Abstract
Many studies have revealed that microRNAs (miRNAs) play crucial roles in cancer development and progression. miRNA-30a (miR-30a), as a member of the miR-30 family, has been implicated in various cancers. However, the role of miR-30a in osteosarcoma remains unclear. In the current study, we found that miR-30a was significantly downregulated in osteosarcoma tissues and cell lines by using quantitative real-time polymerase chain reaction (qRT-PCR). In addition, miR-30a could inhibit cancer cell growth, migration, and invasion in vitro. Furthermore, bioinformatics of miRNA target prediction and luciferase reporter assay indicated that MEF2D is a direct target of miR-30a. miR-30a was able to reduce the mRNA and protein expression of MEF2D as assessed using RT-PCR and Western blotting assay. Interestingly, overexpression of MEF2D partially reversed the miR-30a-reduced cell proliferation, migration, and invasion of osteosarcoma cell, indicating that miR-30a suppresses osteosarcoma cell proliferation and metastasis partially mediated by inhibition of MEF2D. Overall, our study demonstrated that miR-30a functions as a tumor suppressor by targeting MEF2D in osteosarcoma, providing a promising prognostic biomarker and a therapeutic strategy for osteosarcoma.
Keywords: miR-30a, MEF2D, osteosarcoma, proliferation, invasion, migration
Introduction
Osteosarcoma, as the most frequent malignant primary bone tumor, is the leading cause of cancer-related death among children and adolescents.1 Despite the improvements in diagnosis and treatment strategies of osteosarcoma improving the 5-year survival rate of osteosarcoma patients significantly to ~60% over the past several decades, there is still a high risk of distant metastasis and local relapse even after complete surgical resection for osteosarcoma patients.2,3 Thus, it is imperative to elucidate the underlying molecular mechanisms of osteosarcoma and find novel diagnostic and prognostic biomarkers to improve the clinical outcome of osteosarcoma patients.
MicroRNA (miRNA) is an abundant group of small noncoding RNA (with approximately 22 nucleotides) that controls the expression of target genes by binding to the 3′-untranslated region (UTR) of their target mRNAs and plays an important role in a variety of biological processes, including cell proliferation, apoptosis, differentiation, invasion, migration, and so on.4–7 Many studies showed that miRNAs are dysregulated in a variety of cancers and play a critical role in tumorigenesis.8–13 Recent studies demonstrated that miRNAs have been recognized as critical regulators in the development and progression of cancers, including osteosarcoma.14–19 Therefore, identification of novel miRNAs involved in osteosarcoma progression may contribute to the development of a prognostic biomarker and a therapeutic strategy for osteosarcoma.
The miR-30 family contains six distinct mature miRNA sequences: miR-30a/miR-30c-2, miR-30d/miR-30b, and miR-30e/miR-30c-1.20 Accumulating evidences indicate that the deregulation of miR-30a contributes to various malignant tumors, including thyroid cancer, breast cancer, gastric cancer, lung cancer, and colon cancer.21–26 miR-30a suppresses the tumorigenesis processes in these cancers by directly targeting tumor-related proteins. For example, miR-30a suppressed colon cancer cell growth by targeting IRS2 (insulin receptor substrate 2).21 However, the expression and role of miR-30a in the progression of osteosarcoma remain unclear.
In this study, we, for the first time, revealed the deregulated expression of miR-30a in osteosarcoma and investigated the function of miR-30a on osteosarcoma cell proliferation and invasion. Furthermore, we identified MEF2D as a direct target of miR-30a involved in the progression of osteosarcoma. In conclusion, we found that miR-30a acts as a tumor suppressor, and so may serve as a potential therapeutic target in osteosarcoma.
Materials and methods
Human tissue specimens
Paired tissue specimens of osteosarcoma and matched normal tissues were obtained, with written informed consent, from 20 osteosarcoma patients between 2013 and 2014 at the First Affiliated Hospital of Nanchang University. All the tissues were obtained at the time of surgery and immediately stored in liquid nitrogen until use. The Institute Research Medical Ethics Committee of Nanchang University granted approval for this study.
Cell culture and transfection
Osteosarcoma cell lines (MG-63, U2OS, and Saos-2 cells) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in RPMI 1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific). Normal osteoblast cell line NHOst was purchased from the ATCC and was cultured in Dulbecco’s Modified Eagle’s Medium (DMEM)/F12 medium supplemented with 10% fetal bovine. Cultures were maintained at 37°C in a humidified atmosphere with 5% CO2.
MG-63 cells were seeded in 12-well plates. After 12 hours, cells were transiently transfected with the miR-30a (hsa-miR-30a) mimic, miR-30a inhibitor, mimic negative control (NC mimic), and inhibitor negative control (NC inhibitor) sequences using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. The full-length MEF2D cDNA was PCR amplified and cloned into the pcDNA3.1 vector to generate the pcDNA-MEF2D constructs that were used in the rescue assays. MG-63 cells were con-transfected with the miR-30a mimic and pcDNA-MEF2D.
Quantitative real-time polymerase chain reaction
The RNA extracted from cells and human osteosarcoma specimens with the Trizol solution (Sigma-Aldrich Co., St Louis, MO, USA) was subjected to reverse transcription PCR to obtain cDNA (Primescript RT Reagent kit, Takara, Shiga, Japan) according to the manufacturer’s instructions. Quantitative polymerase chain reaction (qPCR) was performed using SYBR Premix Ex Taq II (Takara) with the CFX96™ Real-Time PCR Detection System (Bio-Rad Laboratories Inc., Hercules, CA, USA). The primer sequences of miR-30a and MEF2D were described previously.21,27
Luciferase reporter gene assays
The 3′-UTR of MEF2D containing the putative binding site of miR-30a was amplified and subcloned into pGL3 luciferase promoter vector (Promega Corporation, Fitchburg, WI, USA). The vector was cotransfected with the miR-30a mimics into HEK293 cells for 48 hours. The cells were harvested, and the relative luciferase activity was detected using a dual-luciferase reporter assay kit (Promega Corporation) according to the manufacturer’s instructions. All experiments were performed at least three times.
Western blotting analysis
Whole-cell extracts were prepared with a cell lysis reagent (Sigma-Aldrich Co.) according to the manufacturer’s instruction manual, and the protein was quantified by BCA assay (Pierce, Rockford, IL, USA). Then, the protein samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (10%) and detected by Western blot using polyclonal (rabbit) anti-MEF2D antibody (Santa Cruz Biotechnology Inc., Dallas, TX, USA). Goat anti-rabbit IgG (Pierce) secondary antibody conjugated to horseradish peroxidase and ECL detection systems (SuperSignal West Femto, Pierce) were used for detection.
Cell proliferation assay
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was adopted to assess cell viability as described previously.28
Invasion assay
The cell invasion ability was examined by the transwell invasion assay. Cells were placed in the upper chamber of a 24-well Transwell unit with 8-μm-pore-sized polycarbonate nucleopore filters (Corning Costar, Cambridge, MA, USA). The upper compartment contained serum-free medium while the lower compartment contained medium with 10% fetal bovine serum; the cells were incubated for 48 hours in a humidified atmosphere of 5% CO2 at 37°C. The cells from at least five representative fields were analyzed. The invading cells across the membrane were counted under a light microscope. The cells adhering to the lower surface were fixed and counted. All the experiments were repeated in triplicate.
Wound healing assay
When the transfected and untransfected MG-63 cells had grown to 90% confluence, we used a micropipette tip to produce a scratch in the cell monolayer. The cells were then incubated under standard conditions for 24 hours, and images were captured at different times. The migration potential was estimated by counting the cells that migrated from the wound edge. The migration distance between the leading edge of the migrating cells and the edge of the wound was compared, similar to a previous work.29
Statistical analysis
Each experiment was repeated at least three times. Data were presented as mean ± SD and analyzed using SPSS 19.0 (StataCorp LP, College Station, TX, USA). Statistical comparisons between groups were analyzed using Student’s t-test, and a two-tailed P<0.05 was considered to indicate statistical significance.
Results
Downregulation of miR-30a in osteosarcoma tissues and cell lines
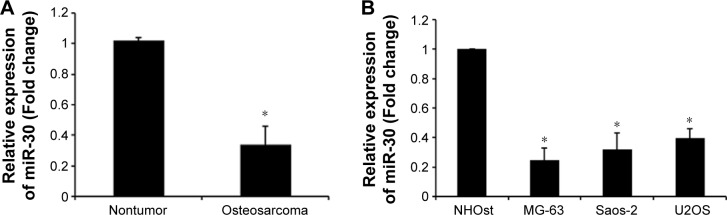
We first detected miR-30a expression in 20 human osteosarcoma and adjacent normal bone tissues using the quantitative real-time PCR (qRT-PCR) assay. As shown in Figure 1A, miR-30a expression was evidently downregulated in osteo-sarcoma tissues compared with the corresponding adjacent normal bone tissues. Furthermore, we investigated the expression of miR-30a in three osteosarcoma cell lines (MG-63, U2OS, and Saos-2 cells) and the human osteoblastic cell line NHOst by qRT-PCR. Similarly, we found that the expression of miR-30a was much less in three osteosarcoma cell lines than that of human osteoblast cell line NHOst (Figure 1B). Collectively, our findings suggest that downregulated miR-30a may be involved in the tumorigenesis of osteosarcoma.
Figure 1.
The expression of miR-30a in human osteosarcoma cell lines and tissues.
Notes: (A) The expression of miR-30a in 20 paired primary osteosarcoma tissues and adjacent nontumor tissues. Data are presented as mean ± SD. *P<0.05 vs nontumor group. (B) The expression of miR-30a in osteosarcoma cell lines (MG-63, Saos-2, and U2OS) and normal osteoblast cell line NHOst. U6 was used as an internal control. Each experiment was repeated three times. Data are presented as mean ± SD. *P<0.05 vs NHOst group. Results are in response to a Student’s t-test.
miR-30a suppresses proliferation, migration, and invasion of osteosarcoma cell
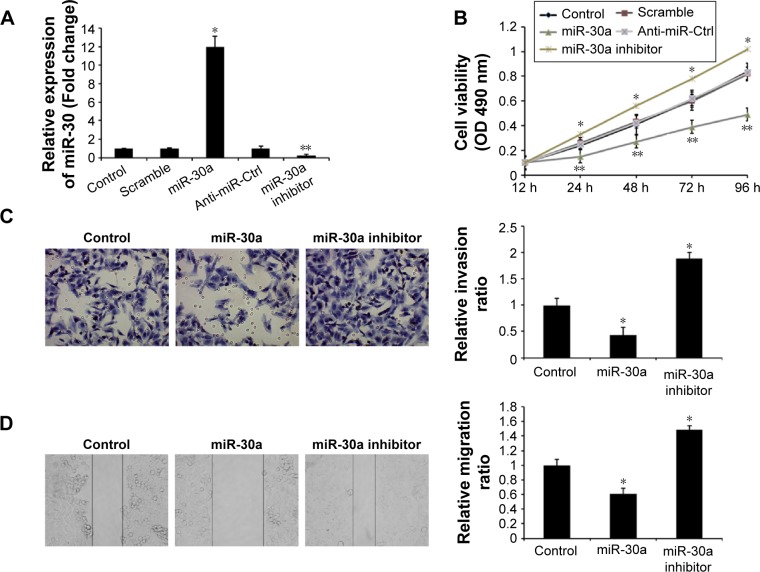
To explore the role of miR-30a in the development of osteosarcoma, we transfected the miR-30a mimic or the miR-30a inhibitor into MG63 cells and then detected proliferation, migration, and invasion of osteosarcoma. As shown in Figure 2A, the expression levels of miR-30a were evidently upregulated by the miR-30a mimic and were evidently downregulated by the miR-30a inhibitor. Next, the MTT assay showed that the proliferation ability of MG-63 cells was significantly decreased by the miR-30a mimic (Figure 2B). Consistent with these results, the proliferation ability of MG63 cells was significantly increased by the miR-30a inhibitor (Figure 2B). Furthermore, the role of miR-30a on MG-63 cell migration and invasion by using the wound healing assay and transwell invasion assay was analyzed. The transwell invasion assay showed that the miR-30a mimic markedly inhibited osteosarcoma cell invasion (Figure 2C), while the miR-30a inhibitor evidently increased osteosarcoma cell invasion (Figure 2C). Wound healing assays showed the inhibition of migration ability of osteosarcoma cells by miR-30a mimics (Figure 2D) and the induction of osteosarcoma cell migration by miR-30a inhibitors (Figure 2D). Taken together, these data demonstrated that miR-30a functions as a tumor suppressor by inhibiting cell proliferation, migration, and invasion of osteosarcoma.
Figure 2.
miR-30a regulates osteosarcoma cell proliferation, migration, and invasion.
Notes: (A) miR-30a expression by transfection with miR-30a mimic and miR-30a inhibitor in MG-63 cells. Data are presented as mean ± SD from three independent experiments. *P<0.01 vs control group and scramble group. **P<0.01 vs control group and anti-miR-Ctrl group. (B) The MTT assay was performed to analyze the effect of miR-30a on cell proliferation of MG-63 cells. *P<0.01 vs control group and scramble group. **P<0.01 vs control group and anti-miR-Ctrl group. Statistical significance was assessed by comparing mean values (±SD) using a Student’s t test for independent groups and was assumed for **P<0.01. (C) The transwell invasion assay was utilized to analyze the effect of miR-30a on cell invasion of MG-63 cells. *P<0.05 vs control group. Magnification ×40. (D) The wound healing assay was used to analyze the effect of miR-30a on cell migration of MG-63 cells. Data are presented as mean ± SD from three independent experiments. *P<0.05 vs control group.
Abbreviations: h, hours; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
miR-30a directly targets MEF2D in osteosarcoma cells
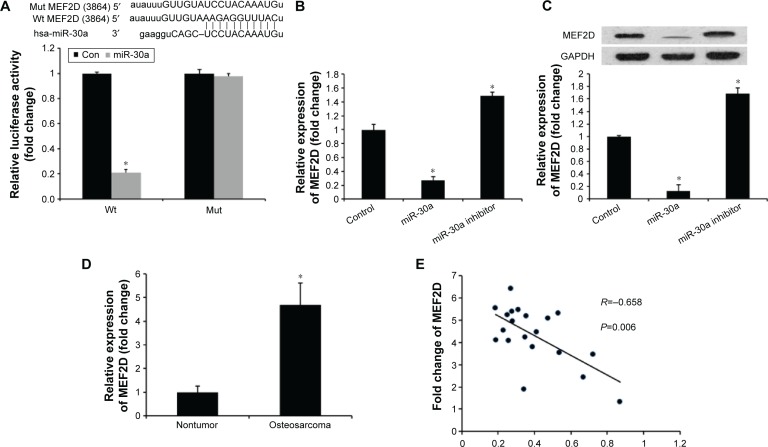
To elucidate the underlying mechanisms of miR-30a on osteosarcoma progression, we predicted targets of miR-30a using the miRNA target prediction websites www.microRNA.org and TargetScan. Here, we identified a conserved miR-30a-binding site in the 3′-UTR of MEF2D mRNA. We then cloned WT or Mut target region sequence of the MEF2D 3′-UTR, which was inserted into a luciferase reporter vector (Figure 3A). Subsequently, these reporter vectors were con-transfected with miR-30a mimics (miR-30a) or mimics control (con) into the HEK293 cell line. As shown in Figure 3A, con-transfection of miR-30a mimics suppressed the luciferase activity of the reporter-containing wild-type MEF2D 3′-UTR sequence, but failed to inhibit that of mutated MEF2D, as assessed by the dual-luciferase reporter assay. These data indicate that MEF2D is one of the direct targets of miR-30a in osteosarcoma cells. To further confirm MEF2D as a direct target of miR-30a, the qRT-PCR and Western blot assay were used to detect the expression of MEF2D in MG-63 cells. As shown in Figure 3B and C, both mRNA and protein expression levels of MEF2D were significantly downregulated by miR-30a mimics and were upregulated by miR-30a inhibitors in MG-63 cells.
Figure 3.
miR-30a negatively regulates MEF2D expression in osteosarcoma cells.
Notes: (A) Sequence alignment of miR-30a and 3′-UTR of MEF2D using mirco-RNA.org and the luciferase reporter assay. MG-63 cells were transiently cotransfected with Wt/Mut 3′-UTR with miRNAs as indicated. The effects of miR-30a on the expression of MEF2D at both mRNA (B) and protein (C) level in MG-63 cells are indicated. Data are presented as mean ± SD from three independent experiments. *P<0.05 vs control group. (D) MEF2D mRNA levels was examined by qRT-PCR in 20 cases of primary osteosarcoma tissues and adjacent nontumor tissues. (E) Correlation of miR-30a levels with MEF2D mRNA levels was examined by qRT-PCR in 20 cases of osteosarcoma tissues (Pearson’s correlation coefficient, R=−0.658). Statistically significant differences are indicated: *P<0.01 vs paired nontumorous tissues.
Abbreviations: GAPDH, glyceraldehyde 3-phosphate dehydrogenase; qRT-PCR, real-time polymerase chain reaction.
The relationship between MEF2D and miR-30a was further analyzed by detecting the expression levels of MEF2D in osteosarcoma tissues and cell lines. We found that MEF2D levels in osteosarcoma tissues were greatly upregulated compared with adjacent nontumor tissues or osteoblast cell line (Figure 3D). Then, we correlated MEF2D with miR-30a expression in the same osteosarcoma specimens. As shown in Figure 3E, the mRNA levels of miR-30a and MEF2D showed a significant inverse correlation when assessed using the Spearman’s correlation analysis (R=−0.658, P<0.001). Taken together, these data strongly supported the speculation that MEF2D is a direct target of miR-30a in osteosarcoma.
Overexpression of MEF2D rescues the miR-30a-induced suppression of osteosarcoma cell proliferation, migration, and invasion
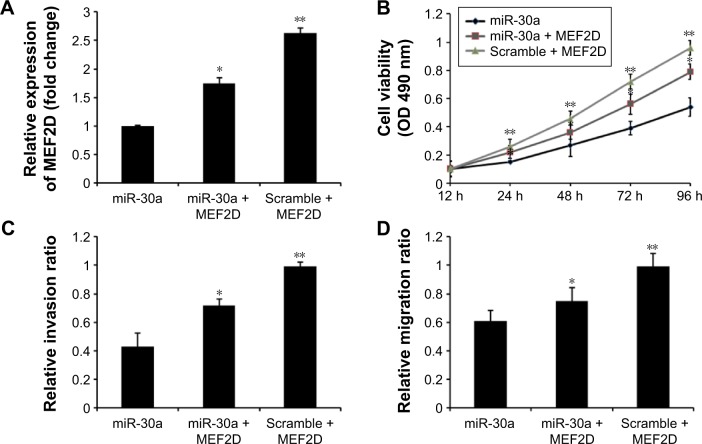
To further confirm that MEF2D is a functional target of miR-30a, MEF2D plasmids were used to restore MEF2D expression. The expression levels of MEF2D were significantly increased in the cells cotransfected with pcDNA-MEF2D and miR-30a mimics compared with the miR-30a mimics-only group (Figure 4A). Moreover, overexpression of MEF2D by transfecting with pcDNA-MEF2D could reverse the inhibition of proliferation, migration, and invasion by miR-30a mimics (Figure 4B–D). In conclusion, these results indicated that MEF2D is involved the miR-30a-suppressed proliferation, migration, and invasion of osteosarcoma cells.
Figure 4.
MEF2D is required for miR-30a-directed osteosarcoma cell proliferation, migration, and invasion.
Notes: (A) MEF2D expression was increased by transfection with MEF2D plasmids in miR-30a mimics osteosarcoma cell. (B) The MTT assay was used to detect MG-63 cells cotransfection with miR-30a mimic and pcDNA-MEF2D plasmid. (C) The wound healing assay was performed to detect the effects on cell migration of MG-63 cells treated as described. (D) The transwell invasion assay was used to detect the effects on cell invasion of MG-63 cells treated as described. Data are presented as mean ± SD from three independent experiments. *P<0.01 vs miR-30a group. **P<0.01 vs miR-30a + MEF2D group.
Abbreviations: h, hours; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; OD, optical density.
Discussion
Increasing evidences have indicated that miRNAs are involved in the tumorigenic processes by targeting a variety of tumor-related genes.14 In this study, we investigated the roles of miR-30a in osteosarcoma cell proliferation, migration, and invasion. We found that miR-30a expression was significantly downregulated in osteosarcoma tissue and cell lines compared with adjacent nontumor tissues and osteoblast cell line NHOst. Further studies indicated that miR-30a was able to regulate cell proliferation, migration, and invasion of osteosarcoma cells by targeting MEF2D. Therefore, our study, for the first time, revealed miR-30a as a tumor suppressor in the progression of osteosarcoma.
The myocyte enhancer factor 2 (MEF2) transcription factors have roles in muscle, cardiac, skeletal, vascular, neural, blood, and immune system cell development through their effects on cell differentiation, proliferation, apoptosis, migration, shape, and metabolism.30 Altered MEF2 activity plays a role in human diseases and has recently been implicated in the initiation and progression of human cancers.31 Recently, it was reported that MEF2D, one member of the MEF2 family, was involved in the progression of several cancers, including hepatocellular carcinoma,31 lung carcinoma,32 and rhabdomyosarcoma.33 In osteosarcoma, MEF2D elevation in osteosarcoma clinical specimens was associated with poor patient prognosis. MEF2D suppression was shown to decrease the proliferation of osteosarcoma cells.27 In our study, the luciferase reporter assay revealed that MEF2D was a direct target of miR-30a in osteosarcoma cells. Overexpression of miR-30a reduced MEF2D mRNA and protein levels in osteosarcoma cells. A significant inverse correlation was observed by the Spearman’s correlation analysis between the mRNA levels of miR-30a and MEF2D in the same osteosarcoma specimens. Furthermore, reexpression of MEF2D rescued the suppressive effect of miR-30a on the proliferation and metastasis of osteosarcoma cells. Our results indicated that miR-30a functions as a tumor suppressor in osteosarcoma by targeting MEF2D. The regulatory mechanisms of miRNAs on deregulated MEF2D in human cancers remain to be explored in the future.
Taken together, our study, for the first time, revealed miR-30a as a tumor suppressor in osteosarcoma, with the expression levels of miR-30a being significantly decreased in tumor tissues and cell lines and its ectopic expression inhibiting cell proliferation and metastasis. The tumor suppressor function of miR-30a was mediated by the regulation of MEF2D expression. These results indicate that miR-30a deregulation may play important roles in tumor progression and that miR-30a may be a potential therapeutic target for the treatment of osteosarcoma.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81460406), the Education Department Science Foundation of the Jiangxi Province (GJJ10329), and the Health Department Science Foundation of the Jiangxi Province (20091057).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 2.Duchman KR, Gao Y, Miller BJ. Prognostic factors for survival in patients with high-grade osteosarcoma using the Surveillance, Epidemiology, and End Results (SEER) Program database. Cancer Epidemiol. 2015;39(4):593–599. doi: 10.1016/j.canep.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Bramer JA, van Linge JH, Grimer RJ, Scholten RJ. Prognostic factors in localized extremity osteosarcoma: a systematic review. Eur J Surg Oncol. 2009;35(10):1030–1036. doi: 10.1016/j.ejso.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 4.Li Z, Lei H, Luo M, et al. DNA methylation downregulated mir-10b acts as a tumor suppressor in gastric cancer. Gastric Cancer. 2015;18(1):43–54. doi: 10.1007/s10120-014-0340-8. [DOI] [PubMed] [Google Scholar]
- 5.Xiao X, Tang C, Xiao S, Fu C, Yu P. Enhancement of proliferation and invasion by MicroRNA-590-5p via targeting PBRM1 in clear cell renal carcinoma cells. Oncol Res. 2013;20(11):537–544. doi: 10.3727/096504013X13775486749335. [DOI] [PubMed] [Google Scholar]
- 6.Yin WZ, Li F, Zhang L, Ren XP, Zhang N, Wen JF. Down-regulation of microRNA-205 promotes gastric cancer cell proliferation. Eur Rev Med Pharmacol Sci. 2014;18(7):1027–1032. [PubMed] [Google Scholar]
- 7.Yang X, Ni W, Lei K. miR-200b suppresses cell growth, migration and invasion by targeting Notch1 in nasopharyngeal carcinoma. Cell Physiol Biochem. 2013;32(5):1288–1298. doi: 10.1159/000354527. [DOI] [PubMed] [Google Scholar]
- 8.Liu Z, Mai C, Yang H, et al. Candidate tumor suppressor CCDC19 regulates miR-184 direct targeting of C-Myc thereby suppressing cell growth in non-small cell lung cancers. J Cell Mol Med. 2014;18(8):1667–1679. doi: 10.1111/jcmm.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Q, Wang Y, Lu X, et al. miR-125b regulates epithelial-mesenchymal transition via targeting Sema4C in paclitaxel-resistant breast cancer cells. Oncotarget. 2015;6(5):3268–3279. doi: 10.18632/oncotarget.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong B, Hu H, Chen J, et al. Caprin-1 is a novel microRNA-223 target for regulating the proliferation and invasion of human breast cancer cells. Biomed Pharmacother. 2013;67(7):629–636. doi: 10.1016/j.biopha.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Raimondo M, Guha S, et al. Circulating microRNAs in pancreatic juice as candidate biomarkers of pancreatic cancer. J Cancer. 2014;5(8):696–705. doi: 10.7150/jca.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duan HF, Li XQ, Hu HY, et al. Functional elucidation of miR-494 in the tumorigenesis of nasopharyngeal carcinoma. Tumour Biol. 2015;36(9):6679–6689. doi: 10.1007/s13277-015-3356-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu J, He ML, Wang L, et al. miR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71(1):225–233. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- 14.Xu JQ, Zhang WB, Wan R, Yang YQ. MicroRNA-32 inhibits osteosarcoma cell proliferation and invasion by targeting Sox9. Tumour Biol. 2014;35(10):9847–9853. doi: 10.1007/s13277-014-2229-x. [DOI] [PubMed] [Google Scholar]
- 15.Ma W, Zhang X, Chai J, Chen P, Ren P, Gong M. Circulating miR-148a is a significant diagnostic and prognostic biomarker for patients with osteosarcoma. Tumour Biol. 2014;35(12):12467–12472. doi: 10.1007/s13277-014-2565-x. [DOI] [PubMed] [Google Scholar]
- 16.Wang G, Li B, Fu Y, et al. miR-23a suppresses proliferation of osteosarcoma cells by targeting SATB1. Tumour Biol. 2015;36(6):4715–4721. doi: 10.1007/s13277-015-3120-0. [DOI] [PubMed] [Google Scholar]
- 17.Wang NG, Wang DC, Tan BY, Wang F, Yuan ZN. Down-regulation of microRNA152 is associated with the diagnosis and prognosis of patients with osteosarcoma. Int J Clin Exp Pathol. 2015;8(8):9314–9319. [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Yang H, Tian Q, Liu Y, Weng Y. Upregulation of microRNA-17-92 cluster associates with tumor progression and prognosis in osteosarcoma. Neoplasma. 2014;61(4):453–460. doi: 10.4149/neo_2014_056. [DOI] [PubMed] [Google Scholar]
- 19.Chong Y, Zhang J, Guo X, et al. MicroRNA-503 acts as a tumor suppressor in osteosarcoma by targeting L1CAM. PLoS One. 2014;9(12):e114585. doi: 10.1371/journal.pone.0114585. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Chang TC, Yu D, Lee YS, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40(1):43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q, Tang Q, Qin D, et al. Role of microRNA 30a targeting insulin receptor substrate 2 in colorectal tumorigenesis. Mol Cell Biol. 2015;35(6):988–1000. doi: 10.1128/MCB.01242-14. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Visone R, Pallante P, Vecchione A, et al. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 2007;26(54):7590–7595. doi: 10.1038/sj.onc.1210564. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Zhang Y, Zhang Y, Ding J, Wu K, Fan D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59(5):579–585. doi: 10.1136/gut.2008.175497. [DOI] [PubMed] [Google Scholar]
- 24.Cheng CW, Wang HW, Chang CW, et al. MicroRNA-30a inhibits cell migration and invasion by downregulating vimentin expression and is a potential prognostic marker in breast cancer. Breast Cancer Res Treat. 2012;134(3):1081–1093. doi: 10.1007/s10549-012-2034-4. [DOI] [PubMed] [Google Scholar]
- 25.Yang C, Pan Y. Fluorouracil induces autophagy-related gastric carcinoma cell death through Beclin-1 upregulation by miR-30 suppression. Tumour Biol. 2015 Jul 25; doi: 10.1007/s13277-015-3775-6. Epub. [DOI] [PubMed] [Google Scholar]
- 26.Kumarswamy R, Mudduluru G, Ceppi P, et al. MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. Int J Cancer. 2012;130(9):2044–2053. doi: 10.1002/ijc.26218. [DOI] [PubMed] [Google Scholar]
- 27.Yu H, Sun H, Bai Y, et al. MEF2D overexpression contributes to the progression of osteosarcoma. Gene. 2015;563(2):130–135. doi: 10.1016/j.gene.2015.03.046. [DOI] [PubMed] [Google Scholar]
- 28.Han K, Zhao T, Chen X, et al. microRNA-194 suppresses osteosarcoma cell proliferation and metastasis in vitro and in vivo by targeting CDH2 and IGF1R. Int J Oncol. 2014;45(4):1437–1449. doi: 10.3892/ijo.2014.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2(2):329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 30.Pon JR, Marra MA. MEF2 transcription factors: developmental regulators and emerging cancer genes. Oncotarget. 2016;7(3):2297–2312. doi: 10.18632/oncotarget.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma L, Liu J, Liu L, et al. Overexpression of the transcription factor MEF2D in hepatocellular carcinoma sustains malignant character by suppressing G2-M transition genes. Cancer Res. 2014;74(5):1452–1462. doi: 10.1158/0008-5472.CAN-13-2171. [DOI] [PubMed] [Google Scholar]
- 32.Song L, Li D, Zhao Y, et al. miR-218 suppressed the growth of lung carcinoma by reducing MEF2D expression. Tumour Biol. 2015 Sep 26; doi: 10.1007/s13277-015-4038-2. Epub. [DOI] [PubMed] [Google Scholar]
- 33.Zhang M, Truscott J, Davie J. Loss of MEF2D expression inhibits differentiation and contributes to oncogenesis in rhabdomyosarcoma cells. Mol Cancer. 2013;12(1):150. doi: 10.1186/1476-4598-12-150. [DOI] [PMC free article] [PubMed] [Google Scholar]