Sidak; data obtained from four analyzed samples). (b) BAY 11-7082 induces “oncomirs” and “tumor suppressor” miRNA expression ratios (with/without inhibitor miRNA levels, normalized to RNU6) in acidic bile–treated murine HM [***P<.0005; ****P<.00005; by one-way ANOVA; Kruskal-Wallis; graph plots, representing miRNA means (middle line), created by GraphPad 6.0].
(B) Schematic representation of NF-κB inhibition by BAY 11-7082 in acidic bile–induced cancer-related mRNA and miRNA phenotypes in murine HM (a) acidic bile–induced NF-κB activation and production of cancer-related mRNA and miRNA phenotypes (b) NF-κB inhibition by topical application of BAY 11-7082 in acidic bile–induced cancer-related mRNA and miRNA phenotypes.
We found that short-term application of acidic bile upregulated the expression of “oncomirs” miR-21, miR-155, and miR-192 and downregulated the expression of “tumor suppressors” miR-451a and miR-375 compared to its corresponding TM (P<.005) (Figure 5A). Acidic bile–treated HM demonstrated significantly higher levels of the analyzed “oncomirs” and lower levels of “tumor suppressor” miRNAs compared to saline-treated HM (P<.005, multiple t test) (Figure 5, A-a).
NF-κB inhibition significantly prevented the acidic bile–induced upregulation of “oncomirs” as well as the downregulation of “tumor suppressor” miRNAs. The application of acidic bile with BAY 11-7082 was observed to induce significantly lower miR-21, miR-155, and miR-192 and higher miR-34a, miR-375, and miR-451a levels in treated HM compared to acidic bile alone (P<.005, multiple t test) (Figure 5, A-a).
Relative expression ratios of the analyzed miRNA markers between acidic bile with BAY 11-7082 and acidic bile alone revealed that “oncomiR” miR-192 and “tumor suppressor” miR-375 were the most affected by NF-κB inhibition (Figure 5, A-b). We observed that miR-192 demonstrated the lowest ratios (with/without inhibitor), with a significant difference compared to miR-21 and miR-155. Similarly, miR-375 showed the highest expression ratios (with/without inhibitor), with a significant difference compared to miR-34a and miR-451a (P<.00005). Additionally, NF-κB inhibition induced significantly higher expression ratios (with/without inhibitor) of miR-451a relative to miR-34a (P<.0005) in acidic bile–treated HM. Finally, we found that the calculated miR-21/375 ratios were significantly lower in HM exposed to acidic bile with BAY 11-7082 compared to HM exposed to acidic bile without BAY 11-7082 (P<.0005) one-way ANOVA, Kruskal-Wallis).
NF-κB Mediates Acidic Bile–Induced Interactions between Cancer-Related miRNA Markers and NF-κB–Related Genes with Oncogenic Function in Acidic Bile–Treated Murine Hypopharyngeal Epithelial Cells In Vitro and In Vivo
We showed that NF-κB inhibitor (BAY 11-7082) prevented the acidic bile–induced transcriptional activation of genes with oncogenic function and deregulation of oncogenic miRNA markers in in vivo murine HM and in cultured murine hypopharyngeal primary cells, as previously shown in cultured normal human hypopharyngeal cells [19]. Figure 5B summarizes the observed effect of BAY 11-7082 in murine hypopharyngeal epithelial cells in vitro and HM in vivo, and demonstrates proposed interactions among cancer-related miRNA markers and NF-κB related genes, supported by the observed statistically significant correlations among their expression changes in cultured MHPC.
BAY 11-7082 Reduces Acidic Bile–Induced Gene Expression for the NF-κB Signaling Pathway in In Vivo Exposed Murine HM
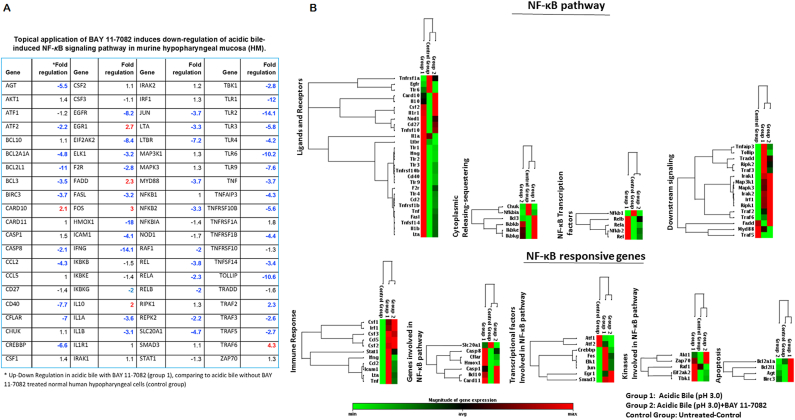
We performed a PCR array for the NF-κB signaling pathway to explore acidic bile–induced gene expression profiling of NF-κB signaling with and without BAY 11-708 compared to saline-treated HM. A significant acceleration of NF-κB signaling in HM was observed by acidic bile relative to control (Figure 6). Specifically, we found 83% of the analyzed genes (70 out of 84) to be upregulated (>two-fold change) in treated HM after short-term exposure to acidic bile (Supplementary Table S3). The effect of BAY 11-7082 reduced the acidic bile–induced transcriptional levels of 48 out of 84 analyzed NF-κB signaling related genes (~60%) (>two-fold change), as similarly observed in human hypopharyngeal primary cells in vitro [19] (Figure 6A).
Figure 6.
Short-term exposure of murine HM to acidic bile upregulates the NF-κB signaling pathway and is reduced by topical application of BAY 11-7082. (A). Table presents the expression changes (upregulation or downregulation by two-fold changes) of 84 genes of NF-κB signaling pathway, analyzed by PCR microarray analysis, in acidic bile–treated HM under the topical application of BAY 11-7082. (B) Heat maps were obtained by RT2-Profiler PCR array analysis for NF-κB signaling. Genes were clustered based on their biological role. Heat maps demonstrated the effect of BAY 11-7082 in gene expression of NF-κB pathway and NF-κB responsive genes (red color for maximum expression and green for minimum). Group 1: acidic bile–treated HM; control group: saline treated-HM, and group 2: acidic bile with BAY 11-7082–treated HM [gene expression has been normalized to two housekeeping genes: GAPDH, glyceraldehyde-3-phosphate dehydrogenase and Hsp90ab1, Heat shock protein 90 alpha (cytosolic), class B member 1].
The effect of acidic bile with and without BAY 11-7082 on the NF-κB signaling pathway is provided in Figure 6B. Among others, NF-κB inhibitor downregulated the acidic bile–induced transcriptional levels of the NF-κB transcription factors Rel (>3.5-fold), RELA(p65) (>2-fold), RELB (>2-fold), and NFkB2 (>3-fold). NF-κB inhibitor also downregulated members of TNF-receptors such as CD40 (>7.5-fold), TNFRSF10B (>5.5-fold), TNFRSF1B (>4-fold), and TNFSF14 (>3-fold). Moreover, BAY 11-7082 reduced the transcriptional levels of receptors and ligands of the innate immune system, like TLR1 (>11-fold), TLR-2 (>14-fold), TLR3 (>5.5-fold), TLR4 (>4-fold), TLR6 (>10-fold), TLR-9 (>7.5-fold), as well as IL-1A (>3.5-fold) and IL1B (>3-fold). Interestingly, BAY 11-7082 increased the transcriptional levels of IL10 (two-fold) known to block NF-κB activity.
BAY 11-7082 also reduced NF-κB downstream signaling, preventing the expression of positive regulators of the NF-κB pathway, such as TNFAIP3 (>4-fold), TRAF3 (>2.5-fold), and Tollip (>10-fold). The effect of BAY 11-7082 reduced the expression of Inhibitor-kappaB kinases IKBKB, IKBE, and IKBKG (~1.5 times), as well as of bcl-3 (>3-fold), which is a co-activator of NF-κB, preventing the cytoplasmic release of NF-κB.
NF-κB inhibition resulted in reduction of NF-κB responsive genes in acidic bile–treated cells. We observed a decrease in the expression of antiapoptotic genes, such as BCL2A1A (>4.5-fold), BCL2L1 (>11-fold), and BIRC3 (>3-fold) genes, and cflar (7-fold), an inhibitor of apoptosis. BAY 11-7082 also downregulated kinases that activate NF-κB pathways, such as EIF2AK2 (>8-fold), and transcriptional factors, such as EGFR (>8-fold), ELK1 (>3-fold), FOS (>2.5-fold), and Jun (>3.5-fold), in line with our data from qPCR analysis. Finally, NF-κB inhibitor affected other NF-κB signaling genes such as Hmox1 (>18-fold), known to exert a protumorigenic role.
Discussion
We present the first report of a topically applied specific NF-κB inhibitor, BAY 11-7082, to HM exposed to acidic bile, considered a potential risk factor in hypopharyngeal carcinogenesis. Our novel findings reveal that topical application of BAY 11-7082 can effectively prevent acidic bile–induced NF-κB activation, its signaling pathway, and its transcriptional activation of NF-κB–related genes previously linked to early neoplastic events in murine HM. The ability of BAY 11-7082 to suppress acidic bile–induced molecular events was successful both in vitro in murine hypopharyngeal primary cells and in vivo in murine HM.
Our data demonstrate that the short-term local effect of acidic bile is very capable of transcriptionally stimulating the NF-κB signaling pathway and associated genes participating in oncogenic pathways of head and neck cancer, including STAT3, EGFR, bcl-2, TNF-α, and WNT5A. The topical application of BAY 11-7082 can diminish the acidic bile–induced NF-κB activation of proliferating cells located at the basal mucosal layer. NF-κB inhibition also reduces the acidic bile–induced upregulation in the majority (~60%) of genes implicated in NF-κB signaling, as similarly identified in human hypopharyngeal primary cells in vitro [19]. These data strongly reveal that the transcriptional activation of molecules involved in antiapoptotic and cell proliferation function, as well as cancer-related cytokines, happens early in HM exposed to acidic bile and that topical application of a pharmacologic NF-κB inhibitor is capable of sufficiently blocking this effect.
The observed early molecular changes, which are prevented by NF-κB inhibitor, also include small miRNA molecules. The effect of BAY 11-7082 in preventing the upregulation of “oncomirs,” such as miR-21, miR-155, and miR-192, and downregulation of “tumor suppressors” miR-34a, miR-375, and miR-451a demonstrates that NF-κB inhibition can restore regulating transcriptional mechanisms that may have originally been activated by acidic bile in vivo. Topical short-term exposure of HM to NF-κB inhibitor characteristically affects the “tumor suppressor” miR-451a and “oncomiR” miR-192, previously linked to hypopharyngeal cancer and gastroesophageal reflux, respectively [32], [35]. Our data are in line with findings from cultured normal human hypopharyngeal cells demonstrating that BAY 11-7082 can reverse the acidic bile–induced miRNA phenotype [unpublished data]. BAY 11-7082 topically applied is also able to inhibit the acidic bile–induced overexpression of the “oncomirs” miR-21 and miR-155, important markers for poor prognosis in head and neck cancer [26], [36], [37], [38]. This view is partly supported by prior studies suggesting that NF-κB directly regulates the expression of miR-21 and miR-155 via their binding promoters [39], [40], [41], [42] and that NF-κB inhibition thereby markedly suppresses upregulation of miR-21 [43].
Our data support previously proposed interactions between acidic bile–induced NF-κB activation and alterations of bcl-2, EGFR, STAT3, TNF-α, and WNT5A, which may directly or indirectly interact with cancer-related miRNA markers, such as “oncomirs” and “tumor suppressor” miRNA markers. Specifically, data demonstrate strong correlations among the analyzed oncogenic miRNA markers, such as “oncomirs” miR-21, and particularly the “tumor suppressors” miR-34a, miR-375, and miR-451a, and NF-κB–related genes, such as NF-κB transcriptional factor RELA(p65), cytokine TNF-α, and the important regulator of cell proliferation STAT3, supporting the role of the studied miRNAs in our proposed acidic bile–related carcinogenic pathway (Figure 5B). These findings are in line with previous studies suggesting interactions of miRNA molecules, such as miR-21 and miR-34a, with other transcriptional factors interacting with NF-κB signaling pathway, such as STAT3, in an NF-κB–dependent manner [39], [44].
According to Tili et al., permanent upregulation of miR-155 may mediate a prolonged inflammatory reaction leading to cancer [45]. Our novel findings show that NF-κB mediating upregulation of “oncomiR” miR-155 and mRNAs of inflammatory molecules, such as TNF-α, IL-6, and IL-1β, can be effectively prevented by BAY 11-7082, suggesting a protective role of NF-κB inhibition in inflammatory events linked to downstream oncogenic pathways.
Whereas the ratio miR-21/miR-375 was previously implicated as a biomarker for poor prognosis in supraglottic cancer [30], we demonstrate that miR-21/miR-375 was impacted by NF-κB inhibition, as similarly observed in human hypopharyngeal primary cells in vitro [unpublished data], thus demonstrating that acidic bile–induced NF-κB activation may interact with both miRNA markers, serving as a potential index for malignant transformation.
Finally, our microscopic examination of treated mucosa revealed that NF-κB inhibitor reduced the acidic bile–induced NF-κB activation not only in superficial layers of hypopharyngeal epithelium but also in its regenerating basal layer. The intense expression of Ki67, a cell proliferation marker, found in the basal and parabasal layers of short-term acidic bile–treated HM supports cell proliferation or regeneration of mucosa under the acidic bile stimulus. The observation that topically applied NF-κB inhibition with BAY 11-7082 successfully decreased Ki67 suggests that BAY 11-7082 is capable of preventing NF-κB–induced cell proliferative events. In this view, the topical use of a suitable NF-κB inhibitor may be central to the search for an effective program of chemo-prevention.
Our data verify the key role of NF-κB in acidic bile–induced early molecular changes promoting malignant transformation and prevented by NF-κB inhibition. However, the link between the acidic bile effect and the NF-κB activation in hypopharyngeal cells is not yet well understood. Dvorak et al. previously showed that bile at low pH could induce oxidative stress and DNA damage in esophageal cells [5], while others suggested that NF-κB could be activated as part of the DNA damage response [46]. Our data show that acidic bile upregulates both NF-κB [9], [17], [19] and phosphorylated H2AX, a marker of DNA damage [33], [34], in treated hypopharyngeal primary cells, suggesting NF-κB activation may be caused by acidic bile–induced DNA damage. However, the exact effect of acidic bile in HM and the promoting events that lead to NF-κB activation should be further investigated using in vitro and in vivo models.
Conclusion
The current study demonstrates the effectiveness of topical BAY 11-7082, an NF-κB inhibitor, in the prevention of early molecular alterations induced by acidic bile in murine HM. The protective role of NF-κB inhibition in acidic bile–induced neoplastic transformation could lead to future translational efforts in head and neck chemoprevention.
Data Availability
The data sets generated during and/or analyzed during the current study are available from the corresponding author.
Author Contributions
-
•
Conception and design: D. P. V., C. T. S., S. G. D.
-
•
Development of methodology: D. P. V., S. G. D.
-
•
Acquisition of data: S. G. D., D. P. V.
-
•
Analysis and interpretation of data: D. P. V., S. G. D., C. T. S.
-
•
Writing, review and/or revision of the manuscript: C. T. S., D. P. V., S. G. D.
-
•
Administrative, technical, or material support: C. T. S.
-
•
Study supervision: D. P. V., C. T. S.
Footnotes
Conflict of interest statement: The authors whose names are listed in this article certify that they have NO affiliations with or involvement in any organization or entity with any financial interest or nonfinancial interest in the subject matter or materials discussed in this manuscript.
Grant support: This study was supported by the Virginia Alden Wright Fund.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2018.02.001.
Appendix A. Supplementary Data
Supplementary material
References
- 1.Galli J, Cammarota G, De Corso E, Agostino McQuaid KR, Laine L, Fennerty MB, Souza R, Spechler SJ. Systematic review: the role of bile acids in the pathogenesis of gastro-oesophageal reflux disease and related neoplasia. Aliment Pharmacol Ther. 2011;34:146–165. doi: 10.1111/j.1365-2036.2011.04709.x. [DOI] [PubMed] [Google Scholar]
- 2.Nehra D, Howell P, Williams CP, Pye JK, Beynon J. Toxic bile acids in gastro-oesophageal reflux disease: influence of gastric acidity. Gut. 1999;44:598–602. doi: 10.1136/gut.44.5.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langevin SM, Michaud DS, Marsit CJ, Nelson HH, Birnbaum AE, Eliot M, Christensen BC, McClean MD, Kelsey KT. Gastric reflux is an independent risk factor for laryngopharyngeal carcinoma. Cancer Epidemiol Biomark Prev. 2013;22:1061–1068. doi: 10.1158/1055-9965.EPI-13-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kauer WK, Peters JH, DeMeester TR, Ireland AP, Bremner CG, Hagen JA. Mixed reflux of gastric and duodenal juices is more harmful to the esophagus than gastric juice alone. The need for surgical therapy re-emphasized. Ann Surg. 1995;222:525–531. doi: 10.1097/00000658-199522240-00010. [discussion 531-33] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dvorak K, Payne CM, Chavarria M, Ramsey L, Dvorakova B, Bernstein H, Holubec H, Sampliner RE, Guy N, Condon A. Bile acids in combination with low pH induce oxidative stress and oxidative DNA damage: relevance to the pathogenesis of Barrett’s oesophagus. Gut. 2007;56:763–771. doi: 10.1136/gut.2006.103697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kauer WK, Stein HJ. Role of acid and bile in the genesis of Barrett's esophagus. Chest Surg Clin N Am. 2002;12:39–45. doi: 10.1016/s1052-3359(03)00064-4. [DOI] [PubMed] [Google Scholar]
- 7.Iftikhar SY, Ledingham S, Steele RJ, Evans DF, Lendrum K, Atkinson M, Hardcastle JD. Bile reflux in columnar-lined Barrett’s oesophagus. Ann R Coll Surg Engl. 1993;75:411–416. [PMC free article] [PubMed] [Google Scholar]
- 8.Kauer WK, Peters JH, DeMeester TR, Feussner H, Ireland AP, Stein HJ, Siewert RJ. Composition and concentration of bile acid reflux into the esophagus of patients with gastroesophageal reflux disease. Surgery. 1997;122:874–881. doi: 10.1016/s0039-6060(97)90327-5. [DOI] [PubMed] [Google Scholar]
- 9.Sasaki CT, Issaeva N, Vageli DP. In vitro model for gastroduodenal reflux-induced nuclear factor-kappaB activation and its role in hypopharyngeal carcinogenesis. Head Neck. 2016;38(Suppl. 1):E1381–91. doi: 10.1002/hed.24231. [DOI] [PubMed] [Google Scholar]
- 10.Sasaki CT, Toman J, Vageli D. The in vitro effect of acidic-pepsin on nuclear factor kappaB activation and its related oncogenic effect on normal human hypopharyngeal cells. PLoS One. 2016;11 doi: 10.1371/journal.pone.0168269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Cho CH. Effect of NF-κB signaling on apoptosis in chronic inflammation-associated carcinogenesis. Curr Cancer Drug Targets. 2010;10:593–599. doi: 10.2174/156800910791859425. [DOI] [PubMed] [Google Scholar]
- 12.Klein JD, Grandis JR. The molecular pathogenesis of head and neck cancer. Cancer Biol Ther. 2010;9:1–7. doi: 10.4161/cbt.9.1.10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nottingham LK, Yan CH, Yang X, Si H, Coupar J, Bian Y, Cheng TF, Allen C, Arun P, Gius D. Aberrant IKKα and IKKβ cooperatively activate NF-κB and induce EGFR/AP1 signaling to promote survival and migration of head and neck cancer. Oncogene. 2014;2014(33):1135–1147. doi: 10.1038/onc.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allen CT, Ricker JL, Chen Z, Van Waes C. Role of activated nuclear factor-kappaB in the pathogenesis and therapy of squamous cell carcinoma of the head and neck. Head Neck. 2007;29:959–971. doi: 10.1002/hed.20615. [DOI] [PubMed] [Google Scholar]
- 15.Loercher A, Lee TL, Ricker JL, Howard A, Geoghegen J, Chen Z, Sunwoo JB, Sitcheran R, Chuang EY, Mitchell JB. Nuclear factor-kappaB is an important modulator of the altered gene expression profile and malignant phenotype in squamous cell carcinoma. Cancer Res. 2004;64:6511–6523. doi: 10.1158/0008-5472.CAN-04-0852. [DOI] [PubMed] [Google Scholar]
- 16.Lee TL, Yang XP, Yan B, Friedman J, Duggal P, Bagain L, Dong G, Yeh NT, Wang J, Zhou J. A novel nuclear factor-kappaB gene signature is differentially expressed in head and neck squamous cell carcinomas in association with TP53 status. Clin Cancer Res. 2007;13:5680–5691. doi: 10.1158/1078-0432.CCR-07-0670. [DOI] [PubMed] [Google Scholar]
- 17.Vageli DP, Prasad ML, Sasaki CT. Gastro-duodenal fluid induced nuclear factor-κappaB activation and early pre-malignant alterations in murine hypopharyngeal mucosa. Oncotarget. 2016;7:5892–5908. doi: 10.18632/oncotarget.6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki CT, Vageli DP. miR-21, miR-155, miR-192, and miR-375 deregulations related to NF-kappaB activation in gastroduodenal fluid-induced early preneoplastic lesions of laryngeal mucosa in vivo. Neoplasia. 2016;18:329–338. doi: 10.1016/j.neo.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vageli DP, Doukas SG, Sasaki CT. Inhibition of NF-κB prevents the acidic bile–induced oncogenic mRNA phenotype in human hypopharyngeal cells. Oncotarget. 2018;9:5876–5891. doi: 10.18632/oncotarget.23143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 21.Lin Y, Bai L, Chen W, Xu S. The NF-κB activation pathways, emerging molecular targets for cancer prevention and therapy. Expert Opin Ther Targets. 2010;14:45–55. doi: 10.1517/14728220903431069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Waes C. Nuclear factor-kappaB in development, prevention, and therapy of cancer (2007) Clin Cancer Res. 2007;13:1076–1082. doi: 10.1158/1078-0432.CCR-06-2221. [DOI] [PubMed] [Google Scholar]
- 23.Pierce JW, Schoenleber R, Jesmok G, Best J, Moore SA, Collins T, Gerritsen ME. Novel inhibitors of cytokine-induced IkappaBalpha phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. J Biol Chem. 1997;272:21096–21103. doi: 10.1074/jbc.272.34.21096. [DOI] [PubMed] [Google Scholar]
- 24.Nakanishi C, Toi M. Nuclear factor-kappaB inhibitors as sensitizers to anticancer drugs. Nat Rev Cancer. 2005;5:297–309. doi: 10.1038/nrc1588. [DOI] [PubMed] [Google Scholar]
- 25.Meng Z, Lou S, Tan J, Xu K, Jia Q, Zheng W. Nuclear factor-kappa B inhibition can enhance apoptosis of differentiated thyroid cancer cells induced by 131I. PLoS One. 2012;7 doi: 10.1371/journal.pone.0033597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allen CT, Ricker JL, Chen Z, Van Waes C. Role of activated nuclear factor-kappaB in the pathogenesis and therapy of squamous cell carcinoma of the head and neck. Head Neck. 2017;29:959–971. doi: 10.1002/hed.20615. [DOI] [PubMed] [Google Scholar]
- 27.Sakurai H, Chiba H, Miyoshi H, Sugita T, Toriumi W. IkappaB kinases phosphorylate NF-kappaB p65 subunit on serine 536 in the transactivation domain. J Biol Chem. 1999;274:30353–30356. doi: 10.1074/jbc.274.43.30353. [DOI] [PubMed] [Google Scholar]
- 28.McQuaid KR, Laine L, Fennerty MB, Souza R, Spechler SJ. Systematic review: the role of bile acids in the pathogenesis of gastro-oesophageal reflux disease and related neoplasia. Aliment Pharmacol Ther. 2011;34:146–165. doi: 10.1111/j.1365-2036.2011.04709.x. [DOI] [PubMed] [Google Scholar]
- 29.Kundu JK, Shin YK, Kim SH, Surh YJ. Reseratrol inhibits phorbol ester-induced expression of COX-2 and activation of NF-kappaB in mouse skin by blocking inhibitor kinase activity. Carcinogenesis. 2006;27:1465–1474. doi: 10.1093/carcin/bgi349. [DOI] [PubMed] [Google Scholar]
- 30.Hu A, Huang JJ, Xu WH, Jin XJ, Li JP, Tang YJ, Huang XF, Cui HJ, Sun GB, Li RL. MiR-21/miR-375 ratio is an independent prognostic factor in patients with laryngeal squamous cell carcinoma. Am J Cancer Res. 2015;5:1775–1785. [PMC free article] [PubMed] [Google Scholar]
- 31.Yan B, Li H, Yang X, Shao J, Jang M, Guan D, Zou S, Van Waes C, Chen Z, Zhan M. Unraveling regulatory programs for NF-kappaB, p53 and microRNAs in head and neck squamous cell carcinoma. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bus P, Siersema PD, Verbeek RE, van Baal JW. Upregulation of miRNA-143, -145, -192, and -194 in esophageal epithelial cells upon acidic bile salt stimulation. Dis Esophagus. 2014;27:591–600. doi: 10.1111/dote.12112. [DOI] [PubMed] [Google Scholar]
- 33.Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothkamm K, Löbrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci U S A. 2003;100:5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukumoto I, Kinoshita T, Hanazawa T, Kikkawa N, Chiyomaru T, Enokida H, Yamamoto N, Goto Y, Nishikawa R, Nakagawa M. Identification of tumour suppressive microRNA-451a in hypopharyngeal squamous cell carcinoma based on microRNA expression signature. Br J Cancer. 2014;111:386–394. doi: 10.1038/bjc.2014.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen D, Cabay RJ, Jin Y, Wang A, Lu Y, Shah-Khan M, Zhou X. MicroRNA deregulations in head and neck squamous cell carcinomas. J Oral Maxillofac Res. 2013;4(1) doi: 10.5037/jomr.2013.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arantes LMRB, Laus AC, Melendez ME, de Carvalho AC, Sorroche BP, De Marchi PR, Evangelista AF, Scapulatempo-Neto C, de Souza Viana L, Carvalho AL. MiR-21 as prognostic biomarker in head and neck squamous cell carcinoma patients undergoing an organ preservation protocol. Oncotarget. 2017;8:9911–9921. doi: 10.18632/oncotarget.14253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hui AB, Bruce JP, Alajez NM, Shi W, Yue S, Perez-Ordonez B, Xu W, O'Sullivan B, Waldron J, Cummings B. Comprehensive microRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res. 2010;16:1129–1139. doi: 10.1158/1078-0432.CCR-09-2166. [DOI] [PubMed] [Google Scholar]
- 39.Niu J, Shi Y, Tan G, Yang CH, Fan M, Pfeffer LM, Wu ZH. DNA damage induces NF-κB-dependent microRNA-21 up-regulation and promotes breast cancer cell invasion. J Biol Chem. 2012;287:21783–21795. doi: 10.1074/jbc.M112.355495. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Yang Z, Fang S, Di Y, Ying W, Tan Y, Gu W. Modulation of NF-κB/miR-21/PTEN pathway sensitizes non-small cell lung cancer to cisplatin. PLoS One. 2015;10 doi: 10.1371/journal.pone.0121547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma X, Becker Buscaglia LE, Barker JR, Li Y. MicroRNAs in NF-κB signaling. J Mol Cell Biol. 2011;3:159–166. doi: 10.1093/jmcb/mjr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerloff D, Grundler R, Wurm AA, Bräuer-Hartmann D, Katzerke C, Hartmann JU, Madan V, Müller-Tidow C, Duyster J, Tenen DG. NF-κB/STAT5/miR-155 network targets PU.1 in FLT3-ITD-driven acute myeloid leukemia. Leukemia. 2015;29:535–547. doi: 10.1038/leu.2014.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y, Xian P-F, Yang L, Wang SX. MicroRNA-21 promotes proliferation of fibroblast-like synoviocytes through mediation of NF-κB nuclear translocation in a rat model of collagen-induced rheumatoid arthritis. Biomed Res Int. 2016;2016 doi: 10.1155/2016/9279078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rokavec M, Öner MG, Li H, Jackstadt R, Jiang L, Lodygin D, Kaller M, Horst D, Ziegler PK, Schwitalla S. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2014;124:1853–1867. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tili E, Croce CM, Michaille JJ. miR-155: on the crosstalk between inflammation and cancer (Review) Int Rev Immunol. 2009;28:264–284. doi: 10.1080/08830180903093796. [DOI] [PubMed] [Google Scholar]
- 46.Janssens S, Tschopp J. Signals from within: the DNA-damage-induced NF-kappaB response. Cell Death Differ. 2006;13:773–784. doi: 10.1038/sj.cdd.4401843. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The data sets generated during and/or analyzed during the current study are available from the corresponding author.