Abstract
Protein Arginine deiminases (PADs) play an important role in the pathogenesis of various diseases, including rheumatoid arthritis, multiple sclerosis, lupus, ulcerative colitis and breast cancer. Therefore, the development of PAD-inhibitors has drawn significant research interest in recent years. Herein, we describe the development of the first photoswitchable PAD-inhibitors. These compounds possess an azobenzene photoswitch to optically control PAD activity. Screening of a series of inhibitors structurally similar to BB-Cl-Amidine afforded compounds 1 and 2 as the most promising candidates for the light-controlled inhibition of PAD2; the cis-isomer of 1 is 10-fold more potent than its trans-isomer, whereas the trans-isomer of 2 is 45-fold more potent than the corresponding cis-isomer. The altered inhibitory potency upon photoisomerization has been confirmed in a competitive activity-based protein profiling (ABPP) assay. Further investigations indicate that the trans-isomer of 2 is an irreversible inhibitor, whereas the cis-isomer acts as a competitive inhibitor. In cells, the trans-isomer of compound 1 is completely inactive, whereas the cis-isomer inhibits histone H3-citrullination in a dose-dependent manner. Taken together, 1 serves as the foundation for developing photopharmaceuticals that can be activated at the desired tissue, using light, to treat diseases where PAD activity is dysregulated.
INTRODUCTION
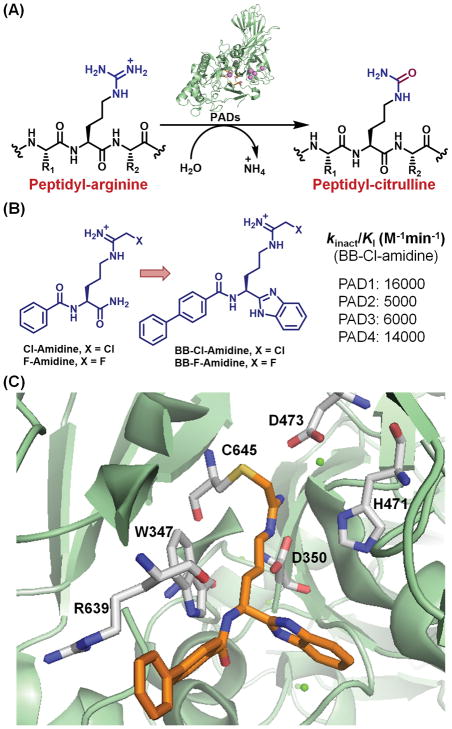
Protein arginine deiminases (PADs) are cysteine hydrolases that mediate the conversion of arginine to citrulline (Figure 1A).1,2 Five isozymes (PAD1, 2, 3, 4 and 6) are known but only four of them (PADs 1–4) are catalytically active.3 The deiminase activity of PADs 1–4 is strictly regulated by the presence of Ca2+ ions, and the holoenzymes are known to bind four (PAD1), five (PADs 3 and 4) and six (PAD2) calcium ions at distinct sites.4,5,6 Although calcium does not play a direct role in catalysis, recent crystal structures indicate that it induces a series of structural rearrangements that leads to the formation of the catalytically competent state. Particularly, the movement of the nucleophilic cysteine (Cys647 and Cys645 in PAD2 and PAD4, respectively) into the active site is triggered by calcium.6 PAD-mediated protein citrullination is known to regulate various cellular processes, including neutrophil extracellular trap (NET) formation, the epigenetic regulation of gene transcription, and the maintenance of pluripotency. Moreover, aberrant protein citrullination is associated with various autoimmune diseases, including rheumatoid arthritis (RA), lupus, multiple sclerosis, ulcerative colitis, and certain cancers.1, 2, 7–14
Figure 1.
(A) PAD-catalyzed citrullination of peptidyl-arginine. (B) Chemical structures of pan-PAD inhibitors and the kinact/KI values for BB-Cl-Amidine. (C) Cocrystal structure of PAD4 bound to BB-F-Amidine indicating the covalent modification of Cys645 by the inhibitor (PDB code 5N0M).15
Given the potential role of PADs in human pathology, the development of PAD inhibitors has attracted significant attention.1, 2, 15 The most widely-used pan-PAD inhibitor is Cl-amidine (Figure 2B), which possesses a chloroacetamidine warhead that mimics the substrate guanidinium and irreversibly inhibits the PADs. Notably, Cl-amidine shows excellent efficacy in animal models of rheumatoid arthritis (RA), lupus, ulcerative colitis, spinal cord injury, breast cancer and atherosclerosis.13, 16–18 Structure-activity relationships ultimately led to the development of BB-Cl-amidine, a second-generation PAD inhibitor in which the phenyl and carboxamide groups of Cl-Amidine are replaced with a biphenyl and benzimidazole group, respectively (Figure 1B).17 BB-Cl-amidine exhibits at least 10-fold more potency than Cl-amidine in cell-based and animal experiments.17, 19, 20 However, the fluoroacetamidine counterparts of Cl-amidine and BB-Cl-amidine, i.e. F-Amidine and BB-F-amidine, exhibit significantly less inhibitory activity, likely due to the lower electrophilicity of the fluoroacetamidine warhead (Figure 1B).21, 22 In agreement with the proposed mechanism of inhibition,23 the cocrystal structure of PAD4 bound to BB-F-amidine indicates that nucleophilic attack of the active site cysteine on the haloacetamidine warhead displaces the halide and irreversibly modifies the enzyme active site (Figure 1C).15 Recently, we showed that BB-F-Yne, an alkyne bearing fluoroacetamidine based PAD inhibitor, is remarkably selective for the PADs in cell based assays.24 By contrast, BB-Cl-yne, possesses a number of off targets including, β-tubulin, β-Actin, clathrin heavy chain 1 (CLTC), bifunctional glutamate/proline-tRNA ligase (EPRS), heterogeneous nuclear ribonucleoprotein U (HNRPU).24 These observations suggest that localized activation or deactivation of chloroacetamidine-containing inhibitors in the target tissue may be an alternative approach to limit potential systemic toxicities.
Figure 2.
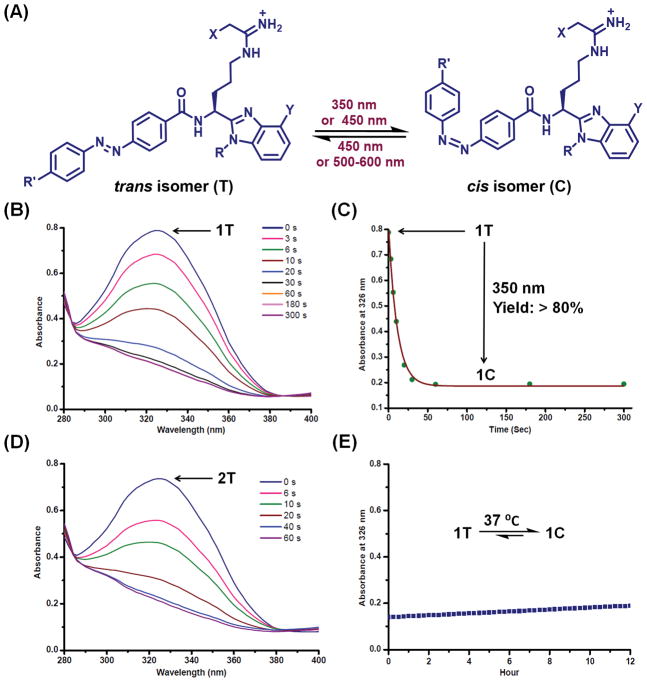
(A) The chemical structures of trans- and cis-isomers of compounds 1-8 (see Scheme 1 for variable groups). (B) The change in the UV-vis spectra of compound 1 upon irradiation with UV-A, demonstrating the photoisomerization of 1T into 1C. (C) Optimisation of UVA-exposure time from the change in absorbance of 1 at 324 nm. (D) Photoisomerization of compound 2 monitored by UV-Vis spectra. (E) Thermal stability of 1C in aqueous buffer at 37 °C. Assay conditions: compound 1 (30 μM), 100 mM TRIS pH 7.4, 50 mM NaCl.
Photopharmacology, which relies on the activation or inactivation of drug molecules in the targeted tissue by light, has led to the development of several light-controlled ion-channel blockers, receptor antagonists, antibiotics, and enzyme inhibitors.25–32 Light serves as a suitable external, noninvasive stimuli to modulate the potency of drug molecules and it can be delivered with high spatiotemporal precision and with a wide range of intensities as well as wavelengths. Photopharmaceuticals are generally developed by incorporating photoresponsive groups into bioactive molecules. The simplest and most widely-used example of such a photoresponsive element is an azobenzene. Azobenzenes undergo a trans-cis reversible isomerisation in the presence of light. While the thermodynamically more stable trans-isomer can be switched to the less stable, bent cis-isomer by UV light (i.e., 350 nm), the reverse process can be achieved by shining blue light (450 nm).29, 30 The excitation wavelengths and the half-life of the photoisomers can also be modulated by incorporating different substitutions on the azobenzene scaffold. The two isomers of azobenzene mainly differ from each other in shape and polarity and consequently, they exhibit different biological activities. Therefore, we hypothesized that the incorporation of azobenzene switches in BB-Cl-amidine, by replacing the biphenyl group, would result in a PAD-inhibitor that is structurally-similar to BB-Cl-amidine but can be activated or inactivated by light. Herein, we report the development of photoresponsive PAD inhibitors that can be used to optically control the inhibition of PADs in vitro and in cell-based assays.
RESULTS AND DISCUSSION
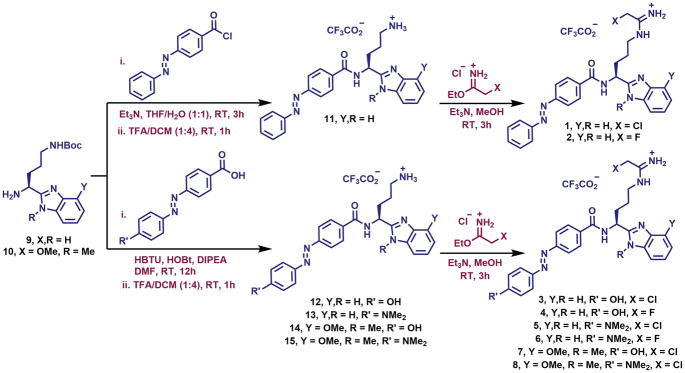
To achieve the optical control of PAD-activity, we synthesized various azobenzene-containing compounds carrying either a fluoro- or chloroacetamidine warhead (compounds 1-8, Scheme 1). While compounds 1 and 2 are the simple azobenzene-modified analogues of BB-Cl-Amidine and BB-F-Amidine, respectively, compounds 3-8 carries various substitutions on the azobenzene as well as the benzimidazole moiety. We hypothesized that the p-hydroxyl and p-dimethylamino substitutions on the terminal aromatic ring of the azobenzene moiety in compounds 3-6 may alter the binding affinity of different photoisomers for the enzyme active site by both electronic and steric influence. The N-methyl and 4-methoxy substitutions on the benzimidazole ring in compounds 7 and 8 were incorporated because we previously showed that these modifications significantly increase the potency and isoform-specificity of benzimidazole-based PAD inhibitors.15 The synthesis of compounds 1-8 is depicted in Scheme 1 and it involves the common precursors 9 and 10, which were synthesized by following a previously reported procedure.15, 17
Scheme 1.
Chemical structures and the synthesis of compounds 1-8.
With this family of the photoresponsive PAD-inhibitors in hand, we next investigated their trans-cis isomerisation by UV-visible spectroscopy (Figure 2A and Figure S1). The thermodynamically more stable trans-isomer of compound 1 (1T) exhibits maximal absorption (λmax) at 324 nm (Figure 2B). However, upon irradiation with UV-A (λ = 320–380 nm), this peak disappears with time, indicating the formation of the cis-isomer of this compound (i.e., 1C). A plot of absorbance at 324 nm versus time indicates that 40–60 s of UV-A exposure is sufficient to convert more than 80 % of 1T into 1C (Figure 2C). A similar phenomenon was also observed for the fluoroacetamidine warhead-containing compound 2 (Figure 2D), indicating that the halogen substitution on the electrophilic warhead does not impact the photoisomerisation of compounds 1 and 2. We also investigated the thermal stability of the cis-isomer by UV-vis spectroscopy, as fast back conversion to the trans-isomer (from the thermodynamically less stable cis-isomer) would limit their utility. Both 1C and 2C exhibit remarkable stability in aqueous buffer at 37 °C as the change in absorbance at 324 nm over 12 h is negligible after irradiating 1T or 2T with UV-A lamps (Figure 2E and Figure S2). Both 1C and 2C can be switched back to their trans-form, i.e. 1T and 2T, respectively, by irradiating with blue light (λ = 400–450 nm) (Figure S1G and H) similarly to other azobenzene-containing photopharmaceuticals.25–32 This phenomenon, however, is not important for our purposes because compounds 1-8 are irreversible PAD-inhibitors and, as such, once the trans- or cis- isomer reacts with the active site cysteine, photoswitching will not turn off the inhibitor to regenerate the active enzyme. The photoswitching properties of compounds 3-8 were also characterised by UV-vis spectroscopy (Figure S1) and the observations are in agreement with the reported photoswitching behaviour of hydroxy- and diaminomethyl-substituted azobenzene photopharmaceuticals.25–32
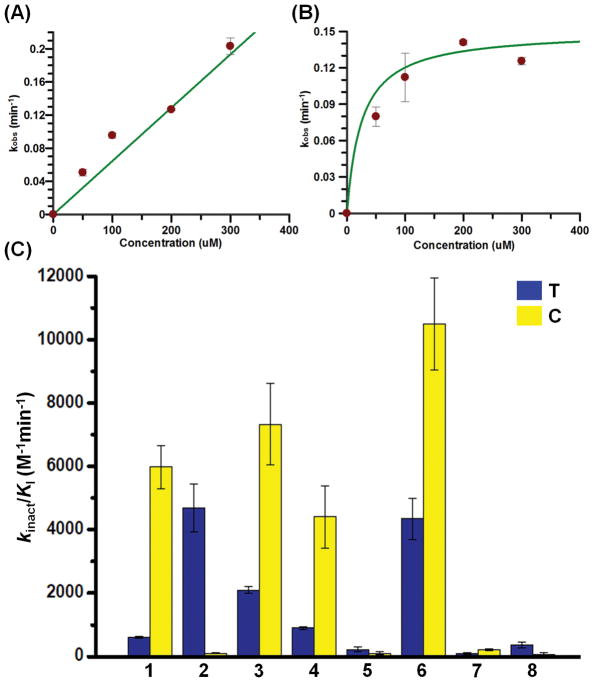
Having established the optimum photoswitching conditions (excitation wavelengths and exposure time), we set out to evaluate the inhibitory potency of the trans- (T) and cis- (C) isomers of the inhibitors in in vitro PAD-inhibition assays. PADs 1–4 were expressed and purified following standard protocols established in the Thompson lab.4, 5 Inhibition of citrulline production was quantified using the COLDER assay (see experimental section). For PAD1, replacement of the biphenyl group in BB-Cl-amidine with the azobenzene decreases the potency of 1T by almost 8-fold, while the fluorinated analogue, 2T exhibits almost 6-fold higher activity than BB-F-amidine for this isozyme (Table 1). Interestingly, compound 3T, with a p-hydroxy substitution, exhibits almost 13-fold higher activity than the unsubstituted analogue 1T, while the fluorinated analogue 4T exhibits 2-fold lower potency than 2T for PAD1. Furthermore, compounds 5T, 7T and 8T are inactive towards PAD1 (Table 1), indicating that the p-dimethylamino substitution on the azobenzene in combination with the N-methyl and 4-methoxy substitutions on the benzimidazole ring disfavors binding of the inhibitor to the PAD1 active site. Next, we evaluated the inhibitory potency of the cis-isomers of 1-8. The changes in the potency upon photoisomerisation of compounds 1-8 were generally found to be within 1–2-fold, although compound 6T exhibits 5-fold higher activity than 6C.
Table 1.
kinact/KI vales for the inhibition of PADs 1–4 by BB-Cl-Amidine, BB-F-Amidine, and trans- (T) and cis- (C) isomers of compounds 1-8 (see Scheme 1 for variable groups).
| Compounds & photoisomers | kinact/KI (M−1min−1) | ||||
|---|---|---|---|---|---|
|
| |||||
| PAD1 | PAD2 | PAD3 | PAD4 | ||
| BB-Cl-Amidine | 16000a | 5000a | 6000a | 14000a | |
| BB-F-Amidine | 900a | 1200a | 3400a | 3750a | |
| 1 | T | 2300 ± 200b | 600 ± 30b | 1000 ± 60b | 10510 ± 590c |
| C | 2000 ± 100b | 5970 ± 670c | 1500 ± 40b | 4900 ± 100b | |
| 2 | T | 4920 ± 760c | 4520 ± 760c | 500 ± 77b | 3880 ± 1320c |
| C | 2850 ± 320c | ≤100 ± 20de | 1100 ± 40b | 4150 ± 420c | |
| 3 | T | 28770 ± 7020c | 2100 ± 100b | 6500 ± 100b | 19920 ± 2170c |
| C | 12000 ± 570c | 7330 ± 1280c | 6300 ± 400b | 27940 ± 180c | |
| 4 | T | 2130 ± 510c | 900 ± 40b | 2020 ± 210c | 3640 ± 100c |
| C | 1030 ± 380c | 4410 ± 980c | 1300 ± 50b | 5580 ± 350c | |
| 5 | T | No Inhibition | 230 ± 70d | No Inhibition | 2440 ± 760c |
| C | No Inhibition | ≤100 ± 40d | No Inhibition | 3070 ± 50c | |
| 6 | T | 870 ± 360c | 4340 ± 650c | 350 ± 50d | 450 ± 40d |
| C | 180 ± 70d | 10490 ± 1460c | 510 ± 50d | 450 ± 40d | |
| 7 | T | ≤40 ± 10d | ≤100 ± 30d | ≤130 ± 30d | 600 ± 30b |
| C | ≤80 ± 30d | 220 ± 30d | 170 ± 20d | 600 ± 100b | |
| 8 | T | No Inhibition | 360 ± 100d | No Inhibition | ≤60 ± 20d |
| C | No Inhibition | 80 ± 40d | No Inhibition | ≤80 ± 20d | |
kinact/KI values were taken from the published results (ref. 15).
kinact/KI was determined from a linear fit of the kobs versus [I] data.
kinact and KI was determined from a nonlinear fit of the kobs versus [I] data.
A single kobs was determined.
Detailed kinetic studies indicate that 2C is actually a reversible inhibitor of PAD2 with a Ki value of 25.2 μM (see Figure S7).
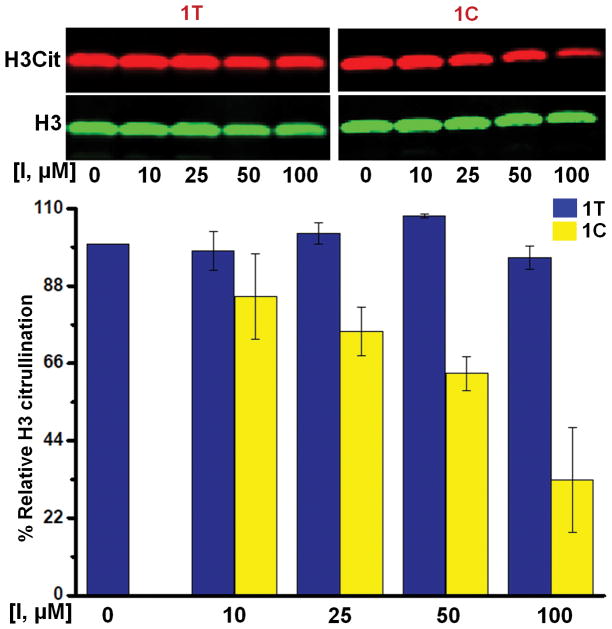
Figure 5.
Inhibition of histone H3 citrullination in HEK293T/PAD2 cells by compound 1. Inhibitor concentrations [I] are given under each lane of the western-blot image. Citrullinated H3 (H3Cit) and H3 are shown in red and green, respectively. Quantification of each band yielded the H3Cit/H3 ratio, from which the % relative H3 citrullination was calculated.
Next, we tested their ability to inhibit PAD3 and PAD4. The potencies of the chloro- and fluoroacetamidine warhead-containing compounds 1-8 for PAD3-inhibition were generally found to be lower than BB-Cl-Amidine and BB-F-Amidine, respectively. Compound 3T exhibited the highest potency in the series towards PAD3 (Table 1). However, upon photoswitching to the cis-isomer (3C), there was no change in potency. For other compounds in the series, photoisomerisation had modest (1.5–2-fold) effects on potency. For PAD4, compounds 1T and 2T exhibit similar potency to BB-Cl-amidine and BB-F-amidine (Table 1). As observed for PAD1 and PAD3, compound 3T exhibited the highest inhibitory activity towards PAD4, which is almost 2-fold higher than that of 1T. However, its fluorinated analogue, 4T, is only just as potent as compound 2T for PAD4 inhibition. While 5T (with a p-dimethylamino substitution) exhibits reasonable activity towards PAD4, the fluorinated analogue 6T is 5-fold less active (Table 1). It should be noted that 5T is completely selective to PAD4 over PAD1 and PAD3, and such selectivity over PAD1 is rare in the literature. However, compounds 7T and 8T exhibit poor inhibitory activity for PAD4, indicating that the benzimidazole-ring substitutions disfavor binding of the inhibitor to the active site of PAD4. Similar to the inhibition of PAD1 and 3, the cis-conformation of these compounds are equipotent to the trans-conformation, e.g. 2C (kinact/KI = 4150 ± 420 M−1min−1) inhibits PAD4 as well as 2T (kinact/KI = 3900 ± 1300 M−1min−1). Moreover, there was only a two-fold change in potency between compounds 1T and 1C (Table 1).
Finally, we investigated the ability of these compounds to inhibit PAD2. Dysregulated PAD2 activity is strongly associated with multiple sclerosis, RA and breast cancer.1, 2 PAD2 is known to be secreted in the synovial fluid of arthritic joints, where it citrullinates numerous extracellular proteins, leading to the development of anticitrullinated protein antibodies (APCA), a diagnostic marker of RA.3, 12 PAD2 is also responsible for the citrullination of histone H3 at R26 position, which triggers the localized decondensation of chromatin and activates gene transcription.13, 33 Notably compound 1T is ~10-times less active than BB-Cl-amidine, indicating that replacement of the biphenyl group with the azobenzene disfavors interactions between the trans-form of the inhibitor and PAD2. Gratifyingly, its potency increases by 10-fold upon excitation to the cis-isomer, 1C (Table 1). Moreover, the kinact/KI value of 1C (6000 ± 680 M−1min−1) is almost identical to that of BB-Cl-amidine (5000 M−1min−1). The increased potency upon photoisomerisation is most likely due to enhanced binding to the PAD2 active site because the plots of kobs versus inhibitor concentration move from linear (1T) to hyperbolic (1C) (Figure 3A), indicative of a lower KI value for the cis-isomer. The kinact and KI values for 1C are 0.15 min−1 and 25.2 μM, respectively (Figure 3B). These results indicate that the less potent compound 1T can be activated to the more potent compound 1C for the inhibition of PAD2 in the presence of UV-A radiation. Surprisingly, 2T, the fluorinated analogue of 1T, is almost 45-fold more active than its cis-isomer 2C (Table 1 and Figure 3C), indicating that the nature and size of the halogen atom in the haloacetamidine warhead plays an important role in the binding and reactivity of the inhibitor. 2T shows saturation kinetics with kinact and KI values of 0.3 min−1 and 66.4 μM, respectively (Figure S4C). By contrast, 2C showed very little, if any, enzyme inactivation, as such a single kobs was used to estimate the kinact/KI of 2C (kinact/KI ≤ 100 ± 20 M−1min−1) (Figure S4D). In comparison to compounds 1 and 2, compounds 3C and 4C are ~ 4- and 5-fold, respectively, more active than 3T and 4T, indicating that the p-hydroxy substitution in the azobenzene ring suppresses the optical control of the inhibition of PAD2 activity (Table 1 and Figure 3C). Compound 6T is also ~2-times more potent than 6C. Interestingly, 6C is ~57-, 20- and 23-fold more selective for PAD2 than PAD1, PAD3 and PAD4, respectively. Such isozyme selectivity has rarely been observed amongst previously developed PAD inhibitors. Furthermore, compound 8T is ~5-times more active than 8C, although the potency of 8T is very low (Table 1 and Figure 3C). Taken together, compounds 1 and 2 are the most promising candidates in the series for the optical control of PAD2 activity – compound 1 can be photo-activated by 10-fold, whereas compound 2 can be photo-deactivated by 45-fold.
Figure 3.
Concentration dependence of pseudo-first-order rate constant of inactivation (kobs) of PAD2 by 1T (A) and 1C (B). (C) Potencies (kinact/KI) of trans- and cis-isomers of compounds 1-8 for the inhibition of PAD2. Assay conditions: PAD2 (2 μM), 100 mM TRIS pH 7.4, 50 mM NaCl, 2 mM DTT, 10 mM CaCl2, and 10 mM BAEE.
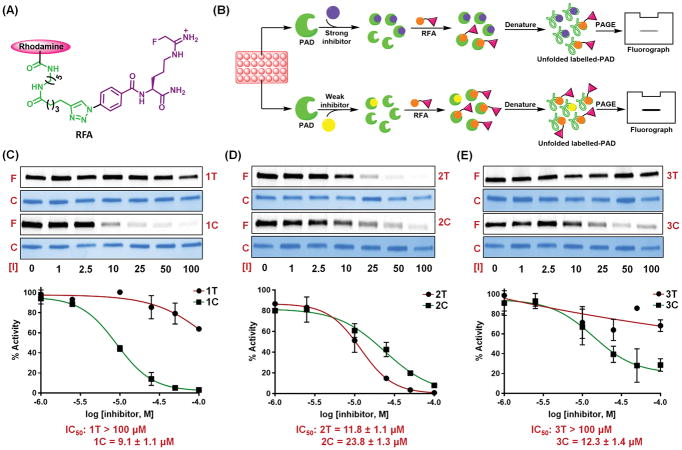
To further confirm the results of our in vitro inhibition assays, we used a previously established competitive activity-based protein profiling (ABPP) assay to visualise the change in inhibitory potency upon photoisomerisation (Figure 4A and B). For these studies, PAD2 was treated with inhibitors in microplates for 30 min prior to the addition of rhodamine-conjugated fluoroamidine (RFA), a PAD-targeted ABPP. For strong inhibitors, RFA-labelling will be blocked, whereas for weak inhibitor a smaller proportion of PAD2 would be occupied, and RFA-labelling is enhanced (Figure 4B).
Figure 4.
(A) Chemical structure of RFA. (B) Workflow of labelling of PAD2 with RFA in the presence of inhibitors. RFA-labelling of PAD2 in the presence of 1T and 1C (C), 2T and 2C (D), 3T and 3C (E). F and C stands for fluorograph and coomassie stain, respectively. Concentrations of inhibitors [I] are given in micromolar (μM). Intensities of each band in the fluorograph were measured using ImageJ software and the relative decrease in the fluorescence intensity has been plotted against the concentration of inhibitor to obtain the IC50 values. Assay conditions: PAD2 (2 μM), 100 mM TRIS pH 7.4, 50 mM NaCl, 2 mM DTT, and 10 mM CaCl2.
Consistent, with the kinact/KI data, 1T is a very weak PAD2 inhibitor (Figure 4C); the IC50 is >100 μM. By contrast, the IC50 of 1C, 9.1 μM, is more than 10-fold lower than that of 1T. Despite the fact that 2T is 45-fold more potent than 2C, these two isomers yielded similar IC50 values, i.e. 11.8 μM and 23.8 μM respectively, suggesting that 2T is only 2-times more active than 2C (Figure 4D). To reconcile this contradiction, we considered the possibility that 2C binds to the PAD2 active site in such a conformation that precludes its reaction with C647, the active site cysteine, but still inhibits the enzyme reversibly. To test this possibility, we determined the Km of Nα-benzoyl-L-arginine ethyl ester (BAEE), a known substrate of PAD2, in the presence of increasing concentrations of 2C. The resulting inhibition patterns indicate that 2C is a reversible, competitive inhibitor, with a Ki value of 25.2 μM (Figure S7). Since 3C is almost 4-fold more potent than 3T (Table 1), we also assayed this compound in this competitive ABPP assay. Interestingly, the IC50 of 3C is >8-fold lower than that of 3T, roughly in agreement with the results obtained using the COLDER assay.
Motivated by these promising inhibition data, we next evaluated their ability to inhibit histone H3 citrullination in cells; histone H3 is citrullinated at multiple positions, including R2, R8, R17 and R26 by PAD2.33–35 For these experiments, HEK293T/PAD2 overexpressing cells36, 37 were incubated with the inhibitor, CaCl2 and ionomycin (a calcium ionophore) for 3 h, and then the cells were lysed, the soluble proteins in the lysate resolved by SDS-PAGE, and transferred to a PVDF membrane. The membrane was then incubated with primary antibodies for histone H3 and citrullinated histone H3 (Cit 2, 8, 17), followed by incubation with appropriate secondary antibodies and imaged by Licor analysis. Consistent with our in vitro data, 1T does not inhibit histone H3 citrullination even at 100 μM. By contrast, the light-activated isomer 1C inhibits citrullination in a dose-dependent manner (Figure 5), suggesting that compound 1 can be photoactivated to inhibit histone H3-citrullination in HEK293T/PAD2 cells. By contrast, 2T only showed modest inhibition (~33% at 100 μM) and 2C did not show any activity (Figure S8A). Similarly, 3C was slightly more active than 3T in inhibiting H3 citrullination by PAD2 (Figure S8B). These results suggest that in addition to inhibitory potency, cell-permeability of the different isomers plays an important role in inhibiting PAD2 activity in the HEK293T/PAD2 cells.
We also evaluated the cytotoxicity of the photoisomers of compounds 1-3 and BB-Cl-amidine in the HEK293T/PAD2 cells. Compound 1, which exhibited the best results for the photoactivation of inhibition of PAD2 was as cytotoxic as BB-Cl-amidine (Figure S9). Notably, the trans- and cis isomers of 1 do not show any difference in the cytotoxicity, suggesting that the cytotoxicity of compound 1 does not originate from the inhibition of PAD2. By contrast, compounds 2 and 3 are less toxic than 1.
Conclusions
Herein, we report the development of the first photoswitchable PAD inhibitor. A series of compounds, 1-8, carrying an electrophilic warhead were evaluated and in vitro inhibition experiments indicate that these compounds can optically control PAD2 activity, with 1 and 2 being the most promising candidates. While the cis-isomer (1C) exhibits 10-fold higher potency than the trans-isomer (1T) of 1, the trans-isomer (2T) exhibits 45-fold more potency than the cis-isomer (2C) of 2. Furthermore, alterations in the inhibitory potency upon photoisomerisation were confirmed using a competitive ABPP assay. This study indicates that the IC50 of 1C is >10-fold lower than that of 1T. In contrast to the kinact/KI data, compound 2C is only 2-fold less potent than 2T in the ABPP-based assay. Detailed enzyme kinetics in the presence of increasing concentrations of 2C revealed that 2C is a reversible, competitive inhibitor of PAD2. Cell-based studies reveal that 1T is inactive, whereas the light-activated isomer 1C inhibits histone H3 citrullination in a dose-dependent manner in HEK293T/PAD2 cells.
In total, the present data sets a foundation for developing photopharmaceuticals to optically control PAD activity. Nevertheless, further research is required to overcome the limitations of the current molecules, including the poor tissue penetration of 350 nm light and modest isozyme selectivity. With regard to the former, introduction of fluoro- or chloro substitutions into the azobenzene scaffold will shift the excitation wavelength from UV to green or red light, respectively.38 With regard to the latter, the PADs generally show tissue or disease specific expression patterns.3 For example, PAD2 is overexpressed in luminal breast cancer cells and its expression correlates with the level of the HER2 protooncogene.13, 33 Thus, compound 1 represents a promising starting point for the development of photopharmaceuticals against diseases in which PAD2 activity is dysregulated. Nevertheless, improvements in potency and interisozyme selectivity will likely be necessary and could be achieved by modifying the chromophore.
EXPERIMENTAL SECTION
Materials and methods
Fmoc-Orn(Boc)-OH was purchased from Chem-Impex International, Inc. 4-(phenylazo)benzoyl chloride, p-(p-dimethylaminophenylazo)benzoate dehydrate, DIPEA, anhydrous methanol, DMF, dichloromethane, piperidine, triethylamine, trifluoroacetic acid, chloroacetonitrile, fluoroacetonitrile and HPLC-grade acetonitrile were obtained from Sigma-Aldrich. 4′-Hydroxyazobenzene-4-carboxylic acid was bought from TCI Chemicals. Precoated silica gel plates were purchased from Merck. Deuterated solvents were acquired from Cambridge Isotope Laboratories. Mouse monoclonal anti-histone H3 (catalogue no. ab10799) and rabbit polyclonal anti-H3 (Citrulline R2, R8 and R17) (catalogue no. ab5103) were purchased from Abcam. NMR spectra were recorded in d4-MeOH or d6-DMSO as solvent. 1H and 13C NMR spectra were recorded using a Bruker 500 MHz NMR spectrometer. Chemical shift values are cited with respect to SiMe4 as the internal standard. Column chromatography was carried out in glass columns. Final compounds were purified by reverse-phase HPLC using a semi-preparative C18 column (Agilent, 21.2 × 250 mm, 10 μm) and water/acetonitrile gradient supplemented with 0.05% trifluoroacetic acid.
Synthesis
Detailed procedures for the synthesis of compounds 1-8 are given in the supporting information. All the compounds were isolated as trifluoroacetate salts and were characterized using 1H, 13C NMR spectroscopy and ESI-Mass spectrometry. The purity of the compounds was analyzed by analytical HPLC.
Inactivation kinetics
Inactivation kinetic parameters for compounds 1-8 were determined by following reported procedures.22, 23 Briefly, PAD1, PAD2, PAD4 (2 μM) or PAD3 (5 μM) was added to a prewarmed (10 min, 37 °C) inactivation mixture (100 mM TRIS pH 7.4, 50 mM NaCl, 10 mM CaCl2, 2 mM DTT, with a final volume of 50 μL) containing various concentrations of inhibitors. Aliquots (6 μL) of the inactivation mixture were removed at various time points and were added to a prewarmed (10 min, 37 °C) reaction mixture (100 mM TRIS pH 7.4, 50 mM NaCl, 10 mM CaCl2, 2 mM DTT, and 10 mM BAEE or 10 mM BAA for PAD3, with a final volume of 60 μL). The reactions were quenched with liquid nitrogen after 15 min and citrulline production was quantified using the COLDER assay.5, 39 The time-dependence of PAD inhibition was fit to equation 1,
| (1) |
using Grafit, version 5.0.11, where ν is velocity, ν0 is initial velocity, k (or kobs is the pseudo-first-order rate constant of inactivation, and t is the time. If the rates of inactivation reached saturation, the concentration dependence of kobs was fit to equation 2,
| (2) |
using Grafit, version 5.0.11, where kinact is the maximal rate of inactivation, KI is the concentration of inhibitor that yields half-maximal inactivation, and [I] is the concentration of inhibitor. If the plot of kobs versus [I] did not saturate and was linear, then the value of kinact/KI was determined from the slope of the line. All the experiments were carried out at least in duplicate.
RFA-labelling
The labeling of PAD2 with RFA in the presence of inhibitor was carried out by following a previously reported procedure.36, 40, 41 Briefly, PAD2 (2 μM final) was added to the reaction mixture (100 mM TRIS pH 7.4, 50 mM NaCl, 10 mM CaCl2, and 2 mM DTT in a final volume of 20 μL) containing DMSO or various concentrations of the inhibitor. The mixture was incubated at 37 °C for 30 min, followed by addition of RFA (10 μL, 2 μM final). Then the reaction mixture was incubated at 37 °C for a further 2 h, quenched with 5X SDS-PAGE dye, incubated at 95 °C for 15 min and then loaded onto a 10% SDS-PAGE gel. After electrophoresis, fluorescence of the protein bands was recorded using a typhoon scanner (excitation/emission maxima of ~546/579, respectively). Fluorescence intensity of the protein bands was quantified using ImageJ software and was plotted against the concentration of the inhibitor to obtain the IC50 values.
Histone H3 Citrullination in HEK293TPAD2 Cells
HEK293T cells stably expressing human PAD2 (HEK293T/PAD2) were cultured as reported earlier.36, 37 Cells were grown to ~90% confluence, trypsinized, quenched with complete media and harvested by centrifugation at 3,000 rpm for 3.5 min. Then the cells were washed with 1X HBS four times and were resuspended in 1X HBS at 8 × 106 cells/mL. 4 × 105 cells were used for subsequent assays with each inhibitor concentration. Calcium chloride (1 mM), ionomycin (10 μM), and either DMSO or various concentrations of inhibitor were incubated with the cells for 3 h. The final concentration of DMSO in each sample was 1%. Triton X-100 (1% final) was then added to each sample and the suspension was sonicated at 4 °C for 1 h, followed by centrifugation at 21,000g for 15 min. Lysates were collected, soluble proteins in the lysates were quantified using the DC-assay (Bio-Rad) and were normalized. 2 μg of protein was loaded onto a 4–15% gradient SDS-PAGE gel, separated by electrophoresis and proteins were then transferred to PVDF membrane (Bio-Rad) at 80 V for 50 min. The membrane was then blocked by treating with phosphate buffered saline containing 0.1% Tween-20 (PBST) and 5% BSA, and the blocked membrane was incubated with primary antibodies for histone H3 (1:1,000) and histone H3Cit 2,8,17 (1:1000) in PBST with 5% BSA for 12 h at 4 °C. After washing the membrane with PBST, it was incubated with anti-mouse and anti-rabbit IgG Licor conjugate (1:5000) in PBST and 5% BSA for 1 h at 25 °C. The membrane was then washed with PBST and imaged by Licor analysis.
Cytotoxicity Studies
HEK293T/PAD2 cells were plated (2 × 104 cells/well) in a 96-well plate and were allowed to grow for 24 h. DMSO or various concentrations of inhibitor were added to the wells and incubated for 24 h. Cell viability was measured using the XTT reagent kit (ATCC) by recording the absorbance at 475 nm and 660 nm. An eight-point dose-response curve was fit to the following equation 3 using GraphPad Prism 7.03 to determine the EC50 values for cell-growth inhibition,
| (3) |
where Top and Bottom are plateaus of the dose-response curve, X is the log of inhibitor-concentration, Hillslope is the slope factor or Hill slope.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grant R35GM118112 (P.R.T.).
ABBREVIATIONS
- RA
Rheumatoid arthritis
- PAD
Protein arginine deiminase
- ACPA
Anti-citrullinated protein antibodies
- NET
Neutrophil extracellular trap
- RFA
Rhodamine-conjugated F-amidine
- ABPP
Activity-based protein profiling
Footnotes
Supporting information. Full synthetic procedures, NMR and ESI-MS characterization, PAD inactivation kinetics, RFA-labelling of PAD2, inhibition of histone H3 citrullination, cytotoxicity studies and Figures S1–S25. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Fuhrmann J, Clancy KW, Thompson PR. Chemical Biology of Protein Arginine Modifications in Epigenetic Regulation. Chem Rev. 2015;115:5413–5461. doi: 10.1021/acs.chemrev.5b00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuhrmann J, Thompson PR. Protein Arginine Methylation and Citrullination in Epigenetic Regulation. ACS Chem Bio. 2016;11:654–668. doi: 10.1021/acschembio.5b00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vossenaar ER, Zendman AJW, van Venrooij WJ, Pruijn GJM. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. BioEssays. 2003;25:1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 4.Knuckley B, Causey CP, Jones JE, Bhatia M, Dreyton CJ, Osborne TC, Takahara H, Thompson PR. Substrate Specificity and Kinetic Studies of PADs 1, 3, and 4 Identify Potent and Selective Inhibitors of Protein Arginine Deiminase 3. Biochemistry. 2010;49:4852–4863. doi: 10.1021/bi100363t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kearney PL, Bhatia M, Jones NG, Yuan L, Glascock MC, Catchings KL, Yamada M, Thompson PR. Kinetic Characterization of Protein Arginine Deiminase 4: A Transcriptional Corepressor Implicated in the Onset and Progression of Rheumatoid Arthritis. Biochemistry. 2005;44:10570–10582. doi: 10.1021/bi050292m. [DOI] [PubMed] [Google Scholar]
- 6.Slade DJ, Fang P, Dreyton CJ, Zhang Y, Fuhrmann J, Rempel D, Bax BD, Coonrod SA, Lewis HD, Guo M, Gross ML, Thompson PR. Protein Arginine Deiminase 2 Binds Calcium in an Ordered Fashion: Implications for Inhibitor Design. ACS Chem Bio. 2015;10:1043–1053. doi: 10.1021/cb500933j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J Exp Med. 2010;207:1853–1862. doi: 10.1084/jem.20100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slade DJ, Subramanian V, Thompson PR. Citrullination unravels stem cells. Nat Chem Bio. 2014;10:327. doi: 10.1038/nchembio.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christophorou MA, Castelo-Branco G, Halley-Stott RP, Oliveira CS, Loos R, Radzisheuskaya A, Mowen KA, Bertone P, Silva JCR, Zernicka-Goetz M, Nielsen ML, Gurdon JB, Kouzarides T. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature. 2014;507:104. doi: 10.1038/nature12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones J, Causey C, Knuckley B, Slack-Noyes JL, Thompson PR. Protein arginine deiminase 4 (PAD4): current understanding and future therapeutic potential. Curr Opin Drug Disc. 2009;12:616–627. [PMC free article] [PubMed] [Google Scholar]
- 11.Moscarello MA, Pritzker L, Mastronardi FG, Wood DD. Peptidylarginine deiminase: a candidate factor in demyelinating disease. J Neurochem. 2002;81:335–343. doi: 10.1046/j.1471-4159.2002.00834.x. [DOI] [PubMed] [Google Scholar]
- 12.Damgaard D, Senolt L, Nielsen MF, Pruijn GJ, Nielsen CH. Demonstration of extracellular peptidylarginine deiminase (PAD) activity in synovial fluid of patients with rheumatoid arthritis using a novel assay for citrullination of fibrinogen. Arthritis Res Ther. 2014;16:498. doi: 10.1186/s13075-014-0498-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McElwee JL, Mohanan S, Griffith OL, Breuer HC, Anguish LJ, Cherrington BD, Palmer AM, Howe LR, Subramanian V, Causey CP, Thompson PR, Gray JW, Coonrod SA. Identification of PADI2 as a potential breast cancer biomarker and therapeutic target. BMC Cancer. 2012;12:500. doi: 10.1186/1471-2407-12-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohanan S, Cherrington BD, Horibata S, McElwee JL, Thompson PR, Coonrod SA. Potential Role of Peptidylarginine Deiminase Enzymes and Protein Citrullination in Cancer Pathogenesis. Biochem Res Int. 2012;2012:11. doi: 10.1155/2012/895343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muth A, Subramanian V, Beaumont E, Nagar M, Kerry P, McEwan P, Srinath H, Clancy K, Parelkar S, Thompson PR. Development of a Selective Inhibitor of Protein Arginine Deiminase 2. J Med Chem. 2017;60:3198–3211. doi: 10.1021/acs.jmedchem.7b00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willis VC, Gizinski AM, Banda NK, Causey CP, Knuckley B, Cordova KN, Luo Y, Levitt B, Glogowska M, Chandra P, Kulik L, Robinson WH, Arend WP, Thompson PR, Holers VM. N-α-benzoyl-N5-(2-chloro-1-iminoethyl)-l-ornithine amide, a Protein Arginine Deiminase Inhibitor, Reduces the Severity of Murine Collagen-Induced Arthritis. J Immunol. 2011;186:4396–4404. doi: 10.4049/jimmunol.1001620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knight JS, Subramanian V, O’Dell AA, Yalavarthi S, Zhao W, Smith CK, Hodgin JB, Thompson PR, Kaplan MJ. Peptidylarginine deiminase inhibition disrupts NET formation and protects against kidney, skin and vascular disease in lupus-prone MRL/lpr mice. Ann Rheum Dis. 2015;74:2199–2206. doi: 10.1136/annrheumdis-2014-205365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knight JS, Luo W, O’Dell AA, Yalavarthi S, Zhao W, Subramanian V, Guo C, Grenn RC, Thompson PR, Eitzman DT, Kaplan MJ. Peptidylarginine Deiminase Inhibition Reduces Vascular Damage and Modulates Innate Immune Responses in Murine Models of Atherosclerosis. Circ Res. 2014;114:947–956. doi: 10.1161/CIRCRESAHA.114.303312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghari F, Quirke A-M, Munro S, Kawalkowska J, Picaud S, McGouran J, Subramanian V, Muth A, Williams R, Kessler B, Thompson PR, Fillipakopoulos P, Knapp S, Venables PJ, La Thangue NB. Citrullination-acetylation interplay guides E2F-1 activity during the inflammatory response. Sci Adv. 2016;2 doi: 10.1126/sciadv.1501257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawalkowska J, Quirke A-M, Ghari F, Davis S, Subramanian V, Thompson PR, Williams RO, Fischer R, La Thangue NB, Venables PJ. Abrogation of collagen-induced arthritis by a peptidyl arginine deiminase inhibitor is associated with modulation of T cell-mediated immune responses. Sci Rep. 2016;6:26430. doi: 10.1038/srep26430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo Y, Knuckley B, Lee Y-H, Stallcup MR, Thompson PR. A Fluoroacetamidine-Based Inactivator of Protein Arginine Deiminase 4: Design, Synthesis, and in Vitro and in Vivo Evaluation. J Am Chem Soc. 2006;128:1092–1093. doi: 10.1021/ja0576233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Y, Arita K, Bhatia M, Knuckley B, Lee Y-H, Stallcup MR, Sato M, Thompson PR. Inhibitors and Inactivators of Protein Arginine Deiminase 4: Functional and Structural Characterization. Biochemistry. 2006;45:11727–11736. doi: 10.1021/bi061180d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knuckley B, Causey CP, Pellechia PJ, Cook PF, Thompson PR. Haloacetamidine-based inactivators of Protein Arginine Deiminase 4 (PAD4): Evidence that General Acid Catalysis Promotes Efficient Inactivation. Chembiochem. 2010;11:161–165. doi: 10.1002/cbic.200900698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nemmara VV, Subramanian V, Muth A, Mondal S, Salinger AJ, Maurais AJ, Tilvawala R, Weerapana E, Thompson PR. The Development of Benzimidazole-Based Clickable Probes for the Efficient Labeling of Cellular Protein Arginine Deiminases (PADs) ACS Chem Biol. 2018 doi: 10.1021/acschembio.7b00957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broichhagen J, Schönberger M, Cork SC, Frank JA, Marchetti P, Bugliani M, Shapiro AMJ, Trapp S, Rutter GA, Hodson DJ, Trauner D. Optical control of insulin release using a photoswitchable sulfonylurea. Nat Comm. 2014;5:5116. doi: 10.1038/ncomms6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Velema WA, van der Berg JP, Hansen MJ, Szymanski W, Driessen AJM, Feringa BL. Optical control of antibacterial activity. Nat Chem. 2013;5:924. doi: 10.1038/nchem.1750. [DOI] [PubMed] [Google Scholar]
- 27.Reisinger B, Kuzmanovic N, Löffler P, Merkl R, König B, Sterner R. Exploiting Protein Symmetry To Design Light-Controllable Enzyme Inhibitors. Angew Chem Int Edit. 2014;53:595–598. doi: 10.1002/anie.201307207. [DOI] [PubMed] [Google Scholar]
- 28.Szymanski W, Ourailidou ME, Velema WA, Dekker FJ, Feringa BL. Light-Controlled Histone Deacetylase (HDAC) Inhibitors: Towards Photopharmacological Chemotherapy. Chem Eur J. 2015;21:16517–16524. doi: 10.1002/chem.201502809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velema WA, Szymanski W, Feringa BL. Photopharmacology: Beyond Proof of Principle. J Am Chem Soc. 2014;136:2178–2191. doi: 10.1021/ja413063e. [DOI] [PubMed] [Google Scholar]
- 30.Broichhagen J, Frank JA, Trauner D. A Roadmap to Success in Photopharmacology. Acc Chem Res. 2015;48:1947–1960. doi: 10.1021/acs.accounts.5b00129. [DOI] [PubMed] [Google Scholar]
- 31.Barber DM, Liu S-A, Gottschling K, Sumser M, Hollmann M, Trauner D. Optical control of AMPA receptors using a photoswitchable quinoxaline-2,3-dione antagonist. Chem Sci. 2017;8:611–615. doi: 10.1039/c6sc01621a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broichhagen J, Podewin T, Meyer-Berg H, von Ohlen Y, Johnston NR, Jones BJ, Bloom SR, Rutter GA, Hoffmann-Röder A, Hodson DJ, Trauner D. Optical Control of Insulin Secretion Using an Incretin Switch. Angew Chem Int Edit. 2015;54:15565–15569. doi: 10.1002/anie.201506384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Bolt M, Guertin MJ, Chen W, Zhang S, Cherrington BD, Slade DJ, Dreyton CJ, Subramanian V, Bicker KL, Thompson PR, Mancini MA, Lis JT, Coonrod SA. Peptidylarginine deiminase 2-catalyzed histone H3 arginine 26 citrullination facilitates estrogen receptor α target gene activation. Proc Natl Acad Sci USA. 2012;109:13331–13336. doi: 10.1073/pnas.1203280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, Kouzarides T. Histone Deimination Antagonizes Arginine Methylation. Cell. 2004;118:545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 35.Clancy KW, Russell A-M, Subramanian V, Nguyen H, Qian Y, Campbell RM, Thompson PR. Citrullination/Methylation Crosstalk on Histone H3 Regulates ER-Target Gene Transcription. ACS Chem Bio. 2017;12:1691–1702. doi: 10.1021/acschembio.7b00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewallen DM, Bicker KL, Madoux F, Chase P, Anguish L, Coonrod S, Hodder P, Thompson PR. A FluoPol-ABPP PAD2 High-Throughput Screen Identifies the First Calcium Site Inhibitor Targeting the PADs. ACS Chem Bio. 2014;9:913–921. doi: 10.1021/cb400841k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewallen DM, Bicker KL, Subramanian V, Clancy KW, Slade DJ, Martell J, Dreyton CJ, Sokolove J, Weerapana E, Thompson PR. Chemical Proteomic Platform To Identify Citrullinated Proteins. ACS Chem Bio. 2015;10:2520–2528. doi: 10.1021/acschembio.5b00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wegener M, Hansen MJ, Driessen AJM, Szymanski W, Feringa BL. Photocontrol of Antibacterial Activity: Shifting from UV to Red Light Activation. J Am Chem Soc. 2017;139:17979–17986. doi: 10.1021/jacs.7b09281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knipp M, Vašák M. A Colorimetric 96-Well Microtiter Plate Assay for the Determination of Enzymatically Formed Citrulline. Anal Biochem. 2000;286:257–264. doi: 10.1006/abio.2000.4805. [DOI] [PubMed] [Google Scholar]
- 40.Luo Y, Knuckley B, Bhatia M, Pellechia PJ, Thompson PR. Activity-Based Protein Profiling Reagents for Protein Arginine Deiminase 4 (PAD4): Synthesis and in vitro Evaluation of a Fluorescently Labeled Probe. J Am Chem Soc. 2006;128:14468–14469. doi: 10.1021/ja0656907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knuckley B, Luo Y, Thompson PR. Profiling Protein Arginine Deiminase 4 (PAD4): A novel screen to identify PAD4 inhibitors. Bioorg Med Chem. 2008;16:739–745. doi: 10.1016/j.bmc.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.