Abstract
The Cre-lox recombination approach is commonly used to generate cell-specific gene inactivation (or activation). We have noticed that the breeding and genotyping sections of papers utilizing Cre-lox techniques are frequently incomplete. While seemingly straightforward, there are important considerations that need to be implemented in the breeding and genotyping to prevent the introduction of experimental confounds. Germline recombination and transient expression of Cre recombinase during development are some examples of the complications that can occur, and conventional genotyping methods may fail to identify these events. In this article, we highlight the importance of testing for unexpected recombination events, suggest strategies to isolate and minimize adverse recombination events, and encourage editors and reviewers to expect more definitive statements regarding the validation of genotyping.
Keywords: transgenic mice, Cre-recombinase, germline recombination, ectopic gene expression
Cre recombinase allows cell-specific manipulation of gene expression
Cre recombinase (Cre) is a 343-amino-acid protein comprised of 4 subunits that recognizes pairs of specific 34 bp DNA sequences called loxP sites. It initially creates a DNA loop and then either excises or inverts the looped segment depending on the orientation of the loxP sites [1–3]. By using cell-specific drivers for Cre, this recombination can be limited to generate conditional knock-out or knock-in animals. This site-specific recombination was initially discovered in bacteriophage P1 and the potential utility for cell-type-specific genetic manipulations was noted [2]. In 1987, the Cre-lox system was utilized for the first time in yeast [3], and subsequently in cultured mammalian cells [4]. With the development of transgenic mice, where novel DNA sequences could be stably introduced into the mouse genome in a heritable manner [5–6], the Cre-lox system was first implemented in T-cells [7], and then in neuronal tissue [8]. Controlling Cre expression using a cell-specific promotor allows selective inactivation, activation, or mutation of genes of interest. The first Cre-driver lines of mice had transgenes with relatively short regulatory regions fused to Cre recombinase, which integrated randomly into the mouse genome. A strategy that gives much more faithful Cre expression than this method involves inserting Cre recombinase into the gene of interest in a bacterial artificial chromosome (BAC) of about 200 kb and using that construct to generate so-called BAC transgenic mice, again with random integration. These transgenic approaches can give cell-specific expression, but regulation of Cre recombinase expression may suffer from chromosomal effects due to the random site of integration and variable numbers of gene copies. A more reliable strategy involves gene targeting (by homologous recombination in embryonic stem cells or CRISPR/Cas9 methodology) such that a single copy of a Cre recombinase gene is expressed from an endogenous gene locus. A cassette including Cre recombinase is typically inserted into the gene of interest either (a) just before the normal initiation codon, (b) after a 2A self-cleaving sequence that is inserted in place of the termination codon, or (c) after an internal ribosome entry site (IRES) sequence inserted just after the normal termination codon. With all the methods described above, the insertion of foreign DNA (the bacteriophage Cre recombinase DNA) can influence expression of both the Cre recombinase and the gene into which it is inserted due to gene silencing (e.g. by DNA methylation). With method (a), insertion of Cre recombinase will likely disrupt expression of the gene, which is useful if one also wants to create a null allele; with method (c) initiation of translation using the IRES will probably be less efficient than initiation of the primary mRNA. It is always necessary to carefully document the expression pattern of new Cre-driver mice. The selective control of gene expression by the Cre-lox method has profoundly impacted many fields by enabling the understanding of gene functions whose global knockout would be lethal or create non-specific results [8–9]. There are now different types of recombinases available and several variants of lox sites that can be used together to create more sophisticated recombination events, expanding the range of applications in which this technology can be implemented.
Unexpected expression of Cre recombinase
Although the Cre-lox system provides remarkable control over gene expression, there are limitations that need to be considered when both the Cre recombinase and lox sites are present in an animal that will then be bred. Cre recombinase is usually targeted to genes that are expressed in a limited number of cells in adult mice. However, there may be unexpected transient expression of Cre recombinase in the germline or during early development. This unexpected gene expression is sometimes referred to as ‘ectopic’ expression suggesting that it is unnatural; however, many genes are normally expressed in the germline or at various stages of development although the significance of this expression is generally unknown. It does not take much expression of Cre-recombinase to act on the loxP sites to generate recombination events. Thus, it is essential to design experiments that can detect these unexpected and unwanted recombination events.
Testing a new Cre-driver line of mice
Most investigators breed a new Cre-recombinase-expressing (Cre-driver) line of mice with a Reporter line of mice in which expression of the Reporter is activated by deleting a ‘lox-stop-lox’ sequence to test for the specificity of the Cre-induced recombination in the offspring. The reporter lines from the Allen Institute are commonly used for this purpose [10–12]. In this scenario, one typically begins with a test cross, in which mice that are heterozygous for Cre recombinase (designated as: Gene XCre/wt) are bred with a heterozygous or homozygous floxed Reporter mice (Reporter lox/wt or Reporter lox/lox, respectively). One expects the Cre-positive offspring to manifest expression of the Reporter in cells that are known to express Gene X, which can be documented by immunohistochemistry or in situ hybridization. It is often the case that normal expression of Gene X during development is not well documented; hence, it is not uncommon to find that expression of the Reporter is more widespread than anticipated because Gene X was expressed transiently during development. The unanticipated Reporter expression can be variable from one offspring to another, presumably depending on the stochastic nature of Cre recombinase expression [10], but genetic background differences may also contribute to this variability. Thus, one needs to examine the expression of several mice, ideally from different parents, to be confident of appropriately restricted expression of the Cre recombinase. If there is unwanted expression observed from the genetic cross of a Cre-driver line with a floxed Reporter line, the investigator must consider how that might influence their results.
Making conditional knockout mice and testing for unexpected recombination
In the experiments described above, the genes carrying Cre recombinase and the floxed Reporter come together at fertilization to turn on a gene of interest through the excision of a STOP cassette in a single copy of the Reporter. However, for conditional gene-knockout experiments, investigators usually want BOTH alleles of the gene of interest (Gene Y) to undergo recombination. As a result, the Cre recombinase and the floxed allele of Gene Y will be together in the germline, where unexpected recombination is common. An easy way to visualize the potential problem of germline expression is to breed mice of both sexes that carry both genes (Gene XCre/wt;Reporterlox/wt mice) that were generated in the test cross (previous paragraph) with wild-type (WT) mice and examine the expression of the Reporter in their offspring. If the expression pattern of these second-generation mice is more widespread than with the first cross, then germline expression of the Cre recombinase is likely. Note that because recombination may occur at the diploid stage of gametogenesis and the resulting haploid daughter cells give rise to the embryo, offspring of this second cross that do not inherit Cre recombinase may still show expression of the Reporter gene (Fig. 1). We highly recommend this second test cross as an easy way to visualize potential problems when generation of conditional, gene-knockout mice is planned.
Figure 1.
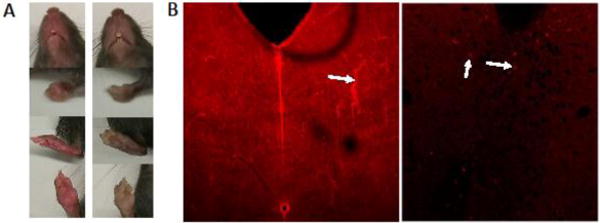
Non-specific expression of a Reporter gene from a genetic cross of Oprk1Cre/wt ; Rosa26lox-stop-lox-tdTomato/wt with a wild-type mouse. (a) Images of facial area, front limb, hind limb, and front paw (from top to bottom) of a Oprk1wt/wt Rosa26lox-stop-lox-tdTomato/wt mouse (left) with nonspecific global expression of tdTomato, and its Oprk1Cre/wt ;Rosa26lox-stop-lox-tdTomato/wt littermate (right). (b) Confocal images of a 40-μm brain section of the dorsal raphe nucleus from the mouse with non-specific expression of tdTomato (left) including in blood vessels (arrow), and its littermate (right) with cell-specific expression of tdTomato (arrows) in kappa opioid receptor expressing neurons taken under the same capture settings.
Generating conditional knock-out animals require two genetic crosses. In the first cross, Gene XCre/wt mice are bred with mice that are either heterozygous or homozygous for the floxed allele of Gene Y (designated as: Gene Ylox/wt or Gene Ylox/lox, respectively) to generate mice with one copy of each gene (Gene XCre/wt;Gene Ylox/wt). In the second cross, Gene XCre/wt;Gene Ylox/wt mice are bred with Gene Ylox/wt or Gene Ylox/lox mice to generate the desired Gene XCre/wt;Gene Ylox/lox mice. If Cre expression is tightly controlled and not expressed in the germline or during development, then one expects Cre-positive offspring of the second cross to include Gene XCre/wt;Gene Ylox/wt and Gene XCre/wt;Gene Ylox/lox. The latter group are the experimental animals that should only have Gene Y recombined on both alleles in cells that express Cre recombinase. However, there is a potential problem with this strategy. After the first cross, there may be recombination of the floxed allele in the germline of Gene XCre/wt;Gene Ylox/wt mice, which will generate a deletion allele (designated here as Δ) due to recombination between loxP sites in some or all of the cells [13]. Hence, the second genetic cross may really be Gene XCre/wt;Gene YΔ/wt bred with Gene Ylox/wt or Gene Ylox/lox. If one uses the conventional, 2-primer PCR strategy to detect the floxed Gene Y, one may not detect this unexpected germline recombination event.
Consider this example: Figure 2a depicts a hypothetical Gene Y with an exon flanked by loxP sites and the location of primers a, b and c. Two of the primers (a forward and b reverse) flank one of the loxP sites and the third primer (c reverse) is designed to lie beyond the other loxP site such that after recombination, PCR with primers a and c produces a product that is longer than the product produced with a and b from the floxed allele. Thus, on a gel, the smallest band (a-b) is the WT allele, the next larger band (a-b which includes the lox site) is the lox allele and the largest band (a-c) is the Δ allele. Depending on the PCR conditions, the very long PCR product from a to c of the lox allele may also be seen at the top of the gel. Figure 2b shows a typical genetic cross used to generate conditional knockout mice. Figure 2c shows a hypothetical gel with the expected PCR products using a 2-primer PCR protocol from tail DNA where recombination is not expected (lanes 1, 2) and the results that would occur with partial (lanes 3, 5) or complete (lanes 4, 6) recombination. Note that with partial recombination the lox allele is less abundant than expected and with complete recombination there is no band representing the lox allele at all. However, if one uses a 3-primer PCR strategy as in Figure 2d, then one can detect the Δ allele and the unexpected recombination becomes obvious. Only the PCR results from the Cre-positive offspring are shown in Figure 2c and d; however, Cre-negative offspring can also reveal evidence of recombination because recombination may occur in diploid stages of gametogenesis and the Cre recombinase gene may not be segregated to haploid gametes. The abundance of each of the primers in the PCR mix should be adjusted so that all three bands are of approximately equal intensity. Also, enough PCR cycles should be used to ensure robust band intensity. However, note that if the PCR reaction is continued beyond the linear phase, then WT/lox heteroduplexes can form and they migrate slower on the gel than the lox allele band, appearing as a doublet. Therefore, when considering the location of primer c, take into account the likely appearance of this heteroduplex band. Obviously, having offspring with global or partial recombination of Gene Y in all cells can affect the interpretation of results. Thus, employing a PCR strategy that can detect the unexpected recombination event (the Δ allele) in all animals, not just the Cre-positive animals, is essential to avoid misinterpreting data.
Figure 2.

Detection of unexpected recombination during generation of conditional knockout mice. (a) Top line, is the wild-type (WT) allele with a hypothetical exon (box); below that is the floxed allele with the exon flanked by loxP sites (arrow heads), and below that is the floxed allele after Cre-mediated recombination. The location of PCR primers, a, b, and c is indicated along with the sizes of the PCR products. (b) A genetic cross between a mouse bearing Gene XCre/wt and Gene Ylox/wt and a mouse homozygous for the floxed gene (Gene Ylox/lox). (c) Hypothetical results from PCR analysis of the offspring that are positive for Cre recombinase using a 2-primer PCR strategy that detects the WT and lox alleles. Lanes 1, 2 show expected results with no unexpected recombination in tail DNA. The WT/lox heteroduplex band may appear after too many PCR cycles. Lanes 3, 5 show the results when there is partial recombination (thin arrow Δ) during early development. Lanes 4, 6 show results when there is complete recombination (thick arrow Δ). Note that with germline or developmental recombination, the lox allele becomes fainter and may even disappear. Recombination can also be observed in Cre-negative offspring (not shown). (d) Hypothetical results using a 3-primer strategy that can detect the Δ allele. Unexpected recombination is revealed by the presence the 200 bp band.
Because of the possibility of variable recombination in the germline, one can deliberately generate the Δ allele by breeding Gene Ylox/wt mice with a ubiquitously expressed Cre-driver line and then generate mice in the first genetic cross that are Gene XCre/wt;Gene YΔ/wt and breed them with Gene Ylox/lox mice. After that cross, the relevant Cre-positive experimental offspring are Gene XCre/wt;Gene YΔ/lox. Figure 3 shows an example of the PCR results expected with this cross using the 3-primer strategy. Even with this cross it is not uncommon to observe offspring where some or all of the floxed alleles have recombined to generate Δ alleles. Genotyping suggests that some mice have all three alleles (Fig. 3c, lane 3). The recombination of the floxed allele in this breeding strategy cannot occur in the germline and must therefore have occured in the early embryo. When Cre recombinase is in the male germline, perhaps a little Cre recombinase is transmitted by the sperm to the egg where the recombination occurs. When Cre recombinase is expressed in oocytes, recombination can occur even after fertilization. Alternatively, the Cre recombinase gene may be expressed transiently after fertilization to allow recombination of the floxed allele in the fertilized egg or early blastomeres. Regardless, the presence of the Δ allele after PCR of DNA from tissues where expression is not expected (e.g., tail or ear DNA) in mice with a WT allele (Fig. 3c, lane 3) is indicative of unexpected recombination. Conversely, unexpected recombination in offspring with the Gene YΔ/lox genotype can be harder to detect unless recombination is robust because the Δ allele is already present (Fig. 3c, lanes 2 and 5). The end result of unexpected recombination with either breeding strategy is the same (compare Figs. 2d and 3c), i.e., it does not matter whether recombination actually occurs in the germline or the early embryo. Figure 3d shows PCR results from a cross between Kiss1Cre/wt; Slc17a6Δ/wt and Slc17a6lox/lox mice, which reveals unexpected recombination.
Figure 3.
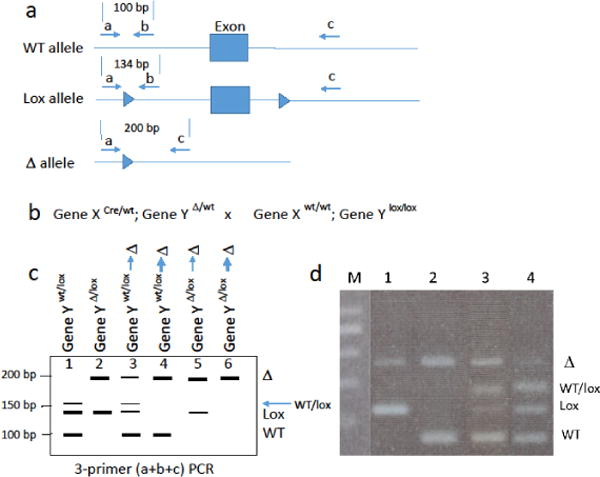
(a) Top line, is the wild-type (WT) allele with a hypothetical exon (box); below that is the floxed allele with the exon flanked by loxP sites (arrow heads), and below that is the floxed allele after Cre-mediated recombination. The location of PCR primers, a, b, and c is indicated along with the sizes of the PCR products. (b) A genetic cross between a mouse bearing Gene XCre and Gene YΔ/wt and a mouse homozygous for the floxed Gene Y (no Cre). Note that in this breeding scheme, recombination cannot occur in the germline because there is no lox allele in the germline. (c) Hypothetical results from PCR analysis of tail DNA (where recombination is not expected) from Cre-positive offspring. Lanes 1, 2 show expected results with no unexpected recombination in tail DNA. The WT/lox heteroduplex band may appear with too many PCR cycles. Lanes 3, 5 show the results when there is partial recombination (thin arrow Δ) during early development. Lanes 4, 6 show results when there is complete recombination (thick arrow Δ) during early development. The results are the same as when the parent with Gene XCre carries Gene Ylox/wt allele (Figure 2D); however, in that case one does not know whether recombination occurred in germline or during early development. Recombination can also be observed in Cre-negative offspring (not shown). (d) PCR results from a genetic cross of the type shown in B; Note the presence of the Δ allele in lanes 2, 3, and 4. In lane 2, there must have been complete recombination of the lox allele during early development because WT and Δ alleles are not possible from this cross; lanes 3 and 4 reveal partial recombination of the lox allele.
It is tempting to maintain colonies of conditional knockout mice by inbreeding Gene XCre/wt;Gene Ywt/lox (or some of the other offspring that were not used for experiments). This can be a very risky strategy in the absence of rigorous surveillance of the Δ allele. With Cre recombinase expressed in both male and female germlines the probability of unexpected recombination increases. Hence, it is easy to produce a colony of mice with complete or partial occurrence of Gene YΔ/Δin most cells, leading to erroneous results.
Appropriate control animals for conditional-knockout experiments
If expression Cre recombinase is not leaky or one is using an inducible Cre recombinase, then the experimental animals can be Gene X Cre/+;Gene Ylox/lox and animals from the same cross that lack Cre can serve as one control. If mice with Cre recombinase have a phenotype (e.g. because animals are heterozygous for loss of function of Gene X), then animals of Gene XCre/+;Gene Y+/+ genotype should be used as controls. If the experimental animals are Gene XCre/+;Gene YΔ/lox, then animals of Gene XCre/+;Gene YΔ/+ genotype are suitable controls. In this case, all animals are heterozygous for Gene X and recombination occurs only where Cre is expressed.
Concluding Remarks and Future Perspectives
The inadvertent generation and interpretation of data of nonspecific or global knock-out animals can be detrimental to an experiment. However, these errors can be tracked and avoided with appropriate genotyping and careful monitoring. Germline expression and developmental gene expression are natural phenomena; consequently, test-crossing new Cre-driver lines with Reporter animals in conjunction with strategies outlined in Box 1 with the 3-primer genotyping system aids in the management of unwanted recombination events. We believe that these proper breeding and genotyping protocols are often overlooked aspects of experiments involving Cre-dependent gene manipulations and errors are often difficult to identify due to incomplete methods sections. Our recommendations for Methods sections are outlined in Box 2 with examples for describing the methods used in Cre-lox studies. There is need for alternative strategies to manipulate gene expression, some of which are outlined in the Outstanding Questions.
Box #1. Categorization of Cre-driver lines and potential solutions when expression pattern is unexpected.
Ideal: Cre is expressed only in adult cells of interest, with no expression in other cells during development or in the germline.
-
Cre is expressed only in adult cells of interest, but there is expression in other cells during development but not in the germline.
Utilize an inducible Cre in the adult, e.g., by using a CreERt, in which the function of Cre is activated by administration of tamoxifen [14], using the Tet system, in which expression of Cre recombinase is dependent on tetracycline or its derivatives such as doxycycline [15–17], or using a Cre recombinase with a nonsense codon that can be suppressed by administration of a nonsense suppressor drug [18].
For neurobiology experiments, one can sometimes achieve regional and temporal control by stereotaxic injection of viruses expressing Cre-dependent genes into the brains of adult mice. Another approach is to inject a virus expressing FLP recombinase into an adult mouse with a FLP recombinase-dependent Cre. FLP-dependent Cre can be made by inserting frt-flanked stop cassette between the promoter and Cre recombinase, or by inserting an intron into Cre, flanking the 2nd exon with double frt sites and inverting it. The action of FLP recombinase inverts the second exon to allow spicing and production of functional Cre recombinase.
-
Cre is expressed only in adult cells of interest, but with expression in the germline.
If there is only partial and variable extents of recombination as revealed by the appearance of the Δ allele, or if the test crosses with a floxed Reporter mouse give variable expression from one mouse to the next (Figs. 2 and 3), then one should test the variability of transmittance of the Cre recombinase gene by male or female parents. After choosing the more conservative donor of the gene, using only those offspring with no obvious Δ allele is a feasible strategy.
If the extent of recombination is low, indicated by variable (stochastic) degrees of recombination, then making a new line with attenuated expression of Cre recombinase (e.g., by removing a nuclear localization signal, not using a codon-optimized Cre, weakening the initiation codon from the ideal RXXATGG sequence, and/or adding sequences that destabilize the Cre recombinase mRNA or protein) is a possible solution. Compromising the activity of Cre recombinase helped reduce unexpected recombination for both AgrpCre and Kiss1Cre constructs, but not CalcaCre (unpublished observations).
If the extent of recombination in the germline is high, then using a mouse with an inducible Cre (see 2a) or using a different strategy (see Outstanding Questions box) is the only solution.
Box #2. Recommendations.
Most published papers (including some of our own) that describe conditional, gene-knockout experiments do not include sufficient information in Materials and Methods sections to evaluate whether the authors were cognizant of the potential problems of unexpected recombination when transmitting Cre recombinase and floxed alleles through the germline and do not fully describe how they dealt with the problem. Investigators using these techniques for the first time may be unaware of the problem because there is no acknowledgment of these genetic complications by established investigators. Therefore, we suggest that all papers involving the use of Cre/lox system (or any of the other recombinase/target sequence approaches) include details in the Material and Methods section (if not in the Results) describing their breeding strategy and the methods that they used to detect unexpected recombination. A typical statement might include the following information.
When breeding the Gene XCre line with a floxed Reporter line (designate line) we (how often) observed expression of the Reporter in (tissue examined) of (number mice examined) in cells of the adults that do not express the Gene X as detected by (method). We also bred (male and/or female) mice carrying both Gene XCre and a floxed Reporter with wild-type mice and the expression pattern was the (same or different) than when breeding the Gene XCre line directly with a floxed Reporter line.
To generate conditional knockout mice we first crossed Gene XCre mice to floxed Gene Y mice and then bred (males or females) carrying both the Cre allele and the floxed allele with (heterozygous or homozygous) floxed Gene Y mice. We carefully looked for unexpected generation of the recombined allele by PCR using primers (identified in Methods) that distinguish all three potential alleles. We (how often) detected evidence of recombination in DNA from tissue where expression is not expected (e.g. tail DNA) and did not use those mice for any experiments.
Highlights.
Cre-lox manipulation is an important tool used widely in mouse experiments to selectively express or knock-out genes of interest in a cell-specific manner. However, germline recombination and transient expression of Cre recombinase in the germline or during development can lead to unexpected expression, which often goes undetected.
It is important to deliberately determine the extent of ‘ectopic’ recombination to avoid misinterpretations of results from animals with unexpected recombination.
New Cre-recombinase lines need to be test-crossed with a Reporter line to identify the expression variability and utilize a strategy that minimizes the adverse effects of unwanted recombination events.
Acknowledgments
We thank Drs. B. Land, G.S. McKnight, L. Zweifel, C. Chavkin, O. Gayol, J. Hidalgo, R. Bluett, and D. Durnam for their feedback on the manuscript. This report was supported by the NIMH Conte Center Grant 5P50MH106428-03.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Yarmolinsky M, Hoess R. The legacy of Nat Sternberg: The genesis of Cre-lox technology. Annu Rev Virol. 2015;2:25–40. doi: 10.1146/annurev-virology-100114-054930. [DOI] [PubMed] [Google Scholar]
- 2.Sternberg N, Hamilton D. Bacteriophage P1 site-specific recombination: I. Recombination between loxP sites. J Mol Biol. 1981;150:467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- 3.Sauer B. Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2087–2096. doi: 10.1128/mcb.7.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmiter RD, Brinster RL. Germ-line transformation of mice. Annu Rev Genet. 1986;20:465–499. doi: 10.1146/annurev.ge.20.120186.002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capecchi MR. The new mouse genetics: altering the genome by gene targeting. Trends Genet. 1989;5:70–76. doi: 10.1016/0168-9525(89)90029-2. [DOI] [PubMed] [Google Scholar]
- 7.Gu M, et al. Deletion of a DNA polymerase Beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 8.Tsien J, et al. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- 9.Joyner A. Gene targeting and development of the nervous system. Curr Opin Neurobiol. 1994;4:37–42. doi: 10.1016/0959-4388(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 10.Feng G, et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 11.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Devel Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spinelli V, et al. Screening strategy to generate cell specific recombination: a case report with the RIP-Cre mice. Transgenic Res. 2015;24:803. doi: 10.1007/s11248-015-9889-1. [DOI] [PubMed] [Google Scholar]
- 14.Metzger D, et al. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci USA. 1995;92:6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gossen M, et al. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;23:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 17.Liu X, et al. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–385. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng F, et al. New inducible genetic method reveals critical roles of GABA in the control of feeding and metabolism. Proc Natl Acad Sci USA. 2016;113:3645–50. doi: 10.1073/pnas.1602049113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dymecki SM, et al. Mapping cell fate and function using recombinase-based intersectional strategies. Meth Enzymol. 2010;477:183–213. doi: 10.1016/S0076-6879(10)77011-7. [DOI] [PubMed] [Google Scholar]
- 20.Swiech L, et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015;33:102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]


