Summary
During cell division, the mitotic kinesin-5 Eg5 generates most of the force required to separate centrosomes during spindle assembly. However, Kif15, another mitotic kinesin, can replace Eg5 function permitting mammalian cells to acquire resistance to Eg5 poisons. Unlike Eg5, the mechanism by which Kif15 generates centrosome separation forces is unknown. Here, we investigated the mechanical properties and force generation capacity of Kif15 at the single molecule level using optical tweezers. We found that the non-motor microtubule-binding tail domain interacts with the microtubule’s E-hook tail with a rupture force higher than the stall force of the motor. This allows Kif15 dimers to productively and efficiently generate forces that could potentially slide microtubules apart. Using an in vitro optical trapping and fluorescence assay, we found that Kif15 slides anti-parallel microtubules apart with gradual force build-up while parallel microtubule bundles remain stationary with a small amount of antagonizing force generated. A stochastic simulation shows the essential role of Kif15’s tail domain for load storage within the Kif15-microtubule system. These results suggest a mechanism for how Kif15 rescues bipolar spindle assembly.
Introduction
A hallmark of the G2/M transition is the conversion of an interphase microtubule cytoskeleton into a bipolar spindle. Bipolarity is essential for spindle function and is established by many microtubule-associated proteins that collectively move centrosomes to opposite cell poles [1]. One factor central to spindle assembly is the homotetrameric kinesin-5, Eg5, that crosslinks and slides anti-parallel microtubules apart [2]. Eg5 inactivation blocks centrosome separation, resulting in a monopolar spindle [3–10]. However, several lines of evidence indicate that kinesin-12s also generate forces relevant for spindle assembly. In C. elegans, a kinesin-12 (KLP-18) rather than kinesin-5 (BMK-1) is essential for spindle formation [11,12]. In humans, kinesin-12 Kif15 prevents [13] or slows [14] kinesin-5 inhibitor (K5I)-triggered collapse of the metaphase spindle. Kif15 overexpression can nullify the cytotoxic effects of K5Is by driving spindle assembly [13,15] and is essential for HeLa cells to acquire K5I resistance [16]. These results suggest that kinesin-5s and kinesin-12s share strong functional homology, but the biophysical underpinnings are unclear because the motors differ in their cell biological and biochemical properties.
In mammalian cells, the mitotic localizations of Kif15 and Eg5 are significantly different. Kif15 enriches on kinetochore microtubules in prometaphase and then distributes uniformly along spindle microtubules at metaphase [17]. In contrast, Eg5 binds microtubules early during spindle assembly irrespective of whether they are attached to kinetochores [3,17] and concentrates near spindle poles at metaphase [3,10,18]. In K5I-resistant cell lines (KIRCs), Kif15 redistributes to non-kinetochore microtubules while microtubule-bound levels of active Eg5 are reduced [16,17]. These localization patterns may indicate that Kif15 can only produce forces sufficient to drive spindle assembly when it is bound to non-kinetochore microtubules in an orientation-dependent manner. Kinetochore microtubules are predominantly parallel-oriented [19], whereas interpolar microtubules are of mixed polarity [20]. Redistribution of Kif15 to non-kinetochore microtubules may lead to significant force production when microtubules are anti-parallel, similar to Eg5.
Elegant biophysical studies have demonstrated that tetrameric Eg5 is optimally designed to drive spindle assembly [2,21–25]. Eg5 only slides anti-parallel microtubules apart [2]. Single molecule and stopped flow experiments have shown that Eg5 resides primarily in a two-head bound state [21], and its catalytic cycle is limited by ATP hydrolysis rather than product release [22]. These features and a second non-motor microtubule-binding site bias Eg5 to remain attached to microtubules [24]. Furthermore, Eg5’s mechanochemical cycle makes force generation scale linearly with motor number within an anti-parallel microtubule overlap [25].
By comparison, our understanding of Kif15 is limited. Some work suggests that Kif15 is tetrameric [26], leading to speculation that Kif15 promotes spindle assembly through a mechanism similar to Eg5, while our previous studies indicate that Kif15 is dimeric [16,27]. As a homodimer, Kif15 can crosslink two microtubules through its motor domains and a second microtubule-binding site (Coil-1) within its coiled-coil stalk [27]. This crosslinking activity enables Kif15 to slide microtubules apart in vitro [27], although the microtubule orientation requirement for this activity is unknown.
Here, we investigate the activity of Kif15 on single microtubules and within microtubule bundles of different filament arrangements. We analyze constructs including full Kif15, a truncated Kif15 motor that does not include the Coil-2 inhibitory domain but includes the non-motor microtubule binding domain Coil-1 (N700), and the isolated microtubule binding domain (Coil-1) (Figure 1A). Single molecule optical tweezers and fluorescence experiments confirm that Kif15 acts as a dimer, takes a large percentage of backward steps, and that Coil-1 sustains forces higher than motor stall. These properties permit build-up of mechanical strain in the microtubule system through forward progress of the motor heads and concomitant resistance or relaxation by Coil-1 rupturing and back stepping. We further show that concerted movement occurs exclusively in anti-parallel microtubules. A stochastic simulation of microtubule sliding by Kif15 reveals the mechanism by which Coil-1 regulates strain build-up. Our work provides insight into how Kif15 generates forces that drive spindle assembly and has implications for the interplay between Eg5 and Kif15 within the spindle.
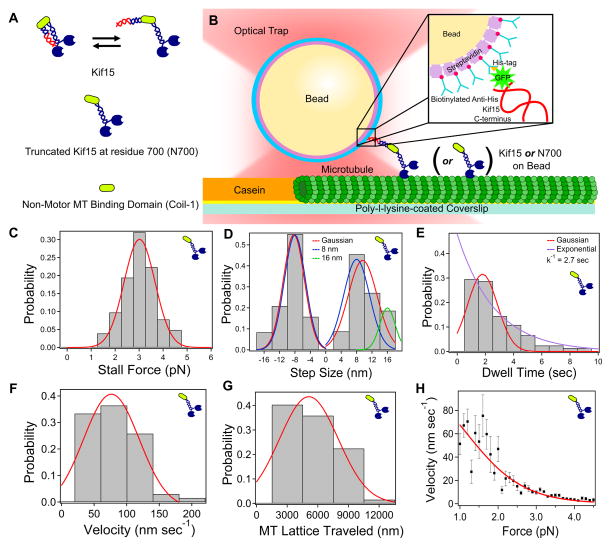
Figure 1. N700 Motility Assay.
(A) Constructs studied: Kif15, N700, Coil-1. (B) Single molecule motility assay (Methods). (C) N700 stall force, averaging 3.0 pN ± 0.6 pN (N=102), which is very similar to Kif15. (D) Step size distribution, averaging 9.6 nm ± 3.8 nm (N=473, forward) and 8.3 nm ± 2.7 nm (N=230, backward). Blue and green: 8 nm and 16 nm constrained fits. (E) Dwell time between steps averaged 1.8 sec ± 1.1 sec. Decay constant (exponential fit, red) is 2.7 sec. (F) Unloaded velocity measured via video tracking was found to be 75.8 nm s−1 ± 41 nm s−1 (N=67). (G) N700 processivity was measured via video tracking the run length as 5120 nm ± 2900 nm, which is limited by microtubule length (N=67). (H) Force-velocity curve. Solid line: fit with a three-state kinetic model (Table 1). Error bars: SEM. See also Figure S1, Movie S1.
Results
Oligomerization state of Kif15
We used the baculovirus expression system to produce and purify Kif15 motor constructs (Kif15 and N700) with a C-terminal GFP-His6-tag (Figure 1B, inset) [27]. Confirmation of the motor’s oligomerization state was necessary to form the foundation of a mechanism regarding Kif15’s ability to generate force during spindle assembly. Single molecule photobleaching of individual motors occurred in two steps (Methods; Figure S1 A–C). This indicates that our construct is a dimer, which agrees with previous rigorous analysis in solution and when expressed in cells [16,27,28].
N700 Generates Force at Single Molecule Level
To evaluate the force generation capabilities of Kif15 and N700, single molecule bead motility assays using optical tweezers were performed (Figure 1B; Methods) [29]. N700, which includes the motor, stalk, and Coil-1 [27], moved robustly, permitting evaluation of motor characteristics such as stall force, step size, backward step probability, and dwell times. Full length Kif15, containing a C-terminal inhibitory domain (Coil-2) [27], moved much less frequently, prompting rigorous analysis of N700.
N700 exhibited a higher frequency of motility events compared to kinesin-1, and traces did not reach a plateauing stall but rather fell off abruptly, similar to sawtooth patterns generated by Eg5 [23]. Its stall force was 3.0 pN ± 0.6 pN (avg±std, N=102; Figure 1C), which is similar to that of Kif15 (see below). N700 took approximately 8 nm steps (size of a tubulin dimer), though some variability was observed (Figure 1D). Backward stepping was observed with a 32% frequency, well above that of kinesin-1 [29–31]. While the vast majority of step sizes are 8 nm, a small population (20%) of 16 nm forward steps were recorded (blue and green constrained fits in Figure 1D) that are likely to be unresolvable rapid 8 nm steps in succession. N700 had 2.7 s dwell times (Figure 1E).
N700 (attached to a bead) moved readily in an unloaded motility assay. Its velocity was 75.8 nm s−1 ± 41 nm s −1 (N=67; Figure 1F), which is similar to previous measurements of Eg5 [23] and fluorescence velocity measurements of N700 and Kif15 [26,27]. The average run length was 5120 nm ± 2900 nm (N=67, Figure 1G). The spread is due to the underlying microtubule length, and values up to 12 μm were measured. Long commitments of Kif15 to motility have been observed previously [26,27]. Upon reaching the end of the microtubule, N700 released 64% of the time. It also has the ability to switch microtubule tracks (Movie S1), as shown previously, but the proposed structure was a tetramer [26]. Instead, N700 can switch tracks likely via its ability to rapidly recommit to motility, as shown by its high run frequency, along with Coil-1 that facilitates attachment and can keep an unbound motor from diffusing away (see below).
Force-velocity analysis of N700
To parameterize the mechanistic underpinnings of Kif15 behavior, a force-velocity curve was generated for N700 (Figure 1H; Methods) [32–34], and was fit with a three-state kinetic model of the form [23,34]
| (1) |
with the force-dependent rate of the mechanochemical cycle
| (2) |
Here, v is the motor velocity under an applied force F; δ1 = 8.2 nm is the average kinesin step size; k1 and k−1 are ATP binding and unbinding rates (globally fit from [23]); k20 is the unloaded mechanical rate; k3 is the ATP hydrolysis rate; kBT (T = 300K) is thermal energy; and δ2 represents the force sensitivity of the three-state model, where larger values indicate a more force-sensitive motor [34]. The measured and fitted parameters for kinesin-1 [34], Eg5 [23], N700, and Kif15 are shown in Table 1, where δ1, k1, and k−1 were held constant. Eg5 and N700 unloaded velocities are similar, but N700 is more processive than Eg5, potentially due to the N700’s ability to recommit to motility upon unbinding from the microtubule. The force sensitivity δ2 is smaller for Eg5 (1.9 nm) than kinesin-1, N700, and Kif15 (4–6 nm). Eg5 has a proline in the middle of the neck linker, resulting in a shorter cover-neck bundle (force generating element) compared to kinesin-1 or Kif15, agreeing with its shorter δ2 [34]. These parameters dictate Kif15’s single molecule and ensemble-level force generation capacity (see modeling results).
Table 1. Three-state kinetic model.
Kinesin-1 and Eg5 values were added for comparison. n/m: not measured.
| Construct | Unloaded Velocity | Run Length | Stall Force | Three-state model | |||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| k1 | k−1 | k20 | k3 | δ2 | vmax | ||||
|
| |||||||||
| (nm s−1) | (nm) | (pN) | (μM−s)−1 | (s−1) | (s−1) | (s−1) | (nm) | (nm s−1) | |
| Kinesin-1[34] | 671 ± 21 | 1370 ± 287 | 4.92 ± 0.08 | 2 | 120 | 12,900 | 76.86 | 5.50 | 630 |
| Eg5[23,34] | 96 ± 2 | 67 ± 7 | 1.5 – 7* | 0.89 | 10 | 86 | 13.5 | 1.9 | 94 |
| N700 | 76 ± 41 | 5120 ± 2900 | 3.0 ± 0.6 | 2 | 120 | 72.2 | 15.1 | 5.4 | 101 |
| Kif15 | n/m | n/m | 2.7 ± 1.0 | 2 | 120 | 88.1 | 16.2 | 4.1 | 111 |
Range of values reported.
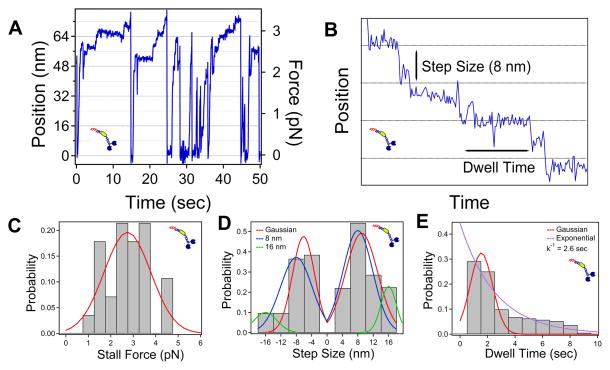
Kif15 and N700 stall at 3 pN
Interestingly, Kif15 is much slower than N700 and has a lower run frequency, or number of run events (walk until maximum force is reached before returning to the trap center) per time, on average 1.2 and 10 runs per 100 sec for Kif15 and N700, respectively (based on run events from raw force/displacement data). This suggests that Coil-2 in Kif15 plays a regulatory role, a feature not fully replicated in the trapped bead assay. Kif15 also shows step size variability in the distribution but shows 8 nm steps near the stall force (Figure 2). Kif15’s stall force is very similar to N700 (Figure 2C). Step size distributions (Figure 2D) also contain 8 nm (blue) and 16 nm (green; 10% of forward and 20% of backward steps) constrained fits.
Figure 2. Kif15 Motility Assay.
(A) Example Kif15 motility trace. (B) Example of 8 nm stepping used for step size and dwell time analysis. (C) Stall force histogram, averaging 2.7 pN ± 1.0 pN (N=27). (D) Step size, averaging 8.9 nm ± 4.1 nm (N=118, forward) and 6.1 nm ± 2.9 nm (N=52, backward). 8 nm (blue) and 16 nm (green) constrained fits. (E) Dwell time, averaging 1.5 sec ± 0.8 sec. Decay constant (exponential fit, purple) is 2.6 sec. See also Movie S2.
Unlike N700, Kif15 unloaded motility on single microtubules was rare. Although Kif15 coated beads bound microtubules, they did not readily move, making it difficult to obtain unloaded measurements. This may be because both Coil-1 and the motor domains bind to the microtubule simultaneously, making motility dependent on whether the motor heads could produce enough force to displace Coil-1 (see below). When motile, traces resembled those of N700 and Eg5. Notably, Kif15 moves processively even with the inhibitory Coil-2 domain present (Movie S2). This could be an effect of the GFP-tag or close association of the C-terminus to the bead, both of which could sequester Coil-2 from association with the motor heads.
Coil-1 Resists Forces Greater Than Stall
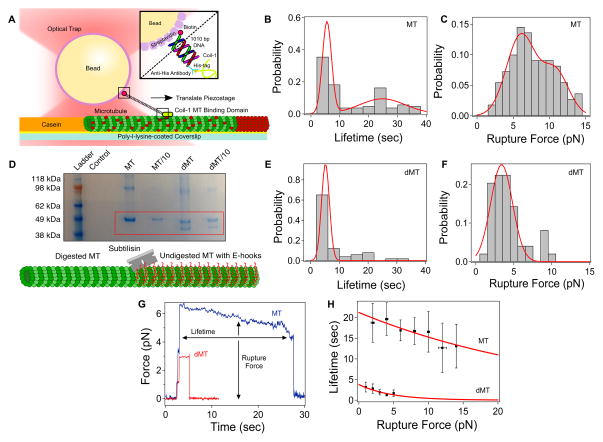
We investigated the Coil-1/microtubule interaction, which would be critical in defining motor function. A Coil-1 rupture force below the stall force implies that motor heads walking along a microtubule would easily rupture Coil-1 bound to an adjacent microtubule, facilitating isolated motor movement with little resistance. A rupture force higher than stall would anchor Coil-1 to the microtubule while the crosslinked motor heads walk along the other, permitting productive, efficient, and collective force build-up within the bundle and concomitant microtubule transport.
We evaluated the strength of the Coil-1/microtubule interaction by measuring the time for the bond to rupture at a load determined by the necessary displacement to fully elongate the DNA tether (Figure 3; Methods). The lifetime distribution was bimodal, which averaged 5.6 sec ± 1.6 sec and 24.8 sec ± 7.9 sec, respectively (N=109; Figure 3B). This potentially reflects binding of Coil-1 to the microtubule’s C-terminal tail (termed E-hook) or to the microtubule lattice. In both cases, the rupture forces are higher than the 3 pN stall force (Figure 3C). The average lifetime is long relative to the dwell times measured for Kif15, suggesting that Kif15 may be able to generate force in microtubule bundles. Interestingly, a significant number of rupture traces show structured motion toward the trap center before rupture (Figure 3G blue and Figure S2). The transitions appear discrete with dwells in between (Figure S2, insets). This structure suggests that Coil-1 may jump to the next available E-hook to readjust its position and maintain the high level of resistance necessary for force generation.
Figure 3. Coil-1 Binding Assay.
(A) Assay design (Methods). (B) Coil-1/microtubule lifetime distribution, averaging 5.6 sec ± 1.6 sec and 24.8 ± 7.9 sec (N=109) (C) Force at rupture, averaging 6.1 pN ± 2.0 pN and 10.81 pN ± 1.8 pN (N=109). (D) E-hook digestion confirmation using SDS-PAGE through separation of alpha and beta tubulin bands (ladder, control (sample buffer and water), microtubules (stock and 10x diluted), digested microtubules (stock and 10x diluted). (E) Coil-1/digested microtubule lifetime, averaging 5.2 sec ± 1.4 sec (N=49), is comparable to the lower value for normal microtubule (panel B). (F) Digested microtubule rupture force, averaging 3.4 pN ± 1.4 pN (N=49). (G) Example ruptures on normal (blue) and digested microtubules (red) (H) Coil-1 lifetime versus rupture force on normal (R2 = 0.77) and digested microtubule (R2 = 0.68). Error bars: SEM. See also Figure S2.
We further investigated the role of the negatively-charged E-hook by cleaving it from microtubules with subtilisin. Digested microtubules were confirmed by fractionation of α- and β-tubulin bands in SDS-PAGE (Figure 3D) [35]. Rupture measurements with digested microtubules revealed a reduced lifetime (5.2 sec ± 1.4 sec, N=49; Figure 3E), and rupture force (3.4 pN ± 1.4 pN, N=49; Figure 3F). The rupture trace on a digested microtubule (Figure 3G, red) was also noticeably flat compared to normal microtubules (Figure 3G, blue). The difference may be due to the lack of available E-hooks to rescue binding once the first binding event fails. The marked lifetime decrease under a given rupture force (Figure 3H) also supports that the E-hook plays a substantial, likely electrostatic, role in binding Coil-1 to microtubules. The dependence on lifetime for Coil-1 on microtubules and digested microtubules was fit to an exponential based on the Bell model [36], which describes the force dependence of bond reaction rates (Figure 3H). Coil-1 lifetime on digested microtubules falls off quickly with increased force in contrast to the relatively flat dependence for native microtubules, revealing the E-hook’s significance in the interaction.
Under physiological conditions, Kif15 targets kinetochore microtubules that contain parallel microtubules [17]. However, in KIRCs, Kif15 also binds interpolar microtubules that are oriented in anti-parallel [16,17]. Thus, Kif15 may somehow discriminate and perform differently between bundle orientations. For example, if Kif15, being a plus end directed motor, can slide anti-parallel microtubules apart, then the Coil-1/microtubule interaction should be strong enough to sustain load as the motor walks. Conversely, if Kif15 transits through parallel bundles, then a weaker Coil-1/microtubule would be advantageous in facilitating motor transport towards the plus end.
To test Coil-1’s orientation sensing ability, a directional pulling assay was performed, similar to the assay in Figure 3A, except using polarity-marked microtubules (Methods) [37]. After the tether was formed in DIC mode, fluorescence visualization unambiguously determined the microtubule orientation. The piezostage was then moved towards the desired microtubule end, and the system was held at a fixed force. The average rupture forces for pulling toward the plus end (7.7 pN ± 2.6 pN, N=26), and the minus end (7.8 pN ± 3.8 pN, N=30) were similar, likely due to the E-hook’s flexibility and reorientation ability. Thus, Coil-1 should respond similarly when Kif15s walk between microtubules in parallel or anti-parallel.
Kif15 Generates Force in Anti-Parallel Microtubule Bundles
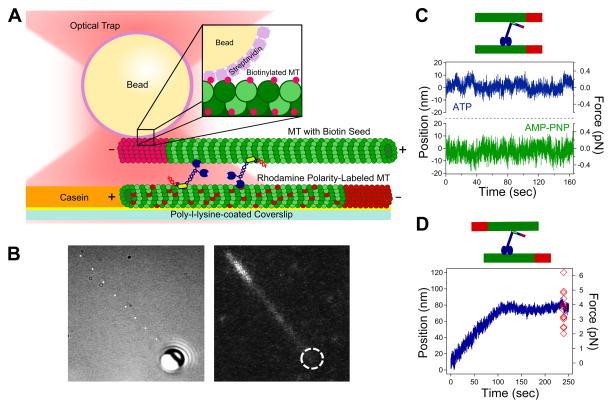
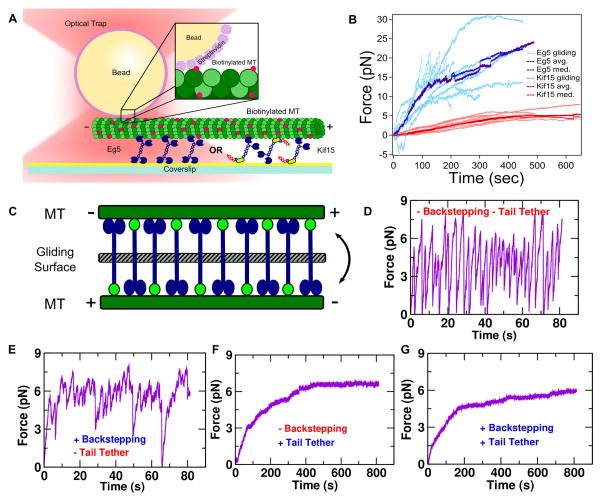
An in vitro optical trapping assay was developed to observe Kif15 behavior in microtubule bundles, a more native environment compared to the bead assay (Figure 4A; Methods, adopted from Shimamoto et al. [25]). After finding a bundle, its orientation (parallel vs. anti-parallel) was determined through fluorescence visualization relative to the known fixed trap position. Since the streptavidin bead only binds the biotinylated tubulin contained in the minus end microtubule seed, the trap center is located at the cargo microtubule minus end (Figure 4B).
Figure 4. Microtubule Bundle Assay.
(A) Assay schematic (Methods). (B) Top microtubule (DIC) with streptavidin bead bound to the biotinylated minus end, bundled to the bottom microtubule (fluorescence). Dashed circle denotes bead location. (C) Parallel bundles yielded baseline force generation. With AMPPNP, only thermal noise is present. Kif15 has a 72% parallel bundle bias (N=43). (D) Anti-parallel bundles yielded a force ramp and plateau at a stall force similar to a single motor (N=17). Red diamonds: individual stall forces. See also Figure S3–4.
Bundles were generated in the presence of ~20 nM Kif15. Parallel bundle traces do not show much force generation but rather exhibit wandering along the baseline with motors antagonizing each other (N=43, Figure 4C). To determine whether this baseline displacement was due to the motors or thermal fluctuations, a control was performed with AMPPNP that locks the motors in place. The trace with ATP present shows distinctive motion in comparison to the AMPPNP trace, revealing that this frustrated motion is due to antagonizing motors (Figure 4C). Anti-parallel bundle traces exhibit a ramp where the motors begin generating force until they stall out against the trap and plateau (Figure 4D). This is similar to Eg5 in microtubule overlaps [25], except the plateau force here remains close to stall forces for individual motors (3.7 pN ± 1.0 pN, N=17). The corresponding translation velocity is 0.44 nm s−1 ± 0.19 nm s−1 (N=17). Also, only 28% of the bundles were anti-parallel in orientation, while the other 72% were parallel. This observation is consistent with Drechsler et al. who showed that Kif15 has a 70% bundling bias for parallel microtubules in the presence of ATP [38].
The same bundling experiment was performed with N700 (Figure S3), behaving similarly to Kif15. There is a 69% bundling bias for the parallel orientation that lacks force generation, whereas in anti-parallel bundles, the force profile shows a ramp followed by a plateau, as with Kif15. Since Kif15 and N700 exhibit similar capabilities, Coil-1’s microtubule bundling role was further investigated through two experiments: (1) bundling with a construct lacking Coil-1 (N420) and (2) with N700 (including Coil-1) but with digested microtubules (Figure S4). Microtubules did not bundle in both cases. Therefore, the Coil-1/E-hook interaction is necessary for bundle formation by Kif15. Together, these results reveal that Kif15 can generate force between anti-parallel microtubules while net progress in parallel microtubules is minimal with frustrated oscillations around baseline.
Kif15 Contains a Force-Feedback Mechanism
The low plateau force in anti-parallel bundles indicates that Kif15 may have a self-governing mechanism that limits its force generating capabilities in large numbers. To test this idea, a combined microtubule gliding/optical tweezers assay was developed (see Methods) to compare Kif15, Eg5 (Figure 5) and N700 (Figure S5). Eg5 behaves similarly to what was found previously, where many motors generate a large amount of force (Figure 5B, blue) [25]. Force plateaus of 20–30 pN were observed (N=8), where some traces reach the stall, fall back briefly with jagged trajectories, and continue to glide persistently. In contrast, Kif15 (N=8) behaves similarly as at very low concentration in the bundle assay. There is a slow and smooth force ramp that finally plateaus minutes later at 5–6 pN. N700 behaved similarly to Kif15 (Figure S5B).
Figure 5. Combined Microtubule Gliding/Optical Tweezer Assay.
(A) Assay schematic (Methods). (B) Loading traces of gliding microtubules on Eg5 (blue, N=8) and Kif15 (red, N=8). Eg5 generates force, sliding faster (0.1 pN/sec ± 0.02 pN/sec) than Kif15 (0.01 pN/sec ± 0.006 pN/sec). Kif15 behaves similarly as in anti-parallel bundles (Figure 4D). (C) Model schematic. Simulation results can be interpreted as a gliding microtubule or anti-parallel bundle (Results). (D–G) Simulated sliding with different model components (Methods). Simulated system contains 5 Kif15 motors. In (F,G), 5 tethers are present with Coil-1 binding to the microtubule at 6.1 pN rupture force while the other end is stationary. Only with tethers are slow ramping and reduced force fluctuation observed. See also Figures S5.
Due to the non-specific nature of motor binding to the glass surface, both the motor heads and Coil-1 can help recruit microtubules to the surface to subsequently glide. The same trace patterns between this gliding assay and the bundle assay reveal that the distributions of motor orientations are likely similar.
To better understand the plateauing behavior, we developed a stochastic model of microtubule gliding by Kif15 (Methods). The trap stiffness, 3-state force-velocity relation, Kif15 stall force, backstep probability, and tail (Coil-1) rupture force were adopted from experimental values. Model parameters unavailable from experiment were adjusted so that resulting behaviors match semi-quantitatively with experiment. We also incorporated “tethered tails” whose opposite motor domain is attached to the substrate and immobile. This mimics the situation where Coil-1 is bound to the microtubule in the gliding assay (Figure 5A). Such scenarios may also arise in microtubule bundles as motor domains waiting for ATP binding are in rigor states (Figure 5C).
We first ran the simulation without backstepping and tail rupturing. The microtubule trajectory followed a sawtooth pattern caused by walking until stall, detachment, and return to the origin (Figure 5D). With backstepping, the force rapidly plateaued, indicating a balance between forward and backward steps was established (Figure 5E). The rapid rise and noisy force trace qualitatively correspond to Eg5 where both ends of the motor walk (hence low chance of forming a tether), which also can backstep (Figure 5B) [24]. On the other hand, when tail rupturing was activated, the force ramped more slowly, and the profile became smoother (Figure 5F,G). For this, backstepping was not essential, but it lowered the plateau force slightly due to the additional force relaxation (Figure 5F,G).
These results provide evidence for Kif15 having a force-feedback mechanism that limits force generation and fluctuation. Tethering by Coil-1 is essential for this feedback. Coil-1 provides support as motor domains walk and transport microtubules. For motor domains awaiting ATP, the corresponding Coil-1, with its high rupture force, serves as a tether that limits microtubule gliding, manifesting a force plateau on the bead.
Discussion
In human cells, it is unknown how Kif15 generates centrosome separation forces necessary to drive spindle assembly [13,17]. Since Eg5 promotes spindle assembly through anti-parallel microtubule-microtubule sliding [2], the prevailing hypothesis is that Kif15 produces outward force through a similar mechanism, prompting several models [13,26,27]. One model posits that Kif15 may exist as a tetramer in solution, and thus is Eg5-like [26]. However, our measurements reported here (Figure S1) and in our previous works [16,27] show that our Kif15 constructs are homodimers in solution and in cells. A recent study from Brouwers et al. supports that Kif15 is a dimer in solution [28]. A second model suggests that Kif15 may complex with TPX2, enabling microtubule-microtubule crosslinking and sliding [13]. Recognizing that Kif15 contains Coil-1, we previously proposed a third model wherein Kif15 dimers drive microtubule-microtubule sliding [27] similarly to fly kinesin-14 (Ncd) [39] and yeast kinesin-8 (Kip3) [40]. This is consistent with the finding that a minimal Kif15 construct (N700) drives microtubule-microtubule sliding [27]. Here, we show that non-truncated Kif15 also catalyzes microtubule-microtubule sliding when anti-parallel. In conjunction with our optical trapping data, our observations provide a biochemical explanation for how Kif15 promotes spindle assembly and yields insight into the functional relationship between Kif15 and Eg5 within the spindle.
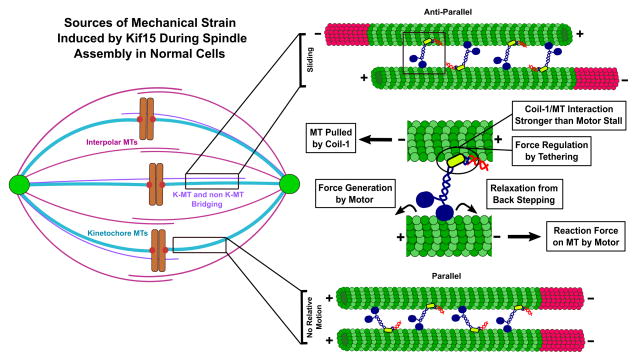
In contrast to a single Eg5 tetramer that walks on both anti-parallel microtubules, Kif15 likely “runs in place” between two microtubules, based on Coil-1 having a rupture force at least two-fold greater than Kif15’s stall. Strong Coil-1/microtubule binding is likely electrostatic, as E-hook removal significantly reduces the strength of the interaction. In an anti-parallel bundle, Kif15 exerts force directed toward the minus end of each microtubule, regardless of whether its motor or Coil-1 domain is bound, causing the microtubules to slide apart (Figure 6). In the parallel case, a Kif15 motor head pushes a microtubule towards its minus end whereas Coil-1 pushes it to the plus end as its motor domain on the other microtubule walks, causing both microtubules to become locked (Figure 6). This model also mirrors the mechanism proposed for fly kinesin-14, a dimeric motor with a non-motor microtubule binding site on its tail [39,41].
Figure 6. Model for Kif15 Function in Microtubule Bundles.
Force generated by Kif15 transmits between microtubule bundles via Coil-1 and the motor head. In the anti-parallel case, the motors collectively slide microtubules apart. In the parallel case, motors yield no relative movement (Discussion).
We also found distinct regulation of force generation between anti-parallel microtubule bundles by Kif15. Force ramps gradually over a period that is much longer than individual motor dwell times to a plateau not significantly larger than the motor’s stall force. Furthermore, force fluctuation is much smaller than observed for Eg5 (Figure 5). Our simulation shows that force regulation by an ensemble of Kif15s is determined mainly by the tethering effect of Coil-1. Although backstepping contributes to force relaxation, its time scale is comparable to the forward stepping time; force fluctuates over a time scale comparable to the motility of individual motors (several seconds). Also, the force fluctuation amplitude is large since it is determined essentially by the motility and detachment of individual motors (Figure 5E). This is the case even with tail rupturing since unbinding of the motor domain occurs more quickly and at lower force. On the other hand, if the motor unbinds from the microtubule mainly via tail rupturing (a tethered case), the tethered tails serve as transient crosslinks between two microtubules, slowing down microtubule sliding and suppressing force fluctuation (Figure 5F,G). A modest level of rupture force is crucial since a very high tether rupture force would correspond to a static crosslink that prevents microtubule sliding.
Force regulation by an ensemble of Kif15 differs significantly from Eg5 [25]. Having motor domains on both ends, force regulation by an Eg5 ensemble occurs on a time scale comparable to that of individual motors. It is, perhaps, for this reason, that Eg5 and Kif15 differ in spindle assembly efficiency. In contrast to Eg5, Kif15 is poor at driving centrosome separation, even under conditions where the motor is overexpressed; spindles remain monopolar for long periods of time, potentially storing energy, and “pop open” when reaching a force threshold [13,17]. The difference in force-producing behaviors of Eg5 and Kif15 also has implications for how the motors distribute within the metaphase spindle. Under normal conditions, Kif15 concentrates on kinetochore microtubules, whereas Eg5 accumulates on spindle microtubules indiscriminately. Ensembles of Eg5 can easily produce forces in excess of 6 pN within anti-parallel overlaps, with single molecules each generating ~1.5 pN [25]. Linear scaling of motors with force implies that small numbers (~4) of Eg5 motors can displace Kif15 molecules from the anti-parallel overlap, a force-feedback mechanism that may cause Eg5 and Kif15 to associate with distinct spindle microtubule geometries [17].
What is the function(s) of Kif15 on kinetochore microtubules? In HeLa cells, Kif15 is required for spindles to remain bipolar at metaphase [13,15]. Similarly, Kif15 slows K5I-triggered collapse of metaphase spindles in RPE-1 cells [42]. These data strongly suggest that Kif15 not only helps bundle microtubules but also somehow generates an outward-directed force within the spindle, even while restricted to kinetochore microtubules. Since Kif15 cannot generate net sliding forces within parallel bundles, we propose that Kif15 motors mechanically link non-kinetochore-microtubules to kinetochore microtubules and slide them apart only when anti-parallel (Figure 6). Near the centromere, such microtubules constitute “bridging fibers”, which link sister kinetochore-fibers and balance inter-kinetochore tension [43]. We speculate that mechanical coupling of non-kinetochore and kinetochore microtubules is more pervasive throughout the spindle, as Kif15 is uniformly distributed along kinetochore microtubules [17]. The distinct function of Kif15 in parallel kinetochore microtubules, its structural and mechanical differences from Eg5, and the jack-knifing effect [17] upon rescuing spindle assembly, demonstrate that Kif15 contributes uniquely to spindle dynamics.
In summary, dimeric Kif15 regulates force generation differently depending on microtubule orientation. Essential to its force-regulating capability is the fine physical balance between the motor head motility properties and the Coil-1/microtubule interaction. Physiologically, Kif15 localizes to kinetochore microtubules to aid in stability and regulate length through plus end tracking, as proposed elsewhere [38], as well as antagonize motor movement that only allows for subtle, well-regulated displacement, as shown here. Kif15 can rescue the function of inhibited Eg5 by utilizing Coil-1 in conjunction with its motor heads to build up mechanical strain and slide anti-parallel microtubules apart, building a bipolar spindle that is necessary for successful mitosis. This mechanism demonstrates the therapeutic importance of Kif15, whose inhibition in tandem with an Eg5 inhibitor could prove to be more effective in the clinic.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Matthew Lang (matt.lang@vanderbilt.edu).
Experimental Model and Subject Details
No cell or animal experiments were conducted.
Method Details
Molecular biology/baculovirus construction
pET15-Kif15-Coil-1 was described previously [27]. pET15-Kif15-N420 was constructed by isothermal assembly. A PCR fragment encompassing Kif15 amino acids 1-420 was generated with Phusion DNA polymerase (Thermo) using the following conditions: 1) 98°C, 30 seconds; 2) 98°C, 10 seconds; 3) 56°C, 30 seconds; 4) 72°C, 90 seconds; 5) repeat steps 2–4 30 times; 6) 72°C, 10 minutes. The fragment was purified and ass embled into pET15 (Novagen) restricted with NdeI and XhoI.
pFASTBAC1-Kif15-GFP (pRO1221) and pFASTBAC1-Kif15-N700-GFP (pRO1222) were constructed by 3 part isothermal assembly into the Bam H1 and Pstl sites of pFASTBAC1 (ThermoFisher Scientific). Kif15 and GFP coding sequences were PCR amplified using conditions described above, but with varying elongation times (step 4). Primers for amplification (see Key Resource Table) included: 1) a (GGS)2 linker in between Kif15 and a 3′-end-positioned GFP; and 2) a hexahistidine tag downstream of GFP. pRO1221 and pRO1222 were used with the Bac-to-Bac system (ThermoFisher Scientific) to create baculoviruses that express Kif15-FL-GFP-His6 and Kif15-N700-GFP-His6, respectively.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| His Tag Antibody, mAb | GenScript | Cat#A00186-100 |
| Penta-His Biotin Conjugate | Qiagen | Cat#34440 |
| Bacterial and Virus Strains | ||
| Bac-to-Bac Baculovirus Expression System | Thermo Scientific | Cat#10359-016 |
| BL21 DE3 | Novagen | Cat#69450 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Kif15 | This paper | n/a |
| N700 | This paper | n/a |
| N420 | This paper | n/a |
| Coil-1 | [27] | n/a |
| Eg5 | [16] | n/a |
| Phusion DNA Polymerase | Thermo Scientific | Cat#F530L |
| Bovine Tubulin | Cytoskeleton | Cat#TL238 |
| PIPES | Sigma | Cat#P-1851 |
| EGTA | Sigma | Cat#E-4378 |
| MgCl2 | Mallinckrodt | Cat#H590 |
| GTP | Cytoskeleton | Cat#BST06 |
| ATP | Sigma | Cat#A26209 |
| Bovine Tubulin | PurSolutions | Cat#1001 |
| Taxol | Cytoskeleton | Cat#TXD01 |
| Subtilisin | Sigma | Cat#P8038 |
| PMSF, phenylmethanesulfonyl fluoride | Sigma | Cat#P7626 |
| GMPCPP | Jena Bioscience | Cat#NU-405L |
| Rhodamine Tubulin | Cytoskeleton | Cat#TL590M |
| Biotinylated Tubulin | Cytoskeleton | Cat#T333P-A |
| Blotting Grade Blocker (casein) | BioRad | Cat#1706404 |
| Glucose Oxidase | Sigma | Cat#G2133 |
| β-D-Glucose | Sigma | Cat#G8270 |
| Catalase | Sigma | Cat#C40 |
| Streptavidin beads | Spherotech | Cat#SVP-05-10 |
| Sulfo-NHS | ThermoScientific | Cat#24510 |
| EDC | ThermoScientific | Cat#22980 |
| Ethanolamine | Sigma | Cat#E9508 |
| Poly-l-lysine | Sigma | Cat#P8920 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| 5′-biotin-TATTGCGTTTCCTCGGTTTC-3′ | IDT | n/a |
| 5′-amine-TTGAAATACCGACCGTGTGA-3′ | IDT | n/a |
| Kif15_N420_5′_IA: GCAGCGGCCTGGTGCCGCGCGGCAGCCATATGGCACCCGGCTGCAAAAC | IDT | n/a |
| Kif15_N420_3′_IA: CGGGCTTTGTTAGCAGCCGGATCCTCGAGCTAAGACTTCTTTTCCTGTTC | IDT | n/a |
| Kif15_pFB_5′: GCAGCGGCCTGGTGCCGCGCGGCAGCCATATGGCACCCGGCTGCAAAAC | IDT | n/a |
| Kif15_FL_(GGS)2_GFP_3′: CTCGCCCTTGCTCACCATTGATCCTCCTGATCCTCCAGATTCACTTCTTTTCTTTTC | IDT | n/a |
| Kif15_FL_(GGS)2_GFP_5′: GAAAAGAAAAGAAGTGAATCTGGAGGATCAGGAGGATCAATGGTGAGCAAGGGCGAG | IDT | n/a |
| GFP_His6_pFB_3′: AGCTTGGTACCGCATGCCTCGAGACTGCAGTCAGTGATGGTGATGGTGATGCTTGTACAGCTCGTCCATG | IDT | n/a |
| Kif15_700_(GGS)2_GFP_3′: CTCGCCCTTGCTCACCATTGATCCTCCTGATCCTCCAATGGCCTCAAAAGCTTG | IDT | n/a |
| Kif15_700_(GGS)2_GFP_5′: CAAGCTTTTGAGGCCATTGGAGGATCAGGAGGATCAATGGTGAGCAAGGGCGAG | IDT | n/a |
| Recombinant DNA | ||
| pET-15 | Novagen | Cat#69661 |
| M13mp18 | [46] | n/a |
| pFASTBAC1 | Thermo Scientific | Cat#10712-024 |
| Software and Algorithms | ||
| Matlab | Mathworks | n/a |
| Stochastic Kinesin Simulation | This paper | n/a |
| Other | ||
| Micro Bio-Spin 30 Column | BioRad | Cat#7326250 |
pFASTBAC-HTc-Eg5-WT was constructed as described previously [16]. The Eg5 coding region from pEGFP-C1-Eg5-WT was amplified and assembled into SalI-EcoRI-restricted pFASTBAC-HTc (ThermoFisher Scientific) by isothermal assembly. This was used with the Bac-to-Bac system (Invitrogen) to create baculoviruses that express His6-Eg5-WT.
Protein expression and purification
His6-Kif15-N420 and His6-Kif15-Coil-1 were expressed in and purified from BL21DE3 cells. Both constructs were expressed for 4 hours in cells cultured at 18°C with 0.4 mM IPTG. For purification, cells were pelleted and resuspended in lysis buffer (PNI [50 mM sodium phosphate, 500 mM NaCl, 20 mM imidazole], 5 mM β-mercaptoethanol, and 1% NP40, and protease inhibitors [1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, and 10 μg/mL each of leupeptin, pepstatin, and chymostatin]). All buffers to purify His6-Kif15-N420 additionally contained 50 μM MgATP. Lysate was incubated with 1 mg/mL lysozyme for 30 minutes, sonicated, and clarified by centrifugation at 35,000 rpm for 1 hour in a Ti 45 rotor (Beckman). ~3 mL of Ni++-NTA agarose (Qiagen) was incubated with the supernatant for 1 hour at 4° C, and then washed with wash buffer (PNI, 5 mM β-ME, 10% glycerol). Protein was eluted with PNI, 5 mM β-ME, and 180 mM imidazole. His6-Kif15-Coil-1 was desalted with a PD10 column (GE Healthcare) equilibrated with 10 mM K-HEPES, pH=7.7, 100 mM KCl, 1 mM DTT, and 20% sucrose. His6-Kif15-N420 was subjected to size exclusion chromatography on a Superdex 200 column equilibrated in 10 mM K-HEPES, pH=7.7, 100 mM KCl, 1 mM DTT, and 0.1 mM MgATP. For both, peak fractions were pooled, aliquoted, frozen in liquid nitrogen, and stored at −80°C.
Kif15-FL-GFP-His6, Kif15-N700-GFP-His6, and His6-Eg5-WT were expressed in Sf9 cells cultured at 27°C for 72 hours. For purification, cells were pelleted and resuspended in lysis buffer (PNI, 5 mM β-mercaptoethanol (β-ME), and 1% NP40, and protease inhibitors [1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, and 10 μg/ml each of leupeptin, pepstatin, and chymostatin]). Lysate was incubated on ice for 15 minutes, sonicated, and clarified by centrifugation at 35,000 rpm for 1 hour in a Ti 45 rotor (Beckman). ~2 ml of Ni++-NTA agarose (Qiagen) was incubated with the supernatant for 1 hour at 4°C, and then washed extensively with wash buffer (PNI, 5 mM β-ME, and 50 μM MgATP). Protein was eluted with PNI, 5 mM β-ME, 0.1 mM MgATP, and 180 mM imidazole, and peak fractions subjected to desalting with a PD10 column (GE Healthcare) equilibrated in 10 mM K-HEPES, pH=7.7, 300 mM KCl, 1 mM DTT, and 0.1 mM MgATP. Protein concentrations were determined using Bradford assays and take into account that Kif15 exists as a dimer in solution and Eg5 exists as a tetramer in solution. Powdered sucrose was added to 20% w/v. Protein was aliquoted, frozen in liquid nitrogen, and stored at −80°C.
Microtubule preparation
Purified bovine tubulin, purchased from Cytoskeleton (TL238), was reconstituted in 25 μL PEM80 buffer (80 mM PIPES (Sigma P-1851), 1 mM EGTA (Sigma E-4378), 4 mM MgCl2 (Mallinckrodt H590), pH adjusted to 6.9 with KOH) supplemented with 1 mM GTP (Cytoskeleton BST06) and kept on ice. Purified tubulin from PurSolutions (bovine, 1001) was also used and reconstituted in the supplied polymerization buffer. 13 μL PEM104 buffer (104 mM PIPES, 1.3 mM EGTA, 6.3 mM MgCl2, pH adjusted to 6.9 with KOH), 2.2 μL 10 mM GTP, and 2.2 μL DMSO were mixed. 4.8 μL of 10 mg/mL tubulin were added to the mixture and allowed to incubate for 40 minutes at 37°C. Subsequently, 2 μL of stabilization solution (STAB, 38.6 μL PEM80, 0.5 μL 100 mM GTP, 4.7 μL 65 g/L NaN3 (Sigma S-8032), 1.2 μL 10 mM Taxol (Cytoskeleton TXD01), 5 uL DMSO (Cytoskeleton)) was added to the stock microtubule solution at room temperature.
Digested microtubules were made by removing the C-terminal E-hook of microtubules with subtilisin. 7.5 μL of pre-formed microtubules were mixed with 0.75 μL of 20 μM subtilisin (Sigma P8038) and was allowed to incubate at 37°C for 40 minutes. To stop digestion, 0.8 μL of 20 mM PMSF (phenylmethanesulfonyl fluoride, Sigma P7626) in DMSO was added to the digested microtubule mixture. 2 μL of STAB solution was then added to the digested microtubules at room temperature.
Polarity-marked microtubules were prepared by making a brightly fluorescent microtubule seed and polymerizing dimmer tubulin from that nucleation point. The microtubule seed was formed using GMPCPP, a non-hydrolysable analog of GTP (Jena Bioscience NU-405L). Rhodamine-labeled tubulin (Cytoskeleton, TL590M) was used in different concentrations to denote the bright seed from the dimmer elongation. First, the seed was polymerized by mixing 13 μL PEM104, 2.2 μL 10 mM GMPCPP, 2.2 μL DMSO, 4 μL non-labeled tubulin (10 mg/mL), and 1 μL rhodamine-labeled tubulin (10 mg/mL). The seed mixture was incubated at 37°C and allowed to incubate for 40 minutes. The elongation solution was made by mixing 13 μL PEM104, 2.2 μL 10 mM GTP, 2.2 μL DMSO, 2 μL non-labeled tubulin (10 mg/mL), and 1.5 μL rhodamine-labeled tubulin (1 mg/mL). The elongation mixture was incubated at 37°C for 1 minute to ensure that the mixture was at least at room temperature. After a minute, 1.5 μL of the seed mixture was added to the elongation mixture and allowed to incubate at 37°C for 40 minutes. Subsequently, 2 μL of STAB solution was added to the polarity-marked microtubules at room temperature.
Microtubule gliding assays
Microtubule gliding assays for Kif15 were performed as described previously [16,27]. Flow cells were constructed with double-stick tape, and motor was added at stock concentration (1.1 μM Kif15; 0.88 μM Eg5) for 3 minutes and X-rhodamine-labeled GMPCPP microtubules (1:9 labeled:unlabeled) at 300 nM tubulin in BRB80 for 3 minutes. Flow cells were washed between each addition with 3 volumes of BRB80, 50 mM KCl, 1 mM MgATP, and 500 μg/mL casein. After the final addition, flow cells were washed with 3 volumes of BRB80, 50 mM KCl, 1 mM MgATP, 500 μg/mL casein, and oxygen scavenging mix (200 μg/mL glucose oxidase, 35 μg/mL catalase, 25 mM glucose, 70 mM β-ME). Microtubule gliding was recorded at 5-s intervals by time-lapse microscopy.
Combined gliding and optical trapping assays were constructed in the same manner except 1:9 biotinylated microtubules were used in place of rhodamine microtubules and 1.25 μm streptavidin beads (Spherotech) were added to the final buffer addition. Beads were trapped in solution and actively bound to the biotinylated microtubules for force measurement.
Bead functionalization
Coil-1 was tethered to 0.44 μm streptavidin polystyrene beads (Spherotech – SVP-05-10) via a 1,010 bp DNA linker functionalized with biotin and a terminal amine. The 1,010 bp DNA linkers were created using PCR and the M13mp18 plasmid template using the following conditions: 1) 98°C, 30 seconds; 2) 98°C, 10 seconds; 3) 49°C, 30 seconds; 4) 72°C, 90 seconds; 5) repeat steps 2–4 35 times; 6) 72°C, 10 minutes. All primers were ordered from Integrated DNA Technologies (IDT). One 5′ biotinylated primer (forward, 5′-biotin-TATTGCGTTTCCTCGGTTTC-3′) and one 5′ amine-functionalized primer (reverse, 5′-amine-TTGAAATACCGACCGTGTGA-3′) were used with the M13mp18 template. After PCR, the amine-functionalized end of the tethers were crosslinked to anti-His tag antibody (GenScript – A00186-100) using sulfo-NHS/EDC chemistry. EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride, Thermo Scientific – 22980), Sulfo-NHS (N-hydroxysulfosuccinimide, Thermo Scientific – 24510), ethanolamine (Sigma – E9508), and PBS (1x, pH 7.4) were used in this reaction. The DNA linkers were then purified using Micro Bio-Spin 30 columns (Biorad). Streptavidin beads, DNA linker (33 ng/μL), and Coil-1 (1 nM) diluted in assay buffer (AB, 80 mM PIPES, 1 mM EGTA, 4 mM MgCl2, 1 mM DTT, 20 μM Taxol, 1 mg/mL casein, 1 mM ATP) were incubated together for 1 hour at 4°C on a rotator to created Coil-1 tethered beads.
Motor functionalized beads were created by binding the C-terminus of the motor to the bead via a His-tag linkage. 0.44 μm streptavidin polystyrene beads (Spherotech – SVP-05-10) were incubated with 0.2 mg/mL biotinylated anti-His tag antibody (Qiagen – 34440) to create anti-His tag coated beads. The beads were washed with PBS (1x, pH 7.4) 4 times by centrifuging at 10,000 rpm for 6 minutes to remove any unreacted biotinylated anti-His. The beads were then incubated with 1 nM of motor diluted in AB for 1 hour at 4°C on a rotator in the presence of a n oxygen scavenging system (5 mg/mL β-D-glucose (Sigma G8270), 0.25 mg/mL glucose oxidase (Sigma G2133), and 0.03 mg/mL catalase (Sigma C40)).
Single molecule optical trapping assays
A flow cell that holds a volume of ~15 μL was assembled using a microscope slide, etched coverslips, and double-sided sticky tape. Before assembly, etched coverslips were incubated in a solution of 100 μL poly-l-lysine (PLL, Sigma P8920) in 30 mL ethanol for 15 minutes. The coverslip was then dried with a filtered air line. After flow cell assembly, microtubules were diluted 150 times from the stock in a solution of PemTax (1 μL 10 mM Taxol in 500 μL PEM80). The diluted microtubules were added to the flow cell via capillary action and allowed to incubate to the PLL surface for 10 minutes. Unbound microtubules were then washed out with 20 μL PemTax. A solution of casein (Blotting-Grade Blocker, Biorad 1706404) diluted in PemTax (1:8 mixture) was then added to the flow cell and allowed to incubate for 10 minutes to block the remainder of the surface to prevent non-specific binding. We found that the assay was very sensitive to the grade of casein used to block the surface and found optimal results with the blotting-grade blocker used here. After the incubation, the flow cell was washed with 50 μL PemTax and 80 μL assay buffer (AB). 20 μL of the bead solution described above (either tethered or with full motor) that had incubated for 1 hour was then added to the flow cell.
Optical trapping measurements were obtained using a custom built instrument with separate trapping and detection systems. The instrument setup and calibration procedures have been described previously [32]. Briefly, beads were trapped with a 1,064 nm laser that was coupled to an inverted microscope with a 100x/1.3 NA oil-immersion objective. Bead displacements from the trap center were recorded at 3 kHz and further antialias filtered at 1.5 kHz. Position calibration and trap stiffness measurements were obtained using custom Matlab (Mathworks) programs.
To ensure that we were at the single molecule limit for both the binding assay and motility assay, the protein-bead ratio was adjusted so that fewer than half of the beads trapped and tested on microtubules showed binding, actually having 5–10% binding the majority of the time.
In the binding assay, beads were trapped in solution and brought close to surface-bound microtubules to allow for binding of Coil-1. Once a tether was confirmed through visual inspection in DIC mode, the tether was centered by an automated two axis piezostage centering routine. Afterward, the bead was again trapped and the piezostage was translated to load the interaction with force. Rupture of the Coil-1-microtubule interaction was confirmed by the bead diffusing away from the microtubule after the trap is turned off once the measurement was complete. A single tether was characterized to have a single break back to baseline in the measurements. This was also the case for the directional pulling assay with polarity-marked microtubules. The exception is that the orientation of the microtubule was checked in fluorescence mode, noted, and then after switching back to DIC mode, the piezostage was translated in the direction of testing (either toward the plus end (dimly fluorescent elongation) or minus end (brightly fluorescent seed) of the microtubule).
In the motor motility assay, a motor-coated bead was trapped in solution and subjected to position calibration and trap stiffness Labview routines. Afterward, the bead was brought close to a surface-bound microtubule to allow for binding. Bead position displacement and force generation were measured for single motor-bound beads.
This is very similar to the unloaded velocity assay except for when the bead is brought close to the microtubule, the trap and detection lasers are turn off, and the bead motion on the microtubule is video-tracked with a DAGE CCD camera. Custom Matlab code based on a cross-correlation method was used to track the bead positions over time [44].
Microtubule bundle assays
Microtubule bundles were generated by adhering a rhodamine-labeled polarity-marked microtubule to a coverslip surface (substrate microtubule) that was further blocked with casein. A motility mixture consisting of a known concentration of motor, polarity-marked microtubules with a biotinylated seed at the minus end (cargo microtubule), and streptavidin coated beads was then added to the flow cell to allow for spontaneous bundle formation through the motors. A free, streptavidin-coated bead was trapped in solution and brought to an apparent microtubule bundle. Bundles appear slightly thicker than normal isolated microtubules under DIC microscopy. Bead binding to an end verifies that a motor-formed bundle was found as beads only bind the biotinylated minus ends of the cargo microtubules. Control experiments in the absence of motor did not bind microtubules.
Single molecule photobleaching assay
Single molecules of FL-Kif15-GFP were imaged at 200 pM in assay buffer through non-specific adsorption to an etched coverslip in a flow cell. The protein was allowed to incubate for 10 minutes, and then unbound motor was washed out with 3 volumes of assay buffer. Images were acquired at 8.66 Hz using TIRF microscopy equipped with an EMCCD camera. GFP was excited with 488 nm at an exposure time of 0.1 sec. Locating individual fluorescent FL-Kif15-GFP molecules for photobleaching analysis was performed from analysis of video frames using custom software written in Matlab (Mathworks) [45].
Data analysis
Nanometer position and piconewton force values were measured using calibration data and trap stiffness measurements from each bead before data acquisition. Those traces were visualized in custom-built Matlab code to determine overall signature of the traces as well as the stall force measurements. Other scripts were used to determine lifetimes, velocities, and local force-velocity relationships. Step-finding code based on a sliding Student’s t-test was used to determine the boundaries of each step to denote a dwell time in between, as well as allow for measuring varied step sizes [46]. A dwell was defined as constant position over time in between steps. In this code, a dwell was measured if the change in the moving average of position was less than 3 nm. This step threshold was chosen due to the defined step sizes of most kinesins (around 8 nm), but could allow for variability without detecting steps from noise (measurements less than 3 nm). Accounting for steps less than 3 nm shows a marked increase in number of steps. However, upon visual inspection of how the trace was been analyze by the code for accuracy, it was found that these steps below 3 nm were indeed noise and therefore not accounted for in averaging.
Force change correlates with the distance the bead has been displaced from the trap center from motor movement. In order to see how velocity correlates with different forces, traces were analyzed over 5 second windows to find the average force, or bead position relative to the trap center. The average velocity that correlates with each window was then calculated using a linear fit. These data sets were then used to construct a force-velocity curve.
Stochastic Simulation
In our computational model, a motor protein contains three elements: motor head, stalk, and non-motor tail. Motor head is a stochastic stepper following the three-state force-velocity relation (Equations 1 and 2). Parameters for the three-state model were taken from experimental data for Kif15 (Table 1). At each time step, forward stepping occurs with probability v(F)dt, where v(F) is the force-dependent speed of the motor, and dt is the time increment per integration step. Under resisting load, a backward step occurs with rate kbefδb, where kb = 0.25/s is the unloaded backward stepping rate, f is the magnitude of the resisting load, and δb = 4 nm is the force sensitivity of the backward steps. Values of kb and δb were chosen so that backstepping occurs about 30% of stepping events, as measured experimentally. Unbinding of a motor head occurs with rate . Here, is the basal off rate, erf(x) is the error function, f is the magnitude of the resisting load, fstall = 2.7 pN is the stall force, and σs = 0.01 pN is the transition width of the error function. An unbound motor head immediately relaxes to its equilibrium position and rebinds to the microtubule with rate 5/s.
For the tail domain, an unbinding event follows a form similar to unbinding of the motor head: , with , ft = 6.1 pN (rupture force of Coil-1), and σt = 0.5 pN. An unbound tail domain immediately relaxes to its equilibrium position and rebinds to the microtubule. The stalk is modeled as a Hookean spring of stiffness 0.5 pN/nm, and force on the motor increases linearly with the distance between its motor head and tail. Choice of model parameters whose values are not experimentally available were made to approximately reproduce the experimental observations in other aspects, such as the motor head stalling and tail rupturing. For the purpose of examining dependence on different model components, in particular, backstepping and tethering (Figure 5D–G), precise values of model parameters are not important.
A microtubule was considered to be rigidly linked to a trapped bead of stiffness 0.04 pN/nm, the same as in the experiment. Time evolution of its position was made via Brownian dynamics method, with a stochastic velocity Verlet integration algorithm. Each run started with five motors. Their motor heads are initially bound to the microtubule and tails are bound to the substrate. After the simulation starts, motors walk along the microtubule and slide it. The microtubule’s position and force were recorded in time. In simulations with tethers (Figure 5F,G), five additional motors were introduced, with their tail domains bound to the cargo microtubule, while their motor domains were affixed to the substrate. For these tethers, rupturing of the tails from the cargo microtubule can happen, but their motor domains are immobile, which mimic the situation that can occur during microtubule gliding (Figure 5A), or in an anti-parallel microtubule bundle where motor heads are in rigor states. A more extensive analysis of this model will be published elsewhere.
Quantification and Statistical Analysis
All experiments were repeated until convergence and high N to ensure that the data were robust. Average and standard deviation are reported throughout the paper and figure legends. Error bars are standard error. The N value for each experiment is reported in each figure. N represents: number of stalls (Figure 1C, Figure 2C), number of steps (Figure 1D, Figure 2D), number of dwells (Figure 1E, Figure 2E), number of velocity traces (Figure 1F), number of processivity measurements (Figure 1G), number of lifetime measurements (Figure 3B, Figure 3E), number of rupture events (Figure 3C, Figure 3F), number of parallel bundle measurements (Figure 4C), number of anti-parallel bundle measurements (Figure 4D), and number of gliding traces (Figure 5). In the custom written algorithms used for analysis, a sliding Student’s t-test was used to determine boundaries (steps, dwells, stalls, change in direction) in each trace.
Supplementary Material
DIC video of a single N700 motor attached to a cargo bead that switches microtubule tracks after a brief pause. The video includes a picture on the left with microtubule orientations and direction of bead movement indicated for clarity.
Kif15 is activated by the addition of GFP on the tail domain, as shown in this microtubule gliding assay. Images were acquired at 2 second intervals and played back at 10 frames per second.
Acknowledgments
This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. 1445197 (D.N.R.). This work was also supported, in part, by the Singapore-MIT Alliance for Research and Technology—BioSyM and NSF Grant No. 1330792 (M.J.L.). R.O. was supported by National Institutes of Health R01 GM086610 and is a scholar of the Leukemia and Lymphoma Society.
Footnotes
Author Contributions
D.N.R. was involved in all aspects of the work including assay development, performing experiments, data analysis, and manuscript preparation. E.G.S., D.N.R., and R.O. expressed and purified the constructs. E.G.S. performed microtubule gliding fluorescence assays. D.K.D. and S.V. aided in assay design. M.S.D. aided in developing the protocol for making digested microtubules. W.H., M.J.L, D.N.R. and R.O designed the simulation. W.H. developed and performed the simulation. D.N.R., M.J.L., R.O., and W.H. prepared the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Heald R, Khodjakov A. Thirty years of search and capture: The complex simplicity of mitotic spindle assembly. J Cell Biol. 2015;211:1103–1111. doi: 10.1083/jcb.201510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapitein LC, Peterman EJG, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- 3.Slangy A, Lane HA, D’Hérin P, Harper M, Kress M, Niggt EA, Blangy A, Lane HA, D’Hérin P, Harper M, et al. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- 4.Enos AP, Morris NR. Mutation of a Gene That Encodes a Kinesin-like Protein Blocks Nuclear Division in A. nidulans. Cell. 1990;60:1019–1027. doi: 10.1016/0092-8674(90)90350-n. [DOI] [PubMed] [Google Scholar]
- 5.Hagan I, Yanagida M. Kinesin-related cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature. 1992;356:74–76. doi: 10.1038/356074a0. [DOI] [PubMed] [Google Scholar]
- 6.Heck MMS, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LSB. The Kinesin-like Protein KLP61F Is Essential for Mitosis in Drosophila. J Cell Biol. 1993;123:665–679. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiae Kinesin-related Gene Products Required for Mitotic Spindle Assembly. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small Molecule Inhibitor of Mitotic Spindle Bipolarity Identified in a Phenotype-Based Screen. Science (80-) 1999;286:971–975. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 9.Roof DM, Meluh PB, Rose MD. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawin KE, LeGuellec K, Philippe M, Mitchison TJ. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 1992;359:540–3. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- 11.Saunders AM, Powers J, Strome S, Saxton WM. Kinesin-5 acts as a brake in anaphase spindle elongation. Curr Biol. 2007;17:453–454. doi: 10.1016/j.cub.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segbert C, Barkus R, Powers J, Strome S, Saxton WM, Bossinger O. KLP-18, a Klp2 Kinesin, Is Required for Assembly of Acentrosomal Meiotic Spindles in Caenorhabditis. Mol Biol Cell. 2003;14:4458–4469. doi: 10.1091/mbc.E03-05-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanenbaum ME, Macůrek L, Janssen A, Geers EF, Alvarez-Fernández M, Medema RH. Kif15 Cooperates with Eg5 to Promote Bipolar Spindle Assembly. Curr Biol. 2009;19:1703–1711. doi: 10.1016/j.cub.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 14.Gayek AS, Ohi R. CDK-1 Inhibition in G2 Stabilizes Kinetochore- Microtubules in the following Mitosis. PLoS One. 2016;11:1–18. doi: 10.1371/journal.pone.0157491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vanneste D, Takagi M, Imamoto N, Vernos I. The Role of Hklp2 in the Stabilization and Maintenance of Spindle Bipolarity. Curr Biol. 2009;19:1712–1717. doi: 10.1016/j.cub.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 16.Sturgill EG, Norris SR, Guo Y, Ohi R. Kinesin-5 inhibitor resistance is driven by kinesin-12. J Cell Biol. 2016;213:213–227. doi: 10.1083/jcb.201507036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sturgill EG, Ohi R. Kinesin-12 differentially affects spindle assembly depending on its microtubule substrate. Curr Biol. 2013;23:1280–1290. doi: 10.1016/j.cub.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharp DJ, Mcdonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, Mitchison TJ, Scholey JM. The Bipolar Kinesin, KLP61F, Cross-links Microtubules within Interpolar Microtubule Bundles of Drosophila Embryonic Mitotic Spindles. J Cell Biol. 1999;144:125–138. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonald KL, O’Toole ET, Mastronarde DN, McIntosh JR. Kinetochore Microtubules in PTK Cells. J Cell Biol. 1992;118:369–383. doi: 10.1083/jcb.118.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mastronarde DN, Mcdonald KL, Ding R, McIntosh JR. Interpolar Spindle Microtubules in PTK Cells. J Cell Biol. 1993;123:1475–1489. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen G, Mickolajczyk KJ, Hancock XWO. The Kinesin-5 Chemomechanical Cycle Is Dominated by a Two-Heads-Bound State. J Biol Chem. 2016;291:20283–20294. doi: 10.1074/jbc.M116.730697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krzysiak TC, Gilbert SP. Dimeric Eg5 Maintains Processivity through Alternating-site Catalysis with Rate-limiting ATP Hydrolysis. J Biol Chem. 2006;281:39444–39454. doi: 10.1074/jbc.M608056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valentine MT, Fordyce PM, Krzysiak TC, Gilbert SP, Block SM. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat Cell Biol. 2006;8:470–6. doi: 10.1038/ncb1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinger JS, Qiu M, Yang G. Report A Nonmotor Microtubule Binding Site in Kinesin-5 Is Required for Filament Crosslinking and Sliding. Curr Biol. 2011;21:154–160. doi: 10.1016/j.cub.2010.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimamoto Y, Forth S, Kapoor TM. Measuring Pushing and Braking Forces Generated by Ensembles of Kinesin-5 Crosslinking Two Article Measuring Pushing and Braking Forces Generated by Ensembles of Kinesin-5 Crosslinking Two Microtubules. Dev Cell. 2015;34:669–681. doi: 10.1016/j.devcel.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drechsler H, McHugh T, Singleton MR, Carter NJ, McAinsh AD. The Kinesin-12 Kif15 is a processive track-switching tetramer. Elife. 2014:e01724. doi: 10.7554/eLife.01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sturgill EG, Das DK, Takizawa Y, Shin Y, Collier SE, Ohi MD, Hwang W, Lang MJ, Ohi R. Report Kinesin-12 Kif15 Targets Kinetochore Fibers through an Intrinsic Two-Step Mechanism. Curr Biol. 2014;24:2307–2313. doi: 10.1016/j.cub.2014.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brouwers N, Mallol Martinez N, Vernos I. Role of Kif15 and its novel mitotic partner KBP in K-fiber dynamics and chromosome alignment. PLoS One. 2017;12:e0174819. doi: 10.1371/journal.pone.0174819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svoboda K, Block SM. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 30.Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct Observation of Kinesin Stepping by Optical Trapping Interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 31.Schnitzer MJ, Visscher K, Block SM. Force production by single kinesin motors. Nat Cell Biol. 2000;2:718–23. doi: 10.1038/35036345. [DOI] [PubMed] [Google Scholar]
- 32.Khalil AS, Appleyard DC, Labno AK, Georges A, Karplus M, Belcher AM, Hwang W, Lang MJ. Kinesin’s cover-neck bundle folds forward to generate force. Proc Natl Acad Sci U S A. 2008;105:19246–19251. doi: 10.1073/pnas.0805147105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carter NJ, Cross RA. Mechanics of the kinesin step. Nature. 2005;435:308–12. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- 34.Hesse WR, Steiner M, Wohlever ML, Kamm RD, Hwang W, Lang MJ. Modular Aspects of Kinesin Force Generation Machinery. Biophysj. 2013;104:1969–1978. doi: 10.1016/j.bpj.2013.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lakämper S, Meyhöfer E. The E-hook of tubulin interacts with kinesin’s head to increase processivity and speed. Biophys J. 2005;89:3223–34. doi: 10.1529/biophysj.104.057505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bell GI. Models for the Specific Adhesion of Cells to Cells. Science (80-) 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 37.Howard J, Hyman AA. Chapter 7 Preparation of Marked Microtubules for the Assay of the Polarity of Microtubule-Based Motors by Fluorescence Microscopy. Methods Cell Biol. 1993;39:105–113. doi: 10.1016/s0091-679x(08)60164-8. [DOI] [PubMed] [Google Scholar]
- 38.Drechsler H, Mcainsh AD. Kinesin-12 motors cooperate to suppress microtubule catastrophes and drive the formation of parallel microtubule bundles. Proc Natl Acad Sci U S A. 2016:E1635–E1644. doi: 10.1073/pnas.1516370113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fink G, Hajdo L, Skowronek KJ, Reuther C, Kasprzak AA, Diez S. The mitotic kinesin-14 Ncd drives directional microtubule – microtubule sliding. Nat Cell Biol. 2009;11:717–23. doi: 10.1038/ncb1877. [DOI] [PubMed] [Google Scholar]
- 40.Su X, Arellano-santoyo H, Portran D, Gaillard J, Vantard M, Thery M, Pellman D. Microtubule-sliding activity of a kinesin-8 promotes spindle assembly and spindle-length control. Nat Cell Biol. 2013;15:948–957. doi: 10.1038/ncb2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braun M, Drummond DR, Cross RA, Mcainsh AD. The kinesin-14 Klp2 organizes microtubules into parallel bundles by an ATP-dependent sorting mechanism. Nat Cell Biol. 2009;11:724–30. doi: 10.1038/ncb1878. [DOI] [PubMed] [Google Scholar]
- 42.Gayek AS, Ohi R. Kinetochore-microtubule stability governs the metaphase requirement for Eg5. Mol Biol Cell. 2014;25:2051–2060. doi: 10.1091/mbc.E14-03-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kajtez J, Solomatina A, Novak M, Polak B, Vukusic K, Rudiger J, Cojoc G, Milas A, Sumanovac Sestak I, Risteski P, et al. Overlap microtubules link sister k-fibres and balance the forces on bi-oriented kinetochores. Nat Commun. 2016;7:10298. doi: 10.1038/ncomms10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gelles J, Schnapp BJ, Sheetz MP. Tracking kinesin-driven movements with nanometre-scale precision. Nature. 1988;331:450–453. doi: 10.1038/331450a0. [DOI] [PubMed] [Google Scholar]
- 45.Shin Y, Davis JH, Brau RR, Martin A, Kenniston JA, Baker TA, Sauer RT, Lang MJ. Single-molecule denaturation and degradation of proteins by the AAA + ClpXP protease. Proc Natl Acad Sci U S A. 2009;106:19340–19345. doi: 10.1073/pnas.0910484106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brady SK, Sreelatha S, Feng Y, Chundawat SPS, Lang MJ. Cellobiohydrolase 1 from Trichoderma reesei degrades cellulose in single cellobiose steps. Nat Commun. 2015;6:1–9. doi: 10.1038/ncomms10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DIC video of a single N700 motor attached to a cargo bead that switches microtubule tracks after a brief pause. The video includes a picture on the left with microtubule orientations and direction of bead movement indicated for clarity.
Kif15 is activated by the addition of GFP on the tail domain, as shown in this microtubule gliding assay. Images were acquired at 2 second intervals and played back at 10 frames per second.