Abstract
Flaviviruses such as dengue (DENV), yellow fever (YFV), West Nile (WNV), and Zika (ZIKV) are human pathogens of global significance. In particular, DENV causes the most prevalent mosquito-borne viral diseases in humans, and ZIKV emerged from obscurity into the spotlight in 2016 as the etiologic agent of congenital Zika syndrome. Owing to the recent emergence of ZIKV as a global pandemic threat, the roles of the immune system during ZIKV infections are as yet unclear. In contrast, decades of DENV research implicate a dual role for the immune system in protection against and pathogenesis of DENV infection. As DENV and ZIKV are closely related, knowledge based on DENV studies has been used to prioritize investigation of ZIKV immunity and pathogenesis, and to accelerate ZIKV diagnostic, therapeutic, and vaccine design. This review discusses the following topics related to innate and adaptive immune responses to DENV and ZIKV: the interferon system as the key mechanism of host defense and viral target for immune evasion, antibody-mediated protection versus antibody-dependent enhancement, and T cell–mediated protection versus original T cell antigenic sin. Understanding the mechanisms that regulate the balance between immune-mediated protection and pathogenesis during DENV and ZIKV infections is critical toward development of safe and effective DENV and ZIKV therapeutics and vaccines.
Keywords: dengue, Zika, adaptive immunity, innate immunity, vaccine
INTRODUCTION
The genus Flavivirus of the family Flaviviridae consists of more than 70 members and includes several medically important viruses, such as dengue (DENV) and Zika (ZIKV). The four serotypes of DENV cause dengue fever and dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS), the most important viral illnesses transmitted by mosquitoes, with a global estimate of 3.6 billion people at risk for infection, 400 million new infections, and 100 million new symptomatic cases annually (1). Over 2 million DHF/DSS cases and 20,000 deaths are estimated to occur each year (2). Dengue fever is characterized by fever, arthralgia, myalgia, abdominal pain, rash, and a viremia that begins 3–4 days following infection by mosquito bite. DHF/DSS manifests, as the fever subsides, with a sudden onset of plasma leakage that can result in hemoconcentration, pleural effusion, ascites, and shock. A small subset of DHF/DSS patients may die because of hypotensive shock. Although coagulation abnormalities and thrombocytopenia are prominent features, plasma leakage is the hallmark of DHF/DSS. In rare cases of DHF/DSS, neurologic abnormalities, including encephalitis, may also occur (3). DENV is transmitted to humans by the mosquitoes Aedes aegypti and Aedes albopictus. Owing to uncontrolled urbanization, globalization, and the spread of DENV-transmitting mosquitoes, multiple DENV serotypes cocirculate, resulting in increased frequency of epidemics and the continued emergence and spread of DHF/DSS. As a result, DENV is a major public health threat throughout the world. Despite the significance of this pathogen, no DENV-specific therapies are available, and a DENV vaccine that elicits protection in people with prior DENV exposure but in not naive individuals and that is not equally protective against all four serotypes has recently begun to be licensed on a country-by-country basis. This is in large part due to an incomplete understanding of the interplay between viral and host factors that contribute to DENV pathogenesis. On the virus side, some DENV lineages are more virologically and epidemiologically fit than others and are thus associated with DHF/DSS manifestations (4–6). On the host side, DENV infection history is the primary determinant associated with development of more severe dengue disease, with potential contributions from genetic variation, age, and sex (7).
Similar to DENV, ZIKV is transmitted by Aedes mosquitoes, and most infected people are asymptomatic or develop a dengue fever—like, self-limiting febrile disease. ZIKV, discovered in 1947, was thus not considered to be a significant human pathogen for 60 years, until large-scale outbreaks started in 2007 in the Pacific Islands, and multiple modes of transmission and new syndromes became recognized during the 2015–2016 outbreak in Latin America. ZIKV is now known to cause fetal infection and congenital Zika syndrome, which includes microcephaly, cerebral malformations, ophthalmological and hearing defects, and arthrogryposis (8). In adults, ZIKV infection may induce Guillain-Barré syndrome, an autoimmune peripheral neuropathy characterized by acute, symmetric limb weakness with decreased or absent deep-tendon reflexes (9–12). Additionally, case reports of ZIKV sexual transmission (13, 14) and ZIKV persistence in semen (15, 16) and vaginal secretions (17) are also mounting. Thus unlike DENV, ZIKV is characterized by transplacental and sexual transmission, life-threatening neurological complications, and viral persistence. Likely owing to its multiple modes of transmission and sharing of DENV’s mosquito vector, ZIKV has now spread to many DENV-endemic regions beyond Latin America, including Southeast Asia and India.
ZIKV-specific therapeutic and in particular vaccine candidates have begun to be developed rapidly (18, 19). However, ZIKV’s unusual transmission modes and features may present many challenges for drug and vaccine development. Moreover, cocirculation of DENV and ZIKV, which share a close antigenic relationship at antibody and T cell levels, may complicate vaccine development efforts for both viruses owing to potential dual roles for antibodies and T cells in protection and pathogenesis. The amino acid sequences of DENV strains within a serotype are approximately 98.1% to 99.0% identical, and the four DENV serotypes share 68.7% to 78.1% identity (20). ZIKV strains (representing both the Asian and African lineages) exist as a single serotype (21) and are 99.2% identical, comparable to sequence identity among different DENV strains, which also exist as multiple lineages. Among pathogenic human flaviviruses, DENV and ZIKV are most closely related to each other, with 55.1% to 56.3% amino acid sequence identity. Accordingly, emerging literature indicates many similarities between these two viruses in terms of interactions between the virus and host immune system. For both viruses, the interferon system is the central mediator of host defense and target of viral counterattack, whereas complex interplays between antibody and T cell responses likely determine the outcome of infection in flavivirus-immune settings. Thus, virus-host interactions related to interferon, antibody, and T cell responses to DENV and ZIKV are the major topics of discussion in this review. A thorough understanding of the immune response to both viruses is paramount for developing urgently needed DENV and ZIKV therapeutics and vaccines that are both safe and effective.
DENV AND ZIKV VIROLOGY
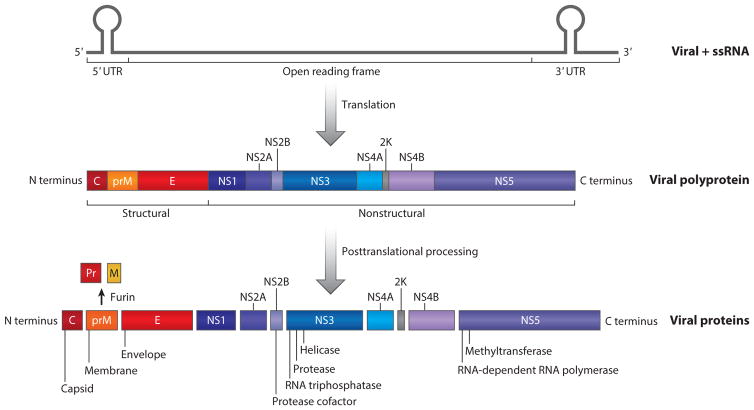
DENV and ZIKV are enveloped, positive-sense, single-stranded RNA viruses. The flaviviral genome is approximately 10.7 kb in length and encodes a polyprotein with three structural proteins [capsid (C)-premembrane (prM)-envelope (E)] at the N terminus and seven nonstructural proteins (NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5) at the C terminus flanked by 5′ and 3′ untranslated regions (UTRs) (22) (Figure 1). The single polyprotein is cleaved into these ten viral proteins by both viral and host proteases. The structural proteins form the virion particle. The C protein surrounds the viral genome, forming a nucleocapsid. The prM protein acts as a chaperone for E during virion assembly and is cleaved by furin to M in the trans-Golgi, resulting in the formation of mature virions than contain E and M (23). However, this cleavage is incomplete, resulting in the release of many immature virions containing prM (24, 25). The M protein regulates fusion of the viral envelope with the host membrane. The E protein is involved in receptor binding, membrane fusion, and viral assembly. E protein is also the major target of neutralizing antibodies and contains each of the four DENV serotype-specific antigenic epitopes. Based on the crystal structure of the DENV E protein ectodomain (26, 27), E is composed of three domains: Domain I (EDI) participates in E protein structural rearrangements required for fusion, domain II (EDII) contains the fusion loop involved in pH-dependent fusion of virus and host cell membranes, and domain III (EDIII) includes the receptor-binding region. The nonstructural proteins are involved in viral polyprotein processing, replication, and innate immune antagonism (28). In particular, NS5, approximately 900 amino acids long, is the largest and most conserved of the flavivirus proteins. It encodes an RNA-dependent RNA polymerase for genome replication, methyltransferase for 5′ RNA cap formation and methylation, and a type I interferon antagonist. NS3 encodes a protease and associates with NS2B (which acts as a cofactor) to form the NS2B3 viral protease complex, and it antagonizes the type I interferon pathway using both proteolytic-dependent and proteolytic-independent activities. In addition to nonstructural proteins, 5′ and 3′ UTRs are part of the viral replication complex, and subgenomic flavivirus RNA (sfRNA) encoded by the 3′ UTR is also involved in viral evasion of the innate immune response (5, 29, 30). During productive infection, DENV binds its cellular receptor, enters cells via receptor-mediated endocytosis, uncoats the viral genome in acidified endosomal vesicles, and releases its RNA into the cytoplasm to initiate viral translation, which is required to generate individual viral proteins, including the viral replicase necessary for subsequent viral RNA replication. Viral particles assemble by budding into the endoplasmic reticulum, pass through the Golgi, where surface glycoprotein modification occurs, and are exocytosed via secretory vesicles.
Figure 1.
The flaviviral genome is approximately 10.7 kb and is a positive-sense, single strand of RNA. Translation results in a polyprotein composed of three structural proteins at the N terminus and seven nonstructural proteins at the C terminus. Posttranslational cleavage and processing produce viral proteins: capsid, premembrane, envelope, NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5. The 2K signal peptide is a sequence of 17 amino acids linking NS4A and NS4B. Abbreviations: C, capsid; E, envelope; NS, nonstructural; prM, premembrane; ssRNA, single-stranded RNA.
Viral Entry Receptors and Cellular Tropism
Central to understanding mechanisms of viral immunity and pathogenesis is the knowledge of viral entry receptors and cellular tropism. Although the E protein has been known to mediate receptor binding and fusion, the precise identity of entry receptors for DENV and ZIKV in humans remains uncertain. DENV appears to use multiple cell surface molecules for binding to and infecting target cells, depending on the cell type. Several candidate molecules—including glycosaminoglycans (31, 32); C-type lectins; dendritic cell–specific ICAM3-grabbing nonintegrin (DC-SIGN) and liver/lymph node–specific ICAM3-grabbing integrin (L-SIGN) (33–35); mannose receptor (36); the phosphatidylserine receptors T cell immunoglobulin and mucin domain (TIM) and Tyro3, Axl, and Mertk (TAM) (37); and the phospholipid receptor CD300a (38)—have been proposed to serve as receptors for DENV based primarily on in vitro studies with cell lines and primary human cells. One TAM family member, Axl, has also been implicated as a ZIKV entry receptor in studies with cell lines and primary human cells. However, recent studies using CRISPR-Cas9-based gene editing in primary human neural progenitor cells and brain organoid cultures (39) and gene-deficient mice lacking Axl or both Axl and Mertk showed no effect of either Axl or Mertk on ZIKV infection (40–42). Therefore, bona fide entry receptors required for DENV and ZIKV infection in humans are as yet unknown, or they may not exist. The exact functions of the various candidate receptors identified to date are unclear. These candidates, individually or in various combinations in a cell type–dependent manner, may serve as true entry receptors that mediate fusion, or they may serve as attachment factors that promote virus adsorption and internalization, leading either to productive or abortive infection, or perhaps influencing the level of productive infection.
Viral entry may also occur via the process known as antibody-dependent enhancement (ADE) of infection, which is discussed in detail in the section titled The Debate Continues: The Role of Adaptive Immunity in Protection Versus Pathogenesis. Owing to incomplete cleavage of the pr domain from the prM protein during viral maturation, DENV and ZIKV populations exist as a mixture of mature, partially mature, and immature virions (43). This structural heterogeneity allows the virion population not only to limit the uniform availability of neutralizing antibody epitopes but also to enter into cells. In particular, immature virions that are not infectious can enter Fcγ receptor (FcγR)-bearing cells through antibody-dependent entry and become infectious (44). Thus, DENV and ZIKV not only counteract but also usurp the host antibody response via FcγR-mediated entry into cells as immune complexes.
In large part owing to incomplete knowledge about DENV entry receptors and the highly heterogeneous and plastic nature of FcγR-expressing cells such as macrophages, the precise cellular tropism of DENV in humans is still not fully defined. Human autopsy studies have reported that cells belonging to the monocyte/macrophage/dendritic cell lineage in the spleen, lymph nodes, lungs, liver, kidney, and stomach are the primary cellular hosts of DENV (45–48). Additionally, endothelial cells, hepatocytes, and neurons in the brain may also support DENV infection (45–51). Likely owing to the use of reagents that might not have distinguished virus-infected cells from those that might have endocytosed or phagocytosed viral antigen, some of these immunohistochemistry- and autopsy-based publications have differed over whether particular cell types are infected by DENV. In peripheral blood samples from DENV-infected patients, monocytes have been identified as the principal DENV hosts (52). In skin biopsy samples from uninfected human donors, Langerhans cells, dendritic cells, and macrophages can serve as DENV cellular hosts (53, 54). Similarly, in spleens from uninfected human donors, macrophages are the target cells for DENV infection in vitro (55). In agreement with human studies, detailed analyses of cellular tropism in highly susceptible mice lacking type I interferon receptor signaling have demonstrated that both dermal dendritic cells and macrophages in the skin (56), but primarily macrophages in other lymphoid (spleen, lymph nodes, thymus, bone marrow, Peyer patches) and nonlymphoid (kidney, heart, lung, and gastrointestinal tract) tissues are the principal cellular hosts of DENV (57, 58). Thus, human and mouse model studies to date indicate cells of the monocyte/macrophage/dendritic cell lineage as the major DENV hosts in the skin and many lymphoid and nonlymphoid organs.
Studies on human clinical samples and cell culture and nonhuman primate and mouse models suggest that ZIKV is pantropic. In clinical samples in situ hybridization has identified neural progenitor cells, glial cells, amniotic epithelial cells, fetal mesenchymal cells (59), and Hofbauer cells (macrophages of the placenta) (60–63) as permissive. Ex vivo and in vitro studies on human cells also suggest that ZIKV replicates in astrocytes (64), trophoblasts (65, 66), fibroblasts (placental, uterine, pulmonary) (67), endothelial cells, various epithelial cells, and peripheral blood mononuclear cells (PBMCs) (68). Numerous other immunohistochemistry-based studies using immune serum or anti-E antibody clone 4G2 on clinical samples have also demonstrated immunoreactivity in neurons, pneumocytes, hepatocytes, and renal epithelial cells (69–71). In vitro studies using 4G2 (39, 72–75) have also shown ZIKV E in glomerular podocytes, mesangial cells (76), endometrial stromal cells (77), retinal pericytes and pigmented epithelial cells (78), dermal fibroblasts and dendritic cells, epidermal keratinocytes (79), and endothelial cells (glomerular, retinal) (80). In rhesus macaques, in situ hybridization and NS2B immunohistochemistry have demonstrated that neurons, macrophages, neutrophils, and dendritic cells are permissive (81–83). Results in macaques have differed in regards to whether ZIKV replicates in B cells (82, 83). Further, independent studies have shown that ZIKV can persist in the macaque central nervous system and lymphoid tissue for over 4 weeks after infection (83, 84). Similar findings in immunocompetent and interferon axis–compromised mouse models suggest pantropism (85, 86). Notably in C57BL/6 mice treated with a monoclonal type I interferon receptor (IFNAR)-blocking antibody, neurons in the retina and optic nerve were permissive to infection (42, 87). In a separate study also using IFNAR blockade, ZIKV was detected in spermatogonia, primary spermatocytes, and Sertoli cells (40). In Ifnar1−/− mice, Leydig cell infection was associated with decreased testosterone production and testicular atrophy (88). Following intravaginal infection in mice, trafficking of permissive round cells (likely macrophages) through the subcapsular sinus of iliac lymph nodes has also been demonstrated (89). Reflecting ZIKV’s broad tropism, viral RNA has been isolated from several human body fluids, including saliva, urine, tears, aqueous humor, breast milk, semen, and vaginal washes (14, 17, 90–93).
Approached from the clinical side, susceptibilities of various placental, blood-brain barrier, and fetal brain cells are most directly relevant to understanding the pathogenesis of microcephaly and fetal Zika syndrome. Permissiveness of cells of the blood-testis barrier and male and female urogenital tracts is most relevant to investigating sexual transmission. The strong association between ZIKV infection and GBS may be related to already-documented susceptibility of neurons to ZIKV replication.
Most pantropic viruses employ a similar dissemination strategy. At the inoculation site virions replicate in tissue macrophages and dendritic cells, which traffic virus to the draining lymph nodes and other lymphoid tissues. In lymphoid tissues, large numbers of macrophages are recruited and amplify the virus. Following a period of mostly cell-associated viremia, the virus spreads to monocytes, macrophages, or dendritic cells in multiple tissues. Assembled virions then spread to infect the remaining local tissues. Although ZIKV tropism studies to date are consistent with this model, many additional questions remain, including how the virus crosses intact epithelia, enters specific body fluids, enters sites of immune privilege, and persists in specific sites and the possible influence of ADE on tropism. Due to the rarity of ZIKV autopsy samples, inability to conduct experiments in human hosts, and the large numbers of immunologic tools available for mouse modeling, further studies in mice and nonhuman primates will accelerate investigations of ZIKV tropism by allowing for phenotyping and genotyping of permissive and restrictive cell populations in stringently controlled experiments.
TYPE I INTERFERONS: A CRITICAL HOST DEFENSE SYSTEM AGAINST DENV AND ZIKV
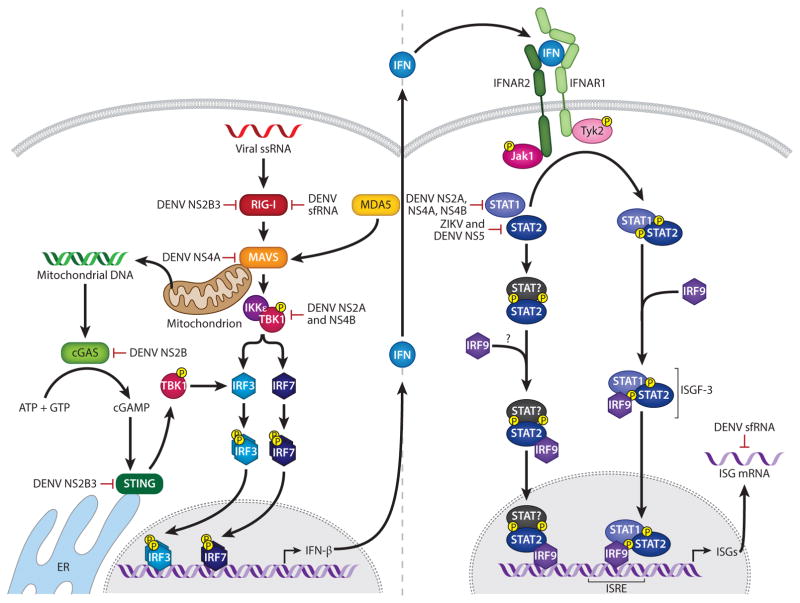
A short course of illness and self-limiting febrile symptoms in most DENV and ZIKV cases implicate a key role for the innate immune system in controlling DENV and ZIKV infections. The interferon system, comprising type I interferons (IFN-α, β), type II interferon (IFN-γ), and type III interferons (IFN-λ1–4), is the primary mechanism by which the innate immune system defends against viruses (94). Type I interferons are encoded by a family of closely related IFN-α genes and one IFN-β gene; type II interferon is encoded by one IFN-γ gene; and type III interferons, IFN-λ1–4, are also known as IL-29, 28A–C. Nearly all nucleated cells can respond to secreted type I interferons and, any nucleated cell can produce type I interferons; thus type I interferons are the primary interferons that are generated in most cell types during viral infections. The type I interferon system is triggered within hours of viral infection, upon recognition of viruses by cellular pattern recognition receptors (PRRs) such as the retinoic acid–inducible gene I (RIG-I)-like receptors (RLRs) and Toll-like receptors (TLRs). Engagement of these PRRs by viral nucleic acids results in the activation of multiple transcription factors, including interferon regulatory factors (IRFs) and NF-κB, that cooperate in inducing interferons and various other inflammatory cytokines and chemokines that orchestrate the innate and adaptive immune responses. RNA viruses that replicate in the cytosol are recognized mainly via the RLRs RIG-I and melanoma-associated differentiation antigen 5 (MDA5). RNA virus replication generates 5′-triphosphorylated single-stranded RNA and double-stranded RNA that are detected by RIG-I and MDA5. Upon RNA binding, RIG-I/MDA5 recruits the adaptor mitochondrial antiviral-signaling protein (MAVS) to trigger activation of the noncanonical IKK-related kinases IKKε and TBK1, which then activate IRF3 and NF-κB to induce type I interferon production (95) (Figure 2). The biological activity of type I interferons is mediated by triggering of their own transcriptional program, which results in expression of hundreds of interferon-stimulated genes (ISGs) (96). Specifically, type I interferons bind to the common type I interferon receptor (IFNAR), a heterodimer consisting of IFNAR1 and IFNAR2 chains, and signals through the Janus kinase–signal transducer and activation of transcription (JAK-STAT) pathway. The JAK kinases JAK1 and TYK2 phosphorylate and activate STAT1 and STAT2, followed by heterodimerization of the activated STAT1 and STAT2 and association with another DNA-binding protein, interferon regulatory factor 9 (IRF-9), to form ISG factor 3 (ISGF-3). The ISGF-3 complex promotes gene expression by binding to interferon-stimulated response elements (ISREs) of type I interferon–dependent genes. These ISGs influence many cellular processes, including RNA processing, protein stability, and cell viability, thereby directly affecting specific steps of the viral life cycle and thus virus replication. ISGs in antigen-presenting cells such as macrophages and dendritic cells are also important for T and B cell activation, affecting the magnitude and quality of the adaptive immune response and eventual virus clearance.
Figure 2.
The interferon system. Following flaviviral infection, viral RNA is sensed by cellular pattern recognition receptors, including RLRs, which induce multiple transcription factors including the IRFs and NF-κB, leading to transcription of interferon. Interferon is secreted and binds the type I interferon receptor to activate the JAK-STAT signal transduction pathway. STAT1 and STAT2 are phosphorylated and form a heterodimer that joins IRF9 to form ISGF-3. The ISGF-3 complex then binds to ISREs in the genome to promote expression of ISGs. DENV may evade the interferon system by multiple mechanisms, including degradation of STAT2, blocking of STAT1 and STAT2 phosphorylation, suppression of STAT1 signaling, blocking of TBK1 phosphorylation, blocking of MAVS binding to RIG-I, induction of mitochondrial elongation linked to viral replication complexes, cleavage of mitochondrial fusion proteins, prevention of RIG-I translocation to mitochondria, binding of TRIM25, cleavage of STING, degradation of cGAS, and inactivation of host RNA-binding proteins. Abbreviations: ER, endoplasmic reticulum; IRF, interferon regulatory factor; ISGF, interferon-stimulated gene factor; ISRE, interferon-stimulated response element; MAVS, mitochondrial antiviral-signaling protein; RLR, RIG-I-like receptor; sfRNA, subgenomic flavivirus RNA; ssRNA, single-stranded RNA.
Several lines of evidence indicate that the type I interferon system is the central mediator of protection against DENV and ZIKV. First, studies with DENV-infected patient samples have shown that, during the early febrile period, patients contain high levels of type I interferons in the serum (97, 98), and the type I interferon signatures are prominent in transcriptional profiling of PBMCs (99–101). Second, cell culture studies have demonstrated that DENV is recognized by both RIG-I and MDA5 (102), and the phosphatases PP1α and PP1γ are required for RIG-I- and MDA5-mediated type I interferon response to DENV (103); type I interferons inhibit DENV infection in a variety of cells (104, 105); autocrine type I interferon action in infected cells, rather than paracrine type I interferon signaling in naive cells, is important for controlling DENV infection in cell cultures (106); and expression of type I interferon–inducible ISGs such as IFITM2/3, viperin, ISG15, ISG20, OAS, BST2, and RyDEN (also known as IRAV) blocks DENV infection (107–113). IFITMs, in particular IFITM3, also inhibit ZIKV infection (114), and IFITM3 interacts with ZMPSTE24 to mediate its antiviral activity against ZIKV (115). Third, mouse models of experimental DENV and ZIKV infection have shown that the interferon system is essential and more important than T and B cell–dependent immunity in controlling DENV infection in mice (40, 116–118); these mouse model data are further described below. Finally, all flaviviruses studied to date must evade the type I interferon system–mediated antiviral defense in order to replicate and cause disease in vertebrate hosts; thus, DENV and ZIKV employ multiple viral mechanisms to antagonize both type I interferon induction and type I interferon signaling (28, 119), underscoring the importance of the type I interferon system in anti-DENV/ZIKV immunity as discussed below.
Murine Studies: Identification of the Type I Interferon Components That Are Required for Protection Against DENV and ZIKV Infection In Vivo
Studies with various gene-deficient mice have revealed the key type I interferon components that are required for mediating protection against DENV in vivo. In particular, MAVS and the combined activities of IRF-3 and IRF-7 are essential for the initial type I interferon induction and restriction of DENV in mice (120, 121), revealing the presence of MAVS-independent and IRF-3-and IRF-7-independent pathways that control DENV infection in vivo in a delayed manner. These late-acting pathways that restrict DENV infection in vivo are as yet to be identified. Based on studies showing that TLR3 and TLR7 can sense DENV infection in cells (122, 123) and that IRF-5 (85, 124, 125) and IRF-1 (126) contribute to host defense against ZIKV and the closely related WNV, MyD88/TRIF and IRF-5/IRF-1 signaling, respectively, are likely candidates for mediating MAVS-independent and IRF-3- and IRF-7-independent antiviral defenses against DENV. At the level of type I interferon signaling, mice lacking IFNAR1 are highly sensitive to DENV infection and thus can succumb to DENV infection depending on the viral challenge strain and dose (127, 128). Dissection of the IFNAR signaling mechanism has revealed that the IFNAR signaling in myeloid cells is essential for protection against DENV (129, 130), and the early STAT1-dependent and the late STAT1-independent pathways can each control DENV infection to a degree but are most effective in concert (131). The later STAT1-independent mechanism of protection against DENV involves the noncanonical IFNAR-STAT2 pathway (132), indicating that the IFNAR-STAT2 pathway, in the absence of STAT1, modulates type I interferon production and ISG response in a delayed manner during DENV infection in vivo. Similarly, studies using mouse models of systemic ZIKV infection have demonstrated that wild-type mice treated with blocking anti-IFNAR monoclonal antibody, Ifnar1−/− mice, Irf3−/−Irf5−/−Irf7−/− triple knockout mice, and Stat2−/− mice, but not wild-type, Irf3−/−, Irf5−/−, Mavs−/−, Tmem173(STING)−/−, and Mb21d1 (cGAS)−/− mice, can succumb to ZIKV infection (85, 125, 133–135). As Stat2−/− mice can succumb to infection with ZIKV (133) but not DENV (132, 136), STAT2-directed signaling may play a more important role against ZIKV than DENV. Additionally, studies using mouse models of intravaginal ZIKV infection have shown a requirement for IFNAR signaling in myeloid cells (89) and combined activities of IRF-3 and -7 or TLR7 and MAVS to restrict local ZIKV infection in the female reproductive tract (118). Another study using direct inoculation of ZIKV into mouse eyes reported a role for ISG15 in controlling ZIKV infection of eyes (87). Collectively, these findings indicate that multiple, redundant pathways at both type I interferon induction and signaling levels contribute to host defense against DENV and ZIKV. They have also set the foundation to determine the precise virus-specific (DENV versus ZIKV), time-specific (early versus late stage of DENV/ZIKV infection, or acute versus persisting ZIKV infection), route-specific (systemic versus mucosal ZIKV infection), tissue-specific (e.g., lymphoid versus nonlymphoid organs/genital organs/placenta), and cell type–specific (e.g., immune versus nonimmune cells, or macrophages versus dendritic cells) mechanisms by which the type I interferon system protects against DENV and ZIKV.
However, some of the signaling molecules involved in the type I interferon system may also provide protective immunity in a type I interferon–independent manner. Based on the following results, PRR-induced, type I interferon–independent, type III interferon–dependent ISGs may be important in protecting against DENV and ZIKV infection depending on the context: (a) The combined activities of IRF-3, -5, and -7 are more important than that of IFNAR in restricting systemic ZIKV infection in mice (125), and IRF-3 and -7 are more important than IFNAR in limiting ZIKV infection in the murine female reproductive tract (118); (b) RIG-I agonists have a part in inhibiting DENV replication in human cells via the RIG-I/MAVS/TBK1/IRF3 signaling axis that is largely type I interferon independent (137); (c) type III interferons have a role in protection against ZIKV infection in human syncytiotrophoblasts from term placenta (138); and (d) the peroxisomal MAVS-based type III interferon–mediated ISG response is protective against DENV infection in various human cells (139). Additionally, based on results revealing the type II interferon receptor–mediated control of DENV infection in the Ifnar1−/− mouse brain (127) and a higher susceptibility of mice lacking both type I and II interferon receptors to DENV and ZIKV infection than mice lacking type I interferon receptors alone (116, 117, 134), type II interferon signaling may be synergistic or nonredundant with the IFNAR pathway in protecting against DENV and ZIKV. Similarly, in terms of virus sensing, TLRs may contribute to host defense against DENV and ZIKV in a context-dependent manner. DENV employs multiple mechanisms to evade the type I interferon system, particularly at the RIG-I-like receptor (RLR) pathway level, as discussed below. The TLR- and plasmacytoid dendritic cell (PDC)-based mechanisms may be key in fighting against DENV and ZIKV infection in settings where the virus inhibits the RLR pathway and in Mavs−/− mice (120), as cell culture studies have suggested roles for TLR3, TLR7, and PDCs in detecting DENV and producing type I interferons (122, 123, 140). However, under certain circumstances, activation of the TLR pathway may contribute to pathogenesis rather than protection. For instance, TLR3 activation following ZIKV infection in human embryonic stem cell–derived cerebral organoids has been associated with attenuated neurogenesis (141), and TLR2, 4, and 6 activation upon binding of DENV NS1 protein to endothelial cells results in proinflammatory cytokine production and loss of endothelial cell integrity (142–144). Deciphering how signaling via type I, II, and III interferons and other inflammatory cytokines intersects and regulates the balance between protection and pathogenesis will be crucial for understanding the precise mechanisms by which the type I interferon system mediates host defense against DENV and ZIKV.
Antagonism of the Type I Interferon System by DENV and ZIKV
To facilitate viral replication/dissemination and cause disease in humans, each pathogenic flavivirus has evolved multiple yet distinct mechanisms for inhibiting the signal transduction pathways that induce type I interferon production and ISG expression. Given the recent emergence of ZIKV, only one mechanism of viral antagonism of the type I interferon signaling has been identified thus far (145). In contrast, the past decade of research has revealed numerous mechanisms by which DENV antagonizes the signal transduction pathways involved in the type I interferon system (28, 119). Multiple DENV proteins as wells as sfRNA are dedicated to inhibition of both the type I interferon induction and signaling axes, further emphasizing a critical role for type I interferons in host defense against DENV.
In particular, DENV NS2A, NS4A, NS4B, NS2B3, and the sfRNA have been observed to target distinct steps of the RIG-I/MDA5 signaling to inhibit type I interferon production. DENV NS2A and NS4B overexpression can inhibit RIG-I/MDA5-dependent type I interferon production by blocking TBK1 phosphorylation (146). DENV NS4A can associate with MAVS, preventing MAVS binding to RIG-I and subsequent IRF3 activation and type I interferon induction (147). DENV NS4B has also been demonstrated to induce the formation of elongated mitochondria that are physically linked with DENV-induced replication factories and favor viral replication by limiting RIG-I-dependent induction of type I and III interferons (148). However, in another study DENV protease NS2B3 was shown to cleave mitochondrial fusion proteins, thereby disrupting mitochondrial fusion and membrane potential and attenuating MAVS activation (149). These two conflicting reports on mitochondrial elongation versus fusion during DENV infection may be specific to the cell line and experimental approach, and both are as yet to be confirmed using relevant primary human cells. The protease-independent function of DENV NS2B3 can also antagonize the RIG-I pathway by binding of NS3 to the adaptor protein 14-3-3ε and thus preventing RIG-I translocation to mitochondria (150). Finally, sfRNA of DENV strains that are associated with greater epidemic potential binds more strongly to the ubiquitin ligase tripartite motif protein 25 (TRIM25), thereby preventing the ubiquitination-dependent activation of RIG-I (5). The existence of so many different mechanisms involving multiple viral proteins and sfRNA to subvert the RIG-I/MDA5 pathway suggests a dominant role for the RLR sensors in inducing a type I interferon response against DENV, consistent with Mavs−/− mouse data demonstrating an essential role for MAVS in mounting the initial antiviral innate immune response against DENV (120). RIG-I and MDA5 function redundantly in detecting DENV in cells (102) and are thus likely to play a similar role in vivo. Based on a study suggesting that overexpression of ZIKV sfRNA limits both RIG-I- and MDA5-directed type I interferon induction, whereas DENV sfRNA inhibited only RIG-I-mediated type I interferon induction (30), ZIKV sfRNA may have a broader antagonistic activity than DENV sfRNA at the level of RLR signaling.
DENV also blocks type I interferon production in human cells via manipulation of the cytosolic DNA receptor cGAS and its adapter STING, an endoplasmic reticulum–resident protein (151–153). cGAS binds to double-stranded DNA and catalyzes the formation of cyclic dinucleotides, which then form cGAMP that activates STING to trigger type I interferon production. cGAS can recognize DNA in various contexts, including microbial and cellular DNA. In DENV-infected cells, cGAS senses mitochondrial DNA (153). DENV NS2B3 protease mediates cleavage of human but not murine STING (151, 152), revealing STING as a species-specific restriction factor against DENV in mice. To counteract NS2B3-mediated cleavage of STING, the endoplasmic reticulum protein SCAP prevents association of NS2B with STING and inhibits the formation of NS2B3 (154), suggesting that STING and SCAP levels may dictate differential susceptibility of cells to DENV infection. The DENV protease also targets cGAS for degradation, but independently of its proteolytic activity and via NS2B rather than the NS2B3 complex (153). DENV targeting of both cGAS and STING (i.e., members of the same pathway) suggests an important role for this pathway in sensing DENV. Thus, redundant targeting of the RLR and cGAS/STING pathways and multiple members of a common pathway may ensure efficient blockade of type I interferon production. In the future, in vivo studies using mice deficient in particular components of the RLR and cGAS pathways and in vitro studies using primary human cells should elucidate the relative importance of the RLR versus cGAS pathways in the host defense against DENV and ZIKV.
DENV also uses multiple viral proteins and sfRNA to antagonize type I interferon signaling. DENV NS2A, NS4A, and NS4B can suppress STAT1 activation and downstream ISG expression (155). To prevent expression of ISGs, DENV sfRNA binds to and inactivates RNA-binding proteins G3BP1, G3BP2, and CAPRIN1, which are required for translation of ISG mRNAs (29). Studies to date indicate that DENV NS5 may be the most potent antagonist of type I interferon signaling. Specifically, DENV NS5 targets human but not murine STAT2 for degradation via interaction with UBR4, a cellular protein with E3 ligase activity responsible for degrading proteins (136, 156). Thus, murine STAT2 is another species-specific restriction factor against DENV besides STING. Recently, ZIKV NS5 was also identified as an antagonist of type I interferon signaling (145). Similarly to DENV NS5, ZIKV NS5 binds and degrades human but not mouse STAT2 and acts as a species-specific restriction factor against ZIKV in mice; however, unlike DENV, ZIKV does not use UBR4, suggesting that ZIKV uses an as yet to be identified mechanism to degrade STAT2. Based on another study showing that ZIKV blocks phosphorylation of both STAT1 and STAT2 in primary human dendritic cells to antagonize type I interferon signaling and limit cytokine production and activation marker expression (157), ZIKV is also likely to use multiple viral proteins and mechanisms to evade type I interferon signaling.
The 5′ methylated cap of the DENV and ZIKV genomes may also be involved in counteracting the interferon system. In general, 2′-O-methylation of the 5′ cap on the virus genomic RNA mimics cellular mRNAs and conceals the viral genome from detection via RLRs (158, 159) and the type I interferon–dependent IFIT family of proteins (160). For DENV, 2′-O-methylation of the 5′ cap is also important for evading an early type I interferon–independent response (161). MAVS localizes to peroxisomes in addition to mitochondria, and peroxisomal MAVS can rapidly induce type III interferon–dependent expression of ISGs that restrict DENV (139). Additionally, DENV capsid, the first viral protein made in flavivirus-infected cells, may be involved in reducing peroxisome numbers and type III interferon production (162). Thus, consistent with murine studies implicating roles for PRR-induced, type I interferon–independent, type III interferon–dependent responses in host defense against DENV and ZIKV infection in vivo, DENV and ZIKV may have evolved strategies to counteract the antiviral effects of early ISGs. These ISGs may be rapidly induced in a type I interferon–independent manner to provide short-term protection, while mitochondrial MAVS activates type I interferon signaling with delayed kinetics to amplify and stabilize the antiviral response. At present, cross talk among various viral proteins and interferon antagonism strategies, and the importance of peroxisome- versus mitochondrion-mediated responses and early type I interferon–independent versus –dependent responses against DENV and ZIKV infection in vivo are as yet to be explored.
Collectively, studies have provided a solid foundation to explore the interactions between the interferon system and virus during DENV/ZIKV infections in animal models, relevant human cell culture models, and humans using state-of-the-art technologies. Despite decades of research, the mechanisms by which the type I interferon system mediates antiviral defense are as yet to be fully understood. Considering their similarity in genome structures and replication strategies, DENV and ZIKV likely share many aspects of their interactions with the type I interferon system. However, due to differences in the biology of DENV and ZIKV and the context-dependent nature of the type I interferon system, many interactions between the type I interferon system and DENV/ZIKV should also vary depending on the virus. Answers to questions related to how ZIKV interactions with the interferon system influence the outcome of infection in barrier tissues such as the placenta, testes, eyes, and brain and during pregnancy in the mother versus the fetus may also provide novel insights into the mechanism of action of the interferon system. Importantly, identifying the specific mechanisms by which DENV and ZIKV activate and evade the interferon system and potentially other components of the innate immune system, including the complement system, γδ T cells, and various types of innate lymphoid cells, is essential for understanding viral pathogenesis and developing effective therapeutics and vaccines against DENV and ZIKV. Particularly in the context of adaptive immunity and vaccine development (discussed below), studies investigating the roles of infected versus bystander cells belonging to the monocyte/macrophage/dendritic cell lineage in regulation of the adaptive immunity are urgently needed.
THE DEBATE CONTINUES: THE ROLE OF ADAPTIVE IMMUNITY IN PROTECTION VERSUS PATHOGENESIS
While primary infection by one DENV serotype confers life-long immunity to that serotype (163), multiple epidemiological studies indicate that DHF/DSS most often occurs in individuals experiencing a second DENV infection by a different (heterotypic) serotype (7, 164). For decades, the two non–mutually exclusive hypotheses of ADE and T cell original antigenic sin have competed to explain why a subset of secondary infections results in more severe dengue disease. Evaluation of these hypotheses is now crucial in the context of infection with not only the four DENV serotypes but also ZIKV, given that DENV and ZIKV share genetic similarity and structural homology with each other, and ZIKV continues to merge into DENV’s established ranges. Emerging evidence indicates that the amino acid sequence conservation and structural similarities among the four DENV serotypes and ZIKV are substantial enough for these viruses to be cross-reactive at both the humoral and the cellular levels (165, 166). In this section, we focus on the potential dual roles of antibodies and T cells in DENV and ZIKV protection and pathogenesis.
Antibody-Mediated Protection
Many years of research have shown that both DENV serotype–specific and DENV serotype–cross-reactive antibodies can inhibit DENV infection (58, 117, 167–169). In natural DENV infections, DENV-specific IgM appears first, followed by IgG (mainly IgG1 and IgG3), which persists despite a steady decline in titer (170). For decades, infected patients have had detectable DENV antibodies that are partially cross-reactive for all four DENV serotypes and for other flaviviruses as well. Primary infection induces long-term immunity to homotypic DENV. However, the cross-reactive antibody response induced by a primary DENV infection tends to be low avidity and weakly neutralizing, whereas the cross-reactive antibody response after a secondary DENV infection can be high avidity and highly neutralizing in vitro (171, 172), and subsequent reinfections are rare. Since the recent emergence of ZIKV, multiple groups have reported cross-reactivity between DENV and ZIKV sera and vice versa, in terms of both binding and neutralization (173–176). Thus, how prior DENV immunity affects the subsequent antibody response to ZIKV and vice versa, in terms of specificity/avidity and binding versus neutralizing titers, is now an important question in the field.
The E protein is the main target of neutralizing antibodies in flavivirus infections, and many recent reviews have discussed epitopes recognized by DENV and ZIKV antibodies (165, 177). In particular, EDIII contains epitopes that are recognized by serotype-specific potently neutralizing antibodies in mice (178, 179). However, these EDIII-specific antibodies represent a small proportion of antibodies in DENV-immune human sera (180); instead, the human antibody response to DENV appears to be dominated by weakly neutralizing and highly cross-reactive antibodies that bind the prM protein or fusion loop epitope (FLE) located in EDII (170). In addition, the anti-DENV antibody response in humans consists of antibodies that recognize complex, quaternary epitopes that are present on intact virions but not recombinant E proteins (181, 182). The most strongly neutralizing DENV monoclonal antibodies identified to date are directed against the conformational quaternary epitopes that span across multiple E proteins (183, 184). Recently, E dimer epitope–specific monoclonal antibodies derived from DENV patients were demonstrated to cross-neutralize ZIKV with high potency (175, 176). Other recent studies showed that EDIII, FLE, and complex quaternary epitopes on E are also targets of monoclonal antibodies isolated from ZIKV-infected patients (174, 185–188). One of these studies also found that preexisting DENV immunity is associated with increased ZIKV neutralizing antibody titers against EDIII (188), setting the stage for investigating the influence of prior DENV exposure on the development of an antibody response to ZIKV and vice versa.
Antibody-Dependent Enhancement
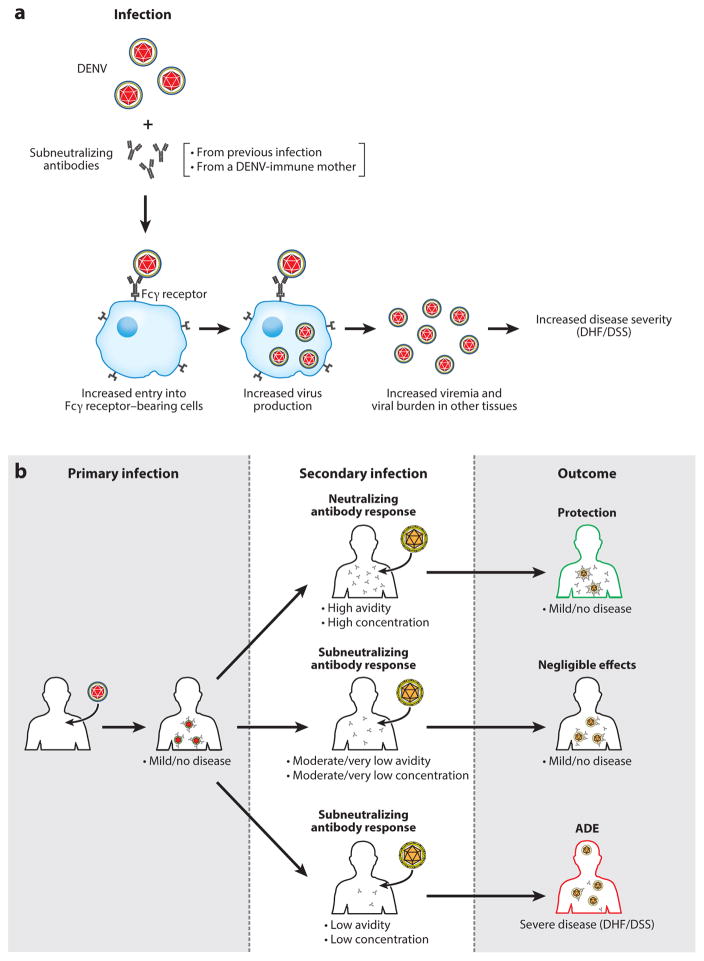
In addition to protection, the antibody response to DENV may contribute to viral replication and disease severity during secondary infection with heterotypic DENV via ADE. According to the ADE hypothesis, DENV-antibody complexes are formed and bind to FcγRs on cells such as macrophages, facilitating viral entry and replication; increased viral loads resulting from ADE then drive the production of inflammatory mediators that increase vascular permeability (Figure 3a). Multiple lines of evidence now support this ADE model. First, mothers of infants with DHF/DSS due to primary infection have DENV-specific antibodies (189), suggesting that antibodies produced during primary infections or passively acquired contribute to DHF/DSS. DHF/DSS has not yet been observed in an infant born to a DENV-naive mother, providing the most compelling argument for the role of anti-DENV antibodies in DENV pathogenesis. Second, studies with nonhuman primates have demonstrated ADE of DENV replication in vivo. Specifically, monkeys develop higher levels and longer duration of viremia in heterologous infections with DENV than in primary infection with the same virus (190), and monkeys passively transferred with DENV-immune human sera (191) or a humanized IgG1 monoclonal antibody have higher viral loads than control monkeys (192). Third, studies with models of DENV infection in mice lacking type I interferon receptor signaling have shown ADE of both DENV replication in vivo and disease severity (58, 193, 194). Recently, a similar passive transfer study using DENV-immune human sera and a model of ZIKV infection in mice lacking STAT2 also demonstrated ADE of ZIKV infection and pathogenesis (195). Collectively, these findings implicate an important role for ADE in both DENV and ZIKV pathogenesis in humans.
Figure 3.
Simplified scheme of outcomes resulting from antibody dynamics: ADE versus protection versus negligible effects. (a) According to the ADE hypothesis, antibodies that are subneutralizing in quality or quantity bind a cross-reactive virion, and this immune complex gains access to permissive cells bearing the Fcγ receptor. Increased viral replication and viremia result in severe disease (DHF/DSS). (b) Antibodies are generated in response to primary infection. At the time of future infection by a cross-reactive virus, four states of antibody CAC result in three categories of clinical outcomes. If CAC is high, then the second infection is neutralized without causing symptoms. If CAC is moderate or minimal, then virus infection is controlled through other mechanisms and Abs have negligible effects. If CAC is low, then antibodies enhance infection and result in severe disease. Abbreviations: Abs, antibodies; ADE, antibody-dependent enhancement; CAC, combined avidity and concentration; DHF/DSS, dengue hemorrhagic fever/dengue shock syndrome.
Studies using a variety of monoclonal antibodies have shown that all neutralizing antibodies can promote ADE in vitro when used at subneutralizing concentrations (196–198). This relates to the concept that a neutralizing antibody can induce ADE if the viral occupancy threshold required for neutralization is not reached (196). Neutralization versus enhancement are simply a function of the stoichiometry of antibody binding to the viral particles (199). For neutralization, an antibody must be of high enough affinity to block correct epitopes on the surface of the virus, and antibody must be present in sufficient concentration. Failure to fulfill either condition will prevent neutralization. Therefore, even neutralizing antibodies will neutralize only if present at sufficiently high concentrations, and they may either mediate ADE if the antibody amount is insufficient for neutralization but sufficient for ADE or exert negligible effect if the antibody amount/condition is insufficient/inappropriate for both ADE and neutralization (Figure 3b). The precise features and molecular mechanisms of ADE are currently unknown. Studies suggest that ADE versus negligible effects of antibodies may depend on engagement of different signaling cascades. For example, signaling-competent and -incompetent forms of FcγR1 and FcγRII induce differential enhancement of DENV immune complex infectivity (200), and FcγRIIB coligation, which requires high antibody concentrations, inhibits ADE (201, 202). ADE may also downregulate activities of key antiviral molecules such as IRF-1 and STAT-1 (203) and increase production of certain cytokines such as TNF, IL-6, and IL-10 (203, 204), and this ADE-mediated evasion of the antiviral response appears to be mediated via coligation of LILRB1, which inhibits expression of ISGs triggered via the activating FcγR pathway (205). Consistent with an important role for FcγR signaling in mediating ADE, a recent study reported a potential role for unfucosylated Fc glycans and particular IgG subclasses in promoting ADE (206). Overall, these results suggest that complex interactions among multiple signaling pathways modulate ADE, providing a framework toward further investigation of ADE mechanisms using relevant human cell culture and animal models.
T Cell Original Antigenic Sin
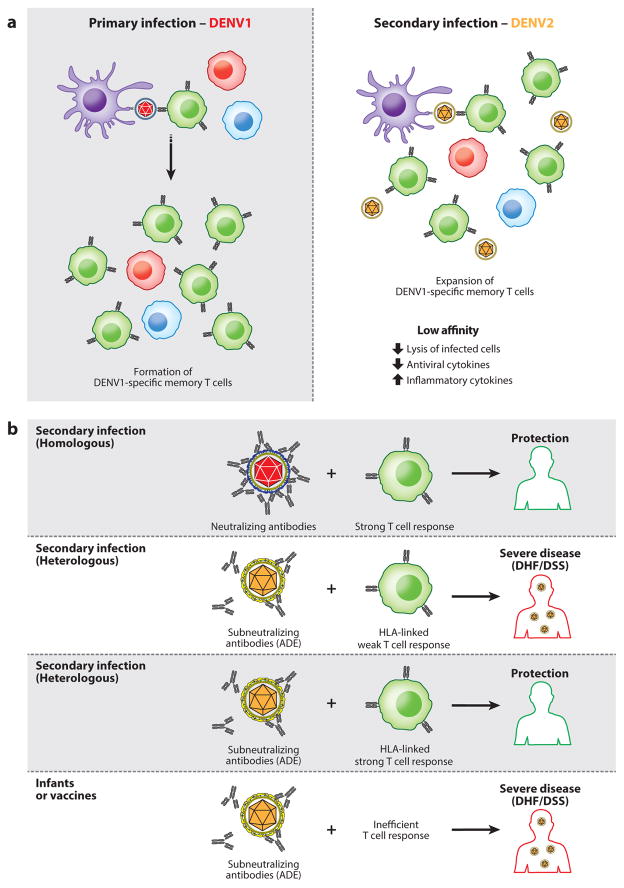
With or without ADE, T cell original antigenic sin postulates that T cells primed against a primary DENV serotype can inappropriately dominate the response to secondary heterotypic infection, resulting in an immune response that can worsen the active secondary infection (Figure 4a). Clinical studies have shown a higher frequency of DENV serotype cross-reactive CD8 T cells with low affinity (207) in the blood of patients experiencing DHF/DSS compared to those with dengue fever, as well as altered cytokine profiles in DENV-specific CD8 T cells from DHF/DSS patients (208, 209). DENV-specific CD4 T cells also produce higher proportions of TNF, a key feature of the cytokine storm in DHF/DSS patients, in response to heterotypic antigen compared to homotypic antigen (210, 211). However, to date, direct evidence for the role of T cells in the pathogenesis of DHF/DSS is lacking.
Figure 4.
T cell original antigenic sin and scheme of outcomes resulting from combined humoral and T cell immunity. (a) T cell original antigenic sin postulates that T cells exposed to a first virus undergo appropriate clonal expansion. However, upon new infection and presentation of cross-reactive viral antigen, the previously expanded T cell populations that have low affinity to the currently infecting virus are activated. This results in a failure to adequately kill infected cells and leads to aberrant, pathogenic immune responses. (b) Evidence better supports a model in which T cells are generally protective against primary infection and secondary infection even under antibody-dependent enhancement (ADE) conditions. However, T cell responses may be attenuated by host factors such as HLA.
Is It Time to Forgive T Cells?
While the evidence suggesting a pathogenic role for T cells is sparse, recent data abound to indicate protective roles for serotype-specific and cross-reactive T cells against DENV infection. The CD8 T cell response to DENV mainly targets the nonstructural proteins NS3, NS4B, and NS5 (212). Studies analyzing epitope-specific CD8 T cell response in blood samples from populations with prior DENV exposure and individuals that have been vaccinated with a live-attenuated tetravalent DENV formulation have revealed that although T cell specificity is skewed toward conserved epitopes, this skew is not associated with impairment of the actual immune response (212–214). In fact, in these populations with prior DENV exposure, the magnitude and breadth of the CD8 T cell response was found to be associated with HLA alleles that correlate with disease susceptibility, suggesting that a high-magnitude/breadth CD8 T cell response is a correlate of protection (212, 215). DENV-specific CD8 T cell responses that were associated with protective HLA alleles were polyfunctional, producing both IFN-γ and TNF-α, and the majority of these cells were effector memory T cells (both CD45RA−CCR7− T effector memory and CD45RA+CCR7− T effector memory RA subsets) that expressed high levels of the programmed death 1 (PD-1) receptor (212, 215). DENV-specific CD8 T cells in the blood may also express CXCR3, CCR5, and the skin-homing marker cutaneous lymphocyte-associated antigen (CLA); these cells can home to the skin during acute DENV infection (216). Finally, higher frequencies of DENV-specific IFN-γ-producing CD8 T cells were also seen in children who developed subclinical infection, compared to those who developed symptomatic secondary DENV infection (217).
Unlike the CD8 T cell response, the anti-DENV CD4 T cell response targets structural proteins (C and E) over nonstructural proteins (218). However, similar to CD8 T cells, CD4 T cells may contribute to protection against DENV. Phase I trials in humans using a live-attenuated vaccine suggested that multifunctional CD4 T cells that produce proinflammatory cytokines might facilitate control of DENV replication or mediate viral clearance (219). In addition, the CD45RA+CCR7− subset of CD4 T cells was expanded in humans with secondary (multiple) DENV infections, particularly in individuals carrying an HLA allele associated with protection from severe disease (220). These CD4 T cells that expand depending on DENV infection history and HLA alleles are likely cytotoxic effectors, based on their expression of granzyme B, perforin, CD107a, and ex vivo DENV-specific cytolytic activity. Thus, both CD4 and CD8 T cells may protect against DENV infection in large part via their cytotoxic effector functions, and HLA alleles seem to dictate the magnitude and breadth of both CD4 and CD8 T cell responses.
In parallel to the evidence in humans, murine studies have demonstrated a critical role for T cells in protection against DENV infection. Specifically, studies using mice lacking type I and/or type II interferon receptors—which support robust DENV replication, unlike their wild-type counterparts, as discussed above—have revealed that CD8 T cells are more important than CD4 T cells in protection against both primary and secondary DENV infections (127, 221–226). In particular, these studies have demonstrated that (a) primary DENV infection elicits polyfunctional effector CD8 T cells that express IFN-γ, TNF, and CD107a and contribute to viral clearance (222), (b) CD8 T cells are required for DENV clearance from the central nervous system (127), (c) CD8 T cells prevent ADE (227), (d) a DENV E protein–based vaccine candidate confers protection via CD8 T cells (226), (e) CD8 T cells are essential for protection against heterotypic reinfection (224), and (f) cross-reactive CD8 T cells that exhibit lower avidity and magnitude of response to heterotypic infection contribute to viral clearance (225). Although CD4 T cells appear not to be essential in controlling DENV infection, they can provide protection in a peptide vaccination setting, also likely through their cytotoxic activities (223). These results for T cell–mediated protection in mice are likely relevant to human infection, as the following observations based on studies with HLA transgenic mice have been validated: (a) The epitopes identified in mice were also recognized by T cells from DENV-exposed humans (225, 228, 229); (b) A dominance of HLA B*0702–restricted responses was detected in both mice and humans (212, 228); (c) CD8 T cell responses target both structural and nonstructural proteins in DENV3 but predominantly nonstructural proteins in the other three DENV serotypes in both mice and humans (214, 229, 230); (d) CD8 T cell responses were broad following primary and homotypic secondary DENV infection in both mice and humans, whereas CD8 T cell responses following heterotypic secondary infection in mice (229) and vaccination with tetravalent live attenuated DENV (214, 231) and natural reinfections in humans (208, 212, 218) were focused toward the conserved nonstructural proteins.
Researchers are also beginning to explore the role of T cells in ZIKV immunity using animal models. T cell responses have been detected in ZIKV-infected nonhuman primates and the peak of activation correlates with ZIKV viral RNA reduction, suggesting protective roles for both CD4 and CD8 T cells in controlling ZIKV replication (232). In all cases, ZIKV rechallenge resulted in complete protection, indicating that primary ZIKV infection induces protective immunity (82, 232). In mice with interferon receptor–competent T cells, CD8 T cells expand and play a critical role in protection against ZIKV infection (233–235). These CD8 T cells express the cytotoxic markers CD107a and granzyme B and exhibit in vivo cytotoxicity. During pregnancy, ZIKV-infected mice have decreased CD8 T cell activation, which could facilitate vertical transmission (235). CD8 T cell responses have also been examined in HLA transgenic mice (166). This study showed that both ZIKV-specific and DENV/ZIKV-cross-reactive CD8 T cells could protect against ZIKV infection. It also revealed that the CD8 T response to ZIKV, similar to the anti-DENV CD8 T cell response, elicits a weak T cell response in HLA-A*0101 transgenic mice compared to HLA-B*0702 transgenic mice; and immunodominance patterns in DENV/ZIKV cross-reactive T cell responses are modulated in a similar manner as heterotypic DENV responses. Collectively, these studies implicate important roles for both ZIKV-specific and DENV/ZIKV-cross-reactive CD8 T cells in protection against ZIKV in humans. Future investigation of both primary and sequential DENV-ZIKV and ZIKV-DENV infections in mice, nonhuman primates, and humans should provide insights into the precise quality and quantity of protective CD8 T cell responses to both viruses, and they may also provide evidence in support of a protective role for CD4 T cells against ZIKV in certain settings.
CONCLUSION
While vaccines have successfully enlisted the antibody response to prevent large-scale morbidity and mortality caused by other viruses, the studies reviewed herein indicate that the four serotypes of DENV present a cross-reactive challenge requiring a different approach. Early studies of DENV-ZIKV cross-immunity further suggest that a more complex, potentially high-risk immunological puzzle is emerging. Two principles to guide vaccine development for these flaviviruses emerge from our investigative experience and assessment of clinical and research studies: Embrace complexity. Do no harm.
Viral species and strain/serotype, host age, prior infection/vaccination status, time since sero-conversion, host genetic factors (in particular HLA), and individual immune status all must be considered when interpreting clinical and research data. While some data would implicate innate factors, antibodies, or T cells in protection or pathogenesis, clinical outcomes reflect the sum of all these factors. The model we present is one of innate immunity backstopped by humoral immunity backstopped by cellular immunity. In most primary and secondary cases of DENV and ZIKV infection, innate immunity is sufficiently backstopped by adaptive immunity to neutralize the virus and result in asymptomatic or negligible infection. However, in a minority of primary infections a weakened innate immune response can result in more severe clinical signs. Secondary infection by a cross-reactive serotype or virus challenges the host to produce an appropriate adaptive response based on memory of a different virus. A strong antibody response will neutralize the second virus and result in negligible disease. A minimal antibody response is insufficient in combined antibody avidity and concentration (CAC) to generate ADE conditions, and a strong T cell response stands behind humoral immunity to neutralize the infection. But if at the time of secondary infection CAC is in a low range conducive to ADE, then a strong T cell response is required to neutralize the infection and abrogate the effects of ADE. If the T cell response is attenuated by host factors such as HLA or inadequate antigen presentation, then ADE will progress unchecked (Figure 4b).
Finally, beginning with epidemiologic observations made nearly 50 years ago of dengue patients in Thailand, enough scientific evidence has been established to indicate that modulation of host flavivirus immunity carries the risk of producing harmful effects. Given this documented risk, flaviviral vaccines should be designed to elicit strong responses from all branches of immunity, and vaccines and therapies should be diligently tested prior to release. Today no animal model matches the mouse for combined capacity to isolate and manipulate single variables, measure in vivo response, repeat and refine experiments, and generate statistical power. Further investigations of flaviviral infection in mouse models, in combination with studies using nonhuman primates and human samples, will prove invaluable to generating safe and effective vaccines and therapeutics.
Acknowledgments
The authors thank Dr. Kenneth Kim for critical review of the manuscript and Anh-Viet Nguyen for preparing the figures. This manuscript was supported by NIH grants (RO1 AI116813, R21 NS100477, and R21 AI127988) and La Jolla Institute for Allergy and Immunology institutional support.
Footnotes
DISCLOSURE STATEMENT
S.S. is listed as an inventor on US patent application 14/800468, “Dengue Virus (DV) Polypeptide Sequences, T Cell Epitopes, and Methods and Uses Thereof.”
LITERATURE CITED
- 1.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, et al. The global distribution and burden of dengue. Nature. 2013;496:504–7. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gubler DJ. The economic burden of dengue. Am J Trop Med Hyg. 2012;86:743–44. doi: 10.4269/ajtmh.2012.12-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solomon T, Dung NM, Vaughn DW, Kneen R, Thao LT, et al. Neurological manifestations of dengue infection. Lancet. 2000;355:1053–59. doi: 10.1016/S0140-6736(00)02036-5. [DOI] [PubMed] [Google Scholar]
- 4.Messer WB, Gubler DJ, Harris E, Sivananthan K, de Silva AM. Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg Infect Dis. 2003;9:800–9. doi: 10.3201/eid0907.030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science. 2015;350:217–21. doi: 10.1126/science.aab3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cologna R, Rico-Hesse R. American genotype structures decrease dengue virus output from human monocytes and dendritic cells. J Virol. 2003;77:3929–38. doi: 10.1128/JVI.77.7.3929-3938.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halstead SB. Dengue. Lancet. 2007;370:1644–52. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. Zika virus and birth defects—reviewing the evidence for causality. N Engl J Med. 2016;374:1981–87. doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- 9.Watrin L, Ghawche F, Larre P, Neau JP, Mathis S, Fournier E. Guillain-Barre syndrome (42 cases) occurring during a Zika virus outbreak in French Polynesia. Medicine. 2016;95:e3257. doi: 10.1097/MD.0000000000003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parra B, Lizarazo J, Jimenez-Arango JA, Zea-Vera AF, Gonzalez-Manrique G, et al. Guillain-Barre syndrome associated with Zika virus infection in Colombia. N Engl J Med. 2016;375:1513–23. doi: 10.1056/NEJMoa1605564. [DOI] [PubMed] [Google Scholar]
- 11.Cao-Lormeau VM, Blake A, Mons S, Lastere S, Roche C, et al. Guillain-Barre syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet. 2016;387:1531–39. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malkki H. CNS infections: Zika virus infection could trigger Guillain-Barre syndrome. Nat Rev Neurol. 2016;12:187. doi: 10.1038/nrneurol.2016.30. [DOI] [PubMed] [Google Scholar]
- 13.D’Ortenzio E, Matheron S, Yazdanpanah Y, de Lamballerie X, Hubert B, et al. Evidence of sexual transmission of Zika virus. N Engl J Med. 2016;374:2195–98. doi: 10.1056/NEJMc1604449. [DOI] [PubMed] [Google Scholar]
- 14.Moreira J, Peixoto TM, Siqueira AM, Lamas CC. Sexually acquired Zika virus: a systematic review. Clin Microbiol Infect. 2017;23:296–305. doi: 10.1016/j.cmi.2016.12.027. [DOI] [PubMed] [Google Scholar]
- 15.Barzon L, Pacenti M, Franchin E, Lavezzo E, Trevisan M, et al. Infection dynamics in a traveller with persistent shedding of Zika virus RNA in semen for six months after returning from Haiti to Italy, January 2016. Eurosurveillance. 2016;21:30316. doi: 10.2807/1560-7917.ES.2016.21.32.30316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Froeschl G, Huber K, von Sonnenburg F, Nothdurft HD, Bretzel G, et al. Long-term kinetics of Zika virus RNA and antibodies in body fluids of a vasectomized traveller returning from Martinique: a case report. BMC Infect Dis. 2017;17:55. doi: 10.1186/s12879-016-2123-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray KO, Gorchakov R, Carlson AR, Berry R, Lai L, et al. Prolonged detection of Zika virus in vaginal secretions and whole blood. Emerg Infect Dis. 2017;23:99–101. doi: 10.3201/eid2301.161394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pierson TC, Graham BS. Zika virus: immunity and vaccine development. Cell. 2016;167:625–31. doi: 10.1016/j.cell.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barouch DH, Thomas SJ, Michael NL. Prospects for a Zika virus vaccine. Immunity. 2017;46:176–82. doi: 10.1016/j.immuni.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X, Vaughan K, Weiskopf D, Grifoni A, Diamond MS, et al. Identifying candidate targets of immune responses in Zika virus based on homology to epitopes in other flavivirus species. PLOS Currents Outbreaks. 2016 Nov 15; doi: 10.1371/currents.outbreaks.9aa2e1fb61b0f632f58a098773008c4b. https://dx.doi.org/10.1371/currents.outbreaks.9aa2e1fb61b0f632f58a098773008c4b. [DOI] [PMC free article] [PubMed]
- 21.Dowd KA, DeMaso CR, Pelc RS, Speer SD, Smith AR, et al. Broadly neutralizing activity of Zika virus-immune sera identifies a single viral serotype. Cell Rep. 2016;16:1485–91. doi: 10.1016/j.celrep.2016.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- 23.Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 24.Zybert IA, van der Ende-Metselaar H, Wilschut J, Smit JM. Functional importance of dengue virus maturation: infectious properties of immature virions. J Gen Virol. 2008;89:3047–51. doi: 10.1099/vir.0.2008/002535-0. [DOI] [PubMed] [Google Scholar]
- 25.Junjhon J, Lausumpao M, Supasa S, Noisakran S, Songjaeng A, et al. Differential modulation of prM cleavage, extracellular particle distribution, and virus infectivity by conserved residues at nonfurin consensus positions of the dengue virus pr-M junction. J Virol. 2008;82:10776–91. doi: 10.1128/JVI.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–19. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 27.Modis Y, Ogata S, Clements D, Harrison SC. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J Virol. 2005;79:1223–31. doi: 10.1128/JVI.79.2.1223-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gack MU, Diamond MS. Innate immune escape by Dengue and West Nile viruses. Curr Opin Virol. 2016;20:119–28. doi: 10.1016/j.coviro.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bidet K, Dadlani D, Garcia-Blanco MA. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLOS Pathog. 2014;10:e1004242. doi: 10.1371/journal.ppat.1004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donald CL, Brennan B, Cumberworth SL, Rezelj VV, Clark JJ, et al. Full genome sequence and sfRNA interferon antagonist activity of Zika virus from Recife, Brazil. PLOS Negl Trop Dis. 2016;10:e0005048. doi: 10.1371/journal.pntd.0005048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Maguire T, Hileman RE, Fromm JR, Esko JD, et al. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med. 1997;3:866–71. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 32.Prestwood TR, Prigozhin DM, Sharar KL, Zellweger RM, Shresta S. A mouse-passaged dengue virus strain with reduced affinity for heparan sulfate causes severe disease in mice by establishing increased systemic viral loads. J Virol. 2008;82:8411–21. doi: 10.1128/JVI.00611-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tassaneetrithep B, Burgess TH, Granelli-Piperno A, Trumpfheller C, Finke J, et al. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J Exp Med. 2003;197:823–29. doi: 10.1084/jem.20021840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navarro-Sanchez E, Altmeyer R, Amara A, Schwartz O, Fieschi F, et al. Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep. 2003;4:1–6. doi: 10.1038/sj.embor.embor866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dejnirattisai W, Webb AI, Chan V, Jumnainsong A, Davidson A, et al. Lectin switching during dengue virus infection. J Infect Dis. 2011;203:1775–83. doi: 10.1093/infdis/jir173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller JL, de Wet BJ, Martinez-Pomares L, Radcliffe CM, Dwek RA, et al. The mannose receptor mediates dengue virus infection of macrophages. PLOS Pathog. 2008;4:e17. doi: 10.1371/journal.ppat.0040017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meertens L, Carnec X, Lecoin MP, Ramdasi R, Guivel-Benhassine F, et al. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe. 2012;12:544–57. doi: 10.1016/j.chom.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carnec X, Meertens L, Dejarnac O, Perera-Lecoin M, Hafirassou ML, et al. The phosphatidylserine and phosphatidylethanolamine receptor CD300a binds dengue virus and enhances infection. J Virol. 2015;90:92–102. doi: 10.1128/JVI.01849-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wells MF, Salick MR, Wiskow O, Ho DJ, Worringer KA, et al. Genetic ablation of AXL does not protect human neural progenitor cells and cerebral organoids from Zika virus infection. Cell Stem Cell. 2016;19:703–8. doi: 10.1016/j.stem.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 40.Govero J, Esakky P, Scheaffer SM, Fernandez E, Drury A, et al. Zika virus infection damages the testes in mice. Nature. 2016;540:438–42. doi: 10.1038/nature20556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hastings AK, Yockey LJ, Jagger BW, Hwang J, Uraki R, et al. TAM receptors are not required for Zika virus infection in mice. Cell Rep. 2017;19:558–68. doi: 10.1016/j.celrep.2017.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miner JJ, Sene A, Richner JM, Smith AM, Santeford A, et al. Zika virus infection in mice causes panuveitis with shedding of virus in tears. Cell Rep. 2016;16:3208–18. doi: 10.1016/j.celrep.2016.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prasad VM, Miller AS, Klose T, Sirohi D, Buda G, et al. Structure of the immature Zika virus at 9 Å resolution. Nat Struct Mol Biol. 2017;24:184–86. doi: 10.1038/nsmb.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodenhuis-Zybert IA, van der Schaar HM, da Silva Voorham JM, van der Ende-Metselaar H, Lei HY, et al. Immature dengue virus: a veiled pathogen? PLOS Pathog. 2010;6:e1000718. doi: 10.1371/journal.ppat.1000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hall WC, Crowell TP, Watts DM, Barros VL, Kruger H, et al. Demonstration of yellow fever and dengue antigens in formalin-fixed paraffin-embedded human liver by immunohistochemical analysis. Am J Trop Med Hyg. 1991;45:408–17. doi: 10.4269/ajtmh.1991.45.408. [DOI] [PubMed] [Google Scholar]
- 46.Bhoopat L, Bhamarapravati N, Attasiri C, Yoksarn S, Chaiwun B, et al. Immunohistochemical characterization of a new monoclonal antibody reactive with dengue virus-infected cells in frozen tissue using immunoperoxidase technique. Asian Pac J Allergy Immunol. 1996;14:107–13. [PubMed] [Google Scholar]
- 47.Jessie K, Fong MY, Devi S, Lam SK, Wong KT. Localization of dengue virus in naturally infected human tissues, by immunohistochemistry and in situ hybridization. J Infect Dis. 2004;189:1411–18. doi: 10.1086/383043. [DOI] [PubMed] [Google Scholar]
- 48.Balsitis SJ, Coloma J, Castro G, Alava A, Flores D, et al. Tropism of dengue virus in mice and humans defined by viral nonstructural protein 3-specific immunostaining. Am J Trop Med Hyg. 2009;80:416–24. [PubMed] [Google Scholar]
- 49.Couvelard A, Marianneau P, Bedel C, Drouet MT, Vachon F, et al. Report of a fatal case of dengue infection with hepatitis: demonstration of dengue antigens in hepatocytes and liver apoptosis. Hum Pathol. 1999;30:1106–10. doi: 10.1016/s0046-8177(99)90230-7. [DOI] [PubMed] [Google Scholar]
- 50.Huerre MR, Lan NT, Marianneau P, Hue NB, Khun H, et al. Liver histopathology and biological correlates in five cases of fatal dengue fever in Vietnamese children. Virchows Arch. 2001;438:107–15. doi: 10.1007/s004280000329. [DOI] [PubMed] [Google Scholar]
- 51.Ramos C, Sanchez G, Pando RH, Baquera J, Hernandez D, et al. Dengue virus in the brain of a fatal case of hemorrhagic dengue fever. J Neurovirol. 1998;4:465–68. doi: 10.3109/13550289809114548. [DOI] [PubMed] [Google Scholar]
- 52.Durbin AP, Vargas MJ, Wanionek K, Hammond SN, Gordon A, et al. Phenotyping of peripheral blood mononuclear cells during acute dengue illness demonstrates infection and increased activation of monocytes in severe cases compared to classic dengue fever. Virology. 2008;376:429–35. doi: 10.1016/j.virol.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu SJ, Grouard-Vogel G, Sun W, Mascola JR, Brachtel E, et al. Human skin Langerhans cells are targets of dengue virus infection. Nat Med. 2000;6:816–20. doi: 10.1038/77553. [DOI] [PubMed] [Google Scholar]
- 54.Cerny D, Haniffa M, Shin A, Bigliardi P, Tan BK, et al. Selective susceptibility of human skin antigen presenting cells to productive dengue virus infection. PLOS Pathog. 2014;10:e1004548. doi: 10.1371/journal.ppat.1004548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blackley S, Kou Z, Chen H, Quinn M, Rose RC, et al. Primary human splenic macrophages, but not T or B cells, are the principal target cells for dengue virus infection in vitro. J Virol. 2007;81:13325–34. doi: 10.1128/JVI.01568-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmid MA, Harris E. Monocyte recruitment to the dermis and differentiation to dendritic cells increases the targets for dengue virus replication. PLOS Pathog. 2014;10:e1004541. doi: 10.1371/journal.ppat.1004541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prestwood TR, May MM, Plummer EM, Morar MM, Yauch LE, Shresta S. Trafficking and replication patterns reveal splenic macrophages as major targets of dengue virus in mice. J Virol. 2012;86:12138–47. doi: 10.1128/JVI.00375-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zellweger RM, Prestwood TR, Shresta S. Enhanced infection of liver sinusoidal endothelial cells in a mouse model of antibody-induced severe dengue disease. Cell Host Microbe. 2010;7:128–39. doi: 10.1016/j.chom.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Eijk AA, van Genderen PJ, Verdijk RM, Reusken CB, Mögling R, et al. Miscarriage associated with Zika virus infection. N Engl J Med. 2016;375:1002–4. doi: 10.1056/NEJMc1605898. [DOI] [PubMed] [Google Scholar]
- 60.Rosenberg AZ, Yu W, Hill DA, Reyes CA, Schwartz DA. Placental pathology of Zika virus: Viral infection of the placenta induces villous stromal macrophage (Hofbauer cell) proliferation and hyperplasia. Arch Pathol Lab Med. 2017;141:43–48. doi: 10.5858/arpa.2016-0401-OA. [DOI] [PubMed] [Google Scholar]
- 61.Schwartz DA. Viral infection, proliferation, and hyperplasia of Hofbauer cells and absence of inflammation characterize the placental pathology of fetuses with congenital Zika virus infection. Arch Gynecol Obstet. 2017;295:1361–68. doi: 10.1007/s00404-017-4361-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhatnagar J, Rabeneck DB, Martines RB, Reagan-Steiner S, Ermias Y, et al. Zika virus RNA replication and persistence in brain and placental tissue. Emerg Infect Dis. 2017;23:405–14. doi: 10.3201/eid2303.161499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miner JJ, Diamond MS. Zika virus pathogenesis and tissue tropism. Cell Host Microbe. 2017;21:134–42. doi: 10.1016/j.chom.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Retallack H, Di Lullo E, Arias C, Knopp KA, Laurie MT, et al. Zika virus cell tropism in the developing human brain and inhibition by azithromycin. PNAS. 2016;113:14408–13. doi: 10.1073/pnas.1618029113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aagaard KM, Lahon A, Suter MA, Arya RP, Seferovic MD, et al. Primary human placental trophoblasts are permissive for Zika virus (ZIKV) replication. Sci Rep. 2017;7:41389. doi: 10.1038/srep41389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tabata T, Petitt M, Puerta-Guardo H, Michlmayr D, Wang C, et al. Zika virus targets different primary human placental cells, suggesting two routes for vertical transmission. Cell Host Microbe. 2016;20:155–66. doi: 10.1016/j.chom.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen JC, Wang Z, Huang H, Weitz SH, Wang A, et al. Infection of human uterine fibroblasts by Zika virus in vitro: implications for viral transmission in women. Int J Infect Dis. 2016;51:139–40. doi: 10.1016/j.ijid.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 68.Hou W, Armstrong N, Obwolo LA, Thomas M, Pang X, et al. Determination of the cell permissiveness spectrum, mode of RNA replication, and RNA-protein interaction of Zika virus. BMC Infect Dis. 2017;17:239. doi: 10.1186/s12879-017-2338-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Azevedo RS, Araujo MT, Martins Filho AJ, Oliveira CS, Nunes BT, et al. Zika virus epidemic in Brazil: I. Fatal disease in adults; clinical and laboratorial aspects. J Clin Virol. 2016;85:56–64. doi: 10.1016/j.jcv.2016.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Noronha L, Zanluca C, Azevedo ML, Luz KG, Santos CN. Zika virus damages the human placental barrier and presents marked fetal neurotropism. Mem Inst Oswaldo Cruz. 2016;111:287–93. doi: 10.1590/0074-02760160085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martines RB, Bhatnagar J, de Oliveira Ramos AM, Davi HP, Iglezias SD, et al. Pathology of congenital Zika syndrome in Brazil: a case series. Lancet. 2016;388:898–904. doi: 10.1016/S0140-6736(16)30883-2. [DOI] [PubMed] [Google Scholar]
- 72.El Costa H, Gouilly J, Mansuy JM, Chen Q, Levy C, et al. ZIKA virus reveals broad tissue and cell tropism during the first trimester of pregnancy. Sci Rep. 2016;6:35296. doi: 10.1038/srep35296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tang H, Hammack C, Ogden SC, Wen Z, Qian X, et al. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell. 2016;18:587–90. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jurado KA, Simoni MK, Tang Z, Uraki R, Hwang J, et al. Zika virus productively infects primary human placenta-specific macrophages. JCI Insight. 2016;1:e88461. doi: 10.1172/jci.insight.88461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quicke KM, Bowen JR, Johnson EL, McDonald CE, Ma H, et al. Zika virus infects human placental macrophages. Cell Host Microbe. 2016;20:83–90. doi: 10.1016/j.chom.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alcendor DJ. Zika virus infection of the human glomerular cells: implications for viral reservoirs and renal pathogenesis. J Infect Dis. 2017;216:162–71. doi: 10.1093/infdis/jix171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pagani I, Ghezzi S, Ulisse A, Rubio A, Turrini F, et al. Human endometrial stromal cells are highly permissive to productive infection by Zika virus. Sci Rep. 2017;7:44286. doi: 10.1038/srep44286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roach T, Alcendor DJ. Zika virus infection of cellular components of the blood-retinal barriers: implications for viral associated congenital ocular disease. J Neuroinflamm. 2017;14:43. doi: 10.1186/s12974-017-0824-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hamel R, Dejarnac O, Wichit S, Ekchariyawat P, Neyret A, et al. Biology of Zika virus infection in human skin cells. J Virol. 2015;89:8880–96. doi: 10.1128/JVI.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu S, DeLalio LJ, Isakson BE, Wang TT. AXL-mediated productive infection of human endothelial cells by Zika virus. Circ Res. 2016;119:1183–89. doi: 10.1161/CIRCRESAHA.116.309866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nguyen SM, Antony KM, Dudley DM, Kohn S, Simmons HA, et al. Highly efficient maternal-fetal Zika virus transmission in pregnant rhesus macaques. PLOS Pathog. 2017;13:e1006378. doi: 10.1371/journal.ppat.1006378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Osuna CE, Lim SY, Deleage C, Griffin BD, Stein D, et al. Zika viral dynamics and shedding in rhesus and cynomolgus macaques. Nat Med. 2016;22:1448–55. doi: 10.1038/nm.4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hirsch AJ, Smith JL, Haese NN, Broeckel RM, Parkins CJ, et al. Zika virus infection of rhesus macaques leads to viral persistence in multiple tissues. PLOS Pathog. 2017;13:e1006219. doi: 10.1371/journal.ppat.1006219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aid M, Abbink P, Larocca RA, Boyd M, Nityanandam R, et al. Zika virus persistence in the central nervous system and lymph nodes of rhesus monkeys. Cell. 2017;169:610–20. e14. doi: 10.1016/j.cell.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li H, Saucedo-Cuevas L, Regla-Nava JA, Chai G, Sheets N, et al. Zika virus infects neural progenitors in the adult mouse brain and alters proliferation. Cell Stem Cell. 2016;19:593–98. doi: 10.1016/j.stem.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brault JB, Khou C, Basset J, Coquand L, Fraisier V, et al. Comparative analysis between flaviviruses reveals specific neural stem cell tropism for Zika virus in the mouse developing neocortex. EBioMedicine. 2016;10:71–76. doi: 10.1016/j.ebiom.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Singh PK, Guest JM, Kanwar M, Boss J, Gao N, et al. Zika virus infects cells lining the blood-retinal barrier and causes chorioretinal atrophy in mouse eyes. JCI Insight. 2017;2:e92340. doi: 10.1172/jci.insight.92340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Uraki R, Hwang J, Jurado KA, Householder S, Yockey LJ, et al. Zika virus causes testicular atrophy. Sci Adv. 2017;3:e1602899. doi: 10.1126/sciadv.1602899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tang WW, Young MP, Mamidi A, Regla-Nava JA, Kim K, Shresta S. A mouse model of Zika virus sexual transmission and vaginal viral replication. Cell Rep. 2016;17:3091–98. doi: 10.1016/j.celrep.2016.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bingham AM, Cone M, Mock V, Heberlein-Larson L, Stanek D, et al. Comparison of test results for Zika virus RNA in urine, serum, and saliva specimens from persons with travel-associated Zika virus disease—Florida, 2016. MMWR. 2016;65:475–78. doi: 10.15585/mmwr.mm6518e2. [DOI] [PubMed] [Google Scholar]
- 91.Tan JJL, Balne PK, Leo YS, Tong L, Ng LFP, Agrawal R. Persistence of Zika virus in conjunctival fluid of convalescence patients. Sci Rep. 2017;7:11194. doi: 10.1038/s41598-017-09479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Furtado JM, Esposito DL, Klein TM, Teixeira-Pinto T, da Fonseca BA. Uveitis associated with Zika virus infection. New Eng J Med. 2016;375:394–96. doi: 10.1056/NEJMc1603618. [DOI] [PubMed] [Google Scholar]
- 93.Colt S, Garcia-Casal MN, Pena-Rosas JP, Finkelstein JL, Rayco-Solon P, et al. Transmission of Zika virus through breast milk and other breastfeeding-related bodily-fluids: a systematic review. PLOS Negl Trop Dis. 2017;11:e0005528. doi: 10.1371/journal.pntd.0005528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.van den Broek MF, Muller U, Huang S, Zinkernagel RM, Aguet M. Immune defence in mice lacking type I and/or type II interferon receptors. Immunol Rev. 1995;148:5–18. doi: 10.1111/j.1600-065x.1995.tb00090.x. [DOI] [PubMed] [Google Scholar]
- 95.Zevini A, Olagnier D, Hiscott J. Crosstalk between cytoplasmic RIG-I and STING sensing pathways. Trends Immunol. 2017;38:194–205. doi: 10.1016/j.it.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–45. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kurane I, Innis BL, Nimmannitya S, Nisalak A, Meager A, Ennis FA. High levels of interferon alpha in the sera of children with dengue virus infection. Am J Trop Med Hyg. 1993;48:222–29. doi: 10.4269/ajtmh.1993.48.222. [DOI] [PubMed] [Google Scholar]
- 98.Becquart P, Wauquier N, Nkoghe D, Ndjoyi-Mbiguino A, Padilla C, et al. Acute dengue virus 2 infection in Gabonese patients is associated with an early innate immune response, including strong interferon alpha production. BMC Infect Dis. 2010;10:356. doi: 10.1186/1471-2334-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sun P, Garcia J, Comach G, Vahey MT, Wang Z, et al. Sequential waves of gene expression in patients with clinically defined dengue illnesses reveal subtle disease phases and predict disease severity. PLOS Negl Trop Dis. 2013;7:e2298. doi: 10.1371/journal.pntd.0002298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Simmons CP, Popper S, Dolocek C, Chau TN, Griffiths M, et al. Patterns of host genome-wide gene transcript abundance in the peripheral blood of patients with acute dengue hemorrhagic fever. J Infect Dis. 2007;195:1097–107. doi: 10.1086/512162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tolfvenstam T, Lindblom A, Schreiber MJ, Ling L, Chow A, et al. Characterization of early host responses in adults with dengue disease. BMC Infect Dis. 2011;11:209. doi: 10.1186/1471-2334-11-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–45. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wies E, Wang MK, Maharaj NP, Chen K, Zhou S, et al. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 2013;38:437–49. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Diamond MS, Roberts TG, Edgil D, Lu B, Ernst J, Harris E. Modulation of dengue virus infection in human cells by alpha, beta, and gamma interferons. J Virol. 2000;74:4957–66. doi: 10.1128/jvi.74.11.4957-4966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Diamond MS, Harris E. Interferon inhibits dengue virus infection by preventing translation of viral RNA through a PKR-independent mechanism. Virology. 2001;289:297–311. doi: 10.1006/viro.2001.1114. [DOI] [PubMed] [Google Scholar]
- 106.Schmid B, Rinas M, Ruggieri A, Acosta EG, Bartenschlager M, et al. Live cell analysis and mathematical modeling identify determinants of attenuation of dengue virus 2′-O-methylation mutant. PLOS Pathog. 2015;11:e1005345. doi: 10.1371/journal.ppat.1005345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jiang D, Weidner JM, Qing M, Pan XB, Guo H, et al. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J Virol. 2010;84:8332–41. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Helbig KJ, Carr JM, Calvert JK, Wati S, Clarke JN, et al. Viperin is induced following dengue virus type-2 (DENV-2) infection and has anti-viral actions requiring the C-terminal end of viperin. PLOS Negl Trop Dis. 2013;7:e2178. doi: 10.1371/journal.pntd.0002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–54. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schoggins JW, Dorner M, Feulner M, Imanaka N, Murphy MY, et al. Dengue reporter viruses reveal viral dynamics in interferon receptor-deficient mice and sensitivity to interferon effectors in vitro. PNAS. 2012;109:14610–15. doi: 10.1073/pnas.1212379109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Suzuki Y, Chin WX, Han Q, Ichiyama K, Lee CH, et al. Characterization of RyDEN (C19orf66) as an interferon-stimulated cellular inhibitor against dengue virus replication. PLOS Pathog. 2016;12:e1005357. doi: 10.1371/journal.ppat.1005357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Balinsky CA, Schmeisser H, Wells AI, Ganesan S, Jin T, et al. IRAV (FLJ11286), an interferon-stimulated gene with antiviral activity against dengue virus, interacts with MOV10. J Virol. 2017;91:e01606–16. doi: 10.1128/JVI.01606-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lin RJ, Yu HP, Chang BL, Tang WC, Liao CL, Lin YL. Distinct antiviral roles for human 2′,5′-oligoadenylate synthetase family members against dengue virus infection. J Immunol. 2009;183:8035–43. doi: 10.4049/jimmunol.0902728. [DOI] [PubMed] [Google Scholar]
- 114.Savidis G, Perreira JM, Portmann JM, Meraner P, Guo Z, et al. The IFITMs inhibit Zika virus replication. Cell Rep. 2016;15:2323–30. doi: 10.1016/j.celrep.2016.05.074. [DOI] [PubMed] [Google Scholar]
- 115.Fu B, Wang L, Li S, Dorf ME. ZMPSTE24 defends against influenza and other pathogenic viruses. J Exp Med. 2017;214:919–29. doi: 10.1084/jem.20161270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shresta S, Kyle JL, Snider HM, Basavapatna M, Beatty PR, Harris E. Interferon-dependent immunity is essential for resistance to primary dengue virus infection in mice, whereas T- and B-cell-dependent immunity are less critical. J Virol. 2004;78:2701–10. doi: 10.1128/JVI.78.6.2701-2710.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Johnson AJ, Roehrig JT. New mouse model for dengue virus vaccine testing. J Virol. 1999;73:783–86. doi: 10.1128/jvi.73.1.783-786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yockey LJ, Varela L, Rakib T, Khoury-Hanold W, Fink SL, et al. Vaginal exposure to Zika virus during pregnancy leads to fetal brain infection. Cell. 2016;166:1247–56. e4. doi: 10.1016/j.cell.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Suthar MS, Aguirre S, Fernandez-Sesma A. Innate immune sensing of flaviviruses. PLOS Pathog. 2013;9:e1003541. doi: 10.1371/journal.ppat.1003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Perry ST, Prestwood TR, Lada SM, Benedict CA, Shresta S. Cardif-mediated signaling controls the initial innate response to dengue virus in vivo. J Virol. 2009;83:8276–81. doi: 10.1128/JVI.00365-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen HW, King K, Tu J, Sanchez M, Luster AD, Shresta S. The roles of IRF-3 and IRF-7 in innate antiviral immunity against dengue virus. J Immunol. 2013;191:4194–201. doi: 10.4049/jimmunol.1300799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang JP, Liu P, Latz E, Golenbock DT, Finberg RW, Libraty DH. Flavivirus activation of plasmacytoid dendritic cells delineates key elements of TLR7 signaling beyond endosomal recognition. J Immunol. 2006;177:7114–21. doi: 10.4049/jimmunol.177.10.7114. [DOI] [PubMed] [Google Scholar]
- 123.Nasirudeen AM, Wong HH, Thien P, Xu S, Lam KP, Liu DX. RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PLOS Negl Trop Dis. 2011;5:e926. doi: 10.1371/journal.pntd.0000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lazear HM, Lancaster A, Wilkins C, Suthar MS, Huang A, et al. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLOS Pathog. 2013;9:e1003118. doi: 10.1371/journal.ppat.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, et al. A mouse model of Zika virus pathogenesis. Cell Host Microbe. 2016;19:720–30. doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Brien JD, Daffis S, Lazear HM, Cho H, Suthar MS, et al. Interferon regulatory factor-1 (IRF-1) shapes both innate and CD8+ T cell immune responses against West Nile virus infection. PLOS Pathog. 2011;7:e1002230. doi: 10.1371/journal.ppat.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Prestwood TR, Morar MM, Zellweger RM, Miller R, May MM, et al. Gamma interferon (IFN-γ) receptor restricts systemic dengue virus replication and prevents paralysis in IFN-α/βreceptor-deficient mice. J Virol. 2012;86:12561–70. doi: 10.1128/JVI.06743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Orozco S, Schmid MA, Parameswaran P, Lachica R, Henn MR, et al. Characterization of a model of lethal dengue virus 2 infection in C57BL/6 mice deficient in the alpha/beta interferon receptor. J Gen Virol. 2012;93:2152–57. doi: 10.1099/vir.0.045088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pinto AK, Brien JD, Lam CY, Johnson S, Chiang C, et al. Defining new therapeutics using a more immunocompetent mouse model of antibody-enhanced dengue virus infection. mBio. 2015;6:e01316–15. doi: 10.1128/mBio.01316-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zust R, Toh YX, Valdes I, Cerny D, Heinrich J, et al. Type I interferon signals in macrophages and dendritic cells control dengue virus infection: implications for a new mouse model to test dengue vaccines. J Virol. 2014;88:7276–85. doi: 10.1128/JVI.03827-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shresta S, Sharar KL, Prigozhin DM, Snider HM, Beatty PR, Harris E. Critical roles for both STAT1-dependent and STAT1-independent pathways in the control of primary dengue virus infection in mice. J Immunol. 2005;175:3946–54. doi: 10.4049/jimmunol.175.6.3946. [DOI] [PubMed] [Google Scholar]
- 132.Perry ST, Buck MD, Lada SM, Schindler C, Shresta S. STAT2 mediates innate immunity to dengue virus in the absence of STAT1 via the type I interferon receptor. PLOS Pathog. 2011;7:e1001297. doi: 10.1371/journal.ppat.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tripathi S, Balasubramaniam VR, Brown JA, Mena I, Grant A, et al. A novel Zika virus mouse model reveals strain specific differences in virus pathogenesis and host inflammatory immune responses. PLOS Pathog. 2017;13:e1006258. doi: 10.1371/journal.ppat.1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rossi SL, Tesh RB, Azar SR, Muruato AE, Hanley KA, et al. Characterization of a novel murine model to study Zika virus. Am J Trop Med Hyg. 2016;94:1362–69. doi: 10.4269/ajtmh.16-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dowall SD, Graham VA, Rayner E, Atkinson B, Hall G, et al. A susceptible mouse model for Zika virus infection. PLOS Negl Trop Dis. 2016;10:e0004658. doi: 10.1371/journal.pntd.0004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ashour J, Morrison J, Laurent-Rolle M, Belicha-Villanueva A, Plumlee CR, et al. Mouse STAT2 restricts early dengue virus replication. Cell Host Microbe. 2010;8:410–21. doi: 10.1016/j.chom.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Olagnier D, Scholte FE, Chiang C, Albulescu IC, Nichols C, et al. Inhibition of dengue and chikungunya virus infections by RIG-I-mediated type I interferon-independent stimulation of the innate antiviral response. J Virol. 2014;88:4180–94. doi: 10.1128/JVI.03114-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bayer A, Lennemann NJ, Ouyang Y, Bramley JC, Morosky S, et al. Type III interferons produced by human placental trophoblasts confer protection against Zika virus infection. Cell Host Microbe. 2016;19:705–12. doi: 10.1016/j.chom.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Odendall C, Dixit E, Stavru F, Bierne H, Franz KM, et al. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat Immunol. 2014;15:717–26. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Decembre E, Assil S, Hillaire ML, Dejnirattisai W, Mongkolsapaya J, et al. Sensing of immature particles produced by dengue virus infected cells induces an antiviral response by plasmacytoid dendritic cells. PLOS Pathog. 2014;10:e1004434. doi: 10.1371/journal.ppat.1004434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dang J, Tiwari SK, Lichinchi G, Qin Y, Patil VS, et al. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell. 2016;19:258–65. doi: 10.1016/j.stem.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Beatty PR, Puerta-Guardo H, Killingbeck SS, Glasner DR, Hopkins K, Harris E. Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Sci Transl Med. 2015;7:304ra141. doi: 10.1126/scitranslmed.aaa3787. [DOI] [PubMed] [Google Scholar]
- 143.Chen J, Ng MM, Chu JJ. Activation of TLR2 and TLR6 by dengue NS1 protein and its implications in the immunopathogenesis of dengue virus infection. PLOS Pathog. 2015;11:e1005053. doi: 10.1371/journal.ppat.1005053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Modhiran N, Watterson D, Muller DA, Panetta AK, Sester DP, et al. Dengue virus NS1 protein activates cells via Toll-like receptor 4 and disrupts endothelial cell monolayer integrity. Sci Transl Med. 2015;7:304ra142. doi: 10.1126/scitranslmed.aaa3863. [DOI] [PubMed] [Google Scholar]
- 145.Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, et al. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe. 2016;19:882–90. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dalrymple NA, Cimica V, Mackow ER. Dengue virus NS proteins inhibit RIG-I/MAVS signaling by blocking TBK1/IRF3 phosphorylation: Dengue virus serotype 1 NS4A is a unique interferon-regulating virulence determinant. mBio. 2015;6:e00553–15. doi: 10.1128/mBio.00553-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.He Z, Zhu X, Wen W, Yuan J, Hu Y, et al. Dengue virus subverts host innate immunity by targeting adaptor protein MAVS. J Virol. 2016;90:7219–30. doi: 10.1128/JVI.00221-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chatel-Chaix L, Cortese M, Romero-Brey I, Bender S, Neufeldt CJ, et al. Dengue virus perturbs mitochondrial morphodynamics to dampen innate immune responses. Cell Host Microbe. 2016;20:342–56. doi: 10.1016/j.chom.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yu CY, Liang JJ, Li JK, Lee YL, Chang BL, et al. Dengue virus impairs mitochondrial fusion by cleaving mitofusins. PLOS Pathog. 2015;11:e1005350. doi: 10.1371/journal.ppat.1005350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chan YK, Gack MU. A phosphomimetic-based mechanism of dengue virus to antagonize innate immunity. Nat Immunol. 2016;17:523–30. doi: 10.1038/ni.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, et al. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLOS Pathog. 2012;8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yu CY, Chang TH, Liang JJ, Chiang RL, Lee YL, et al. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLOS Pathog. 2012;8:e1002780. doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Aguirre S, Luthra P, Sanchez-Aparicio MT, Maestre AM, Patel J, et al. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat Microbiol. 2017;2:17037. doi: 10.1038/nmicrobiol.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liu H, Zhang L, Sun J, Chen W, Li S, et al. Endoplasmic reticulum protein SCAP inhibits dengue virus NS2B3 protease by suppressing its K27-linked polyubiquitylation. J Virol. 2017;91:e02234–16. doi: 10.1128/JVI.02234-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. PNAS. 2003;100:14333–38. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Morrison J, Laurent-Rolle M, Maestre AM, Rajsbaum R, Pisanelli G, et al. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLOS Pathog. 2013;9:e1003265. doi: 10.1371/journal.ppat.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bowen JR, Quicke KM, Maddur MS, O’Neal JT, McDonald CE, et al. Zika virus antagonizes type I interferon responses during infection of human dendritic cells. PLOS Pathog. 2017;13:e1006164. doi: 10.1371/journal.ppat.1006164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Schuberth-Wagner C, Ludwig J, Bruder AK, Herzner AM, Zillinger T, et al. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1-2′O-methylated self RNA. Immunity. 2015;43:41–51. doi: 10.1016/j.immuni.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12:137–43. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–56. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chang DC, Hoang LT, Mohamed Naim AN, Dong H, Schreiber MJ, et al. Evasion of early innate immune response by 2′-O-methylation of dengue genomic RNA. Virology. 2016;499:259–66. doi: 10.1016/j.virol.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.You J, Hou S, Malik-Soni N, Xu Z, Kumar A, et al. Flavivirus infection impairs peroxisome biogenesis and early antiviral signaling. J Virol. 2015;89:12349–61. doi: 10.1128/JVI.01365-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sabin AB. Research on dengue during World War II. Am J Trop Med Hyg. 1952;1:30–50. doi: 10.4269/ajtmh.1952.1.30. [DOI] [PubMed] [Google Scholar]
- 164.Guzman MG, Kouri G, Valdes L, Bravo J, Alvarez M, et al. Epidemiologic studies on dengue in Santiago de Cuba, 1997. Am J Epidemiol. 2000;152:793–99. doi: 10.1093/aje/152.9.793. discussion 804. [DOI] [PubMed] [Google Scholar]
- 165.Fernandez E, Diamond MS. Vaccination strategies against Zika virus. Curr Opin Virol. 2017;23:59–67. doi: 10.1016/j.coviro.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wen J, Tang WW, Sheets N, Ellison J, Sette A, et al. Identification of Zika virus epitopes reveals immunodominant and protective roles for dengue virus cross-reactive CD8+ T cells. Nat Microbiol. 2017;2:17036. doi: 10.1038/nmicrobiol.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kaufman BM, Summers PL, Dubois DR, Cohen WH, Gentry MK, et al. Monoclonal antibodies for dengue virus prM glycoprotein protect mice against lethal dengue infection. Am J Trop Med Hyg. 1989;41:576–80. doi: 10.4269/ajtmh.1989.41.576. [DOI] [PubMed] [Google Scholar]
- 168.Kaufman BM, Summers PL, Dubois DR, Eckels KH. Monoclonal antibodies against dengue 2 virus E-glycoprotein protect mice against lethal dengue infection. Am J Trop Med Hyg. 1987;36:427–34. doi: 10.4269/ajtmh.1987.36.427. [DOI] [PubMed] [Google Scholar]
- 169.Kyle JL, Balsitis SJ, Zhang L, Beatty PR, Harris E. Antibodies play a greater role than immune cells in heterologous protection against secondary dengue virus infection in a mouse model. Virology. 2008;380:296–303. doi: 10.1016/j.virol.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wahala WM, Silva AM. The human antibody response to dengue virus infection. Viruses. 2011;3:2374–95. doi: 10.3390/v3122374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tsai WY, Lai CY, Wu YC, Lin HE, Edwards C, et al. High-avidity and potently neutralizing cross-reactive human monoclonal antibodies derived from secondary dengue virus infection. J Virol. 2013;87:12562–75. doi: 10.1128/JVI.00871-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tsai WY, Durbin A, Tsai JJ, Hsieh SC, Whitehead S, Wang WK. Complexity of neutralizing antibodies against multiple dengue virus serotypes after heterotypic immunization and secondary infection revealed by in-depth analysis of cross-reactive antibodies. J Virol. 2015;89:7348–62. doi: 10.1128/JVI.00273-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Priyamvada L, Quicke KM, Hudson WH, Onlamoon N, Sewatanon J, et al. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. PNAS. 2016;113:7852–57. doi: 10.1073/pnas.1607931113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Stettler K, Beltramello M, Espinosa DA, Graham V, Cassotta A, et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science. 2016;353:823–26. doi: 10.1126/science.aaf8505. [DOI] [PubMed] [Google Scholar]
- 175.Swanstrom JA, Plante JA, Plante KS, Young EF, McGowan E, et al. Dengue virus envelope dimer epitope monoclonal antibodies isolated from dengue patients are protective against Zika virus. mBio. 2016;7:e01123–16. doi: 10.1128/mBio.01123-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nat Immunol. 2016;17:1102–8. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Priyamvada L, Hudson W, Ahmed R, Wrammert J. Humoral cross-reactivity between Zika and dengue viruses: implications for protection and pathology. Emerg Microbes Infect. 2017;6:e33. doi: 10.1038/emi.2017.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sukupolvi-Petty S, Austin SK, Purtha WE, Oliphant T, Nybakken GE, et al. Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J Virol. 2007;81:12816–26. doi: 10.1128/JVI.00432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Shrestha B, Brien JD, Sukupolvi-Petty S, Austin SK, Edeling MA, et al. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLOS Pathog. 2010;6:e1000823. doi: 10.1371/journal.ppat.1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wahala WM, Kraus AA, Haymore LB, Accavitti-Loper MA, de Silva AM. Dengue virus neutralization by human immune sera: role of envelope protein domain III-reactive antibody. Virology. 2009;392:103–13. doi: 10.1016/j.virol.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Edeling MA, Austin SK, Shrestha B, Dowd KA, Mukherjee S, et al. Potent dengue virus neutralization by a therapeutic antibody with low monovalent affinity requires bivalent engagement. PLOS Pathog. 2014;10:e1004072. doi: 10.1371/journal.ppat.1004072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.de Alwis R, Smith SA, Olivarez NP, Messer WB, Huynh JP, et al. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. PNAS. 2012;109:7439–44. doi: 10.1073/pnas.1200566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, et al. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol. 2015;16:170–77. doi: 10.1038/ni.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Fibriansah G, Tan JL, Smith SA, de Alwis R, Ng TS, et al. A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins. Nat Commun. 2015;6:6341. doi: 10.1038/ncomms7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sapparapu G, Fernandez E, Kose N, Bin C, Fox JM, et al. Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature. 2016;540:443–47. doi: 10.1038/nature20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wang Q, Yang H, Liu X, Dai L, Ma T, et al. Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus. Sci Transl Med. 2016;8:369ra179. doi: 10.1126/scitranslmed.aai8336. [DOI] [PubMed] [Google Scholar]
- 187.Hasan SS, Miller A, Sapparapu G, Fernandez E, Klose T, et al. A human antibody against Zika virus crosslinks the E protein to prevent infection. Nat Commun. 2017;8:14722. doi: 10.1038/ncomms14722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Robbiani DF, Bozzacco L, Keeffe JR, Khouri R, Olsen PC, et al. Recurrent potent human neutralizing antibodies to Zika virus in Brazil and Mexico. Cell. 2017;169:597–609. e11. doi: 10.1016/j.cell.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Halstead SB. Pathogenesis of dengue: challenges to molecular biology. Science. 1988;239:476–81. doi: 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- 190.Halstead SB, Shotwell H, Casals J. Studies on the pathogenesis of dengue infection in monkeys: II. Clinical laboratory responses to heterologous infection. J Infect Dis. 1973;128:15–22. doi: 10.1093/infdis/128.1.15. [DOI] [PubMed] [Google Scholar]
- 191.Halstead SB. In vivo enhancement of dengue virus infection in rhesus monkeys by passively transferred antibody. J Infect Dis. 1979;140:527–33. doi: 10.1093/infdis/140.4.527. [DOI] [PubMed] [Google Scholar]
- 192.Goncalvez AP, Engle RE, St Claire M, Purcell RH, Lai CJ. Monoclonal antibody-mediated enhancement of dengue virus infection in vitro and in vivo and strategies for prevention. PNAS. 2007;104:9422–27. doi: 10.1073/pnas.0703498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Balsitis SJ, Williams KL, Lachica R, Flores D, Kyle JL, et al. Lethal antibody enhancement of dengue disease in mice is prevented by Fc modification. PLOS Pathog. 2010;6:e1000790. doi: 10.1371/journal.ppat.1000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ng JK, Zhang SL, Tan HC, Yan B, Martinez JM, et al. First experimental in vivo model of enhanced dengue disease severity through maternally acquired heterotypic dengue antibodies. PLOS Pathog. 2014;10:e1004031. doi: 10.1371/journal.ppat.1004031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Bardina SV, Bunduc P, Tripathi S, Duehr J, Frere JJ, et al. Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science. 2017;356:175–80. doi: 10.1126/science.aal4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Pierson TC, Xu Q, Nelson S, Oliphant T, Nybakken GE, et al. The stoichiometry of antibody-mediated neutralization and enhancement of West Nile virus infection. Cell Host Microbe. 2007;1:135–45. doi: 10.1016/j.chom.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yamanaka A, Kosugi S, Konishi E. Infection-enhancing and -neutralizing activities of mouse monoclonal antibodies against dengue type 2 and 4 viruses are controlled by complement levels. J Virol. 2008;82:927–37. doi: 10.1128/JVI.00992-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Morens DM, Halstead SB. Disease severity-related antigenic differences in dengue 2 strains detected by dengue 4 monoclonal antibodies. J Med Virol. 1987;22:169–74. doi: 10.1002/jmv.1890220208. [DOI] [PubMed] [Google Scholar]
- 199.Pierson TC, Diamond MS. Molecular mechanisms of antibody-mediated neutralisation of flavivirus infection. Expert Rev Mol Med. 2008;10:e12. doi: 10.1017/S1462399408000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Rodrigo WW, Jin X, Blackley SD, Rose RC, Schlesinger JJ. Differential enhancement of dengue virus immune complex infectivity mediated by signaling-competent and signaling-incompetent human FcγRIA (CD64) or FcgγRIIA (CD32) J Virol. 2006;80:10128–38. doi: 10.1128/JVI.00792-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Boonnak K, Slike BM, Donofrio GC, Marovich MA. Human FcγRII cytoplasmic domains differentially influence antibody-mediated dengue virus infection. J Immunol. 2013;190:5659–65. doi: 10.4049/jimmunol.1203052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Chan KR, Zhang SL, Tan HC, Chan YK, Chow A, et al. Ligation of Fc gamma receptor IIB inhibits antibody-dependent enhancement of dengue virus infection. PNAS. 2011;108:12479–84. doi: 10.1073/pnas.1106568108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Chareonsirisuthigul T, Kalayanarooj S, Ubol S. Dengue virus (DENV) antibody-dependent enhancement of infection upregulates the production of anti-inflammatory cytokines, but suppresses anti-DENV free radical and pro-inflammatory cytokine production, in THP-1 cells. J Gen Virol. 2007;88:365–75. doi: 10.1099/vir.0.82537-0. [DOI] [PubMed] [Google Scholar]
- 204.Boonnak K, Slike BM, Burgess TH, Mason RM, Wu SJ, et al. Role of dendritic cells in antibody-dependent enhancement of dengue virus infection. J Virol. 2008;82:3939–51. doi: 10.1128/JVI.02484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Chan KR, Ong EZ, Tan HC, Zhang SL, Zhang Q, et al. Leukocyte immunoglobulin-like receptor B1 is critical for antibody-dependent dengue. PNAS. 2014;111:2722–27. doi: 10.1073/pnas.1317454111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wang TT, Sewatanon J, Memoli MJ, Wrammert J, Bournazos S, et al. IgG antibodies to dengue enhanced for FcγRIIIA binding determine disease severity. Science. 2017;355:395–98. doi: 10.1126/science.aai8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Mongkolsapaya J, Dejnirattisai W, Xu XN, Vasanawathana S, Tangthawornchaikul N, et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med. 2003;9:921–27. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- 208.Duangchinda T, Dejnirattisai W, Vasanawathana S, Limpitikul W, Tangthawornchaikul N, et al. Immunodominant T-cell responses to dengue virus NS3 are associated with DHF. PNAS. 2010;107:16922–27. doi: 10.1073/pnas.1010867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Rothman AL. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat Rev Immunol. 2011;11:532–43. doi: 10.1038/nri3014. [DOI] [PubMed] [Google Scholar]
- 210.Mangada MM, Rothman AL. Altered cytokine responses of dengue-specific CD4+ T cells to heterologous serotypes. J Immunol. 2005;175:2676–83. doi: 10.4049/jimmunol.175.4.2676. [DOI] [PubMed] [Google Scholar]
- 211.Mangada MM, Endy TP, Nisalak A, Chunsuttiwat S, Vaughn DW, et al. Dengue-specific T cell responses in peripheral blood mononuclear cells obtained prior to secondary dengue virus infections in Thai schoolchildren. J Infect Dis. 2002;185:1697–703. doi: 10.1086/340822. [DOI] [PubMed] [Google Scholar]
- 212.Weiskopf D, Angelo MA, de Azeredo EL, Sidney J, Greenbaum JA, et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. PNAS. 2013;110:E2046–53. doi: 10.1073/pnas.1305227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Weiskopf D, Cerpas C, Angelo MA, Bangs DJ, Sidney J, et al. Human CD8+ T-cell responses against the 4 dengue virus serotypes are associated with distinct patterns of protein targets. J Infect Dis. 2015;212:1743–51. doi: 10.1093/infdis/jiv289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Weiskopf D, Angelo MA, Bangs DJ, Sidney J, Paul S, et al. The human CD8+ T cell responses induced by a live attenuated tetravalent dengue vaccine are directed against highly conserved epitopes. J Virol. 2015;89:120–28. doi: 10.1128/JVI.02129-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.de Alwis R, Bangs DJ, Angelo MA, Cerpas C, Fernando A, et al. Immunodominant dengue virus-specific CD8+ T cell responses are associated with a memory PD-1+ phenotype. J Virol. 2016;90:4771–79. doi: 10.1128/JVI.02892-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Rivino L, Kumaran EA, Thein TL, Too CT, Gan VC, et al. Virus-specific T lymphocytes home to the skin during natural dengue infection. Sci Transl Med. 2015;7:278ra35. doi: 10.1126/scitranslmed.aaa0526. [DOI] [PubMed] [Google Scholar]
- 217.Hatch S, Endy TP, Thomas S, Mathew A, Potts J, et al. Intracellular cytokine production by dengue virus-specific T cells correlates with subclinical secondary infection. J Infect Dis. 2011;203:1282–91. doi: 10.1093/infdis/jir012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Rivino L, Kumaran EA, Jovanovic V, Nadua K, Teo EW, et al. Differential targeting of viral components by CD4+ versus CD8+ T lymphocytes in dengue virus infection. J Virol. 2013;87:2693–706. doi: 10.1128/JVI.02675-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lindow JC, Borochoff-Porte N, Durbin AP, Whitehead SS, Fimlaid KA, et al. Primary vaccination with low dose live dengue 1 virus generates a proinflammatory, multifunctional T cell response in humans. PLOS Negl Trop Dis. 2012;6:e1742. doi: 10.1371/journal.pntd.0001742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Weiskopf D, Bangs DJ, Sidney J, Kolla RV, De Silva AD, et al. Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity. PNAS. 2015;112:E4256–63. doi: 10.1073/pnas.1505956112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zompi S, Santich BH, Beatty PR, Harris E. Protection from secondary dengue virus infection in a mouse model reveals the role of serotype cross-reactive B and T cells. J Immunol. 2012;188:404–16. doi: 10.4049/jimmunol.1102124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Yauch LE, Zellweger RM, Kotturi MF, Qutubuddin A, Sidney J, et al. A protective role for dengue virus-specific CD8+ T cells. J Immunol. 2009;182:4865–73. doi: 10.4049/jimmunol.0801974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Yauch LE, Prestwood TR, May MM, Morar MM, Zellweger RM, et al. CD4+ T cells are not required for the induction of dengue virus-specific CD8+ T cell or antibody responses but contribute to protection after vaccination. J Immunol. 2010;185:5405–16. doi: 10.4049/jimmunol.1001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zellweger RM, Tang WW, Eddy WE, King K, Sanchez MC, Shresta S. CD8+ T cells can mediate short-term protection against heterotypic dengue virus reinfection in mice. J Virol. 2015;89:6494–505. doi: 10.1128/JVI.00036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Elong Ngono A, Chen HW, Tang WW, Joo Y, King K, et al. Protective role of cross-reactive CD8 T cells against dengue virus infection. EBioMedicine. 2016;13:284–93. doi: 10.1016/j.ebiom.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zellweger RM, Miller R, Eddy WE, White LJ, Johnston RE, Shresta S. Role of humoral versus cellular responses induced by a protective dengue vaccine candidate. PLOS Pathog. 2013;9:e1003723. doi: 10.1371/journal.ppat.1003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zellweger RM, Eddy WE, Tang WW, Miller R, Shresta S. CD8+ T cells prevent antigen-induced antibody-dependent enhancement of dengue disease in mice. J Immunol. 2014;193:4117–24. doi: 10.4049/jimmunol.1401597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Weiskopf D, Yauch LE, Angelo MA, John DV, Greenbaum JA, et al. Insights into HLA-restricted T cell responses in a novel mouse model of dengue virus infection point toward new implications for vaccine design. J Immunol. 2011;187:4268–79. doi: 10.4049/jimmunol.1101970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Weiskopf D, Angelo MA, Sidney J, Peters B, Shresta S, Sette A. Immunodominance changes as a function of the infecting dengue virus serotype and primary versus secondary infection. J Virol. 2014;88:11383–94. doi: 10.1128/JVI.01108-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Weiskopf D, Cerpas C, Angelo MA, Bangs DJ, Sidney J, et al. Human CD8+ T-cell responses against the 4 dengue virus serotypes are associated with distinct patterns of protein targets. J Infect Dis. 2015;212:1743–51. doi: 10.1093/infdis/jiv289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Chu H, George SL, Stinchcomb DT, Osorio JE, Partidos CD. CD8+ T-cell responses in flavivirus-naive individuals following immunization with a live-attenuated tetravalent dengue vaccine candidate. J Infect Dis. 2015;212:1618–28. doi: 10.1093/infdis/jiv258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Dudley DM, Aliota MT, Mohr EL, Weiler AM, Lehrer-Brey G, et al. A rhesus macaque model of Asian-lineage Zika virus infection. Nat Commun. 2016;7:12204. doi: 10.1038/ncomms12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Elong Ngono A, Vizcarra EA, Tang WW, Sheets N, Joo Y, et al. Mapping and role of the CD8+ T cell response during primary Zika virus infection in mice. Cell Host Microbe. 2017;21:35–46. doi: 10.1016/j.chom.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Pardy RD, Rajah MM, Condotta SA, Taylor NG, Sagan SM, Richer MJ. Analysis of the T cell response to Zika virus and identification of a novel CD8+ T cell epitope in immunocompetent mice. PLOS Pathog. 2017;13:e1006184. doi: 10.1371/journal.ppat.1006184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Winkler CW, Myers LM, Woods TA, Messer RJ, Carmody AB, et al. Adaptive immune responses to Zika virus are important for controlling virus infection and preventing infection in brain and testes. J Immunol. 2017;198:3526–35. doi: 10.4049/jimmunol.1601949. [DOI] [PMC free article] [PubMed] [Google Scholar]