Abstract
STUDY QUESTION
Are there novel hyaladherins in human sperm?
SUMMARY ANSWER
Zona pellucida-binding protein 2 (ZPBP2), containing a Link-like hyaluronic acid (HA)-binding domain, and several other proteins containing BX7B motifs, such as ADAM32 and Midkine, may be novel hyaladherins with HA-binding properties.
WHAT IS KNOWN ALREADY
HA-binding proteins (hyaladherins), which can bind HA surrounding the cumulus-oophorus complex, are distinct from hyases such as PH 20 (SPAM1) and are expressed by mature spermatozoa. Although HABP1 and CD44 are reasonably well characterized hyaladherins and the former has been implicated in sperm-oocyte interactions, the overall significance of sperm hyaladherins for male fertility is still poorly understood.
STUDY DESIGN, SIZE, DURATION
This was a laboratory-based investigation into human sperm hyaladherins undertaken as part of a three year PhD programme sponsored by the EU Marie Curie Training network, Reprotrain.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Protein homogenates of sperm obtained from young men of unknown fertility (N = 4) were partitioned into HA-binding and non-binding fractions by a protein affinity ‘panning’ method; their subsequent characterization was by liquid chromatography-tandem mass spectrometry (LC-MS-MS) and partitioning behaviour was confirmed by western blotting. Sequences of proteins from both fractions were submitted to PDBsum to look for orthologous entries (PDB codes) and all returned codes were queried against the matching protein using SAS (Sequences Annotated by Structure) looking for structural similarities between them. A systematic search for other common features of hyaladherins was also undertaken.
MAIN RESULTS AND THE ROLE OF CHANCE
The presence of BX7B sequence motifs found in several well-described hyaladherins including RHAMM was used to assess efficacy of potential hyaladherin partitioning by the HA substrate. The data showed that 50% (14/28) and 34.5% (28/81) of proteins in the bound and unbound fractions, respectively, contained these motifs (one-tailed Z-score = 1.45; P = 0.074), indicating weak discrimination by the substrate. Querying PDBsum with sequences for all bound proteins returned several PDB codes matching ZPBP2 with the HA-binding Link domain of the hyaladherin, CD44. Western blot analysis confirmed the affinity partitioning of proteins indicated by the LC-MS/MS results, with ADAM32 (containing two BX7B motifs) and ZPBP2 (containing a Link-like HA-binding domain) present only in the binding fraction. There remains the possibility that the putative hyaladherins uncovered by this study were coincidentally enriched by HA-binding.
LARGE SCALE DATA
The full proteomics data set is available on request.
LIMITATIONS REASONS FOR CAUTION
The protein extraction methods or the HA substrate used to pan them in this study were probably not ideal, as hyaladherins expected to be present in sperm homogenates (such as CD44 and RHAMM) were not detected.
WIDER IMPLICATIONS OF THE FINDINGS
The results provide evidence that ZPBP2, found only in the bound fraction, may have hyaladherin-like properties, which could reflect the evolutionary background context of contemporary sperm-oocyte interaction mechanisms.
STUDY FUNDING AND COMPETING INTEREST(S)
An EU Marie Curie Sklodowska Initial Training Network Scholarship, supporting Ms Torabi, is gratefully acknowledged. This project was also supported and funded by the Efficacy and Mechanism Evaluation Programme, a UK MRC and NIHR partnership (Grant No 11/14/ 34). There is no conflict of interest in relation to this work.
Keywords: Hyaluronic acid, Hyaluronic Acid-Binding Protein, CD44, Link module, BX7B motif, ZPBP2, ADAM32
Introduction
In natural reproductive cycles, ejaculated spermatozoa encounter physiological selection pressures during their journey across the female reproductive tract, where sperm encounter several biological ‘checkpoints’, ensuring that only the ‘fitter’ cells reach the oocyte (Suarez and Pacey, 2006; Ikawa et al., 2010). In ART, the success of embryo development and pregnancy outcome also depends on sperm quality (Sakkas et al., 2000) and the selection of viable sperm, including sperm with high DNA integrity and chromatin maturity is crucial (Hekmatdoost et al., 2009). To a large extent, sperm selection for ART particularly with ICSI depends on the embryologists’ experience of picking the best sperm and is based on microscopic parameters of sperm motility, viability and morphology. Sperm abnormalities, however, particularly those occurring at the molecular level cannot always be detected by microscopic observation alone (Palermo et al., 1992). Celik-Ozenci et al. (2004) showed that sperm with normal motility and morphology may have chromosomal abnormalities and ICSI provides no barrier preventing sperm with chromatin and other defects from participating in the fertilization process. Negative effects on ICSI outcomes of the use of poor quality sperm may include miscarriage, an increased risk of congenital abnormalities and childhood cancer (Celik-Ozenci et al., 2004; Gopalkrishnan et al., 2000; Halliday, 2012; Jaleel and Khan, 2013; Larsen et al., 2013). These issues have led to the development of alternative methods of sperm selection for ICSI based on functional properties, potentially mimicking the natural processes occurring in the reproductive tract of healthy individuals and sperm binding to hyaluronic acid (HA) is one such method.
HA is a non-sulphated glycosaminoglycan with numerous biological functions in the extracellular matrix and on the cell surface (Amemiya et al., 2005). HA is abundant in the female reproductive tract and may be involved in sperm-oocyte interactions (Ghosh et al., 2007). The cumulus-oophorus complex (COC) is also HA-rich (Zhuo and Kimata, 2001) and HA permeates the zona pellucida and the perivitelline space of mammalian oocytes (Vandevoort et al., 1997). During in vivo fertilization, mature spermatozoa bind HA in the extracellular matrix of the COC via hyaladherins and subsequently release unrelated hyases (for example, PH20/SPAM1) facilitating HA digestion and penetration of the cumulus mass. Immature spermatozoa, however, do not bind to HA or may bind it more weakly (Huszar et al., 2003; Nasr-Esfahani et al., 2008).
Hyaladherins including CD44 (Bajorath et al., 1998; Underhill, 1992) and RHAMM (Hardwick et al., 1992; Yang et al., 1994), both present in ejaculate spermatozoa (Amaral et al., 2014), are categorized by whether they contain a Link domain or module (a sequence of ~100 amino acids composed of two alpha-helices, two triple-stranded anti-parallel beta-sheets and two disulphide bonds (Barta et al., 1993)) or a BX7B motif (where the ‘B’s are arginine (R) or lysine (K) residues and the ‘X’ is a sequence of seven non-acidic and at least one basic amino acid with a covalent bond) or combinations thereof (Amemiya et al., 2005; Day and Prestwich, 2002; Yang et al., 1994). CD44, for example, contains both a Link module and a BX7B motif.
Investigating sperm hyaladherins is justified in relation to understanding the fertilization potential of sperm and the causes of male infertility. A previous study in our laboratory provided evidence for the complexity of hyaladherin expression in ejaculated human spermatozoa and showed that sperm binding to HA is enhanced by capacitation (Torabi et al., 2017) and may involve unknown hyaladherins. To improve our understanding, proteins extracted from washed and homogenized ejaculate human spermatozoa were subjected to affinity panning on a HA-coated surface (Amemiya et al., 2005). Adherent (binding) and non-binding proteins were recovered and subsequently characterized by tandem mass spectrometry (LC-MS/MS).
Materials and Methods
Reagents used
EDTA, Triton-X-100, Acrylamide/Bis-acrylamide, 30% solution, Sodium dodecyl sulphate (SDS), Ammonium persulphate (APS) and N,N,N′,N′-Tetramethylethylenediamine (TEMED) were obtained from Sigma-Aldrich (UK). PVDF membranes and Amicon Ultra-0.5 and 15 ml centrifugal filter units were obtained from Millipore (UK). Pierce BCA protein assay kit and Pierce™ protein-free (PBS/TBS) blocking buffer were obtained from Thermo Fisher Scientific (UK). Tris Base and NaCl were purchased from Fisher Scientific (UK). Protease inhibitor cocktail was purchased from Cell Signalling Technology (UK). ColorPlus prestained protein ladder was acquired from New England Biolabs (UK). Clarity™ western ECL substrate and Bradford protein assay reagent were purchased from Bio-Rad (UK). HA-coated dishes were purchased from Biocoat (USA). The monoclonal anti-ZPBP2 antibody was purchased from antibodies-online. The monoclonal anti-ADAM32 (sc-376 738) and anti-Alpha-tubulin (SC-5286) antibodies were obtained from Santa Cruz Biotechnology. Water for mass spectrometry containing 0.1% formic acid (LC-MS Chromasolv®) was from Honeywell (Seelze, Germany) and the acetonitrile from Fluka. The rest of the reagents were supplied by Sigma-Aldrich (St. Louis, MO) unless otherwise stated.
Ethical approval
The study was considered and nationally approved by the relevant UK Integrated Research Application System (IRAS) Ethics Committee (NRES 12_NE_0192) on 13 January 2013 and locally approved by the University of Leeds’ School of Medicine Research Ethics Committee (SoMREC/13/017) on 28 November 2013.
Semen analysis
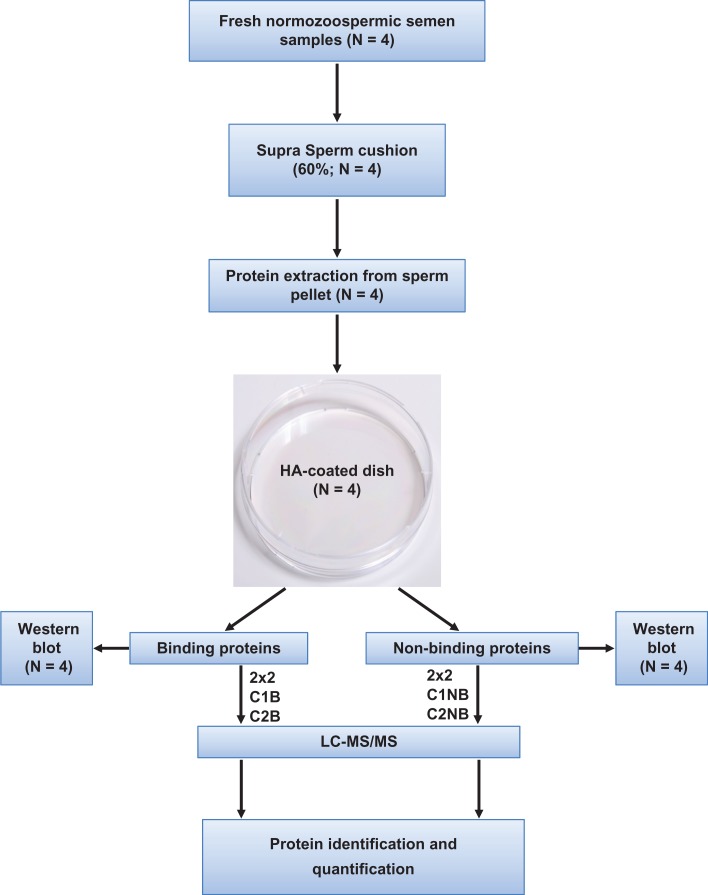
A schematic of the experimental plan is shown in Fig. 1. Human semen samples were obtained ethically from young male volunteers (18–25 years of age) of unproven fertility by masturbation into sterile, tissue culture grade universal containers after three days abstinence. Collected semen samples were immediately liquefied for 30 min at 37°C. The semen parameters related to each sample are shown in Table I. After liquefaction, all samples were checked for volume, sperm concentration, sperm morphology and the number of round cells according to the WHO (World Health Organisation) criteria (WHO, 2010) and following, where appropriate, the guidelines recommended by Bjorndahl et al. (2016). Only those with normal semen parameters as defined by WHO criteria were included in the study.
Figure 1.
Flow chart of experimental design.
Table I.
Semen parameters of young male volunteers (unproven fertility) participating in the study.
| Sample | Volunteer Age | Sperm concentration (million/ml) | Total sperm count (×106) | Semen volume (ml) | Sperm motility (%) |
|---|---|---|---|---|---|
| D1 | 28 | 233 | 349.5 | 1.5 | 92 |
| D2 | 36 | 200 | 300 | 1.5 | 90 |
| D3 | 19 | 251 | 1204.8 | 4.8 | 87 |
| D4 | 20 | 179 | 716 | 4 | 61 |
| Mean ± SD | 26 ± 8 | 216 ± 32.34 | 643 ± 418 | 3 ± 1.70 | 83 ± 14.5 |
Sperm preparation
To maximize the recovery of sperm, liquefied semen samples were centrifuged through a 60% density cushion of SupraSperm™ using special inserts (ProInsert-Nidacon, Sweden) to remove any round cell contamination at 300 × g for 20 min. A pellet retrieval pipette (ProInsert kit) was used to aspirate sperm pellets which were then resuspended and washed in PBS (pH: 7.2) followed by centrifugation at 300 × g for 10 min (two repeated washes). Sperm motility and morphology were assessed using a Leitz Laborlux 12 light microscope and the sperm pellets were used for further protein extraction.
Protein extraction and affinity panning of sperm proteins
To reduce costs while as far as possible retaining sample heterogeneity, two aliquots of each sample were pooled into two (~2 × 108 sperm in total, each) combined samples (C1 and C2) and extracted separately in 500 μl of a mild lysis buffer (150 mM NaCl, 20 mM Tris-HCl (pH: 8), 2 mM EDTA, 1× protease inhibitor cocktail, 0.5% Triton X-100) (D’Cruz et al., 1993). The suspensions were sonicated on ice for 15 s on, 40 s off, at an amplitude of 10 microns. The sonication was repeated four times. Samples were incubated on ice for 1 h with constant shaking. To recover extracted soluble proteins, samples were centrifuged at 16 000 × g for 20 min and supernatants were aspirated and transferred into fresh tubes. Pellets were discarded. Protein quantification was performed using the Pierce BCA protein assay kit according to manufacturer’s instruction (Thermo Fisher Scientific, UK). HA-coated dishes (Biocoat, USA) were used to pan for proteins in the sample homogenates (C1 and C2) with an affinity for HA. To block non-specific binding, dishes were treated beforehand with 1.5 ml of Pierce™ protein-free (PBS) blocking buffer (Thermo Fisher Scientific, UK) for 30 min at room temperature (RT) with gentle shaking. Dishes were then washed twice with 2 ml of phosphate buffered saline (PBS). Extracted proteins from the sperm of two different men were used and loaded on to separate HA-coated dishes at a concentration of 1.5 mg/ml. Dishes were incubated for 75 min at RT with gentle shaking after which, non-binding proteins were decanted for frozen storage at −80°C.
To minimize non-specific contamination of the binding fractions, dishes were washed with 1.5 ml PBS (four times for 1 min each wash). Bound proteins were then recovered by incubating the plate surface with 400 μl of a pre-heated (95°C) Laemmli-based buffer containing 0.125 M Tris-HCl (pH: 6.8), 4%w/v SDS and 10%v/v 2-mercaptoethanol for 5 min with gentle shaking. Bound proteins were aspirated using a pipette. Both protein fractions (HA-binding and non-binding) were processed through Amicon Ultra-15 ml centrifugal desalting filters (3 kDa) according to manufacturer’s instruction (Millipore, UK). The proteins were then concentrated using Amicon’s Ultra-0.5 ml centrifugal filters according to manufacturer’s instruction (Millipore, UK). A final volume (40 μl) was obtained after concentration and 30 μl of each was precipitated using TCA-acetone (see below). Ten (10 μl) aliquots were stored at −80°C for western blot analysis (see below).
Protein precipitation
To precipitate proteins, one volume of 100% (w/v) TCA was added to four volumes of the protein solution (final conc of 20% TCA) from both HA-binding and non-binding samples. All samples were incubated for 20 min at 4°C and centrifuged at 19 000×g for 5 min. Supernatants were discarded. Pellets were washed with 200 μl of ice-cold acetone and centrifuged at 19 000×g for 5 min at 4°C (twice, with washing between). Pellets were dried and stored at −80°C before sequencing by LC-MS/MS as outlined below.
Liquid chromatography-tandem mass spectrometry
Proteins were digested with trypsin (Promega, Madison, WI) at 37°C overnight according to the manufacturer’s recommendations. Tryptic peptides were separated by means of nano-liquid chromatography using an Eksigent NanoLC AS2 ultra (AB SCIEX) with a flow rate of 400 nl/min, an EASY C18 trap column (5 μm, 120 Å, 100 μm inner diameter × 2 cm in length), and an EASY C18 analytical column (3 μm, 120 Å, 75 μm inner diameter × 10 cm in length). The following linear gradient, using solvent B (97% acetonitrile, 0.1% formic acid) and solvent A (3% acetonitrile, 0.1% formic acid), was employed: 5–40% buffer B (65 min); 40–100% buffer B (5 min). MS/MS analysis was performed using an LTQ Orbitrap Velos (Thermo Fisher Scientific) with a nanoelectrospray ion source with precursor ion selection in the Orbitrap at 30.000 of resolution, selecting the 15 most intense precursor ions, with a collision energy of 35 in positive ion mode. MS/MS data acquisition was completed using Xcalibur 2.1 (Thermo Fisher Scientific). Figure 1 shows a schematic illustration of the study plan.
Protein identification, quantification and analysis
Proteome Discoverer 1.4 (Thermo Fisher Scientific) was used to characterize all proteins. Database searching included all entries from the Homo sapiens UniProtKB/Swiss-Prot database (release 02–2015) using SEQUEST version 28.0 (Thermo Fisher Scientific). A 1% false discovery rate (FDR) and at least one (≥1) unique peptide per protein were the criteria used for protein identification. The dissociated or ‘ungrouping’ of proteins from their respective families was used during the quantification process in order to avoid possible ambiguities associated with different isoforms of the same protein.
Identified proteins in binding and non-binding fractions from the two combined samples were compared and checked against the whole human sperm proteome (Amaral et al., 2014) and only those present in this database were included for further analysis. Additional information about the identified proteins was obtained using UniProtKB/Swiss-Prot, and BioMart - Ensembl. DAVID Bioinformatics Resources 6.8 was used for functional ontological annotation clustering (Huang et al., 2007).
PDBsum was used to align amino acid sequences of identified proteins with sequences for which structural information is available (Laskowski, 2001, 2009; Laskowski et al., 2005). The SAS (Sequences annotated by structure) server was then used (http://www.ebi.ac.uk/thornton-srv/databases/sas/) to determine the structural similarities between identified proteins in HA-binding and non-binding fractions with the HA-binding domain of CD44 (highlighted by PDBsum). SAS annotates protein residues according to residue type (polar, non-polar, aliphatic and aromatic), secondary structure (alpha-helix, beta-strand, turn and coil), inter-molecular contacts (number of hydrogen bonds, total contacts, nucleic acid contacts and metal ions contacts to ligand), active sites and residue similarity (Milburn et al., 1998). Sequence similarities were computed using the PAM250 log odds scores matrix with FASTA used as the sequence search method and presented as a simple pairwise comparisons. The colouring of residue similarity signifies identical amino acid types (red) down to conservative, semi-conservative and dissimilar residues using a spectrum ranging from orange through to blue. In addition, a Z-score and an E-value for sequence similarity were computed (Pagni and Jongeneel, 2001).
Western blot analysis
To verify the results of LC-MS/MS experiments, 15 μg of binding and non-binding proteins from each combined sample were resolved by SDS PAGE and western blotted. Samples were dissolved and diluted 1:1 with 2× Laemmli buffer (4% SDS, 10% 2-mercaptoethanol, 20% glycerol, 0.004% bromophenol blue, 0.125 M Tris-HCl (pH: 6.8)) and boiled for 5 min at 95°C. The proteins were separated on a 10% gel at 120 V for 100 min in 1× SDS running buffer (25 mM Tris-base, 190 mM Glycine, 0.1% SDS). Proteins were then transferred to PVDF membranes at 250 mA for 90 min in 1× transfer buffer (25 mM Tris-base, 190 mM Glycine, and 20% methanol). The membrane was blocked with Pierce™ protein-free (TBS) blocking buffer (Thermo Fisher Scientific, UK) for 1 h at RT with constant shaking and then probed with antibodies to two proteins specific to the binding fraction, zona pellucida-binding protein 2 (ZPBP2) and ADAM32 (200 μg/ml) at a primary antibody concentration of 1/500 and 1/200, respectively overnight at 4°C. An alpha tubulin antibody (200 μg/ml at a final concentration of 1/2000) was used as a control for proteins present in both fractions and as a loading control. Membranes were washed three times with TBS containing 0.1% Tween-20 for 15 min each and incubated with 1:1000 dilution of relevant HRP-conjugated secondary antibodies for 1 h at RT (all primary and secondary antibodies were diluted in Pierce™ protein-free (TBS)). Membranes were washed as described above and the Clarity western ECL substrate was used to visualize protein bands (Bio-Rad, UK).
Results
Identification of proteins in HA-binding and non-binding fractions
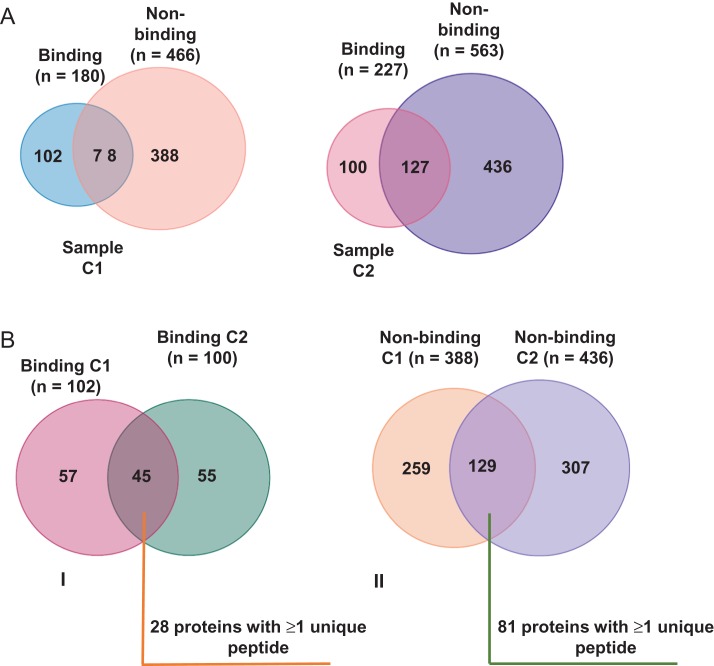
LC-MS/MS data resulted in the identification of 180 (sample C1) and 227 (sample C2) proteins in HA-binding fractions and 466 (sample C1) and 563 (sample C2) proteins in non-binding fractions, respectively (Fig. 2A). As shown in Fig. 2BI, 45 proteins were common and specific (no overlaps with non-binding fractions) to both HA-binding fractions and had records in the human sperm proteome database (Amaral et al., 2014), of which 28 had ≥1 unique peptides per protein. There were 129 proteins common to both non-binding fractions with 81 recorded in the human sperm proteome database and represented by ≥1 unique peptides (Fig. 2BII). A list of all mass spectrometry data can be found in Supplementary Tables I–IV. Additional information about the identified proteins was obtained using UniProtKB/Swiss-Prot, and BioMart - Ensembl. Table II lists proteins in the binding fractions. The equivalent list from the non-binding fractions is shown in Supplementary Table V.
Figure 2.
Venn diagrams illustrating overlaps between (A) HA- and non-binding proteins in two (CI and C2) sperm samples from four volunteer donors of unproven fertility and (B) overlaps between combined HA-binding (I) and non-binding proteins (II) from these samples.
Table II.
Proteins in both (C1 and C2) binding fractions with ≥1 unique identifying peptide and with no overlap with proteins in the non-binding fractions.
| UniProt/Swiss-Prot Accession | Description | UniProt Gene name | Potential HA-binding domain (N) |
|---|---|---|---|
| A4D1T9 | protease, serine, 37 | PRSS37 | BX7B (1) |
| P03973 | Antileukoproteinase, secretory leucocyte peptidase inhibitor | SLPI | BX7B (1) |
| P05109 | S100 calcium binding protein A8 | S100A8 | BX7B (1) |
| P13647 | Keratin, type II cytoskeletal 5 | KRT5 | N/D |
| P20155 | serine protease inhibitor, Kazal type 2 (acrosin-trypsin inhibitor) | SPINK2 | N/D |
| P21741 | midkine (neurite growth-promoting factor 2) | MDK | BX7B (4) |
| P35663 | cylicin, basic protein of sperm head cytoskeleton 1 | CYLC1 | BX7B (6) |
| P62987 | ubiquitin A-52 residue ribosomal protein fusion product 1 | UBA52 | BX7B (1) |
| P99999 | cytochrome c, somatic | CYCS | BX7B (1) |
| Q00796 | sorbitol dehydrogenase | SORD | N/D |
| Q13618 | cullin 3 | CUL3 | BX7B (2) |
| Q16568 | CART prepropeptide | CARTPT | N/D |
| Q16836 | hydroxyacyl-CoA dehydrogenase | HADH | N/D |
| Q6NUT2 | Probable C-mannosyltransferase | DPY19L2 | BX7B (2) |
| Q6P4A8 | phospholipase B domain-containing 1 | PLBD1 | BX7B (1) |
| Q6UWM5 | GLIPR1-like protein 1 | GLIPR1L1 | N/D |
| Q6X784 | zona pellucida-binding protein 2 | ZPBP2 | Link-like domain |
| Q86YZ3 | hornerin | HRNR | N/D |
| Q8N5Q1 | family with sequence similarity 71, member E2 | FAM71E2 | BX7B (3) |
| Q8TC27 | ADAM metallopeptidase domain 32 | ADAM32 | BX7B (2) |
| Q8WZ59 | transmembrane protein 190 | TMEM190 | N/D |
| Q96KX0 | lysozyme-like 4 | LYZA | N/D |
| Q96QH8 | sperm acrosome associated 5B | SPACA5 | N/D |
| Q9BVA1 | tubulin, beta 2B class IIb | TUBB2B | N/D |
| Q9BWH2 | FUN14 domain-containing 2 | FUNDC2 | BX7B (1) |
| Q9NPJ3 | acyl-CoA thioesterase 13 | ACOT13 | N/D |
| Q9UII2 | ATPase inhibitory factor 1 | ATPIF1 | N/D |
| Q9Y6A4 | cilia and flagella associated protein 20 | CFAP20 | BX7B (3) |
Numbers in parenthesis indicate the number of BX7B motifs found in the protein sequence. N/D is not detected.
An ontology analysis was undertaken to determine whether binding and non-binding proteins carried distinct functional signatures indicating a potential HA affinity effect. Following submission to DAVID, results suggested distinct signatures with the bound fraction carrying a weak enrichment for secreted proteins although a stronger enrichment in the unbound fraction was found for chaperone and chaperone-like proteins (Supplementary Tables VI and VII).
Alignment of the sequences of HA-binding and non-binding proteins using PDBsum and SAS
The amino acid sequences of the 28 proteins in the HA-binding fractions were submitted to PDBsum, which generates lists of structural features (PDB codes) conserved between proteins. One protein (ZPBP2) contained a sequence motif bearing similarity to the Link module of CD44, while 14 others (50% in total; Table II) contained BX7B sequences. There were 28 proteins (34.5%; Supplementary Table V) from the non-binding fraction which also contained the BXB7 motif, indicating a slight enrichment for proteins containing the motif in the binding fractions (one-tailed Z-score = 1.45; P = 0.074). No proteins containing a Link-like sequence were present in the non-binding fraction. Proteins with multiple (≥2) BX7B motifs, including ADAM32 (Uniprot QBTC27) with two and Midkine (Uniprot P21741) with four, of which the latter is known to bind HA and other glycosaminoglycans (http://www.ebi.ac.uk/QuickGO/GProtein?ac=P21741), were more abundant in the binding (25%) versus the non-binding (15%) fractions although this difference was not significant (one-tailed Z-score = 1.22; P = 0.111). Hence, the trends suggest that HA-binding motifs may have influenced the partitioning of the two populations. Charge differences between proteins were also explored as an alternative partitioning mechanism. Net charge is determined by proteins’ pI and since HA carries a net negative charge at pH 8.0 (the pH of the extraction buffer), basic proteins with a net positive charge could have electrostatically interacted with the substrate. There were 20 (71.4%) proteins in the binding fraction, compared with 21 (26%) in the non-binding fraction, likely to carry a net positive charge, suggesting that charge effects may have influenced the partitioning of proteins in the fractions (one-tailed Z-score = 4.28; P < 0001).
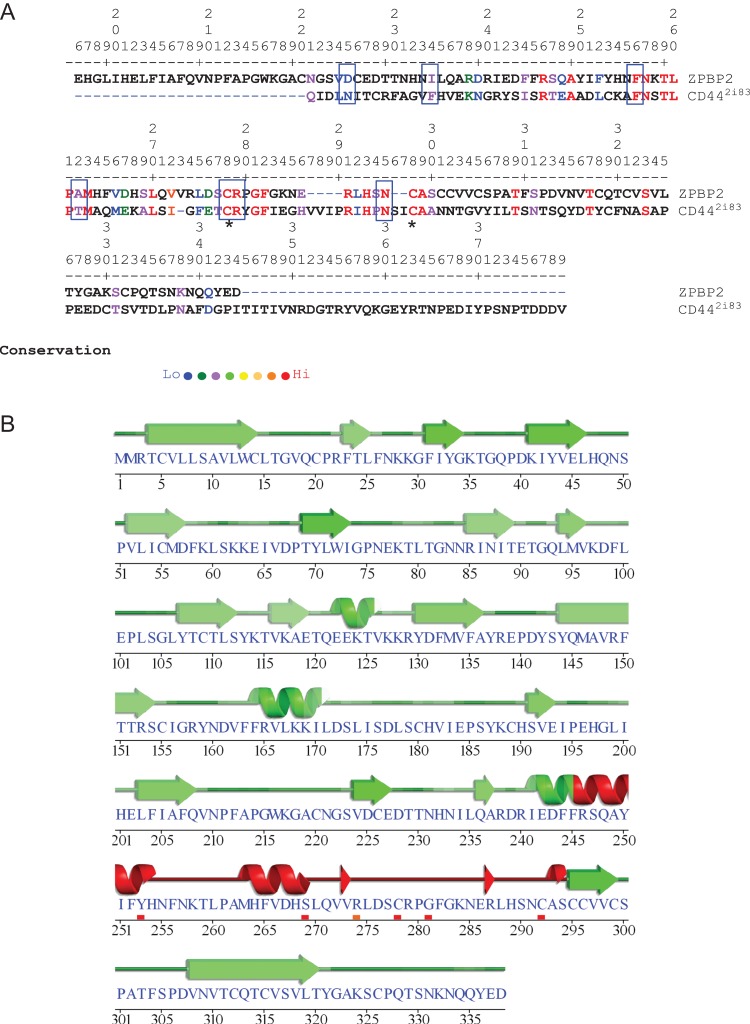
Of all the proteins in the binding fractions, ZPBP2 which contains a sequence motif with similarities to the Link module found in CD44 was flagged as one of the more interesting. ZPBP2 (also known as ZPBPL) was first reported in 2003 from an in silico search of a region on human chromosome 17q12 that is frequently amplified in breast and stomach cancers (Katoh and Katoh, 2003). Figure 3A shows the alignment between ZPBP2 and CD44 (PDB code 2i83, residues 21–178) with a 32.7% sequence identity and 69% overall similarity covering a 55 amino acid overlap. Unlike CD44 where the Link module is close to the amino-terminal end, the equivalent sequence in ZPBP2 is located towards the carboxyl-terminal end. There were no proteins with a sequence resembling the Link module in the non-binding fractions. The 2i83 PDB code comparison is shown because it corresponds with the CD44’s HA-binding form although other valid codes representing a similar residue overlap were also flagged (Table III) (Liu and Finzel, 2014; Takeda et al., 2006; Teriete et al., 2004). As ZPBP2 may have hyaladherin properties based on sequence similarities with the Link domain, further structural similarities were investigated using the Sequences Annotated by Structure (SAS) server (Milburn et al., 1998). This resource checks for the number of contacts between the protein and its presumptive ligand based on secondary structure prediction and was used to scan the homologous HA-binding motif on ZPBP2. The PROSITE secondary structure alignment (PS01241) for 2i83 (http://prosite.expasy.org/cgi-bin/prosite/prosite-search-ac?PS01241) with ZPBP2 is shown in Fig. 3B. Residues in ZPBP2 conserved with those in CD44 within the HA-binding domain, included Arg46/246, Ala49/249, Phe56/256, Asn57/257, Thr59/259, Leu60/260, Pro61/261, Met63/263, Leu70/270, Cys78/278, Arg79/279, Gly81/281, Phe82/282, Arg91/287, His93/289, Asn95/291, Cys98/292, Ala99/293, Thr109/303, Thr117/311, Ser123/317). In Fig. 3A, boxes indicate conserved residues involved in ligand binding and the stars indicate conserved cysteine residues.
Figure 3.
Homology between the ligand (HA) bound form of CD44 (PDB code 2i83, 158 amino acids) and ZPBP2; 338 amino acids). Panel A shows the best residue alignment of ZPBP2 with 2i83. Blue dashed lines indicate alignment adjustment spacing of 2i83 which means the residue numbers in panels A and B will not necessarily correspond. Boxes border residues implicated in ligand binding. Residue conservation is denoted by colours ranging from red (identical) to least conserved (blue). Panel B shows the predicted secondary structure (helices and strands) of ZPBP2 with the confidence for helix and strand predictions denoted by green shading (darker shade means higher confidence). Red shading signifies the Link domain signature of CD44 with the small shaded bars indicating the strongest drivers of predicted secondary structure. Stars in Panel A indicate the cysteines thought to be critical to the HA-binding configuration of the Link module.
Table III.
PDB codes for sequences matching ZPBP2 (http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=index.html).
| PDB code | Model | Length | %-identity | AA overlap | z-score | Ligands | Protein name |
|---|---|---|---|---|---|---|---|
| 4pz3(A) | X-ray 1.08 Å | 150 | 32.7 | 55 | 116.4 | ALA-ALA-ALA-VAL, PEG, EDO, MES. | High-resolution crystal structure of the human cd44 hyaluronan domain complex with undefined peptides. |
| 1uuh(A) | X-ray 2.20 Å | 150 | 32.7 | 55 | 116.4 | Hyaluronan-binding domain of human cd44. | |
| 4pz4(A) | X-ray 1.60 Å | 154 | 32.7 | 55 | 116.2 | SO4, PEG, GOL, EDO, DMS. | High-resolution crystal structure of the human cd44 hyaluronan domain in new space group. |
| 2i83(A) | NMR | 158 | 32.7 | 55 | 116.1 | Hyaluronan-binding domain of cd44 in its ligand-bound form. | |
| 1 poz(A) | NMR | 159 | 32.7 | 55 | 116.0 | Solution structure of the hyaluronan-binding domain of human cd44. |
PDB was also used to check whether the Link module that could influence the partitioning of proteins into binding and non-binding fractions was present in any of the 81 proteins common to the non-binding fractions. None returned a PDB code. We also aligned the sequences of ten putative HA-binding proteins containing the Link module (Table IV*) and 11 randomly selected proteins from the non-binding fraction (Table IV‡) without evidence for a Link module. SAS was then used to look for any existing structural similarities between them and the CD44 motifs flagged by the PDB codes (4pz3, 4pz4, 1uuh, 1 poz and 2i83; Table III). When aligned with these sequences, the first seven proteins in the list had the highest Z-scores (123–321) and lowest E-values (4.5e−16−2.2e−5). The next three proteins had lower Z-scores (50.6−112.1) and higher E-values (1.9e−4−0.38). The 11 proteins in the non-binding fraction (Table IV‡) occasionally showed higher percentage identities with the CD44 HA-binding domain (PLIN3 for example). Similarly, human axonemal dynein light intermediate polypeptide 1 (O14645) also showed a high % identity with sequences represented by PDB codes 4pz3 and 1uuh but with a far lower Z-score and high E-value. Relative Z-scores and E-values give more accurate predictions of structural relationships between proteins than percentage identity and although not reported to contain a Link module hitherto, ZPBP2 is included in Table IV because it was flagged by PDB / SAS and has a relatively high Z-score and low E-value.
Table IV.
List of 10 HA-binding proteins containing and 11 randomly selected proteins from the HA-non-binding fraction without the Link module.
| PDB code | Smith Waterman Score | % Identity | a.a Overlap | Seq length | Z-score | E-value | Protein name/UniProt accession number |
|---|---|---|---|---|---|---|---|
| 4pz4 (a.a 18–171) | 227 | 35 | 120 | 227 | 279.5 | 9.2e-14 | TSG6_Human tumour necrosis factor-inducible gene 6 protein (P98066)* |
| 4pz3(a.a 18–170) | 227 | 35 | 120 | 227 | 279.9 | 8.8e-14 | |
| 1uuh(a.a 20–178) | 227 | 35 | 120 | 227 | 279.9 | 8.8e-14 | |
| 2i83 (a.a 21–178) | 227 | 35 | 120 | 227 | 279.2 | 9.6e-14 | |
| 1 poz (a.a 20–178) | 227 | 35 | 120 | 227 | 279.1 | 9.7e-14 | |
| 4pz4 (a.a 18–171) | 259 | 32.6 | 138 | 322 | 317.6 | 7e-16 | LYVE1_Human Lymphatic vessel endothelial HA receptor 1 (Q9Y5Y7)* |
| 4pz3(a.a 18–170) | 259 | 32.6 | 138 | 322 | 318 | 6.7e-16 | |
| 1uuh(a.a 20–178) | 259 | 32.6 | 138 | 322 | 318 | 6.7e-16 | |
| 2i83 (a.a 21–178) | 262 | 32.3 | 158 | 322 | 321 | 4.5e-16 | |
| 1 poz (a.a 20–178) | 262 | 32.3 | 158 | 322 | 320 | 4.6e-16 | |
| 4pz4 (a.a 18–171) | 148 | 26.2 | 103 | 862 | 173 | 7.9e-08 | Human putative uncharacterized protein DKFZp434E0321 (Fragment) (CAB61358) (Q9UF98)* |
| 4pz3(a.a 18–170) | 148 | 26.2 | 103 | 862 | 173.4 | 7.5e-08 | |
| 1uuh(a.a 20–178) | 148 | 26.2 | 103 | 862 | 173.4 | 7.5e-08 | |
| 2i83 (a.a 21–178) | 148 | 26.2 | 103 | 862 | 172.7 | 8.3e-08 | |
| 1 poz (a.a 20–178) | 148 | 26.2 | 103 | 862 | 172.6 | 8.4e-08 | |
| 4pz4 (a.a 18–171) | 154 | 29.8 | 94 | 2415 | 173.1 | 7.8e-08 | Human Aggrecan core protein (P16112)* |
| 4pz3(a.a 18–170) | 154 | 29.8 | 94 | 2415 | 173.5 | 7.4e-08 | |
| 1uuh(a.a 20–178) | 154 | 29.8 | 94 | 2415 | 173.5 | 7.4e-08 | |
| 2i83 (a.a 21–178) | 154 | 29.8 | 94 | 2415 | 172.6 | 8.3e-08 | |
| 1 poz (a.a 20–178) | 154 | 29.8 | 94 | 2415 | 172.5 | 8.4e-08 | |
| 4pz4 (a.a 18–171) | 123 | 26.1 | 88 | 340 | 149.6 | 1.6e-06 | Human Hyaluronan and proteoglycan link protein 2 (BRAL1) (Q9GZV7)* |
| 4pz3(a.a 18–170) | 123 | 26.1 | 88 | 340 | 150 | 1.5e-06 | |
| 1uuh(a.a 20–178) | 123 | 26.1 | 88 | 340 | 150 | 1.5e-06 | |
| 2i83 (a.a 21–178) | 123 | 26.1 | 88 | 340 | 123 | 1.7e-06 | |
| 1 poz (a.a 20–178) | 123 | 26.1 | 88 | 340 | 149.2 | 1.7e-06 | |
| 4pz4 (a.a 18–171) | 182 | 31.4 | 102 | 2570 | 207.2 | 9.9e-10 | HUMAN Stabilin-1 (Q9NY15)* |
| 4pz3(a.a 18–170) | 182 | 31.4 | 102 | 2570 | 207.6 | 9.3e-19 | |
| 1uuh(a.a 20–178) | 182 | 31.4 | 102 | 2570 | 207.6 | 9.3e-10 | |
| 2i83 (a.a 21–178) | 182 | 31.4 | 102 | 2570 | 206.7 | 1e-09 | |
| 1 poz (a.a 20–178) | 182 | 31.4 | 102 | 2570 | 206.7 | 1e-09 | |
| 4pz4 (a.a 18–171) | 115 | 25.5 | 94 | 1321 | 129.3 | 2.2e-05 | Human Neurocan core protein (O14594)* |
| 4pz3(a.a 18–170) | 115 | 25.5 | 94 | 1321 | 129.7 | 2e-05 | |
| 1uuh(a.a 20–178) | 115 | 25.5 | 94 | 1321 | 129.7 | 2e-05 | |
| 2i83 (a.a 21–178) | 115 | 25.5 | 94 | 1321 | 128.9 | 2.3e-05 | |
| 1 poz (a.a 20–178) | 115 | 25.5 | 94 | 1321 | 128.8 | 2.3e-05 | |
| 4pz4 (a.a 18–171) | 123 | 24.3 | 107 | 911 | 65.4 | 0.075 | Human Brevican core protein (Q96GW7)* |
| 4pz3(a.a 18–170) | 123 | 24.3 | 107 | 911 | 65.8 | 0.071 | |
| 1uuh(a.a 20–178 | 123 | 24.3 | 107 | 911 | 65.8 | 0.071 | |
| 2i83 (a.a 21–178) | 123 | 24.3 | 107 | 911 | 65.1 | 0.078 | |
| 1 poz (a.a 20–178) | 123 | 24.3 | 107 | 911 | 65 | 0.079 | |
| 4pz4 (a.a 18–171) | 123 | 27.8 | 108 | 3396 | 111.6 | 0.00021 | Human Versican core protein (P13611)* |
| 4pz3(a.a 18–170) | 123 | 27.8 | 108 | 3396 | 112.1 | 0.00019 | |
| 1uuh(a.a 20–178 | 123 | 27.8 | 108 | 3396 | 112.1 | 0.00019 | |
| 2i83 (a.a 21–178) | 123 | 27.8 | 108 | 3396 | 111.2 | 0.00022 | |
| 1 poz (a.a 20–178) | 123 | 27.8 | 108 | 3396 | 111.2 | 0.00022 | |
| 4pz4 (a.a 18–171) | 61 | 26.2 | 61 | 629 | 51 | 0.39 | Human Sushi domain-containing protein 5 (KIAA0527) (O60279)* |
| 4pz3(a.a 18–170) | 61 | 26.2 | 61 | 629 | 51.4 | 0.38 | |
| 1uuh(a.a 20–178 | 61 | 26.2 | 61 | 629 | 51.4 | 0.38 | |
| 2i83 (a.a 21–178) | 61 | 26.2 | 61 | 629 | 50.6 | 0.4 | |
| 1 poz (a.a 20–178) | 61 | 26.2 | 61 | 629 | 50.6 | 0.41 | |
| 4pz4 (a.a 18–171) | 88 | 32.7 | 55 | 316 | 107 | 3.7e-04 (0.00037) | Zona pellucida-binding protein 2 (ZPBP2) (Q6 × 784)*? |
| 4pz3(a.a 18–170) | 88 | 32.7 | 55 | 316 | 107.5 | 3.5e-4 | |
| 1uuh(a.a 20–178) | 88 | 32.7 | 55 | 316 | 107.5 | 3.5e-4 | |
| 2i83 (a.a 21–178) | 88 | 32.7 | 55 | 316 | 106.8 | 3.8e-04 | |
| 1 poz (a.a 20–178) | 88 | 32.7 | 55 | 316 | 106.7 | 3.9e-04 | |
| 4pz4 (a.a 18–171) | 40 | 20.7 | 29 | 215 | 38 | 0.93 | Human sperm acrosome membrane-associated protein 3 (Q81XA5)‡ |
| 4pz3(a.a 18–170) | 40 | 20.7 | 29 | 215 | 38.3 | 0.92 | |
| 1uuh(a.a 20–178) | 40 | 20.7 | 29 | 215 | 38.3 | 0.92 | |
| 2i83 (a.a 21–178) | 40 | 20.7 | 29 | 215 | 37.7 | 0.93 | |
| 1 poz (a.a 20–178) | 40 | 20.7 | 29 | 215 | 37.6 | 0.94 | |
| 4pz4 (a.a 18–171) | 58 | 25 | 92 | 548 | 39.8 | 0.88 | Human T-complex protein 1 subunit theta (P50990)‡ |
| 4pz3(a.a 18–170) | 58 | 25 | 92 | 548 | 40.1 | 0.86 | |
| 1uuh(a.a 20–178) | 58 | 25 | 92 | 548 | 40 | 0.86 | |
| 2i83 (a.a 21–178) | 58 | 225 | 92 | 548 | 39.4 | 0.89 | |
| 1 poz (a.a 20–178) | 58 | 25 | 92 | 548 | 39.3 | 0.89 | |
| 4pz4 (a.a 18–171) | 46 | 23.4 | 64 | 170 | 52.7 | 0.33 | CAMP_Human Cathelicidin antimicrobial peptide (P49913)‡ |
| 4pz3(a.a 18–170) | 46 | 23.4 | 64 | 170 | 53 | 0.32 | |
| 1uuh(a.a 20–178) | 46 | 23.4 | 64 | 170 | 53 | 0.32 | |
| 2i83 (a.a 21–178) | 46 | 23.4 | 64 | 170 | 52.4 | 0.34 | |
| 1 poz (a.a 20–178) | 46 | 23.4 | 64 | 170 | 52.3 | 0.34 | |
| 4pz4 (a.a 18–171) | 39 | 23.5 | 34 | 258 | 42.4 | 0.77 | Human Axonemal dynein light intermediate polypeptide 1 (O14645)‡ |
| 4pz3(a.a 18–170) | 34 | 55.6 | 9 | 258 | 42.7 | 0.76 | |
| 1uuh(a.a 20–178) | 34 | 55.6 | 9 | 258 | 42.7 | 0.76 | |
| 2i83 (a.a 21–178) | 39 | 23.5 | 34 | 258 | 42.1 | 0.79 | |
| 1 poz (a.a 20–178) | 39 | 23.5 | 34 | 258 | 42 | 0.79 | |
| 4pz4 (a.a 18–171) | 49 | 38.9 | 18 | 434 | 33 | 0.99 | PLIN3_HUMAN Perilipin-3 O60664‡ |
| 4pz3(a.a 18–170) | 49 | 38.9 | 18 | 434 | 33.4 | 0.99 | |
| 1uuh(a.a 20–178) | 49 | 38.9 | 18 | 434 | 33.4 | 0.99 | |
| 2i83 (a.a 21–178) | 49 | 38.9 | 18 | 434 | 32.7 | 0.99 | |
| 1 poz (a.a 20–178) | 49 | 38.9 | 18 | 434 | 32.6 | 0.99 | |
| 4pz4 (a.a 18–171) | 53 | 23.9 | 67 | 148 | 71.5 | 0.035 | LYZL6_HUMAN Lysozyme-like protein 6 (O75951)‡ |
| 4pz3(a.a 18–170) | 50 | 28.1 | 64 | 148 | 63.2 | 0.099 | |
| 1uuh(a.a 20–178) | 50 | 28.1 | 64 | 148 | 63.2 | 0.099 | |
| 2i83 (a.a 21–178) | 50 | 28.1 | 64 | 148 | 62.6 | 0.11 | |
| 1 poz (a.a 20–178) | 50 | 28.1 | 64 | 148 | 62.5 | 0.11 | |
| 4pz4 (a.a 18–171) | 38 | 25 | 32 | 150 | 52.8 | 0.32 | Human Cytochrome c oxidase subunit 5 A, mitochondrial (P20674)‡ |
| 4pz3(a.a 18–170) | 38 | 25 | 32 | 150 | 53.1 | 0.31 | |
| 1uuh(a.a 20–178) | 38 | 25 | 32 | 150 | 53.1 | 0.31 | |
| 2i83 (a.a 21–178) | 38 | 25 | 32 | 150 | 52.6 | 0.33 | |
| 1 poz (a.a 20–178) | 38 | 25 | 32 | 150 | 52.5 | 0.33 | |
| 4pz4 (a.a 18–171) | 45 | 23.3 | 43 | 381 | 50.1 | 0.43 | Human Putative heat shock protein HSP 90-beta 2 (Q58FF8)‡ |
| 4pz3(a.a 18–170) | 45 | 23.3 | 43 | 381 | 50.4 | 0.41 | |
| 1uuh(a.a 20–178) | 45 | 23.3 | 43 | 381 | 50.4 | 0.41 | |
| 2i83 (a.a 21–178) | 45 | 23.3 | 43 | 381 | 49.8 | 0.44 | |
| 1 poz (a.a 20–178) | 45 | 23.3 | 43 | 381 | 49.7 | 0.44 | |
| 4pz4 (a.a 18–171) | 33 | 17.4 | 46 | 369 | 24.5 | 1 | Human Hsc70-interacting protein (P50502)‡ |
| 4pz3(a.a 18–170) | 33 | 17.4 | 46 | 369 | 24.8 | 1 | |
| 1uuh(a.a 20–178) | 33 | 17.4 | 46 | 369 | 24.8 | 1 | |
| 2i83 (a.a 21–178) | 36 | 19.4 | 36 | 369 | 40.2 | 0.86 | |
| 1 poz (a.a 20–178) | 36 | 19.4 | 36 | 369 | 40.1 | 0.86 | |
| 4pz4 (a.a 18–171) | 41 | 25.4 | 59 | 378 | 30.4 | 1 | Human Hsp90 co-chaperone Cdc37 (Q16543)‡ |
| 4pz3(a.a 18–170) | 41 | 25.4 | 59 | 378 | 30.8 | 1 | |
| 1uuh(a.a 20–178) | 41 | 25.4 | 59 | 378 | 30.8 | 1 | |
| 2i83 (a.a 21–178) | 41 | 25.4 | 59 | 378 | 30.1 | 1 | |
| 1 poz (a.a 20–178) | 41 | 25.4 | 59 | 378 | 30 | 1 | |
| 4pz4 (a.a 18–171) | 41 | 24 | 75 | 507 | 24.4 | 1 | ARSA_HUMAN Arylsulfatase A (P15289)‡ |
| 4pz3(a.a 18–170) | 38 | 22 | 50 | 507 | 39.5 | 0.88 | |
| 1uuh(a.a 20–178) | 38 | 22 | 50 | 507 | 39.5 | 0.88 | |
| 2i83 (a.a 21–178) | 42 | 23.7 | 76 | 507 | 40 | 0.87 | |
| 1 poz (a.a 20–178) | 42 | 23.7 | 76 | 507 | 39.9 | 0.87 |
SAS was used to look for any existing structural similarities with the PDB flagged CD44 motifs (Table III). The first 10 proteins in the table (*) are putative HA-binding proteins known to contain the Link module. A set of 11 randomly selected proteins from the experimentally derived non-binding fractions that do not contain a Link module (‡) are also shown for comparison. Apart from containing or not containing a Link module, proteins are listed in no particular order. ZPBP2 (*?) is included in Table IV because it was flagged by PDB/SAS and has relatively high Z-scores and low E-values.
Western blotting to confirm the results of proteomics
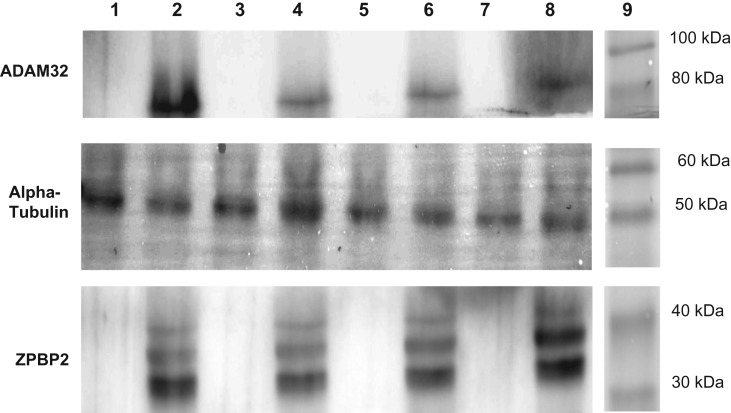
To confirm the results obtained by LC-MS/MS, western blotting was performed using antibodies against ZPBP2, ADAM32 and alpha tubulin on binding and non-binding fractions from the four samples individually and independently processed prior to combining for proteomic analysis. ADAM32 was chosen because it has two BX7B motifs. Antibodies against ZPBP2 and ADAM32 detected protein bands at approximately 38 kDa and 85 kDa, respectively in binding fractions (Lanes 2, 4, 6, 8), consistent with our sequencing and partitioning data (Fig. 4). Additional bands at ~36 kDa and 42 kDa were revealed using the antibody to ZPBP2. No signals were detected in non-binding fractions (Lanes 1, 3, 5, 7). The antibody against alpha-tubulin (present in both fractions and used here as a loading control) detected a single band at ~50 kDa in all samples.
Figure 4.
Western blotting on binding (lanes 2, 4, 6, 8) and non-binding (lanes 1, 3, 5, 7) proteins using antibodies directed against ZPBP2 and ADAM32. Alpha-tubulin, present in both binding and non-binding fractions, was used as a loading control. Western blot was run on samples before combining.
Discussion
The present study was conducted to search for novel hyaladherins in human sperm by allowing the soluble proteins extracted from homogenized sperm to interact with a HA-coated surface before their recovery and characterization by tandem mass spectrometry (LC-MS/MS). This simple affinity panning process (Amemiya et al., 2005) resulted in the identification and selection for further analysis of 28 proteins from the binding fractions, including one containing a sequence related to the HA-binding domain of CD44 (Liu and Finzel, 2014; Takeda et al., 2006; Teriete et al., 2004) that was identified as ZPBP2. Based on the proportion of positively charged proteins in the binding versus non-binding fractions of extracted sperm proteins, electrostatic attraction is also likely to have played a role in the outcomes of the panning process (Lenormand et al., 2008; Purcell et al., 2014). Nonetheless, a conformational change in conjunction with the overall charge of a protein could also favour interaction with HA, which is known to be transitory in nature (Kohda et al., 1996; Liu and Finzel, 2014; Misra et al., 2015).
SAS determines and applies unique identifiers from different crystallographic (X-ray or Nuclear Magnetic Resonance) submissions, which in the case of the ZPBP2 included five records (PDB codes 4pz3, 1uuh, 4pz4, 2i83 and 1 poz) relating to the HA-binding (Link) domain of CD44 (Liu and Finzel, 2014; Takeda et al., 2006; Teriete et al., 2004). CD44 is one of the better understood hyaladherins and is a Type I transmembrane protein located on the surface of many different cell types including mammalian sperm as reported previously (Bains et al., 2002). Its location on the sperm surface has also been shown to alter during and following capacitation and the acrosome reaction (Torabi et al., 2017). CD44 has many different isoforms due to alternative splicing in the proximal region of the extracellular domain and also shows evidence of post-translational modifications (Oliferenko et al., 2000; Toole, 2004). CD44 probably binds HA through a disulphide-bond stabilized HA-binding domain (HABD) located within the Link module (Takeda et al., 2006) and the peptide sequence in ZPBP2 bearing similarity to the Link domain of CD44 is based on the latter’s ligand-bound configuration (Bajorath et al., 1998) and retains two of the cysteine residues thought to be responsible for stabilizing the HABD.
Both ZPBP1 and ZPBP2 are located on the acrosome and the role of the former in binding the zona pellucida is already documented (Mori et al., 1993, 1995). Both proteins belong to the immunoglobulin like domain-containing family (Lin et al., 2007) and are co-designated by sequence similarity, sharing a similar, amino-terminal signal sequence and a carboxyl-terminal zona-binding homologous domain (ZBHD) with 15 conserved cysteine residues (Katoh and Katoh, 2003). Loss of either protein leads to disruption of acrosome formation and sperm morphology (ZPBP1/2) and to male mouse sterility (ZPBP1) or subfertility (ZPBP2; Lin et al., 2007). The ZPHD and putative Link domains, overlap (Fig. 3A) and share common features, notably the two conserved cysteines (Fig. 3A; stars) supporting the latter’s disulphide linked HA-binding configuration (Teriete et al., 2004).
Excepting teleosts, which have no ZPBPs and use a completely distinct process for fertilization (Morisawa, 1999), the ZPHD domain is conserved across all vertebrate species (Lin et al., 2007). Amphibians express just one ZPBP, which is more similar to mammalian ZPBP2 than to ZPBP1 (Lin et al., 2007), explaining PDPsum’s ‘failure’ to report homology with ZPBP1. These data suggest that the recognition of zona pellucida protein 2 (ZP2) by either of the ZPBPs and of HA by hyaladherins containing the Link module like CD44, may utilize similar mechanisms. Hence, zona recognition by amphibian sperm ZPBPs could have arisen via an earlier HA-binding (or lectin binding) property that was partially conserved during the likely gene duplication event that gave rise to ZPBP1 (Lin et al., 2007; Kohda et al., 1996). The original ZPBPs of primitive vertebrates may not have had time to evolve the more specific sperm-oocyte interactions seen in modern mammals. Alongside CD44, however, they may still have hyaladherin-like properties. In the human, ZPBP2 has at least two known isoforms of 338 (38.6) kDa and 316 (36.2 kDa) amino acids, respectively, both of which contain the ZPHD sequence. These and an additional band at ~42 kDa, which may represent a novel isoform of this protein (Lin et al., 2007), were detected only in the binding fractions by western blotting (Fig. 4).
The Link module present in CD44 is also present in several other hyaladherins including TSG-6, versican, link protein, brevican, aggrecan and neurican, which were not detected in either binding or non-binding fractions. The BX7B motif has been reported in RHAMM, CD44, and the hyaluronidase, HABP1 (C1QBP), among others (Day and Prestwich, 2002; Yang et al., 1994). Midkine (MK) is a secreted growth factor rich in basic amino acids and cysteine with a molecular mass of ~13 kDa (Jono and Ando, 2010). The two BX7B motifs in ADAM32 and the four in Midkine as well as the net positive charge of the proteins may help explain their presence in the binding fraction. ADAM32 has been identified as a type I non-catalytic metalloprotease-like transmembrane protein (Wolfsberg et al., 1995). The protein is expressed during spermatogenesis and processed during epididymal maturation to become localized on the sperm surface (Kim et al., 2006). Western blot analysis confirmed that ADAM32 was confined to the binding fractions. To date 29 ADAMs have been identified of which 15 are expressed in testis (Primakoff and Myles, 2000). This suggests a specific relationship between ADAM function, spermatogenesis and fertilization. ADAM2 (fertilin beta) and ADAM3 (cyritestin) interact with integrin in the plasma membrane of mouse oocytes (Wassarman et al., 2001).
The absence of known sperm hyaladherins, including CD44, RHAMM and C1QBP, in any of the extract fractions as well as the presence of BX7B motifs in some proteins from the non-binding fractions was surprising and warrants further consideration (Bajorath et al., 1998; Hardwick et al., 1992; Underhill, 1992; Yang et al., 1994). One possibility is that the transient and dynamic binding reported between hyaladherins and HA that facilitates cell adhesion and migration relating to conformational changes in protein structure was responsible (Kohda et al., 1996; Liu and Finzel, 2014; Misra et al., 2015). HA-binding proteins without the required conformational configuration may well have been extracted by our methods but were in the wrong conformational arrangement to permit efficient enrichment by HA-binding. Alternatively, the missing hyaladherins may not have been sufficiently abundant in either fraction to be detectable by the LC-MS/MS method used. CD44, for example, is not listed in the most recent and comprehensive human sperm proteome (Amaral et al., 2014). Moreover, the mild solvent used in the extraction process, designed to preserve tertiary structure, may have failed to access these proteins, which were lost in the insoluble pellets following sperm homogenization. Membrane anchoring could also have rendered many proteins inaccessible under the extraction conditions used here (Gupta et al., 1991). Finally, BX7B motifs may not be universally involved in HA-binding, with only RHAMM having been shown to have an absolute requirement for them to date (Day, 2000; Yang et al., 1994). Work is in progress to isolate proteins from sperm permitted to undergo capacitation beforehand where conformational or localization changes to hyaladherins, increasing access to the extraction solvent, may have occurred (Torabi et al., 2017). Modification of the HA substrate may also improve its capacity to recognize and bind hyaladherins and experiments are also underway exploring this possibility.
Just one protein (ZPBP2) present only in HA-binding fractions contained a Link-like module. Regardless of the efficacy of the panning process itself, the uncovering of a structural relationship between ZPBP2 and CD44 is noteworthy and suggests that the general HA-sperm recognition process occurring in the COC may predate the appearance of more specialized protein–protein, sperm-zona interactions in this context. The synthesis of larger quantities of both isoforms of ZPBP2 is warranted to more fully study their structural and possible HA-binding characteristics, their relationship to hyaladherins and their involvement in male infertility.
Supplementary data
Supplementary data are available at Molecular Human Reproduction online.
Authors’ roles
Ms F.T. contributed to the concept and design of the study, undertook the majority of the work reported in the manuscript including semen sample processing, sperm protein extraction, panning, preparation for LC-MS.MS, data analysis, writing of the manuscript and correction checking. F.T. is also an ‘Early Stage Researcher’ in the Marie Curie ITN, ‘Reprotrain’ network. Dr O.A.B. contributed to the concept and design of the study, assisted with preparation for LC-MS/MS, data analysis and critical reviewing of the manuscript. Dr J.M.E. contributed to the concept and design of the study, undertook the LC-MS/MS experiments, provided critical insight with the proteomic output and critically reviewed the manuscript. Prof R.O. contributed to the concept and design of the study, supervised and assisted in the laboratory work, provided critical insight with the proteomic output and critically reviewed the manuscript. R.O. is also the programme manager for the Marie Curie ITN, ‘Reprotrain.’ Dr D.M. contributed to the concept and design of the study, supervised the laboratory work, and co-wrote and provided critical reading of the manuscript. D.M. is also a participant in the Marie Curie ITN, ‘Reprotrain’ and is a NIHR-EME grant holder.
Funding
The authors gratefully acknowledge a scholarship of the EU Marie Curie Sklodowska Reproductive Biology Early Research Training network (REPRO-TRAIN) Project ID 289880, supporting Ms Torabi. This project was also supported and funded by the Efficacy and Mechanism Evaluation Programme, a Medical research Council (MRC) and National Institute of Health Research partnership (Grant No 11/14/ 34). The views expressed in this publication are those of the author(s) and not necessarily those of the MRC, National Health Service, NIHR or the Department of Health.
Conflict of interest
On behalf of all the authors, Dr Miller declares that we have no conflict of interest in relation to this work.
Supplementary Material
References
- Amaral A, Castillo J, Ramalho-Santos J, Oliva R. The combined human sperm proteome: cellular pathways and implications for basic and clinical science. Hum Reprod Update 2014;20:40–62. [DOI] [PubMed] [Google Scholar]
- Amemiya K, Nakatani T, Saito A, Suzuki A, Munakata H. Hyaluronan-binding motif identified by panning a random peptide display library. Biochim Biophys Acta 2005;1724:94–99. [DOI] [PubMed] [Google Scholar]
- Bains R, Adeghe J, Carson RJ. Human sperm cells express CD44. Fertil Steril 2002;78:307–312. [DOI] [PubMed] [Google Scholar]
- Bajorath J, Greenfield B, Munro SB, Day AJ, Aruffo A. Identification of CD44 residues important for hyaluronan binding and delineation of the binding site. J Biol Chem 1998;273:338–343. [DOI] [PubMed] [Google Scholar]
- Barta E, Deak F, Kiss I. Evolution of the hyaluronan-binding module of link protein. Biochem J 1993;292:947–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorndahl L, Barratt CL, Mortimer D, Jouannet P. ‘How to count sperm properly’: checklist for acceptability of studies based on human semen analysis. Hum Reprod 2016;31:227–232. [DOI] [PubMed] [Google Scholar]
- Celik-Ozenci C, Jakab A, Kovacs T, Catalanotti J, Demir R, Bray-Ward P, Ward D, Huszar G. Sperm selection for ICSI: shape properties do not predict the absence or presence of numerical chromosomal aberrations. Hum Reprod 2004;19:2052–2059. [DOI] [PubMed] [Google Scholar]
- D’Cruz OJ, Haas GG Jr., Lambert H. Heterogeneity of human sperm surface antigens identified by indirect immunoprecipitation of antisperm antibody bound to biotinylated sperm. J Immunol 1993;151:1062–1074. [PubMed] [Google Scholar]
- Day AJ. Understanding hyaluronan-protein interactions. Glycoforum 2000, Japan.
- Day AJ, Prestwich GD. Hyaluronan-binding proteins: tying up the giant. J Biol Chem 2002;277:4585–4588. [DOI] [PubMed] [Google Scholar]
- Ghosh I, Chattopadhaya R, Kumar V, Chakravarty BN, Datta K. Hyaluronan binding protein-1: a modulator of sperm-oocyte interaction. Soc Reprod Fertil Suppl 2007;63:539–543. [PubMed] [Google Scholar]
- Gopalkrishnan K, Padwal V, Meherji PK, Gokral JS, Shah R, Juneja HS. Poor quality of sperm as it affects repeated early pregnancy loss. Arch Androl 2000;45:111–117. [DOI] [PubMed] [Google Scholar]
- Gupta S, Batchu RB, Datta K. Purification, partial characterization of rat kidney hyaluronic acid binding protein and its localization on the cell surface. Eur J Cell Biol 1991;56:58–67. [PubMed] [Google Scholar]
- Halliday J. Outcomes for offspring of men having ICSI for male factor infertility. Asian J Androl 2012;14:116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick C, Hoare K, Owens R, Hohn HP, Hook M, Moore D, Cripps V, Austen L, Nance DM, Turley EA. Molecular cloning of a novel hyaluronan receptor that mediates tumor cell motility. J Cell Biol 1992;117:1343–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmatdoost A, Lakpour N, Sadeghi MR. Sperm chromatin integrity: etiologies and mechanisms of abnormality, assays, clinical importance, preventing and repairing damage. Avicenna J Med Biotechnol 2009;1:147–160. [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 2007;8:R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huszar G, Ozenci CC, Cayli S, Zavaczki Z, Hansch E, Vigue L. Hyaluronic acid binding by human sperm indicates cellular maturity, viability, and unreacted acrosomal status. Fertil Steril 2003;79:1616–1624. [DOI] [PubMed] [Google Scholar]
- Ikawa M, Inoue N, Benham AM, Okabe M. Fertilization: a sperm’s journey to and interaction with the oocyte. J Clin Invest 2010;120:984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaleel R, Khan A. Paternal factors in spontaneous first trimester miscarriage. Pak J Med Sci 2013;29:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jono H, Ando Y. Midkine: a novel prognostic biomarker for cancer. Cancers (Basel) 2010;2:624–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M, Katoh M. Identification and characterization of human ZPBP-like gene in silico. Int J Mol Med 2003;12:399–404. [PubMed] [Google Scholar]
- Kim T, Oh J, Woo JM, Choi E, Im SH, Yoo YJ, Kim DH, Nishimura H, Cho C. Expression and relationship of male reproductive ADAMs in mouse. Biol Reprod 2006;74:744–750. [DOI] [PubMed] [Google Scholar]
- Kohda D, Morton CJ, Parkar AA, Hatanaka H, Inagaki FM, Campbell ID, Day AJ. Solution structure of the link module: a hyaluronan-binding domain involved in extracellular matrix stability and cell migration. Cell 1996;86:767–775. [DOI] [PubMed] [Google Scholar]
- Larsen EC, Christiansen OB, Kolte AM, Macklon N. New insights into mechanisms behind miscarriage. BMC Med 2013;11:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res 2001;29:221–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA. PDBsum new things. Nucleic Acids Res 2009;37:D355–D359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Chistyakov VV, Thornton JM. PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res 2005;33:D266–D268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand H, Deschrevel B, Tranchepain F, Vincent JC. Electrostatic interactions between hyaluronan and proteins at pH 4: how do they modulate hyaluronidase activity. Biopolymers 2008;89:1088–1103. [DOI] [PubMed] [Google Scholar]
- Lin YN, Roy A, Yan W, Burns KH, Matzuk MM. Loss of zona pellucida binding proteins in the acrosomal matrix disrupts acrosome biogenesis and sperm morphogenesis. Mol Cell Biol 2007;27:6794–6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LK, Finzel B. High-resolution crystal structures of alternate forms of the human CD44 hyaluronan-binding domain reveal a site for protein interaction. Acta Crystallogr F Struct Biol Commun 2014;70:1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn D, Laskowski RA, Thornton JM. Sequences annotated by structure: a tool to facilitate the use of structural information in sequence analysis. Protein Eng 1998;11:855–859. [DOI] [PubMed] [Google Scholar]
- Misra S, Hascall VC, Markwald RR, Ghatak S. Interactions between Hyaluronan and Its Receptors (CD44, RHAMM) Regulate the Activities of Inflammation and Cancer. Front Immunol 2015;6:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori E, Baba T, Iwamatsu A, Mori T. Purification and characterization of a 38-kDa protein, sp38, with zona pellucida-binding property from porcine epididymal sperm. Biochem Biophys Res Commun 1993;196:196–202. [DOI] [PubMed] [Google Scholar]
- Mori E, Kashiwabara S, Baba T, Inagaki Y, Mori T. Amino acid sequences of porcine Sp38 and proacrosin required for binding to the zona pellucida. Dev Biol 1995;168:575–583. [DOI] [PubMed] [Google Scholar]
- Morisawa S. Fine structure of micropylar region during late oogenesis in eggs of the hagfish Eptatretus burgeri (Agnatha). Dev Growth Differ 1999;41:611–618. [DOI] [PubMed] [Google Scholar]
- Nasr-Esfahani MH, Razavi S, Vahdati AA, Fathi F, Tavalaee M. Evaluation of sperm selection procedure based on hyaluronic acid binding ability on ICSI outcome. J Assist Reprod Genet 2008;25:197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliferenko S, Kaverina I, Small JV, Huber LA. Hyaluronic acid (HA) binding to CD44 activates Rac1 and induces lamellipodia outgrowth. J Cell Biol 2000;148:1159–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagni M, Jongeneel CV. Making sense of score statistics for sequence alignments. Brief Bioinform 2001;2:51–67. [DOI] [PubMed] [Google Scholar]
- Palermo G, Joris H, Devroey P, Van Steirteghem AC. Pregnancies after intracytoplasmic injection of single spermatozoon into an oocyte. Lancet 1992;340:17–18. [DOI] [PubMed] [Google Scholar]
- Primakoff P, Myles DG. The ADAM gene family: surface proteins with adhesion and protease activity. Trends Genet 2000;16:83–87. [DOI] [PubMed] [Google Scholar]
- Purcell BP, Kim IL, Chuo V, Guinen T, Dorsey SM, Burdick JA. Incorporation of sulfated hyaluronic acid macromers into degradable hydrogel scaffolds for sustained molecule delivery. Biomater Sci 2014;2:693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakkas D, Manicardi G, Bizzaro D, Bianchi PG. Possible consequences of performing intracytoplasmic sperm injection (ICSI) with sperm possessing nuclear DNA damage. Hum Fertil (Camb) 2000;3:26–30. [DOI] [PubMed] [Google Scholar]
- Suarez SS, Pacey AA. Sperm transport in the female reproductive tract. Hum Reprod Update 2006;12:23–37. [DOI] [PubMed] [Google Scholar]
- Takeda M, Ogino S, Umemoto R, Sakakura M, Kajiwara M, Sugahara KN, Hayasaka H, Miyasaka M, Terasawa H, Shimada I. Ligand-induced structural changes of the CD44 hyaluronan-binding domain revealed by NMR. J Biol Chem 2006;281:40089–40095. [DOI] [PubMed] [Google Scholar]
- Teriete P, Banerji S, Noble M, Blundell CD, Wright AJ, Pickford AR, Lowe E, Mahoney DJ, Tammi MI, Kahmann JD et al. Structure of the regulatory hyaluronan binding domain in the inflammatory leukocyte homing receptor CD44. Mol Cell 2004;13:483–496. [DOI] [PubMed] [Google Scholar]
- Toole BP. Hyaluronan: from extracellular glue to pericellular cue. Nat Rev Cancer 2004;4:528–539. [DOI] [PubMed] [Google Scholar]
- Torabi F, Binduraihem A, Miller D. Sedimentation properties in density gradients correspond with levels of sperm DNA fragmentation, chromatin compaction and binding affinity to hyaluronic acid. Reprod Biomed Online 2017;34:298–311. [DOI] [PubMed] [Google Scholar]
- Underhill C. CD44: the hyaluronan receptor. J Cell Sci 1992;103:293–298. [DOI] [PubMed] [Google Scholar]
- Vandevoort CA, Cherr GN, Overstreet JW. Hyaluronic acid enhances the zona pellucida-induced acrosome reaction of macaque sperm. J Androl 1997;18:1–5. [PubMed] [Google Scholar]
- Wassarman PM, Jovine L, Litscher ES. A profile of fertilization in mammals. Nat Cell Biol 2001;3:E59–E64. [DOI] [PubMed] [Google Scholar]
- Wolfsberg TG, Primakoff P, Myles DG, White JM. ADAM, a novel family of membrane proteins containing A Disintegrin and Metalloprotease domain: multipotential functions in cell-cell and cell-matrix interactions. J Cell Biol 1995;131:275–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Yang BL, Savani RC, Turley EA. Identification of a common hyaluronan binding motif in the hyaluronan binding proteins RHAMM, CD44 and link protein. EMBO J 1994;13:286–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo L, Kimata K. Cumulus oophorus extracellular matrix: its construction and regulation. Cell Struct Funct 2001;26:189–196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.