KIF20B is in the Kinesin-6 family that includes KIF23/MKLP1 and KIF20A/MKLP2. Previously we showed that mouse Kif20b regulates cerebral cortex growth and neural stem cell midbodies. Now we show KIF20B has a cell-autonomous role in regulating cytokinetic furrowing and abscission. KIF20B may coordinate late midbody maturation before abscission.
Abstract
Cytokinesis requires the cooperation of many cytoskeletal and membrane regulators. Most of the major players required for cytokinesis are known, but the temporal regulation and adaptations for different cell types are less understood. KIF20B (previously called MPHOSPH1 or MPP1) is a member of the Kinesin-6 family, which also includes the better-known members KIF23/MKLP1 and KIF20A/MKLP2. Previously, we showed that mouse Kif20b is involved in cerebral cortex growth and midbody organization of neural stem cells. Here, using siRNA-mediated knockdown of KIF20B in a human cell line and fixed and live imaging, we show that KIF20B has a cell-autonomous role in cytokinesis. KIF20B depletion affects the speed of both furrow ingression and abscission. It localizes to microtubules of the central spindle and midbody throughout cytokinesis, at sites distinct from the other Kinesin-6 family members. KIF20B is not required for midbody assembly, but may accelerate or coordinate midbody maturation. In particular, KIF20B appears to regulate late steps of maturation including anillin dispersal, ESCRT-III recruitment, and the formation of microtubule constriction sites.
INTRODUCTION
Cytokinesis is fundamentally important to building and renewing tissues, but it is still poorly understood. It consists of two sequential processes: cleavage furrow ingression, which takes minutes, and abscission, which can last more than an hour (for a review, see Green et al., 2012; Mierzwa and Gerlich, 2014). Moreover, cytokinesis has different spatial and temporal regulation in different cell types or at different times in development (Singh and Pohl, 2014; Lenhart and DiNardo, 2015). Although many of the major players of cytokinesis have been defined, how the temporal control of furrowing and abscission is regulated is not well understood. Furthermore, how abscission regulation influences tissue development is only beginning to be explored (Dionne et al., 2015).
After chromosome segregation, interpolar microtubules are bundled in an antiparallel manner to form the central spindle, which is involved in specifying and focusing the plane of cleavage. As the cleavage furrow ingresses to form a thin intercellular bridge, the central spindle microtubules are compacted into a dense structure called the midbody. The midbody serves as a platform to mediate the final severing event, abscission. It contains more than 150 proteins and lipids (Skop et al., 2004; Atilla-Gokcumen et al., 2014). During midbody assembly, these proteins are partitioned into distinct subdomains (Elia et al., 2011; Green et al., 2012; Hu et al., 2012). The midbody core or “dark zone” is at the center where microtubules of the two spindle halves overlap in an antiparallel arrangement and are surrounded by electron-dense material (Mullins and Biesele, 1977). PRC1 organizes the antiparallel microtubules in the central spindle and then localizes to the midbody core (Mollinari et al., 2002; Hu et al., 2012) Around the midbody core is a bulge that appears as a ring by light microscopy. The ring contains the scaffolding protein anillin. Tightly packed parallel microtubules emanate from either side of the midbody core and form the midbody flanks. The flanks are labeled by an AuroraB kinase signal that terminates near the eventual abscission site(s). After midbody assembly, CEP55 is recruited to the midbody center where it helps form the bulge and scaffolds sequential recruitment of ESCRT-I and -III components on either side of the bulge (Zhao et al., 2006; Morita et al., 2007; Lee et al., 2008). As the time for abscission gets nearer, constriction sites (also called secondary ingressions) form on one or both sides of the midbody bulge. There the microtubules are even more tightly packed (Mullins and Biesele, 1977). How the constriction sites are positioned and formed is not understood, but ESCRT-III filaments and endosomes are thought to be involved (Elia et al., 2011; Guizetti et al., 2011; Schiel et al., 2012). Together the ESCRT disassembly factor VPS4 and the microtubule-depolymerizing enzyme spastin are thought to complete abscission by coordinating membrane scission and microtubule severing (Morita et al., 2007; Elia et al., 2011; Guizetti et al., 2011). In HeLa cells and MDCK cells, a second abscission occurs on the other flank to release the midbody (Elia et al., 2011; Guizetti et al., 2011). Other reports have observed abscission on only one side, and this may depend on cell type or daughter fates (Ettinger et al., 2011; Kuo et al., 2011).
Microtubule motors are crucial for mediating the cytoskeletal reorganizations that take place during cell division. The Kinesin-6 family members are thought to have roles in cytokinesis and cancer (Baron and Barr, 2015). The family is defined by homology in the motor domain; the stalks and tails are divergent (Dagenbach and Endow, 2004; Miki et al., 2005). They are distinguished by a long insertion in loop-6 of the motor domain, and a relatively long neck domain that may enable the two heads in a homodimer to bridge longer distances than adjacent tubulin binding sites. They homodimerize through their coiled-coil domains in the stalks. Caenorhabditis elegans has one member of this gene family, zen-4. Drosophila has two members, pavarotti and subito. Vertebrates have three members of the Kinesin-6 family: KIF23/MKLP1, KIF20A/MKLP2, and KIF20B (previously called Mphosph1 or Mpp1). KIF23/MKLP1 partners with MgcRacGAP to promote central spindle assembly (Glotzer, 2005; Nishimura and Yonemura, 2006). Later, it localizes to the midbody bulge and is required for a stable midbody to form (Matuliene and Kuriyama, 2002; Zhu et al., 2005a,b). KIF20A/MKLP2 localizes to the midbody flanks, and is required to recruit AuroraB kinase there (Neef et al., 2003; Gruneberg et al., 2004; Zhu et al., 2005a). KIF20B apparently evolved in the vertebrate lineage to become structurally divergent, with an extraordinarily long stalk (∼1000 amino acids) containing three hinges that may allow increased flexibility (Kamimoto et al., 2001; Dagenbach and Endow, 2004; Miki et al., 2005). In cell-free assays, KIF20B acted as a slow, plus-end directed microtubule motor, and was sufficient to slide and bundle microtubules (Abaza et al., 2003). When knocked down in human HCT116 cells, a subset of cells failed cytokinesis, and sometimes underwent apoptosis (Abaza et al., 2003). However, the role of KIF20B in cytokinesis is not known.
We previously isolated a Kif20b mouse mutant in a genetic screen for genes involved in cerebral cortex development (Dwyer et al., 2011; Janisch, Vock, et al., 2013). The mutant embryos have small brains (microcephaly) with increased apoptosis. Mutant neurons have wider, more branched axons (McNeely, Cupp, et al., 2017). In the embryonic neural stem cells in mutant brains, no changes in mitotic indices or cleavage angles were evident, but abnormalities in cytokinetic midbodies were significant. Midbodies had altered shape and organization at the apical membrane (Janisch, Vock, et al., 2013; Janisch and Dwyer, 2016). We hypothesized that defective or failed abscission in a subset of neural stem cells caused them to undergo apoptosis, thus depleting the progenitor pool, reducing neurogenesis, and leading to a small brain.
Motivated by a desire to understand mechanisms of abscission in polarized neural stem cells, but hindered by the dearth of data about KIF20B’s role in cytokinesis, we set out to investigate it in a simpler more tractable system, to generate hypotheses that can then be tested in primary mouse neural cells and intact brain tissue. In the mutant brains, the observed phenotypes could be due to non-cell-autonomous roles of KIF20B, or secondary effects from previous defective divisions. We chose the HeLa human cancer cell line because the cells proliferate as isolated cells, and have a flat morphology, enabling easier imaging of cytokinesis and of the cytoskeleton. They are readily transfected, tolerant of live imaging, and many more antibodies are made against human proteins than mouse. Finally, since the majority of studies of abscission proteins have used HeLa cells, it is an essential baseline cell type for comparison.
RESULTS
KIF20B protein localizes to microtubules in the central spindle and midbody throughout cytokinesis, in a pattern distinct from that of other Kinesin-6 family members
Before analyzing the specific role of KIF20B in cytokinesis in HeLa cells, we first sought to determine its detailed subcellular localization in relation to the microtubule cytoskeleton during cytokinesis. To do this, we coimmunostained HeLa cells for endogenous KIF20B along with α-tubulin. KIF20B is a low abundance protein, but it is readily detected in dividing cells when concentrated in cytokinetic structures. In cells fixed in anaphase, KIF20B signal appears as puncta on the microtubules of the central spindle (Figure 1A). In cells with more deeply ingressed furrows, KIF20B is more concentrated in the center of the central spindle as a band (Figure 1B). In early midbodies, KIF20B signal is detected in two disks on either side of the midbody core or “dark zone” (arrowhead; Figure 1, C and D). In late midbodies, KIF20B signal is more extended along the midbody flanks (Figure 1E). If microtubule constriction sites are visible, KIF20B surrounds them, in this case appearing as four distinct spots on the midbody (Figure 1F). In cells that appear to have undergone abscission (with a large gap in tubulin staining on one side of the midbody), KIF20B can be detected still surrounding the dark zone (arrowheads), and on both flanks on either side of the abscission site (Figure 1, G–I). In a few cases where a microtubule strand remained connecting the sister cells, they were decorated with dots of KIF20B (arrows in Figure 1, G–G″). Overexpression of a GFP-tagged full-length KIF20B in cell lines or neurons usually causes cell death, as previously reported by our group and others (Abaza et al., 2003; McNeely, Cupp, et al., 2017). However, a small number of surviving cells expressing GFP-KIF20B showed a similar localization as endogenous KIF20B detected by immunostaining, flanking the dark zone (Figure 1, J–J″). Together these data suggest that KIF20B first associates with central spindle microtubules starting in early anaphase, then accumulates at microtubule plus ends that coincide with the plane of furrow ingression, and finally remains concentrated at the narrowest portions of the midbody surrounding the dark zone and constriction sites throughout the abscission process.
FIGURE 1:
KIF20B localizes to the central spindle and midbody throughout cytokinesis in HeLa cells. Panels A–I show immunofluorescence staining for endogenous KIF20B (green) and tubulin (red), at sequential phases of cytokinesis from anaphase furrowing (A) to postabscission (I). Arrowheads point to the central dark zone in all pictures. (A–B″) During anaphase, KIF20B starts to accumulate as speckles along the microtubules of the central spindle (A′), and in later furrows forming a dense band in the middle of the central spindle (B′). (C–D″) In early midbodies, KIF20B accumulates on the inner flanks of the midbody surrounding the dark zone (arrowhead), forming a cap-like structure. (E–F″) In late midbody stage, KIF20B spreads out on the midbody flanks surrounding the constriction sites, resulting in four distinct spots of KIF20B localization. (G–I″) Near abscission in very thin midbodies, small spots of KIF20B can be seen still surrounding the central dark zone (arrowhead). The small arrows in the G panels point to KIF20B dots localizing along a strand of microtubules. (J–J″) GFP-KIF20B expressed in HeLa cell shows the same localization within the midbody as detected by antibodies to KIF20B. Scale bars represent 5 µm for A–I″ and 10 µm for panel J.
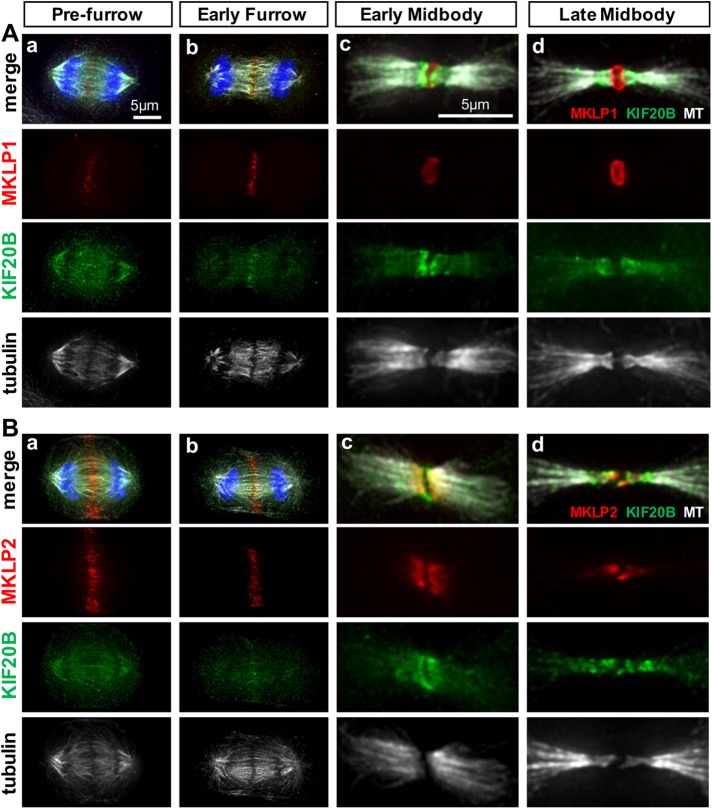
To compare the subcellular localization of endogenous KIF20B with those of the related Kinesin-6 family members MKLP1/KIF23 and MKLP2/KIF20A, we performed double immunolabeling and imaged cells at four stages of cytokinesis: postanaphase prefurrow, early furrow, early midbody, and late midbody (Figure 2). Interestingly, in cells that have segregated chromosomes but not yet begun furrow ingression (prefurrow), KIF20B signal is widely distributed on the central spindle and mitotic spindle microtubules, whereas both MKLP1 and MKLP2 are enriched at the central spindle midzone (Figure 2, Aa and Ba). In early furrows, all three family members are accumulated at the central spindle midzone where microtubule plus ends overlap (Figure 2, Ab and Bb). In the early midbody, MKLP1 is seen as a ring around the midbody core (Figure 2Ac), whereas MKLP2 and KIF20B are similarly localized on the inner flanks adjacent to the core, with KIF20B extending more widely than MKLP2 (Figure 2Bc). Late midbodies display this difference even more clearly. MKLP1 remains strictly at the midbody center (Figure 2Ad). KIF20B overlaps MKLP2 on the inner flanks, but shows additional enriched signal distally around the microtubule constriction sites (Figure 2Bd). These data show that KIF20B localizes differently than its other subfamily members, accumulating at the midzone slightly later in anaphase than either MKLP1 or MKLP2, and localizing more widely on the midbody flanks than MKLP2, surrounding the constriction sites. These data concur with our previous data in mouse cells showing that Kif20b protein localization overlapped with that of Aurora B kinase in the midbody flanks, but was more enriched on the outer flanks of Aurora B signal (Janisch, Vock, et al., 2013). In addition, these data support the notion that the three Kinesin-6 family members have different functions in abscission.
FIGURE 2:
KIF20B protein shows overlapping but distinct localization from MKLP1/KIF23 and MKLP2/KIF20A during cytokinesis. (A) Immunostaining HeLa cells for endogenous MKLP1 (red) and KIF20B (green) with α-tubulin (white) shows distinct localizations at prefurrow (a), early furrow (b), early midbody (c), and late midbody (d) stages. (B) Immunostaining for endogenous MKLP2 (red) and KIF20B (green) with α-tubulin (white) shows substantial overlap in the early furrow (b) and early midbody (c) stages, but that KIF20B is more broadly distributed on the central spindle in the prefurrow stage (a) and is enriched on the outer flanks of the constriction sites in the late midbody stage (d).
Knockdown of KIF20B increases multinucleate cells
To investigate the cell-autonomous requirement and primary role(s) of KIF20B in cell division, we depleted endogenous KIF20B from HeLa cells using small interfering RNA (siRNA) transfections. We used a previously published KIF20B-specific siRNA sequence (“siRNA1” from Abaza et al., 2003), and confirmed that it depletes endogenous KIF20B to undetectable levels in 98% of HeLa cell midbodies by 24 h posttransfection and did not deplete other Kinesin-6 family members (see Materials and Methods and Supplemental Figure 1 for details). First, we compared general mitotic and cytokinesis parameters between asynchronous cells transfected with control siRNA (siLUC) or siKIF20B (siKIF), and fixed 24 or 48 h after transfection. Depletion of KIF20B did not significantly change the mitotic index or midbody index, but only slightly reduced the fraction of mitotic cells in telophase detected at 48 h (Figure 3, A–C). However, KIF20B depletion resulted in a 2.5-fold increase in the occurrence of multinucleated cells (with two or more clearly distinct nuclei) at 24 h, and a striking increase in multilobed nuclei at 48 h posttransfection (Figure 3, D–H).Two additional siRNAs targeting different KIF20B sequences also increased the rates of multinucleation and multi-lobed nuclei (Supplemental Figure 1, D and E). Multilobed nuclei are likely a later, secondary consequence of cytokinesis failure—the fusion of two or more nuclei (Neumann et al., 2010). Finally, KIF20B knockdown caused a small but significant increase in apoptosis at 24 h (Figure 3I). Together these data support the conclusion that in dissociated human cells as well as in the developing mouse brain, KIF20B has a role in cytokinesis.
FIGURE 3:
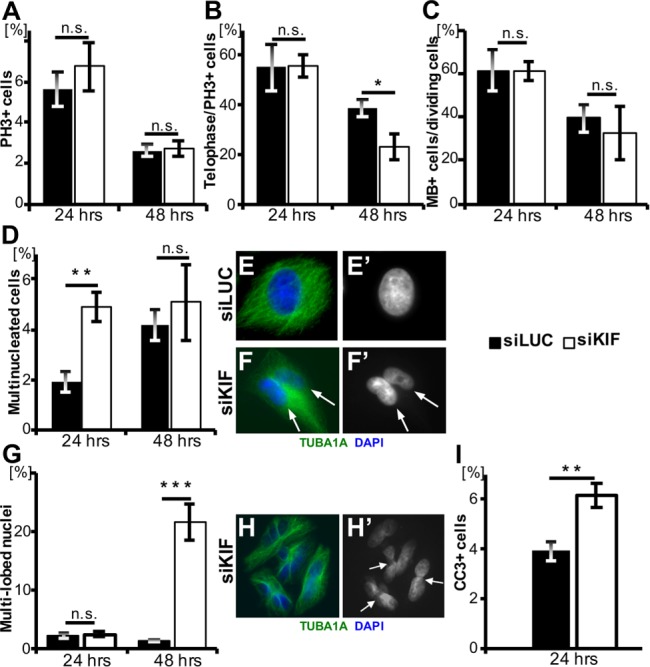
Cytokinesis defects in KIF20B-depleted asynchronous HeLa cell cultures. (A) The average mitotic index was not altered by KIF20B depletion at 24 h (n = 5 coverslips/treatment, siLUC = 1192 cells, siKIF = 1408 cells) or 48 h posttransfection (n = 7 coverslips/treatment, siLUC = 1870 cells, siKIF = 2513 cells). Mitotic index was defined as the number of phosphohistone H3 positive (PH3+) cells divided by the total cell count by DAPI+ nuclei. PH3 immunostaining signal is strong in prophase, metaphase, and anaphase; weak in telophase; and absent in posttelophase late midbody stage cells. (B) Average percentage of telophase cells out of mitotic cells (PH3+) was not changed at 24 h but decreased at 48 h posttransfection in siKIF knockdown cells (p = 0.038). Telophase was characterized by the presence of condensed chromatin. For 24 h, n = 5 coverslips/treatment (siLUC = 67 cells; siKIF = 95 cells); for 48 h posttransfection, n = 7 coverslips/treatment (siLUC = 49 cells; siKIF = 69 cells). (C) The average percentage of midbody stage cells out of all dividing cells (PH3+ or PH3− with a midbody) was not significantly different in siKIF-treated cells. At 24 h, n = 8 coverslips/treatment, with 2556 total siLUC cells, and 3048 total siKIF cells; at 48 h posttransfection, n = 6 coverslips/treatment, with 952 total siLUC cells, and 1176 total siKIF cells). (D) The percentage of multinucleate cells out of total cells was significantly increased in the KIF20B-depleted cultures at 24 h posttransfection (p = 0.005). For 24 h, n = 5 coverslips/treatment with 1192 total siLUC cells and 1408 total siKIF cells. For 48 h, n = 6 coverslips/treatment, with 1870 total siLUC cells and 2513 total siKIF cells). (E, E′) A control siLUC HeLa cell stained with α-tubulin (TUBA1A) and DAPI with a single nucleus. (F, F′) siKIF transfected cell stained with TUBA1A and DAPI showing two nuclei within the same cell (white arrows). (G) The percentage of cells with multi-lobed nuclei was significantly greater in the knockdown cells than in the control cells (p = 4.77 × 10−5) at 48 h posttransfection. For 24 h, n = 5 coverslips/treatment, with 1192 total siLUC cells and 1408 total siKIF cells. For 48 h, n = 6 coverslips/treatment, with 1870 total siLUC cells and 2513 siKIF cells. (H, H′) siKIF transfected cells stained with TUBA1A and DAPI to show multi-lobed phenotype. White arrows point at constrictions within single nuclei. (I) Average apoptotic index (cleaved-caspase-3 CC3+ out of total cells) was increased after siKIF treatment (p = 0.006). n = 8 coverslips/treatment, with 1878 siLUC cells and 1486 siKIF cells. Coverslips were prepared from three independent siRNA transfection experiments. p Values stated for Student’s t test.
Furrow ingression is slower in KIF20B-depleted cells
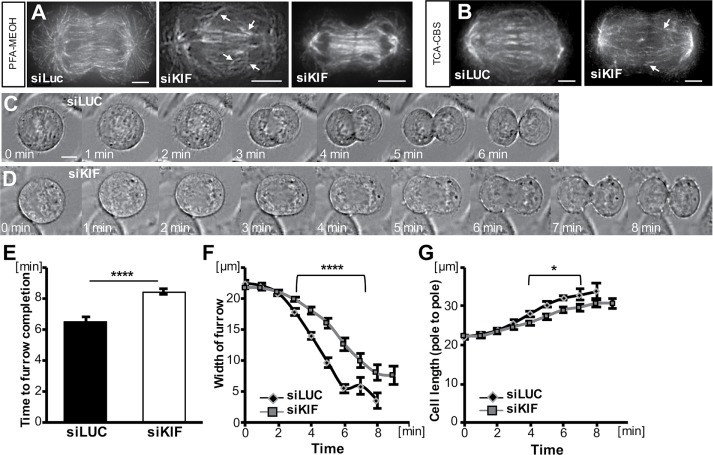
Next, we sought to analyze cytokinesis in KIF20B-depleted cells in more detail. Analyses were done at 24 h posttransfection of siRNA, since KIF20B was depleted and phenotypes were already observed. First, we examined the cleavage furrowing stage. In fixed cell images, we noticed that the central spindles of anaphase cells in the siKIF20B-treated cultures sometimes appeared disorganized, with nonparallel microtubule bundles or asymmetric gaps (Figure 4, A and B, arrowheads), but often appeared symmetric (Figure 4A, right cell, and Supplemental Figure 2). In blinded scoring, 7 of 18 (39%) siKIF20B-treated furrowing cells had discernible disorganization in their central spindles, whereas only 6 of 31 (19%) siLUC-treated furrowing cells did. Although these sample sizes are too small and the microtubule spacing too variable during furrow ingression for statistical significance, these data suggest that KIF20B-depleted anaphase cells may have increased rates of irregularities in the central spindle. Together with the KIF20B localization on the central spindle microtubules shown in Figures 1 and 2, it seems plausible that KIF20B could help organize or stabilize microtubule bundles during anaphase to telophase.
FIGURE 4:
Cleavage furrow ingression is slower in KIF20B-depleted cells. (A, B) Example images of central spindles of furrowing cells fixed with either PFA-MeOH (A) or TCA-CBS (B) and stained for α-tubulin (TUBA1A). Central spindles of some KIF20B-depleted cells appear to have irregularities in the angles or left-right symmetry of the microtubule bundles (arrowheads). (C, D) Representative brightfield time-lapse live images of an siLUC-treated cell (C) that completed furrowing in 6 min and an siKIF-treated cell (D) that completed furrowing in 8 min. (E) Average total time from anaphase onset (chromosome segregation onset) to completion of cleavage furrowing ingression was increased in siKIF cells (****: p = 3 × 10−7). (F) Furrow widths of siKIF cells decreased steadily but more slowly than those of siLUC cells (for points under bracket; ****: p = 3 × 10−5). (G) Cell lengths (pole to pole) of siKIF20B-treated cells also increased steadily but more slowly than in siLUC cells (*: p ≤ 0.05). For E–G, nsiLUC = 20 cells and nsiKIF = 24 cells across five independent imaging sessions. t = 0 was the last time point before anaphase onset (chromosome segregation) was detectable. Scale bars represent 10 µm.
Because the anaphase spindle regulates cleavage furrow positioning and ingression (Green et al., 2012), this notion prompted us to examine the kinetics of furrow ingression in live cell time-lapse imaging. By collecting images every 1 min as control or KIF20B-depleted mitotic cells progressed through cleavage, we observed in each case that furrows appeared qualitatively normal, occurring only at the cell equator, ingressing steadily, and never regressing (Figure 4, C and D). However, there was a quantitative difference. The average total duration of furrow ingression from anaphase onset to completion was significantly increased in KIF20B-depleted cells, from 6.5 min to 8.4 min (Figure 4E). Plotting furrow width over time demonstrates that the onset of furrow ingression is similar, but ingression proceeds at a slower rate in siKIF-treated cells (Figure 4F). A smaller but similar reduction was seen in the rate of cell lengthening from pole to pole (Figure 4G). Together these data show that, although KIF20B is not required for central spindle formation or furrow ingression, it may contribute to central spindle organization and promote rapid furrow ingression.
KIF20B loss alters midbody width but not subdomain structure
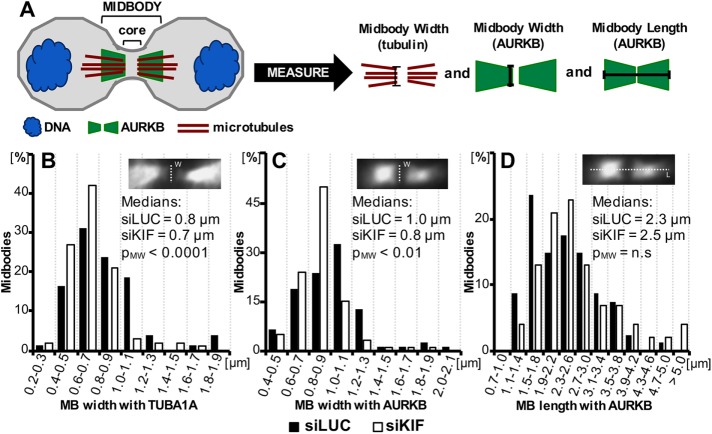
We previously showed that in embryonic mouse brains, loss of Kif20b disrupted the shapes and positioning of neural stem cell midbodies. Midbodies still formed at the apical membrane of the neuroepithelium, but were more often misaligned, and had an altered distribution of axis ratios, primarily due to increased width (Janisch, Vock, et al., 2013; Janisch and Dwyer, 2016). To test whether this phenotype reflects a cell-autonomous primary requirement for KIF20B in abscission, and whether it occurs in cells not contained in a polarized epithelium, we measured the lengths and widths of midbodies of HeLa cells treated with control or KIF20B siRNA (Figure 5A). Indeed, we found that siKIF20B-treated cells did have a significantly shifted distribution of midbody widths, but surprisingly there were more thin midbodies and fewer wide midbodies. This was observed with either tubulin or Aurora B kinase immunostaining (Figure 5, B and C). Midbodies also tended to be longer when KIF20B was depleted, but the shift was not statistically significant (Figure 5D). Together with our previously published midbody shape analyses in embryonic mouse brains, these data show that loss of KIF20B causes changes in the abscission stage of cytokinesis in a cell-autonomous manner.
FIGURE 5:
Midbodies tend to be thinner in KIF20B-depleted HeLa cells. 24 h after transfection with control (siLUC) or KIF20B (siKIF) siRNA, cells were fixed and immunostained for α-tubulin (TUBA1A) and Aurora B kinase (AURKB). Midbodies were imaged and measured for width and length as shown in A and insets. (A) Schematic of midbody (not to scale) and measurements. The midbody core “dark zone” contains dense overlapping microtubules, but does not stain with tubulin or Aurora B antibodies due to density. Midbody widths were measured adjacent to the dark zone by both tubulin and AURKB signal. Lengths were measured by AURKB signal only. The midbody “edge” was considered to be where no signal was detected. (B, C) Plots of the distributions of midbody widths for KIF20B-depleted cells (siKIF, white bars) and controls (siLUC, black bars) show that KIF20B-depleted cells more often have thin midbodies when measured by TUBA1A or AURKB signal (medians: siLUC = 0.8 µm, siKIF = 0.7 µm, pM-W < 0.0001, and distribution shape pK-S = 0.0205; and siLUC = 1.0 µm, siKIF = 0.8 µm, pM-W < 0.01, and distribution shape pK-S = 0.000419, respectively). (D) Median midbody length, as measured with AURKB signal, shows a trend to be increased in siKIF-treated cells, but does not reach statistical significance (pM-W = 0.28; pK-S = 0.85). p Values (pM-W) for medians are calculated with Mann-Whitney U test; p values (pK-S) for distribution shape are calculated with a Kolmogorov-Smirnov test; n.s., not significant. nsiLUC = 80 midbodies and nsiKIF = 100 midbodies, from three independent siRNA transfections.
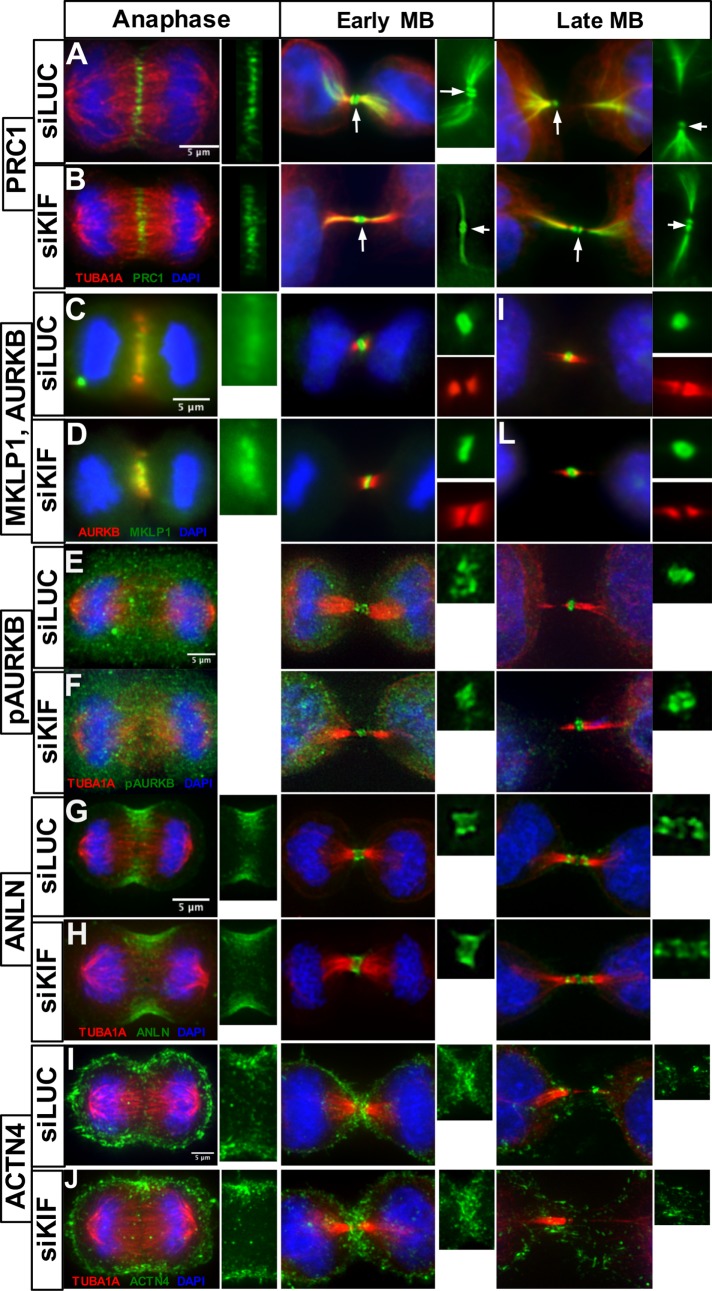
We hypothesized that KIF20B may regulate midbody shape directly or indirectly by binding microtubules or by localizing effectors to midbody microtubules. To further investigate midbody structure and protein recruitment when KIF20B is depleted, we took advantage of the wider variety of antibodies that work for immunofluorescence on human cells than on mouse cells. We tested whether the major subdomains of the midbody form when KIF20B is depleted, and whether several key regulators of cytokinesis are localized properly in the subdomains as well as in furrows (Figure 6). Protein regulator of cytokinesis 1 (PRC1), which is required for formation of the central spindle (Mollinari et al., 2002; Zhu et al., 2006) and was shown to interact with KIF20B (Kanehira et al., 2007), shows normal localization at the center of the central spindle in anaphase, and in the midbody in two disks in the dark zone, as well as the flanks. This pattern was not disrupted in KIF20B-depleted cells (Figure 6, A and B). The Kinesin-6 family member KIF23/MKLP1, required for midbody formation and abscission (Matuliene and Kuriyama, 2002; Zhu et al., 2005a) localizes normally to the central spindle and to the midbody center in KIF20B-depleted cells (Figure 6, C and D). Aurora B kinase (AURKB) localizes to the central spindle during furrowing, and on the flanks of the midbody in both control and siKIF20B-treated cells (Figure 6, C and D). Activated Aurora B kinase phosphorylated at Thr-232 (pAURKB), which was shown to regulate the abscission checkpoint (Steigemann et al., 2009; Carlton et al., 2012), localizes to the center of the midbody in both control and KIF20B-depleted cells (Figure 6, E and F). Anillin (ANLN), a scaffold that cross-links filaments in the contractile furrow membrane, does not require KIF20B for recruitment. In both control and siKIF-treated cells, it localizes to the furrowing membrane, remains in the midbody as a ring around the central bulge at early stages, and later localizes to both the central bulge and constriction sites (Figure 6, G and H). α-Actinin-4 (ACTN4), which cross-links actin and regulates cytokinetic furrowing (Mukhina et al., 2007), was pulled down as a candidate binding protein with KIF20B (Maliga et al., 2013). However, it appears indistinguishable in the furrows of KIF20B-depleted cells, and as expected, is not enriched in midbodies of control or KIF20B-depleted cells (Figure 6, I and J). These data demonstrate that KIF20B is not required for recruitment of several key proteins to the midbody, and moreover that when KIF20B is depleted, the primary midbody subdomains of central dark zone, bulge, and flanks are specified.
FIGURE 6:
Midbody assembly appears normal in KIF20B-depleted cells. (A, B) PRC1 localizes to the central spindle in anaphase in both siLUC and siKIF cells. In early and late midbodies (MB), PRC1 lines the microtubules of the midbody flanks and extends into the cell on the microtubule network. It also forms two distinct disks around the core of the midbody (white arrow). (C, D) MKLP1/KIF23 localizes to the central spindle in anaphase in siLUC and siKIF cells. In early and late midbodies, it is found in the center of the midbody (dark zone) in control or KIF20B depleted cells. By contrast, Aurora kinase B (AURKB) localizes to the flanks of the midbody in early and late stages in both siLUC and siKIF cells. (E, F) Phospho-T232-Aurora B (pAURKB), representing “activated” Aurora B kinase, is diffusely localized during anaphase in both siLUC and siKIF cells. In early midbodies, pAURKB localizes to the center dark zone and inner flanks as a diffuse blob, but appears as a more compact disk in later midbodies, similar in siLUC and siKIF cells. (G, H) Anillin (ANLN) localizes to the furrowing cell cortex during anaphase in siLUC and siKIF cells. In the early midbody stage, ANLN forms a wide ring around the center of the midbody. In late midbodies, ANLN can be found in the center of the midbody as well as at the constriction sites. These localizations were not disrupted in KIF20B-depleted cells. (I, J) α-Actinin-4 (ACTN4) is distributed on the entire cell cortex but clearly accumulates in the cleavage furrow during anaphase in both siLUC and siKIF cells. In early midbody stage, ACTN4 enriches in a half-circle shape at the edges of both daughter cells underneath the midbody. In the late midbody stage, there is no longer enrichment of ACTN4 around the midbody, in both siLUC and siKIF cells. Images in panels A and B and E–J were taken with a DeltaVision deconvolution microscope and those in C and D with a wide-field microscope. At least 15 midbody-stage and five anaphase cells imaged for each marker and condition. Scale bars are 5 µm for all images.
KIF20B depletion disrupts late midbody maturation
Because the higher proportion of thin and long midbodies in KIF20B-depleted cells suggested a delay in abscission (Figure 5), but the major midbody subdomains appear to assemble normally (Figure 6), we sought to further characterize the late-stage midbodies. First, we quantified midbodies showing evidence of a late event—microtubule constriction sites. In fixed midbodies immunostained for tubulin, constriction sites appear as pinches or gaps in microtubule staining a few microns away from the central bulge, and are the presumed abscission sites (Guizetti et al., 2011; Hu et al., 2012). These constriction sites (also called secondary ingressions) may be visible on one or both sides of the midbody center (Figure 7, A and B). Interestingly, we found that in the KIF20B-depleted cells, a smaller percentage of midbodies had a detectable constriction site (Figure 7C). This suggests a defect in the structure or formation of constrictions. As a second marker of late midbodies, we analyzed the localization of CEP55, a key abscission regulator that accumulates in late midbodies, starting ∼50–60 min after anaphase onset in HeLa cells (Bastos and Barr, 2010; Guizetti et al., 2011). It localizes to and maintains the structure of the central bulge of the midbody, and then recruits ESCRT proteins to mediate abscission (Zhao et al., 2006; Lee et al., 2008). Accordingly, we observed that in midbodies without constriction sites, CEP55 immunostaining signal appeared in two disks perpendicular to the microtubules inside the central dark zone (Figure 7D, left). In late midbodies with at least one microtubule constriction site, the CEP55 disks appeared closer together, usually still resolvable with deconvolution in most control cells, but less often in KIF20B-depleted cells (Figure 7D, right; arrowheads point to constriction sites). Interestingly, in KIF20B-depleted midbodies with at least one constriction, the CEP55 signal had a greater maximum intensity, although occupying the same area (Figure 7, E and F). Together, the decreased frequency of midbodies with constriction sites, and increased CEP55 intensity in the midbodies that do have constrictions, suggest that KIF20B depletion causes a defect or delay in a late stage of midbody maturation, between CEP55 recruitment and the formation of constriction sites.
FIGURE 7:
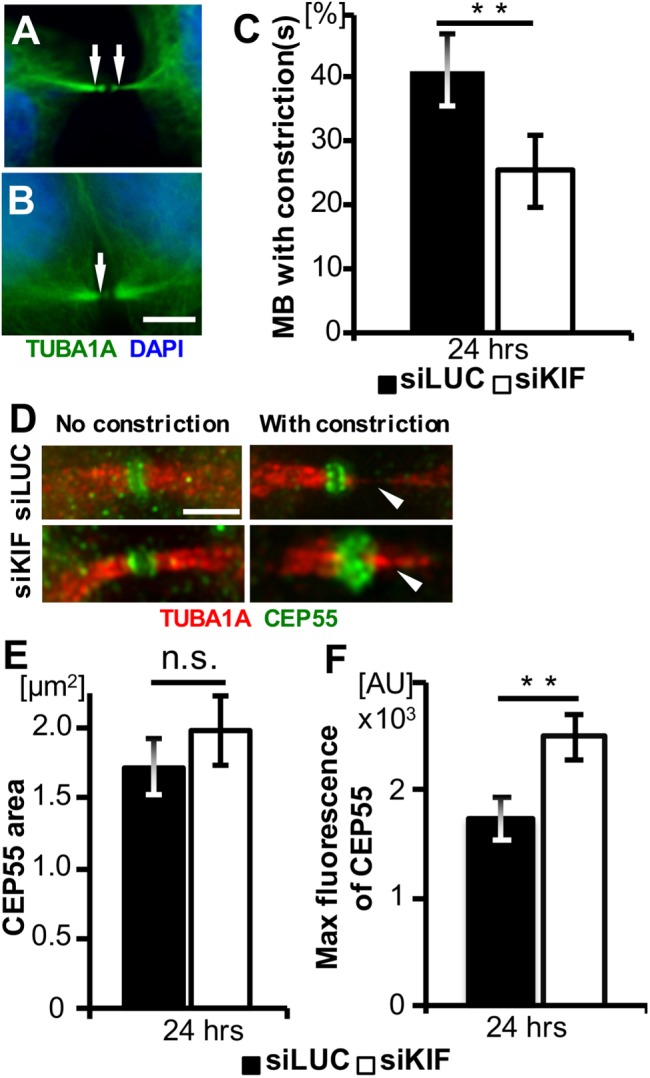
Midbodies of KIF20B-depleted cells show reduced frequency of constriction sites and increased CEP55 intensity. (A, B) Representative wide-field images of tubulin in midbodies with one or two constriction sites visible (arrows). (C) The average percentage of midbodies having at least one visible constriction site was reduced in KIF20B-depleted cells at 24 h posttransfection (**: p = 0.01, n = 5 experiments with two coverslips each). (D) Representative deconvolved images of endogenous CEP55 localization to the midbody bulge in both siLUC- and siKIF20B-treated cells. In midbodies without constriction sites, CEP55 appears in two distinct disks. In late midbodies having constriction sites (arrowheads), CEP55 is denser but can still be resolved into two disks by deconvolution in control cells, but not in siKIF-treated cells. (E) The average area of CEP55 signal in the core of late midbodies with constriction sites is not significantly different between siLUC- and siKIF-treated cells. (F) The average maximum fluorescence intensity of CEP55 is significantly higher in late midbodies with constriction sites after KIF20B depletion. **: p = 0.01, n = 26 siLUC and 17 siKIF late midbodies. p Values calculated with a two-tailed Student’s t test, except that in C a paired t test was used. Scale bars for A and B, 5 µm; for D–K, 2.5 µm.
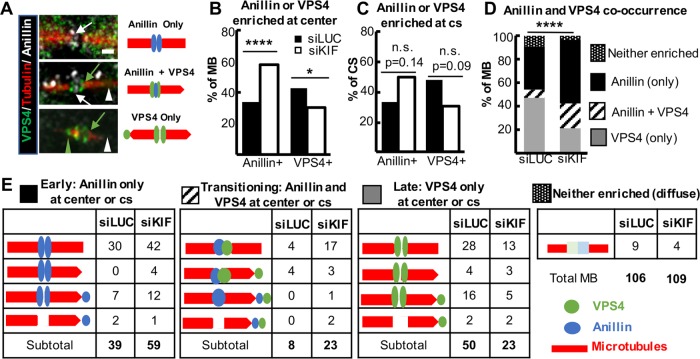
It is not well understood how late-stage midbodies form constriction sites in preparation for final abscission, but may involve both membrane constriction by ESCRT-III filament assembly and turnover, and localized microtubule severing by spastin (Connell et al., 2009; Elia et al., 2011; Guizetti et al., 2011; Schiel et al., 2011; Mierzwa et al., 2017). Spastin is recruited to late midbodies first to the central bulge and then to constriction sites (Gershony et al., 2014). This recruitment pattern of spastin does not appear to require KIF20B, as it was observed in most control and KIF20B-depleted midbodies (Supplemental Figure 3). To analyze ESCRT-III recruitment, we performed double immunostaining with anillin and VPS4, the most downstream component of the ESCRT-III machinery that mediates filament formation and turnover, and is enriched just before abscission (Elia et al., 2011; Mierzwa et al., 2017). It was shown that in late midbodies, anillin and ESCRT-III both localize to the midbody center and constriction sites, but that anillin dissipates and disappears as ESCRT-III accumulates (Renshaw et al., 2014). Accordingly, we observed by double immunostaining that midbodies variously showed enrichment of anillin only (early stage), both anillin and VPS4 (transitional), or VPS4 only (latest stage; Figure 8A). Interestingly, we found that among KIF20B-depleted midbodies, almost twice as many had anillin enriched at the center bulge compared with control midbodies, and a higher percentage had anillin enrichment at constriction sites also (Figure 8, B, C, and E). However, the frequency of observations of VPS4 enrichment was reversed, with a smaller proportion of midbodies showing VPS4 at the center or constrictions when KIF20B was depleted (Figure 8, B, C, and E). Categorizing the midbodies by whether they had enriched anillin only, VPS4 only, or both anillin and VPS4, we found that the population was shifted in the KIF20B-depleted cells to the earlier and transitional stage categories (Figure 8, D and E). The KIF20B-depleted midbodies were 50% more likely to be anillin-only enriched, more than twice as likely to be transitional, with both anillin and VPS4 enrichment, but only half as likely to be late-stage VPS4 only. These data suggest that KIF20B is not required for VPS4 recruitment to the midbody bulge and constriction sites (nor presumably for upstream ESCRT-III components), but that its activity may enhance the speed or extent of VPS4 recruitment. Alternatively, KIF20B function may aid anillin dispersal. In any case, the data support the notion that KIF20B functions to accelerate or coordinate late-stage midbody maturation processes.
FIGURE 8:
Pattern of anillin and VPS4 recruitment is consistent with late-stage maturation defect in KIF20B-depleted midbodies. (A) Left, example deconvolution images of endogenous VPS4 and anillin localization in representative midbodies of HeLa cells. Right, schematic representations of staining in images on left. Arrows point to the central bulge region, and arrowheads point to constriction sites. White, anillin; green, VPS4; red, tubulin. Scale bar, 1 µm. (B) The percentage of midbodies with anillin enriched in the center is increased, and the percentage with VPS4 enriched in the center is decreased. n = 106 siLUC-treated cells, and 109 siKIF-treated cells from two coverslips each of two independent experiments. (C) The percentage of constriction sites (cs) that have anillin enrichment was increased, while the percentage of constriction sites that have VPS4 enrichment was decreased, in KIF20B depleted midbodies, but did not reach statistical significance. n = 48 siLUC constriction sites in 38 midbodies, and n = 46 siKIF constriction sites in 34 midbodies. (D) Bar plot of anillin and VPS4 co-occurrence in midbodies shows KIF20B-depleted midbodies are significantly shifted out of the latest-stage category (VPS4-only) and into the early (anillin-only) and transitional (anillin plus VPS4) categories. n = 106 siLUC-treated midbodies, n = 109 siKIF-treated midbodies. (E) Detailed schematic representations and raw tallies of subcategories of anillin and VPS4 enrichment data plotted in bar graphs in B–D. White space in microtubules symbolizes the central dark zone, and pointed ends symbolize constriction sites (cs). Anillin (blue) or VPS4 (green) enrichment was scored at midbody centers or constriction sites. *p < 0.05; ****p < 0.0001; n.s., not significant (Fisher’s test for B and C, Chi-square test for D).
KIF20B depletion disrupts abscission timing
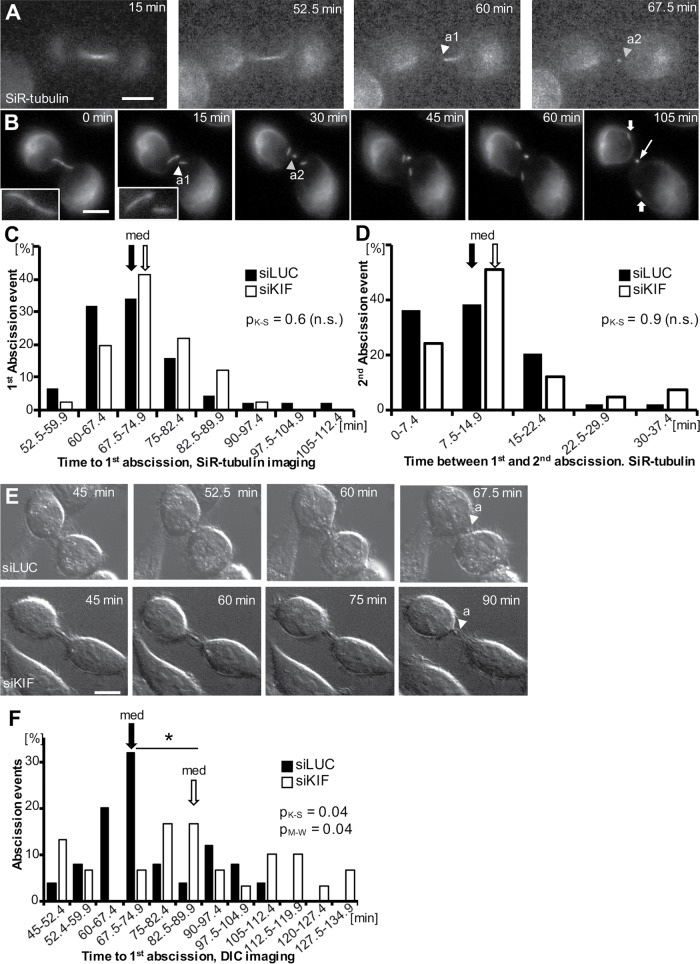
On the basis of the late-stage midbody defects we observed, we decided to directly test whether loss of KIF20B causes delays or failures in abscission by performing live cell time-lapse imaging with siRNA-treated HeLa cells. First, the cell-permeable tubulin dye silicon-rhodamine tubulin (SiR-tubulin) was used to label midbodies and monitor abscission (Figure 9, A–D, and Supplemental Movies 1 and 2). This far-red dye shows a more than 10-fold increase in fluorescence upon binding to polymerized microtubules (Lukinavicius et al., 2014). Figure 9, A and B show example time-lapse images of abscission collected using wide-field microscopy with a 20× dry or 63× oil objective (Figure 9, A and B, and Supplemental Movies 1 and 2). In both cases, the first abscission (a1, white arrowhead) and second abscission (a2, yellow arrowhead) can be observed, and the midbody remnant can be followed for at least 1 h after abscission (Figure 9B, thin arrow at 105 min). Interestingly, at the higher magnification, the severed midbody flanks can also still be resolved as coherent microtubule bundles more than 1 h after the second abscission (Figure 9B, wide arrows at 105 min).
FIGURE 9:
KIF20B depletion delays and dysregulates abscission. (A, B) Example wide-field time-lapse imaging of abscission in a HeLa cell labeled with SiR-tubulin at 20× (A) and 63× (B) (arrowhead a1: abscission 1, arrowhead a2: abscission 2; filled arrow: midbody flanks, open arrow: midbody remnant). Images captured every 7.5 min (A) and every 15 min (B). Insets in B show the intact midbody and the first abscission. (C) Frequency distribution of time from anaphase to first abscission shows a similar median time (67.5 min, arrows, pM-W = 0.13) and similar distribution shape (pK-S = 0.6) for siLUC (black bars) and siKIF-treated cells (white bars) imaged and scored using SiR-tubulin. n = 44 siLUC cells and 41 siKIF20B cells from four experiments. (D) The median time between first and second abscissions (arrows, 7.5 min, pM-W = 0.09) imaged with SiR-tubulin was not different in KIF20B-depleted cells (siKIF) and controls (siLUC). The distributions were not significantly different between siLUC- and sKIF-treated cells (pK-S = 0.9). The sample is the same as in C. (E) Representative selected planes from time-lapse z-stack images of abscission using differential interference contrast (DIC) microscopy, captured every 7.5 min (arrowhead with “a” signifies abscission). Top, in this siLUC-treated cell, abscission was observed 67.5 min after anaphase onset. Bottom, in this siKIF-treated cell, abscission was observed 90 min after anaphase onset. The second abscission event was not discernible by DIC imaging. (F) Increased median time (67.5 vs. 82.5 min) and altered distribution of time to abscission after KIF20B depletion, shown in a frequency graph of time from anaphase to first abscission discerned using DIC time-lapse microscopy in siKIF- (white bars) compared with siLUC-treated cells (black bars). For distribution, *pK-S = 0.04; for medians, *pM-W = 0.04. n = 25 siLUC cells and 30 siKIF cells from four experiments. K-S: Kolmogorov-Smirnov test; M-W: Mann-Whitney U test. Scale bars, 10 µm.
Movie S1.
Example of live cell abscission imaging from Figure 9A using widefield time-lapse imaging with 20x dry objective of aHeLa cell treated with siKIF20B, and labeled with SiR-tubulin to visualize microtubules. Image stacks were collected every 7.5 minutes and maximum intensity projections used to score abscission. Abscission occurred on both sides of the midbody sequentially.
Movie S2.
Example of live cell widefield time-lapse imaging shown in Figure 9Busing a 63x oil objective to image a control HeLa cell stained with SiR-tubulin undergoing abscission on both sides of the midbody sequentially. Images stacks were collected every 15 minutes and converted to maximum intensity projections.
By imaging every 7.5 min, we found that control cells had a median time from anaphase onset to first abscission of 67.5 min (Figure 9C, black bars). This is similar to the timing observed previously in HeLa cells using either GFP-tubulin or photoactivatable dye transfer (Steigemann et al., 2009; Guizetti et al., 2011), suggesting that SiR-tubulin is a valid tool for abscission studies. Surprisingly, however, in KIF20B-depleted cells, the median time to first abscission was not significantly different from controls (Figure 9C, white bars). Because midbody inheritance or release has been proposed to influence daughter cell fate in stem cell divisions (Dubreuil et al., 2007; Ettinger et al., 2011; Kuo et al., 2011; Salzmann et al., 2014), we also wanted to test whether KIF20B regulates the time between first and second abscissions. We found that the second abscission usually occurred within 15 min of the first abscission (∼75% of the cells), in both siLUC- and siKIF-treated cells (Figure 9D). This is similar to but slightly faster than what was reported using GFP-tubulin (Guizetti et al., 2011; Gershony et al., 2017). Interestingly, the cells that had the longest times to first abscission were not the ones that had the longest times between first and second abscissions, suggesting they are independent events (unpublished data).
The lack of a detectable change in abscission timing using SiR-tubulin imaging after KIF20B depletion was surprising, given the late midbody defects we had observed. SiR-tubulin is a derivative of the taxane docetaxel, which stabilizes microtubules, but it is much less toxic and does not arrest mitosis at the dosage we used (Lukinavicius et al., 2014). However, we hypothesized that SiR-tubulin might be slightly stabilizing microtubules and thereby rescuing the effect of loss of KIF20B. To test this hypothesis, we employed a different method of scoring abscission—differential interference contrast (DIC) imaging—without SiR-tubulin (Figure 9E and Supplemental Movies 3 and 4). With this method, only the first abscission event could be discerned with confidence. The median time to first abscission in control siRNA cells was 67.5 min, the same as observed with SiR-tubulin (Figure 9, C and F, black arrows). Remarkably, however, we found that in siKIF20B-treated cells, abscission timing was significantly dysregulated, with a wider distribution of times, and a median increase of 15 min (Figure 9F, white bars, white arrow). Together, these live cell time-lapse imaging experiments show that KIF20B loss has a small but significant effect on the timing of abscission, and further suggest that KIF20B does so by stabilizing microtubules.
Movie S3.
DIC live time-lapse imaging movie from Figure 9E (top) of an siLUC-treated HeLa cell undergoing abscission, with images collected every 7.5 minutes. Single planes at the level of the midbody bulge were chosen from z-stacks. All time points starting from the last image prior to anaphase onset are included here.
Movie S4.
DIC live time-lapse imaging movie shown in Figure 9E (bottom) of an siKIF20B-treated HeLa cell undergoing abscission, with images collected every 7.5 minutes. Single planes at the level of the midbody bulge were chosen from z-stacks. All time points starting from the last image prior to anaphase onset are included here.
DISCUSSION
With the major players required for cytokinesis largely identified, there is a growing need to understand the roles of proteins that regulate the temporal aspects of cytokinesis, or play specialized roles in different cell types. We have demonstrated here that KIF20B, a Kinesin-6 family member required for normal brain size, has cell-autonomous roles in cytokinesis, temporally regulating both cytokinetic furrow ingression and abscission. We found that KIF20B localizes to microtubules of the central spindle and midbody throughout cytokinesis, particularly enriched near the midbody core and also around constriction sites, in patterns distinct from the other two Kinesin-6 family members. KIF20B depletion resulted in slower furrow ingression, and dysregulated abscission timing. Analyses of midbody structure and markers suggest that KIF20B is not required for midbody assembly or specification of subdomains, but may promote efficient midbody maturation and timely abscission. KIF20B may act in part by stabilizing microtubules. In the context of our previous work, these data suggest that seemingly subtle defects in the midbody and temporal control of cytokinesis can have devastating consequences for brain development.
Midbody maturation is an evolving concept in the cytokinesis field, describing a series of events during 1 h or more that prepare the intercellular bridge to be cut by the abscission machinery. It includes sequential recruitment of many proteins to the midbody bulge and their formation into rings, thinning of the midbody flanks, shedding of membrane, formation of constriction sites, and relocalization of some proteins (e.g., anillin, spastin, VPS4) to the constriction sites (Hu et al., 2012; Mierzwa and Gerlich, 2014; Renshaw et al., 2014). Our data suggest that KIF20B helps accelerate or coordinate midbody maturation. Its localization around the core of early midbodies and around the constriction sites of late midbodies suggests that it may aid maturation by keeping microtubules tightly packed or preventing them from sliding.
Multiple pieces of evidence suggest that KIF20B functions to organize microtubule bundles in cells by cross-linking. First, KIF20B is sufficient to cross-link microtubules in a cell-free assay (Abaza et al., 2003). Second, we previously showed that in axons of Kif20b mouse mutant neurons, microtubules are less tightly packed and more frequently invade growth cone filopodia, suggesting increased sliding (McNeely, Cupp, et al., 2017). Similarly, another Kinesin-6, Pavarotti/MKLP1, inhibits microtubule sliding in Drosophila neurons (del Castillo et al., 2015). Third, here we showed that depletion of KIF20B caused increased disorganization in central spindle microtubules, and altered midbody widths. It could be that KIF20B helps bundle and anchor microtubules at the center of the midzone, and any irregularities in anaphase are compounded as the midbody forms and matures. Although KIF20B is not required for constriction sites to form, or for spastin localization to the constrictions, an attractive possibility is that KIF20B stabilizes microtubules in a particular arrangement to facilitate formation of constriction sites, or the activity of severing enzymes like spastin.
The finding that SiR-tubulin appears to rescue the abscission timing defect also supports the idea that KIF20B promotes efficient abscission by stabilizing microtubules. More work is needed to determine the utility and limitations of using SiR-tubulin to live image abscission. At a minimum, it provides a way to label endogenous microtubules in live cells with low background and without the need to transfect plasmids or inject labeled protein. Of note, SiR-tubulin was recently used to label midbodies in a study of ESCRT recruitment during abscission in HeLa cells (Mierzwa et al., 2017). However, careful comparisons of microtubule dynamics and the kinetics of cytokinesis measured with varying dosages of SiR-tubulin versus other tubulin reagents, such as GFP-tubulin, are important for the field.
The phenotypes caused by depletion of KIF20B from HeLa cells are subtle compared with those caused by depletion of other midbody proteins such as CEP55 or the other Kinesin-6 family members. HeLa cells depleted of KIF23/MKLP1 usually fail to assemble a proper midbody core, regress their furrow, and become binucleate (Matuliene and Kuriyama, 2002; Zhu et al., 2005b). HeLa cells depleted of KIF20A/MKLP2 may either fail to complete furrow ingression (Neef et al., 2003; Kitagawa et al., 2013), or assemble a midbody but fail to complete abscission (Zhu et al., 2005b). Knockdown of CEP55 causes cells to be stuck in abscission for hours, and manifests as a large increase in midbody index in fixed cell populations (Zhao et al., 2006). But knockdown of KIF20B caused only a 2-min delay in furrowing and a 15-min delay in abscission in our experiments; roughly 25% increases. This small effect size explains why we did not find an increased mitotic index or midbody index in fixed cells. This may also explain why KIF20B was not identified in high-throughput RNA interference screens for genes involved in cell division (Kittler et al., 2004, 2007; Neumann et al., 2010), or for motor protein knockdown phenotypes (Zhu et al., 2005b). In retrospect, the small magnitude of KIF20B phenotypes probably did not meet the thresholds applied in those screens, but are revealed only upon focused quantitative analysis. Our genetic screen for developmental phenotypes, rather than a single-cell screen, revealed the importance of this Kinesin-6 (Dwyer et al., 2011).
Our previous analyses showed that Kif20b is essential for normal brain size in the mouse, even though it is not absolutely required for cytokinesis. The Kif20b mutant embryos form most organs normally, and do grow a small brain. This demonstrates that some tissues do not require Kif20b, and that many neural stem cells divisions occur successfully without it. However, the mutant neural stem cells show abnormal midbody shapes and organization, and undergo increased apoptosis (Janisch, Vock, et al., 2013). What we do not yet know is whether the temporal changes in cytokinesis observed upon KIF20B loss in HeLa cells are increased in severity in neuroepithelial stem cells lacking Kif20b, or whether the neural cells are simply much more sensitive to delays in cytokinesis. Polarized neuroepithelial stem cells are very tall and thin, and furrow ingression proceeds from the basal side to the apical. In addition, furrowing and abscission must be coordinated with the inheritance of cell fate determinants and apical membrane junctions. Interestingly, Kif20b may help link midbodies to the apical membrane (Janisch, Vock, et al., 2013). These factors suggest that cytokinesis in the neuroepithelium is much more challenging than in a HeLa cell and requires both spatial and temporal precision (Dwyer et al., 2016; Johnson et al., 2017). Thus, small defects in cytokinesis could trigger apoptosis or premature differentiation, depleting the progenitor pool. HeLa cells, the most commonly used mammalian system to study cytokinesis, cannot fully model the cytokinesis mechanisms and phenotypes of developing tissues like the early brain.
Nevertheless, despite such limitations, the cell line work herein has generated valuable high-resolution data and hypotheses about KIF20B’s functions to test in the developing brain. Future experiments will examine whether similar defects in midbody maturation and abscission occur in neural stem cells in culture and in intact brain explants, as well as how the timing of furrowing and abscission are coordinated with tissue polarity and daughter fates. This work underscores the need for more studies of cytokinesis in developing tissues in addition to isolated single cells. There may be many important players that regulate the timing or precision of cytokinesis that have not yet been identified from single-cell screens. Mutations in these genes may be discovered as causing developmental or functional defects in animal models or human clinical diseases.
MATERIALS AND METHODS
Cell culture
HeLa cells (human cervical carcinoma cells) were obtained from the American Type Culture Collection; we did not reauthenticate. HeLa cells were grown in 10-cm Petri dishes with DMEM (Life Technologies, Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals), 15 mM HEPES (Life Technologies, Thermo Fisher Scientific), 1X glutamine (Life Technologies, Thermo Fisher Scientific), 1X nonessential amino acids (Life Technologies, Thermo Fisher Scientific) and 1X Penicillin/Streptomycin (Life Technologies, Thermo Fisher Scientific). Cells were grown at 37°C and 5% CO2 until they reached ∼70% confluency before subculturing for use in experiments. Coverslips were examined for Mycoplasma contamination monthly by 4’6-diamidino-2-phenylindole (DAPI) staining of nuclei. For siRNA transfections, unsynchronized HeLa cells were harvested, counted, and plated onto 22-mm-diameter glass coverslips with a density of 50,000 cells per coverslip, unless otherwise stated.
Transfection with GFP-KIF20B
GFP-KIF20B plasmid was provided by F. Pirollet (Grenoble Institut des Neurosciences) (Abaza et al., 2003), and verified by sequencing. Cells were used when ∼70% confluent and transfected using a PolyJet transfection kit (SignaGen Laboratories) according to the manufacturer’s instructions. Cells were cotransfected with mCherry to monitor transfection efficiency. After 24 h, cells were fixed with 4% paraformaldehyde (PFA) followed by ice-cold methanol before staining and imaging.
Transfection with siRNA
For knockdown experiments, we independently tested two custom-made siRNAs (Invitrogen) previously reported to knock down human KIF20B (siRNA#1: 5′AAAGGACAGAGUCGUCUGAUUUU, siRNA#2: 5′AAUGGCAGUGAAACACCCUGGUU from Abaza et al., 2003). Because we observed that siRNA#2 depleted KIF23/MKLP1 as well as KIF20B, we discontinued use of siRNA#2 and used siRNA#1 for most experiments. Additional siRNAs were purchased from Life Technologies/Thermo Fisher: s18420 (called siRNA#3) 5′GCAAGUUAAAGAAUAUCGAtt and s18421 (siRNA#4) 5′CAAACGUAUUAGUUCAGCAtt. siRNAs #1, #3, and #4 caused similarly complete depletion of KIF20B to levels undetectable in 98–100% of midbodies at 24 h posttransfection (Supplemental Figure 1, C and D), and caused increased multinucleate and multi-lobed cells (Supplemental Figure 1E). A standard negative control siRNA to firefly luciferase (siLUC) was designed by Invitrogen. Cells were transfected using Lipofectamine RNAiMAX (Thermo Fisher) transfection reagent according to the manufacturer’s instructions. The final concentrations of siRNA were 10 nM. Transfection efficiency was found to be 98–100% using fluorescently tagged siRNA (Block-It from Invitrogen and siGlo from Dharmacon). Cells were fixed either 24 or 48 h (with medium change) after transfection and analyzed by immunofluorescence staining. For live cell imaging, cells were plated at a density of 50,000 cells per chamber in a two-chamber coverglass (Nunc Lab-Tek; Thermo Fisher Scientific) before transfection and imaged 22–24 h after transfection.
Immunocytochemistry
The standard fixation for most antibodies used was 4% PFA/PBS (phosphate-buffered saline) for 2 min at room temperature, followed with −20°C methanol for 10 min. Cells were then washed three times with PBS to remove any residual methanol and stored at 4°C until usage. For phospho-AURORA kinase B (pAURK) and spastin staining, cells were fixed with −20°C methanol for 10 min followed by three washes with PBS. For staining for ANILLIN (ANLN) and α-ACTININ 4 (ACTN4), cells were fixed with 10% trichloroacetic acid (TCA) in cytoskeleton buffer with sucrose (CBS; 10 mM MES, pH 6.1, 138 mM KCl, 3 mM MgCl2, 2 mM EGTA [ethylene glycol-bis-N,N,N’,N’-tetraacetic acid], 0.32 M sucrose). Briefly, 10% TCA in CBS was added to the cells and incubated on ice for 15 min. Cells were permeabilized with 0.2% Triton X-100, 50 mM glycine in PBS for 2 min on ice, then quenched with 50 mM glycine in PBS for 20 min.
Cells were blocked with 2% normal goat serum (NGS) in PBS with 0.1% Triton X-100 (PBST) for 1 h at room temperature. After three washes with PBS for 10 min each, primary antibodies diluted in blocking buffer were applied for 3 h at room temperature. Appropriate secondary antibodies diluted in blocking buffer were applied after three washes with PBS for 5 min each and incubated for 30 min at room temperature in the dark. Cells were mounted with Fluoromount (Diagnostic BioSystems) after a nuclear counterstain with DAPI (Fisher Scientific) and two washes with PBS for 10 min.
Antibodies
Primary antibodies used were as follows: mouse monoclonal DM1α (α-tubulin; 1:500) was from Abcam; rat anti-TUBA1A (clone YL ½; 1:750) was from Novus Biologicals; mouse polyclonal anti-CEP55 (1:200) was from Abnova; mouse anti-Aurora kinase B (AURKB; 1:300) was from BD Biosciences; rabbit anti-phospho-T232-Aurora kinase B (pAURKB; 1:200) was from Rockland; rabbit anti-KIF20A (A300-879A; 1:100) was from Bethyl Labs; goat anti-anillin (ANLN; 1:300), mouse monoclonal anti-ANLN (1:100), rabbit anti-MKLP1 (sc-867; 1:100), rabbit anti-PRC1 (1:50), mouse monoclonal anti-human-spastin (3G11/1; 1:50), and mouse anti-human-MPP1(KIF20B; 1:300) were from Santa Cruz; rabbit anti-mouse-Kif20b (1:500) was custom-made by Covance (Janisch, Vock, et al., 2013); rabbit anti-cleaved-caspase 3 (CC3; 1:200) and rabbit anti-phosphohistone H3 (PH3, Alexa Fluor 647 conjugated; 1:400) were from Cell Signaling; rabbit anti-α-ACTININ4 (ACTN4; 1:250) was from Millipore; and rabbit anti-VPS4 (1:500) was from Sigma Aldrich. Both the monoclonal mouse anti-human MPP1 (KIF20B) antibody (shown in Figures 1 and 2 and Supplemental Figure 1) and the polyclonal rabbit anti-mouse-Kif20b (shown in Supplemental Figure 1B) were validated by verifying that the midbody staining was lost in Kif20b mutant mouse cells. All other primary antibodies were validated by verifying that the staining patterns matched multiple published reports. Secondary antibodies were goat or donkey polyclonal immunoglobulin G (H+L) conjugated to Alexa fluorophores against according species and were used at 1:200 (Life Technologies).
Imaging and data analysis
Fixed images were either collected with a Zeiss AxioVision ImagerZ1 wide-field microscope with 40×/1.3 or 100×/1.25 Oil M27 APO objectives or a DeltaVision Elite with TrueLight deconvolution microscope with 60×/1.42 Oil Plan APO objective. For comparisons between siLUC and siKIF20B knockdown cells, exposure times were kept constant for each treatment and images were taken on the same day. For DeltaVision images, deconvolved maximum intensity projections of z-stacks are displayed unless otherwise specified. For image analysis and data acquisition of images, we used Fiji/ImageJ (http://imagej.net/Fiji/Downloads).
Live cell imaging was done on an inverted Zeiss AxioObserver microscope equipped with a temperature and CO2-controlled chamber set at 5% CO2 and 37°C, multipoint acquisition and DefiniteFocus. Illumination and exposure times were kept to minimum practical levels. Images were acquired with an AxioCam MRm camera and Zeiss Zen software. Cells in metaphase were chosen in brightfield based on their shape and chromatin appearance. siKIF20B cells were imaged first, followed by siLUC cells. Time 0 was the last frame when chromatin was aligned at the metaphase plate. Cells were unsynchronized. After each live imaging experiment, cells were immunostained for KIF20B and checked for expression; in each case 0/20 midbodies examined had detectable KIF20B.
Live imaging of furrow ingression was done by collecting brightfield images with a 63× oil objective. Image stacks with 0.75 μm step size were captured every minute. Furrow was considered completely ingressed when no further ingression could be observed using raw z-stacks or projections. In no case was furrow regression observed.
Abscission data
Brightfield.
Image stacks with 0.5 μm step size were collected every 7.5 min with a 63× oil objective and DIC for 4–6 h. Abscission was scored in the z-plane where the midbody bulge was visible, and was defined as the separation of the midbody from one or both cells (i.e., movement of the bulge away from a cell, and connection no longer detectable). Twenty-five siLUC cells and 30 siKIF20B cells were analyzed.
SiR-tubulin fluorescence imaging.
Cells were treated with 100 nm silicon-rhodamine tubulin (SiR-tubulin) according to the manufacturer’s instructions (Cytoskeleton) at least 6 h before imaging. Cells were confirmed to have bipolar spindles using the SiR-tubulin. Cells with more than two spindles (observed in both control and depleted cultures) were not used for measurements. Image stacks with 1.0 μm step size were captured with a 20× objective and a Cy5 (far-red) filter, and maximum intensity projections of selected planes were used for analysis. The first abscission was defined as the first microtubule break, with no visible fluorescence between the flank and the bulge. The second abscission was defined as the last break of the microtubules allowing the release of the midbody remnant into the medium. SiR-tubulin enabled the visualization of both abscission events. DIC images were blinded for analysis. Live cell images were collected and analyzed using Zen Blue software (Zeiss).
Other data analysis and statistics were done with Microsoft Excel, GraphPad Prism, and PAST (http://palaeo-electronica.org/2001_1/past/issue1_01.htm). Unless otherwise indicated, p values were calculated with two-tailed Student’s t test, and error bars are SEM.
Supplementary Material
Acknowledgments
We thank Fabienne Pirollet for the GFP-KIF20B plasmid. We are grateful to Todd Stukenberg, Jim Casanova, Bettina Winckler, Jing Yu, and Xiaowei Lu, and members of their laboratories, and Jessica Little for advice and discussions. This work was supported by the National Institutes of Health (Grant No. R01 NS076640 to N.D.D.), an American Cancer Society seed grant (Grant No. ACS-IRG-81-001-26 to N.D.D.), and a Harrison Undergraduate Research Award to J.M.D.
Abbreviations used:
- CBS
cytoskeleton buffer with sucrose
- cs
constriction site
- DAPI
4’6-diamidino-2-phenylindole
- DIC
differential interference contrast
- EGTA
ethylene glycol-bis-N,N,N’,N’-tetraacetic acid
- FBS
fetal bovine serum
- HCT116
human colon tumor 116
- HEPES
4-(2-hydroxyl)-1-piperazineethanesulfonic acid
- LUC
luciferase
- MB
midbody
- MDCK
Madin–Darby canine kidney
- MES
2-(N-morpholino)ethanesulfonic acid
- NGS
normal goat serum
- PBS
phosphate-buffered saline
- PBST
phosphate-buffered saline with Triton X-100
- PFA
paraformaldehyde
- SiR
silicon-rhodamine
- TCA
trichloroacetic acid.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-08-0495) on November 22, 2017.
REFERENCES
Boldface denotes co–first authors.
- Abaza A, Soleilhac JM, Westendorf J, Piel M, Crevel I, Roux A, Pirollet F. M phase phosphoprotein 1 is a human plus-end-directed kinesin-related protein required for cytokinesis. J Biol Chem. 2003;278:27844–27852. doi: 10.1074/jbc.M304522200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atilla-Gokcumen GE, Muro E, Relat-Goberna J, Sasse S, Bedigian A, Coughlin ML, Garcia-Manyes S, Eggert US. Dividing cells regulate their lipid composition and localization. Cell. 2014;156:428–439. doi: 10.1016/j.cell.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron RD, Barr FA. The kinesin-6 members MKLP1, MKLP2 and MPP1. In: Kozielski F.S.B.F., editor. Kinesins and Cancer. Dordrecht, Netherlands: Springer; 2015. pp. 193–222. [Google Scholar]
- Bastos RN, Barr FA. Plk1 negatively regulates Cep55 recruitment to the midbody to ensure orderly abscission. J Cell Biol. 2010;191:751–760. doi: 10.1083/jcb.201008108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Caballe A, Agromayor M, Kloc M, Martin-Serrano J. ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science. 2012;336:220–225. doi: 10.1126/science.1217180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell JW, Lindon C, Luzio JP, Reid E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 2009;10:42–56. doi: 10.1111/j.1600-0854.2008.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagenbach EM, Endow SA. A new kinesin tree. J Cell Sci. 2004;117:3–7. doi: 10.1242/jcs.00875. [DOI] [PubMed] [Google Scholar]
- del Castillo U, Lu W, Winding M, Lakonishok M, Gelfand VI. Pavarotti/MKLP1 regulates microtubule sliding and neurite outgrowth in Drosophila neurons. Curr Biol. 2015;25:200–205. doi: 10.1016/j.cub.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne LK, Wang XJ, Prekeris R. Midbody: from cellular junk to regulator of cell polarity and cell fate. Curr Opin Cell Biol. 2015;35:51–58. doi: 10.1016/j.ceb.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil ., Marzesco AM, Corbeil D, Huttner WB, Wilsch-Brauninger M. Midbody and primary cilium of neural progenitors release extracellular membrane particles enriched in the stem cell marker prominin-1. J Cell Biol. 2007;176:483–495. doi: 10.1083/jcb.200608137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer ND, Chen B, Chou SJ, Hippenmeyer S, Nguyen L, Ghashghaei HT. Neural stem cells to cerebral cortex: emerging mechanisms regulating progenitor behavior and productivity. J Neurosci. 2016;36:11394–11401. doi: 10.1523/JNEUROSCI.2359-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer ND, Manning DK, Moran JL, Mudbhary R, Fleming MS, Favero CB, Vock VM, O’Leary DD, Walsh CA, Beier DR. A forward genetic screen with a thalamocortical axon reporter mouse yields novel neurodevelopment mutants and a distinct Emx2 mutant phenotype. Neural Dev. 2011;6:3. doi: 10.1186/1749-8104-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci USA. 2011;108:4846–4851. doi: 10.1073/pnas.1102714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger AW, Wilsch-Brauninger M, Marzesco AM, Bickle M, Lohmann A, Maliga Z, Karbanova J, Corbeil D, Hyman AA, Huttner WB. Proliferating versus differentiating stem and cancer cells exhibit distinct midbody-release behaviour. Nat Commun. 2011;2:503. doi: 10.1038/ncomms1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershony O, Pe’er T, Adar S, Noach-Hirsh M, Elia N, Tsur A. Cytokinetic abscission is an acute G1 event. Cell Cycle. 2014;13:3436–3441. doi: 10.4161/15384101.2014.956486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershony O, Sherman S, Adar S, Segal I, Nachmias D, Goliand I, Elia N. Measuring abscission spatiotemporal dynamics using quantitative high-resolution microscopy. Methods Cell Biol. 2017;137:205–224. doi: 10.1016/bs.mcb.2016.03.032. [DOI] [PubMed] [Google Scholar]
- Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- Green RA, Paluch E, Oegema K. Cytokinesis in animal cells. Annu Rev Cell Dev Biol. 2012;28:29–58. doi: 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- Gruneberg U, Neef R, Honda R, Nigg EA, Barr FA. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J Cell Biol. 2004;166:167–172. doi: 10.1083/jcb.200403084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizetti J, Schermelleh L, Mantler J, Maar S, Poser I, Leonhardt H, Muller-Reichert T, Gerlich DW. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 2011;331:1616–1620. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- Hu CK, Coughlin M, Mitchison TJ. Midbody assembly and its regulation during cytokinesis. Mol Biol Cell. 2012;23:1024–1034. doi: 10.1091/mbc.E11-08-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janisch KM, Dwyer ND. Imaging and quantitative analysis of cytokinesis in developing brains of Kinesin-6 mutant mice. Methods Cell Biol. 2016;131:233–252. doi: 10.1016/bs.mcb.2015.06.008. [DOI] [PubMed] [Google Scholar]
- Janisch KM, Vock VM, Fleming MS, Shrestha A, Grimsley-Myers CM, Rasoul BA, Neale SA, Cupp TD, Kinchen JM, Liem KF, Dwyer ND. The vertebrate-specific Kinesin-6, Kif20b, is required for normal cytokinesis of polarized cortical stem cells and cerebral cortex size. Development. 2013;140:4672–4682. doi: 10.1242/dev.093286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CA, Wright CE, Ghashghaei HT. Regulation of cytokinesis during corticogenesis: focus on the midbody. FEBS Lett. 2017;591:4009–4026. doi: 10.1002/1873-3468.12676. [DOI] [PubMed] [Google Scholar]
- Kamimoto T, Zama T, Aoki R, Muro Y, Hagiwara M. Identification of a novel kinesin-related protein, KRMP1, as a target for mitotic peptidyl-prolyl isomerase Pin1. J Biol Chem. 2001;276:37520–37528. doi: 10.1074/jbc.M106207200. [DOI] [PubMed] [Google Scholar]
- Kanehira M, Katagiri T, Shimo A, Takata R, Shuin T, Miki T, Fujioka T, Nakamura Y. Oncogenic role of MPHOSPH1, a cancer-testis antigen specific to human bladder cancer. Cancer Res. 2007;67:3276–3285. doi: 10.1158/0008-5472.CAN-06-3748. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Fung SY, Onishi N, Saya H, Lee SH. Targeting Aurora B to the equatorial cortex by MKlp2 is required for cytokinesis. PLoS One. 2013;8:e64826. doi: 10.1371/journal.pone.0064826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Putz G, Pelletier L, Poser I, Heninger AK, Drechsel D, Fischer S, Konstantinova I, Habermann B, Grabner H, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- Kittler R, Surendranath V, Heninger AK, Slabicki M, Theis M, Putz G, Franke K, Caldarelli A, Grabner H, Kozak K, et al. Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat Methods. 2007;4:337–344. doi: 10.1038/nmeth1025. [DOI] [PubMed] [Google Scholar]
- Kuo TC, Chen CT, Baron D, Onder TT, Loewer S, Almeida S, Weismann CM, Xu P, Houghton JM, Gao FB, et al. Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat Cell Biol. 2011;13:1214–1223. doi: 10.1038/ncb2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Elia N, Ghirlando R, Lippincott-Schwartz J, Hurley JH. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322:576–580. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhart KF, DiNardo S. Somatic cell encystment promotes abscission in germline stem cells following a regulated block in cytokinesis. Dev Cell. 2015;34:192–205. doi: 10.1016/j.devcel.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinavicius G, Reymond L, D’Este E, Masharina A, Gottfert F, Ta H, Guther A, Fournier M, Rizzo S, Waldmann H, et al. Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat Methods. 2014;11:731–733. doi: 10.1038/nmeth.2972. [DOI] [PubMed] [Google Scholar]
- Maliga Z, Junqueira M, Toyoda Y, Ettinger A, Mora-Bermudez F, Klemm RW, Vasilj A, Guhr E, Ibarlucea-Benitez I, Poser I, et al. A genomic toolkit to investigate kinesin and myosin motor function in cells. Nat Cell Biol. 2013;15:325–334. doi: 10.1038/ncb2689. [DOI] [PubMed] [Google Scholar]
- Matuliene J, Kuriyama R. Kinesin-like protein CHO1 is required for the formation of midbody matrix and the completion of cytokinesis in mammalian cells. Mol Biol Cell. 2002;13:1832–1845. doi: 10.1091/mbc.01-10-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeely KC, Cupp TD, Little JN, Janisch KM, Shrestha A, Dwyer ND. Mutation of Kinesin-6 Kif20b causes defects in cortical neuron polarization and morphogenesis. Neural Dev. 2017;12:5. doi: 10.1186/s13064-017-0082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzwa BE, Chiaruttini N, Redondo-Morata L, von Filseck JM, Konig J, Larios J, Poser I, Muller-Reichert T, Scheuring S, Roux A, Gerlich DW. Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat Cell Biol. 2017;19:787–798. doi: 10.1038/ncb3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzwa B, Gerlich DW. Cytokinetic abscission: molecular mechanisms and temporal control. Dev Cell. 2014;31:525–538. doi: 10.1016/j.devcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Mollinari C, Kleman JP, Jiang W, Schoehn G, Hunter T, Margolis RL. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J Cell Biol. 2002;157:1175–1186. doi: 10.1083/jcb.200111052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhina S, Wang YL, Murata-Hori M. α-Actinin is required for tightly regulated remodeling of the actin cortical network during cytokinesis. Dev Cell. 2007;13:554–565. doi: 10.1016/j.devcel.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins JM, Biesele JJ. Terminal phase of cytokinesis in D-98s cells. J Cell Biol. 1977;73:672–684. doi: 10.1083/jcb.73.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef R, Preisinger C, Sutcliffe J, Kopajtich R, Nigg EA, Mayer TU, Barr FA. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J Cell Biol. 2003;162:863–875. doi: 10.1083/jcb.200306009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Yonemura S. Centralspindlin regulates ECT2 and RhoA accumulation at the equatorial cortex during cytokinesis. J Cell Sci. 2006;119:104–114. doi: 10.1242/jcs.02737. [DOI] [PubMed] [Google Scholar]
- Renshaw MJ, Liu J, Lavoie BD, Wilde A. Anillin-dependent organization of septin filaments promotes intercellular bridge elongation and Chmp4B targeting to the abscission site. Open Biol. 2014;4:130190. doi: 10.1098/rsob.130190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzmann V, Chen C, Chiang CY, Tiyaboonchai A, Mayer M, Yamashita YM. Centrosome-dependent asymmetric inheritance of the midbody ring in Drosophila germline stem cell division. Mol Biol Cell. 2014;25:267–275. doi: 10.1091/mbc.E13-09-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiel JA, Park K, Morphew MK, Reid E, Hoenger A, Prekeris R. Endocytic membrane fusion and buckling-induced microtubule severing mediate cell abscission. J Cell Sci. 2011;124:1411–1424. doi: 10.1242/jcs.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiel JA, Simon GC, Zaharris C, Weisz J, Castle D, Wu CC, Prekeris R. FIP3-endosome-dependent formation of the secondary ingression mediates ESCRT-III recruitment during cytokinesis. Nat Cell Biol. 2012;14:1068–1078. doi: 10.1038/ncb2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Pohl C. A function for the midbody remnant in embryonic patterning. Commun Integr Biol. 2014;7:e28533. doi: 10.4161/cib.28533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop AR, Liu H, Yates J, Meyer BJ, Heald R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science. 2004;305:61–66. doi: 10.1126/science.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steigemann P, Wurzenberger C, Schmitz MH, Held M, Guizetti J, Maar S, Gerlich DW. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136:473–484. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]
- Zhao WM, Seki A, Fang G. Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol Biol Cell. 2006;17:3881–3896. doi: 10.1091/mbc.E06-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Bossy-Wetzel E, Jiang W. Recruitment of MKLP1 to the spindle midzone/midbody by INCENP is essential for midbody formation and completion of cytokinesis in human cells. Biochem J. 2005a;389:373–381. doi: 10.1042/BJ20050097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Lau E, Schwarzenbacher R, Bossy-Wetzel E, Jiang W. Spatiotemporal control of spindle midzone formation by PRC1 in human cells. Proc Natl Acad Sci USA. 2006;103:6196–6201. doi: 10.1073/pnas.0506926103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan JB, Abraham RT, Jiang W. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol Cell. 2005b;16:3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.