Abstract
The Arabidopsis IMMUTANS gene encodes a plastid homolog of the mitochondrial alternative oxidase, which is associated with phytoene desaturation. Upon expression in Escherichia coli, this protein confers a detectable cyanide-resistant electron transport to isolated membranes. In this assay this activity is sensitive to n-propyl-gallate, an inhibitor of the alternative oxidase. This protein appears to be a plastid terminal oxidase (PTOX) that is functionally equivalent to a quinol:oxygen oxidoreductase. This protein was immunodetected in achlorophyllous pepper (Capsicum annuum) chromoplast membranes, and a corresponding cDNA was cloned from pepper and tomato (Lycopersicum esculentum) fruits. Genomic analysis suggests the presence of a single gene in these organisms, the expression of which parallels phytoene desaturase and ζ-carotene desaturase gene expression during fruit ripening. Furthermore, this PTOX gene is impaired in the tomato ghost mutant, which accumulates phytoene in leaves and fruits. These data show that PTOX also participates in carotenoid desaturation in chromoplasts in addition to its role during early chloroplast development.
Carotenoids have well known biological properties that include light harvesting and protection against photo-oxidation. Carotenoids can be further metabolized for instance to abscisic acid in plant cells and to vitamin A in animal cells (for review, see Cunningham and Gantt, 1998). In plant cells carotenoids are produced in plastids. The main carotenoids in chloroplasts are β-carotene and xanthophylls. In non-photosynthetic chromoplasts the distribution of carotenoids is subject to considerable variation from one species to another (for review, see Bramley, 1997). This ranges from the accumulation of an intermediate in the pathway such as lycopene in ripe tomato (Lycopersicum esculentum) fruits to the production of species-specific carotenoids as in pepper (Capsicum annuum) fruits.
However, all of these molecules are derived from a common precursor, namely phytoene, a C40 acyclic molecule. Phytoene is subjected to four consecutive desaturation (dehydrogenation) reactions that lead to the formation of lycopene. This latter compound contains the typical chromophore of 11 conjugated double bonds. Phytoene desaturation to ζ-carotene via phytofluene is catalyzed by phytoene desaturase (PDS) (Bartley et al., 1991), and ζ-carotene desaturation to lycopene via neurosporene is catalyzed by ζ-carotene desaturase (ZDS; Albrecht et al., 1995). These enzymes catalyze similar reactions and share significant sequence similarity (Albrecht et al., 1995).
Although these enzymes are active in an heterologous environment, e.g. when produced in Escherichia coli cells (Fraser et al., 1993; Bartley et al., 1999; Breitenbach et al., 1999), they require a number of co-factors in plastids. The first evidence for co-factor requirement was obtained using daffodil flower chromoplasts where it was shown that molecular oxygen and a redox mediator acting between the desaturase and O2 (Beyer et al., 1989) as well as quinones (Mayer et al., 1992; Schultz et al., 1993) are required. Genetic evidence for quinone requirement was also obtained using Arabidopsis mutants accumulating phytoene (Norris et al., 1995). Furthermore, phytoene desaturation is linked to a respiratory redox chain in daffodil flowers, which surprisingly involves the 23-kD protein from the chloroplastic oxygen-evolving complex (Nievelstein et al., 1995).
It has been recent that the cloning of the Arabidopsis gene IMMUTANS has identified a new important factor for phytoene desaturation (Carol et al., 1999; Wu et al., 1999). Inactivation of this gene results in reduced phytoene desaturation and consequently reduced carotenoid content leading to photo-oxidative damage, which results in a variegated phenotype consisting of white and green sectors. White sectors arise from irreversible photo-oxidative damage at an early stage of chloroplast formation (Carol et al., 1999) when carotenoid synthesis is normally increased (Corona et al., 1996). Green sectors originate from cells that avoided irreversible photo-oxidative damage. This clarifies previous data postulating that the IMMUTANS gene product is active during an early phase of plant development (Wetzel et al., 1994). The IMMUTANS gene product shows limited similarity with mitochondrial alternative oxidases (AOX; for reviews, see Day et al., 1995; Siedow and Umbach, 1995; Vanlerberghe and McIntosh, 1997; Wagner and Moore, 1997), suggesting that it may function as a terminal oxidase located within plastids (Carol et al., 1999; Wu et al., 1999).
In this report we monitored the terminal oxidase activity of the IMMUTANS gene product after expression in E. coli. In addition we examined the involvement of this protein during a particular developmental stage, namely during chromoplast differentiation in pepper and tomato fruits when carotenoid production is enhanced. These data are reinforced by our demonstration that the tomato ghost mutant, the phenotype of which resembles immutans in leaves but also accumulates phytoene in fruits (Mackinney et al., 1956; Scolnik et al., 1987), is impaired in the corresponding gene.
RESULTS
Catalytic Properties of the IMMUTANS Gene Product Expressed in E. coli
We tested the potential plastid terminal oxidase (PTOX) activity of the IMMUTANS gene product after expression in E. coli cells (see “Materials and Methods”). After induction of this chimeric gene by isopropylthio-β-galactoside, cells were harvested and their membranes were isolated. The overexpressed polypeptide (see below) was visible in Coomassie Blue-stained protein gels and could be immunodetected in these membranes (not shown) but not in control membranes (from cells transformed with the cloning vector alone).
To assay for PTOX enzymatic activity, oxygen consumption by these membranes was measured. As shown in Figure 1a, NADH addition initiates oxygen consumption in membranes from both control cells and from cells expressing the putative PTOX. An addition of cyanide (KCN; 2 mm) progressively, but strongly, inhibited oxygen consumption in control membranes. In contrast a significant cyanide-resistant oxygen consumption was consistently observed (in 12 experiments) in membranes from the IMMUTANS-expressing cells (Fig. 1, a and b).
Figure 1.
Oxygen consumption in isolated E. coli membranes from cells expressing the Arabidopsis IMMUTANS gene product (PTOX) and control cells. a, Oxygen uptake was measured using an O2 electrode after addition of NADH as an electron donor, KCN, and n-propyl-gallate (nPG). Dithionite (Na2S2O4) was added at the end of the experiment to verify that the reactions did not proceed to complete oxygen depletion. Numbers refer to oxygen consumption in nmol O2 min−1 mg−1 protein in the linear zone of the traces (when full effect of the added compound was obtained). b, Relative respiration rate after sequential addition of NADH (set as 100), KCN, and nPG. Means of 12 experiments and se are shown. c, Same as a except that nPG was added prior to KCN. Traces are of the same scale in a given panel.
To examine the possibility that this cyanide-resistant oxygen consumption could be due to the induction of cytochrome bd oxidase, which is more resistant to cyanide than cytochrome bo oxidase, these experiments were repeated in the presence of 10 mm KCN. Cyanide resistance was also observed in this case (data not shown). Therefore, a possible rise in cytochrome bd oxidase level, induced indirectly by PTOX, does not appear to explain our data.
Furthermore, this cyanide-resistant oxygen consumption was abolished by addition of nPG, an inhibitor of the mitochondrial AOX (Siedow and Umbach, 1995). In the latter case the residual rate of oxygen consumption was similar to that observed in control membranes treated with KCN alone or KCN plus nPG. An addition of nPG to control membranes prior to KCN had little effect (Fig. 1c), which is in agreement with other data (Berthold, 1998). An addition of nPG to PTOX membranes prior to KCN also had little effect (Fig. 1c), suggesting that PTOX cannot efficiently compete with the E. coli cytochrome path when the latter is active. This is not unexpected considering that PTOX is not a normal constituent of these membranes. In addition it should be remembered that the engagement of the AOX path in mitochondria is often largely influenced by the inhibition of the cytochrome path (Day et al., 1995).
These biochemical data suggest that the IMMUTANS gene product is inserted in E. coli membranes where it functions as a terminal oxidase, which by-passes the E. coli cytochrome path when this path is inhibited by cyanide. Thus this plastid protein behaves similarly to the mitochondrial AOX (a quinol:oxygen oxidoreductase) when expressed in E. coli (Kumar and Söll, 1992; Berthold, 1998).
PTOX Polypeptide Is Present in Fruit Chromoplasts
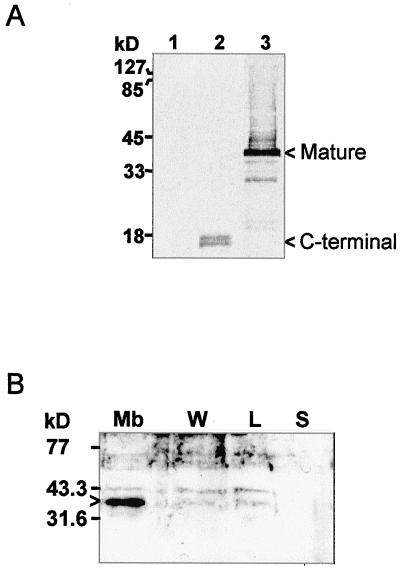
Antibodies raised against the mature IMMUTANS gene product expressed in E. coli (as described in “Materials and Methods”) immunodetected the PTOX polypeptide expressed in E. coli (Fig. 2A, lane 3), but no polypeptide in control (non-recombinant) E. coli cells (lane 1). The specificity of the antibodies is also shown by the fact that they recognize a polypeptide of shorter size in E. coli cells expressing a truncated version (missing the N-terminal coding region) of the gene construct (Fig. 2A, lane 2).
Figure 2.
Immunodetection of the IMMUTANS polypeptide after expression in E. coli (A) and after sub-fractionation of purified chromoplasts from a ripening pepper fruit (B). A, Three E. coli strains were used: control (lane 1), expressing the 130 C-terminal amino acids (lane 2), or expressing the full mature polypeptide (lane 3). E. coli cells were grown and total protein recovered as described in “Materials and Methods.” In lane 3, the smear above the mature 41-kD band is due to incomplete resolubilization of the IMMUTANS polypeptide from inclusion bodies. B, Achlorophyllous membranes (Mb), membrane-wash fraction (W), low-density lipid fraction (L), and stroma (S) were fractionated as described in “Materials and Methods.” Protein samples were separated by SDS/12.5% (v/v) PAGE and transferred to nitrocellulose membranes. Position of size markers is shown on the left. The primary antibody was raised as described in “Materials and Methods.” A horseradish peroxidase-coupled secondary antibody was used. Detection was performed colorimetrically (A) or by enhanced chemiluminescence (B). Bands discussed in the text are indicated by arrowheads.
Using this antibody, we investigated the presence of PTOX in fruit chromoplasts as a preliminary step to examine whether PTOX participates also in carotenoid biosynthesis during this developmental stage. Protein gel-blot analysis revealed a polypeptide of approximately 41 kD in chromoplasts isolated from red pepper fruits and purified on Suc gradients. After further sub-fractioning of chromoplasts on Suc gradients, the protein was found in the membrane fraction (consisting of the internal achlorophyllous membrane and the envelope) but not in the soluble fraction (Fig. 2B). The PTOX protein was detected in only very faint amounts (which can be due to contamination) in the membrane wash fraction and in the low-density lipid fraction that contains globules and other light membranes structures.
Cloning and Characterization of PTOX cDNAs from Pepper and Tomato Fruits
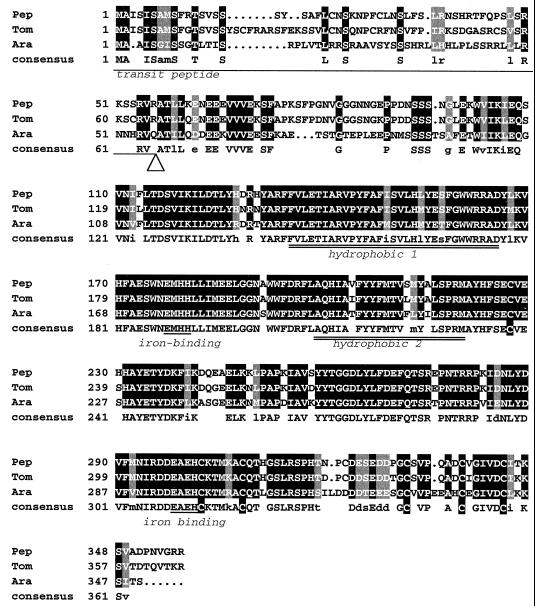
A portion of the Arabidopsis IMMUTANS cDNA encoding the mature PTOX protein was used as a hybridization probe to screen a cDNA library from green pepper fruit under non-stringent conditions. All positive clones that were analyzed appeared to be derived from the same gene, as suggested by identical sequences in the 3′-untranslated region. The DNA sequence of a full-length clone has been deposited in the databases (accession no. AF177981). This pepper cDNA was subsequently used to isolate the corresponding cDNA from a red tomato fruit library (accession no. AF177980). Figure 3 compares the deduced amino acid sequences from pepper, tomato, and Arabidopsis.
Figure 3.
Comparison of the deduced amino acid sequences of pepper, tomato, and Arabidopsis PTOX. Amino acids identical in all sequences are boxed in black and shown in uppercase in the consensus line (where Cys are boxed). Residues identical in two sequences but substituted by an equivalent amino acid in the third are boxed in gray and shown in lowercase in the consensus line. The transit peptide, two hydrophobic domains, and two putative iron-binding sites (ExxH) are underlined. The arrowhead shows the likely cleavage site of the transit peptide.
As expected, the plastid targeting transit peptides showed limited sequence similarity, except for the N-terminal region and the region around the proposed cleavage site (ATR/Q-AT). The mature PTOX polypeptides share 91% identity (95% similarity) between pepper and tomato and 75% identity (85% similarity) between pepper and Arabidopsis. The identity between PTOX and AOX polypeptides is approximately 25%. Sequence alignment of PTOX sequences reveals the presence of two conserved hydrophobic regions separated by a highly conserved hydrophilic segment, an organization also found in AOXs. The N-terminal domain is mainly hydrophilic and contains a long stretch of poorly conserved amino acids. The C-terminal domain is also mainly hydrophilic and contains one conserved motif (EAEH) that matches a putative iron-binding site (ExxH) that is also conserved in AOXs. No other potential iron-binding site is present in this region, but instead one is present in the hydrophilic region separating the two hydrophobic regions as recently proposed for AOXs (Andersson and Nordlung, 1999). In addition the C-terminal region contains six conserved cysteines in PTOX, whereas the rest of the polypeptide is devoid of Cys. This is in contrast with AOX structures in which two conserved cysteines are present in the N-terminal region.
Genomic Structure of PTOX in Tomato
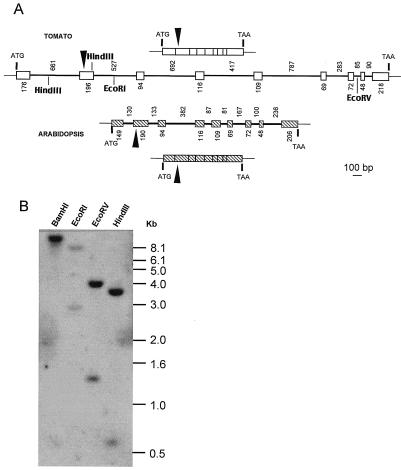
A genomic clone of the tomato PTOX gene was isolated after PCR amplification of total genomic DNA using oligonucleotide primers based on the PTOX cDNA sequence. Tomato PTOX spans approximately 4.8 kb (accession no. AF177979), whereas the Arabidopsis homolog (Carol et al., 1999) is located on a 2.5-kb genomic DNA fragment. This difference is essentially due to intron sizes (Fig. 4A). Both genes have eight introns the relative distribution of which is identical among the two species. The first five introns are longer in tomato than Arabidopsis, however, the last two are shorter in tomato. All introns follow the GT/AG splicing rule. Blast analysis of the different introns indicated that in tomato, 350 bp of the fifth intron show 82% identity with a portion of a DNA fragment of unknown function located on chromosome 6 in tomato between markers APS and GP 79 (accession no. LEU81378).
Figure 4.
Genomic organization of PTOX from tomato and Arabidopsis. A, Schematic representation of cDNAs and genomic fragments from PTOX. Upper and lower lines represent transcripts, whereas intermediate lines represent genomic structures. Exons are indicated by boxes, introns by lines. Numbers indicate the length in base pairs of exons (below) and introns (above). Transcription start site (ATG) and stop codon (TAA) are indicated. Putative processing site of the targeting sequence is indicated with a black arrowhead. Cleavage sites for restriction enzymes used in B are shown for the tomato gene. B, Southern-blot analysis of tomato PTOX. Tomato DNA was digested with the indicated restriction endonucleases and hybridized with the radiolabeled tomato full-length cDNA probe. Size markers (kilobase pairs) are indicated on the right.
Southern genomic analysis under stringent conditions indicates that PTOX is present as a single gene per haploid genome in tomato (Fig. 4B) and in pepper (data not shown). When these experiments were repeated under low stringent conditions, no additional bands were revealed (data not shown).
Expression of PTOX, PDS, and ZDS Genes during Fruit Ripening in Pepper and Tomato
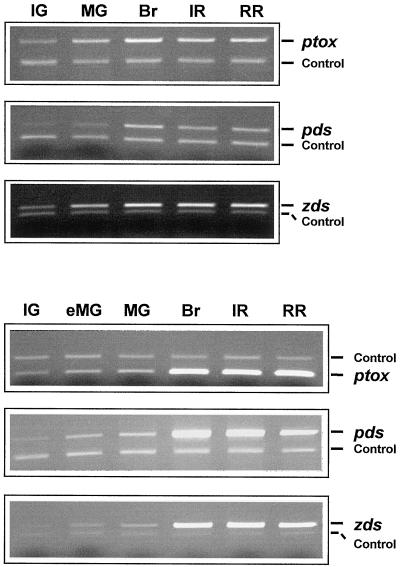
To assess whether PTOX is expressed during ripening, total RNA was extracted from pepper and tomato fruit at different ripening stages. Transcripts of PTOX, PDS, and ZDS were detected following reverse transcriptase (RT)-PCR in the presence of specific oligonucleotides. Appropriate controls were included to verify that the amount of RT-PCR product reflects proportionally the amount of the corresponding transcript in the RNA samples (see “Materials and Methods”).
In pepper all three genes have a similar pattern of expression during ripening. As shown in Figure 5 (top), the transcript levels show a 4- to 6-fold increase between the immature green stage and the breaker stage (early visible signs of color change). The levels of transcript then remain relatively constant, but with a slight decrease during the red stages. It is interesting that in tomato all three genes when compared with each other also show a similar expression pattern (Fig. 5, bottom). However, this expression pattern is different in comparison with the expression pattern in pepper. In tomato the transcript levels show a 2- to 4-fold increase between the immature green and the mature green stage (fruit of adult size) but dramatically increase at the breaker stage and remain high throughout ripening, showing only a slight decrease in the later stages of ripening.
Figure 5.
Expression of PTOX, PDS, and ZDS genes during pepper (top) and tomato (bottom) fruit development. mRNA levels were determined by RT-PCR amplification of total cellular RNA. Equal amounts of total RNA were used in each reaction. The PCR products were separated by 1.5% (v/v) agarose gel electrophoresis and visualized by ethidium bromide staining. Amplification of globin mRNA (added to the RT reaction mix) was used as a control for the RT-PCR reaction (see also “Materials and Methods”). IG, Immature green; eMG, early mature green (adult size); MG, mature green; Br, breaker; IR, intermediate red (4 d after breaker); RR, red ripe (14 d after breaker).
The Tomato ghost Mutant Is Affected in the PTOX Gene
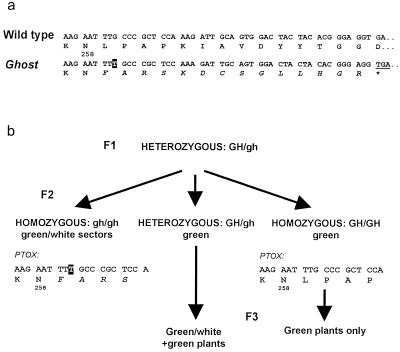
Because the tomato ghost mutant accumulates phytoene in fruits (Mackinney et al., 1956; Scolnik et al., 1987), we decided to investigate whether PTOX is mutated in this line. We cloned a PTOX cDNA from two plants homozygous for ghost, and two wild-type plants of the same cultivar. Whereas the sequences from the wild-type plants were identical to the sequence reported above, the sequences from the ghost plants showed a frame shift after the Asn-258 codon (Fig. 6a). This cDNA potentially encodes a truncated polypeptide lacking part of the C-terminal region containing the second potential iron-binding motif (Fig. 3). To further confirm that the ghost locus corresponds to PTOX, 20 seeds from plants heterogyzous for ghost were germinated. The segregation of this PTOX frame-shift mutation and the ghost phenotype was compared. As shown in Figure 6b, the PCR-amplified genomic fragment from 100% of the plants showing the ghost phenotype (homozygous for ghost) contained the frame-shift. In contrast, all plants homozygous for the wild-type GHOST gene (showing no ghost phenotype in the next generation) yielded PTOX genomic fragments identical to the wild-type sequence.
Figure 6.
Identification of the tomato ghost gene. a, Partial sequence of the PTOX cDNA showing a T insertion (boxed) in ghost plants when compared to wild-type and deduced amino acid sequences. The amino acid sequence created by the mutation is shown in italics. b, Cosegregation of the ghost recessive phenotype and the mutation in the PTOX genomic sequence from a heterozygous F1 plant to the F2 generation. Homozygous F2 wild-type plants were discriminated from heterozygous plants in the F3 generation.
DISCUSSION
The Arabidopsis IMMUTANS Gene Product Behaves Like a Quinol:Oxygen Oxidoreductase
The identification of the IMMUTANS gene product as a polypeptide showing limited sequence similarity with mitochondrial AOX was previously taken as an indication that it fulfils the role of a PTOX associated with phytoene desaturation (Carol et al., 1999; Wu et al., 1999). We show here using a functional assay, following expression in E. coli, that PTOX is capable of conferring weak but significant cyanide-resistant electron transport in isolated membranes. The fact that this activity is weak (although the polypeptide is visible in Coomassie Blue-stained gels) and only clearly detectable when the cytochrome path is inhibited is not surprising in such an heterologous assay based on E. coli electron transport chain. It is important to mention that these results are highly reproducible (Fig. 1b). This activity is sensitive to nPG, which is an inhibitor of the cyanide-resistant mitochondrial AOX. Therefore, PTOX appears to be functionally equivalent to a quinol:oxygen oxidoreductase. It should be mentioned that salicylhydroxamic acid, another commonly used AOX inhibitor, was not used in this assay since unlike nPG it has been shown to also inhibit components of the E. coli electron transport chain (Berthold, 1998). It should also be mentioned that an effect of nPG on carotenoid accumulation, in developing seedlings for example, could not be assessed since under our experimental conditions nPG delayed germination at 0.1 mm (with no effect on carotenoid accumulation) and prevented germination at higher concentrations (not shown).
Structural Features of PTOX
Hydropathy analysis indicates a similar structure for all three PTOX polypeptides studied here and AOX polypeptides, namely three hydrophilic regions separated by two hydrophobic domains. A model for AOX structure (Siedow and Umbach, 1995) proposes that these hydrophobic regions are helical transmembrane domains. Searches for secondary structures in PTOX sequences consistently predicted helical structures matching almost perfectly these two potential transmembrane domains and most of the hydrophilic region separating them. Additional helical structures can be postulated after the second hydrophobic domain. Although different predictions were obtained in this C-terminal region using different programs, predictions for AOX and PTOX were again quite similar, despite limited primary sequence similarity.
Therefore, by analogy with the structure proposed by Siedow et al. (1995) for AOX, which is based on structures of binuclear iron proteins such as methane mono-oxygenase, a four-helix structure forming a binuclear iron center can be postulated for the C-terminal region. A weak point for this model in the case of PTOXs is that they do not contain two iron-binding motifs (ExxH) in this region but only the second one. It is interesting that a more recent alternative model has been proposed by Andersson and Nordlung (1999) for AOXs taking into account recent AOX sequences in which the first iron-binding motif in the C-terminal region is not conserved either (like in PTOX). This model proposes that the hydrophobic regions are not membrane spanning. This allows a spatial proximity between the remaining conserved iron-binding motif and a similar ExxH motif in the hydrophilic region separating the hydrophobic regions. It is striking that such an ExxH motif is also conserved in the corresponding position (position 177 in the pepper sequence in Fig. 3) in all PTOX sequences.
It is also notable that conserved cysteines are present in the PTOX C-terminal region, which in AOXs is devoid of Cys. In contrast, plant AOXs have two conserved Cys in their N-terminal region, the first one being involved in the formation of the less active disulfide-linked dimer of the protein and α-keto-acid (e.g. pyruvate) activation (Rhoads et al., 1998; Vanlerberghe et al., 1998). Whether PTOX is subject to redox control or activation by pyruvate will be examined using the functional assay described here. This could provide some indication as to whether the C-terminal conserved cysteines in PTOXs are functionally equivalent to the N-terminal conserved cysteines in AOXs.
Involvement of the PTOX in Carotenoid Biosynthesis in Chromoplasts
The PTOX polypeptide could be immunodetected in plastid fractions from pepper (Fig. 2) and Arabidopsis (not shown) as a polypeptide of approximately 41 kD. This is larger than the theoretical molecular mass of the mature polypeptide (34.3 kD) but is similar to the apparent molecular mass of the mature polypeptide expressed in E. coli. This difference may be due to the partially hydrophobic nature of PTOX.
Ripening pepper fruits were chosen because isolation of highly purified chromoplasts is feasible in this case. The PTOX polypeptide was mainly found in the achlorophyllous membrane fraction of these chromoplasts, which is known to contain carotenoid biosynthetic enzymes (Bouvier et al., 1994). Further evidence for the involvement of PTOX in carotenoid biosynthesis in chromoplasts is provided by the cloning of related PTOX cDNAs from pepper and tomato fruits. Since PTOX is encoded by a single gene in all species studied here, it seems likely that the same polypeptide operates during early stages of plastid development as well as late stages, namely chromoplast formation. Gene expression studies have shown that these PTOX genes are induced during fruit ripening. Furthermore, when compared with the PDS and ZDS genes, a similar profile of induction during fruit ripening was observed in the same organism. The expression of PDS obtained here is in agreement with previous reports (Ronen et al., 1999 and references therein). The expression of ZDS during ripening has not to our knowledge been reported to date.
Genetic evidence for the involvement of PTOX in carotenoid biosynthesis in chromoplasts is provided by our data showing that PTOX corresponds to the tomato GHOST gene. Like immutans, the ghost mutant shows variegated green/white leaves and phytoene accumulation in white sectors. In addition, ghost is interesting since it possesses poorly colored petals (compared with the yellow carotenoid-containing wild-type petals) and fruits that do not accumulate the red lycopene pigment but phytoene instead (Mackinney et al., 1956; Scolnik et al., 1987). Despite this lack of carotenoid desaturation, ghost fruits exhibit other ripening features such as softening. These data show that PTOX has a preponderant role in carotenoid accumulation in petal and fruit chromoplasts in addition to its role during early chloroplast differentiation (Wetzel et al., 1994; Carol et al., 1999; Wu et al., 1999).
Does PTOX Act with Both PDS and ZDS Reactions?
In contrast with PDS, no data are currently available demonstrating the involvement of PTOX with ZDS activity. However, this is most likely the case since (a) PDS and ZDS share considerable sequence similarity, (b) they catalyze similar enzymatic reactions, (c) ZDS also uses quinones as cofactors (Breitenbach et al., 1999), (d) no evidence was obtained for a second PTOX gene, and (e) the PDS, ZDS, and PTOX genes are co-expressed when carotenoid biosynthesis is enhanced during fruit ripening.
The data presented here and the availability of antibodies against PTOX represent a new step toward the elucidation of carotenoid desaturation complexes and their mode of action.
MATERIALS AND METHODS
Plant Materials
Pepper (Capsicum annuum cv Yolo Wonder) and tomato (Lycopersicum esculentum cv Ailsa Craig) plants were grown under greenhouse conditions. The ghost mutant (accession no. LA0295) and corresponding wild-type line (San Marzano, LA0180) were obtained from the Tomato Genetic Resource Center (University of California, Davis) and grown in shaded conditions.
Chromoplast Isolation and Sub-Fractionation
A pepper fruit at an early ripening stage was ground in extraction buffer (1.65 m sorbitol, 5 mm EDTA, 5 mm β-mercaptoethanol, and 0.25 m Tris [tris(hydroxymethyl)-aminomethane], pH 7.6). The extract was filtered, centrifuged for 5 min at 2,000g, resuspended in extraction buffer, and loaded onto a discontinuous Suc gradient (0.5 m/0.84 m/1.45 m). After 15 min of centrifugation at 60,000g, intact chromoplasts were recovered at the 0.84-m/1.45-m interface. Chromoplasts were lysed by resuspension in 5 mm EDTA, 5 mm β-mercaptoethanol, and 0.25 m Tris, pH 7.6, and homogenized in a Potter homogenizer. The stromal fraction was recovered by a 100,000g centrifugation. Alternatively, to fractionate the insoluble fraction, the lysed chromoplasts were loaded onto a 0.5-m/0.9-m Suc step gradient and centrifuged 15 min at 60,000g. The low-density lipid fraction was recovered on top of the gradient, whereas the membrane fraction was recovered at the interface. The latter fraction was diluted and repurified by 1 h of centrifugation on a linear 0.5-m/0.9-m Suc gradient. All insoluble fractions were washed and recentrifuged at 100,000g.
Library Screening and Southern-Blot Analysis
A ripening tomato fruit (Kausch and Handa, 1997) and a green pepper fruit cDNA library (Matsui et al., 1996) were screened essentially as described (Albrecht et al., 1995). Tomato genomic DNA was extracted according to Tieman et al. (1992). For Southern analysis, 25 μg of DNA were digested with restriction endonucleases, electrophoresed on 0.8% (w/v) agarose gel, and transferred to nitrocellulose (Optitrans BA-S 85, Schleicher & Schull, Keene, NH) following standard procedures. Filters were hybridized with a radiolabeled full-length cDNA probe of tomato PTOX. Hybridization was performed at 62°C (high stringency) or 47°C (low stringency) with 106 cpm/mL hybridization solution. After hybridization filters were washed in 2×, 1×, and then 0.1× SSC containing 0.1% (w/v) SDS at 62°C or 47°C.
PCR Amplification of Tomato PTOX Gene
Two sets of primers were used to produce overlapping fragments of the entire coding region (cv Ailsa Craig). One set, T5F (5′-CTAACAACTTTCCCACTTTGG-3′) and T5R, (5′-CAATTTATCGTAAGTCTCGTATGC-3′) amplifies 800 bp in the 5′ side of the cDNA and 3,910 bp from tomato genomic DNA. A second set, T3F (5′-ATGGCATATCAT-TTCTCTGAATGTGTGGA-3′) and T3R, (5′-GTATATACA-AGTATAGTTGTCCGC-3′) amplifies 540 bp from the 3′-end of the cDNA and 915 bp from genomic DNA. PCR reactions were performed in a 25-μL reaction volume with 1.8-μg genomic DNA, 0.4 μm of primer, 2 mm MgCl2, and Elongase enzyme mixture (Bethesda Research Laboratory [BRL], Gaithersburg, MD). After 3 min at 95°C, PCR amplification was performed by 35 cycles consisting of 30 s at 94°C, 40 s at 50°C, and 2 min at 68°C, followed by a 10-min extension at 68°C.
Amplification of a PTOX fragment from ghost plants was performed as above using primers T3F and T5R. For cloning the entire cDNA from this mutant, 1 μL of a reverse transcriptase reaction mixture (see below) was submitted to PCR amplification in the presence of 0.1 μm of primers T5F and T3R, 1 mm dNTP, and 1.5 mm MgCl2 and Elongase. Amplification conditions were as above with an annealing temperature raised to 52°C.
PCR products were cloned in the pGEM-T easy vector (Promega, Madison, WI) according to manufacturer's instructions. Sequence analyses were performed using the GCG package (Genetic Computer Group, Madison, WI) and software from Infobiogen (www.infobiogen.fr) or the Pasteur Institute (www.pasteur.fr).
Extraction of RNA
Fruits were ground in a coffee grinder cooled with liquid nitrogen. Ground material was added to a mixture (preheated to 80°C) of extraction buffer (0.1 m Tris, pH 8.0, 10 mm EDTA, 0.1 m LiCl, and 1% [w/v] SDS) and water-saturated phenol and vortexed. Samples were centrifuged, and the aqueous phase was re-extracted with chloroform. The aqueous phase was collected, and RNA was precipitated with 0.5 volumes 6 m LiCl. RNA samples were treated with proteinase K in 10 mm Tris, pH 7.5, and 0.4% (w/v) SDS at 50°C for 30 min, and re-extracted with phenol/chloroform. Leaf RNA was extracted in a similar manner and routinely treated with DNase.
RNA concentration and purity were determined by spectrophotometry and visualized by electrophoresis on formaldehyde/agarose gels stained with ethidium bromide. Samples were checked for DNA contamination by PCR using a 4-fold excess of RNA with respect to the concentration routinely used in the RT-PCR reactions.
Measurement of mRNA by RT-PCR
Reverse transcription was carried out using 250 ng of total RNA. Linearity of the RT reaction was established for RNA amounts between 125 and 500 ng. The reaction mixture included 1 mm dNTPs, 0.5 μm oligo(dT), 20 units RNaseOut (BRL), 0.1 pg of control RNA (rabbit globin mRNA from reticulocyte polyribosomes; BRL), 10 mm dithiothreitol, 1× RT buffer (BRL), and 100 units Moloney murine leukemia virus reverse transcriptase (BRL) in total volume of 20 μL. Each reaction was carried out in duplicate. The reaction mixture was incubated for 10 min at 20°C, for 35 min at 37°C, and then for 15 min at 42°C. Duplicate samples were pooled to give a final volume of 40 μL, and aliquots were taken for several parallel PCR amplification.
The PCR mix contained 0.6 to 2.0 μg of each primer (based on sequences from the 3′ portion of cDNAs), 1.4× Taq polymerase buffer, 5 mm MgCl, 0.25 mm dNTPs, 1.5 units Taq polymerase, and 8 μL of RT reaction mixture (corresponding to the original amount of 100 ng of RNA) in a total volume of 100 μL. The final concentration of cDNA in the PCR reaction mix therefore corresponded to 1 ng/μL of the original RNA, and the linearity of the PCR amplification was verified for concentration between 0.5 and 2 ng/μL (using a sample from the developmental stage that gave the strongest signal). The amplification consisted of 24 cycles of 30 s at 94°C, 20 s at 55°C (50°C for pepper PTOX), and 20 s at 72°C.
Gene Construct and Protein Production
The portion of the Arabidopsis IMMUTANS cDNA coding for the entire mature peptide was PCR-amplified using oligonucleotides based on the cDNA sequence and extended by a BamHI (upstream oligonucleotide) or a PstI (downstream oligonucleotide) restriction site. After restriction digestion, this PCR fragment was in-frame inserted in the Escherichia coli expression vector pQE31 (Qiagen, Valencia, CA) and cleaved using the same enzymes. The recombinant protein that possesses a 6×-His-tag was produced in E. coli, purified according to supplier recommendations, and used to raise polyclonal antibodies in rabbit.
Immunodetection
Protein samples were fractionated by SDS/PAGE and electroblotted onto nitrocellulose. Immunodetection was performed using either the horseradish peroxidase conjugate substrate kit (Bio-Rad Laboratories, Hercules, CA) or the enhanced chemiluminescence western-blotting kit (Amersham, Buckinghamshire, UK) as recommended by the suppliers.
Measurement of Oxygen Consumption
E. coli cells (strain XL-1 Blue) were grown in M9/glycerol medium until OD600 = 0.3. Isopropylthio-β-galactoside was then added (final concentration 40 μm) to induce expression of the recombinant gene during 3 h. The control strain was grown in parallel. After lysis and elimination of the debris, membranes were recovered upon centrifugation at 100,000g for 1 h. Pelleted membranes were resuspended in 0.2 m Tris-HCl, pH 7.5 and 0.75 m Suc. Oxygen consumption was measured in a Clark O2 electrode chamber (Hansatech, King's Lynn, UK). A typical assay contained 100 μg of membrane protein in the following buffer: 50 mm Tris-maleate, pH 7.5, 0.2 mm decyl-plastoquinone, 10 mm KCl, 5 mm MgCl2, and 1 mm EDTA.
ACKNOWLEDGMENTS
We wish to thank E. Charpentier for technical assistance, J.P. Alcaraz for sequencing, and G. Clabault for constant aid. We are grateful to Dr. A.J. Dorne (University of Grenoble/Commissariat à l'Energie Atomique) for helpful discussions, to Prof. P.M. Bramley and co-workers (Royal Holloway, University of London) for sharing unpublished sequence information, to Prof. A.K. Handa (Purdue University, West Lafayette, IN) and Prof. K. Matsui (Yamaguchi University, Japan) for their gift of a cDNA library, and to R. Curtis and colleagues (Tomato Genetic Resource Center) for providing tomato seeds.
Footnotes
This work was supported by the European Commission DGXII Biotechnology Programme (contract BIO4–96–2077).
LITERATURE CITED
- Albrecht M, Klein A, Hugueney P, Sandmann G, Kuntz M. Molecular cloning and functional expression in E. coli of a novel plant enzyme mediating ζ-carotene desaturation. FEBS Lett. 1995;372:199–202. doi: 10.1016/0014-5793(95)00978-i. [DOI] [PubMed] [Google Scholar]
- Andersson ME, Nordlung P. A revised model for the active site of alternative oxidase. FEBS Lett. 1999;449:17–22. doi: 10.1016/s0014-5793(99)00376-2. [DOI] [PubMed] [Google Scholar]
- Bartley GE, Scolnik PA, Beyer P. Two Arabidopsis thaliana carotene desaturase, expressed in Escherichia coli, catalyze a poly-cis pathway to yield pro-lycopene. Eur J Biochem. 1999;259:396–403. doi: 10.1046/j.1432-1327.1999.00051.x. [DOI] [PubMed] [Google Scholar]
- Bartley GE, Vitanen PE, Pecker I, Chamovitz D, Hirschberg J, Scolnik PA. Molecular cloning and expression in photosynthetic bacteria of a soybean cDNA coding for phytoene desaturase, an enzyme of the carotenoid biosynthetic pathway. Proc Natl Acad Sci USA. 1991;88:6532–6536. doi: 10.1073/pnas.88.15.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold DA. Isolation of mutants of the Arabidopsis thaliana alternative oxidase (ubiquinol:oxygen oxidoreductase) resistant to salicylhydroxamic acid. Biochim Biophys Acta. 1998;1364:73–83. doi: 10.1016/s0005-2728(98)00015-2. [DOI] [PubMed] [Google Scholar]
- Beyer P, Mayer MP, Kleinig H. Molecular oxygen and the state of geometric isomerism of intermediates are essential in the carotene desaturation and cyclisation reactions in daffodil chromoplasts. Eur J Biochem. 1989;184:141–150. doi: 10.1111/j.1432-1033.1989.tb15000.x. [DOI] [PubMed] [Google Scholar]
- Bouvier F, Hugueney P, d'Harlingue A, Kuntz M, Camara B. Xanthophyll biosynthesis in chromoplasts: isolation and molecular characterization of an enzyme catalyzing the conversion of 5,6-epoxycarotenoid into ketocarotenoid. Plant J. 1994;6:45–54. doi: 10.1046/j.1365-313x.1994.6010045.x. [DOI] [PubMed] [Google Scholar]
- Bramley PM. Isoprenoid metabolism. In: Dey PM, Harborne JB, editors. Plant Biochemistry. San Diego: Academic Press; 1997. pp. 417–437. [Google Scholar]
- Breitenbach J, Kuntz M, Takaichi S, Sandmann G. Catalytic properties of an expressed and purified higher plant type ζ-carotene desaturase from Capsicum annuum. Eur J Biochem. 1999;265:376–383. doi: 10.1046/j.1432-1327.1999.00746.x. [DOI] [PubMed] [Google Scholar]
- Carol P, Stevenson D, Bisanz C, Breitenbach J, Sandmann G, Mache R, Coupland G, Kuntz M. Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell. 1999;11:57–68. doi: 10.1105/tpc.11.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona V, Aracri B, Kosturkova G, Bartley GE, Pitto L, Giorgetti L, Scolnik PA, Giuliano G. Regulation of a carotenoid biosynthesis gene promoter during plant development. Plant J. 1996;9:505–512. doi: 10.1046/j.1365-313x.1996.09040505.x. [DOI] [PubMed] [Google Scholar]
- Cunningham FX, Gantt E. Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:557–583. doi: 10.1146/annurev.arplant.49.1.557. [DOI] [PubMed] [Google Scholar]
- Day DA, Whelan J, Millar AH, Siedow JN, Wiskich JT. Regulation of the alternative oxidase in plants and fungi. Aust J Plant Physiol. 1995;22:497–509. [Google Scholar]
- Fraser PD, Linden H, Sandmann G. Purification and reactivation of recombinant Synechococcus phytoene desaturase from an overexpressing strain of Escherichia coli. Biochem J. 1993;291:687–692. doi: 10.1042/bj2910687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kausch KD, Handa AK. Molecular cloning of a ripening-specific lipoxygenase and its expression during wild-type and mutant tomato fruit development. Plant Physiol. 1997;113:1041–1050. doi: 10.1104/pp.113.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar AM, Söll D. Arabidopsis alternative oxidase sustains Escherichia coli respiration. Proc Natl Acad Sci USA. 1992;89:10842–10846. doi: 10.1073/pnas.89.22.10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinney G, Rick CM, Jenkins JA. The phytoene content of tomatoes. Proc Natl Acad Sci USA. 1956;42:404–408. doi: 10.1073/pnas.42.7.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Shibutani M, Hase T, Kajiwara T. Bell pepper fruit fatty acid hydroperoxide lyase is a cytochrome P450 (CYP74B) FEBS Lett. 1996;394:21–24. doi: 10.1016/0014-5793(96)00924-6. [DOI] [PubMed] [Google Scholar]
- Mayer MP, Nievelstein V, Beyer P. Purification and characterization of a NADPH dependent oxidoreductase from chromoplasts of Narcissus pseudonarcissus: a redox-mediator possibly involved in carotenoid desaturation. Plant Physiol Biochem. 1992;30:389–398. [Google Scholar]
- Nievelstein V, Vandekerckove J, Tadros MH, vonLintig J, Nitschke W, Beyer P. Carotene desturation is linked to a respiratory redox pathway in Narcissus pseudonarcissus chromoplast membranes: involvement of a 23-kDa oxygen-evolving-complex-like protein. Eur J Biochem. 1995;233:864–872. doi: 10.1111/j.1432-1033.1995.864_3.x. [DOI] [PubMed] [Google Scholar]
- Norris SR, Barrette TR, DellaPenna D. Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell. 1995;7:2139–2149. doi: 10.1105/tpc.7.12.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads DM, Umbach AL, Sweet CR, Lennon AM, Rauch GS, Siedow JN. Regulation of the cyanide-resistant alternative oxidase of plant mitochondria: identification of the cysteine residue involved in α-keto acid stimulation and intersubunit disulfide bond formation. J Biol Chem. 1998;273:30750–30756. doi: 10.1074/jbc.273.46.30750. [DOI] [PubMed] [Google Scholar]
- Ronen G, Cohen M, Zamir D, Hirschberg J. Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. Plant J. 1999;17:341–351. doi: 10.1046/j.1365-313x.1999.00381.x. [DOI] [PubMed] [Google Scholar]
- Schultz A, Ort O, Beyer P, Kleinig H. SC-0051, A 2-benzoyl-cyclohexane-1,3-dione bleaching herbicide, is a potent inhibitor of the enzyme p-hydroxyphenylpyruvate dioxygenase. FEBS Lett. 1993;318:161–166. doi: 10.1016/0014-5793(93)80013-k. [DOI] [PubMed] [Google Scholar]
- Scolnik PA, Hinton P, Greenblatt IM, Giuliano G, Delanoy MR, Spector DL, Pollock D. Somatic instability of carotenoid biosynthesis in the tomato ghost mutant and its effect on plastid development. Planta. 1987;171:11–18. doi: 10.1007/BF00395063. [DOI] [PubMed] [Google Scholar]
- Siedow JN, Umbach AL. Plant mitochondrial electron transfer and molecular biology. Plant Cell. 1995;7:821–831. doi: 10.1105/tpc.7.7.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedow JN, Umbach AL, Moore AL. The active site of the cyanide-resistant oxidase from plant mitochondria contains a binuclear iron center. FEBS Lett. 1995;362:10–14. doi: 10.1016/0014-5793(95)00196-g. [DOI] [PubMed] [Google Scholar]
- Tieman DM, Harriman RW, Ramamohan G, Handa AK. An antisense pectin methylesterase gene alters pectin chemistry and soluble solids, in tomato fruits. Plant Cell. 1992;4:667–679. doi: 10.1105/tpc.4.6.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L. Alternative oxidase: from gene to function. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:703–734. doi: 10.1146/annurev.arplant.48.1.703. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L, Yip JYH. Molecular localization of a redox-modulated process regulating plant mitochondrial electron transport. Plant Cell. 1998;10:1551–1560. doi: 10.1105/tpc.10.9.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AM, Moore AL. Structure and function of the plant alternative oxidase: its putative role in the oxygen defense mechanism. Biosci Rep. 1997;17:319–333. doi: 10.1023/a:1027388729586. [DOI] [PubMed] [Google Scholar]
- Wetzel CM, Jiang C-Z, Meehan LJ, Voytas DF, Rodermel SR. Nuclear-organelle interactions: the immutans variegation mutant of Arabidopsis is plastid autonomous and impaired in carotenoid biosynthesis. Plant J. 1994;6:161–175. doi: 10.1046/j.1365-313x.1994.6020161.x. [DOI] [PubMed] [Google Scholar]
- Wu D, Wright DA, Wetzel C, Voytas DF, Rodermel S. The IMMUTANS variegated locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell. 1999;11:43–55. doi: 10.1105/tpc.11.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]