Abstract
STUDY QUESTION
Could the follicle proteome be mapped by identifying specific proteins that are common or differ between three developmental stages from the secondary follicle (SF) to the antrum-like stage?
SUMMARY ANSWER
From a total of 1401 proteins identified in the follicles, 609 were common to the three developmental stages investigated and 444 were found uniquely at one of the stages.
WHAT IS KNOWN ALREADY
The importance of the follicle as a functional structure has been recognized; however, up-to-date the proteome of the whole follicle has not been described. A few studies using proteomics have previously reported on either isolated fully-grown oocytes before or after meiosis resumption or cumulus cells.
STUDY DESIGN, SIZE, DURATION
The experimental design included a validated mice model for isolation and individual culture of SFs. The system was chosen as it allows continuous evaluation of follicle growth and selection of follicles for analysis at pre-determined developmental stages: SF, complete Slavjanski membrane rupture (SMR) and antrum-like cavity (AF). The experiments were repeated 13 times independently to acquire the material that was analyzed by proteomics.
PARTICIPANTS/MATERIALS, SETTING, METHODS
SFs (n = 2166) were isolated from B6CBA/F1 female mice (n = 42), 12 days old, from 15 l. About half of the follicles isolated as SF were analyzed as such (n = 1143) and pooled to obtain 139 μg of extracted protein. Both SMR (n = 359) and AF (n = 124) were obtained after individual culture of 1023 follicles in a microdrop system under oil, selected for analysis and pooled, to obtain 339 μg and 170 μg of protein, respectively. The follicle proteome was analyzed combining isoelectric focusing (IEF) fractionation with 1D and 2D LC-MS/MS analysis to enhance protein identification. The three protein lists were submitted to the ‘Compare gene list’ tool in the PANTHER website to gain insights on the Gene Ontology Biological processes present and to Ingenuity Pathway Analysis to highlight protein networks. A label-free quantification was performed with 1D LC-MS/MS analyses to emphasize proteins with different expression profiles between the three follicular stages. Supplementary western blot analysis (using new biological replicates) was performed to confirm the expression variations of three proteins during follicle development in vitro.
MAIN RESULTS AND THE ROLE OF CHANCE
It was found that 609 out of 1401 identified proteins were common to the three follicle developmental stages investigated. Some proteins were identified uniquely at one stage: 71 of the 775 identified proteins in SF, 181 of 1092 in SMR and 192 of 1100 in AF. Additional qualitative and quantitative analysis highlighted 44 biological processes over-represented in our samples compared to the Mus musculus gene database. In particular, it was possible to identify proteins implicated in the cell cycle, calcium ion binding and glycolysis, with specific expressions and abundance, throughout in vitro follicle development.
LARGE SCALE DATA
Data are available via ProteomeXchange with identifier PXD006227.
LIMITATIONS, REASONS FOR CAUTION
The proteome analyses described in this study were performed after in vitro development. Despite fractionation of the samples before LC-MS/MS, proteomic approaches are not exhaustive, thus proteins that are not identified in a group are not necessarily absent from that group, although they are likely to be less abundant.
WIDER IMPLICATIONS OF THE FINDINGS
This study allowed a general view of proteins implicated in follicle development in vitro and it represents the most complete catalog of the whole follicle proteome available so far. Not only were well known proteins of the oocyte identified but also proteins that are probably expressed only in granulosa cells.
STUDY FUNDING/COMPETING INTEREST(S)
This study was supported by the Portuguese Foundation for Science and Technology, FCT (PhD fellowship SFRH/BD/65299/2009 to A.A.), the Swedish Childhood Cancer Foundation (PR 2014-0144 to K.A.R-.W.) and Stockholm County Council to K.A.R-.W. The authors of the study have no conflict of interest to report.
Keywords: ovarian follicle, follicle development, in vitro growth, mouse, proteomics
Introduction
The development of successful systems for in vitro growth of ovarian follicles from the earliest stage until maturity is predicted to have an enormous impact on assisted reproductive treatments. In particular, the method is necessary for the increasing number of young cancer patients undergoing cryopreservation of ovarian tissue in clinical programs for fertility preservation, but for whom the risk of reintroducing of malignant cells precludes transplantation (Telfer and Zelinski, 2013). The best developed systems applied to mammalian follicles that can be translated to human follicle-culture systems have been achieved using mice, which have proved to be an excellent model for translational reproductive research, and live offspring have been obtained (Smitz and Cortvrindt, 2002; Xu et al., 2006; Eppig et al., 2009). However, the current culture systems still need to be improved to increase the possibility of obtaining fully competent oocytes in humans (Telfer and Zelinski, 2013).
A series of biological processes is needed to achieve oocyte developmental competence for fertilization and embryo development (Picton et al., 1998). This competence is gained gradually during folliculogenesis, which is a highly complex process with several mechanisms and steps, which are not yet clear. After the activation of a primordial follicle, important changes occur such as granulosa and theca cell differentiation and proliferation. Simultaneously, the oocyte volume increases and it grows until it reaches the appropriate size to induce nuclear maturation, which finally allows fertilization and the development of a viable embryo (Gilchrist et al., 2004). In these processes, several critical steps, involving a cascade of molecular interactions, have been described with high transcriptional activity for synthesis of new proteins (Picton et al., 1998; Gilchrist et al., 2004; Eppig et al., 2005). However, not all mechanisms have been elucidated, and expanding our knowledge of protein expression during the early stages may add important information about the requirements for successful follicle development. Proteomic approaches allow the identification and characterization of proteins that are expressed in a cell. These approaches present several advantages over mRNA expression profiling since the presence of mRNA in a cell is not strictly correlated with protein expression (Ma et al., 2008).
Over the last decade, proteomic studies of female reproduction have essentially focused on oocyte maturation and early embryonic development of several mammalian species. In mice, the proteome of fully-grown oocytes obtained by superovulation before and after meiosis resumption has been reported, and the list of proteins identified is increasing (Ma et al., 2008; Zhang et al., 2009; Wang et al., 2010; Pfeiffer et al., 2011; Demant et al., 2012). Nonetheless, these studies report on proteins identified from isolated oocytes despite the essential role played by the bidirectional communication between oocyte and its surrounding cells in the follicle, which is required for oocyte growth and maturation (Matzuk et al., 2002). In mice, Meng et al. (2007) have investigated the surrounding follicle cells using mature cumulus-oocyte complexes (COC). However, at this stage of development, the cumulus cells are further differentiated from the previous granulosa cells due to the disruption of intercellular connections and consequent expansion occurring during ovulation (Hawkins and Matzuk, 2010).
To date, no study of the whole follicle proteome has been reported in any species. Our study was designed to identify proteins expressed in the follicular unit during its growth and development. For this purpose, an experimental model for mouse in vitro follicle development was used, which allows the growth and maturation of oocytes from secondary stage follicles up to fertilizable oocytes (Cortvrindt et al., 1996). This system was preferred over several newly developed culture systems as it has been validated by obtaining live offspring and because it permits continuous follicle observation (Cortvrindt et al., 1996; Smitz and Cortvrindt, 2002; Picton et al., 2008), which makes it possible to detect even slight morphological changes indicative of follicle development at the three pre-determined morphological stages we attempted to investigate.
Material and Methods
Animals
All animal experimentation described was previously approved by the University Pierre et Marie Curie and conducted in accordance with accepted standards of humane animal care.
B6CBAF1/J female mice (n = 42), 12 days old, obtained from Charles River®, Lyon, France were sacrificed by cervical dislocation. The ovaries were collected to a 37°C warmed dissection medium (Leibovitz L15 (Gibco®, Cergy-Pontoise, Fance) enriched with 10% fetal bovine serum (FBS, Gibco®), 100 UI/ml penicillin (Sigma-Aldrich®, Saint Quentin Fallavier, France) and 100 μg/ml streptomycin (Sigma-Aldrich®).
Preantral ovarian follicle isolation, culture and selection of follicles for analysis
Ovarian follicles were isolated by mechanical dissection under stereomicroscope (Nikon®, France) using 27 gauge needles (Sherwood Medical®, Evry, France), ensuring that the follicular structure remained intact. Secondary follicles (SFs) with a diameter between 100 and 130 μm, a clearly visible round and central oocyte, two or more granulosa-cell layers, and some adhering theca cells were analyzed as such or put in culture to obtain follicles with a complete Slavjanski membrane rupture (SMR) and antrum-like follicles (AF) (Fig. 1a). The SMR stage was obtained between Days 6 and 9 of culture, when a complete rupture of the Slavjanski membrane and the expansion of granulosa could be observed, as signs of growing follicles. The AF stage was collected at the end of the culture (Day 12) when the follicles reached a diameter of at least 500 μm and a space within the granulosa-cell layers was visible, corresponding to the antrum formation in vivo.
Figure 1.
Schematic chart indicating (a) Type and numbers of animals used in the study and numbers of secondary follicles (SFs) that were isolated and cultured in repeated independent experiments. From culture, follicles were collected when Slavjsanki Membrane Rupture (SMR) was observed or antrum-like cavity (AF). Morphological characteristics of the follicles selected at each stage are illustrated indicating: size, visible oocyte (O), granulosa cells (GC) theca cells (TC) and the presence of antrum (black arrow). The quantity of pooled protein obtained from selected follicles at each stage is also indicated. (b) Schematic description of how the total protein extract (TPE) from each follicle stage was fractionated by isoelectric focusing (IEF) with stacking in SDS-PAGE gel and manual band excision for digestion.
Isolated SFs were cultured individually in 20 μl microdrops, 10 per Petri dish, covered by mineral oil (Sigma-Aldrich®, Saint Quentin Fallavier, France) in an atmosphere containing 5% CO2 at 37°C. An α-minimal essential medium with glutaMAX (α-MEM GlutaMAX, Gibco®) supplemented with 5% FBS (Gibco®), 10 μg/ml transferrin (Boehringer Mannheim®, Meylan, France), 5 μg/ml insulin (Boehringer Mannheim®) and 100 mIU/ml recombinant follicle stimulating hormone (rFSH, MSD®, Puteaux, France) was used. The follicles were observed daily using an inverted microscope (Nikon®) at a magnification of ×100 or ×200 to observe morphological characteristics and follicular size. Two perpendicular diameters were taken using a calibrated ocular micrometer (Nikon®). Follicular diameter was estimated by measuring the granulosa-cell mass, but not the theca cells. From Day 2 onwards, half of the culture medium was refreshed.
All follicles recovered for the study were washed three times in phosphate-buffered saline (PBS, Gibco-BRL®), dried and stored at −196°C until needed.
Proteomic analysis
The chemicals and material used for proteomic analysis were purchased from Sigma-Aldrich® (Saint Quentin Fallavier, France) and GE Healthcare® (Vélizy, France), unless otherwise indicated.
Protein extraction
Protein extraction was performed by adding 15 μl/vial of lysis buffer containing 7 M urea, 2 M thiourea, 1% 3-[3-cholamidopropyl)dimethylamonio]-1-propanesulfonate (CHAPS), 0.5% 3-(Decyldimethylammonio) propanesulfonate inner salt (SB3–10), 0.5% Triton × 100, 10% isobutanol, 50 mM dithiothreiol (DTT) and 0.5% ampholyte 3–10. After 2 × 5 min of sonication and 45 min of 20–000 × g centrifugation at 4°C, the supernatants were collected and identified as total protein extract (TPE). Protein concentration was measured three times for each sample using the Bradford assay (Bradford, 1976) (Biorad®, France), and samples were stored at −80°C until use.
Protein fractionation
For each group, 100 μg of TPE was loaded into Immobilized pH gradient (IPG) Immobiline DryStrips (7 cm, pH 4–7) (Fig. 1b). Isoelectrofocusing (IEF) migration was performed on an Ettan IPGphor II isoelectric focusing (IEF) system, with a multistep program: 50 V for 1 h, 200 V for 1 h, gradient 200–1000 V in 45 min, 1000 V for 45 min, gradient 1000–4000 V in 1 h and 4000 V for 3 h. Following isoelectric migration, the strips were treated in a reduction buffer (6 M Urea, 75 mM Tris pH 8.8, 30% glycerol, 2% SDS, 65 mM DTT) for 15 min, followed by 15 min in an alkylation buffer (6 M Urea, 75 mM Tris pH 8.8, 30% glycerol, 2% SDS, 135 mM iodoacetamide). Each strip was then cut into five equal pieces using a scalpel blade (1 cm of the acidic end of the strip was discarded). Proteins from each strip fraction were extracted with a short stacking migration (1 h at 10 mA) in an SDS-PAGE gel, 12% acrylamide. Proteins from TPEs (10 μg) were stacked in the same way. Gels were stained with Coomassie Blue R-250, using Imperial Protein Stain (Pierce, Thermo Fischer Scientific®, Courtaboeuf, France) (Fig. 1b). Thus, for each group, six fractions (five obtained by protein fractionation and one corresponding to TPE) were treated and analyzed in parallel.
In-gel tryptic digestion
For each fraction, a unique large band was excised and cut into several 1 mm3 cubes (Fig. 1b). Gel pieces were destained with a 25 mM ammonium bicarbonate (AmBic) in 50% ethanol solution, dehydrated in acetonitrile (ACN) and dried at room temperature. Gel slices were then rehydrated with 70 μl of trypsin solution (200 ng/μl in 50 mM AmBic) on ice for 30 min and incubated overnight at 37°C. Supernatants were collected in new tubes, and the gel pieces were incubated twice with 60% ACN in 0.1% trifluoroacetic acid to extract the remaining peptides. The extracts were concentrated and dried in a Speed-Vac (Thermo Fischer Scientific®), and then resuspended in 40 μl of 30% ACN with 0.1% formic acid (FA) and stored at −80°C until MS analysis.
LC-MS/MS analysis
Peptide samples were desalted using Zip-Tip C18 pipette tips (Millipore®, Fontenay sous Bois, France), following the manufacturer’s instructions. Two configurations of high-performance liquid chromatography (HPLC) were used for LC-MS/MS analysis: 1D LC-MS/MS and 2D LC-MS/MS. The nanoHPLC (Ultimate 3000, Thermo Fischer Scientific®) was coupled to an ion trap mass spectrometer (MS) (HCTultra, Bruker® (Bremen, Germany)) used in the positive mode with the selection of eight precursors from each MS spectrum for fragmentation by collision-induced dissociation. Six serial injections were performed for each fraction of each group, four in 1D LC-MS/MS and two in 2D LC-MS/MS.
In the 1D configuration, peptide mixtures were concentrated and desalted for 5 min on a precolumn C18 (5 mm, 300 μm i.d., 100 Å, Dionex) with a mobile phase A1 (2% ACN/0.1% FA) at a flow rate of 20 μl/min. Then, the peptide mixtures were separated and eluted on an analytical column RP-C18 (15 cm, 75 μm i.d., 100 Å, Dionex) at a flow rate of 200 nl/min, using 2–30% gradient buffer 1 (95% ACN, 0.1% FA).
In the 2D configuration, peptides were first separated by sequential elution from a silica capillary column strong cation exchanger (SCX, 15 mm, 300 μm i.d., Dionex) with 20 μl of 0, 20, 30, 50, 100 and 1000 mM of ammonium acetate. Each of these salt steps was then treated as was the 1D approach.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD006227.
Data analysis
Protein identification
Tandem mass spectra data were searched against the Uniprot KB mouse databases (June 2012) using X!tandemPipeline V 3.3.2 (http://pappso.inra.fr/bioinfo/xtandempipeline/), taking into consideration trypsin as the cleavage enzyme, carbamidomethylation of cysteine residues as a fixed modification and methionine oxidation as a variable modification, a peptide mass tolerance of 0.5 Da and a charge of +2 or +3.
Proteins identified were only validated if they had at least one unique peptide, a P-value < 0.05 for peptides and a P-value < 0.003 for proteins, and a false discovery rate lower than 1%.
GO analysis
Gene Ontology (GO) was performed using the PANTHER 8.1 (Protein ANalysis THrough Evolutionary Relationship) classification system (http://www.pantherdb.org). Data were divided into protein classes and biological processes. A statistical over-representation test was performed to highlight biological processes over-represented in our samples. The Bonferroni correction for multiple testing was used.
Label-free quantification
For label-free quantitation, Bruker® raw data files from 1D LC-MS/MS acquisitions were imported into the Progenesis LC-MS software version 4.1.4832.42146 (Nonlinear Dynamics, Newcastle upon Tyne, UK) for feature detection, alignment and quantification.
For each group, the acquisitions were aligned according to retention time, using a combination of automatic and manual alignment. Peptide peak picking was undertaken with an absolute ion-intensity threshold of 100 000, a maximal ion charge state of 3 and a retention time ranging from 25 to 100 min. MS/MS peak lists were generated with the 100 most intense fragment-ion peaks. For a given peptide, only the five most intense MS/MS spectra were used to generate MS/MS peak lists, which were exported as mgf files. Protein identification was conducted using in-house Mascot software version 2.2.07 (Matrix Science, London, UK) against the Mus musculus proteome set (release 2013–05, containing 50 807 sequences) from SwissProt/Trembl database, and using the same parameters as above for protein identification. Mascot search results were imported back into Progenesis software to link the identified peptides to the detected abundances of those peptides. Any peptides identified with a Mascot score lower than 33 (significance threshold of P < 0.05) were removed. The protein quantitation was made using non-conflicting peptides. Only proteins with a fold-change >2 between the groups (ANOVA P-value < 0.05), and with at least two peptides with significant changes (ANOVA q-value < 0.05 and fold-change >2), were accepted.
Bioinformatics analysis
To identify biological pathways and networks that were significantly represented in our proteomic datasets, Ingenuity Pathways Analysis (IPA; Ingenuity Systems, Qiagen Company, Redwood City, CA, USA) and Pathway Studio (Elsevier, BV, New York, NY, USA) were used.
The IPA maps protein networks using an algorithm based on molecular function, cellular function and functional group. Path Designer completes the research workflow from data analysis through the creation of images able to suggest a summary of networks selected. Fisher’s exact test was used to calculate the P-value determining the probability of each biological function or pathway being assigned by chance. In the test, P ≤ 0.05 was used, and only networks scoring ≥2, which have >99% confidence of not being generated by chance, were selected.
In Pathway Studio, the proteins identified were submitted to the 9.0 ResNEt Mammal database from Elsevier, which concerns biological and ontological interactions and pathways.
Western blotting
Western blotting was performed in a new biological replicate for each group and repeated three times for each protein. Protein extraction and quantification were performed as described above. From the TPE, 5 μg of protein was loaded per well in NuPAGE®4–12% Bis-Tris Gel (Life Technologies™, Carlsbad, CA, USA). After running the gel according to the XCell SureLock® Mini-Cell protocol, proteins were transferred to a PVDF membrane. The membrane was then blocked for at least 30 min with StartingBlock Blocking Buffer (Pierce®, Carlsbad, CA, USA). The blots were then probed with monoclonal mice PKA, (ab75996, 1:1000 dilution), monoclonal mice LDHB (ab85319 1:10 000 dilution) and monoclonal rabbit p27 (ab92741, 1:1000 dilution) all from Abcam® (Cambridge, UK) overnight at 4oC. The blots were then washed and probed with the secondary antibody (anti-rabbit or anti-mouse HRP conjugated, 1:10 000 dilutions, GE Healthcare®, Uppsala, Sweden). West Femto Maximum Sensitivity Substrate (Pierce®) was used to detect the signal from the blot. Band intensity was calculated on a recurring basis using Image Lab Software from BioRad®.
Results
Proteome of ovarian follicles grown in vitro
Fractionation of the TPE from the AF stage allowed the identification of 3.3 times more proteins than when only the TPE was analyzed (Fig. 2a). The quality of the fractionation method was demonstrated by the low overlap of proteins identified in several fractions, with 79% of the proteins being only identified in one fraction (Fig. 2b). The four serial injections performed in the 1D LC-MS/MS configuration allowed the identification of 600 proteins in SF, 737 in SMR and 710 in AF. The two serial injections in the 2D LC-MS/MS configuration allowed an increment in the number of proteins found in follicles of all stages. The increases were thus substantial and 25.2, 40.2 and 47.5% more proteins could be identified in SF, SMR and AF, respectively.
Figure 2.
Fractionation was performed to increase the likelihood of obtaining a higher number of identified proteins in the samples. The figure illustrates this by showing: (a) the number of proteins identified in AF stage analyzing either only TPE or only the Fractions, and the combination of both; (b) the percentage of proteins identified in one single fraction, or in 2, 3, 4 or 5 fractions and (c) a Venn diagram of proteins identified in ovarian follicles. The blue circle contains proteins identified at the SF stage; the green circle contains proteins identified at the SMR stage; and the purple circle contains proteins identified at the AF stage.
The combination of these two configurations made it possible to further increase the number of proteins identified and therefore to obtain a more complete protein profile in each stage, with 775 proteins identified in SF, 1092 in SMR and 1100 in AF (Supplementary Table S1).
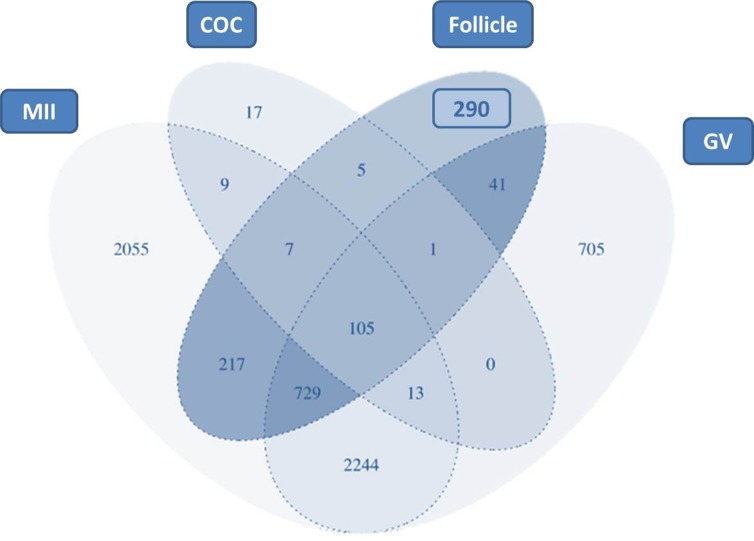
Of the identified proteins, 609 (43.4%) were common to the three developmental stages, whereas 348 (24.8%) were common to two stages of development and 444 proteins were identified in a single stage: 71 (5.1%) in SF, 181 (12.9%) in SMR and 192 (13.7%) in AF (Fig. 2c). From a total of 1401 different proteins identified in the follicle, 290 proteins had not been previously described in proteomic studies of metaphase II oocyte (MII), germinal vesicle oocyte (GV) or COC (Fig. 3). The list of the proteins herein obtained can be consulted in Supplementary Table S2.
Figure 3.
Venn diagram comparing the mouse follicle proteome found in this study and the proteome of metaphase II oocytes (MII) (Ma et al., 2008; Zhang et al., 2009; Wang et al., 2010; Pfeiffer et al., 2011), germinal vesicle oocytes (GV) (Wang et al., 2010; Demant et al., 2012) and cumulus-oocyte complex (COC) (Meng et al., 2007).
GO analysis
Proteins identified in each stage were assigned to protein class and biological process using PANTHER GO analysis.
For the three stages, GO analysis distributed the hit genes in 22 protein classes (Fig. 4a). Most proteins identified in the three stages were classified as nucleic binding proteins (18–22%). Thereafter, the four protein classes with more proteins attributed were enzymes: hydrolases, oxidoreductases, transferases and enzyme modulators. These classes together corresponded to ~45% of the proteins identified at SF and SMR, and to 40% of the proteins identified at AF. Cytoskeletal proteins represented 8–9% of the proteins identified in the three stages, and chaperones, transcription factors and proteases represented about 5% each. The, remaining 13 classes each had <5% proteins assigned in all groups (Fig. 4a).
Figure 4.
Gene Ontology (GO) analysis of the proteins identified at the three stages. (a) Distribution of the proteins identified for each group according to PANTHER protein class. (b) Functional classification based on Biological Processes. *Signifies biological process over-represented in samples when compared to the whole mouse genome.
Concerning biological processes, 487 proteins from SF, 701 from SMR and 670 from AF were assigned to the metabolic process cluster. This represented more than 60% of the proteins at every stage of development. Cellular process, transport and specifically protein transport, cell cycle and cell communication were also clusters with numerous proteins assigned (Fig. 4b).
In spite of similar biological-process distribution between the stages, some were shown to be predominant in a given stage when compared to the whole gene Mus musculus database (Supplementary Table S3).
At the SF stage, a total of 77 identified proteins could be matched to the cluster cellular component organization, wherein 57 and 7 proteins were assigned to cellular component morphogenesis and mitochondrial transport, respectively. These subclusters were over-represented only in the SF stage, along with anatomical structure morphogenesis and RNA localization.
Within metabolic processes, ferredoxin and pyrimidine metabolic processes were over-represented in SMR, with 15 and 6 proteins assigned, respectively, while the vitamin metabolic process, with 10 proteins identified, was most prominent in AF. Metabolic processes associated to glycogen, fatty acids and DNA—in particular, DNA replication—were over-represented at SMR and AF stages, but not at SF stage.
Over 20 other biological processes were significantly over-represented in the three follicle groups of the study, which included: cell cycle, more specifically mitosis within the cellular process; generation of precursor metabolites and energy; protein metabolic processes, including protein-folding; carbohydrate metabolic processes; translation; and nuclear and protein transport (Supplementary Table S3).
Protein-network analysis
During follicular development, granulosa-cell proliferation and differentiation are essential to achieve oocyte maturity and competence. Culture conditions directly affect granulosa-cell health and proliferation, as well as cell stress and apoptosis levels (Eppig et al., 2000; Picton et al., 2008). In the samples, proteins with apoptosis or DNA repair were not over-represented in any of the groups. However, metabolic processes related to reactive oxygen species (ROS) were over-represented in all three stages. Pathway Studio analysis associated 112 proteins identified in this study with ROS generation (Fig. 5b). From these, 59 were identified at SF (52.7%), 98 at SMR (87.5%) and 96 at AF (85.7%). For the 95 proteins attributed by this bioinformatics analyzer to DNA repair (Fig. 5a), the distribution between the three stages was similar to that found for proteins associated to ROS generation. For apoptosis and DNA repair biological processes, fewer proteins were identified at SF stage compared to the other two stages, and more proteins were identified in SMR stage.
Figure 5.
Follicle proteins identified related to (a) DNA repair and (b) ROS generation. The images were generated by Pathway Studio. Proteins identified are shown using white background and non-identified using color background.
Through IPA analysis, proteins involved in cell-cycle processes, with a particular focus on a cyclin-dependent kinases (CDKs) network, were highlighted. Indeed, 12 proteins, including CDKs and related proteins, exhibited a particular expression in the three stages (Fig. 6). At the SF stage, four of those proteins, such as S-phase kinase-associated protein 1 (SKP1), F-box/WD repeat-containing protein 12 (FBXW12), breast cancer type 2 susceptibility protein homolog (BRCA2) and Ubiquitin-associated protein 2-like (UBAP2L), were identified, even if cyclins were not (Fig. 6a). At the SMR stage, three of the four cyclins from this network were identified, namely, CDK 1, 4 and 6. Four other proteins that are directly or indirectly associated with those CDKs were also detected at the SMR stage, such as CDK5 regulatory subunit-associated protein 3 (CDK5RAP3), Forkhead box protein O1 (FOXO1), UDP-N-acetylglucosamine-peptide N-acetylglucosaminyltransferase 110 kDa subunit (OGT) and fibronectin (FN1), in addition to the four proteins already detected at SF stage (Fig. 6b). At the AF stage, a cyclin-dependent kinase inhibitor 1B (CDKN1B) was identified. Consequently, other proteins connected with this inhibitor and identified in the previous stages were no longer observed, such as the three CDKs, SKP1/SKP2, FBXW12, BRCA2, FOXO1, OGT and UBAP2L. This illustrated a complete reversal of the CDK network protein profile at this stage (Fig. 6c).
Figure 6.
CDKs and related protein interactions underlined by Ingenuity Pathway Analysis (IPA). Proteins in gray were detected in the samples, and those in white were not. This network showed 12 proteins whose expressions were different between the three stages: (a) SF stage, (b) SMR stage and (c) AF stage. Proteins surrounded in green indicate they were identified at that stage; proteins surrounded in red were not identified.
Analysis of protein-abundance in the three developmental stages
Label-free quantification was performed on 1D LC-MS/MS data (incompatible with 2D LC-MS/MS data) in order to emphasize protein-abundance variations during in vitro follicle development. Differential levels of expression were found to be significant for 57 proteins between the three stages (Table I) and were distributed into five distinct abundance profiles (Fig. 7).
Table I.
Proteins differently expressed in the three stages.
| Accession number | Protein name | Gene name | Protein score | ANOVA P-value | Fold-change | Max. | Min. | Profile Type (Fig. 7) |
|---|---|---|---|---|---|---|---|---|
| Q8K3V4 | Protein-arginine deiminase type-6 | Padi6 | 1103.2 | 1.9E-07 | 3.6 | SF | AF | P1 |
| P16125 | L-lactate dehydrogenase B chain | Ldhb | 879.7 | 7.0E-06 | 4.9 | SF | AF | P1 |
| F8WIV2 | Protein Serpinb6a | Serpinb6a | 648.1 | 3.4E-07 | 2.4 | SF | AF | P1 |
| Q9CWU5 | KH domain-containing protein 3 | Khdc3 | 569.8 | 5.9E-05 | 3.2 | SF | SMR/AF | P1 |
| Q9R0P9 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | Uchl1 | 509.7 | 6.9E-05 | 4.4 | SF | AF | P1 |
| P24549 | Retinal dehydrogenase 1 | Aldh1a1 | 418.6 | 8.0E-04 | 2.0 | SF | SMR | P1 |
| P10761 | Zona pellucida sperm-binding protein 3 | Zp3 | 346.3 | 7.2E-05 | 4.9 | SF | AF | P1 |
| P54869 | Hydroxymethylglutaryl-CoA synthase, mitochondrial | Hmgcs2 | 335.2 | 1.0E-02 | 2.1 | SF | SMR | P1 |
| Q9DAW9 | Calponin-3 | Cnn3 | 308.2 | 2.3E-04 | 2.5 | SF | AF | P1 |
| Q9WVB3 | Transducin-like enhancer protein 6 | Tle6 | 239.0 | 1.9E-06 | 3.5 | SF | AF | P1 |
| P10518 | Delta-aminolevulinic acid dehydratase | Alad | 230.6 | 2.0E-04 | 2.7 | SF | AF | P1 |
| P20239 | Zona pellucida sperm-binding protein 2 | Zp2 | 189.8 | 7.0E-05 | 2.6 | SF | AF | P1 |
| Q08EC7 | Pla2g4c protein | Pla2g4c | 188.0 | 2.4E-04 | 3.1 | SF | AF | P1 |
| P24472 | Glutathione S-transferase A4 | Gsta4 | 153.6 | 3.3E-04 | 2.7 | SF | SMR | P1 |
| Q9CWE6 | Oocyte-expressed protein homolog | Ooep | 94.9 | 2.0E-02 | 4.8 | SF | SMR/AF | P1 |
| P14211 | Calreticulin | Calr | 276.5 | 1.3E-05 | 4.6 | SMR | SF | P2 |
| Q60817 | Nascent polypeptide-associated complex subunit alpha | Naca | 270.9 | 1.6E-04 | 2.3 | SMR | AF | P2 |
| Q60605 | Myosin light polypeptide 6 | Myl6 | 174.3 | 1.6E-03 | 3.4 | SMR | SF/AF | P2 |
| G3UWR0 | Calumenin | Calu | 161.2 | 5.1E-05 | 3.1 | SMR | SF | P2 |
| Q4VAA2 | Protein CDV3 | Cdv3 | 106.5 | 2.1E-04 | 2.5 | SMR | AF | P2 |
| P20029 | 78 kDa glucose-regulated protein | Hspa5 | 2951.4 | 2.6E-05 | 2.7 | AF | SF | P3 |
| P17182 | Alpha-enolase | Eno1 | 1548.7 | 4.5E-07 | 5.0 | AF | SF | P3 |
| P05213 | Tubulin alpha-1B chain | Tuba1b | 1527.4 | 7.0E-04 | 2.7 | AF | SF | P3 |
| P09103 | Protein disulfide-isomerase | P4hb | 1416.4 | 4.3E-06 | 2.7 | AF | SF | P3 |
| P16858 | Glyceraldehyde-3-phosphate dehydrogenase | Gapdh | 701.4 | 1.6E-07 | 4.0 | AF | SF | P3 |
| P14152 | Malate dehydrogenase, cytoplasmic | Mdh1 | 462.0 | 4.8E-06 | 2.1 | AF | SF | P3 |
| P48036 | Annexin A5 | Anxa5 | 445.8 | 5.5E-04 | 2.0 | AF | SF | P3 |
| Q99KV1 | DnaJ homolog subfamily B member 11 | Dnajb11 | 420.4 | 1.1E-03 | 2.2 | AF | SF | P3 |
| G5E8N5 | L-lactate dehydrogenase | Ldha | 412.5 | 3.1E-06 | 3.4 | AF | SF | P3 |
| P24815 | 3 beta-hydroxysteroid dehydrogenase/Delta 5-- > 4-isomerase type 1 | Hsd3b1 | 394.6 | 6.4E-03 | 3.1 | AF | SF | P3 |
| E9PXX7 | Thioredoxin domain-containing protein 5 | Txndc5 | 380.2 | 4.7E-03 | 2.1 | AF | SF | P3 |
| P57759 | Endoplasmic reticulum resident protein 29 | Erp29 | 361.6 | 3.1E-05 | 2.5 | AF | SF | P3 |
| O08807 | Peroxiredoxin-4 | Prdx4 | 358.2 | 4.0E-05 | 2.2 | AF | SF | P3 |
| P29758 | Ornithine aminotransferase, mitochondrial | Oat | 354.1 | 5.3E-04 | 2.1 | AF | SF | P3 |
| P51656 | Estradiol 17-beta-dehydrogenase 1 | Hsd17b1 | 351.2 | 3.6E-03 | 2.4 | AF | SF | P3 |
| P53994 | Ras-related protein Rab-2A | Rab2a | 330.6 | 3.3E-03 | 2.0 | AF | SF | P3 |
| Q9DBJ1 | Phosphoglycerate mutase 1 | Pgam1 | 329.4 | 6.5E-04 | 2.2 | AF | SF | P3 |
| P17742 | Peptidyl-prolyl cis-trans isomerase A | Ppia | 325.5 | 5.8E-03 | 2.2 | SMR | SF | P3 |
| P09411 | Phosphoglycerate kinase 1 | Pgk1 | 300.5 | 2.8E-03 | 2.7 | AF | SF | P3 |
| Q60715 | Prolyl 4-hydroxylase subunit alpha-1 | P4ha1 | 271.1 | 7.0E-05 | 2.2 | AF | SF | P3 |
| Q9QXT0 | Protein canopy homolog 2 | Cnpy2 | 254.6 | 3.8E-03 | 2.3 | SMR | SF | P3 |
| A2A7Q5 | Prolyl 3-hydroxylase 1 | Lepre1 | 225.9 | 2.7E-03 | 2.3 | AF | SF | P3 |
| G5E850 | Cytochrome b-5, isoform CRA_a | Cyb5 | 222.6 | 4.9E-05 | 3.4 | AF | SF | P3 |
| Q3U4W8 | Ubiquitin carboxyl-terminal hydrolase | Usp5 | 216.6 | 5.5E-03 | 2.2 | AF | SF | P3 |
| G3XA14 | Protein Akr1cl | Akr1cl | 212.7 | 7.5E-03 | 3.4 | SMR | SF | P3 |
| P34884 | Macrophage migration inhibitory factor | Mif | 210.1 | 1.6E-05 | 2.1 | AF | SF | P3 |
| P22935 | Cellular retinoic acid-binding protein 2 | Crabp2 | 130.9 | 2.2E-04 | 2.6 | AF | SF | P3 |
| Q9CYA0 | Cysteine-rich with EGF-like domain protein 2 | Creld2 | 107.5 | 2.0E-02 | 2.9 | AF | SF | P3 |
| P63158 | High-mobility group protein B1 | Hmgb1 | 267.9 | 2.0E-03 | 2.5 | SF | SMR | P4 |
| Q9JKB1 | Ubiquitin carboxyl-terminal hydrolase isozyme L3 | Uchl3 | 226.8 | 1.1E-03 | 2.6 | SF | SMR | P4 |
| Q9ERD7 | Tubulin beta-3 chain | Tubb3 | 1342.8 | 1.6E-03 | 3.3 | AF | SMR | P5 |
| P31324 | cAMP-dependent protein kinase type II-beta regulatory subunit | Prkar2b | 1129.2 | 1.7E-04 | 3.7 | AF | SF | P5 |
| Q05816 | Fatty acid-binding protein, epidermal | Fabp5 | 781.9 | 9.1E-05 | 3.1 | AF | SF | P5 |
| P17751 | Triosephosphate isomerase | Tpi1 | 627.6 | 5.7E-05 | 2.3 | AF | SF | P5 |
| Q9D0F9 | Phosphoglucomutase-1 | Pgm1 | 403.2 | 1.2E-04 | 2.3 | AF | SF/SMR | P5 |
| Q99JF5 | Diphosphomevalonate decarboxylase | Mvd | 161.0 | 9.7E-05 | 3.6 | AF | SF | P5 |
| Q62159 | Rho-related GTP-binding protein RhoC | Rhoc | 136.3 | 2.1E-04 | 2.4 | AF | SMR | P5 |
Fold-change and ANOVA were calculated between the maximal and the minimal abundance obtained for each protein. The protein score is the sum of Mascot scores of the identified peptides. Proteins were validated when at least two peptides were significant, with an ANOVA P-value 0.05 and a fold-change ≥2. Profile Type (P1–P5) refers to the normalized abundance behavior of the protein (see Fig. 7).
Figure 7.
Standardized Normalized Abundance profiles of proteins differently expressed between developmental stages. (P1) 15 proteins with highest abundance in SF stage. (P2) 5 proteins with maximal abundance at SMR stage. (P3) 28 proteins corresponding to increasing abundance throughout developmental stages. (P4) 2 proteins with a significant decrease at SMR stage compared to the other two stages. (P5) 7 proteins with a significant variation between the two first stages (SF and SMR) and the AF stage, which is the stage with the highest abundance.
Fifteen proteins had their maximal abundance at the SF stage (Fig. 7 P1). Of these, 11 had significant variations between this stage and the other stages, including 7 proteins associated with the oocyte, such as zona pellucida sperm-binding proteins 2 and 3 (ZP2 and ZP3), oocyte-expressed protein homolog (OOEP), KH domain-containing protein 3 (Filia), transducin-like enhancer protein 6 (TLE6), protein-arginine deiminase type-6 (PADI6) and phospholipase A2, group IVC (Pla2g4c). In this same profile type, it was also possible to observe two proteins with a significant variation between SF and SMR stages, but not in AF stage. The two remaining proteins had a significantly lower abundance at the AF stage compared to the SF and SMR stages (Table I).
From the five proteins with the highest abundance at the SMR stage, which characterize profile 2 (Fig. 7 P2), three calcium ion-binding proteins, calreticulin (CALR), calumenin (CALU) and myosin light polypeptide 6 (MYL6), were significantly less abundant in the other stages. The other two proteins varied between the SMR and AF stages.
A gradual increase in protein-abundance throughout the three stages was the most frequent profile observed, with 28 fitting proteins (Fig. 7 P3). Conversely, the profile with a lower abundance at SMR stage compared to SF and AF stages, with two proteins, was the less frequent profile (Fig. 7 P4).
At AF stage, seven proteins had their maximal abundance, with a significant variation between this stage and the other two (Fig. 7 P5). Two of these proteins are enzymes implicated in glycolysis: alpha-enolase (ENOA) and phosphoglucomutase-1 (PGM1), and a third, fatty acid-binding protein (FABP5), is a glucose transporter. A regulatory subunit of the cAMP-dependent protein kinases involved in cAMP signaling in cells (named PKAR2B) was also particularly abundant at this stage.
Moreover, by analyzing the list of 57 differentially expressed proteins, IPA suggested that glycolysis was the top canonical pathway, with nine proteins involved in this pathway. Eight showed a regular increase during follicle development, with maximal expression at AF stage. Conversely, the remaining protein, L-lactate dehydrogenase chain B (LDHB), had a maximal expression at SF stage and decreased during development (Fig. 8).
Figure 8.
Abundance profile of proteins implicated in glycolysis, with a significant variation throughout follicle development in vitro. In red, eight proteins with increasing abundance during the stages. In blue, one protein, LDHB, with decreasing abundance during follicle development in vitro.
Protein-abundance results were validated by western blot analysis of selected proteins, namely PKAR2B and LDHB (Fig. 9).
Figure 9.
Western blot demonstrating variable expression of PRKAR2B, LDHB and CDKN1B at the three stages. It is possible to observe an increasing expression of PRKAR2B and CDKN1B during the culture, with a higher expression in the two later stages of development. The decreasing expression of LDHB during follicle development is also shown by western blot, with similar abundance in the SMR and AF stages.
Discussion
Previous studies of ovarian proteomics have used isolated oocytes obtained after superovulation and mainly investigated oocyte maturation. The oocytes thus obtained are fully grown and often present the maturity signs of meiosis resumption (Metaphase II oocytes). However, oocyte competence for maturation, fertilization and embryo development is slowly acquired during oocyte development, and it is highly dependent on the interactions between the oocyte and its surrounding cells, their specialization and functioning (Picton et al., 1998; Gilchrist et al., 2004); hence, there is interest in investigating the growth of the complete follicle unit at early stages and following its development.
The choice of the three follicle stages analyzed in this study was based on morphological characteristics previously described when a microdrop culture system is used. The culture system chosen in this study has been previously validated, as live offspring have been obtained, and it allows continuous observation of follicle growth to select morphologically appropriate follicles for the study from the SFs up to the stage of pre-ovulatory follicle (Cortvrindt et al., 1996; Picton et al., 2008).
Follicle proteome analyses and biological processes identified
We attempted to seek potential biological developmental markers of the entire functional follicle structure, rather than to study different cell types individually. By applying a combined 1D and 2D LC-MS/MS analysis, preceded by in-gel IEF fractionation in this study, it was possible to increase the number of proteins identified in follicles. Hence, 1401 different proteins were successfully identified, representing, to the best of our knowledge, the first protein profile of the whole ovarian follicle unit. Interestingly, the qualitative and quantitative analysis of the data pointed out not only biological processes but also proteins which seem to have a particular behavior throughout follicle growth.
GO analysis showed a similar distribution in all three stages for protein classes and biological processes. As in previous studies concerning the oocyte proteome, most proteins identified were assigned to metabolic processes, followed by cellular processes, transport and cell communication (Meng et al., 2007; Ma et al., 2008; Zhang et al., 2009; Wang et al., 2010; Pfeiffer et al., 2011; Demant et al., 2012). However, it was observed that, compared to the whole Mus musculus genome, 44 biological processes were over-represented in the current study’s samples. More than 20 biological processes were over-represented simultaneously in all three stages, with special emphasis on the cell cycle, particularly mitosis. Indeed, IPA emphasized the variable expression of twelve proteins involved in the cell cycle throughout the three stages. We observed that at the SF stage only 4 of the 12 proteins, with expression variation in the CDK network suggested by IPA, were identified. In particular, CDKs themselves were not detected. This is consistent with the fact that, at this stage, the follicle has only a few granulosa-cell layers, and consequently, the mitotic activity is lower than in other stages of development.
Conversely, the highest mitotic activity happened at the SMR stage, with an extremely high level of proliferation of granulosa and theca cells. This was underlined by identification of eleven of the 12 proteins associated with the cell cycle. The identification of the CDKs only at this stage sustains the intense mitotic activity of somatic cells in the growing follicle, because these proteins, when connected with cyclins, are responsible for cell-cycle progression and consequently granulosa-cell proliferation (Sherr, 1994; Coqueret, 2002; Shimizu et al., 2013). FOXO1, also only identified at SMR stage, was previously described as being specifically expressed in the granulosa cells of growing follicles (Tarnawa et al., 2013), with lower levels of expression in antral and Graafian follicles (Shi and LaPolt, 2003). It was also described as regulating the gene expression associated with proliferation, apoptosis and homeostasis of granulosa cells in culture (Park et al., 2005; Liu et al., 2009, 2013). Furthermore, CDKs 4/6 and FOXO1 have not been previously reported in the proteome of isolated oocytes or COC, suggesting that they are only expressed in granulosa cells, or at least they are more abundant before ovulation.
A reduction in granulosa-cell proliferation at Day 9 of ovarian follicle culture was previously described (Pesty et al., 2007), indicating that even if the follicle is still increasing in size, mitotic activity decreases from that day onward. In the present study, the same behavior was highlighted by the identification of cyclin-dependent kinase inhibitor 1B (CDKN1B), also known as p27Kip1, only at the AF stage. This protein prevents the formation of cyclin E-CDK2 and cyclin D-CDK4/CDK6 complexes, resulting in inhibition of cell-cycle progression and cell proliferation (Bayrak and Oktay, 2003; Rajareddy et al., 2007). In addition, the SPK1/SPK2 complex, which is known to be involved in CDKN1B degradation, was not identified at this stage, although it was detected in previous stages analyzed. Since CDKN1B has been described as being detected in granulosa cells at all stages of follicular development, the current study’s results showed that this protein is more abundant at AF stage compared to the other stages (Fig. 9). Moreover, previous studies showed the important role of this protein in follicle luteinization and ovulation (Fero et al., 1996; Kiyokawa et al., 1996; Nakayama et al., 1996), so it could be suggested as a marker for culture and follicle quality.
Nuclear and protein transport, generation of precursor metabolites and energy, carbohydrate metabolic process, protein-folding and translation were other biological processes over-represented in the three stages. These results are consistent with the major role of interaction between the granulosa cells and the oocyte, as well as the high levels of transcription, inducing expression of proteins needed for follicular and oocyte growth and acquisition of molecular capacities for future events.
Otherwise, some biological processes seemed to be more relevant to a specific stage of development since they were only over-represented in one of the development stages chosen for this study. In particular, the biological processes related to ferredoxins were only over-represented in SMR stage, and it was also only at this stage that adrenoxin (FDX1), called ferredoxin 1 in humans, was identified. Ferredoxin 1 is described as being involved in steroidogenesis (Miller, 2005) and it seems that transcription of this protein increases with the level of cAMP (Imamichi et al., 2013). This could be an indicator of steroidogenesis activity increasing in developing follicles. These results are consistent with estradiol secretion studies in ovarian follicles in culture, which associated secretion with cellular proliferation, describing a stabilization or decrease of estradiol secretion associated with a reduction in granulosa-cell proliferation (Boland et al., 1993; Liu et al., 2002; Dorphin et al., 2012).
The over-representation of metabolic processes related to glycogen and fatty acids at both SMR and AF stages supported the information from a previous study suggesting that these metabolic processes have an increasing role throughout follicular development (Collado-Fernandez et al., 2012).
Analysis of protein-abundance in the three developmental stages
Different levels of expression between the three stages were observed for 57 proteins. The majority of the proteins that were more abundant in SF stage are expressed in the oocyte, such as the glycoproteins ZP2 and ZP3 (Epifano et al., 1995), oocyte- and embryo-abundant PAD (PADI6) (Wright et al., 2003) and three protein members of the subcortical maternal complex (SCMC): OOEP, Filia (KHDC3) and TLE6 (Li et al., 2008). Although these proteins have been shown to increase during oocyte development (Epifano et al., 1995; Wright et al., 2003; Li et al., 2008; Zheng and Dean, 2009; Yurttas et al., 2010), their expression decreased when looking at the entire follicle during in vitro development. This is consistent with the higher representation of oocyte proteins in SF stage follicles when compared to the later stages where granulosa-cell growth is predominant and their proteins become much more abundant.
At the SMR stage, quantification analysis pointed out calcium-associated proteins, which is one of the most ubiquitous signaling molecules and which controls a wide variety of cellular processes. Calreticulin (CARL) is the major Ca2+-binding chaperone in oocytes (Zhang et al., 2010). Accumulating evidence has found that intracellular free Ca2+ plays an important role in regulation of the meiotic maturation of oocytes and early embryonic development (Homa et al., 1993; Sousa et al., 1997; Whitaker, 2006). In the current study, quantitative analysis of the proteome revealed that this protein, as well as other calcium-binding proteins, such as calumenin (CALU) and myosin light protein 6 (MYL6), were more abundant at the SMR stage. This suggests that calcium exchanges in the follicle may have a relevant role at this point in development.
Quantitative analysis also highlighted carbohydrate metabolism, particularly glycolysis, which is described as an important pathway for glucose utilization by larger follicles, as an important biological process in follicle development (Harris et al., 2007; Xu et al., 2009; Collado-Fernandez et al., 2012). Glycolysis is required for follicle growth and estradiol secretion, and it is the main source of ATP in mouse preantral follicles in vitro (Boland et al., 1994b; a). It was possible to identify several proteins involved in this pathway, such as pyruvate kinase M2 (KPYM), PGM1, L-lactate dehydrogenase chains A and B (LDHA, LDHB), fructose-bisphosphate aldolase A (ALDOA), alpha and gamma enolase (ENOA, ENOG), ADP glucokinase (ADPGK), GAPDH, triosephosphate isomerase (TPIS), 6-phosphofructokinase type C (K6PP), glucose-6-phosphate (G6PD1) and phosphoglycerate mutase 1 (PGAM1). The abundance for nine of these proteins was found to vary throughout follicle development (Table I). The expression of LDHB reached a maximum at the SF stage, while the other eight exhibited an increase in abundance throughout follicle development, achieving a maximum at AF stage. These results illustrate a strong need for energy supply at this stage of in vitro development. Previous studies showed that LDHB and associated mRNA increased in the oocyte during follicular development, maturation and the beginning of embryonic development (Brinkworth and Masters, 1978; Roller et al., 1989; Winger et al., 2000; Li et al., 2006). Yet, our results showed a lower abundance during follicular development, which could be interpreted as a protein specifically localized in the oocyte.
Another protein of relevant interest, and abundant in AF stage, was PRKAR2B, which is a regulatory subunit of the cAMP-dependent protein kinase (PKA) involved in cAMP signaling. Indeed, high levels of cAMP within the oocyte are essential to keep the meiotic cycle on hold. Nevertheless, the source of cAMP is still controversial since one theory suggests that the oocyte itself generates the inhibitory cAMP (Mehlmann et al., 2002; Horner et al., 2003; Mehlmann, 2005) whereas another theory suggests that meiotic arrest is dependent on cumulus or granulosa cells supplying this inhibitor (Sela-Abramovich et al., 2006). Higher levels of a protein implicated in cAMP signaling at AF stage suggest that this protein is likely to be produced in the granulosa cells since at this stage, in the current samples there is a higher number of granulosa cells compared to the other stages. It would be of great interest to further investigate the regulation of PKA during follicle development in order to assess its potential role in folliculogenesis.
Apoptosis, ROS and DNA repair
During in vivo folliculogenesis, a large number of follicles will not grow and will degenerate, mainly due to proliferation arrest of granulosa cells and consequent apoptosis. In the ovary, almost 99% of follicles that begin development will die; therefore apoptosis is expected to be a major biological process in the ovary proteome (He et al., 2014). In the current study, only 5% of the proteins identified were attributed to that biological process. Since apoptosis was not over-represented at any stage of development, it can be concluded that in vitro culture conditions differ from those in vivo, as there is not increased expression of proteins related to apoptosis, taking account of appropriate culture conditions.
In vivo, the follicles constantly compete for developmental resources, leading to the death of the weakest. Nevertheless, in the type of culture that we describe here, the follicles grew individually without competition for resources. The lack of competition between follicles lead to a larger number of follicles reaching ovulatory stage and a reduced apoptosis rate compared to in vivo conditions.
However, Pathway Studio analysis demonstrated that at SMR and AF stages, twice as many proteins involved in ROS production and DNA repair were identified compared to SF stage. This could be justified by the culture conditions since the SF stage was exposed to those conditions for a shorter time. Nevertheless, at the end of the culture, the AF stage had fewer proteins associated with those biological processes compared to SMR stage. These results could be justified by the change of follicular structure in this type of culture, passing from a tridimensional to a bidimensional structure, as the follicle grows attached to the culture surface (Cortvrindt et al., 1996; Pesty et al., 2007; Picton et al., 2008). However, if increased transcription of proteins involved in ROS production and DNA repair is induced by this structural change, it seems that it does not have a significant negative impact in follicular development since these types of protein was reduced again in the last culture phase.
Conclusion
Follicular integrity is crucial for oocyte development from follicle activation until maturation. In this study, it was possible to identify 1401 proteins expressed in the follicle during its growth in vitro, representing the first catalog of a follicle proteome. Although this protein list is not exhaustive, it shows the most abundant proteins expressed in the growing follicle, and it can be used as a baseline for further studies about the role of a protein in follicle development.
The comparative analysis of the three distinct morphological stages of follicles made it possible to highlight several functional groups of proteins exhibiting differential expression during in vitro development, suggesting different roles at specific stages. The combination of qualitative and quantitative analyses highlighted specific expression patterns for proteins associated with the CDK network, glycolysis, calcium binding and regulation, as well as a regulatory subunit of PKA. Further complementary studies should be carried out to understand the specific activation of developmental pathways that allow follicle development and maturation.
This study contributes to the identification of several mechanisms involved in the ovarian follicles development in vitro. The resulting data may potentially serve future improvements of current and still-suboptimal mammalian follicle-culture systems, which is particularly interesting as alternatives for female patients treated for cancer are urgently needed.
Supplementary data
Supplementary data are available at Molecular Human Reproduction online.
Authors’ roles
A.A., K.A.R-.W., T.A.S. and C. Poirot designed the study. A.A., K.A.R-.W., S.C. and C. Pionneau conducted the experiments, with A.A. having the main responsibility. C.F. realized the Ingenuity Pathways Analysis (IPA) and Pathway Studio analysis. A.A., S.C. and C. Pionneau analyzed the data, with A.A. having the main responsibility. K.A.R-.W. and C. Poirot provided economic and administrative support. A.A., K.A.R-.W., S.C., C. Pionneau, C.F., T.A.S. and C. Poirot participated in the writing of the manuscript and approved the final version.
Funding
The Portuguese Foundation for Science and Technology, FCT (PhD fellowship SFRH/BD/65299/2009 to A.A.), the Swedish Childhood Cancer Foundation (PR 2014-0144 to K.A.R-.W.) and Stockholm County Council (to K.A.R-.W.).
Conflict of interest
The authors of the study have no conflict of interest to report.
Supplementary Material
Acknowledgements
The authors wish to thank Karine Reynaud (Alfort Veterinary School, Maisons Alfort, France) and Karthik Govindasamy Muralidharan, Arian Lundberg and Jonas Bergh (Department of Oncology-Pathology, Cancer Center Karolinska, Karolinska Institutet) for laboratory assistance and administrative support. We also thank the anonymous reviewers whose comments and suggestions helped improve and clarify this manuscript.
References
- Bayrak A, Oktay K. The expression of cyclin-dependent kinase inhibitors p15, p16, p21, and p27 during ovarian follicle growth initiation in the mouse. Reprod Biol Endocrinol 2003;1:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland NI, Humpherson PG, Leese HJ, Gosden RG. Pattern of lactate production and steroidogenesis during growth and maturation of mouse ovarian follicles in vitro. Biol Reprod 1993;48:798–806. [DOI] [PubMed] [Google Scholar]
- Boland NI, Humpherson PG, Leese HJ, Gosden RG. Characterization of follicular energy metabolism. Hum Reprod 1994. a;9:604–609. [DOI] [PubMed] [Google Scholar]
- Boland NI, Humpherson PG, Leese HJ, Gosden RG. The effect of glucose metabolism on murine follicle development and steroidogenesis in vitro. Hum Reprod 1994. b;9:617–623. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248–254. [DOI] [PubMed] [Google Scholar]
- Brinkworth RI, Masters CJ. On the localization of lactate dehydrogenase in the ovaries and reproductive tracts of rats and mice. Mech Ageing Dev 1978;8:299–310. [DOI] [PubMed] [Google Scholar]
- Collado-Fernandez E, Picton HM, Dumollard R. Metabolism throughout follicle and oocyte development in mammals. Int J Dev Biol 2012;56:799–808. [DOI] [PubMed] [Google Scholar]
- Coqueret O. Linking cyclins to transcriptional control. Gene 2002;299:35–55. [DOI] [PubMed] [Google Scholar]
- Cortvrindt R, Smitz J, Van Steirteghem AC. In-vitro maturation, fertilization and embryo development of immature oocytes from early preantral follicles from prepuberal mice in a simplified culture system. Hum Reprod 1996;11:2656–2666. [DOI] [PubMed] [Google Scholar]
- Demant M, Trapphoff T, Frohlich T, Arnold GJ, Eichenlaub-Ritter U. Vitrification at the pre-antral stage transiently alters inner mitochondrial membrane potential but proteome of in vitro grown and matured mouse oocytes appears unaffected. Hum Reprod 2012;27:1096–1111. [DOI] [PubMed] [Google Scholar]
- Dorphin B, Prades-Borio M, Anastacio A, Rojat P, Coussieu C, Poirot C. Secretion profiles from in vitro cultured follicles, isolated from fresh prepubertal and adult mouse ovaries or frozen-thawed prepubertal mouse ovaries. Zygote 2012;20:181–192. [DOI] [PubMed] [Google Scholar]
- Epifano O, Liang LF, Familari M, Moos MC Jr, Dean J. Coordinate expression of the three zona pellucida genes during mouse oogenesis. Development 1995;121:1947–1956. [DOI] [PubMed] [Google Scholar]
- Eppig JJ, Hosoe M, O’Brien MJ, Pendola FM, Requena A, Watanabe S. Conditions that affect acquisition of developmental competence by mouse oocytes in vitro: FSH, insulin, glucose and ascorbic acid. Mol Cell Endocrinol 2000;163:109–116. [DOI] [PubMed] [Google Scholar]
- Eppig JJ, O’Brien MJ, Wigglesworth K, Nicholson A, Zhang W, King BA. Effect of in vitro maturation of mouse oocytes on the health and lifespan of adult offspring. Hum Reprod 2009;24:922–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JJ, Pendola FL, Wigglesworth K, Pendola JK. Mouse oocytes regulate metabolic cooperativity between granulosa cells and oocytes: amino acid transport. Biol Reprod 2005;73:351–357. [DOI] [PubMed] [Google Scholar]
- Fero ML, Rivkin M, Tasch M, Porter P, Carow CE, Firpo E, Polyak K, Tsai LH, Broudy V, Perlmutter RM et al. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell 1996;85:733–744. [DOI] [PubMed] [Google Scholar]
- Gilchrist RB, Ritter LJ, Armstrong DT. Oocyte-somatic cell interactions during follicle development in mammals. Anim Reprod Sci 2004;82–83:431–446. [DOI] [PubMed] [Google Scholar]
- Harris SE, Adriaens I, Leese HJ, Gosden RG, Picton HM. Carbohydrate metabolism by murine ovarian follicles and oocytes grown in vitro. Reproduction 2007;134:415–424. [DOI] [PubMed] [Google Scholar]
- Hawkins SM, Matzuk MM. Oocyte-somatic cell communication and microRNA function in the ovary. Ann Endocrinol 2010;71:144–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Teng H, Zhou T, Guo Y, Wang G, Lin M, Sun Y, Si W, Zhou Z, Guo X et al. Unravelling the proteome of adult rhesus monkey ovaries. Mol Biosyst 2014;10:653. [DOI] [PubMed] [Google Scholar]
- Homa ST, Carroll J, Swann K. The role of calcium in mammalian oocyte maturation and egg activation. Hum Reprod 1993;8:1274–1281. [DOI] [PubMed] [Google Scholar]
- Horner K, Livera G, Hinckley M, Trinh K, Storm D, Conti M. Rodent oocytes express an active adenylyl cyclase required for meiotic arrest. Dev Biol 2003;258:385–396. [DOI] [PubMed] [Google Scholar]
- Imamichi Y, Mizutani T, Ju Y, Matsumura T, Kawabe S, Kanno M, Yazawa T, Miyamoto K. Transcriptional regulation of human ferredoxin 1 in ovarian granulosa cells. Mol Cell Endocrinol 2013;370:1–10. [DOI] [PubMed] [Google Scholar]
- Kiyokawa H, Kineman RD, Manova-Todorova KO, Soares VC, Hoffman ES, Ono M, Khanam D, Hayday AC, Frohman LA, Koff A. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1). Cell 1996;85:721–732. [DOI] [PubMed] [Google Scholar]
- Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell 2008;15:416–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SS, Liu YH, Tseng CN, Singh S. Analysis of gene expression in single human oocytes and preimplantation embryos. Biochem Biophys Res Commun 2006;340:48–53. [DOI] [PubMed] [Google Scholar]
- Liu HC, He Z, Rosenwaks Z. In vitro culture and in vitro maturation of mouse preantral follicles with recombinant gonadotropins. Fertil Steril 2002;77:373–383. [DOI] [PubMed] [Google Scholar]
- Liu Z, Castrillon DH, Zhou W, Richards JS. FOXO1/3 depletion in granulosa cells alters follicle growth, death and regulation of pituitary FSH. Mol Endocrinol 2013;27:238–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Rudd MD, Hernandez-Gonzalez I, Gonzalez-Robayna I, Fan HY, Zeleznik AJ, Richards JS. FSH and FOXO1 regulate genes in the sterol/steroid and lipid biosynthetic pathways in granulosa cells. Mol Endocrinol 2009;23:649–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Guo X, Wang F, Zhao C, Liu Z, Shi Z, Wang Y, Zhang P, Zhang K, Wang N et al. Protein expression profile of the mouse metaphase-II oocyte. J Proteome Res 2008;7:4821–4830. [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Burns KH, Viveiros MM, Eppig JJ. Intercellular communication in the mammalian ovary: oocytes carry the conversation. Science 2002;296:2178–2180. [DOI] [PubMed] [Google Scholar]
- Mehlmann LM. Stops and starts in mammalian oocytes: recent advances in understanding the regulation of meiotic arrest and oocyte maturation. Reproduction 2005;130:791–799. [DOI] [PubMed] [Google Scholar]
- Mehlmann LM, Jones TL, Jaffe LA. Meiotic arrest in the mouse follicle maintained by a Gs protein in the oocyte. Science 2002;297:1343–1345. [DOI] [PubMed] [Google Scholar]
- Meng Y, Liu XH, Ma X, Shen Y, Fan L, Leng J, Liu JY, Sha JH. The protein profile of mouse mature cumulus-oocyte complex. Biochim Biophys Acta 2007;1774:1477–1490. [DOI] [PubMed] [Google Scholar]
- Miller WL. Minireview: regulation of steroidogenesis by electron transfer. Endocrinology 2005;146:2544–2550. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Ishida N, Shirane M, Inomata A, Inoue T, Shishido N, Horii I, Loh DY, Nakayama K. Mice lacking p27(Kip1) display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell 1996;85:707–720. [DOI] [PubMed] [Google Scholar]
- Park Y, Maizels ET, Feiger ZJ, Alam H, Peters CA, Woodruff TK, Unterman TG, Lee EJ, Jameson JL, Hunzicker-Dunn M. Induction of cyclin D2 in rat granulosa cells requires FSH-dependent relief from FOXO1 repression coupled with positive signals from Smad. J Biol Chem 2005;280:9135–9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesty A, Miyara F, Debey P, Lefevre B, Poirot C. Multiparameter assessment of mouse oogenesis during follicular growth in vitro. Mol Hum Reprod 2007;13:3–9. [DOI] [PubMed] [Google Scholar]
- Pfeiffer MJ, Siatkowski M, Paudel Y, Balbach ST, Baeumer N, Crosetto N, Drexler HC, Fuellen G, Boiani M. Proteomic analysis of mouse oocytes reveals 28 candidate factors of the ‘reprogrammome’. J Proteome Res 2011;10:2140–2153. [DOI] [PubMed] [Google Scholar]
- Picton H, Briggs D, Gosden R. The molecular basis of oocyte growth and development. Mol Cell Endocrinol 1998;145:27–37. [DOI] [PubMed] [Google Scholar]
- Picton HM, Harris SE, Muruvi W, Chambers EL. The in vitro growth and maturation of follicles. Reproduction 2008;136:703–715. [DOI] [PubMed] [Google Scholar]
- Rajareddy S, Reddy P, Du C, Liu L, Jagarlamudi K, Tang W, Shen Y, Berthet C, Peng SL, Kaldis P et al. p27kip1 (cyclin-dependent kinase inhibitor 1B) controls ovarian development by suppressing follicle endowment and activation and promoting follicle atresia in mice. Mol Endocrinol 2007;21:2189–2202. [DOI] [PubMed] [Google Scholar]
- Roller RJ, Kinloch RA, Hiraoka BY, Li SS, Wassarman PM. Gene expression during mammalian oogenesis and early embryogenesis: quantification of three messenger RNAs abundant in fully grown mouse oocytes. Development 1989;106:251–261. [DOI] [PubMed] [Google Scholar]
- Sela-Abramovich S, Edry I, Galiani D, Nevo N, Dekel N. Disruption of gap junctional communication within the ovarian follicle induces oocyte maturation. Endocrinology 2006;147:2280–2286. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. G1 phase progression: cycling on cue. Cell 1994;79:551–555. [DOI] [PubMed] [Google Scholar]
- Shi F, LaPolt PS. Relationship between FoxO1 protein levels and follicular development, atresia, and luteinization in the rat ovary. J Endocrinol 2003;179:195–203. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Hirai Y, Miyamoto A. Expression of cyclins and cyclin-dependent kinase inhibitors in granulosa cells from bovine ovary. Reprod Domest Anim 2013;48:e65–e69. [DOI] [PubMed] [Google Scholar]
- Smitz JE, Cortvrindt RG. The earliest stages of folliculogenesis in vitro. Reproduction 2002;123:185–202. [DOI] [PubMed] [Google Scholar]
- Sousa M, Barros A, Silva J, Tesarik J. Developmental changes in calcium content of ultrastructurally distinct subcellular compartments of preimplantation human embryos. Mol Hum Reprod 1997;3:83–90. [DOI] [PubMed] [Google Scholar]
- Tarnawa ED, Baker MD, Aloisio GM, Carr BR, Castrillon DH. Gonadal expression of Foxo1, but not Foxo3, is conserved in diverse Mammalian species. Biol Reprod 2013;88:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer EE, Zelinski MB. Ovarian follicle culture: advances and challenges for human and nonhuman primates. Fertil Steril 2013;99:1523–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Kou Z, Jing Z, Zhang Y, Guo X, Dong M, Wilmut I, Gao S. Proteome of mouse oocytes at different developmental stages. Proc Natl Acad Sci USA 2010;107:17639–17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker M. Calcium at fertilization and in early development. Physiol Rev 2006;86:25–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winger QA, Hill JR, Shin T, Watson AJ, Kraemer DC, Westhusin ME. Genetic reprogramming of lactate dehydrogenase, citrate synthase, and phosphofructokinase mRNA in bovine nuclear transfer embryos produced using bovine fibroblast cell nuclei. Mol Reprod Dev 2000;56:458–464. [DOI] [PubMed] [Google Scholar]
- Wright PW, Bolling LC, Calvert ME, Sarmento OF, Berkeley EV, Shea MC, Hao Z, Jayes FC, Bush LA, Shetty J et al. ePAD, an oocyte and early embryo-abundant peptidylarginine deiminase-like protein that localizes to egg cytoplasmic sheets. Dev Biol 2003;256:73–88. [DOI] [PubMed] [Google Scholar]
- Xu M, Barrett SL, West-Farrell E, Kondapalli LA, Kiesewetter SE, Shea LD, Woodruff TK. In vitro grown human ovarian follicles from cancer patients support oocyte growth. Hum Reprod 2009;24:2531–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Kreeger PK, Shea LD, Woodruff TK. Tissue-engineered follicles produce live, fertile offspring. Tissue Eng 2006;12:2739–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurttas P, Morency E, Coonrod SA. Use of proteomics to identify highly abundant maternal factors that drive the egg-to-embryo transition. Reproduction 2010;139:809–823. [DOI] [PubMed] [Google Scholar]
- Zhang DX, Li XP, Sun SC, Shen XH, Cui XS, Kim NH. Involvement of ER-calreticulin-Ca2+ signaling in the regulation of porcine oocyte meiotic maturation and maternal gene expression. Mol Reprod Dev 2010;77:462–471. [DOI] [PubMed] [Google Scholar]
- Zhang P, Ni X, Guo Y, Guo X, Wang Y, Zhou Z, Huo R, Sha J. Proteomic-based identification of maternal proteins in mature mouse oocytes. BMC Genomics 2009;10:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Dean J. Role of Filia, a maternal effect gene, in maintaining euploidy during cleavage-stage mouse embryogenesis. Proc Natl Acad Sci USA 2009;106:7473–7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.