Abstract
We calculate the solubility limit of pentapeptides in water by simulating the phase separation in an oversaturated aqueous solution. The solubility limit order followed by our model peptides (GGRGG > GGDGG > GGGGG > GGVGG > GGQGG > GGNGG > GGFGG) is found to be the same as that reported for amino acid monomers from experiment (R > D > G > V > Q > N > F). Investigation of dynamical properties of peptides shows that the higher the solubility of a peptide is, the lower the time spent by the peptide in the aggregated cluster is. We also demonstrate that fluctuations in conformation and hydration number of peptide in monomeric form are correlated with the solubility of the peptide. We considered energetic mechanisms and dynamical properties of interbackbone CO–CO and CO···HN interactions. Our results confirm that CO–CO interactions more than the interbackbone H-bonds are important in peptide self-assembly and association. Further, we find that the stability of H-bonded peptide pairs arises mainly from coexisting CO–CO and CO···HN interactions.
Graphical Abstract
INTRODUCTION
Solubility is an important factor which drives a range of chemical and biological phenomena.1 The solubility limit of a solute is defined by the concentration of the solution at saturation. In such a solution both the free energy of solvation as well as the effective interactions among solutes determine the properties of the system including conformations and phase behavior. Cellular cytoplasm is a concentrated, crowded mixture with many components near their solubility limits.2 The phase behaviors of proteins and protein–RNA mixtures establish the formation and activity of nonenveloped organelles, also referred to as membraneless organelles or biomolecular condensates, in cells.3,4 These structures are known to play a role in the regulation of many biological processes.
Aggregation of proteins and peptides, driven by solubility, has been shown to share molecular mechanisms similar to the collapse of the polypeptide chain.5,6 Many efforts have been made to understand the role of side-chains in the collapse and folding of proteins.7–9 The role of the backbone in this process is understood to be of similar importance to that of the side-chains from transfer free energy measurements.10 A number of experimental and theoretical efforts have worked to parse the effects of the side-chains from the backbone.11,12 The backbone has often been modeled as either an alanine or a glycine chain.13 Here we use oligoglycine as our reference to understand the effect that various side-chains have on the solubility limit of peptides in solution.
Although the free energy of hydration is more favorable for dilute solutions of longer oligoglycines, they collapse in water to unstructured globules.6,14–17 Experimentally, the solubility of oligoglycines in water reduces with length.18 This suggests that self-association of backbone amide (CONH) groups and backbone–water interactions are enhanced at different rates with respect to the length of the peptide leading to backbone collapse. Simulation was employed recently to model the saturation concentration of pentaglycine in water.5 The purpose of the present study is to extend solubility limit modeling to aid in the interpretation of side-chain effects on peptide aggregation.
Here, we probe the role of various interactions involving backbone and side-chains involved in aggregation and hence solubility of peptides. Backbone hydrogen bonding (H-bonding) interactions have long been proposed to control collapse of proteins into their folded states.19,20 Interactions that govern peptide molecular recognition via aggregation and protein collapse share similar features. Significant numbers of H-bonds are observed in the reported well-ordered structures produced from self-assembly of oligoglycines (polyglycines I and II).21,22 In polyglycine I which resembles a β-structure, each chain forms H-bonds with neighboring parallel or antiparallel chains.21 Similarly, oligoglycine chains are parallel and hexagonally packed in polyglycine II crystals, and each of these chains forms a single H-bond with each of its six identical neighbors.22 Because of competition with water molecules in a solution, a reduction in the contribution of H-bonds to the stability of peptide clusters is expected in water as observed in simulations of pentaglycines in water.5 We note that each H-bond between amide groups in water has been shown to contribute 1–2 kcal/mol favorably of free energy which is significantly less than the contribution (>3 kcal/mol) upon burial and dehydration.23–31
In addition to backbone H-bonds, other dipolar interactions among groups in the main-chain occur as well. Backbone interactions between dipoles, especially of the carbonyl groups, CO–CO, with a contact distance less than 4 Å are common in helices and β-sheets.32,33 These contacts arise from interactions between the carbonyl (CO) dipoles,34,35 and have been ascribed to the so-called n → π* interaction.36–43 In classical terms, the attraction between the oppositely charged atoms is given angular dependence by excluded volume and the like charge repulsions which constitute a major part of the amide dipole–dipole interactions.32,43,44 It has been argued that interactions between CO groups modulate conformations of peptides, peptoids, and related small molecules.32–41,44–48 Relevant to these findings, perturbation theory (IMPT) applied to a perpendicular propanone dimer shows that the attractive energy of a nonbonded CO–CO interaction is comparable to the CO···HN H-bond in water.49 In their computational analyses of helices and β-sheets in native proteins, Maccallum et al. found that CO–CO interactions are occasionally more attractive than CO···HN H-bonds, and backbone H-bonds usually deviate from linearity and adopt a geometry that maximizes the favorable CO–CO interaction.32
Collapsed states as well as the aggregated structure of oligoglycines in water have been found to have more numerous CO–CO interactions than backbone H-bonds.5,6,15,17 Such systems are quite disordered compared to folded proteins or crystalline peptides. Despite the high abundance of short-range CO–CO interactions and their similarity in magnitude of attractive strength to H-bonds in water, they have received less attention in protein folding research relative to that of H-bonds due to the central importance of secondary structure. Motivated by the necessity of understanding the nature of backbone CO–CO and CO···HN interactions and their relative importance in self-association of backbone amide groups, we present an analysis of these interactions for a variety of concentrated peptide solutions.
Side-chains play important roles in the solubility of a peptide as well as protein folding.10,50 As in the case of oligoglycines, the solubility limit of an amino acid in water cannot be predicted from its hydration free energy alone; an amino acid with more favorable dilute solution hydration free energy is not necessarily more soluble in water. For example, despite its much more favorable hydration free energy, Gln is less soluble than Val in water.50–54 Some glycine-rich proteins have been found to exhibit globular-like conformations55 but without a hydrophobic side-chain core. These systems violate the premise that folding is driven by the hydrophobic side-chains gravitating to the interior, avoiding the solvent, but rather highlights our hypothesis that folding is a cooperative and correlated process between the solute–solvent and solute–solute enthalpic and entropic energies. The backbone dihedrals, the solute inter- and intramolecular interactions, including the H-bond network, and the solute–solvent interactions work with or against each other to drive the system to a collapsed or folded state. We conduct our investigation on model pentaglycines substituted with six different amino acid side-chains using unsubstituted pentaglycine as a reference. Structural, energetic, and dynamical properties of various interactions involving backbone and side-chains are investigated to probe the nature of these interactions and their relative importance in self-assembly of our model peptides. Preferential interaction coefficients and transfer free energies have also been used to decipher protein and peptide interactions in unfolding and aggregation,56,57 but have not been included as part of this study.
The rest of this study is organized into three sections. After the description of the systems, models, and methods employed in this study, we present the results obtained from simulations. We first address aggregation and the solubility limit of our model peptides in water. Stability of peptides in the aggregated cluster as well as in saturated solution are compared thereafter. This is followed by a presentation of structural, energetic, and dynamic properties. We summarize the findings and draw our conclusions in the last section.
MODELS AND METHODS
We wish to interpret the side-chain effects on the solubility of pentapeptides and to understand the role of various interactions involving backbone and side-chains behind aggregation. To consider the solubility limit of peptides, we performed MD simulations of peptides with uncharged sequences GGGGG, GGNGG, GGQGG, GGVGG, and GGFGG, as well as charged side-chains GGRGG and GGDGG denoted here, respectively, as gly, asn, gln, val, phe, arg, and asp at finite concentration in explicit water. The peptides were initially built in extended conformation with the ends capped with acetyl (CH3CO) and N-methyl (NHCH3) groups using the AMBER58 software package. There were 625 peptides and ~90 000 water molecules then arranged on a lattice in a cubic box (see Table SI-I for overview of simulations) giving a total concentration of around 0.3 M, which is expected to be well above the solubility limit. For those with charged side-chains, arg and asp, we included 0.1 M NaCl and neutralizing ions. Due to the nature of the systems with net charge, some analyses (e.g., dipole correlations, etc.) are only performed and compared for the uncharged side-chains chosen.
Our protocol to avoid the metastable parts of the phase diagram and kinetic traps follows our previous work.5 After energy minimization, simulations were performed first in the isothermal–isobaric (NpT) ensemble for 2 ns with targeted temperature and pressure of 300 K and 1 atm, respectively, and then in the canonical (NVT) ensemble for 1 ns with the average box volume obtained from the last 1 ns of the previous NpT simulation run. Finally, 140 ns of MD trajectories were generated in the microcanonical (NVE) ensemble, the last 40 ns of which were considered for equilibrium analysis. This choice of ensemble in a large system ensures that thermostats and barostats are not affecting the phase behavior.
The NAMD59 package was employed. The simulations used the TIP3P60 water with the peptide forces described by the AMBER ff12SB force field. We employed an integration time step of 2 fs and a spherical cutoff distance of 12 Å for nonbonding interactions. The long-range electrostatic interactions were treated with the particle mesh Ewald method. All bonds involving hydrogen were constrained using the SHAKE algorithm.
In earlier work in this laboratory61 a comparative analysis of force fields for short, Gly3, and long, Gly10, peptides in solution as a function of three all-atom molecular mechanics force fields was performed. Those long simulations, 300 ns and 1 μs, for 2 different peptide lengths allowed evaluation of the structural metrics, such as the end-to-end distance, radius of gyration, and the distribution of conformations to be evaluated as a function of force field. The results showed different structural tendencies based on the force field. In particular, the CHARMM 36 (C36) force field tended to produce more extended conformations. Others have discussed the ability of the molecular mechanics force field to represent the protein–solvent interactions, in particular those of intrinsically disordered systems.62–66 These involve modifications to existing terms, use of alternate solvent parameters, or entirely new force field parameter sets. In this study we are not comparing the force field parameters but, instead, choose to evaluate the details of aggregation using one of the commonly used molecular mechanics force fields.
RESULTS AND DISCUSSION
Solubility Limit of Peptides
All of the uncharged systems studied spontaneously phase separated in tens of nanoseconds from initiation, similar to our previous study of GGGGG using the CHARMM potential.5 After the saturated solution was equilibrated, we performed clustering analyses of our model peptides. A cutoff distance of 4 Å between heavy atoms of different peptides was applied for this clustering. The time-averaged, bimodal distribution of the peptides in clusters of different sizes, shown in Figure 1, demonstrates the presence of the expected phase separation. All systems had a dilute phase with a few peptide monomers and small clusters (up to 5) along with a concentrated phase of large clusters of size greater than 570. As expected, the phase separated systems avoid clusters of intermediate sizes when far from metastability.
Figure 1.
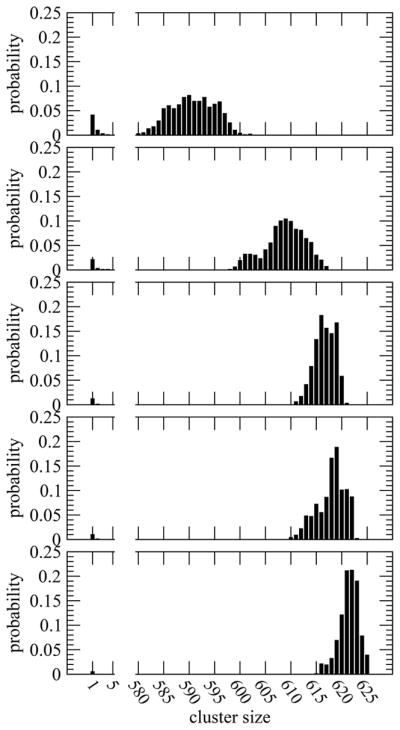
Probability (normalized to one) of finding uncharged peptides in clusters of different sizes for systems gly, val, gln, asn, and phe (from top to bottom, respectively).
Our simulations display the effects of side-chains in peptide aggregation. We find that, in the case of gly, the average number of peptide monomers in the saturated solution is around 26, and clusters in the concentrated phase average 590.7 ± 0.8. For the other uncharged compounds we find peptide aggregation increases and the solubility limit decreases in the presence of the side-chains chosen. We note that these span a range of classical hydrophobicity scales.67 The number of monomers or small clusters with these side-chains decreases, and the distribution shifts in favor of large clusters. The average size of the large uncharged cluster is greatest in the phe system, 621.4 ± 0.4. The cluster size is slightly higher for asn (618.0 ± 0.6) than that for gln (616.7 ± 0.4) which, in turn, is higher than that for val (608.8 ± 0.9).
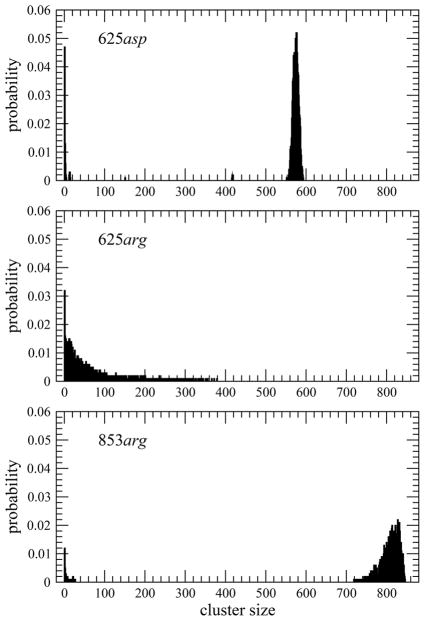
For the charged systems arg and asp, we tried the same procedure. We found that the asp system followed the expected phase separation pattern of the uncharged side-chain solutions. The arg system did not. We expected it to be more soluble, but we had issues with metastability and the system did not phase separate after 200 ns. This was reminiscent of what was seen in an earlier simulation with a solution of the gly system roughly 10 times more dilute,5 while above the solubility limit the metastability issues led to no discernible phase separation in 200 ns. This is evident in Figure 2. Thus, for arg we raised the initial concentration by more than a third to include 853 arg peptides. This system phase separated and produced stable averages without further issues.
Figure 2.
Probability (normalized to 1) of finding charged peptides in clusters of different sizes. In the top two panels, both systems had 625 GGXGG peptides. The failure of the arg system to clearly phase separate led us to increase the initial concentration to 853 GGRGG peptides shown in the bottom panel.
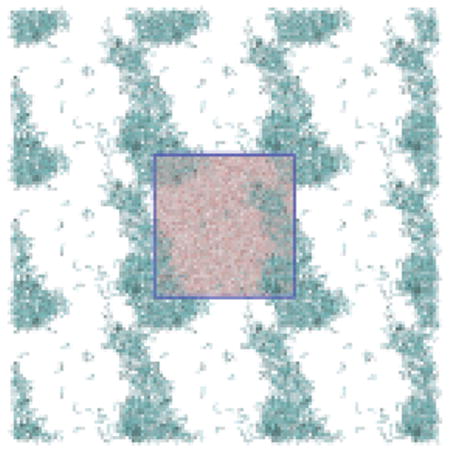
The large cluster formed by the solutes after phase separation is often continuous across the periodic boundary box (Figure SI-1). This arrangement, which is stable due to interpeptide interactions, is similar to that seen in the aggregation of hydrophobic particles and other phase separations.68,69 Each solution here separates into organic-rich and water-rich (peptide saturated) phases (Figure SI-1). The peptide concentration in the saturated solution was measured by subtracting the number of peptides in the large cluster from the total divided by the solvent excluded volume remaining after removing that cluster. We used the software package DAlpha-ball70 to compute solvent excluded volume of the large cluster.
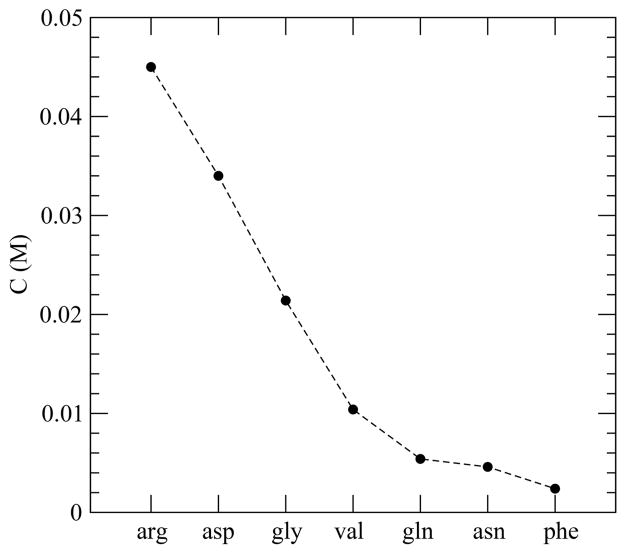
In Figure 3, we present the concentration of peptides in the saturated solution or the solubility limit for each system. For pentaglycine, we find that the concentration of the saturated solution is 0.021 ± 0.001 M, which is higher than that measured (0.016 ± 0.003 M) with another force field previously.5 The experimentally18 measured solubility of pentaglycine is 0.005 M at pH 5.4 and temperature 298.15 K which is 4 times lower than the force field dependent estimate here. We remark that none of the current force fields were parametrized to fit this property, nor were they parameterized at pH values comparable to experimental ones.
Figure 3.
Peptide concentration in saturated solution for all the systems studied.
As expected, the amount of peptide in the saturated solution, or in other words the solubility of the peptide, decreases in the presence of nonpolar side-chains relative to gly and increases for those systems studied with charge. For instance, we find that substitution of a H-atom in a glycine unit with a benzyl (C6H5CH2–) group causes more reduction in solubility than that with an iso-propyl ((CH3)2CH2–) group. That is in agreement with the experimental finding that aqueous solubility of amino acid molecules decreases on enhancing the nonpolar portion.53,54 Beyond classically hydrophobic residues, we also find that substitution of a glycine unit with an asparagine or a glutamine, both of which have polar side-chains, also gives a solubility less than gly. The charged side-chains produce a strong increase in solubility over the gly system. Overall, solubility of our model peptides follows an order (arg > asp > gly > val > gln > asn > phe) which is the experimental aqueous solubility of amino acid molecules.50,53,54
Stability of Peptides in Organic-Rich and Water-Rich Phases
Each monomer in the peptide saturated water-rich phase has 40–45 (depending on the peptide considered) water molecules within 4 Å of peptide heavy atoms (see Figure SI-3). Considering the neutral side-chain systems, computation of the total stabilization energy of each monomer arising from its interaction energies with these water molecules suggests a trend (gln > asn > phe > gly > val) for the systems studied which is similar to the trend in solvation free energy of neutral Gly, Val, Gln, Asn, and Phe units in water as reported from Monte Carlo simulations.51 In a more recent free energy simulation study of N-acetyl-X-methylamide amino acids, Boresch and co-workers observed a slightly altered trend in dilute solution solvation free energy with Gln > Asn > Gly > Phe > Val.52 The solvation free energy values and their ordering in these previous studies51,52 were found to depend strongly on whether they were capped or zwitterionic. These results in light of the current study indicate how solvation free energy is only one factor determining the solubility limit and phase behavior.
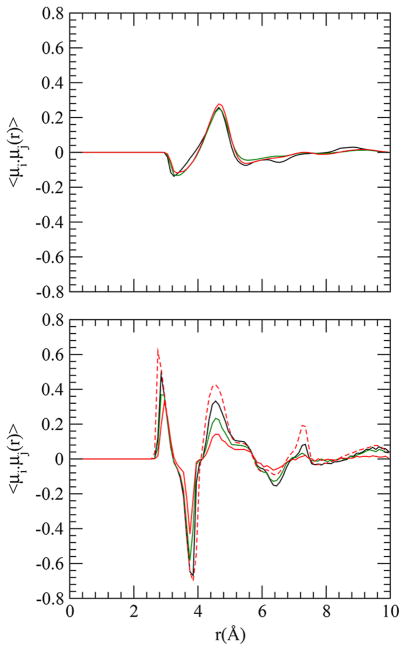
We considered both intramolecular and intermolecular correlations between amides. Previous simulations showed a distinctive correlation between amide dipoles within an oligoglycine with a strong positive correlation (parallel dipoles) near 3 Å and a strong negative correlation (antiparallel dipoles) near 4 Å.5,6 Such distinctive correlations arise from constraints created by bonded peptide backbone and hence should be dependent on conformational fluctuation of the peptide. We computed the correlation between amide dipoles i and j at a distance r, 〈μi·μj(r)〉, similarly to that described previously.5
The curves for pentaglycine (both intrapeptide and interpeptide), presented in Figure 4, are very similar to those found for another force field.5 However, the intrapeptide correlation declines with larger uncharged side-chains; the correlation almost vanishes in the case of F-substituted pentaglycine. Larger uncharged side-chain substitution reduces the conformational space available in solution. The trend of decreasing correlation gly < val < gln < asn < phe is similar to that of aggregation propensity of the peptides studied. Also, in line with the expectation that conformational fluctuation of a peptide decreases on moving from monomeric form to a large cluster, we find that the intrapeptide dipole correlation is stronger for a peptide in the cluster than that in monomeric form (Figure 4).
Figure 4.
Interpeptide (top) and intrapeptide (bottom) dipole correlations as a function of distance in systems gly (black), gln (green), and phe (red). Solid and dashed lines in the bottom panel are, respectively, for monomeric peptides and for all peptides in the system (in the cluster as well as in saturated solution).
The behavior of the hydration layer for the uncharged peptides was examined by calculating a water number fluctuation function, Nw(t), for isolated monomers and peptides in the organic-rich phase. The function was defined as
| (1) |
where nw(t) represents the number of waters of hydration (contact waters) at time t and nw is the average number of hydration water. The average hydration number at time t and for the entire simulation was estimated separately for peptides in clusters and in monomeric form. A cutoff distance of 4 Å between peptide heavy atoms and water oxygen was used to define a water molecule in the hydration layer.
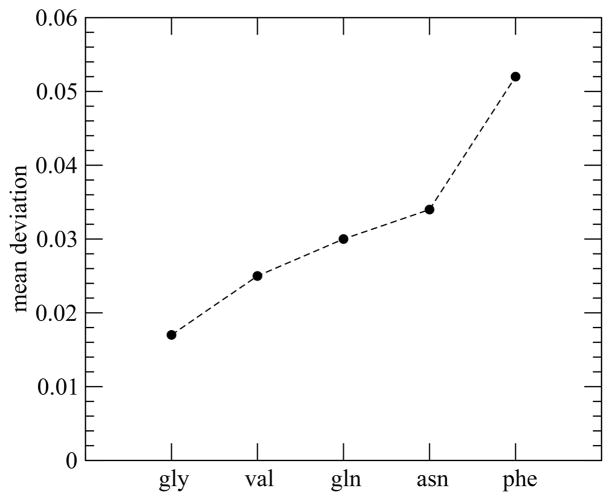
The uncharged side-chain differences affect the fluctuation in hydration number of monomeric peptides (Figure SI-2). The greater the fluctuation in the hydration number is, the higher the aggregation propensity of the peptide is. Fluctuation of Nw(t) for phe, the highest among the peptides studied, is nearly 3 times that for pentaglycine. This is in accord with modern theories of the hydrophobic effect related to the fluctuation of the first solvation shell water.71,72 We note that the order for the fluctuations (Figure 5) is the same as that for the solubility limits but has differences with many hydrophobicity scales.67,73 These findings are also in accord with previous work on the hydration layer of helices facilitating the actin binding process of protein HP-36.74
Figure 5.
Mean deviation of water number fluctuation function.
The interaction energy of an uncharged peptide with a water does not depend strongly on size of the cluster to which the peptide belongs although the total stabilization energy from water does (Figure SI-3). However, because clustering increases the local density of peptides, the total energetic gain from interpeptide interactions increases significantly in a large cluster. This stabilization is higher in magnitude than the energetic loss from the reduction in the number of water molecules interacting directly with a tagged peptide in large cluster. We find a decrease in energy of 26–35 kcal/mol on moving from its monomeric state to a large cluster, which provides a noteworthy energetic contribution to peptide clustering.
We calculated a free energy change, ΔGs, for peptides between the saturated solution and the large cluster using the relation ΔGs = −RT ln Ks, where Ks is the equilibrium constant defined as
| (2) |
In this definition, [LC] and [SS] represent molar concentrations of peptides in the large clusters and saturated solution, respectively. The estimated values of Ks and ΔGs for the uncharged systems are in Table 1. Since we are well beyond the solubility limit, peptide aggregation is spontaneous and favored in all the systems studied. The relative stability of the cluster is smallest for gly and greatest for phe. We find the relative cluster stability of the uncharged peptides to be gly < val < gln < asn < phe.
Table 1.
Cluster Stabilitya
| system | Ks | ΔGs | τ | non | noff |
|---|---|---|---|---|---|
| gly | 17.2 | −1.7 | 965 | 77.8 | 77.9 |
| val | 37.6 | −2.2 | 1314 | 55.3 | 55.6 |
| gln | 74.3 | −2.6 | 1416 | 26.2 | 26.2 |
| asn | 88.3 | −2.7 | 1665 | 23.7 | 23.8 |
| phe | 172.6 | −3.1 | 1703 | 23.1 | 23.0 |
Ks, ΔGs, and τ represent, respectively, equilibrium constant, relative stability of large cluster (in kcal/mol), and lifetime (in ps) of a peptide in large cluster. non and noff, respectively, are the number of peptides that reach and leave the cluster per nanosecond.
During the simulations, a cluster changes its size by collecting or releasing one or more monomers. Therefore, following Rashke et al.,68 we compute the free energy (ΔGm) of formation of a cluster of size n + 1 (Sn+1) by addition of a monomer (S1) to a cluster of size n (Sn) using the relation where Km is the equilibrium constant for the monomer addition process and C0 is the reference state. Figure 6 presents the average molar concentration of each cluster used in the free energy of cluster formation. We find that formation of a large cluster is more favorable than that of a dimer or trimer. On average, the free energy of formation of a large cluster of pentaglycine by monomer addition is around −2.6 kcal/mol. The cluster formation becomes more favorable for the uncharged substituted pentaglycines. The average free energy of cluster formation is −3.0 kcal/mol in val, −3.3 kcal/mol in gln, −3.6 kcal/mol in asn, and −3.8 kcal/mol in phe.
Figure 6.
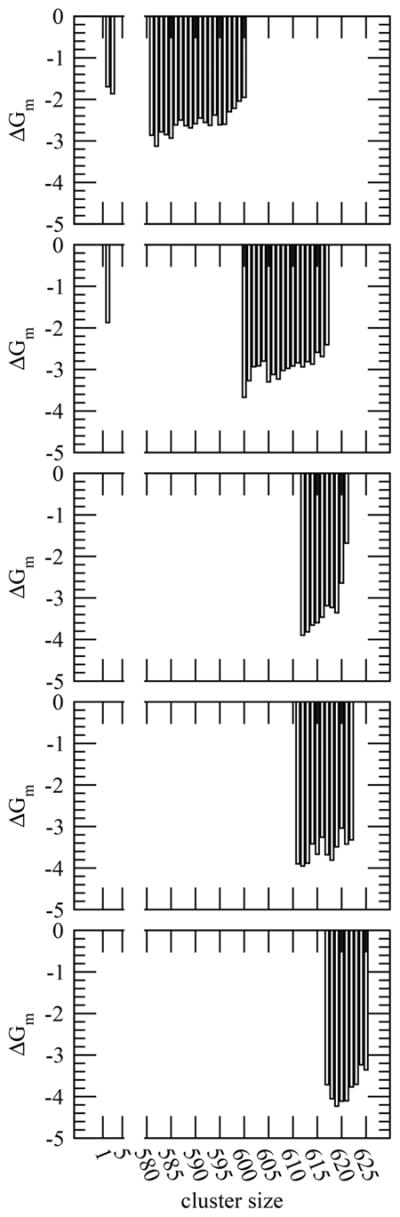
Free energy (in kcal/mol) of cluster formation by monomer addition for systems gly, val, gln, asn, and phe (from top to bottom, respectively). Clusters with very low concentrations are excluded in the free energy calculations.
Dynamical Properties of Peptide Clusters
To compare the kinetic stability of our model uncharged peptide clusters, we calculated the average lifetime of a peptide in a large cluster as well as the flux to leave the cluster. This would be complicated by charge neutrality for the charged systems, and so, we restrict the analysis to the neutral solutes. We use a survival function, S(t), to calculate lifetime.
| (3) |
In this relation, the variable h is equal to unity when the tagged peptide makes contact with a large cluster and is zero otherwise. The other variable H takes a value of unity if the tagged peptide remains continuously connected to the large cluster to time t and is zero otherwise.
In all the neutral systems studied, the survival function decays rapidly during the initial periods. However, there are some peptides that remain in the large cluster for almost 40 ns before leaving or exchanging with those in solution. We find that the relaxation of the function for our neutral peptides is the fastest for gly and the slowest in system phe, indicating relative kinetic stability. We show in Table 1 the average lifetime of a peptide in the large cluster as estimated by time integration of S(t). For the neutral system it can be seen that gly spends the shortest time in the large cluster, whereas time spent by phe is nearly 2 times longer. This is further reflected in the average number of peptides that leave the cluster per nanosecond (noff). We find that noff is at a maximum for gly and at a minimum for phe (Table 1), despite the presence of the smallest cluster in the former and the largest cluster in the latter. As a check of equilibrium, the number of peptides that leave the cluster per nanosecond was found to be essentially equal to the number of peptides that join the cluster per nanosecond (non).
Interactions on Neutral Peptide Aggregation
To develop an assessment of forces that stabilize the large cluster formed by our model peptides, we investigated structural, energetic, and dynamical properties of CO–CO and CO···HN H-bonding interactions. Following earlier work,75 a H-bond was identified by imposing cutoff distances for N–O and O–H (of 3.4 and 2.4 Å, respectively, here), and a simultaneous cutoff angle (of 45°) for H–N–O. We considered that a CO–CO interaction exists if the O–C separation does not exceed 4 Å. The stabilization energies of a backbone CO involved in either CO–CO or CO···HN H-bonding interactions are simply obtained from the nonbonded energy function.
Only the CO-to-CO and CO-to-NH interactions (rather than the whole peptide moiety) were used to calculate stabilization energy. Note that H-bond strength in the DSSP algorithm76 for assigning protein secondary structure elements is measured by considering only CO-to-NH interactions as in our study, but instead of employing the full nonbonded potential, it employs only the electrostatic part and hence overestimates attraction between close oppositely charged atoms. Finally, the average lifetimes of these interactions were computed by time integration of the survival function, S(t), as defined in eq 3. The variable h in this case is equal to unity when the tagged CO–CO or CO···HN pair exists and is zero otherwise. The other variable H takes a value of unity if the tagged pair remains continuously paired to time t and is zero otherwise.
In all the systems studied, the number of H-bonds formed by a peptide with surrounding peptides with this definition is 1.9–2.4 with the current potential model (Table SI–II). With a different potential and H-bond definition, previous studies found that number to be around 2 for the aggregation of pentaglycines.5 A β10–40 peptide collapse in water has been found to be driven by a small number of H-bonds.77 Given that pentapeptides with the blocking groups could make 6 donor and 6 acceptor H-bonds by either definition, the number found in these relatively unstructured clusters is far less than that expected from maximally H-bonded regular structures familiar from secondary structural elements.
We find that the number of CO–CO interactions as defined here is more than twice the number of H-bonds. It is notable here that, for a strict cutoff distance of 3.6 Å, which is the reported32 average distance between C- and O-atoms of H-bonded CONH groups in α-helices and antiparallel β-sheets (a criteria which dismisses half the distribution), the number of CO–CO interactions is close to the number of H-bonds as obtained from our highly relaxed H-bond definition. While this finding could give the impression that all of these short COCO interactions are an inevitable consequence of H-bonds between CONH pairs, our data in Table 2 clearly suggest that short CO–CO interactions do exist even in the absence of H-bonds. This shows the importance of CO–CO interactions in aggregation of our model peptides. As expected, side-chains sterically reduce the ability of backbone carbonyls to interact with other groups. The formation of a H-bond gives about 1.4 kcal/mol energetic stabilization (Table SI–II). This value, which is much lower than the attractive strength of a H-bond in the gas phase, agrees well with the generally accepted strength of a H-bond in water (1–2 kcal/mol).23–31,78 On the other hand, the CO–CO backbone interaction gives an energetic stabilization of 1.8 kcal/mol in system gly, and the gain increases slightly more in the presence of side-chains. Notably, using IMPT on a perpendicular propanone dimer, Allen et al. obtained an attractive energy of 1.8 kcal/mol for CO–CO interactions at a C–O separation of 3.02 Å.49
Table 2.
Probability (p) of Observing Backbone CO in Different Groups Considered and Their Lifetime (τ) and Energetic Stability (E) in System glya
| gCO | gOH | gCOH | gfree | |
|---|---|---|---|---|
| p | 0.267 (0.134) | 0.013 (0.168) | 0.349 (0.194) | 0.371 (0.504) |
| E (kcal/mol) | −7.276 | −5.938 | −8.557 | −7.063 |
| τ (fs) | 126 | 29 | 235 |
Numbers within parentheses are for O–C cutoff distance of 3.6 Å. For other systems, see Supporting Information.
We find that a CO–CO contact relaxes relatively slowly (Figure SI-4) and thus has a longer lifetime (Table SI–II) than a H-bond. Other work found that collapsed states of oligoglycines in water are stabilized by the more numerous CO–CO interactions than H-bonds.6,15,17 Similarly, short CO–CO contacts are common in proteins,32,33 and these interactions alter conformational preferences of peptides, peptoids, and other related small molecules.32–41,44–48
Coexisting CO–CO and CO···HN Interactions in Neutral Systems
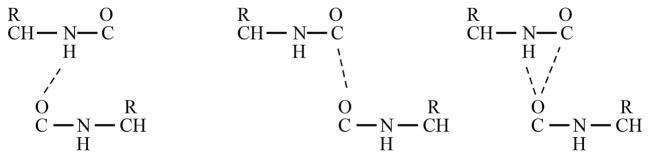
We now decompose backbone carbonyl interactions into four different groups, gi, based on the type of interbackbone interactions the considered O-atom makes: (a) CO–CO interaction exists but not a H-bond (gCO), (b) H-bond exists but not a CO–CO interaction (gOH), (c) CO–CO and H-bonding interactions exist simultaneously (gCOH), and (d) neither CO–CO nor H-bonding interactions exists (gfree). This classification is illustrated in Figure 7.
Figure 7.
Decomposition of backbone CO into different groups based on the interbackbone interactions. Left: a H-bond exists but not a CO–CO interaction (group gOH). Middle: a CO–CO interaction exists but not a H-bond (group gCO). Right: CO–CO and H-bonding interactions exist simultaneously (group gCOH).
Table 2 presents the probability of observing the classified interaction groups for gly. We find a significant number of backbone COs that interact directly with identical neighbors but do not form H-bonds (gCO). Among the backbone carbonyls involved in CO–CO interactions, ~60% simultaneously form interbackbone H-bonds. With these conventions, the relative population of backbone carbonyls that form H-bonds without interacting directly with identical neighbors (gOH) is negligible, <2%. Of the backbone carbonyls that form H-bonds, more than 94% interact simultaneously with neighboring carbonyls (gCOH) as well. Propensities of the interactions have previously been observed in computational and crystallographic analyses of protein α-helices and β-sheets.32,33
The energetic contribution of backbone carbonyls to the stability was calculated by considering CO group interactions (with C–O separation less than 4 Å), NH, Cα groups, and water. The survival function, S(t), used to compute lifetime is similar to that defined in eq 3. The variable h in this case is equal to unity when the tagged CO belongs to the particular group considered and is zero otherwise. The other variable H takes a value of unity if the tagged CO remains continuously in the same considered group to time t and is zero otherwise.
Figure SI-9 displays the relaxation of S(t), and the estimated lifetimes are listed in Table 2. We observe dramatic differences in the relaxation behavior of S(t) for the three groups considered. The survival function for group gOH decays at a much faster rate. A backbone CO belonging to this group has the shortest lifetime (~30 fs), suggesting that a H-bond without any contribution from the CO–CO interaction in these unstructured systems is not kinetically very stable. Relaxation of S(t) for group gCO is noticeably slower than that for group gOH, resulting in a longer (~4 times) lifetime for the former. Among the groups considered, relaxation is the slowest for interaction group gCOH. It is clear that a backbone CO is the most stable in gCOH and the least stable in gOH with an energy difference of more than 2.4 kcal/mol. Interestingly, the energy of a backbone carbonyl in gOH is less favorable than that in gfree by more than 0.8 kcal/mol. On the other hand, the stability of a carbonyl is slightly higher in gCO than that in gfree. Thus, consideration of Figure SI-9 and Table 2 suggests that the coexistence of the CO–CO contact is correlated with survival of a backbone H-bond. A backbone CO in gCO provides some stability to the cluster, holding the peptides together in a large cluster for some time without any help from a H-bond. Nonetheless, the coexistence of H-bonds and CO–CO interactions increases stability.
Water interacts significantly with the backbone carbonyls with a maximum in gfree for gly of 6.8 kcal/mol and minimum in gCOH of only ~1.3 kcal/mol. Importantly, it is not only the obvious reduction in number of water molecules near backbone carbonyls in gCOH but also the decreased strength of interactions between them that cause the significant reduction (Figure SI-5).
In Table 3, we present structural, energetic, and dynamical properties of CO–CO and CO···HN interactions for backbone COs in different groups to demonstrate the influence of neighboring groups on these interactions. The survival function, S(t), used here is similar to that defined in eq 3. The variable h is equal to unity when the tagged CO–CO or CO···HN interaction exists with backbone CO belonging to the particular group considered and is zero otherwise. The other variable H takes a value of unity if the tagged CO–CO or CO···HN interaction remains continuous in the same considered group to time t and is zero otherwise.
Table 3.
Decomposition of Interpeptide CO-to-CO and CO-to-HN Interactions into Groups gCO, gOH, and gCOH in System glya
| CO–CO | CO···HN | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| gCO | gOH | gCOH | gCO | gOH | gCOH | |
| n | 1.298 | 0 | 1.772 | 0 | 1.013 | 1.098 |
| E | −1.422 | −1.988 | −1.507 | −1.437 | ||
| τ | 99 | 153 | 29 | 195 | ||
For other systems, see Supporting Information. n, E, and τ represent, respectively, number, energy (in kcal/mol), and lifetime (in fs) of interactions.
We see that backbone COs that form H-bonds gain more stability through interactions with identical groups than do backbone COs with no H-bonds (gCO). Slower relaxation of CO–CO interactions in gCOH as compared to that in gCO is also noticed (Figure SI-6). Additionally, for a backbone CO in gCOH, the energetic stabilization from a CO–CO interaction is higher than that from a H-bond. Total energetic stabilization of backbone COs from CO–CO interactions is higher than that from H-bonding interactions by more than 2 times.
In agreement with previous suggestions that backbone H-bonds in α-helices and β-sheets usually deviate from linearity to maximize favorable backbone CO–CO interaction,32 our simulations capture slight destabilization (by ~0.1 kcal/mol) of H-bonds on moving from gOH to gCOH. The relaxation of H-bonds slows down noticeably with coexisting CO–CO interactions (Figure SI-6). This dynamic slowing is a reflection of the stability of a backbone CO in gCOH as compared to that in gOH and indicates the advantage of coexisting CO–CO interactions for survival of a backbone H-bond. It is notable here that the much faster relaxation of H-bonds in gOH as compared to that of CO–CO contacts in gCO causes the differential relaxation behaviors of CO–CO and CO···HN interactions, shown in Figure SI-4.
Figure 8 shows the probability of an interacting carbonyl converting from one group state to another. Table 4 presents the pairwise energies for CO–CO and CO···HN interactions prior to conversion from one group to another. A backbone carbonyl almost always converts to either gCOH or gfree both from groups gCO and gOH. The rate of conversion from gOH is much faster than that from gCO (Table SI–V).
Figure 8.
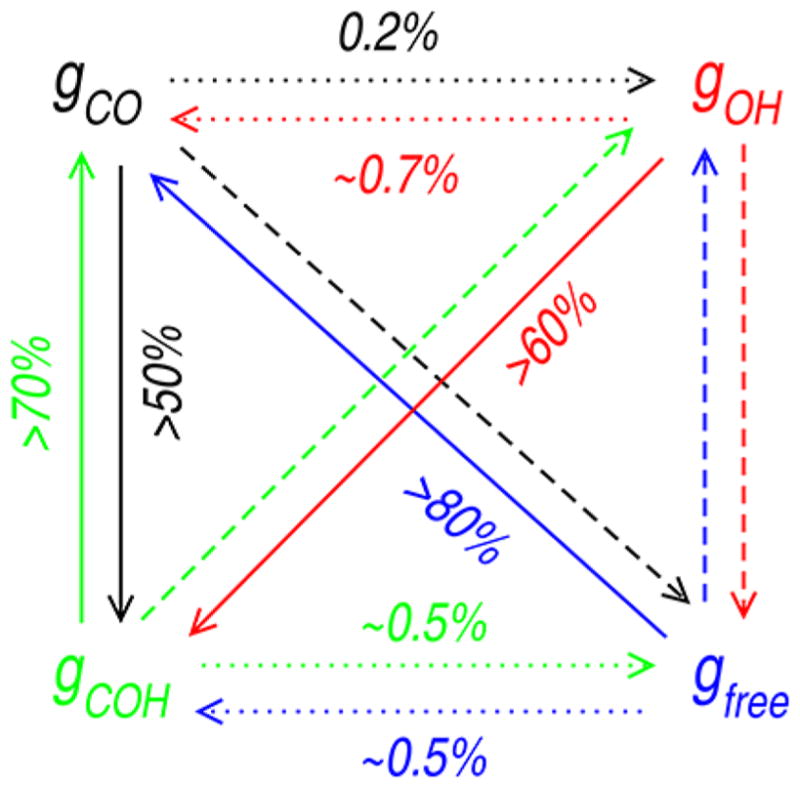
Schematic representation of probability of conversion of backbone CO from one group to another. Probability is at a maximum for the solid line and at a minimum for the dotted line. For details, see Table SI–V.
Table 4.
Interaction Energy (in kcal/mol) for CO–CO and CO···HN Interactions Prior to Conversion from Groups gCO, gOH, and gCOH to Other Groups in System glya
| to ↓ | from → | |||
|---|---|---|---|---|
|
| ||||
| gCO | gOH | gCOH | ||
| CO–CO | gCO | −1.912 | ||
| gOH | −1.408 | −1.680 | ||
| gCOH | −1.909 | |||
| gfree | −0.877 | −1.425 | ||
| CO···HN | gCO | −1.143 | −0.704 | |
| gOH | −1.616 | |||
| gCOH | −1.622 | |||
| gfree | −1.168 | −1.302 | ||
For other systems, see Figure SI-8.
We find that not only is the probability of conversion from gCOH to gOH significantly lower than that to gCO but also the time needed is noticeably shorter for conversion to the latter, indicating a shorter time required to break the H-bond than that to break the CO–CO contact for a backbone CO in gCOH (Table SI–V). It is also important to note that a backbone CO that is free in solution (gfree) shows a much higher (more than 4 times) preference to interact with another backbone CO than with backbone NH. We find the CO–CO interaction initiates the formation of a new H-bond. On the other hand, although a H-bond between backbone amide groups can lead to the formation of a new CO–CO contact, the effect is less significant.
Not surprisingly, the choice of cutoff distance influences these findings. We note that a reduction of O–C cutoff distance from 4.0 to 3.6 Å (the average) (a) increases probability of conversion from gCO to gfree, gOH to gCOH, gCOH to gOH, and gfree to gOH, and (b) decreases the probability of conversion from gCO to gCOH, gOH to gfree, gCOH to gCO, and gfree to gCO (data not presented).
To understand the geometric criteria for conversion of a backbone CO from gCOH to either gCO or gOH, we considered the CO–CO and CO···HN distances as well as the CO–C and CO···H angles in system gly when backbone CO interacts simultaneously with NH and identical groups in the main-chain of surrounding peptides. For CO–CO interactions, we find that the mean contact distance (dOC) is 3.6 Å and the mean CO–C contact angle (θOC) is 129°. The mean value of dOC found here is essentially the same as the reported average distance between C and O-atoms of H-bonded CONH groups in proteins (3.6 Å for α-helices and antiparallel β-sheets and 3.7 Å for parallel β-sheets).32 We find that disruption of CO–CO contacts leading to conversion of backbone CO from gCOH to gOH is expected for CO–CO contact distances more than 3.8 Å or CO–C contact angles less than 120°.
For H-bonds, the calculated mean distance (dOH) and the mean CO–H contact angle (θOH) are 2.0 Å and 138°, respectively. In the region dOH ≃ 1.9 Å and 110° ≤ θOH ≤ 170°, contribution of CO···HN interaction to stability of backbone CO in gCOH is at a maximum (>1.6 kcal/mol). A higher frequency of H-bond disruption leading to conversion of backbone CO from gCOH to gCO is expected for a H-bond distance of more than 2.2 Å or for a CO···H contact angle less than 100°.
Interactions of Uncharged Side-Chains
To probe the interactions of side-chains in the large cluster, we consider the radial distributions among side-chains and the backbone about side-chains (Figure SI-7). We find that the side-chain Asn shows the highest tendency to interact directly with the backbone. From the location of the first peak (<3 Å), most of these noncovalent interactions between side-chain and backbone in asn can be assigned to H-bonding interactions. The tendency of the iso-propyl group in Val to interact directly is the lowest both with the backbone and side-chains. We also find a close packing of phenyl rings in the arrangement of peptides. Among the uncharged side-chains considered, the benzyl group in Phe shows the highest tendency to cluster. Interactions between phenyl groups are common in proteins, and the high tendency of aromatic groups to be surrounded by identical groups in solution is often considered to drive protein folding. Our calculations also show that the angle between the ring planes of interacting phenyl pair is usually ~90°. In other words, phenyl groups prefer to interact perpendicularly which is in accord with solution,79 crystal structures of benzene (Cambridge database), and analyses of proteins.80
To further characterize the interactions of side-chains in asn and gln, we find that side-chain N interacts more with backbone O of other peptides than does side-chain O with backbone N (Figure SI-7). The number of interpeptide H-bonds between side-chain N and backbone O in the large cluster is more than 2 times higher than that between side-chain O and backbone N. The probability of a side-chain to form H-bonds with side-chains is significantly lower than with a backbone. This is due in part to the much higher population of backbone H-bonding sites than those of side-chains. We find the side-chain O of Asn interacts more than that of Gln with backbone N of same peptide, whereas it is the side-chain N of Gln that interacts more than that of Asn with backbone oxygens of the same peptide.
Comparison between Charged and Uncharged Aggregates
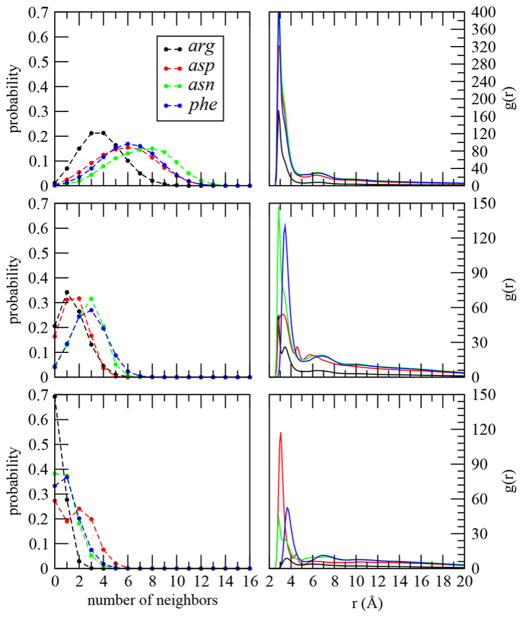
The mechanism of association leading to phase separation depends on the chemical nature of the side-chain. In Figure 9 we compare our acidic, asp, and basic, arg, peptides with both polar, asn, and nonpolar, phe, systems. The interactions both among and between side-chains and backbone in the phase separated cluster can be characterized in different ways. On the left side of Figure 9 we show the backbone–backbone, side-chain–backbone and side-chain–side-chain probability of neighbors within 4 Å. On the right-hand side are the corresponding radial distribution functions. As expected the backbones of the pentapeptides can have contacts with a more substantial number of other backbones than with side-chains. The side-chain–side-chain neighbor distribution is the narrowest.
Figure 9.
Comparison between charged and uncharged peptide associations. Left: Probability distribution of the number of backbone within 4 Å of backbone (top), backbone within 4 Å of a side-chain (middle), and side-chain within 4 Å of a side-chain (bottom) for peptides in the large cluster. Right: Site–site radial distribution functions (based on shortest heavy atom distance) between the heavy atoms of peptide backbones (top), between the heavy atoms of peptide side-chains and backbones (middle), and between the heavy atoms of peptide side-chains (bottom) for peptides in the large cluster.
The cationic and anionic side-chains cluster quite differently. The arg system generally interacts with fewer partners regardless of whether they are backbone or side-chain. Interestingly, we find asp to make more side-chain to side-chain contacts than the other systems. Proteins are known to hold like charged groups together in active sites for catalysis,81 but here we do not have any polymer architecture leading to constraint. Accounting for differences in side-chain volume we see more similarity between the uncharged groups than the charged ones.
CONCLUSION
We analyzed the effect of side-chains on aggregation/association of peptides by directly calculating the solubility limit of classical molecular models. Spontaneous aggregation separated the solution into organic-rich and peptide saturated water-rich phases. Correlated with the changes in peptide aggregation were the fluctuations in the waters of the first hydration shell. It was found that the greater the fluctuations are, the higher the aggregation propensity of the peptide is, which is reflected in modern theories of hydrophobicity.71,72,82,83 However, we note that our fluctuation trend also predicted the clustering propensity of Asn and Gln side-chains which are classically hydrophilic. Yet, these residues are well-known to play a role in peptide aggregation (plaque forming) diseases.84,85
Our preliminary assessment of forces that hold the disordered peptides together in aggregates showed a greater contribution of CO–CO interactions than that of H-bonds. The relative importance of CO–CO and H-bonding interactions in peptide aggregation was then examined closely by decomposing backbone CO into different interactions the considered O-atom made. Backbone COs that participate in H-bonding interactions with backbone NH are almost always surrounded by other CO groups from the main-chains.
Our simulations also captured the high tendency of the benzyl group in Phe to be surrounded by identical groups in solution, the tendency often considered to be the driving force of protein folding. In contrast, the tendency of the side-chain –CONH2 group in Asn and Gln, which readily aggregate with a low solubility limit, was not seen to form a significant number of H-bonds with identical groups. Instead, the side-chain N was observed forming H-bonds mainly with the backbone.
The H-bond has long been considered to be the most important contributor to forces that hold backbone amide groups together and stabilize proteins in their folded states. Disordered systems such as the peptides considered here show different behaviors. Our study suggests that a backbone amide pair in a disordered peptide connected by a H-bond gains maximum energetic and dynamic stability only if the pair can make use of other CO–CO interactions as well. Our results thus bring clarity to the role of CO–CO interaction in formation and stability of disordered peptide aggregates. Extended to unstructured proteins, backbone H-bonds are unlikely to form or survive for a long time without surrounding backbone carbonyls.
Supplementary Material
Acknowledgments
We gratefully acknowledge the Robert A. Welch Foundation (H-0037), the National Science Foundation (CHE-1152876), and the National Institutes of Health (GM-037657) for partial support of this work. B.M.P. thanks Dr. Dilip Asthagiri for comments. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation Grant ACI-1548562. The authors also acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing HPC resources that have contributed to the research results reported within this paper. URL: http://www.tacc.utexas.edu.
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpcb.7b10734.
Detailed description of how the fluctuations in the peptide conformations were determined; tables of system overview parameters, H-bonding interactions, probabilities, lifetimes, and energetic stabilities; and figures showing the system at the end of equilibration, time evolution, variation in peptide interactions, survival functions, decompositions, rdf’s, and interaction energies (PDF)
References
- 1.Kauzmann W. Some Factors in the Interpretation of Protein Denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 2.Ellis RJ. Macromolecular Crowding: Obvious but Under-appreciated. Trends Biochem Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 3.Lin Y, Protter DSW, Rosen M, Parker R. Formation and Maturation of Phase- Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shin Y, Brangwynne CP. Liquid Phase Condensation in Cell Physiology and Disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 5.Karandur D, Wong KY, Pettitt BM. Solubility and Aggregation of Gly5 inWater. J Phys Chem B. 2014;118:9565–9572. doi: 10.1021/jp503358n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karandur D, Harris RC, Pettitt BM. Protein Collapse Driven Against Solvation Free Energy Without H-bonds. Protein Sci. 2016;25:103–110. doi: 10.1002/pro.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanford C. Contribution of Hydrophobic Interactions to the Stability of the Globular Conformation of Proteins. J Am Chem Soc. 1962;84:4240–4247. [Google Scholar]
- 8.Baldwin RL. How Does Protein Folding Get Started? Trends Biochem Sci. 1989;14:291–294. doi: 10.1016/0968-0004(89)90067-4. [DOI] [PubMed] [Google Scholar]
- 9.Spassov VZ, Yan L, Flook PK. The Dominant Role of Side-Chain Backbone Interactions in Structural Realization of Amino Acid Code. ChiRotor: A Side-Chain Prediction Algorithm Based on Side-Chain Backbone Interactions. Protein Sci. 2007;16:494–506. doi: 10.1110/ps.062447107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auton M, Rösgen J, Sinev M, Holthauzen LM, Bolen DW. Osmolyte Effects on Protein Stability and Solubility: A Balancing Act Between Backbone and Side-Chains. Biophys Chem. 2011;159:90–99. doi: 10.1016/j.bpc.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moeser B, Horinek D. Unified Description of Urea Denaturation: Backbone and Side Chains Contribute Equally in the Transfer Model. J Phys Chem B. 2014;118:107–114. doi: 10.1021/jp409934q. [DOI] [PubMed] [Google Scholar]
- 12.Canchi DR, García AE. Backbone and Side-Chain Contributions in Protein De-naturation by Urea. Biophys J. 2011;100:1526–1533. doi: 10.1016/j.bpj.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Street TO, Bolen DW, Rose GD. A Molecular Mechanism for Osmolyte-Induced Protein Stability. Proc Natl Acad Sci U S A. 2006;103:13997–14002. doi: 10.1073/pnas.0606236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu CY, Kokubo H, Lynch GC, Bolen DW, Pettitt BM. Backbone Additivity in the Transfer Model of Protein Solvation. Protein Sci. 2010;19:1011–1022. doi: 10.1002/pro.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu CY, Lynch GC, Kokubo H, Pettitt BM. TMAO Influence on the Backbone of: an Oligoglycine Model. Proteins: Struct, Funct Genet. 2010;78:695–704. doi: 10.1002/prot.22598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teufel DP, Johnson CM, Lum JK, Neuweiler H. Backbone-Driven Collapse in Unfolded Protein Chains. J Mol Biol. 2011;409:250–262. doi: 10.1016/j.jmb.2011.03.066. [DOI] [PubMed] [Google Scholar]
- 17.Tran HT, Mao A, Pappu RV. Role of Backbone-Solvent Interactions in Deter- mining Conformational Equilibria of Intrinsically Disordered Proteins. J Am Chem Soc. 2008;130:7380–7392. doi: 10.1021/ja710446s. [DOI] [PubMed] [Google Scholar]
- 18.Lu J, Wang XJ, Yang X, Ching CB. Solubilities of Glycine and Its Oligopeptides in Aqueous Solutions. J Chem Eng Data. 2006;51:1593–1596. [Google Scholar]
- 19.Pauling L, Corey RB, Branson HR. The Structure of Proteins: Two Hydrogen-Bonded Helical Configurations of the Polypeptide Chain. Proc Natl Acad Sci U S A. 1951;37:205–211. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolen DW, Rose GD. Structure and Energetics of the Hydrogen-Bonded Backbone in Protein Folding. Annu Rev Biochem. 2008;77:339–362. doi: 10.1146/annurev.biochem.77.061306.131357. [DOI] [PubMed] [Google Scholar]
- 21.Bamford CH, Brown L, Cant EM, Elliott A, Hanby WE, Malcolm BR. Structure of Polyglycine. Nature. 1955;176:396–397. [Google Scholar]
- 22.Crick FHC, Rich A. Structure of Polyglycine II. Nature. 1955;176:780–781. doi: 10.1038/176780a0. [DOI] [PubMed] [Google Scholar]
- 23.Scholtz JM, Marqusee S, Baldwin RL, York EJ, Stewart JM, Santoro M, Bolen DW. Calorimetric Determination of the Enthalpy Change for the α-helix to Coil Transition of an Alanine Peptide in Water. Proc Natl Acad Sci U S A. 1991;88:2854–2858. doi: 10.1073/pnas.88.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myers JK, Pace CN. Hydrogen Bonding Stabilizes Globular Proteins. Biophys J. 1996;71:2033–2039. doi: 10.1016/S0006-3495(96)79401-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takano K, Yamagata Y, Funahashi J, Hioki Y, Kuramitsu S, Yutani K. Contribution of Intra- and Intermolecular Hydrogen Bonds to the Conformational Stability of Human Lysozyme. Biochemistry. 1999;38:12698–12708. doi: 10.1021/bi9910169. [DOI] [PubMed] [Google Scholar]
- 26.Ben-Tal N, Sitkoff D, Topol IA, Yang AS, Burt SK, Honig B. Free Energy of Amide Hydrogen Bond Formation in Vacuum, in Water, and in Liquid Alkane Solution. J Phys Chem B. 1997;101:450–457. [Google Scholar]
- 27.Williams DH, Searle MS, Mackay JP, Gerhard U, Maplestone RA. Toward an Estimation of Binding Constants in Aqueous Solution: Studies of Associations of Vancomycin Group Antibiotics. Proc Natl Acad Sci U S A. 1993;90:1172–1178. doi: 10.1073/pnas.90.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fersht AR, Shi JP, Knill-Jones J, Lowe DM, Wilkinson AJ, Blow DM, Brick P, Carter P, Waye MMY, Winter G. Hydrogen Bonding and Biological Specificity Analysed by Protein Engineering. Nature. 1985;314:235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- 29.Sheu SY, Yang DY, Selzle HL, Schlag EW. Energetics of Hydrogen Bonds in Peptides. Proc Natl Acad Sci U S A. 2003;100:12683–12687. doi: 10.1073/pnas.2133366100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shirley BA, Stanssens P, Hahn U, Pace CN. Contribution of Hydrogen Bonding to the Conformational Stability of Ribonuclease T1. Biochemistry. 1992;31:725–732. doi: 10.1021/bi00118a013. [DOI] [PubMed] [Google Scholar]
- 31.Arora N, Jayaram B. Strength of Hydrogen Bonds in α Helices. J Comput Chem. 1997;18:1245–1252. [Google Scholar]
- 32.Maccallum PH, Poet R, Milner-White EJ. Coulombic Interactions Between Partially Charged Main-Chain Atoms not Hydrogen-Bonded to Each Other Influence the Conformations of α-helices and Antiparallel β-sheets. A New Method for Analysing the Forces Between Hydrogen Bonding Groups in Proteins Includes All the Coulombic Interactions. J Mol Biol. 1995;248:361–373. doi: 10.1016/s0022-2836(95)80056-5. [DOI] [PubMed] [Google Scholar]
- 33.Bartlett GJ, Choudhary A, Raines RT, Woolfson DN. n → π* Interactions in Proteins. Nat Chem Biol. 2010;6:615–620. doi: 10.1038/nchembio.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paulini R, Müller K, Diederich F. Orthogonal Multipolar Interactions in Structural Chemistry and Biology. Angew Chem, Int Ed. 2005;44:1788–1805. doi: 10.1002/anie.200462213. [DOI] [PubMed] [Google Scholar]
- 35.Fischer FR, Wood PA, Allen FH, Diederich F. Orthogonal Dipolar Interactions Between Amide Carbonyl Groups. Proc Natl Acad Sci U S A. 2008;105:17290–17294. doi: 10.1073/pnas.0806129105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bretscher LE, Jenkins CL, Taylor KM, DeRider ML, Raines RT. Conformational Stability of Collagen Relies on a Stereoelectronic Effect. J Am Chem Soc. 2001;123:777–778. doi: 10.1021/ja005542v. [DOI] [PubMed] [Google Scholar]
- 37.DeRider ML, Wilkens SJ, Waddell MJ, Bretscher LE, Weinhold F, Raines RT, Markley JL. Collagen Stability: Insights from NMR Spectroscopic and Hybrid Density Functional Computational Investigations of the Effect of Electronegative Substituents on Prolyl Ring Conformations. J Am Chem Soc. 2002;124:2497–2505. doi: 10.1021/ja0166904. [DOI] [PubMed] [Google Scholar]
- 38.Hinderaker MP, Raines RT. An Electronic Effect on Protein Structure. Protein Sci. 2003;12:1188–1194. doi: 10.1110/ps.0241903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hodges JA, Raines RT. Energetics of an n → π* Interaction that Impacts Protein Structure. Org Lett. 2006;8:4695–4697. doi: 10.1021/ol061569t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shoulders MD, Guzei IA, Raines RT. 4-Chloroprolines: Synthesis, Conformational Analysis, and Effect on the Collagen Triple Helix. Biopolymers. 2008;89:443–454. doi: 10.1002/bip.20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choudhary A, Gandla D, Krow GR, Raines RT. Nature of Amide Carbonyl-Carbonyl Interactions in Proteins. J Am Chem Soc. 2009;131:7244–7246. doi: 10.1021/ja901188y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newberry RW, Raines RT. A Prevalent Intraresidue Hydrogen Bond Stabilizes Proteins. Nat Chem Biol. 2016;12:1084. doi: 10.1038/nchembio.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newberry RW, Raines RT. The n → π* Interaction. Acc Chem Res. 2017;50:1838–1846. doi: 10.1021/acs.accounts.7b00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deane CM, Allen FH, Taylor R, Blundell TL. Carbonyl-Carbonyl Interactions Stabilize the Partially Allowed Ramachandran Conformations of Asparagine and Aspartic Acid. Protein Eng, Des Sel. 1999;12:1025–1028. doi: 10.1093/protein/12.12.1025. [DOI] [PubMed] [Google Scholar]
- 45.Horng JC, Raines RT. Stereoelectronic Effects on Polyproline Conformation. Protein Sci. 2006;15:74–83. doi: 10.1110/ps.051779806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao J, Kelly JW. Toward Quantification of Protein Backbone-Backbone Hydrogen Bonding Energies: An Energetic Analysis of an Amide-to-Ester Mutation in an α-helix within a Protein. Protein Sci. 2008;17:1096–1101. doi: 10.1110/ps.083439708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorske BC, Stringer JR, Bastian BL, Fowler SA, Blackwell HE. New Strategies for the Design of Folded Peptoids Revealed by a Survey of Noncovalent Interactions in Model Systems. J Am Chem Soc. 2009;131:16555–16567. doi: 10.1021/ja907184g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jakobsche CE, Choudhary A, Miller SJ, Raines RT. n → π* Interaction and n → π Pauli Repulsion Are Antagonistic for Protein Stability. J Am Chem Soc. 2010;132:6651–6653. doi: 10.1021/ja100931y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen FH, Baalham CA, Lommerse JPM, Raithby PR. Carbonyl-Carbonyl Interactions can be Competitive with Hydrogen Bonds. Acta Crystallogr, Sect B: Struct Sci. 1998;54:320–329. [Google Scholar]
- 50.Auton M, Holthauzen LMF, Bolen DW. Anatomy of Energetic Changes Accompanying Urea-Induced Protein Denaturation. Proc Natl Acad Sci U S A. 2007;104:15317–15322. doi: 10.1073/pnas.0706251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang J, Lenhoff AM, Sandler SI. Solvation Free Energy of Amino Acids and Side-Chain Analogues. J Phys Chem B. 2007;111:2098–2106. doi: 10.1021/jp0620163. [DOI] [PubMed] [Google Scholar]
- 52.König G, Bruckner S, Boresch S. Absolute Hydration Free Energies of Blocked Amino Acids: Implications for Protein Solvation and Stability. Biophys J. 2013;104:453–462. doi: 10.1016/j.bpj.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohn EJ, Mcmeekin TL, Edsall JT, Weare JH. Studies in the Physical Chemistry of Amino Acids, Peptides and Related Substances. II. The Solubility of α- Amino Acids in Water and in Alcohol-Water Mixtures. J Am Chem Soc. 1934;56:2270–2282. [Google Scholar]
- 54.Needham TE, Jr, Paruta AN, Gerraughty RJ. Solubility of Amino Acids in Pure Solvent Systems. J Pharm Sci. 1971;60:565–567. doi: 10.1002/jps.2600600410. [DOI] [PubMed] [Google Scholar]
- 55.Gates ZP, Baxa MC, Yu W, Riback JA, Li H, Roux B, Kent SBH, Sosnick TR. Perplexing Cooperative Folding and Stability of a Low-Sequence Complexity, Polyproline 2 Protein Lacking a Hydrophobic Core. Proc Natl Acad Sci U S A. 2017;114:2241–2246. doi: 10.1073/pnas.1609579114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Record M, Anderson C. Interpretation of Preferential Interaction Coefficients of Non-electrolytes and of Electrolyte Ions in Terms of a Two-Domain Model. Biophys J. 1995;68:786–794. doi: 10.1016/S0006-3495(95)80254-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Courtenay E, Capp M, Saecker R, Record M. Thermodynamic Analysis of Interactions Between Denaturants and Protein Surface Exposed on Unfolding: Interpretation of Urea and Guanidinium Chloride m-Values and their Correlation with Changes in Accessible Surface Area (ASA) Using Preferential Interaction Coefficients and the Local-Bulk Domain Model. Proteins: Struct, Funct Genet. 2000;41:72–85. doi: 10.1002/1097-0134(2000)41:4+<72::aid-prot70>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 58.Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber Biomolecular Simulation Programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable Molecular Dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 61.Drake JA, Pettitt BM. Force Field-Dependent Solution Properties of Glycine Oligomers. J Comput Chem. 2015;36:1275–1285. doi: 10.1002/jcc.23934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, de Groot BL, Grubmuller H, MacKerell AD., Jr CHARMM36m: An Improved Force Field for Folded for Intrinsically Disordered Proteins. Nat Methods. 2017;14:71. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Piana S, Donchev AG, Robustelli P, Shaw DE. Water Dispersion Interactions Strongly Influence Simulated Structural Properties of Disordered Protein States. J Phys Chem B. 2015;119:5113–5123. doi: 10.1021/jp508971m. [DOI] [PubMed] [Google Scholar]
- 64.Best RB, Zheng W, Mittal J. Balanced Protein-Water Interactions Improve Properties of Disordered Proteins and Non-Specific Protein Association. J Chem Theory Comput. 2014;10:5113–5124. doi: 10.1021/ct500569b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Best RB, Zheng W, Mittal J. Correction to Balanced Protein-Water Interactions Improve Properties of Disordered Proteins and Non-Specific Protein Association. J Chem Theory Comput. 2015;11:1978–1978. doi: 10.1021/acs.jctc.5b00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mercadante D, Wagner JA, Aramburu IV, Lemke EA, Gräter F. Sampling Long-versus Short-Range Interactions Defines the Ability of Force Fields To Reproduce the Dynamics of Intrinsically Disordered Proteins. J Chem Theory Comput. 2017;13:3964–3974. doi: 10.1021/acs.jctc.7b00143. [DOI] [PubMed] [Google Scholar]
- 67.White SH, Wimley WC. Membrane Protein Folding and Stability: Physical Principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 68.Raschke TM, Tsai J, Levitt M. Quantification of the Hydrophobic Interaction by Simulations of the Aggregation of Small Hydrophobic Solutes in Water. Proc Natl Acad Sci U S A. 2001;98:5965–5969. doi: 10.1073/pnas.111158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wei D, Patey GN. Ferroelectric Liquid-Crystal and Solid Phases Formed by Strongly Interacting Dipolar Soft Spheres. Phys Rev A: At, Mol, Opt Phys. 1992;46:7783–7792. doi: 10.1103/physreva.46.7783. [DOI] [PubMed] [Google Scholar]
- 70.Edelsbrunner H, Koehl P. The Weighted-Volume Derivative of a Space-Filling Diagram. Proc Natl Acad Sci U S A. 2003;100:2203–2208. doi: 10.1073/pnas.0537830100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patel AJ, Varilly P, Chandler D. Fluctuations of Water Near Extended Hydrophobic and Hydrophilic Surfaces. J Phys Chem B. 2010;114:1632–1637. doi: 10.1021/jp909048f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sarupria S, Garde S. Quantifying Water Density Fluctuations and Compressibility of Hydration Shells of Hydrophobic Solutes and Proteins. Phys Rev Lett. 2009;103:037803-1–037803-4. doi: 10.1103/PhysRevLett.103.037803. [DOI] [PubMed] [Google Scholar]
- 73.Eisenberg D, McLachlan AD. Solvation Energy in Protein Folding and Binding. Nature. 1986;319:199–203. doi: 10.1038/319199a0. [DOI] [PubMed] [Google Scholar]
- 74.Bandyopadhyay S, Chakraborty S, Bagchi B. Secondary Structure Sensitivity of Hydrogen Bond Lifetime Dynamics in the Protein Hydration Layer. J Am Chem Soc. 2005;127:16660–16667. doi: 10.1021/ja054462u. [DOI] [PubMed] [Google Scholar]
- 75.Sarma R, Paul S. Exploring the Molecular Mechanism of Trimethylamine-N-oxide’s Ability to Counteract the Protein Denaturing Effects of Urea. J Phys Chem B. 2013;117:5691–5704. doi: 10.1021/jp401750v. [DOI] [PubMed] [Google Scholar]
- 76.Kabsch W, Sander C. Dictionary of Protein Secondary Structure: Pattern Recognition of Hydrogen-Bonded and Geometrical Features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 77.Kim S, Takeda T, Klimov DK. Mapping Conformational Ensembles of Aβ Oligomers in Molecular Dynamics Simulations. Biophys J. 2010;99:1949–1958. doi: 10.1016/j.bpj.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davis AM, Teague SJ. Hydrogen Bonding, Hydrophobic Interactions, and Failure of the Rigid Receptor Hypothesis. Angew Chem, Int Ed. 1999;38:736–749. doi: 10.1002/(SICI)1521-3773(19990315)38:6<736::AID-ANIE736>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 79.Lowden LJ, Chandler D. Theory of Intermolecular Pair Correlations for Molecular Liquids. Applications to the Liquids Carbon Tetrachloride, Carbon Disulfide, Carbon Diselenide, and Benzene. J Chem Phys. 1974;61:5228–5241. [Google Scholar]
- 80.Singh J, Thornton JM. The Interaction Between Phenylalanine Rings in Proteins. FEBS Lett. 1985;191:1–6. [Google Scholar]
- 81.Brooks CL, III, Karplus M, Pettitt BM. Advances in Chemical Physics. Vol. 71. John Wiley and Sons; New York: 1988. Proteins: A Theoretical Perspective of Dynamics, Structure, and Thermodynamics; p. 259. [Google Scholar]
- 82.Lu X, Murphy RM. Asparagine Repeat Peptides: Aggregation Kinetics and Comparison with Glutamine Repeats. Biochemistry. 2015;54:4784–4794. doi: 10.1021/acs.biochem.5b00644. [DOI] [PubMed] [Google Scholar]
- 83.Murphy RM, Kendrick BS. Protein Misfolding and Aggregation. Biotechnol Prog. 2007;23:548–552. doi: 10.1021/bp060374h. [DOI] [PubMed] [Google Scholar]
- 84.Perutz MF, Pope BJ, Owen D, Wanker EE, Scherzinger E. Aggregation of Proteins with Expanded Glutamine and Alanine Repeats of the Glutamine-Rich and Asparagine-rich Domains of Sup35 and of the Amyloid β-peptide of Amyloid Plaques. Proc Natl Acad Sci U S A. 2002;99:5596–5600. doi: 10.1073/pnas.042681599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tsai HHG, Reches M, Tsai CJ, Gunasekaran K, Gazit E, Nussinov R. Energy Landscape of Amyloidogenic Peptide Oligomerization by Parallel-Tempering Molecular Dynamics Simulation: Significant Role of Asn Ladder. Proc Natl Acad Sci U S A. 2005;102:8174–8179. doi: 10.1073/pnas.0408653102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.