Abstract
Digital diffusion tensor imaging (DTI) templates of the adult human brain are commonly used in neuroimaging research, and their characteristics influence the accuracy of the application. However, a systematic evaluation of the characteristics and performance of standardized and study-specific DTI templates has not been conducted. The purpose of this work was to compare eight available standardized DTI templates to each other (ICBM81, ENIGMA, FMRIB58, SRI24, IIT2, NTU-DSI-122-DTI, IIT v.3.0, Eve), as well as to study-specific templates, in terms of template characteristics (image sharpness, ability to identify small brain structures, artifacts, mean values, noise properties) and performance in spatial normalization and detection of small inter-group FA differences. The IIT v.3.0 template was shown to combine a number of desirable characteristics: includes full-tensor information, is population-based, has high image sharpness, shows no visible artifacts, has low noise levels, has diffusion tensor properties and spatial features representative of data from the average individual adult brain. Furthermore, the IIT v.3.0 template was shown to allow higher inter-subject DTI spatial normalization accuracy, and detection of smaller inter-group FA differences, compared to all other templates, including study-specific templates. These findings were consistent when evaluating the templates in younger as well as older adult cohorts.
Keywords: Diffusion, DTI, MRI, template, atlas, comparison
1. Introduction
Digital diffusion tensor imaging (DTI) templates of the adult human brain are indispensable neuroimaging tools. The most common applications of DTI templates are in voxel-wise analyses, where they are used as references for inter-subject spatial normalization (Zhang and Arfanakis, 2013)(Smith et al., 2006), and in region of interest analyses, where they are used for conventional or skeletonized atlas-based segmentation (Zhang and Arfanakis, 2014). DTI templates are also used in automated seed selection for fiber-tracking (Y. Zhang et al., 2010)(Nucifora et al., 2012)(Suarez et al., 2012), as spatial maps forming the basis of brain atlases (Peng et al., 2009)(Mori et al., 2008), and as standards for algorithm evaluation (Shi et al., 2013)(Zhou et al., 2015). The fundamental assumption in all the above applications is that the characteristics of the DTI template are representative of those of DTI data from the average individual adult brain. The degree to which this is true influences the success of the application (Van Hecke et al., 2011)(Zhang and Arfanakis, 2013).
The adult DTI templates that have been presented and used in the literature include A) both single-person and population-based templates (referring to the number of individuals used in their construction), as well as B) templates that contain full tensor information and others containing only tensor-derived scalar quantities, such as fractional anisotropy (FA) and mean diffusivity. A) In theory, a notable drawback of a single-person template is that a single brain’s characteristics may not be representative of the general population, and therefore, such a template may introduce a bias (Joshi et al., 2004). This limitation is also true, though to a lesser degree, for a population-based DTI template constructed with a relatively small number of participants. As the number of participants increases, a population-based DTI template becomes more representative of the average characteristics of the general population. Specifically, it has been demonstrated that population-based DTI templates constructed using a bootstrap approach and a small number of participants are characterized by highly variable diffusion characteristics, and that this variability decreases asymptotically as the number of participants increases, with more than approximately 60 persons resulting to a highly stable template (S. Zhang et al., 2010). B) Templates that contain full tensor information clearly have additional functionality compared to those containing only tensor-derived scalar quantities. Furthermore, in theory, tensor-based registration to a template containing full tensor information allows more accurate inter-subject spatial normalization of DTI data than registration to a template of a scalar quantity (e.g. FA) (Alexander and Gee, 2000)(Park et al., 2003). Among population-based full tensor templates of the adult human brain, those that have spatial features and diffusion characteristics most similar to those of the DTI data from individual adults should theoretically lead to highest accuracy in various template-dependent applications (Zhang et al., 2011).
Adult DTI templates can be divided into two main categories: study-specific (Van Hecke et al., 2011)(Zhang et al., 2007) and standardized (Mori et al., 2008)(Peng et al., 2009). The former are constructed in each study separately based on the data collected specifically for that study, and the latter are constructed once and used in multiple investigations. Study-specific DTI templates can only be used as references for inter-subject spatial normalization for the purposes of voxel-wise analyses, since they typically lack complementary resources, such as labels and other features of an atlas, which limits their functionality (Van Hecke et al., 2011). Nonetheless, study-specific templates are in theory most representative of the data under investigation and are thought to result in more accurate spatial normalization than standardized templates (Van Hecke et al., 2011). However, when the requirements for building an optimal study-specific template are not satisfied due to various limitations, the resulting study-specific template is suboptimal and may not be representative of the individual data under investigation, thus leading to low spatial normalization accuracy (Van Hecke et al., 2011). Additionally, differences across study-specific templates complicate integration of results across studies. In contrast, a high-quality, demographically representative, standardized DTI template may allow consistently high accuracy across studies, minimize inefficiencies and complexities associated with template construction, enable a variety of applications (when it is part of a comprehensive atlas), and facilitate integration of findings across modalities and across studies. Several standardized DTI templates have become available over the last decade (Mori et al., 2008)(Peng et al., 2009)(Oishi et al., 2009)(Rohlfing et al., 2010)(Zhang et al., 2011)(Jahanshad et al., 2013)(Hsu et al., 2015) and have been used extensively. However, a systematic evaluation of the characteristics and performance of available standardized and study-specific DTI templates has not been conducted. Only isolated pair-wise comparisons of standardized templates have been presented in (Peng et al., 2009)(Zhang et al., 2011)(Hsu et al., 2015), and a comparison of a study-specific template to a single standardized template has been presented in (Van Hecke et al., 2011). This is a critical gap in literature, as template selection may have important implications in the accuracy of DTI investigations (Van Hecke et al., 2011).
The purpose of this work was to compare available standardized DTI templates to each other, as well as to study-specific templates, in terms of template characteristics and performance. Eight standardized DTI templates and two study-specific templates constructed based on publicly available data on younger and older adults, were compared in terms of image sharpness, ability to identify small brain structures, artifacts, mean values, and noise properties. Since DTI templates most commonly serve as references for spatial normalization, all templates were also compared in terms of the accuracy of spatial matching achieved when they are used as references for normalization of younger and older adults, separately. Finally, power analysis was conducted to assess the impact of differences in spatial normalization accuracy across templates on the ability to detect small inter-group FA differences in younger and older adults, separately.
2. Materials and Methods
2.1 MRI Data
Two brain MRI datasets were used in this work. All participants provided written informed consent in accordance with procedures approved by the local institutional committees for the protection of human subjects.
Dataset 1 included DTI and T1-weighted data from 72 healthy young and middle-aged adults (22 male, 31 ± 13 years of age, 18 – 59 years of age) (Appendix 1), obtained from the Enhanced Nathan Kline Institute Rockland Sample (http://fcon_1000.projects.nitrc.org/indi/enhanced/sharing.html) and collected on a 3 T Siemens MRI scanner (Erlangen, Germany). The DTI data were acquired using a single-shot spin-echo echo-planar diffusion imaging sequence with the following imaging parameters: TR = 2,400 ms, TE = 85 ms, field-of-view 21.2 cm × 18 cm, 2 mm slice thickness, 64 axial slices, 106 × 90 image matrix, b = 1,500 s/mm2 for 128 diffusion gradient directions, nine b = 0 s/mm2 volumes, and a multi-band acceleration factor of 4. The T1-weighted data were acquired using an MPRAGE sequence with the following imaging parameters: TR = 1,900 ms, TE = 2.52 ms, preparation time = 900 ms, flip angle 9 degrees, field of view 25 cm × 25 cm, 176 sagittal slices, 1 mm slice thickness, 256 × 256 image matrix, and an acceleration factor of 2.
Dataset 2 included DTI and T1-weighted data from 72 healthy older adults (17 male, 72 ± 5 years of age, 65 - 85 years of age) (Appendix 1), also obtained from the Enhanced Nathan Kline Institute Rockland Sample and collected with the same protocol as Dataset 1.
2.2 Pre-processing
For the DTI data of both Datasets 1 and 2, the brain was extracted from the raw DTI data, and corrections for eddy-current distortions as well as bulk motion were accomplished by affine registration to the first volume with no diffusion weighting (b=0 sec/mm2). Distortions due to magnetic field non-uniformities were corrected by non-linear registration to the corresponding T1-weighted MPRAGE data. The b-matrix was reoriented and the diffusion tensor was estimated in each brain voxel. Maps of FA and trace were generated from the diffusion tensors. All DTI data pre-processing steps were accomplished using TORTOISE (www.tortoisedti.org) (Pierpaoli et al., 2010).
2.3 Comparison of Eight Standardized and Two Study-Specific DTI Brain Templates
Eight standardized DTI templates of the adult human brain and two study-specific templates were compared as described in the following sections. The two study-specific diffusion tensor templates were constructed based on Datasets 1 and 2, separately, and are referred to here as SS-y (for younger) and SS-o (for older), respectively. The template construction process followed the popular DTI-TK approach recommended in: http://dti-tk.sourceforge.net/pmwiki/pmwiki.php?n=Documentation.Registration. The eight standardized templates included five providing full tensor information: IIT v.3.0 (based on 72 persons, 20–40 years of age) (Appendix 2 provides details on the construction of this template) (Varentsova et al., 2014), IIT2 (67 persons, 20–40 years of age) (Zhang et al., 2011), ICBM81 (81 persons, 18–59 years of age) (Mori et al., 2008), Eve (1 person, 32 years of age) (Oishi et al., 2009), and NTU-DSI-122-DTI (122 persons, 19–40 years of age) (Hsu et al., 2015); and three providing only scalar quantities derived from the diffusion tensor: ENIGMA (400 persons, 18–85 years of age) (Jahanshad et al., 2013), FMRIB58 (58 persons, 20–50 years of age) (FMRIB, Oxford, UK), and SRI24 (24 persons, 19–84 years of age) (Rohlfing et al., 2010). These standardized templates were selected because they are publicly available and commonly used in neuroimaging research.
2.3.1 Comparison of Template Characteristics
First, FA maps of all templates were compared in terms of image sharpness assessed by means of the normalized power spectra for the anterior-posterior, left-right, and inferior-superior axes, separately (Zhang et al., 2011). FA maps were also compared in terms of the user’s ability to identify certain small brain structures based on visual inspection. Furthermore, FA maps of all templates were evaluated with regard to image artifacts content.
FA values for selected white matter regions of interest (ROI) were extracted and compared across templates. The ROIs were located in the genu and splenium of the corpus callosum, forceps minor, forceps major, superior cingulum, anterior and posterior limb of the internal capsule, external capsule, parahippocampal white matter, and cortico-pontine white matter in the brainstem, and included homologous structures from both hemispheres (Wakana et al., 2004). ROIs were drawn in the FA maps of each template separately. Care was taken to sample homologous brain tissue across templates, and to sample the central portion of the structures listed above in order to minimize partial volume effects. FA values in the selected ROIs were compared across templates using ANOVA with Tukey’s honestly significant difference (HSD) post hoc test (Tukey, 1949). Only differences with p<0.05 were considered significant.
The noise characteristics of the IIT v.3.0 template were extracted using a bootstrap approach described in (Zhang et al., 2011), and compared to those of IIT2 (noise characteristics of IIT2 were extracted previously using the same approach). More specifically, maps of the total variance of the diffusion tensor (TVDT) (Papadakis et al., 1999) (Appendix 3), standard deviation of FA (FAstd) and trace (tracestd) (Zhang et al., 2011), and 95% cone of uncertainty (COU) (Jones, 2003) (Appendix 3) were compared between IIT v.3.0 and IIT2.
2.3.2 Comparison of Inter-subject DTI Spatial Normalization Accuracy
The accuracy of inter-subject DTI spatial normalization achieved when using each of the templates as reference was compared across templates, for normalization of younger and older adults, separately. For that purpose, the DTI data from Datasets 1 and 2 were registered to the standardized and corresponding study-specific templates. For templates providing full tensor information (IIT v.3.0, IIT2, ICBM81, Eve, NTU-DSI-122-DTI, SS-y, SS-o), tensor-based registration was conducted using DTI-TK (Zhang et al., 2006), which is shown to be one of the top-performing DTI registration tools (Wang et al., 2015)(Wang et al., 2011)(Zhang and Arfanakis, 2013)(Kochunov et al., 2015)(Irfanoglu et al., 2016). For templates providing only tensor-derived scalar quantities (ENIGMA, FMRIB58, SRI24), the FA maps of Datasets 1 and 2 were first registered to the template FA maps using ART (Ardekani et al., 2005), one of the top-performing registration tools in its class (Klein et al., 2009), and the resulting transformations were applied to the diffusion tensors of Datasets 1 and 2 using finite strain tensor reorientation (Alexander et al., 2001). A number of metrics were used to assess the accuracy of inter-subject spatial normalization achieved for each template by measuring similarity of whole tensors, or similarity in portions of the diffusion tensor contents across participants. These metrics have been defined previously and are also described in Appendix 3. The average Euclidean distance of tensors (DTED) (Zhang et al., 2011), the average Euclidean distance of the deviatoric tensors (DVED), the average log-Euclidean tensor distance (LETD) (Arsigny et al., 2006), and the average overlap of eigenvalues-eigenvectors between tensors (OVL) (Basser and Pajevic, 2000), over all possible pairs of participants (separately for Datasets 1 and 2), were estimated in each voxel. In addition, the coherence of primary eigenvectors (COH) (Zhang et al., 2011), the COU (Jones, 2003) and the TVDT (Papadakis et al., 1999) were estimated in each voxel. White matter was then segmented for each template through K-means clustering of the mean FA maps of the corresponding normalized data, and histograms of the above quantities in white matter were generated for Datasets 1 and 2, separately (Zhang and Arfanakis, 2013). Each histogram was normalized by the corresponding total number of white matter voxels. Histograms of the metrics listed above were compared across templates using the one-sided two-sample Kolmogorov-Smirnov (KS) test. Differences were considered significant at p<0.05.
2.3.3 Impact of Spatial Normalization Accuracy on the Ability to Detect Small Inter-group FA Differences
Power analysis was used to assess the impact of spatial normalization accuracy achieved for each of the templates on the ability to detect small inter-group FA differences. More specifically, FA standard deviation maps generated after the registration of Dataset 1 to each of the templates were used in power analyses to assess the minimum FA differences that can be detected in each white matter voxel across two groups, assuming 100 participants per group, significance at p<0.05, and power>0.95. This was accomplished using the “sampsizepwr” function in Matlab (Mathworks, Natick, Massachusetts) for one-sided t-tests. White matter was defined after registration of Dataset 1 to each template as described above. Maps of the minimum detectable inter-group FA differences were generated for all templates. Cumulative distributions of the values presented in these maps were compared across templates using the one-sided two-sample Kolmogorov-Smirnov (KS) test. Differences were considered significant at p<0.05. The same power analysis on the ability to detect small inter-group FA differences was repeated for data from older adults, using FA standard deviation maps generated after the registration of Dataset 2 to each of the templates.
3. Results
3.1 Comparison of Template Characteristics
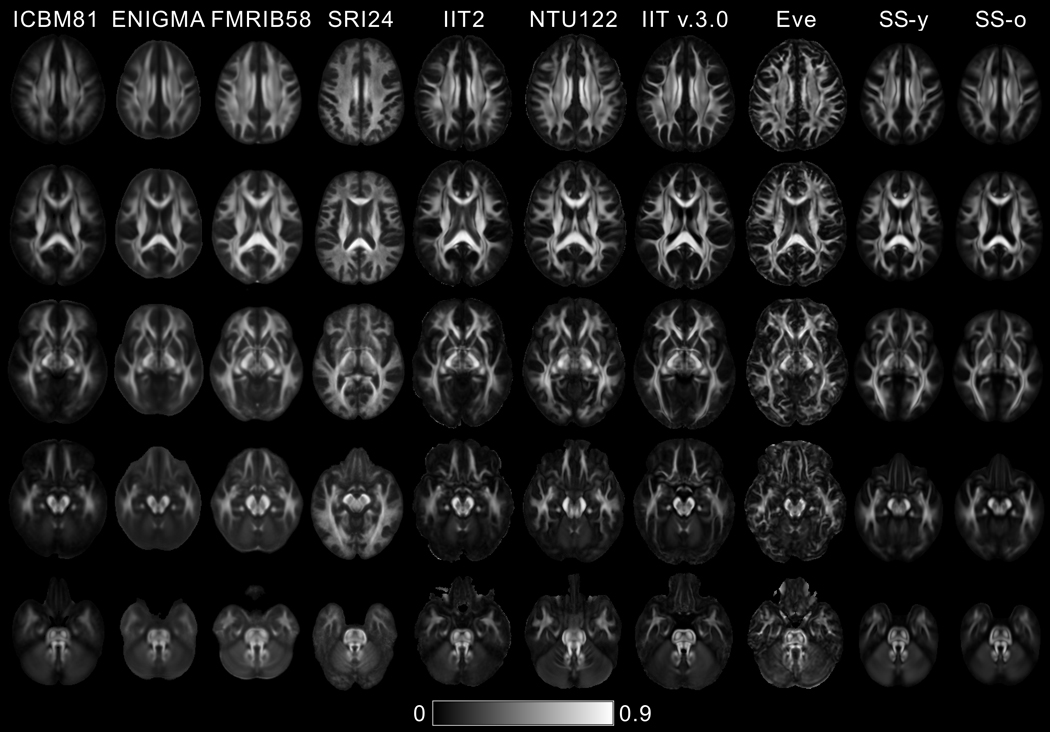
According to visual inspection, image sharpness was higher in the FA maps of Eve and IIT v.3.0 compared to all other templates (Fig. 1). This finding was supported by a quantitative comparison of the normalized power spectra of FA maps from different templates (Fig. 2). The energy at high spatial frequencies was higher in the normalized power spectra of FA maps from Eve and IIT v.3.0 compared to other DTI templates, on average over all axes (Fig. 2).
Figure 1.
FA maps of five axial brain slices of the ICBM81, ENIGMA, FMRIB58, SRI24, IIT2, NTU-DSI-122-DTI, IIT v.3.0, Eve, SS-y and SS-o templates.
Figure 2.
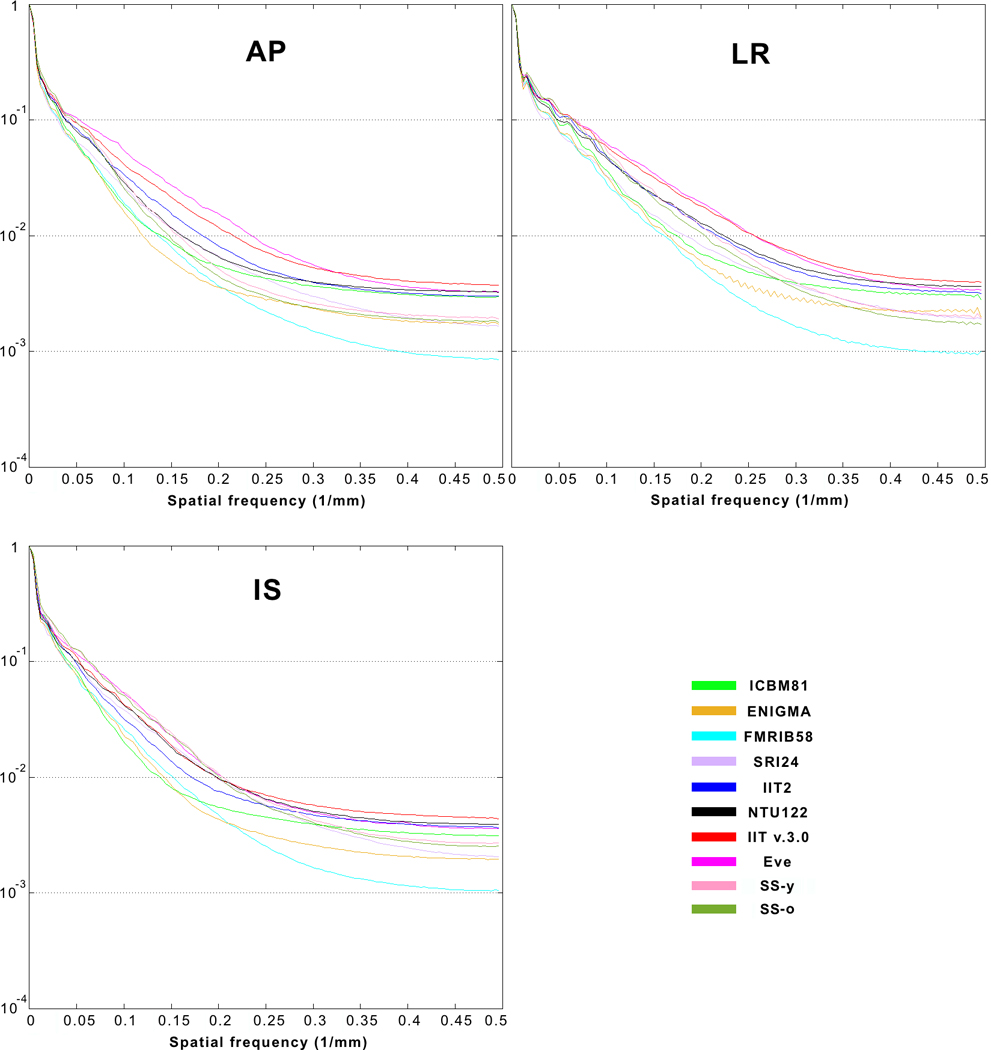
Normalized power spectra of FA maps for the anterior-posterior (AP), left-right (LR), and inferior-superior (IS) axes, separately, from the ICBM81, ENIGMA, FMRIB58, SRI24, IIT2, NTU-DSI-122-DTI, IIT v.3.0, Eve, SS-y and SS-o templates.
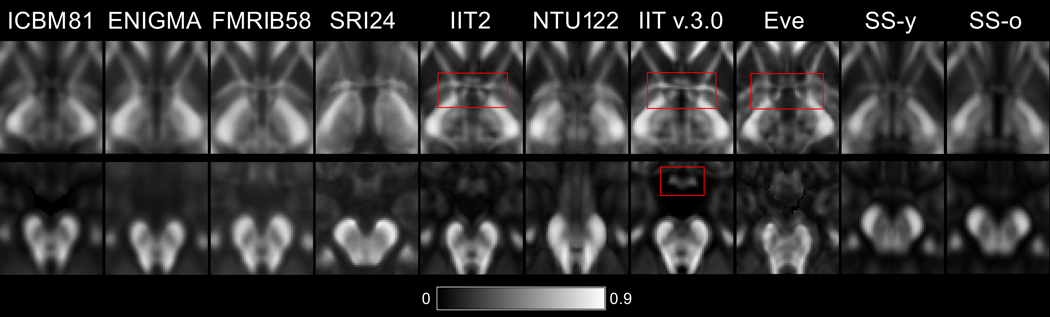
In general, fine white matter details were observed throughout the brain in IIT v.3.0 and Eve templates, even in areas near the cortex, while the same information was often lost due to blurring in other templates, especially in ICBM81, ENIGMA, and FMRIB58 (Fig. 1). Fine white matter details in the area of the anterior commissure (Fig. 3 top row) were visible in IIT v.3.0, Eve and IIT2 templates, while, in contrast, the anterior commissure was considered indistinguishable from the column and pre-commissural part of the fornix in ENIGMA, SRI24, SS-y, and SS-o, and not present in ICBM81 and NTU-DSI-122-DTI. The optic chiasm (Fig. 3 bottom row) was considered to be well-defined in IIT v.3.0, while it was not discernible in NTU-DSI-122-DTI, ENIGMA or SS-o.
Figure 3.
Magnified axial FA maps through the anterior commissure (top row) and optic chiasm (bottom row) of the ICBM81, ENIGMA, FMRIB58, SRI24, IIT2, NTU-DSI-122-DTI, IIT v.3.0, Eve, SS-y, SS-o templates. Fine white matter details in the area of the anterior commissure (top row) are visible in IIT v.3.0, Eve and IIT2 templates, while, in contrast, the anterior commissure is indistinguishable from the column and pre-commissural part of the fornix in ENIGMA, SRI24, SS-y, and SS-o, and not present in ICBM81 and NTU-DSI-122-DTI. The optic chiasm (bottom row) is well-defined in IIT v.3.0, while it is not discernible in NTU-DSI-122-DTI, ENIGMA, or SS-o.
In terms of image artifacts, visual inspection revealed eddy-current-induced artifacts mainly in the frontal lobe in ICBM81 and Eve (Fig. 4). NTU-DSI-122-DTI suffered by pronounced ghosting of the pons anterior to the actual structure (Fig. 4). The FA maps of SRI24 were deemed atypical since they exhibited low FA values in a substantially smaller portion of the brain than typical FA maps (Figs. 1,4).
Figure 4.
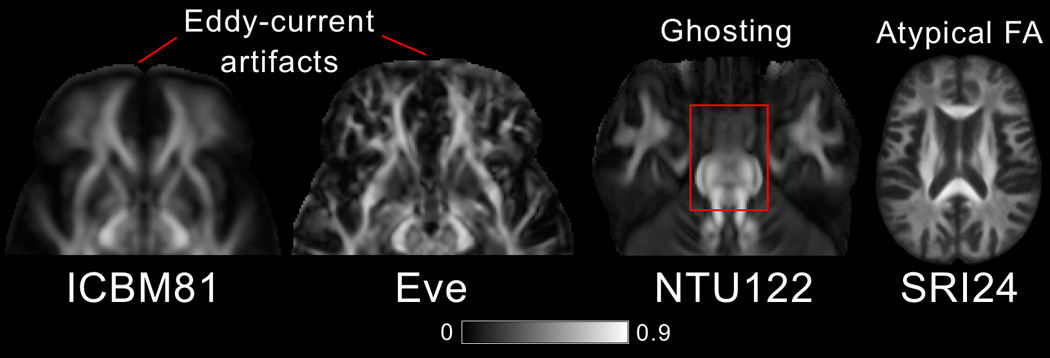
Artifacts. Eddy-current-induced artifacts in the frontal lobe of ICBM81 and Eve. Ghosting of the pons in NTU-DSI-122-DTI. Atypical FA map in SRI24.
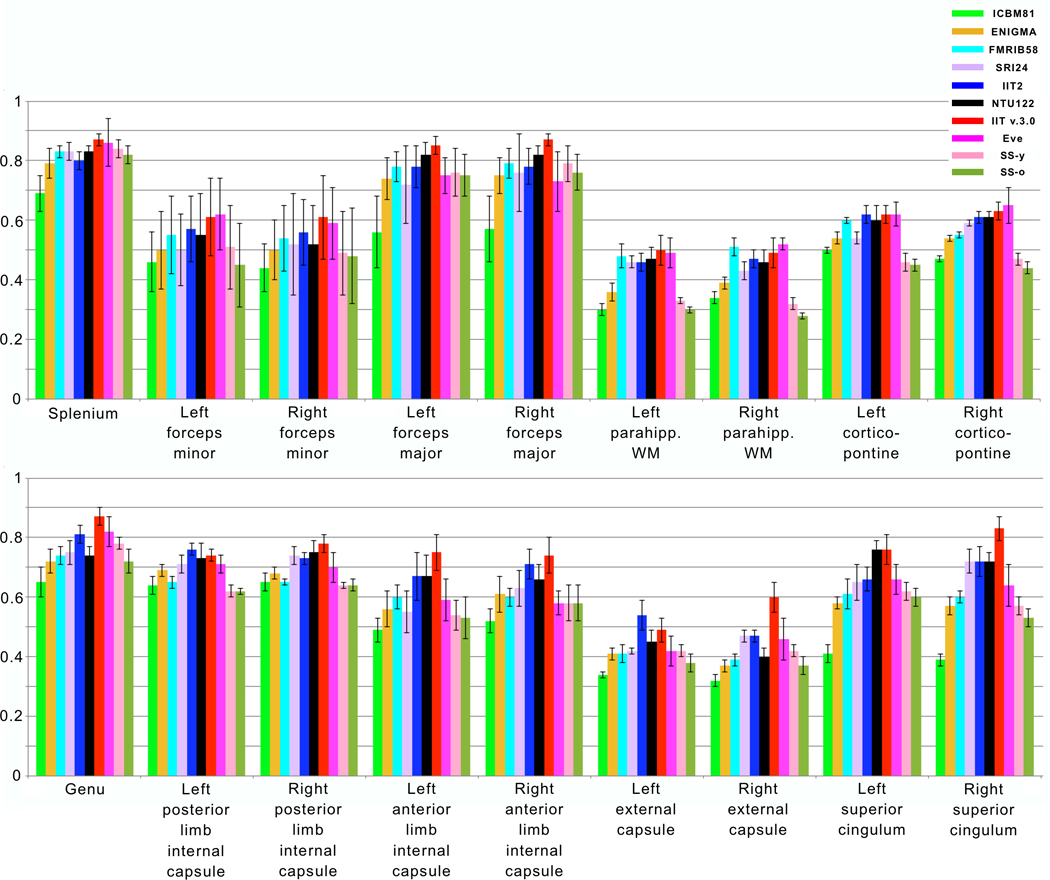
Mean and standard deviation of FA values from the selected white matter ROIs are presented in Figure 5 for all 10 templates (also tabulated in Appendix 4). ANOVA showed significant differences in mean FA values of all regions across templates. Post hoc comparisons using Tukey’s honestly significant difference test are listed in Appendix 4. According to these post hoc comparisons, in most ROIs, mean FA values of the ICBM81, ENIGMA, FMRIB58, SRI24, SS-y, and SS-o templates were significantly lower than those of the IIT2, NTU-DSI-122-DTI, IIT v.3.0, and Eve templates (p<0.05) (Appendix 4). In addition, even though FA values in gray matter were not quantified since most templates lack the images necessary for precise definition of gray matter, visual inspection of FA maps revealed elevated gray matter FA values in ENIGMA and FMRIB58 templates (Fig. 1).
Figure 5.
Mean FA values for homologous white matter regions of interest in the ICBM81, ENIGMA, FMRIB58, SRI24, IIT2, NTU-DSI-122-DTI, IIT v.3.0, Eve, SS-y, SS-o templates. Error bars denote standard deviation of FA values in the selected regions.
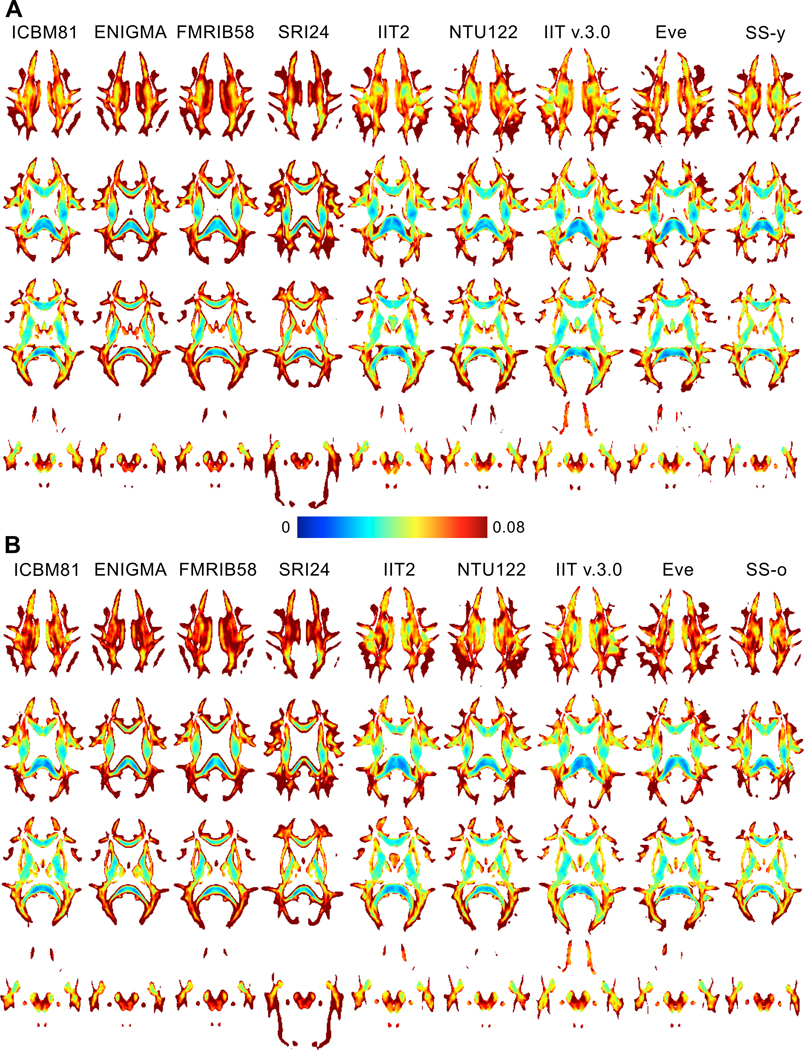
Maps of FAstd, tracestd, TVDT and COU showed reduced noise in white matter of IIT v.3.0 compared to IIT2 (Fig. 6). The improvement in FAstd was most visible at the edges of white matter structures (Fig. 6A), and in tracestd at the interface between brain tissue and CSF-filled spaces (Fig. 6B). The improvements in TVDT and COU were present throughout white matter (Fig. 6C,D).
Figure 6.
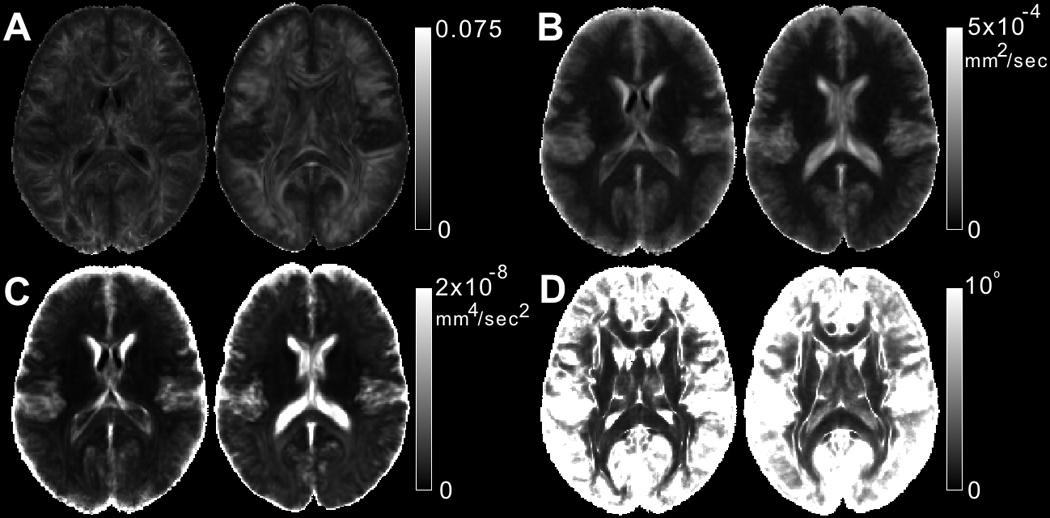
Maps of A) FAstd, B) tracestd, C) TVDT and D) COU for one slice of the IIT v.3.0 (left image in each image-pair) and IIT2 (right image in each image-pair) DTI templates.
3.2 Comparison of Inter-subject DTI Spatial Normalization Accuracy
The accuracy of inter-subject spatial normalization of DTI data from Datasets 1 and 2 achieved when using each of the standardized and study-specific templates as reference was compared across templates. It was demonstrated that, for Dataset 1 (younger adults), use of the IIT v.3.0 template resulted in a significantly higher number of white matter voxels with high COH and OVL, and low COU, DTED, DVED, LETD, and TVDT, compared to all other templates (including the corresponding study-specific template, SS-y), suggesting higher inter-subject DTI spatial normalization accuracy when using the IIT v.3.0 template (p<10−8 in all cases, one-sided two-sample KS test) (Fig. 7A). For Dataset 2 (older adults), higher inter-subject DTI spatial normalization accuracy was again achieved when using the IIT v.3.0 template compared to all other templates (including the corresponding study-specific template, SS-o), (p<10−8 in all cases, one-sided two-sample KS test) (Fig. 7B).
Figure 7.
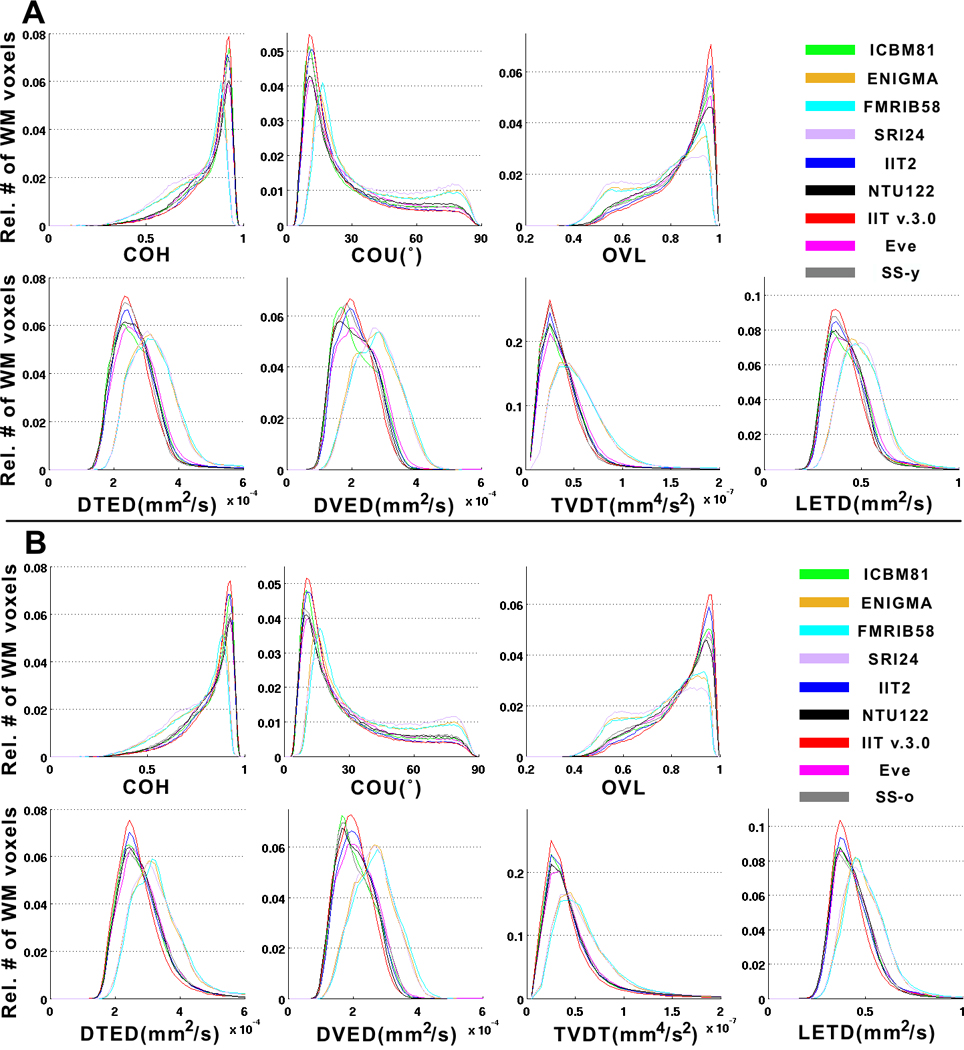
A) Histograms of the relative number of white matter voxels corresponding to different values of COH, COU, TVDT, average OVL, DTED, DVED, LETD, for registration of DTI data from Dataset 1 (younger adults) to the ICBM81, ENIGMA, FMRIB58, SRI24, IIT2, NTU-DSI-122-DTI, IIT v.3.0, Eve, SS-y templates. B) The same histograms for registration of DTI data from Dataset 2 (older adults) to the standardized templates and corresponding study-specific template, SS-o. Use of the IIT v.3.0 template for spatial normalization of DTI data from Datasets 1 and 2 (separately) results in a significantly higher number of white matter voxels with high COH and OVL, and low COU, DTED, DVED, LETD, and TVDT, compared to other templates (including the corresponding study-specific templates, SS-y and SS-o) (p<10−8 in all cases, one-sided two-sample KS test). This shows that, for both Datasets 1 and 2, inter-subject DTI spatial normalization accuracy is higher when using the IIT v.3.0 template compared to other standardized templates or the corresponding study-specific templates.
3.3 Impact of Spatial Normalization Accuracy on the Ability to Detect Small Inter-group FA Differences
Power analysis showed that, on average over all white matter, use of the IIT v.3.0 template allowed detection of smaller inter-group FA differences compared to other templates (including the study-specific templates), for both younger (Fig. 8A) and older adults (Fig. 8B). This was demonstrated in Figure 8 as a higher number of white matter voxels with cooler colors and a lower number of white matter voxels with warmer colors for IIT v.3.0. Also, the cumulative distribution of the minimum detectable inter-group FA differences in white matter was significantly higher for IIT v.3.0 compared to other templates (p<10−8 in all cases, one-sided two-sample KS test), for both younger (Fig. 9A) and older adults (Fig. 9B). (Appendix 5 presents an additional power analysis assessing the impact of spatial normalization accuracy achieved for each of the templates on the ability to detect a reduction of white matter FA in aging).
Figure 8.
Maps of the minimum detectable FA differences across A) two younger adult groups and B) two older adult groups, when using different templates as reference for spatial normalization (results are based on power analyses assuming 100 participants per group, p<0.05 and power>0.95). The color bar shows the correspondence of colors to minimum detectable FA differences. On average over all white matter, when using the IIT v.3.0 template, a higher number of voxels with cooler colors and a lower number of voxels with warmer colors are obtained, suggesting that the IIT v.3.0 template allows detection of smaller inter-group FA differences compared to other templates, for both younger and older adults.
Figure 9.
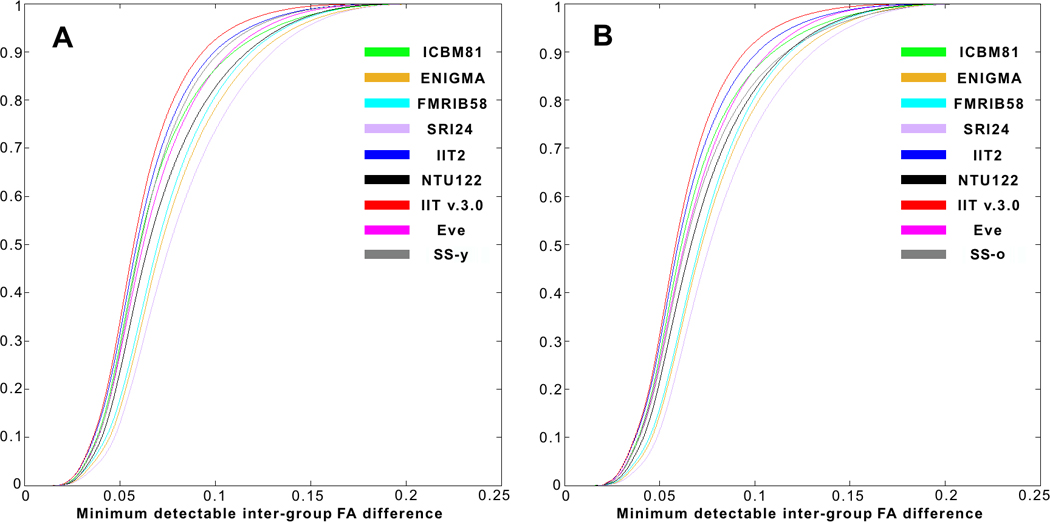
Cumulative distributions of the minimum detectable inter-group FA differences in white matter, when using different templates as reference for spatial normalization. Use of the IIT v.3.0 template allows detection of smaller inter-group FA differences compared to other templates, for both A) younger and B) older adults.
4. Discussion
A number of DTI templates of the adult human brain are commonly used in neuroimaging research, and their characteristics influence the accuracy of the application. In this work, available standardized DTI templates were compared to each other, as well as to study-specific templates, in terms of their characteristics and performance in spatial normalization and detection of small inter-group FA differences. According to this analysis, the IIT v.3.0 template was shown to combine a number of desirable characteristics over other templates: includes full-tensor information, is population-based, has high image sharpness, shows no visible artifacts, has low noise levels, has diffusion tensor properties and spatial features representative of data from the average individual adult brain. It was also demonstrated that the IIT v.3.0 template allows higher inter-subject DTI spatial normalization accuracy, and detection of smaller inter-group FA differences, compared to other templates (including study-specific templates), for both younger and older adults.
4.1 Comparison of Image Sharpness and Ability to Distinguish Small White Matter Features Across DTI Templates
Both Eve and IIT v.3.0 templates were shown to have higher image sharpness compared to all other DTI templates and provided the ability to distinguish small white matter structures. Although, this is straightforward for Eve since it is a single subject template, it is more difficult to achieve in a population-based template due to brain structural differences across participants. The high image sharpness and preservation of fine white matter details in the IIT v.3.0 template was probably due to more accurate spatial normalization of data from individual participants compared to other population-based templates. This, in turn, was due to the use of a top-performing tensor-based registration algorithm (Zhang et al., 2006) on data with no visible artifacts (Gui et al., 2008) (Appendix 2 provides details on the construction of the IIT v.3.0 template).
A template in which fine white matter details typical of data from individual adult brains are either missing or blurred, may negatively impact template applications. For example, the fact that the anterior commissure is not present in the ICBM81 and NTU-DSI-122-DTI templates (Fig. 3) means that when these templates are used as references for inter-subject spatial normalization, spatial matching of this particular structure across subjects may be inaccurate since the location of the target is unknown. The same problem exists in several of the standardized and study-specific templates for numerous other white matter structures that are relatively small yet play important roles in the brain (e.g. posterior commissure, optic chiasm, decussation of the superior cerebellar peduncle, and others less known). Similarly, white matter structures that are blurred in a template (most blurring was seen in ICBM81, ENIGMA, FMRIB58) (Figs. 1,2) constitute imprecisely-defined targets, and may also reduce inter-subject spatial normalization accuracy. Other template applications, such as localization of semantic labels for atlas construction based on template-derived maps, or algorithm evaluation on a template that is assumed to represent data from the average individual adult brain, may also be negatively impacted by templates with low image sharpness and lacking fine white matter details typically present in individual datasets. Enhancing the accuracy of various template applications requires a template with spatial features that are representative of those of data from the average individual adult brain. From the available DTI templates, Eve has the characteristics of an individual dataset, since it is based on data from a single subject. However, this also means that the features in Eve may not be representative of those in data from the average individual adult brain. In contrast, IIT v.3.0 has similar sharpness to Eve (Figs. 1,2), preserves fine white matter details seen in typical individual data (Figs. 1,3), and as a population-based template, it may be more representative of the features in data from the average individual adult brain.
4.2 Comparison of Image Artifacts Content Across DTI Templates
Image artifacts were visible in at least four of the DTI templates. The eddy-current induced artifacts in ICBM81 and Eve were probably due to residual eddy-current artifacts present in the echo-planar imaging data used to develop these templates (Mori et al., 2008)(Oishi et al., 2009). The white matter ghosting in NTU-DSI-122-DTI might be a result of inaccuracies in spatial normalization of individual datasets. The atypical FA maps in SRI24 were probably due to inaccurate spatial matching of individual DTI datasets caused by the exclusively T1-driven registration (Rohlfing et al., 2010). In addition to the above visible artifacts, less visible errors may have also contaminated the DTI templates. More specifically, eddy-current artifacts not only introduce bright bands at the edges of the brain, but also errors in tensors throughout the brain, and thus, the uncorrected eddy-current artifacts in individual datasets used to develop the ICBM81 and Eve templates suggest that tensor errors inside the brain have been carried over to the final templates. The inaccuracies in spatial normalization that may have led to the white matter ghosting in NTU-DSI-122-DTI may have also affected neighboring brain regions. Similarly, the inaccuracies in spatial matching of diffusion information that probably led to the atypical FA maps in SRI24 may have negatively influenced diffusion information throughout that template. Furthermore, all templates excluding IIT2 and IIT v.3.0, were based on data collected with spin-echo echo-planar DTI and suffered from different degrees of signal loss and signal pileup due to magnetic field non-uniformities. Even though these magnetic field-related artifacts may have been averaged out and may not be visible in the final templates, their effects remain in the form of errors in diffusion characteristics in the affected brain regions (mainly frontal and temporal lobes). Templates with any of the artifacts described above may negatively impact the accuracy of various template applications. In contrast, IIT2 and IIT v.3.0 are the only templates based on artifact-free DTI data (Appendix 2) (Gui et al., 2008). Furthermore, for IIT v.3.0, spatial normalization of individual datasets was conducted using one of the top-performing DTI registration tools (Zhang et al., 2006)(Wang et al., 2015)(Wang et al., 2011)(Zhang and Arfanakis, 2013)(Kochunov et al., 2015)(Irfanoglu et al., 2016). Consequently, the approach followed in the construction of IIT v.3.0 (Appendix 2) minimized the artifacts that were shown to contaminate other DTI templates.
4.3 Comparison of Mean FA Values Across DTI Templates
Mean FA values in white matter were lower on average in ICBM81, ENIGMA, FMRIB58, SRI24, SS-y, and SS-o templates compared to IIT2, NTU-DSI-122-DTI, IIT v.3.0, and Eve templates (Fig. 5, Appendix 4). Furthermore, mean FA values in gray matter were higher in ENIGMA and FMRIB58. These FA findings may in part be due to the use of less effective registration algorithms leading to less accurate spatial normalization of data from individual participants during construction of ICBM81, ENIGMA, FMRIB58, and SRI24 templates compared to IIT2, NTU-DSI-122-DTI, IIT v.3.0, and Eve templates (Peng et al., 2009). Another element that may have contributed to the above FA findings may be the fact that ICBM81, ENIGMA, FMRIB58, SRI24, SS-y, and SS-o included older adults (age range 18–85 years) compared to IIT2, NTU-DSI-122-DTI, IIT v.3.0, and Eve (age range 19–40 years), and FA is known to decrease with age (Pfefferbaum et al., 2000). Finally, the hardware and software used for data acquisition may have also contributed to the FA differences observed across templates.
4.4 Comparison of Noise Characteristics between IIT2 and IIT v3.0 Templates
The noise in various tensor properties was lower in IIT v.3.0 compared to IIT2 (Fig. 6). This comparison included only IIT2 and IIT v.3.0 because other templates do not provide noise estimates. Since the data used in the construction of IIT2 constituted 93% of the data used to build IIT v.3.0, the improvement in noise characteristics in IIT v.3.0 was exclusively due to improved tensor matching across individual participants and suggests higher confidence in the information presented in IIT v.3.0 compared to IIT2.
4.5 Comparison of Inter-subject DTI Spatial Normalization Accuracy
The IIT v.3.0 template resulted in the highest inter-subject DTI spatial normalization accuracy when used as a reference, compared to all other templates (including study-specific templates), for both younger and older adults (Fig. 7). IIT2, SS-y, SS-o, NTU-DSI-122-DTI, ICBM81 and Eve showed average performance, while FMRIB58, ENIGMA and SRI24 resulted in the lowest inter-subject spatial normalization accuracy. All three templates in the last group were templates containing only FA information, preventing tensor-based registration, which may have contributed to the low performance (Alexander and Gee, 2000)(Park et al., 2003). FMRIB58 and ENIGMA were in the group of templates with substantial blurring. Furthermore, SRI24 included artifacts (discussed earlier). The above factors may have contributed to the low inter-subject spatial normalization accuracy for FMRIB58, ENIGMA and SRI24. In contrast, all templates with average spatial normalization accuracy, IIT2, SS-y, SS-o, NTU-DSI-122-DTI, ICBM81, Eve, included full tensor information and allowed tensor-based registration. They did not however reach top performance probably due to bias (in the case of the single-subject Eve template), blurring (mainly in ICBM81, SS-y, SS-o), and artifacts (in the case of ICBM81 and NTU-DSI-122-DTI). In contrast, IIT v.3.0 is a full-tensor, population-based template with similar features and diffusion characteristics to those of typical datasets from individual adult brains, low noise, and no visible artifacts, and these factors may have contributed to the higher inter-subject spatial normalization accuracy when using IIT v.3.0 as reference, compared to other templates.
4.6 IIT v.3.0 vs. Study-Specific Templates
At first glance, the above results showing higher inter-subject DTI spatial normalization accuracy when using a standardized instead of a study-specific template appear to be in conflict with the work by (Van Hecke et al., 2011), which has established the use of study-specific templates as the modus operandi in DTI investigations. However, van Hecke et al. only compared spatial normalization accuracy between a study-specific template and a single standardized template, namely ICBM81, and showed that the coefficient of variation (COV) of FA and mean diffusivity values across subjects was lower when using the study-specific template instead of ICBM81. The results of the present work are actually in agreement with this finding by van Hecke et al. More specifically, Figure 7A of the present work also showed that use of the study-specific template instead of the ICBM81 resulted in lower DTED, TVDT, and LETD, suggesting higher spatial normalization accuracy when using the former instead of the latter (Note #1: the DTED, TVDT and LETD are more related to the COV of FA and mean diffusivity used by van Hecke et al. compared to the other measures included in Fig. 7A; Note #2: Only the results of Fig. 7A, and not Figure 7B, can be compared to those of van Hecke et al., since the data in Fig. 7B correspond to a different age-range). Furthermore, the same figure of the present work shows that IIT v.3.0 achieves even lower DTED, TVDT, and LETD than both templates, probably due to the enhanced characteristics of IIT v.3.0 compared to other templates. In addition, van Hecke et al. used FA-based instead of tensor-based registration, which leads to suboptimal spatial matching. Overall, the scope of the paper by van Hecke et al. was limited by the fact that several of the templates considered in the present work were simply not available at that time, and tensor-based registration was not yet widely used. Similar limitations as in (Van Hecke et al., 2011) were also true for (Zhang and Arfanakis, 2013) who also compared spatial normalization accuracy between a study-specific template and only two standardized templates (ICBM81, IIT2), and showed that a study-specific template may outperform the standardized templates in terms of spatial normalization accuracy. The present investigation substantially extends the work by (Van Hecke et al., 2011) and (Zhang and Arfanakis, 2013), and demonstrates that IIT v.3.0, a standardized template, can lead to higher spatial normalization accuracy than study-specific templates constructed with state-of-the-art approaches. This result is of high significance as it pertains to the template selection strategy used in many DTI studies. In agreement with the findings of the present work, Cabeen et al. (Cabeen et al., 2017) recently showed that, use of the IIT v.3.0 template in various voxel-based analysis approaches (VBA, TBSS etc.) resulted in better fitting (higher R2) of models relating age to FA, compared to study-specific templates.
Although the relative performance in spatial normalization across standardized templates remained relatively consistent for both younger (Fig. 7A) and older (Fig. 7B) adult data, the performance of SS-o relative to standardized templates (Fig. 7B) was lower than that of SS-y compared to the same standardized templates (Fig. 7A). This more pronounced difference in spatial normalization accuracy of older adult data between the top-performing standardized template and the corresponding study-specific template may reflect the difficulty in constructing a study-specific template that is representative of individual brain data when dealing with data from older adults, which are known to have higher variability compared to data from younger adults. This finding does not suggest that data from older adult brains register better to young instead of older adult templates. It simply means that until a high-quality older adult template is constructed, spatial normalization of older adult data is more accurate when using the IIT v.3.0 template than a study-specific template constructed with state-of-the-art approaches. The above has significant implications for DTI studies on older adults.
In addition to higher spatial normalization accuracy, the IIT v.3.0 standardized template offers several other important benefits to DTI investigations over study-specific templates. First, it eliminates the complexities and delays associated with constructing separate templates for different studies. It should be noted here that care and time must be invested when generating study-specific templates, since a poorly constructed study-specific template may result in even lower spatial normalization accuracy than that shown in Figure 7. Second, adoption of a standardized template by the community may facilitate integration and comparison of findings across studies. Third, a standardized template provides a space in which semantic labels and other resources can be developed, which may enhance functionality.
4.7 Impact of Spatial Normalization Accuracy on the Ability to Detect Small Inter-group FA Differences
Inter-subject spatial normalization is a key element of DTI voxel-wise investigations, among others, and high normalization accuracy is important to ensure high sensitivity and specificity in statistical analyses. Since the IIT v.3.0 template resulted in the highest inter-subject DTI spatial normalization accuracy when used as a reference, it also allowed detection of smaller inter-group FA differences on average over all white matter compared to all other templates, for both younger and older adult data (Figs. 8,9). The difference between IIT v.3.0 and study-specific templates was more pronounced for data from older adults (Fig. 9). Although the present investigation on the ability to detect small inter-group differences used a conventional voxel-wise analytic approach, the results are also relevant for analyses using tract-based spatial statistics (TBSS) (Smith et al., 2006), which projects information from the central portion of imperfectly aligned tracts onto a white matter skeleton to reduce the effects of misregistration. TBSS has been shown to address only a small portion of the effect of residual misalignment across subjects (Zalesky, 2011). Therefore, maximizing normalization accuracy through the use of a high-quality template continues to be crucial even in TBSS-type analyses.
4.8 Caveats
A few caveats must be considered when evaluating the findings of this work. Regarding the evaluation of inter-subject spatial normalization accuracy when using different templates as reference, it should be stressed that registration results depend not only on the template, but also on the quality of the individual data, as well as the registration algorithm. In the present work, data with typical quality from a popular publicly available database, and state-of-the-art publicly available registration methods, were used for the assessment of normalization accuracy. The findings of the present work might not generalize to other conditions. Also, maps of mean diffusivity and other tensor-derived quantities were not compared in this work since they are not available for most templates, and are not commonly used in template-based applications.
5. Conclusion
Standardized and study-specific DTI templates of the adult human brain were evaluated in this work. The DTI template of the IIT Human Brain Atlas (v.3.0) was shown to combine a number of desirable characteristics over other templates: includes full-tensor information, is population-based, has high image sharpness, shows no visible artifacts, has low noise levels, and has diffusion tensor properties and spatial features representative of data from the average individual adult brain. It was also demonstrated that the IIT v.3.0 template allows higher inter-subject DTI spatial normalization accuracy, and detection of smaller inter-group FA differences, for both younger and older adults, compared to other available standardized templates as well as study-specific templates constructed with state-of-the-art techniques. The IIT v.3.0 DTI template and associated resources are available for download at www.nitrc.org/projects/iit.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute of Biomedical Imaging and Bioengineering (NIBIB) (R21EB006525), the National Institute of Neurological Disorders and Stroke (NINDS) (R21NS076827), and the National Institute of Aging (NIA) (R01AG052200).
References
- Alexander DC, Gee JC. Elastic Matching of Diffusion Tensor Images. Comput Vis Image Underst. 2000;77:233–250. doi: 10.1006/cviu.1999.0817. [DOI] [Google Scholar]
- Alexander DC, Pierpaoli C, Basser PJ, Gee JC. Spatial transformations of diffusion tensor magnetic resonance images. IEEE Trans Med Imaging. 2001;20:1131–9. doi: 10.1109/42.963816. [DOI] [PubMed] [Google Scholar]
- Ardekani BA, Guckemus S, Bachman A, Hoptman MJ, Wojtaszek M, Nierenberg J. Quantitative comparison of algorithms for inter-subject registration of 3D volumetric brain MRI scans. J Neurosci Methods. 2005;142:67–76. doi: 10.1016/j.jneumeth.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Arsigny V, Fillard P, Pennec X, Ayache N. Log-Euclidean metrics for fast and simple calculus on diffusion tensors. Magn Reson Med. 2006;56:411–21. doi: 10.1002/mrm.20965. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Pajevic S. Statistical artifacts in diffusion tensor MRI (DT-MRI) caused by background noise. Magn Reson Med. 2000;44:41–50. doi: 10.1002/1522-2594(200007)44:1<41::aid-mrm8>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Cabeen RP, Bastin ME, Laidlaw DH. A Comparative evaluation of voxel-based spatial mapping in diffusion tensor imaging. Neuroimage. 2017;146:100–112. doi: 10.1016/j.neuroimage.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M, Peng H, Carew JD, Lesniak MS, Arfanakis K. A tractography comparison between turboprop and spin-echo echo-planar diffusion tensor imaging. Neuroimage. 2008;42:1451–62. doi: 10.1016/j.neuroimage.2008.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-C, Lo Y-C, Chen Y-J, Wedeen VJ, Isaac Tseng W-Y. NTU-DSI-122: A diffusion spectrum imaging template with high anatomical matching to the ICBM-152 space. Hum Brain Mapp. 2015;36:3528–41. doi: 10.1002/hbm.22860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irfanoglu MO, Nayak A, Jenkins J, Hutchinson EB, Sadeghi N, Thomas CP, Pierpaoli C. DR-TAMAS: Diffeomorphic Registration for Tensor Accurate Alignment of Anatomical Structures. Neuroimage. 2016;132:439–54. doi: 10.1016/j.neuroimage.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad N, Kochunov PV, Sprooten E, Mandl RC, Nichols TE, Almasy L, Blangero J, Brouwer RM, Curran JE, de Zubicaray GI, Duggirala R, Fox PT, Hong LE, Landman BA, Martin NG, McMahon KL, Medland SE, Mitchell BD, Olvera RL, Peterson CP, Starr JM, Sussmann JE, Toga AW, Wardlaw JM, Wright MJ, Hulshoff Pol HE, Bastin ME, McIntosh AM, Deary IJ, Thompson PM, Glahn DC. Multi-site genetic analysis of diffusion images and voxelwise heritability analysis: a pilot project of the ENIGMA-DTI working group. Neuroimage. 2013;81:455–69. doi: 10.1016/j.neuroimage.2013.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DK. Determining and visualizing uncertainty in estimates of fiber orientation from diffusion tensor MRI. Magn Reson Med. 2003;49:7–12. doi: 10.1002/mrm.10331. [DOI] [PubMed] [Google Scholar]
- Joshi S, Davis B, Jomier M, Gerig G. Unbiased diffeomorphic atlas construction for computational anatomy. Neuroimage. 2004;23(Suppl 1):S151–60. doi: 10.1016/j.neuroimage.2004.07.068. [DOI] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Jahanshad N, Marcus D, Winkler A, Sprooten E, Nichols TE, Wright SN, Hong LE, Patel B, Behrens T, Jbabdi S, Andersson J, Lenglet C, Yacoub E, Moeller S, Auerbach E, Ugurbil K, Sotiropoulos SN, Brouwer RM, Landman B, Lemaitre H, den Braber A, Zwiers MP, Ritchie S, van Hulzen K, Almasy L, Curran J, deZubicaray GI, Duggirala R, Fox P, Martin NG, McMahon KL, Mitchell B, Olvera RL, Peterson C, Starr J, Sussmann J, Wardlaw J, Wright M, Boomsma DI, Kahn R, de Geus EJC, Williamson DE, Hariri A, van ‘t Ent D, Bastin ME, McIntosh A, Deary IJ, Hulshoff Pol HE, Blangero J, Thompson PM, Glahn DC, Van Essen DC. Heritability of fractional anisotropy in human white matter: a comparison of Human Connectome Project and ENIGMA-DTI data. Neuroimage. 2015;111:300–11. doi: 10.1016/j.neuroimage.2015.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Oishi K, Jiang H, Jiang L, Li X, Akhter K, Hua K, Faria AV, Mahmood A, Woods R, Toga AW, Pike GB, Neto PR, Evans A, Zhang J, Huang H, Miller MI, van Zijl P, Mazziotta J. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. Neuroimage. 2008;40:570–82. doi: 10.1016/j.neuroimage.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucifora PGP, Wu X, Melhem ER, Gur RE, Gur RC, Verma R. Automated diffusion tensor tractography: implementation and comparison to user-driven tractography. Acad Radiol. 2012;19:622–9. doi: 10.1016/j.acra.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Faria A, Jiang H, Li X, Akhter K, Zhang J, Hsu JT, Miller MI, van Zijl PCM, Albert M, Lyketsos CG, Woods R, Toga AW, Pike GB, Rosa-Neto P, Evans A, Mazziotta J, Mori S. Atlas-based whole brain white matter analysis using large deformation diffeomorphic metric mapping: application to normal elderly and Alzheimer’s disease participants. Neuroimage. 2009;46:486–99. doi: 10.1016/j.neuroimage.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadakis NG, Xing D, Huang CL, Hall LD, Carpenter TA. A comparative study of acquisition schemes for diffusion tensor imaging using MRI. J Magn Reson. 1999;137:67–82. doi: 10.1006/jmre.1998.1673. [DOI] [PubMed] [Google Scholar]
- Park HJ, Kubicki M, Shenton ME, Guimond A, McCarley RW, Maier SE, Kikinis R, Jolesz FA, Westin CF. Spatial normalization of diffusion tensor MRI using multiple channels. Neuroimage. 2003;20:1995–2009. doi: 10.1016/j.neuroimage.2003.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Orlichenko A, Dawe RJ, Agam G, Zhang S, Arfanakis K. Development of a human brain diffusion tensor template. Neuroimage. 2009;46:967–980. doi: 10.1016/j.neuroimage.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferbaum A, Sullivan EV, Hedehus M, Lim KO, Adalsteinsson E, Moseley M. Age-related decline in brain white matter anisotropy measured with spatially corrected echo-planar diffusion tensor imaging. Magn Reson Med. 2000;44:259–68. doi: 10.1002/1522-2594(200008)44:2<259::aid-mrm13>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Pierpaoli C, Walker L, Irfanoglu MO, Barnett A, Basser P, Chang L-C, Koay C, Pajevic S, Rohde G, Sarlls J, Wu M. TORTOISE: an integrated software package for processing of diffusion MRI data. ISMRM Annual Meeting Proceedings; 2010. p. 1597. [Google Scholar]
- Rohlfing T, Zahr NM, Sullivan EV, Pfefferbaum A. The SRI24 multichannel atlas of normal adult human brain structure. Hum Brain Mapp. 2010;31:798–819. doi: 10.1002/hbm.20906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Roger G, Vachet C, Budin F, Maltbie E, Verde A, Hoogstoel M, Berger J-B, Styner M. Software-based Diffusion MR Human Brain Phantom for Evaluating Fiber-tracking Algorithms. Proc SPIE--the Int Soc Opt Eng. 2013:8669. doi: 10.1117/12.2006113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols TE, Mackay CE, Watkins KE, Ciccarelli O, Cader MZ, Matthews PM, Behrens TEJ. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage. 2006;31:1487–505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Suarez RO, Commowick O, Prabhu SP, Warfield SK. Automated delineation of white matter fiber tracts with a multiple region-of-interest approach. Neuroimage. 2012;59:3690–700. doi: 10.1016/j.neuroimage.2011.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey JW. Comparing individual means in the analysis of variance. Biometrics. 1949;5:99–114. [PubMed] [Google Scholar]
- Van Hecke W, Leemans A, Sage CA, Emsell L, Veraart J, Sijbers J, Sunaert S, Parizel PM. The effect of template selection on diffusion tensor voxel-based analysis results. Neuroimage. 2011;55:566–73. doi: 10.1016/j.neuroimage.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Varentsova A, Zhang S, Arfanakis K. Development of a high angular resolution diffusion imaging human brain template. Neuroimage. 2014;91:177–186. doi: 10.1016/j.neuroimage.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakana S, Jiang H, Nagae-Poetscher LM, van Zijl PCM, Mori S. Fiber Tract–based Atlas of Human White Matter Anatomy. Radiology. 2004;230:77–87. doi: 10.1148/radiol.2301021640. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gupta A, Liu Z, Zhang H, Escolar ML, Gilmore JH, Gouttard S, Fillard P, Maltbie E, Gerig G, Styner M. DTI registration in atlas based fiber analysis of infantile Krabbe disease. Neuroimage. 2011;55:1577–86. doi: 10.1016/j.neuroimage.2011.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yu Q, Liu Z, Lei T, Guo Z, Qi M, Fan Y. Evaluation on diffusion tensor image registration algorithms. Multimed Tools Appl. 2015 doi: 10.1007/s11042-015-2727-x. [DOI] [Google Scholar]
- Zalesky A. Moderating registration misalignment in voxelwise comparisons of DTI data: a performance evaluation of skeleton projection. Magn Reson Imaging. 2011;29:111–25. doi: 10.1016/j.mri.2010.06.027. [DOI] [PubMed] [Google Scholar]
- Zhang H, Yushkevich PA, Alexander DC, Gee JC. Deformable registration of diffusion tensor MR images with explicit orientation optimization. Med Image Anal. 2006;10:764–85. doi: 10.1016/j.media.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Zhang H, Yushkevich PA, Rueckert D, Gee JC. Unbiased white matter atlas construction using diffusion tensor images. Med Image Comput Comput Assist Interv. 2007;10:211–8. doi: 10.1007/978-3-540-75759-7_26. [DOI] [PubMed] [Google Scholar]
- Zhang S, Arfanakis K. White matter segmentation based on a skeletonized atlas: effects on diffusion tensor imaging studies of regions of interest. J Magn Reson Imaging. 2014;40:1189–98. doi: 10.1002/jmri.24445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Arfanakis K. Role of standardized and study-specific human brain diffusion tensor templates in inter-subject spatial normalization. J Magn Reson Imaging. 2013;37:372–381. doi: 10.1002/jmri.23842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Carew JD, Arfanakis K. Variability of diffusion tensor characteristics in human brain templates: Effect of the number of subjects used for the development of the templates. ISMRM Annual Meeting Proceedings; 2010. p. 1637. [Google Scholar]
- Zhang S, Peng H, Dawe RJ, Arfanakis K. Enhanced ICBM diffusion tensor template of the human brain. Neuroimage. 2011;54:974–84. doi: 10.1016/j.neuroimage.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Oishi K, Faria AV, Jiang H, Li X, Akhter K, Rosa-Neto P, Pike GB, Evans A, Toga AW, Woods R, Mazziotta JC, Miller MI, van Zijl PCM, Mori S. Atlas-guided tract reconstruction for automated and comprehensive examination of the white matter anatomy. Neuroimage. 2010;52:1289–301. doi: 10.1016/j.neuroimage.2010.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou MX, Yan X, Xie HB, Zheng H, Xu D, Yang G. Evaluation of non-local means based denoising filters for diffusion kurtosis imaging using a new phantom. PLoS One. 2015;10:e0116986. doi: 10.1371/journal.pone.0116986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.