Abstract
Despite widespread use of the Bacille Calmette-Guérin (BCG) vaccine, tuberculosis (TB) remains a leading cause of global mortality from a single infectious agent (Mycobacterium tuberculosis or Mtb). Here, over two independent Mtb challenge studies, we demonstrate that subcutaneous vaccination of rhesus macaques (RM) with Rhesus Cytomegalovirus vectors encoding Mtb antigen (Ag) inserts (RhCMV/TB), which elicit and maintain highly effector-differentiated, circulating and tissue-resident Mtb-specific CD4+ and CD8+ memory T cell responses, can reduce the overall (pulmonary and extra-pulmonary) extent of Mtb infection and disease by 68% compared to unvaccinated controls after intra-bronchial Erdman strain Mtb challenge, with challenge occurring at ~1 year after first vaccination. Fourteen of 34 RhCMV/TB-vaccinated RM (41%) across both studies showed no TB disease by computed tomography (CT) scan or at necropsy after challenge (compared with 0 of 17 unvaccinated controls) and 10 of these RM were Mtb culture-negative in all tissues, an exceptional long-term vaccine effect in the Erdman strain Mtb RM challenge model. These results suggest that complete vaccine-mediated immune control of highly pathogenic Mtb is possible if immune effector responses can intercept Mtb infection at its earliest stages.
INTRODUCTION
The natural immune response to Mycobacterium tuberculosis (Mtb), characterized pathologically by necrotizing granulomatous inflammation, limits the progression of infection in most subjects, but at the same time provides a haven for pathogen persistence and promotes host-to-host transmission1–4. An emerging consensus in the field suggests that over many thousands of years of evolution as an obligate human pathogen, Mtb has developed the ability to manipulate the human immune response so as to achieve the optimal balance between pathogenicity and immunity that provides maximal penetrance in the human population1,2,5–7. Natural Mtb immunity would therefore include a mixture of protective and anti-protective components, the latter of which would contribute to Mtb persistence and transmission. In keeping with this hypothesis, the Mycobacterium bovis-derived BCG vaccine, which enhances natural adaptive anti-mycobacterial immunity, is not completely efficacious. BCG protects vaccinated individuals, particularly children, from severe miliary TB, but fails to substantially reduce the incidence of pulmonary TB in adolescents and adults, a requirement for vaccine-mediated control of the global TB epidemic4,8,9. A corollary of this hypothesis is that for a vaccine to have sufficient potency to prevent pulmonary TB and thereby impact the TB epidemic, it will likely need to possess the following characteristics: 1) the ability to elicit effective, durable anti-Mtb immune responses that lack the components of natural Mtb immunity that are advantageous to the microbe, and 2) the ability to control/eliminate any nascent Mtb infection prior to the microbe’s establishment of an immune environment conducive to persistence (likely including mature granulomas), and/or be potent enough to overcome any Mtb-associated immunoregulation contributing to persistence. Studies in the mouse TB model have shown that accelerating effector cell production and delivery to lung in primary Mtb infection improves outcome10. Consistent with this concept of enhanced Mtb control with earlier effector cell delivery, vaccination of nonhuman primates (NHP) via aerosol delivery of either BCG or an attenuated Mtb vaccine, which potentiates both innate and adaptive cellular immune responses in the lung, has been shown to enhance protection against Mtb challenge11–13. Heterologous viral vaccine vectors would presumably have advantages over mycobacteria-based vaccines as these agents would likely lack the anti-protective immunoregulatory manipulation of mycobacteria, yet would potentially have an ability to elicit Mtb-specific CD4+ and CD8+ T cell responses with relevant functional profiles (for example, the ability to synthesize effector cytokines such as IFN-γ and TNF which are associated with Mtb control1,2,9). To date, however, TB vaccines based on poxvirus and adenovirus vectors have shown only modest enhancement of Mtb control in NHP models, and a Phase 2b trial of a Modified Vaccinia Ankara/Ag85A vaccine failed to show clinical benefit14–18.
The reason for the limited success of poxvirus- and adenovirus-based vectors as TB vaccines is unknown, but we hypothesize that these particular viral vectors simply failed to elicit, maintain and/or deliver sufficient numbers of functionally appropriate, effector-differentiated, Mtb-specific T cells at relevant tissue sites such as the bronchial mucosa, alveolar tissue and lung-draining lymph nodes (LNs) to effectively control Mtb challenge. In this regard, we and others have previously shown that Cytomegalovirus (CMV) is unique among microbial agents that infect humans and NHP in its ability to elicit and maintain robust, life-long, circulating and tissue-resident, effector-differentiated CD4+ and CD8+ memory T cell responses19,20. Indeed, CMV-specific CD4+ and CD8+ T cells constitute an average of 10% of the entire circulating memory T cell population in CMV-infected people21. We also have previously demonstrated that RhCMV-based vectors can be exploited as a uniquely effective “effector memory” vaccine against simian immunodeficiency virus (SIV) in RM22–24, and have established that these vectors can elicit and maintain circulating and tissue-resident CD4+ and CD8+ effector-differentiated T cells (transitional effector memory T cells – TTrEM and fully differentiated effector memory T cells – TEM) to any exogenous Ag insert22–25, including those encoding Mtb proteins25. Since RhCMV vector-elicited TTrEM/TEM both circulate in blood and accumulate in lung, and can produce copious amounts of TNF and IFN-γ upon Ag recognition22–24, the RhCMV/TB vaccine provides a unique opportunity to experimentally test the hypothesis that such high frequency, preformed, in situ or rapidly recruited, Mtb-specific TNF-and IFN-γ-producing TTrEM/TEM would be able to abrogate or markedly reduce TB progression after highly pathogenic Mtb challenge.
RESULTS
Relative Immunogenicity of RhCMV/TB and BCG (Study 1)
RM are highly susceptible to Mtb infection (particularly the highly pathogenic Erdman strain), even with very low doses of challenge inoculum, and show very similar immunology and pathology to humans, with the exception that whereas most (~85–90%) human TB infections are non-progressive over the lifetime of the infected individual, the vast majority of Mtb-infected RM progress to disease following experimental exposure4,26–30. Mtb infection of RM therefore represents a close-to-human, yet extremely stringent, model for testing TB vaccine activity. Indeed, the protection afforded by intradermally administered (i.d.) BCG vaccine in this model is variable and modest8,13,16,31, allowing vaccine developers to target enhancement of protection over that of i.d. BCG in this model as the goal of any new TB vaccine concept.
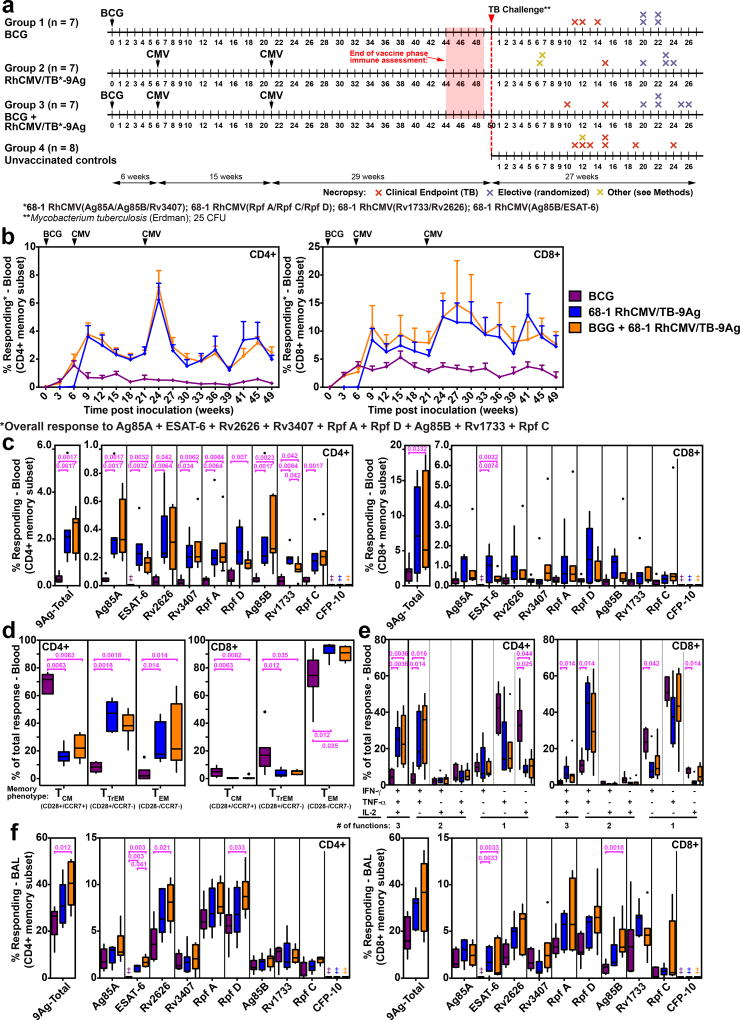
To test the hypothesis that vaccination with RhCMV/TB would elicit more effective immunity than i.d. BCG, and/or enhance the protection of i.d. BCG vaccination, we vaccinated 3 groups of RM (n=7/group; all naturally RhCMV+ at study assignment) with the following vaccines: 1) RhCMV/TB alone (subcutaneous administration of a set of 4 RhCMV vectors based on the 68-1 strain that together express 9 different Mtb proteins: ESAT-6, Ag85A, Ag85B, Rv3407, Rv1733, Rv2626, Rpf A, Rpf C, Rpf D; Supplementary Fig. 1a), 2) i.d. BCG alone, and 3) i.d. BCG followed (6 weeks later) by subcutaneous RhCMV/TB vaccination (Fig. 1a). As expected, the RhCMV/TB vaccine elicited and maintained high frequency CD4+ and CD8+ T cell responses in blood to all 9 Mtb inserts, as measured by overlapping 15mer peptide mix-induced expression of intracellular TNF and/or IFN-γ by flow cytometric intracellular cytokine (ICS) analysis (Fig. 1b–c). During the plateau phase, RhCMV vector-elicited Ag85A-specific responses were predominantly effector-differentiated, manifesting an almost exclusive TEM phenotype for CD8+ T cell responses, and a mixed TTrEM and TEM phenotype for CD4+ T cell responses (Fig. 1d). Approximately half of the RhCMV/TB-elicited Ag85A-specific CD4+ and CD8+ T cells responding in the ICS assays produced both TNF and IFN-γ (with or without IL-2), with the remainder predominantly producing TNF alone (Fig. 1e). BCG induced circulating CD4+ and CD8+ T cell responses to 8 of the 9 insert Ags (all except ESAT-6, which is not expressed by BCG32). In peripheral blood, the overall magnitude of the BCG-elicited T cell responses to these 8 Ags was considerably less than observed in RhCMV/TB-vaccinated RM (Fig. 1b,c). BCG-elicited Ag85A-specific responses predominantly exhibited a central memory (TCM) phenotype for the CD4+ T cells, and a TEM phenotype for the CD8+ T cells (Fig. 1d). The majority of these Ag85A-specific T cells produced either TNF or IL-2 alone (CD4+) or TNF or IFN-γ alone (CD8+), but not both TNF and IFN-γ (Fig. 1e). Notably, the BCG-induced CD4+ and CD8+ T cell response to the 8 BCG-expressed Mtb Ags was not large enough to significantly change the plateau-phase magnitude, phenotype, or function of the peripheral blood Mtb Ag-specific responses in the RM that received both BCG and RhCMV/TB relative to the RM that received RhCMV/TB vaccination alone (Fig. 1b–e). Differences in response magnitude between BCG- and RhCMV/TB-vaccinated RM were less apparent in bronchoalveolar lavage (BAL) fluid, with the responses in the latter group only modestly higher than in the former group (Fig. 1f). None of the 3 vaccinations used in Study 1 elicited significant antibody (Ab) responses to the 9 TB Ags in the RhCMV vector inserts (Supplementary Fig. 1b).
Figure 1. Immunogenicity of RhCMV/TB and i.d. BCG vaccines (Study 1).
(a) Schematic of the RM groups, (n=29 biologically independent animals), vaccination and challenge protocols, and necropsy time points of Study 1. (b) Longitudinal analysis of the overall CD4+ and CD8+ T cell response to the 9 Mtb insert proteins after vaccination. The background-subtracted frequencies of cells producing TNF and/or IFN-γ by flow cytometric ICS assay to peptide mixes comprising each of the Mtb proteins within the memory CD4+ or CD8+ T cell subsets were summed with the figure showing the mean (+ SEM; n=7 per group) of these overall (summed) responses at each time point. (c) Boxplots compare the individual Mtb protein (each of the 9 Mtb inserts plus the non-insert CFP-10)-specific and overall (summed) Mtb-specific CD4+ and CD8+ T cell response frequencies (defined by TNF and/or IFN-γ production) in peripheral blood between the same vaccine groups as in b at the end of the vaccine phase (each data point is the mean of response frequencies in 3 separate samples from weeks 44–49; ‡ indicates no response detected). (d) Boxplots compare the memory differentiation of the vaccine-elicited CD4+ and CD8+ memory T cells in peripheral blood responding to Ag85A with TNF and/or IFN-γ production at the end of vaccine phase (week 47). Memory differentiation state was based on CD28 vs. CCR7 expression, delineating central memory (TCM), transitional effector memory (TTREM), and effector memory (TEM), as designated. (e) Boxplots compare the frequency of vaccine-elicited CD4+ and CD8+ memory T cells in peripheral blood responding to Ag85A with TNF, IFN-γ and IL-2 production, alone and in all combinations at the end of vaccine phase (week 49). (f) Boxplots compare the individual Mtb protein-specific and overall (summed) Mtb-specific CD4+ and CD8+ T cell response frequencies (defined by TNF and/or IFN-γ production) in bronchoalveolar lavage (BAL) fluid between the vaccine groups at the end of the vaccine phase (weeks 46–47). In c–f, plots show a box from 1st to 3rd quartiles (IQR) and a line at the median, with whiskers extending to the farthest data point within 1.5*IQR above and below the box respectively; all data points outside of the whiskers are plotted individually; the Kruskal-Wallis (KW) test was used to determine the significance of differences between vaccine groups with the Wilcoxon rank sum test used to perform pair-wise analysis if KW p-values were ≤0.05; brackets indicate pair-wise comparisons with Holm-adjusted, two-sided Wilcoxon p-values ≤0.05 (with actual p-values shown; n=7 per group).
Effect of BCG, RhCMV/TB, and BCG + RhCMV/TB vaccination on Mtb challenge (Study 1)
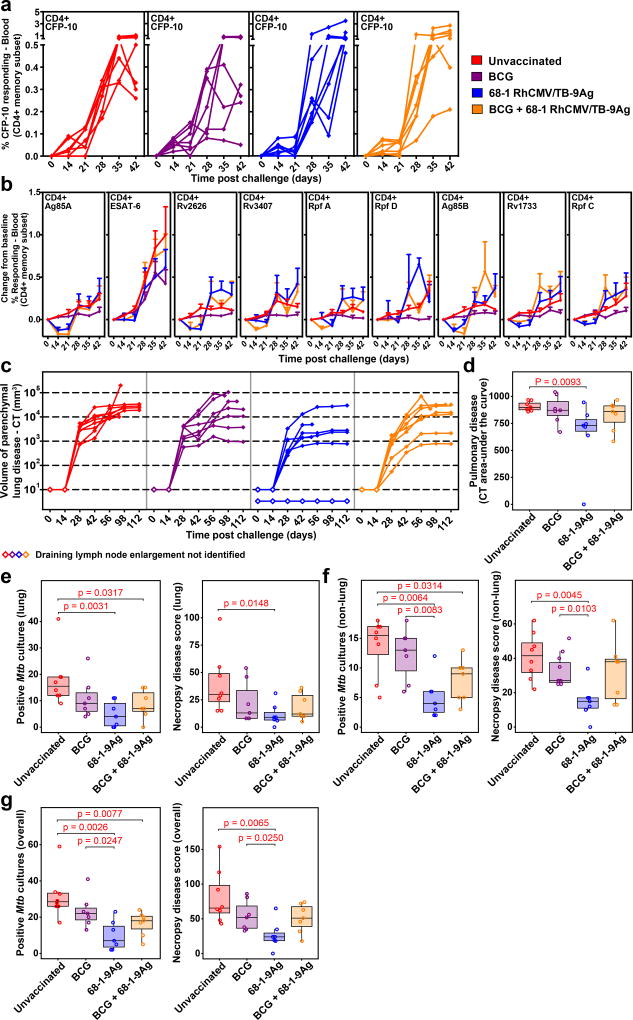
Fifty weeks after initial vaccination, the 3 groups of vaccinated RM and a control group of unvaccinated RM (n=8; all naturally RhCMV+) were challenged by intrabronchial instillation of 25 colony-forming units (CFUs) of Erdman strain Mtb bacteria into the right lower lobe. The effectiveness of challenge was confirmed by de novo development of CD4+ and CD8+ T cell responses to the CFP-10 Ag (Fig. 2a, Supplementary Fig. 2a; note that the Mtb-expressed CFP-10 Ag was not included in the RhCMV/TB vectors, and, like ESAT-6, is not expressed by BCG32). The unvaccinated RM manifested de novo CD4+ and CD8+ T cell responses to all 9 TB vaccine-insert Ags, including strong responses to ESAT-6 (Fig. 2b; Supplementary Fig. 2b). Only a very modest increase in the magnitude of CD4+ and CD8+ T cell responses to the TB vaccine-insert Ags was detected after Mtb challenge in vaccinated RM from all 3 vaccine groups (shown as the change in %responding from baseline; Fig. 2b; Supplementary Fig. 2b). The observation that the magnitude of this change-from-baseline after challenge in the vaccinated RM was not significantly different from the magnitude of the de novo responses to challenge in unvaccinated RM suggests that none of the vaccines used in this study were able to prime for an anamnestic Mtb-specific T cell response (i.e., a secondary, systemic immune response of substantially greater magnitude than the primary response). Initially, we planned to monitor post-challenge pulmonary immune responses by BAL fluid analysis, but BAL was discontinued when 2 RM were euthanized and taken to necropsy due to acute cardiopulmonary complications associated with the BAL procedure (see Methods). The development of pulmonary disease after challenge was monitored every two weeks by CT scan assessment of lesion volume, and both pulmonary and extra-pulmonary disease was determined at necropsy, with necropsy targeted to be performed either at clinical endpoint, or after 20 weeks post-infection (pi), by randomization (Fig. 1a). The extent of Mtb infection and TB disease at necropsy was quantified by 1) detailed pathologic scoring (based on comprehensive gross and microscopic pathologic examination; see Supplementary Fig. 3 and Methods), 2) by the frequency of recovery of culturable Mtb from a standard set of tissue samples, including 40 lung punch biopsies (randomly sampled using stereology to standardize sample collection across all study RM), as well as representative samples of trachea, all lung-draining and other chest LNs (hilar, carinal, paratracheal, mediastinal), selected peripheral LNs (axillary, inguinal, mesenteric) and other selected organs (spleen, liver, kidney, pancreas), and 3) by the amount of culturable Mtb in lung-draining LNs (CFU/gm), a nearly universal site of Mtb infection in RM with progressive TB disease26.
Figure 2. Outcome of Mtb challenge (Study 1).
(a) Development of peripheral blood CD4+ T cell responses to the peptide mixes comprising the non-vaccine insert Mtb protein CFP-10 after Mtb challenge by flow cytometric ICS analysis (response defined by TNF and/or IFN-γ production after background subtraction in memory subset; CFP-10-specific CD8+ T cell responses shown in Supplementary Fig. 2a). (b) Mean (+ SEM; n=7 per vaccine group; n=6 unvaccinated RM) change in peripheral blood CD4+ T cell response frequencies to peptide mixes comprising the vaccine insert Mtb proteins after Mtb challenge from pre-challenge baseline by flow cytometric ICS analysis (response defined as in a; analogous CD8+ T cell responses shown in Supplementary Fig. 2b). There were no significant differences in post-challenge change-from-baseline response dynamics between unvaccinated RM and any of the vaccine groups (see Methods). (c) CT quantification of disease volume in the pulmonary parenchyma after Mtb challenge (presence or absence of draining LN enlargement indicated by closed vs. open symbols). (d) Boxplots compare the AUC of CT-determined pulmonary lesional volume (day 0–112; see Methods) of the 4 RM groups. (e–g) Boxplots compare the extent of TB at necropsy measured by Mtb recovery with mycobacterial culture and by pathologic disease score (see Methods) in lung parenchyma (e), all non-lung parenchymal tissues (f) and all tissues (g). In c–g, n=7 per vaccine group and n=8 unvaccinated RM; in d–g, plots show jittered points with a box from 1st to 3rd quartiles (IQR) and a line at the median, with whiskers extending to the farthest data point within 1.5*IQR above and below the box respectively; unadjusted two-sided Wilcoxon p-values ≤0.05 are shown (see also Supplementary Fig. 4a).
Pulmonary disease developed rapidly in unvaccinated control RM with progression to severe (≥10,000 mm3 total lesional volume) lung parenchymal disease by CT scan in 7 of 8 RM by day 56pi and all RM by day 98pi (Fig. 2c). In both RM groups that received i.d. BCG, the development of pulmonary disease was more variable, but 5 of 7 RM in each group developed severe disease by day 98pi. In contrast, 5 of the 7 RM vaccinated with RhCMV/TB-alone developed only mild pulmonary disease (<3,000 mm3 total lesional volume, n=4) or no disease (n=1). The overall area-under-the-curve (AUC) of pulmonary lesion volume of the RhCMV/TB-alone group during the first 16 weeks pi (using imputed AUC values for monkeys taken to necropsy prior to week 16 pi, see Methods) was significantly reduced compared with the unvaccinated control group (Fig. 2d; Supplementary Fig. 4a), whereas there was no significant reduction in this parameter in either the i.d. BCG-alone or i.d. BCG + RhCMV/TB groups. Across all groups, the CT-determined lesional AUC through week 16 closely correlated with pulmonary parenchymal disease at necropsy as measured by both pathologic scoring and extent of Mtb recovery by culture (Supplementary Fig. 5a), and consistent with this, the extent of pulmonary TB at necropsy was also significantly reduced in the RhCMV/TB vector group compared to unvaccinated controls by both these necropsy measures (Fig. 2e; Supplementary Fig. 4a). No significant pulmonary disease reduction was observed in the i.d. BCG-only vaccinated cohort by either necropsy measure, and the combination i.d. BCG + RhCMV/TB vaccine regimen resulted in a significant reduction in Mtb burden, but not in pathologic score.
TB is typically not restricted to pulmonary parenchyma in RM infected with Erdman strain Mtb26, and necropsy analysis revealed extensive extra-pulmonary disease in the unvaccinated RM, including both lung-associated LN involvement and extra-thoracic spread (Supplementary Fig. 6). Importantly, the extent of overall disease as measured by pathologic score was also closely correlated with the recovery of viable Mtb in pulmonary and non-pulmonary parenchymal tissues (Supplementary Fig. 5b), and by both pathologic score and Mtb recovery criteria, extra-pulmonary and overall disease were significantly reduced in the RhCMV/TB-vaccinated cohort relative to unvaccinated RM (Fig. 2f,g; Supplementary Fig. 4a). We also found a >2 log reduction in the density of culturable Mtb (CFU/gm) in all lung-draining LNs in RhCMV/TB-vaccinated RM compared to unvaccinated control RM (P=0.0003; Supplementary Fig. 5e). By Poisson modeling, the overall (pulmonary + extra-pulmonary) extent of disease was reduced in the RhCMV/TB-vaccinated group by 68.7% by extent of Mtb recovery (P<0.0001) and 67.3% by pathologic score (P<0.0001) relative to unvaccinated controls (Supplementary Fig. 4b). In contrast, the extent of extra-pulmonary and overall disease in i.d. BCG-only vaccinated RM was not significantly different from unvaccinated RM by either criterion (Fig. 2f,g; Supplementary Fig. 4a). The density of viable mycobacteria in lung-draining LNs was also not different in i.d. BCG-vaccinated RM relative to unvaccinated controls (Supplementary Fig. 5e). In keeping with these results, the extent of extra-pulmonary and overall disease, as well as the density of viable Mtb in lung-draining LNs, in RM receiving RhCMV/TB alone, was also significantly reduced when directly compared to the i.d. BCG-only vaccinated RM (Fig. 2f,g; Supplementary Figs. 4a and 5e). Using the same Poisson modeling, RhCMV/TB vaccination reduced overall disease relative to i.d. BCG vaccination alone by 57.7% (P=0.0007) and 51.4% (P=0.01) for extent of Mtb recovery and pathologic score, respectively (Supplementary Fig. 4b). Vaccination of RM with i.d. BCG 6 weeks before the initial RhCMV/TB vaccination appeared to reduce both the pulmonary and extra-pulmonary protection mediated by the RhCMV/TB vaccination alone, as the i.d. BCG + RhCMV/TB group did not manifest a significant reduction in pathologic score relative to unvaccinated controls (Fig. 2e–g and Supplementary Fig. 4a). However, the extent of Mtb recovery in both pulmonary and extra-pulmonary tissues and the density of viable Mtb in lung-draining LNs was still significantly reduced in the RM receiving i.d. BCG + RhCMV/TB relative to unvaccinated controls (Fig. 2e–g; Supplementary Figs. 4a and 5e). Thus, RhCMV/TB-vaccinated RM manifested superior overall outcomes compared to RM vaccinated with i.d. BCG alone, and the i.d. administration of BCG 6 weeks prior to RhCMV/TB vaccination appeared to reduce, but not abolish, the protection afforded by the RhCMV/TB vaccine.
Immunogenicity and protective capacity of RhCMV/TB variants (Study 2)
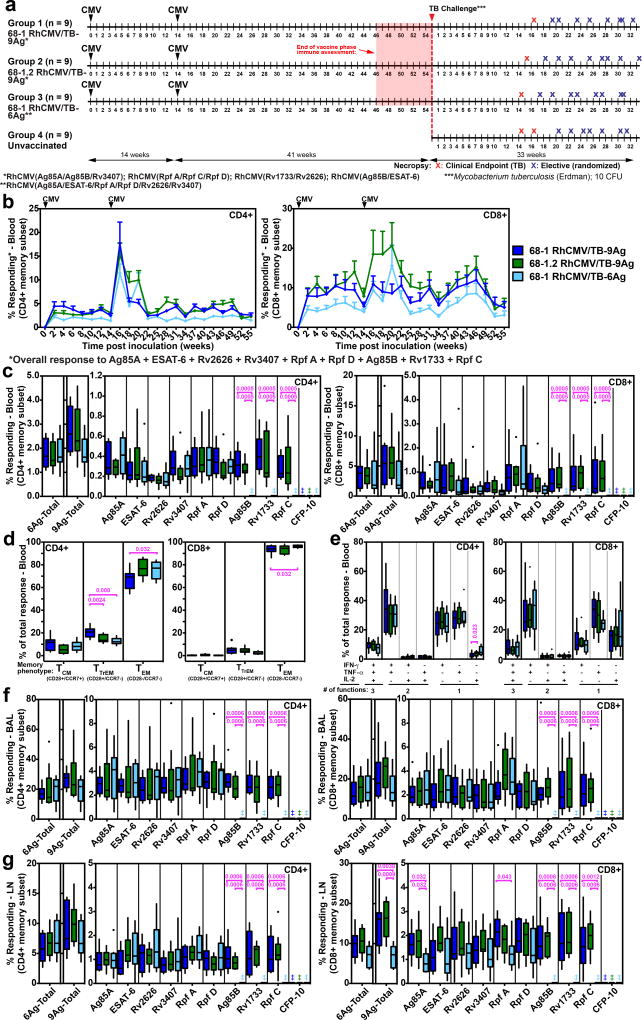
To confirm and further characterize the protective effect of RhCMV/TB vaccination, we performed a second larger Mtb challenge study (n=9/group; all RM naturally RhCMV+ at assignment) in which we compared the same 68-1 RhCMV/TB-9 Ag vector set used in Study 1 (group 1) with an analogous RhCMV/TB-9 Ag vector set based on the 68-1.2 RhCMV backbone (group 2), and with a 68-1 RhCMV/TB-6 Ag vector expressing a single 6 Ag Mtb polyprotein (Ag85A; ESAT-6; Rv3407; Rv2626; Rpf A; Rpf D) (group 3) (Fig. 3a; Supplementary Fig. 1). Strain 68-1 RhCMV vectors elicit unconventional CD8+ T cell responses that are restricted by MHC-II and MHC-E, whereas the Rh157.5/Rh157.4 gene-repaired 68-1.2 RhCMV vectors elicit CD8+ T cells that target conventionally MHC-Ia-restricted epitopes25,33. The comparison of group 1 to group 2 therefore allows determination of the contribution of unconventionally restricted CD8+ T cells to RhCMV/TB-mediated protection. In the group 1 vs. group 3 comparison, we sought to determine whether the vaccine effect observed in Study 1 with the 4 RhCMV/TB vector set encoding 9 Mtb Ags (3 each in the acute phase, latency and resuscitation Ag types) can be recapitulated by a single RhCMV/TB vector expressing 6 Mtb Ags (2 each from these Ag types) as a single polyprotein, a simpler vaccine configuration which is better suited for clinical translation. As expected, the magnitude of the overall Mtb-specific and individual Mtb insert-specific CD4+ and CD8+ T cell responses elicited by the 68-1 and 68-1.2 RhCMV/TB-9 Ag vectors were comparable in blood throughout the vaccination phase, and in BAL fluid and peripheral LN at the end of vaccination phase, as was the memory differentiation and functional phenotype of the Mtb-specific response in blood (Fig. 3b–g). However, also as expected25, the CD8+ T cells elicited by the 68-1 RhCMV/TB vaccine were unconventionally (MHC-II and MHC-E) restricted, whereas those elicited by 68-1.2 RhCMV/TB vaccine were conventionally (MHC-Ia) restricted (Supplementary Fig. 7). The T cell responses recognizing the 6 Ags common to both the 68-1 RhCMV/TB-6 Ag and RhCMV/TB-9 Ag vaccines were similar between groups 1 and 3 with respect to magnitude, phenotype and (unconventional) MHC restriction, except for slightly different levels of CD8+ T cell responses in peripheral LN (Fig. 3b–g; Supplementary Fig. 7). Thus, the 3 different RhCMV-based vaccines used in Study 2 generated CD4+ and CD8+ T cell responses to insert Ags that were of comparable magnitude, function and phenotype, but differed, as intended, in the epitope targeting and restriction of the CD8+ T cell responses (68-1 vs. 68-1.2), or in the number of Mtb Ags targeted (6 Ag vector vs. 9 Ag vector sets). As in Study 1, none of the 3 RhCMV/TB vaccines used in Study 2 elicited a specific Ab response to any of the TB Ags expressed by these vectors (Supplementary Fig. 1b).
Figure 3. Immunogenicity of RhCMV/TB vaccines (Study 2).
(a) Schematic of RM groups, vaccination and challenge protocols, and necropsy time points of Study 2 (n=36 biologically independent animals). (b) Longitudinal analysis of the overall CD4+ and CD8+ T cell response to the 9 Mtb Ags after vaccination, as described in Fig. 1b (n=9 per group). (c) Boxplots compare the individual Mtb protein (each of the 9 Mtb inserts plus the non-insert CFP-10)-specific and overall (summed) Mtb-specific CD4+ and CD8+ T cell response frequencies (defined by TNF and/or IFN-γ production) in peripheral blood between the vaccine groups at the end of the vaccine phase (each data point is the mean of response frequencies in 3 separate samples from weeks 49–55; ‡ indicates no response detected). (d) Boxplots compare the memory differentiation phenotypes (see Fig. 1d) of the vaccine-elicited CD4+ and CD8+ memory T cells in peripheral blood responding to Ag85A with TNF and/or IFN-γ production at the end of vaccine phase (weeks 51–52). (e) Boxplots compare the frequency of vaccine-elicited CD4+ and CD8+ memory T cells in peripheral blood responding to Ag85A with TNF, IFN-γ and/or IL-2, alone and in all combinations at the end of vaccine phase (weeks 49–50). (f,g) Boxplots compare the individual Mtb protein (the 9 Mtb inserts plus the non-insert CFP-10)-specific and overall (summed) Mtb-specific CD4+ and CD8+ T cell response frequencies (defined by TNF and/or IFN-γ production) in BAL (f) and in peripheral LN (g) between the vaccine groups at the end of vaccine phase (weeks 46–47). In c–g, plots show a box from 1st to 3rd quartiles (IQR) and a line at the median, with whiskers extending to the farthest data point within 1.5*IQR above and below the box respectively; all data points outside of the whiskers are plotted individually. Statistics were performed as described in Fig. 1 with brackets indicating pair-wise comparisons with Holm-adjusted, two-sided Wilcoxon p-values ≤0.05 (with actual p-values shown; n=9 per group).
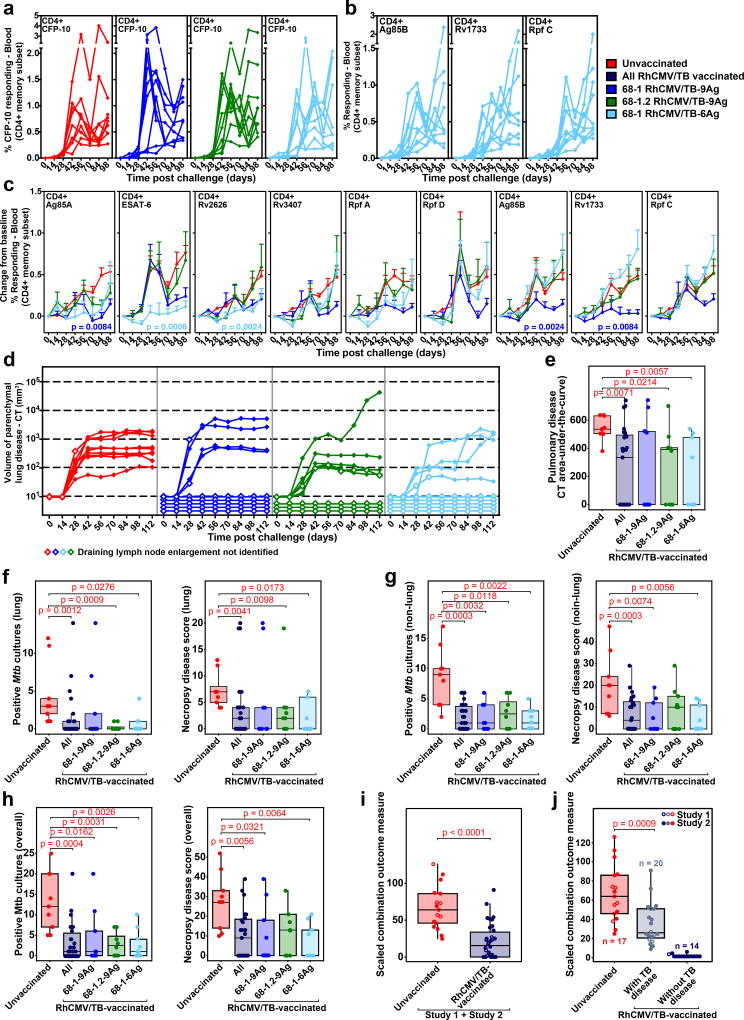
After a 56-week vaccination period, all vaccinated RM (groups 1–3) and unvaccinated control RM (group 4) were intrabronchially challenged with ~10 CFUs of Erdman strain Mtb bacteria – the reduction in dose relative to Study 1 was intended to slow TB progression in Study 2 RM to more closely resemble the course of human Mtb infection. In addition, BAL was not performed post-challenge in this experiment to prevent procedure-related mortality and the possibility of artificial enhancement of bacteria spread within the lung. All Study 2 RM developed de novo CFP-10-specific T cell responses in blood following challenge, and the RhCMV/TB-6 Ag-vaccinated RM (group 3) also developed de novo T cell responses in blood to the Ag85B, Rpf C, and Rv1733 Ags, which were not included in their vaccine (Fig. 4a,b; Supplementary Fig. 2c,d and 8). As observed in Study 1, the change-from-baseline in the magnitude of the post-challenge TB insert-specific CD4+ and CD8+ T cell responses in RhCMV/TB-vaccinated RM was less than or equal to the magnitude of the de novo responses to these Ags in the unvaccinated RM, confirming lack of a true anamnestic response in the RhCMV/TB-vaccinated animals (Fig. 4c; Supplementary Fig. 2e). All 9 unvaccinated (group 4) RM developed TB lesions on CT scans by day 28pi, but as anticipated, the disease progression in this study was slower and reached a lower plateau earlier than in Study 1 (Fig. 4d), with only 2 unvaccinated (group 4) control RM developing endpoint TB disease over the course of observation (Fig. 3a). Remarkably, 13 of the 27 vaccinated RM (5 RM each in group 1 and group 3 and 3 RM in group 2) did not develop any radiologic signs of pulmonary TB (including no CT-detectable hilar adenopathy) at any time point through to random elective necropsy at >16 weeks pi, and the average CT-determined lesional AUC in lung parenchyma of the overall cohort of vaccinated RM was significantly reduced compared to the unvaccinated group 4 (Fig. 4d,e; Supplementary Fig. 4c).
Figure 4. Outcome of Mtb challenge (Study 2 and Overall).
(a,b) Development of peripheral blood CD4+ T cell responses to the peptide mixes comprising the non-vaccine insert Mtb protein CFP-10 in all Study 2 RM (a), and comprising the Ag85B, Rv1733 and Rpf C proteins in group 3 RM only (b; RM vaccinated with the single 68-1 RhCMV/TB-6Ag vector lacking these 3 inserts) after Mtb challenge by flow cytometric ICS analysis, as described in Fig. 2a (peripheral blood CD8+ T cell responses and tissue CD4+ and CD8+ T cell responses to these same Ags are shown in Supplementary Figs. 2c,d and 8, respectively). (c) Mean (+ SEM; n=9 per group) change in peripheral blood CD4+ T cell response frequencies to peptide mixes comprising the vaccine insert Mtb proteins after Mtb challenge from pre-challenge baseline by flow cytometric ICS analysis, as described in Fig. 2b (analogous CD8+ T cell responses shown in Supplementary Fig. 2e). Holm-adjusted Wilcoxon p-values ≤0.05 are shown for change-from-baseline AUC values of post-challenge responses in any of the vaccinated groups versus the unvaccinated controls. (d) CT scan-based quantification of disease volume in the pulmonary parenchyma after Mtb challenge (presence or absence of draining LN enlargement indicated by closed and open symbols, respectively; n=9 per group). (e) Boxplots compare the AUC values of CT scan-determined pulmonary lesional volume (day 0–112) of the unvaccinated RM vs. all RhCMV/TB-vaccinated RM vs. RM in each individual RhCMV/TB vaccine group (n=9 per group). (f–h) Boxplots compare the extent of TB at necropsy measured by Mtb recovery with mycobacterial culture and by pathologic disease score in lung parenchyma (f), all non-lung parenchymal tissues (g) and all tissues (h) in the same RM groups as in e. (i,j) The boxplots and unadjusted two-sided Wilcoxon test p-values compare the outcome of Mtb challenge in all unvaccinated RM vs. all RhCMV/TB only-vaccinated RM across both Study 1 (n=8 vs. n=7) and Study 2 (n=9 vs. n=27) using a scaled outcome measure that combines both Mtb culture and pathologic score data (see Methods). In (j), the RhCMV/TB-vaccinated RM are divided into 2 groups based on presence (n=20) or absence (n=14) of granulomatous disease at necropsy (necropsy pathologic score ≥4 vs. =0, respectively) vs. n=17 unvaccinated RM. In e–j, plots show jittered points with a box from 1st to 3rd quartiles (IQR) and a line at the median, with whiskers extending to the farthest data point within 1.5*IQR above and below the box respectively (unadjusted, two-sided Wilcoxon p-values ≤0.05 are shown; see also Supplementary Fig. 4).
At necropsy, none of the 13 CT lesion-negative RM from vaccine groups 1–3 had any macroscopic granulomatous disease, and 10 of these 13 were culture-negative in all tissue samples tested (the remaining 3 were Mtb+ in lung-draining LNs only; Supplementary Figs. 5c,d and 9). Moreover, despite the development of CD4+ and CD8+ T cell responses to Mtb proteins not present within their vaccine in lung, lung-draining and peripheral LNs, and spleen (Supplementary Fig. 8), extensive histopathologic analysis of lung and lung-draining LNs from these 13 TB disease-free RM showed no microscopic granulomatous inflammation. As in Study 1, the pathologic scores at necropsy (same as Study 1) and the extent of Mtb recovery (same as Study 1 except for expansion of analysis to retropharyngeal, submandibular, iliosacral LNs and tonsils) were again strongly correlated (Supplementary Fig. 5d). The overall extent of disease by both measures was significantly reduced in the overall (pooled groups 1–3) RhCMV/TB-vaccinated group compared to the unvaccinated control group, with no significant difference in protection between individual groups 1–3, and similar protection in both pulmonary and extra-pulmonary tissues (Fig. 4f–h; Supplementary Figs. 4c and 9). For the pooled vaccinated group, the overall reduction in disease extent relative to the unvaccinated control group was 74.5% by Mtb culture (P=0.0024) and 61.4% by pathologic score (P=0.0011) using Poisson modeling (Supplementary Fig. 4d).
To assess the effect of RhCMV/TB vaccination (given alone) on Mtb challenge outcome, relative to no vaccination across Study 1 and Study 2, we used a scaled (normalized) combination outcome parameter based on both Mtb culture and pathologic score (see Methods). As shown in Fig. 4i, the difference in overall (Study 1 + 2) outcome between unvaccinated and RhCMV/TB (only)-vaccinated RM was highly significant, corresponding to a 68% reduction in this combined measure of Mtb infection and disease in the vaccinated RM by Poisson modeling (Supplementary Fig. 4e). Of note, despite the fact that the average extent of TB progression in the unvaccinated control RM was quite different in Studies 1 and 2, the relative reduction in infection and disease associated with RhCMV/TB vaccination was similar for each study (Supplementary Fig. 4). Across both Study 1 and 2, 14 of 34 RhCMV/TB (only)-vaccinated RM (41%; 95% confidence interval: 26%-58%) manifested no detectable granulomatous disease at necropsy (vs. 0 of 17 unvaccinated controls; P=0.0018), and 10 of these 14 were culture-negative for Mtb (Fig. 4j; Supplementary Figs. 6 and 9). Importantly, the extent of disease in the remaining RhCMV/TB (only) vaccinees (e.g., excluding the RhCMV/TB vaccinated RM with no detectable disease and including only those vaccinated RM with detectable disease at necropsy; n = 20) was still significantly reduced compared with the unvaccinated controls (41.5% reduction by Poisson modeling; Fig. 4j and Supplementary Fig. 4e), indicating that in contrast to the “all or none” protection of the RhCMV/SIV vaccine against SIV challenge23,24, the vaccine effect of the RhCMV/TB vaccine includes both complete and partial protection. Of note, 32 of the 34 RM vaccinated with RhCMV/TB alone (94%), manifested an extent of disease at necropsy less than the median value in the unvaccinated controls, and 24 of the 34 vaccinated RM (71%) manifested an extent of disease at or below the lowest level of disease extent in the unvaccinated controls (Fig. 4j).
Innate response to Mtb challenge is modified in RhCMV/TB protected RM
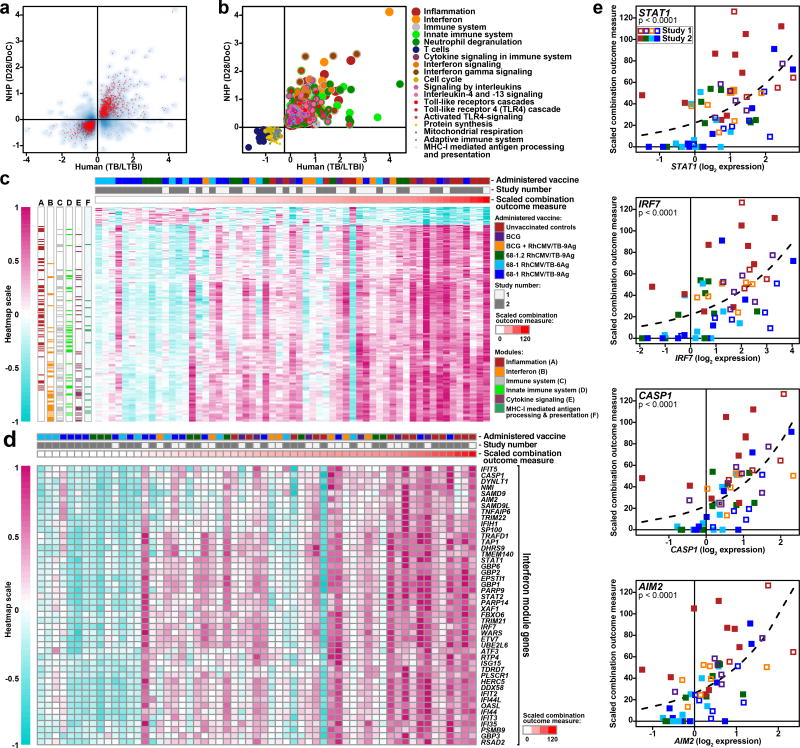
Development of active TB after Mtb infection in humans is associated with profound changes in the expression of immune-related and inflammatory genes in whole blood34–37. To determine whether our study RM manifested analogous transcriptional changes after Mtb challenge, and whether vaccine-associated protection modified this signature, we performed RNA-sequencing (RNA-Seq) transcriptome analysis of whole blood RNA collected immediately prior to, and 28 days after Mtb challenge. We first determined whether genes that were significantly up- or down-regulated in active TB patients compared to controls without active TB in two African cohorts37 were similarly regulated in the blood of the unvaccinated RM after challenge. As shown in Fig. 5a,b, the unvaccinated control RM in our study displayed a whole blood transcriptional response to Mtb infection that was highly congruent with the response in humans, involving similar and robust up- and down-regulation of a common set of genes in immune-associated modules and pathways. We next tested whether the magnitude of induction of this whole blood transcriptional signature at 28 days Mtb post-challenge was associated with outcome at necropsy across all Study 1 and Study 2 RM, as measured by the scaled combination outcome parameter described above. As shown in Fig. 5c–e, a large number of genes that were highly enriched for the interferon transcriptional module exhibited expression patterns that strongly correlated with extent of disease of Study 1 and 2 RM at necropsy. These interferon response-associated genes include members of a signature associated with progression to active TB disease in humans34, classical antiviral responses (e.g., IRF7, IFIH1, DDX58, RSAD2) and inflammasome components (AIM2 and STAT1). Notably, this TB disease-associated transcriptional signature was profoundly reduced in the vast majority of RhCMV/TB-vaccinated RM with complete Mtb control (e.g., no detectable granulomatous disease at necropsy), indicating that in these RM, protective vaccine-elicited immune responses countered the spread of Mtb early enough after challenge to ameliorate the potent generalized effect of Mtb infection on innate immune signaling.
Figure 5. Transcriptional response to Mtb challenge reduced in protected RM.
(a) The median longitudinal log2 gene expression fold-change between 28 days after challenge and the day of challenge in unvaccinated RM from Study 1 and 2 (n=13; y-axis) is plotted against the average median log2 expression difference computed between HIV-negative active TB patients and latent TB infection controls from South Africa (TB: n=46, LTBI: n=48) and Malawi (TB: n=51, LTBI: n=35) (x axis) for 3406 significantly regulated genes identified in the human study37. Genes significantly regulated in Mtb-challenged, unvaccinated RM are shown in red (one-sided, paired Wilcoxon rank sum test, p<0.05; FDR<0.33; complete list of p-values shown in Supplementary Table 1). The overall Spearman rank correlation coefficient between expression fold-changes in the human study and RM for all 3406 genes is 0.58. (b) Commonly up-regulated (n=793) or down-regulated (n=689) genes in humans with TB disease and unvaccinated Study 1 + 2 RM (n=13) 28 days after Mtb infection from panel a were tested for significant over-representation of transcriptional modules53 and immune pathways (Reactome)54 (p<0.002, FDR<0.01;one-sided Fisher’s Exact Test), which were then color-coded according to significance and pathway membership. Enrichment p-values ranged from p<10−40 for inflammation and interferon, to p=2×10−3 for Class I MHC (complete list of p-values shown in Supplementary Table 2). (c) The heatmap depicts scaled expression fold-changes for all 214 genes (rows) for all Study 1 and 2 RM (columns; n=59) ordered by scaled combination outcome measure (red bar at top). Post-challenge expression fold-changes for 214/1482 commonly regulated genes in panel a were strongly associated with the scaled combination outcome measure as indicated by Poisson modeling (p<0.05, FDR<0.1; complete list of p-values shown in Supplementary Table 3). Genes with fold-changes positively associated with outcome are significantly enriched for the interferon transcriptional module (FDR=3×10−27) and other pathways (Supplementary Table 4). Day 28/Day of challenge log2 fold-changes were scaled by the maximum absolute log2 fold-change observed. Annotations of genes to significantly over-represented pathways are shown in colored boxes at the left side of the heatmap. (d) Heatmap as in panel c showing expression profiles of 46 genes from the interferon transcriptional module. (e) Scatterplots depicting scaled combination outcome measure (y axis) as a function of Day 28/Day of challenge log2 fold-changes for representative genes from the interferon transcriptional module. The dashed line indicates the best-fit Poisson model. P-values indicate the significance of the regression coefficient in a sandwich-adjusted Poisson model (n=59) for the association between gene expression and the scaled combination outcome measure.
Correlates of protection
Given the effector memory nature of RhCMV/TB vaccine-elicited T cell responses, and the lack of anamnestic T cell or relevant Ab responses, the variable level of protection observed in RhCMV/TB vaccinated RM (ranging from complete to none) would likely reflect a composite integration of the frequency and function of RhCMV/TB vaccine-elicited T cells, as well as that of innate immune effectors, immediately after challenge, at the local sites of early Mtb replication in lung and lung-draining LNs. Since direct immunologic interrogation of all these sites is not possible, we asked whether analysis of accessible, surrogate sites would provide insight into the immunologic differences among RhCMV/TB vaccinated RM (with or without i.d. BCG) that determine subsequent control or non-control of Mtb challenge. In this regard, the outcome heterogeneity among all RhCMV/TB-vaccinated RM in Study 1 and 2, as measured by the scaled combination outcome parameter described in Fig. 4, was not predicted by the ICS-determined magnitudes of the RhCMV/TB-elicited, TNF/IFN-γ-defined, CD4+ and/or CD8+ T cell responses in blood or BAL fluid (or Study 2 only, peripheral LN) immediately prior to challenge (Supplementary Fig. 10a–c). In addition, in Study 2, there were no significant differences in the magnitudes of the AUC values of the post-challenge Mtb Ag-specific CD4+ and CD8+ T cell responses between the 13 RhCMV/TB-vaccinated RM that were completely protected after challenge and the 14 RhCMV/TB-vaccinated RM with TB disease (Supplementary Fig. 10d,e), except for reduced ESAT-6- and CFP-10-specific CD4+ T cell responses in the protected group (likely reflecting lower antigenic loads of these two related acute-phase Ags in the protected RM after challenge).
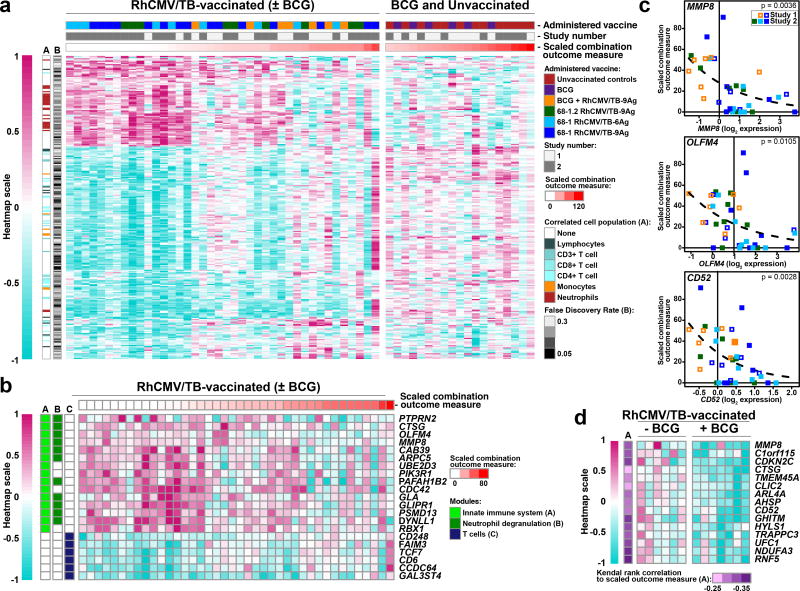
These data suggest that the overall Mtb-specific T cell responses measured in these surrogate sites may not accurately reflect the relevant memory T cell responses in lung tissue and lung-draining LN and/or that the measured responses are not the only, or not the major, limiting immunologic factors that determine outcome. To more globally determine whether other peripheral readouts of immune and inflammatory state are associated with differential outcomes after Mtb challenge in RhCMV/TB-vaccinated RM, we analyzed the pre-challenge whole blood transcriptomes to identify gene expression associations with the scaled combination outcome measure. This analysis revealed a set of 280 genes with pre-challenge expression profiles that were significantly associated with eventual outcome (p<0.05, FDR<0.33), out of which 258 (92%; Fig. 6a) exhibited no significant pre-challenge association with outcome for unvaccinated and i.d. BCG-vaccinated RM – suggesting that these signatures are associated with outcome only in the context of RhCMV/TB vaccination. Transcriptional module and pathway enrichment analysis revealed significant (p<0.05, FDR<0.15) over-representation of neutrophil degranulation and innate immunity functions amongst genes that were expressed higher in RM that would ultimately be protected (“protection signature”), whereas genes that were expressed higher in RM that would ultimately develop severe disease (“susceptibility signature”) were significantly enriched for the T cell transcriptional module (Fig. 6b,c). Although these enrichment analysis results raise the possibility that relative abundances of specific leukocyte populations prior to Mtb challenge would be associated with eventual outcome, no significant associations between outcome and leukocyte population counts were identified. Nevertheless, genes significantly correlated with neutrophil counts were over-represented within the protection signature, whereas genes significantly correlated with T cell counts (and to a lesser extent, monocytes) were over-represented within the susceptibility signature (Fig. 6a), consistent with the module and pathway analysis results. Visual inspection of the heatmap for the protection and susceptibility signatures (Fig 6a) suggested that the association with protection was predominantly binary, with strongly protected RM exhibiting a distinct expression pattern compared to other RM. Logistic regression modeling of the data supported this interpretation, with 38/77 (49%) and 70/181 (39%) of protection and susceptibility genes, respectively, demonstrating reasonable potential to discriminate the 14 protected RM from the remaining RhCMV/TB-vaccinated RM. Since the i.d. BCG + RhCMV/TB-vaccinated group from Study 1 exhibited a trend towards impaired protection compared to the RhCMV/TB-only vaccinated group alone (despite having similar RhCMV/TB-elicited T cell responses; Fig. 1b–f), we assessed whether expression of the protection signature was impaired in Study 1 i.d. BCG + RhCMV/TB RM compared to RhCMV/TB-only vaccinated RM on the day of challenge (Fig. 6d). This analysis revealed that a subset of protection signature genes, including MMP8, CTSG, and CD52, were suppressed in the i.d. BCG + RhCMV/TB-vaccinated group relative to the RhCMV/TB-only vaccinated group, suggesting that BCG-driven modulation of inflammation may have countered a protective innate immune response in RhCMV/TB vaccinated RM, and thereby contributed to the reduced protection observed with the combined vaccine regimen.
Figure 6. Pre-challenge transcriptional profiles correlate with post-challenge outcome in RhCMV/TB-vaccinated RM.
(a) Heatmap visualization of the relative gene expression (log2 expression fold-change) of genes expressed in the whole blood immediately prior to challenge of RhCMV/TB-vaccinated RM (with or without i.d. BCG) from Study 1 + 2 that are significantly associated with the scaled combination outcome measure by Poisson modeling (p<0.05, FDR<0.33; FDR values are shown in the heatmap and all p-values are shown in Supplementary Table 5; n=40 RM), and are not associated with outcome for the unvaccinated and i.d. BCG-vaccinated RM (p>0.2; n=19 RM). The left and right sides of the heatmap show expression of the genes in RhCMV/TB-vaccinated vs. unvaccinated or i.d. BCG-vaccinated RM, respectively. The heatmap also delineates whether the expression of each gene correlates with the absolute counts of any of the designated leukocyte populations in the same blood sample (if more than one population correlates, the strongest association is shown; complete results are shown in Supplementary Table 7). Note that genes positively associated with eventual disease are significantly enriched for genes associated with T cell counts (FDR<0.05; one-sided Fisher’s Exact Test; p-values values provided in Supplementary Table 8), whereas genes negatively associated with eventual disease (positively correlated with RhCMV/TB-mediated protection) are significantly enriched for genes associated with neutrophil counts (FDR=3×10−3). (b) The testing of genes in panel a (protection signature: n=77 genes; susceptibility signature: n=181 genes) for over-representation of transcriptional modules and pathways revealed three significant results (p<0.05, FDR<0.15, one-sided Fisher’s Exact Test), indicated by the colored bars on the left of the heatmap (p-values provided in Supplementary Table 6). The heatmap shows the relative expression of specific genes (same scale as in panel a) with pre-challenge associations with the scaled combination outcome measure that belong to the significantly enriched modules and pathways. (c) Scatterplots depicting scaled combination outcome measure (y-axis) as a function of pre-challenge expression of representative protection-associated genes from the neutrophil degranulation module (MMP8 and OLFM4, the latter expressed by a neutrophil subset55), as well as CD52, a broadly expressed gene that is also expressed by neutrophils56. The dashed line indicates the best-fit Poisson model (The p-values shown indicate the sandwich-adjusted Poisson p-values for the association between pre-challenge gene expression and the scaled combination outcome measure; n=40 RM). (d) Heatmap visualization of 15 genes from panel a with protection-associated pre-challenge expression levels which exhibit significantly reduced expression on the day of challenge (one-sided Wilcoxon tests p<0.05, FDR<0.33) in RM vaccinated with i.d. BCG prior to RhCMV/TB vaccination (n=7) compared to RM vaccinated with RhCMV/TB only (n=6) from Study 1. See Methods for details of data preparation for visualization.
DISCUSSION
In this report, we demonstrate that subcutaneously administered RhCMV-based TB vaccines are able to elicit and maintain immune effector responses that can control Mtb at the earliest stages of infection, and that the protection afforded by this vaccine can be complete, if not sterilizing. To our knowledge, this is the first demonstration of complete prevention of TB disease in a substantial fraction of RM by a peripherally administered, long-acting vaccine after highly pathogenic (Erdman strain) Mtb challenge. Although Kaushal, et al.12 have reported impressive protection in RM against a less pathogenic Mtb strain (CDC1551) with high dose, aerosol “vaccination” of a live, attenuated mycobacterium-based TB vaccine (MtbΔSigH), in their studies, Mtb challenge occurred only 8 weeks after aerosol vaccine administration, a time when their replicating MtbΔSigH would still be present in lungs of vaccinated RM and both innate and adaptive immunity would presumably be at or near peak levels. In contrast, the potent RhCMV/TB vaccine-mediated protection demonstrated here against low-dose, but highly pathogenic, Erdman strain Mtb challenge occurred as long as 55 weeks following initial vaccination, and 40 weeks following boost of the subcutaneously administered vaccine, consistent with establishment of long-term protection.
The working hypothesis upon which CMV vaccine vector development is based relates to the ability of these persistent vectors to elicit and maintain effector-differentiated (“effector memory”), insert-specific CD4+ and CD8+ T cell responses in mucosal effector sites and secondary lymphoid tissues, as well as circulating in the blood, pre-positioned to intercept mucosally-acquired pathogens at the very onset of infection. While it is difficult to definitively establish mechanism-of-action in a vaccine protection study, the available data are consistent with the operation of this “effector memory” hypothesis in the RhCMV/TB vaccine-mediated control of Mtb infection. In keeping with previous work on RhCMV/SIV vaccines22–24, RhCMV/TB vectors elicit robust circulating and tissue-based CD4+ and CD8+ T cell responses (but little to no insert-specific Ab responses), with both the CD4+ and CD8+ T cell responses in blood being highly effector memory-biased (TTREM + TEM for CD4+; TEM for CD8+). Although there are other semi-innate and innate cell types in the lung and lung-draining LNs that could potentially participate in protection against Mtb38, the effector-differentiated, Mtb-specific CD4+ and CD8+ T cells are essentially the only adaptive immune responses detected in RhCMV/TB-vaccinated RM. Moreover, protection appeared very early in many RM – before the onset of detectable pathology and early enough to reduce and even abrogate the potent interferon-driven innate immune response to Mtb infection. This protection also occurred in the absence of a true anamnestic response, consistent with the concept of protection being mediated by established effectors already “in place” or rapidly recruited at the time of challenge. It is also noteworthy that the most pronounced adaptive immune difference between RhCMV/TB-elicited and the (non-protective) i.d. BCG-elicited T cell responses was in the Mtb-specific CD4+ T cell responses prior to challenge, in particular their differentiation and functional potential, with the i.d. BCG vaccine eliciting predominantly monofunctional, central memory responses vs. polyfunctional (TNF- + IFN-γ-producing) effector-differentiated (TEM + TTREM) responses by the RhCMV/TB vaccine. With respect to Mtb-specific CD8+ T cell responses, the protection afforded by 68-1.2 RhCMV/TB vaccination indicates the unconventional MHC-II and MHC-E-restricted CD8+ T cells are not required for protection, and suggests that either conventional or unconventional CD8+ T cell responses can contribute to protection, or that protection is independent of CD8+ T cells altogether.
The observation that outcome heterogeneity among RhCMV/TB vaccinated RM was not predicted by the pre-challenge magnitude of RhCMV/TB-elicited, TNF and/or IFN-γ-defined, CD4+ or CD8+ T cell responses in blood, BAL fluid or LN does not refute the “effector memory” hypothesis, as these sampling sites and the ICS assay read-out may not accurately reflect the frequency and function of resident memory responses in lung and lung-draining LN that are likely the relevant population for earliest immune control of infection and thus for protection39. In contrast to T cell responses that either continuously recirculate (central memory responses) or have a required blood-borne component (classic anamnestic memory responses), effector memory responses are regulated locally, are not well represented by blood measurements, are difficult to characterize ex vivo, and are not uniform across tissue compartments40,41. It should also be noted that a correlation between pre-challenge Mtb insert-specific T cell response magnitude and subsequent protection would be expected only if this T cell response magnitude was the major limiting component of the protective immune response. Since CD4+ and CD8+ T cells are not thought to directly kill Mtb, but rather to work through innate immune (primarily phagocytic) effectors, it is possible that a required innate immune co-effector component is the limiting factor that primarily determines whether an individual RhCMV/TB vaccinee will be completely protected, partially protected, or not at all protected after challenge. In this regard, the gene expression levels identified in pre-challenge transcriptomic analysis of whole blood as predictive of the RhCMV/TB vaccine effect preferentially included genes expressed by neutrophils and associated with innate immunity and neutrophil degranulation gene pathways, including genes that encode neutrophil granule effector molecules (MMP8, CTSG) (Fig. 6). This protection signature is somewhat surprising since in overt TB disease, neutrophils have been primarily associated with TB pathogenesis, mediating tissue destruction, necrosis, and cavitation, and providing a pro-Mtb milieu, but not effective Mtb killing42–45. However, some studies have demonstrated an ability of neutrophils to kill, growth-restrict, or trap Mtb (preventing its spread), raising the question of whether the role of the neutrophil in TB immunity and pathogenesis is both timing and context dependent7,46–49. Indeed, one study demonstrated an inverse correlation between neutrophil counts and the risk of TB infection in contacts of patients diagnosed with pulmonary TB47, consistent with an early protective role for this cell population.
Our demonstration that neutrophil gene expression patterns, but not neutrophil counts themselves, predict outcome in RhCMV/TB-vaccinated RM, but not in unvaccinated and i.d. BCG-only vaccinated RM suggests that neutrophils might be important co-effectors for RhCMV/TB vaccine-elicited T cell responses, and that their differentiation or activation state determines, at least in part, their ability to contribute to RhCMV/TB vaccine-mediated protection. Interestingly, it appears that the protection-associated transcriptomic signature in neutrophils primarily delineates RhCMV/TB-vaccinated RM destined for complete or nearly complete protection from those with partial or no protection (Fig. 6), suggesting that the contribution of “primed” neutrophils to protection occurs very early after challenge and wanes thereafter. The extent to which this protective neutrophil differentiation/activation state is related to RhCMV/TB vaccination, monkey genetics, or immune events at the time of challenge remains to be determined, but it is notable that i.d. BCG vaccination 6 weeks before RhCMV/TB vaccination appears to deleteriously modify the expression of several of the protection-associated genes (Fig. 6), suggesting that innate immune modulation might account for the apparent anti-protective effect of giving i.d. BCG before RhCMV/TB vaccination. Additional studies to determine the effect of remote (neonatal) BCG vaccination on RhCMV/TB vector-mediated protection in adolescent and adult RMs will be required to clarify the potential clinical significance of this putative antagonistic effect in at risk human populations who have previously received the BCG vaccine at birth.
In summary, the data presented here demonstrate that RhCMV-based vaccines manifest significant protection against aggressive Erdman-strain TB in highly susceptible RM. Although delineation of the specific immunologic mechanisms responsible for protection in RhCMV/TB-vaccinated individuals will require additional study, the nature of RhCMV/TB vector immunogenicity and the dynamics of protection after challenge suggests that protection is based on both the ability of this persistent viral vector to elicit and maintain Mtb-specific effector-differentiated resident memory T cell responses, and on a conducive innate immune state at the time of Mtb challenge that might also be vaccine-related50,51. Importantly, it has been documented in natural CMV infection that the robust TTrEM/TEM responses elicited by this virus are maintained for life19,20. To our knowledge, CMV is unique in this ability, and recent data indicates that strategically attenuated CMV vectors, potentially safe enough for clinical translation, maintain this unique immunogenicity (Hansen, Früh and Picker, unpublished data). How the above described putative interaction between CMV vaccine-elicited TTrEM/TEM and BCG-/Mtb-elicited immune regulation will play out over clinically relevant timeframes in people remains to be determined. However, the ability of RhCMV/TB vaccination to completely prevent development of TB disease in more than 40% of vaccinated RM and to provide a nearly 70% overall vaccine effect offers promise that a human CMV/TB vaccine might be effective in preventing pulmonary TB in adolescents and adults52, and thereby contribute to ending the global TB epidemic.
METHODS
Rhesus Macaques
Sixty-five purpose-bred, pedigreed, male RM (Macaca mulatta) of Indian genetic background were used in the described studies. At assignment, these RM were specific-pathogen free (SPF) as defined by being free of Macacine herpesvirus 1, D-type simian retrovirus, simian T-lymphotrophic virus type 1, simian immunodeficiency virus, and Mtb, but were all naturally RhCMV-infected. All RM used in this study were housed at the Oregon National Primate Research Center (ONPRC) in Animal Biosafety level (ABSL)-2 (vaccine phase) and ABSL-3 rooms (challenge phase) with autonomously controlled temperature, humidity, and lighting. RM were single cage-housed due to the infectious nature of the study and had visual, auditory and olfactory contact with other animals. Because the RM were single cage-housed, an enhanced enrichment plan was designed and overseen by nonhuman primate behavior specialists. RM were fed commercially prepared primate chow twice daily and received supplemental fresh fruit or vegetables daily. Fresh, potable water was provided via automatic water systems. All RM were observed twice daily to assess appetite, attitude, activity level, hydration status and evidence of disease (tachypnea, dyspnea, coughing). Physical exams including body weight and complete blood counts were performed at all protocol time points. RM care and all experimental protocols and procedures were approved by the ONPRC Institutional Animal Care and Use Committee (IUCUC). The ONPRC is a Category I facility. The Laboratory Animal Care and Use Program at the ONPRC is fully accredited by the American Association for Accreditation of Laboratory Animal Care (AAALAC), and has an approved Assurance (#A3304-01) for the care and use of animals on file with the NIH Office for Protection from Research Risks. The IACUC adheres to national guidelines established in the Animal Welfare Act (7 U.S.C. Sections 2131–2159) and the Guide for the Care and Use of Laboratory Animals (8th Edition) as mandated by the U.S. Public Health Service Policy.
Animal Procedures
RM were sedated with ketamine HCl or Telazol® for intradermal and subcutaneous vaccine administration, venipuncture, bronchoalveolar lavage, LN biopsy, intrabronchial Mtb inoculation and computed tomography (CT) procedures. Mtb Erdman (kindly provided by Dr. J. Flynn, U. Pittsburgh) was diluted in saline, lightly sonicated and bacteria were delivered to a segmental bronchus in the right caudal lung lobe using a bronchoscope. The RM in Studies 1 and 2 received 25 and 10 colony forming units (CFU), respectively, in a volume of 1 ml57. Pre- and post-challenge axial CT scans (2.5 mm slices) were obtained using a multi-section CT scanner using helical technique, collimation 3 mm and pitch 1.5 (CereTom, Neurologica Corp., Danvers, MA) and reconstructed as 1.25 mm slices to improve detection sensitivity58,59. Nonionic iodinated contrast (Isovue 370, 1–2 ml/kg, Bracco Diagnostics, Princeton, NJ) was administered IV at a rate of 1–2 ml/s. CT scans were obtained with 120 kVp and 200 mA. All RM were imaged pre-challenge, at two-week intervals for the duration of the studies and immediately prior to necropsy. Scans were interpreted by a veterinarian who was blinded to the study group of the subject. Lesion area in sequential scans was determined from transverse slices through entire lung fields using the IMPAX 6.5.5.3020 software area tool (AGFA HealthCare N.V., Mortsel, Belgium) and lesion volume determined by multiplying area by 1.25. One RM developed extensive bilateral miliary disease with estimated lesion volume >200,000 mm3 and no further attempts to estimate lesion volume were made in this animal.
Necropsy
The humane criteria for removing RM with end-stage TB from the studies are as follows: 1) marked lethargy, 2) severe dyspnea at rest and/or failure to maintain adequate oxygenation (85%) based on pulse oximetry or blood gas analysis, 3) hemoptysis, 4) weight loss (>15% in 2 weeks; >25% over any time course in an adult animal), 5) hypothermia <96°F with supplemental heating, 6) persistent anemia (<20% for 2 weeks), 7) dehydration unresponsive to oral rehydration therapy for 3 days, 8) non-responsiveness to therapy for spontaneous disease conditions, 9) poor appetite, requiring more than 3 orogastric tube feedings in 7 days, 10) obtundation, 11) neurologic deficits, and 12) persistent self-injurious behavior unresponsive to a change in location or enrichment. RM that manifested one or more of these end-stage criteria were immediately euthanized and taken to necropsy. RM that remained clinically well after post-infection week 16 were randomized and scheduled for euthanasia and necropsy at the rate of two per week. There were 3 exceptions to this general rule of necropsy initiated by end-stage disease criteria or randomization (3 necropsies that are designated as “other” in Fig. 1a). One non-end-stage RM (D1) in Study 1 was euthanized on the same day as an end-stage RM because the IACUC does not permit housing a single RM alone in a room. Two additional RM (N1, N3) in Study 1 were euthanized because of failure to maintain adequate oxygenation following a bronchoalveolar lavage procedure that was not attributable to end-stage Mtb disease. To avoid this issue in Study 2, BALs were not performed after challenge.
At the humane or scheduled endpoint RM were euthanized with sodium pentobarbital overdose (>50 mg/kg) and exsanguinated via the distal aorta. The necropsy procedure included complete gross pathologic evaluation of abdominal organs and tissues and the brain prior to entering the thoracic cavity to avoid contamination. Macroscopic granulomas in liver, spleen, kidney and the brain were counted, measured and photographed in serial 5 mm tissue slices whereas granulomas occurring in extra-thoracic LNs, the gastrointestinal tract and soft tissues were collected, measured and photographed with minimum sectioning and given a numeric point value score using a semi-quantitative grading system (Supplementary Fig. 3). Granulomas (≤10) occurring in these tissues were bisected and one half collected in sterile media tubes for Mtb culture (see below) and the remaining half immersed in 10% neutral-buffered formalin for histologic analysis. Representative granulomas were selected for Mtb culture and histology from tissues with >10 granulomas. Single small granulomas (≤1 mm) were utilized entirely for bacteriology. If no granulomas were identified, representative samples of selected tissues (see below) were submitted for Mtb culture. Representative samples of all abdominal and mediastinal organs and associated major LN groups, and both the brain and spinal cord were collected and then fixed in 10% neutral-buffered formalin for histologic analysis. Additional samples from these tissues were collected for mononuclear isolation. The pleura and thoracic wall were examined on entering the thoracic cavity and macroscopic granulomas and adhesions were collected, counted, measured, photographed, scored as described (Supplementary Fig. 3) and sampled for bacteriology and histology. The thoracic viscera were removed en bloc, and then transferred to a sterile cutting board for examination and dissection. Extreme care was taken to avoid Mtb contamination of thoracic tissues by spillage of granuloma contents during dissection, and this was accomplished in all but 1 RM in which gross spillage of granuloma contents was observed upon opening the thoracic cavity. The heart was removed and examined, and pulmonary and mediastinal LNs and individual lung lobes were dissected free, weighed and photographed. LNs were divided into samples for Mtb culture, histopathology, and mononuclear cell isolation (with Mtb culture prioritized if tissue was limiting). Macroscopic granulomas in individual lung lobes were counted, measured, photographed and scored in serial 5 mm tissue slices. Samples for Mtb culture from the right and left lung slices were harvested using a nonbiased, stereologic sampling method60. Briefly, an opaque plastic 96 hole strip-tube rack was placed over the adjacent lung slices and individual 5 mm lung tissue cores (30 from the right lung and 10 from the left lung) were collected through these the openings in the rack (irrespective of any gross pathology) in a consistent pattern across all study monkeys. These stereologically-obtained (essentially random) lung cores were bisected and one-half processed for bacteriology and the remaining half immersed in 10% neutral-buffered formalin for histologic analysis. In addition, as indicated above, extensive representative samples of all lung lobes and lung-draining LN, all abdominal and mediastinal organs/tissues and all major non-lung-draining LN groups were collected and immersed in 10% neutral-buffered formalin for histologic analysis. Tissues for histologic analysis were routinely processed and embedded in paraffin. Sections (6 mm) were stained with hematoxylin and eosin. All told, between 100 and 120 tissue blocks were examined by a veterinary pathologist for each monkey necropsy, ~50 of which focused on the lung, trachea/major bronchi, and lung draining LNs, and the remainder all the other non-pulmonary tissues. Selected tissues were stained by the Ziehl-Neelsen method for acid-fast bacteria.
Vaccines
The 68-1 and 68-1.2 RhCMV/TB vectors were constructed by bacterial artificial chromosome (BAC) recombineering and were reconstituted and amplified into vector preparations as previously described22–24,33. The Mtb Ags to be included in these vectors were selected by a bioinformatics selection criteria starting with the scoring of 4000 Mtb open reading frames (ORF) by 11 criteria61. Those criteria included immunogenicity, vaccine efficacy, expression in granulomas, secretion, and role in hypoxic survival. The top candidates were then further screened by a deeper bioinformatics analysis of predicted T cell epitopes and curated to include antigens that are active during different stages of TB infection. In addition, all antigens had been shown to be at least partially protective in a mouse challenge model. The final choice of 9 Mtb proteins for the vaccine inserts included 3 representative proteins from so-called acute phase (85A, 85B, ESAT-6), latency (Rv1733, Rv3407, Rv2626) and resuscitation (Rpf A, Rpf C and Rpf D) classes of Mtb Ags. These 9 Ags were expressed in 4 different RhCMV/TB vectors (to be used in combination) for both the 68-1 and 68-1.2 backbones, as follows: 1) Ag85A/Ag85B/Rv3407 (GenBank #KY611401), 2) Rv1733/Rv2626 (GenBank #KY611402), 3) Rpf A/Rpf C/Rpf D (GenBank #KY611403), and 4) Ag85B/ESAT-6 (GenBank #KY611404). The GenBank Accession # corresponds to the sequences as they are found in the final vectors. Polyprotein #s 1–3 were inserted into the nonessential Rh211 ORF under the control of the murine CMV IE promotor. Polyprotein #4 was inserted in the same region of the RhCMV genome but under the control of the EF1a promotor (Supplementary Fig. 1a). For the single 6 Ag-expressing, 68-1 RhCMV/TB vector, a single polyprotein insert consisting of 2 Ags from each of 3 classes described above (acute: ESAT-6, Ag85A; latency: Rv3407, Rv2626; resuscitation: RpfA, RpfD; GenBank #KY611405) was used to replace the nonessential Rh107 gene, placing its expression under the control of the endogenous Rh107 promoter (Supplementary Fig. 1a). All BACs were analyzed by restriction digestion to confirm genomic integrity and were further examined by next generation sequencing (NGS) on an Illumina MiSeq sequencer to ensure the absence of any unintended mutations in the transgene. Full genome sequences of viral DNA isolated from vaccine stocks used in the presented study [68-1 RhCMV/85A/85B/Rv3407 (MF468139); 68-1.2 RhCMV/85A/85B/Rv3407 (MF468141); 68-1.2 RhCMV/85B/ESAT-6 (MF468142); 68-1.2 RhCMV/Rpf A/Rpf C/Rpf D (MF468143); 68-1.2 RhCMV/Rv1733c/Rv2626c (MF468144); 68-1 RhCMV/Rpf A/Rpf C/Rpf D (MF468145) and 68-1 RhCMV/Rv1733c/Rv2626c (MF468146)] or full genome sequences of the BAC of the mutant used to derive the vaccine stocks [68-1 RhCMV/85B/ESAT-6 (MF468140) and 68-1 ΔRh107 RhCMV/TB-6Ag (MF468147)] were submitted to GenBank. To reconstitute the vaccine vectors, the BACs were electroporated into telomerized or primary rhesus fibroblasts and kept in culture until full cytopathic effect was achieved. At this point, transgene expression was confirmed by immunoblot of infected cell lysates and vaccine stocks were generated by the ONPRC Molecular Virology Support Core. Overall genomic integrity and transgene expression of the final vaccine stocks were confirmed by immunoblots, NGS, and pilot immunogenicity studies in RM. Expression of the ORFs neighboring the TB Ag insertion site was confirmed by RT-PCR. RhCMV/TB vector stocks were titered using primary rhesus fibroblasts in a TCID50 assay. Study 1 and 2 RM were vaccinated by subcutaneous administration of 5 × 106 pfu of each of the designated RhCMV/TB vectors. For the RM receiving the 4 vector set, each vector was administered in a separate limb (right arm, left arm, right leg, left leg). RM were given the test vaccines twice, with the second dose (homologous boost) administered 15 weeks (Study 1) or 14 weeks (Study 2) after the first dose. The BCG vaccine (Danish strain 1331; Batch #111005A) was obtained from the Statens Serum Institute (Copenhagen, Denmark) and was reconstituted per the manufacturer’s instructions (Diluent Batch #386587B). RM were BCG-vaccinated by the intradermal administration of 100 µl of vaccine containing 5.5 × 105 CFUs into the mid-back.
Immunologic Assays
Mtb-specific CD4+ and CD8+ T cell responses were measured in blood, BAL fluid and tissues by flow cytometric ICS, as previously described in detail22,25,33. Briefly, mononuclear cell preparations from blood, BAL fluid or tissue were incubated at 37°C in a humidified 5% CO2 atmosphere with overlapping, consecutive 15mer peptide mixes (11 amino acid overlap) comprising these proteins, or individual 15mer peptides from these proteins, and the co-stimulatory molecules CD28 and CD49d (eBioscience) for 1 hour, followed by addition of Brefeldin A (Sigma-Aldrich) for an additional 8 hrs. Co-stimulation without antigenic peptides served as a background control. As previously described25, the MHC restriction (MHC-Ia, MHC-E, MHC-II) of a peptide-specific response was determined by pre-incubating isolated mononuclear cells for 1 hr at room temperature (prior to adding peptides and incubating per the standard ICS assay) with the following blockers: 1) the pan anti-MHC-I mAb W6/32 (10mg/ml), 2) the MHC-II-blocking CLIP peptide (MHC-II-associated invariant chain, amino acids 89–100; 20µM), and 3) the MHC-E-blocking VL9 peptide (VMAPRTLLL; 20µM). Blocking reagents were not washed, but remain throughout the assay. Following incubation, stimulated cells were fixed, permeabilized and stained as previously described22,25,33 using combinations of the following fluorochrome-conjugated mAbs: SP34-2 (CD3; Pacific Blue, Alexa700), L200 (CD4; AmCyan, BV510), SK-1 (CD8α; PerCP-Cy5.5), MAB11 (TNF-α; FITC, PE), B27 (IFN-γ; APC), FN50 (CD69; PE, PE-TexasRed), B56 (Ki-67; FITC), and in polycytokine analyses, JES6-5H4 (IL-2; PE, PE Cy-7). To determine the cell surface phenotype of Mtb-specific CD4+ and CD8+ T cells, mononuclear cells were stimulated as described above, except that the CD28 co-stimulatory mAb was used as a fluorochrome conjugate to allow CD28 expression levels to be later assessed by flow cytometry, and in these experiments, cells were surface-stained after incubation for lineage markers CD3, CD4, CD8, CD95 and CCR7 (see below for mAb clones) prior to fixation/permeabilization and then intracellular staining for response markers (CD69, IFN-γ, TNF-α; note that Brefeldin A treatment preserves the pre-stimulation cell-surface expression phenotype of phenotypic markers examined in this study). Data was collected on an LSR-II (BD Biosciences). Analysis was performed using FlowJo software (Tree Star). In all analyses, gating on the lymphocyte population was followed by the separation of the CD3+ T cell subset and progressive gating on CD4+ and CD8+ T cell subsets. Antigen-responding cells in both CD4+ and CD8+ T cell populations were determined by their intracellular expression of CD69 and one or more cytokines (either or both of the IFN-γ and TNF; ± IL-2 in polycytokine analyses). After subtracting background, the raw response frequencies above the assay limit of detection were “memory-corrected” (e.g., % responding out of the memory population), as previously described22,23 using combinations of the following fluorochrome-conjugated mAbs to define the memory vs. naïve subsets SP34-2 (CD3; Alexa700, PerCP-Cy5.5), L200 (CD4; AmCyan), SK-1 (CD8a; APC, PerCP-cy-5.5), MAB11 (TNF-α; FITC), B27 (IFN-γ; APC), FN50 (CD69; PE), CD28.2 (CD28; PE-TexasRed), DX2 (CD95; PE), 15053 (CCR7; Pacific Blue), and B56 (Ki-67; FITC). As previously described21, assay background is determined by measurement of the net response to the Ags of interest in a cohort of monkeys that have never been exposed to these Ags. We, conservatively, use 4 standard deviations above the mean of these background values as our cut-off value or 0.05%, whichever is higher (responses below this cut-off are considered negative or response = 0). For this study, the net responses to the 10 Mtb Ags in blood in all the quantiferon-negative RM pre-vaccination, was determined to be 0.004% ± 0.004% for CD4+ T cells (cut-off = 0.02%) and 0.003% ± 0.004 for CD8+ T cell responses (cut-off = 0.02%). For BAL, the mean background + SD was determined to be 0.004% ± 0.008% for both CD4+ and CD8+ T cells (cut-off = 0.036%). Since these cut-off levels were all <0.05%, we used our conservative default cut-off of 0.05% for all responses. For memory phenotype and polycytokine analysis of Mtb Ag-specific T cells, all cells expressing CD69 plus one or more cytokines were first Boolean gated, and then this overall Ag-responding population was subdivided into the subsets of interest on the basis of surface phenotype or cytokine production pattern22,23.
A customized Luminex subclass assay63 was used to quantify the relative concentration of each Immunoglobulin (Ig) isotype among Mtb antigen-specific Abs. Carboxylated microspheres (Luminex) were coupled with Mtb protein Ags by covalent NHS-ester linkages via the cross-linker EDC and NHS (Thermo Scientific) per the manufacturer’s instructions. Ag-coated microspheres (5000 per well) were added to a 96-well filter plate (Millipore). Each Ig sample (5 µg) was added to five replicate wells of a 96-well plate and incubated for 16 hrs at 4°C. The microspheres were washed and the Ig isotype-specific detection reagents were added (Southern Biotech) for 2 hr at room temperature. The beads were then washed and read on a Bio-Plex 200 System. The background signal, defined as mean fluorescent intensity (MFI) of microspheres incubated with PBS, was subtracted. Each sample was tested twice.
Mycobacterial (Mtb) Culture
Tissues routinely collected at necropsy for Mtb burden analysis in both Study 1 and 2 included: 30 stereologic punches from right lung lobes, 10 punches from left lung lobes, trachea, left hilar LN, right hilar LN, left carinal LN, right carinal LN, paratracheal LN, mediastinal LN, axillary LN, inguinal LN, mesenteric LN, spleen, pancreas, left medial lobe of liver, right medial lobe of liver, left lateral lobe of liver, right lateral lobe of liver, liver caudate, left kidney, and right kidney. In Study 2 we also collected and cultured retropharyngeal LN, tonsil, submandibular LN, and iliosacral LN. In one RM in Study 2, Mtb culture analysis was not reported due to gross contamination of thoracic tissues with granuloma contents. Tissues were collected in HBSS (Hyclone, SH30031.3) and were then homogenized in an IKA grinder tube with an IKA Ultra-Turrax Tube Drive homogenizer. The tissue homogenate was then filtered over a 70µm wire screen to remove debris and 200µl of this material was plated neat and in serial dilutions (1/10, 1/100) on 7H11 agar plates (Remel). All plates were incubated at 37°C and M. tuberculosis growth was enumerated 28 and 42 days later. A tissue was considered Mtb+ if any colonies with the correct morphologic features were identified. Selected cultures were analyzed by the Ziehl-Neelsen method for acid-fast bacteria to confirm colony morphology. Bacterial density in lung-draining LNs was calculated in CFU per gram of tissue.
Data Preparation (Immunology and Vaccine-Mediated Protection)
Area under the curve of log(CT-scan-determined pulmonary disease volume)
We computed the AUC of the log-transformed CT scan-determined pulmonary disease volume measurements from time 0 (set to 0) to day 112. Next, we imputed missing values for monkeys taken to necropsy before the full series of scheduled CT scan time points. We computed the AUC of this augmented data. The imputation procedure that we employed used linear regression to estimate missing values from previous time points. As a sensitivity analysis, we also imputed the missing values using a more conservative rule (replacing missing values with the largest non-missing value at the same time point among monkeys receiving the same treatment, excluding for further conservatism one high-valued outlier unvaccinated RM); the resulting AUCs were highly insensitive to this procedure (Pearson correlation over 0.99 for both studies).
Necropsy score data
The non-negative count-valued necropsy scores are amenable to Poisson modeling. Model evaluations supported inclusion of the additional parameter for overdispersion in the negative binomial model. In TB Study 2 the estimated extra parameter was zero, so this model was equivalent to a simple Poisson model. For this Poisson modeling we employed sandwich-based estimates of variance-covariance matrices as an alternative method to account for overdispersion, using the vcovHC function in the sandwich package in R62–64. We also considered and rejected employing the zero-inflated Poisson (ZIP) model.
Necropsy culture data
Necropsy culture inputs were quantitative measures of culture growth with multiple replicates per tissue. We treated these data as binary indicators of a culture being positive vs. negative (zero), and evaluated the total number of positive cultures. Model evaluations of necropsy culture outcome data favored the more expressive negative binomial models over Poisson models, and did not support using the ZIP model. Note that one animal in TB Study 2 (I3; see Supplementary Fig. 9) was missing necropsy culture data but did have necropsy score data; the analyses of the culture data therefore excluded this RM. Thus, for the left panels of Fig. 4f–h (“Positive Mtb cultures”), the sample size for the 68-1.2-9Ag (green-colored) group is n = 8. As described below, we imputed the missing value for use in computing the scaled (normalized) combination outcome measure.
Scaled (normalized) combination outcome measure
As shown in Supplementary Fig. 5, we found strong correlation between the necropsy score and necropsy culture outcome measures within each study, although the scales were different: necropsy cultures are about one-third as large as necropsy scores in both studies. We created the intermediate, study-specific scaled combination outcome measure to gain measurement precision by averaging these two very similar outcome measures. To ensure that each receives equal weight in the combined measure, and to maintain discreteness in support of Poisson analysis of the statistic, we first scaled the inputs by multiplying the necropsy culture values by 3 and then adding these to the necropsy scores. For Study 2, as indicated above, one RM (I3) had a missing necropsy culture value, so for this animal we computed the scaled combination outcome measure using an imputed necropsy culture value, which we obtained by multiplying the observed necropsy score value by the estimated coefficient from a simple linear regression model relating the two values. This monkey’s necropsy score value was 29, its imputed necropsy culture value was 8, and its scaled combination outcome measure value was 53. We employed a negative binomial regression model of these study-specific combined measures versus treatment and study, and used the estimated coefficient on study (0.3796) to further scale the TB Study 1 scaled combination outcome values. Therefore, the scaled combination outcome variable for TB Study 1 RM is the study-specific value multiplied by 0.3796 and then rounded to maintain the discreteness of the final variable for Poisson analysis.
Challenge phase LN cell-forming units per gram
We computed the mean of log10-transformed measures of CFU per gram across five LN tissue samples (carinal left and right, and hilar left and right; paratracheal). The few missing values were first imputed using the mean value for the LN.
End-of-vaccine-phase T cell response data
We also evaluated longitudinal flow cytometric ICS measures of CD4+ and CD8+ immune responses targeting the 9 individual Mtb proteins in the RhCMV vaccines, and CFP-10. Our primary summary of these data was a measure of the immune response at the end of vaccine phase. These pre-challenge baseline immunogenicity values are geometric means of three independent measurements over the time periods shown in Figs. 1a and 3a. Totals over 6 or 9 antigens were computed prior to log-transformation and normalization. Normalization shifts and scales these values to have mean zero, standard deviation 1 so that units have the interpretation of z-scores measuring the number of standard deviations that an immune measurement (on the log scale) is from the overall (study-specific) mean of that measurement.
Change-from-baseline T cell responses after Mtb challenge
We computed areas under the curve of time-varying differences from the pre-challenge baseline immunogenicity values for each of the 9 insert antigens over 42 or 98 days post-challenge (for TB Studies 1 and 2, respectively), on the natural (pre-log-transformation and pre-normalization) scale.
Antibody responses to vaccination
We computed log 10-transformed baseline-subtracted MFI values after replacing differences less than 1 by 1 (such that after log10-transformation these values are zero). For plotting purposes, we computed positivity thresholds as the median among unvaccinated RM of the baseline-subtracted measure prior to truncation and log transformation, plus three standard deviations. For TB antigens, we computed these threshold values separately for Study 1 and 2. For the SIV gp120 control Ag, which was used as a positive control using Ig obtained from 5 SIV-infected RM (SIVIG), we computed this background threshold using the unvaccinated RM from both studies (these values are plotted along with the SIVIG values in Supplementary Fig. 1b inset).
Statistical Analysis (Immunology and Vaccine-Mediated Protection)
All statistical analyses were conducted in R64. [See also the Life Sciences Reporting Summary.]
Sample size and treatment assignment
For both studies, the primary constraint on sample size was availability of RM subjects and BSL-3 facilities. Groups were balanced for monkey traits and treatments assigned non-randomly, but arbitrarily.
Vaccine-mediated protection
As described above, we evaluated three primary outcome measures for evidence of a difference across treatment arms within each study: CT scan AUC from challenge to day 112 post-challenge, pathologic score at necropsy, and extent of Mtb infection at necropsy (# Mtb+ tissues). The equivalence of the three Study 2 vaccine groups by these outcome measures (Supplementary Fig. 4) justified a pooled analysis. We employed non-parametric tests for primary comparisons and used parametric models for estimating confidence intervals of treatment effects. For comparisons of outcome measures across pairs of groups, we used two-sided Wilcoxon tests. Boxplots in Figs. 2 and 4 show unadjusted p-values of only the pairwise comparisons that are significant at the 0.05 level. We employed Holm adjustment65 for the specified primary non-parametric comparisons: between unvaccinated and vaccinated TB Study 1 groups, and separately between BCG-only and other vaccinated groups. For TB Study 2, we similarly applied Holm-adjustment within groups of comparisons between unvaccinated and vaccinated TB Study 2 groups individually, and between the RM administered the original 68-1 9Ag vector and the two modifications (68-1 6Ag, and 68-1.2 9Ag). Boxplots show unadjusted p-values; Holm-adjusted p-values are shown in Supplementary Fig. 4. Vaccine-mediated protection is reported as 100%-W, where W is 100 times the estimated rate (or confidence limit) of the Poisson or negative binomial model representing a count-valued outcome measure (necropsy score, necropsy culture, or the scaled combination outcome measure) among a vaccine group, as a fraction of the rate for unvaccinated RM. For analysis of correlations across outcome variables, we used Spearman’s rank-transformed correlation statistic (r) and test. For Spearman’s test, p-values were computed via the asymptotic t approximation using the cor.test method in R. Binary comparisons of the dichotomous endpoint defined by any evidence of TB disease at necropsy were conducted using Fisher’s exact test, and confidence intervals for the dichotomous endpoint were estimated using Wilson’s score method66.
T cell and antibody immunogenicity and immune correlates analysis
For comparisons of T cell immunogenicity across vaccine-receiving treatment groups at pre-challenge baseline, for comparisons of changes from baseline after challenge, and for comparisons of pre-challenge Ab responses across treatment groups, we first employed Kruskal-Wallis (KW) tests. The boxplots indicate significance of pairwise Wilcoxon tests if the KW test had a p-value ≤0.05 and the Wilcoxon test had a Holm-adjusted p-value ≤0.05. Due to missingness of blood immunology data from week 41 of RM U6 (Study 1; BCG-only group), for Fig. 1d, n = 6 for the BCG group; and for Fig. 1c, the data point for this RM used only 2, rather than 3, time points for averaging. Due to missingness of immunology data of RM D1 and D2 (Study 1; Unvaccinated group), n = 6 for the unvaccinated group. For analysis of immune response correlations with the scaled combination outcome measure (defined above) among RM receiving RhCMV and no BCG, we computed Spearman’s statistic and test, and in the scatterplots we also show the curve of the best-fitting negative binomial model. We confirmed through sensitivity analyses that the low correlations (Supplementary Fig. 10) were also found in each study separately and in each outcome measure, so the lack of significance is not an artifact of these analysis choices. Extensive non-parametric analyses as well as parametric analyses employing negative binomial models to estimate single-parameter and multi-parameter associations between this scaled combination measure and pre-challenge immune responses to RhCMV immunogens revealed no statistically supported ICS-determined T cell response magnitude correlate of that outcome, or of other related outcomes.
Transcriptomic Analysis
RNA processing and sequencing
RM whole blood was drawn directly into PAXgene blood RNA tubes (PreAnalytiX, Hombrechtikon, Switzerland), which were stored at –20°C. RNA was extracted from the blood in the PAXgene tubes according to the manufacturer’s instructions. Globin transcript depletion (GlobinClear, ThermoFisher Scientific, MA, USA) was performed on the extracted RNA, followed by cDNA library preparation using the Illumina TruSeq Stranded mRNA sample preparation kit (Illumina, CA, USA) for Study 1, and Eukaryote Transcriptome Library Construction Protocol (SOP-TRA J005, Beijing Genomics Institute, Shenzhen, China) for Study 2. For Study 1, globin transcript depletion, cDNA library preparation and RNA sequencing were performed by Expression Analysis/Q2 Solutions, Inc. (Durham, NC). For Study 2, these services were provided by Beijing Genomics Institute (Shenzhen, China). For Study 1, the sequencing strategy was 51bp paired-end reads at an average sequencing depth of 40M reads per sample. For Study 2, the sequencing strategy was 49bp paired-end reads at an average sequencing depth of 70M reads per sample. RNA extraction failures for one Study 1 BCG group RM (U2), one Study 1 68-1 RhCMV/TB group RM (N1), and four unvaccinated group RM (L1, L2, D1, and D2) precluded generation of RNA-Seq data for these animals, giving a total of 23 RM analyzed by RNA-Seq for Study 1 (n=4 unvaccinated; n=6 RhCMV/TB vaccinated; n=6 BCG vaccinated, n=7 BCG+RhCMV/TB vaccinated). All 36 Study 2 RM were analyzed by RNA-Seq (n=9 for each group). For Study 1, RNA samples collected on the day of challenge and 14 and 28 days after challenge were analyzed by RNA-Seq. For Study 2, RNA samples collected prior to vaccination (for RhCMV/TB-vaccinated groups), on the day of challenge, and 28 days after challenge were analyzed by RNA-Seq.
RNA-Seq data processing
Read pairs were preprocessed using scripts that adjust base calls with phred scores <5 to “N” and remove read pairs for which either end has fewer than 30 unambiguous (non-N) base calls. Processed read pairs were aligned using STAR (v.2.3.0.1)67 to the MacaM assembly68. Read pairs in which both ends mapped concordantly were used as input to the HTseq program69 (run in stranded, intersection-strict mode) to assign gene counts. Of the 16,050 gene models, 11,977 and 12,517 had at least 10 counts in at least 5% of the samples from Study 1 and 2, respectively, with 11,678 gene models meeting these criteria in both studies. Log2-transformed values of counts normalized by trimmed mean of M-values (TMM)70 adjusted library counts were computed for Study 1 and 2 using the calcNormFactors function of the edgeR package71 and the voom function of the package limma72. This widely-applied transformation seeks to normalize estimated gene expression levels for differences in both library size and overall transcriptome diversity and will help to minimize the effect of technical differences between samples or protocols between Study 1 and 2.
Log2-transformed counts for Study 1 and 2 were then merged into a unified dataset that was used for all subsequent RNA-Seq-based correlates analyses.
Identification of commonly regulated genes in TB patients and unvaccinated RM from Study 1 and 2 after Mtb challenge
Normalized Illumina microarray data for HIV-uninfected South African and Malawi patients with TB (n = 46 from South Africa; n = 51 from Malawi) or control individuals with latent TB infection (LTBI) (n = 48 from South Africa; n = 35 from Malawi) from a previously published human TB study37 was downloaded from the Gene Expression Omnibus along with the Entrez Gene annotations for the microarray platform. For cases where multiple probes targeted a common gene, the probe with the maximum 90% quantile of gene expression was selected. The human microarray and macaque RNA-Seq datasets were then matched in terms of gene symbols, yielding 10,075 genes that were assayed by both platforms. TB-associated genes in the human study were identified by two-sided Wilcoxon tests comparing TB patients to controls for the South African and Malawi sites. For each gene, the maximum Wilcoxon p-value for the two sites was false discovery rate (FDR)-adjusted for multiple comparisons using the Benjamini-Hochberg FDR adjustment procedure, after having first assigned p=1 for any genes with discordant patterns between the cohorts. The purpose of using the maximum p-value from the two sites as a summary statistic was to ensure that only genes were selected that were robustly and consistently associated with TB disease irrespective of any site-associated contributions of genetics or environment to transcriptional levels. At a significance threshold of p<0.05 and FDR<0.33, 3406 TB-responsive genes were identified from the human study. These genes were then tested for responses in the unvaccinated RM from Study 1 (n=4) and Study 2 (n=9) by one-sided paired Wilcoxon tests comparing expression at 28 days post-challenge to pre-challenge levels, using the directionality from the response in humans. At a significance threshold of p<0.05 and FDR<0.33, 1482/3406 TB-responsive genes from the human study were validated in the RM model, and median fold-changes were correlated between both systems (Fig. 5a–b; Supplementary Table 1).
Identification of associations between transcriptional responses and outcome
Poisson regression modeling with sandwich adjustment, as described above (Necropsy score data), was used to test whether expression levels or expression fold-changes were significant predictors for outcome after challenge, as indicated by the scaled combination outcome measure. Briefly, Wald tests (R function: waldtest(); library: lmtest)73 with the regression coefficient covariance matrix being provided by heteroskedasticity-consistent covariance matrix estimation (R function: vcovHC(), library: sandwich62,63) were performed comparing two Poisson regression models that were generated for each gene:
Model 1: outcome ~ gene + Study
Model 2: outcome ~ Study
(where: “outcome” = the scaled combination outcome measure, “gene” = log 2-transformed expression level or fold change for a specific gene, and “Study” = an additive factor with two levels: 0 or 1, with “0” corresponding to Study 1 and “1” corresponding to Study 2.)
While the scaled combination outcome measure is harmonized between Study 1 and 2, minor technical differences between Study 1 and 2 RNA-Seq analysis (e.g. sequencing depth, library protocol, or service provider) may give rise to small shifts in expression between Study 1 and 2 that could confound the modeling; including the additive “Study” term in the model addresses this possible effect. Wald test p-values from comparing Model 1 with Model 2 were FDR-adjusted for multiple comparisons using the Benjamini-Hochberg procedure. This overall strategy was applied to identify the post-challenge (Fig. 5) and pre-challenge (Fig. 6) transcriptional correlates with outcome as described below:
Post-challenge correlates with outcome
When modeling outcome as a function of the log2 fold-change post challenge (day 28/day of challenge), data from all RM were included (all RhCMV/TB groups, BCG, and unvaccinated; n=59) from Study 1 and 2. To focus the analysis on the TB disease signature that is common between humans and RM, the set of genes analyzed was restricted to the 1482 TB-responsive genes that were discovered in the human TB cohort and validated in the unvaccinated RM (Fig. 5a). At the thresholds applied in the other analyses (p<0.05, FDR<0.33) a large number of significant associations were detected (1007 genes). For this reason, an additional filtering step was applied to focus the results on the strongest associations. Requiring that the absolute value of the overall Kendall tau rank correlation coefficient between the scaled combination outcome measure and gene expression fold-changes exceeded a value of 0.4 yielded the final set 214 genes shown in Fig. 5c (Supplementary Table 3).
Pre-challenge correlates with outcome
When modeling outcome as a function of the log2 expression levels measured immediately before challenge, two analyses were performed: (i) using data from all RM that received RhCMV/TB vaccines (all RhCMV/TB groups and the BCG + RhCMV/TB group from Study 1; n=40), and (ii) using data from the unvaccinated and BCG (only) vaccine groups (n=19; Fig. 6a–c). The objective of (i) was to identify pre-challenge correlates of eventual protection or susceptibility that were operational within the context of RhCMV/TB-induced immune responses (note that RhCMV/TB-elicited Mtb-specific T cell responses were similar regardless of prior BCG vaccination; Fig. 1b–f). The objective of (ii) was to confirm that the associations in (i) were restricted to the context of RhCMV/TB vaccination and less likely to be due to monkey-intrinsic differences. Prior to Poisson regression, the average pre-challenge log2 expression levels for the unvaccinated RM was subtracted from the pre-challenge log2 expression levels of all RM within the same study.
For the RhCMV/TB-focused analysis (i), the complete transcriptome (11,678 genes) was analyzed. At the thresholds applied in the other analyses (p<0.05, FDR<0.33), 475 genes with significant associations were identified. This initial set was reduced to 315 genes by requiring that the sign of the Kendall tau rank correlation coefficient between gene expression and outcome be consistent for Study 1 and Study 2 (different correlation signs could arise from the difference in sample numbers between Study 1 and Study 2; requiring consistent directionality penalized associations driven predominantly by Study 2 in favor of shared associations). We noted that genes with overall negative correlations with outcome exhibited much greater sign consistency between Study 1 and Study 2 than genes with overall positive correlations with outcome (94/107 consistent genes negatively correlated with outcome; 221/368 consistent genes positively correlated with outcome). The genes with the strongest associations between log2 expression and outcome were then identified by ranking the absolute value of the overall Kendall tau rank correlation coefficient between the scaled combination outcome measure and log2 gene expression levels (using “study adjusted” values for genes with nominally significant study terms – defined below). Requiring the absolute value of the Kendall tau coefficient to exceed 0.2 yielded a set of 280 genes for subsequent analysis. All arbitrary thresholds including this Kendall tau threshold were determined once and held fixed thereafter to ensure interpretability of resulting statistics. To determine whether these candidate signatures were potentially driven by the RhCMV/TB vaccine, we tested whether these genes exhibited significant associations with outcome for the BCG and unvaccinated groups by Poisson modeling as described above. Of the 280 genes, 7 were nominally significant (p<0.05) and 22 exhibited p<0.2, indicating an overall absence of evidence for an association between the expression of the gene set and outcome for RM that did not receive RhCMV/TB vaccination. The 22 genes with p<0.2 for analysis (ii) were excluded, yielding the 258 genes shown in Figure 6a (Supplementary Table 5). Of these, 181 exhibited pre-challenge expression levels that were positively correlated with values of the scaled combination outcome measure and were termed the “susceptibility signature,” meaning that higher expression of these genes prior to Mtb exposure was associated with a tendency to ultimately develop more severe disease. Seventy-seven of the 258 genes exhibited pre-challenge expression levels that were negatively correlated with values of the scaled combination outcome measure and were termed the “protection signature,” meaning that higher expression of these genes prior to Mtb exposure was associated with a tendency to ultimately develop less severe disease or even to have complete protection.
As described in the Results, inspection of the heatmap visualizations of protection and susceptibility signatures (Fig. 6a–b) suggested that the association between pre-challenge expression levels and outcome was predominantly binary, with strongly protected RMs exhibiting a distinct expression pattern compared to all other RM. This conjecture was explored by logistic regression modeling in which the scaled combination outcome measure was discretized into two levels: “completely protected” (14 RM that exhibited no signs of disease), and “non-protected” (all other RM that received RhCMV/TB vaccine). The modeling was essentially as above, replacing non-negative discrete numeric “outcome” with binary “protection”, and employing Chi-squared tests to compare models in lieu of the Wald test. Also, instead of performing the modeling by including a “Study” term (Model 1 and Model 2), the “Study” term was removed and the modeling was performed using the “study adjusted” expression values for instances where the “Study” term was statistically significant based on the full Poisson models (Model 1 and Model 2). This is because there was only one protected RM in Study 1, and it is therefore not possible to reliably estimate a “Study” term for the logistic regression models. In addition to evaluating the statistical significance of the binary models, discriminatory potential of the models was evaluated using receiver operating curve (ROC) metrics (R package: pROC) to assess ROC AUC and stratified accuracy for Study 1 and 2 at the optimal ROC operating point. As described, results of this analysis supported the binary interpretation of the signatures, with 49% and 39% of protection and susceptibility genes, respectively, exhibiting “reasonable discriminatory ability” (Supplementary Table 5), which we defined as p<0.05, ROC AUC lower 95% confidence interval >0.5, and having a ROC operating point that simultaneously achieves at least 50% sensitivity and specificity in both Study 1 and Study 2.
Heatmap visualizations
Heatmap visualizations were generated in R using the pheatmap library.
Post-challenge correlates with outcome
The “Study” term in Model 1 was not nominally significant for any of the 214 genes identified (i.e., p>0.05), and for this reason, the log2 fold-change data shown in Fig. 5c–e is presented without any additional transformations beyond the overall scaling for the heatmaps (dividing the values for each gene by the maximum absolute fold-change observed for that gene).
Pre-challenge correlates with outcome
The “Study” term in Model 1 was nominally significant for about ½ of the genes identified, indicating small differences in pre-challenge gene expression that did not influence associations with outcome but complicate visualizations. For these genes, “study adjusted” log2-transformed expression profiles were computed by adding to Study 2 log2-transformed expression levels the estimate of the “Study” coefficient divided by the estimate of the “gene” coefficient from Model 1. This transformation aligns the Study 1 and 2 log2 expression levels to a common trajectory and facilitates visualization of the associations with outcome identified by the modeling. In the heatmaps, log2 expression profiles or “study adjusted” log2 expression profiles are shown after scaling the values for each gene by the maximum absolute fold-change observed for that gene.
Module and pathway enrichment analysis
Fisher’s exact test was employed to determine whether genes belonging to pre-defined transcriptional modules or immune pathways were significantly over-represented within sets of genes that were identified as associated with outcome or other characteristics. This analysis utilized two external sources for association of specific genes with transcriptional modules or pathways. The first was the set of blood transcriptional co-expression modules defined by Obermoser, et al53. These modules are advantageous because they define sets of similarly annotated genes that exhibit concordant expression patterns across an extensive set of immune-related blood transcriptional profiles. Enrichment analysis of these modules was restricted to the annotated modules, and modules with common annotations were merged. Definition and annotation of the modules is provided at: http://www.biir.net/public_wikis/module_annotation/G2_Trial_8_Modules. The transcriptional modules were complemented by curated pathway definitions from Reactome54, which are not based on co-expression, but rather extensive signaling pathway knowledge from the literature. Enrichment analysis of these pathways was restricted to those within the “Immune System” branch of the overall hierarchy. Each enrichment analysis required a “reference” gene set that constitutes the relevant compendium of candidate genes from which the gene set of interest was derived. The reference gene sets for each enrichment analysis are defined below. To exclude spurious enrichment results, modules and pathways with fewer than 50 members within the reference gene set were excluded. Gene sets for enrichment analysis were defined according to the association of interest and directionality (i.e., up-regulated genes were tested for module and pathway over-representation separately from down-regulated genes). Multiple comparisons were addressed by performing the Benjamini-Hochberg adjustment to the Fisher’s exact test p-values, with p-values for gene sets derived from a common analysis but exhibiting opposite directionality being adjusted together. Enrichment significance thresholds were set at p<0.05 and FDR<0.15.
Enrichment analysis of TB-responsive genes in humans and RM (Fig. 5b and Supplementary Table 2)
The reference gene sets were the 10,075 genes commonly detected in the human and NHP platforms and the gene sets of interest were the 793 up- and 689 down-regulated genes from the 1482 genes regulated in common. Enriched modules and pathways with fewer than 20 members within the up- and down-regulated genes are not shown.
Enrichment analysis of TB-responsive genes with post-challenge fold-changes associated with outcome in RM (Figs. 5c–d and Supplementary Table 4)
The reference gene sets were the 1482 genes regulated in common between human and NHP and the gene sets of interest were the 196 genes positively associated and 18 genes negatively associated from the 214 with post-challenge fold-changes associated with outcome. Enriched modules and pathways with fewer than 2 members within the positively- and negatively-associated genes are not shown.
Enrichment analysis of genes with pre-challenge expression profiles that are associated with outcome in RhCMV/TB-vaccinated RM (Fig. 6a–b and Supplementary Table 6)
The reference gene sets were the 11,678 genes detected in common between Study 1 and 2 and the gene sets of interest were the 181 genes positively associated (susceptibility signature) and 77 genes negatively associated (protection signature) from the 258 with pre-challenge expression profiles associated with outcome. Enriched modules and pathways with <2 members within the positively- and negatively-associated genes are not shown.
Integration of transcriptional measurements with leukocyte population counts
To gain insight into the contribution of leukocyte trafficking to the pre-challenge protection and susceptibility signatures, we integrated the transcriptome data with whole blood leukocyte percentages. Bulk lymphocyte, monocyte, and neutrophil population percentages in whole blood on the day of challenge were determined by Coulter Counter (Study 1) and Horiba blood cell counter (Study 2). Basophil and eosinophil counts were excluded because they were marginal in both studies and not likely to contribute to the whole blood transcriptome readouts. The contribution of T cells (CD3+) and T cell subsets (CD4+, CD8+) to the bulk lymphocyte count was further determined for Study 1 and 2 by flow cytometry. Leukocyte counts were utilized in three exploratory analyses: (i) testing leukocyte populations themselves for pre-challenge associations with outcome, (ii) identifying the leukocyte population most associated with specific genes, and (iii) determining whether leukocyte population differences may be responsible for the pre-challenge transcriptional associations with outcome. Both untransformed and log10-transformed leukocyte % were investigated in the exploratory analyses.
Testing leukocyte populations alone for pre-challenge associations with outcome
In this analysis, the Poisson regression analysis (described above) for identifying pre-challenge transcriptional correlates with outcome was repeated exactly, but substituting leukocyte population counts (and log transformations thereof) for genes. No single leukocyte population, nor ratios between monocytes and lymphocytes, neutrophils to lymphocytes, or neutrophils to monocytes, achieved nominally significant associations with outcome (i.e., p>0.05 for all populations) or nominally significant Kendall rank correlations with outcome (i.e., p>0.05 for all populations).
Identifying the leukocyte population most associated with specific genes
Given the non-transitive nature of correlations, it is possible that genes that are significantly associated with outcome may be significantly associated with certain leukocyte populations, even if those leukocyte populations themselves are not significantly associated with outcome. This scenario could arise, for example, if the activation state of a certain leukocyte subpopulation, as indicated by expression of a specific gene set, was associated with outcome. We determined the leukocyte population associated with each gene by performing robust linear regression (R: rlm, package: MASS) using the following model structure:
Model 3: Cell ~ gene + Study
(where “Cell” = the leukocyte population percentage of interest (neutrophils, monocytes, lymphocytes, T cells, CD4+ T cells, or CD8+ T cells and log transformations thereof), and “gene” and “Study” are as defined as for models 1 and 2).
As the focus of the analysis was for the interpretation of the pre-challenge protection and susceptibility signatures (Fig. 6a), robust linear regression modeling was restricted to leukocyte counts and transcriptome data for RM that received RhCMV/TB vaccination. The significance of the association between the gene and leukocyte population was determined by robust F-tests on the “gene” coefficient from the model (R: f.robftest, package: sfsmisc). Plausible leukocyte/cell associations were defined as those for which the model coefficient was nominally significant (p<0.05) and the coefficient was positive (indicating a positive relationship between transcription and the cells). Of the genes with more than one plausible association meeting these criteria, the cell population with the smallest p-value was selected.
To determine whether genes with plausible associations with specific leukocyte populations were over-represented in the pre-challenge protection or susceptibility signatures, Fisher’s exact tests were performed as described above, except replacing the transcriptional module and pathway annotations with the associations between genes and cell populations. Given the hierarchical relationship between CD4+ and CD8+ T cells, T cells, and lymphocytes, genes/lymphocyte associations detected lower in the hierarchy were also assigned to upper branches of the hierarchy (i.e., a gene associated with CD4+ T cells would also be counted as a gene associated with T cells and lymphocytes). Similarly, genes most associated with actual CD4+ T cell percentages or log-transformed CD4+ T cell percentages were simply regarded as “CD4+ T cell-associated.” The full list of gene by leukocyte populations is provided in Supplementary Table 7. Enrichment results for association with leukocyte populations are provided in Supplementary Table 8.
Determining whether leukocyte population differences may be responsible for the pre-challenge transcriptional associations with outcome
Additional Poisson regression analyses were performed to determine whether the association between pre-challenge expression of a certain gene and outcome were driven by the leukocyte population with which the gene was most associated. This followed the identical sandwich-adjusted Poisson modeling and Wald test for model comparison as described above, except including new terms for the leukocyte population associated with the gene being tested:
Model 4: outcome ~ gene + Cell + Study
Model 5: outcome ~ Cell + Study
(where: “Cell” = the leukocyte population most associated with the gene being analyzed, and “gene” and “Study” are as defined above).
If variation in the leukocyte population was driving the association between outcome and the gene, then the gene would not contribute to the model fit and there would be no statistically significant difference revealed by the Wald test comparing Model 4 and Model 5. As above, this analysis was performed only for RM that received RhCMV/TB vaccine, only at the pre-challenge time point, and only for those genes from the protection and susceptibility signatures (Fig. 6a) for which there was a plausibly associated leukocyte population. Of the 280 genes, 82 were associated with at least one leukocyte population, and four of these (AMIGO1, DDX39B, GMEB2, and TYSND1, all from the susceptibility signature) lost nominal significance in the comparison between Model 4 and 5 (p values ranged from 0.05 to 0.08 for these genes) (Supplementary Table 5). This result suggests that the protection and susceptibility signatures do not likely result from nor are driven by trafficking differences in bulk leukocyte populations.
Testing for differences between RhCMV/TB and BCG + RhCMV/TB pre-challenge transcriptional patterns
One-sided Wilcoxon rank-sum tests were performed to compare the expression levels of genes from the protection signature in the RhCMV/TB group (n=6) to the BCG + RhCMV/TB(n=7) group from Study 1. Given the tendency within the BCG + RhCMV/TB group towards reduced protection, we tested whether pre-challenge expression of the protection-associated genes was reduced in this group compared to the group that received RhCMV alone. At p<0.05 and FDR<0.33, 15 genes were identified that meet this criterion (Fig. 6d and Supplementary Table 9).
DATA AND COMPUTER CODE AVAILABILITY
The datasets generated and/or analyzed during the current study, as well as the computer code used to perform statistical analysis, are available from the corresponding authors on reasonable request. Raw and processed RNA-Seq data for Study 1 and 2 have been deposited into the Gene Expression Omnibus under accession #GSE102440.
Supplementary Material
Acknowledgments
This work was supported by AERAS, the Bill and Melinda Gates Foundation (OPP1087783 to A.A. and D.Z.), and the National Institutes of Health (U19 AI106761 to A.A.; P51 OD011092 to ONPRC; U42 OD010426 to ONPRC). We thank C. Kahl, S. Hagen, J. Bae, I. Pelletier, Y. Guo, EM Borst, LS Uebelhoer, and J. Womack for technical assistance, C. Scanga and J. Flynn for guidance on the RM TB model, including sharing of NHP protocols and provision of Mtb challenge stocks, P. Barry (UC, Davis) and T. Shenk (Princeton University) for the 68-1 and 68-1.2 BACs, respectively, W. Hanekom, L. Stuart, and D. Barber for helpful discussions, D. Casimiro for manuscript review, J. Strussenberg for BSL3 management, L. Boshears for administrative assistance, and A. Townsend for figure preparation.
Footnotes
AUTHOR CONTRIBUTIONS
S.G.H. planned animal experiments and supervised all immunologic and virologic studies and data analysis. G.X. and J.C.F. processed monkey samples and tissues and performed immunologic and bacteriologic analysis, assisted by R.M.G., C.M.H., A.B.V., E.A., K.T.R., A.N.S., P.R., L.H., H.P. and M.S.L. A.W.S. performed assay development and supervised flow cytometry. K.F., D.M., E.E.M., M.M. and M.A.J. designed, constructed, and/or validated the RhCMV/TB vectors used in the study. A.W.L. supervised all animal procedures, including CT scanning, assisted by S.L.P., J.M.T, M.F., C.A., and R.C.Z. M.K.A. planned and provided overall supervision of monkey protocols, interpreted CT scans, performed all necropsies and interpreted both gross pathology and histopathology. D.J.L. designed experimental approaches and reviewed data. M.S., A.B., and T.G.E. contributed to data interpretation and/or study design. L.L. performed the Ab assays under the supervision of G.A. J.V., J.M.B., and S.S. processed samples and data for transcriptomic analysis. D.E.Z. planned, executed, and interpreted the transcriptomic analysis, assisted by L.M.A, S.S., and A.A. P.T.E. planned and performed all statistical analyses. L.J.P. conceived the RhCMV vector strategy, planned and supervised all experiments and data analysis, and wrote the manuscript, assisted by S.G.H., P.T.E., D.E.Z. and M.K.A.
COMPETING FINANCIAL INTERESTS STATEMENT
OHSU and Drs. Picker, Hansen, Malouli, and Früh have a significant financial interest in Vir Biotechnology, Inc., a company that may have a commercial interest in the results of this research and technology. The potential individual and institutional conflicts of interest have been reviewed and managed by OHSU. Dr. Evans has served as a clinical consultant to Vir Biotechnology and also has a significant financial interest in that company.
References
- 1.Cambier CJ, Falkow S, Ramakrishnan L. Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell. 2014;159:1497–1509. doi: 10.1016/j.cell.2014.11.024. [DOI] [PubMed] [Google Scholar]
- 2.Orme IM, Robinson RT, Cooper AM. The balance between protective and pathogenic immune responses in the TB-infected lung. Nat Immunol. 2015;16:57–63. doi: 10.1038/ni.3048. [DOI] [PubMed] [Google Scholar]
- 3.Shaler CR, Horvath CN, Jeyanathan M, Xing Z. Within the Enemy’s Camp: contribution of the granuloma to the dissemination, persistence and transmission of Mycobacterium tuberculosis. Front Immunol. 2013;4:30. doi: 10.3389/fimmu.2013.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pai M, et al. Tuberculosis. Nat Rev Dis Primers. 2016;2:16076. doi: 10.1038/nrdp.2016.76. [DOI] [PubMed] [Google Scholar]
- 5.Comas I, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42:498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comas I, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 2013;45:1176–1182. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorhoi A, Kaufmann SH. Pathology and immune reactivity: understanding multidimensionality in pulmonary tuberculosis. Semin Immunopathol. 2016;38:153–166. doi: 10.1007/s00281-015-0531-3. [DOI] [PubMed] [Google Scholar]
- 8.Hawn TR, et al. Tuberculosis vaccines and prevention of infection. Microbiol Mol Biol Rev. 2014;78:650–671. doi: 10.1128/MMBR.00021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaufmann SH. Future vaccination strategies against tuberculosis: thinking outside the box. Immunity. 2010;33:567–577. doi: 10.1016/j.immuni.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 10.Griffiths KL, et al. Targeting dendritic cells to accelerate T-cell activation overcomes a bottleneck in tuberculosis vaccine efficacy. Nat Commun. 2016;7:13894. doi: 10.1038/ncomms13894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barclay WR, et al. Protection of monkeys against airborne tuberculosis by aerosol vaccination with bacillus Calmette-Guerin. Am Rev Respir Dis. 1973;107:351–358. doi: 10.1164/arrd.1973.107.3.351. [DOI] [PubMed] [Google Scholar]
- 12.Kaushal D, et al. Mucosal vaccination with attenuated Mycobacterium tuberculosis induces strong central memory responses and protects against tuberculosis. Nat Commun. 2015;6:8533. doi: 10.1038/ncomms9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verreck FAW, et al. Variable BCG efficacy in rhesus populations: Pulmonary BCG provides protection where standard intra-dermal vaccination fails. Tuberculosis (Edinb) 2017;104:46–57. doi: 10.1016/j.tube.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Jeyanathan M, et al. AdHu5Ag85A Respiratory Mucosal Boost Immunization Enhances Protection against Pulmonary Tuberculosis in BCG-Primed Non-Human Primates. PLoS One. 2015;10:e0135009. doi: 10.1371/journal.pone.0135009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darrah PA, et al. Aerosol vaccination with AERAS-402 elicits robust cellular immune responses in the lungs of rhesus macaques but fails to protect against high-dose Mycobacterium tuberculosis challenge. J Immunol. 2014;193:1799–1811. doi: 10.4049/jimmunol.1400676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verreck FA, et al. MVA.85A boosting of BCG and an attenuated, phoP deficient M. tuberculosis vaccine both show protective efficacy against tuberculosis in rhesus macaques. PLoS One. 2009;4:e5264. doi: 10.1371/journal.pone.0005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tameris MD, et al. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial. Lancet. 2013;381:1021–1028. doi: 10.1016/S0140-6736(13)60177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tameris M, et al. The candidate TB vaccine, MVA85A, induces highly durable Th1 responses. PLoS One. 2014;9:e87340. doi: 10.1371/journal.pone.0087340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarvis MA, Hansen SG, Nelson JA, Picker LJ, Früh K. Vaccine Vectors Using the Unique Biology and Immunology of Cytomegalovirus. In: Reddehase MJ, editor. Cytomegaloviruses: From Molecular Pathogenesis to Intervention. Vol. 2. Caister Academic Press; 2013. [Google Scholar]
- 20.Cicin-Sain L, et al. Cytomegalovirus-specific T cell immunity is maintained in immunosenescent rhesus macaques. J Immunol. 2011;187:1722–1732. doi: 10.4049/jimmunol.1100560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sylwester AW, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med. 2005;202:673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen SG, et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med. 2009;15:293–299. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen SG, et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 2011;473:523–527. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen SG, et al. Immune clearance of highly pathogenic SIV infection. Nature. 2013;502:100–104. doi: 10.1038/nature12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen SG, et al. Broadly targeted CD8(+) T cell responses restricted by major histocompatibility complex E. Science. 2016;351:714–720. doi: 10.1126/science.aac9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scanga CA, Flynn JL. Modeling tuberculosis in nonhuman primates. Cold Spring Harb Perspect Med. 2014;4:a018564. doi: 10.1101/cshperspect.a018564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharpe S, et al. Ultra low dose aerosol challenge with Mycobacterium tuberculosis leads to divergent outcomes in rhesus and cynomolgus macaques. Tuberculosis (Edinb) 2016;96:1–12. doi: 10.1016/j.tube.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Gormus BJ, Blanchard JL, Alvarez XH, Didier PJ. Evidence for a rhesus monkey model of asymptomatic tuberculosis. J Med Primatol. 2004;33:134–145. doi: 10.1111/j.1600-0684.2004.00062.x. [DOI] [PubMed] [Google Scholar]
- 29.Sibley L, et al. Route of delivery to the airway influences the distribution of pulmonary disease but not the outcome of Mycobacterium tuberculosis infection in rhesus macaques. Tuberculosis (Edinb) 2016;96:141–149. doi: 10.1016/j.tube.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Mothe BR, et al. The TB-specific CD4(+) T cell immune repertoire in both cynomolgus and rhesus macaques largely overlap with humans. Tuberculosis (Edinb) 2015;95:722–735. doi: 10.1016/j.tube.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langermans JA, et al. Divergent effect of bacillus Calmette-Guerin (BCG) vaccination on Mycobacterium tuberculosis infection in highly related macaque species: implications for primate models in tuberculosis vaccine research. Proc Natl Acad Sci U S A. 2001;98:11497–11502. doi: 10.1073/pnas.201404898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu T, et al. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci U S A. 2003;100:12420–12425. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansen SG, et al. Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science. 2013;340:1237874. doi: 10.1126/science.1237874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zak DE, et al. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet. 2016;387:2312–2322. doi: 10.1016/S0140-6736(15)01316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cliff JM, Kaufmann SH, McShane H, van Helden P, O’Garra A. The human immune response to tuberculosis and its treatment: a view from the blood. Immunol Rev. 2015;264:88–102. doi: 10.1111/imr.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berry MP, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaforou M, et al. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS Med. 2013;10:e1001538. doi: 10.1371/journal.pmed.1001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Libero G, Singhal A, Lepore M, Mori L. Nonclassical T cells and their antigens in tuberculosis. Cold Spring Harb Perspect Med. 2014;4:a018473. doi: 10.1101/cshperspect.a018473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rayner EL, et al. Early lesions following aerosol infection of rhesus macaques (Macaca mulatta) with Mycobacterium tuberculosis strain H37RV. J Comp Pathol. 2013;149:475–485. doi: 10.1016/j.jcpa.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 40.Steinert EM, et al. Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell. 2015;161:737–749. doi: 10.1016/j.cell.2015.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thome JJ, Farber DL. Emerging concepts in tissue-resident T cells: lessons from humans. Trends Immunol. 2015;36:428–435. doi: 10.1016/j.it.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dallenga T, Schaible UE. Neutrophils in tuberculosis--first line of defence or booster of disease and targets for host-directed therapy? Pathog Dis. 2016;74 doi: 10.1093/femspd/ftw012. [DOI] [PubMed] [Google Scholar]
- 43.Mishra BB, et al. Nitric oxide prevents a pathogen-permissive granulocytic inflammation during tuberculosis. Nat Microbiol. 2017;2:17072. doi: 10.1038/nmicrobiol.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ong CW, et al. Neutrophil-Derived MMP-8 Drives AMPK-Dependent Matrix Destruction in Human Pulmonary Tuberculosis. PLoS Pathog. 2015;11:e1004917. doi: 10.1371/journal.ppat.1004917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mattila JT, Maiello P, Sun T, Via LE, Flynn JL. Granzyme B-expressing neutrophils correlate with bacterial load in granulomas from Mycobacterium tuberculosis-infected cynomolgus macaques. Cell Microbiol. 2015;17:1085–1097. doi: 10.1111/cmi.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lyadova IV. Neutrophils in Tuberculosis: Heterogeneity Shapes the Way? Mediators Inflamm. 2017;2017:8619307. doi: 10.1155/2017/8619307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martineau AR, et al. Neutrophil-mediated innate immune resistance to mycobacteria. J Clin Invest. 2007;117:1988–1994. doi: 10.1172/JCI31097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warren E, Teskey G, Venketaraman V. Effector Mechanisms of Neutrophils within the Innate Immune System in Response to Mycobacterium tuberculosis Infection. J Clin Med. 2017;6 doi: 10.3390/jcm6020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seiler P, et al. Early granuloma formation after aerosol Mycobacterium tuberculosis infection is regulated by neutrophils via CXCR3-signaling chemokines. Eur J Immunol. 2003;33:2676–2686. doi: 10.1002/eji.200323956. [DOI] [PubMed] [Google Scholar]
- 50.Jeyanathan M, et al. Differentially imprinted innate immunity by mucosal boost vaccination determines antituberculosis immune protective outcomes, independent of T-cell immunity. Mucosal Immunol. 2013;6:612–625. doi: 10.1038/mi.2012.103. [DOI] [PubMed] [Google Scholar]
- 51.Beverley PC, et al. A novel murine cytomegalovirus vaccine vector protects against Mycobacterium tuberculosis. J Immunol. 2014;193:2306–2316. doi: 10.4049/jimmunol.1302523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knight GM, et al. Impact and cost-effectiveness of new tuberculosis vaccines in low- and middle-income countries. Proc Natl Acad Sci U S A. 2014;111:15520–15525. doi: 10.1073/pnas.1404386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Obermoser G, et al. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013;38:831–844. doi: 10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fabregat A, et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016;44:D481–487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clemmensen SN, et al. Olfactomedin 4 defines a subset of human neutrophils. J Leukoc Biol. 2012;91:495–500. doi: 10.1189/jlb.0811417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ambrose LR, Morel AS, Warrens AN. Neutrophils express CD52 and exhibit complement-mediated lysis in the presence of alemtuzumab. Blood. 2009;114:3052–3055. doi: 10.1182/blood-2009-02-203075. [DOI] [PubMed] [Google Scholar]
- 57.Capuano SV, 3rd, et al. Experimental Mycobacterium tuberculosis infection of cynomolgus macaques closely resembles the various manifestations of human M. tuberculosis infection. Infect Immun. 2003;71:5831–5844. doi: 10.1128/IAI.71.10.5831-5844.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rumboldt Z, Huda W, All JW. Review of portable CT with assessment of a dedicated head CT scanner. AJNR Am J Neuroradiol. 2009;30:1630–1636. doi: 10.3174/ajnr.A1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rubin GD. Lung nodule and cancer detection in computed tomography screening. J Thorac Imaging. 2015;30:130–138. doi: 10.1097/RTI.0000000000000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luciw PA, et al. Stereological analysis of bacterial load and lung lesions in nonhuman primates (rhesus macaques) experimentally infected with Mycobacterium tuberculosis. Am J Physiol Lung Cell Mol Physiol. 2011;301:L731–738. doi: 10.1152/ajplung.00120.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zvi A, Ariel N, Fulkerson J, Sadoff JC, Shafferman A. Whole genome identification of Mycobacterium tuberculosis vaccine candidates by comprehensive data mining and bioinformatic analyses. BMC Med Genomics. 2008;1:18. doi: 10.1186/1755-8794-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeileis A. Econometric computing with HC and HAC covariance matrix estimators. Journal of Statistical Software. 2004;11:1–17. [Google Scholar]
- 63.Zeileis A. Object-oriented computation of sandwich estimators. Journal of Statistical Software. 2006;16:1–16. [Google Scholar]
- 64.R Core Team. R. R Foundation for Statistical Computing. Vol. 2015. Vienna; Austria: 2015. A language and environment for statistical computing. [Google Scholar]
- 65.Holm S. A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics. 1979;6:65–70. [Google Scholar]
- 66.Agresti A, Coull BA. Approximate is better than “exact” for interval estimation of binomial proportions. The American Statistician. 1998;52:119–126. [Google Scholar]
- 67.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zimin AV, et al. A new rhesus macaque assembly and annotation for next-generation sequencing analyses. Biol Direct. 2014;9:20. doi: 10.1186/1745-6150-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeileis A, Hothorn T. Diagnostic Checking in Regression Relationships. R News. 2002;2:7–10. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.