Abstract
mRNA synthesis is one of the earliest readouts of the activity of a transcribed gene, which is of particular interest in the context of metazoan cell fate specification. These processes are intrinsically dynamic and stochastic, which makes in vivo single-cell measurements inevitable. Here, we present the application of a technology that has been widely used in single celled organisms to measure transcriptional activity in developing embryos of the fruit fly Drosophila melanogaster. The method allows quantification of instantaneous polymerase occupancy of active gene loci and thereby enables the development and testing of models of gene regulation in development.
Keywords: nascent mRNA, transcription activity, quantitative live imaging, fluorescence, Drosophila
1 Introduction
The observation of mRNA transcript production in real time has become a familiar routine in the context of single cells.[1–3] This method entails the use of an mRNA tagging system in which nascent transcripts are tagged with multiple repeats of a stem loop sequence that is recognized by a cognate binding protein (Fig. 1A). The latter is constitutively expressed and fused to a fluorescent protein that can be visualized using standard live microscopy techniques.[4–6] The stem loop cluster binds multiple fluorescent proteins resulting in spatially localized fluorescence at the gene locus, which is further enhanced by each additional polymerase that is engaged in transcriptional elongation (Fig. 1B). In the fruit fly Drosophila melanogaster, this MS2 system has been implemented to study maternal mRNA transport in oocytes [7] and transcription [8–13].
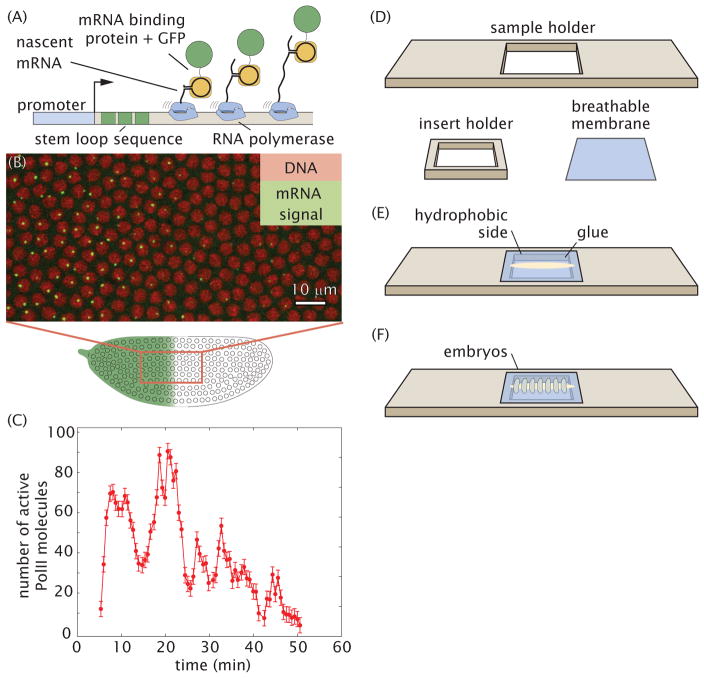
Figure 1. Quantifying transcriptional dynamics in live fly embryos.
(A) 24 repeats of the MS2 loop sequence is added to a gene that, when transcribed, folds into a stem loop that is recognized by an mRNA binding protein fused to GFP; fluorescence is proportional to transcriptional activity. (B) Typical field of view showing sites of transcription in single nuclei. (C) Number of actively transcribing Pol II molecules as a function of time for a labeled site of nascent transcript formation. (D,E,F) Sample holder and breathable membrane for embryo mounting.
Here we present a detailed protocol to visualize and quantify nascent transcripts in living fly embryos using the MS2 stem loop system (Fig. 1C). Details are provided regarding the transgenic construct design and implementation, embryo handling for live imaging, and various suitable microscopy techniques. We discuss confocal and two-photon imaging conditions for optimal high quality images for quantitative analysis. In particular we discuss calibration of the fluorescence signal to obtain absolute units in terms of numbers of actively transcribing RNA Polymerase II (PolII) molecules.
2 Materials
2.1. Creating MS2 reporter constructs and transgenic fly lines
Plasmid pCR4-24XMS2SL-stable [4] (Addgene #31865) to make a new MS2 reporter construct
pIB-hbP2-P2P-lacZ-αTUb3′UTR [14]
pBϕ-eve2-MS2-yellow [10] for plasmids ready for transgenesis using recombination mediated cassette exchange (RMCE) [15] or single attP site integration [16], respectively.
Transformation competent E. Coli (e.g. XL1-Blue)
Stbl2 competent E. Coli cells (Life Technologies 10268-019).
Bloomington Drosophila stock center fly lines #27388 (for RMCE) or #9750 (single attP).
In-house microinjection setup to generate transgenics or third-party company such as Bestgene (www.thebestgene.com) or Rainbow Transgenics (www.rainbowgene.com).
2.2. Fly lines
2.3. Embryo collection
Apple juice agar plates.
Active dry yeast (e.g. Fleischmann’s ADY 2192).
Halocarbon oil 27 (e.g. Sigma-Aldrich).
Absorbent paper towels.
20 % Bleach.
Distilled or reverse osmosis water.
2.4. Embryo mounting
Breathable Lumox Film (e.g. Sarstedt 94.6077.317)
Double-sided sticky tape.
50 mL conical tubes.
1.5 mL Eppendorf tubes.
Heptane.
Platform rocker (Nutator).
Table top centrifuge.
Scintillation vial.
Embryo and membrane slide holder (http://www.sculpteo.com/en/gallery/public/hggarcia/).
Dumont #5 forceps.
Round brush (size 0 or smaller).
2.5. Imaging
Custom-built 2-photon microscope [17] with a Zeiss 25x 0.8NA objective, GaAsP photomultipliers (Hamamatsu H10770PA-40 SEL), Pockel Cell or neutral density filters to modulate laser output, Coherent power meter #1159770 and ScanImage control software (www.scanimage.org)
Confocal microscope with high sensitivity photodetectors. We use a Leica SP8 with 63x, 1.4 NA objective, Argon and Helium-Neon lasers, or white light laser and HyD detectors and a Zeiss 780 with 40x, 1.4 NA objective, Argon and Helium-Neon lasers and GaAsP detectors.
Diagnostic slides for measuring flat field (Chroma #92001)
2.6. Data analysis
Mathworks Matlab
FlyRNAQuant (github.com/PrincetonUniversity/FlyRNAQuant)
3. Methods
The protocol begins with the creation of transgenic Drosophila lines bearing stem loop-tagged reporter constructs. The mounting of embryos onto a custom-made slide sample holder (Fig. 1D–F) is described. This sample holder makes it possible to flatten the embryos and have as many nuclei as possible in the same focal plane. Lastly, the imaging protocol for the measurement of transcriptional activity in live embryos is described.
3.1. Creating DNA reporter molecules
Use the unique restriction sites in pIB-hbP2-P2P-lacZ-αTUb3′UTR (RMCE) or pBϕ-eve2-MS2-yellow (single attP insertion) to insert new regulatory regions using regular cutting-and-pasting with restriction enzymes or using Gibson assembly (vector maps available at benchling.com/garcialab).
Transform the newly generated plasmids into regular cloning bacterial strains such as XL1-Blue due to their high transformation efficiency.
Some of the MS2 stem loops can be lost. Before sequencing colonies screen them using restriction enzymes such as EcoRV. Send for sequencing with primers for the new insert and for the stem loops.
Once sequencing is confirmed transform the plasmid into Stbl2 cells for archival purposes.
Generate transgenic flies by injecting the newly generated vector in-house or through a company.
3.2. Embryo glue
Densely pack a 50 mL conical tube with strips of double-sided sticky tape.
Fill the tube with heptane.
Mix the tube overnight on a nutator.
Using forceps extract the tape and pipette the heptane into Eppendorf tubes.
Spin down the tubes at maximum speed on a table-top centrifuge.
Pipette the supernatant into a scintillation vial and store at 4 °C.
3.3. Preparing the sample holder for live imaging of embryos
Identify the hydrophobic side of the permeable membrane using a sharpie: it will be harder to write on the hydrophobic side.
Cut a 2 cm x 2 cm square.
Mount the membrane into the membrane holder as shown in Figs. 1D and E
Apply a thin line of glue to the membrane (Fig. 1E). This glue will avoid embryo rolling during sample preparation.
3.4. Preparing transgenic embryos for live imaging
Two days before imaging prepare a cage of flies containing 50–100 virgin females of His-RFP;MCP-GFP flies and 20–40 males containing the reporter construct.
90 minutes before preparing the embryos replace the plate in the cage for a new one with fresh yeast.
After the waiting time replace the plate again.
Cover plate with halocarbon oil and image embryos using a dissecting scope with trans-illumination (see Note 1).
Using the forceps pick early embryos [18] and transfer them to a small (1 cm x 1 cm) cutout of a paper towel.
Cover with bleach for 1 minute.
Absorb bleach using a paper towel and wash with water for 1 minute.
Absorb water and transfer paper towel to a petri dish with water. Undamaged embryos will float.
Using the brush transfer embryos to a small dry cutout of paper towel.
Pick embryos one by one in the right orientation using the brush and place them on the glue on the membrane (Fig. 1F) (see Note 2).
Put two drops of halocarbon oil on embryos and place an 18 x 18 mm 1.5 coverglass on them. Let the sample sit for one minute.
Inspect the samples for embryo flattening. If embryos need further flattening use the side of a Kimwipe to absorb oil from the side of the coverglass. Monitor coverglass so that it doesn’t slide and the embryos so that they do not explode!
This sample can be used both on an upright or inverted configuration.
3.5. Setting up the microscopes for imaging
Turn on the microscope system and lasers at least 90 minutes before imaging the embryos.
3.5.1. 2-photon microscopy for live imaging
Set the TiSa laser wavelength to 970 nm.
Set up power meter at the objective exit.
Park scanner or set it to a high zoom of 80 or larger and scan.
Measure power and use the Pockel Cell or the neutral density filter to adjust it to 10 mW.
Set scanning to a pixel size of 220 nm, 512 pixels per line, 256 lines per frame and a frequency of 250 Hz per line (4 ms/line).
Set Z-stack continuous acquisition to 10 slices separated by 1 μm, each slice is averaged 3 times.
3.5.2. Confocal microscopy for live imaging
Use a 488 nm and a 561 nm laser excitation lines.
Set up power meter at the 10x objective exit, zoom to 40x, and find the right laser power settings for a 35 μW output power in 488nm and 20 μW output power in 561 nm.
Set scanning to a pixel size of 220 nm, 512 pixels per line, 256 lines per frame and a frequency of 400 Hz per line and bidirectional scanning.
Set Z-stack continuous acquisition to 21 slices separated by 0.5 μm, each slice is averaged 3 times.
3.6. Live imaging
Find the embryos using brightfield illumination and save their positions using an automated XY stage.
Using fluorescence illumination look for an embryo whose nuclei are just migrating to the surface.
Find the nuclei and transcription spots, and start a continuous acquisition by centering the Z-stack on the middle of the nuclei (see Notes 3 and 4).
The sample should be monitored regularly as nuclei and their transcription spots tend to drift during acquisition. Adjust the center of the Z-stack as needed throughout the experiment.
Once done with the acquisition take an image of the full embryo image in order to determine the correct AP position of the zoomed-in image and to check for photobleaching (see Note 5).
Using the same acquisition parameters of the Z-stack measure the flat field using diagnostic slides for measuring flat field (see Note 6).
3.7. Live data analysis
Download the FlyRNAQuant software and manual.
Install the software as instructed in the manual and download a sample data set.
Follow the analysis of the sample data set all the way to the end to ensure the full functionality of the code.
Transfer the obtained data to the analysis computer with analysis software.
Go through the different steps detailed in the manual for the automated analysis.
If single cell information is required particular attention needs to be paid to the curation step of the traces.
3.8. Calibration of absolute number of active PolII molecules
Measure the transcriptional activity of reporter fly line P2P-MS2-lacZ as described in Methods 3.3., 3.4., 3.6. and 3.7.
Integrate the fluorescence over nuclear cycle 13 to infer the number of total number of mRNA molecules produced as described in [8].
Measure the number of mRNA molecules produced by the reporter line P2P-lacZ in the cytoplasm during mitosis 12 and mitosis 13 using the single-molecule FISH (smFISH) protocol described in Chapter 8 of this book.[19]
Take the difference of mRNA molecules obtained by smFISH during mitosis 12 and 13 and compare it to the quantity obtained by live imaging during the same developmental stage in order to calibrate the arbitrary fluorescence units of the MS2-MCP-EGFP reporter.[8]
Acknowledgments
The authors are grateful through support provided by the Princeton Dickie Fellowship (HG), the Burroughs Wellcome Fund Career Award at the Scientific Interface (HG), The Sloan Research Foundation (HG), The Searle Scholars Program (TG and HG), the Shurl & Kay Curci Foundation (HG), the National Institutes of Health Grants P50 GM071508 and R01 GM097275 (TG).
Footnotes
The halocarbon oil allows to easily image through the embryos and score their developmental time.
Mounting a typical amount of 16 embryos usually ensures finding several in the right orientation and developmental stage for imaging.
If no spots or low fluorescent signal is detected, check and/or increase laser power. Image the flat field slide as a positive control of the detectors.
If the imaging conditions are checked and the problem of no/low signal persists, it is possible that the MS2 stem loops were lost during the cloning procedure. Check the number of loops as in step 3 of Method 3.1.If necessary, rescreen bacterial colonies.
If pronounced photobleaching on the MCP-GFP channel is detected reduce the laser power and/or reduce the frequency of acquisition.
The laser power will have to be reduced dramatically so as to not saturate and damage the detectors.
Contributor Information
Hernan Garcia, Department of Molecular and Cell Biology, Department of Physics, and Biophysics Graduate Group; University of California, Berkeley, CA, USA.
Thomas Gregor, Joseph Henry Laboratory of Physics and Lewis-Sigler Institute for Integrative Genomics; Princeton University, Princeton, NJ, USA.
References
- 1.Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol. 2003;13(2):161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123(6):1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 3.Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011;332(6028):475–478. doi: 10.1126/science.1202142. 332/6028/475 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2(4):437–445. doi: 10.1016/s1097-2765(00)80143-4. S1097-2765(00)80143-4 [pii] [DOI] [PubMed] [Google Scholar]
- 5.Yunger S, Rosenfeld L, Garini Y, Shav-Tal Y. Single-allele analysis of transcription kinetics in living mammalian cells. Nat Methods. 2010;7(8):631–633. doi: 10.1038/nmeth.1482. nmeth.1482 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods. 2011 doi: 10.1038/nmeth.1551. nmeth.1551 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol. 2003;13(14):1159–1168. doi: 10.1016/s0960-9822(03)00451-2. S0960982203004512 [pii] [DOI] [PubMed] [Google Scholar]
- 8.Garcia HG, Tikhonov M, Lin A, Gregor T. Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Curr Biol. 2013;23(21):2140–2145. doi: 10.1016/j.cub.2013.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucas T, Ferraro T, Roelens B, De Las Heras Chanes J, Walczak AM, Coppey M, Dostatni N. Live imaging of bicoid-dependent transcription in Drosophila embryos. Curr Biol. 2013;23(21):2135–2139. doi: 10.1016/j.cub.2013.08.053. [DOI] [PubMed] [Google Scholar]
- 10.Bothma JP, Garcia HG, Esposito E, Schlissel G, Gregor T, Levine M. Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. Proc Natl Acad Sci U S A. 2014;111(29):10598–10603. doi: 10.1073/pnas.1410022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bothma JP, Garcia HG, Ng S, Perry MW, Gregor T, Levine M. Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo. eLife. 2015:4. doi: 10.7554/eLife.07956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferraro T, Esposito E, Mancini L, Ng S, Lucas T, Coppey M, Dostatni N, Walczak AM, Levine M, Lagha M. Transcriptional Memory in the Drosophila Embryo. Curr Biol. 2016;26(2):212–218. doi: 10.1016/j.cub.2015.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukaya T, Lim B, Levine M. Enhancer Control of Transcriptional Bursting. Cell. 2016 doi: 10.1016/j.cell.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H, Xu Z, Mei C, Yu D, Small S. A system of repressor gradients spatially organizes the boundaries of bicoid-dependent target genes. Cell. 2012;149(3):618–629. doi: 10.1016/j.cell.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bateman JR, Lee AM, Wu CT. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics. 2006;173(2):769–777. doi: 10.1534/genetics.106.056945. genetics.106.056945 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314(5806):1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- 17.Liu F, Morrison AH, Gregor T. Dynamic interpretation of maternal inputs by the Drosophila segmentation gene network. Proc Natl Acad Sci U S A. 2013;110(17):6724–6729. doi: 10.1073/pnas.1220912110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Figard L, Sokac AM. Imaging cell shape change in living Drosophila embryos. J Vis Exp. 2011;(49) doi: 10.3791/2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Little SC, Tikhonov M, Gregor T. Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell. 2013;154(4):789–800. doi: 10.1016/j.cell.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]