Abstract
Background
Pediatric PAX:FOXO1 fusion-negative rhabdomyosarcoma represents a diverse spectrum of tumors with marked differences in histology, myogenic differentiation and clinical behavior.
Methods
This study sought to evaluate the clinical and mutational spectrum of 24 pediatric PAX:FOXO1 fusion-negative human rhabdomyosarcomas with high levels of myogenic differentiation. Tumors were sequenced using OncoPanel v.2 (OPv2), a panel consisting of the coding regions of 504 genes, previously linked to human cancer.
Results
Most (19 of 24) tumors arose at head/ neck or genitourinary sites, and overall survival was 100% with a median followup time of 4.6 years (range: 1.4 to 8.6 years). RAS pathway gene mutations were the most common mutations in PAX:FOXO1 fusion-negative, highly differentiated rhabdomyosarcomas. In addition, Hedgehog (Hh) and mechanistic Target of Rapamycin (mTOR) gene mutations with evidence for functional relevance (high-impact) were identified in subsets of tumors. The presence of Hh and mTOR pathway gene mutations was mutually exclusive, and associated with high-impact RAS pathway gene mutations in 3 of 4 Hh-mutated and 1 of 6 mTOR-mutated tumors.
Conclusions
Interestingly, Hh and mTOR gene mutations were previously associated with rhabdomyomas, which are also known to preferentially arise at head/neck and genitourinary sites. Findings from this study further support that PAX:FOXO1 fusion-negative, highly differentiated rhabdomyosarcoma and rhabdomyoma may represent a continuous spectrum of tumors.
Keywords: rhabdomyosarcoma, rhabdomyoma, sequencing, RAS, Hedgehog signaling, mTOR signaling
Introduction
Rhabdomyosarcomas (RMS) are cancers with skeletal muscle differentiation. Exclusive chromosomal translocations at t(2;13)(q35;q14) or t(1;13)(p36;q14) distinguish two disease categories1, 2. PAX:FOXO1 fusion-positive (PF+) tumors exhibit alveolar histology, aggressive clinical behavior and unfavorable outcomes. In contrast, PAX:FOXO1 fusion-negative (PF−) RMS represents a diverse spectrum of tumors with marked differences in histology, myogenic differentiation, clinical behavior and outcome1, 2. The most common oncogenic mutations in human PF- RMS are in RAS pathway genes3, 4.
Genetically distinct mouse models of RMS reflect the clinicopathological heterogeneity of the human disease5–7. For example, mice that are haploinsuffcient for Patched1 (Ptch1)8 or express a constitutively hyperactive Smoothened (Smo) allele9, 10 resulting in activated Hedgehog (Hh) signaling, develop highly differentiated tumors with muscle differentiation, resembling highly differentiated RMS or fetal rhabdomyomas (FRM) in humans5, 8, 10. Interestingly, humans with Nevoid Basal Cell Carcinoma Syndrome (NBCCS), which is associated with PTCH1 germline mutations, also are susceptible to develop highly differentiated RMS and FRM11. Finally, Hedgehog (Hh) pathway gene mutations were detected in FRM tumors9.
The association between Hh pathway gene mutations and highly differentiated myogenic tumors9, 11 led us to hypothesize that, similar to human FRM, highly-differentiated human RMS may carry Hh gene mutations and may be part of a continuous spectrum of highly differentiated myogenic tumors12. Several published studies have evaluated sequence variants in the human RMS genome. PTCH1 mutations were detected in a few non-alveolar RMS cases by fluorescence in-situ hybridization13, 14; however, the RMS sequencing cohorts published by Chen et al. and Shern et al. did not include any tumors with Hh pathway gene mutations (Table S1)3, 4. In this study, we demonstrate that RAS pathway gene mutations were the most common mutations in PF- RMS tumors with high, rhabdomyoma-like levels of skeletal muscle differentiation. In addition, we identified Hedgehog (Hh) and mechanistic Target of Rapamycin (mTOR) gene mutations in subsets of tumors.
Materials & Methods
Highly differentiated RMS and normal human muscle
Forty-one RMS cases labeled “highly differentiated” or “rhabdomyoma-like” were identified by LT on central pathology review of tumors of children/ adolescents enrolled in Children’s Oncology Group (COG) RMS trials. Hematoxylin and Eosin-stained sections were re-reviewed by LT to confirm the diagnosis of rhabdomyosarcoma as well as high levels of skeletal muscle differentiation (D3). In highly differentiated RMS tumors, myoblasts with ample eosinophilic, fibrillary cytoplasm were seen throughout tumor tissue, as previously described by Kodet et al15. Two cases were PAX:FOXO1 fusion-positive. Ultimately, D3 histology was confirmed in 24 PF- cases (Figure 1). Corresponding normal tissue was not available. Data on IRS stage, group, histology, primary tumor location, tumor size, age at diagnosis, gender, event-free and overall survival were obtained from COG records (Table 1). Discarded normal human skeletal muscle, obtained from 7 donors without tumor history served as control samples. Two PF- RMS tumors, which exhibited only occasional, scattered myoblasts throughout tumor tissue and did not meet criteria for highly differentiated RMS, were obtained from the National Development & Research Institutes, Inc. (NDRI). All studies involving human tissue samples were approved by the relevant institutional review boards (DFCI protocol No 12–496; Joslin Diabetes Center, CHS#06-42 and #08-21; Charité EA1/056/14). Informed consent was obtained from each participant or each participant’s guardian.
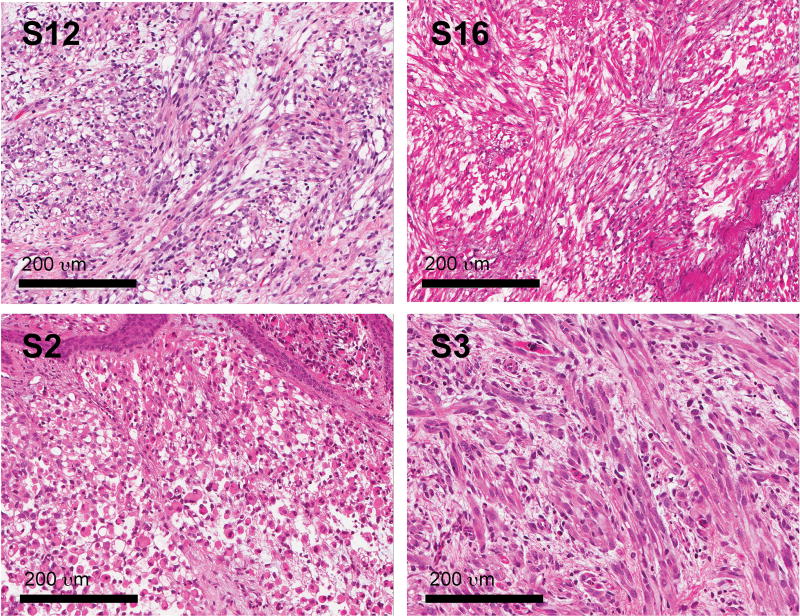
Figure 1. Histology of PF- D3 RMS tumors.
D3 histology is defined by a predominance of myoblasts, including elongated, multi-nucleated cells with abundant cytoplasm throughout tumor tissue as previously described by Kodet et al15.
Table 1.
Clinical characteristics of 24 F- D3 RMS tumors.
| F- D3 RMS (n=24) |
|
|---|---|
| Original histology assignment | |
| Embryonal RMS | 19 (79%) |
| Spindle cell RMS | 3 (13%) |
| Botryoid RMS | 1 (4%) |
| Alveolar RMS | 0 (0%) |
| RMS NOS | 1 (4%) |
| Age at diagnosis | |
| ≤ 1year | 3 (13%) |
| 1–10 years | 12 (50%) |
| 10–21 years | 9 (37%) |
| Gender | |
| Female | 8 (33%) |
| Male | 16 (67%) |
| Primary site | |
| Genitourinary | 11 (46%) |
| Head/neck | 8 (33%) |
| Extremities | 0 (0%) |
| Other | 5 (21%) |
| IRS stage | |
| Stage 1 | 7 (29%) |
| Stage 2 | 7 (29%) |
| Stage 3 | 8 (33%) |
| Stage 4 | 2 (8%) |
| IRS group | |
| Group I | 7 (29%) |
| Group II | 4 (17%) |
| Group III | 11 (46%) |
| Group IV | 2 (8%) |
| IRS risk | |
| Low | 12 (50%) |
| Intermediate | 10 (42%) |
| High | 2 (8%) |
| Primary tumor size | |
| ≥5cm | 15 (63%) |
| <5cm | 9 (37%) |
Immunohistochemistry
Formalin-fixed, paraffin-embedded (FFPE) tissue sections were baked, deparaffinized, subjected to heat-induced antigen retrieval in 10mM sodium citrate buffer, pH6 and stained against actin (1:200, Dako M0635) as previously described6, 9. Slides were lightly counterstained with haematoxylin.
DNA isolation and sequencing analysis
FFPE tumor tissue was scraped off 3–4 glass slides per tumor and skeletal muscle sample. All tissue samples were obtained prior to the initiation of chemotherapy and/ or radiotherapy. DNA was isolated using the Covaris protocol, and DNA concentration was determined using PicoGreen dsDNA reagent (Invitrogen). DNA yield was adequate (> 200 ng) for all skeletal muscle, 17 PF− tumor and 2 PF+ tumor samples. Three PF− tumor samples were included in the sequencing run despite suboptimal DNA yields between 121.9 – 196.8ng. Two tumors (S25, S26) were excluded from sequencing analyses due to insufficient DNA yield below 40ng.
Sequencing analysis
Sequencing was performed using OncoPanel v.2 (OPv2), an Agilent SureSelect custom designed bait set consisting of the coding regions of 504 genes (Table S2), previously linked to human cancer. DNA from 22 PF- D3 RMS tumors and 8 normal human muscle samples was sequenced; 2 PF- D3 RMS tumors were excluded from the sequencing analysis because of insufficient DNA availability.
Samples were prepared for sequencing as previously described16. Briefly, prior to library preparation, DNA was fragmented (Covaris sonication) to 250 bp and further purified using Agentcourt AMPure XP beads. Size-selected DNA was ligated to specific adaptors during library preparation (Illumina TruSeq). For all samples (including those with insufficient DNA yield), size distribution was within the expected range after DNA fragmentation and library preparation. Each library was made with sample-specific barcodes and quantified using qPCR. Libraries were pooled in equal mass to a total of 500ng for OPv2 enrichment using the Agilent SureSelect hybrid capture kit. The captures were then sequenced on a total of 5 lanes of the HiSeq 2500 in Rapid Run Mode. BAMF files are available for download at the European Genome Archives (EGA; accession EGAD00001003746 and EGAD00001003887).
Pooled sample reads were de-convoluted (de-multiplexed) and sorted using the Picard tools (http://broadinstitute.github.io/picard/command-line-overview.html#Overview). Next, reads were aligned to the reference sequence b37 edition from the Human Genome Reference Consortium using bwa (http://bio-bwa.sourceforge.net/bwa.shtml). Duplicate reads were identified and removed using the Picard tools17. Alignments were further refined using the GATK tool for localized realignment around indel sites (http://gatkforums.broadinstitute.org/discussion/38/local-realignment-around-indels). For muscle samples, 94.1% to 98.1% of target bases were covered at > 30×. We achieved > 30× coverage in 89.2% to 99% of target bases in 14 PF− tumor samples, 11.5% to 77.5% of target bases in 8 PF− samples and 95.1% to 97.8% of target bases in 2 PF+ tumors.
Mutation analysis for single nucleotide variants (SNV) was performed using MuTect v1.1.4 in paired mode with the CEPH cell line as the project normal18. Variants were annotated using Oncotator. We considered only SNVs that were non-synonymous, detected in a tumor fraction of > 30%, absent in normal skeletal muscle specimens and listed in the Exome Variant Server (ESP), NHLBI Exome Sequencing Project (6,500 exome release) with a population frequency ≤1% in either the African-American or European-American subset. Insertions and deletions (InDels) were called using Indel Locator (http://www.broadinstitute.org/cancer/cga/indelocator). We only considered those indels found in an allele fraction of > 25%. Finally, copy number analysis was performed on all samples using Nexus7.1 (BioDiscovery Inc) after calculating the sequencing coverage using PICARD. All samples were compared to normal CEPH DNA and one of the skeletal muscle samples as reference.
Alamut was used to perform in silico pathogenicity analysis of RAS, Hh, mTOR and NOTCH pathway gene mutations by 3 different algorithms (Sorting Intolerant From Tolerant (SIFT), MutationTaster (MuT) and Polymorphism Phenotyping v2 (PolyPhen)) and evaluate population frequency as per the Exome Aggregation Consortium browser (ExAc). Sequence variations in Hh and mTOR pathway genes were verified by PCR. Mutations that were predicted to be damaging in silico were considered high-impact and included in Figure 5.
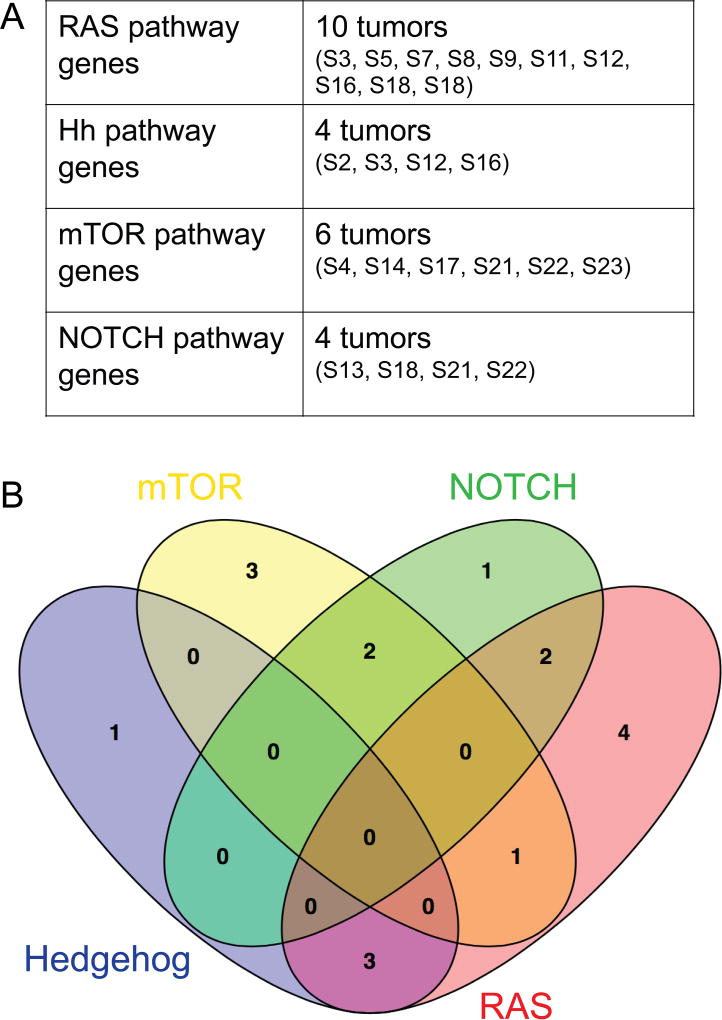
Figure 5. RAS, NOTCH, Hh and mTOR pathway gene mutations in PF- D3 RMS.
Mutations were considered likely damaging, if one of 3 different algorithms predicted pathogenicity in silico (Table S5). (A) Such high-impact mutations with evidence for functional relevance were detected in RAS genes in 10 tumors, in Hh genes in 4 tumors, in mTOR genes in 6 tumors and in NOTCH genes in 4 tumors. (B) High-impact mutations with evidence for functional relevance in Hh and mTOR gene mutations in PF- D3 RMS were mutually exclusive. One of 6 mTOR-mutated tumors and 3 of 4 Hh-mutated tumors exhibited concurrent RAS pathway mutations.
Statistical analysis
Event-free survival (EFS) was defined as the time from study entry to the first occurrence of disease progression, disease relapse, or death. For those not experiencing one of these events, EFS was censored at last contact. Estimates of overall survival (OS) and EFS were calculated using the Kaplan-Meier method.
Results
Terminal muscle differentiation in PF- D3 RMS
Our cohort included 24 PF- highly differentiated human RMS tumors. These tumors contained many elongated, multi-nucleated myoblasts with abundant eosinophilic, fibrillary cytoplasm (Figure 1), which were identified on Hematoxylin and Eosin stained tissue sections and consistent with highly differentiated D3 histology as described by Kodet et al15. Marked cellular atypia, as evidenced by nuclear size/ polymorphism and hyperchromasia, confirmed a diagnosis of rhabdomyosarcoma in all cases; rhabdomyoma histology was excluded. All 24 tumors were initially classified and treated as embryonal RMS; three tumors were further described as spindle cell RMS (13%), one as botryoid RMS (4%) and one as RMS not otherwise specified (NOS) (Table 1).
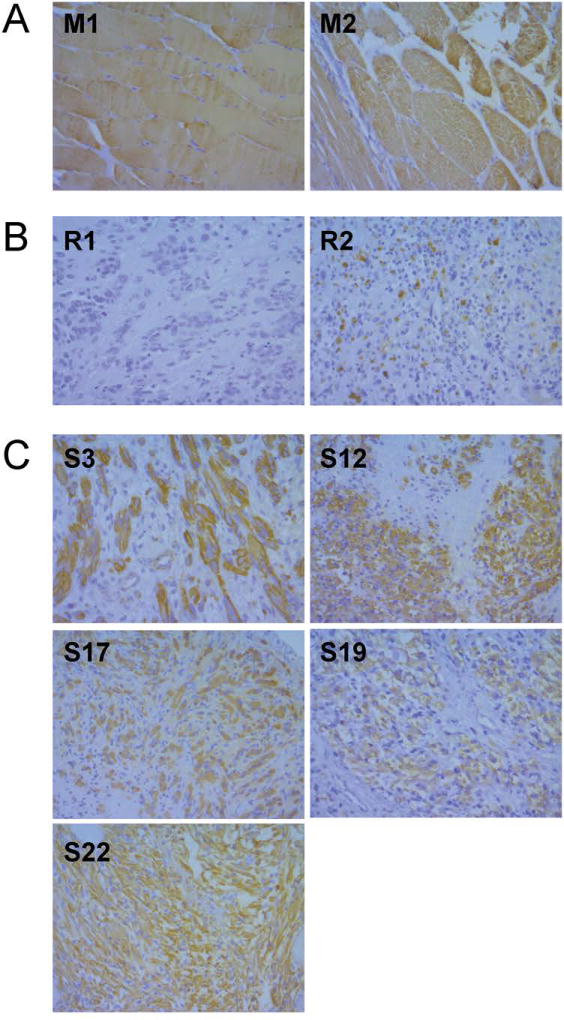
To further explore the presence of terminal muscle differentiation features, actin expression was evaluated in 3 human skeletal muscle samples, 2 random PF- human RMS tumors obtained from the NDRI and 5 PF- D3 human RMS tumors. As expected, there was strong cytoplasmatic actin staining in normal muscle fibers (Figure 2A) and minimal actin staining in the 2 random RMS specimens (Figure 2B). Consistent with their highly differentiated morphology, 4 of 5 PF- D3 human RMS tumors (S3, S12, S17, S22) evaluated exhibited strong actin staining (Figure 2C). Of note, the level of actin staining was not used to select P3F- D3 RMS samples that were sequenced.
Figure 2. Actin expression in skeletal muscle and RMS tissue.
The myofilament actin marks terminal muscle differentiation. (A) Normal muscle exhibits strong cytoplasmatic actin staining. (B) Random RMS samples demonstrate minimal actin staining (C) Actin expression in PF- D3 RMS tumors evidences terminal muscle differentiation.
Clinical characteristics of PF- D3 RMS
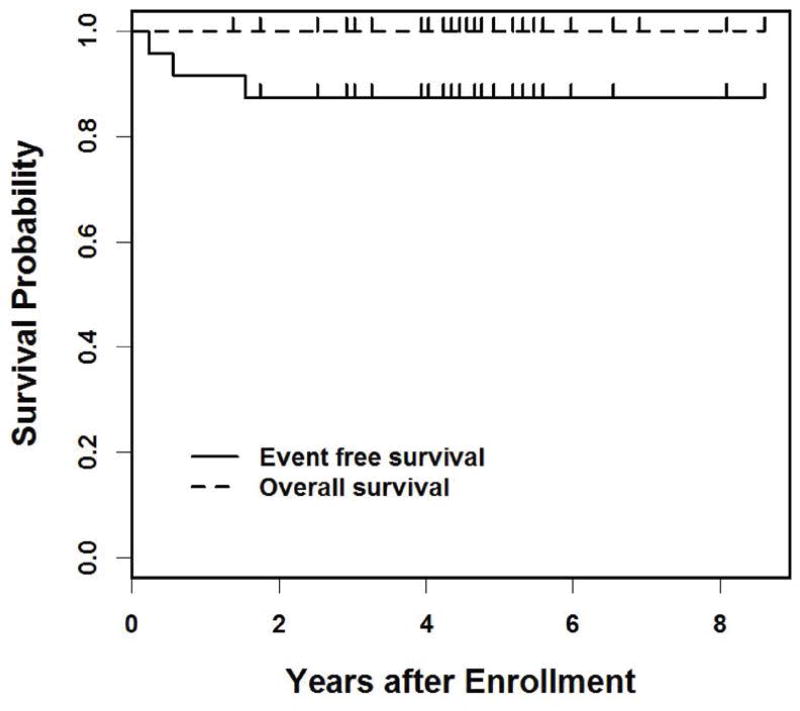
For the 24 PF- D3 RMS tumors included in this analysis, clinical and survival data were obtained from COG records. Eleven (46%) of these tumors originated in the genitourinary system, and 8 (33%) tumors arose at head and neck sites (Table 1). Other primary sites were the retroperitoneum, mediastinum, chest well, pelvis and paraspinal region (one tumor each). Mean age at diagnosis was 8.1 years (± S.D. 6.3 years). Two cases (8%) were associated with distant metastases (stage 4). Twenty-two (92%) tumors were classified as stage 1–3 and low/ intermediate IRS risk (Table 1). Two patients with stage 3 disease experienced relapse/progression after 80 and 202 days, respectively. Another patient with stage 1 disease relapsed/ progressed after 1.6 years. Of note, one of 3 recurrent cases was excluded from the sequencing analysis because of insufficient DNA availability (Figure 4). Overall survival was 100% with a median followup time of 4.6 years (range: 1.4 to 8.6 years, Figure 3).
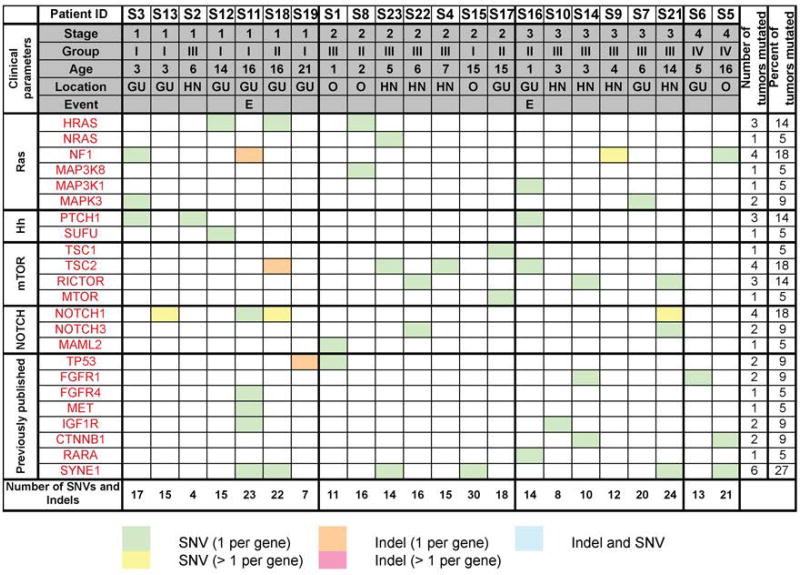
Figure 4. Mutational spectrum of PF- D3 RMS.
Mutations in RAS, NOTCH, Hh and mTOR pathway genes and in genes previously found mutated in RMS3, 4. SNVs were considered if they were non-synonymous, detected in a tumor fraction >30%, not present in normal muscle samples and listed in the Exome Variant Server, NHLBI Exome Sequencing Project (ESP), Seattle, WA, USA, with a population frequency < 1%). InDels were considered if allele frequency was > 25%. The number of SNVs and Indels refers to the total number of aberrations observed in the tumor. Of note, only 2 of 3 recurrent cases were included in the sequencing analysis; one recurrent case was excluded because of insufficient DNA availability.
Figure 3. Survival of patients diagnosed with PF- D3 RMS.
Overall survival and event-free survival was determined for 24 individuals diagnosed with PF- D3 RMS (median followup time of 1.8 years; range: 0.4 to 4.5 years).
Frequent RAS pathway gene mutations in PF- D3 RMS
The genetic underpinnings of 22 PF- D3 RMS tumors and 8 normal human muscle samples were examined by targeted Illumina sequencing (OncoPanel v. 2). We identified a total of 4 to 30 SNVs (non-synonymous, not present in normal muscle samples, detected in a tumor fraction >30%, listed in the Exome Variant Server, NHLBI Exome Sequencing Project (ESP), Seattle, WA, USA, with a population frequency < 1%) and InDels (allele frequency > 25%) (Figure 4, Tables S3–S4) per tumor. CNV analyses were performed for all tumors; however, the Nexus quality score was insufficient, and it was not possible to call any CNVs except for high amplifications that were well above the noise level at the Insulin Like Growth Factor 1 receptor (IGF1R) locus in 2 samples (S1, S19; data not shown). Figure S1 includes all mutated genes (n=16) detected in 4 or more PF- D3 RMS tumors. The most frequent aberrations in individual genes were detected in Lysine (K)-Specific Methyltransferase 2C (MLL3; 18 tumors mutated), Spectrin Repeat Containing Nuclear Envelope Protein 1 (SYNE1; 6 tumors mutated) and LDL Receptor Related Protein 2 (LRP2; 7 tumors mutated).
We also detected aberrations in a number of genes, which were previously found mutated in RMS3, 4, including NOTCH and RAS pathway genes (Figure 4, Table S1). Of note, the frequency of RAS gene mutations in PF- D3 tumors (45%) was comparable to that noted in previous sequencing cohorts of random PF- RMS tumors (29–41%, Table S1,3, 4). The frequency of NOTCH gene mutations in PF- D3 RMS tumors (27%) was higher than that observed in previous cohorts of random PF- RMS (0–7%, Table S1,3, 4).
Specifically, 10 NOTCH pathway gene mutations were detected in 6 tumors (Figure 4). Eight of these mutations (detected in 4 tumors) were predicted to be damaging in silico (Figure 5A, Table S5); the NOTCH1 mutation in S11, and the Mastermind Like Transcriptional Coactivator 2 (MAML2) mutation in S1 were predicted to be benign aberrations (Table S5).
Twelve RAS pathway gene mutations were detected in 10 of 22 (45%) PF- D3 RMS tumors sequenced (Figure 4). Mutated RAS pathway genes in PF- D3 RMS included Neurofibromin 1 (NF1), HRas Proto-Oncogene, GTPase (HRAS), Neuroblastoma RAS Viral Oncogene Homolog (NRAS), Mitogen-Activated Protein Kinase Kinase Kinase 8 (MAP3K8), Mitogen-Activated Protein Kinase Kinase Kinase 1 (MAP3K1) and Mitogen-Activated Protein Kinase 3 (MAPK3). In silico pathogenicity analysis predicted that 10 of 11 RAS pathway gene mutations were damaging as per at least one of three algorithms applied (Figure 5A, Table S5). The NF1 mutation detected in tumor S3 was predicted to be a benign aberration (Table S5).
Hh and mTOR pathway gene mutations in PF- D3 RMS
In addition to RAS and NOTCH, two other signaling pathways were highly represented among the genes mutated in PF- D3 RMS. Mutations in Hh and mTOR genes were not seen in previous sequencing cohorts of random PF- RMS tumors (Table S1,3,4). Firstly, 35 variants in Hh pathway genes were detected in 19 PF- D3 RMS tumors (Table S6). Four of these 35 variants were non-synonymous, not present in normal muscle samples and listed in the Exome Variant Server, NHLBI Exome Sequencing Project (ESP), Seattle, WA, USA, with a population frequency < 1% (Figure 4). The SUFU Negative Regulator Of Hedgehog Signaling (SUFU) mutation and 2 of 3 PTCH1 mutations were subsequently confirmed by PCR (Table S5). PCR did not confirm the PTCH1 mutation (c.4048C>T) detected in S2, but identified an alternative PTCH1 mutation (c.4225T>C) in the same tumor (Table S5). Also, all 4 Hh pathway gene mutations detected in PF- D3 RMS were predicted to be pathogenic by at least one algorithm in silico (Figure 5A, Table S5). Secondly, 9 mTOR pathway gene mutations were identified in 8 PF- D3 RMS tumors (Figure 4). Mutated mTOR pathway genes included Tuberous Sclerosis 1 (TSC1), Tuberous Sclerosis 2 (TSC2), RPTOR Independent Companion Of MTOR Complex 2 (RICTOR) and Mechanistic Target Of Rapamycin (MTOR). All 9 mTOR pathway gene mutations were confirmed by PCR except for the RICTOR mutations in S14 and S21 (insufficient amount of DNA available, Table S5). Six of these mutations (detected in 6 tumors) were predicted to be damaging in silico (Figure 5A, Table S5); the TSC2 mutations in S16 and S18 and the TSC1 mutation in S17 were predicted to be benign aberrations (Table S5). It was not determined if the observed Hh and mTOR pathway gene mutations activated or inactivated the function of the respective gene products.
In summary, we identified high-impact, likely damaging mutations with evidence for functional relevance in Hh genes in 4 tumors, in mTOR gene in 6 tumors, in NOTCH genes in 4 tumors and in RAS genes in 10 tumors (Figure 5A). Interestingly, both Hh and mTOR gene mutations were previously associated with rhabdomyoma development9, 11, 19–21. All mTOR- and Hh-mutated PF- D3 RMS tumors arose at head/ neck or GU sites, were diagnosed at stages 1–3 and in patients aged 1–10 years (Table S7). There were no tumors carrying concurrent mTOR and Hh pathway gene mutations (Figure 5B). Of 6 mTOR pathway gene mutated tumors, one carried concurrent RAS and 2 contained concurrent NOTCH gene mutations. Of 4 Hh gene mutated tumors, 3 exhibited concurrent RAS pathway gene mutations.
Discussion
PF- D3 RMS tumors contained frequent, high-impact RAS, NOTCH, Hh and mTOR pathway gene mutations with in silico evidence for functional relevance. The presence of Hh and mTOR pathway gene mutations were mutually exclusive, and associated with high-impact RAS pathway gene mutations in 3 of 4 Hh-mutated and 1 of 6 mTOR-mutated tumors.
Previous studies established that RAS gene mutations are the most common mutations detected in all PF- RMS tumors3, 4. Hh gene aberrations in human RMS are rare, and mTOR pathway gene mutations have not been identified in human RMS thus far3, 4, 22, 23. On the other hand, RAS gene mutations were not found in a previously published cohort of fetal rhabdomyomas9, and Hh and mTOR pathway gene mutations have been clearly associated with fetal and cardiac rhabdomyomas9, 11, 19–21. Highly differentiated RMS tumors and rhabdomyomas appear to share mutations in Hh and mTOR pathway genes. These are associated with frequent mutations in RAS pathway genes in highly differentiated RMS, but not rhabdomyomas.
Kodet et al. previously noted that highly differentiated RMS and fetal rhabdomyomas were difficult to distinguish histologically, and suggested that fetal cellular rhabdomyomas and differentiated RMS may be variant manifestations of the same disease12. In our series described here, two thirds of PF- D3 RMS tumors arose at head/neck or genitourinary sites, and overall survival was substantially better than expected in children with RMS1. Interestingly, head/neck or genitourinary locations also are known to be common sites of origin of rhabdomyomas, which never metastasize and have excellent outcomes, but exhibit locally aggressive growth and occasional multifocality24–26. As previously suggested by Kodet12, 15 and ourselves9, 11, highly differentiated RMS tumors and rhabdomyomas might form a continuous spectrum of tumors. This could have important implications for treatment. Similar to the therapeutic requirements of rhabdomyomas, cure of highly differentiated RMS may depend on wide surgical resection rather than radiation and/ or systemic chemotherapy.
Hh and mTOR signaling effects on skeletal muscle differentiation were previously evaluated. Activation of mTOR signaling stimulated muscle differentiation/ regeneration27, 28. Similarly, activation of Hh signaling was shown to promote muscle differentiation29, 30, whereas Hh blockade impeded muscle regeneration post injury31. The differentiating effects of activated Hh and mTOR signaling in normal skeletal muscle could be consistent with a highly differentiated phenotype of Hh- and mTOR-mutated myogenic tumors. Also, Hh pathway activation in mice resulted in the formation of highly differentiated myogenic tumors8, 10, and PTCH1 inactivating mutations (resulting in Hh activation) were found in human fetal rhabdomyomas9. Finally, in human PF- RMS, expression of the Hh effectors GLI Family Zinc Finger 1 (GLI1), GLI Family Zinc Finger 3 (GLI3) and PTCH1, was previously correlated with low expression of the early myoblast marker Myod1 by Zibat et al32. Of note, this same study reported that Hh pathway activation in PF- RMS was associated with reduced survival32, whereas all 4 patients with Hh-mutated PF- D3 RMS tumors in the series first reported here survived. It is possible that the varied spectrum of PF- RMS comprises clinically distinct tumors with canonical Hh activation due to activating aberrations in Hh genes as well as tumors with non-canonical Hh activation due to aberrations in RMS-relevant upstream effectors.
PF- RMS represents a remarkably heterogeneous group of tumors. This series focuses specifically on those tumors that exhibit high, rhabdomyoma-like levels of differentiation. These tumors are rare. While our study does not allow an estimate on what proportion of PF- RMS tumors are highly differentiated, Kodet et al previously reviewed the histology of 163 non-alveolar RMS tumors arising in the periorbital area. Fourteen (9%) of these 163 orbital tumors met D3 criteria for highly differentiated RMS15. Our study highlights the presence of frequent mutations in RAS pathway and rhabdomyoma-relevant genes in these tumors. Data should be interpreted within the context of the study’s limitations: (i) Our sequencing cohort of 22 PF-, D3 RMS cases is not necessarily representative, but represents a convenience cohort; (ii) our targeted sequencing approach included 504 cancer-relevant genes, but not the entire genome; (iii) lack of paired germline samples prevents distinction of germline versus somatic mutations; and (iv) it’s possible that certain mutations were missed due to moderate target base coverage. Nevertheless, our findings further support that highly differentiated RMS and rhabdomyomas might form a continuous spectrum of tumors and share certain molecular and clinical characteristics. It will be important to further dissect the molecular and clinicopathological heterogeneity of PF- RMS in future studies evaluating larger, representative cohorts of tumors.
Supplementary Material
Figure S1. Most frequent mutations in PF- D3 RMS. Figure S1 summarizes the most frequent aberrations in individual genes in PF- D3 RMS. SNVs were considered if they were non-synonymous, detected in a tumor fraction >30%, not present in normal muscle samples and listed in the Exome Variant Server, NHLBI Exome Sequencing Project (ESP), Seattle, WA, USA, with a population frequency < 1%). InDels were considered if the allele frequency was > 25%. Also, of note, aberrations in major histocompatibility complex genes were discarded from the list in Figure S1.
Table S1. Frequency of RAS (NRAS, KRAS, HRAS, NF1, BRAF, MAP3K8, MAP3K1, MAPK3), Hh (PTCH1, SUFU), mTOR (TSC1, TSC2, RICTOR, MAML) and Notch (NOTCH1, NOTCH2, NOTCH3, MAML2) gene mutations in PF- RMS.
Table S2. CCGD Oncopanel v2 gene list.
Table S3. SNVs detected in 22 PF- D3 RMS tumors (non-synonymous, not present in normal muscle samples, listed in the Exome Variant Server, NHLBI Exome Sequencing Project (ESP), Seattle, WA, USA, with a population frequency < 0.01/1% in European Americans (EA) or African Americans (AA)).
Table S4. Indels detected in 22 PF- D3 RMS (allele frequency > 25%).
Table S5. In silico pathogenicity analysis of Hh, mTOR, NOTCH and RAS pathway mutations detected in 22 PF- D3 RMS tumors.
Table S6. Hh pathway gene variants in 22 PF- D3 RMS.
Table S7. Clinical characteristics of RAS-, Hh-, mTOR- and NOTCH-mutated PF- D3 RMS tumors (total n=22) analyzed.
Acknowledgments
This work was funded in part by P.A.L.S. Bermuda/St. Baldrick’s (to SH), by the Joslin Diabetes Center (to AJW) and by U10CA180886, U10CA180999, U10CA98543 and U10CA98413 (National Cancer Institute, Bethesda, MD). Sequencing was performed by the Center for Cancer Genome Discovery at the Dana-Farber Cancer Institute, Boston, MA. The authors are grateful to Victor Pastor for help with in silico prediction of pathogenicity of mutations.
Footnotes
Disclosures
The authors declare no conflicts of interests.
The authors also declare no competing financial interests. Content is solely the responsibility of the authors.
Author contributions: Lisa A. Teot: conceptualization, data curation, formal analysis, investigation; Michaela Schneider: formal analysis; Aaron R. Thorner: formal analysis, methodology; Jing Tian: formal analysis; Yueh-Yun Chi: formal analysis; Matthew Ducar: formal analysis; Ling Lin: formal analysis; Marcin Wlodarski: data curation; methodology; Holcombe E Grier: investigation; Christopher DM Fletcher: investigation, writing; Paul van Hummelen: formal analysis, methodology; Stephen X Skapek: data curation, investigation, funding acquisition; Douglas S Hawkins: data curation, investigation, funding acquisition, writing; Amy J Wagers: investigation, funding acquisition, writing; Carlos Rodriguez-Galindo: conceptualization, data curation, investigation, funding acquisition, writing; Simone Hettmer: conceptualization, data curation, formal analysis, methodology, investigation, funding acquisition, writing.
References
- 1.Hettmer S, Li Z, Billin AN, et al. Rhabdomyosarcoma: current challenges and their implications for developing therapies. Cold Spring Harb Perspect Med. 2014;4:a025650. doi: 10.1101/cshperspect.a025650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parham DM. Pathologic classification of rhabdomyosarcomas and correlations with molecular studies. Mod Pathol. 2001;14:506–514. doi: 10.1038/modpathol.3880339. [DOI] [PubMed] [Google Scholar]
- 3.Chen X, Stewart E, Shelat AA, et al. Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer Cell. 2013;24:710–724. doi: 10.1016/j.ccr.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shern JF, Chen L, Chmielecki J, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014;4:216–231. doi: 10.1158/2159-8290.CD-13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hettmer S, Bronson RT, Wagers AJ. Distinct malignant behaviors of mouse myogenic tumors induced by different oncogenetic lesions. Front Oncol. 2015;5:50. doi: 10.3389/fonc.2015.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hettmer S, Liu J, Miller CM, et al. Sarcomas induced in discrete subsets of prospectively isolated skeletal muscle cells. Proc Natl Acad Sci U S A. 2011;108:20002–20007. doi: 10.1073/pnas.1111733108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubin BP, Nishijo K, Chen HI, et al. Evidence for an unanticipated relationship between undifferentiated pleomorphic sarcoma and embryonal rhabdomyosarcoma. Cancer Cell. 2011;19:177–191. doi: 10.1016/j.ccr.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hahn H, Nitzki F, Schorban T, Hemmerlein B, Threadgill D, Rosemann M. Genetic mapping of a Ptch1-associated rhabdomyosarcoma susceptibility locus on mouse chromosome 2. Genomics. 2004;84:853–858. doi: 10.1016/j.ygeno.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Hettmer S, Teot LA, van Hummelen P, et al. Mutations in Hedgehog pathway genes in fetal rhabdomyomas. J Pathol. 2013;231:44–52. doi: 10.1002/path.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao J, Ligon KL, Rakhlin EY, et al. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 2006;66:10171–10178. doi: 10.1158/0008-5472.CAN-06-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hettmer S, Teot LA, Kozakewich H, et al. Myogenic tumors in nevoid Basal cell carcinoma syndrome. J Pediatr Hematol Oncol. 2015;37:147–149. doi: 10.1097/MPH.0000000000000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kodet R, Fajstavr J, Kabelka Z, Koutecky J, Eckschlager T, Newton WA., Jr Is fetal cellular rhabdomyoma an entity or a differentiated rhabdomyosarcoma? A study of patients with rhabdomyoma of the tongue and sarcoma of the tongue enrolled in the intergroup rhabdomyosarcoma studies I, II, and III. Cancer. 1991;67:2907–2913. doi: 10.1002/1097-0142(19910601)67:11<2907::aid-cncr2820671133>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 13.Bridge JA, Liu J, Weibolt V, et al. Novel genomic imbalances in embryonal rhabdomyosarcoma revealed by comparative genomic hybridization and fluorescence in situ hybridization: an intergroup rhabdomyosarcoma study. Genes Chromosomes Cancer. 2000;27:337–344. doi: 10.1002/(sici)1098-2264(200004)27:4<337::aid-gcc1>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 14.Tostar U, Malm CJ, Meis-Kindblom JM, Kindblom LG, Toftgard R, Unden AB. Deregulation of the hedgehog signalling pathway: a possible role for the PTCH and SUFU genes in human rhabdomyoma and rhabdomyosarcoma development. J Pathol. 2006;208:17–25. doi: 10.1002/path.1882. [DOI] [PubMed] [Google Scholar]
- 15.Kodet R, Newton WA, Jr, Hamoudi AB, Asmar L, Wharam MD, Maurer HM. Orbital rhabdomyosarcomas and related tumors in childhood: relationship of morphology to prognosis--an Intergroup Rhabdomyosarcoma study. Med Pediatr Oncol. 1997;29:51–60. doi: 10.1002/(sici)1096-911x(199707)29:1<51::aid-mpo10>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 16.Sholl LM, Do K, Shivdasani P, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight. 2016;1:e87062. doi: 10.1172/jci.insight.87062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benyounes N, Fohlen M, Devys JM, Delalande O, Moures JM, Cohen A. Cardiac rhabdomyomas in tuberous sclerosis patients: a case report and review of the literature. Arch Cardiovasc Dis. 2012;105:442–445. doi: 10.1016/j.acvd.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 20.Choi JE, Chae JH, Hwang YS, Kim KJ. Mutational analysis of TSC1 and TSC2 in Korean patients with tuberous sclerosis complex. Brain Dev. 2006;28:440–446. doi: 10.1016/j.braindev.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Elawabdeh N, Sobol S, Blount AC, Shehata BM. Unusual presentation of extracardiac fetal rhabdomyoma of the larynx in a pediatric patient with tuberous sclerosis. Fetal Pediatr Pathol. 2013;31:43–47. doi: 10.3109/15513815.2012.659416. [DOI] [PubMed] [Google Scholar]
- 22.Calzada-Wack J, Schnitzbauer U, Walch A, et al. Analysis of the PTCH coding region in human rhabdomyosarcoma. Hum Mutat. 2002 Sep;20(3):233–4. doi: 10.1002/humu.9056. 2002;20:233–234. [DOI] [PubMed] [Google Scholar]
- 23.Pressey JG, Anderson JR, Crossman DK, Lynch JC, Barr FG. Hedgehog pathway activity in pediatric embryonal rhabdomyosarcoma and undifferentiated sarcoma: a report from the Children's Oncology Group. Pediatr Blood Cancer. 2011;57:930–938. doi: 10.1002/pbc.23174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapadia SB, Meis JM, Frisman DM, Ellis GL, Heffner DK. Fetal rhabdomyoma of the head and neck: a clinicopathologic and immunophenotypic study of 24 cases. Hum Pathol. 1993;24:754–765. doi: 10.1016/0046-8177(93)90013-7. [DOI] [PubMed] [Google Scholar]
- 25.Kapadia SB, Meis JM, Frisman DM, Ellis GL, Heffner DK, Hyams VJ. Adult rhabdomyoma of the head and neck: a clinicopathologic and immunophenotypic study. Hum Pathol. 1993;24:608–617. doi: 10.1016/0046-8177(93)90240-h. [DOI] [PubMed] [Google Scholar]
- 26.de Trey LA, Schmid S, Huber GF. Multifocal adult rhabdomyoma of the head and neck manifestation in 7 locations and review of the literature. Case Rep Otolaryngol. 2013;2013:758416. doi: 10.1155/2013/758416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erbay E, Chen J. The mammalian target of rapamycin regulates C2C12 myogenesis via a kinase-independent mechanism. J Biol Chem. 2001;276:36079–36082. doi: 10.1074/jbc.C100406200. [DOI] [PubMed] [Google Scholar]
- 28.Ge Y, Wu AL, Warnes C, et al. mTOR regulates skeletal muscle regeneration in vivo through kinase-dependent and kinase-independent mechanisms. Am J Physiol Cell Physiol. 2009;297:C1434–1444. doi: 10.1152/ajpcell.00248.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hettmer S, Lin MM, Tchessalova D, et al. Hedgehog-driven myogenic tumors recapitulate skeletal muscle cellular heterogeneity. Exp Cell Res. 2016;340:43–52. doi: 10.1016/j.yexcr.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elia D, Madhala D, Ardon E, Reshef R, Halevy O. Sonic hedgehog promotes proliferation and differentiation of adult muscle cells: Involvement of MAPK/ERK and PI3K/Akt pathways. Biochim Biophys Acta. 2007;1773:1438–1446. doi: 10.1016/j.bbamcr.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 31.Straface G, Aprahamian T, Flex A, et al. Sonic hedgehog regulates angiogenesis and myogenesis during post-natal skeletal muscle regeneration. J Cell Mol Med. 2009;13:2424–2435. doi: 10.1111/j.1582-4934.2008.00440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zibat A, Missiaglia E, Rosenberger A, et al. Activation of the hedgehog pathway confers a poor prognosis in embryonal and fusion gene-negative alveolar rhabdomyosarcoma. Oncogene. 2010;29:6323–6330. doi: 10.1038/onc.2010.368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Most frequent mutations in PF- D3 RMS. Figure S1 summarizes the most frequent aberrations in individual genes in PF- D3 RMS. SNVs were considered if they were non-synonymous, detected in a tumor fraction >30%, not present in normal muscle samples and listed in the Exome Variant Server, NHLBI Exome Sequencing Project (ESP), Seattle, WA, USA, with a population frequency < 1%). InDels were considered if the allele frequency was > 25%. Also, of note, aberrations in major histocompatibility complex genes were discarded from the list in Figure S1.
Table S1. Frequency of RAS (NRAS, KRAS, HRAS, NF1, BRAF, MAP3K8, MAP3K1, MAPK3), Hh (PTCH1, SUFU), mTOR (TSC1, TSC2, RICTOR, MAML) and Notch (NOTCH1, NOTCH2, NOTCH3, MAML2) gene mutations in PF- RMS.
Table S2. CCGD Oncopanel v2 gene list.
Table S3. SNVs detected in 22 PF- D3 RMS tumors (non-synonymous, not present in normal muscle samples, listed in the Exome Variant Server, NHLBI Exome Sequencing Project (ESP), Seattle, WA, USA, with a population frequency < 0.01/1% in European Americans (EA) or African Americans (AA)).
Table S4. Indels detected in 22 PF- D3 RMS (allele frequency > 25%).
Table S5. In silico pathogenicity analysis of Hh, mTOR, NOTCH and RAS pathway mutations detected in 22 PF- D3 RMS tumors.
Table S6. Hh pathway gene variants in 22 PF- D3 RMS.
Table S7. Clinical characteristics of RAS-, Hh-, mTOR- and NOTCH-mutated PF- D3 RMS tumors (total n=22) analyzed.