Abstract
Sleep is an evolutionarily conserved behavior that is increasingly recognized as important for human health. While its precise function remains controversial, sleep has been suggested to play a key role in a variety of biological phenomena, ranging from synaptic plasticity to metabolic clearance. Although it is clear that sleep is regulated by the circadian clock, how this occurs remains enigmatic. Here, we examine the genetic mechanisms by which the circadian clock regulates sleep, drawing upon recent work in fruit flies, zebrafish, mice, and humans. These studies reveal that central and local clocks utilize diverse mechanisms to regulate different aspects of sleep, and a better understanding of this multilayered regulation may lead to a better understanding of the function(s) of sleep.
Keywords: Sleep, Circadian, Genes
From clockwork gears to sleep behavior
The biological clocks ticking within essentially all animals coordinate diverse and widespread physiological processes and behaviors across the 24 hr day. Starting with the seminal identification of the period mutant by Benzer and Konopka in 1971 and culminating with the recent awarding of the 2017 Nobel Prize in Physiology or Medicine to Drs. Hall, Rosbash, and Young [1], there has been intense focus on unraveling the molecular genetic basis of the core circadian oscillator. As our understanding of the mechanisms that mediate intrinsic rhythmicity and phase resetting of the core oscillator has matured, there has been increasing attention to how these circadian clock oscillations regulate various physiological functions and behaviors, such as sleep. In this review, after providing a general introduction to sleep, we will focus on recent advances in our understanding of the genetic mechanisms by which it is regulated by the circadian clock. These studies suggest that circadian orchestration of multiple genetic pathways enables multilayered control of different aspects of sleep, and a deeper understanding of these processes may ultimately yield insights into the fundamental functions of sleep.
Sleep is an ancient, conserved behavior with multiple proposed functions
Sleep is a vital, phylogenetically conserved phenomenon that has been identified in animals ranging from worms and jellyfish to birds and mammals [2–7]. Despite its near ubiquity, sleep remains a poorly understood process. While its function(s) remain an intensely debated topic, sleep has been proposed to be important for energetic and metabolic processes [8, 9], immune system function [10], clearance of neuronal waste products and maintaining ionic balance [11–13], synaptic homeostasis and neural plasticity [14–17], and learning and memory [18–20]. How a single behavioral entity plays a key role in such diverse processes is unclear, but may relate to the notion that sleep is not a unitary phenomenon, but rather comprises multiple distinct sub-states [21]. In more primitive organisms, such as jellyfish, it may be that sleep serves a single primordial function, and that various other functions for sleep have been added during the evolution of more complex animals. Sleep in humans, for example, can be measured by electroencephalography (EEG) and be broadly categorized into rapid-eye movement (REM) sleep, and “light” non-REM (NREM) vs “deep” NREM sleep (slow wave sleep), based largely upon waveform frequency/amplitude patterns. These distinct sleep sub-states preferentially occur during different times during the night, are differentially regulated, and have been proposed to be important for distinct processes [22]. Beyond these canonical sleep sub-states, there is growing recognition that these basic categories do not adequately describe the full spectrum of potential sleep sub-states [23]. Distinct sleep sub-states associated with different electrophysiological signatures, arousal thresholds, and/or behaviors have also been described in invertebrates, suggesting that the diversity of sleep states is an evolutionarily conserved phenomenon [24–27].
Studies of the regulation and function of sleep have been greatly aided by the use of model organisms. In this review, in addition to research in humans, we focus on circadian/sleep work performed in fruit flies, zebrafish, and mice. How is sleep measured in these model organisms? Sleep can be identified using electrophysiological measures such as EEG, or by behavioral criteria. The latter includes sustained periods of quiescence occurring in a circadian pattern, reduced sensitivity to stimuli (increased arousal threshold), and homeostatic rebound following prolonged wakefulness [28]. Thus, characterization of sleep phenotypes in model organisms utilizes electrophysiological or behavioral (consolidated immobility associated with an increased arousal threshold) measures. Sleep is regulated by a homeostatic process (which reflects sleep need) and a circadian process (which reflects endogenous circadian time) [29, 30]. Here, we will mainly discuss the latter, but will first discuss the molecular and circuit basis of the core circadian clock.
Tick-Tock… Canonical Clocks Coordinating Rhythms
Forward genetic screens in animal models have identified canonical circadian clock genes which form a transcriptional-translational feedback loop (TTFL), turning over at a rate which approximates the solar day (~24hrs) [for review see 31]. The molecular basis of the core circadian oscillator and its basic functions are largely conserved from flies to humans (Fig. 1). In mammals, the basic TTFL comprises transcriptional activators (CLOCK/NPAS2 and BMAL1) that drive the expression of transcriptional repressors (PERIOD and CRYPTOCHROME), which then feedback to inhibit CLOCK/BMAL activity and suppress their own expression [32]. The oscillatory activity of these transcription factors and their regulators (clock genes, CGs) lead to the rhythmic expression of a large network of genes (clock-controlled genes, CCGs), which has been estimated to comprise up to 43% of the transcriptome across the entire body [33]. In this way, the molecular clocks found in almost every single cell in our bodies can locally coordinate oscillatory gene expression [34].
Figure 1. Conservation of canonical circadian clock from invertebrates (A) to vertebrates (B).
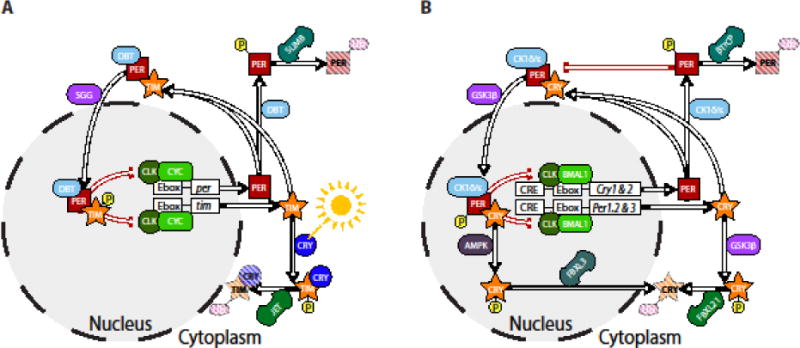
A) In a highly simplified model of the invertebrate circadian clock Clock and cycle genes drive transcription of period and timeless via enhancer boxes (Ebox) found upstream of the genes. PER and TIM proteins form heterodimers which translocate back to the nucleus to inhibit their own translation. While CRY-mediated ubiquitination and subsequent proteosomal degradation of TIM drives daily resetting by light, clock speed is controlled by the rate of degradation of PER proteins. B) Similar feedback and cycling occurs in mammals with the Cryptochrome genes substituting for timeless in the core oscillator. Cytosolic PER and CRY proteins are ubiquitinated by two separate mechanisms when not heterodimerized. PER is flagged for degradation by the doubletime orthologues CK1δ/ε leading to ubiquitination by βTrCP, while phase-resetting occurs via intracellular signaling pathways AMPK (Cry1) and GSK3β (Cry2) triggering FBXL3 ubiquitination and proteosomal degradation mediated intracellular signaling. In these figures, several accessory loops in both flies and mammals are omitted to focus on the balance of synthesis and degradation of clock proteins, which has been implicated in the circadian timing of sleep in humans. Indeed, all mammalian genes presented here, except Clk and Bmal1, have at least one allelic variant associated with a sleep-wake disorder. Abbreviations: AMPK - AMP-activated protein kinase; BMAL1 – Brain and muscle arntl-like protein 1; βTrCP – β-transducin repeat-containing protein; CK1δ/ε – Casein kinase 1 delta/epsilon; CLK – Clock; CRE – cAMP response element; CRY – Cryptochrome; CYC – Cycle; Ebox – promoter enhancer box; FBXL – F-box and leucine rich repeat protein ; GSK3β – Glycogen synthase kinase-3; PER – Period; SGG – Shaggy; TIM – Timeless; Ub – post-translational ubiquitination.
These cell-autonomous clocks throughout the brain and body are generally synchronized with each other and the external environment, which is achieved by coordination by a central pacemaker and integration with environmental cues [35, 36]. In fruit flies and mice, circadian clock neurons (~150 distributed neurons in flies and the densely packed suprachiasmatic nucleus in mice) receive environmental inputs (“zeitgebers”) and set the pace of the circadian rhythms of the animal by directly or indirectly inducing release of secreted signals including neurotransmitters, neuropeptides, and hormones [37–40]. The crucial role of these circadian pacemaker neurons in sleep regulation is demonstrated by loss of cycling of sleep/wake behavior in flies and mice when these neurons are ablated [41–43]. Zebrafish also have a region in the hypothalamus containing several neuropeptide-expressing cell populations, similar to that seen in the mammalian SCN; however, it is not required for pineal rhythmicity in larval zebrafish and thus may not play a key circadian pacemaking role [44]. Instead, it may be that light serves as a master organizer of circadian clocks in zebrafish [45].
Mutations in Core Clock Genes Cause Human Circadian Rhythm Sleep Disorders
The genetic pathways by which the circadian clock regulates sleep has been addressed at both the levels of the CGs themselves, as well as CCGs. Work over the past two decades has shown that knockout of canonical CGs in mice and flies leads to marked and varying effects on sleep amount and timing [46–53]. However, perhaps the most compelling evidence for an important role for CGs in regulating sleep comes from the analysis of the genetic basis of human circadian rhythm sleep disorders, specifically Familial Advanced Sleep Phase Syndrome (FASPS) and more recently Delayed Sleep Phase Syndrome (DSPS). Over the past ~15 years, several studies have identified a number of clock genes (Casein Kinase 1 delta/epsilon, Period2, Cryptochrome2, Period3) that lead to FASPS [54–57], where affected individuals exhibit markedly earlier bedtimes and wake times, compared to the general population. On the other hand, DSPS is characterized by delayed sleep initiation and trouble awakening at an appropriate hour. The prevalence of DSPS in the general population is significantly higher than that for ASPS, which raised the possibility that DSPS mainly results from voluntary behaviors (e.g, staying up at night with electronic devices or media). However, mutations in 2 clock genes, Period3 [58] or Cryptochrome1 [59], have recently been shown to cause this disorder.
In contrast to clock null mutants in animal models, which result in loss of rhythmic cycling of sleep behavior under constant conditions [49], the mutations causing ASPS and DSPS alter the phase of the consolidated sleep period of the individual. In addition, these mutations generally do not affect sleep quantity, although clinically, these disorders can restrict sleep amount because of forced wake-up times or bed times from social or work-related cues. At a molecular level, the ASPS mutations destabilize core clock genes. For example, the P415A/H417R variants of Per3 induce more rapid degradation of the PER3 protein[57], which facilitates degradation of PER1 and PER2 proteins, shortening the cycle length of the core oscillator. Similarly, a shortened period was observed in human subjects and transgenic mice carrying the point mutation A260T in Cry2 [56]. This genetic variant leads to enhanced affinity of CRY2 for FBXL3, a key ubiquitin ligase which targets CRY2 for degradation.
The recently identified 1657+3A>C Cry1 variant [59] causing DSPS illustrates another way by which the clock machinery may be perturbed: by slowing down rather than speeding up the core oscillator. Instead of affecting stability, this variant, which results in the skipping of exon 11 and a truncation of the C-terminus, enhances the repressor activity of CRY. This significantly lengthens the timing of feedback onto the CLOCK:BMAL1 heterodimer, and hence lengthens the circadian period. Because humans with these ASPS and DSPS mutations have intact circadian entrainment, the changes in their period length do not lead to non-24 hour sleep/wake disorder, where their wake-up times free-run, shifting progressively each day until misaligned with the environment. Instead, external cues such as light serve to reset their phase, inducing a consistent advance or delay of their sleep time each day, making them appear as extreme ‘early birds’ or ‘night owls’.
Clock-Controlled Genes Regulate Sleep via Extracellular Signaling Pathways
Compared with our understanding of the molecular basis of the core circadian oscillator, significantly less is known about the genetic mechanisms by which the circadian clock regulates rhythmic expression of behaviors such as sleep. Starting with classical SCN transplantation studies in the 1990s, a prevailing model for how the SCN exerts controls over behaviors is via the release of diffusible factors [60, 61]. In addition, circadian pacemaker neurons in flies and mice also project, directly or indirectly, to structures that regulate sleep (e.g., the arousal-promoting pars intercerebralis in flies and the sleep-promoting ventrolateral preoptic nucleus in mice) [62–64]. In this section, we will focus on the former process and discuss how circadian clocks regulate sleep via release of diffusible factors. A few such factors like Prokineticin2 (Prok2), transforming growth factor-α, and cardiotrophin-like cytokine have been previously suggested to act as circadian output molecules that regulate sleep timing in mammals [65–71]. However, functional evidence in support of this notion is relatively sparse, and recent evidence from zebrafish suggests instead that it acts to mediate light-dependent regulation of sleep [72]. Here, we discuss several recent studies that have re-examined the roles of melatonin and histamine, and identified a function for DH31 (a neuropeptide in Drosophila) in regulating the circadian timing of sleep.
Melatonin is required for circadian timing of sleep
The role of melatonin in sleep regulation has been somewhat controversial. Melatonin, the so-called “darkness hormone”, is released at night [73] in both diurnal and nocturnal animals [74] . Its synthesis in the pineal gland is gated by the circadian system in mammals and potently inhibited by light [75]. Although the timing of melatonin release is strongly linked to circadian phase [76, 77], via autonomic innervation by the SCN [78], studies in humans have generally found mild effects of exogenous melatonin administration on nighttime sleep behavior [79, 80]. Moreover, melatonin is not considered soporific in nocturnal rodents, and the most widely used mouse background for sleep/circadian studies, C57BL/6, lacks melatonin owing to loss of a key biochemical synthesis enzyme [81]. These observations raise the question of whether melatonin is dispensable for sleep regulation.
To address this issue, one study examined zebrafish larvae that lack melatonin due to a mutation in arylalkylamine-N-acetyltransferase 2 (aanat2) [82]. aanat2 mutants exhibit a significant reduction in night-time sleep, consistent with a sleep-promoting role for melatonin. Moreover, under constant darkness conditions, the normal rhythmic sleep behavior observed in wild-type zebrafish larvae is lost in aanat2 mutants. This phenotype is reminiscent of loss of rhythmic sleep behavior observed with loss of core clock molecules [46, 48–49], suggesting that melatonin plays a key role in the circadian regulation of sleep behavior. How do we reconcile the divergent findings regarding melatonin and sleep from zebrafish, mice, and humans? The weak sleep-promoting effects of melatonin in humans may reflect reduced effectiveness of exogenous melatonin due to the presence of significant endogenous melatonin signaling at night. Indeed, when given during the subjective day (when endogenous levels are low), melatonin has a more robust sleep-promoting effect in both fish and humans [83, 84]. Melatonin may be dispensable for circadian regulation of sleep in mice, because they are nocturnal animals and tend to sleep during the day.
The DH31 neuropeptide promotes wakefulness at the end of the night
In Drosophila, a recent study has identified a neuropeptide that is secreted from clock neurons to specifically regulate sleep [85]. Diuretic hormone 31 (DH31) is the fly homolog of calcitonin-related peptide in vertebrates, and loss-of-function mutations in this gene result in increased late-night sleep, without affecting circadian locomotor rhythmicity. These data suggest that DH31 normally acts to promote wakefulness towards the end of the night. This phenotype depends upon the expression of DH31 in a subset of clock neurons in Drosophila, the DN1 subgroup, and functional imaging experiments reveal that the activity of the DN1 neurons peaks towards the end of the night. Together, these data support a role for DH31 as a clock output molecule of DN1 neurons that acts specifically in the late night to promote arousal, in preparation for morning awakening.
Histamine signaling is regulated by local clocks to rhythmically promote arousal
In addition to the secretion of signaling molecules under control of central pacemakers described above, the cycling of local cell-endogenous circadian clocks also tunes the signaling of independent circuits to amplify information received from central pacemaker cells. Histaminergic cells of the tuberomammillary nucleus (TMN) in mice promote arousal, firing rapidly after wake onset but quieting during sleep. Expression of histidine decarboxylase (HDC), the rate limiting enzyme for histamine biosynthesis, cycles within these cells in phase with their electrical activity. Interestingly, selectively abolishing the local core clock via conditional BMAL1 knockout in only these cells eliminates HDC cycling and results in increased histamine levels during the daytime rest period. These animals exhibit significantly decreased daytime NREM sleep, in addition to greater sleep fragmentation and increased transitions between REM and NREM sleep. Although whole-animal measures of circadian rhythmicity remained unaffected, the decrease in regular and recovery SWA induced learning and memory deficits [86]. Thus, in histaminergic TMN cells, local cycling of the circadian transcriptional translation feedback loop directly influences the biosynthetic pathway of an arousal promoting neurotransmitter, and in this manner regulates various aspects of sleep in vivo.
Clock-Controlled Genes Regulate Sleep By Intrinsic Modulation of Neuronal Excitability
Wide Awake and Fbxl4 coordinate rhythmic GABA sensitivity in clock neurons to regulate sleep
Another process by which the circadian clock modulates sleep is via tuning of electrical activity of sleep/wake circuits. In Drosophila, a subset of clock neurons has been shown to promote arousal (l-LNvs), and a clock output molecule, named WIDE AWAKE (WAKE) acts in these cells to regulate the timing of sleep onset [87]. WAKE levels cycle under circadian clock control in the l-LNvs, rising in the early night to increase the levels and membrane targeting of a GABAA receptor (RDL). This enhanced GABA sensitivity of the arousal-promoting l-LNvs at dusk inhibits spontaneous firing and evoked excitability of these neurons to facilitate the switch from wakefulness to sleep. The absence of WAKE (as seen in wake mutants) results in greater l-LNv firing at dusk and specifically prolongs sleep latency (time from lights out to first sleep bout) [87], similar to the phenotype observed in patients with sleep-onset insomnia. The clock-dependent cycling of l-LNv excitability in sleep regulation not only depends on WAKE, but also a specific E3 ubiquitin ligase (Fbxl4), whose expression is also CLOCK-dependent [88]. Fbxl4 acts in an opponent manner to WAKE, by rhythmically degrading RDL at dawn to promote arousal, with mutant flies exhibiting significantly increased sleep and reduced sleep onset latency [88]. Strikingly, there is a single homolog of WAKE in mice and humans that is correspondingly enriched in the suprachiasmatic nucleus in mice [87], suggesting that these mechanisms are conserved in mammals.
A Clock-regulated microRNA regulates sleep by tuning excitability of sleep/wake circuits
The core circadian oscillator modulates the expression of many downstream genes, by not only directly regulating gene transcription, but also indirectly via control of non-coding RNAs, including microRNAs (miRs) [89–91]. Recently, one of these miRs, miR-92a, was shown to regulate neural excitability and also sleep in Drosophila [92]. Levels of miR-92a in fly clock neurons are elevated at night, and, like WAKE, miR-92a acts to suppress neural excitability of these cells. In arousal-promoting dopaminergic neurons, overexpression and knockdown of miR-92a levels led to an increase and decrease in sleep-bout duration, respectively, exclusively during the daytime. Conversely, overexpression of miR-92a in a sleep-promoting circuit increased daytime sleep. miR-92a therefore is an example of a post-transcriptional mechanism by which the circadian clock tunes the excitability of sleep/wake circuits to regulate both sleep and arousal across 24 hours.
Concluding Remarks and Future Perspectives
Our review of recent literature updates the varied mechanisms by which circadian clocks regulate sleep: from actions of the central clock and local clocks on rhythmic release of secreted molecules to cyclical tuning of intrinsic neural excitability. Given the paucity of knowledge regarding clock output mechanisms, it is likely that many more such genes will be identified in the future. Why are so many different clock-dependent genetic mechanisms necessary for regulating sleep? Sleep has been proposed to serve multiple functions [93]. These different sleep-related processes likely require spatial and temporal segregation. Related to the latter point, recent work using next generation sequencing technologies [for review see 31] demonstrates that, by using a variety of genetic regulatory mechanisms, the transcriptional network governed by the circadian clock is able to temporally segregate distinct cellular functions across nearly any phase of the 24 hr cycle. This fine-grained temporal resolution potentially enables different clock output genes to regulate the timing of various sleep processes. As an example, in Drosophila, WAKE promotes sleep onset in the early night, while DH31 promotes arousal in the late night. Further work is needed to not only delineate the functions of sleep, but also how timing of specific sleep-related processes may support these functions.
Forward genetic approaches in model organisms have been a fruitful avenue to identify genes involved in circadian rhythms and sleep [94–97], and will likely continue to be so into the future. However, the circuit mechanisms by which master circadian clocks regulate outputs such as sleep remain poorly understood. Advances in optogenetic and chemogenetic interrogation, as well as whole-brain functional imaging, should accelerate progress in these areas. These studies are also being coupled with single-cell sequencing methods to further refine our understanding of the circuit mechanisms regulating sleep [98]. Delineation of these output circuits will in the future not only establish the neural basis for these behaviors, but will also be critical for understanding the precise functions of the clock-regulated genes that impact sleep.
Table 1.
Genes implicated in circadian regulation of sleep.
| Gene | Function | Manipulation | Organism | Phenotype | Refs |
|---|---|---|---|---|---|
| Aanat2 | Melatonin biosynthesis | Null | Fish | ↓ Nighttime sleep (L:D), Arrythmic (D:D) | [82] |
|
| |||||
| Bmal1 | Transcription factor | KO | Mouse | ↑ Total sleep | [28] |
| cycle | Transcription factor | Null | Fly | ↑ Sleep latency, Locomotion | [52, 87] |
|
| |||||
| βTrcp2 | F-box WD40 repeat | KO | Mouse | Destabilized rest/activity timing | [99] |
| slimb | F-box WD40 repeat | Overexpression | Fly | ↓ Rest/activity amplitude, ↑ % arrhythmic | [100] |
|
| |||||
| CK1δ/ε | Kinase | Ck1δ T44A | Human | ASPS | [54] |
| Ck1δ T44A | Mouse | Shortened period | [54] | ||
| Ck1δ T44A | Fly | Lengthened period | [54] | ||
| Ck1ε tau | Mouse | ↑ REM sleep time | [101] | ||
|
| |||||
| Clc | Cytokine | Infusion | Mouse | Suppressed locomotor activity | [71] |
| Cntfr | CLC receptor | Ab infusion | Mouse | ↑ Daytime locomotion | [71] |
|
| |||||
| Clock | Transcription factor | 3111C allele | Human | “Night owl” tendency | [102] |
| Antimorph | Mouse | ↓ Total sleep time | [46] | ||
| Null | Fly | ↓ sleep, ↑ locomotion | [52, 87] | ||
| Npas2 | Transcription factor | KO | Mouse | ↓ Nighttime SWS+REM, ↓ Rebound sleep | [103, 47] |
|
| |||||
| Cryptochrome | Transcriptional repressor | Cry1 1657+3A>C | Human | DSPS | [59] |
| Cry2 A260T | Human | ASPS, ↑ Cry degradation by Fbxl3 | [56] | ||
| Cry1/2 KO | Mouse | ↑ Homeostatic sleep drive | [50] | ||
|
| |||||
| Dec2 | Transcriptional repressor | P385R | Human | ↓ Total sleep, ↓ NREM and REM sleep | [104] |
| P385R | Mouse | ↓ Total sleep, ↓ NREM and REM sleep | [104] | ||
| Y362H | Human | ↓ Baseline sleep, ↓ sleep | [105] | ||
| Morpholino | Fish | ↓ Sleep time, ↑ Sleep fragmentation | [106] | ||
| Sharp | Transcriptional repressor | Sharp1/2 KO | Mouse | ↑ Nighttime NREM, ↓ daytime REM | [107] |
|
| |||||
| Dh31 | Neuropeptide | Null | Fly | ↑ Sleep, ↓ Arousal threshold | [85] |
| Overexpression | Fly | ↓ Nighttime sleep | [85] | ||
|
| |||||
| Fbxl | F-box leucine-rich repeat | fbxl4Δ45 Null | Fly | ↓ sleep latency, ↑ total sleep | [88] |
|
| |||||
| miR-92a | Neural excitability | Overexpression | Fly | ↑ Daytime sleep | [92] |
| KO | Fly | ↓ Daytime sleep | [92] | ||
|
| |||||
| Period | Transcriptional repressor | Per2 hypomorph | Human | ASPS | [55] |
| Per3 haplotype | Human | DSPS | [58] | ||
| Per3 P415A/H417R | Human | ASPS | [57] | ||
| Per3 P415A/H417R | Mouse | Period lengthening in constant light | [57] | ||
| Per3 P415A/H417R | Fly | Advanced activity offset, Shortened period | [57] | ||
| Per1/2 KO | Mouse | ↓ NREM sleep (L:D) | [49] | ||
| Null | Fly | ↓ Sleep | [87] | ||
|
| |||||
| Prok2 | Neuropeptide | Infusion | Mouse | ↓ Rest/activity amplitude | [65] |
| Overexpression | Fish | ↓ Rest/activity amplitude | [72] | ||
| Prokr2 | PRK2 Receptor | KO | Mouse | ↓ Nighttime locomotion | [68] |
|
| |||||
| Rev-ERB α/β | Transcriptional repressor | Rev-ERBα KO | Mouse | ↓ Homeostatic sleep, Advanced sleep/wake | [53] |
|
| |||||
| TGFα | Neuropeptide | Infusion | Mouse | ↓ Locomotion | [69, 70] |
| Egfr | TGFα receptor | Null | Mouse | ↑ Daytime activity | [69] |
| rho | EGF receptor | Receptor activation | Fly | ↑ Sleep | [38] |
|
| |||||
| wide awake | Neural excitability | Null | Fly | ↑ Sleep latency, ↓ Total sleep | [87] |
Highlights.
Work over the past few decades has led to detailed understanding of the genetic mechanisms underlying the core circadian clock, culminating in the Nobel Prize in Physiology or Medicine in 2017
Much less is known about how this core clock regulates output behaviors, such as sleep
Emerging data in fruit flies, zebrafish, mice, and humans reveal that central and local clocks use diverse mechanisms to regulate distinct aspects of sleep
Outstanding Questions Box.
What is the role of local clocks within specific sleep/wake circuits in sleep regulation? Do local clocks regulate transcriptional networks altering electrical activity, metabolism, or even sleep need in these neurons in a circuit-specific manner?
Astrocytes express circadian clock genes from flies to mammals. Do local clocks in these cells impact cortical function and sleep need or sleep quality?
Recent GWAS studies have identified human genomic loci associated with sleep and circadian rhythms phenotypes. Do these sequences contain new genes and/or non-coding RNAs which are relevant to the circadian regulation of sleep? Can model organisms help clarify the role of these molecules?
Given the increasing recognition that disordered sleep and circadian rhythms may contribute to the development or progression of neurodegenerative diseases in humans, do alterations in the mechanisms mediating the circadian regulation of sleep play a role in diseases of aging in humans?
Acknowledgments
This work was supported by NIH grants NS079584 and NS094571 (M.N.W.)
Glossary Box – Key sleep- and circadian-related terms
- Circadian Clock/Core Oscillator
The central molecular mechanism, consisting of a transcriptional-translational feedback loop (defined below), by which animals endogenously keep time, and align their physiological and behavioral patterns to the 24-hour day
- Transcriptional-Translational Feedback Loop (TTFL)
See Figure 1. Core clock transcription factors (CLOCK/NPAS2 and BMAL) drive expression of repressors (PERIOD and CRYPTOCHROME), which translocate back into the nucleus to inhibit the core clock transactivators, thus suppressing their own expression. In this manner, many genes regulated by these transcription factors are subject to ~24-hour oscillation in expression levels
- Circadian Period
The length of time required to complete one full cycle of rhythmic molecular oscillations or behavior. Circadian periods (‘circa’ = about, ‘diem’ = day), occur with approximately 24-hour regular intervals
- Circadian Phase
The phase of a circadian rhythm is defined relative to a reference point of the rhythm, such as the temperature nadir in mammals
- Circadian Rhythms
Biological rhythms that oscillate with a ~24 hr period that are regulated by the core circadian clock. Examples include cycling of sleep/wake behavior, feeding behavior, core body temperature, and blood cortisol levels
- Entrainment
The ability of the circadian clock to adjust its phase, based upon environmental cues, including light, temperature, and food availability
- Zeitgeber
German for “time-giver.” Environmental cues that adjust the phase of the circadian clock. Light is the most powerful zeitgeber, but food access, temperature, and social cues also serve to entrain the clock
- Electroencephalography (EEG)
An assay that measures brain activity, in which voltage changes resulting from the synchronized firing of thousands of cortical neurons are detected at the scalp, amplified, and recorded. EEG is a commonly used method for measuring sleep in mammals, including humans
- Non-REM (NREM) Sleep
A state of sleep defined by characteristic EEG waveforms with increasing cohesiveness and amplitude and reduced frequency. In deep NREM sleep, high amplitude delta waves (0.5–4 Hz) are prominent (Slow-Wave Activity, SWA), and their spectral power correlates with sleep need
- Rapid Eye Movement (REM) Sleep
also known as ‘Paradoxical Sleep,’ REM is defined by low amplitude, mixed frequency EEG waveforms, similar to that seen in wakefulness, coupled with muscle atonia and characteristic horizontal eye movements. In humans, REM is the sleep sub-state associated with vivid dreams, but its specific function remains contested
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ibanez C. The 2017 Nobel Prize in Physiology or Medicine - Advanced Information: Discoveries of Molecular Mechanisms Controlling the Circadian Rhythm. Nobel Media AB; 2017. Nobelprize.org. [Google Scholar]
- 2.Raizen DM, et al. Lethargus is a Caenorhabditis elegans sleep-like state. Nature. 2008;451(7178):569–572. doi: 10.1038/nature06535. [DOI] [PubMed] [Google Scholar]
- 3.Nath RD, et al. The Jellyfish Cassiopea Exhibits a Sleep-like State. Curr Biol. 2017;27(19):2984–2990. doi: 10.1016/j.cub.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vorster APA, et al. Characterization of sleep in Aplysia californica. Sleep. 2014;37(9):1453–1463. doi: 10.5665/sleep.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tisdale RK, et al. Sleep-Related Electrophysiology and Behavior of Tinamous (Eudromia elegans): Tinamous Do Not Sleep Like Ostriches. Brain Behav Evol. 2017;89(4):249–261. doi: 10.1159/000475590. [DOI] [PubMed] [Google Scholar]
- 6.Lesku JA, et al. History and future of comparative analyses in sleep research. Neurosci Biobehav Rev. 2009;33(7):1024–36. doi: 10.1016/j.neubiorev.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Saper CB, Fuller PM. Wake–sleep circuitry: an overview. Current Opinion in Neurobiology. 2017;44:186–192. doi: 10.1016/j.conb.2017.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benington JH, Craig Heller H. Restoration of brain energy metabolism as the function of sleep. Progress in Neurobiology. 1995;45(4):347–360. doi: 10.1016/0301-0082(94)00057-o. [DOI] [PubMed] [Google Scholar]
- 9.Spiegel K, et al. Impact of sleep debt on metabolic and endocrine function. The Lancet. 1999;354(9188):1435–1439. doi: 10.1016/S0140-6736(99)01376-8. [DOI] [PubMed] [Google Scholar]
- 10.Besedovsky L, et al. Sleep and immune function. Pflügers Archiv - European Journal of Physiology. 2012;463(1):121–137. doi: 10.1007/s00424-011-1044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellesi M, et al. Sleep Loss Promotes Astrocytic Phagocytosis and Microglial Activation in Mouse Cerebral Cortex. The Journal of Neuroscience. 2017;37(21):5263–5273. doi: 10.1523/JNEUROSCI.3981-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie L, et al. Sleep Drives Metabolite Clearance from the Adult Brain. Science. 2013;342(6156):373–377. doi: 10.1126/science.1241224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding F, et al. Changes in the composition of brain interstitial ions control the sleep-wake cycle. Science. 2016;352(6285):550–555. doi: 10.1126/science.aad4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tononi G, Cirelli C. Sleep and the price of plasticity: from synaptic and cellular homeostasis to memory consolidation and integration. Neuron. 2014;81(1):12–34. doi: 10.1016/j.neuron.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watson BO, et al. Network Homeostasis and State Dynamics of Neocortical Sleep. Neuron. 2016;90(4):839–52. doi: 10.1016/j.neuron.2016.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diering GH, et al. Homer1a drives homeostatic scaling-down of excitatory synapses during sleep. Science. 2017;355(6324):511–515. doi: 10.1126/science.aai8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Vivo L, et al. Ultrastructural evidence for synaptic scaling across the wake/sleep cycle. Science. 2017;355(6324):507–510. doi: 10.1126/science.aah5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall L, et al. Boosting slow oscillations during sleep potentiates memory. Nature. 2006;444(7119):610–613. doi: 10.1038/nature05278. [DOI] [PubMed] [Google Scholar]
- 19.Rasch B, et al. Odor cues during slow-wave sleep prompt declarative memory consolidation. Science. 2007;315(5817):1426–9. doi: 10.1126/science.1138581. [DOI] [PubMed] [Google Scholar]
- 20.Boyce R, et al. REM sleep and memory. Current Opinion in Neurobiology. 2017;44:167–177. doi: 10.1016/j.conb.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Levenstein D, et al. Sleep regulation of the distribution of cortical firing rates. Curr Opin Neurobiol. 2017;44:34–42. doi: 10.1016/j.conb.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buzsaki G. Rhythms of the Brain. Oxford University Press; USA: 2006. [Google Scholar]
- 23.Putilov AA. Prospects of using electroencephalographic signatures of the chronoregulatory processes for meaningful, parsimonious and quantitative description of the sleep–wake sub-states. Biological Rhythm Research. 2011;42(3):181–207. [Google Scholar]
- 24.Frank MG, et al. A Preliminary Analysis of Sleep-Like States in the Cuttlefish Sepia officinalis. PLOS ONE. 2012;7(6):e38125. doi: 10.1371/journal.pone.0038125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Alphen B, et al. A Dynamic Deep Sleep Stage in Drosophila. The Journal of Neuroscience. 2013;33(16):6917–6927. doi: 10.1523/JNEUROSCI.0061-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Swinderen B, Andretic R. Dopamine in Drosophila: setting arousal thresholds in a miniature brain. Proc Biol Sci. 2011;278:906–913. doi: 10.1098/rspb.2010.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Swinderen B, et al. Uncoupling of Brain Activity from Movement Defines Arousal States in Drosophila. Current Biology. 2004;14(2):81–87. [PubMed] [Google Scholar]
- 28.Zimmerman JE, et al. Conservation of sleep: insights from non-mammalian model systems. Trends in Neurosciences. 2008;31(7):371–376. doi: 10.1016/j.tins.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borbély AA, et al. The two-process model of sleep regulation: a reappraisal. J Sleep Res. 2016;25(2):131–43. doi: 10.1111/jsr.12371. [DOI] [PubMed] [Google Scholar]
- 30.Borbély AA. A two process model of sleep regulation. Hum Neurobiol. 1982;1(3):195–204. [PubMed] [Google Scholar]
- 31.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18(3):164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ueda HR, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37(2):187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 33.Zhang R, et al. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci U S A. 2014;111(45):16219–24. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoo SH, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101(15):5339–46. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schibler U. Interaction Between Central and Peripheral Clocks in Mammals. In: Kumar V, editor. Biological Timekeeping: Clocks, Rhythms and Behaviour. Springer; India: 2017. pp. 337–363. [Google Scholar]
- 36.Buijs RM, Kalsbeek A. Hypothalamic integration of central and peripheral clocks. Nat Rev Neurosci. 2001;2(7):521–526. doi: 10.1038/35081582. [DOI] [PubMed] [Google Scholar]
- 37.Selcho M, et al. Central and peripheral clocks are coupled by a neuropeptide pathway in Drosophila. Nat Comm. 2017;8:15563. doi: 10.1038/ncomms15563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown TM, Piggins HD. Electrophysiology of the suprachiasmatic circadian clock. Prog Neurobiol. 2007;82(5):229–55. doi: 10.1016/j.pneurobio.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Maywood ES, et al. Minireview: The Circadian Clockwork of the Suprachiasmatic Nuclei Analysis of a Cellular Oscillator that Drives Endocrine Rhythms. Endocrinology. 2007;148(12):5624–5634. doi: 10.1210/en.2007-0660. [DOI] [PubMed] [Google Scholar]
- 40.Dubowy C, Sehgal A. Circadian Rhythms and Sleep in Drosophila melanogaster. Genetics. 2017;205(4):1373–1397. doi: 10.1534/genetics.115.185157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renn SCP, et al. A pdf Neuropeptide Gene Mutation and Ablation of PDF Neurons Each Cause Severe Abnormalities of Behavioral Circadian Rhythms in Drosophila. Cell. 1999;99(7):791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- 42.Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci U S A. 1972;69(6):1583–6. doi: 10.1073/pnas.69.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rusak B. The role of the suprachiasmatic nuclei in the generation of circadian rhythms in the golden hamster, Mesocricetus auratus. J Comp Physiol. 1977;118(2):145–164. [Google Scholar]
- 44.Noche RR, et al. Circadian rhythms in the pineal organ persist in zebrafish larvae that lack ventral brain. BMC Neuroscience. 2011;12(1):7. doi: 10.1186/1471-2202-12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitmore D, et al. Light acts directly on organs and cells in culture to set the vertebrate circadian clock. Nature. 2000;404:87–91. doi: 10.1038/35003589. [DOI] [PubMed] [Google Scholar]
- 46.Naylor E, et al. The circadian clock mutation alters sleep homeostasis in the mouse. J Neurosci. 2000;20(21):8138–43. doi: 10.1523/JNEUROSCI.20-21-08138.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franken P, et al. NPAS2 as a transcriptional regulator of non-rapid eye movement sleep: genotype and sex interactions. Proc Natl Acad Sci U S A. 2006;103(18):7118–23. doi: 10.1073/pnas.0602006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laposky A, et al. Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation. Sleep. 2005;28(4):395–409. doi: 10.1093/sleep/28.4.395. [DOI] [PubMed] [Google Scholar]
- 49.Shiromani PJ, et al. Sleep rhythmicity and homeostasis in mice with targeted disruption of mPeriod genes. Am J Physiol Regul Integr Comp Physiol. 2004;287:R47–57. doi: 10.1152/ajpregu.00138.2004. [DOI] [PubMed] [Google Scholar]
- 50.Wisor JP, et al. A role for cryptochromes in sleep regulation. BMC Neurosci. 2002;3:20. doi: 10.1186/1471-2202-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hendricks JC, et al. Rest in Drosophila Is a Sleep-like State. Neuron. 2000;25(1):129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 52.Hendricks JC, et al. Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J Biol Rhythms. 2003;18(1):12–25. doi: 10.1177/0748730402239673. [DOI] [PubMed] [Google Scholar]
- 53.Mang GM, et al. Altered Sleep Homeostasis in Rev-erb α Knockout Mice. Sleep. 2016;39(3):589–601. doi: 10.5665/sleep.5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Y, et al. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434(7033):640–4. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 55.Toh KL, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291(5506):1040–3. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 56.Hirano A, et al. A Cryptochrome 2 mutation yields advanced sleep phase in humans. eLife. 2016;5:e16695. doi: 10.7554/eLife.16695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang L, et al. A PERIOD3 variant causes a circadian phenotype and is associated with a seasonal mood trait. Proc Natl Acad Sci U S A. 2016;113(11):E1536–E1544. doi: 10.1073/pnas.1600039113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ebisawa T, et al. Association of structural polymorphisms in the human period3 gene with delayed sleep phase syndrome. EMBO reports. 2001;2(4):342–346. doi: 10.1093/embo-reports/kve070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patke A, et al. Mutation of the Human Circadian Clock Gene CRY1 in Familial Delayed Sleep Phase Disorder. Cell. 2017;169(2):203–215 e13. doi: 10.1016/j.cell.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ralph MR, et al. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247(4945):975–8. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- 61.Silver R, et al. A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature. 1996;382(6594):810–3. doi: 10.1038/382810a0. [DOI] [PubMed] [Google Scholar]
- 62.Chou TC, et al. Afferents to the ventrolateral preoptic nucleus. J Neurosci. 2002;22(3):977–90. doi: 10.1523/JNEUROSCI.22-03-00977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cavanaugh DJ, et al. Identification of a circadian output circuit for rest:activity rhythms in Drosophila. Cell. 2014;157(3):689–701. doi: 10.1016/j.cell.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cavey M, et al. Circadian rhythms in neuronal activity propagate through output circuits. Nat Neurosci. 2016;19:587–595-81. doi: 10.1038/nn.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng MY, et al. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature. 2002;417(6887):405–10. doi: 10.1038/417405a. [DOI] [PubMed] [Google Scholar]
- 66.Cheng M, et al. Regulation of prokineticin 2 expression by light and the circadian clock. BMC Neurosci. 2005;6:17. doi: 10.1186/1471-2202-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li JD, et al. Attenuated circadian rhythms in mice lacking the prokineticin 2 gene. J Neurosci. 2006;26(45):11615–23. doi: 10.1523/JNEUROSCI.3679-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prosser HM, et al. Prokineticin receptor 2 (Prokr2) is essential for the regulation of circadian behavior by the suprachiasmatic nuclei. Proc Natl Acad Sci U S A. 2007;104(2):648–53. doi: 10.1073/pnas.0606884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kramer A, et al. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science. 2001;294(5551):2511–5. doi: 10.1126/science.1067716. [DOI] [PubMed] [Google Scholar]
- 70.Kramer A, et al. A Screen for Secreted Factors of the Suprachiasmatic Nucleus. In: Michael WY, editor. Methods in Enzymology. Academic Press; 2005. pp. 645–663. [DOI] [PubMed] [Google Scholar]
- 71.Kraves S, Weitz CJ. A role for cardiotrophin-like cytokine in the circadian control of mammalian locomotor activity. Nat Neurosci. 2006;9(2):212–9. doi: 10.1038/nn1633. [DOI] [PubMed] [Google Scholar]
- 72.Chen S, et al. Light-Dependent Regulation of Sleep and Wake States by Prokineticin 2 in Zebrafish. Neuron. 2017;95(1):153–168.e6. doi: 10.1016/j.neuron.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Utiger RD. Melatonin — The Hormone of Darkness. New England Journal of Medicine. 1992;327(19):1377–1379. doi: 10.1056/NEJM199211053271909. [DOI] [PubMed] [Google Scholar]
- 74.Reiter RJ. The pineal-1977. Eden Press; 1977. [Google Scholar]
- 75.Cardinali DP. Melatonin. A Mammalian Pineal Hormone. Endocr Rev. 1981;2(3):327–346. doi: 10.1210/edrv-2-3-327. [DOI] [PubMed] [Google Scholar]
- 76.Lewy AJ, Sack RL. The dim light melatonin onset as a marker for circadian phase position. Chronobiol Int. 1989;6(1):93–102. doi: 10.3109/07420528909059144. [DOI] [PubMed] [Google Scholar]
- 77.Boivin DB, et al. Dose-response relationships for resetting of human circadian clock by light. Nature. 1996;379(6565):540–542. doi: 10.1038/379540a0. [DOI] [PubMed] [Google Scholar]
- 78.Kalsbeek A, et al. A network of (autonomic) clock outputs. Chronobiol Int. 2006;23(3):521–35. doi: 10.1080/07420520600651073. [DOI] [PubMed] [Google Scholar]
- 79.Scheer FAJL, Czeisler CA. Melatonin, sleep, and circadian rhythms. Sleep Med Rev. 9(1):5–9. doi: 10.1016/j.smrv.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 80.Brzezinski A, et al. Effects of exogenous melatonin on sleep: a meta-analysis. Sleep Med Rev. 2005;9(1):41–50. doi: 10.1016/j.smrv.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 81.Kasahara T, et al. Genetic variation of melatonin productivity in laboratory mice under domestication. Proc Natl Acad Sci U S A. 2010;107(14):6412–7. doi: 10.1073/pnas.0914399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gandhi AV, et al. Melatonin is required for the circadian regulation of sleep. Neuron. 2015;85(6):1193–9. doi: 10.1016/j.neuron.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhdanova IV, et al. Melatonin promotes sleep-like state in zebrafish. Brain Res. 2001;903(1):263–268. doi: 10.1016/s0006-8993(01)02444-1. [DOI] [PubMed] [Google Scholar]
- 84.Wyatt JK, et al. Sleep-Facilitating Effect of Exogenous Melatonin in Healthy Young Men and Women Is Circadian-Phase Dependent. Sleep. 2006;29(5):609–618. doi: 10.1093/sleep/29.5.609. [DOI] [PubMed] [Google Scholar]
- 85.Kunst M, et al. Calcitonin Gene-Related Peptide Neurons Mediate Sleep-Specific Circadian Output in Drosophila. Curr Biol. 2014;24(22):2652–2664. doi: 10.1016/j.cub.2014.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu X, et al. Circadian Factor BMAL1 in Histaminergic Neurons Regulates Sleep Architecture. Curr Biol. 2014;24(23):2838–2844. doi: 10.1016/j.cub.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu S, et al. WIDE AWAKE Mediates the Circadian Timing of Sleep Onset. Neuron. 2014;82(1):151–166. doi: 10.1016/j.neuron.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li Q, et al. Fbxl4 serves as a clock output molecule that regulates sleep through promotion of rhythmic degradation of the GABAA receptor. Curr Biol. 2017;27(23):3616–3625. doi: 10.1016/j.cub.2017.10.052. [DOI] [PubMed] [Google Scholar]
- 89.Alvarez-Saavedra M, et al. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum Mol Genet. 2011;20(4):731–751. doi: 10.1093/hmg/ddq519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cheng HYM, et al. microRNA Modulation of Circadian-Clock Period and Entrainment. Neuron. 2007;54(5):813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luo W, Sehgal A. Regulation of Circadian Behavioral Output via a MicroRNA-JAK/STAT Circuit. Cell. 2012;148(0):765–79. doi: 10.1016/j.cell.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen X, Rosbash M. MicroRNA-92a is a circadian modulator of neuronal excitability in Drosophila. Nat Comm. 2017;8:14707. doi: 10.1038/ncomms14707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Krueger JM, et al. Sleep function: Toward elucidating an enigma. Sleep Med Rev. 2016;28:46–54. doi: 10.1016/j.smrv.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68(9):2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vitaterna MH, et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264(5159):719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Koh K, et al. Identification of SLEEPLESS, a sleep-promoting factor. Science. 2008;321(5887):372–6. doi: 10.1126/science.1155942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Funato H, et al. Forward-genetics analysis of sleep in randomly mutagenized mice. Nature. 2016;539(7629):378–383. doi: 10.1038/nature20142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chung S, et al. Identification of preoptic sleep neurons using retrograde labelling and gene profiling. Nature. 2017;545(7655):477–481. doi: 10.1038/nature22350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.D’Alessandro M, et al. Stability of Wake-Sleep Cycles Requires Robust Degradation of the PERIOD Protein. Curr Biol. 2017;27(22):3454–3467. doi: 10.1016/j.cub.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ko HW, et al. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- 101.Zhou L, et al. The circadian clock gene Csnk1e regulates rapid eye movement sleep amount, and nonrapid eye movement sleep architecture in mice. Sleep. 2014;37(4):785–93. doi: 10.5665/sleep.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Katzenberg D, et al. A CLOCK polymorphism associated with human diurnal preference. Sleep. 1998;21(6):569–76. doi: 10.1093/sleep/21.6.569. [DOI] [PubMed] [Google Scholar]
- 103.Dudley CA, et al. Altered Patterns of Sleep and Behavioral Adaptability in NPAS2-Deficient Mice. Science. 2003;301(5631):379–383. doi: 10.1126/science.1082795. [DOI] [PubMed] [Google Scholar]
- 104.He Y, et al. The transcriptional repressor DEC2 regulates sleep length in mammals. Science. 2009;325(5942):866–70. doi: 10.1126/science.1174443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pellegrino R, et al. A Novel BHLHE41 Variant is Associated with Short Sleep and Resistance to Sleep Deprivation in Humans. Sleep. 2014;37(8):1327–1336. doi: 10.5665/sleep.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Srdanović S, et al. Transient knock-down of kcna2 reduces sleep in larval zebrafish. Behav Brain Res. 2017;326:13–21. doi: 10.1016/j.bbr.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 107.Baier PC, et al. Mice Lacking the Circadian Modulators SHARP1 and SHARP2 Display Altered Sleep and Mixed State Endophenotypes of Psychiatric Disorders. PLOS ONE. 2014;9(10):e110310. doi: 10.1371/journal.pone.0110310. [DOI] [PMC free article] [PubMed] [Google Scholar]


