Summary
The Toxoplasma inner membrane complex (IMC) is a specialized organelle underlying the parasite’s plasma membrane that consists of flattened rectangular membrane sacs that are sutured together and positioned atop a supportive cytoskeleton. We have previously identified a novel class of proteins localizing to the transverse and longitudinal sutures of the IMC, which we named ISCs. Here we have used proximity-dependent biotin identification (BioID) at the sutures to better define the composition of this IMC subcompartment. Using ISC4 as bait, we demonstrate biotin-dependent labeling of the sutures and have uncovered two new ISCs. We also identified five new proteins that exclusively localize to the transverse sutures which we named TSCs, demonstrating that components of the IMC sutures consist of two groups, those that localize to the transverse and longitudinal sutures (ISCs) and those residing only in the transverse sutures (TSCs). In addition, we functionally analyze the ISC protein ISC3 and demonstrate that ISC3-null parasites have morphological defects and reduced fitness in vitro. Most importantly, Δisc3 parasites exhibit a complete loss of virulence in vivo. These studies expand the known composition of the IMC sutures and highlight the contribution of ISCs to the ability of the parasite to proliferate and cause disease.
Introduction
Members of the phylum Apicomplexa are obligate intracellular parasites that cause medically and economically important diseases in humans and animals. Plasmodium species are the causative agents of malaria, which results in 200 million clinical cases and more than half a million deaths annually, while Cryptosporidium infections are a significant contributor of diarrheal disease in children in the developing world (Kotloff et al., 2013, WHO, 2014). Toxoplasma gondii, the etiological agent of toxoplasmosis, is the most experimentally tractable apicomplexan and serves as a model system for studying how this group of pathogens infect their hosts and cause disease.
Hallmarks of apicomplexan biology include a distinct mode of replication whereby the progeny are assembled within the mother parasite, and invasion via the formation of a ring-shaped tight junction interface between the parasite and host plasma membranes, through which the parasite gains entry into the host cell (Carruthers et al., 2007, Shen et al., 2012, Francia et al., 2014). Central to these two unique biological processes is an organelle called the inner membrane complex (IMC), which consists of a series of flattened membrane sacs called alveoli that are sutured together and positioned atop a supportive cytoskeletal network of filamentous coiled-coil proteins (D’Haese et al., 1977, Porchet et al., 1977, Mann et al., 2001). The IMC serves structural roles as the anchor for the parasite’s actin-myosin motor that powers gliding motility and invasion and also as the scaffold for daughter cell formation during replication (Harding et al., 2014). Despite the critical roles of the IMC, the protein composition of this organelle is still being elucidated and the functions of these constituents remain largely unexplored.
We have previously utilized an in vivo biotinylation approach called BioID to uncover new proteins in the IMC (Chen et al., 2015). This techniques relies on the proximity-dependent biotinylation of interacting partners and proximal proteins by the promiscuous biotin ligase BirA* that is fused to a protein of interest (Roux et al., 2012). Using the IMC proteins ISP3 and AC2 as bait, we demonstrated targeted enrichment of novel labeled proteins from their respective subcompartments and greatly expanded the known IMC proteome (Chen et al., 2015). The most notable subset of proteins identified in these experiments is the IMC sutures components (ISCs), which localize to the transverse and longitudinal sutures that demark the junctions between the alveolar sacs. The IMC sutures had been observed by electron microscopy for ~40 years, but the proteins that comprise these structures had remained elusive prior to our BioID experiments (Porchet et al., 1977). We identified four ISCs and showed that they associate with either the IMC membranes (ISCs 2/3) or the underlying cytoskeleton (ISCs 1/4). Most of these ISCs lack identifiable domains that would suggest function and the organization of the ISCs within the sutures is unknown.
However, ISC3 is predicted to be a choline transporter-like protein (CTL) and contains ten transmembrane domains, which is consistent with CTLs in other organisms (Michel et al., 2006). Choline is an essential nutrient that serves as a precursor in the biogenesis of lipid components such as phosphotidylcholine (PtdCho) and sphingomyelin for incorporation into cellular membranes (Zeisel et al., 1994). CTLs are expressed in many different cell types and mediate choline uptake across plasma membranes for phospholipid synthesis (Michel et al., 2006). For example, PtdCho can be synthesized de novo via the CDP-choline arm of the Kennedy pathway: the imported choline is initially phosphorylated by an enzyme called choline kinase, and after subsequent conversion to CDP-choline, the modified choline headgroup is combined with a diacylglycerol (DAG) backbone to form PtdCho (Zeisel et al., 1994). T. gondii encodes a functional choline kinase that is refractory to genetic ablation, suggesting de novo PtdCho synthesis via the CDP-choline arm of the Kennedy pathway is essential for parasite growth and replication, although T. gondii is also capable of scavenging phospholipids from the host cell (Charron et al., 2002, Sampels et al., 2012).
In this report, we expand upon our previous BioID studies by using an ISC4-BirA* fusion protein to identify additional IMC sutures components. Using this approach, we uncovered two new ISCs as well as a novel subset of sutures components that specifically localize to the transverse but not the longitudinal junctions of the IMC plates. We demonstrate that similar to the ISCs, this new class contains IMC membrane or cytoskeleton-associated components. In addition, we functionally evaluate ISC3 using a gene knockout approach and demonstrate that Δisc3 parasites have a reduced fitness in vitro and exhibit morphological defects within the parasitophorous vacuole as well as in the extracellular environment. Surprisingly, disruption of ISC3 results in a complete loss of virulence, demonstrating that this protein is absolutely crucial for establishing an infection and causing disease in vivo. Taken together, these studies show that proteins in the IMC sutures segregate into two groups (ISCs and TSCs), demonstrate the importance of ISCs to parasite fitness and virulence, and provide a foundation for assessing the functional contribution of this IMC subcompartment to parasite biology.
Results
ISC4-BirA* biotinylates novel IMC sutures components
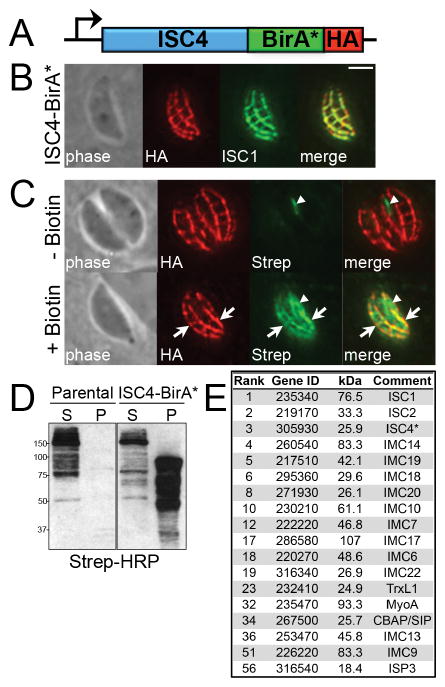
To identify novel ISCs by BioID, we generated parasites expressing an ISC4-BirA* fusion protein, with a C-terminal 3×HA tag, by endogenous gene tagging in the RHΔhptΔku80 strain (Fig. 1A). We assessed the localization of the fusion by immunofluorescence assays (IFA) and observed staining in the transverse and longitudinal sutures of the IMC (Fig. 1B), a pattern consistent with endogenous ISC4 (Chen et al., 2015). ISC4-BirA* also co-localized with ISC1, confirming correct targeting of the fusion protein to the IMC sutures (Fig. 1B).
Figure 1. ISC4-BirA* biotinylates proteins in the IMC sutures.
A) Diagram of the endogenous locus encoding ISC4 fused to BirA*, plus a C-terminal 3xHA epitope tag, driven by its own promoter. B) IFA demonstrating correct targeting of the ISC4-BirA* fusion protein to the IMC sutures, as determined by co-localization with ISC1. Red: mouse anti-HA antibody. Green: rat anti-ISC1 antibody. Scale bar = 2 μm. C) IFA of ISC4-BirA* expressing parasites, grown for 24 hours +/- biotin. ISC4-BirA* biotinylates proteins in the transverse and longitudinal sutures in a biotin-dependent manner (arrows). Endogenously biotinylated proteins in the apicoplast are detectable as background in the absence of biotin (arrowheads). Red: mouse anti-HA antibody. Green: streptavidin-Alexa Fluor 488. D) Western blot showing TX-100 fractionation of insoluble, biotinylated proteins in the IMC cytoskeleton (P, pellet) from soluble background proteins (S, supernatant) in ISC4-BirA* parasite lysates. Note the similar profile of endogenously biotinylated background proteins extracted in the soluble fractions of parental (left panel) and ISC4-BirA* parasite lysates (right). E) Proteins localizing to the IMC sutures and/or cytoskeleton are selectively enriched among the highest-scoring ISC4-BioID hits. The bait protein (ISC4) is marked by an asterisk. Gene ID numbers shown are from ToxoDB. The complete listing of ISC4-BioID hits is provided in Table S1.
To determine whether the ISC4-BirA* fusion could biotinylate interacting or proximal proteins upon trafficking to the IMC sutures, we assessed labeled proteins in the parasite by IFA using fluorophore-conjugated streptavidin. In the absence of biotin in the growth medium, we detected only the background of endogenously biotinylated proteins in the parasite apicoplast and mitochondria (Jelenska et al., 2001). However, in parasites grown in media supplemented with biotin, we observed robust streptavidin staining along the transverse and longitudinal sutures that overlapped with the fusion (Fig. 1C), demonstrating that ISC4-BirA* is catalytically active and labels proteins in the IMC sutures.
We then analyzed ISC4-BirA* parasite lysates by Western blot with streptavidin-HRP to determine if multiple proteins are biotinylated by the fusion protein. To reduce background from apicoplast and mitochondrial proteins, we exploited the fact that ISC4 is associated with the IMC cytoskeleton and thus insoluble in the detergent TX-100 (Chen et al., 2015). Using this approach, we were able to release the majority of the background proteins into the soluble fraction of both parental (RHΔhptΔku80) and ISC4-BirA* lysates (Fig. 1D). Importantly, we detected an array of distinct bands that were specifically enriched in the insoluble fraction of ISC4-BirA* lysates, indicating multiple proteins were labeled by the fusion (Fig. 1D). The insoluble fraction from each line was then solubilized in SDS and biotinylated proteins were purified using streptavidin affinity chromatography for downstream identification (data not shown).
Identification of novel ISCs by BioID
To identify the targets labeled by ISC4-BirA*, the biotinylated proteins isolated from fractionated lysates of parental (RHΔhptΔku80) and ISC4-BirA* parasites grown in biotin were subjected to MuDPIT mass spectrometric analysis. Proteins that were unique to ISC4-BirA* samples were scored as hits based on the number of identified spectra and unique peptides (Table S1). Notably, the top three hits identified by mass spectrometry were ISCs 1/2/4, which is consistent with preferential labeling of proteins in the transverse and longitudinal sutures of the IMC and suggests these ISCs are organized in close proximity (Fig. 1E). As expected, our top hits were also significantly enriched for IMC cytoskeletal proteins, including IMCs 6/7/9/10/13/14/17/18/19/20/22 (Fig. 1E), as well as a series of hypothetical proteins (Table S1). These top hits were filtered for proteins with signature cyclical expression patterns similar to known IMC proteins and selected proteins were localized within parasites by endogenous gene tagging (Behnke et al., 2010).
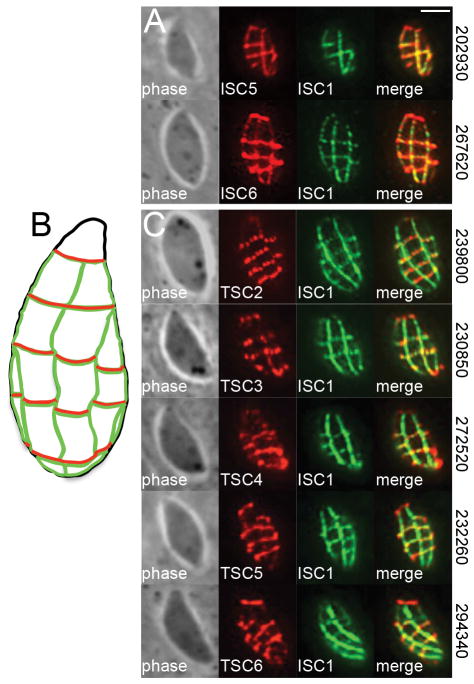
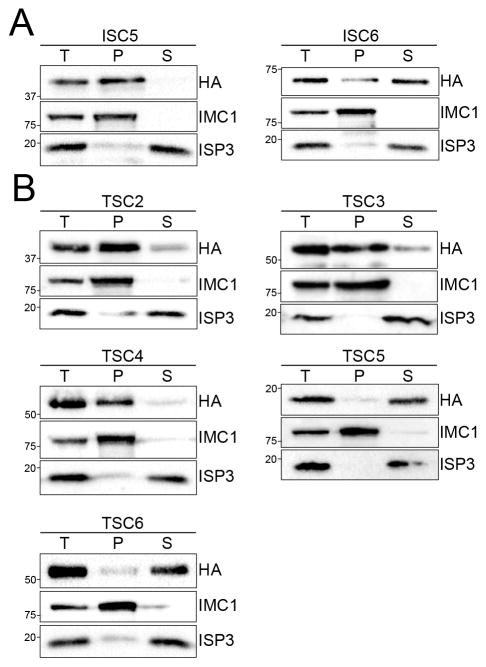
Using this approach, we identified two new proteins that localize to the transverse and longitudinal sutures of the IMC: TgGT1_202930 (designated as ISC5) and TgGT1_267620 (ISC6). These new ISCs, which have no recognizable domains by BLAST analysis, stain the junctions of the IMC plates, overlapping completely with the IMC sutures marker ISC1 by IFA (Fig. 2A). We next used detergent extraction assays to determine whether these ISCs are associated with the IMC membrane sacs or underlying cytoskeleton. ISC5 was completely resistant to detergent extraction and co-fractionated with the cytoskeletal marker IMC1, indicating that it is embedded in the IMC cytoskeletal meshwork (Fig. 3A). In contrast, ISC6 appears to be associated with the IMC membranes as it was mostly solubilized under these conditions, which is consistent with the presence of three predicted transmembrane domains at the C-terminus of the protein (Fig. 3A). These extraction data are in agreement with our previous findings that the IMC sutures contain both membrane and cytoskeleton-associated components (Chen et al., 2015).
Figure 2. Identification of novel IMC sutures proteins by ISC4-BioID.
A) Endogenous gene tagging of hypothetical proteins from the ISC4-BioID data set yielded two additional ISCs (ISC 5/6) which stain the transverse and longitudinal sutures demarked by ISC1. Red: mouse anti-HA antibody. Green: rat anti-ISC1 antibody. Scale bar = 2 μm. B,C) Diagram (B) and IFA (C) showing localization of TSCs (TSCs 2–6) which specifically stain the transverse sutures of the IMC. Note the lack of co-localization between the TSCs and ISC1 along the longitudinal sutures. ToxoDB gene ID numbers for each protein are shown on the right. Red: mouse anti-HA antibody. Green: rat anti-ISC1 antibody.
Figure 3. ISC4-BioID hits contain both membrane and cytoskeleton-associated sutures components.
A,B) Western blots showing TX-100 detergent extraction analyses of new ISCs (A) and TSCs (B) identified from endogenous tagging of ISC4-BioID hits. The total lysate (T) was partitioned into the insoluble pellet (P) or soluble supernatant (S) fractions. Fractionation was monitored using IMC1 (insoluble) and ISP3 (soluble) controls.
The TSCs constitute a new class of IMC sutures components
From these ISC4-BioID studies, we also discovered a novel subset of proteins that exclusively stain the transverse junctions of the IMC plates (Fig. 2B,C). Their localization at the transverse but not the longitudinal sutures was confirmed by co-staining with ISC1 (Fig. 2C). This staining pattern is similar to a previously identified protein in the transverse sutures called CBAP/SIP (Lentini et al., 2014, Tilley et al., 2014), which also ranked highly in the ISC4-BioID data set (Fig. 1E). Based on their localization, we named this new class of proteins Transverse Sutures Components or TSCs (CBAP/SIP was designated as TSC1). In total, we identified five new TSCs in our data set: TgGT1_239800 (hereafter referred to as TSC2), TgGT1_230850 (TSC3), TgGT1_272520 (TSC4), TgGT1_232260 (TSC5), and TgGT1_294340 (TSC6). Detergent extraction analyses indicated that like the ISCs, the TSCs contain proteins associated with either the cytoskeletal (TSCs 2/3/4) or membrane (TSCs 5/6) elements of the IMC (Fig. 3B). The solubility of TSC5 and TSC6 in these extraction assays is consistent with the presence of three and one transmembrane domains, respectively, that likely anchor these proteins to the IMC membrane sacs.
Identification of novel IMC proteins by ISC4-BioID
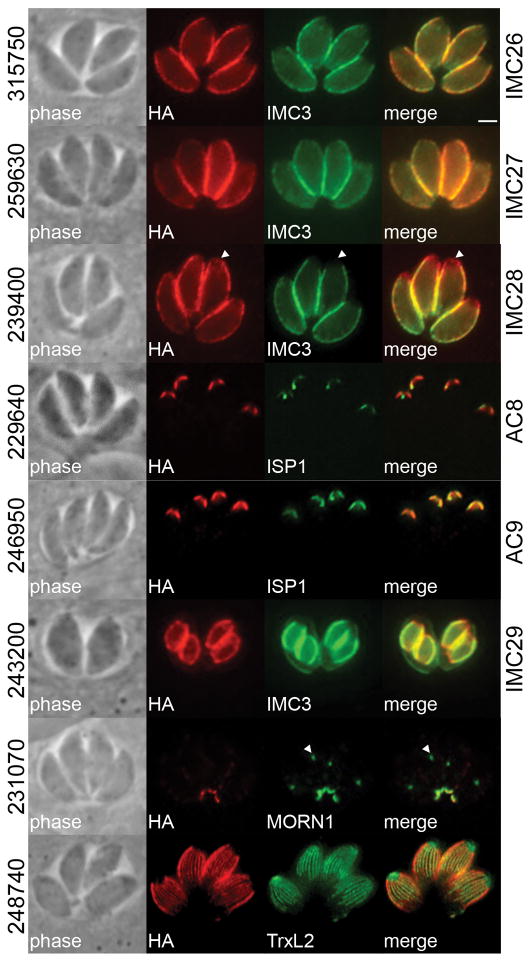
Endogenous gene tagging of ISC4-BioID hits also yielded a number of novel proteins in the IMC or the associated subpellicular microtubules (Fig. 4). From this data set, we uncovered two proteins that localize to the central and basal subcompartments but not the apical cap, TgGT1_315750 (hereafter designated as IMC26) and TgGT1_259630 (IMC27) and one that is present in all three IMC subcompartments, TgGT1_239400 (IMC28). We also identified two proteins residing in the apical cap, TgGT1_229640 (AC8) and TgGT1_246950 (AC9), and a protein that appears to be present exclusively on the daughter cell IMC, TgGT1_243200 (IMC29). Finally, we also identified a protein in the basal complex (TgGT1_231070) and another that co-localized with the subpellicular microtubules of the cortical cytoskeleton (TgGT1_248740). The labeling of known and novel IMC proteins demonstrates that, as expected, ISC4-BirA* biotinylates targets in a variety of IMC subcompartments.
Figure 4. Identification of novel IMC proteins by ISC4-BioID.
Endogenous gene tagging of hypothetical proteins from the ISC4-BioID data set yielded proteins from various IMC subcompartments or the subpellicular microtubules: TgGT1_315750 (designated as IMC26) and TgGT1_259630 (IMC27) reside in the center and base of the IMC, demarked by IMC3; TgGT1_239400 (IMC28) is present throughout the entire IMC, note lack of co-localization between TgGT1_239400 and IMC3 at the apical cap (arrowheads); TgGT1_229640 (AC8) and TgGT1_246950 (AC9) localize to the apical cap demarked by ISP1; TgGT1_243200 (IMC29) appears to primarily stain the daughter cell IMC; TgGT1_231070 is present in the basal complex as co-localized with MORN1 (which also stains the parasite centrocone, arrowheads); and TgGT1_248740 co-localizes with the subpellicular microtubules, visualized by expression of mEmeraldFP-TrxL2 (Liu et al., 2013). Red: mouse or rabbit anti-HA antibody. Green: rat anti-IMC3, mouse anti-ISP1, or rabbit anti-MORN1 antibody. Scale bar = 2 μm.
Generation of ISC3 knockout and complemented strains
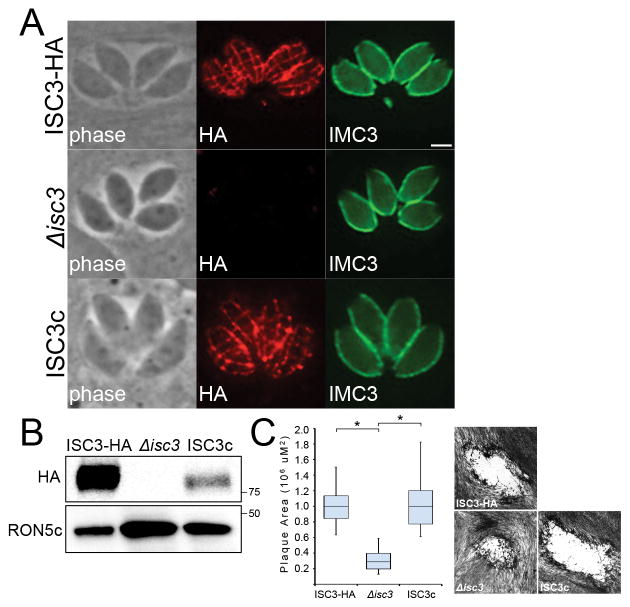
To begin to understand the functional contribution of IMC sutures components, we chose to investigate the role of ISC3 in parasite fitness using a gene knockout approach. ISC3 is a putative choline transporter-like protein that associates with the IMC membranes via ten predicted transmembrane domains (Chen et al., 2015). Since choline is utilized for the synthesis of membrane components such as PtdCho and sphingomyelin, we hypothesized that ISC3 may have a role in the import of this critical micronutrient across the IMC membranes (Zeisel et al., 1994). Thus, we disrupted ISC3 in parental ISC3-HA parasites using CRISPR/Cas9 and loss of the HA-tagged protein in these mutants was monitored by IFA and Western blot (Fig. 5A,B) (Sidik et al., 2014). Using this approach, we successfully generated Δisc3 parasites, demonstrating that this gene is not absolutely required for parasite survival in vitro.
Figure 5. Δisc3 parasites have a reduced fitness in vitro.
A,B) Disruption of ISC3 as shown by loss of the HA-tagged ISC3 protein using IFA (A) and Western blot (B). Δisc3 parasites were complemented with a wild-type ISC3-HA transgene to generate the ISC3c line. Red: mouse anti-HA antibody. Green: rat anti-IMC3 antibody. Scale bar = 2 μm. C) Quantification and example of plaques produced by ISC3-HA, Δisc3, or ISC3c parasites after 9 days of growth. Plaque areas are depicted as a box-whisker plot, with the middle line corresponding to the median, the bottom and top boxes representing the 25th and 75th percentiles, respectively, and whiskers corresponding to the smallest and largest plaques. Representative plaques for each parasite line are shown. Data were analyzed by one-way ANOVA followed by Bonferroni post hoc tests. *p<0.001
We subsequently complemented this Δisc3 strain with a wild-type copy of ISC3-HA that was stably integrated into the remote UPRT locus (hereafter referred to as ISC3c) (Donald et al., 1995). This transgene is driven by the RON5 promoter (similar to IMC proteins, rhoptry proteins are synthesized de novo during endodyogeny and also have cyclical expression patterns) (Behnke et al., 2010). In ISC3c parasites, we observed HA staining along the transverse and longitudinal sutures by IFA, indicating expression from the RON5 promoter did not perturb ISC3 targeting to the proper IMC subcompartment (Fig. 5A). Interestingly, Western blot analysis showed ISC3 expression in the ISC3c strain was much lower compared to wild-type parasites (Fig. 5B). This is most likely due to differences in RON5 promoter strength compared to the ISC3 endogenous promoter but expression could also be impacted by accessibility of the expression cassette at the UPRT locus.
Δisc3 parasites have a reduced fitness in vitro
To determine whether disruption of ISC3 has an effect on parasite fitness, we compared the ability of ISC3-HA, Δisc3, and ISC3c parasites to form plaques on human foreskin fibroblast (HFF) monolayers. These plaque assays measure overall parasite fitness (host cell invasion, growth and replication, and egress) over multiple successive lytic cycles. We found that Δisc3 parasites produced significantly smaller plaques (~70% reduction in size) compared to the parental strain (Fig. 5C). This decrease in plaque size formation was not due to defects in parasite invasion, as parental and Δisc3 parasites exhibited similar capacities to invade host cells in red-green invasion assays (data not shown). Importantly, complementation with the ISC3-HA transgene completely rescued the small plaque phenotype observed in Δisc3 parasites (Fig. 5C). ISC3c parasites produced nearly identical-sized plaques compared to the wild-type strain, demonstrating that the lower levels of ISC3-HA expression are still sufficient for normal parasite growth in culture. Collectively, these data show that Δisc3 parasites have a reduced fitness in vitro that is specifically due to absence of ISC3.
Δisc3 parasites exhibit morphological defects within host cells
To assess the basis for the reduced fitness of Δisc3 parasites, we examined this strain more closely for defects in parasite replication by IFA with IMC markers. Within most vacuoles, some of the parasites appeared normal but others within the vacuole were aberrant and misshapen (Fig. 6A, arrowheads). Other vacuoles were highly disordered (containing parasites with gross morphological defects) or appeared to be abortive, with vacuolized parasites (Fig. 6B). We also stained Δisc3 parasites with an array of organellar markers (e.g. the mitochondria, apicoplast, nucleus, and rhoptries) and sometimes observed missegregated nuclei (Fig. 6B, middle panel) or complete breakdown of the mitochondria (Fig. 6B, bottom panel), consistent with severe replication defects and/or parasite death. Other organelles such as the apicoplast appeared mostly normal, although this was difficult to ascertain in more disordered vacuoles (not shown). Importantly, these morphological defects were observed in two Δisc3 strains generated by completely independent CRISPR/Cas9 knockout experiments and were rescued by complementation. These data suggest that the reduced fitness of ISC3 knockouts in vitro occurs at the level of parasite replication.
Figure 6. Δisc3 parasites exhibit morphological defects within host cells.
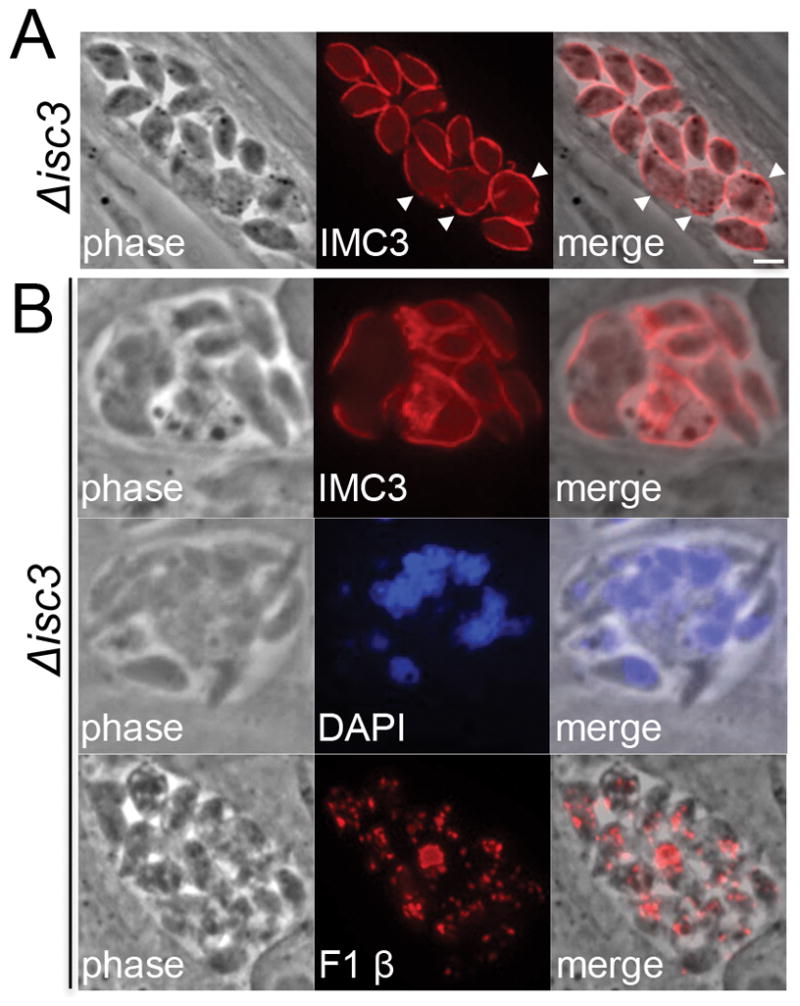
A) Representative IFA of Δisc3 parasites stained with the IMC marker IMC3. Parasites with aberrant morphology can be observed within the majority of knockout vacuoles (arrowheads). Red: rat anti-IMC3 antibody. Scale bar = 2 μm. B) Some Δisc3 vacuoles contain more severe replication defects, as assessed by staining for the IMC (top panel), nuclei (middle), and mitochondria (bottom). Red: rat anti-IMC3 or mouse anti-F1 ATP synthase β subunit antibody. Blue: DAPI.
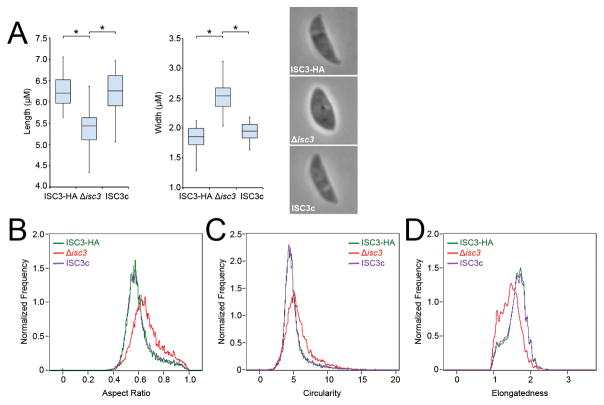
Extracellular Δisc3 parasites display an irregular morphology
To determine if ISC3 plays a role in parasite morphology, we examined extracellular ISC3 knockouts using light microscopy, as a similar function has been described for CBAP/SIP/TSC1 (Lentini et al., 2014, Tilley et al., 2014). We found that disruption of ISC3 results in shorter and wider parasites, giving these knockouts a bloated appearance compared to the parental strain (Fig. 7A). This phenotype was specifically due to loss of ISC3 as these morphological changes are largely abrogated in the complemented strain. We next utilized ImageStream flow cytometry to assess and quantitate parasite morphology in a high-throughput, population-scale analysis. In agreement with measurements obtained by light microscopy, we found that Δisc3 parasites generally have rounder cell shapes, as reflected by shifts towards higher aspect ratio and circularity values compared to wild-type and complemented parasites (Fig. 7B,C). Furthermore, these knockout parasites exhibit shorter and wider cell shapes, as indicated by a shift towards lower elongatedness values (Fig. 7D). Taken together with previous studies of CBAP/SIP/TSC1 mutants, these findings support a role for IMC sutures components in the establishment and/or maintenance of parasite shape.
Figure 7. Analysis of extracellular Δisc3 parasite morphology.
A) Measurements of the length and width of extracellular ISC3-HA, Δisc3, or ISC3c parasites. Representative images of each parasite line are shown. Loss of ISC3 results in shorter and wider parasites, a phenotype that is rescued by complementation. Parasite lengths and widths are depicted as box-whisker plots, with the middle lines corresponding to the median, the bottom and top boxes representing the 25th and 75th percentiles, respectively, and whiskers corresponding to the smallest and largest measurements. Data were analyzed by one-way ANOVA followed by Bonferroni post hoc tests. *p<0.001 B-D) ImageStream flow cytometry analysis of extracellular ISC3-HA, Δisc3, or ISC3c parasite morphology using aspect ratio (B), circularity (C), and elongatedness (D) parameters. Aspect ratio measures the ratio of the transverse to longitudinal axis of a cell and more circular cells will have an aspect ratio value closer to 1. Circularity measures the average distance of the cell boundary from its center, divided by the variation of this distance; more circular cells have a lower variation and thus a higher circularity value. Elongatedness measures the ratio of height to width of a cell’s bounding box; long and narrow cells have a higher elongatedness value. Δisc3 parasites (red) display a shift towards higher values of aspect ratio (B) and circularity (C) and lower values of elongatedness (D) compared to parental ISC3-HA parasites (green). These shifts are completely reversed in the ISC3c strain (purple). At least 10,000 images were analyzed for each parasite line.
The ISC3 paralog TgGT1_212990 is dispensable for parasite growth in vitro
To determine whether other predicted CTL proteins could have functional roles at the IMC and/or sutures, we determined the localization of the ISC3 paralog, TgGT1_212990, using endogenous gene tagging. We localized this protein to a compartment that mostly overlaps with the VAC/PLV, an acidified vacuole that is part of the parasite’s endolysosomal system (Fig. S1) (Miranda et al., 2010, Parussini et al., 2010). This localization suggests TgGT1_212990 likely has IMC-independent functions. We then deleted TgGT1_212990 using CRISPR/Cas9 and homology-dependent repair (HDR) by replacing the entire gene and the downstream HXGPRT marker (used for endogenous gene tagging) with a DHFR cassette (Fig. S1A). Knockout parasites that had undergone the desired recombination event were readily isolated by selection as shown by PCR, IFA, and Western blot analysis (Fig. S1A–C) and had no detectable fitness defects in vitro as measured by plaque assays (Fig. S1D). We also examined compensation in Δisc3 parasites (by TgGT1_212990) and Δtggt1_212990 parasites (by ISC3) and observed no gross changes in TgGT1_212990 and ISC3 localization, respectively (Fig. S1E,F), demonstrating that these CTL proteins likely do not compensate for each other in these mutants. Collectively, these data demonstrate that TgGT1_212990 is dispensable for parasite growth in culture.
TgGT1_249640 is a predicted phosphaditic acid phosphatase (PAP) that localizes to the IMC
As ISC3 may play a role in PtdCho transport across the IMC membranes, we also assessed whether other enzymes in the Kennedy pathway of PtdCho biosynthesis are present in the IMC. We identified a putative PAP encoded by TgGT1_249640 with a cyclical expression pattern similar to IMC proteins (Behnke et al., 2010). Using endogenous gene tagging, we localized TgGT1_249640 to the apical and central subcompartments of the IMC but not the base (Fig. 8). TgGT1_249640 contains a PAP2 and four predicted transmembrane domains, which may anchor the protein into the IMC membranes. In the Kennedy pathway, PAP enzymes convert phosphaditic acid (PA) to diacylglycerol, which upon the addition of a CDP-choline headgroup forms PtdCho (Carman et al., 2006). However, it is unclear whether TgGT1_249640 functions in this capacity at the IMC or is involved in the PA signaling pathways at the parasite periphery that regulate microneme exocytosis (Bullen et al., 2016). We were unable to successfully isolate TgGT1_249640-null clones in repeated gene knockout experiments with CRISPR/Cas9, suggesting this protein is essential (data not shown).
Figure 8. A putative PAP localizes to the parasite IMC.
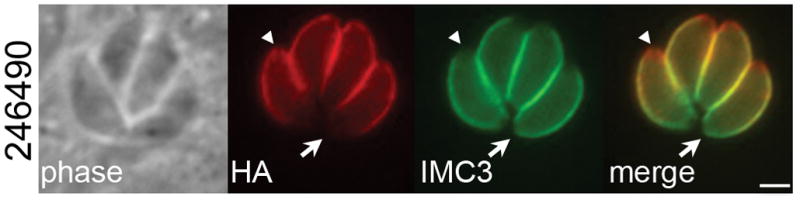
IFA showing localization of TgGT1_246490, which contains a predicted PAP2 domain, to the apical and central but not basal subcompartments of the IMC. Note the lack of co-localization between TgGT1_246490 and IMC3 (present in the center and the base) at the apical cap (arrowheads) and base (arrows). Red: mouse anti-HA antibody. Green: rat anti-IMC3 antibody. Scale bar = 2 μm.
Disruption of ISC3 results in a complete loss of virulence in vivo
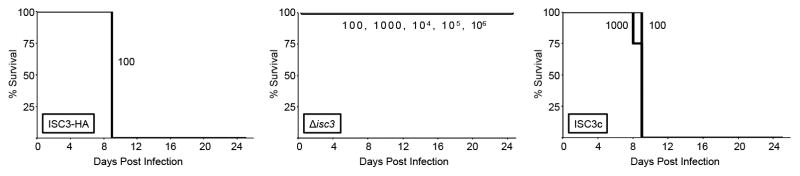
To determine if the reduced fitness of Δisc3 parasites in vitro translates to defects in vivo, we examined the virulence of these parasites in a mouse model. Since we disrupted ISC3 in the hypervirulent type I strain (LD100= ~1), C57BL/6 mice were injected intraperitoneally with a low dose of 100 ISC3-HA, Δisc3, or ISC3c parasites. While all mice infected with the wild-type strain succumbed to infection by day 9, all mice subjected to this dose of Δisc3 parasites survived and failed to display visible signs of infection (Fig. 9). We then infected mice with increasing doses of Δisc3 parasites varying over several orders of magnitude. To our surprise, we observed a 100% survival rate in mice at all doses tested, even up to 106 parasites per mouse, demonstrating that Δisc3 parasites are essentially avirulent (Fig. 9). Importantly, this loss of virulence was completely reversed in the ISC3c strain, demonstrating this phenotype is specifically due to the loss of ISC3. We also examined whether infection with Δisc3 parasites was protective against a subsequent T. gondii challenge. We found that all seropositive mice injected with the lowest dose (100) of knockout parasites were resistant to a lethal challenge of 104 RHΔhptΔku80 (wild-type) tachyzoites (not shown). Finally, we also determined whether Δisc3 parasites could be recovered from brain homogenates (at 30 days post-infection) of mice injected with doses of 1000, 104, 105, or 106 parasites. We were unable to recover parasites after culturing these homogenates on HFF monolayers, suggesting that the knockout parasites were unable to survive in these hosts. These in vivo experiments demonstrate that ISC3 is required for T. gondii to establish an infection and cause disease in mice.
Figure 9. Disruption of ISC3 results in a complete loss of parasite virulence in vivo.
Kaplan-Meier survival curves for C57BL/6 mice infected with 100 ISC3-HA (left), 100, 1000, 104, 105, or 106 Δisc3 (middle), or 100 or 1000 ISC3c parasites (right panel). Each group of 4 mice was injected intraperitoneally and monitored for 25 days. The viability of injected parasites was monitored in parallel using plaque assays (~15–20% plaquing efficiency across all strains/doses, not shown).
Discussion
As new components of the IMC are discovered and characterized, it is becoming clear that the structure is composed of a number of distinct subcompartments, each containing a unique array of protein constituents that likely play specialized roles in IMC organization and function. These subcompartments include the membranous alveolar sacs and the underlying cytoskeletal meshwork, which are further divided into the apical cap, the central and basal rows of alveolar sacs, and the basal complex (Porchet et al., 1977, Mann et al., 2001, Gubbels et al., 2006, Beck et al., 2010). The IMC sutures delineate these regions and subdivide the organelle into its discrete subcompartments (Porchet et al., 1977, Chen et al., 2015). In this report, we have expanded the repertoire of known proteins in the IMC sutures and show that they segregate into ISCs that reside in the transverse and longitudinal sutures and TSCs that localize exclusively to the transverse sutures. From this study and previous reports, a total of six ISCs and six TSCs have been identified, which provides a framework for investigating the functional contribution of the IMC sutures to parasite biology (Chen et al., 2015). In addition, we provide the first functional assessment of an ISC, ISC3, and demonstrate the importance of the IMC sutures in parasite growth in vitro and virulence in vivo. Finally, in addition to novel ISCs and TSCs, we also identified eight new proteins in the IMC or associated subpellicular microtubules, further broadening the known IMC proteome.
We have previously shown the utility of BioID for identifying IMC proteins that are present in the alveolar sacs or the underlying cytoskeleton (Chen et al., 2015). To reduce the amount of background proteins contaminating our ISC4-BioID experiments, we implemented a fractionation step prior to the purification of biotinylated proteins. Using detergent extractions of the IMC cytoskeleton, we were able to release the background of endogenously biotinylated proteins, present in the apicoplast and mitochondria, into the detergent soluble fraction. Although our enrichment for cytoskeletal proteins using this approach potentially came at the expense of membrane IMC sutures components, our ISC4-BioID data set included detergent-soluble proteins such as ISC3, ISC6, and TSCs 5/6 (albeit in some cases as lower-ranked hits in the dataset), indicating we were still able to retain at least a subset of these proteins in our fractionated samples. This is consistent with our detergent extraction analyses of ISC6 and TSCs 5/6, which demonstrate that a small fraction of these membrane-associated components remains in the insoluble pellet after extraction (Fig. 3). However, it is formally possible that less abundant IMC membrane proteins may have been lost during the fractionation step and thus were excluded from our ISC4-BioID data set. Importantly, the top three hits from this experiment were ISCs 1/2/4, which is consistent with these proteins being interactors or in close proximity to each other (Table S1).
The identification of a larger number of ISCs and TSCs enables us to examine these proteins for potential common features (e.g. trafficking determinants, functional domains) and assess their conservation throughout the Apicomplexa. Preliminary examination of the amino acid sequences of the six identified ISCs or TSCs did not reveal any obvious conserved motifs or sequences that could potentially target these proteins to the transverse and longitudinal sutures, or exclusively to the transverse sutures in the case of TSCs. In addition, the new ISCs and TSCs reported here lack conserved domains or features that could suggest common structural or functional roles at the IMC sutures, although ISC6 and TSCs 5/6 contain predicted transmembrane domains at their C-termini that presumably anchor these proteins into the IMC membranes (Fig. 3). Thus, more systematic mutagenesis or deletion studies of the ISCs and TSCs are required to elucidate trafficking determinants and structural and functional features of these proteins.
Interestingly, there appears to be limited conservation of T. gondii ISCs and TSCs in more distantly related apicomplexans such as P. falciparum. Of the ISCs, only ISC1 (PF3D7_1341500) and ISC3 (PF3D7_1431900) have homologs in P. falciparum by BLAST analyses (data not shown). In contrast, homologs to most of the ISCs (except for ISC4) are present in the more closely related parasite Eimeria tenella, which belongs to the same subgroup (Coccidia) as T. gondii. The conservation of TSCs shares a similar pattern as only TSC1/CBAP/SIP and TSC2 appear to be conserved in P. falciparum but coccidians such as E. tenella and Sarcocystis neurona retain homologs to most of the TSCs. Conversely, the only previously identified IMC sutures protein in P. falciparum, MAL13P1.228, appears specific to the genus (Kono et al., 2012). Thus, at least a subset of ISCs and TSCs appear to be subgroup-specific and may have specialized functions in the IMC.
The difference in protein composition of the IMC sutures between T. gondii and P. falciparum is consistent with differences in the arrangement of the IMC sutures in these two apicomplexans. The T. gondii IMC in tachyzoites is composed of several rows of rectangular plates that are organized into a quilt-like arrangement, with multiple transverse and longitudinal sutures forming the junctions between these plates (Porchet et al., 1977, Chen et al., 2015). This arrangement is presumably conserved in bradyzoites, which retain a similar IMC architecture as observed by time-lapse microscopy of differentiating parasites (Dzierszinski et al., 2004). While the sutures are thought to be absent in most stages of P. falciparum in which the IMC consists of a single vesicle, the gametocyte IMC consists of up to thirteen ring-shaped plates that encircle and form segments along the parasite (Meszoely et al., 1987, Bannister et al., 1995, Kono et al., 2012). These segments are delineated by transverse sutures, which have been observed by ultrastructural analyses and IFA by staining for the IMC sutures protein MAL13P1.228 (Meszoely et al., 1987, Kono et al., 2012). In addition, only one longitudinal suture-like structure has been observed (by staining for MAL13P1.228), further highlighting the different arrangements of IMC sutures between these two distantly related parasites (Kono et al., 2012).
Our functional analysis of ISC3 provides the first evidence demonstrating the importance of ISCs to parasite biology. Parasites lacking ISC3 have a reduced fitness in vitro and display an array of morphological defects within host cells and in the extracellular environment. It is unclear whether these defects are due to the loss of ISC3 as a choline transporter or as a structural component of the IMC sutures, or a combination of both factors. It is possible that a decrease in ISC3-mediated choline import in these mutants results in an altered IMC membrane composition and integrity, leading to the aberrant morphology of Δisc3 parasites. Consistent with this hypothesis, we also localized a putative PAP, TgGT1_249640, to the IMC (Fig. 8). PAP proteins catalyze the conversion of PA to DAG, which can be combined with CDP-choline to form PtdCho as part of the Kennedy pathway. However, PAP enzymes also provide DAG for cell signaling pathways and in T. gondii, the interconversion of PA/DAG at the parasite periphery by unidentified PAP(s) regulates efficient microneme secretion (Bullen et al., 2016). Initial experiments to generate knockout parasites lacking TgGT1_249640 using CRISPR/Cas9 were unsuccessful, suggesting that this PAP is essential for parasite survival.
Alternatively, ISC3 may instead serve a strictly structural role at the IMC sutures, a function that has been described for the transverse sutures component CBAP/SIP/TSC1 (Lentini et al., 2014, Tilley et al., 2014). Mutants lacking CBAP/SIP/TSC1 have a shorter, “stumpy” morphology compared to wild-type parasites and are impaired in gliding motility, host cell invasion and virulence in mice. While disruption of ISC3 also results in changes in parasite shape, the IMC defects observed in Δisc3 parasites appear to be limited to replication as these mutants have similar invasion capacities compared wild-type parasites. This lack of an effect on invasion was somewhat surprising given the morphological defects observed in Δisc3 parasites, however it is possible that the more aberrant or misshapen parasites fail to attach to the host cells and thus are removed by washing during our red-green invasion assays. In addition, our observation that only a fraction of ISC3 protein (compared to wild-type expression levels) is sufficient to rescue the fitness defects of Δisc3 parasites may also suggest an enzymatic rather than structural function for ISC3. A dissection of the molecular underpinnings of the Δisc3 mutant phenotypes described here will be the subject of future studies. Genetic studies of additional ISCs and TSCs will help elucidate the components that are critical for maintaining parasite morphology and/or structural integrity of the IMC membrane sacs.
Surprisingly, Δisc3 parasites are completely avirulent in vivo, as mice were completely resistant to dosages of up to 106 knockout parasites, whereas the type I parental strain has an LD100=1 in mice. This is in contrast to various other type I knockouts which have fitness defects and produce significantly smaller plaques in vitro yet are still able to kill mice with low doses of parasites, albeit sometimes with a delay in the time of death (Blume et al., 2009, Shen et al., 2014, Hammoudi et al., 2015, El Bissati et al., 2016, Pszenny et al., 2016). This discrepancy between the in vitro and in vivo phenotypes of Δisc3 parasites may reflect a reduced capacity of these knockouts to establish a productive infection upon intraperitoneal injection into mice. Our inability to recover Δisc3 parasites from mouse brain homogenates after 30 days of infection supports this hypothesis and may suggest the utility of Δisc3 parasites as a vaccine strain (although the ability to maintain a chronic infection would best be evaluated with ISC3 knockouts generated in a cystogenic type II strain). This avirulent phenotype is most likely the result of the replication defects observed in vitro, but we cannot exclude the possibility that extracellular survival, invasion, or egress may also be affected in these cell types. The dramatic loss of virulence in vivo observed in Δisc3 parasites points to this IMC protein as a new potential therapeutic target for T. gondii and related pathogens.
Experimental Procedures
Toxoplasma and host cell culture
T. gondii RHΔku80Δhpt and modified strains were grown on confluent monolayers of human foreskin fibroblast (HFF) host cells in DMEM supplemented with 10% fetal bovine serum, as previously described (Donald et al., 1996).
Antibodies
The following previously described primary antibodies were used in immunofluorescence (IFA) or Western blot assays: anti-IMC1 (mAb 45.15) (Wichroski et al., 2002), anti-ISP1 (mAb 7E8) (Beck et al., 2010), anti-F1 ATP synthase β subunit (mAb 5F4) (Jacot et al., 2013), mouse anti-ISP3 (Beck et al., 2010), rabbit anti-CPL (Larson et al., 2009), rabbit anti-MORN1 (Gubbels et al., 2006) and rat anti-IMC3 (Anderson-White et al., 2011). The hemagglutinin (HA) epitope was detected with mouse anti-HA (mAb HA.11) (Covance) or rabbit anti-HA (Invitrogen). Rabbit RON5c antibody was generated using recombinant 6xHis-tagged RON5c protein that was purified as previously described (Straub et al., 2009) for polyclonal antibody production (Cocalico Biologicals). To generate rat anti-ISC1, the full coding region of ISC1 was PCR-amplified (Table S2) and cloned into the pET28a vector between EcoRI and SalI sites. The resulting 6xHis-tagged ISC1 was overexpressed and purified as previously described (Straub et al., 2009) for polyclonal antibody production (Cocalico Biologicals).
Immunofluorescence assays (IFA) and Western blots
For IFA, HFFs were grown to confluency on coverslips and infected with T. gondii parasites. After 18–36 hours, the coverslips were fixed and processed for indirect immunofluorescence as previously described (Bradley et al., 2005). Primary antibodies were detected by species-specific secondary antibodies conjugated to Alexa Fluor 594/488 dyes. The coverslips were mounted in Vectashield (Vector Labs) and viewed with an Axio Imager.Z1 fluorescent microscope (Zeiss) as previously described (Beck et al., 2010).
For Western blot, parasites were lysed in Laemmli sample buffer (50 mM Tris-HCl [pH 6.8], 10% glycerol, 2% SDS, 1% 2-mercaptoethanol, 0.1% bromophenol blue) and lysates were resolved by SDS-PAGE and transferred onto nitrocellulose membranes. Blots were probed with the indicated primary antibodies, followed by secondary antibodies conjugated to horse radish peroxidase (HRP). Target proteins were visualized by chemiluminescence.
Generation of ISC4-BirA* parasites and epitope tagging of BioID hits
To generate parasites expressing the ISC4-BirA* fusion protein, the 3’ region of ISC4 was PCR-amplified (using the designated primers in Table S2) and inserted into pBirA*-3×HA-LIC-DHFR using a ligation-independent cloning approach (Chen et al., 2015). 100 μg of the construct was linearized and transfected into RHΔku80Δhpt parasites. Transgenic parasites were selected in 1 μM pyrimethamine and cloned by limiting dilution. Clones that had undergone the intended recombination event were screened by IFA and Western blot against the 3×HA tag. A clone expressing the correct fusion protein was selected and designated ISC4-BirA*.
For endogenous tagging of genes from the ISC4-BioID data set, we used the plasmids p3×HA-LIC-DHFR, p3×HA-LIC-HPT, or p3×HA-LIC-CAT (Fung et al., 2012, Beck et al., 2014). A 3′ portion of each gene was PCR-amplified (Table S2) and inserted into the respective plasmid to generate a 3×HA epitope tag fusion prior to the stop codon of each gene. 100 μg of each construct was linearized and transfected into RHΔku80Δhpt parasites and HA-positive clones were isolated as described above following selection with 1 μM pyrimethamine, 50 μg/ml mycophenolic acid/xanthine, or 1 μM chloramphenicol.
Detergent extraction assays
Extracellular parasites were washed in PBS, pelleted, and lysed in 1 mL of 1% Triton X-100 lysis buffer (50mM Tris-HCl [pH 7.4], 150mM NaCl) supplemented with Complete Protease Inhibitor Cocktail (Roche) for 30 min on ice. Lysates were centrifuged for 15 min at 14,000 × g. Equivalent fractions of the total lysate, supernatant, and pellet were separated by SDS-PAGE and analyzed by Western blot.
Detergent fractionation and affinity capture of biotinylated proteins
HFF monolayers infected with parasites expressing the ISC4-BirA* fusion or the parental (RHΔhptΔku80) line were grown in media containing 150 μM biotin for 24 hours prior to parasite egress. Extracellular parasites were collected, washed in PBS, and lysed in 1% Triton X-100 lysis buffer supplemented with Complete Protease Inhibitor Cocktail (Roche) for 30 minutes on ice. Lysates were centrifuged for 15 min at 14,000 × g to pellet the insoluble IMC cytoskeletal fraction (Mann et al., 2001). The insoluble pellet was then solubilized using 1% SDS buffer (50 mM Tris [pH 7.5], 150 mM NaCl) and sonication, diluted to a final concentration of 0.1% SDS, and incubated with Streptavidin Plus UltraLink resin (Pierce) at room temperature for 4 hours under gentle agitation. Beads were collected by centrifugation and washed five times in 0.1% SDS buffer (50mM Tris-HCl [pH 7.4], 150mM NaCl), followed by three washes in 8M urea buffer (50mM Tris-HCl [pH 7.4], 150mM NaCl). 10% of each sample was boiled in Laemmli sample buffer and eluted proteins were analyzed by Western blot by streptavidin-HRP prior to mass spectrometry.
Mass spectrometry of biotinylated proteins
Purified proteins bound to streptavidin beads were reduced, alkylated and digested by sequential addition of lys-C and trypsin proteases (Kaiser et al., 2005, Wohlschlegel, 2009). The peptide mixture was desalted using C18 tips and fractionated online using a 75 μM inner diameter fritted fused silica capillary column with a 5 μM pulled electrospray tip and packed in-house with 15 cm of Luna C18(2) 3 μM reversed phase particles. The gradient was delivered by an easy-nLC 1000 ultra high-pressure liquid chromatography (UHPLC) system (Thermo Scientific). MS/MS spectra was collected on a Q-Exactive mass spectrometer (Thermo Scientific) (Michalski et al., 2011, Kelstrup et al., 2012). Data analysis was performed using the ProLuCID and DTASelect2 implemented in the Integrated Proteomics Pipeline - IP2 (Integrated Proteomics Applications, Inc., San Diego, CA) (Tabb et al., 2002, Xu et al., 2006, Cociorva et al., 2007). Protein and peptide identifications were filtered using DTASelect and required minimum of two unique peptides per protein and a peptide-level false positive rate of less than 5% as estimated by a decoy database strategy (Elias et al., 2007). Normalized spectral abundance factor (NSAF) values were calculated as described (Florens et al., 2006).
Generation of Δisc3 and Δtggt1_212990 parasites using CRISPR/Cas9
CRISPR/Cas9-mediated gene disruption of ISC3 or TgGT1_212990 was performed using the pU6-Universal system as previously described (Sidik et al., 2014). 20 bp protospacer sequences targeting the coding region of ISC3 or TgGT1_212990 (see Table S2) were designed using Protospacer Workbench software (MacPherson et al., 2015). Selected 20 bp protospacer oligos (Table S2) were annealed and ligated into the BsaI-digested pU6-Universal plasmid to generate pU6-ISC3 or pU6-212990.
ISC3 was disrupted using a double-stranded 79 bp oligo (Table S2) that serves as a template for HDR at the Cas9-induced double-stranded break and harbors a premature stop codon along with a mutation in the protospacer adjacent motif (PAM) to prevent further Cas9 targeting of the locus after recombination. ISC3-HA parasites co-transfected with the 79 bp oligos and pU6-ISC3 were cloned immediately by limiting dilution (Chen et al., 2015). Clones that had undergone the desired gene editing event were screened by IFA and Western blot for loss of the 3×HA tag. A positive clone was designated Δisc3.
To delete the entire locus of TgGT1_212990 and the downstream HXGPRT drug marker used for endogenous gene tagging, we PCR-amplified a DHFR cassette (Table S2) using a 5′ primer containing the 40 bp sequence immediately upstream of the TgGT1_212990 start codon and a 3′ primer in the 3′ UTR of the DHFR cassette (which is identical to that of the HXGPRT marker, thus the entire UTR serves as the 3′ homology arm during HDR). TgGT1_212990-HA parasites were transfected with this modified DHFR cassette and pU6-212990. Following selection with 1 μM pyrimethamine, transgenic parasites that had undergone the intended recombination event were screened by IFA and Western blot for loss of the 3xHA tag. A positive clone was designated Δtggt1_212990. Deletion of the TgGT1_212990 locus and recombination of the DHFR cassette in this clone was verified by PCR (see Table S2 and Fig. S1A). Loss of the HXGPRT marker was confirmed by sensitivity to 50 μg/ml mycophenolic acid/xanthine.
Complementation of Δisc3 parasites with ISC3-HA
The entire coding region of ISC3 was PCR-amplified (see Table S2) and cloned into the pUPRTKO-RON5-HA plasmid using NotI and blunted BglII sites to generate pUPRTKO-ISC3-HA (Beck et al., 2014). The plasmid was linearized and transfected into Δisc3 parasites, followed by selection with 5 μg/ml 5-fluorodeoxyuridine to facilitate targeted replacement of the UPRT locus as previously described (Donald et al., 1995). ISC3-HA expressing clones were screened by IFA and Western blot and a HA-positive clone was designated ISC3c.
Plaque assays
Freshly lysed, extracellular ISC3-HA, Δisc3, and ISC3c parasites were allowed to infect confluent HFF monolayers and form plaques for 9 days. Cells were fixed with ice-cold methanol and stained with 0.4% Crystal Violet. 30 plaques from each line were visualized with an Axio Imager.Z1 microscope (Zeiss) and plaque areas were measured using ZEN imaging software (Zeiss) to generate plaque size means and interquartile ranges.
Analysis of extracellular parasite morphology
Freshly lysed, extracellular ISC3-HA, Δisc3, and ISC3c parasites were washed in PBS, fixed in solution with 4% formaldehyde, and allowed to settle and attach to glass coverslips. Coverslips were washed with three exchanges of PBS to remove unattached parasites and mounted in ProLong Gold (Molecular Probes). A total of 50 parasites from each line were visualized with an Axio Imager.Z1 microscope (Zeiss) and parasite lengths and widths were measured using ZEN imaging software (Zeiss) to generate means and interquartile ranges.
For ImageStream flow cytometry analysis, freshly lysed, extracellular parasites were washed in PBS, fixed in solution with 4% formaldehyde, and processed for indirect immunofluorescence with anti-IMC1 and Alexa Fluor 488-conjugated secondary antibodies as previously described (Bradley et al., 2005). At least 50,000 images per parasite line were acquired on an ImageStreamX Mark II imaging flow cytometer (Amnis) using 60× magnification and a core width of 6 μM. IMC1 staining was detected in channel 2 (with the 488 nm laser set to 100 mW) and bright-field images were collected in channel 5. To select parasites were image analyses, we gated for cells in focus (on bright-field), followed by single cells, and finally IMC1-positive cells. Analysis of parasite morphological features (aspect ratio, circularity, and elongatedness) were performed on IDEAS software using bright-field images and the erode mask function. At least 10,000 images per parasite strain were analyzed.
Mouse virulence assays
Intracellular ISC3-HA, Δisc3, and ISC3c parasites were mechanically liberated (via syringe lysis) from infected HFF monolayers and resuspended in Opti-MEM medium (Thermo Fisher Scientific) prior to intraperitoneal injection into female C57BL/6 mice at the following dosages: 100 ISC3-HA parasites; 100, 1000, 104, 105, or 106 Δisc3 parasites; or 100 or 1000 ISC3c parasites (4 mice per dose). In parallel, the viability of resuspended parasites for injection was determined by plaque assays (~15–20% plaquing efficiency across all strains/doses, data not shown). Mice were monitored for symptoms of infection and weight loss for 25 days and survival for 30 days. Surviving mice were bled and seroconversion was assessed via Western blot with RHΔhptΔku80 (wild-type) lysates. Mice previously injected with a dose of 100 Δisc3 parasites were challenged with intraperitoneal injection of 104 RHΔhptΔku80 tachyzoites and monitored for an additional 30 days. Mice previously injected with a dosage of 1000, 104, 105, or 106 Δisc3 parasites were sacrificed and brains were harvested and homogenized in PBS. Brain homogenates were cultured on HFF monolayers and monitored for the presence of parasites for at least an additional 30 days.
Statistical Analysis
All statistical analyses were performed using SPSS statistics software (IBM). Data were analyzed by one-way ANOVA followed by Bonferroni post hoc tests unless otherwise indicated. Differences were considered significant at p<0.001 and denoted with an asterisk.
Supplementary Material
Figure S1. The ISC3 paralog TgGT1_212990 is dispensable for parasite growth. A) Strategy for deleting the entire TgGT1_212990 locus and the downstream HXGPRT marker used for endogenous gene tagging (left). The DHFR cassette used for recombination at the CRISPR/Cas9-induced double-stranded break has a 5’ homology arm containing the 40 bp sequence (in red) immediately upstream of the TgGT1_212990 start codon. The 3′ UTRs of the DHFR and HXGPRT markers (green) are identical so the entire sequence functions as the 3′ homology arm during homology-dependent repair. Using this approach, only the 5′ primer is gene-specific for the PCR amplification of a drug cassette targeting an endogenously tagged gene of interest. Loss of the TgGT1_212990 locus and recombination of the DHFR cassette was verified by PCR using parental TgGT1_212990-HA parasites as a control (right). B,C) IFA (B) and Western blot (C) showing loss of the HA-tagged TgGT1_212990 protein (co-localized with the VAC marker CPL). Red: mouse anti-HA antibody. Green: rabbit anti-CPL antibody. Scale bar = 2 μm. D) Quantification of plaques produced by TgGT1_212990-HA and Δtggt1_212990 parasites after 9 days of growth. Plaque areas are depicted as a box-whisker plot, with the middle line corresponding to the median, the bottom and top boxes representing the 25th and 75th percentiles, respectively, and whiskers corresponding to the smallest and largest plaques. Data were analyzed with an independent samples test and no significant differences were detected. E,F) IFA showing no gross changes in TgGT1_212990 localization in Δisc3 parasites (E) and ISC3 localization in Δtggt1_212990 parasites (F). Red: mouse anti-HA antibody. Green: rabbit anti-CPL or rat anti-ISC1 antibody.
Table S1. List of top ISC4-BioID hits identified by mass spectrometry. Proteins present in both experimental samples but not the controls are shown. Hypothetical proteins localized by endogenous gene tagging in this study are highlighted in blue. Data is from two experimental replicates. Gene IDs and corresponding descriptions are from ToxoDB.
Table S2. Oligonucleotide primers used in this study. All primer sequences are shown in the 5′ to 3′ orientation.
Acknowledgments
We thank the following for generously providing reagents: Marc-Jan Gubbels (Boston College) for anti-IMC3 and anti-MORN1 antibodies, Vern Carruthers (Michigan University) for anti-CPL antibodies, Sebastian Lourido (Whitehead Institute, Massachusetts Institute of Technology) for the pU6-Universal plasmid, and Ke Hu (Indiana University) for the pTub-mEmeraldFP-TrxL2 plasmid. This work was supported by NIH grants #AI064616 to P.J.B. and #GM089778 to J.A.W. and Ruth L. Kirschstein National Research Service Awards #AI007323 to A.L.C. and #GM007185 to C.P.C. A.H.L. was supported by the Arnold and Mabel Beckman Scholars Program and UCLA Undergraduate Research Fellowship Program; A.S.H. was supported by the UCLA Undergraduate Research Scholars Program; E.W.K. was supported by the Philip Whitcome Pre-doctoral Fellowship in Molecular Biology; and H.N.B. was supported by the Amgen Scholars Program, UCLA Undergraduate Research Fellowship Program, and the Howard Hughes Undergraduate Research Fellowship. ImageStream flow cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility that is supported by NIH awards P30 #CA016042 and 5P30 #AI028697, and by the JCCC, the UCLA AIDS Institute, the David Geffen School of Medicine at UCLA, the UCLA Chancellor’s Office, and the UCLA Vice Chancellor’s Office of Research.
References
- Anderson-White BR, Ivey FD, Cheng K, Szatanek T, Lorestani A, Beckers CJ, et al. A family of intermediate filament-like proteins is sequentially assembled into the cytoskeleton of Toxoplasma gondii. Cell Microbiol. 2011;13:18–31. doi: 10.1111/j.1462-5822.2010.01514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister LH, Mitchell GH. The role of the cytoskeleton in Plasmodium falciparum merozoite biology: an electron-microscopic view. Ann Trop Med Parasitol. 1995;89:105–111. doi: 10.1080/00034983.1995.11812940. [DOI] [PubMed] [Google Scholar]
- Beck JR, Chen AL, Kim EW, Bradley PJ. RON5 is critical for organization and function of the Toxoplasma moving junction complex. PLoS Pathog. 2014;10:e1004025. doi: 10.1371/journal.ppat.1004025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck JR, Rodriguez-Fernandez IA, Cruz de Leon J, Huynh MH, Carruthers VB, Morrissette NS, Bradley PJ. A novel family of Toxoplasma IMC proteins displays a hierarchical organization and functions in coordinating parasite division. PLoS Pathog. 2010:6. doi: 10.1371/journal.ppat.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke MS, Wootton JC, Lehmann MM, Radke JB, Lucas O, Nawas J, et al. Coordinated progression through two subtranscriptomes underlies the tachyzoite cycle of Toxoplasma gondii. PLoS One. 2010;5:e12354. doi: 10.1371/journal.pone.0012354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume M, Rodriguez-Contreras D, Landfear S, Fleige T, Soldati-Favre D, Lucius R, Gupta N. Host-derived glucose and its transporter in the obligate intracellular pathogen Toxoplasma gondii are dispensable by glutaminolysis. Proc Natl Acad Sci U S A. 2009;106:12998–13003. doi: 10.1073/pnas.0903831106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley PJ, Ward C, Cheng SJ, Alexander DL, Coller S, Coombs GH, et al. Proteomic analysis of rhoptry organelles reveals many novel constituents for host-parasite interactions in Toxoplasma gondii. J Biol Chem. 2005;280:34245–34258. doi: 10.1074/jbc.M504158200. [DOI] [PubMed] [Google Scholar]
- Bullen HE, Jia Y, Yamaryo-Botte Y, Bisio H, Zhang O, Jemelin NK, et al. Phosphatidic Acid-Mediated Signaling Regulates Microneme Secretion in Toxoplasma. Cell Host Microbe. 2016;19:349–360. doi: 10.1016/j.chom.2016.02.006. [DOI] [PubMed] [Google Scholar]
- Carman GM, Han GS. Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem Sci. 2006;31:694–699. doi: 10.1016/j.tibs.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers V, Boothroyd JC. Pulling together: an integrated model of Toxoplasma cell invasion. Curr Opin Microbiol. 2007;10:83–89. doi: 10.1016/j.mib.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Charron AJ, Sibley LD. Host cells: mobilizable lipid resources for the intracellular parasite Toxoplasma gondii. J Cell Sci. 2002;115:3049–3059. doi: 10.1242/jcs.115.15.3049. [DOI] [PubMed] [Google Scholar]
- Chen AL, Kim EW, Toh JY, Vashisht AA, Rashoff AQ, Van C, et al. Novel components of the Toxoplasma inner membrane complex revealed by BioID. MBio. 2015;6:e02357–02314. doi: 10.1128/mBio.02357-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cociorva D, DLT, Yates JR. Validation of tandem mass spectrometry database search results using DTASelect. Current protocols in bioinformatics / editoral board, Andreas D. Baxevanis ... [et al.] 2007;Chapter 13(Unit 13):14. doi: 10.1002/0471250953.bi1304s16. [DOI] [PubMed] [Google Scholar]
- D'Haese J, Mehlhorn H, Peters W. Comparative electron microscope study of pellicular structures in coccidia (Sarcocystis, Besnoitia and Eimeria) Int J Parasitol. 1977;7:505–518. doi: 10.1016/0020-7519(77)90014-5. [DOI] [PubMed] [Google Scholar]
- Donald RG, Carter D, Ullman B, Roos DS. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J Biol Chem. 1996;271:14010–14019. doi: 10.1074/jbc.271.24.14010. [DOI] [PubMed] [Google Scholar]
- Donald RG, Roos DS. Insertional mutagenesis and marker rescue in a protozoan parasite: cloning of the uracil phosphoribosyltransferase locus from Toxoplasma gondii. Proc Natl Acad Sci U S A. 1995;92:5749–5753. doi: 10.1073/pnas.92.12.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierszinski F, Nishi M, Ouko L, Roos DS. Dynamics of Toxoplasma gondii differentiation. Eukaryot Cell. 2004;3:992–1003. doi: 10.1128/EC.3.4.992-1003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Bissati K, Suvorova ES, Xiao H, Lucas O, Upadhya R, Ma Y, et al. Toxoplasma gondii Arginine Methyltransferase 1 (PRMT1) Is Necessary for Centrosome Dynamics during Tachyzoite Cell Division. MBio. 2016;7:e02094–02015. doi: 10.1128/mBio.02094-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, Workman JL, Washburn MP. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40:303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia ME, Striepen B. Cell division in apicomplexan parasites. Nat Rev Microbiol. 2014;12:125–136. doi: 10.1038/nrmicro3184. [DOI] [PubMed] [Google Scholar]
- Fung C, Beck JR, Robertson SD, Gubbels MJ, Bradley PJ. Toxoplasma ISP4 is a central IMC sub-compartment protein whose localization depends on palmitoylation but not myristoylation. Mol Biochem Parasitol. 2012;184:99–108. doi: 10.1016/j.molbiopara.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbels MJ, Vaishnava S, Boot N, Dubremetz JF, Striepen B. A MORN-repeat protein is a dynamic component of the Toxoplasma gondii cell division apparatus. J Cell Sci. 2006;119:2236–2245. doi: 10.1242/jcs.02949. [DOI] [PubMed] [Google Scholar]
- Hammoudi PM, Jacot D, Mueller C, Di Cristina M, Dogga SK, Marq JB, et al. Fundamental Roles of the Golgi-Associated Toxoplasma Aspartyl Protease, ASP5, at the Host-Parasite Interface. PLoS Pathog. 2015;11:e1005211. doi: 10.1371/journal.ppat.1005211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding CR, Meissner M. The inner membrane complex through development of Toxoplasma gondii and Plasmodium. Cell Microbiol. 2014;16:632–641. doi: 10.1111/cmi.12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacot D, Daher W, Soldati-Favre D. Toxoplasma gondii myosin F, an essential motor for centrosomes positioning and apicoplast inheritance. EMBO J. 2013;32:1702–1716. doi: 10.1038/emboj.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenska J, Crawford MJ, Harb OS, Zuther E, Haselkorn R, Roos DS, Gornicki P. Subcellular localization of acetyl-CoA carboxylase in the apicomplexan parasite Toxoplasma gondii. Proc Natl Acad Sci U S A. 2001;98:2723–2728. doi: 10.1073/pnas.051629998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser P, Wohlschlegel J. Identification of ubiquitination sites and determination of ubiquitin-chain architectures by mass spectrometry. Methods in enzymology. 2005;399:266–277. doi: 10.1016/S0076-6879(05)99018-6. [DOI] [PubMed] [Google Scholar]
- Kelstrup CD, Young C, Lavallee R, Nielsen ML, Olsen JV. Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole orbitrap mass spectrometer. Journal of proteome research. 2012;11:3487–3497. doi: 10.1021/pr3000249. [DOI] [PubMed] [Google Scholar]
- Kono M, Herrmann S, Loughran NB, Cabrera A, Engelberg K, Lehmann C, et al. Evolution and architecture of the inner membrane complex in asexual and sexual stages of the malaria parasite. Mol Biol Evol. 2012;29:2113–2132. doi: 10.1093/molbev/mss081. [DOI] [PubMed] [Google Scholar]
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- Larson ET, Parussini F, Huynh MH, Giebel JD, Kelley AM, Zhang L, et al. Toxoplasma gondii cathepsin L is the primary target of the invasion-inhibitory compound morpholinurea-leucyl-homophenyl-vinyl sulfone phenyl. J Biol Chem. 2009;284:26839–26850. doi: 10.1074/jbc.M109.003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentini G, Kong-Hap M, El Hajj H, Francia M, Claudet C, Striepen B, et al. Identification and characterization of Toxoplasma SIP, a conserved apicomplexan cytoskeleton protein involved in maintaining the shape, motility and virulence of the parasite. Cell Microbiol. 2014 doi: 10.1111/cmi.12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wetzel L, Zhang Y, Nagayasu E, Ems-McClung S, Florens L, Hu K. Novel thioredoxin-like proteins are components of a protein complex coating the cortical microtubules of Toxoplasma gondii. Eukaryot Cell. 2013;12:1588–1599. doi: 10.1128/EC.00082-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson CR, Scherf A. Flexible guide-RNA design for CRISPR applications using Protospacer Workbench. Nat Biotechnol. 2015;33:805–806. doi: 10.1038/nbt.3291. [DOI] [PubMed] [Google Scholar]
- Mann T, Beckers C. Characterization of the subpellicular network, a filamentous membrane skeletal component in the parasite Toxoplasma gondii. Mol Biochem Parasitol. 2001;115:257–268. doi: 10.1016/s0166-6851(01)00289-4. [DOI] [PubMed] [Google Scholar]
- Meszoely CA, Erbe EF, Steere RL, Trosper J, Beaudoin RL. Plasmodium falciparum: freeze-fracture of the gametocyte pellicular complex. Exp Parasitol. 1987;64:300–309. doi: 10.1016/0014-4894(87)90040-3. [DOI] [PubMed] [Google Scholar]
- Michalski A, Damoc E, Hauschild JP, Lange O, Wieghaus A, Makarov A, et al. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol Cell Proteomics. 2011;10:M111011015. doi: 10.1074/mcp.M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel V, Yuan Z, Ramsubir S, Bakovic M. Choline transport for phospholipid synthesis. Exp Biol Med (Maywood) 2006;231:490–504. doi: 10.1177/153537020623100503. [DOI] [PubMed] [Google Scholar]
- Miranda K, Pace DA, Cintron R, Rodrigues JC, Fang J, Smith A, et al. Characterization of a novel organelle in Toxoplasma gondii with similar composition and function to the plant vacuole. Mol Microbiol. 2010;76:1358–1375. doi: 10.1111/j.1365-2958.2010.07165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parussini F, Coppens I, Shah PP, Diamond SL, Carruthers VB. Cathepsin L occupies a vacuolar compartment and is a protein maturase within the endo/exocytic system of Toxoplasma gondii. Mol Microbiol. 2010;76:1340–1357. doi: 10.1111/j.1365-2958.2010.07181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porchet E, Torpier G. Freeze fracture study of Toxoplasma and Sarcocystis infective stages (author's transl) Zeitschrift fur Parasitenkunde. 1977;54:101–124. doi: 10.1007/BF00380795. [DOI] [PubMed] [Google Scholar]
- Pszenny V, Ehrenman K, Romano JD, Kennard A, Schultz A, Roos DS, et al. A Lipolytic Lecithin:Cholesterol Acyltransferase Secreted by Toxoplasma Facilitates Parasite Replication and Egress. J Biol Chem. 2016;291:3725–3746. doi: 10.1074/jbc.M115.671974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. The Journal of cell biology. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampels V, Hartmann A, Dietrich I, Coppens I, Sheiner L, Striepen B, et al. Conditional mutagenesis of a novel choline kinase demonstrates plasticity of phosphatidylcholine biogenesis and gene expression in Toxoplasma gondii. J Biol Chem. 2012;287:16289–16299. doi: 10.1074/jbc.M112.347138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Buguliskis JS, Lee TD, Sibley LD. Functional analysis of rhomboid proteases during Toxoplasma invasion. MBio. 2014;5:e01795–01714. doi: 10.1128/mBio.01795-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Sibley LD. The moving junction, a key portal to host cell invasion by apicomplexan parasites. Curr Opin Microbiol. 2012;15:449–455. doi: 10.1016/j.mib.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidik SM, Hackett CG, Tran F, Westwood NJ, Lourido S. Efficient genome engineering of Toxoplasma gondii using CRISPR/Cas9. PLoS One. 2014;9:e100450. doi: 10.1371/journal.pone.0100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub KW, Cheng SJ, Sohn CS, Bradley PJ. Novel components of the Apicomplexan moving junction reveal conserved and coccidia-restricted elements. Cell Microbiol. 2009;11:590–603. doi: 10.1111/j.1462-5822.2008.01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. Journal of proteome research. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilley LD, Krishnamurthy S, Westwood NJ, Ward GE. Identification of TgCBAP, a novel cytoskeletal protein that localizes to three distinct subcompartments of the Toxoplasma gondii pellicle. PLoS One. 2014;9:e98492. doi: 10.1371/journal.pone.0098492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. World Malaria Report 2014. WHO; Geneva, Switzerland: 2014. [Google Scholar]
- Wichroski MJ, Melton JA, Donahue CG, Tweten RK, Ward GE. Clostridium septicum alpha-toxin is active against the parasitic protozoan Toxoplasma gondii and targets members of the SAG family of glycosylphosphatidylinositol-anchored surface proteins. Infection and immunity. 2002;70:4353–4361. doi: 10.1128/IAI.70.8.4353-4361.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel JA. Identification of SUMO-conjugated proteins and their SUMO attachment sites using proteomic mass spectrometry. Methods in molecular biology. 2009;497:33–49. doi: 10.1007/978-1-59745-566-4_3. [DOI] [PubMed] [Google Scholar]
- Xu T, Venable JT, Kyu Park S, Cociorva D, Lu B, Liao L, et al. ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Molecular & Cellular Proteomics. 2006;5:S174–S174. [Google Scholar]
- Zeisel SH, Blusztajn JK. Choline and human nutrition. Annu Rev Nutr. 1994;14:269–296. doi: 10.1146/annurev.nu.14.070194.001413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The ISC3 paralog TgGT1_212990 is dispensable for parasite growth. A) Strategy for deleting the entire TgGT1_212990 locus and the downstream HXGPRT marker used for endogenous gene tagging (left). The DHFR cassette used for recombination at the CRISPR/Cas9-induced double-stranded break has a 5’ homology arm containing the 40 bp sequence (in red) immediately upstream of the TgGT1_212990 start codon. The 3′ UTRs of the DHFR and HXGPRT markers (green) are identical so the entire sequence functions as the 3′ homology arm during homology-dependent repair. Using this approach, only the 5′ primer is gene-specific for the PCR amplification of a drug cassette targeting an endogenously tagged gene of interest. Loss of the TgGT1_212990 locus and recombination of the DHFR cassette was verified by PCR using parental TgGT1_212990-HA parasites as a control (right). B,C) IFA (B) and Western blot (C) showing loss of the HA-tagged TgGT1_212990 protein (co-localized with the VAC marker CPL). Red: mouse anti-HA antibody. Green: rabbit anti-CPL antibody. Scale bar = 2 μm. D) Quantification of plaques produced by TgGT1_212990-HA and Δtggt1_212990 parasites after 9 days of growth. Plaque areas are depicted as a box-whisker plot, with the middle line corresponding to the median, the bottom and top boxes representing the 25th and 75th percentiles, respectively, and whiskers corresponding to the smallest and largest plaques. Data were analyzed with an independent samples test and no significant differences were detected. E,F) IFA showing no gross changes in TgGT1_212990 localization in Δisc3 parasites (E) and ISC3 localization in Δtggt1_212990 parasites (F). Red: mouse anti-HA antibody. Green: rabbit anti-CPL or rat anti-ISC1 antibody.
Table S1. List of top ISC4-BioID hits identified by mass spectrometry. Proteins present in both experimental samples but not the controls are shown. Hypothetical proteins localized by endogenous gene tagging in this study are highlighted in blue. Data is from two experimental replicates. Gene IDs and corresponding descriptions are from ToxoDB.
Table S2. Oligonucleotide primers used in this study. All primer sequences are shown in the 5′ to 3′ orientation.