Abstract
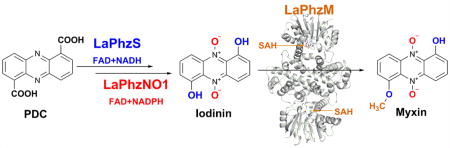
Myxin is a well-known antibiotic that had been used for decades. It belongs to the phenazine natural products that exhibit various biological activities, which are often dictated by the decorating groups on the heteroaromatic three-ring system. The three rings of myxin carry a number of decorations, including an unusual aromatic N5,N10-dioxide. We previously showed that phenazine 1,6-dicarboxylic acid (PDC) is the direct precursor of myxin, and two redox enzymes (LaPhzS and LaPhzNO1) catalyze the decarboxylative hydroxylation and aromatic N-oxidations of PDC to produce iodinin (1.6-dihydroxy-N5,N10-dioxide phenazine). In this work, we identified the LaPhzM gene from Lysobacter antibioticus OH13 and demonstrated that LaPhzM encodes a SAM-dependent O-methyltransferase converting iodinin to myxin. The results further showed that LaPhzM is responsible for both monomethoxy and dimethoxy formation in all phenazine compounds isolated from strain OH13. LaPhzM exhibits relaxed substrate selectivity, catalyzing O-methylation of phenazines with non-, mono-, or di-N-oxide. In addition, we demonstrated a one-pot biosynthesis of myxin by in vitro reconstitution of the three phenazine-ring decorating enzymes. Finally, we determined the X-ray crystal structure of LaPhzM with a bound cofactor at 1.4 Å resolution. The structure provided molecular insights into the activity and selectivity of the first characterized phenazine O-methyltransferase. These results will facilitate future exploitation of the thousands of phenazines as new antibiotics through metabolic engineering and chemoenzymatic syntheses.
Graphical abstract
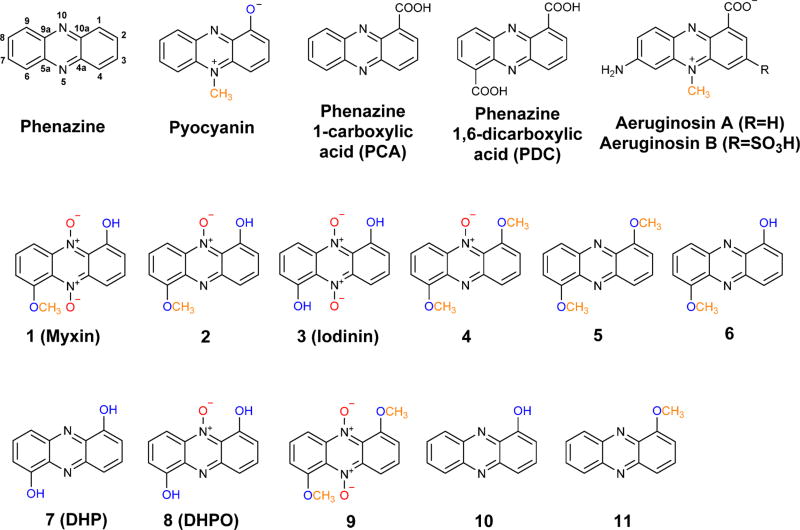
Phenazines are a large group of heterotricyclic N-containing aromatic compounds. Over 6000 phenazines have been isolated from living organisms or chemically synthesized during the last one and half centuries.1–4 Among them, several hundreds were reported to possess biological activities, including antimicrobial, antimalarial, and anticancer functions. The first phenazine, pyocyanin (1-hydroxy-N5-methylphenazine), was isolated from the Gram negative opportunistic pathogen Pseudomonas aeruginosa in the late 19th century (Figure 1).1 This bacterial pathogen uses pyocyanin as a virulence factor for infection,5, 6 and recent work has shown that pyocyanin is a redox-active metabolite facilitating extracellular electron transfer and survival in anoxic environments, which is critical for biofilm development.7–11
Figure 1.
Chemical structure of selected phenazine natural products isolated from Pseudomonas aeruginosa and Lysobacter antibioticus. Some of the functional groups on the phenazine ring are color highlighted to indicate the decorations relevant to this research.
All phenazines share a three-ring core (dibenzopyrazine) but differ at the decorations on the ring system. The decorating groups, including carboxylate, hydroxy, amide, N-methyl, O-methyl (methyoxy), and prenyl groups, dictate the difference in phenazine biological activities.12 Recent studies also showed that phenazines of P. aeruginosa could be selectively modified by surrounding organisms as a means to control the pathogen’s biofilms.13 The biosynthetic mechanism for the phenazine core has been extensively investigated by several groups.1–4, 14, 15 Five to seven genes are required to convert chorismic acid to phenazine 1-carboxylic acid (PCA) and/or phenazine-1,6-dicarboxylic acid (PDC; Figure 1). In contrast, relatively little has been done on the molecular mechanism underlying the decorations on the rings. Phenazine natural products with a hydroxy group at the six modifiable carbons of the phenazine core have been isolated from Pseudomonas species.1 Interestingly, the hydroxy groups are rarely methylated, although several well-known Pseudomonas products contain an N-methyl group, such as pyocyanin and aeruginosins (Figure 1). In L. antibioticus OH13, we previously isolated six phenazines (1–6), including the well-known antibiotic myxin (1-hydroxy-6-methoxyphenazine-N5,N10-dioxide).16 Myxin possesses potent antimicrobial activity with a unique mode of action and was manufactured as a copper(II) complex, cuprimyxin, which had been used for decades as a topical broad spectrum veterinary antibiotic and antifungal drug.12 Five of the six compounds isolated from L. antibioticus contain one or two O-methyl groups, but no N-methyl group (Figure 1).16 The O-methylation is important for the antibiotic activity of phenazines. For example, myxin and iodinin (1,6-dihydroxyphenazine N5,N10-dioxide) have an identical structure, except that none of the hydroxy groups is methylated in iodinin (Figure 1), but myxin has much higher antimicrobial activity than iodinin in all the tests we conducted.16 While O-methylation is very common in natural product biosynthesis, the O-methylation in myxin and other phenazines had not been investigated.
In pyocyanin, two enzymes are involved in the decoration of the phenazine ring during biosynthesis in P. aeruginosa.17–20 The first enzyme (PhzM) is an S-adenosylmethionine (SAM) dependent N-methyltransferase, converting PCA to N5-methyl-PCA. Curiously, PhzM was shown to be active only in the physical presence of PhzS, a flavin-dependent hydroxylase catalyzing a decarboxylative hydroxylation in pyocyanin biosynthesis. The N-methylation product, N5-methyl-PCA, was not isolated in the PhzM reaction. It was proposed that N5-methyl-PCA could be unstable and served as an enzyme-bound intermediate that was further processed by the second enzyme, PhzS, to produce the final product, pyocyanin. The result suggested that a protein–protein interaction between PhzM and PhzS might be required in pyocyanin production, as this interaction could ensure an efficient production of pyocyanin by preventing the formation and release of the unstable intermediate N5-methyl-PCA. The crystal structure of both PhzM and PhzS had been reported.17–20 The 1.8 Å dimeric structure of PhzM showed three domains per monomer characteristic of many small molecule methyltransferase but did not contain any ligand. The 2.4 Å crystal structure of PhzS showed that the enzyme belongs to the family of aromatic hydroxylases.
Phenazine ring decorations are usually specific to each producing organism. In strain OH13, the LaPhz gene cluster contains four genes that were predicted to be responsible for phenazine ring decoration.16 One gene codes for the FAD-dependent enzyme LaPhzS that catalyzes twice the decarboxylative hydroxylation of PDC, the common precursor of all phenazines in strain OH13 (Figure 1). This reaction converts PDC to 1,6-dihydroxyphenazine (7). The next gene encodes LaPhzNO1, a Baeyer–Villiger-like flavin enzyme that catalyzes N-oxidation at both N5 and N10 of the phenazine core. This enzyme eventually converts 1,6-dihydroxyphenazine (7) to 1,6-dihydroxyphenazine N5,N10-dioxide (iodinin, 3). Within the LaPhz gene cluster, there remain two genes (LaPhzM and LaPhzX) that have not been investigated for their function. LaPhzM shows a sequence similarity to genes coding for methyltransferases including pyocyanin PhzM,18 and LaPhzX is similar to genes encoding small monooxygenases that catalyze oxidation of phenolic compounds to corresponding quinones and do not require any metal ion or other cofactors.21, 22 Interestingly, although LaPhzM is homologous to pyocyanin PhzM (Figure S1), no N-methyl containing phenazines have been identified from strain OH13. In this work, we showed that LaPhzM is responsible for O-methylation in the biosynthesis of myxin and other phenazine natural products isolated from strain OH13. We characterized the SAM-dependent O-methyltransferase LaPhzM through heterologous expression, in vitro assay of the enzyme activity and selectivity, one-pot reconstitution of myxin biosynthesis in vitro, and determination of the crystal structure of LaPhzM complexed with cofactor.
RESULTS AND DISCUSSION
LaPhzM Encodes an O-Methyltransferase for Production of Both Monomethylated and Dimethylated Phenazines in L. antibioticus OH13
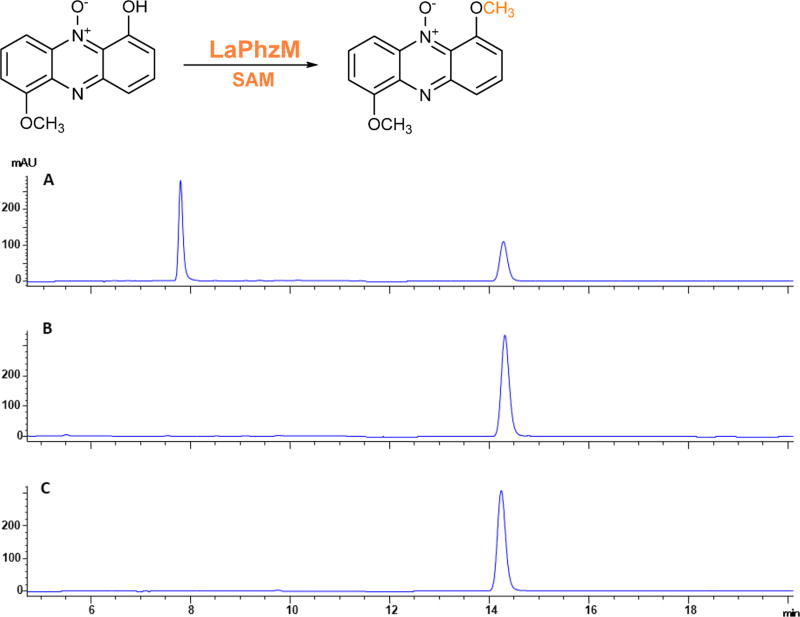
The LaPhzM gene within the phenazine biosynthetic gene cluster LaPhz in strain OH13 was heterologously expressed in E. coli as a 36.7 kDa protein (Figure S2). To study the role of LaPhzM in the biosynthesis of myxin and other phenazines, we incubated LaPhzM with 6-hydroxy-1-methoxyphenazine N5-oxide (2), the main phenazine compound produced by strain OH13.16 The substrate eluted at a retention time of 14.3 min on HPLC, and the reaction yielded one new product with a retention time of 6.8 min (Figure 2 and Figure S3). The production of this product was dependent on LaPhzM and SAM, as neither the boiled LaPhzM nor the lack of SAM produced this compound. The new product had the same retention time as 1,6-dimethoxyphenazine N5-oxide (4), which had previously been isolated from strain OH13.16 Mass spectrometry analysis of the new product gave an m/z of 257.07 (Figure S4A), which is also consistent with [M + H]+ for 1,6-dimethoxyphenazine N5-oxide (calculated mass 256.08). The result demonstrates that LaPhzM is a SAM-dependent O-methyltransferase for phenazines. A kinetics study of the enzyme gave a KM of 33.6 µM and kcat of 0.6 min−1, with a Vmax of 2.9 µM min−1, when 6-hydroxy-1-methoxyphenazine N5-oxide (2) was the substrate to produce 1,6-dimethoxyphenazine N5-oxide (4; Figure S3).
Figure 2.
HPLC analysis of the reaction product of LaPhzM with 6-hydroxy-1-methoxyphenazine N5-oxide (2) as substrate. (A) LaPhzM + substrate with SAM. (B) Boiled LaPhzM + substrate with SAM. (C) Substrate 2.
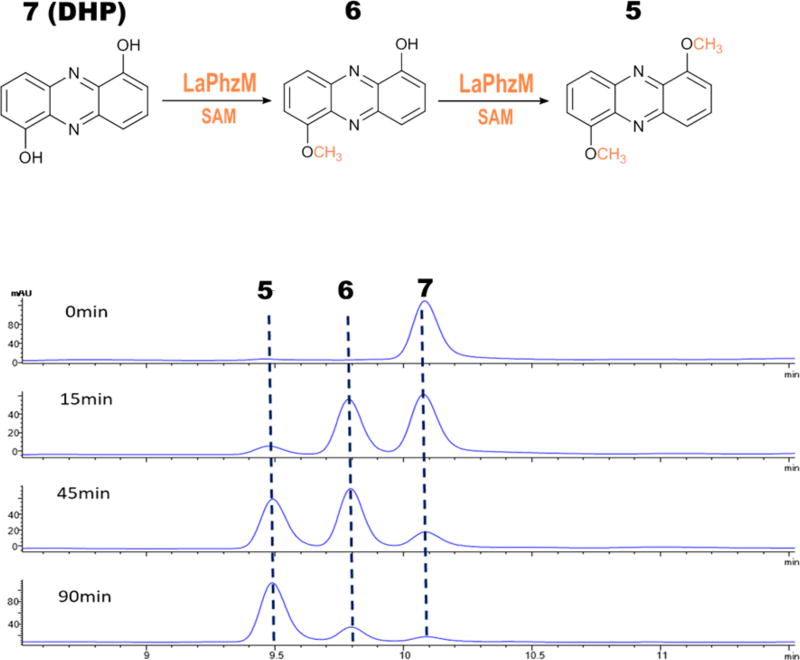
In addition to compounds 2 and 4, strain OH13 produces three other O-methyl containing phenazines, including 1-hydroxy-6-methoxyphenazine (6), 1,6-dimethoxyphenazine (5), and myxin (1).16 To further understand the role and substrate selectivity of LaPhzM, we tested the enzyme’s activity with dihydroxyphenazine as a substrate (Figure 3). 1,6-Dihydroxyphenazine (DHP, 7) gave a retention time of 10.1 min and an m/z of 213.40 for [M + H]+ (Figure 3 and Figure S4B), which is also consistent with the calculated mass of the compound (calculated mass 212.06). After 15 min of reaction, a major peak at 9.8 min appeared. This product comigrated with 1-hydroxy-6-methoxyphenazin (6) and gave an m/z of 227.02 for [M + H]+ (Figure 3 and Figure S4C), which is consistent with the calculated mass of 6 (calculated mass 226.07). As the reaction continued, the peaks for the substrate (7) and the first product (6) decreased and a third peak at 9.5 min became the predominant product in reaction mixture (Figure 3). This peak had the same retention time as 1,6-dimethoxyphenazine (5) and gave an m/z of 241.02 for [M + H]+ (Figure S4D), which is consistent with the calculated mass of 5 (calculated mass 240.09). The results show that LaPhzM is responsible for O-methylation of both the monomethylated and dimethylated phenazines produced by strain OH13.16
Figure 3.
HPLC analysis of reaction products of LaPhzM with 1,6-dihydroxyphenazine (DHP, 7) as substrate. From top to bottom, the HPLC traces are for reactions incubated for 0, 15, 45, and 90 min, respectively.
Additionally, we compared the relative reaction rate of LaPhzM toward different substrates (Figure S5). The enzyme exhibited the highest relative activity when the dihydroxy-containing phenazine DHP (7) was the substrate, followed by monohydroxy-containing phenazine (10), monohydroxy-monomethoxy-containing phenazine (2), and nonphenazine substrates (8-hydroxyquinoline and 6-hydroxyquinoline).
Reconstitution of Myxin Biosynthesis in Vitro
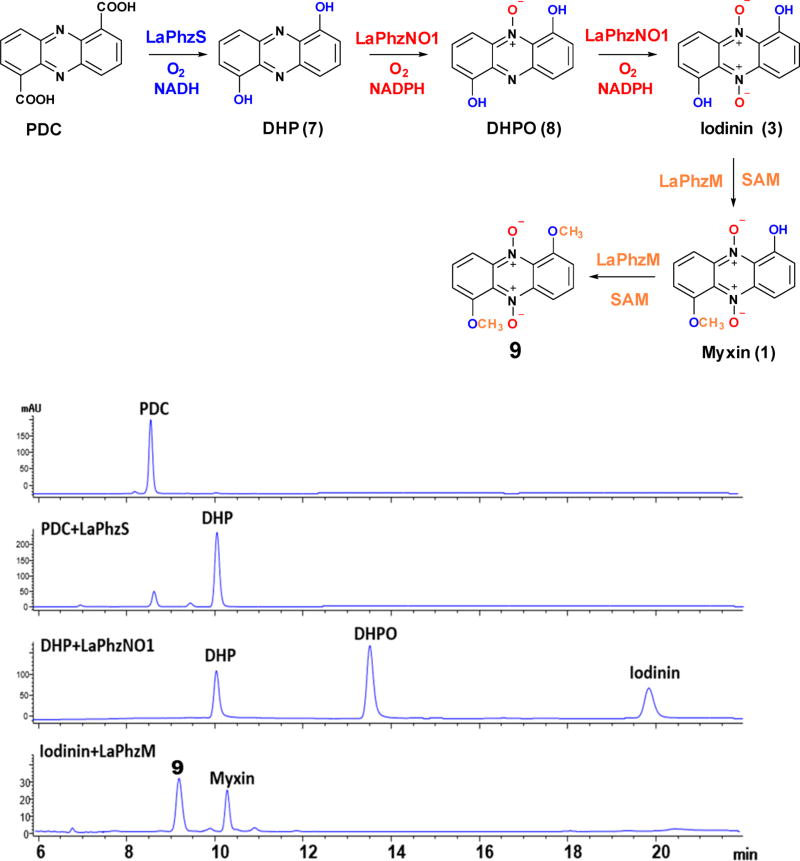
Myxin is perhaps the most interesting phenazine natural product due to its potent antibiotic activity and rich decorations on the tricyclic system. Although the biosynthetic pathway from shikimate to PDC is known, the steps from PDC to myxin had not been completely established. With the heterologous expression and purification of LaPhzM, LaPhzS, and LaPhzNO1, we set out to reconstitute the myxin biosynthetic pathway in vitro. PDC gave a retention time of 8.6 min on HPLC and was converted to 1,6-dihydroxyphenazine (DHP, 7) by LaPhzS in the presence of the cofactors FAD and NADH (Figure 4). DHP ran at 10.0 min and gave an m/z of 213.40 for [M + H]+ (Figure S4B). When the second enzyme LaPhzNO1 and another cofactor NADPH were added in the reaction mixture, two new peaks appeared at 13.5 and 19.8 min, in addition to DHP (Figure 4). The peak at 13.5 min comigrated with 1,6-dihydroxyphenazine N5-mono-oxide (DHPO, 8) on HPLC and gave an m/z of 229.08 for [M + H]+ (Figure S4H), whereas the peak at 19.8 min showed the same retention time as iodinin (1,6-dihydroxyphenazine N5,N10-dioxide).16 Finally, when the third enzyme LaPhzM and the cofactor SAM were added to the reaction mixture, the peaks for PDC, DHP, DHPO, and iodinin all disappeared. Instead, two new peaks at 9.2 and 10.3 min showed up on HPLC (Figure 4). The peak at 10.3 min comigrated with myxin on HPLC and gave an m/z of 259.39 for [M + H]+ (Figure 4 and Figure S4I), whereas the peak at 9.2 min gave an m/z of 273.26 for [M + H]+, which is consistent with 1,6-dimethoxyphenazine N5,N10-dioxide (9; Figure 4 and Figure S4J). This dimethoxy and dioxide phenazine is a new compound that had not been observed in the previous studies of strain OH13.16 Together, the results demonstrate that three enzymes, LaPhzS, LaPhzNO1, and LaPhzM, are sufficient to decorate the shikimate-derived PDC into the long-time used antibiotic myxin. In this pathway, the first step is the double decarboxylative hydroxylation of PDC catalyzed by LaPhzS, which provides substrates for both LaPhzNO1 and LaPhzM. The N-oxidation activity of LaPhzNO1 is relatively selective, as it requires a free hydroxy group at carbon-1 and/or carbon-6 of the phenazine ring.16 In contrast, the O-methylation activity of LaPhzM is relatively flexible. LaPhzM can use nonoxide, mono-oxide, or dioxide phenazines as substrates. This flexibility leads to a variety of phenazine derivatives as found in strain OH13 (Figure 1).
Figure 4.
In vitro reconstitution of myxin biosynthesis. In addition to myxin and other known phenazines, the in vitro reactions produced a new compound (9), a phenazine with dimethoxy and di-N-oxide, which had not been isolated from strain OH13.
Test of LaPhzS and LaPhzM for Pyocyanin Biosynthesis in Vitro
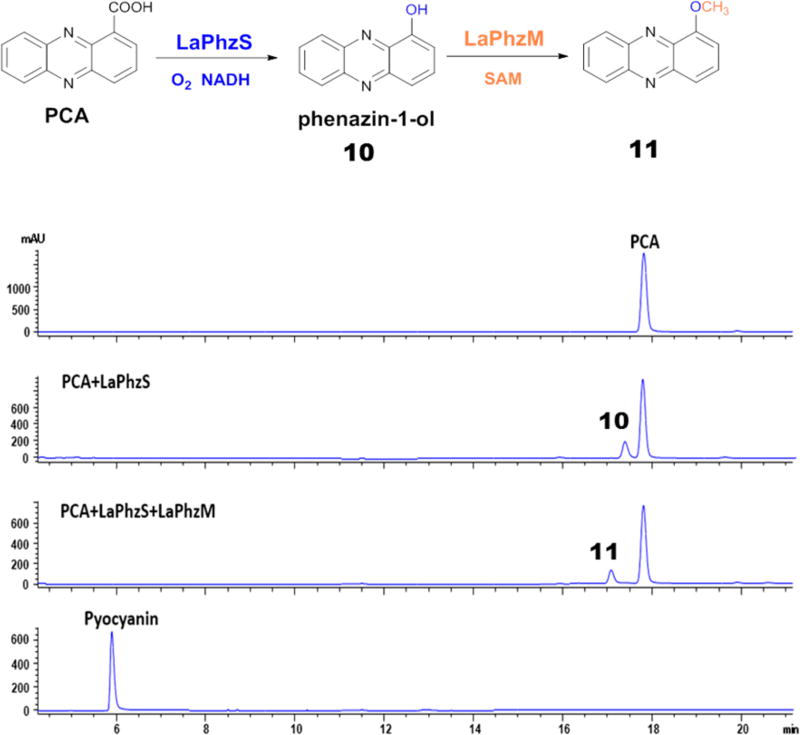
In addition to myxin, pyocyanin is another well-known phenazine natural product that has been subject to many studies.1–4 Pyocyanin contains a methyl group at N5, but no O-methyl group (Figure 1). LaPhzS and LaPhzM for myxin biosynthesis in L. antibioticus are the respective homologues of PhzS and PhzM for pyocyanin biosynthesis in P. aeruginosa. LaPhzS and LaPhzM, together with LaPhzNO1, are the decorating enzymes that convert the dicarboxylic PDC to the antibiotic myxin, whereas PhzS and PhzM are the decorating enzymes that convert the monocarboxylic PCA to the virulence factor pyocyanin.17–20 PhzM catalyzes the N-methylation step in pyocyanin biosynthesis, producing the intermediate N5-methyl-PCA. However, the N-methylated product has not been isolated. This is in contrast to the reactions catalyzed by LaPhzM, which generated a variety of methylated products as described above. One reason could be due to the unstable nature of N5-methyl-PCA, as suggested previously.18 On the other hand, there could be other possibilities. For example, PCA might not be the direct substrate of PhzM; instead, 1-hydroxyphenazine (10), the product of PCA through a decarboxylative hydroxylation catalyzed by PhzS,17 could be the substrate of PhzM. This hypothesis is in agreement with the previous observation that PhzM’s function required the presence of PhzS in the reaction.18 To test if LaPhzM had any N-methylation activity, we tested if the enzymes from strain OH13 were able to convert PCA to pyocyanin. When LaPhzS was incubated with PCA in the presence of FAD and NADH, a small peak at 17.4 min was produced from PCA, which ran at 17.8 min on HPLC (Figure 5). This product gave an m/z of 197.05 for [M + H]+, which is consistent with the mass of 1-hydroxyphenazine (10; Figure S4E). When both LaPhzS and LaPhzM were incubated with PCA in the presence of FAD, NADH, and SAM, another small peak at 17.1 min was produced, while the peak at 17.4 min disappeared on HPLC (Figure 5). This product gave an m/z of 211.03 for [M + H]+ (Figure S4F), which is coincident with the mass of pyocyanin. However, none of the two small peaks comigrated with standard pyocyanin on HPLC, which had a retention time of 5.9 min, although it also gave an m/z of 211.09 for [M + H]+ (Figure 5 and Figure S4G). Therefore, the product at 17.4 min is 1-hydroxyphenazine (10), and the product at 17.1 min is 1-methoxyphenazine (11). The results show that, like PhzS, LaPhzS is able to catalyze the decarboxylative hydroxylation of PCA to produce 1-hydroxyphenazine, but LaPhzM can only convert 1-hydroxyphenazine to 1-methoxyphenazine, rather than 1-hydroxy-N5-methylphenazine (pyocyanin). LaPhzM does not function as an N-methyltransferase when PCA is the substrate. Thus, LaPhzM is an O-methyltransferase, whereas PhzM is an N-methyltransferase. Finally, it should be pointed out that only a small fraction of PCA was converted to products 10 and 11 by LaPhzS and LaPhzM, suggesting that the phenazine monocarboxylic acid is not an optimal substrate for the Lysobacter enzymes.
Figure 5.
HPLC analysis of reaction products of LaPhzS and LaPhzM with PCA as a substrate. Pyocyanin was also included as a reference.
Overview of the Three-Dimensional Structure of LaPhzM
In order to shine light on the structural basis of the enzyme activity and substrate selectivity of LaPhzM, we determined a crystallographic structure of LaPhzM in complex with the cofactor at 1.42 Å, using the mitomycin 7-O-methyltransferase MmcR from Streptomyces lavendulae as an initial search model for phasing.23 An overview of the LaPhzM structure is shown in Figure S6. Similar to many other O-methyltransferases, LaPhzM forms a dimeric structure with a dimerization domain in the N terminus and a catalytic domain with the α/β Rossmann fold in the C terminus of each monomer. The overall fold of LaPhzM mirrors that of MmcR despite the low sequence identity between the two proteins (Figure S7A).23 All the secondary structural elements are also conserved in PhzM (Figure S7B) with a larger structural variation in comparison to that of MmcR.
The Active Site
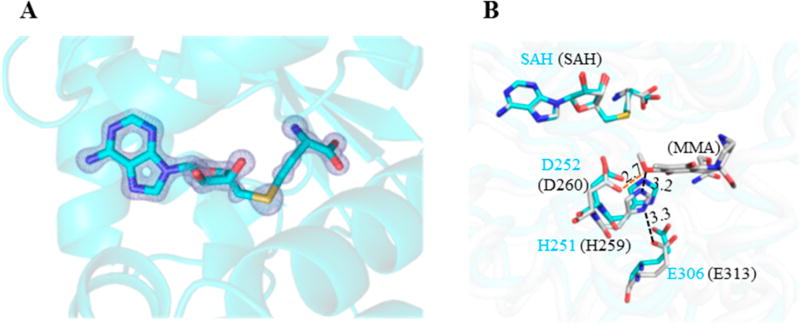
The electron density around the cofactor binding site clearly indicates an S-adenosyl-l-homocysteine (SAH), instead of a SAM, in the LaPhzM crystal structure (Figure 6A). Since SAM was incubated with LaPhzM during crystallization, this observation indicates that LaPhzM is capable of catalyzing the demethylation of SAM in the absence of the phenazine substrate. The surrounding environment at the cofactor binding site of LaPhzM closely resembles that of MmcR and other similar SAM-dependent O-methyltransferases (Figure S8).23, 24 The proteinaceous environment at the active site of LaPhzM possesses the structural characteristics for the general acid/base-mediated transmethylation of class I O-methyltransferases.24 A 3D structural alignment shows that the His251-Glu306 pair takes similar geometric arrangement as their corresponding residues (His259 and Glu313, respectively) in MmcR (Figure 6B), which have been suggested to form a hydrogen bond network and be involved in deprotonating the hydroxyl group of the substrate.23 Interestingly, Asp252 (Asp260 in MmcR) adjacent to His251 and SAH is also within the hydrogen bonding distance with the substrate near the active site. Sequence alignments indicate that His251, Asp252, and Glu306 are highly conserved in the LaPhzM homologues and other similar O-methyltransferases. It remains to be tested whether Asp252 plays a crucial role in positioning the substrate and/or proton shuttling during the catalytic process.24
Figure 6.
Active site of the LaPhzM. (A) The 2Fo–Fc electron density map around the cofactor binding site. The map is illustrated in light blue color and contoured at 3σ. The cofactor product S-adenosyl-l-homocysteine (SAH) is represented by sticks. (B) The stick representation of the active site in LaPhzM (cyan carbon atoms) overlaid with the MmcR-SAH-mitomycin A (MMA) complex (PDB ID: 3GXO, gray carbon atoms). The residues in LaPhzM are labeled in cyan fonts, and the corresponding ones in MmcR are labeled in black fonts in the round bracket. The nitrogen atoms are in blue, oxygen in red, and sulfur in yellow.
Modeling of the LaPhzM-Substrate Complex
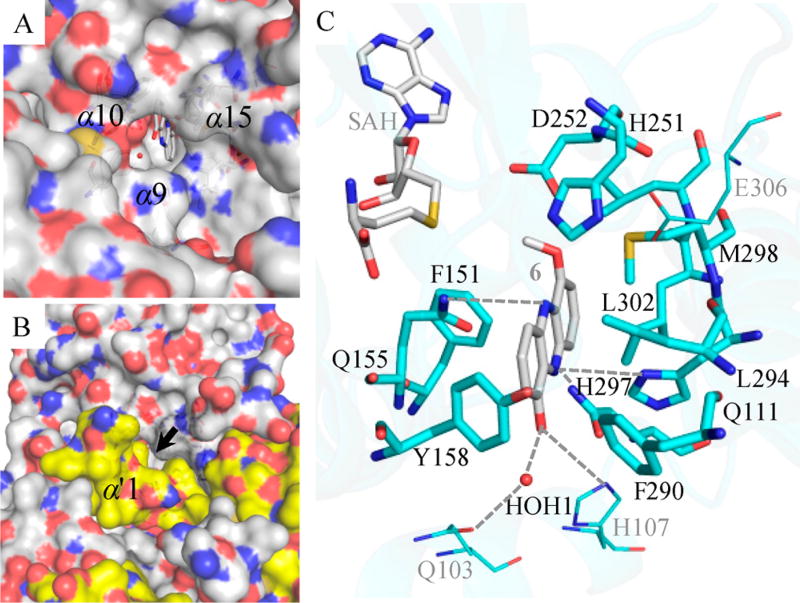
Although the LaPhzM structure determined in this study is in the substrate-free form, cavity analysis by CAVER and structural comparison with the MmcR-SAH-mitomycin A complex (PDB ID: 3GXO) suggest that the characterized LaPhzM-SAH complex is in an open state for substrate binding.23, 25 Indeed, 1-hydroxy-6-methoxyphenazine (6), the intermediate methylated product from DHP, can be fit well into the substrate binding pocket of LaPhzM by ligand docking (Figure 7). In this model, the substrate entry is surrounded by helices α9, α10, and α15 in one monomer and is capped by helix α′1 from the second monomer, which leaves a narrow channel for substrate entry (Figure 7A and B). At the substrate binding site, 6 is surrounded by 11 residues in a largely hydrophobic environment (Figure 7C). Particularly, two aromatic residues Tyr158 and Phe290 located in helices α10 and α15, respectively, are at the surface of LaPhzM facing the solvent environment, presumably serving as the gate of the substrate entry. These two residues are conserved only in the close homologues of LaPhzM with high sequence identity and likely play an important role in defining the substrate preference of LaPhzM. Tyr158 and Phe290 are absent in MmcR, which leaves a wider opening gate to accommodate the large polar tail of mitomysin A. Apart from the hydrophobic and van der Waals interactions, and the hydrogen bond network involving the three residues mentioned above at the active site, a few polar residues including Gln103, His107, Gln155, and His297 may also form hydrogen bonds with the 1-hydroxyl group, N5, and N10, respectively, in DHP and thus prepare the substrate for the optimal orientation for transmethylation (Figure 7).
Figure 7.
Substrate binding pocket of LaPhzM with 6 fit into the structure by autodocking. (A and B) Surface representations of LaPhzM viewed from the protein–solvent interface toward the substrate binding pocket with and without the second monomer of the dimer, respectively. The black arrow indicates the proposed narrow substrate entry tunnel between helices α9, α10, and α15 from one monomer and helix α′1 from the second monomer. The surface presentation is colored by the underlying atoms with nitrogen in blue and oxygen in red. The surface around carbon atoms is colored white for one monomer and yellow for the second monomer for clarity. The local environment at the substrate binding pocket is shown in C. The carbon atoms are colored cyan for the amino acid residues and gray for SAH and 6. The water molecule is shown in the red sphere. SAH, 6, and the amino acid residues within 4 Å of 6 are represented by sticks.
Overall, the local environment at the substrate binding pocket of LaPhzM strongly suggests that proximity may also be an important mechanism for determining the enzymatic activity and phenazine selectivity as observed in the enzymatic activity assay. The higher activity of LaPhzM to 1,6-dihydroxyphenazine (DHP) in comparison to 1-hydroxyphenazine is likely due to the hydrogen bonding interactions between one hydroxy group of DHP and Glu103/His107 that facilitate the orientation of the second hydroxy optimal for methyl transfer (Figure 7). Among the three phenazine substrates tested, LaPhzM shows the lowest activity to 6-hydroxy-1-methoxyphenazine N5-oxide (2; Figure S5). The proton in the 6-hydroxyl group of 2 has been proposed to form a stable hydrogen bond with oxidized N5 in a pseudo-six-member ring (Figure S9).12 The presence of an intramolecular hydrogen bond network in 2 may reduce the efficiency of deprotonation on the 6-hydroxyl group mediated by a hydrogen bonding with His251 and thus reduce the transmethylation rate.
Structural Comparison of the Substrate Binding Pocket in LaPhzM and PhzM
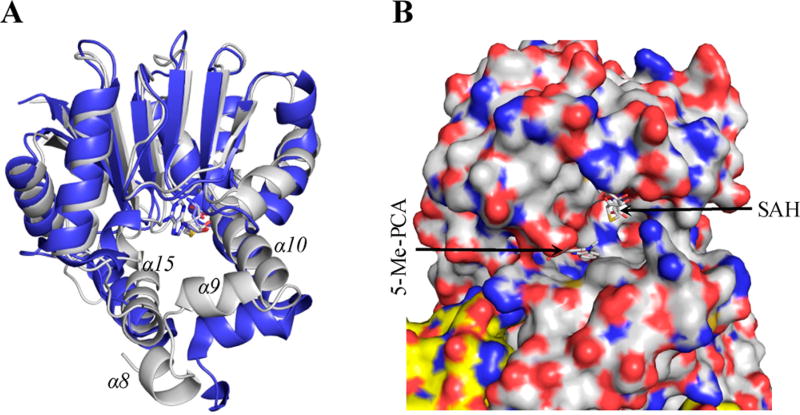
The absence of the cofactor and substrate in the reported PhzM structure makes it difficult for a direct comparison of the substrate binding pocket between the two structures. Nonetheless, SAH can be modeled into the active site of PhzM using its homologue IOMT (PDB ID: 1FP2) as a model, as demonstrated previously owing to the high structural similarity in this region.18 When overlaying the two structures around the SAH binding site, the most noticeable difference is the solvent accessibility at the substrate binding pocket. The helices α9, α10, and α15 in PhzM are further away from the active site (up to 7 Å relative to the corresponding Cα atoms in LaPhzM). In addition, different from LaPhzM, helix α′1 in PhzM does not cover the opening of the active site. These differences in the local structural arrangement result in a much more solvent exposed active site in PhzM (Figure 8), which is not optimal for SAM and/or substrate binding and the reaction of transmethylation. These observations may explain the difficulties in cocrystallizing PhzM with a SAM/SAH.18
Figure 8.
Comparison of the substrate binding pocket of PhzM (PDB ID: 2IP2) and LaPhzM. (A) A ribbon presentation of PhzM (blue) overlaid with LaPhzM (light gray). Only the residues from AA121 to the end of the C terminus are included for clarity. (B) A surface presentation around the active site of PhzM viewed from the protein–solvent interface. The surface presentation is colored by the underlying atoms with nitrogen in blue and oxygen in red. The surface around carbon atoms is colored white for one monomer and yellow for the second monomer for clarity. SAH was modeled to the active site of PhzM guided by its homologue IOMT (PDB ID: 1FP2); 5-Me-PCA (N5-methylphenazine carboxylic acid) was manually placed near SAH for the purpose of demonstration.
As discussed above in pyocyanin biosynthesis, it was proposed that failing to detect the product when incubating PhzM with SAM and the substrate PCA in the absence of PhzS was probably due to the instability of the product N5-methyl-PCA.18 However, no SAM consumption was reported either in the study, while this would be expected if the transmethylation did occur. The results from the structural comparison between PhzM and LaPhzM provide structural insights in supporting that the alternative explanation may exist, since a free PhzM as in the reported structure is not optimal for catalyzing the transmethylation reaction. The interaction of PhzM with PhzS may be required for desolvation of the active site, which is necessary for the stable cofactor/substrate binding and efficient transmethylation. By interacting with PhzM, PhzS could either cap the substrate binding pocket directly by itself, or trigger conformational changes in PhzM and lead to a similar structural arrangement at the substrate binding pocket as observed in LaPhzM.
In conclusion, we have characterized the LaPhzM gene from strain OH13 and demonstrated that LaPhzM is a SAM-dependent O-methyltransferase responsible for all methoxy formation in phenazine natural products isolated from strain OH13, including the well-known antibiotic myxin. For the first time, we demonstrated the biosynthesis of myxin from PDC, one of the two common precursors for all phenanzine natural products, through in vitro reconstitution of the ring-decorating steps using purified enzymes. Finally, we determined the cofactor-bound X-ray crystal structure of LaPhzM at 1.42 Å resolution, which provides molecular insights into the catalytic activity and O-methyl selectivity of the enzyme. These findings will facilitate future efforts to improve myxin production in strain OH13 through metabolic pathway engineering, and the identification of the phenazine-decorating enzymes will find uses in chemoenzymatic syntheses of new phenazine antibiotics.
METHODS
Cloning and Expression of LaPhzM in E. coli
LaPhzM was amplified from the genomic DNA of strain OH13 (CGMCC No.7561), using primers LaPhzM-PF/LaPhzM-PR (Table S1). The product was cloned into the EcoRI/HindIII sites in plasmid pET28a (Novagen) and confirmed by DNA sequencing. This construct was introduced into E. coli BL21(DE3) for expression. LaPhzM was purified using a Ni-NTA column (GE Life Science) and a size exclusion column (Sephadex 200; Figure S2). A typical stock solution contained LaPhzM of 30 mg mL−1, in 50 mM Tris buffer at pH 8, 200 mM NaCl, and 1 mM DTT and was stored at −80 °C. In the protein preparation for LaPhzM cyrstalization, the thrombin cleavage site was removed from the LaPhzM expression construct.
Enzyme Activity Assays
Compound 2 was prepared from strain OH13 culture.16 PDC and PCA were purchased from Alligator Reagent Inc., China, and pyocyanin from Sigma-Aldrich. DHP (7) was purified by HPLC from the reaction mixture of PDC and LaPhzS.16 The LaPhzS reaction was conducted in a 100 µL mixture containing LaPhzS (1 µM), the substrate (PDC or PCA, 100 µM), NADH (100 µM), and FAD (100 µM) in a phosphate buffer (KH2PO4/K2HPO4, 100 mM, pH 7.0). The reaction mixture was incubated at RT for 30 min. The reaction was quenched with an equal volume of ethyl acetate, and the organic phase containing the product was collected and evaporated to dryness. The residues were dissolved in methanol (100 µL), and the supernatant was obtained by centrifugation at 13 200g for 10 min. A fraction (20 µL) of the supernatant was subjected to reversed-phase HPLC analysis or LC-MS as described previously.16 The LaPhzNO1-catalyzed N-oxidation was carried out in the same conditions as that for LaPhzS, except NADH was replaced by NADPH (100 µM). The LaPhzM-catalyzed methyltransfer reaction was conducted in a 100 µL mixture containing LaPhzM (5 µM), substrate (50 µM), and SAM (180 µM), in a Tris buffer (50 mM, pH 7.8). The mixture was incubated at RT for 30 min, and the reaction was stopped by adding ethyl acetate. To quantify the selectivity, kinetics analysis was performed with compound 2 and SAM. Initial velocity of the reaction was measured at a variety of substrate concentrations. The reaction was quantified by measuring the SAM product, SAH, using reversed-phase HPLC (254 nm). A calibration curve of SAH was prepared prior to the enzyme assays. These data were globally fitted to the Michaelis–Menten equation, yielding KM values. In the reactions to compare various potential substrates for LaPhzM, the relative conversion rate was calculated based on the conversion rate of compound 2 to compound 4, which was set as 100%. The relative rate of other substrates converted by LaPhzM was expressed as the percent of that of compound 2 converted to compound 4.
In reactions with PCA as the substrate to test the activity of LaPhzS and LaPhzM in pyocyanin biosynthesis in vitro, the procedures were the same as above, but with the following modifications for HPLC analysis. A sample (20 µL) was injected into a reversed-phase column (Cosmosil, 4.6 ID × 250 mm) in an Agilent HPLC (1220 Infinity LC). Water/0.05% formic acid (solvent A) and methanol 0.05% formic acid (solvent B) were used as the mobile phases with a flow rate of 1.0 mL/min. The HPLC program was as follows: 5–25% B in the first 5 min, 25–80% B in 5–25 min, 100% B at 26–35 min, back to 5% B at 36 min, and maintained up to 40 min. The phenazines were detected at 254 nm on a UV detector. LCQ-MS was used to verify the mass of the peak of PCA and products.
Crystallization
SAM (Biolab Inc.) was added to the purified LaPhzM at a molar ratio of 1.5:1 before concentrating the protein to 20 mg mL−1. LaPhzM was crystallized by sitting-drop vapor diffusion at RT (~295 K). The protein solution at 20 mg mL−1 was mixed in 1:1 volume ratio with the reservoir solution containing 0.2 M ammonium formate and 18–22% w/v PEG 3350. Crystals typically reached the maximum size in about 1 week. Santovac 5 Cryo Oil (Hampton Research) was used as the cryoprotectant when harvesting the crystal.
X-Ray Diffraction Data Collection, Processing, and Refinement
X-ray diffraction data were collected on Beamline 9–2 at the Stanford Synchrotron Radiation Lightsource (SSRL), with a 6 M Pixel Array Detector. A set of 1200 diffraction images was collected at 12 658 eV with an oscillation angle of 0.15°. Diffraction data were integrated with the XDS program package26 and subsequently processed using programs of the CCP4 suite.27 The phase was determined by molecular replacement using the structure of the mitomycin 7-O-methyltransferase MmcR from Streptomyces lavendulae (PDB code 3gwz, 34% of sequence identity)23 as the search model and employing the BALBES automated molecular replacement pipeline server.28 Further refinement and structural validation were carried out by using REFMAC5, COOT, and MolProbity.29–31 The diffraction data and refinement statistics are listed in Tables S2 and S3, respectively. All but the 6×His tag and the first nine residues in the N terminus of the native protein have been included in the model. The programs COOT and PyMOL (the PyMOL Molecular Graphics System, version 1.7.1 Schrödinger) were used for structural analysis and for generating figures. The atomic coordinates and experimental data (code 6C5B) have been deposited in the Protein Data Bank (www.wwpdb.org).
Molecular Docking
The prediction of LaPhzM–substrate binding was performed with Autodock Vina employing default parameters using a grid with dimensions of 26 × 20 × 20 Å centered at the substrate binding cavity.32 Water molecules were removed from the LaPhzM structure in preparation of docking. The coordinates for methylated phenazines were modified from the structures downloaded from the PubChem database. The Gasteiger charges and hydrogens were added to the receptor structure and phenazines with AutoDocktools.33 The receptor structure was treated as a rigid body during docking analysis to exclude the uncertainty caused by the structural flexibilities.
Supplementary Material
Acknowledgments
This work was supported in part by the National Institutes of Health (R01AI097260), UNL Redox Biology Center, a Faculty Seed Grant, and a Revision Award from UNL. We thank Y. Zhao for helpful discussion at the beginning of this project, R. Cerny and K. Wulser for technical assistance, and H. Li for the insightful discussion on the charge distribution for molecular docking.
Footnotes
ASSOCIATED CONTENT
- Details of primer sequences, statistics for data processing and refinement, multiple amino acid sequence alignment, SDS-PAGE of proteins, enzyme kinetics, protein secondary structural elements, models of cofactor and substrate binding environments in protein structures, and spectroscopic data for identification of phenazines (PDF)
Author Contributions
L.D. and L.Z. conceived and designed experiments. J.J., S.W., and D.B. carried out experiments. L.D., L.Z., J.J., and A.S. analyzed data. J.J., L.D., and L.Z. wrote the manuscript.
The authors declare no competing financial interest.
References
- 1.Laursen JB, Nielsen J. Phenazine natural products: biosynthesis, synthetic analogues, and biological activity. Chem. Rev. 2004;104:1663–1686. doi: 10.1021/cr020473j. [DOI] [PubMed] [Google Scholar]
- 2.Cimmino A, Evidente A, Mathieu V, Andolfi A, Lefranc F, Kornienko A, Kiss R. Phenazines and cancer. Nat. Prod. Rep. 2012;29:487–501. doi: 10.1039/c2np00079b. [DOI] [PubMed] [Google Scholar]
- 3.Mavrodi DV, Blankenfeldt W, Thomashow LS. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu. Rev. Phytopathol. 2006;44:417–445. doi: 10.1146/annurev.phyto.44.013106.145710. [DOI] [PubMed] [Google Scholar]
- 4.Mavrodi DV, Peever TL, Mavrodi OV, Parejko JA, Raaijmakers JM, Lemanceau P, Mazurier S, Heide L, Blankenfeldt W, Weller DM, Thomashow LS. Diversity and evolution of the phenazine biosynthesis pathway. Appl. Environ. Microbiol. 2010;76:866–879. doi: 10.1128/AEM.02009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pirnay JP, De Vos D, Cochez C, Bilocq F, Vanderkelen A, Zizi M, Ghysels B, Cornelis P. Pseudomonas aeruginosa displays an epidemic population structure. Environ. Microbiol. 2002;4:898–911. doi: 10.1046/j.1462-2920.2002.00321.x. [DOI] [PubMed] [Google Scholar]
- 6.Selezska K, Kazmierczak M, Musken M, Garbe J, Schobert M, Haussler S, Wiehlmann L, Rohde C, Sikorski J. Pseudomonas aeruginosa population structure revisited under environmental focus: impact of water quality and phage pressure. Environ. Microbiol. 2012;14:1952–1967. doi: 10.1111/j.1462-2920.2012.02719.x. [DOI] [PubMed] [Google Scholar]
- 7.Price-Whelan A, Dietrich LE, Newman DK. Rethinking ’secondary’ metabolism: physiological roles for phenazine antibiotics. Nat. Chem. Biol. 2006;2:71–78. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- 8.Dietrich LE, Price-Whelan A, Petersen A, Whiteley M, Newman DK. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Mol. Microbiol. 2006;61:1308–1321. doi: 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Wilks JC, Danhorn T, Ramos I, Croal L, Newman DK. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J. Bacteriol. 2011;193:3606–3617. doi: 10.1128/JB.00396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glasser NR, Kern SE, Newman DK. Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol. Microbiol. 2014;92:399–412. doi: 10.1111/mmi.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dietrich LE, Teal TK, Price-Whelan A, Newman DK. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science. 2008;321:1203–1206. doi: 10.1126/science.1160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chowdhury G, Sarkar U, Pullen S, Wilson WR, Rajapakse A, Fuchs-Knotts T, Gates KS. DNA strand cleavage by the phenazine di-N-oxide natural product myxin under both aerobic and anaerobic conditions. Chem. Res. Toxicol. 2012;25:197–206. doi: 10.1021/tx2004213. [DOI] [PubMed] [Google Scholar]
- 13.Costa KC, Glasser NR, Conway SJ, Newman DK. Pyocyanin degradation by a tautomerizing demethylase inhibits Pseudomonas aeruginosa biofilms. Science. 2017;355:170–173. doi: 10.1126/science.aag3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pierson LS, 3rd, Gaffney T, Lam S, Gong F. Molecular analysis of genes encoding phenazine biosynthesis in the biological control bacterium Pseudomonas aureofaciens 30–84. FEMS Microbiol. Lett. 1995;134:299–307. doi: 10.1111/j.1574-6968.1995.tb07954.x. [DOI] [PubMed] [Google Scholar]
- 15.Mavrodi DV, Ksenzenko VN, Bonsall RF, Cook RJ, Boronin AM, Thomashow LS. A seven-gene locus for synthesis of phenazine-1-carboxylic acid by Pseudomonas fluorescens 2–79. J. Bacteriol. 1998;180:2541–2548. doi: 10.1128/jb.180.9.2541-2548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Y, Qian G, Ye Y, Wright S, Chen H, Shen Y, Liu F, Du L. Heterocyclic aromatic N-oxidation in the biosynthesis of phenazine antibiotics from Lysobacter antibioticus. Org. Lett. 2016;18:2495–2498. doi: 10.1021/acs.orglett.6b01089. [DOI] [PubMed] [Google Scholar]
- 17.Greenhagen BT, Shi K, Robinson H, Gamage S, Bera AK, Ladner JE, Parsons JF. Crystal structure of the pyocyanin biosynthetic protein PhzS. Biochemistry. 2008;47:5281–5289. doi: 10.1021/bi702480t. [DOI] [PubMed] [Google Scholar]
- 18.Parsons JF, Greenhagen BT, Shi K, Calabrese K, Robinson H, Ladner JE. Structural and functional analysis of the pyocyanin biosynthetic protein PhzM from Pseudomonas aeruginosa. Biochemistry. 2007;46:1821–1828. doi: 10.1021/bi6024403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gohain N, Thomashow LS, Mavrodi DV, Blankenfeldt W. The purification, crystallization and preliminary structural characterization of FAD-dependent monooxygenase PhzS, a phenazine-modifying enzyme from Pseudomonas aeruginosa. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 2006;62:989–992. doi: 10.1107/S1744309106034464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gohain N, Thomashow LS, Mavrodi DV, Blankenfeldt W. The purification, crystallization and preliminary structural characterization of PhzM, a phenazine-modifying methyltransferase from Pseudomonas aeruginosa. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 2006;62:887–890. doi: 10.1107/S1744309106029149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen B, Hutchinson CR. Tetracenomycin F1 monooxygenase: oxidation of a naphthacenone to a naphthacenequinone in the biosynthesis of tetracenomycin C in Streptomyces glaucescens. Biochemistry. 1993;32:6656–6663. doi: 10.1021/bi00077a019. [DOI] [PubMed] [Google Scholar]
- 22.Kendrew SG, Hopwood DA, Marsh EN. Identification of a monooxygenase from Streptomyces coelicolor A3(2) involved in biosynthesis of actinorhodin: purification and characterization of the recombinant enzyme. J. Bacteriol. 1997;179:4305–4310. doi: 10.1128/jb.179.13.4305-4310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh S, Chang A, Goff RD, Bingman CA, Gruschow S, Sherman DH, Phillips GN, Jr, Thorson JS. Structural characterization of the mitomycin 7-O-methyltransferase. Proteins: Struct., Funct., Genet. 2011;79:2181–2188. doi: 10.1002/prot.23040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liscombe DK, Louie GV, Noel JP. Architectures, mechanisms and molecular evolution of natural product methyltransferases. Nat. Prod. Rep. 2012;29:1238–1250. doi: 10.1039/c2np20029e. [DOI] [PubMed] [Google Scholar]
- 25.Chovancova E, Pavelka A, Benes P, Strnad O, Brezovsky J, Kozlikova B, Gora A, Sustr V, Klvana M, Medek P, Biedermannova L, Sochor J, Damborsky J. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput. Biol. 2012;8:e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabsch W. XDS. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 28.Long F, Vagin AA, Young P, Murshudov GN. BALBES: a molecular-replacement pipeline. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. COOT: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.