Abstract
We investigated plasma microRNA (miRNA) profiles associated with variation of hyperglycemia, measured as hemoglobin A1c (HbA1c), in two panels of patients with type 1 diabetes (T1D). Using the HTG Molecular Diagnostics EdgeSeq platform, 2,083 miRNAs were measured in plasma from 71 patients included in a screening panel. Quantitative real-time PCR was used to measure the candidate miRNAs in plasma from 95 patients included in an independent replication panel. We found 10 miRNAs replicated in both panels and 4 with high statistical significance. The strongest positive correlations with HbA1c were found with miR-125b-5p (rs = 0.40, P = 6.0 × 10−5) and miR-365a-3p (rs = 0.35, P = 5.9 × 10−4). The strongest negative correlations were found with miR-5190 (rs = –0.30, P = 0.003) and miR-770-5p (rs = –0.27, P = 0.008). Pathway analysis revealed that 50 Kyoto Encyclopedia of Genes and Genomes pathways were significantly enriched by genes targeted by these four miRNAs. The axon guidance signaling pathway was enriched (P < 1 × 10−7) by genes targeted by all four miRNAs. In addition, three other pathways (Rap1 signaling, focal adhesion, and neurotrophin signaling) were also significantly enriched but with genes targeted by only by three of the identified miRNAs. In conclusion, our study identified four circulating miRNAs that were influenced by variation in hyperglycemia. Dysregulation of these miRNAs, which are associated with hyperglycemia in patients with T1D, may contribute to the development of diabetes complications. However, there are multitudes of possible mechanisms/pathways through which dysregulation of these miRNAs may impact risk of diabetes complications.
Introduction
Diabetes is characterized by chronic hyperglycemia. Clinical trials in type 1 diabetes (T1D) and type 2 diabetes (T2D) have demonstrated that the degree of hyperglycemia is a major risk factor for the development of late diabetes complications (1,2). Various mechanisms have been proposed through which hyperglycemia may impact the development of complications such as retinopathy, nephropathy, and neuropathy (3). These mechanisms include nonenzymatic glycation of proteins, generation of oxidative stress, activation of the renin-angiotensin system, DNA methylation, and others (3). Dysregulation of microRNA (miRNA) expression resulting from hyperglycemia was recently proposed to be another such mechanism (4).
miRNAs are endogenous short noncoding RNA molecules that regulate gene expression at the posttranslational level and modulate a variety of physiological processes in both health and disease (5). More than 2,500 human miRNAs are known, and new ones are continuously being discovered. Mature miRNAs bind to their target mRNA(s) and interfere with their translation (6). As such, elevated levels of miRNAs result in lower expression levels of their targeted gene(s) and likely decrease protein levels as well. Conversely, lower levels of miRNAs should result in higher levels of their target gene(s)/protein(s). At least 60% of human protein-coding genes are targeted/regulated by miRNAs (7).
miRNA profiles associated with hyperglycemia in diabetes in humans have not yet been studied in a comprehensive way. Some previous reports investigated associations between circulating miRNAs and hyperglycemia in patients with T2D (8–10). However, these studies were limited in the scope of profiling and used variable normalization methods, and their results remain controversial. Additionally, few reports focused on the effect of hyperglycemia on specific miRNA profiles. For example let-7c-5p and let-7a-5p were shown to be negatively correlated with HbA1c in serum obtained from children with T1D (11), and miR-375 was reported to be increased in plasma from patients with T1D compared with individuals with normal glucose tolerance (12,13).
The aim of this study was to examine profiles of circulating miRNAs according to variation of hyperglycemia measured by HbA1c levels in patients with T1D. Using the HTG Molecular Diagnostics EdgeSeq platform that measured the majority of known circulating miRNAs, we determined concentration of 2,083 miRNAs in plasma obtained from patients included in a screening panel. miRNAs correlated with HbA1c in this panel were examined further for similar associations in plasma obtained from patients included in the replication panel using quantitative real-time PCR (qPCR). The new miRNAs strongly associated with variation in HbA1c were subjected to bioinformatics analysis to identify genes and pathways targeted by these miRNAs.
Research Design and Methods
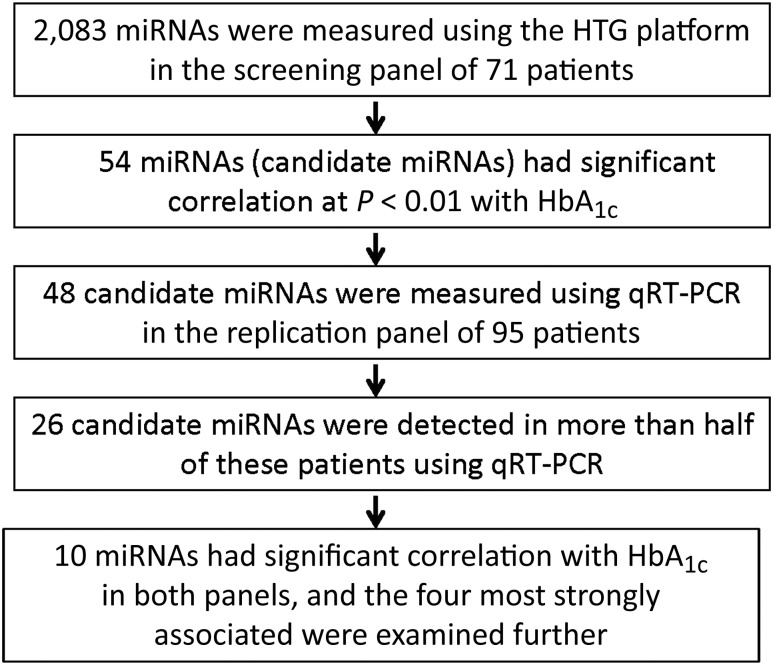
The study design and methods used are outlined in Fig. 1. All subjects included in this research were recruited as part of the Joslin Kidney Study. Study protocols on recruitment and examination of patients in the Joslin Kidney Study and related consent procedures were approved by the Joslin Diabetes Center institutional review board.
Figure 1.
Outline of the study design to identify miRNAs associated with HbA1c.
Screening Panel
Our screening panel consisted of 71 patients randomly selected from the participants in the second Joslin Kidney Study with baseline clinical data, baseline measurements of HbA1c, and baseline specimens of plasma available for use in this study (14). Briefly, the second Joslin Kidney Study is a longitudinal investigation on the natural history of early diabetic nephropathy in nonproteinuric patients (n = 660) with T1D attending the Joslin Clinic between 2003 and 2006. More information about study enrollment and protocols to examine these patients can be found in previous publications (14,15).
miRNA Expression Profiling in the Screening Panel
To determine the concentration of 2,083 known miRNAs in plasma from patients in our screening panel, we applied HTG EdgeSeq technology (HTG Molecular Diagnostics, Inc., Tucson, AZ), a new next-generation sequencing–based miRNA profiling platform (16,17). Fifteen-microliter aliquots of plasma from 71 patients were submitted to HTG Molecular Diagnostics for analysis. The samples were run on an EdgeSeq processor using the EdgeSeq miRNA Whole Transcriptome Assay. After the processor step, samples were individually barcoded by adding sequence adaptors and molecular barcodes to each plasma sample. Barcoded samples were pooled, individually purified, and sequenced on the Illumina NextSeq 500 sequencing platform using a High Output v2 kit (75 cycles) with two-index read. Data were retrieved from the sequencer in the form of FASTQ files and processed using the EdgeSeq parser software (HTG Molecular Diagnostics, Inc.).
Normalization of miRNA Expression Data in the Screening Panel
Normalization of miRNA expression data was performed using the edgeR (version 3.12.1) (18) and limma (version 3.26.9) (19) packages from Bioconductor in R. We considered miRNAs detectable if they had expression levels of >1 counts per million (CPM) in more than half of our samples (>36). We then applied quantile normalization, a nonscaling approach that forces the distribution of read counts in all experimental samples to be equivalent and assumes that 1) most target miRNAs are not differentially expressed and 2) that the true expression distribution of miRNAs is similar across all samples (20).
Replication Panel
We randomly selected 95 patients from among those participating in the Joslin Proteinuria Cohort as a replication panel. Briefly, the Joslin Proteinuria Cohort comprised 424 patients who developed proteinuria while attending the Joslin Clinic between 1991 and 2004 (21). These patients were enrolled into a long-term follow-up study (22). For the replication panel, only patients with chronic kidney disease stages 1 and 2 were considered. For those randomly selected, baseline plasma specimens together with baseline clinical data and baseline HbA1c measurements were used. For comparison of plasma concentration of candidate miRNAs in the replication panel and in individuals without diabetes, a group of 30 healthy parents of T1D patients were used. More information about protocols of enrollment and baseline examination can be found in previous publications (21,22).
RNA Isolation and qRT-PCR
Total RNA was isolated from 60-μL plasma samples from members of the replication panel using the QIAGEN miRNeasy Serum/Plasma Kit (QIAGEN, Valencia, CA) according to the manufacturer’s protocol and as already published (23). Candidate miRNAs identified in the screening panel were used to design a custom miScript miRNA PCR Array (QIAGEN). As some assays were not available on this platform, our custom miScript miRNA PCR Array included 48 miRNAs identified in the screening panel as well as several positive control miRNAs.
Reverse transcription of RNA isolated from plasma was performed using the miScript II RT Kit with miScript HiSpec Buffer (QIAGEN). One and a half microliters of isolated RNA were used to prepare a 10-μL reverse transcription reaction as specified by the manufacturer. The preamplification was performed using miScript PreAMP PCR Kit and a custom miScript PreAMP Primer Mix according to the manufacturer’s protocol. Following the reverse transcription and preamplification, the levels of the candidate miRNAs included on the custom miRNA PCR Array were assayed by SYBR Green–based qRT-PCR using 0.25 μL of diluted cDNA in a 10-μL reaction on an Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). Amplification results were analyzed with the SDS 2.4 software (Applied Biosystems). Ct values >35 were considered as negative amplification. Global mean normalization, as described by Mestdagh et al. (24), was used to normalize the resulting qRT-PCR data. Relative quantification values were calculated using the ΔΔCt method.
The above protocols were applied also to measure the four miRNAs strongly associated with hyperglycemia in a group of 30 control subjects without diabetes.
Determination of HbA1c in Study Patients
Baseline HbA1c was measured during routine clinic visits or for research purposes in the Joslin clinical laboratory. The methods used over time were calibrated according to Diabetes Control and Complications Trial (DCCT) standards: in the 1990s, Bio-Rad HPLC analyzer (Bio-Rad, Hercules, CA); in 2001–2005, Tosoh 2+2 HPLC analyzer; and in 2006 and beyond, Tosoh G7 HPLC analyzer (Tosoh Bioscience, South San Francisco, CA).
Pathway Analysis
To predict the target genes for the candidate miRNAs, we used the open-access web server miRWalk2.0 (zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2) that integrates 12 target prediction algorithms (25). To increase prediction accuracy, we required that putative target genes be identified by at least 6 of the 12 algorithms included in miRWalk2.0. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed for overlapped genes using Database for Annotation, Visualization, and Integrated Discovery (DAVID) (david.ncifcrf.gov) (26). A Fisher exact test P value <0.01 was used to identify significantly targeted pathways in KEGG and enriched gene target pathways obtained from these databases.
Statistical Analysis
All statistical analyses were conducted using SAS for Windows, version 9.4 (SAS Institute, Cary, NC) or R (version 3.2.4). Correlations between HbA1c and candidate miRNA expression levels were estimated by Spearman rank correlation test.
For further assessment of levels of candidate miRNAs, patients were divided into tertiles based on HbA1c levels and data were compared across tertiles. Groupwise comparisons of differences in miRNA levels were assessed by nonparametric one-way ANOVA and Dwass-Steel-Critchlow-Fligner test as appropriate. P values <0.05 were considered statistically significant.
Results
Clinical Characteristics of Study Panels
Clinical characteristics for patients included in the screening and replication panels are presented in Table 1. Importantly, both studies included patients with a very broad range of hyperglycemia. The interquartile range of HbA1c was <7.9% and >9.2% in the screening panel and <8.3% and >10.1% in the replication panel. Relative to patients in the screening panel, the replication panel patients were more often male, were younger, had a shorter duration of diabetes, had slightly higher systolic and diastolic blood pressures, and had lower BMI; the majority also had microalbuminuria or proteinuria. In addition, patients in both panels had normal estimated glomerular filtration rate (eGFR), although it was slightly lower in those included in the replication panel.
Table 1.
Clinical characteristics of participants in screening panel and replication panel
| Screening panel (study 1) | Replication panel (study 2) | |
|---|---|---|
| N | 71 | 95 |
| Male/female, n | 31/40 | 47/48 |
| Age (years) | 43 [31, 52] | 37 [29, 44] |
| Duration of diabetes (years) | 27 [15, 33] | 22 [16, 30] |
| HbA1c (%) | 8.3 [7.9, 9.2] | 9.3 [8.3, 10.1] |
| HbA1c (mmol/mol) | 68 [63, 77] | 78 [67, 87] |
| eGFR (mL/min/1.73 m2) | 108 [98, 120] | 104 [83, 117] |
| ACR (mg/g) | 20 [12, 42] | 335 [16, 944] |
| Systolic blood pressure (mmHg) | 120 [114, 130] | 127 [116, 144] |
| Diastolic blood pressure (mmHg) | 70 [66, 74] | 78 [70, 86] |
| BMI (kg/m2) | 26.6 [24, 31] | 24.8 [23, 29] |
Data are expressed as median [first quartile, third quartile] unless otherwise indicated. All clinical characteristics are from baseline examinations.
Screening for Candidate miRNAs Using the EdgeSeq Platform
Using the EdgeSeq platform, expression profiles were measured for 2,083 miRNAs in patients included in the screening panel. After filtering out nondetectable miRNAs, we normalized the data using quantile normalization with sample weights and examined correlations between these miRNA expression levels and HbA1c levels using Spearman rank correlation test. A total of 54 miRNAs were found to be correlated with HbA1c at P value <0.01 (Supplementary Table 1).
Examination of the Candidate miRNAs in the Replication Panel Using qRT-PCR
Among the 54 candidate miRNAs identified in the screening panel, 48 were assayed in the replication panel. Among these 48 candidate miRNAs, only 26 miRNAs were detected in more than half of the study patients (>48 out of 95 patients). The rest of the miRNAs were detected in a fewer number of patients or not at all (Supplementary Table 1). We performed Spearman rank correlation testing of these 26 miRNAs and HbA1c levels. Ten miRNAs were correlated with HbA1c at P < 0.05 (Table 2). Five miRNAs were positively correlated with HbA1c, and five were negatively correlated with HbA1c. miR-125b-5p (rs = 0.40, P = 6.0 × 10−5) and miR-365a-3p (rs = 0.35, P = 5.9 × 10−4) had the strongest and most significant positive correlation with HbA1c. miR-5190 (rs = −0.30, P = 0.003) and miR-770-5p (rs = −0.27, P = 0.008) had the strongest and most significant negative correlation with HbA1c. Interestingly, plasma levels of miR-125b-5p and miR-365a-3p were strongly and positively correlated with each other as were miR-5190 and miR-770-5p (rs > 0.65, P < 10−6). On the other hand, correlation among the first two and the two others was negative and much weaker (rs = −0.20 and −0.40, P < 0.05) (Supplementary Table 2).
Table 2.
Spearman correlation between HbA1c and the 10 miRNAs with P values <0.01 in HTG platform and with P values <0.05 in qRT-PCR
| Screening panel
(by HTG platform) |
Replication panel
(by qRT-PCR) |
|||
|---|---|---|---|---|
| miRNA | Coefficient | P | Coefficient | P |
| miR-125b-5p | 0.32 | 0.0072 | 0.4 | 0.00006 |
| miR-365a-3p | 0.33 | 0.0046 | 0.35 | 0.00059 |
| miR-7-1-3p | 0.33 | 0.0047 | 0.26 | 0.0107 |
| miR-193a-5p | 0.32 | 0.0059 | 0.25 | 0.014 |
| miR-200c-3p | 0.32 | 0.0064 | 0.24 | 0.0173 |
| miR-5190 | −0.36 | 0.0019 | −0.3 | 0.0028 |
| miR-770-5p | −0.31 | 0.0076 | −0.27 | 0.0075 |
| miR-6799-3p | −0.36 | 0.0023 | −0.21 | 0.0398 |
| miR-6793-5p | −0.36 | 0.0023 | −0.21 | 0.0423 |
| miR-1228-3p | −0.32 | 0.0074 | −0.2 | 0.0466 |
Results for other miRNAs are provided in Supplementary Table 1. Spearman correlation coefficients for the four candidate miRNA with strongest correlation with HbA1c after adjustment for relevant clinical covariates are shown in the legend of Supplementary Table 2.
The correlation between HbA1c and these four candidate miRNAs remained statistically significant after adjustment for age, BMI, systolic blood pressure, and albumin-to-creatinine ratio (ACR). Spearman correlation coefficients were as follows: rs = 0.34, P = 0.0025 for miR-125b-5p; rs = 0.31, P = 0.005 for miR-365a-3p; rs = −0.26, P = 0.019 for miR-5190; and rs = −0.30, P = 0.0074 for miR-770-5p. The other six miRNAs were less correlated with HbA1c in the replication study and were not analyzed further.
Distribution of Plasma Levels of the Four Candidate miRNAs Across Tertiles of HbA1c
To further examine the relationship between the four candidate miRNAs and hyperglycemia, the distribution of each of these miRNAs was plotted across tertiles of HbA1c levels in the screening and in the replication panels separately. The panel-specific tertiles of HbA1c were <8.1%, 8.1–8.9%, and >8.9% for the screening panel and <8.6%, 8.6–9.7%, and >9.7% for the replication panel. Clinical characteristics for tertile subgroups are shown in Supplementary Table 3. In the screening panel, the highest tertile subgroup was slightly older and had slightly higher eGFR than the lowest tertile subgroup. However, these two covariates were not significantly different among the tertiles in the replication panel.
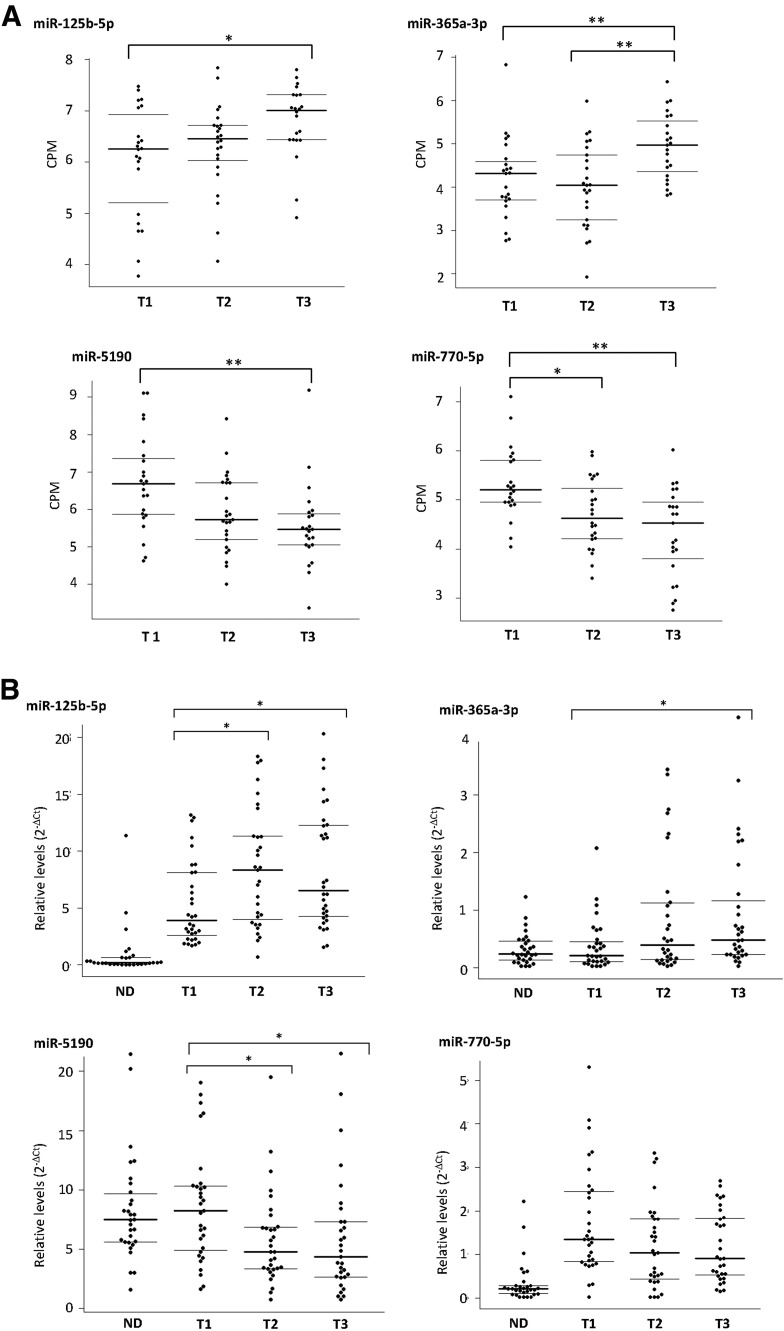
As shown in Fig. 2A, among patients in the screening panel, plasma levels of miR-125b-5p and miR-365a-3p increased with tertiles of HbA1c. Whereas the increase of plasma levels of miR-125b-5p appears to be dose dependent, miR-365a-3p levels were only significantly increased in the highest HbA1c tertile (HbA1c >8.9%). In contrast, plasma levels of miR-5190 and miR-770-5p decreased in a dose-dependent manner with increasing tertiles of HbA1c (i.e., worsening hyperglycemia).
Figure 2.
A: Distribution of concentration of four candidate miRNAs according to tertiles of HbA1c in the screening panel of T1D patients. The plasma concentration of the candidate miRNAs was measured by HTG platform and is expressed as CPM with quantile normalization. The HbA1c tertile cut points were as follows: tertile 1 (T1), HbA1c <8.1%; tertile 2 (T2), 8.1 ≤ HbA1c ≤ 8.9%; and tertile 3 (T3), HbA1c >8.9%. B: Distribution of concentration of four candidate miRNAs according to tertiles of HbA1c in the replication panel of T1D patients and in control subjects without diabetes (ND). The plasma concentrations of the candidate miRNAs were measured by the qRT-PCR method and are expressed as levels relative to the average concentrations of all detected miRNAs. The HbA1c tertile cut points were as follows: T1, HbA1c <8.6%; T2, 8.6 ≤ HbA1c ≤ 9.7%; and T3, HbA1c >9.7%. Horizontal bars indicate median (bold) and first and third quartile in each group. Groupwise comparison between tertiles was assessed by Dwass-Steel-Critchlow-Fligner test. *P < 0.05, **P < 0.01.
As shown in Fig. 2B, among patients in the replication panel, despite a very different method of determining concentrations of miRNAs, the patterns of association of the four candidate miRNAs according to the tertiles of HbA1c were similar to those in patients in the screening panel. Figure 2B also shows the distribution of plasma levels of the four candidate miRNAs obtained from 30 control subjects without diabetes using qRT-PCR. Plasma levels of miR-125b-5p were very low in the control subjects (at detection limits) compared with increasing levels of HbA1c in patients with T1D (P < 0.00001). Plasma levels of miR-365a-3p were low but well detected in control subjects without diabetes and similar to those in the first tertile of HbA1c. On the other hand, plasma levels of miR-5190 were high in control subjects without diabetes and similar to those in the first tertile of HbA1c. The levels of this miRNA were significantly lower in the second and third tertile of HbA1c. Plasma levels of miR-770-5p in control subjects without diabetes were the most contrasting. They were very low, whereas levels of this miRNA were high in the first tertile (best glycemic control) and decreased in a dose-dependent manner with increasing HbA1c.
Correlation of Plasma Levels of the Four Candidate miRNAs With Clinical Characteristics
The four candidate miRNAs showed strong correlation with HbA1c levels in both panels, as shown in Table 2 and Fig. 2. In the screening panel, the levels of miRNAs were not correlated at all with other clinical characteristics such as BMI, eGFR, urinary albumin excretion measured as ACR, and systolic blood pressure (data not shown). In the replication panel, variation of BMI and eGFR were not associated with variation in miRNAs. In contrast, all four miRNAs were significantly associated with ACR and systolic blood pressure (Supplementary Table 3). These associations, however, did not change the correlations of these miRNAs with HbA1c (Supplementary Table 3).
Target Genes and Target Pathways for the Four Candidate miRNAs
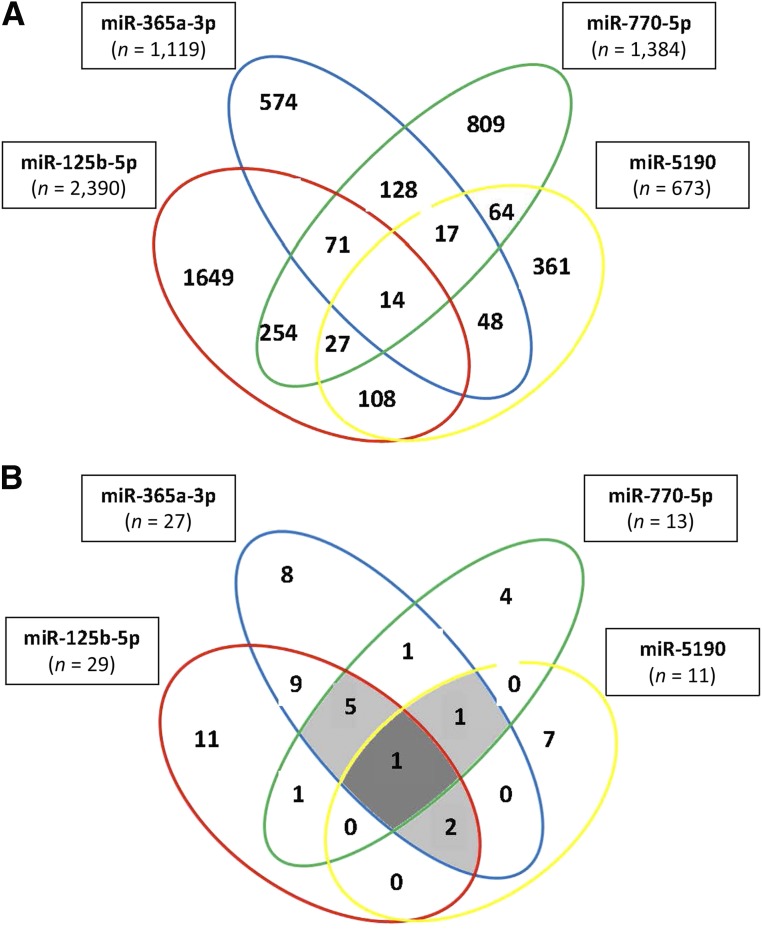
Using miRWalk2.0 software, 5,566 predicted genes were identified by at least 6 prediction algorithms that were targeted by the 4 candidate miRNAs highly correlated with HbA1c. The numbers of genes targeted by each of the miRNAs are shown in boxes in Fig. 3A. Since many of the genes were targeted by multiple miRNAs, the total of unique genes was only 4,391. The multiple-color Venn diagram presented in Fig. 3A shows the distribution of these genes according to the candidate miRNAs that targeted them.
Figure 3.
A: Venn diagram of genes targeted by each of the four candidate miRNAs. The genes were selected as targets if they had overlapped in at least in 6 of 12 prediction algorithms (MicroT4, miRWalk, miRBridge, miRanda, miRDB, miRMap, Pictar2, PITA, miRNAMap, RNAhybrid, RNA22, and TargetScan). B: Venn diagram of pathways enriched with genes targeted by the four candidate miRNAs with P < 0.01. Nineteen pathways related to cancer were eliminated. In the middle of the diagram, there is the axon guidance signaling pathway, which was enriched by genes targeted by all four miRNAs. Other enriched pathways are listed in Table 3 and Supplementary Tables 4 and 5.
To identify pathways enriched with these genes, a KEGG pathway analysis was performed. After eliminating cancer-related pathways, this analysis identified 29 pathways enriched with genes targeted by miR-125b-5p, 27 by miR-365a-3p, 13 by miR-770-5p, and 13 by miR-5190 (a total of 82 pathways) (Supplementary Table 5). Many of the pathways, however, overlapped, so in total there were 50 unique pathways enriched at statistical significance of P < 0.01.
Figure 3B shows distribution of the enriched pathways as a Venn diagram according to the candidate miRNAs, and Table 3 provides the name of the pathways with information about degree of enrichment with targeted genes. The pathway of the axon guidance signaling was enriched with genes targeted by all four miRNAs at very high statistical significance (P < 1.0 × 10−7). There were eight other pathways enriched with genes targeted by three miRNAs, the most statistically significant of which were the Rap1 signaling pathway (P < 7.5 × 10−8), focal adhesion pathway (P < 1.6 × 10−7), and neurotrophin signaling pathway (P < 6.3 × 10−8). Eleven pathways were enriched with genes that were targeted by two miRNAs. The remaining 30 pathways were enriched with genes targeted only by one miRNA, and the degree of enrichment was less statistically significant (Supplementary Table 6).
Table 3.
KEGG pathways enriched by genes targeted by the four candidate miRNAs (cancer pathways are not included)
| miRNA(s) | KEGG pathway | Number of genes in pathway |
|||
|---|---|---|---|---|---|
| Total | Targeted | Expected numbers | P | ||
| All four miRNAs | Axon guidance | 127 | 57 | 29.3 | 1.0E-07* |
| miR-125b-5p, miR-365a-3p, and miR-770-5p | Rap1 signaling | 210 | 78 | 44 | 7.5E-08* |
| Focal adhesion | 206 | 76 | 43.2 | 1.6E-07* | |
| Thyroid hormone signaling | 114 | 46 | 23.9 | 4.20E-06 | |
| MAPK signaling | 255 | 83 | 53.4 | 1.30E-05 | |
| Cholinergic synapse | 111 | 41 | 23.3 | 1.60E-04 | |
| miR-125b-5p, miR-365a-3p, and miR-5190 | Neurotrophin signaling | 120 | 49 | 22.8 | 6.3E-08* |
| cGMP-PKG signaling | 166 | 56 | 31.6 | 8.70E-06 | |
| miR-365a-3p, miR-770-5p, and miR-5190 | Oxytocin signaling | 158 | 43 | 22.9 | 4.20E-05 |
| miR-125b-5p and miR-365a-3p | ErbB signaling | 87 | 34 | 14.3 | 1.10E-06 |
| Sphingolipid signaling | 120 | 42 | 19.8 | 1.40E-06 | |
| Insulin resistance | 108 | 38 | 17.8 | 4.30E-06 | |
| Ras signaling | 226 | 64 | 37.3 | 8.00E-06 | |
| Hepatitis B | 145 | 46 | 23.9 | 8.40E-06 | |
| Insulin signaling | 138 | 44 | 22.7 | 1.20E-05 | |
| Wnt signaling | 138 | 44 | 22.7 | 1.20E-05 | |
| HIF-1 signaling | 98 | 34 | 16.2 | 2.10E-05 | |
| PI3K-Akt signaling | 345 | 84 | 56.9 | 1.30E-04 | |
| miR-125b-5p and miR-770-5p | Adherens junction | 71 | 29 | 12.7 | 1.50E-05 |
| miR-365a-3p and miR-770-5p | HTLV-I infection | 256 | 55 | 30.2 | 1.10E-05 |
HTLV-I, human T-cell leukemia virus type I.
*Pathways with strongest statistical support for enrichment with genes targeted by the four candidate miRNAs.
Genes Targeted by the Four Candidate miRNAs in the Axon Guiding Signaling Pathway
To illustrate the complexity of the possible effects of the candidate miRNAs, we analyzed genes targeted by these miRNAs in the axon guiding signaling pathway. In the KEGG database, this pathway consists of 129 distinct genes. Among these genes, 57 are predicted to be targeted and regulated by the 4 candidate miRNAs, while only 29.3 genes were expected if there was no enrichment (P < 1.0 × 10−7 for difference between targeted and expected) (Table 3). Table 4 lists target genes grouped according to candidate miRNAs. It is striking that a large proportion of these genes had similar names disregarding the grouping. Furthermore, the majority of these target genes belonged to the same gene families such as EPHRINs, SEMPAPHORINs, NETRINs, and SLITs. The target genes included ligands and receptors as well as activators and activated genes.
Table 4.
List of genes in the axon guidance pathway targeted by each of the four candidate miRNAs
| Candidate miRNA | Function | Ephrins | Semaphorins | Netrins | Slits | Other genes |
|---|---|---|---|---|---|---|
| miR-125b-5p specific |
Ligand |
EFNA5 |
SEMA3E
SEMA4B
SEMA4C |
SLIT1 |
DPYSL5
GSK3B
LIMK1
MAPK3
PAK6
PPP3R2 |
|
| Receptor |
EPHA2
EPHA8
EPHB3 |
PLXNA1 |
ROBO2 |
|||
| Activator |
SRGAP2 |
|||||
| Activated |
RAC3 |
|||||
| miR-365a-3p specific |
Ligand |
EFNA3 |
SEMA6D |
ARHGEF12
RND1 |
||
| Activated |
RAC1 |
|||||
| miR-125b and miR-365a |
Ligand |
SEMA5A |
NTN1 |
NFATC4 |
||
| miR-770-5p |
Ligand |
EFNB2
EFNB3 |
SEMA6A |
ABL1
LIMK2
NCK2
NFATC3
PPP3CA
RGS3 |
||
| Receptor |
EPHA3
EPHB1 |
NRP1 |
||||
| miR-5190 |
Ligand |
SEMA3C |
MET |
|||
| Receptor |
PLXNC1 |
|||||
| miR-770 and miR-125b, or miR-365a |
Ligand |
SEMA4D
SEMA5B |
KRAS
MAPK1 |
|||
| Receptor |
EPHA7 |
UNC5C |
||||
| Activator |
SRGAP1 |
|||||
| Activated |
RAC2 |
|||||
| miR-5190 and miR-125b, and/or miR-365a | Ligand |
SLIT3 |
ABLIM3 CXCL12 NFATC2 | |||
| Receptor |
EPHA4 |
UNC5B
UNC5D |
||||
| Activator | SRGAP3 |
Discussion
Our study is the first to comprehensively examine nearly all known circulating miRNAs for association with variation in hyperglycemia in T1D. Using a new next-generation sequencing–based miRNA platform, we measured plasma levels of 2,083 known miRNAs in exploratory panel and found that 54 miRNAs correlated with hyperglycemia. Among these miRNAs, 10 were confirmed by qRT-PCR in an independent replication panel and 4 of them—miR-125b-5p, miR-365a-3p, miR-5190, and miR-770-5p—showed very significant correlation with HbA1c levels. These correlations remained significant disregarding adjustment for various clinical characteristics including presence or absence of kidney complications. Bioinformatics analyses showed that these four candidate miRNAs target the expression of more than 4,000 genes/proteins that may impact 50 KEGG pathways. Four of these pathways had the highest statistical support for enrichment with genes targeted by these putative hyperglycemia-regulated miRNAs. These include the axon guidance signaling pathway, Rap1 signaling pathway, focal adhesion pathway, and neurotrophin signaling pathway. Overall, our study showed that hyperglycemia impacts expression of a relatively few specific circulating miRNAs that target thousands of genes and many dozens of pathways. The interpretation and experimental validation of our findings creates a formidable challenge considering that our study measured circulating miRNAs and not intracellular miRNAs in humans. Furthermore, not all human miRNAs are present in animals. Below, we discuss our findings in the context of the limited literature regarding the pathways and miRNAs involved.
The axon guidance signaling pathway contains 127 genes/proteins, and in our study, 57 of these genes were predicted to be targeted by the 4 candidate miRNAs (only 29 genes would be expected by chance). This pathway is involved in the formation of neuronal network, and it is guided by several guidance factors/proteins, including such ligands as semaphorins, ephrins, slits, and netrins and their corresponding receptors neurophilin, Eph, Robo1-4, and UNC5. Singh et al. (27) reported that mRNAs for genes encoding axon guidance signaling pathway were upregulated in the hyperglycemic state in human endothelial cells, and recent reports implicated some of these proteins and this pathway in the development of diabetic retinopathy and nephropathy (28,29).
The Rap1 signaling pathway contains 210 genes/proteins; 78 of these genes are predicted to be targeted by 3 putative hyperglycemia-regulated miRNAs identified in our study (44 would have been expected by chance). Rap1 is a small GTPase that controls diverse processes, including cell adhesion, cell–cell junction formation, and cell polarity. Rap1 plays a dominant role in the control of cell–cell and cell–matrix interactions by regulating the function of integrins and other adhesion molecules in various cell types (30). Rap1 also regulates MAPK activity in a manner highly dependent on the context of cell types (31). Hyperglycemia inhibits Rap1 expression and its activity, which leads to tubular cell injury in patients with diabetic nephropathy and STZ-induced diabetic animal models (32).
The focal adhesion pathway contains 206 genes/proteins. A total of 76 genes in this pathway are predicted to be targeted by 3 putative hyperglycemia-regulated miRNAs (43 would be expected by chance). Cell–matrix adhesions play important roles in biological processes including cell motility, proliferation, differentiation, regulation of gene expression, and cell survival. Focal adhesion is also related to growth factor–mediated signaling in similar morphological alterations and modulation of gene expressions, suggesting considerable cross talk between adhesion and growth factor–mediated signaling as well as the Rap1 signaling pathway (33).
The neurotrophin signaling pathway contains 120 genes/proteins, and 3 putative hyperglycemia-regulated miRNAs identified in our study are predicted to target 49 of the genes in this pathway (23 would have been expected by chance). Neurotrophin/Trk signaling plays an important role for neural development and additional higher-order activities, including learning and memory. A recent study indicates that diabetes-induced alterations in neurotrophin expression play multiple roles in steering events such as neurodegeneration, inflammation, and vascular dysfunction in the diabetic retina (34).
In contrast to the results of our pathway analysis that showed that all the putative hyperglycemia-regulated miRNAs might target a large number of genes in multiple pathways, many of the previous publications aimed at studying single miRNAs and focused on limited downstream effects. For example, studies on miR-125b have focused on its impact on the regulation of specific downstream gene targets. Villeneuve et al. (35) reported that miR-125b levels were upregulated in diabetic db/db vascular smooth muscle cells compared with those in control mice and that this miRNA was able to target and downregulate Suv39h1, a histone methyltransferase that mediates histone H3-lysine-9 trimethylation (H3K9me3). The authors also found that histone H3K9me3 levels were decreased at the promoters of key inflammatory genes interleukin-6 and monocyte chemoattractant protein 1. Other studies have reported that miR-125b-5p expression is involved in the immune responses to viral infection (36). Interestingly, plasma miR-125b-5p levels and hyperglycemia were considered risk factors for poor prognosis in children with viral encephalitis (37). No similar research has been done regarding the biology of the other putative hyperglycemia-regulated miRNAs identified in our study, i.e., miR-365a-3p, miR-5190, and miR-770-3p.
Among six other miRNAs that were correlated with HbA1c in our study, three (miR-193a-5p, miR-200c-3p, and miR-7-1-3p) were positively correlated with HbA1c and three (miR-1228-3p, miR-6793-5p, and miR-6799-3p) were negatively correlated. Because these miRNAs were only nominally significant, the role of these miRNAs in the context of biological pathways was not explored. However, the biology of some of these has been previously investigated. Yu et al. (38) reported that miR-193a-5p suppresses the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. They also found that miR-193a-3p targeted PIK3R3 and mTOR by binding its 3′ untranslated region directly in non-small-cell lung cancer. Zhang et al. (39) showed that miR-200c-3p was upregulated in aortas from db/db mice and renal arteries from humans with diabetes. They also demonstrated that overexpression of miR-200c-3p impaired endothelium-dependent relaxations in nondiabetic mouse aortas, whereas suppression of miR-200c by anti–miR-200c enhanced endothelium-dependent relaxations in diabetic db/db mice. In addition, hyperglycemia upregulated miR-200c-3p expression levels through reactive oxygen species elevation and leads to endothelial dysfunction via the miR-200c/ZEB1/COX-2 signaling cascade. Also, miR-200c is involved in extracellular matrix regulation (such as collagens) through Zeb1 (40) and Akt activation through FOG2 (41,42) in diabetic nephropathy. In a human study, Bhatt et al. (43) reported that serum miR-200 family expression was significantly elevated in patients with long-standing T1D with severe complications. Hence, these two miRNAs may be involved in glucose metabolism or pathogenesis of diabetes complications. Yan and Zhao (44) reported that miR-1228-3p was downregulated in apoptotic cells and that overexpression of miR-1228-3p reduced MOAP1 expression and delayed the progression of stress-induced cell apoptosis. miR-6799-3p and miR-6793-5p are new miRNAs and have not been investigated.
Osipova et al. (45) reported diabetes-associated miRNAs in 68 pediatric patients with T1D. They demonstrated that miR-21 and miR-210 were significantly upregulated in the plasma of T1D patients compared with subjects without diabetes. Erener et al. (11) reported that let-7c-5p and let-7a-5p were negatively correlated with HbA1c in serum from 19 children with T1D. miR-375 was reported to be increased in plasma from patients with T1D compared with individuals with normal glucose tolerance (12,13). None of these findings were confirmed in our study. The discrepancies can likely be due to the lack of replication studies in the previous research, the methods used to measure miRNAs in these studies, and the analysis implemented in these studies, including normalization that relied on the use of an exogenous spike-in control (i.e., Caenorhabditis elegans miR-39) as the only reference.
The mechanisms of increasing or decreasing circulating miRNA levels in the hyperglycemic state are still unknown. Simionescu et al. (46) reported that hyperglycemia increased the levels of Drosha, DGCR8, and Dicer expression and several miRNAs (miR-223, miR-92a, miR-486, miR-122, miR-125a, and miR-146a) in human macrophages. Nishikawa et al. (47) reported that hyperglycemia causes pathological changes in small vessels and results in tissue damage in endothelial cells. This damage may result in secretion or leakage of the miRNAs into circulation that then communicates with distant cell/organ target and targeted genes/proteins/pathways. Although we could not determine whether the putative hyperglycemia-regulated miRNAs identified in our study were secreted from certain cells or whether they are derived from damaged cells, circulating miRNA profiles can be useful not only to better understand the pathogenesis of diabetes but also to identify novel therapeutic targets to prevent or treat diabetes complications.
Finally, some strengths and limitations of our study need to be considered. To increase the reliability of our findings, we implemented a two-stage study design. In the first stage, candidate miRNAs were searched in a comprehensive way using the EdgeSeq platform to sequence and quantify 2,083 miRNAs in the screening panel. In the second stage, due to limited resources, we used a qRT-PCR to validate the initial findings using the replication panel. Apparently, the latter approach was not sensitive enough to detect nearly half of the candidate miRNAs detected in the first stage. At this time, we cannot establish the reasons for the discrepancies. However, it is possible that extraction of RNA from plasma to run qRT-PCR may result in a loss of certain species of miRNAs. This would suggest that more candidate miRNAs may have been regulated by hyperglycemia than our study identified. Furthermore, by using qRT-PCR and studying control subjects without diabetes, we showed that two out of four candidate miRNAs examined had very low expression compared with that in patients with diabetes. The biology for these differences is unknown but was most likely unrelated to level of hyperglycemia. Additionally, our study is a clinical observation, and we could not investigate the mechanisms of regulation of miRNA expression, regulation of expression at the locus of each miRNA, and regulation through degradation/modification of these miRNAs. Further studies are necessary to investigate these issues.
Supplementary Material
Article Information
Acknowledgments. The authors would like to thank Jonathan Dreyfuss and Hui Pan of the Bioinformatics Core of the Joslin Diabetes Center for advice regarding the normalization of miRNA data.
Funding. This research was supported by National Institutes of Health grant DK041526-23 and Novo Nordisk Foundation grant NNF14OC0013659 (A.S.K.) and the Mary K. Iacocca Fellowship; the Sunstar Foundation, Japan (Hiroo Kaneda Scholarship); and the Foundation for Growth Science, Japan (E.S.).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. E.S. designed the study, performed the experiments, analyzed data, and wrote the manuscript. M.G.P. contributed to analysis of data and edited the manuscript. Z.I.M.D. contributed to implementation of experiments and analysis of data. A.M.S. was responsible for data management of clinical and experimental data and provided statistical analysis of data. M.A.N. contributed to the editing of the manuscript. A.S.K. developed hypotheses for the study, supervised implementation of the study and analyses of data, and contributed to the writing and editing of the manuscript. A.S.K. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db17-1207/-/DC1.
References
- 1.Nathan DM, Genuth S, Lachin J, et al.; Diabetes Control and Complications Trial Research Group . The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N Engl J Med 1993;329:977–986 [DOI] [PubMed] [Google Scholar]
- 2.Stratton IM, Adler AI, Neil HA, et al. Association of glycaemia with macrovascular and microvascular complications of type 2 diabetes (UKPDS 35): prospective observational study. BMJ 2000;321:405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett EJ, Liu Z, Khamaisi M, et al. Diabetic microvascular disease: an endocrine society scientific statement. J Clin Endocrinol Metab 2017;102:4343–4410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexandru N, Badila E, Weiss E, Cochior D, Stępień E, Georgescu A. Vascular complications in diabetes: microparticles and microparticle associated microRNAs as active players. Biochem Biophys Res Commun 2016;472:1–10 [DOI] [PubMed] [Google Scholar]
- 5.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010;11:597–610 [DOI] [PubMed] [Google Scholar]
- 6.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 2009;10:126–139 [DOI] [PubMed] [Google Scholar]
- 7.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zampetaki A, Kiechl S, Drozdov I, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 2010;107:810–817 [DOI] [PubMed] [Google Scholar]
- 9.Kong L, Zhu J, Han W, et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol 2011;48:61–69 [DOI] [PubMed] [Google Scholar]
- 10.Pescador N, Pérez-Barba M, Ibarra JM, Corbatón A, Martínez-Larrad MT, Serrano-Ríos M. Serum circulating microRNA profiling for identification of potential type 2 diabetes and obesity biomarkers. PLoS One 2013;8:e77251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erener S, Marwaha A, Tan R, Panagiotopoulos C, Kieffer TJ. Profiling of circulating microRNAs in children with recent onset of type 1 diabetes. JCI Insight 2017;2:e89656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latreille M, Herrmanns K, Renwick N, et al. miR-375 gene dosage in pancreatic β-cells: implications for regulation of β-cell mass and biomarker development. J Mol Med (Berl) 2015;93:1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seyhan AA, Nunez Lopez YO, Xie H, et al. Pancreas-enriched miRNAs are altered in the circulation of subjects with diabetes: a pilot cross-sectional study. Sci Rep 2016;6:31479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krolewski AS, Niewczas MA, Skupien J, et al. Early progressive renal decline precedes the onset of microalbuminuria and its progression to macroalbuminuria. Diabetes Care 2014;37:226–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosolowsky ET, Ficociello LH, Maselli NJ, et al. High-normal serum uric acid is associated with impaired glomerular filtration rate in nonproteinuric patients with type 1 diabetes. Clin J Am Soc Nephrol 2008;3:706–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma P, Retz M, Siefker-Radtke A, et al. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single-arm, phase 2 trial. Lancet Oncol 2017;18:312–322 [DOI] [PubMed] [Google Scholar]
- 17.Lizarraga D, Huen K, Combs M, Escudero-Fung M, Eskenazi B, Holland N. miRNAs differentially expressed by next-generation sequencing in cord blood buffy coat samples of boys and girls. Epigenomics 2016;8:1619–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014;15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003;19:185–193 [DOI] [PubMed] [Google Scholar]
- 21.Rosolowsky ET, Skupien J, Smiles AM, et al. Risk for ESRD in type 1 diabetes remains high despite renoprotection. J Am Soc Nephrol 2011;22:545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skupien J, Warram JH, Smiles AM, et al. The early decline in renal function in patients with type 1 diabetes and proteinuria predicts the risk of end-stage renal disease. Kidney Int 2012;82:589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pezzolesi MG, Satake E, McDonnell KP, Major M, Smiles AM, Krolewski AS. Circulating TGF-β1–regulated miRNAs and the risk of rapid progression to ESRD in type 1 diabetes. Diabetes 2015;64:3285–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mestdagh P, Van Vlierberghe P, De Weer A, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol 2009;10:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods 2015;12:697. [DOI] [PubMed] [Google Scholar]
- 26.Dennis G Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:3. [PubMed] [Google Scholar]
- 27.Singh KK, Mantella LE, Pan Y, et al. A global profile of glucose-sensitive endothelial-expressed long non-coding RNAs. Can J Physiol Pharmacol 2016;94:1007–1014 [DOI] [PubMed] [Google Scholar]
- 28.Cerani A, Tetreault N, Menard C, et al. Neuron-derived semaphorin 3A is an early inducer of vascular permeability in diabetic retinopathy via neuropilin-1. Cell Metab 2013;18:505–518 [DOI] [PubMed] [Google Scholar]
- 29.Aggarwal PK, Veron D, Thomas DB, et al. Semaphorin3a promotes advanced diabetic nephropathy. Diabetes 2015;64:1743–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim C, Ye F, Ginsberg MH. Regulation of integrin activation. Annu Rev Cell Dev Biol 2011;27:321–345 [DOI] [PubMed] [Google Scholar]
- 31.York RD, Yao H, Dillon T, et al. Rap1 mediates sustained MAP kinase activation induced by nerve growth factor. Nature 1998;392:622–626 [DOI] [PubMed] [Google Scholar]
- 32.Xiao L, Zhu X, Yang S, et al. Rap1 ameliorates renal tubular injury in diabetic nephropathy. Diabetes 2014;63:1366–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner CE. Paxillin and focal adhesion signalling. Nat Cell Biol 2000;2:E231–E236 [DOI] [PubMed] [Google Scholar]
- 34.Mysona BA, Shanab AY, Elshaer SL, El-Remessy AB. Nerve growth factor in diabetic retinopathy: beyond neurons. Expert Rev Ophthalmol 2014;9:99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villeneuve LM, Kato M, Reddy MA, Wang M, Lanting L, Natarajan R. Enhanced levels of microRNA-125b in vascular smooth muscle cells of diabetic db/db mice lead to increased inflammatory gene expression by targeting the histone methyltransferase Suv39h1. Diabetes 2010;59:2904–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun YM, Lin KY, Chen YQ. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol 2013;6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao QL, Ma YX, Yuan DW, Zhang QC, Zeng J, Li H. MicroRNA-125b in peripheral blood: a potential biomarker for severity and prognosis of children with viral encephalitis. Neurol Sci 2017;38:1437–1444 [DOI] [PubMed] [Google Scholar]
- 38.Yu T, Li J, Yan M, et al. MicroRNA-193a-3p and -5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene 2015;34:413–423 [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, Liu J, Qu D, et al. Inhibition of miR-200c restores endothelial function in diabetic mice through suppression of COX-2. Diabetes 2016;65:1196–1207 [DOI] [PubMed] [Google Scholar]
- 40.Kato M, Arce L, Wang M, Putta S, Lanting L, Natarajan R. A microRNA circuit mediates transforming growth factor-β1 autoregulation in renal glomerular mesangial cells. Kidney Int 2011;80:358–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hyun S, Lee JH, Jin H, et al. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell 2009;139:1096–1108 [DOI] [PubMed] [Google Scholar]
- 42.Park JT, Kato M, Yuan H, et al. FOG2 protein down-regulation by transforming growth factor-β1-induced microRNA-200b/c leads to Akt kinase activation and glomerular mesangial hypertrophy related to diabetic nephropathy. J Biol Chem 2013;288:22469–22480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhatt S, Gupta MK, Khamaisi M, et al. Preserved DNA damage checkpoint pathway protects against complications in long-standing type 1 diabetes. Cell Metab 2015;22:239–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan B, Zhao JL. miR-1228 prevents cellular apoptosis through targeting of MOAP1 protein. Apoptosis 2012;17:717–724 [DOI] [PubMed] [Google Scholar]
- 45.Osipova J, Fischer DC, Dangwal S, et al. Diabetes-associated microRNAs in pediatric patients with type 1 diabetes mellitus: a cross-sectional cohort study. J Clin Endocrinol Metab 2014;99:E1661–E1665 [DOI] [PubMed] [Google Scholar]
- 46.Simionescu N, Niculescu LS, Carnuta MG, et al. Hyperglycemia determines increased specific microRNAs levels in sera and HDL of acute coronary syndrome patients and stimulates microRNAs production in human macrophages. PLoS One 2016;11:e0161201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishikawa T, Edelstein D, Du XL, et al. Normalizing mitochondrial superoxide production blocks three pathways of hyperglycaemic damage. Nature 2000;404:787–790 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.