Abstract
Peritoneal carcinomatosis and peritoneal sarcomatosis (PC/PS) is a potential complication of nearly all solid tumors and results in profoundly increased morbidity and mortality. Despite the ubiquity of PC/PS, there are no clinically relevant targeted therapies for either its treatment or prevention. To identify potential therapies, we developed in vitro models of PC/PS using tumor cell lines and patient-derived spheroids (PDS) that recapitulate anoikis resistance and spheroid proliferation across multiple cancer types. Epithelial- and mesenchymal-derived cancer cell lines (YOU, PANC1, HEYA8, CHLA10, TC71) were used to generate spheroids and establish growth characteristics. Differential gene expression analyses of these spheroids to matched adherent cells, revealed a consensus spheroid signature. This spheroid signature discriminates primary tumor specimens from tumor cells found in ascites of ovarian cancer patients and in our PDS models. Key in this gene expression signature is BNIP3 and BNIP3L, known regulators of autophagy and apoptosis. Elevated BNIP3 mRNA expression is associated with poor survival in ovarian cancer patients and elevated BNIP3 protein, as measured by immunohistochemistry (IHC), is also associated with higher-grade tumors and shorter survival. Pharmacological induction of autophagy with rapamycin significantly increased spheroid formation and survival while decreasing the induction of apoptosis. In contrast, the autophagy inhibitor hydroxychloroquine abrogated spheroid formation with a clear increase in apoptosis. Modulation of BNIP3 and the critical autophagy gene Beclin-1 (BECN1) also caused a significant decrease in spheroid formation. Combined, these data demonstrate how modulation of BNIP3-related autophagy, in patient-derived spheroids and in vitro spheroid models, alters the survival and morphology of spheroids.
Background
There is an unmet need to develop effective and novel treatment strategies to prevent and treat local metastasis such as peritoneal carcinomatosis and sarcomatosis (PC/PS). Multiple cancers, including gastrointestinal, gynecological and sarcomatous malignancies, have the potential to grow and disseminate throughout the peritoneal cavity [1, 2]. In up to 50% of gastric cancer, >80% of ovarian cancer, 10–20% of pancreatic cancers, and 20% of sarcomas can present with peritoneal surface dissemination or isolated peritoneal recurrence [3]. PC is the second most common site of recurrence in colorectal cancer [4]. Median survival for patients with colorectal cancer and PC is less than 6 months if untreated and approximately one year if treated with systemic chemotherapy. This is significantly shorter than patients with non-PC metastases treated with systemic chemotherapy [5–7]. Regardless of tumor origin, the prognosis of patients with peritoneal surface malignancies is poor.
Existing therapies for peritoneal disease
A biological understanding of dissemination through intraperitoneal spread has prompted the concept that PC is a locoregional disease and therefore may respond to locoregional therapy. Multiple randomized trials have demonstrated the improved efficacy of intraperitoneal chemotherapy vs. systemic chemotherapy in optimally debulked ovarian cancer [8, 9]. Two large meta-analyses in gastric cancer, each with >1,000 cases included, have demonstrated the benefit of surgical debulking followed by heated intraperitoneal chemotherapy (HIPEC) when compared to surgery +/− systemic chemotherapy [10, 11]. Recently published large retrospective analysis of patients with appendiceal, colon and rectal cancer diagnosed with PC and treated with aggressive surgical and peritoneal chemotherapy found survival benefits that compare favorably to recent data for systemic chemotherapy in metastatic colorectal cancer [12, 13]. Likewise, published data in children with isolated peritoneal sarcomatosis also revealed significant survival benefit for patients who were able to undergo a complete cytoreduction and intraperitoneal chemotherapy [14]. Similar data have also been observed in adults with isolated peritoneal sarcomatosis although samples sizes in various trials have been small [15, 16]. However, the number of patients who achieve long term benefit from this approach is limited and in most diseases the availability of randomized clinical trial data is scarce. The biological mechanisms involved in this very aggressive cancer phenotype are poorly understood.
Biology of peritoneal disease
When cells lose their attachment and are forced to suspend in a fluid environment, they undergo a detachment-induced apoptosis termed anoikis [17, 18]. Anoikis has been demonstrated to be an important mediator for acinar lumen formation in normal breast tissue and anoikis resistance has been demonstrated to be critical for mammary tumor formation and metastases [19]. Anoikis resistance is a hallmark of all cancer cells [20, 21] but may be especially important in the biology of PC/PS as the primary mode of tumor dissemination. We have isolated circulating tumorspheres from the ascites fluid of patients with ovarian cancer and shown morphologic similarities to tumorspheres grown from cell lines derived from multiple primary sites that can be grown on Poly-2-hydroxyethyl methacrylate (polyHEMA).
Similarly, recent studies with selective in vivo passaging of a syngeneic ovarian cancer cells resulted in enhanced anoikis resistance and a higher rate of metabolism and autophagy in more aggressive cells [22]. The intrinsic pathway for apoptosis activation has been shown to be important in anoikis sensitivity and resistance. Pro-apoptotic and anti-apoptotic members of the Bcl-2 family modulate the release of the death-promoting proteins from the mitochondria into the cytoplasm. In non-transformed intestinal cells, anoikis was triggered by the detachment-induced down-regulation of Bcl-XL [10]. Ras-transformed cells display significant anoikis resistance in part to decreased Bak and the failure to down-regulate Bcl-XL [10–12]. The regulatory mechanisms that control the delicate balance between autophagy, anoikis resistance and anoikis mediated apoptosis have yet to be elucidated. Interestingly, Bcl-2/E1B 19kDa interacting protein (BNIP3) is a BH3-only Bcl-2 family member that has been shown to be a potent inducer of both apoptosis and autophagy [23, 24]. Many groups have shown BNIP3 is up-regulated under hypoxic conditions [25, 26] and other situations that induce cellular stress [24, 27, 28]. The exact role of BNIP3 in the regulation of autophagy and apoptosis is poorly understood. Under various conditions, BNIP3 has been alternately shown to induce apoptosis [29–31], autophagy [32–34] or autophagocytic cell death [35, 36].
We show in this paper that spheroid formation across multiple cancers demonstrate similar, patterned activation of BNIP3-linked autophagy. We specifically report on the role of BNIP3 and autophagy in anoikis resistance. In our patient-derived spheroids and in vitro spheroid models, modulation of autophagy altered the survival of the spheroids and may serve as the basis for future treatments.
Materials & Methods
Western Blotting
Cells were isolate from described experiments and were lysed using lysis buffer containing 100mM Tris, 4% SDS, pH 7.2 with added protease and phosphatase inhibitors (Cat# 11836153001 and 4906845001 respectively from Sigma). Equal amounts (10–30ug) of cell extracts were resolved using SDS-polyacrylamide gel electrophoresis and transferred on a pretreated PVDF membrane for 1 hour at 4C. Membranes were incubated overnight at 4C using Rabbit monoclonal antibodies of, LC3B (NB100-22200, Novus), Beclin (sc11427, Santa Cruz), Cleaved Caspase 3 (sc22171-R, Santa Cruz), Atg1/Ulk1(A7481, Sigma) GAPDH (5174P, Cell Signaling) and Mouse monoclonal antibodies of BNIP3 (MAB4147, R&D Systems) at optimized dilutions and then probed with rabbit polyclonal antibodies (7074, Cell Signaling) or mouse polyclonal antibodies (7076, Cell Signaling). Proteins were visualized using enhanced chemiluminescence (ECL) detection agents (Advansta, CA).
3D spheroid culture
Cells from early passage (<10 from thaw) were seeded in triplicates at 570,000 cells/well in a six-well plate pre-coated with poly-2-hydroxyethyl methacrylate (Poly-HEMA, Sigma cat# P3932) dissolved at 12% w/v in 95% ethanol. Cells were grown in RPMI1640 or DMEM with l-glutamine supplemented with 10%FBS and Pen-Strep. Culture medium with spheroids were collected and centrifuged @ 500 rpm for 5’. After discarding supernatant, spheroids were disrupted gently with 1X TBS-Tween20 (50mM Tris-Cl, pH 7.6; 150mM NaCl; 0.1% Tween 20 for a 30s. Viable cells were manually counted after addition of Trypan Blue using a Hemacytometer.
Apoptosis assay
Caspase activity was measured using CaspaseGlo 3/8 System (Promega Inc., cat# G8090) or cleaved caspase by Western blotting as a surrogate for apoptosis. For CaspaseGlo experiments, spheroid cultures were grown as above and at the indicated time points caspase activity was measured on a microplate reader. Caspase activity was normalized to the absolute cell count in the sample being measured prior to comparisons.
Genetic perturbation
BNIP3 (cat# TR316465), BCLN1 (TL314484V), and scrambled (SCR) shRNA were obtained from Origene. Viral particles were generated with 293T cells and used to infect HEYA8 cells according to manufacturer instructions. Infected cells were selected with puromycin (Gibco, Cat# A11138-03) at 5mcg/ml. Genetic knockdown was confirmed by western blotting prior to use in spheroid assays and during the course of each spheroid experiment.
Spheroid signature generation and refinement
Total RNA from triplicate cell culture pellets was isolated using the Qiagen RNeasy kit as per manufacturer’s directions. RNA quality was confirmed using Nanodrop. RNA was then profiled using Illumina HT-12 bead arrays. To minimize batch effect, similar cell lines were grouped on the same chip. Using the BRB-tools suite using BRB-ArrayTools developed by Dr. Richard Simon and the BRB-ArrayTools Development Team resultant data were normalized using RMA. Probes were subsequently 1.25 fold-change filtered. Based on the phenotypes of interest, differential gene expression was calculated using Significance of Analysis of Microarrays (SAM) that incorporates appropriate adjustments for multiplicity. Genes that met a false discovery rate (FDR) of 5% of were retained. For each cell line analyzed, differentially expressed genes with an FDR of 5% were retained and then compared to one another. After adjusting and accounting for directionality, a consensus overlap gene signature was generated termed “spheroid signature”. Resultant spheroid signatures were also evaluated in GSE33874 (ovarian cancer solid tumor) and GSE44104 (ovarian cancer ascites).
Pathway enrichment and network analysis
Gene Set Enrichment Analysis was performed using the Java version of GSEA v2.2 [37]. Relevant gene signatures were downloaded from MSigDB. Ingenuity Pathway Analysis (IPA, www.ingenuity.com) was used to prioritize upstream and downstream regulators of the spheroid signature via the Molecule Activity Predictor tool. Only direct interactions and experimentally determined relationships were considered for this analysis. Resultant network was visualized using IPA’s Path Designer tool.
Genomic profiling of patient ascites samples grown as spheroids
Ascites samples were collected from patients with ovarian cancer scheduled to undergo therapeutic paracentesis and frozen. Samples were subsequently thawed and expanded in a 2D model. Using the 3D spheroid culture described above, these patient samples were grown as spheroids and total RNA was collected using the Qiagen RNeasy kit and profiled using similar methodologies as above.
Tissue Microarray Analysis
Sections (4–6 µm thick) of formalin-fixed paraffin-embedded tissue arrays containing ovarian tumors at various stages were purchased from US Biomax Inc. (Rockville, MD). Slides were baked at 65°C for one hour and immunostaining was performed on the fully automated Bond RX autostaining system (Leica Biosystems, Buffalo Grove, IL). Briefly, heat-induced antigen retrieval was done using ER1 (citrate buffer) for 20 minutes, slides were stained with a rabbit monoclonal antibody to BNIP3 (clone EPR4034 Cat. ab109362, Abcam, Cambridge, MS) at a 1:200 dilution for 30 minutes and the Bond Polymer Refine (DAB) detection system (Leica Biosystems, Buffalo Grove, IL) was used. A pathologist specializing in gynecological malignancies read and scored the stained slides blinded to outcomes data. Survival analysis was performed using Kaplan-Meier estimation using the Graphpad PRISM 6.0 software.
Results
In vitro spheroid model demonstrates anoikis resistance and proliferation across multiple cell lines
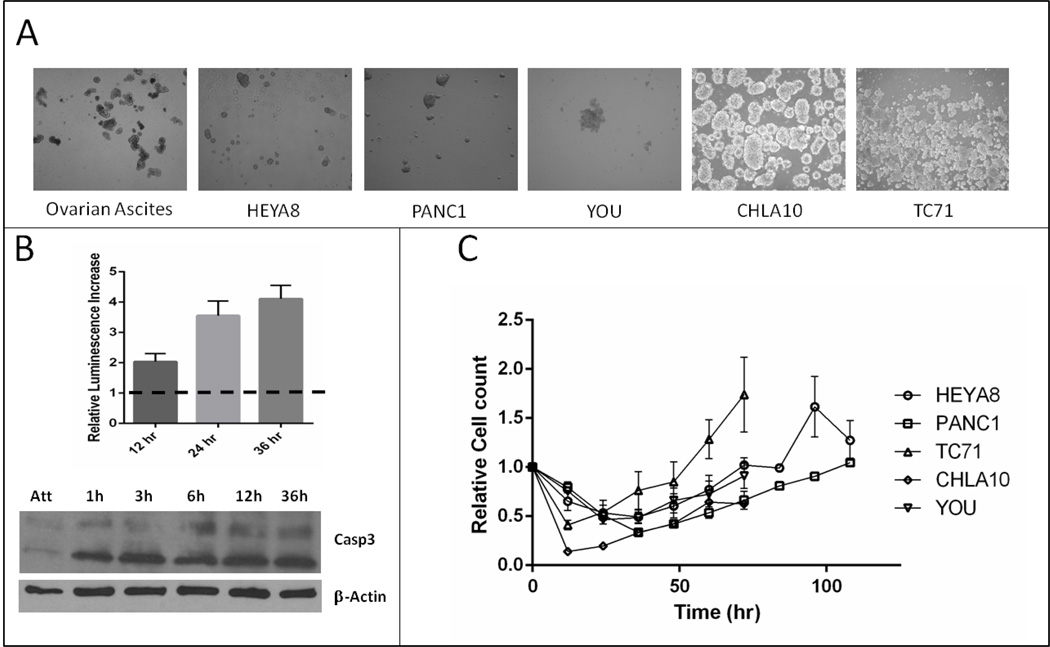
Five cancer cell lines representing one mesothelial (YOU), two epithelial (PANC1, HEYA8) and two sarcomatoid malignancies (CHLA10, TC71) that can spread via peritoneal metastasis, were grown in attachment free conditions to produce free floating tumor spheroids (Figure 1A). These spheroids showed a similar size and morphology to patient-derived spheroids (PDS) isolated from ascites of ovarian cancer patients (Figure 1A). An increase in apoptosis was detected upon culturing in attachment free conditions consistent with previous published data on anoikis (Figure 1B). All in vitro cell lines tested showed a similar temporal pattern of cell death, spheroid development, followed by tumor spheroid growth of an anoikis resistant population after 18–36hrs (Figure 1C).
Figure 1.
A. Demonstration of spheroids from primary ascites and multiple tumor cell lines grown in plyHEMA coated plates. B, Increase in apoptosis over time as HEYA8 cells are plated in attachment free conditions on polyHEMA plates. Top panel demonstrates relative caspase activity as measured by CaspaseGlo 3/7 and bottom increase in cleaved Caspase-3 by WB. C, Cell viability of multiple cell lines grown on polyHEMA plates.
Transcriptomic analysis of spheroids demonstrates common genes
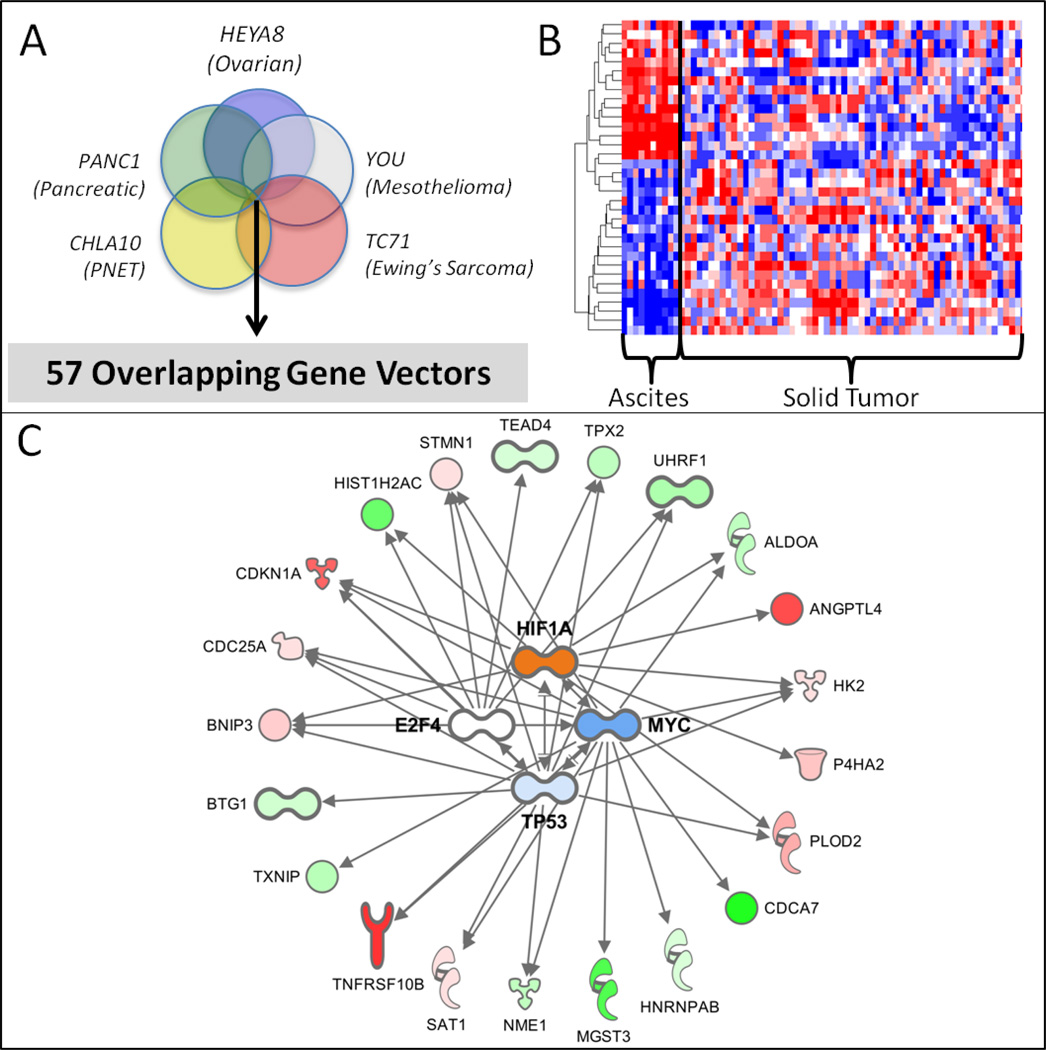
To determine if there were significant similarities between the different tumor cell lines and their corresponding spheroids, each cell line was grown in attachment-free conditions and total RNA from the resultant mature spheroids were isolated at 72 hours. Differential gene expression between the attached and spheroid cultures were performed for each cell line. Genes that met a false discovery rate (FDR) of 5% were retained resulting in a per cell line spheroid signature. The number of unique differentially expressed genes was all in a similar range except for the CHLA10 cell line which had roughly half of the others. The unique gene counts per cell line are as follows: CHLA10 (792 genes), YOU (1,793 genes), HEYA8 (1,269 genes), TC71 (1,483 genes), PANC1 (1,298 genes).
To remove tumor subtype specific effects, we then generated a consensus spheroid signature by taking the intersection of the differentially expressed genes after carefully controlling for directionality (Figure 2A). This in vitro consensus spheroid signature resulted in a set of 57 genes (Table 1). BNIP3, a gene with reported roles in both autophagy induction and apoptosis, was one of the top transcriptomic alterations in all gene sets.
Figure 2.
A. Venn diagram representing significant overlap in genes from multiple cultured cancer cell lines. This cell line signature was further refined against a gene signature associated with primary tumor cells isolated from ovarian cancer patients ascites. B. Heatmap indicating the discriminatory power of a subset of the cell line derived spheroid signature to identify primary ovarian tumors from ascites compared to solid tumor samples. C.
Table 1.
Directionality adjusted overlapping spheroid signature from across five cell lines
| Symbol | Entrez Gene Name | Direction Spheroid |
|---|---|---|
| AK4 | adenylate kinase 4 | UP |
| ALDOA | aldolase, fructose-bisphosphate A | UP |
| ANGPTL4 | angiopoietin like 4 | UP |
| ARMCX3 | armadillo repeat containing, X-linked 3 | UP |
| BCL6 | B-cell CLL/lymphoma 6 | UP |
| BNIP3 | BCL2/adenovirus E1B 19kDa interacting protein 3 | UP |
| BNIP3L | BCL2/adenovirus E1B 19kDa interacting protein 3-like | UP |
| BTG1 | B-cell translocation gene 1, anti-proliferative | UP |
| CCDC34 | coiled-coil domain containing 34 | DOWN |
| CDC25A | cell division cycle 25A | DOWN |
| CDCA4 | cell division cycle associated 4 | DOWN |
| CDCA7 | cell division cycle associated 7 | DOWN |
| CDKN1A | cyclin-dependent kinase inhibitor 1A | UP |
| DHRS7 | dehydrogenase/reductase (SDR family) member 7 | UP |
| DLGAP5 | discs large homolog associated protein 5 | DOWN |
| DNAJB9 | DnaJ heat shock protein family (Hsp40) member B9 | UP |
| DNAJC9 | DnaJ heat shock protein family (Hsp40) member C9 | DOWN |
| ENO2 | enolase 2 | UP |
| EPB41L4A-AS1 | EPB41L4A antisense RNA 1 | UP |
| GABARAPL1 | GABA type A receptor associated protein like 1 | UP |
| GBP2 | guanylate binding protein 2 | UP |
| GINS2 | GINS complex subunit 2 | DOWN |
| HIST1H2AC | histone cluster 1, H2ac | UP |
| HIST1H2BD | histone cluster 1, H2bd | UP |
| HK2 | hexokinase 2 | UP |
| HLA-A | major histocompatibility complex, class I, A | UP |
| HLA-E | major histocompatibility complex, class I, E | UP |
| HLA-F | major histocompatibility complex, class I, F | UP |
| HLA-H | major histocompatibility complex, class I, H | UP |
| HNRNPA1 | heterogeneous nuclear ribonucleoprotein A1 | DOWN |
| HNRNPAB | heterogeneous nuclear ribonucleoprotein A/B | DOWN |
| KIF20A | kinesin family member 20A | DOWN |
| MGST3 | microsomal glutathione S-transferase 3 | UP |
| MKRN1 | makorin ring finger protein 1 | UP |
| MXD4 | MAX dimerization protein 4 | UP |
| NME1 | NME/NM23 nucleoside diphosphate kinase 1 | DOWN |
| NOP16 | NOP16 nucleolar protein | DOWN |
| NRBP2 | nuclear receptor binding protein 2 | UP |
| NREP | neuronal regeneration related protein | UP |
| P4HA2 | prolyl 4-hydroxylase subunit alpha 2 | UP |
| PGK1 | phosphoglycerate kinase 1 | UP |
| PLOD2 | procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 | UP |
| SAT1 | spermidine/spermine N1-acetyltransferase 1 | UP |
| SLC2A1 | solute carrier family 2 member 1 | UP |
| SLC2A6 | solute carrier family 2 member 6 | UP |
| STMN1 | stathmin 1 | DOWN |
| TEAD4 | TEA domain transcription factor 4 | DOWN |
| TNFRSF10B | tumor necrosis factor receptor superfamily member 10b | UP |
| TNIP1 | TNFAIP3 interacting protein 1 | UP |
| TOMM40 | translocase of outer mitochondrial membrane 40 | DOWN |
| TPX2 | TPX2, microtubule nucleation factor | DOWN |
| TSC22D1 | TSC22 domain family member 1 | UP |
| TXNIP | thioredoxin interacting protein | UP |
| UHRF1 | ubiquitin like with PHD and ring finger domains 1 | DOWN |
| VASN | vasorin | UP |
| WBP2 | WW domain binding protein 2 | UP |
| ZNF467 | zinc finger protein 467 | UP |
Tumor spheroid signature effectively discriminates patient primary tumor from ascites
To validate our in vitro spheroid signatures, we evaluated their utility in discriminating ovarian ascites from primary ovarian tumors using both the HEYA8 (ovarian cell line) and the consensus spheroid signature. As a validation set, we used the gene expression profiles of 60 primary ovarian tumors (GSE33874) as compared to 10 ovarian ascites samples (GSE44104). For each available gene in the signature, we marked whether it was capable of discerning ascites from primary tumor. Of the 1,223 genes of the HEYA8 spheroid signature (Supplemental Table 1), 767 (nominal p <= 0.05) were discriminatory. Using GSEA, the ovarian cell line spheroid signature was highly discriminatory (nominal p < 0.0001). Similarly, of the 54 genes available for evaluation from the consensus spheroid signature, the majority of the genes (34) were discriminatory (nominal p <= 0.05) (Figure 2B) between ascites and primary ovarian cancer individually and using GSEA (nominal p = 0.1). Similarly, fresh tumor cells isolated from ascites of patients with ovarian cancer were grown as a monolayer and in attachment free conditions and the differences in the transcriptome were compared to our previously generated signature. As before, the majority (34 genes) of the in vitro consensus signature genes were highly discriminatory in our patient-derived spheroids (nominal p<=0.05, Supplementary Figure 1). In contrast, when we evaluated whether this spheroid signature recapitulated that of a stemness signature by Saxena et al [38], there was little significant overlap (2 genes) (Supplementary Figure 1).
Gene enrichment analysis highlights the autophagy pathway and the genes BNIP3/BNIP3L
Examination of the spheroid signature revealed an enrichment in p53, cMYC, HIF1alpha and E2F4 related genes (Figure 2C). BNIP3, a gene that has been shown to be central to cellular fate decisions regarding apoptosis and autophagy was one of the most highly altered gene across all datasets (top 5% in all signatures). To better understand what pathways the consensus spheroid signature represented, we performed enrichment analyses using IPA and by GSEA. Noteworthy, pathways related to autophagy (Table 2) were highly represented in our consensus spheroid signature (Bonferonni p << 0.001). In contrast, using the individual ovarian spheroid cell line signature prioritized numerous pathways relating to proliferation, apoptosis, cell death, and necrosis (Supplemental Table 2). We observe a similar pattern of alterations using GSEA (MSigDB H: Hallmarks of Cancer signatures) over our in vitro data comparing spheroid versus attached conditions. In this case, we see alterations in lipid and glucose metabolism along with hypoxia and the p53 pathway alterations. It is worth noting that the Hallmarks of Cancer do not yet specifically contain an autophagy pathway (Supplemental Table 3). Examining both our patient ascites-derived spheroid data and that from retrospective cinical samples, BNIP3/BNIP3L were found to be up regulated in spheroids or malignant ovarian tissue compared to attached cells or normal ovarian samples (Figure 4C).
Table 2.
Enrichment Analysis of Spheroid Signature (Bonferonni p = 0.05)
| Diseases of Functions Annotation | # Molecules | adjusted p-value |
|---|---|---|
| proliferation of tumor cell lines | 14 | 0.0001605 |
| autophagy of prostate cancer cell lines | 3 | 0.000272 |
| proliferation of cells | 20 | 0.00185 |
| autophagy of colon cancer cell lines | 3 | 0.00329 |
| metastasis of melanoma cell lines | 3 | 0.00655 |
| polyploidization of cells | 3 | 0.0105 |
| cell death of breast cancer cell lines | 6 | 0.01135 |
| cell death of muscle cells | 6 | 0.0155 |
| apoptosis of melanoma cell lines | 4 | 0.0191 |
| proliferation of erythroid progenitor cells | 3 | 0.02575 |
| lymphocytic cancer | 7 | 0.0346 |
| apoptosis of hepatoma cell lines | 4 | 0.0361 |
| growth of tumor | 8 | 0.0395 |
| metastasis | 8 | 0.0395 |
| lifespan of fibroblast cell lines | 2 | 0.0412 |
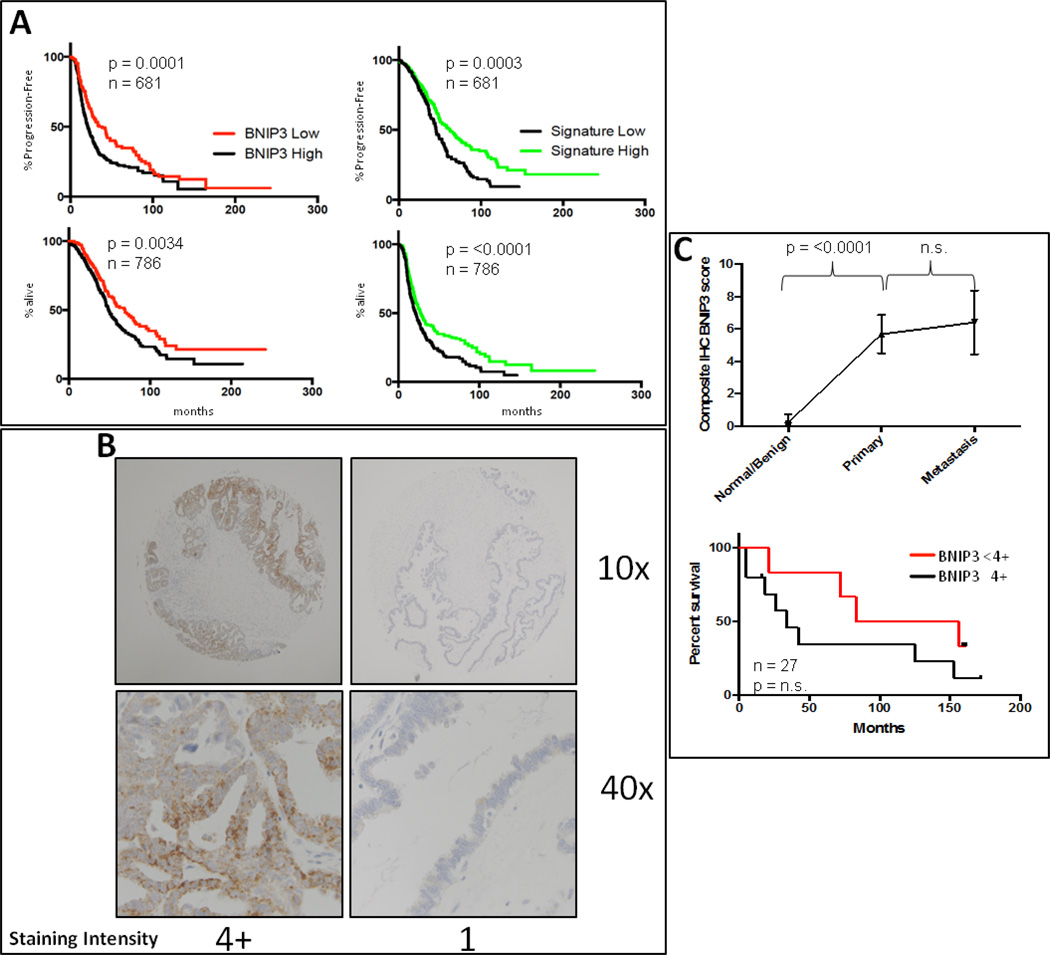
Figure 4.
A. BNIP3 or consensus spheroid signature transcript levels correlate with survival in ovarian cancer patients. B. Representative IHC staining of ovarian cancer tissue showing high (4+) and low (1) staining of BNIP3. C. A significant difference in BNIP3 staining was noted between benign ovarian tissue and malignant tissue in the array, independent of primary or metastatic site. Level of BNIP3 in tumor tissue also correlated with survival in this cohort.
Examining the role of BNIP3 and BECN1 in anoikis resistance
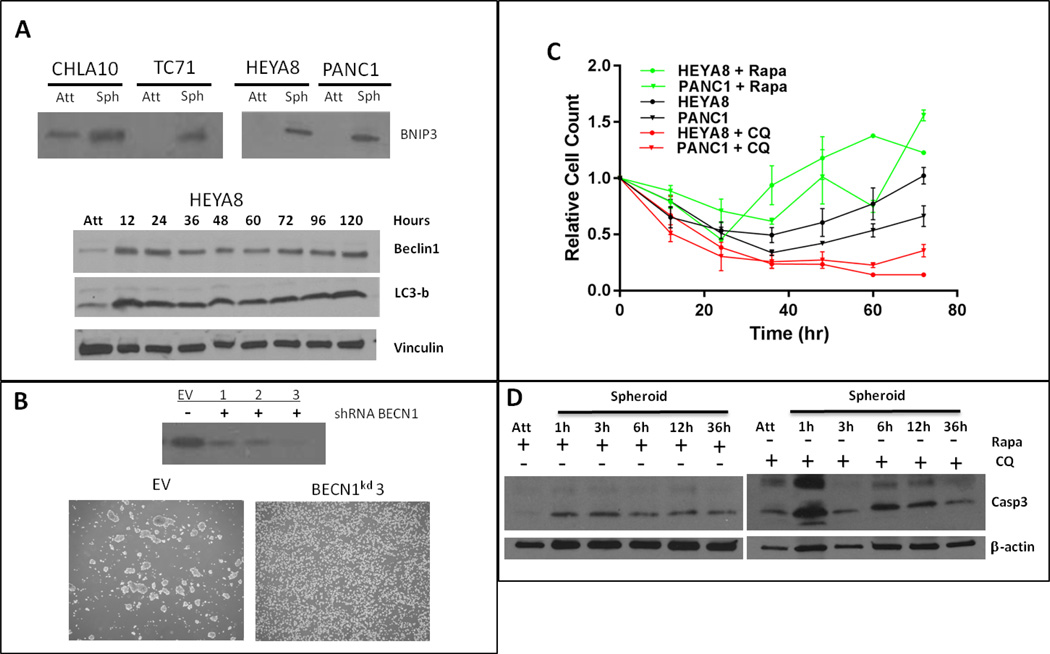
Given the previously published relationship between BNIP3 and beclin-1, Bcl-XL and Bcl-2, and the significant up-regulation of BNIP3 across all cell lines in our spheroid model, we examined the importance of autophagy in spheroid formation. BNIP3 up-regulation was confirmed at the protein level in multiple cell lines under attachment free conditions (Figure 3A). BECN1 and LC3b also increased almost immediately with detachment and stayed elevated throughout spheroid development (Figure 3A), indicating activation of autophagy in response to attachment free conditions. Modulation of autophagy by shRNA mediated depletion of BECN1 demonstrated effective inhibition of spheroid formation in attachment free conditions compared to scrRNA.
Figure 3.
A. BNIP3 increases in all cell lines in response to attachment independence. Beclin-1 and LC3b are increased in HEYA8 cells 12h after attachment loss and remain elevated throughout the spheroid time course. B. Spheroid morphology is changed in BECN1 knockdown compared to scramble shRNA. C. Pharmacologic perturbation of autophagy with rapamycin or hydroxychloroquine modulates the ability of HEYA8 and PANC1 cells to survive in attachment free conditions. D. Apoptosis increases with CQ treatment compared to rapamycin treated cells after loss of attachment.
Activation of autophagy induces whereas inhibition of autophagy inhibits spheroid formation
We next examined whether pharmacologic modulation of autophagy would alter the ability of cell lines to form spheroids. Rapamycin, a known inducer, and hydroxychloroquine, an inhibitor, were used to pharmacologically modulate autophagy. In HEYA8 and PANC1 cell lines, rapamycin leads to enhanced cell survival compared to control at non-cytotoxic concentrations (Figure 3C). However, treatment with hydroxychloroquine abrogated cell survival in those same cell lines (Figure 3C). Similarly, HEYA8 cells treated with hydroxychloroquine showed an increase in apoptosis upon loss of attachment compared to rapamycin treated or untreated cells (Figure 3D).
Increased expression of BNIP3 in patients with ovarian cancer correlates with worsened PFS at both RNA and protein levels
To determine whether the presence of elevated BNIP3 in human samples led to earlier recurrence in previously non-metastatic ovarian cancer patients, we used transcriptomic and clinical data from a repository of ovarian cancer datasets. The data included treatment and response information that was set up using the GEO and TCGA banks [39]. From this curated set, we identified 75 women with serous ovarian, stage I or stage II optimally debulked cancer. When stratified by high versus low BNIP3 expression (split at the median), we noted that women with high BNIP3 expression had a shorter median PFS (logrank p=0.0001) and OS (logrank p=0.0034) than those with low BNIP3 RNA expression based on Kaplan-Meier analysis (Figure 4A). A similar trend was seen when the consensus spheroid signature was used to stratify patients (Figure 4A).
We then determined the level of expression of BNIP3 in human tumor samples from an ovarian tissue microarray. Samples were scored by a blinded gynecology/oncology pathologist on a scale of 1–4+ based on intensity of staining in tumor cells. When tumor samples were compared to control normal ovarian tissue there was a significant difference in the amount of BNIP staining (Figure 4B). We then determined the PFS in patients stratified based on BNIP levels in tumor tissue. Using Kaplan-Meier analysis, we noted a trend towards decreased overall survival in patients whose tumor samples scored 4+ when compared to those who scored <4+ (Figure 4C).
Discussion
Peritoneal carcinomatosis and sarcomatosis (PC/PS) are a devastating complication of multiple malignancies. We have used an in vitro model to determine common biological mechanisms that define anoikis resistance and spheroid formation in multiple cell types from varying cancers. Previous work has shown that tumor cells cultured in attachment free conditions undergo an EMT-like phenotypic change however the role of autophagy in this process has not been previously determined.
Here, we approach anoikis resistance from the novel view that autophagy is a critical driver of this PC/PS. Indeed, if this relationship is confirmed, it has very immediate implications. Rapamycin and its clinically relevant derivatives are FDA approved medications in a host of cancers and many have been tested in many diseases that can present with peritoneal disease. This class of drugs is a known up-regulator of autophagy and therefore may be potentially harmful in patients whose primary mode of tumor dissemination is through carcinomatosis or sarcomatosis. STAT3 has recently been shown to be an upstream modulator of BNIP3 induced autophagy, either through induction and stabilization of HIF1a or Concanavalin A induced activation of membrane type-1 matrix metalloproteinase and subsequent activation of STAT3 [40, 41]. While the exact mechanisms of STAT3 regulation of BNIP3 are still under investigation, the clinical availability of STAT3 inhibitors may prove useful in clinical trials of patients with peritoneal dissemination of ovarian and other cancers. Thus, it is key to better understand the regulatory mechanisms of anoikis and to identify biomarkers in patients where targeted therapeutics aimed at inhibiting this passive dissemination of tumor cells may improve survival.
We first determined the optimal conditions for growth of tumor cell spheroids and that the size and morphology was consistent with tumorspheres isolated from patients with peritoneal carcinomatosis. While there are many different procedures to generate tumorspheres in vitro, many rely on matrix interaction and growth in either matrigel, soft agar, or culturing of single spheres in attachment independent round bottom flasks. Unfortunately, mechanical forces may play a significant role in 3D culture morphology [42] and therefore we chose to culture cells in polyHEMA coated plates in serum containing media to mimic the conditions encountered by tumor cells within the peritoneal compartment. We have shown that culturing cells derived from varying tumor types under these conditions yield an expected amount of apoptotic cell death through the process of anoikis as well as a population of cells that are able to survive and proliferate under these conditions.
We then constructed a genomic signature of these attachment independent replicating tumor cells from various tumor types. The members of this signature were able to accurately differentiate between tumor samples from primary sites compared to tumor cells derived from ascites of patients with ovarian cancer. We furthermore demonstrate that through signature refinement, a core set of autophagy related genes emerges from background pathways seen in the larger sets. Indeed, others have shown enrichment of tumor initiating or tumor stem cells when selecting under attachment independent conditions. Our anoikis resistance signature demonstrated little similarity when compared to published ovarian stem cell signatures indicating a unique ascites/anoikis resistance signature. Unfortunately, clinically annotated genetic expression data for patients with peritoneal disease from other tumor sites is unavailable for validation, but current protocols will prospectively collect this data and be analyzed accordingly.
We hypothesized that BNIP3 may play a central role in spheroid survival due to shunting of cells between apoptosis and cell death and survival through autophagy. We demonstrated that knockdown of one of the central mediators of autophagy, BECN1, inhibited anoikis resistance and spheroid production. Chemical modulation with rapamycin or hydroxychloroquine also demonstrated a central role of autophagy to this process.
Previous studies have highlighted the importance of EMT factors in anoikis resistance. Ko et al. demonstrated the importance of PKCK2 as a mediator of EMT and cadherin switching in early anoikis resistance (<48hrs) in [43]. We did not see similar changes in PKCK2 or N-cadherin transcript level at a FDR of 5% in our experimental design. This may be attributed to the fact that our spheroids were profiled at the 72 hour timepoint that these EMT changes may be an early effect.
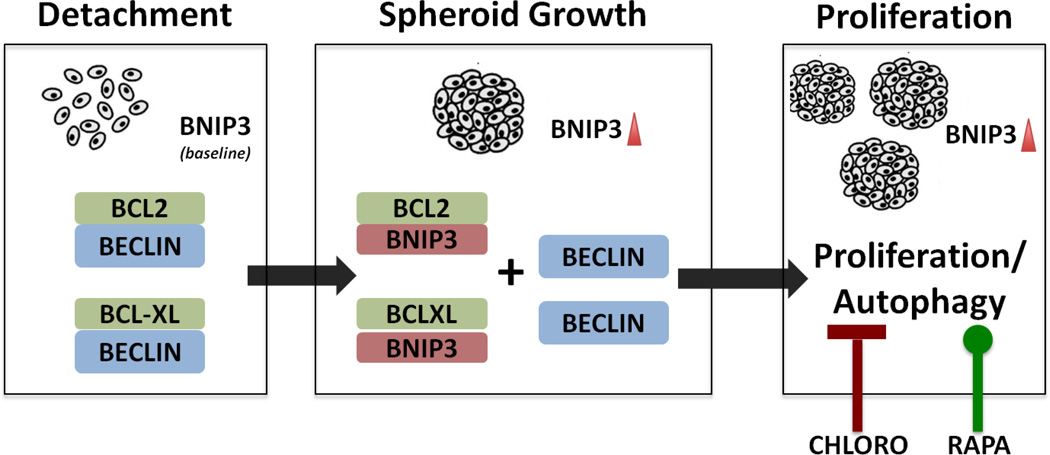
We demonstrate that in peritoneal metastasis, BNIP3-driven autophagy resistance may play a pivotal role in disease progression. Figure 5 summarizes our findings. The ability to determine which early stage ovarian cancer patient might be at risk for further developing peritoneal spread is clinically important. We further demonstrate that activation of BNIP3 clinically is prognostic of a poor outcome. Whether this is indicative that tumors are more prone to autophagy and thus forming spheroids developing local metastasis will require further studies. Nevertheless, our finding provides a cautionary tale for using an unopposed TORC1 inhibitor and potentially a druggable pathway that may alter the natural history of spheroid formation and thus peritoneal metastasis.
Figure 5.
When cell lose attachment, elevated BNIP3 allows dissociation of BECN1/BCL-2 complexes and allows for initiation of autophagy and inhibition of apoptosis to promote cell growth in tumorspheres. Autophagy modulators rapamycin and chloroquine can alter this response and cell survival in attachment free conditions.
Supplementary Material
Implications.
Development of BNIP3/BNIP3L targeting agents or autophagy targeting agents may reduce morbidity and mortality associated with peritoneal carcinomatosis and sarcomatosis.
References
- 1.de Cuba EM, et al. Understanding molecular mechanisms in peritoneal dissemination of colorectal cancer : future possibilities for personalised treatment by use of biomarkers. Virchows Arch. 2012;461(3):231–243. doi: 10.1007/s00428-012-1287-y. [DOI] [PubMed] [Google Scholar]
- 2.Zhu L, McManus MM, Hughes DP. Understanding the Biology of Bone Sarcoma from Early Initiating Events through Late Events in Metastasis and Disease Progression. Front Oncol. 2013;3:230. doi: 10.3389/fonc.2013.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel R, et al. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 4.Elferink MA, et al. Metachronous metastases from colorectal cancer: a population-based study in North-East Netherlands. Int J Colorectal Dis. 2015;30(2):205–212. doi: 10.1007/s00384-014-2085-6. [DOI] [PubMed] [Google Scholar]
- 5.Verwaal VJ, et al. Randomized trial of cytoreduction and hyperthermic intraperitoneal chemotherapy versus systemic chemotherapy and palliative surgery in patients with peritoneal carcinomatosis of colorectal cancer. J Clin Oncol. 2003;21(20):3737–3743. doi: 10.1200/JCO.2003.04.187. [DOI] [PubMed] [Google Scholar]
- 6.Franko J, et al. Treatment of colorectal peritoneal carcinomatosis with systemic chemotherapy: a pooled analysis of north central cancer treatment group phase III trials N9741 and N9841. J Clin Oncol. 2012;30(3):263–267. doi: 10.1200/JCO.2011.37.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klaver YL, et al. Population-based survival of patients with peritoneal carcinomatosis from colorectal origin in the era of increasing use of palliative chemotherapy. Ann Oncol. 2011;22(10):2250–2256. doi: 10.1093/annonc/mdq762. [DOI] [PubMed] [Google Scholar]
- 8.Tewari D, et al. Long-term survival advantage and prognostic factors associated with intraperitoneal chemotherapy treatment in advanced ovarian cancer: a gynecologic oncology group study. J Clin Oncol. 2015;33(13):1460–1466. doi: 10.1200/JCO.2014.55.9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hess LM, et al. A meta-analysis of the efficacy of intraperitoneal cisplatin for the front-line treatment of ovarian cancer. Int J Gynecol Cancer. 2007;17(3):561–570. doi: 10.1111/j.1525-1438.2006.00846.x. [DOI] [PubMed] [Google Scholar]
- 10.Yan TD, et al. A systematic review and meta-analysis of the randomized controlled trials on adjuvant intraperitoneal chemotherapy for resectable gastric cancer. Ann Surg Oncol. 2007;14(10):2702–2713. doi: 10.1245/s10434-007-9487-4. [DOI] [PubMed] [Google Scholar]
- 11.Sun J, et al. Meta-analysis of randomized controlled trials on laparoscopic gastrectomy vs. open gastrectomy for distal gastric cancer. Hepatogastroenterology. 2012;59(118):1699–1705. doi: 10.5754/hge12259. [DOI] [PubMed] [Google Scholar]
- 12.Ung L, Chua TC, David LM. Peritoneal metastases of lower gastrointestinal tract origin:a comparative study of patient outcomes following cytoreduction and intraperitoneal chemotherapy. J Cancer Res Clin Oncol. 2013;139(11):1899–1908. doi: 10.1007/s00432-013-1507-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elias D, et al. A comparative study of complete cytoreductive surgery plus intraperitoneal chemotherapy to treat peritoneal dissemination from colon, rectum, small bowel, and nonpseudomyxoma appendix. Ann Surg. 2010;251(5):896–901. doi: 10.1097/SLA.0b013e3181d9765d. [DOI] [PubMed] [Google Scholar]
- 14.Hayes-Jordan A, et al. Cytoreductive surgery and Hyperthermic Intraperitoneal Chemotherapy (HIPEC) for children, adolescents, and young adults: the first 50 cases. Ann Surg Oncol. 2015;22(5):1726–1732. doi: 10.1245/s10434-014-4289-y. [DOI] [PubMed] [Google Scholar]
- 15.Baumgartner JM, et al. Aggressive locoregional management of recurrent peritoneal sarcomatosis. J Surg Oncol. 2013;107(4):329–334. doi: 10.1002/jso.23232. [DOI] [PubMed] [Google Scholar]
- 16.Sommariva A, et al. Cytoreductive surgery and hyperthermic intraperitoneal chemotherapy in patients with peritoneal sarcomatosis: long-term outcome from a single institution experience. Anticancer Res. 2013;33(9):3989–3994. [PubMed] [Google Scholar]
- 17.Paoli P, Giannoni E, Chiarugi P. Anoikis molecular pathways and its role in cancer progression. Biochim Biophys Acta. 2013;1833(12):3481–3498. doi: 10.1016/j.bbamcr.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 18.Tan K, et al. Uncovering a key to the process of metastasis in human cancers: a review of critical regulators of anoikis. J Cancer Res Clin Oncol. 2013;139(11):1795–1805. doi: 10.1007/s00432-013-1482-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fung C, et al. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell. 2008;19(3):797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding XF, et al. 13,14-bis(cis-3,5-dimethyl-1-piperazinyl)-beta-elemene, a novel beta-elemene derivative, shows potent antitumor activities via inhibition of mTOR in human breast cancer cells. Oncol Lett. 2013;5(5):1554–1558. doi: 10.3892/ol.2013.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagaprashantha LD, et al. The sensors and regulators of cell-matrix surveillance in anoikis resistance of tumors. Int J Cancer. 2011;128(4):743–752. doi: 10.1002/ijc.25725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai Q, Yan L, Xu Y. Anoikis resistance is a critical feature of highly aggressive ovarian cancer cells. Oncogene. 2015;34(25):3315–3324. doi: 10.1038/onc.2014.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vasagiri N, Kutala VK. Structure, function, and epigenetic regulation of BNIP3: a pathophysiological relevance. Mol Biol Rep. 2014;41(11):7705–7714. doi: 10.1007/s11033-014-3664-x. [DOI] [PubMed] [Google Scholar]
- 24.Sun L, et al. Upregulation of BNIP3 mediated by ERK/HIF-1alpha pathway induces autophagy and contributes to anoikis resistance of hepatocellular carcinoma cells. Future Oncol. 2014;10(8):1387–1398. doi: 10.2217/fon.14.70. [DOI] [PubMed] [Google Scholar]
- 25.Kothari S, et al. BNIP3 plays a role in hypoxic cell death in human epithelial cells that is inhibited by growth factors EGF and IGF. Oncogene. 2003;22(30):4734–4744. doi: 10.1038/sj.onc.1206666. [DOI] [PubMed] [Google Scholar]
- 26.Mellor HR, Harris AL. The role of the hypoxia-inducible BH3-only proteins BNIP3 and BNIP3L in cancer. Cancer Metastasis Rev. 2007;26(3–4):553–566. doi: 10.1007/s10555-007-9080-0. [DOI] [PubMed] [Google Scholar]
- 27.Awan MU, et al. Neuroprotective role of BNIP3 under oxidative stress through autophagy in neuroblastoma cells. Mol Biol Rep. 2014;41(9):5729–5734. doi: 10.1007/s11033-014-3444-7. [DOI] [PubMed] [Google Scholar]
- 28.Rubiolo JA, et al. Yessotoxin Induces Er-Stress Followed By Autophagic Cell Death In Glioma Cells Mediated By mTOR and BNIP3. Cell Signal. 2013 [PubMed] [Google Scholar]
- 29.Vande Velde C, et al. BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol Cell Biol. 2000;20(15):5454–5468. doi: 10.1128/mcb.20.15.5454-5468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubli DA, Ycaza JE, Gustafsson AB. Bnip3 mediates mitochondrial dysfunction and cell death through Bax and Bak. Biochem J. 2007;405(3):407–415. doi: 10.1042/BJ20070319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi Y, et al. PTEN induces apoptosis and cavitation via HIF-2-dependent Bnip3 upregulation during epithelial lumen formation. Cell Death Differ. 2015;22(5):875–884. doi: 10.1038/cdd.2014.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tracy K, Macleod KF. Regulation of mitochondrial integrity, autophagy and cell survival by BNIP3. Autophagy. 2007;3(6):616–619. doi: 10.4161/auto.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tracy K, et al. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol. 2007;27(17):6229–6242. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinsay MN, et al. Bnip3-mediated mitochondrial autophagy is independent of the mitochondrial permeability transition pore. Autophagy. 2010;6(7):855–862. doi: 10.4161/auto.6.7.13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daido S, et al. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res. 2004;64(12):4286–4293. doi: 10.1158/0008-5472.CAN-03-3084. [DOI] [PubMed] [Google Scholar]
- 36.Han B, et al. A prolyl-hydroxylase inhibitor, ethyl-3,4-dihydroxybenzoate, induces cell autophagy and apoptosis in esophageal squamous cell carcinoma cells via up-regulation of BNIP3 and N-myc downstream-regulated gene-1. PLoS One. 2014;9(9):e107204. doi: 10.1371/journal.pone.0107204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vathipadiekal V, et al. Identification of a potential ovarian cancer stem cell gene expression profile from advanced stage papillary serous ovarian cancer. PLoS One. 2012;7(1):e29079. doi: 10.1371/journal.pone.0029079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Penzvalto Z, et al. MEK1 is associated with carboplatin resistance and is a prognostic biomarker in epithelial ovarian cancer. BMC Cancer. 2014;14:837. doi: 10.1186/1471-2407-14-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.You L, et al. The role of STAT3 in autophagy. Autophagy. 2015;11(5):729–739. doi: 10.1080/15548627.2015.1017192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pratt J, Annabi B. Induction of autophagy biomarker BNIP3 requires a JAK2/STAT3 and MT1-MMP signaling interplay in Concanavalin-A-activated U87 glioblastoma cells. Cell Signal. 2014;26(5):917–924. doi: 10.1016/j.cellsig.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 42.Pedersen JA, Swartz MA. Mechanobiology in the third dimension. Ann Biomed Eng. 2005;33(11):1469–1490. doi: 10.1007/s10439-005-8159-4. [DOI] [PubMed] [Google Scholar]
- 43.Ko H, et al. Protein kinase casein kinase 2-mediated upregulation of N-cadherin confers anoikis resistance on esophageal carcinoma cells. Mol Cancer Res. 2012;10(8):1032–1038. doi: 10.1158/1541-7786.MCR-12-0261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.