Abstract
Cell-to-cell variation in gene expression creates a need for techniques that characterize expression at the level of individual cells. This is particularly true for rare circulating tumor cells (CTCs), in which subtyping and drug resistance are of intense interest. Here we describe a method for cell analysis – single-cell mRNA cytometry – that enables the isolation of rare cells from whole blood as a function of target mRNA sequences. This approach uses two classes of magnetic particles that are labelled to selectively hybridize with different regions of the target mRNA. Hybridization leads to the formation of large magnetic clusters that remain localized within the cells of interest, thereby enabling the cells to be magnetically separated. Targeting specific intracellular mRNAs enables sorting of CTCs from normal hematopoietic cells. No PCR amplification is required to determine RNA expression levels and genotype at the single-cell level, and minimal cell manipulation is required. To demonstrate this approach we use single-cell mRNA cytometry to detect clinically-important sequences in prostate cancer specimens.
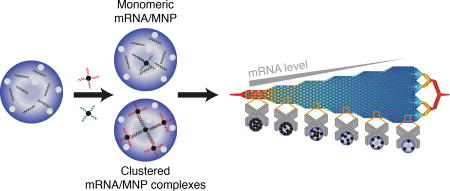
Graphical abstract
Introduction
Gene expression is a stochastic process, and, as a result, mRNA levels exhibit heterogeneity even within a population of isogenic cells1. Studies of gene expression are typically carried out via bulk transcriptome measurement approaches, wherein cells are pooled together and their average gene expression is determined. This strategy generates a transcriptional signature of the bulk population of cells.
The desire to instead study cellular heterogeneity in detail has motivated the development of assays that are capable of characterizing gene expression at the single-cell level2. Most single-cell transcriptional analysis methods are based on RNA sequencing3, quantitative reverse transcription PCR (RT-qPCR) combined with microfluidics4,5, or techniques based on fluorescence hybridization6,7. Unfortunately, RNA sequencing requires mRNA isolation and pre-amplification using PCR, and this may result in amplification bias as well as a significant loss of transcripts8. RT-qPCR combined with microfluidics may provide a closer look at RNA expression within single cells; however, a large percentage of mRNA species can be lost during the purification and processing steps. In addition, the reverse transcription step may introduce artifacts due to template-switching, primer-independent cDNA synthesis, and DNA-dependent DNA polymerase activity9. Fluorescence in situ hybridization10,11 and other techniques based on nanoparticle probes12 do not require pre-amplification, and several of these methods are semi-quantitative for individual cells analyzed in situ. However, often the target mRNA must be labeled with several fluorescent probes to achieve sufficient signal strength, and this precludes accurate quantitation. Moreover, for the analysis of rare cells such as circulating tumour cells (CTCs), cells must first be captured from whole blood, identified, and then subjected to expression analysis. This introduces uncertainty about how the analysis workflow affects the results obtained.
Measurements at the single cell level are particularly important for the study of cancer cells and tumors. Tumors are inherently heterogeneous: different regions of a tumor may experience different levels of exposure to oxygen, chemotherapeutics and other biochemical factors. CTCs are rare tumour cells shed from primary and metastatic tumor sites into the circulation as viable and apoptotic cells, and may exhibit even greater heterogeneity because of dynamic changes correlated to their presence in the bloodstream13.
Here, we report a novel approach – single cell mRNA cytometry – that utilizes nanoparticle-mediated profiling of cancer cells at the single-cell level based on the expression of specific mRNA sequences. Cellular mRNAs are targeted with pairs of probes appended to magnetic nanoparticles (MNPs). Hybridization of mRNA sequences with the tagged MNPs triggers the formation of microscale magnetic clusters that become trapped within the cells. The clusters enhance the magnetic susceptibility of the cells and facilitate their capture within a fluidic device (Figure 1 and Supplementary Figure 1). The device features different capture zones that trap cells with differing magnetic susceptibilities, and after immunostaining, individual cells are then readily visualized within the device to determine their RNA levels. This mRNA profiling approach does not require enzymatic amplification, and is therefore free of amplification bias. It is quantitative when benchmarked against PCR, and is amenable to the analysis of low (~10) numbers of cells, an important feature for the analysis of low levels of bloodborne cells like CTCs. While numerous methods have been applied to the capture and analysis of CTCs14–28, this is the first to do so by targeting intracellular mRNAs.
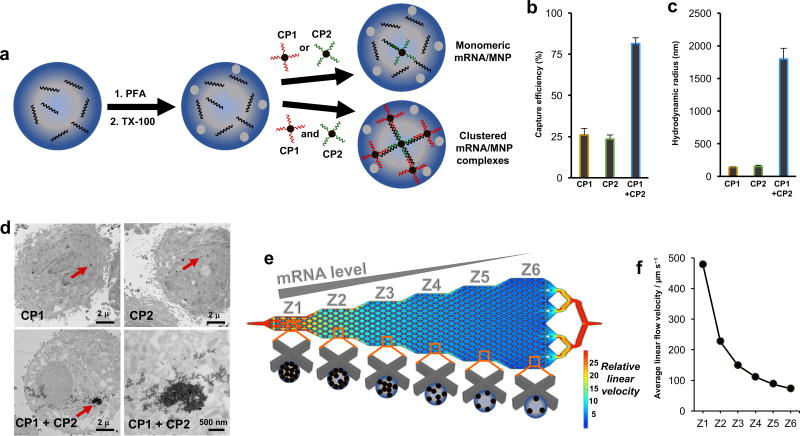
Figure 1. Cellular mRNA analysis approach.
(A) Cells are fixed with 4% paraformaldehyde (PFA) and permeabilized with 0.3% Triton X-100 (TX-100). The cells are incubated with two capture probes (CPs), which are composed of magnetic nanoparticles (MNPs) conjugated to DNA sequences complementary to the target mRNA. Clusters of MNPs are formed and trapped within the cells if two independent CPs are used. (B) When PC3 cells are subjected to magnetic capture based on the targeting of survivin mRNA, only low levels of cell capture are observed if single capture probes are used, while when two capture probes are coincubated with the cells, capture efficiency is increased significantly. (C) DLS measurements of the hydrodynamic radius of magnetic nanoparticles subsequent to hybridization of survivin RNA with either individual capture probes or the combined probes. Statistical analyses of data are provided in Supplementary tables S2−S7. (D) TEM images of PC3 cells after targeting survivin mRNA with CP1, CP2, or CP1+CP2. (E) Device featuring six sequential zones that feature different average linear flow velocities (1×, 0.47×, 0.31×, 0.23×. 0.18×, 0.15×) to facilitate capturing cells with different magnetic content. Cells with high magnetic content are captured in the first zone, whereas cells with medium to low magnetic content are captured in later zones. (F) Distribution of linear velocities at a flow rate of 600 µL h−1 for Zone 1 – 6.
Results and Discussion
Single-cell mRNA cytometry based on intracellular assembly of magnetic clusters
Our approach to mRNA cytometry at the single-cell level is based on magnetic capture of cells using iron oxide nanoparticles functionalized with DNA capture probes complementary to a mRNA sequence of interest (Figure 1A). The capture probes are designed to be specific for a target mRNA. To allow the nanoparticles to penetrate the cells, cells are fixed, permeabilized, and then incubated with the particles.
We found that the use of single nanoparticle-tethered capture probes was not sufficient for high levels of magnetic capture. In proof-of-concept studies monitoring the capture efficiency of a model cell line, low capture efficiencies were observed when a single capture probe was used (Figure 1B).
In contrast, when a combination of two capture probes were used, capture efficiency increased significantly. DLS measurements revealed that combining the two capture probes produced large aggregates in the presence of the complementary target strand, indicating that the dual probe strategy triggered the self-assembly of large magnetic clusters. (Figure 1C and Supplementary Figure 2). These clusters are likely retained within the permeabilized cells, while the single nanoparticles could diffuse out of the cells even after binding a target sequence. TEM studies confirmed the presence of nanoparticle clusters within cells containing a target sequence (Figure 1D).
The cells bearing internalized MNPs are trapped within a fluidic device that features six zones exhibiting differing linear velocities to allow differential sorting of cells with varying levels of bound MNPs (Figure 1E and 1F). Because MNPs have low magnetic susceptibilities, the fluidic channel contains X-shaped microfabricated structures to create localized subzones of low flow velocity and favorable capture dynamics. The first zone has a high linear velocity and thus retains cells with high magnetic content since the retaining magnetic force overcomes the drag force created by the locally high flow velocity. The following five zones exhibit gradually reduced linear velocities (see Figure 1F and Supplementary Information for simulation information). This design allows cells with high magnetic content (i.e., high mRNA expression) to be trapped in the first zone, whereas cells with lower mRNA expression become trapped in later zones based on their mRNA level. This device design has been used to perform high-resolution qualitative profiling of extracellular proteins29; however, it is the first report applied to nucleic acid-based capture.
Profiling of a mRNA sequence using single cell cytometry
In the first suite of experiments, we assessed the capture efficiency of a device designed to facilitate mRNA cytometry and its ability to sort cells bearing different numbers of MNPs. Cultured PC3 cells, a prostate cancer cell line, were labeled with two MNPs targeting the mRNA for survivin, a gene sequence that has been explored as a potential cancer biomarker. Survivin promotes cell division and supresses apoptosis in many human cancers. The antiapoptotic effect is related to its ability to inhibit caspases either directly or indirectly30. The transcription of the survivin gene is higher in tumors compared to normal tissues and is often correlated with metastasis and poor prognosis in cancer patients31.
The cell trapping profiles obtained by targeting the survivin mRNA approach were visualized by immunostaining cells with epithelial markers (EpCAM, CK) and also by confirming the presence of well-defined cell nuclei using the nuclear stain DAPI (Figure 2A). CD45 was also included in the immunostaining protocol to enable the identification of white blood cells when whole blood samples were processed. When CP1 and CP2 were used separately, very low levels of cells were captured (Figure 2A). A non-specific capture probe (NSP) was also used and did not produce significant levels of trapped cells. However, when CP1 and CP2 were used together, much higher levels of cells were observed in the capture device, and the cells were visualized primarily in the first zone of the capture device, indicating high levels of expression. The capture profile and efficiency were unaffected if the PC3 cells were spiked into whole blood. To provide a means to gauge the overall capture efficiency, capture was also carried out with an anti-EpCAM antibody conjugated to MNPs. EpCAM is an epithelial marker found on the surface of tumor cells, and therefore is a standard protein marker to target particularly when cancer cells are isolated from blood. In all trials in which cellular mRNA was tagged with MNPs, a separate sample aliquot was analyzed using anti-EpCAM to provide an overall cell or CTC count.
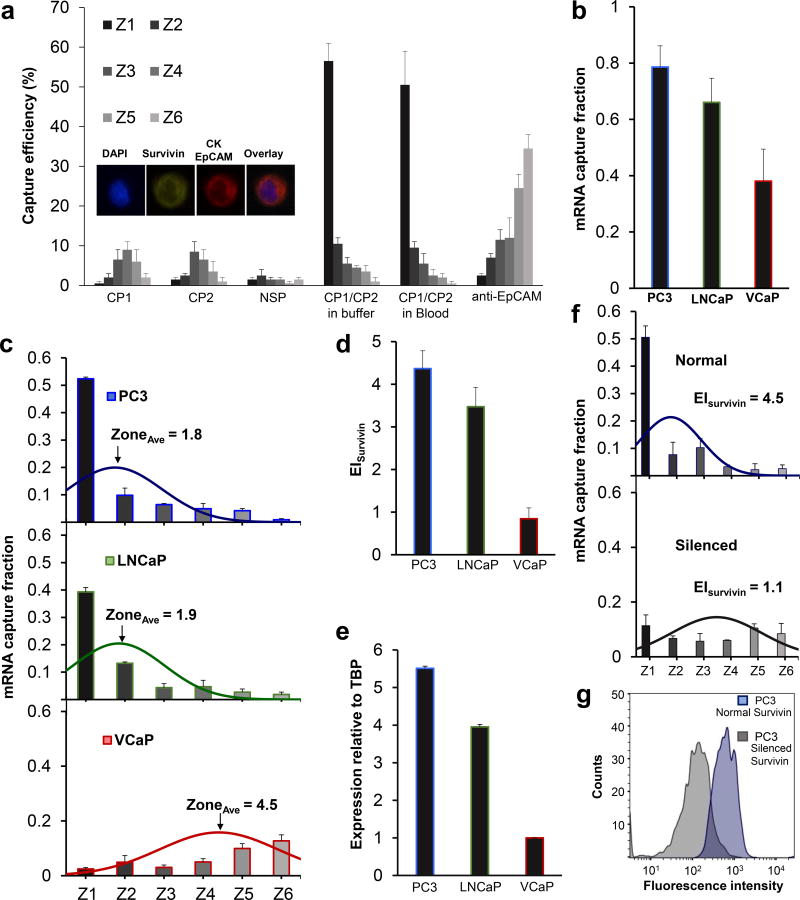
Figure 2. Cell capture and profiling mediated by mRNA-directed magnetic nanoparticles.
(A) Capture efficiency of PC3 cells after incubation with CP1 (complementary to survivin mRNA), CP2 (complementary to survivin mRNA), a non-specific probe (NSP), and a combination of CP1 and CP2 in a buffer solution and blood. A control experiment was carried out in which PC3 cells were captured using magnetic nanoparticles tagged with anti-EpCAM. One hundred cells were used in these trials. Inset shows immunostaining combination used to identify cancer cells. (B) Cellular analysis of survivin mRNA in PC3, LNCaP, and VCaP cell lines. Two hundred cells were used in these trials. The curves represent the normal distribution fit to the capture data. The mRNA capture fraction reflects the capture using mRNA-targeted nanoparticles relative to those labelled with anti-EpCAM. (C) Overall mRNA capture fraction for PC3, LNCaP, and VCaP cells, which compares the number of cells captured with mRNA-targeted nanoparticles versus anti-EpCAM targeted nanoparticles. (D) Expression index, which reflects the mRNA capture fraction divided by the average capture zone. (E) Survivin expression levels determined by RT-qPCR. (F) EISurvivin in PC3 cells before and after silencing the survivin gene with LY2181308 siRNA. Two hundred cells were used in these trials. The curves represent the normal distribution fit to the data. (G) Flow cytometric analysis of survivin protein in PC3 cells before and after silencing the survivin gene. Statistical analyses of data are provided in Supplementary tables S8−S11.
Three prostate cancer cell lines (PC3, LNCaP, and VCaP) were tested in parallel to compare capture efficiencies and the profiles collected using mRNA cytometry (Figure 2B, Supplementary Figure 3). The cells were spiked into blood to ensure that heterogeneous samples were compatible with the approach. The number of cells captured using anti-EpCAM was compared to the number captured using the mRNA-directed approach to determine the overall mRNA capture fraction. For each of the cell lines tested, the overall, EpCAM-mediated capture efficiencies were high (VCaP 92±4%, LNCaP 95±3%, PC3 92±6%), but for the mRNA-targeted trials, the capture levels varied (VCaP 38±11%, LNCaP 66±9%, PC3 79±8%), reflecting the varied expression of survivin in these cell lines. The comparison of the levels of capture when mRNA-targeting was used compared to EpCAM-targeting allowed us to estimate the capture fractions (Figure 2C). Levels of nonspecific capture were taken into account in the calculation of capture fraction (see supplementary information). These studies were conducted with 200 cells spiked into one milliliter of blood; comparable results were obtained with 15 and 50 cells in the same volume (Supplementary Figure 4). While a low level of non-specific capture of white blood cells was observed (Supplementary Figure 5), these cells do not cause false positives because of their distinct staining profiles.
For each cell line, the median zone of capture was determined to provide a parameter that could be used to refine the calculation of relative RNA expression for the cell lines. The PC3 and LNCaP cells were primarily captured in the early zones of the device and had average zone values of 1.8 and 1.9, respectively. The VCaP cells, in addition to having a much lower overall capture efficiency, had a much larger average zone value of 4.5. An expression index (EI) for the survivin mRNA was then calculated for each cell line; values are shown in Figure 2D. The EI was calculated by dividing the capture fraction by the average zone parameter as described in the supporting information. For example, for PC3 cells, the average zone value is 1.8 (Figure 2B), and the overall mRNA-mediated capture efficiency relative to anti-EpCAM mediated capture is 0.79 (Figure 2C). The EI is therefore calculated to be 4.4 (Figure 2D). For VCaP cells the capture efficiency is 0.38 (Figure 2C) and the average zone is 4.5 (Figure 2B). The EI is therefore calculated to be 0.84 (Figure 2B).
Reverse transcription and quantitative PCR was performed using the same cell lines to evaluate the relative expression of survivin. The TATA-box binding protein, TBP, was used as a standard, and the expression levels of survivin were compared to TBP for each cell line (Figure 2E). The levels of expression measured using mRNA cytometry (Figure 2D) and PCR (Figure 2E) are comparable, indicating that the method offers a quantitative approach to monitoring gene expression. For example, the EI values for PC3 and VCaP calculated using single cell mRNA cytometry are 4.4 and 0.84, respectively, and the relative expression levels measured using PCR are 5.5 and 1. The values measured using the two methods agree within measurement error. The concordance of our expression index measurements with PCR-based RNA quantitation provides support to the notion that mRNA cytometry is quantitative, which could not be assumed because cells with a given number of nanoparticles could settle in a number of different adjacent zones (see simulations in Supplementary Information). These results also support the notion that the uptake of magnetic nanoparticles by the different cell types does not influence the expression profiling capability of this mRNA-targeted approach.
We then proceeded to demonstrate the selectivity of the approach by analyzing survivin mRNA in PC3 cells before and after silencing the survivin gene with a small interfering RNA (siRNA). PC3 cells were transfected with LY2181308, a previously characterized siRNA directed against survivin.32 We found that the transfected PC3 cells have exhibited lower EIsurvivin compared to control cells (Figure 2F and Supplementary Figure 6). Flow cytometric analysis of the survivin protein revealed that the protein level decreased by ~83% (Figure 2G). The results corroborated the mRNA expression data obtained using our approach. In these measurements, the overall mRNA-mediated capture efficiency decreases, and the average capture zone also changes, consistent with siRNA knocking down expression. However, the cell capture performed using anti-EpCAM remains constant (Supplementary Figure 7). Therefore, the cells not captured by targeting survivin mRNA are still visible in these trials and we can conclusively determine that RNA expression has decreased.
The sensitivity and dynamic range of the mRNA cytometry approach was also assessed (Supplementary Figure 4). Analysis of as few as 10 cells in a milliliter of cells could be reproducibly achieved, and the EI values were constant between 10 and 500 cells. When 1000 cells were analyzed, the early zones of the device appeared to saturate and this effect then shifted the EI to lower values. However, most clinical specimens would not contain such a high cell count. Specimens – especially from early-stage cancer patients - could contain fewer than 10 cells, in which case a full 10 ml blood sample would need to be processed rather than the 1 ml samples utilized here. The throughput of the analysis – a sample can be processed in ~ 100 minutes (600 µL/hr) – is suitable for clinical applications.
The performance of single cell mRNA cytometry was benchmarked against flow cytometry and fluorescence in situ hybridization (FISH) in order to assess sensitivity relative to these methods. Cells were stained (Supplementary Figure 5) with fluorescent probes and analyzed using flow cytometry could be visualized at low cell counts when suspended in buffered solution, but when spiked into blood, over 1000 cells were required for detection. Even after the depletion of red and white blood cells, residual cells caused a significant background signal that obscured the signal emitted from the RNA probes.
RNA FISH was performed on cells captured in the fluidic capture device (Figure 1E). Cells were incubated with probes with attached MNPs and fluorophores, and then their fluorescence was imaged after cell capture. The level of fluorescence was higher in the earlier zones than later zones, providing independent confirmation that the number of nanoparticles in cells captured in different zones differed.
Analysis of clinically-relevant mRNAs in rare cells
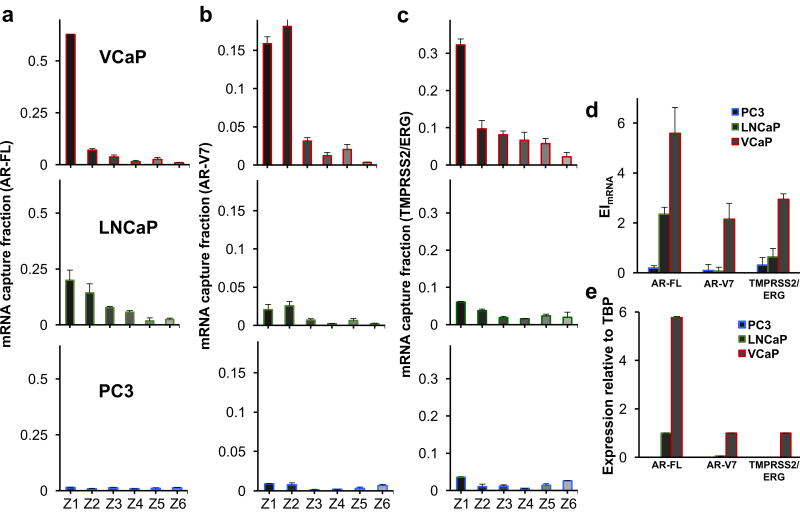
We used this approach to analyze three prostate cancer-specific mRNAs, including full-length androgen receptor (AR-FL), AR splice variant 7 (AR-V7), and TMPRSS2/ERG in VCaP, LnCAP, and PC3 cells. Notably, expression of the androgen receptor is considered a key oncogenic driver at various stages of prostate cancer development and progression33. AR-V7 mRNA is the most abundantly expressed variant that drives prostate cancer during androgen deprivation therapy34. It was recently identified as a predictive biomarker for the resistance to abiraterone and enzalutamide in metastatic castrate-resistant prostate cancer patients35. The TMPRSS2(Exon 1)/ERG(Exon 4) fusion is the most frequent gene fusion in prostate cancer, appearing in about 50% of prostate cancer patients and representing 90% of all prostate cancer gene fusions36. In addition, the presence of TMPRSS2/ERG has been correlated with cancer aggression and metastatic potential37.
The expression pattern of each mRNA was analyzed using our single-cell cytometry approach (Figure 3, Supplementary Figures 8 – 10). The expression index was calculated for each mRNA (Figure 3D), and RT-qPCR was used to analyze the mRNAs in the three cell lines (Figure 3E). The two methods produced comparable profiles, again indicating that single cell mRNA cytometry can be used to quantify gene expression levels.
Figure 3. Analysis of clinically-relevant mRNAs.
Analysis of (A) AR-FL, (B) AR-V7, and (C) TMPRSS2/ERG, in PC3, LNCaP, and VCaP cell lines using single-cell mRNA cytometry. Two hundred cells were used in these trials. The overall mRNA expression was determined using (D) the magnetic ranking approach and (E) RT-qPCR. Statistical analyses of data are provided in Supplementary tables S12−S20. The agreement between the EI values measured with magnetic ranking cytometry and the relative expression levels calculated using RT-qPCR indicates that the new single-cell level technique is quantitative.
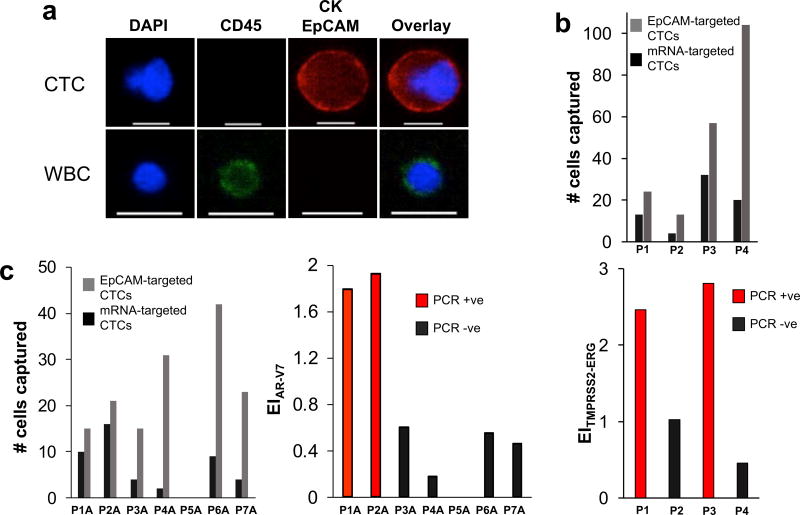
To investigate whether the approach to mRNA analysis in CTCs offers an avenue to clinical utility, we analyzed the TMPRSS2/ERG and AR-V7 mRNAs in blood samples collected from a small cohort of patients undergoing treatment for metastatic castration-resistant prostate cancer. An average of 12 mL of blood was analyzed per patient and CTCs were identified using immunofluorescence and either mRNA- or EpCAM-based capture (Figure 4). Representative images of a CTC captured from patient samples versus a white blood cell are shown in Figure 4A. A patient sample was considered positive for the target mRNA when the EITMPRSS2/ERG was at least 1.5. Samples that tested positive for TMPRSS2/ERG or AR-V7 by mRNA cytometry exhibited significantly higher expression than those that tested negative as measured by PCR (Figure 4B and 4C).
Figure 4. Analysis of clinical samples.
(A) Representative image of a CTC captured from a prostate cancer patient blood sample versus a white blood cell (WBC). The cells were stained with APC-labeled anti-CK, APC-labeled anti-EpCAM, AF488-labeled anti-CD45, and DAPI. Only CK+/EpCAM+/CD45−/DAPI+ cells are counted as CTC. The scale bar is 15 µm. (B) Analysis of blood samples collected from prostate cancer patients for the TMPRSS2-ERG gene fusion. Samples that tested positive for the gene fusion (see Supplementary Figure 11) exhibited significantly higher expression indices than those that tested negative. (C) Analysis of blood samples collected from prostate cancer patients for the androgen receptor splice variant AR-V7. Samples that tested positive for AR-V7 (see Supplementary Figure 12) exhibited significantly higher expression indices than those that tested negative.
In each analysis of a patient sample, the mRNA-based measurement was conducted along with a total CTC count obtained using anti-EpCAM labeled beads. In 10 of the 11 patients tested, CTCs were visualized, but only 4 of the patients exhibited either of the targeted mRNAs. This mRNA analysis method, therefore can provide both a CTC count and information concerning the absence or presence of clinically-relevant mRNAs.
In future, expanded studies of clinically-relevant mRNAs will be needed to establish the detection algorithm for each sequence; however, the initial results presented herein support to the idea that single cell mRNA cytometry can report on the presence or absence of clinically-relevant sequences in patient blood samples. The technique will also need to be tested on early-stage cancer patients – whose samples typically exhibit much lower levels of CTCs – to investigate its utility for non-metastatic patients.
CONCLUSIONS
The single-cell mRNA cytometry method described here provides a new amplification-free means to characterize genotypes and gene expression patterns in intact, bloodborne cancer cells and is broadly applicable to other cell types. This approach relies on microscale clusters of magnetic nanoparticles formed in response to the presence of a specific mRNA inside human cells. While the self-organization and sensing applications of many different types of nanoparticles has been studied38–42, this is the first example of an analysis approach that reports on the self-assembly of magnetic nanoparticles.
While flow cytometry can be used to detect intracellular RNAs labeled with fluorescent probes, high cell numbers are required. The approach reported here allows the study of mRNA expression at the single cell level. It is quantitative, and the expression levels measured using mRNA cytometry correlate closely with those measured using reverse transcription and enzymatic amplification. The method probes RNA levels directly rather than relying on serial enzymatic reactions and isolation steps. It is of interest in the analysis of specific mRNAs in circulating tumor cells that are relevant for therapeutic decision-making. Future improvements to the approach described here will be required to extend this method to the analysis of samples from patients with early-stage cancers, and to resolve bimodal populations of cells with differing expression levels.
METHODS
Chip fabrication
Chips were fabricated using Poly(dimethoxysilane) (PDMS, Dow Chemical, US) soft-lithography. Masters were fabricated on silicon substrates and patterned in SU-8 3050 (Microchem, US). Prior to use, devices were conditioned with 1% Pluronic F68 (Sigma-Aldrich, US) in phosphate-buffered saline (PBS) for 1 h, to reduce the nonspecific adsorption. Each device was sandwiched between two arrays of N52 Nd FeB magnets (K&J Magnetics, US, 1.5 mm by 8 mm) with alternating polarity.
Cell culture
VCaP cells (ATCC CRL-2876) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, ATCC 30-2002). PC3 cells were cultured in F-12K Medium (ATCC 30-2004). LNCaP cells were cultured in RPMI-1640 medium (ATCC 30-2001). All media were supplemented with 10% FBS and 1% penicillin-streptomycin and cells were cultured at 37 °C and 5% CO2 in T75 flasks.
Preparation of the magnetic nanoparticles-labeled capture probes
Briefly, 100 µL of 20 µM of the antisense oligonucleotide solution in Dulbecco's phosphate-buffered saline (DPBS, Sigma-Aldrich, US), were heated for 5 min at 60 °C for deaggregation. Afterward, the solution was transferred to a microtitre plate and incubated with 1.5 µL of 10 mg mL−1 streptavidin-coated magnetic nanoparticles (100 nm, Chemicell, US) for 30 min at room temp. Subsequently, the magnetic nanoparticles-labeled capture probes (MNPs-CPs) were pelleted using a magnetic-ring stand (Thermofisher Scientific, US) and washed three times with DPBS, containing 1 mM dithiothreitol (DPBS/DTT).
Cellular mRNA analysis
Cancer cells (200 cells in 100 µL DPBS) were fixed with 100 µL of 8% paraformaldehyde (PFA, Sigma-Aldrich, US) solution in DPBS/DTT for 15 min at 37 °C. After centrifugation, the cells were incubated with 100 µL of 0.3% Triton X-100 (TX-100, Sigma-Aldrich, US) in DPBS/DTT for 10 min at room temp. Then, 100 µL of labeled MNPs in DPBS/DTT were added and the suspension was gently shaken for 3 h at room temp. The cells were loaded into the microfluidic device at a flow rate of 600 µL h−1.
Cell staining and imaging
Captured cells were counted using fluorescence microscopy. Prior to staining, captured cells were fixed inside the chip using 100 µL of 4% PFA in DPBS/DTT followed by 100 µL of 0.2% TX-100 in DPBS/DTT for permeabilization. Captured cells were immunostained with a mixture of 3% allophycocyanin-labeled anti-cytokeratin antibody (APC-CK, GTX80205, Genetex, US), 3% APC-labeled anti-EpCAM antibody (APC-EpCAM, Miltenyi Biotec Inc., US), and 3% alexafluor 488-labeled anti-CD45 antibody (AF488-CD45, MHCD4520, Invitrogen, US) in 100 µL PBS containing 1% bovine serum albumin (BSA, Sigma-Aldrich, US) and 0.1% Tween-20 (Sigma-Aldrich, US). Chips were scanned using a Nikon Ti-E Eclipse microscope with an automated stage controller and a CMOS Camera (Andor Neo).
Calculation of capture fraction and expression index
The mRNA capture fraction is calculated from formula 1:
| (1) |
NCP denotes the number of cancer cells captured using the capture probe, NNSP represents the number of cells captured by the nonspecific probe, and NAb is the total number of cells in the sample captured by anti-EpCAM. The percentage of cells captured in each zone is multiplied by the mRNA capture fraction to demonstrate the distribution of cell populations bearing different mRNA expression levels and generate a normal distribution fit from which the average capture zone (ZoneAve) is determined.
The mRNA expression index (EImRNA) can then be calculated from formula 2:
| (2) |
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the Canadian Institutes of Health Research (Grant #FDN-148415), the Natural Sciences and Engineering Research Council of Canada (Grant #2016-06090), the Province of Ontario though the Ministry of Research, Innovation and Science (Grant #RE05-009), and the National Cancer Institute of the National Institutes of Health (Grant # 1R33CA204574). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the other funding agencies. We also thank Dr. Anthony Joshua at the Princess Margaret Hospital for supplying clinical specimens.
Footnotes
DATA AVAILABILITY
The authors declare that data supporting the findings of this study are available within the paper and its supplementary information file.
Author contributions. M.L., S.O.K., and E.H.S. conceived and designed the experiments; M.L., R.M.M., M.P. S.U.A., I.I., C.-L. H., and M.M. performed the experiments and analyzed the data: All authors discussed the results, and contributed to the preparation and editing of the manuscript.
Competing financial interest statement. The authors do not have any competing financial interests.
Supplementary information available. Supplementary information is available in the online version of the paper.
References
- 1.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 2.Bendall SC, Nolan GP. From single cells to deep phenotypes in cancer. Nat. Biotechnol. 2012;30:639–647. doi: 10.1038/nbt.2283. [DOI] [PubMed] [Google Scholar]
- 3.Yu M, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalinich M, et al. An RNA-based signature enables high specificity detection of circulating tumor cells in hepatocellular carcinoma. Proc. Natl. Acad. Sci. U.S.A. 2017;114:1123–1128. doi: 10.1073/pnas.1617032114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark IC, Abate AR. Finding a helix in a haystack: nucleic acid cytometry with droplet microfluidics. Lab Chip. 2017;17:2032–2045. doi: 10.1039/c7lc00241f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briley WE, Bondy MH, Randeria PS, Dupper TJ, Mirkin CA. Quantification and real-time tracking of RNA in live cells using Sticky-flares. Proc. Natl. Acad. Sci. U.S.A. 2015;112:9591–95955. doi: 10.1073/pnas.1510581112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 8.Deng Q, Ramskold D, Reinius B, Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 9.Livak KJ, et al. Methods for qPCR gene expression profiling applied to 1440 lymphoblastoid single cells. Methods. 2013;59:71–79. doi: 10.1016/j.ymeth.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyubimova A, et al. Single-molecule mRNA detection and counting in mammalian tissue. Nat. Protoc. 2013;8:1743–1758. doi: 10.1038/nprot.2013.109. [DOI] [PubMed] [Google Scholar]
- 11.Itzkovitz S, van Oudenaarden A. Validating transcripts with probes and imaging technology. Nat. Methods. 2011;8:S12–19. doi: 10.1038/nmeth.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halo TL, et al. NanoFlares for the detection, isolation, and culture of live tumor cells from human blood. Proc. Natl. Acad. Sci. U.S.A. 2014;111:17104–17109. doi: 10.1073/pnas.1418637111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alix-Panabieres C, Pantel K. Challenges in circulating tumour cell research. Nat. Rev. Cancer. 2014;14:623–631. doi: 10.1038/nrc3820. [DOI] [PubMed] [Google Scholar]
- 14.Lang JM, Casavant BP, Beebe DJ. Circulating tumor cells: getting more from less. Sci. Transl. Med. 2012;4:141ps113. doi: 10.1126/scitranslmed.3004261. [DOI] [PubMed] [Google Scholar]
- 15.Green BJ, et al. Beyond the capture of circulating tumor cells: next-generation devices and materials. Angew. Chem. Int. Ed. Engl. 2016;55:1252–1265. doi: 10.1002/anie.201505100. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Chen K, Fan ZH. Circulating tumor cell isolation and analysis. Adv. Clin. Chem. 2016;75:1–31. doi: 10.1016/bs.acc.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Sheng W, Fan ZH. An ensemble of aptamers and antibodies for multivalent capture of cancer cells. Chem. Commun. 2014;50:6722–6725. doi: 10.1039/c4cc02002b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu X, et al. Marker-specific sorting of rare cells using dielectrophoresis. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15757–15761. doi: 10.1073/pnas.0507719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams AA, et al. Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor. J. Am. Chem. Soc. 2008;130:8633–8641. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talasaz AH, et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3970–3975. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stott SL, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schiro PG, et al. Sensitive and high-throughput isolation of rare cells from peripheral blood with ensemble-decision aliquot ranking. Angew. Chem. Int. Ed. Engl. 2012;51:4618–4622. doi: 10.1002/anie.201108695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao W, et al. Bioinspired multivalent DNA network for capture and release of cells. Proc. Natl. Acad. Sci. U. S. A. 2012;109:19626–19631. doi: 10.1073/pnas.1211234109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozkumur E, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci. Transl. Med. 2013;5:179ra147. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Wu M, Han X, Wang P, Qin L. High-throughput, label-free isolation of cancer stem cells on the basis of cell adhesion capacity. Angew. Chem. Int. Ed. Engl. 2015;54:10838–10842. doi: 10.1002/anie.201505294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Zhou L, Qin L. High-throughput 3D cell invasion chip enables accurate cancer metastatic assays. J. Am. Chem. Soc. 2014;136:15257–15262. doi: 10.1021/ja5072114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon HJ, et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat. Nanotechnol. 2013;8:735–741. doi: 10.1038/nnano.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poudineh M, et al. Tracking the dynamics of circulating tumour cell phenotypes using nanoparticle-mediated magnetic ranking. Nat. Nanotechnol. 2017;12:274–281. doi: 10.1038/nnano.2016.239. [DOI] [PubMed] [Google Scholar]
- 30.Altieri DC. Validating survivin as a cancer therapeutic target. Nat. Rev. Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 31.Fulda S, Vucic D. Targeting IAP proteins for therapeutic intervention in cancer. Nat. Rev. Drug Discov. 2012;11:109–124. doi: 10.1038/nrd3627. [DOI] [PubMed] [Google Scholar]
- 32.Carrasco RA, et al. Antisense inhibition of survivin expression as a cancer therapeutic. Mol. Cancer. Ther. 2011;10:221–232. doi: 10.1158/1535-7163.MCT-10-0756. [DOI] [PubMed] [Google Scholar]
- 33.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer. 2015;15:701–711. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson D, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antonarakis ES, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 37.Tomlins SA, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci. Transl. Med. 2011;3:94ra72. doi: 10.1126/scitranslmed.3001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edwardson TG, Lau KL, Bousmail D, Serpell CJ, Sleiman HF. Transfer of molecular recognition information from DNA nanostructures to gold nanoparticles. Nat. Chem. 2016;8:162–170. doi: 10.1038/nchem.2420. [DOI] [PubMed] [Google Scholar]
- 39.Park SY, et al. DNA-programmable nanoparticle crystallization. Nature. 2008;451:553–556. doi: 10.1038/nature06508. [DOI] [PubMed] [Google Scholar]
- 40.Aldaye FA, Sleiman HF. Dynamic DNA templates for discrete gold nanoparticle assemblies: control of geometry, modularity, write/erase and structural switching. J. Am. Chem. Soc. 2007;129:4130–4131. doi: 10.1021/ja070017i. [DOI] [PubMed] [Google Scholar]
- 41.Chuah K, et al. Ultrasensitive electrochemical detection of prostate-specific antigen (PSA) using gold-coated magnetic nanoparticles as 'dispersible electrodes'. Chem. Commun. 2012;48:3503–3505. doi: 10.1039/c2cc30512g. [DOI] [PubMed] [Google Scholar]
- 42.Farlow J, et al. Formation of targeted monovalent quantum dots by steric exclusion. Nat. Methods. 2013;10:1203–1205. doi: 10.1038/nmeth.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.