Abstract
The study of Drosophila muscle development dates back to the middle of the last century. Since that time, Drosophila has proved to be an ideal system for studying muscle development, differentiation, function, and disease. As in humans, Drosophila muscle forms via a series of conserved steps, starting with muscle specification, myoblast fusion, attachment to tendon cells, interactions with motorneurons, and sarcomere and myofibril formation. The genes and mechanisms required for these processes share striking similarities to those found in humans. The highly tractable genetic system and imaging approaches available in Drosophila allow for an efficient interrogation of muscle biology and for application of what we learn to other systems. In this article, we review our current understanding of muscle development in Drosophila, with a focus on myoblast fusion, the process responsible for the generation of syncytial muscle cells. We also compare and contrast those genes required for fusion in Drosophila and vertebrates.
Keywords: Drosophila, muscle development, membrane fusion, actin cytoskeleton
Introduction
From Drosophila to human, skeletal muscles develop and mature through similar steps: cells of the mesoderm are first specified to be skeletal muscle cells, which then undergo cell-cell fusion to add mass to these cells. After fusion is completed, sarcomeres assemble to provide contractile force, and the muscle cell builds connections with its surrounding tissues, such as tendons and motor neurons. Many of the genes that are critical for these steps are conserved across species. This also holds true for nautilus, the fly ortholog of vertebrate MYOD, which is a key regulator of myogenic differentiation in Drosophila (Michelson et al., 1990; reviewed by (Abmayr and Keller, 1998)). As discussed below, Nautilus interacts with other transcription and chromatin regulators, and determines muscle identity, morphology, and size (reviewed in (Dobi et al., 2015)).
During Drosophila’s life cycle, two periods of myoblast fusion occur to form the larval muscle system and adult muscle system, respectively (Figure 1). The larval body wall muscles form during the embryonic stage. These muscles have been intensively studied as they represent muscle at its simplest: a single muscle fiber forms a single muscle. The adult muscle system forms during the pupal stage of development. In contrast to the larva, each single adult muscle is composed of multiple muscle fibers and thus more closely resembles the muscle organization found in vertebrate skeletal muscle (reviewed by Dobi et al., 2015, Taylor, 2006). This pertains in particular to the indirect flight muscles, the largest and best-studied muscle set of the adult fly. In this review, we first give a brief overview of muscle specification (Figure 2) and then focus on a discussion of the current studies in myoblast fusion both in Drosophila (Figure 3, Table 1) and vertebrates (Table 2).
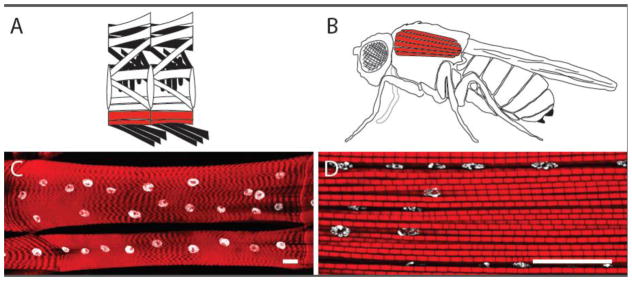
Figure 1. Larval and adult Drosophila muscles.
A) Schematic representation of the larval muscles. The Ventral Longitudinal (VL) muscles 3 (top) and 4 (bottom) are highlighted in red. B) Schematic representation of the adult muscles. The Dorsal Longitudinal Muscles (DLMs) that compose the major portion of the Indirect Flight Muscles are highlighted in red. C) Representative image of the 3rd instar larval VL muscles 3 and 4. D) Representative image of the DLMs where it is possible to observe multiple myofibrils and nuclei. C and D) Actin labeling in sarcomere, red; Nuclei, white. Scale bars, 25μm.
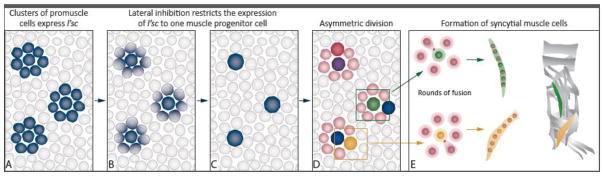
Figure 2. Specification, Fusion and Differentiation of Muscle Cells.
A) Expression of l’sc occurs in clusters of mesodermal cells with competency to become muscle progenitor cells. B and C) The activity of the Notch and RTK-Ras pathways restricts the expression of l’sc to a single muscle progenitor cell of a particular identity. D) The remaining cells from the initial cluster of promuscle cells become FCMs (pink). The muscle progenitor cells undergo asymmetric division to form either two founder cells (red and purple) or one founder cell (green or yellow) and an adult muscle progenitor (blue). E) Founder cells shown on 2D express different identity genes and undergo several rounds of fusion (actin foci, red) to form syncytial muscle cells with different characteristics. The green founder cell expresses apterous and forms muscle LT1 and the yellow founder cell expresses nautilus to form muscle VA1 (modified from (Dobi et al., 2015)). The myotubes maturate by attaching to tendon cells and developing sarcomeres (not shown). The final position of these muscles is highlighted in the hemisegment schematic on the right.
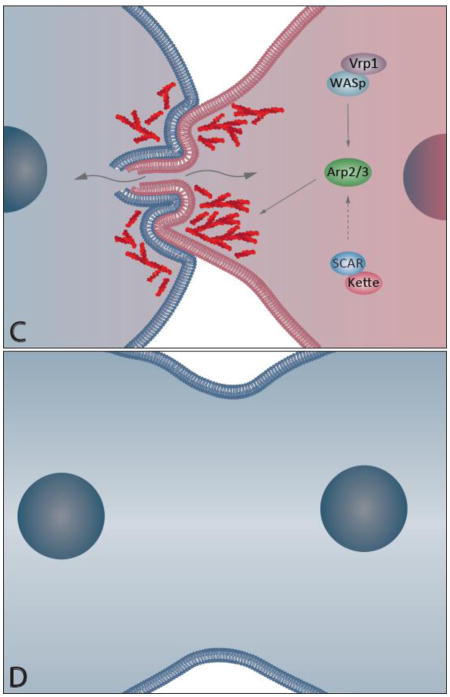
Figure 3. Schematic representation of myoblast fusion.
In the Drosophila embryo, fusion of an individual founder cell (FC) with multiple fusion competent myoblasts (FCMs) gives rise to a single multinucleated muscle cell; the number of FCM fusion events determines the number of nuclei in the mature muscle cell. A) Recognition and Adhesion: FC and FCM express different membrane proteins that allow the two cells to recognize and adhere to each other. When in contact, each cell initiates the fusion process by recruiting several proteins to the fusion site. B) Actin Focus Formation: Through the activity of the fusion machinery on both the FC and the FCM, actin (red) monomers are assembled into filaments and form a dense F-actin focus on the FCM side and a thin layer on the FC side. The FCM actin focus invades the FC with multiple finger-like protrusions. N-cadherin is removed from the membrane to allow for the next steps of fusion. C) Pore Formation. The formation of a fusion pore in the membrane occurs, allowing for the exchange of cytoplasm between both cells. The repression of the FCM transcriptional profile begins (FCM pink nucleus turns blue). D) Post Fusion. After one fusion event, the resulting cell has one additional nucleus that has the transcriptional profile of the FC (blue nuclei). At this moment, the cell either prepares for another round of fusion or stops fusing with FCMs.
Table 1.
List of gene products required in Drosophila myoblast fusion
| Protein | Localization | Function | Reference(s) |
|---|---|---|---|
| Arp2/3 | Cytoplasm | Actin polymerization | (Richardson et al., 2007) |
| Blown Fuse (Blow) | Cytoplasm | Regulation of WASp activity | (Doberstein et al., 1997), (Jin et al., 2011) |
| Crk | Cytoplasm | Adaptor protein | (Balagopalan et al., 2006; Galletta et al., 1999) |
| Diaphanous (Dia) | Cytoplasm | Actin polymerization | (Deng et al., 2015) |
| Dumbfounded (Duf) | FC - Membrane | Recognition and Adhesion | (Ruiz Gomez et al., 2000) |
| Hibris (Hbs) | FCM - Membrane | Recognition and Adhesion | (Artero et al., 2001) |
| Kette | Cytoplasm | SCAR activation | (Richardson et al., 2007) |
| Schizo/Loner | Cytoplasm | Rac localization and N-cadherin removal | (Chen et al., 2003), (Dottermusch-Heidel et al., 2012) |
| Myoblast City (Mbc) | Cytoplasm | Regulation of Rac activity | (Erickson et al., 1997) |
| Myosin II (Myo II) | FC - Cytoplasm | Cortical tension generation on the FC/myotube | (Kim et al., 2015) |
| N-cadherin | Membrane | Adhesion | (Dottermusch-Heidel et al., 2012) |
| PI(4,5)P2 | Membrane | Actin cytoskeleton remodeling | (Bothe et al., 2014) |
| Rac | Cytoplasm | SCAR activation | (Hakeda-Suzuki et al., 2002) |
| Rolling pebbles (Rols) | Cytoplasm | Adaptor protein involved with Duf recycling | (Chen and Olson, 2001), (Rau et al., 2001) |
| Rst | FC - Membrane | Recognition and Adhesion | (Strünkelnberg et al., 2001) |
| SCAR | Cytoplasm | Arp2/3 activation | (Richardson et al., 2007) |
| Singles Bar (Sing) | Membrane | Vesicle trafficking | {Estrada:2007hg} |
| Sticks and Stones (Sns) | FCM - Membrane | Recognition and Adhesion | (Bour et al., 2000) |
| Verprolin1 (Vrp1) | Cytoplasm | Regulation WASp activity | (Massarwa et al., 2007), (Jin et al., 2011) |
| WASp | Cytoplasm | Arp2/3 activation | (Schäfer et al., 2007) |
| WHAMY | Cytoplasm | Actin polymerization | (Brinkmann et al., 2016) |
Table 2.
List of gene products required for vertebrate myoblast fusion
| Protein | Location and function | Animal model | Reference(s) |
|---|---|---|---|
| Adam12 | Membrane Mediates myoblast adhesion Interaction with the actin-binding protein α-actinin-2 promotes myoblast fusion |
Mouse | (Galliano et al., 2000; Zolkiewska, 1999) |
| BAI1 | Membrane Mediates recognition of phosphatidylserine on apoptotic cells Regulates myoblast fusion through Dock1/Rac1 pathway |
Mouse | (Hochreiter- Hufford et al., 2013) |
| BAI3 | Membrane Interacts with the scaffold protein ELMO to regulate myoblast fusion, possibly through the ELMO/DOCK/Rac1 pathway |
Chicken | (Hamoud et al., 2014) |
| Caveolin-3 | Membrane Involved in myoblast fusion (unclear role) Contradictory fusion phenotypes in different experimental models |
Mouse | (Galbiati et al., 1999; Volonte et al., 2003) |
| Cdc42 | Cytoplasm Required for actin rearrangement at the fusion site |
Mouse | (Vasyutina et al., 2009) |
| CKIP-1 | Cytoplasm Regulates myoblast fusion by interacting with both phosphoinositides on the membrane and Arp2/3 subunit |
Mouse Zebrafish |
(Baas et al., 2012) |
| Crk Crkl |
Cytoplasm Adaptor proteins Crk/Crkl genetically interact with DOCK1/DOCK5 to regulate myoblast fusion |
Zebrafish | (Moore et al., 2007) |
| DOCK1 | Cytoplasm Required for myoblast fusion by activating Rac1 |
Zebrafish Mouse |
(Laurin et al., 2008; Moore et al., 2007) |
| DOCK5 | Cytoplasm Functions redundantly with DOCK1 Also involved in post-fusion myofiber development |
Zebrafish Mouse |
(Laurin et al., 2008; Moore et al., 2007) |
| EHD2 | Membrane and cytoplasm Regulates myoblast fusion by mediating endocytic recycling of membrane proteins, such as Myoferlin |
Mouse | (Doherty et al., 2008) |
| Gm7325 Myomixer Minion Myomerger |
Membrane Interacts with Myomaker to induce fusion Functions in cell recognition (proposed) |
Mouse | (Bi et al., 2017; Quinn et al., 2017; Zhang et al., 2017) |
| JAM-B JAM-C |
Membrane Both physically interact to mediate fusion Facilitate myocytes recognition and adhesion (proposed) |
Zebrafish | (Powell and Wright, 2011) |
| Kirrel | Membrane Required for fast muscle precursor fusion |
Zebrafish | (Srinivas et al., 2007) |
| M-cadherin | Membrane Mediates myoblast adhesion Promotes fusion by activating the Rac1 pathway |
Mouse | (Charrasse et al., 2007; 2006) |
| MOR23 | Membrane Regulates cell adhesion during myoblast fusion |
Mouse | (Griffin et al., 2009) |
| Myoferlin | Membrane Regulates myoblast fusion through unclear mechanism, potentially by acting as a scaffold protein that links fusion proteins with the membrane |
Mouse | (Doherty et al., 2005) |
| Myomaker | Membrane Muscle-specific protein that induces fusion during muscle development and repair Mediate cell recognition (proposed) |
Mouse Zebrafish Chicken |
(Landemaine et al., 2014; Luo et al., 2015; Millay et al., 2013; Zhang and Roy, 2017) |
| N-WASp | Cytoplasm Required in both fusing partners |
Mouse | (Gruenbaum-Cohen et al., 2012) |
| Rac1 | Cytoplasm Required for proper recruiting of Arp2/3 to the contact site Regulates actin remodeling during fusion Unlike in Drosophila, constitutively active Rac1 in zebrafish leads to myoblasts hyperfusion |
Mouse Zebrafish Chicken |
(Sieiro et al., 2017; Srinivas et al., 2007; Vasyutina et al., 2009) |
| Stab2 | Membrane Involved in the phosphatidylserine-dependent fusion of myoblasts during muscle development and regeneration |
Mouse | (Park et al., 2016) |
| Talin 1/2 | Cytoplasm Regulates myoblast fusion, potentially by mediating the interaction between integrins and cytoskeleton |
Mouse | (Conti et al., 2009) |
| VLA4 VCAM1 |
Membrane VLA-4 and its counter receptor VCAM-1 are involved in secondary myogenesis |
Mouse | (Rosen et al., 1992) |
| β-1 integrin | Membrane Involved with membrane breakdown during myoblast fusion |
Mouse | (Schwander et al., 2003) |
Preparing for fusion: specification of muscle cells
Muscle Specification in the Drosophila embryo forms the larval muscles
The body wall or skeletal muscles are derived from somatic (Drosophila) or paraxial (mammals) mesoderm, respectively. In Drosophila, the allocation of somatic mesoderm depends on the crosstalk between the mesoderm and the ectoderm (Azpiazu et al., 1996; Frasch, 1995; Staehling-Hampton et al., 1994), as well as the combination of transcription factors that are expressed in the mesoderm (reviewed in Baylies et al., 1998; Dobi et al., 2015). After the allocation of the somatic mesoderm, the bHLH protein Lethal of Scute (L’sc) is expressed in clusters of promuscle cells (Figure 2A), under the control of the Receptor Tyrosine Kinase (RTK) pathway (Carmena et al., 1995). In each cluster, a combination of the Notch and RTK-Ras signaling pathway activities restricts the expression of L’sc to one cell—the muscle progenitor cell (Figure 2B, C) (Carmena et al., 1995; 1998b; Fuerstenberg and Giniger, 1998). Each progenitor cell then undergoes an asymmetric division to form either two founder cells (FC) or a single FC and an adult muscle progenitor (AMP) (Figure 2D) (Carmena et al., 1998a; Ruiz Gomez and Bate, 1997). The cytoplasmic membrane-associated protein Numb plays a critical role in determining muscle fate. Numb antagonizes Notch-mediated lateral inhibition and allows the expression of muscle identity proteins in the muscle progenitor cells (Ruiz Gomez and Bate, 1997). Different combinations of identity transcription factors expressed in each muscle progenitor, such as Slouch, Eve, Krüppel, Apterous, and the MyoD ortholog, Nautilus, in conjunction with chromatin regulators such as Sin3a (Dobi et al., 2014) determine the different cell identities of each FC, and hence, the final muscle pattern (Figure 2E, Dobi et al., 2015). After L’sc is restricted to the progenitor cell, the remaining cells in the promuscle cell cluster adopt a fusion competent myoblast (FCM) fate. The specification of FCMs is controlled by Notch-mediated lateral inhibition as well as by the transcription factors Lame duck and Tramtrack (Boyle and Berg, 2009).
Completion of FC and FCM specification allows for fusion between the two cell types, generating syncytial muscle cells (Figures 2E and 3). Throughout the period of fusion, the transcriptional profiles in both FCs and FCMs are dynamically regulated (Bataillé et al., 2017; Bourgouin et al., 1992; Dohrmann et al., 1990; Jagla et al., 1998). With the completion of each round of fusion, the fused FCM nuclei undergo progressive reprogramming, such that FCM-specific genes are suppressed, and the transcriptional pattern of the FC nucleus is adopted. The nucleus from the FC retains the expression of identity transcription factors after fusion. Additional experimental evidence suggests that the nucleus from the FC remains transcriptionally different from other nuclei in the same muscle fiber (Bataillé et al., 2017). Fusion events between FCs and FCMs are completed at late stage 15 (~13.5h AEL) (Beckett and Baylies, 2007; Richardson et al., 2008). Myotubes derived from myoblast fusion differentiate into fully striated, functional larval muscles (Figure 1A, C). These muscles grow without additional fusion events throughout the larval stage.
Muscle specification in the Drosophila pupa forms the adult muscles
Compared to the larval muscle, less is known about the specification process of adult Drosophila muscles. The prominent muscle groups in the adult Drosophila include the Direct Flight Muscles (DFM), the Indirect Flight Muscles (IFM), the jump muscles, the abdominal muscles, and the leg muscles. Based on morphology and gene expression, these adult somatic muscles can be divided into two types: tubular and fibrillar muscles. The Drosophila abdominal, jump, leg and direct flight muscles are tubular muscles with aligned myofibrils that rely on glycolytic metabolism, while the Indirect Flight Muscles (IFMs) are fibrillar muscles with distinct, unaligned myofibrils (Figure 1D) and are primarily oxidative (Peckham et al., 1990). The IFMs are composed of 7 pairs of dorsal-ventral muscles (DVMs) and 6 pairs of dorsal longitudinal muscles (DLMs) (Figure 1B). DVMs and DLMs have different orientation and attachment sites, and are generated from fusion between myoblasts and templates with different origins (reviewed in Dobi et al., 2015). The specification of DFMs requires the homeodomain transcription factors Cut and Apterous (Ghazi et al., 2000). The specification of IFMs depends on the transcriptional regulators Vestigial (Sudarsan et al., 2001) and Scalloped (Deng et al., 2009). In addition, Spalt major in the IFMs directs the fibrillar muscle fate (Schönbauer et al., 2011). Readers are directed to reviews on the specification of adult muscle for more detail (Dobi et al., 2015; Maqbool et al., 2006; Roy and Vijayraghavan, 1999).
Muscle Specification in vertebrates
Similar to Drosophila, the specification of muscle progenitor cells in vertebrates also requires a combination of signals from adjacent tissues (reviewed in Munsterberg et al., 1995; Bryson-Richardson and Currie, 2008). Extracellular signals induce the expression of Paired Box (Pax) 3 and Pax7 in the somites (Duprez et al., 1998). Pax genes have dual roles: they enhance the proliferation of muscle precursor cells (Relaix et al., 2005), and they activate the expression of MyoD and Myf5, two members of the myogenic regulatory factor (MRF) family (Maroto et al., 1997) (Sato et al., 2010), reviewed in (Bryson-Richardson and Currie, 2008). MRFs are a family of four transcription factors: MyoD, Myf5, myogenin, and MRF4. MRFs activate the expression of genes required for muscle differentiation. MyoD and Myf5 are the first MRFs expressed in muscles and function redundantly to trigger myoblast specification (Rudnicki et al., 1993). Myogenin functions after muscle progenitor formation and regulates terminal differentiation of committed myoblasts (Hasty et al., 1993). Similar to Myogenin, MRF4 also regulates terminal differentiation of myoblasts and functions upstream of MyoD to direct the specification of myogenic cells (Kassar-Duchossoy et al., 2004). In addition to MRFs, other transcription factors are required for muscle differentiation, such as the Myocyte enhancer factor 2 (Mef2), which plays a conserved role in muscle cell differentiation from Drosophila to human (Dodou et al., 2003; Lilly et al., 1994; Molkentin et al., 1995). After the specification of myogenic cells, two waves of myogenesis take place to form the final vertebrate muscle pattern. The first wave generates primary myofibers. The primary fibers function similarly to FCs in Drosophila, and determine the shape and identity of muscles (slow or fast). The second wave generates secondary myofibers that align alongside the primary myofibers and adds mass to the muscles (Harris et al., 1989). The formation of both types of myofibers requires fusion of myoblasts. In vitro experiments suggest that the fusion process occurs in two periods: the first involves the fusion of individual myoblasts to form the nascent myotubes; the second period involves fusion between the myotube and additional myoblasts (reviewed in Rochlin et al., 2010).
Making Larval muscle: Myoblast Fusion in the Drosophila Embryo
Recognition and Adhesion
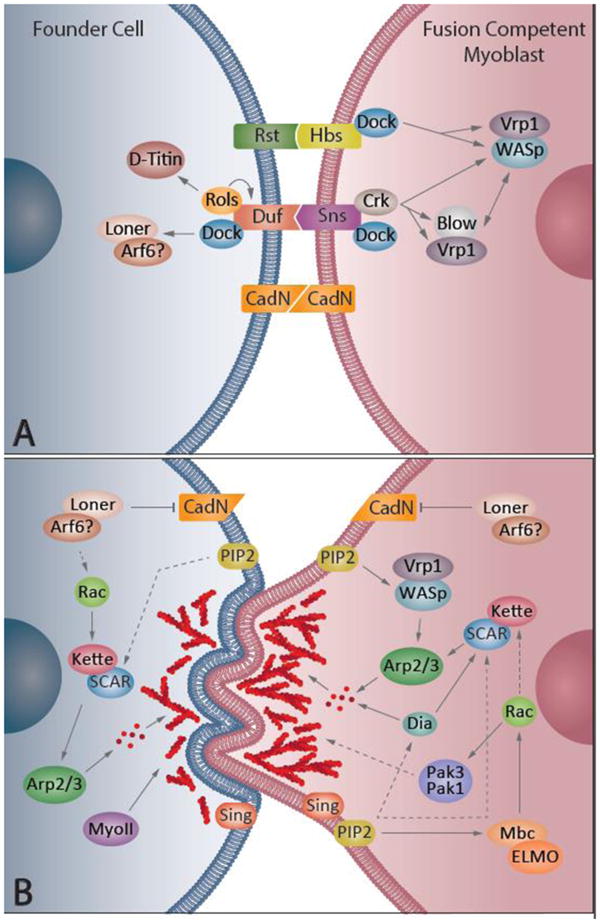
Myoblast fusion begins with cell-cell recognition and adhesion (Figure 3A, Table 1). In Drosophila, recognition and adhesion between the FC and FCMs are mediated by cell-specific immunoglobulin super family (IgSF) members. The IgSF members expressed in the FC are Dumbfounded (Duf) and Roughest (Rst) (Ruiz Gomez et al., 2000). Duf and Rst are transmembrane proteins that function redundantly to mediate recognition and adhesion (Strünkelnberg et al., 2001). In the FCMs, Sticks and Stones (Sns) and Hibris (Hbs) are the counterparts of Duf and Rst (Artero et al., 2001; Bour et al., 2000). Hbs can partially compensate for Sns function, but its activity is less efficient than Sns (Shelton et al., 2009). Expression of Rst is also detected in a subset of FCMs; however, its function in the FCM is unclear (Strünkelnberg et al., 2001). The interaction between Duf-Sns and Rst-Hbs is required for cell recognition and adhesion. Ectopic expression of Duf induces myoblast aggregation and adhesion, but is insufficient to trigger fusion (Ruiz Gomez et al., 2000). The interaction between Duf/Rst-Sns/Hbs is also required in other developmental events such axon guidance and eye development (Bao et al., 2010; Ramos et al., 1993), and this “cassette” of transmembrane proteins is referred to as an “adhesion module” in these contexts. If the FC/myotube and FCMs are not in direct contact before adhesion, cell migration is required to bring the two types of cells into close proximity (Richardson et al., 2007; Rochlin et al., 2010). An intriguing recent development in the understanding of fusion-related adhesion is the identification of N-cadherin as a contributor to myoblast fusion in the Drosophila embryo. N-cadherin is a member of the cadherin family of adhesion proteins. N-cadherin is present, but not restricted at the fusion site (Dottermusch-Heidel et al., 2012). One possible function of N-cadherin is to mediate cell adhesion through the formation of cellular junction-like structures at the fusion site (Hamp et al., 2016). For membrane fusion to proceed, however, N-cadherin needs to be removed from the fusion site (Hamp et al., 2016). Kette and Schizo/Loner, upstream regulators of Arp2/3-mediated actin rearrangements (see Actin rearrangement section below), are reported to facilitate removal of N-cadherin (Hamp et al., 2016, Dottermusch-Heidel et al., 2012, Onel et al., 2014). Notably, N-cadherin loss of function does not result in fusion defects in the Drosophila embryo (Charlton et al., 1997; Dottermusch-Heidel et al., 2012), suggesting potential redundant functions with other members of the cadherin family.
Actin rearrangements at the fusion site
After the cells adhere to one another, signals from these transmembrane recognition and adhesion molecules cause the actin cytoskeleton in both the FC and FCM to undergo remodeling, resulting in a filamentous-actin (F-actin)-based focus structure in the FCM and a thin sheath of F-actin on the opposing inner membrane in the FC (Figure 2B). The actin focus in the FCM mediates the formation of invasive finger-like membrane protrusions (Sens et al., 2010). This actin structure in the FCM is thought to provide the necessary force to push the lipid bilayers of the cells into close proximity. In response to the mechanical forces generated by the invasive protrusions from the FCM, non-muscle Myosin II (MyoII) is recruited to the fusion site in the FC/myotube. The accumulation of MyoII generates cortical tension, facilitates cell membrane juxtaposition, and promotes fusion pore formation (see below) (Kim et al., 2015).
A major regulator of the actin focus during myoblast fusion is Arp2/3. Arp2/3 is a multi-subunit protein complex that binds to actin filaments and polymerizes new actin filaments at a 70-degree angle from the pre-existing actin filament. Arp2/3 is activated by the nucleation-promoting factors (NPFs), Wiskott-Aldrich Syndrome protein (WASp) and SCAR (also known as WAVE). Signal transduction from the membrane to these actin cytoskeleton regulators is partially mediated by the activity of Crk, as well as Dreadlock (Dock), the Drosophila homolog of Nck. Both Dock and Crk are adapter proteins with src homology 2 (SH2)/SH3 domains. Dock can interact with all four IgSF transmembrane proteins that have SH2 or SH3 domains, but it only genetically interacts with Duf, Sns, and Hbs (Kaipa et al., 2013). In addition, rescue experiments using a series of sns deletions and site-directed mutations suggest that Sns functions through interacting with a complex array of proteins (Kocherlakota et al., 2008). In the FCMs, the Dock SH3 domain biochemically and genetically interacts with WASp, as well as the WASp regulator, Verprolin 1 (Vrp1, also known as Solitary (Sltr) and D-Wip) (Kaipa et al., 2013). In both the FC and FCMs, Dock biochemically and genetically interacts with the Arp2/3 NPF SCAR. Through a similar mechanism as Dock, Crk biochemically interacts with Sns. In a FCM, Crk physically interacts with the WASp regulatory proteins Vrp1 and Blow (Kim et al., 2007). Thus, the adaptor proteins Dock and Crk function as links that relay signals from the membrane to the actin cytoskeleton.
In the FC, another adaptor protein that links activity at the membrane to the actin cytoskeleton is Rolling pebbles (Rols, also known as Antisocial (Ants)/Rols). Rols is recruited to the fusion site by Duf. In a positive feedback loop, Rols replenishes Duf at the membrane, and thus enables additional rounds of fusion (Menon et al., 2005). Rols interacts with two proteins that regulate the actin cytoskeleton organization: Myoblast city (Mbc) (Chen and Olson, 2001; Rau et al., 2001) and D-titin (Menon and Chia, 2001). Mbc is the Drosophila homolog of the mammalian protein DOCK1, which mediates actin rearrangements by acting as a guanine nucleotide exchange factor (GEF) for the small GTPase Rac. However, recent data suggest that the downstream target of Rols is not Mbc: rescue experiments suggest that the activity of Mbc is not required in the FC for the initial fusion event (Haralalka et al., 2011). Another actin cytoskeletal protein that can bind to Rols is D-titin (Menon and Chia, 2001). D-titin is recruited to the fusion site by Rols and mediates myoblast fusion in a mechanism that remains unclear. D-titin is also involved in maintaining myotube morphology (Menon and Chia, 2001; Zhang et al., 2000).
While Mbc appears not to be required by the FCs initially, Schizo/Loner, another GEF that regulates actin rearrangements, is required in FCs to mediate fusion. Schizo/Loner is recruited to the fusion site by Duf and Rst (Bulchand et al., 2010). Domain analysis suggests that the function of Schizo/Loner is to recruit and activate the small GTPase dARF6 at the fusion site. Schizo/Loner and dARF6 control the membrane localization of Rac (Chen et al., 2003). However, arf6 maternal/zygotic null mutants do not have a myoblast fusion defect (Dyer et al., 2007), and the fusion phenotype in a dominant negative dARF6 background is not as severe as that in the schizo/loner mutant. Together these data suggest that the downstream target of Schizo/Loner is another GTPase or that there is redundancy with other dARF family members such as dARF1 (Chen et al., 2003).
Once activated, the small GTPase Rac regulates the function of SCAR and is critically important in both FCs and FCMs to mediate fusion. The activity of SCAR is controlled by the pentameric WRC complex (the WAVE/SCAR regulatory complex). The WRC is composed of Kette/Nap1, Sra1, Abi, Hspc300, and SCAR itself (Eden et al., 2002). Rac activity is essential for the localization of the WRC to the fusion site, as well as activation of SCAR via the release of the SCAR VCA domain from an inhibited state (Chen et al., 2010; Gildor et al., 2009). As part of the WRC, Kette is also essential for myoblast fusion as it controls the localization and stability of SCAR (Richardson et al., 2007). It has not been reported whether loss of function in the other WRC components, Sra1, Abi and Hspc300, lead to a fusion phenotype. This may be due to a lack of appropriate mutant alleles, as well as a maternal supply of protein/mRNA in the embryo that masks the role of these proteins during the time period in which fusion takes place. Nevertheless, overexpression of WRC components in muscle cells results in fusion block and suggests a potential role of other WRC components during fusion (Deng et al., 2016).
In the FC, SCAR activates its downstream target Arp2/3, and rearranges the actin cytoskeleton into a thin sheath along the fusion site (Sens et al., 2010). In the FCM, SCAR activity is also regulated by Rac. However, Rac localization and activation is regulated through Mbc (Haralalka et al., 2011). Mbc is recruited to the fusion site by the phospholipid PI(4,5)P2 (PIP2). When PIP2 activity is sequestered via overexpression of the pleckstrin homology domain of phospholipase C-gamma (PHplcγ), Mbc is concentrated in the cytoplasm instead of at the fusion site, leading to mislocalization of active Rac (Bothe et al., 2014). Mbc interacts with ELMO using its SH3 domain and forms the Mbc/ELMO complex. The GEF activity of Mbc/ELMO is tightly regulated during myoblast fusion, and it controls the level of active Rac. Both overexpression and loss of function of Mbc/ELMO cause defects in myoblast fusion, which are reminiscent of constitutively active Rac and Rac mutants, respectively (Geisbrecht et al., 2008). When activated by Mbc/ELMO, Rac, in turn, binds to the WRC and regulates Arp2/3 activity at the fusion site.
In addition to SCAR, the other NPF, WASp, also regulates Arp2/3 at the fusion site. Unlike SCAR, which functions both in FCs and FCMs, WASp activity is only required in the FCM (Jin et al., 2011). The activity of WASp is regulated by Blown Fuse (Blow) and Vrp1. The WASp-homology-1 (WH1) domain in WASp can bind to the WASp-binding domain (WBD) in Vrp1 (Ramesh and Geha, 2009). The interaction between WASp and Vrp1 is required to stabilize and localize WASp at the fusion site (Jin et al., 2011). This interaction is disrupted when Blow competes with WASp for Vrp1 WBD binding (Jin et al., 2011). Thus, after cell recognition and adhesion, a signal is transduced from the membrane via the adaptor protein Crk, which recruits Blow and Vrp1 to the fusion site. At the fusion site, the WASp-Vrp1 complex promotes actin polymerization by regulating Arp2/3 activity. Blow suppresses WASp activity by competing with WASp for Vrp1 binding and by dissociating the WASp-Vrp1 complex. Since the binding affinity between Blow-Vrp1 is lower than WASp-Vrp1, dissociated WASp interacts with Vrp1 again and promotes another round of branched actin polymerization (Jin et al., 2011). It has been reported that in FCs, the function of WASp and SCAR is redundant, as overexpression of WASp in the FCs can partially restore fusion in kette mutant background (Hamp et al., 2016). FCM-specific reduction of the WASp level in a kette mutant background rescues fusion defects. It has been proposed that a particular ratio of active WASp and SCAR needs to be maintained for fusion to occur (Hamp et al., 2016).
Recent studies have indicated that in addition to the branched actin network generated by Arp2/3, the linear actin filament nucleated by Diaphanous (Dia) also plays a role during myoblast fusion (Deng et al., 2015; Haralalka et al., 2014). Dia is a member of the formin family of actin polymerization proteins. Dia is enriched at the actin focus during the formation of Drosophila somatic musculature and dorsal pharyngeal musculature. Dia loss of function blocks myoblast fusion (Deng et al., 2015). Interestingly, overexpression of active Dia also leads to fusion defects (Deng et al., 2015; Haralalka et al., 2014), suggesting that the activity of Dia is highly regulated. Notably, in addition to Dia, the WASp family member WHAMY can also promote the assembly of linear actin filament. It has been reported that WHAMY synergizes with WASp during myoblast fusion (Brinkmann et al., 2016). Altogether, these data suggest that a balance between linear and branched actin filament is required at the fusion site for fusion to occur.
Fusion pore formation
A critical step in cell fusion is the formation of fusion pores that allow for cytoplasmic continuity between the fusing cells (Figure 3C). Studies from virus-cell fusion and C. elegans epithelial cell fusion have identified fusogens that mediate fusion pore formation (Mohler et al., 2002; reviewed in Sapir et al., 2008). A fusogen is a transmembrane protein that localizes to the fusion site and its presence is essential and sufficient to trigger membrane fusion (Sapir et al., 2008). Despite the identification of many gene products that are required for myoblast fusion in both Drosophila and vertebrates, no fusogen has been identified in these systems to date. Therefore, the mechanism that brings the membranes in close proximity to allow pore formation during myoblast membrane fusion remains a critical, open question.
It has been proposed that the actin cytoskeleton plays a crucial role in forcing the juxtaposition of the FC/FCM cell membranes, leading to the formation and expansion of fusion pores (reviewed by (Onel and Renkawitz-Pohl, 2009; Schejter, 2016; Schulman et al., 2015). During Drosophila embryonic myoblast fusion, the F-actin focus in the FCM forms podosome-like structures at the fusion site, which provide an invasive force on the apposing FC and pushes the cell membranes of the FCM and FC into closer proximity. The F-actin focus later evolves into a single-channel fusion pore on the membrane of the fusing cells (Sens et al., 2010). Studies aimed at investigating the function of actin regulators during pore formation have been performed. In these studies, the exchange of fluorescent proteins between the FCM and the FC were measured as a reporter of cytoplasmic continuity in various fusion mutant backgrounds. From these experiments, it has been proposed that SCAR-regulated actin remodeling is required for the initiation of fusion pore formation (Gildor et al., 2009). The function of WASp and its regulators is required for the formation of invasive structures (Jin et al., 2011; Kim et al., 2007; Sens et al., 2010). Therefore it is believed that WASp-regulated actin remodeling is required for pore formation (Jin et al., 2011; Kim et al., 2007; Sens et al., 2010) and expansion (Gildor et al., 2009). Notably, the actin focus resolves prior to fusion, suggesting that actin and other proteins located at the fusion site need to be removed, allowing fusion to occur. Thus far, the removal of proteins associated with the actin cytoskeleton at the fusion site has not been well studied. One potential protein involved in this process is Schizo/Loner, which has been reported to remove N-cadherin from the fusion site to permit fusion (Dottermusch-Heidel et al., 2012). In addition, the transmembrane protein singles bar (sing) is required during myoblast fusion. sing mutant embryos display a cell fusion block with accumulation of vesicles at the fusion site, suggesting a potential role of Sing in vesicle trafficking during fusion (Estrada et al., 2007).
Other studies suggest alternative models of fusion pore formation (Dhanyasi et al., 2015; Doberstein et al., 1997; Onel and Renkawitz-Pohl, 2009). Electron microscopy images have shown the presence of prefusion complexes at the fusion site. These prefusion complexes contain electron-dense, paired vesicles at the apposed plasma membranes. It has been suggested that these paired vesicles resolve into membrane plaques. Subsequently, the juxtaposed plasma membranes vesiculate and form sacs that enclose the extracellular space. Multiple fusion pores are observed in these situations (Doberstein et al., 1997). Similar membrane structures and multiple fusion pores at the fusion site have also been reported during myoblast fusion which leads to the formation of the Drosophila adult flight muscle (Dhanyasi et al., 2015). These data emphasize the requirement for the delivery and the clearing of vesicles at the fusion site as critical for membrane breakdown (Doberstein et al., 1997). These data also suggest that instead of one fusion pore, multiple pores are formed to facilitate fusion. It is still unclear whether these differences of fusion pore number arise from different fixation methods, context or other variables (reviewed in (Schejter, 2016)).
Making Adult Muscle: Myoblast Fusion in the Drosophila Pupa
The Drosophila adult muscles form during metamorphosis in the pupal stages. During the first few hours of metamorphosis, most of the Drosophila larval body wall muscles are destroyed through histolysis, but a few muscles, such as the Dorsal Oblique (DO1-3) muscles, escape degradation and serve as templates for some adult muscles, particularly the Dorsal Longitudinal Flight muscles (Fernandes et al., 1991). The growth of adult muscles is achieved through hundreds of cell-cell fusion events, in which a pool of myoblasts fuse with either template myotubes or muscle precursor cells (Fernandes et al., 1991).
The pathways that regulate myoblast fusion to form the Drosophila adult muscles are not fully understood. Based on our current knowledge, the mechanisms that regulate myoblast fusion during adult myogenesis are similar to those that regulate embryonic myogenesis. Fusion occurs between the adult muscle progenitors (AMPs) and myoblasts that migrate from the imaginal discs to form the adult abdominal muscles, the leg muscles, and the DVMs. The DLMs are formed through fusion between myoblasts and the muscle templates that are preserved during histolysis (Fernandes et al., 1991). The AMPs associated with the wing imaginal disc go through two phases of amplification during adult muscle development. The initial phase is a symmetrical division which is regulated by Notch signaling and leads to increased AMP numbers. The second phase is an asymmetrical division that is regulated by Notch and Wnt signaling and results in AMP self-renewal and the generation of myoblast progeny required for the later fusion events (Gunage et al., 2014). It has been proposed that during muscle injury, the AMPs could function similarly to satellite cells, whose progeny fuse with myotubes to repair muscles (Gunage et al., 2014). A recent study has shown that dIlp6 signals from a muscle niche can activate dormant AMPs through the Insulin-Notch-dMyc cascade (Aradhya et al., 2015).
Similar to embryonic myoblast fusion, the transmembrane proteins Duf, Rst, Sns, and Hbs are required to mediate cell recognition and adhesion (Gildor et al., 2012). After adhesion, the fusing myoblast flattens on the myotube, such that the cell membranes are in tight apposition (Dhanyasi et al., 2015). Actin accumulates primarily at the myoblast side of the fusion site, after receiving signals from the cell membrane, and forms a focus-like structure. As with embryonic myogenesis, actin rearrangements during adult myogenesis require the function of Arp2/3, as well as the actin nucleation-promoting factors (NPFs) WASp and SCAR. Loss of function studies revealed that WASp is required for fusion pore initiation. (Mukherjee et al., 2011). Similar to embryonic myoblast fusion, the MARVEL domain protein Sing is expressed in adult muscles, and its activity is required for adult myoblast fusion (Brunetti et al., 2015).
Despite these similarities, however, there are differences between Drosophila embryonic and adult myogenesis. In embryos, FCMs and FCs are positioned relatively close to one another prior to fusion. The FCMs in contact with FCs/myotubes are responsible for the early fusion events. The FCMs that are located more internally migrate prior to making contact and fusing with the FCs/myotubes (Beckett and Baylies, 2007; Richardson et al., 2008). During adult myogenesis, myoblasts often need to migrate long distances from the imaginal discs to the muscle template/myotubes. More importantly, during migration, these myoblasts maintain a semi-differentiated state via the Notch signaling pathway. Each myoblast expresses the Notch ligand Delta and represses the differentiation of its neighboring cells. Notch signaling decays when myoblasts reach the vicinity of the myotubes, and this allows for the terminal differentiation of the myoblasts and the expression of FCM markers such as Sns (Gildor et al., 2012).
During adult muscle formation, the adhesion between myoblasts and myotubes also requires filopodia that emanate from the myotube surface. The filopodia are actin-based structures that bring the myoblasts and myotubes into close proximity. The formation and maintenance of filopodia requires the activity of Enabled and IRSp53 (Segal et al., 2016). The requirement for these two proteins during embryonic myoblast fusion has not been reported. In addition, reducing WASp and SCAR activity simultaneously in embryos abolishes actin focus formation (Bothe et al., 2014). However, double knockdown of WASp and SCAR during adult muscle formation results in enlarged actin foci, suggesting novel roles of the NPFs during Drosophila adult myoblast fusion (Mukherjee et al., 2011).
Making muscle in Vertebrates: Myoblast Fusion
Compared to Drosophila, the vertebrate muscle is a more complex system, both in terms of its structure and the regulatory mechanisms required during development. Over the past few decades, researchers have identified numerous genes that are required for myoblast fusion in vertebrates (reviewed in Rochlin et al., 2010; Demonbreun et al., 2015). Nevertheless, how these different proteins coordinate fusion remains unclear. The morphological steps that are required for myoblast fusion and many of the genes necessary for the process are conserved between Drosophila and vertebrates. Therefore, we review recent updates in vertebrate fusion-related genes based on their localization and which steps they are involved in during fusion (Table 2).
Similar to the process of Drosophila myoblast fusion, the myotube-myoblast fusion during muscle development in vertebrates is also asymmetric and requires recognition molecules located on the cell membrane. One pair of membrane proteins required for fusion is Integrin vLA-4 and its receptor VCAM-1. Integrin vLA-4 is expressed in myotubes and VCAM-1 in myoblasts. Interactions between vLA-4 and VCAM-1 are required for the alignment of the secondary myofiber with the primary myofiber, as well as for the second phase of muscle cell fusion (Rosen et al., 1992). In addition to vLA-4 and VCAM-1, the transcription factor NFAT2C also mediates cell recognition by regulating the expression of IL-4. IL-4 is a cytokine that functions as a secreted myoblast recruitment factor (Horsley et al., 2003). Another pair of membrane proteins required for fusion is the immunoglobin superfamily proteins Jamb and Jamc. In zebrafish, Jamb and Jamc are expressed in fast-muscle myoblasts. The physical interaction between Jamb and Jamc is required for myoblast fusion during the formation of fast-twitch muscles. Loss of function of either protein severely impairs myoblast fusion (Powell and Wright, 2011; Sedwick, 2011). In addition, it has been shown that Nephrin, an ortholog of the Drosophila transmembrane protein Sns, localizes on myoblasts and mediates cell recognition (Sohn et al., 2009). In zebrafish, Kirrel, an ortholog of Nephrin, is localized to membranes of myoblasts and is required for fusion (Srinivas et al., 2007). After recognition, adhesion molecules such as M-cadherin (Hollnagel et al., 2002), N-cadherin (Radice et al., 1997), and integrin family members (Schwander et al., 2003) mediate the adhesion and alignment of the myoblasts. It has been reported that the G-protein coupled receptors BAI1 and BAI3 also play a role during fusion, through signal transduction from the membrane to the cytoskeleton via the Dock1/Rac1 pathway (Hamoud et al., 2014; Hochreiter-Hufford et al., 2013).
Unlike in Drosophila, an actin focus structure has not been reported during vertebrate myoblast fusion. Nevertheless, actin cytoskeletal rearrangements play a critical role in vertebrate muscle formation. Similar to Drosophila mbc mutants, Dock1-null mice embryos exhibit severely impaired myoblast fusion and skeletal muscle content is reduced (Laurin et al., 2008). Dock1 is the mammalian ortholog of Mbc, and it functions as a GEF protein that activates Rac GTPase. In a conditional Rac1 knockout mouse model, the recruitment of Arp2/3 and F-actin to the cell-contact site is reduced, resulting in impaired myoblast migration and fusion (Vasyutina et al., 2009). Experiments using C2C12 mouse myoblast cell culture suggest that Nap1, the mammalian ortholog of Kette and the downstream target of Rac1, is also required for regulating WAVE dependent actin remodeling during myoblast fusion (Nowak et al., 2009). Moreover, a recent study reported the requirement of the non-muscle myosin IIA (NM-MHC-IIA) during myoblast fusion. NM-MHC-IIA is involved in the formation of the cortical actin wall in the FC/myotube in the rat L6 myoblast cell culture, providing structural and mechanical support to facilitate membrane alignment and fusion pore formation (Duan and Gallagher, 2009). Together, these data suggest a conserved role of actin during muscle development.
It has been a major challenge to identify muscle-specific genes that ensure that fusion occurs only between myoblasts rather than other cell types. The first such gene found in vertebrates is Myomaker/Tmem8c. Myomaker is a transmembrane protein that is specifically expressed in the muscles during muscle development and regeneration (Landemaine et al., 2014; Luo et al., 2015; Millay et al., 2013; 2014; Zhang and Roy, 2017). In humans, reduced Myomaker function is linked to Carey-Fineman-Ziter syndrome (Di Gioia et al., 2017), a congenital myopathy. Myomaker is required for fusion and is localized to the adhesion site. Expressing Myomaker in fibroblasts is sufficient to induce fusion between fibroblasts and myoblasts, but it is not sufficient to induce fibroblast-fibroblast fusion (Millay et al., 2013). These reports suggest that additional proteins on the myoblasts are required to interact with Myomaker to induce fusion.
Recently, Gm7325, the potential counterpart of Myomaker, has been identified by three different labs, and was given the name Myomixer (Bi et al., 2017), Myomerger (Quinn et al., 2017) and Minion (Zhang et al., 2017). Gm7325 is an 84 amino acid microprotein with a similar expression pattern to Myomaker. The function of Gm7325 is required for Myomaker-induced fusion, as simultaneous expression of Myomaker and Gm7325 induces fusion between non-muscle cells (Bi et al., 2017; Quinn et al., 2017; Zhang et al., 2017). The mechanism by which Myomaker and Gm7325 regulates fusion is not well understood. A physical interaction between Myomaker and Gm7325 has been reported (Bi et al., 2017), but remains controversial (Quinn et al., 2017; Zhang and Roy, 2017). In Gm7325- and Myomaker-deficient myoblasts, cell fusion is blocked at different fusion stages. Based on whether the cell membranes align in the deficient myoblasts, it has been proposed that Myomaker is necessary for cell recognition, while Gm7325 is required for downstream events such as actin cytoskeleton remodeling (Zhang et al., 2017).
Cell fusion in vertebrates is required not only during early embryonic myogenesis, but also during muscle regeneration after damage. As with Drosophila adult muscle progenitors (AMPs), a pool of myoblasts remains undifferentiated during vertebrate myogenesis. These myoblasts are satellite cells that associate with myofibers. Satellite cells contribute to post-natal muscle growth, as well as to muscle repair. The specification of satellite cells is critically dependent on Pax7, as no satellite cells are found in Pax7 mutant mice (Seale et al., 2000). With intact muscles, satellite cells are mitotically quiescent and express Pax7 and Myf5, but not Myogenin or MyoD (reviewed in (Yin et al., 2013)). Upon muscle injury, myofiber necrosis triggers inflammatory responses, which induce satellite cells to proliferate (Tidball, 1995). After proliferation, the majority of satellite cells differentiate and start to express MyoD and myogenin (Rantanen et al., 1995). These cells later fuse with damaged myofibers or fuse with each other to generate new muscle fibers (Grounds and Yablonka-Reuveni, 1993). Thus far, the process of satellite cell fusion is mechanistically similar to myoblast fusion during primary and secondary myofiber formation.
Conclusion
Myoblast fusion in organisms ranging from the fly to human occurs through conserved steps: cell recognition, membrane apposition, actin rearrangement, pore formation, and cytoplasmic continuity. The investigation of myoblast fusion in Drosophila continues to provide valuable insights to vertebrate muscle development. However, there are many questions that remain unanswered: How do muscles control the number of fusion events during development and during repair? How do the nuclei added to a myotube adopt the program of the growing myotube? How are movement and transcriptional diversity among the added myonuclei controlled? And when considering the mechanisms of fusion, how are the ratios of linear and branched actin filaments controlled at the actin focus? What are the actin depolymerization factors that resolve the actin focus after fusion pore formation? What causes pore formation between the engaged cells? The knowledge gained from answering these questions in model systems such as Drosophila will illuminate how the key steps of fusion are controlled and how the syncytial muscle cell develops and is maintained.
Acknowledgments
We thank the members of the Baylies lab for helpful discussions, Iliona Wolfowicz for the fly drawing in Figure 1, and our funding agencies SFRH/BD/52041/2012 to MA, NIH [GM078318, AR108981] to MKB, and National Cancer Institute [P30 CA 008748] core grant to MSKCC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aradhya R, Zmojdzian M, Da Ponte JP, Jagla K. Muscle niche-driven Insulin-Notch-Myc cascade reactivates dormant Adult Muscle Precursors in Drosophila. Elife. 2015;4:2609. doi: 10.7554/eLife.08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artero RD, Castanon I, Baylies MK. The immunoglobulin-like protein Hibris functions as a dose-dependent regulator of myoblast fusion and is differentially controlled by Ras and Notch signaling. Development. 2001;128:4251–4264. doi: 10.1242/dev.128.21.4251. [DOI] [PubMed] [Google Scholar]
- Azpiazu N, Lawrence PA, Vincent JP, Frasch M. Segmentation and specification of the Drosophila mesoderm. Genes Dev. 1996;10:3183–3194. doi: 10.1101/gad.10.24.3183. [DOI] [PubMed] [Google Scholar]
- Baas D, Caussanel-Boude S, Guiraud A, Calhabeu F, Delaune E, Pilot F, Chopin E, Machuca-Gayet I, Vernay A, Bertrand S, et al. CKIP-1 regulates mammalian and zebrafish myoblast fusion. J Cell Sci. 2012;125:3790–3800. doi: 10.1242/jcs.101048. [DOI] [PubMed] [Google Scholar]
- Balagopalan L, Chen MH, Geisbrecht ER, Abmayr SM. The CDM superfamily protein MBC directs myoblast fusion through a mechanism that requires phosphatidylinositol 3,4,5-triphosphate binding but is independent of direct interaction with DCrk. Mol Cell Biol. 2006;26:9442–9455. doi: 10.1128/MCB.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Fischbach KF, Corbin V, Cagan RL. Preferential adhesion maintains separation of ommatidia in the Drosophila eye. Dev Biol. 2010;344:948–956. doi: 10.1016/j.ydbio.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataillé L, Boukhatmi H, Frendo JL, Vincent A. Dynamics of transcriptional (re)-programming of syncytial nuclei in developing muscles. BMC Biol. 2017;15:48. doi: 10.1186/s12915-017-0386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylies MK, Bate M, Ruiz Gomez M. Myogenesis: a view from Drosophila. Cell. 1998;93:921–927. doi: 10.1016/s0092-8674(00)81198-8. [DOI] [PubMed] [Google Scholar]
- Beckett K, Baylies MK. 3D analysis of founder cell and fusion competent myoblast arrangements outlines a new model of myoblast fusion. Dev Biol. 2007;309:113–125. doi: 10.1016/j.ydbio.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi P, Ramirez-Martinez A, Li H, Cannavino J, McAnally JR, Shelton JM, Sánchez-Ortiz E, Bassel-Duby R, Olson EN. Control of muscle formation by the fusogenic micropeptide myomixer. Science. 2017;356:323–327. doi: 10.1126/science.aam9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothe I, Deng S, Baylies M. PI(4,5)P2 regulates myoblast fusion through Arp2/3 regulator localization at the fusion site. Development. 2014;141:2289–2301. doi: 10.1242/dev.100743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour BA, Chakravarti M, West JM, Abmayr SM. Drosophila SNS, a member of the immunoglobulin superfamily that is essential for myoblast fusion. Genes Dev. 2000;14:1498–1511. [PMC free article] [PubMed] [Google Scholar]
- Bourgouin C, Lundgren SE, Thomas JB. Apterous is a Drosophila LIM domain gene required for the development of a subset of embryonic muscles. Neuron. 1992;9:549–561. doi: 10.1016/0896-6273(92)90192-g. [DOI] [PubMed] [Google Scholar]
- Boyle MJ, Berg CA. Control in time and space: Tramtrack69 cooperates with Notch and Ecdysone to repress ectopic fate and shape changes during Drosophila egg chamber maturation. Development. 2009;136:4187–4197. doi: 10.1242/dev.042770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann K, Winterhoff M, Onel SF, Schultz J, Faix J, Bogdan S. WHAMY is a novel actin polymerase promoting myoblast fusion, macrophage cell motility and sensory organ development in Drosophila. J Cell Sci. 2016;129:604–620. doi: 10.1242/jcs.179325. [DOI] [PubMed] [Google Scholar]
- Brunetti TM, Fremin BJ, Cripps RM. Identification of singles bar as a direct transcriptional target of Drosophila Myocyte enhancer factor-2 and a regulator of adult myoblast fusion. Dev Biol. 2015;401:299–309. doi: 10.1016/j.ydbio.2015.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson-Richardson RJ, Currie PD. The genetics of vertebrate myogenesis. Nat Rev Genet. 2008;9:632–646. doi: 10.1038/nrg2369. [DOI] [PubMed] [Google Scholar]
- Bulchand S, Menon SD, George SE, Chia W. The intracellular domain of Dumbfounded affects myoblast fusion efficiency and interacts with Rolling pebbles and Loner. PLoS ONE. 2010;5:e9374. doi: 10.1371/journal.pone.0009374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena A, Bate M, Jiménez F. Lethal of scute, a proneural gene, participates in the specification of muscle progenitors during Drosophila embryogenesis. Genes Dev. 1995;9:2373–2383. doi: 10.1101/gad.9.19.2373. [DOI] [PubMed] [Google Scholar]
- Carmena A, Gisselbrecht S, Harrison J, Jiménez F, Michelson AM. Combinatorial signaling codes for the progressive determination of cell fates in the Drosophila embryonic mesoderm. Genes Dev. 1998a;12:3910–3922. doi: 10.1101/gad.12.24.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena A, Murugasu-Oei B, Menon D, Jiménez F, Chia W. Inscuteable and numb mediate asymmetric muscle progenitor cell divisions during Drosophila myogenesis. Genes Dev. 1998b;12:304–315. doi: 10.1101/gad.12.3.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton CA, Mohler WA, Radice GL, Hynes RO, Blau HM. Fusion competence of myoblasts rendered genetically null for N-cadherin in culture. J Cell Biol. 1997;138:331–336. doi: 10.1083/jcb.138.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrasse S, Comunale F, Fortier M, Portales-Casamar E, Debant A, Gauthier-Rouvière C. M-cadherin activates Rac1 GTPase through the Rho-GEF trio during myoblast fusion. Mol Biol Cell. 2007;18:1734–1743. doi: 10.1091/mbc.E06-08-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrasse S, Comunale F, Grumbach Y, Poulat F, Blangy A, Gauthier-Rouvière C. RhoA GTPase regulates M-cadherin activity and myoblast fusion. Mol Biol Cell. 2006;17:749–759. doi: 10.1091/mbc.E05-04-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EH, Olson EN. Antisocial, an intracellular adaptor protein, is required for myoblast fusion in Drosophila. Dev Cell. 2001;1:705–715. doi: 10.1016/s1534-5807(01)00084-3. [DOI] [PubMed] [Google Scholar]
- Chen EH, Pryce BA, Tzeng JA, Gonzalez GA, Olson EN. Control of myoblast fusion by a guanine nucleotide exchange factor, loner, and its effector ARF6. Cell. 2003;114:751–762. doi: 10.1016/s0092-8674(03)00720-7. [DOI] [PubMed] [Google Scholar]
- Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, Umetani J, Billadeau DD, Otwinowski Z, Rosen MK. Structure and control of the actin regulatory WAVE complex. Nature. 2010;468:533–538. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti FJ, Monkley SJ, Wood MR, Critchley DR, Müller U. Talin 1 and 2 are required for myoblast fusion, sarcomere assembly and the maintenance of myotendinous junctions. Development. 2009;136:3597–3606. doi: 10.1242/dev.035857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demonbreun AR, Biersmith BH, McNally EM. Membrane fusion in muscle development and repair. Semin Cell Dev Biol. 2015;45:48–56. doi: 10.1016/j.semcdb.2015.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Hughes SC, Bell JB, Simmonds AJ. Alternative requirements for Vestigial, Scalloped, and Dmef2 during muscle differentiation in Drosophila melanogaster. Mol Biol Cell. 2009;20:256–269. doi: 10.1091/mbc.E08-03-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S, Bothe I, Baylies M. Diaphanous regulates SCAR complex localization during Drosophila myoblast fusion. Fly (Austin) 2016;10:178–186. doi: 10.1080/19336934.2016.1195938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S, Bothe I, Baylies MK. The Formin Diaphanous Regulates Myoblast Fusion through Actin Polymerization and Arp2/3 Regulation. PLoS Genet. 2015;11:e1005381. doi: 10.1371/journal.pgen.1005381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanyasi N, Segal D, Shimoni E, Shinder V, Shilo BZ, Vijayraghavan K, Schejter ED. Surface apposition and multiple cell contacts promote myoblast fusion in Drosophila flight muscles. J Cell Biol. 2015;211:191–203. doi: 10.1083/jcb.201503005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gioia SA, Connors S, Matsunami N, Cannavino J, Rose MF, Gilette NM, Artoni P, de Macena Sobreira NL, Chan WM, Webb BD, et al. A defect in myoblast fusion underlies Carey-Fineman-Ziter syndrome. Nat Commun. 2017;8:16077. doi: 10.1038/ncomms16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doberstein SK, Fetter RD, Mehta AY, Goodman CS. Genetic analysis of myoblast fusion: blown fuse is required for progression beyond the prefusion complex. J Cell Biol. 1997;136:1249–1261. doi: 10.1083/jcb.136.6.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobi KC, Halfon MS, Baylies MK. Whole-genome analysis of muscle founder cells implicates the chromatin regulator Sin3A in muscle identity. Cell Rep. 2014;8:858–870. doi: 10.1016/j.celrep.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobi KC, Schulman VK, Baylies MK. Specification of the somatic musculature in Drosophila. Wiley Interdiscip Rev Dev Biol. 2015;4:357–375. doi: 10.1002/wdev.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodou E, Xu SM, Black BL. mef2c is activated directly by myogenic basic helix-loop-helix proteins during skeletal muscle development in vivo. Mech Dev. 2003;120:1021–1032. doi: 10.1016/s0925-4773(03)00178-3. [DOI] [PubMed] [Google Scholar]
- Doherty KR, Cave A, Davis DB, Delmonte AJ, Posey A, Earley JU, Hadhazy M, McNally EM. Normal myoblast fusion requires myoferlin. Development. 2005;132:5565–5575. doi: 10.1242/dev.02155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty KR, Demonbreun AR, Wallace GQ, Cave A, Posey AD, Heretis K, Pytel P, McNally EM. The endocytic recycling protein EHD2 interacts with myoferlin to regulate myoblast fusion. J Biol Chem. 2008;283:20252–20260. doi: 10.1074/jbc.M802306200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann C, Azpiazu N, Frasch M. A new Drosophila homeo box gene is expressed in mesodermal precursor cells of distinct muscles during embryogenesis. Genes Dev. 1990;4:2098–2111. doi: 10.1101/gad.4.12a.2098. [DOI] [PubMed] [Google Scholar]
- Dottermusch-Heidel C, Groth V, Beck L, Onel SF. The Arf-GEF Schizo/Loner regulates N-cadherin to induce fusion competence of Drosophila myoblasts. Dev Biol. 2012;368:18–27. doi: 10.1016/j.ydbio.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Duan R, Gallagher PJ. Dependence of myoblast fusion on a cortical actin wall and nonmuscle myosin IIA. Dev Biol. 2009;325:374–385. doi: 10.1016/j.ydbio.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprez D, Fournier-Thibault C, Le Douarin N. Sonic Hedgehog induces proliferation of committed skeletal muscle cells in the chick limb. Development. 1998;125:495–505. doi: 10.1242/dev.125.3.495. [DOI] [PubMed] [Google Scholar]
- Dyer N, Rebollo E, Domínguez P, Elkhatib N, Chavrier P, Daviet L, González C, González-Gaitán M. Spermatocyte cytokinesis requires rapid membrane addition mediated by ARF6 on central spindle recycling endosomes. Development. 2007;134:4437–4447. doi: 10.1242/dev.010983. [DOI] [PubMed] [Google Scholar]
- Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- Erickson MR, Galletta BJ, Abmayr SM. Drosophila myoblast city encodes a conserved protein that is essential for myoblast fusion, dorsal closure, and cytoskeletal organization. J Cell Biol. 1997;138:589–603. doi: 10.1083/jcb.138.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada B, Maeland AD, Gisselbrecht SS, Bloor JW, Brown NH, Michelson AM. The MARVEL domain protein, Singles Bar, is required for progression past the pre-fusion complex stage of myoblast fusion. Dev Biol. 2007;307:328–339. doi: 10.1016/j.ydbio.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J, Bate M, Vijayraghavan K. Development of the indirect flight muscles of Drosophila. Development. 1991;113:67–77. doi: 10.1242/dev.113.1.67. [DOI] [PubMed] [Google Scholar]
- Frasch M. Induction of visceral and cardiac mesoderm by ectodermal Dpp in the early Drosophila embryo. Nature. 1995;374:464–467. doi: 10.1038/374464a0. [DOI] [PubMed] [Google Scholar]
- Fuerstenberg S, Giniger E. Multiple roles for notch in Drosophila myogenesis. Dev Biol. 1998;201:66–77. doi: 10.1006/dbio.1998.8944. [DOI] [PubMed] [Google Scholar]
- Galbiati F, Volonte D, Engelman JA, Scherer PE, Lisanti MP. Targeted down-regulation of caveolin-3 is sufficient to inhibit myotube formation in differentiating C2C12 myoblasts. Transient activation of p38 mitogen-activated protein kinase is required for induction of caveolin-3 expression and subsequent myotube formation. J Biol Chem. 1999;274:30315–30321. doi: 10.1074/jbc.274.42.30315. [DOI] [PubMed] [Google Scholar]
- Galletta BJ, Niu XP, Erickson MR, Abmayr SM. Identification of a Drosophila homologue to vertebrate Crk by interaction with MBC. Gene. 1999;228:243–252. doi: 10.1016/s0378-1119(99)00010-4. [DOI] [PubMed] [Google Scholar]
- Galliano MF, Huet C, Frygelius J, Polgren A, Wewer UM, Engvall E. Binding of ADAM12, a marker of skeletal muscle regeneration, to the muscle-specific actin-binding protein, alpha -actinin-2, is required for myoblast fusion. J Biol Chem. 2000;275:13933–13939. doi: 10.1074/jbc.275.18.13933. [DOI] [PubMed] [Google Scholar]
- Geisbrecht ER, Haralalka S, Swanson SK, Florens L, Washburn MP, Abmayr SM. Drosophila ELMO/CED-12 interacts with Myoblast city to direct myoblast fusion and ommatidial organization. Dev Biol. 2008;314:137–149. doi: 10.1016/j.ydbio.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazi A, Anant S, Vijayraghavan K. Apterous mediates development of direct flight muscles autonomously and indirect flight muscles through epidermal cues. Development. 2000;127:5309–5318. doi: 10.1242/dev.127.24.5309. [DOI] [PubMed] [Google Scholar]
- Gildor B, Massarwa R, Shilo BZ, Schejter ED. The SCAR and WASp nucleation-promoting factors act sequentially to mediate Drosophila myoblast fusion. EMBO Rep. 2009;10:1043–1050. doi: 10.1038/embor.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildor B, Schejter ED, Shilo BZ. Bidirectional Notch activation represses fusion competence in swarming adult Drosophila myoblasts. Development. 2012;139:4040–4050. doi: 10.1242/dev.077495. [DOI] [PubMed] [Google Scholar]
- Griffin CA, Kafadar KA, Pavlath GK. MOR23 promotes muscle regeneration and regulates cell adhesion and migration. Dev Cell. 2009;17:649–661. doi: 10.1016/j.devcel.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grounds MD, Yablonka-Reuveni Z. Molecular and cell biology of skeletal muscle regeneration. Mol Cell Biol Hum Dis Ser. 1993;3:210–256. doi: 10.1007/978-94-011-1528-5_9. [DOI] [PubMed] [Google Scholar]
- Gruenbaum-Cohen Y, Harel I, Umansky KB, Tzahor E, Snapper SB, Shilo BZ, Schejter ED. The actin regulator N-WASp is required for muscle-cell fusion in mice. Proc Natl Acad Sci USa. 2012;109:11211–11216. doi: 10.1073/pnas.1116065109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunage RD, Reichert H, Vijayraghavan K. Identification of a new stem cell population that generates Drosophila flight muscles. Elife. 2014;3:180. doi: 10.7554/eLife.03126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakeda-Suzuki S, Ng J, Tzu J, Dietzl G, Sun Y, Harms M, Nardine T, Luo L, Dickson BJ. Rac function and regulation during Drosophila development. Nature. 2002;416:438–442. doi: 10.1038/416438a. [DOI] [PubMed] [Google Scholar]
- Hamoud N, Tran V, Croteau LP, Kania A, Côté JF. G-protein coupled receptor BAI3 promotes myoblast fusion in vertebrates. Proc Natl Acad Sci USa. 2014;111:3745–3750. doi: 10.1073/pnas.1313886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamp J, Löwer A, Dottermusch-Heidel C, Beck L, Moussian B, Flötenmeyer M, Onel SF. Drosophila Kette coordinates myoblast junction dissolution and the ratio of Scar-to-WASp during myoblast fusion. J Cell Sci. 2016;129:3426–3436. doi: 10.1242/jcs.175638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralalka S, Shelton C, Cartwright HN, Guo F, Trimble R, Kumar RP, Abmayr SM. Live imaging provides new insights on dynamic F-actin filopodia and differential endocytosis during myoblast fusion in Drosophila. PLoS ONE. 2014;9:e114126. doi: 10.1371/journal.pone.0114126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralalka S, Shelton C, Cartwright HN, Katzfey E, Janzen E, Abmayr SM. Asymmetric Mbc, active Rac1 and F-actin foci in the fusion-competent myoblasts during myoblast fusion in Drosophila. Development. 2011;138:1551–1562. doi: 10.1242/dev.057653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AJ, Duxson MJ, Fitzsimons RB, Rieger F. Myonuclear birthdates distinguish the origins of primary and secondary myotubes in embryonic mammalian skeletal muscles. Development. 1989;107:771–784. doi: 10.1242/dev.107.4.771. [DOI] [PubMed] [Google Scholar]
- Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- Hochreiter-Hufford AE, Lee CS, Kinchen JM, Sokolowski JD, Arandjelovic S, Call JA, Klibanov AL, Yan Z, Mandell JW, Ravichandran KS. Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature. 2013;497:263–267. doi: 10.1038/nature12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollnagel A, Grund C, Franke WW, Arnold HH. The cell adhesion molecule M-cadherin is not essential for muscle development and regeneration. Mol Cell Biol. 2002;22:4760–4770. doi: 10.1128/MCB.22.13.4760-4770.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V, Jansen KM, Mills ST, Pavlath GK. IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell. 2003;113:483–494. doi: 10.1016/s0092-8674(03)00319-2. [DOI] [PubMed] [Google Scholar]
- Jagla T, Bellard F, Lutz Y, Dretzen G, Bellard M, Jagla K. ladybird determines cell fate decisions during diversification of Drosophila somatic muscles. Development. 1998;125:3699–3708. doi: 10.1242/dev.125.18.3699. [DOI] [PubMed] [Google Scholar]
- Jin P, Duan R, Luo F, Zhang G, Hong SN, Chen EH. Competition between Blown fuse and WASP for WIP binding regulates the dynamics of WASP-dependent actin polymerization in vivo. Dev Cell. 2011;20:623–638. doi: 10.1016/j.devcel.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaipa BR, Shao H, Schäfer G, Trinkewitz T, Groth V, Liu J, Beck L, Bogdan S, Abmayr SM, Onel SF. Dock mediates Scar- and WASp-dependent actin polymerization through interaction with cell adhesion molecules in founder cells and fusion-competent myoblasts. J Cell Sci. 2013;126:360–372. doi: 10.1242/jcs.113860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Gayraud-Morel B, Gomès D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- Kim JH, Ren Y, Ng WP, Li S, Son S, Kee YS, Zhang S, Zhang G, Fletcher DA, Robinson DN, et al. Mechanical tension drives cell membrane fusion. Dev Cell. 2015;32:561–573. doi: 10.1016/j.devcel.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Shilagardi K, Zhang S, Hong SN, Sens KL, Bo J, Gonzalez GA, Chen EH. A critical function for the actin cytoskeleton in targeted exocytosis of prefusion vesicles during myoblast fusion. Dev Cell. 2007;12:571–586. doi: 10.1016/j.devcel.2007.02.019. [DOI] [PubMed] [Google Scholar]
- Kocherlakota KS, Wu JM, McDermott J, Abmayr SM. Analysis of the cell adhesion molecule sticks-and-stones reveals multiple redundant functional domains, protein-interaction motifs and phosphorylated tyrosines that direct myoblast fusion in Drosophila melanogaster. Genetics. 2008;178:1371–1383. doi: 10.1534/genetics.107.083808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landemaine A, Rescan PY, Gabillard JC. Myomaker mediates fusion of fast myocytes in zebrafish embryos. Biochem Biophys Res Commun. 2014;451:480–484. doi: 10.1016/j.bbrc.2014.07.093. [DOI] [PubMed] [Google Scholar]
- Laurin M, Fradet N, Blangy A, Hall A, Vuori K, Côté JF. The atypical Rac activator Dock180 (Dock1) regulates myoblast fusion in vivo. Proc Natl Acad Sci USa. 2008;105:15446–15451. doi: 10.1073/pnas.0805546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly B, Galewsky S, Firulli AB, Schulz RA, Olson EN. D-MEF2: a MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages during Drosophila embryogenesis. Proc Natl Acad Sci USa. 1994;91:5662–5666. doi: 10.1073/pnas.91.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Li E, Nie Q, Zhang X. Myomaker, Regulated by MYOD, MYOG and miR-140-3p, Promotes Chicken Myoblast Fusion. Int J Mol Sci. 2015;16:26186–26201. doi: 10.3390/ijms161125946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maqbool T, Soler C, Jagla T, Daczewska M, Lodha N, Palliyil S, Vijayraghavan K, Jagla K. Shaping leg muscles in Drosophila: role of ladybird, a conserved regulator of appendicular myogenesis. PLoS ONE. 2006;1:e122. doi: 10.1371/journal.pone.0000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroto M, Reshef R, Münsterberg AE, Koester S, Goulding M, Lassar AB. Ectopic Pax-3 activates MyoD and Myf-5 expression in embryonic mesoderm and neural tissue. Cell. 1997;89:139–148. doi: 10.1016/s0092-8674(00)80190-7. [DOI] [PubMed] [Google Scholar]
- Massarwa R, Carmon S, Shilo BZ, Schejter ED. WIP/WASp-based actin-polymerization machinery is essential for myoblast fusion in Drosophila. Dev Cell. 2007;12:557–569. doi: 10.1016/j.devcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Menon SD, Chia W. Drosophila rolling pebbles: a multidomain protein required for myoblast fusion that recruits D-Titin in response to the myoblast attractant Dumbfounded. Dev Cell. 2001;1:691–703. doi: 10.1016/s1534-5807(01)00075-2. [DOI] [PubMed] [Google Scholar]
- Menon SD, Osman Z, Chenchill K, Chia W. A positive feedback loop between Dumbfounded and Rolling pebbles leads to myotube enlargement in Drosophila. J Cell Biol. 2005;169:909–920. doi: 10.1083/jcb.200501126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millay DP, O’Rourke JR, Sutherland LB, Bezprozvannaya S, Shelton JM, Bassel-Duby R, Olson EN. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature. 2013;499:301–305. doi: 10.1038/nature12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millay DP, Sutherland LB, Bassel-Duby R, Olson EN. Myomaker is essential for muscle regeneration. Genes Dev. 2014;28:1641–1646. doi: 10.1101/gad.247205.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler WA, Shemer G, del Campo JJ, Valansi C, Opoku-Serebuoh E, Scranton V, Assaf N, White JG, Podbilewicz B. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev Cell. 2002;2:355–362. doi: 10.1016/s1534-5807(02)00129-6. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- Moore CA, Parkin CA, Bidet Y, Ingham PW. A role for the Myoblast city homologues Dock1 and Dock5 and the adaptor proteins Crk and Crk-like in zebrafish myoblast fusion. Development. 2007;134:3145–3153. doi: 10.1242/dev.001214. [DOI] [PubMed] [Google Scholar]
- Mukherjee P, Gildor B, Shilo BZ, Vijayraghavan K, Schejter ED. The actin nucleator WASp is required for myoblast fusion during adult Drosophila myogenesis. Development. 2011;138:2347–2357. doi: 10.1242/dev.055012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak SJ, Nahirney PC, Hadjantonakis AK, Baylies MK. Nap1-mediated actin remodeling is essential for mammalian myoblast fusion. J Cell Sci. 2009;122:3282–3293. doi: 10.1242/jcs.047597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onel SF, Renkawitz-Pohl R. FuRMAS: triggering myoblast fusion in Drosophila. Dev Dyn. 2009;238:1513–1525. doi: 10.1002/dvdy.21961. [DOI] [PubMed] [Google Scholar]
- Onel SF, Rust MB, Jacob R, Renkawitz-Pohl R. Tethering membrane fusion: common and different players in myoblasts and at the synapse. J Neurogenet. 2014;28:302–315. doi: 10.3109/01677063.2014.936014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Yun Y, Lim JS, Kim MJ, Kim SY, Kim JE, Kim IS. Stabilin-2 modulates the efficiency of myoblast fusion during myogenic differentiation and muscle regeneration. Nat Commun. 2016;7:10871. doi: 10.1038/ncomms10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckham M, Molloy JE, Sparrow JC, White DC. Physiological properties of the dorsal longitudinal flight muscle and the tergal depressor of the trochanter muscle of Drosophila melanogaster. J Muscle Res Cell Motil. 1990;11:203–215. doi: 10.1007/BF01843574. [DOI] [PubMed] [Google Scholar]
- Powell GT, Wright GJ. Jamb and jamc are essential for vertebrate myocyte fusion. PLoS Biol. 2011;9:e1001216. doi: 10.1371/journal.pbio.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn ME, Goh Q, Kurosaka M, Gamage DG, Petrany MJ, Prasad V, Millay DP. Myomerger induces fusion of non-fusogenic cells and is required for skeletal muscle development. Nat Commun. 2017;8:15665. doi: 10.1038/ncomms15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radice GL, Rayburn H, Matsunami H, Knudsen KA, Takeichi M, Hynes RO. Developmental defects in mouse embryos lacking N-cadherin. Dev Biol. 1997;181:64–78. doi: 10.1006/dbio.1996.8443. [DOI] [PubMed] [Google Scholar]
- Ramesh N, Geha R. Recent advances in the biology of WASP and WIP. Immunol Res. 2009;44:99–111. doi: 10.1007/s12026-008-8086-1. [DOI] [PubMed] [Google Scholar]
- Ramos RG, Igloi GL, Lichte B, Baumann U, Maier D, Schneider T, Brandstätter JH, Fröhlich A, Fischbach KF. The irregular chiasm C-roughest locus of Drosophila, which affects axonal projections and programmed cell death, encodes a novel immunoglobulin-like protein. Genes Dev. 1993;7:2533–2547. doi: 10.1101/gad.7.12b.2533. [DOI] [PubMed] [Google Scholar]
- Rantanen J, Hurme T, Lukka R, Heino J, Kalimo H. Satellite cell proliferation and the expression of myogenin and desmin in regenerating skeletal muscle: evidence for two different populations of satellite cells. Lab Invest. 1995;72:341–347. [PubMed] [Google Scholar]
- Rau A, Buttgereit D, Holz A, Fetter R, Doberstein SK, Paululat A, Staudt N, Skeath J, Michelson AM, Renkawitz-Pohl R. rolling pebbles (rols) is required in Drosophila muscle precursors for recruitment of myoblasts for fusion. Development. 2001;128:5061–5073. doi: 10.1242/dev.128.24.5061. [DOI] [PubMed] [Google Scholar]
- Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- Richardson BE, Beckett K, Baylies MK. Live imaging of Drosophila myoblast fusion. Methods Mol Biol. 2008;475:263–274. doi: 10.1007/978-1-59745-250-2_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson BE, Beckett K, Nowak SJ, Baylies MK. SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development. 2007;134:4357–4367. doi: 10.1242/dev.010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochlin K, Yu S, Roy S, Baylies MK. Myoblast fusion: when it takes more to make one. Dev Biol. 2010;341:66–83. doi: 10.1016/j.ydbio.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen GD, Sanes JR, LaChance R, Cunningham JM, Roman J, Dean DC. Roles for the integrin VLA-4 and its counter receptor VCAM-1 in myogenesis. Cell. 1992;69:1107–1119. doi: 10.1016/0092-8674(92)90633-n. [DOI] [PubMed] [Google Scholar]
- Roy S, Vijayraghavan K. Muscle pattern diversification in Drosophila: the story of imaginal myogenesis. Bioessays. 1999;21:486–498. doi: 10.1002/(SICI)1521-1878(199906)21:6<486::AID-BIES5>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Ruiz Gomez M, Bate M. Segregation of myogenic lineages in Drosophila requires numb. Development. 1997;124:4857–4866. doi: 10.1242/dev.124.23.4857. [DOI] [PubMed] [Google Scholar]
- Ruiz Gomez M, Coutts N, Price A, Taylor MV, Bate M. Drosophila dumbfounded: a myoblast attractant essential for fusion. Cell. 2000;102:189–198. doi: 10.1016/s0092-8674(00)00024-6. [DOI] [PubMed] [Google Scholar]
- Sapir A, Avinoam O, Podbilewicz B, Chernomordik LV. Viral and developmental cell fusion mechanisms: conservation and divergence. Dev Cell. 2008;14:11–21. doi: 10.1016/j.devcel.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Rocancourt D, Marques L, Thorsteinsdóttir S, Buckingham M. A Pax3/Dmrt2/Myf5 regulatory cascade functions at the onset of myogenesis. PLoS Genet. 2010;6:e1000897. doi: 10.1371/journal.pgen.1000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer G, Weber S, Holz A, Bogdan S, Schumacher S, Müller A, Renkawitz-Pohl R, Onel SF. The Wiskott-Aldrich syndrome protein (WASP) is essential for myoblast fusion in Drosophila. Dev Biol. 2007;304:664–674. doi: 10.1016/j.ydbio.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Schejter ED. Myoblast fusion: Experimental systems and cellular mechanisms. Semin Cell Dev Biol. 2016;60:112–120. doi: 10.1016/j.semcdb.2016.07.016. [DOI] [PubMed] [Google Scholar]
- Schönbauer C, Distler J, Jährling N, Radolf M, Dodt HU, Frasch M, Schnorrer F. Spalt mediates an evolutionarily conserved switch to fibrillar muscle fate in insects. Nature. 2011;479:406–409. doi: 10.1038/nature10559. [DOI] [PubMed] [Google Scholar]
- Schulman VK, Dobi KC, Baylies MK. Morphogenesis of the somatic musculature in Drosophila melanogaster. Wiley Interdiscip Rev Dev Biol. 2015;4:313–334. doi: 10.1002/wdev.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwander M, Leu M, Stumm M, Dorchies OM, Ruegg UT, Schittny J, Müller U. Beta1 integrins regulate myoblast fusion and sarcomere assembly. Dev Cell. 2003;4:673–685. doi: 10.1016/s1534-5807(03)00118-7. [DOI] [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Sedwick C. Jamb and jamc muscle in on myoblast fusion. PLoS Biol. 2011;9:e1001217. doi: 10.1371/journal.pbio.1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal D, Dhanyasi N, Schejter ED, Shilo BZ. Adhesion and Fusion of Muscle Cells Are Promoted by Filopodia. Dev Cell. 2016;38:291–304. doi: 10.1016/j.devcel.2016.07.010. [DOI] [PubMed] [Google Scholar]
- Sens KL, Zhang S, Jin P, Duan R, Zhang G, Luo F, Parachini L, Chen EH. An invasive podosome-like structure promotes fusion pore formation during myoblast fusion. J Cell Biol. 2010;191:1013–1027. doi: 10.1083/jcb.201006006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton C, Kocherlakota KS, Zhuang S, Abmayr SM. The immunoglobulin superfamily member Hbs functions redundantly with Sns in interactions between founder and fusion-competent myoblasts. Development. 2009;136:1159–1168. doi: 10.1242/dev.026302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieiro D, Véron N, Marcelle C. The chicken embryo as an efficient model to test the function of muscle fusion genes in amniotes. PLoS ONE. 2017;12:e0177681. doi: 10.1371/journal.pone.0177681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn RL, Huang P, Kawahara G, Mitchell M, Guyon J, Kalluri R, Kunkel LM, Gussoni E. A role for nephrin, a renal protein, in vertebrate skeletal muscle cell fusion. Proc Natl Acad Sci USa. 2009;106:9274–9279. doi: 10.1073/pnas.0904398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas BP, Woo J, Leong WY, Roy S. A conserved molecular pathway mediates myoblast fusion in insects and vertebrates. Nat Genet. 2007;39:781–786. doi: 10.1038/ng2055. [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton K, Hoffmann FM, Baylies MK, Rushton E, Bate M. dpp induces mesodermal gene expression in Drosophila. Nature. 1994;372:783–786. doi: 10.1038/372783a0. [DOI] [PubMed] [Google Scholar]
- Strünkelnberg M, Bonengel B, Moda LM, Hertenstein A, de Couet HG, Ramos RG, Fischbach KF. rst and its paralogue kirre act redundantly during embryonic muscle development in Drosophila. Development. 2001;128:4229–4239. doi: 10.1242/dev.128.21.4229. [DOI] [PubMed] [Google Scholar]
- Sudarsan V, Anant S, Guptan P, Vijayraghavan K, Skaer H. Myoblast diversification and ectodermal signaling in Drosophila. Dev Cell. 2001;1:829–839. doi: 10.1016/s1534-5807(01)00089-2. [DOI] [PubMed] [Google Scholar]
- Taylor MV. Comparison of Muscle Development in Drosophila and Vertebrates. In: Sink H, editor. Muscle Development in Drosophila. New York, NY: Springer New York; 2006. pp. 169–203. [Google Scholar]
- Tidball JG. Inflammatory cell response to acute muscle injury. Med Sci Sports Exerc. 1995;27:1022–1032. doi: 10.1249/00005768-199507000-00011. [DOI] [PubMed] [Google Scholar]
- Vasyutina E, Martarelli B, Brakebusch C, Wende H, Birchmeier C. The small G-proteins Rac1 and Cdc42 are essential for myoblast fusion in the mouse. Proc Natl Acad Sci USa. 2009;106:8935–8940. doi: 10.1073/pnas.0902501106. [DOI] [PMC free article] [PubMed] [Google Scholar]