Abstract
STUDY QUESTION
Do paternal exposures to folic acid deficient (FD), and/or folic acid supplemented (FS) diets, throughout germ cell development adversely affect male germ cells and consequently offspring health outcomes?
SUMMARY ANSWER
Male mice exposed over their lifetimes to both FD and FS diets showed decreased sperm counts and altered imprinted gene methylation with evidence of transmission of adverse effects to the offspring, including increased postnatal-preweaning mortality and variability in imprinted gene methylation.
WHAT IS KNOWN ALREADY
There is increasing evidence that disruptions in male germ cell epigenetic reprogramming are associated with offspring abnormalities and intergenerational disease. The fetal period is the critical time of DNA methylation pattern acquisition for developing male germ cells and an adequate supply of methyl donors is required. In addition, DNA methylation patterns continue to be remodeled during postnatal spermatogenesis. Previous studies have shown that lifetime (prenatal and postnatal) folic acid deficiency can alter the sperm epigenome and increase the incidence of fetal morphological abnormalities.
STUDY DESIGN, SIZE, DURATION
Female BALB/c mice (F0) were placed on one of four amino-acid defined diets for 4 weeks before pregnancy and throughout pregnancy and lactation: folic acid control (Ctrl; 2 mg/kg), 7-fold folic acid deficient (7FD; 0.3 mg/kg), 10-fold high FS (10FS, 20 mg/kg) or 20-fold high FS (20FS, 40 mg/kg) diets. F1 males were weaned to their respective prenatal diets to allow for diet exposure during all windows of germline epigenetic reprogramming: the erasure, re-establishment and maintenance phases.
PARTICIPANTS/MATERIALS, SETTINGS, METHODS
F0 females were mated with chow-fed males to produce F1 litters whose germ cells were exposed to the diets throughout embryonic development. F1 males were subsequently mated with chow-fed female mice. Two F2 litters, unexposed to the experimental diets, were generated from each F1 male; one litter was collected at embryonic day (E)18.5 and one delivered and followed postnatally. DNA methylation at a global level and at the differentially methylated regions of imprinted genes (H19, Imprinted Maternally Expressed Transcript (Non-Protein Coding)—H19, Small Nuclear Ribonucleoprotein Polypeptide N—Snrpn, KCNQ1 Opposite Strand/Antisense Transcript 1 (Non-Protein Coding)—Kcnq1ot1, Paternally Expressed Gene 1—Peg1 and Paternally Expressed Gene 3-Peg3) was assessed by luminometric methylation analysis and bisulfite pyrosequencing, respectively, in F1 sperm, F2 E18.5 placenta and F2 E18.5 brain cortex.
MAIN RESULTS AND THE ROLE OF CHANCE
F1 males exhibited lower sperm counts following lifetime exposure to both folic acid deficiency and the highest dose of folic acid supplementation (20FS), (both P < 0.05). Post-implantation losses were increased amongst F2 E18.5 day litters from 20FS exposed F1 males (P < 0.05). F2 litters derived from both 7FD and 20FS exposed F1 males had significantly higher postnatal-preweaning pup death (both P < 0.05). Sperm from 10FS exposed males had increased variance in methylation across imprinted gene H19, P < 0.05; increased variance at a few sites within H19 was also found for the 7FD and 20FS groups (P < 0.05). While the 20FS diet resulted in inter-individual alterations in methylation across the imprinted genes Snrpn and Peg3 in F2 E18.5 placenta, ≥50% of individual sites tested in Peg1 and/or Peg3 were affected in the 7FD and 10FS groups. Inter-individual alterations in Peg1 methylation were found in F2 E18.5 day 10FS group brain cortex (P < 0.05).
LARGE SCALE DATA
Not applicable.
LIMITATIONS REASONS FOR CAUTION
The cause of the increase in postnatal-preweaning mortality was not investigated post-mortem. Further studies are required to understand the mechanisms underlying the adverse effects of folic acid deficiency and supplementation on developing male germ cells. Genome-wide DNA and histone methylome studies as well as gene expression studies are required to better understand the links between folic acid exposures, an altered germ cell epigenome and offspring outcomes.
WIDER IMPLICATIONS OF THE FINDINGS
The findings of this study provide further support for paternally transmitted environmental effects. The results indicate that both folic acid deficiency and high dose supplementation can be detrimental to germ cell development and reproductive fitness, in part by altering DNA methylation in sperm.
STUDY FUNDING AND COMPETING INTERESTS
This study was supported by a grant to J.M.T. from the Canadian Institutes of Health Research (CIHR #89944). The authors declare they have no conflicts of interest.
Keywords: folic acid, paternal effects, DNA methylation, developmental programming, epigenetics, intergenerational effects, sperm, male-mediated, folate
Introduction
Paternal transmission of disease resulting from exposures of male germ cells to environmental factors such as under- and over-nutrition is receiving increased attention due to the emergence of new mechanisms to explain male-mediated effects and the potential for intergenerational consequences (Lane et al., 2014). In particular, environmental exposures can impact the epigenome, including DNA methylation, and result in heritable alterations in gene expression. Folate (vitamin B9) plays a critical role in 1-carbon metabolism involving DNA synthesis and biological methylation reactions such as DNA, histone and protein methylation. Low folate intake and variants in folate metabolism pathway enzymes can alter the availability of cellular methyl groups. Conversely, supplements of folic acid, the synthetic form of folate, are given to pregnant women to prevent neural tube defects (NTDs) and other birth defects and to subfertile men to improve semen parameters including sperm counts (Ebisch et al., 2007; Wilson et al., 2015). DNA methylation patterning is particularly dynamic in developing male germ cells, which could make them particularly susceptible to low or high-folate status during either gestational (in utero) or adult exposures. While there is recent evidence that low paternal folate may alter the sperm epigenome and result in adverse pregnancy outcomes (Lambrot et al., 2013), the potential for male-mediated effects on the offspring mediated by high dietary folate intake has not been examined.
Folate metabolism generates the universal methyl donor S-adenosyl methionine, the primary source of methyl groups for several cellular reactions including epigenetic modifications such as DNA and histone methylation. It is well established that low peri-conceptional folate status is associated with certain pregnancy complications and NTDs (Czeizel and Dudas, 1992; Gordon, 1995; Czeizel et al., 2011). As such, supplements containing 0.4 mg folic acid per day or more are recommended for women considering pregnancy (Wilson et al., 2015). Since the late 1990s, many countries have introduced programs to fortify the food supply with folic acid (Miller and Ulrich, 2013) and rates of NTDs have subsequently decreased (De-Regil et al., 2010). Among others, epigenetic mechanisms have been suggested to underlie the effect of folic acid in preventing birth defects.
Higher folic acid doses of 4–5 mg/day (i.e. 10-fold the pregnancy supplement dose) are recommended, for instance, in certain high-risk pregnancies (Wilson et al., 2015), and there are ongoing clinical trials assessing the use of 4 mg/day folic acid for the prevention of birth defects other than NTDs, and pre-eclampsia (Wen et al., 2013; Bortolus et al., 2014). Based on the results of small scale clinical trials showing that high dose folic acid resulted in improved sperm concentrations in semen (Wong et al., 2002; Ebisch et al., 2007), men with idiopathic infertility are often given supplements containing 5 mg/day of folic acid. However, use of higher doses of folic acid, in the range of 1–5 mg, is controversial due in part to questions regarding the effects of circulating un-metabolized folic acid (UFA) (Smith et al., 2008; Reynolds, 2016). High dose folic acid results in the presence of significant levels of UFA in blood, which is then reduced by dihydrofolate reductase (DHFR) to a bioactive form. For instance, in elderly men given 5 mg/day over 3 weeks, UFA was found in 100% of the group versus 26% at baseline (Obeid et al., 2011). It has been suggested that due to the relatively low activity of DHFR in human liver, increasing the dose of folic acid above a maximum of 1 mg/day may not continue to increase the folate pool available for biological methylation reactions (Bailey and Ayling, 2009). Consistent with this, a few recent studies have shown unexpected decreases in DNA methylation associated with the use of folic acid; increasing doses of a methyl donor would have been predicted to result in increases in DNA methylation. For example, a large epigenome-wide study linked higher maternal plasma folate concentrations with lower DNA methylation levels in cord blood (Joubert et al., 2016). Similarly, DNA hypomethylation was observed in the sperm of infertile men treated with 5 mg/day of folic acid for 6 months (Aarabi et al., 2015).
Evidence from rodent models also indicates that there can be deleterious developmental effects of high dose folic acid supplements. In rodent studies the baseline diets usually contain the recommended daily intake of folic acid (2 mg folic acid/kg diet), with the supplemented diets ranging from moderate (2.5-fold baseline levels), to high (10-fold) or very high (20-fold), with the 10-fold diet roughly equivalent to the 4–5 mg/day human dose. Both 10-fold and 20-fold doses of folic acid given to female mice before and during gestation resulted in embryonic loss and delay, growth retardation and birth defects (Pickell et al., 2011; Mikael et al., 2013). In mouse and rat studies, gestational moderate to high dose (2.0–10-fold), with or without postnatal folic acid supplementation, has been linked to altered DNA methylation patterns in offspring somatic tissues, including the brain (Schaible et al., 2011; Sie et al., 2013; Barua et al., 2016). Here, we chose to study 10-fold and 20-fold folic acid supplements as we were interested in determining the intergenerational effects of clinically relevant doses rather than doses that could be achieved through diet or over the counter supplements.
During their development starting in the prenatal gonad, male germ cells undergo major changes in epigenetic programming involving DNA methylation, histone modifications and non-coding RNAs (ncRNAs). For instance, in mice, DNA methylation at most of the ~20 million CpG sites in the genome is erased at mid-gestation in the primordial germ cells and then re-established for the most part in the prospermatogonia of the fetal gonad prior to birth. During postnatal spermatogenesis, DNA methylation is maintained during cell divisions in spermatogonia and most increases and decreases in DNA methylation occur prior to spermiogenesis (Ly et al., 2015). Certain sequences, such as differentially methylated regions (DMRs) of imprinted genes, have characteristic sperm-specific patterns of DNA methylation, which are transmitted to the offspring. Imprinted genes play critical roles in normal fetal growth and development as well as placental function (Tunster et al., 2013). Histone modifications, some of which are transmitted to the offspring, are also remodeled in prenatal and postnatal male germ cells with histone methylation, in particular, a modification potentially sensitive to methyl donor supply (Ly et al., 2015).
The availability of methyl donors, such as folate, is critical for normal DNA methylation patterning in male germ cells. In humans, early studies reported associations between low blood folate concentrations and male subfertility (Bentivoglio et al., 1993; Forges et al., 2007). Also, infertility and aberrant DNA methylation in sperm have been associated with variants in key enzymes involved in folate metabolism such as methylenetetrahydrofolate reductase (MTHFR) (Forges et al., 2007; Gong et al., 2005). In mice, folic acid deficiency resulted in decreased sperm counts (Swayne et al., 2012) and lifetime (in utero and postnatal exposures) dietary folic acid deficiency was associated with evidence of alterations to the sperm epigenome (Lambrot et al., 2013). In the Lambrot et al. (2013) study, embryos fathered by the exposed males showed an increased incidence of malformations. As evidence of its critical role in the testis, MTHFR is expressed at high levels in both prenatal and postnatal male germ cells (Garner et al., 2013) and loss of MTHFR is associated with DNA methylation defects in sperm and infertility (Kelly et al., 2005; Chan et al., 2010). Mutation in the gene for another enzyme necessary for the utilization of methyl groups from the folate cycle, methionine synthase reductase (MTRR), was associated with epigenetic instability and the inheritance of birth defects across a number of generations (Padmanabhan et al., 2013).
While both low and high folate can impact the epigenome of germ cells and cord blood, it is not clear if the epigenetic alterations will translate into effects on the offspring. Here, our aim was to determine the effects of lifetime folic acid deficient and supplemented diets in male mice to better understand the physiological impact of folate status on male germ cell development, intergenerational offspring outcomes, and global and imprinted gene DNA methylation profiles.
Materials and Methods
Mice and diets
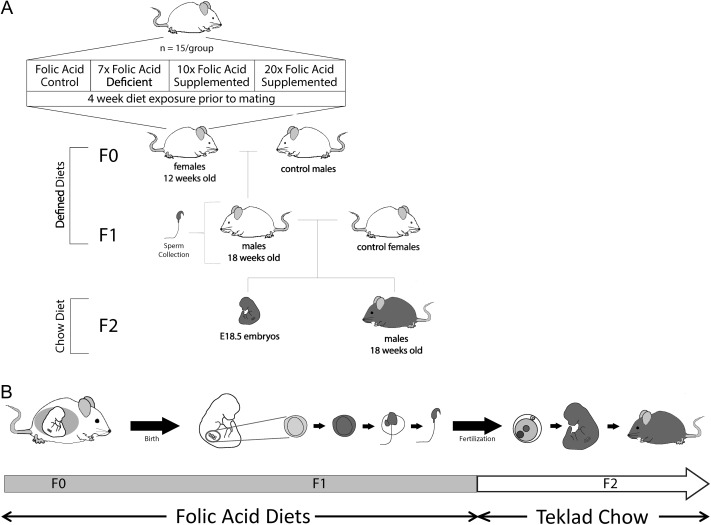
All procedures were carried out in accordance with the Canadian Council on Animal Care and the study was approved by the McGill University Animal Care Committee. Mice were housed at the Montreal Children's Hospital Research Institute's pathogen-free animal facility under a 12 Light: 12 Dark cycle in a temperature and humidity controlled environment with access to food and water ad libitum. The breeding and study scheme is outlined in Fig. 1. Eight-week old (F0) female BALB/c mice (n = 15/group) (Charles River, Canada) were fed one of four amino-acid defined diets (Harlan Teklad, USA) for 4 weeks prior to mating: folic acid control (Ctrl; 2 mg/kg diet) containing the recommended daily intake of folic acid for mice (Reeves et al., 1997), 7-fold folic acid deficient (7FD; 0.3 mg/kg diet), 10-fold folic acid supplemented (10FS; 20 mg/kg diet) or 20-fold FS (20FS; 40 mg/kg diet) diets. All diets contained 1% succinylsulfathiazole in order to prevent de novo synthesis of folate by intestinal bacteria. These diets have been used in several previous mouse studies of the developmental and epigenetic effects of folate deficiency and supplementation (e.g. Pickell et al., 2011; Mikael et al., 2013; Lambrot et al., 2013; Christensen et al., 2015). Following these 4 weeks, females were mated with sexually mature, 10-week-old (F0) male BALB/c mice that were fed regular mouse chow diets. Throughout mating, gestation and lactation, the females were fed their diets and the F1 male offspring were weaned at postnatal day (PND) 20 onto the same diets as their mothers and fed the diets until being killed at PND~200.
Figure 1.
Intergenerational reproductive effects of lifetime folic acid deficiency and supplementation. A) Mating scheme for intergenerational exposures to diet. Eight-week-old BALB/c F0 females were fed either a Ctrl, 7FD, 10FS or 20FS diet, (n = 15 for each) for 4 weeks prior to breeding with BALB/c males fed with regular rodent chow. Females were killed, and F1 male pups received the same experimental diet as their mother. At 18 weeks of age, one F1 male from each litter was mated with a female fed with rodent chow. Females were maintained on the rodent chow through pregnancy and lactation. From weaning until death, F2 male pups received rodent chow. B) Time line of exposure to folic acid defined diets. (Ctrl, folic acid control diet; 7FD, 7× folic acid deficient; 10FS, 10× FS; 20FS, 20× FS). FS, folic acid supplemented; FD, folate deficient.
At 18 weeks of age (approximately three rounds of spermatogenesis), 54 F1 males representative of 10 Ctrl, 11 FD, 10 10FS and 9 20FS different original F0 litters per diet group were mated with 10-week-old female BALB/c mice fed regular mouse chow. Throughout mating, gestation and lactation, the females were fed the regular 18% protein rodent chow diets (Harlan Teklad, USA). F2 male offspring were weaned to and maintained on the regular mouse chow diets.
Breeding studies
In order to assess the outcome in offspring of paternal lifetime folic acid exposures, F1 male BALB/c mice (n = 11–15 per diet group representing 8–10 F0 independent litters) were each mated with a second 10-week-old female BALB/c mouse. The presence of a vaginal plug on the morning after mating was designated 0.5 days post coitum (d.p.c.). Isolated pregnant females were fed regular mouse chow until 18.5 d.p.c., at which point they were killed. Ovaries were collected to determine the number of ovulation sites by counting corpora lutea (CL). Uterine horns were removed and opened to assess implantation by counting viable embryos and resorptions. Preimplantation loss was calculated for each female as the difference between the number of CL and implantation sites. The difference between the numbers of implantation sites and resorptions was used as a measure of post-implantation loss. Placentas, viable embryos and resorptions were removed and weighed. Embryos were sexed according to ano-genital distance (and later verified by PCR to ensure accuracy), their crown-rump length measured and evaluation of gross morphological abnormalities performed. Referring to ‘The Atlas of Mouse Development’ (Kaufman, 1992), embryos and late resorptions were examined for developmental delay and malformations such as cleft palate, closed eyelids, pointy nose, thick neck, curved tail, back and limb malformations. Embryos that deviated by two SDs from the means of the mean embryo weights, calculated per litter, across the Ctrl, were considered either growth restricted (two SDs below mean of means) or growth enhanced (two SDs above mean of means).
Sperm and tissue collection
F1 male mice (n = 17–20 per group, representative of 10 Ctrl, 11 7FD, 10 10FS and 9 20FS independent F0 litters) were killed upon generation of male F2 pups, at PND ~200 and weighed. Paired testes, epididymides and emptied seminal vesicles were removed and weighed. The right testis was frozen for testicular sperm counts. Mature spermatozoa from paired cauda epididymides were collected as previously described (Chan et al., 2012) and kept frozen at −80°C until use.
Sperm counts
Frozen testes (n = 6 per diet group) of F1 males in each diet group were used for hemocytometric testicular sperm counts as described by Robb et al. (1978), with modifications (Kelly et al., 2003). To prepare for counting, a weighed portion of the left testis was homogenized (Polytron, setting 5; Brinkmann Instruments Inc, Westbury, NY, USA) for 2 × 15-second periods, separated by a 30-second interval, in 5 ml of 0.9% NaCl, 0.1% thimerosal and 0.5% Triton X-100. Elongated spermatid nuclei with a shape characteristic of step 17–19 spermatids and resistant to homogenization were counted.
Plasma and red blood cell total folate
Whole blood (n = 5/group) from F0 females and F1 and F2 males was taken by cardiac puncture, maintained on ice and plasma separated and processed within an hour of sampling. Samples were shipped frozen to the Health Canada Nutrition Laboratory for analysis. The Lactobacillus casei microbiological assay was used to measure plasma and red blood cell (RBC) folate as previously described (Horne and Patterson, 1988). RBC folate content was normalized to total protein, which was determined using the modified Lowry assay (Bensadoun and Weinstein, 1976).
DNA methylation analyses
Frozen tissues were homogenized using a mortar and pestle. DNA was isolated using the DNeasy Blood and Tissue kit (Qiagen, Germany) as per the manufacturer's protocol from less than ~10 mg of homogenized frozen placenta and brain frontal cortex tissues. To control for the heterogeneity of placental and brain frontal cortex samples, whole tissues were homogenized by mortar and pestle to produce a uniform powder representative of the full tissue of each type for DNA isolation. Sperm DNA was extracted using the QIAmp DNA Microkit (Qiagen) with modifications. Sperm samples were incubated overnight at 56°C in sperm lysis buffer containing EDTA, Tris, dithiothreitol and proteinase K. Quantitative measurement of the DNA methylation levels on CpG dinucleotides was accomplished on isolated genomic DNA that was subjected to bisulfite treatment using the EpiTect Bisulfite kit (Qiagen, Germany). Imprinted germline differentially methylated domains were amplified using primers specific to pyrosequencing applications. Pyrosequencing was performed as previously described by Ronaghi et al., 1998. Amplified sequences were sequenced using the PyroMark Q24 kit (Qiagen) and the PyroMark Q24 Vacuum Workstation (Qiagen) using the manufacturer's protocol. Designed primers for assessment of H19, Imprinted Maternally Expressed Transcript (Non-Protein Coding)—H19, Small Nuclear Ribonucleoprotein Polypeptide N—Snrpn, KCNQ1 Opposite Strand/Antisense Transcript 1 (Non-Protein Coding)—Kcnq1ot1, Paternally Expressed Gene 1—Peg1 and Paternally Expressed Gene 3 Peg3 are listed in Supplementary Table S1.
Genome-wide DNA methylation was assessed using the luminometric methylation assay (LUMA) as previously described (Luttropp et al., 2015) with a few modifications using the PyroMark Q24. Briefly, duplicate digestion was carried out for both restriction enzymes HpaII and MspI (Thermo Fisher Scientific, USA). For each digestion, 500 ng of DNA diluted in 10 μl of DNase-free water was digested in 10 μl of enzyme restriction master mix [7 μl DNase-free water, 2 μl 10× Tango Buffer, 0.5 μl EcoRI (10 U/μl) and 0.5 μl restriction enzyme (HpaII or MspI) (10 U/ μl)]. Following the 4 h digestion at 37°C, samples were analyzed on a Pyrosequencer Q24 from Qiagen under the ‘AQ’ mode. To do so, 20 μl of Pyrosequencing Annealing Buffer was added to each digested DNA sample for a total of 40 μl, and 30 μl (375 ng of DNA) was loaded onto a Pyrosequencing plate. The nucleotides were not diluted and the Pyrosequencing reagents were added to the cartridge according to the volumes determined from the run preparation using the PyroMark Q24 software. Percentage methylation was calculated using the normalized peak ratios of HpaII over MspI as in the following formula: % Methylation = 100 [1-(HapII/EcoRI/MspI/EcoRI)].
Statistical analysis
Unless otherwise specified, results are expressed as the mean ± SEM. Data were graphed and analyzed with Prism 5 (GraphPad Software Incorporated, USA). Comparisons were made by Fisher's exact test, ANOVA followed by the Dunnett's multiple comparison test compared to control, or one-way ANOVA. Increased variance at individual CpGs was calculated by an F-test between deficient or supplemented versus control diet groups. Increased variance across an imprinted gene locus was calculated by ANOVA followed by the Dunnett's multiple comparison test on the mean of the all CpG variances across the imprinted locus, supplemented or deficient versus control. A level of significance for all analyses was set at P < 0.05.
Results
Serum and RBC folate concentrations
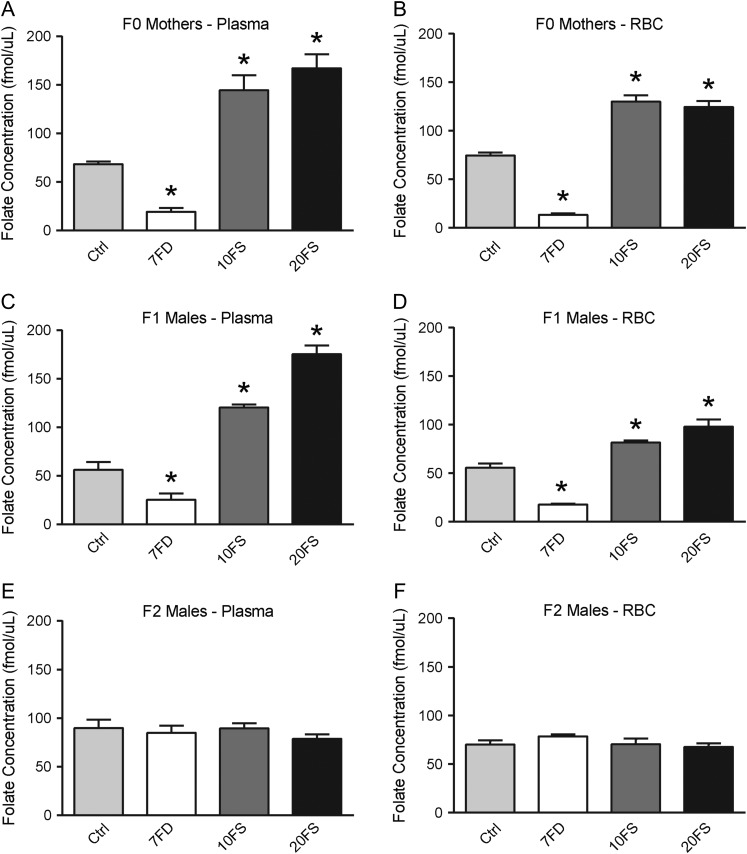
To determine effects of the different diets on folate status of the males in each of the study generations (Fig. 1), plasma and RBC total folate concentrations were evaluated in the F0 mothers (as a measure of in utero exposure of male pups), F1 and F2 adult males. Relative to the control diet (Ctrl), the deficient 7FD diet resulted in significantly lower RBC and plasma folate concentrations in the F0 mothers and F1 males (Fig. 2). In contrast, compared to the Ctrl F0 mothers and F1 males, both the 10FS and 20FS diets resulted in significantly higher RBC and plasma folate concentrations (Fig. 2A–D). All F2 males were fed regular chow based diets throughout their lives from in utero to postnatal stages of development (Fig. 1). No differences were found in the RBC or plasma folate concentrations of the F2 males, regardless of the paternal diet group (Fig. 2E and F). It should be noted that the plasma folate concentrations were higher in F2 chow-fed mice compared to Ctrl fed F1 male mice, which likely reflects the higher folate content of regular rodent chow compared to the defined diet.
Figure 2.
Plasma and RBC folate concentrations. Plasma and RBC folate concentrations in F0 dams (A–B), F1 male progeny (C–D) and F2 male progeny (E–F) when killed (n = 5). *P < 0.05 by one-way ANOVA with Dunnett's multiple comparisons test versus Ctrl. RBC, red blood cell.
Health and reproductive effects of lifetime exposure to folic acid deficient or supplemented diets
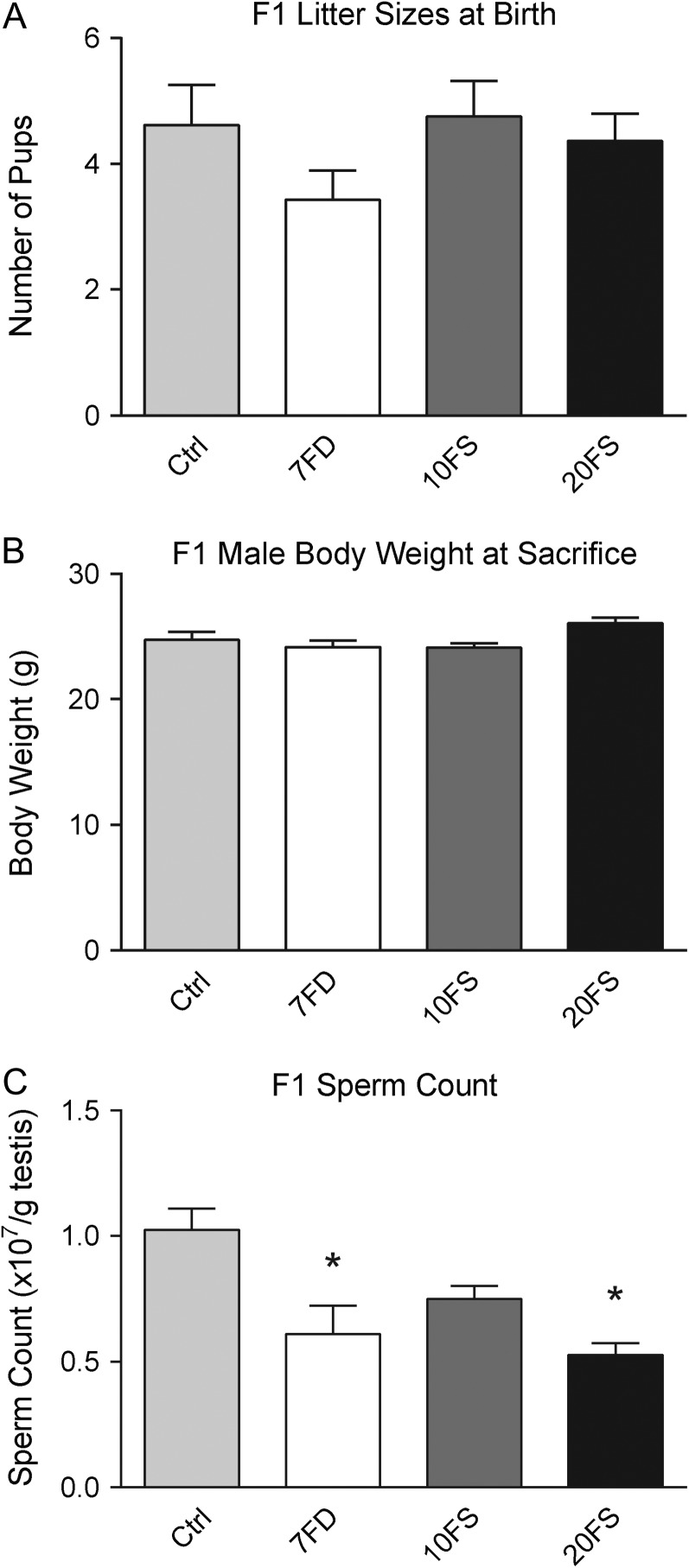
We next assessed whether in utero and postnatal exposures to folic acid deficiency and supplementation affected the general and reproductive health of the F1 males. F1 litter sizes at birth did not differ between folic acid control and supplementation diet groups (Fig. 3). Adult F1 male body weights at necropsy (18 weeks of age) were similar among the groups (Fig. 3B). In addition, there were no differences in reproductive organ weights (paired epididymides, paired testes and seminal vesicle weights) among the groups (Supplementary Fig. 1). As an indicator that germ cells were affected by the diets, testicular sperm counts were measured and found to be significantly lower in the 7FD and 20FS groups (Fig. 3C) compared to the control. Thus although the F1 males showed no signs of compromised general or reproductive health, the decreased sperm counts indicated adverse effects of the in utero and postnatal exposure to the 7FD and 20FS folic acid diets on developing germ cells.
Figure 3.
Lifetime folic acid deficiency and 20FS supplementation decrease sperm count. Effect of lifetime folic acid deficiency and supplementation on F1 litter sizes at birth (A; n = 11–15 litters/group), F1 adult male body weight (B; n = 17–20/group) and F1 sperm count (C; n = 6/group). *P < 0.05 by one-way ANOVA with Dunnett's multiple comparisons test.
Differential effects of paternal lifetime folic acid deficiency and supplementation on offspring (F2) outcomes
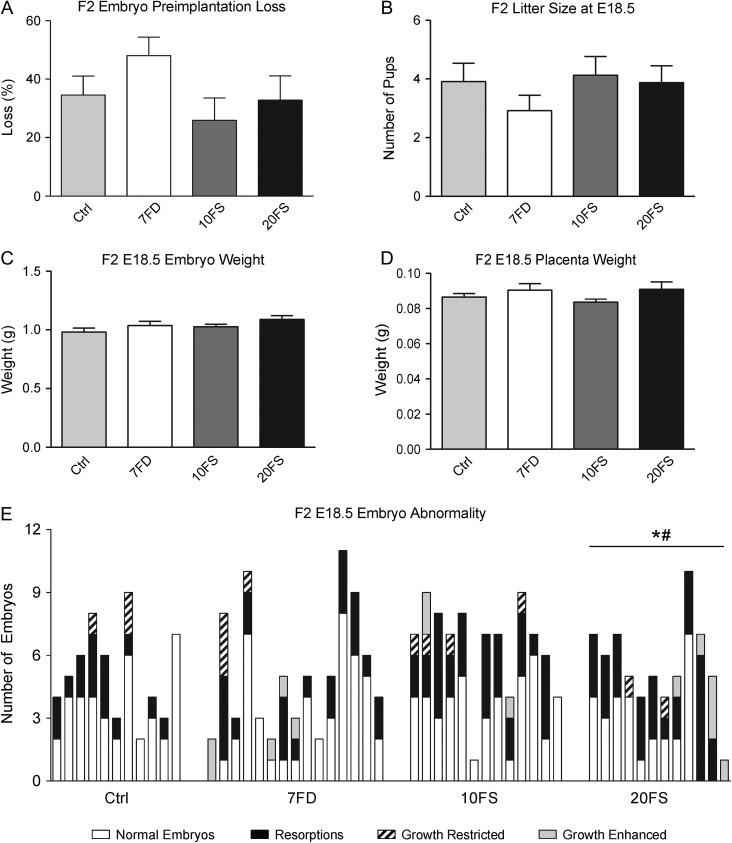
To determine whether the decreased sperm counts seen in F1 males resulted in compromised reproductive outcomes, lifetime diet-exposed F1 males were mated to chow-fed females and effects of the paternal diets on pregnancy outcomes determined at E18.5 for the F2 generation (Fig. 4). Preimplantation loss did not differ between the groups (Fig. 4A). At E18.5, mean F2 litter sizes were similar for the four groups (Fig. 4B). There were no differences among diet groups in embryo and placenta weights (Fig. 4C and D). F2 embryo outcomes for individual litters from at least 6–7 different F1 males per group are shown in Fig. 4E. The incidence of resorptions, as well as the incidence of embryos with any abnormality, including resorption, growth restriction or growth enhancement, were significantly increased in the litters sired by 20FS exposed F1 males (Fig. 4E). Congenital malformations such as craniofacial and limb abnormalities and cleft palate were not found in any of the groups. The E18.5 results gave a preliminary indication of adverse reproductive outcomes in the 20FS group.
Figure 4.
Effects of lifetime folic acid deficiency and supplementation on pregnancy outcomes at E18.5. Preimplantation loss of F2 at embryonic day (E)18.5 (A; n = 11–15 F2 litters, representing n = 7–9 original F1 litters), F2 litter sizes at E18.5 (B), F2 embryo weights at day 18.5 (C; n = 25–38 embryos, representing n = 7–9 original F1 litters), and F2 placental weights at embryonic day 18.5 (D). Incidence of fetal abnormalities at E18.5 per litter (E; n = 11–15 litters); growth restriction and enhancement are defined as a 2-fold SD difference of embryo weight to the group mean of litter mean weights. *P < 0.05 by one-way ANOVA with Dunnett's multiple comparisons test.
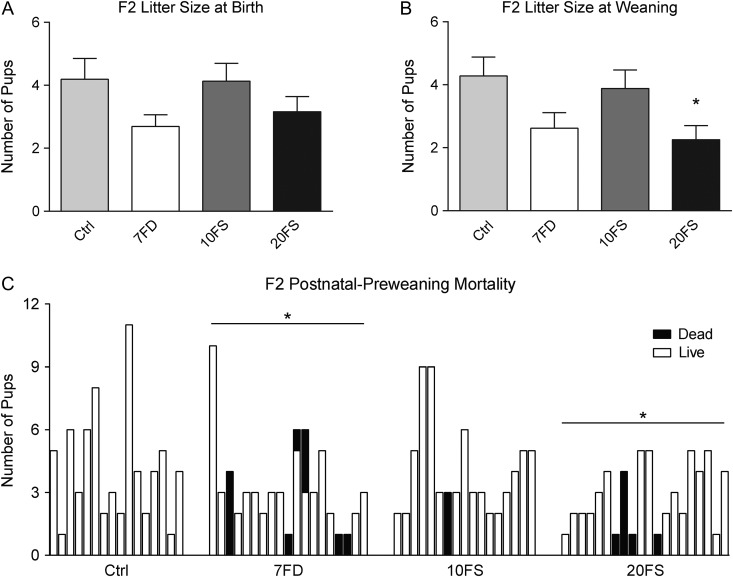
The F2 offspring was followed up in more detail by examining pups after birth in the early postnatal period (Fig. 5). Litter sizes did not differ significantly between the groups (Fig. 5A). Corresponding F2 litter sizes were re-assessed at the time of weaning on PND21. On PND21, a significantly lower litter size was observed in F2 litters from 20FS fathers (Fig. 5B); a trend but not significant change (P = 0.07) was seen for the 7FD group. To further examine the effects of paternal diet on the occurrence of pup mortality in early postnatal life, the proportion of pups that died prior to weaning was compared to those that survived to adulthood for each litter (Fig. 5C). While no evidence of postnatal mortality was found in litters sired by Ctrl males, significant increases in postnatal mortality were found in the 7FD and 20FS litters. For the F2 7FD litters, postnatal-preweaning death affected some or all pups in affected litters. Taken together, the results indicate that both paternal (F1) folic acid deficiency and the 20FS level of supplementation have intergenerational consequences, adversely affecting the F2 offspring.
Figure 5.
Lifetime folic acid deficiency and 20FS supplementation result in postnatal-preweaning mortality. Effect of lifetime folic acid deficiency and supplementation on F2 litter sizes at birth (A; n = 16–20 F2 litters, representing n = 9–11 original F1 litters), F2 litter sizes at weaning (PND21) (B), and incidence of pup postnatal mortality per litter (C). *P < 0.05 by one-way ANOVA with Dunnett's multiple comparisons test or by Fisher's exact test.
Effects of folic acid diets on sperm DNA methylation patterns
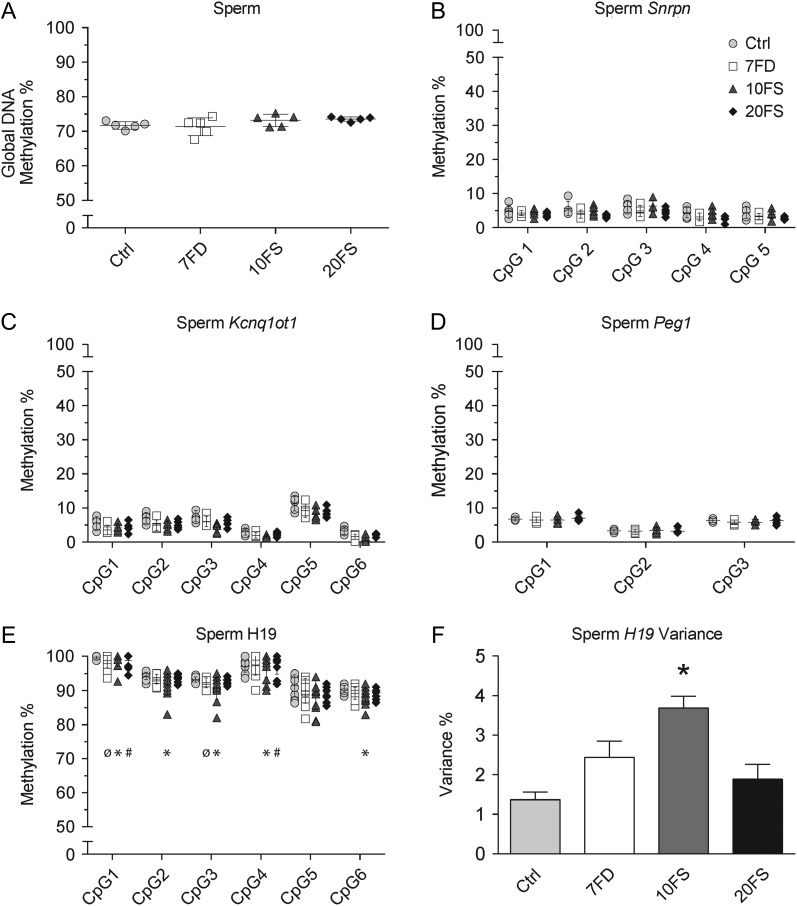
Based on the role of folic acid as a methyl donor and the proposed roles of epigenetic abnormalities in mediating intergenerational effects, DNA methylation was assessed in the sperm of the F1 males. Both whole genome (global methylation) and sequence-specific approaches were used. Sperm from F1 males from at least five original F0 litters per group were chosen for DNA methylation analysis. Using LUMA, DNA methylation levels were assessed at CCGG sites (~20% of all CpG sites) across the genome (Fig. 6). Sperm DNA methylation levels of 70–75% were similar for all four groups of F1 males (Fig. 6A). Next, imprinted genes were targeted since they possess distinct sperm-specific patterns that escape preimplantation reprogramming, are critical for normal fetal and placental development, and are affected in men with low sperm counts. The germline DMRs of maternally and paternally methylated imprinted genes are expected to have low (0–10%) and high (90–100%) levels of methylation, respectively, in sperm. Bisulfite pyrosequencing was used to examine the methylation of the DMRs of the three maternally methylated imprinted genes Snrpn, Kcnq1ot1 and Peg1 (Figs. 6B–D). All three maternally methylated imprinted loci possessed normal low levels of DNA methylation (1–11%) in sperm of the F1 males and were not affected by the diets. The paternally methylated imprinted gene H19 had high levels of DNA methylation (85–99%) in sperm in all F1 males, regardless of diet group (Fig. 6E). However, whereas sperm H19 DNA methylation values for the control Ctrl sperm clustered closely together, more variation in sperm DNA methylation was noted in some of the diet groups. Significant increases in variances at a few individual sites were observed in two groups, at CpGs 1 and 3 for sperm from 7FD F1 males and at CpGs 1 and 4 for sperm from the 20FS F1 males; in contrast, increases in variances were seen at all but CpG5 for sperm from 10FS F1 males. In particular, a significant increase in methylation variance across the H19 DMR locus was found across the six assayed CpGs following 10FS exposure in F1 males (Fig. 6F). Thus, subtle but potentially important effects of the diets on the paternally methylated imprinted gene H19 were found in the sperm of the F1 males.
Figure 6.
F1 sperm global DNA methylation and DMR methylation at imprinted genes uncovers variance of H19 methylation. Global DNA methylation was measured by LUMA (A). Imprinted gene methylation was quantified using bisulfite pyrosequencing at maternally methylated genes Snrpn (B; n = 5/group), Kcnq1ot1 (C; n = 5/group) and Peg1 (D; n = 5/group) and paternally methylated gene H19 (E; n = 10–13/group). H19 variance (F) was measured as a mean of variances of all six CpGs. For individual CpG variance (panel E), ø (7FD), * (10FS), # (20FS) = P < 0.05 by F-test between deficient or supplemented versus control diet groups. For across-locus variance analysis (panel F), *P < 0.05 by one-way ANOVA with Dunnett's multiple comparisons test. LUMA, luminometric methylation analysis; DMR, differentially methylated region.
Epigenetic effects in the F2 offspring
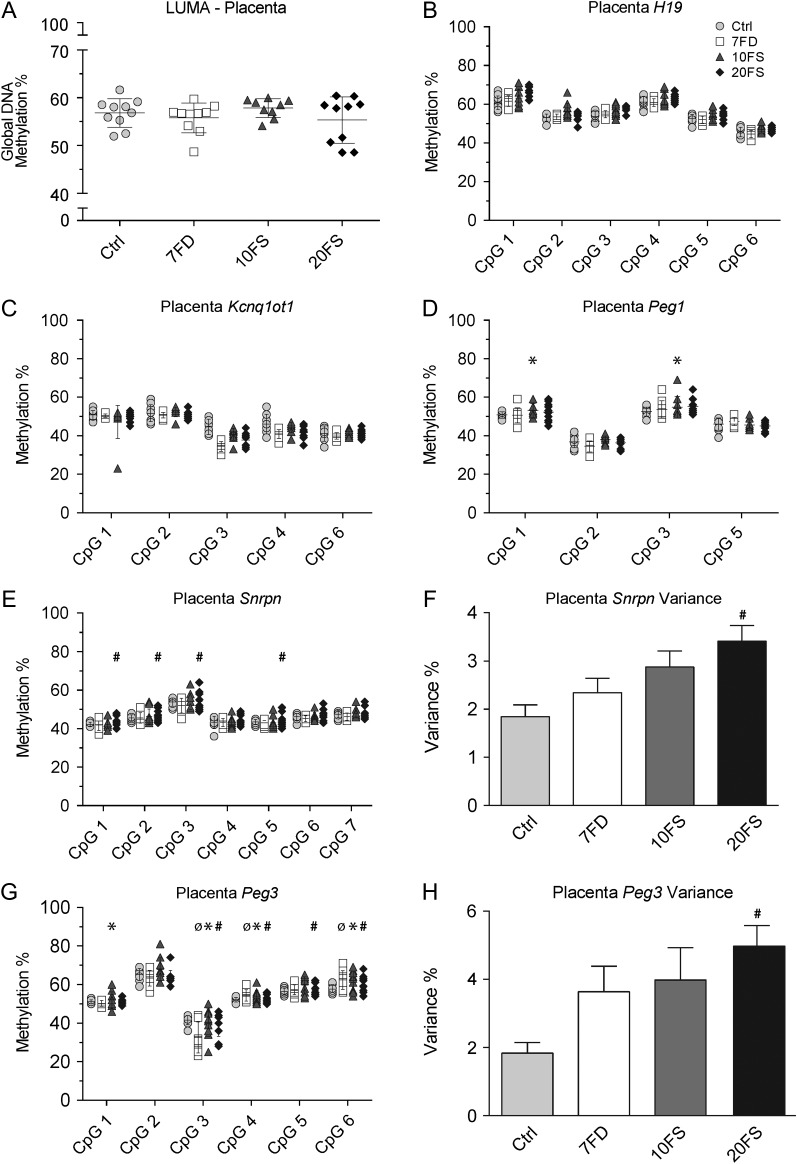
To determine if increased variance of DNA methylation in F1 sperm could escape epigenetic reprogramming during preimplantation development and be inherited in the offspring, global and imprinted gene DNA methylation were examined in F2 E18.5 placentas (Fig. 7) and embryonic tissue (Fig. 8). E18.5 F2 litters were chosen to represent offspring from F1 males of five different F0 females per group; DNA methylation levels were assessed for a male and female from each litter. Mean global methylation in placentas was lower than that for sperm, ranging from 55–57% (Fig. 7A). For the F2 placentas, no differences in global mean DNA methylation were observed between the groups (Fig. 7A). However, the 20FS group showed two distinct clusters, one within the expected range and one of lower global methylation; males and females were equally distributed between the two clusters. Interestingly, we observed that two of four embryos in the lower 20FS cluster were sired by F1 males that also sired litters with postnatal death; in contrast none of those in the upper cluster were sired by F1 males siring litters with postnatal deaths (Fig. 7A).
Figure 7.
F2 E18.5 placenta global DNA methylation and DMR methylation at imprinted genes uncovers variance of Snrpn and Peg3 methylation in F2 pups sired by 20FS exposed males. Global DNA methylation was measured using LUMA (A; n = 10/group). Imprinted gene methylation was quantified using bisulfite pyrosequencing at paternally methylated gene H19 (B) and maternally methylated genes Kcnq1ot1 (C), Peg1 (D), Snrpn (E) and Peg3 (G), (n = 5/group). Snrpn and Peg3 variances were measured as a mean of variances of all seven and six CpGs, respectively. For individual CpG variance (panels D, E and G), ø (7FD), * (10FS), # (20FS) = P < 0.05 by F-test between deficient or supplemented versus control diet groups. For across-locus variance analysis (panels F and H) *P < 0.05 by one-way ANOVA with Dunnett's multiple comparisons test.
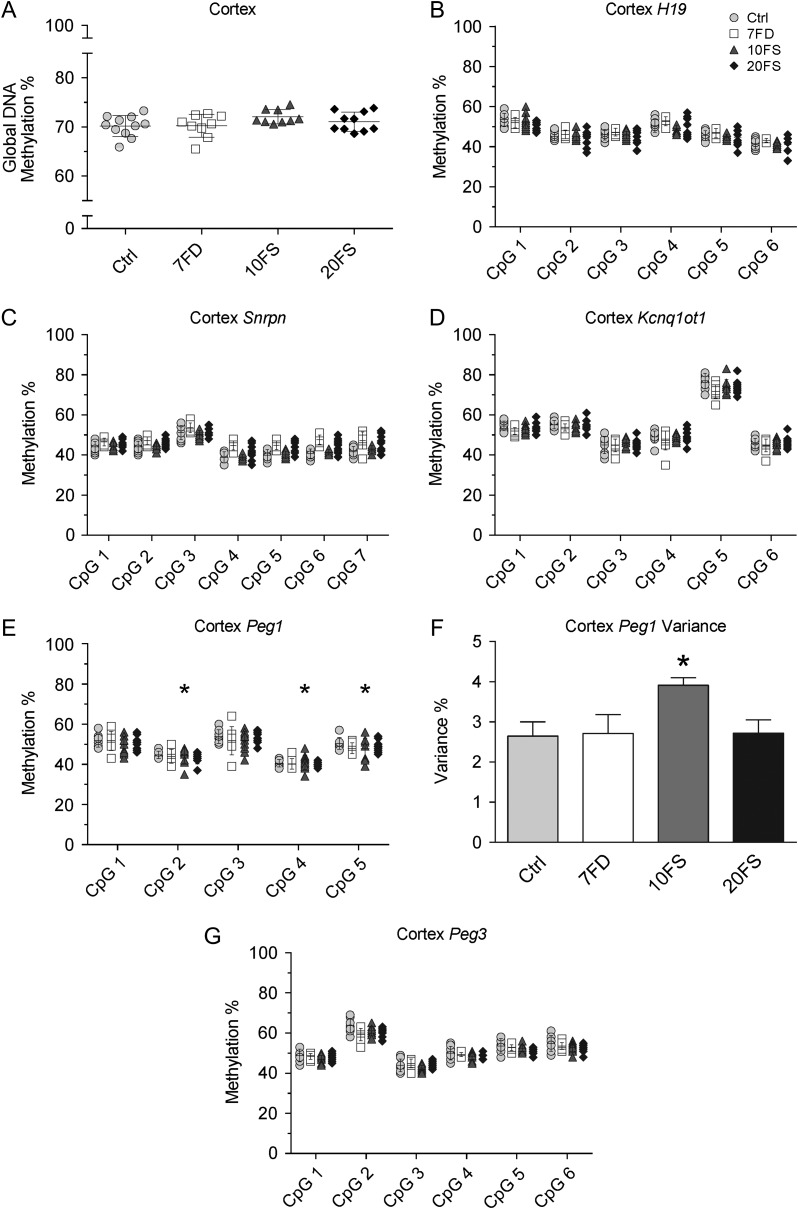
Figure 8.
F2 E18.5 cortex global DNA methylation and DMR methylation at imprinted genes uncovers variance of Peg1 methylation in F2 pups sired by 10FS exposed males. Global DNA methylation was measured using LUMA (A, n = 10/group). Loci of paternally methylated gene H19 (B) and maternally methylated genes Snrpn (C), Kcnq1ot1 (D), Peg1 (E) and Peg3 (G) methylation levels were quantified by pyrosequencing (n = 5/group). Peg1 variance was measured as a mean of variances of all five CpGs. For individual CpG variance (panel E), * (10FS) = P < 0.05 by F-test between deficient or supplemented versus control diet groups. For across-locus variance analysis (panels F) *P < 0.05 by one-way ANOVA with Dunnett's multiple comparisons test.
The possibility of aberrant DNA methylation inheritance from F1 males to F2 offspring was assessed at a gene-specific level in placentas by examining F2 imprinted gene methylation at the paternally methylated imprinted gene H19 and the maternally methylated imprinted genes Snrpn, Kcnq1ot1, Peg1 and Peg3. Methylation levels averaged ~50% for all imprinted genes as would be expected for a somatic or non-germ cell tissue. No differences between groups in overall mean placental DNA methylation were observed at any of the imprinted loci (Fig. 7B–E and G). Placental H19 methylation values were tightly clustered at most CpGs examined for all groups. In contrast, for the maternally methylated imprinted genes, there were significant increases in the variance of DNA methylation levels at individual CpGs across ≥50% of CpGs in a given locus for Peg1 (10FS group-CpGs 1 and 3), Snrpn (20FS group-CpGs 1, 2, 3 and 5) and Peg3 (7FD group-CpGs 3, 4 and 6; 10FS group-CpGs 1, 3, 4 and 6 and 20FS group-CpGs 3, 4, 5 and 6) in the placentas of F2 embryos (Fig. 7C–E, G). Additionally, there were significant increases in the variance of DNA methylation levels across the Snrpn and Peg3 loci in the placentas of F2 embryos from the 20FS group (Fig. 7 F and H).
Global and gene-specific DNA methylation were also assessed in the brains of the embryos corresponding to the placentas examined (Fig. 8). Brain tissue was chosen because it is a key embryonic tissue that depends on normal DNA methylation patterning for its development and was found to be susceptible to periconception exposures (e.g. Ecker et al., 2004; deWaal et al., 2015). Global DNA methylation levels in the cortex averaged 70–72% and did not differ significantly between the groups or show evidence of variance between embryos (Fig. 8A). Significant increases were seen in the variance of DNA methylation levels at individual CpGs across ≥50% of CpGs in a given locus for Peg1 (10FS group-CpGs 2, 4 and 5; Fig. 8E). No increases were found in the variance of DNA methylation levels at individual CpGs across the other imprinted genes H19, Snrpn, Kcnq1ot1 and Peg3 loci in the cortices of F2 embryos from the 7FD, 10FS and 20FS groups (Fig. 8B–D, G). While the H19, Snrpn, Kcnq1ot1 and Peg3 imprinted genes showed normal ~50% DNA methylation levels and little evidence of variance across the entire locus between embryos in all groups, a significant increase in DNA methylation variance across the Peg1 locus was observed for the 10FS group compared to the Ctrl group (Fig. 8F).
Taken together, there was evidence of inter-individual DNA methylation variation in the F2 offspring, most notable in the placentas, associated with paternal folic acid deficiency and supplementation.
Paternal lifetime exposure to folic acid deficient or supplemented diets: effects on surviving male offspring health
To determine if paternal lifetime folic acid deficiency or supplementation could affect postnatal development of F2 pups that survived post-weaning, we investigated the body weights, reproductive organ weights and sperm counts of adult males. Adult F2 male body weights (Supplementary Fig. 2A) at necropsy (~20 weeks of age) and male reproductive organ weights (paired testes, paired epididymides and seminal vesicles, Supplementary Figs. 2B–D) were similar for all the groups. Furthermore, unlike in the F1 generation, testicular sperm counts of the F2 males did not differ between the groups. Thus, adult F2 males did not show signs of male reproductive abnormalities persisting beyond the F1 generation.
Discussion
Our data indicate that a lifetime exposure of male mice to both folic acid deficient and highly supplemented diets result in decreased sperm counts, adverse outcomes in his offspring and evidence of epigenetic alterations (see model in Fig. 9). While folic acid deficient diets have previously been linked to paternally mediated intergenerational effects in a different strain of mouse, C57BL/6 (Lambrot et al., 2013) rather than the BALB/c strain used in the current study, it was unexpected that a FS diet could produce similar outcomes. This is potentially concerning since, although the treatment duration was longer than that clinically used, the doses of folic acid supplements used in the current study,(10- and 20-fold the daily recommended intake of folic acid for mice) are within the range used clinically in high-risk pregnancies and for the treatment of male subfertility. Male germ cells undergo extensive epigenomic reprogramming, including that of DNA methylation, during their development from primordial germ cells in the fetal gonad through postnatal spermatogenesis, including mitotic, meiotic and postmeiotic phases. Folic acid is an important source of methyl groups needed for DNA methylation and thus decreases and increases in folic acid intake have the potential to impact DNA methylation programs. Our study was designed to expose male germ cells to low-, high- and very high-folate levels throughout male germ cell development.
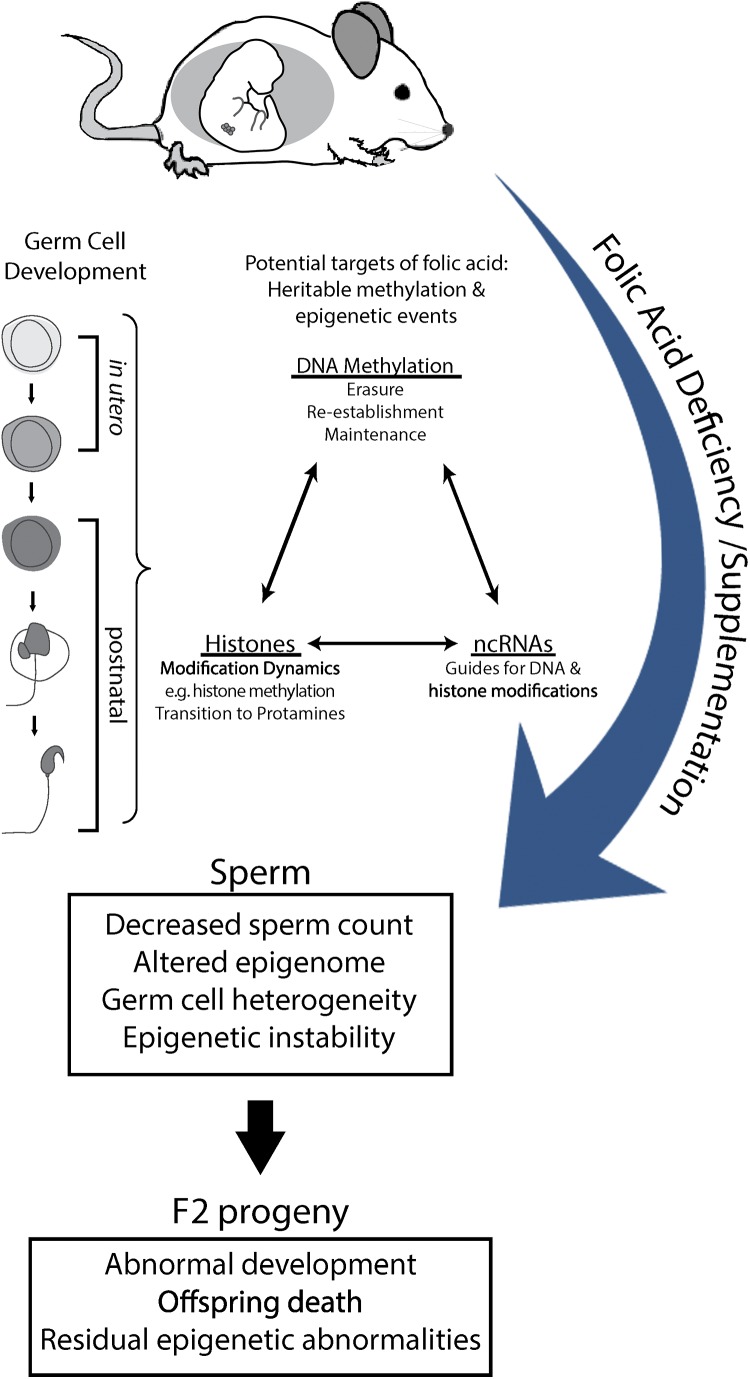
Figure 9.
Model of postulated epigenetic mechanisms and interactions underlying the adverse outcomes in offspring of males exposed to FD and FS diets, starting from fertilization. Lifetime exposure to both FD and FS spans two vulnerable windows of epigenetic reprogramming during germ cell development: in utero and postnatal. Here, in each window of development, both window-specific (e.g. DNA methylation erasure and histone-protamine transition) and development-long (e.g. DNA methylation re-establishment/maintenance and histone modification dynamics) events are at risk of being influenced by external exposures of folic acid. Interactions between DNA methylation, histone modifications and ncRNA expression all have the potential to contribute to the decreased reproductive fitness of sperm, observed as decreased sperm count, altered epigenome (DNA methylation variance), germ cell heterogeneity and epigenetic instability. Ultimately, the cumulative and interacting epigenetic effects of lifetime exposures on the sperm manifest in the F2 progeny as abnormal development, increased offspring death and residual epigenetic abnormalities. non-coding RNA, ncRNA.
Plasma and RBC folate concentrations reflected the folic acid content of the experimental diets consumed by the mice. For example, F0 dams and F1 sires that consumed a folic acid deficient diet had lower plasma and RBC folate concentrations, whereas F0 and F1 mice consuming the supplemented diets had higher plasma and RBC folate concentrations. All F2 males fed the chow diet had a similar folate status regardless of parental or grand-maternal diet. The effects of the defined diets on plasma folate concentrations are in keeping with previous studies utilizing the same diets, with the 7FD diet resulting in a near 4-fold decrease and the 10FS and 20FS diets leading to ~2-fold increases in plasma folate concentrations (Li et al., 2006; Pickell et al., 2011; Mikael et al., 2013). The relationship between folic acid intake and RBC folate concentration is nonlinear suggesting a threshold for folate accumulation in tissues. In relation to our study, the germ cells of the F1 males were exposed to higher or lower folate both in utero (F0) and postnatally during periods in which they were developing from primordial germ cells to mature spermatozoa and reprogramming their DNA methylation levels. Since F2 males were not exposed directly to the diets, it can be proposed that differences in their placenta and tissue methylation profiles are due to exposure of the F1 germ cells to the different diets.
The lifetime folic acid diets did not appear to affect the general health of the F1 males, as indicated by a lack of difference in body and reproductive organ weights. However, we observed a decrease in sperm counts in the testes of F1 males fed the 7FD and 20FS diets. This is similar to findings by Swayne et al. (2012) in which male BALB/c mice fed a folic acid deficient diet for 15 weeks during the post-weaning period resulted in a ~40% reduction in cauda sperm number. In contrast, Lambrot et al. (2013) did not find a decrease in sperm counts in C57BL/6 male mice exposed over a lifetime to the same 7FD diet we used. The discrepancy may be due to the different mouse strains, BALB/c versus C57BL/6, used in the various studies. For instance, testes of BALB/c mice appear to be more susceptible to perturbations in the folate metabolic pathway (Chan et al., 2010). The folate dependent enzyme MTHFR is found in particularly high levels in the prenatal and postnatal testis (Chen et al., 2001; Garner et al., 2013). The MTHFR deficient BALB/c strain of mice have very few germ cells in their testes and are infertile in contrast to the MTHFR deficient C57BL/6 strain mice that have decreased sperm counts (50% of normal) and are fertile (Kelly et al., 2005; Chan et al., 2010). A previous study characterized differences in folate metabolizing and DNA repair enzymes between the two strains and reported that BALB/c mice are more prone than C57BL/6 mice to accumulating DNA damaged cells (resulting in cell death) when one-carbon donors are low (Knock et al., 2011). Folic acid is required for nucleotide synthesis and biological methylation reactions through the folate metabolic pathway. The folic acid deficient diet could affect either of these pathways leading to the decreased sperm counts. These findings are consistent with observations in humans where low serum folate has been correlated with lower sperm counts (De Sanctis et al., 2011)
The decrease in F1 sperm count associated with the 20FS diet, which was similar to that observed in the 7FD group, was unexpected and its basis may provide clues to the adverse pregnancy outcomes in the offspring of both groups of males. A few lines of evidence indicate that excess folic acid results in functional folate deficiency. Down regulation of folate metabolic pathway enzymes may be related to the presence of circulating UFA in individuals taking high dose folic acid supplements (Kelly et al., 1997; Bailey et al., 2010). UFA in turn results in the accumulation of cellular dihydrofolate, an inhibitor of folate metabolism pathway enzymes such as MTHFR (Matthews and Daubner, 1982). In in vitro studies, excess folic acid inhibits MTHFR activity in brain extracts (Hollinger et al., 1982). In addition, in mouse studies excess dietary folic acid has recently been shown to result in decreased MTHFR protein levels as well as production of a less active form of MTHFR in liver (Christensen et al.,. 2015). Together, our data suggest that both folic acid deficiency and high dose folic acid supplements may impact male germ cells in a similar manner either by directly (7FD) or indirectly (e.g. MTHFR inhibition) reducing the provision of methyl groups from folic acid.
In addition to the reduction in sperm number, we also have evidence of adverse outcomes in the offspring (F2) of the F1 males exposed to the folic acid deficient and supplemented diets. One of the most striking findings of this study was an increase in postnatal-preweaning mortality in F2 litters from F1 males fed the 7FD and 20FS. To the best of our knowledge, this is the first study to examine the intergenerational implications of paternal lifetime exposure to both folic acid deficiency and folic acid supplementation in the postnatal period of F2 offspring. The increased mortality in litters (F2 generation) from FD exposed fathers could be a result of aberrant development, similar to findings by Lambrot et al. (2013) in which embryos demonstrated developmental delay and abnormalities. Unfortunately, since the pups died at different times and were in most cases not recovered before the mothers cannibalized them, we were unable to perform autopsies to determine the cause of death. Interestingly, another study on male-mediated effects (sperm based) of drugs used to treat testicular cancer also reported postnatal mortality of unknown cause in the offspring of the exposed males (Bieber et al., 2006). The increased mortality in the postnatal window in the current study suggests that paternal lifetime low- or high-folic acid exposures affected male germ cells and that the effects were transmitted to offspring that survived beyond the embryonic period.
As DNA methylation patterns are erased and reset in the developing male germ cells, we postulated that altered methyl donor supply caused by the folic acid diets would impact epigenetic programming in sperm. Global DNA methylation levels measured using the LUMA assay provided an initial assessment of DNA methylation levels at ~20 million sites across the mouse genome. In previous human studies, we have shown decreases in overall sperm DNA methylation associated with folic acid supplement consumption and MTHFR deficiency (Aarabi et al., 2015). In the current study, sperm global DNA methylation levels were unaffected by the diets, giving a preliminary indication that there were no major global alterations in male germ cell DNA methylation reprogramming. However, as the bulk of DNA methylation takes place in repeat sequences, more subtle effects at developmentally important sequences would be missed with a global assay. We were particularly interested in imprinted genes, since germ cell processing of these sequences is unique. In both male and female primordial germ cells methylation is erased on all imprinted sequences. In the male germline, DNA methylation is re-established on the DMRs of paternally methylated genes, such as H19, while maternally methylated genes, such as Snrpn, remain unmethylated. Lower or higher intakes of folate could alter the availability of methyl co-factors and thus potentially interfere with the erasure or re-establishment of methylation on imprinted genes. In a mouse study of the effects of gestational folic acid supplements on fetal (F1) brain, alteration in imprinted gene methylation was reported, suggesting that germ cells might also be susceptible (Barua et al., 2014).
Overall, mean imprinted gene methylation levels were unaffected in sperm. Other studies have similarly reported that mean methylation of imprinted genes in spermatozoa is unaffected by lifetime folic acid deficiency in mice or postnatal folic acid supplementation in human studies (Lambrot et al., 2013; Aarabi et al., 2015). However, by examining only overall mean imprinted gene methylation on millions of sperm, inter-individual differences may be missed. An initial indication of perturbed epigenetic marks associated with the diet was seen in the F1 sperm at the paternally methylated imprinted gene H19. We observed a significant increased variance of DNA methylation levels at five out of six assayed individual CpGs within the H19 locus, as well as across the locus, between individual animals in the 10FS group; individual H19 CpGs were also affected in the 7FD and 20FS groups. Interestingly, the most marked sperm H19 variability was found in the 10FS group when it might have been expected in the 7FD and 20FS groups. It is possible that, as reflected by the decrease in sperm counts, the sperm epigenome in the 7FD and 20FS groups was more severely affected than that in the 10FS group. In a previous study on enriched populations of mouse spermatogonial stem cells in vitro, a similar variability in male germ cell DNA methylation, in response to Mthfr haploinsufficiency and varying concentrations of methyl groups, was found (Garner et al., 2013). It is possible that such inter-individual variability in germ cell DNA methylation is a common response to folate metabolic pathway and methyl donor perturbations or that it is secondary to effects on other epigenetic modulators, such as histone methylation, which undergoes significant remodeling during male germ cell development (Fig. 9). The results in the current study suggest the possibility that DNA methylation alterations may contribute to the observed folate diet related intergenerational effects.
Another way to examine the significance of epigenetic abnormalities residing in sperm is to examine the resulting offspring for epigenetic defects. We sampled both the extra-embryonic and embryonic tissues. The placenta is particularly useful for assessing epigenetic effects of peri-conceptional exposures including germ cell exposures, as it typically shows higher levels of epigenetic perturbation than the corresponding embryo; this is thought to be due to the retention or tolerance of epigenetically abnormal cells in the placenta but not in the embryo (Rivera et al., 2008; Fortier et al., 2008). Thus the placenta is thought to provide a more sensitive indicator of epigenetic perturbations than the embryo. Increases in the variance of both individual CpGs as well as pan-locus imprinted gene methylation were noted for maternally methylated imprinted genes but not for the paternally methylated gene H19 in the E18.5 day placentas of all the F2 diet groups. Alterations in variance but not in mean DNA methylation of imprinted genes suggests that a subset of F1 aberrant gametes, not detected in whole sperm studies (Fig. 6), from FD and FS exposed males, develop into functional spermatozoa and can result in viable embryos that survive at least until the end of gestation. The observed increased placental DNA methylation variability for some but not all imprinted genes examined could also reflect underlying epigenetic instability induced by the diets and perhaps based on an epigenetic modification other than DNA methylation; this possibility is supported by the fact that DNA methylation of maternally methylated imprinted genes was not affected in the F1 sperm.
The paternal diets resulted in less evidence of epigenetic effects on global DNA and imprinted gene methylation in the brain cortex as compared to the placenta. While the placenta is thought to provide a more sensitive indicator of epigenetic perturbations, somatic tissues remain equally important to investigate as they may result in abnormal tissue structure or function in the offspring. Owing to its complexity, the brain has been postulated to be particularly sensitive to germ cell or early embryo perturbations in DNA methylation patterns. Previous studies have shown that gestational folic acid supplementation can result in altered gene expression and DNA methylation in the cerebellum of the offspring (Barua et al., 2014, 2016); these studies employed genome-wide approaches to DNA methylation profiling. In the current study, there was little evidence of altered global or imprinted gene methylation in the cortex with the exception of effects on Peg1 in F2 brain cortex from the 10FS group. It is unclear why the 10FS group was the only one affected, although as discussed above for the sperm H19 methylation results, it is possible that affected sperm are more viable in the 10FS groups as compared to the two other diet groups. It is clear that much more detailed profiling will be required in our model to determine the extent and type of epigenetic effects in brain and other somatic tissues of the F2 offspring.
Lambrot et al. (2013) also identified aberrant DNA methylation in precursors of spermatozoa associated with the administration of folic acid deficient diets. However, these differentially methylated loci were no longer present is mature spermatozoa, possibly through mechanisms of correction or elimination of affected gametes by apoptosis. Instead, the authors noted abnormal patterns of chromatin marks persisting in mature spermatozoa. Together, results from the Lambrot and the current study suggest that lifetime folic acid deficiency and folic acid supplementation may not only transiently affect DNA methylation of sperm but also affect other epigenetic marks such as chromatin modifications. Subsequently, the inheritance of an altered epigenome or perhaps epigenetic instability may result in abnormal development in the offspring leading to increased morphological abnormalities and/or death (see model in Fig. 9).
Our findings do not support a simple model of diet-induced altered DNA methylation as the sole basis for the prenatal and postnatal F2 progeny death. We thus propose a more complex model (Fig. 9) on which to base further studies. The decreased F1 sperm counts demonstrate that reproductive fitness has been affected and is possibly impaired. The observed postnatal death in the F2 generation suggests that exposed F1 male germ cells are functional but abnormal. Finally, increased imprinted gene DNA methylation variance in F1 sperm and in F2 somatic tissues hints that there is underlying epigenetic dysregulation or instability as a result of the paternal diets. The mild variability in imprinted gene methylation is unlikely to be the basis of the F2 offspring death. DNA methylation alterations may still play a role if genome-wide effects, not examined here, are present at susceptible regions such as regulatory elements of developmental genes. It is equally likely that other epigenetic modifications, such as histone methylation, are affected by the diets. We suggest that the folate diets may affect more than one epigenetic mechanism in exposed developing male germs and result in epigenetic instability. In the Padmanabhan et al. (2013) study, a mutation in the folate metabolism gene Mtrr in the first generation caused congenital defects in the mutant progeny as well as in wild-type descendants for up to four generations; it is postulated that the Mtrr mutation resulted in epigenetic instability, however, the underlying germline inherited epimutations have not been identified.
It is clear that further studies will be required to understand the basis of, and more fully explore the mechanisms underlying, the adverse effects of folic acid deficiency supplementation on developing male germ cells and the offspring sired by the exposed males. Our results demonstrate that the epigenetic phenotypes can be highly variable and underline the fact that numerous offspring will need to be examined to better understand the spectrum of epigenetic defects induced in developing male germ cells by folate diets. Genome-wide DNA methylome studies, histone modification profiles, as well as gene expression studies to get at functional outcomes will be required. It will also be important to separate out effects of prenatal (gestational) and postnatal exposures to male germ cells, as is being done for somatic exposures (e.g. Sie et al., 2013). It is possible that gestational exposures, when the major phase of epigenetic reprogramming of male germ cells takes place, may represent a particularly susceptible window for the induction of DNA methylation defects in male germ cells.
Supplementary data
Supplementary data are available at Molecular Human Reproduction online.
Supplementary Material
Acknowledgements
We thank Dr Marisa Bartolomei for providing primer information before publication along with advice on the LUMA and pyrosequencing assays. J.M.T. is a James McGill Professor. The Research Institute, McGill University Health Centre is supported in part by the Fonds de la recherche du Québec-santé (FRQ-S).
Authors’ roles
L.L. performed the majority of experiments and took the lead in manuscript preparation, D.C. aided in study design and data analysis, M.A. aided in study design and pyrosequencing, M.L. performed LUMA assays and aided in pyrosequencing, N.A.B. performed RBC and plasma folate measurements assays, A.M. aided in folate measurement assays, and manuscript editing, J.T. oversaw completion of the study, was responsible for the study design, and finalized the manuscript.
Funding
The Canadian Institutes of Health Research (CIHR #89944 to J.M.T). L.L. is a recipient of studentships from the Montreal Children's Hospital-RI-MUHC and FRQ-S.
Conflict of interest
None declared.
References
- Aarabi M, San Gabriel MC, Chan D, Behan NA, Caron M, Pastinen T, Bourque G, MacFarlane AJ, Zini A, Trasler J. High-dose folic acid supplementation alters the human sperm methylome and is influenced by the MTHFR C677T polymorphism. Hum Mol Genet 2015;24:6301–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey RL, Mills JL, Yetley EA, Gahche JJ, Pfeiffer CM, Dwyer JT, Dodd KW, Sempos CT, Betz JM, Picciano MF. Unmetabolized serum folic acid and its relation to folic acid intake from diet and supplements in a nationally representative sample of adults aged > or =60 y in the United States. Am J Clin Nutr 2010;92:383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SW, Ayling JE. The extremely slow and variable activity of dihydrofolate reductase in human liver and its implications for high folic acid intake. Proc Natl Acad Sci USA 2009;106:15424–15429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua S, Kuizon S, Brown WT, Junaid MA. DNA methylation profiling at single-base resolution reveals gestational folic acid supplementation influences the epigenome of mouse offspring cerebellum. Front Neurosci 2016;10:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua S, Kuizon S, Chadman KK, Flory MJ, Brown WT, Junaid MA. Single-base resolution of mouse offspring brain methylome reveals epigenome modifications caused by gestational folic acid. Epigenetics Chromatin 2014;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensadoun A, Weinstein D. Assay of proteins in the presence of interfering materials. Anal Biochem 1976;70:241–250. [DOI] [PubMed] [Google Scholar]
- Bentivoglio G, Melica F, Cristoforoni P. Folinic acid in the treatment of human male infertility. Fertil Steril 1993;60:698–701. [DOI] [PubMed] [Google Scholar]
- Bieber AM, Marcon L, Hales BF, Robaire B. Effects of chemotherapeutic agents for testicular cancer on the male rat reproductive system, spermatozoa, and fertility. J Androl 2006;27:189–200. [DOI] [PubMed] [Google Scholar]
- Bortolus R, Blom F, Filippini F, van Poppel MN, Leoncini E, de Smit DJ, Benetollo PP, Cornel MC, de Walle HE, Mastroiacovo P. Prevention of congenital malformations and other adverse pregnancy outcomes with 4.0 mg of folic acid: community-based randomized clinical trial in Italy and the Netherlands. BMC Pregnancy Childbirth 2014;14:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D, Cushnie DW, Neaga OR, Lawrance AK, Rozen R, Trasler JM. Strain-specific defects in testicular development and sperm epigenetic patterns in 5,10-methylenetetrahydrofolate reductase-deficient mice. Endocrinology 2010;151:3363–3373. [DOI] [PubMed] [Google Scholar]
- Chan D, Delbes G, Landry M, Robaire B, Trasler JM. Epigenetic alterations in sperm DNA associated with testicular cancer treatment. Toxicol Sci 2012;125:532–543. [DOI] [PubMed] [Google Scholar]
- Chen Z, Karaplis AC, Ackerman SL, Pogribny IP, Melnyk S, Lussier-Cacan S, Chen MF, Pai A, John SW, Smith RS et al. Mice deficient in methylenetetrahydrofolate reductase exhibit hyperhomocysteinemia and decreased methylation capacity, with neuropathology and aortic lipid deposition. Hum Mol Genet 2001;10:433–443. [DOI] [PubMed] [Google Scholar]
- Christensen KE, Mikael LG, Leung KY, Levesque N, Deng L, Wu Q, Malysheva OV, Best A, Caudill MA, Greene ND et al. High folic acid consumption leads to pseudo-MTHFR deficiency, altered lipid metabolism, and liver injury in mice. Am J Clin Nutr 2015;101:646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeizel AE, Dudas I. Prevention of the first occurrence of neural-tube defects by periconceptional vitamin supplementation. N Eng J Med 1992;327:1832–1835. [DOI] [PubMed] [Google Scholar]
- Czeizel AE, Dudas I, Paput L, Banhidy F. Prevention of neural-tube defects with periconceptional folic acid, methylfolate, or multivitamins. Ann Nutr Metab 2011;58:263–271. [DOI] [PubMed] [Google Scholar]
- De Sanctis V, Candini G, Giovannini M, Raiola G, Katz M. Abnormal seminal parameters in patients with thalassemia intermedia and low serum folate levels. Pediatr Endocrinol Rev 2011;8(Suppl 2):310–313. [PubMed] [Google Scholar]
- de Waal E, Vrooman LA, Fischer E, Ord T, Mainigi MA, Coutifaris C, Schultz RM, Bartolomei MS. The cumulative effect of assisted reproduction procedures on placental development and epigenetic perturbations in a mouse model. Hum Mol Genet 2015;24:6975–6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-Regil LM, Fernandez-Gaxiola AC, Dowswell T, Pena-Rosas JP. Effects and safety of periconceptional folate supplementation for preventing birth defects. Cochrane Database Syst Rev 2010: CD007950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebisch IM, Thomas CM, Peters WH, Braat DD, Steegers-Theunissen RP. The importance of folate, zinc and antioxidants in the pathogenesis and prevention of subfertility. Hum Reprod Update 2007;13:163–174. [DOI] [PubMed] [Google Scholar]
- Ecker DJ, Stein P, Xu Z, Williams CJ, Kopf GS, Bilker WB, Abel T, Schultz RM. Long-term effects of culture of preimplantation mouse embryos on behavior. Proc Natl Acad Sci USA 2004;101:1595–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forges T, Monnier-Barbarino P, Alberto JM, Gueant-Rodriguez RM, Daval JL, Gueant JL. Impact of folate and homocysteine metabolism on human reproductive health. Hum Reprod Update 2007;13:225–238. [DOI] [PubMed] [Google Scholar]
- Fortier AL, Lopes FL, Darricarrere N, Martel J, Trasler JM. Superovulation alters the expression of imprinted genes in the midgestation mouse placenta. Hum Mol Genet 2008;17:1653–1665. [DOI] [PubMed] [Google Scholar]
- Garner JL, Niles KM, McGraw S, Yeh JR, Cushnie DW, Hermo L, Nagano MC, Trasler JM. Stability of DNA methylation patterns in mouse spermatogonia under conditions of MTHFR deficiency and methionine supplementation. Biol Reprod 2013;89:125. [DOI] [PubMed] [Google Scholar]
- Gong M, Dong W, He T, Shi Z, Huang G, Ren R, Huang S, Qiu S, Yuan R. MTHFR 677 C>T polymorphism increases the male infertility risk: a meta-analysis involving 26 studies. PLoS One 2015;10:e0121147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon N. Folate metabolism and neural tube defects. Brain Dev 1995;17:307–311. [DOI] [PubMed] [Google Scholar]
- Hollinger JL, Hommes OR, van de Wiel TJ, Kok JC, Jansen MJ. In vitro studies of 5, 10-methylenetetrahydrofolate reductase: inhibition by folate derivatives, folate antagonists, and monoamine derivatives. J Neurochem 1982;38:638–642. [DOI] [PubMed] [Google Scholar]
- Horne DW, Patterson D. Lactobacillus casei microbiological assay of folic acid derivatives in 96-well microtiter plates. Clin Chem 1988;34:2357–2359. [PubMed] [Google Scholar]
- Joubert BR, den Dekker HT, Felix JF, Bohlin J, Ligthart S, Beckett E, Tiemeier H, van Meurs JB, Uitterlinden AG, Hofman A et al. Maternal plasma folate impacts differential DNA methylation in an epigenome-wide meta-analysis of newborns. Nat Commun 2016;7:10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman MH. The Atlas of Mouse Development. San Diego, CA: Academic Press, INC, 1992:337–370. [Google Scholar]
- Kelly P, McPartlin J, Goggins M, Weir DG, Scott JM. Unmetabolized folic acid in serum: acute studies in subjects consuming fortified food and supplements. Am J Clin Nutr 1997;65:1790–1795. [DOI] [PubMed] [Google Scholar]
- Kelly TL, Li E, Trasler JM. 5-aza-2’-deoxycytidine induces alterations in murine spermatogenesis and pregnancy outcome. J Androl 2003;24:822–830. [DOI] [PubMed] [Google Scholar]
- Kelly TL, Neaga OR, Schwahn BC, Rozen R, Trasler JM. Infertility in 5,10-methylenetetrahydrofolate reductase (MTHFR)-deficient male mice is partially alleviated by lifetime dietary betaine supplementation. Biol Reprod 2005;72:667–677. [DOI] [PubMed] [Google Scholar]
- Knock E, Deng L, Krupenko N, Mohan RD, Wu Q, Leclerc D, Gupta S, Elmore CL, Kruger W, Tini M et al. Susceptibility to intestinal tumorigenesis in folate-deficient mice may be influenced by variation in one-carbon metabolism and DNA repair. J Nutr Biochem 2011;22:1022–1029. [DOI] [PubMed] [Google Scholar]
- Lambrot R, Xu C, Saint-Phar S, Chountalos G, Cohen T, Paquet M, Suderman M, Hallett M, Kimmins S. Low paternal dietary folate alters the mouse sperm epigenome and is associated with negative pregnancy outcomes. Nat Commun 2013;4:2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane M, Robker RL, Robertson SA. Parenting from before conception. Science 2014;345:756–760. [DOI] [PubMed] [Google Scholar]
- Li D, Pickell L, Liu Y, Rozen R. Impact of methylenetetrahydrofolate reductase deficiency and low dietary folate on the development of neural tube defects in splotch mice. Birth Defects Res A Clin Mol Teratol 2006;76:55–59. [DOI] [PubMed] [Google Scholar]
- Luttropp K, Sjoholm LK, Ekstrom TJ. Global Analysis of DNA 5-Methylcytosine Using the Luminometric Methylation Assay, LUMA. Methods Mol Biol 2015;1315:209–219. [DOI] [PubMed] [Google Scholar]
- Ly L, Chan D, Trasler JM. Developmental windows of susceptibility for epigenetic inheritance through the male germline. Semin Cell Dev Biol 2015;43:96–105. [DOI] [PubMed] [Google Scholar]
- Matthews RG, Daubner SC. Modulation of methylenetetrahydrofolate reductase activity by S-adenosylmethionine and by dihydrofolate and its polyglutamate analogues. Adv Enzyme Regul 1982;20:123–131. [DOI] [PubMed] [Google Scholar]
- Mikael LG, Deng L, Paul L, Selhub J, Rozen R. Moderately high intake of folic acid has a negative impact on mouse embryonic development. Birth Defects Res A Clin Mol Teratol 2013;97:47–52. [DOI] [PubMed] [Google Scholar]
- Miller JW, Ulrich CM. Folic acid and cancer—where are we today. Lancet 2013;381:974–976. [DOI] [PubMed] [Google Scholar]
- Obeid R, Kirsch SH, Kasoha M, Eckert R, Herrmann W. Concentrations of unmetabolized folic acid and primary folate forms in plasma after folic acid treatment in older adults. Metabolism 2011;60:673–680. [DOI] [PubMed] [Google Scholar]
- Padmanabhan N, Jia D, Geary-Joo C, Wu X, Ferguson-Smith AC, Fung E, Bieda MC, Snyder FF, Gravel RA, Cross JC et al. Mutation in folate metabolism causes epigenetic instability and transgenerational effects on development. Cell 2013;155:81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickell L, Brown K, Li D, Wang XL, Deng L, Wu Q, Selhub J, Luo L, Jerome-Majewska L, Rozen R. High intake of folic acid disrupts embryonic development in mice. Birth Defects Res A Clin Mol Teratol 2011;91:8–19. [DOI] [PubMed] [Google Scholar]
- Reeves PG. Components of the AIN-93 diets as improvements in the AIN-76A diet. J Nutr 1997;127:838S–841S. [DOI] [PubMed] [Google Scholar]
- Reynolds EH. What is the safe upper intake level of folic acid for the nervous system? Implications for folic acid fortification policies. Eur J Clin Nutr 2016;70:537–540. [DOI] [PubMed] [Google Scholar]
- Rivera RM, Stein P, Weaver JR, Mager J, Schultz RM, Bartolomei MS. Manipulations of mouse embryos prior to implantation result in aberrant expression of imprinted genes on day 9.5 of development. Hum Mol Genet 2008;17:1–14. [DOI] [PubMed] [Google Scholar]
- Robb GW, Amann RP, Killian GJ. Daily sperm production and epididymal sperm reserves of pubertal and adult rats. J Reprod Fertil 1978;54:103–107. [DOI] [PubMed] [Google Scholar]
- Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science 1998;281:363–365. [DOI] [PubMed] [Google Scholar]
- Schaible TD, Harris RA, Dowd SE, Smith CW, Kellermayer R. Maternal methyl-donor supplementation induces prolonged murine offspring colitis susceptibility in association with mucosal epigenetic and microbiomic changes. Hum Mol Genet 2011;20:1687–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sie KK, Li J, Ly A, Sohn KJ, Croxford R, Kim YI. Effect of maternal and postweaning folic acid supplementation on global and gene-specific DNA methylation in the liver of the rat offspring. Mol Nutr Food Res 2013;57:677–685. [DOI] [PubMed] [Google Scholar]
- Smith AD, Kim YI, Refsum H. Is folic acid good for everyone. Am J Clin Nutr 2008;87:517–533. [DOI] [PubMed] [Google Scholar]
- Swayne BG, Kawata A, Behan NA, Williams A, Wade MG, Macfarlane AJ, Yauk CL. Investigating the effects of dietary folic acid on sperm count, DNA damage and mutation in Balb/c mice. Mutat Res 2012;737:1–7. [DOI] [PubMed] [Google Scholar]
- Tunster SJ, Jensen AB, John RM. Imprinted genes in mouse placental development and the regulation of fetal energy stores. Reproduction 2013;145:R117–R137. [DOI] [PubMed] [Google Scholar]
- Wen SW, Champagne J, Rennicks White R, Coyle D, Fraser W, Smith G, Fergusson D, Walker MC. Effect of folic acid supplementation in pregnancy on preeclampsia: the folic acid clinical trial study. J Pregnancy 2013;2013:294312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RD, Wilson RD, Audibert F, Brock JA, Carroll J, Cartier L, Gagnon A, Johnson JA, Langlois S, Murphy-Kaulbeck L et al. Pre-conception folic acid and multivitamin supplementation for the primary and secondary prevention of neural tube defects and other folic acid-sensitive congenital anomalies. J Obstet Gynaecol Can 2015;37:534–552. [DOI] [PubMed] [Google Scholar]
- Wong WY, Merkus HM, Thomas CM, Menkveld R, Zielhuis GA, Steegers-Theunissen RP. Effects of folic acid and zinc sulfate on male factor subfertility: a double-blind, randomized, placebo-controlled trial. Fertil Steril 2002;77:491–498. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.