Abstract
Ethylene perception in Arabidopsis is controlled by a family of five genes, including ETR1, ERS1 (ethylene response sensor 1), ERS2, ETR2, and EIN4. ERS1, the most highly conserved gene with ETR1, encodes a protein with 67% identity to ETR1. To clarify the role of ERS1 in ethylene sensing, we biochemically characterized the ERS1 protein by heterologous expression in yeast. ERS1, like ETR1, forms a membrane-associated, disulfide-linked dimer. In addition, yeast expressing the ERS1 protein contains ethylene-binding sites, indicating ERS1 is also an ethylene-binding protein. This finding supports previous genetic evidence that isoforms of ETR1 also function in plants as ethylene receptors. Further, we used the ethylene antagonist 1-methylcyclopropene (1-MCP) to characterize the ethylene-binding sites of ERS1 and ETR1. We found 1-MCP to be both a potent inhibitor of the ethylene-induced seedling triple response, as well as ethylene binding by yeast expressing ETR1 and ERS1. Yeast expressing ETR1 and ERS1 showed nearly identical sensitivity to 1-MCP, suggesting that the ethylene-binding sites of ETR1 and ERS1 have similar affinities for ethylene.
Ethylene responses in Arabidopsis are mediated by a small family of receptors, including ETR1. The ETR1 gene encodes a His kinase of the two-component class prevalent in bacterial and some eukaryotic systems (Chang et al., 1993). Four ETR1-like genes have been cloned from Arabidopsis (for review, see Johnson and Ecker, 1998) and cluster into two subfamilies based on sequence similarity and overall gene structure. Subfamily I consists of ETR1 and ERS1 (ethylene response sensor 1), whereas subfamily II consists of ETR2, ERS2, and EIN4 (Hua et al., 1997).
ETR1, the first ethylene receptor gene cloned (Chang et al., 1993), was identified in a screen to identify mutants lacking the ethylene-mediated “triple response” phenotype (Bleecker et al., 1988). The N-terminal hydrophobic region of the ETR1 protein contains three putative membrane-spanning subdomains, which form the ethylene-binding site (Schaller and Bleecker, 1995; Rodriguez et al., 1999). Two Cys residues, also in the N terminus of ETR1, mediate disulfide linkage of ETR1 monomers (Schaller et al., 1995). The C terminus of ETR1 is likely involved in transmitting the ethylene signal, as this region contains both His kinase and response regulator domains. Although His kinase activity of ETR1 has been demonstrated (Gamble et al., 1998), the role of kinase activity in signaling is still unclear. Two hybrid and in vitro binding assays have shown that C-terminal regions of both ETR1 and ERS1 interact with CTR1 (Clark et al., 1998), a raf-like kinase that negatively regulates ethylene responses (Kieber et al., 1993). Therefore, CTR1 may be directly modulated by the ethylene receptors.
Biochemical analysis has shown that yeast transformed with the ETR1 gene contain high-affinity ethylene-binding sites and that binding is saturable (Schaller and Bleecker, 1995). A structural model of the ethylene-binding domain of ETR1 predicts that a copper ion, coordinated by amino acids within the N-terminal hydrophobic domain, mediates ethylene binding to ETR1 (Rodriguez et al., 1999). Mutant forms of ETR1 have been expressed in yeast and tested for ethylene-binding activity (Schaller and Bleecker, 1995; Hall et al., 1999; Rodriguez et al., 1999). These studies have shown that some of the dominant mutations in ETR1 abolish ethylene binding by the receptor, whereas other mutations do not affect ethylene binding but may affect receptor signaling (Hall et al., 1999). Mutational analysis so far has implicated the first two transmembrane domains in forming the ethylene-binding site, as all mutations that abolish ethylene binding are localized to these domains (Schaller and Bleecker, 1995; Hall et al., 1999).
All mutations that have been isolated in the ETR1 gene family that cause an ethylene-insensitive phenotype are genetically dominant. Although no dominant ethylene-insensitive mutants of ERS1 or ERS2 have been isolated, mutant forms of these genes introduced transgenically into plants also confer dominant ethylene insensitivity (Hua et al., 1995; Hua et al., 1998). These experiments, as well as the observation that single loss-of-function mutants in four of the five ETR1 family members show normal sensitivity to ethylene, suggest that the ETR1 family members may at least partially possess overlapping functions in ethylene perception and signaling (Hua and Meyerowitz, 1998). Double and triple loss-of-function mutants show a constitutive ethylene-response phenotype, consistent with a model in which the ethylene receptors are negative regulators of the ethylene-response pathway (Hua and Meyerowitz, 1998).
However, a question remaining unresolved is how each ethylene receptor isoform contributes to ethylene perception and signaling. Although the genetic evidence indicates the proteins are functionally redundant, several lines of evidence suggest the five isoforms may not possess entirely equivalent activities. For example, the degeneracy of the kinase domains in ETR2, EIN4, and ERS2 and lack of response regulator domains in ERS1 and ERS2 indicate that each protein may play a slightly different role in the plant. Furthermore, the mRNA expression patterns of the ETR1 family members in Arabidopsis on the whole overlap, but there are some differences in their mRNA expression patterns (Hua et al., 1998). In addition, mRNA expression levels of ERS1, ETR2, and ERS2 are up-regulated by ethylene (Hua et al., 1998), but the significance of this up-regulation is unknown.
In this study we sought to clarify the role of ERS1 in ethylene signaling through a biochemical characterization of the ERS1 protein. Because ERS1 is most highly conserved with ETR1, we focused on determining if ERS1 is an ethylene-binding protein, and if so, how its binding characteristics compared with the ETR1 ethylene receptor.
RESULTS
Analysis of Transgenic Yeast Expressing ERS1
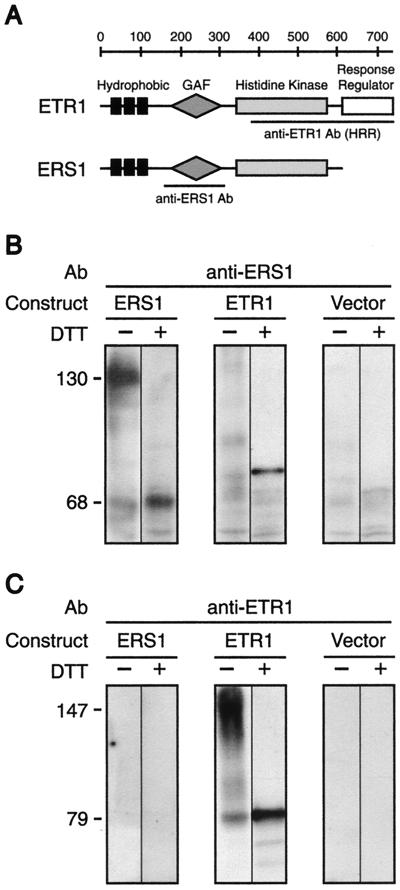
Of the four ETR1-like genes in Arabidopsis, ERS1 is the most closely related to ETR1. As shown in Figure 1A, ERS1 shares structural similarities to ETR1, containing an N-terminal hydrophobic region followed by a His kinase domain, but lacks a C-terminal response regulator domain. Similar to ETR1, the amino-terminal region of ERS1 is predicted to contain three membrane-spanning domains.
Figure 1.
Expression of ETR1 and ERS1 in yeast. A, Structure of ETR1 and ERS1. Hydrophobic, GAF, His kinase, and response regulator domains are indicated. Regions used for generating domain-specific antibodies are also indicated. B, Western blot with ERS1 antibody. Membrane fractions were isolated from yeast expressing ETR1, ERS1, or transformed with vector alone. Samples were incubated in the absence (−) or presence (+) of 100 mm dithiothreitol (DTT) for 1 h at 37°C, then subjected to SDS-PAGE. Positions of the ERS1 monomer and dimer are indicated. C, Western blot with ETR1 antibody. The same membrane fractions used in B were probed with the ETR1-HRR antibody. Positions of the ETR1 monomer and dimer are indicated.
To begin a biochemical characterization of ERS1, a cDNA clone containing the full-length ERS1 gene was isolated from Arabidopsis and expressed in yeast under the control of a constitutive ADH1 promoter. This system has previously been used to biochemically characterize the ethylene-binding site of the ETR1 protein (Schaller and Bleecker, 1995; Hall et al., 1999). ERS1 protein expression was then analyzed by western blot. As shown in Figure 1B, a polyclonal antibody generated against an Escherichia coli-expressed portion of ERS1 (amino acids 133–332) specifically recognizes a polypeptide in membrane fractions isolated from yeast transformed with the ERS1 construct. The calculated molecular mass of the polypeptide identified is 68 kD, consistent with the predicted size of the ERS1 protein based on amino acid sequence. This polypeptide is not present in control yeast transformed with vector alone or yeast transformed with a similar construct containing the ETR1 gene.
We found that the antibodies generated against ERS1 showed some cross-reactivity with ETR1, detecting a 79-kD protein in extracts prepared from yeast expressing ETR1 (Fig. 1B). The anti-ERS1 antibodies were generated against a region of ERS1 that shows 70% identity with ETR1. We did not see any cross-reactivity with the anti-ETR1 antibodies (Fig. 1C) generated against a region of ETR1 that shows 53% identity with ERS1.
The ERS1 protein migrated on SDS-PAGE gels at two different molecular masses, depending on treatment with reducing agent (Fig. 1B). In the presence of the reducing agent DTT, ERS1 migrated at 68 kD, whereas in the absence of reducing agent, the protein migrated at 130 kD. The sensitivity of the ERS1 protein to reducing agent is similar to ETR1, which forms a 147-kD disulfide-linked dimer in both plant and yeast membranes and can be converted to a 79-kD monomer in the presence of DTT (Schaller et al., 1995). These data indicate that ERS1, like ETR1, is capable of forming a disulfide-linked dimer.
Yeast Expressing the ERS1 Protein Binds Ethylene
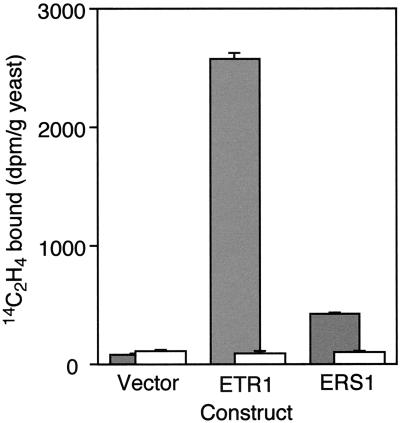
To determine if ERS1 is an ethylene-binding protein, in vivo [14C]ethylene-binding assays were carried out with transgenic yeast cells expressing the full-length ERS1 protein. As shown in Figure 2, ethylene binding was detected in yeast expressing ERS1. Much of the [14C]ethylene was displaceable by competition with unlabeled [12C]ethylene, indicating that the binding was saturable. Control yeast transformed with vector alone showed no saturable binding sites for ethylene. The level of ethylene binding by yeast expressing ERS1 was significantly above background levels but less than that detected in yeast expressing ETR1. The finding that both yeast expressing ETR1 and ERS1 bind ethylene indicates that both ETR1 and ERS1 could serve as ethylene receptors in Arabidopsis.
Figure 2.
Ethylene binding by yeast expressing ETR1 or ERS1. Transgenic yeast was incubated with 0.07 μL L−1 [14C]ethylene (gray bars), or with 0.07 μL L−1 [14C]ethylene and 1,000 μL L−1 [12C]ethylene (white bars). The difference between these two values represents the saturable binding. Samples were run in triplicate, and sd are shown. Yeast transformed with empty vector were used as a control.
Comparison of Protein Expression Levels between Yeast Expressing ERS1-Glutathione S-Transferase (GST) and ETR1-GST
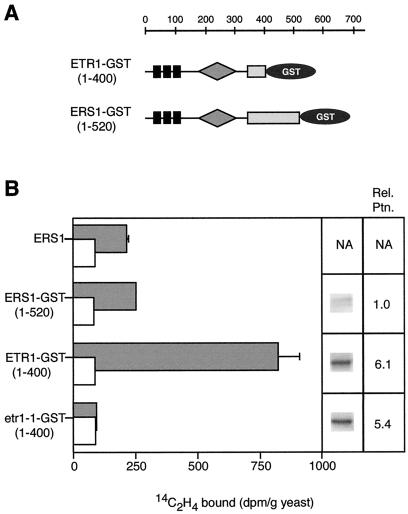
Differences in the amount of ethylene binding by yeast expressing ETR1 and ERS1 (Fig. 2) could reflect either a difference in affinity for ethylene or different expression levels of the proteins. In order to distinguish between these possibilities, we examined the expression levels of ETR1 and ERS1 in the transgenic yeast system. To directly compare protein expression levels on the same western blots, GST fusions of both ETR1 and ERS1 were constructed, and anti-GST antibodies were used for immunodetection. As shown in Figure 3A, the ETR1-GST(1-400) construct included the first 400 amino acids of ETR1, followed by a C-terminal GST tag. The ERS1-GST(1-520) construct included the first 520 amino acids of ERS1 also fused to a C-terminal GST tag. Constructs encoding a truncation of the ETR1 protein fused to a C-terminal GST tag have been previously shown to retain the ability to bind ethylene and localize to yeast membrane fractions, which is similar to full-length ETR1 (Rodriguez et al., 1999).
Figure 3.
Protein expression level comparisons between ETR1-GST(1-400) and ERS1-GST(1-520). A, Domain structure of the ETR1-GST(1-400) and ERS1-GST(1-520) constructs. Black boxes represent transmembrane domains; gray triangles represent the GAF domain; light gray boxes represent the kinase domain; and dark circles represent the GST protein. B, Transgenic yeast was incubated with 0.07 μL L−1 [14C]ethylene (gray bars), or with 0.07 μL L−1 [14C]ethylene and 1,000 μL L−1 [12C]ethylene (white bars). The difference between these two values represents the saturable binding. Samples were run in triplicate and sd are shown. Yeast expressing a mutant form of the ETR1 gene (etr1-1-GST[1-400]) was used as a control. Equal amounts of yeast membranes from yeast used in the binding assays were analyzed by western blots probed with anti-GST antibodies. Protein levels were quantified by chemifluorescence imaging on a Storm system and are reported as relative levels compared with the ERS1-GST(1-520) protein level. NA, Not applicable.
As shown in Figure 3B, yeast expressing ERS1-GST(1-520) bound ethylene at levels similar to yeast expressing the full-length ERS1 protein. Control yeast expressing ETR1-GST(1-400) with a mutation equivalent to etr1-1(C65Y) showed no ethylene binding above background, confirming that the presence of the GST tag did not affect ethylene-binding levels.
Membranes isolated from yeast used in the ethylene-binding assays were analyzed by western blot. Western blots probed with anti-GST antibodies and quantified with chemifluorescent imaging indicated that the ETR1-GST(1-400) protein was expressed at levels approximately six times that of ERS1-GST(1-520). The ERS1-GST(1-520) protein migrated as a doublet, possibly due to proteolysis, and both bands were included in the quantification. These data indicate that ERS1-GST(1-520) is expressed at lower levels than ETR1-GST(1-400) and may account for the lower levels of ethylene binding detected in yeast expressing ERS1 protein.
Effect of 1-Methylcyclopropene (1-MCP) on Ethylene Perception
After determining that yeast expressing ETR1 and ERS1 bind ethylene, we sought to obtain preliminary evidence comparing their relative affinities for ethylene. To do so, we examined the effects of the competitive inhibitor 1-MCP upon ethylene binding to yeast expressing ETR1 and ERS1 and upon ethylene-induced changes in Arabidopsis growth. Recently, 1-MCP has been shown to act at very low concentrations to inhibit ethylene-induced ripening and senescence. Its effectiveness as a competitive inhibitor is at least an order of magnitude better than that of other unsaturated cyclic olefins, such as trans-cyclooctene and 2,5-norbornadiene (Sisler et al., 1996a, 1996b; Sisler and Serek, 1999).
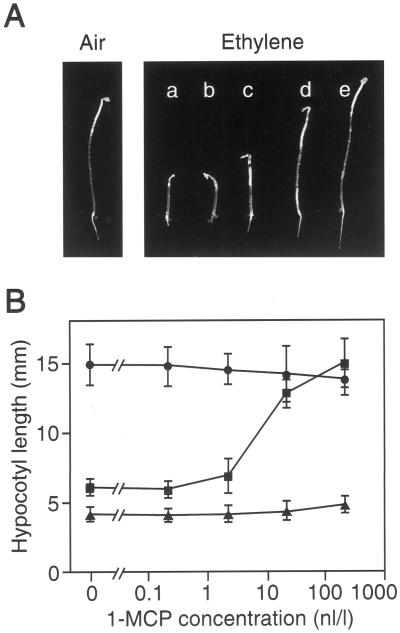
To test the effect of 1-MCP upon ethylene responses in Arabidopsis we analyzed the ability of 1-MCP to reverse the triple response of seedlings to ethylene. The ethylene-mediated triple response is characterized by an inhibition of hypocotyl and root elongation, radial swelling, and the formation of an exaggerated apical hook. As shown in Figure 4A, ethylene at a concentration of 1 μL L−1 induces the triple response in dark-grown Arabidopsis seedlings. Seedlings grown in the presence of 1 μL L−1 ethylene were treated with various concentrations of 1-MCP (Fig. 4). At a concentration of 2.2 nL L−1 the effect of 1-MCP upon ethylene responses first becomes apparent, with complete reversal of ethylene effects at 220 nL L−1 1-MCP. The effect of 1-MCP on the seedling-growth response was fit to the Hill equation to determine the concentration of inhibitor required to reduce growth response to 50% of its value in the absence of inhibitor (IC50). This analysis yielded an IC50 of 10.6 nL L−1 for the effect of 1-MCP. Given that the apparent dissociation constant (Kr) of ethylene for the hypocotyl-growth response is 0.11 μL L−1 (Chen and Bleecker, 1995), the apparent inhibition constant (KI) of 1-MCP for the hypocotyl-growth response is 1.05 nL L−1.
Figure 4.
1-MCP as an antagonist of ethylene responses in dark-grown Arabidopsis seedlings. A, Effect of 1-MCP on the triple response to ethylene. Representative seedlings are shown following growth in air or 1 μL L−1 ethylene. Seedlings labeled a through e were treated with 0, 0.22, 2.2, 22, and 220 nL L−1 1-MCP, respectively. B, Effect of 1-MCP on hypocotyl growth. Seedlings for wild-type (█), etr1-1 (●), and ctr1-2 (▴) mutants, were grown in 1 μL L−1 ethylene with the indicated amounts of 1-MCP. The mean hypocotyl length at each 1-MCP concentration is shown. At least 20 seedlings were used for measurements at each data point, and sd are shown.
Wild-type seedlings grown in the presence of 1-MCP look very much like seedlings grown in air, indicating 1-MCP does not appear to have deleterious effects upon the seedlings at the concentrations tested. The effect of 1-MCP was also tested upon Arabidopsis seedlings carrying the ctr1-2 mutation, which leads to a constitutive ethylene response in the presence or absence of ethylene. 1-MCP was not able to reverse the effects of the ctr1-2 mutation (Fig. 4B); ctr1-2 plants still showed the triple response phenotype at concentrations of 1-MCP that reversed the ethylene effects upon wild-type plants. This result is consistent with a model in which the effects of 1-MCP occur upstream of ctr1-2 in the ethylene signal transduction pathway with a direct effect upon the receptor.
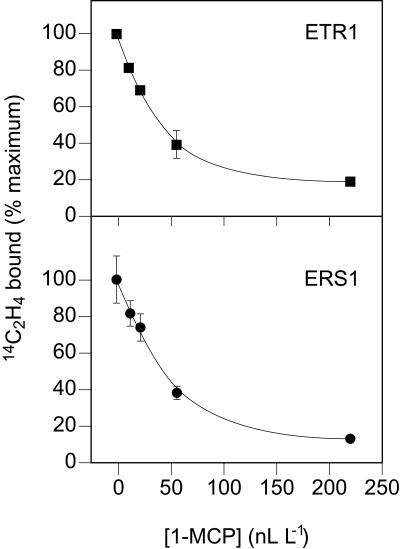
We examined the effects of 1-MCP upon [14C]ethylene binding to transgenic yeast expressing either the ETR1 or the ERS1 genes. As shown in Figure 5, 1-MCP proved to be an effective inhibitor of ethylene binding in both cases with maximal inhibition of ethylene binding reached at 220 nL L−1. Yeast expressing either ETR1 or ERS1 demonstrated almost identical kinetics for inhibition of ethylene binding by 1-MCP with an IC50 of 31.6 nL L−1 and 37.9 nL L−1, respectively. The dissociation constant (Kd) for binding of ethylene to transgenic yeast expressing the ETR1 gene is 0.036 μL L−1 (Schaller and Bleecker, 1995). Thus, 1-MCP has a KI of 10.7 nL L−1 in the ethylene-binding assay with ETR1. A Kd value for binding of ethylene to transgenic yeast expressing ERS1 could not be determined due to the low level of ethylene binding.
Figure 5.
1-MCP as an antagonist of ethylene binding in yeast expressing ETR1 or ERS1. Saturable ethylene binding of yeast expressing ETR1 or ERS1 was determined at 0.07 μL L−1 [14C]ethylene in the presence of the indicated levels of 1-MCP. In the absence of 1-MCP, the ETR1 sample bound 3,028 dpm/g yeast, and the ERS1 sample bound 341 dpm/g yeast. Samples were run in triplicate, and sd are shown.
DISCUSSION
Studies of ethylene signaling in Arabidopsis indicate that receptor gene families in plants may function similar to many of their animal counterparts, increasing their flexibility at responding to complex environments. In Arabidopsis, genetic evidence initially suggested that ethylene responses were mediated by a gene family. Dominant point mutations in at least three genes resulted in ethylene-insensitive plants all showing similar phenotypes although to different degrees. Subsequent cloning of ETR1 (Chang et al., 1993), ETR2 (Sakai et al., 1998), and EIN4 (Hua et al., 1998) confirmed that these genes constituted a gene family similar to prokaryotic two-component His kinase regulators.
The observation that loss-of-function mutants of ETR1, ETR2, EIN4, and ERS2 show normal ethylene responsiveness has provided further genetic evidence that the ETR1 receptor family possess at least partially overlapping functions (Hua and Meyerowitz, 1998). A constitutive ethylene-response phenotype at the seedling stage only emerges in the double mutants and is accentuated in triple and quadruple mutants. This phenotype is consistent with a model for ethylene signaling in which the ethylene receptors in an unbound state are negative regulators of the ethylene response pathway, and ethylene binding relieves this repression (Hua and Meyerowitz, 1998). However, not all double mutant combinations show the same alterations in ethylene responses, suggesting that the five receptor isoforms may differ qualitatively or quantitatively in their contribution to the seedling triple response.
In this paper we have begun to define the biochemical characteristics of the ERS1 protein to understand its specific role in ethylene signaling and identify biochemical similarities and differences between the ethylene receptor isoforms. Using a yeast expression system and in vivo ethylene-binding assays we have shown that ERS1, like ETR1, encodes an ethylene-binding protein. This is consistent with the high amino acid conservation between the proteins, including all residues known to be essential for ethylene binding to ETR1. It will be interesting to determine if the members of subfamily II (ETR2, ERS2, and EIN4) also are able to bind ethylene and if they do so with similar kinetics to ETR1 and ERS1. The subfamily II genes contain a fourth hydrophobic segment at their N termini, and it is unclear if this hydrophobic stretch of amino acids serves as a signal sequence or a fourth transmembrane domain, which could have implications for altering the ethylene-binding site.
In addition we have shown that the ERS1 protein forms a membrane-associated disulfide-linked dimer, which is another characteristic shared with ETR1. ETR1 forms a dimer in both plant and yeast membranes, and this linkage is mediated by two Cys residues in the extreme N terminus of the protein (Cys-4 and Cys-6) (Schaller et al., 1995). These two cysteines are conserved in ERS1, as well as in the other three ETR1-like genes. However, neighboring residues are not conserved, further suggesting these two cysteines play an important role in receptor structure or function. The finding that both ETR1 and ERS1 form dimers is significant because this indicates that they may function similarly to the bacterial sensor proteins in which the kinase domain of one monomer phosphorylates a conserved His residue in trans on the other monomer (Parkinson, 1993). In addition since both ERS1 and ETR1 form homodimers, the possibility exists that they might form heterodimers as well. Heterodimerization is a means by which other receptors, such as growth factor receptors, fine-tune signaling.
In the case of ETR1 and ERS1 several lines of data are consistent with the two proteins containing ethylene-binding sites with similar affinities for ethylene. Ethylene-binding assays carried out near the Kd for ethylene binding to ETR1 (Schaller and Bleecker, 1995) indicated a similar stoichiometry of ethylene binding per unit expressed protein. The virtually identical sensitivity of yeast expressing ERS1 and ETR1 to the competitive inhibitor 1-MCP also provides evidence that the two receptor isoforms have similar binding affinities for ethylene.
The inhibitory effects of a compound on ethylene binding and action can be quantified by the determination of an apparent KI (Sisler et al., 1990; Sisler, 1991; Abeles et al., 1992). We determined an apparent KI for 1-MCP for its effects on the ethylene-growth response (KI = 1.05 nL L−1, gas) and for its effects on ethylene binding to ETR1 in transgenic yeast (KI = 10.7 nL L−1, gas). The KI values calculated for 1-MCP in this or any study must be viewed with caution, given that they are based on estimated dissociation constants for ethylene that were obtained using in vivo assays. For example, synthesis and turnover of receptor protein over the course of the assays could have a direct influence on the Kd values obtained in both the plant and yeast systems. The apparent dissociation constant for the response (Kr) obtained from seedling dose-response analysis is even more subject to deviation from the true Kd for ethylene at the receptor due to the number of possible rate-limiting steps between ethylene perception and physiological response (for discussion, see Chen and Bleecker, 1995). Incorrect estimates of the Kd for ethylene would lead to incorrect values for the KI for 1-MCP.
These caveats notwithstanding, the 10-fold lower apparent KI for 1-MCP in the ethylene-mediated growth response, relative to the KI value calculated for 1-MCP in the ethylene binding to yeast expressing ETR1, indicates that 1-MCP is a much more effective inhibitor of ethylene responses than is predicted by its inhibitory effect on ethylene binding to the receptor. The greater sensitivity of the plant response to inhibition by 1-MCP is consistent with the effects of dominant mutations in the ethylene receptors on ethylene responses. According to the inverse agonist model for ethylene action, dominant point mutations in any one of the ethylene receptor genes are apparently sufficient to keep enough receptors consitutively active to completely repress ethylene responses. If the mechanism of inhibition by 1-MCP worked in a similar manner, binding of 1-MCP to a subset of receptors would be sufficient to constitutively activate enough receptors to keep the system repressed and ethylene responses off.
Although our data indicate ETR1 and ERS1 share similar biochemical properties at the level of signal perception, they do have important structural differences that may result in quantitative differences in signaling. ERS1 is lacking the response regulator domain found in ETR1, ETR2, and EIN4. This structural difference may certainly have consequences on signaling to downstream effectors, since the response regulator domain is often involved in phosphate transfer in other well-characterized signaling pathways. Whereas yeast two hybrid experiments have shown that the kinase domain of ERS1 interacts with CTR1, it does so with less affinity than ETR1 (Clark et al., 1998). This difference may also have implications on downstream signaling, for example, by altering the kinetics of signaling.
Another difference between ERS1 and ETR1 is that mRNA expression of ERS1 is ethylene inducible, whereas ETR1 mRNA expression is not (Hua et al., 1998). ERS1 orthologs in tomato (NR; Wilkinson et al., 1997) and Rumex (RP-ERS1; Vriezen et al., 1997) are also ethylene inducible, whereas the corresponding ETR1 orthologs are not. In Rumex and tomato, the increase in ERS1 mRNA parallels increasing tissue sensitivity to ethylene, and in melon it parallels fruit enlargement (Sato-Nara et al., 1999). The negative regulator model of ethylene receptor function leads to the prediction that up-regulation of ERS1 mRNA expression by ethylene would down-regulate ethylene responses. Given that the half-life for release of ethylene bound to yeast expressing ETR1 is 11 h (Schaller and Bleecker, 1995), new receptor synthesis may be a means to reactivate CTR1 and thus attenuate ethylene responses when ethylene levels decrease.
Many questions still remain to be answered to fully understand how ethylene responses are coordinately controlled by the five ethylene receptor isoforms. The isolation of an ERS1 loss-of-function mutant will help clarify the role of ERS1 in ethylene signaling and will be useful in determining if ERS1 has any ethylene-independent functions in Arabidopsis. In addition it will be interesting to determine if ERS1 is indeed an active His kinase, how this activity contributes to its function, and the consequences a lack of a response regulator has on ERS1 activity.
MATERIALS AND METHODS
Plasmid Constructions
To obtain a cDNA clone of ERS1, a probe was generated by PCR using Arabidopsis genomic DNA as template and primers specific for the first exon of ERS1. The 5′ primer was GAGACGCATGTGAATCAAGATGA and the 3′ primer was GAGGTATGTGCATAGCTTGAG. A cDNA clone containing the complete coding sequence for ERS1 (cERS1-6A) was isolated from an Arabidopsis cDNA library in Lambda ZAP II (Stratagene, La Jolla, CA; Kieber et al., 1993). This was converted into the pBluescript SK+ (Stratagene) plasmid and used for subsequent ERS1 constructs.
For expression in yeast, the vector pYcDE-2 was used (Hadfield et al., 1986). This vector has a constitutive ADH1 promoter, an EcoRI cloning site, and allows for Trp selection in yeast. A variant of the pYcDE-2 vector was prepared by addition of a 10-mer NotI linker to the end-filled EcoRI site, effectively replacing the EcoRI site with a NotI cloning site. To remove the 5′-non-coding sequence of ERS1, PCR was performed using the cERS1-6A clone as template, with a 5′ primer containing a NotI site at the end (GAGCGGCCGCAATGGAGTCATGCGATTGTTTT) and M13-reverse primer. The PCR product for ERS1 was digested with NotI and cloned into pYcDE-2. Cloning of ETR1 into pYcDE-2 has been previously described (Schaller and Bleecker, 1995).
For construction of the ERS1(1-520)-GST fusion protein, the GST gene (Smith and Johnson, 1988) was amplified using PCR from the PGEX-4T1 vector (Amersham-Pharmacia Biotech, Uppsala), introducing an EcoRI site in the 5′ primer (GGGAATTCTATTCATGTCCCCTATACTAGG). A stop codon was included in the 3′ primer, followed by an EcoRI site (GGGACTTAAGGAGTTCCACGCGGAACCGG). The GST fragment was digested with EcoRI and cloned into the EcoRI site of the ERS1 gene within the pYcDE-2 yeast shuttle vector. For construction of the ETR1-GST(1-400) fusion protein, the GST gene was PCR amplified from PGEX-4T2 (Amersham-Pharmacia Biotech), introducing an BglII site at the 5′ end and EcoRI and BamHI sites at the 3′ end. The PCR product was digested with BglII and BamHI and cloned into the BglII site of the ETR1 gene (cETR1-5.2; Schaller and Bleecker, 1995). The resulting construct was digested with EcoRI, and the 1.9-kb fusion of ETR1(1-400) with the GST gene was gel purified and cloned into the EcoRI site of pYcDE-2 for yeast expression.
For expression of a 6-His-tagged ERS1 fusion protein in Escherichia coli, the vector pET-15b (Novagen, Madison, WI) was used. PCR was carried out using the ERS1 cDNA clone as template, a 5′ primer with an NdeI site (TAGATAGACATATGGGTCTTATTTTAA) and a 3′ primer with a BamHI site (GCCTCTTGAGGATCCTTGTCTAAAGC). The PCR product was digested with NdeI and BamHI and cloned into pET-15b. The cloned portion of ERS1 represents amino acids 133 to 332 of the protein.
Yeast Transformation and Growth
Constructs were transformed into yeast Saccharomyces cerevisiae (Schiestl and Gietz, 1989), strain LRB520 (MAT his3leu2trp1 ura3-52yck2-1::HIS3), and standard media and procedures used for growth (Ausubel et al., 1994).
Arabidopsis Growth
Arabidopsis seedlings were grown on agar plates containing 0.5× Murashige and Skoog basal salt mixture, pH 5.7 (Murashige and Skoog, 1962), 0.8% (w/v) agar, and B5 vitamins consisting of 100 mg/mL inositol, 1 mg/mL nicotinic acid, 1 mg/mL pyridoxin HCl, and 10 mg/mL thiamine HCl. Seeds were stratified for 4 d at 4°C, then moved to 22°C (time 0). For light-grown seedlings, plates were kept under continuous fluorescent light. For dark-grown seedlings, plates were light-treated for 12 h before being moved to the dark.
Protein Isolation
For yeast membrane protein isolation, 3.0-g aliquots of yeast were resuspended in 3.0 mL of extraction buffer (50 mm Tris [tris(hydroxymethyl)aminomethane], pH 7.5, 100 mm NaCl, 10% [v/v] glycerol, and 1% [v/v] dimethyl sulfoxide; Ausubel et al., 1994). Three volumes of chilled glass beads were added to the yeast cells. Cells were vortexed seven times (30-s intervals) with 1-min intervals on ice between vortexing. The supernatant was spun at 10,000g for 10 min at 4°C to remove cellular debris. Membranes were pelleted at 30,000g for 30 min and resuspended with a homogenizer (Wheaton, Millville, NJ ) in 1.2-mL assay buffer (10 mm MES [2-(N-morpholino)-ethanesulfonic acid], pH 5.5, 20% [w/v] Suc, and 1% [v/v] dimethyl sulfoxide). This extract was then directly added to 2× SDS-PAGE sample buffer. Protein concentrations were determined by the Bradford method (Bradford, 1976), using bovine serum albumin as a standard.
Antibody Production
The ERS1-6-His fusion protein was expressed in E. coli from the pET-15b vector according to the manufacturer (Novagen). Inclusion bodies were purified by SDS-PAGE on 12% (w/v) polyacrylamide gels. The protein band was excised, electroeluted, and concentrated with Centricon 10 microconcentrators (Amicon, Beverly, MA). Antisera were prepared from recombinant protein by the University of Wisconsin Medical Facility, and cleared against yeast proteins as described (Chang et al., 1992). Preparation of the ETR1-HRR antibody was described previously (Schaller and Bleecker, 1995).
SDS-PAGE and Western Blotting
Protein samples were mixed with SDS-PAGE loading buffer (125 mm Tris, pH 6.8, 20% [v/v] glycerol, 4% [w/v] SDS, and 0.01% [w/v] bromphenol blue) in the presence or absence of 100 mm DTT as indicated. Samples were incubated at 37°C for 1 h and then subjected to SDS-PAGE using 8% (w/v) gels. Proteins were electrotransferred (12 V, 30 min) from gels to Immobilon membrane (Millipore, Milford, MA), and the membrane was then blocked with non-fat dry milk.
For western blotting, the anti-ERS1 antibody was used at a 1:5,000 dilution, the anti-ETR1(HRR) antibody was used at a 1:5,000 dilution, and the anti-GST (Sigma, St. Louis) antibody used at a 1:5,000 dilution. Immunodecorated proteins were visualized by chemiluminescence according to the manufacturer's instructions (Kirkegaard and Perry, Gaithersburg, MD). For protein quantification experiments, immunodecorated proteins were visualized by chemifluorescence according to the manufacturer's instructions (Amersham-Pharmacia Biotech). Protein bands were quantified using the ImageQuant software on a STORM Phosphorimager (Molecular Dynamics, Sunnyvale, CA).
Ethylene Binding to Transgenic Yeast
Ethylene binding experiments were performed as previously described (Schaller and Bleecker, 1995; Hall et al., 1999), using a modification of the method originally described by Sisler (1979). Curve-fitting was performed using SIGMAPLOT. IC50 values were determined by fitting data to the Hill equation using SIGMAPLOT. KI values were determined from the equation KI = IC50/(1 + [S]/Kd).
Ethylene Treatment of Arabidopsis
For the experiment shown in Figure 4, 1-L gas-tight jars were used. Ethylene (final concentration = 1 μL L−1) and 1-MCP (final concentrations as indicated) were introduced through an injection port. Seedlings were grown in the dark on vertically oriented plates and removed after 4 d of growth. Hypocotyl length was determined using the program NIH Image (version 1.6) after first scanning the plates using Adobe Photoshop (version 5.5) and a LaCie scanner. Dose response data were fitted to the Hill equation (Weyers et al., 1987) using SIGMAPLOT. Ethylene concentration was determined by gas chromatography using a column of Carboxen 1000, 45/60 mesh size (Supelco, Bellefonte, PA), with ethylene as the calibration standard.
Preparation and Use of 1-MCP
1-MCP was prepared (Magid et al., 1971) and stored in the gas phase in a container with saturated (NH4)2SO4 as a seal. Concentration was determined by gas chromatography using a column of Carbopak B (80/120 mesh size), 3% (v/v) SP1500 (Supelco) with butane as the calibration standard.
ACKNOWLEDGMENTS
We thank the University of Wisconsin-Madison Pathology department for the use of their Storm system and Magdaly Cintron for assistance making the 6-His-tagged ERS1 construct. We thank Anita Klein and the Bleecker laboratory for comments on the manuscript and Rick Cote for assistance with SIGMAPLOT.
Footnotes
This work was supported by the National Science Foundation (grant nos. MCB–9603679 to G.E.S. and MCB–9513463 to A.B.B.), by the HATCH/U.S. Department of Agriculture project (grant no. 386 to G.E.S.), by the U.S. Department of Energy (grant no. DE–FG02–91ER20029 to A.B.B.), and by the U.S. Department of Energy-National Science Foundation-U.S. Department of Agriculture Collaborative Research in Plant Biology Program (grant no. BIR92–20311 to A.E.H.).
LITERATURE CITED
- Abeles FB, Morgan PW, Salveit ME. Ethylene in Plant Biology. San Diego: Academic Press; 1992. [Google Scholar]
- Ausubel FM, Brent R, Kingston R, Moore DD, Seidman JG, Smith JA, Strohl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1994. [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H. Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science. 1988;241:1086–1089. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Chang C, Schaller GE, Patterson SE, Kwok SF, Meyerowitz E, Bleecker AB. The TMK1 gene from Arabidopsis codes for a protein with structural and biochemical characteristics of a receptor protein kinase. Plant Cell. 1992;4:1263–1271. doi: 10.1105/tpc.4.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QG, Bleecker AB. Analysis of ethylene signal transduction kinetics associated with seedling-growth responses and chitinase induction in wild-type and mutant Arabidopsis. Plant Physiol. 1995;108:597–607. doi: 10.1104/pp.108.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA. 1998;95:5401–5406. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble RL, Coonfield ML, Schaller GE. Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc Natl Acad Sci USA. 1998;95:7825–7829. doi: 10.1073/pnas.95.13.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield C, Cashmore AM, Meacock PA. An efficient chloramphenicol-resistance marker for Saccharomyces cerevisiae and Escherichia coli. Gene. 1986;45:149–158. doi: 10.1016/0378-1119(86)90249-0. [DOI] [PubMed] [Google Scholar]
- Hall AE, Chen QG, Findell JL, Schaller GE. The relationship between ethylene binding and dominant insensitivity conferred by mutant forms of the ETR1 ethylene receptor. Plant Physiol. 1999;121:291–300. doi: 10.1104/pp.121.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Hua J, Sakai H, Meyerowitz EM. The ethylene receptor family in Arabidopsis. In: Kanellis K, Kende H, Grierson D, editors. Biology and Biotechnology of the Plant Hormone Ethylene. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 71–76. [Google Scholar]
- Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM. EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell. 1998;10:1321–1332. doi: 10.1105/tpc.10.8.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PR, Ecker JR. The ethylene gas signal transduction pathway: a molecular perspective. Annu Rev Genet. 1998;32:227–254. doi: 10.1146/annurev.genet.32.1.227. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Magid RM, Clarke TC, Duncan DD. An efficient and convenient synthesis of 1-methylcyclopropene. J Org Chem. 1971;36:1320–1321. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Parkinson JS. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- Rodriguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB. A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science. 1999;283:996–998. doi: 10.1126/science.283.5404.996. [DOI] [PubMed] [Google Scholar]
- Sakai H, Hua J, Chen QG, Chang C, Medrano LJ, Bleecker AB, Meyerowitz EM. ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:5812–5817. doi: 10.1073/pnas.95.10.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato-Nara K, Yuhashi K-I, Higashi K, Hosoya K, Mitsuru K, Ezura H. Stage- and tissue-specific expression of ethylene receptor homolog genes during fruit development in Muskmelon. Plant Physiol. 1999;119:321–329. doi: 10.1104/pp.120.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Bleecker AB. Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science. 1995;270:1809–1811. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- Schaller GE, Ladd AN, Lanahan MB, Spanbauer JM, Bleecker AB. The ethylene response mediator ETR1 from Arabidopsis forms a disulfide-linked dimer. J Biol Chem. 1995;270:12526–12530. doi: 10.1074/jbc.270.21.12526. [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Sisler EC. Measurement of ethylene binding in plant tissue. Plant Physiol. 1979;64:538–542. doi: 10.1104/pp.64.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisler EC. Ethylene-binding components in plants. In: Suttle JC, editor. The Plant Hormone Ethylene. Boca Raton, FL: CRC Press; 1991. pp. 81–99. [Google Scholar]
- Sisler EC, Blankenship SM, Guest M. Competition of cyclooctenes for ethylene binding activity in plants. Plant Growth Regul. 1990;12:125–132. [Google Scholar]
- Sisler EC, Dupille E, Serek M. Effect of 1-methylcyclopropene and methylenecyclopropane on ethylene binding and ethylene action on cut carnations. Plant Growth Regul. 1996a;18:79–86. [Google Scholar]
- Sisler EC, Serek M. Compounds controlling the ethylene receptor. Bot Bull Acad Sin. 1999;40:1–7. [Google Scholar]
- Sisler EC, Serek M, Dupille E. Comparison of cyclopropene, 1-methylcyclopropene, and 3,3-dimethyl-cyclopropene as ethylene antagonists in plants. Plant Growth Regul. 1996b;18:169–174. [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Vriezen WH, van Rijn CP, Voesenek LA, Mariani C. A homolog of the Arabidopsis thaliana ERS gene is actively regulated in Rumex palustris upon flooding. Plant J. 1997;11:1265–1271. doi: 10.1046/j.1365-313x.1997.11061265.x. [DOI] [PubMed] [Google Scholar]
- Weyers JDB, Paterson NW, Brook A. Towards a quantitative definition of plant hormone sensitivity. Plant Cell Environ. 1987;14:1–12. doi: 10.1111/j.1365-3040.1987.tb02073.x. [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Clark DG, Bleecker AB, Chang C, Meyerowitz EM, Klee HJ. A dominant mutant receptor from Arabidopsis confers ethylene insensitivity in heterologous plants. Nat Biotechnol. 1997;15:444–447. doi: 10.1038/nbt0597-444. [DOI] [PubMed] [Google Scholar]