Abstract
For decades, RNA served in a supporting role between the genetic carrier (DNA) and the functional molecules (proteins). Finally, it is time for RNA to take the center stage in all aspects of biology. The retina provides a unique opportunity to dissect molecular underpinnings of neuronal diversity and disease. Transcriptome profiles of the retina and its resident cell types have unraveled unique features of the RNA landscape. Discovery of distinct RNA molecules in the retina and recognition of RNA processing as a major cause of retinal neurodegeneration have prompted design of biomarkers and novel therapeutic paradigms. Here, we have reviewed RNA biology as it pertains to the retina, emphasizing new avenues for investigations in development and disease.
RNA: The messenger and beyond
“Unstable intermediate carrying information from genes to ribosomes for protein synthesis”
Sydney Brenner, 1961
With this provocative title for his ground-breaking discovery, Sydney Brenner announced the birth of a new era in the nascent field of molecular biology; and, the messenger RNA (mRNA) was proposed as the linkage molecule between genes and proteins [1, 2]. RNA was discovered in the late 1890’s, but another 60 years will go by before a function for RNA in protein synthesis will be put forth. Around the same time, transfer RNA (tRNA) was identified in bacteria [3], bringing the number of known RNA species to three (ribosomal, transfer and messenger RNA, named after their function). The focus in early 60s was on the “central dogma” (see Glossary), and all attention centered on the transfer of genetic information from the double helical structure of DNA to proteins, the drivers of cellular function. RNA was perceived as the support molecule and did not attract as much immediate scrutiny from scientists. However, the focus began to shift in late 60s with the realization of critical roles of RNA in protein translation, the discovery of hetero-nuclear (hn) and small-nuclear (sn) RNAs [4–6], and later the critical role of RNA as an enzyme, e.g., ribonuclease-P [7, 8]. “RNA world” theory emerged thereafter to explain the origin of life [9]. Yet, for the most part, and despite the knowledge that mRNA is a small fraction of the transcribed sequences in any cell, RNA remained as an intermediate between DNA and protein. With the identification of long non-coding RNAs H19 [10] and XIST [11, 12], associated with imprinting and X-inactivation, respectively, RNA acquired its due as more than just an “unstable intermediate” or part of “junk DNA”. At least 15 distinct types of RNA molecules have been identified to date, in diverse biological contexts, in nucleus, cytoplasm, or even circulating fluids.
Modern technologies of probing RNA, including next generation sequencing, and computational tools have given a new meaning to the term “RNA world”. We can now catalog and monitor the RNA landscape, yet we know woefully little about the function of most transcribed sequences. RNA is capable of performing a plethora of functions; these include catalysis of biochemical reactions (ribozymes) [8], response to stimuli (riboswitch) [13], carrying genetic information (viral genomes), formation of complex organization (with self and other organic molecules) [14], gene regulation by epigenetic modifications [15], and guiding cellular localization and compartmentalization [16, 17]. In this review, we cover the role of RNA and RNA biology in the context of mammalian retina, our window to the world, and describe how we can use this knowledge for elucidating complexities associated with retinal development and disease.
The retina
Of the five senses, humans rely most heavily on the sense of sight to navigate the world. In a recent nationwide poll [18], most individuals surveyed (87.5%) said good vision is essential to overall health, and about 50% stated that losing their vision is the worst possible health outcome. Vision impairment associated with retinal dysfunction is a major cause of incurable blindness worldwide. Innovations in technology and relative ease in clinical ascertainment have unraveled an array of retinal phenotypes. With over 250 known disease-causing genes (https://sph.uth.edu/retnet/), extensive biological insights, and relative ease in manipulation, the retina offers a unique opportunity to investigate neuronal diversity, functional organization, and disease mechanisms.
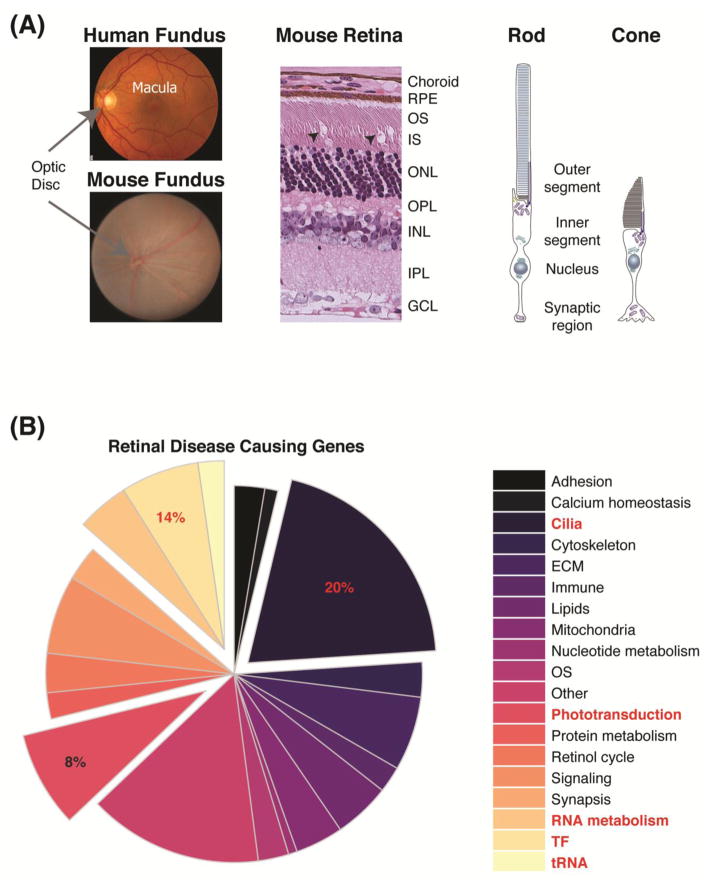
The retina is the photosensitive part of the eye, containing five major types of neurons organized in a laminar structure (similar to the cerebral cortex), which is optimized for capture, processing and transmission of the visual signal [19, 20]. During embryogenesis, the retina originates from the neuroectoderm, differentiating into three cellular layers (outer, inner and ganglion nuclear layers; ONL, INL and GCL, respectively) and two synaptic layers (outer and inner plexiform layer; OPL and IPL, respectively). The photoreceptors in the ONL initiate the phototransduction cascade. Most mammalian retina are dominated by rod photoreceptors; the human retina contains roughly 105 million rods that mediate dim light vision and about 6 million cone photoreceptors that are critical for high visual acuity and color vision. Visual information is then transmitted to and processed by bipolar, horizontal and amacrine cells in the INL, and finally sent to the brain by ganglion cells through the optic nerve.
Photoreceptors possess four morphologically-distinct domains: the outer segment (OS), the inner segment (IS), the cell body including the nucleus, and the synaptic region (Figure 1A). The OS is a modified sensory cilium that is formed by hundreds of stacked membranous discs carrying components of the phototransduction cascade, including the visual pigment (opsin). The type of opsin expressed defines the sensitivity and identity of the photoreceptor; based on the maximal spectral sensitivity of the opsin, the human cones are broadly classified into three sub-types, L- (long, 564 nm), M- (medium, 533 nm) and S- (short, 437 nm) cones. A single type of rod photoreceptor, expressing the rhodopsin visual pigment, is present in the vertebrate retina. In a “steady state”, opsin is covalently bound to 11-cis retinal chromophore. Upon exposure to light photons, 11-cis retinal isomerizes to all-trans retinal, resulting in conformational change of the opsin molecule. Phototransduction comprises of the biochemical chain of events that lead to hyperpolarization of the photoreceptor and subsequent transmission of the visual signal to inner retina neurons. All-trans retinal is then recycled back to 11-cis via retinoid cycle involving retinal pigment epithelium (RPE). Defects in photoreceptor or RPE function are the primary cause of visual impairment in retinal and macular degeneration; of these, 14% of disease is attributed to mutations in genes associated with RNA life cycle, secondary only to cilia biogenesis and transport (Figure 1B).
Figure 1.
Retinal structure and disease-causing genes. (A) Fundus images of a healthy human and mouse retinas, depicting the blood vessels, optic nerve head, and macula (in human). Histology of mouse retina, showing the laminar structure of the retina. Arrow heads point to the unique cone OS. A schematic representation of photoreceptors, highlighting the different morphological and functional domains of the cell. (B) Pie-chart of reported retinal degeneration genes (RetNet; https://sph.uth.edu/retnet), organized by the functional groups. The top three groups are indicated in red and pulled out of the pie. Abbreviations: RPE, retinal pigment epithelium; OS, outer segment; IS, inner segment; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer; ECM, extracellular matrix.
Retinal transcriptome
The RNA profile (transcriptome) provides the correct blueprint for each cell’s morphological and functional identity. Acquisition of a specific cell fate depends not only on expression of the correct repertoire of genes, but also on suppression of genes that may interfere with desired cellular functions. Transcriptional networks guiding retinal development have been the subject of intense investigations [21–24]; this is especially true for photoreceptors with detailed transcriptome profiling of the dynamic changes and gene regulatory networks associated with morphogenesis and functional maturation [25]. A combinatorial (and rather incomplete) network of transcription factors, including members of the basic helix-loop-helix, homeodomain, and fork-head families, tightly regulate temporal transcriptional events in the retinal progenitors. Transcriptional landscape in the developing vertebrate retina corresponds to the birth order of cells; the initial wave is enriched for genes associated with differentiation of ganglion cells, followed by amacrine and horizontal cells, cone photorecetors and 10–15% of rods, in the embryonic stage (Brooks and Swaroop, manuscript in preparation)[26–28]. Postnatally-expressed genes correlate highly to rod photoreceptors and their functional maturation because of their abundance in the mouse retina, with bipolar neurons and Muller glia being the other postnatal differentiating cells [29–31]. Several aspects of the retinal transcriptome make it especially interesting; the retina displays a high prevalence of alternative splicing and alternative promotor usage, and a plethora of non-coding transcripts [32–35]. Retinal development in humans proceeds in two distinct waves; the first corresponds to differentiation of the foveal region and the second to expansion and subsequently differentiation of the peripheral retina [36]. Extensive tissue- and cell-type specific heterogeneity (as much as 85%) has been observed in protein-coding transcripts by transcriptome profiling of human tissues [37]; this is especially true for the retina [33, 36]. As in brain, retinal photoreceptors exhibit unique set of transcript isoforms generated by alternate promoter usage or alternate splicing [25].
Alternative splicing
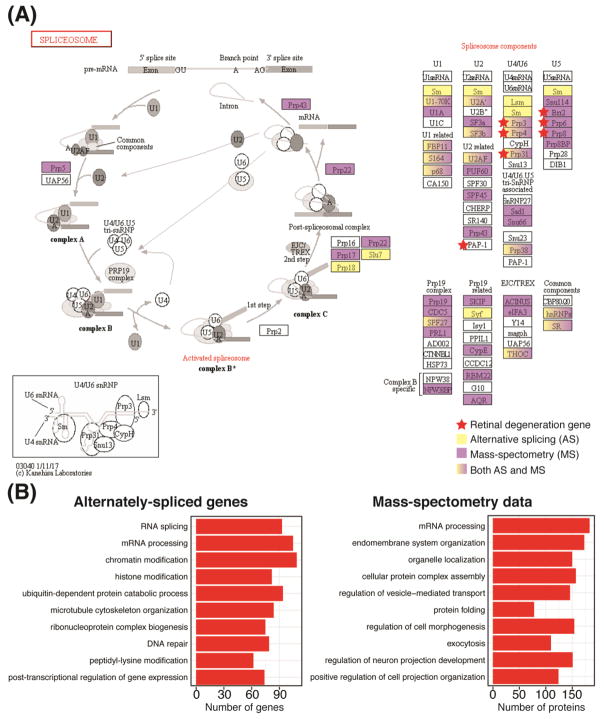
Transcriptome analyses have unraveled an “unprecedented” level of alternative splicing (AS) events in the mammalian retina, especially in the photoreceptors [25, 32, 33, 38]. Over 50% of genes in the human retina exhibit altered exon composition compared to the reference sequence [33, 36]. Why is AS so predominant in the retina? Is splicing evolutionarily linked to light perception? In Arabidopsis, mutations in splicing factors (e.g., SKIP and RRC1) show a defect in the plant’s ability to respond to light [39]. AS is proposed to facilitate tissue diversity, and is essential to cell fate determination [40]. This is beautifully exemplified in the retina, where AS contributes to diverse protein functions [41], including gene regulation [25] and modulation of disease phenotype [42]. Photoreceptors are especially sensitive to global splicing defects as mutations in splicing factors can lead to non-syndromic retinal degeneration; for example, heterozygous mutations in the SNRNP200 gene specifically lead to autosomal dominant retinitis pigmentosa (RP) [43]. Notably, pathways involving RNA processing/binding are significantly enriched in over 2000 genes that exhibit 10,000 independent alternately-spliced events, with components of the spliceosome machinery itself undergoing extensive alternative splicing (Figure 2A, B), as indicated by Gene Ontology (GO) analysis of the photoreceptor developmental transcriptome data [25]. Splicing in mouse photoreceptors is shown to exhibit atypical neuronal patterns that involve “high inclusion” of specific microexons (hallmark of neuronal cells) and are guided, to a significant extent, by the Musashi1 protein [44]. Microexons appear to module tissue-specific protein-protein interaction networks, and their dysregulation has been observed in autistic brains [45]. RNA-binding proteins (RBPs) tightly regulate the inclusion of these exons during neurogenesis, via splicing enhancers located in close proximity to 5′ and 3′ splice acceptor sites [46]. A network of unique RPBs and splicing factors with specific microexon inclusion could have a dramatic effect on the retinal protein interactome. Mass-spectrometry analysis of the retinal proteome during development and disease is necessary to validate transcriptome data; our early proteomic analysis (unpublished) suggests an enrichment of proteins associated with the splicing machinery (Figure 2A, B) and may yield insights as to why retina invests significant resources to maintain its unique RNA landscape.
Figure 2.
Alternative splicing in the retina. (A) Schematic representation of the spliceosome pathway (from KEGG database; http://www.genome.jp/kegg/pathway.html). Each component of the spliceosome is marked in purple if it was enriched in mass-spectrometry data obtained from trypsinized retinal lysate or in yellow if it was enriched in alternatively-spliced gene analysis of developing photoreceptors. The lysate from P28 C57BL/6 mouse retina was separated by high-pH reversed phase liquid chromatography and concatenated into 15 fractions for LC MS/MS analysis using Orbitrap Lumos Tribrid mass spectrometer (Thermo Fisher Scientific) (Zelinger, unpublished data). Retinal disease genes are indicated by red star. Spliceosome components listed in KEGG pathway use an alias nomenclature; e.g., PRPF genes in NCBI database (https://www.ncbi.nlm.nih.gov/gene) are designated as PRP here. (B) Top ten Gene Ontology (GO) terms enriched in either alternatively-spliced genes or total protein mass-spectrometry data from the retina. Both analyses identified RNA processing as one of the top biological processes.
Splicing defects and retinal degeneration
Since the first documentation of rhodopsin defects in autosomal dominant RP [47], thousands of mutations have now been identified in over 250 genes, making retinal diseases among the most prolific in genetics research. In 2001, mutations in splicing factor genes were first reported to cause autosomal dominant retinitis pigmentosa [48, 49]. Genetic analyses of families and patients with retinal degeneration have now identified mutations in a number of splicing factor genes including PRPF3, PRPF4, PRPF6, PRPF8, PRPF31 and PAP1 (RP9) (https://sph.uth.edu/retnet). Notably, PRPF6, PRPF8 and PRPF31 splicing factors have also been linked to cilia biogenesis [50]. Given the prevalence of alternately-spliced transcripts in the retina, it is not surprising that dysregulation of RNA processing is among the key defects associated with retinal degenerative diseases [51, 52]. A spectrum of mutations in Stargardt disease are associated with abberrant splicing events in ABCA4 [53]. Splicing-associated defects in the RPE gene Bestrophin 1 cause autosomal dominant vitreoretinochoroidopathy [54]. However, one needs to be careful in interpreting in vitro strategies as no splicing defect was detected in patient stem cell-derived RPE cells carrying the splicing mutation c.704T > C, p.V235A in the Bestrophin gene [55].
Non-coding RNAs
Next generation sequencing has revealed transcription of almost three-quarters of the human genome, even though only 1% of the transcripts are protein-coding [56]. Not surprisingly, an array of non-coding (nc) RNAs are detected in the mammalian retina, especially photoreceptors [57]. Here we briefly discuss three main classes of ncRNAs.
Conditional ablation of Dicer1, a key enzyme in miRNA processing, in the mouse retina demonstrated critical functions of miRNAs in retinal development, especially affecting the differentiation of late-born cells such as rods and Müller glia [58, 59]. Furthermore, reduction or loss of Dicer1 in the RPE has been implicated in Alu RNA toxicity in age-related macular degeneration (AMD) and RPE degeneration in mice [60, 61]. Twenty miRNAs dominate the retinal miRNAome [62]; of these, miR-184 and miR204 are shown to be involved in inherited diseases, resulting in either ocular [63] or ocular and retinal phenotypes [64], respectively. Loss of miR-155 and its target gene CCN1 is reported to result in vascular changes in the mouse retina [65]. Several miRs expressed in retinal and RPE are suggested to contribute to pathology in AMD and diabetic retinopathy; however, we have limited understanding of their precise functions at this stage.
The highly-expressed miRNAs, originating in the miR-183/96/182 cluster, are linked to formation of the retinal laminar structure and photoreceptor outer segment and synapses [66–68]. The studies on this miRNA cluster previously demonstrated that “microRNA metabolism in neurons is higher than in most other cells types and linked to neuronal activity” [69]. These miRNAs are transcriptionally upregulated by light in mouse retinal neurons, and are rapidly decayed during dark adaptation. In dark conditions, photoreceptors are depolarized and release the neurotransmitter, glutamate. Interestingly, voltage-dependent glutamate transporter Slc1a1 is a target of miR-183/96/182 cluster, and under low glutamic load, Slc1a1 could help clear glutamate from the photoreceptor synaptic region. MiR-183/96/182 cluster is also implicated in cone outer segment maintenance [70] by targeting genes associated with membrane trafficking, lipid metabolism, and cilia formation. The rapid turnover of miRNAs in retinal neurons, and consequently, rapid changes in expression of their targets could facilitate accurate coding of the visual stimulus in a rapidly changing environment.
As many as 50,000 lncRNA genes have been annotated in the human genome [71], and about 40% of these are expressed in neuronal tissues [72]. Over 2300 distinct lncRNAs are reportedly expressed in developing photoreceptors, and their functional relevance was suggested based on bioinformatic analysis [57]. LncRNA RNCR2 is involved in retinal cell fate acquisition [73], whereas RNCR4 and other noncoding RNAs are implicated in organization of the retinal architecture [67]. Search of taurine-upregulated genes identified a potential ncRNA, TUG1, and its knockdown by RNAi resulted in abnormal photoreceptor outer segments [74]. RNCR2 and RNCR3 are also implicated in microvascular dysfunction [75, 76]. A circadian-regulated lncRNA ENSMUST00000138486 (human gene: MIR4435-2HG and Morrbid) that is highly expressed in RPE/choroid may regulate the expression of phagocytosis-associated gene Mertk [77]. Several antisense RNAs (a subgroup of lncRNAs) originate from eye transcription factor loci [57, 78] and appear to modulate gene expression [79, 80].
Another class of ncRNAs, tRNAs, has received much less scrutiny. Nonetheless, defects in mitochondrial tRNA genes are associated with complex syndromic conditions [81, 82] and retinal degeneration, usually in combination with sensorineural hearing loss [83, 84]. Curiously, a homozygous variant in histidyl-tRNA synthetase (HARS) appear to be associated with Usher syndrome (USH3B) [85]. Mutations in TRNT1, a nucleotidyltransferase critical for tRNA processing, lead to RP with erythrocytic microcystosis [86].
RNA based diagnosis and therapy of retinal diseases
The ability of antisense oligonucleotide to prevent replication and translation of Rous sarcoma virus was the first demonstration of RNA-based strategies to alter gene expression [87]. Oligonucleotides are commonly used in research, and clinical studies are being performed to assess their utility in a clinical setting. RNA can also be considered as a biomarker for retinal diseases, especially since dysregulation of a subset of miRNAs has been consistently recognized in retina, vitreous or serum of AMD patients and in diabetic retinopathy blood samples [88, 89]. In addition, detection of a specific isoform of TUBD1 in blood can potentially serve as a biomarker for diabetic retinopathy [90].
RNA has emerged as a potential therapeutic target as well. RNA interference (RNAi) technology involves the use of a RNA molecule complementary to a sequence in the target mRNA for gene silencing either by inhibition of translation or by degradation of the target [91, 92]. Six different RNA-based drugs, including an RNA aptamer, are currently available or in clinical trials for treatment of neovascular AMD and their targets include vascular endothelial growth factor [93]. RNAi is also shown to be effective in treating dominant RP in a mouse model carrying a rhodopsin mutation [94]. Two adeno-associated virus vectors were used to deliver RNAi against the endogenous mutated rhodopsin and a codon-modified rhodopsin replacement gene resistant to suppression. This combination of RNAi and gene replacement can provide the broad direction for developing “mutation independent” treatment for genetic diseases [95]. Ribozymes, which are RNAs that possess a catalytic activity to cleave either itself or other RNAs, have been used to target several genetic diseases [96]. Treatment of retinal degeneration caused by rhodopsin mutations [97] provided “proof of concept” for this strategy. Antisense oligonucleotide-based strategies have been developed to restore correct splicing patterns for USH2A [98], CEP290 [99] and OPA1 [100]. However, additional research is needed to assess the effectiveness of RNA-based therapies.
Concluding Remarks and Future Directions
RNA is no longer just an intermediate molecule; however, the true functional potential of diverse RNA entities is far from clear. The retina provides a unique opportunity to study RNA biology during development and disease. Here, we highlight a few interesting avenues for future research.
RNA modifications (epitranscriptome)
At least 150 different chemical modifications have been described for RNA, and the epitranscriptome is emerging as a key regulator of transcriptional landscape [101, 102]. The most common type of RNA modification, is N6-methyladenosine (m6A), which might mirror miRNA functions, by upregulation or rapid degradation of RNA during developmental transitions [103]. RNA methylation can modulate most steps of RNA processing [104], localization [105], secondary structure and interaction with proteins [106], and is even implicated in regulation of the circadian clock [107]. Little is known about RNA modifications in the retina.
Polyadenylation is another RNA modification that is getting a second look, especially since many miRNAs and RNA-binding proteins bind RNA at the 3′-untranslated region. Polyadenylation changes are reported during retinal development [108]. A key feature of polyadenylation dynamics in the retina is preferential usage of weak proximal polyadenylation sites in early development, indicating a distinct regulatory mechanism.
RNA-based cellular communication
RNA-based communication has origins in the early days of life on earth (the “RNA world”) [109]. Curiously, though not validated yet, secretion of proteins and RNAs by exosomes at neuronal synapses can serve as a possible mode of communication in the nervous system [110]. Cargo of secreted exosomes from prion-infected neurons includes diverse RNA species that may correlate to disease progression [111]. Analysis of purified exosomes by next generation sequencing may unravel the role of small ncRNAs in cell-cell communication in the retina and possibly in retinal neurodegeneration.
RNA binding proteins (RBP) and Cellular Metabolism
We are beginning to realize that efficient organization of cellular pathways and/or effective response to distinct stimuli may be facilitated by rapid assembly (or disassembly) of specialized subcellular membraneless organelles and/or macromolecular structures. Ribonucleoprotein complexes exhibit unique and highly dynamic properties, behaving like liquid droplets, with RNA molecule(s) regulating the biophysical characteristics of such membraneless organelles [112]. One of the RP genes, Ceramide kinase like (CERKL), is a RNA binding protein that co-localizes with stress granules and processing (P)-bodies [113]. The exact function of CERKL and the RNAs it binds to are currently unknown; however, CERKL’s ability to bind poly A-binding protein PABP, elongation factor eIF3B and heat shock protein HSP70 suggests its possible role in modulating mRNA translation and stability. In the mammalian retina, the RNA-binding protein RBPMS is selectively expressed in ganglion cells though its role is not understood [114].
Several metabolic enzymes are now being reported as RNA-binding proteins with unexpected role in feedback regulation [115]. Recently, hypoxia-induced formation of non-membrane “glycolytic bodies” (G-bodies) was shown to require RNA [116]. This observation has significant implications for retinal function and degenerative disease because the photoreceptors display high energy requirements, with reliance on aerobic glycolysis [117, 118] and because of low mitochondrial reserve capacity [119].
As the field of RNA-biology continues to grow, novel and evolving roles of RNA in distinct cellular contexts are expected to have far-reaching consequences during development, homeostasis and disease pathogenesis.
Trends box.
After decades of playing a second fiddle, RNA has now acquired its coveted role in all aspects of biological research.
Transcriptome profiling has identified key features unique to the retina, including non-coding RNAs that are likely linked to both development and disease pathology.
Alternative splicing is a hallmark of transcriptomes of developing mammalian retina, and especially photoreceptors. At least 50% of the retina-expressed genes exhibit an altered structure compared to the reference.
Diverse and functionally characterized neurons in the retina provide a unique model to dissect the function of distinct RNA molecules in biological processes and evaluate novel therapeutic paradigms.
Outstanding Questions Box.
How does the landscape of the RNA epitranscriptome in the retina impact cell function during development and aging?
Do the lncRNAs interact directly and predominantly with the genome or form complexes with other RNAs and proteins?
Are all lncRNAs truly non-coding, or do the small ORFs (present in many lncRNAs) have biological function?
What are membraneless RNA-protein complexes in retinal neurons and what is their role in cellular homeostasis?
What is the biological significance of developmentally-controlled alternative splicing in the retina, and especially photoreceptors, and what is the role of splicing in retinal neurodegeneration?
Can we use RNA biomarkers to monitor progression of retinal disease and in the design of novel RNA-based therapies for retinal disease?
Acknowledgments
We thank Angel Aponte and Marjan Gucek (NHLBI Proteomics core facility) for obtaining the MS data and Vijender Chaitankar, Matthew Brooks and D. Thad Whitaker for advice. Our research is supported by Intramural Research Program of the National Eye Institute (EY000450 and EY000546).
Glossary
- Age-related macular degeneration (AMD)
a common multifactorial retinal disease, which is a leading cause of vision impairment among people over 50.
- Alternative splicing
a highly-regulated RNA processing mechanism, resulting in generation of distinct RNA transcripts from a single gene.
- Antisense oligonucleotide
polymer of 15–20 deoxyribonucleotides or ribonucleotides with complementary sequence orientation of 3′ → 5′ to the sense sequence of a target molecule.
- Bipolar, horizontal and amacrine cells
inner retina neurons that transmit visual information from photoreceptors to ganglion cells after integration and processing.
- Central dogma
the two-step process of transcription and translation for information flow from DNA to protein via RNA.
- Choroid
vascular layer of the eye between the neural retina and the sclera, providing nutrients, oxygen and removing waste.
- Circadian clock
daily rhythms of biological activity. All cells have an intrinsic clock. The master clock in the suprachiasmatic nucleus is reset by light-dark cycles and controls production of melatonin.
- Dicer1
a member of the ribonuclease III family that contains RNA helicase motif. It cleaves dsRNAs and pre-miRNAs into fragments of 22 nucleotides, producing short interfering RNAs.
- Epitranscriptome
a collection of biochemical modifications of RNAs, which have an effect on the function of the transcript without changing the sequence.
- Ganglion cells
retinal neurons that receive visual information from the bipolar and amacrine cells. Axons of the ganglion cells form the optic nerve that transmits information to the brain.
- Long non-coding RNAs
a diverse class of RNAs of > 200 nucleotides that presumably lack protein-coding potential.
- Microexons
exons of <51 nucleotides, especially common in neuronal tissues. These exons appear to have a role in regulating protein interaction networks.
- miRNAs
non-coding RNAs of 22 nucleotides that drive post-transcriptional regulation of gene expression.
- Müller glia cells
glial cells that extend through all retinal cell layers, playing roles in maintaining retinal structure and signaling.
- Retinal pigment epithelium (RPE)
pigmented cells adjacent to the photoreceptors. RPE creates blood-retina barrier between choroid and neural retina and performs retinoid recycling, phagocytosis of shed outer segment discs, and transport of nutrients and oxygen.
- Retinitis pigmentosa (RP)
heterogeneous group of inherited disorders that lead to dysfunction or death of photoreceptors. Initial clinical symptoms are night-blindness and visual field constriction, progressing to complete blindness.
- Ribonuclease-P
essential endonuclease that catalyzes the cleavage of 5′ leader sequence from precursor tRNAs.
- Stress granules and P-bodies
membraneless organelles comprising of RNAs and proteins. While stress granules are transient, P-bodies are present constantly in cells. Both play a major role in regulation of RNA metabolism, via degradation or silencing.
- Taurine
A sulfur amino acid that is not incorporated into proteins. Taurine has diverse biological functions, including in neurotransmission, stabilization of cell membranes and ion transport.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brenner S, et al. An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature. 1961;190:576–581. doi: 10.1038/190576a0. [DOI] [PubMed] [Google Scholar]
- 2.Gros F, et al. Unstable ribonucleic acid revealed by pulse labelling of Escherichia coli. Nature. 1961;190:581–5. doi: 10.1038/190581a0. [DOI] [PubMed] [Google Scholar]
- 3.Hoagland MB, et al. A soluble ribonucleic acid intermediate in protein synthesis. J Biol Chem. 1958;231(1):241–57. [PubMed] [Google Scholar]
- 4.Weinberg RA, Penman S. Small molecular weight monodisperse nuclear RNA. J Mol Biol. 1968;38(3):289–304. doi: 10.1016/0022-2836(68)90387-2. [DOI] [PubMed] [Google Scholar]
- 5.Warner JR, et al. Rapidly labeled HeLa cell nuclear RNA. I. Identification by zone sedimentation of a heterogeneous fraction separate from ribosomal precursor RNA. J Mol Biol. 1966;19(2):349–61. doi: 10.1016/s0022-2836(66)80009-8. [DOI] [PubMed] [Google Scholar]
- 6.Holmes DS, et al. Chromosomal RNA: its properties. Science. 1972;177(4043):72–4. doi: 10.1126/science.177.4043.72. [DOI] [PubMed] [Google Scholar]
- 7.Robertson HD, et al. Purification and properties of a specific Escherichia coli ribonuclease which cleaves a tyrosine transfer ribonucleic acid presursor. J Biol Chem. 1972;247(16):5243–51. [PubMed] [Google Scholar]
- 8.Guerrier-Takada C, et al. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35(3 Pt 2):849–57. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 9.Gilbert W. Origin of life: The RNA world. Nature. 1986;319:618. [Google Scholar]
- 10.Brannan CI, et al. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10(1):28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brockdorff N, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71(3):515–26. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 12.Brown CJ, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527–42. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 13.Winkler WC, et al. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol. 2003;10(9):701–7. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 14.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris KV, et al. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305(5688):1289–92. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 16.Lecuyer E, et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131(1):174–87. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Wong HH, et al. RNA Docking and Local Translation Regulate Site-Specific Axon Remodeling In Vivo. Neuron. 2017;95(4):852–868. e8. doi: 10.1016/j.neuron.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott AW, et al. Public Attitudes About Eye and Vision Health. JAMA Ophthalmol. 2016;134(10):1111–1118. doi: 10.1001/jamaophthalmol.2016.2627. [DOI] [PubMed] [Google Scholar]
- 19.Lamb TD, et al. Evolution of the vertebrate eye: opsins, photoreceptors, retina and eye cup. Nat Rev Neurosci. 2007;8(12):960–76. doi: 10.1038/nrn2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoon M, et al. Functional architecture of the retina: development and disease. Prog Retin Eye Res. 2014;42:44–84. doi: 10.1016/j.preteyeres.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siegert S, et al. Transcriptional code and disease map for adult retinal cell types. Nat Neurosci. 2012;15(3):487–95. S1–2. doi: 10.1038/nn.3032. [DOI] [PubMed] [Google Scholar]
- 22.He J, et al. How variable clones build an invariant retina. Neuron. 2012;75(5):786–98. doi: 10.1016/j.neuron.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bassett EA, Wallace VA. Cell fate determination in the vertebrate retina. Trends Neurosci. 2012;35(9):565–73. doi: 10.1016/j.tins.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 24.Gregory-Evans CY, et al. Gene networks: dissecting pathways in retinal development and disease. Prog Retin Eye Res. 2013;33:40–66. doi: 10.1016/j.preteyeres.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Kim JW, et al. NRL-Regulated Transcriptome Dynamics of Developing Rod Photoreceptors. Cell Rep. 2016;17(9):2460–2473. doi: 10.1016/j.celrep.2016.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang SS, et al. A biphasic pattern of gene expression during mouse retina development. BMC Dev Biol. 2006;6:48. doi: 10.1186/1471-213X-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swaroop A, et al. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010;11(8):563–76. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aavani T, et al. Temporal profiling of photoreceptor lineage gene expression during murine retinal development. Gene Expr Patterns. 2017;23–24:32–44. doi: 10.1016/j.gep.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Wang S, et al. A gene regulatory network controls the binary fate decision of rod and bipolar cells in the vertebrate retina. Dev Cell. 2014;30(5):513–27. doi: 10.1016/j.devcel.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ueki Y, et al. A transient wave of BMP signaling in the retina is necessary for Muller glial differentiation. Development. 2015;142(3):533–43. doi: 10.1242/dev.118745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ueno K, et al. Analysis of Muller glia specific genes and their histone modification using Hes1-promoter driven EGFP expressing mouse. Sci Rep. 2017;7(1):3578. doi: 10.1038/s41598-017-03874-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan J, et al. Dynamic usage of alternative splicing exons during mouse retina development. Nucleic Acids Res. 2011;39(18):7920–30. doi: 10.1093/nar/gkr545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinelli M, et al. An atlas of gene expression and gene co-regulation in the human retina. Nucleic Acids Res. 2016;44(12):5773–84. doi: 10.1093/nar/gkw486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Popova EY, et al. Identification and prediction of alternative transcription start sites that generate rod photoreceptor-specific transcripts from ubiquitously expressed genes. PLoS One. 2017;12(6):e0179230. doi: 10.1371/journal.pone.0179230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li M, et al. RNA expression in human retina. Hum Mol Genet. 2017;26(R1):R68–R74. doi: 10.1093/hmg/ddx219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoshino A, et al. Molecular Anatomy of the Developing Human Retina. Dev Cell. 2017;43:763–779. doi: 10.1016/j.devcel.2017.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Consortium G. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farkas MH, et al. Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC Genomics. 2013;14:486. doi: 10.1186/1471-2164-14-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu SH. Gene expression regulation in photomorphogenesis from the perspective of the central dogma. Annu Rev Plant Biol. 2014;65:311–33. doi: 10.1146/annurev-arplant-050213-040337. [DOI] [PubMed] [Google Scholar]
- 40.Baralle FE, Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol. 2017;18(7):437–451. doi: 10.1038/nrm.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai G, et al. Alternative splicing governs cone cyclic nucleotide-gated (CNG) channel sensitivity to regulation by phosphoinositides. J Biol Chem. 2014;289(19):13680–90. doi: 10.1074/jbc.M114.562272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy D, et al. Alternative Splicing Shapes the Phenotype of a Mutation in BBS8 To Cause Nonsyndromic Retinitis Pigmentosa. Mol Cell Biol. 2015;35(10):1860–70. doi: 10.1128/MCB.00040-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao C, et al. Autosomal-dominant retinitis pigmentosa caused by a mutation in SNRNP200, a gene required for unwinding of U4/U6 snRNAs. Am J Hum Genet. 2009;85(5):617–27. doi: 10.1016/j.ajhg.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy D, et al. The Musashi 1 Controls the Splicing of Photoreceptor-Specific Exons in the Vertebrate Retina. PLoS Genet. 2016;12(8):e1006256. doi: 10.1371/journal.pgen.1006256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irimia M, et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 2014;159(7):1511–23. doi: 10.1016/j.cell.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li YI, et al. RBFOX and PTBP1 proteins regulate the alternative splicing of micro-exons in human brain transcripts. Genome Res. 2015;25(1):1–13. doi: 10.1101/gr.181990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dryja TP, et al. Mutations within the rhodopsin gene in patients with autosomal dominant retinitis pigmentosa. N Engl J Med. 1990;323(19):1302–7. doi: 10.1056/NEJM199011083231903. [DOI] [PubMed] [Google Scholar]
- 48.McKie AB, et al. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13) Hum Mol Genet. 2001;10(15):1555–62. doi: 10.1093/hmg/10.15.1555. [DOI] [PubMed] [Google Scholar]
- 49.Vithana EN, et al. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11) Mol Cell. 2001;8(2):375–81. doi: 10.1016/s1097-2765(01)00305-7. [DOI] [PubMed] [Google Scholar]
- 50.Wheway G, et al. An siRNA-based functional genomics screen for the identification of regulators of ciliogenesis and ciliopathy genes. Nat Cell Biol. 2015;17(8):1074–87. doi: 10.1038/ncb3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu MM, Zack DJ. Alternative splicing and retinal degeneration. Clin Genet. 2013;84(2):142–9. doi: 10.1111/cge.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bacchi N, et al. Splicing-correcting therapeutic approaches for retinal dystrophies: where endogenous gene regulation and specificity matter. Invest Ophthalmol Vis Sci. 2014;55(5):3285–94. doi: 10.1167/iovs.14-14544. [DOI] [PubMed] [Google Scholar]
- 53.Sangermano R, et al. ABCA4 midigenes reveal the full splice spectrum of all reported noncanonical splice site variants in Stargardt disease. Genome Res. 2017 doi: 10.1101/gr.226621.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yardley J, et al. Mutations of VMD2 splicing regulators cause nanophthalmos and autosomal dominant vitreoretinochoroidopathy (ADVIRC) Invest Ophthalmol Vis Sci. 2004;45(10):3683–9. doi: 10.1167/iovs.04-0550. [DOI] [PubMed] [Google Scholar]
- 55.Carter DA, et al. Mislocalisation of BEST1 in iPSC-derived retinal pigment epithelial cells from a family with autosomal dominant vitreoretinochoroidopathy (ADVIRC) Sci Rep. 2016;6:33792. doi: 10.1038/srep33792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zelinger L, et al. Regulation of Noncoding Transcriptome in Developing Photoreceptors by Rod Differentiation Factor NRL. Invest Ophthalmol Vis Sci. 2017;58(11):4422–4435. doi: 10.1167/iovs.17-21805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.La Torre A, et al. Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proc Natl Acad Sci U S A. 2013;110(26):E2362–70. doi: 10.1073/pnas.1301837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sundermeier TR, et al. DICER1 is essential for survival of postmitotic rod photoreceptor cells in mice. FASEB J. 2014;28(8):3780–91. doi: 10.1096/fj.14-254292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaneko H, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471(7338):325–30. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohana R, et al. MicroRNAs are essential for differentiation of the retinal pigmented epithelium and maturation of adjacent photoreceptors. Development. 2015;142(14):2487–98. doi: 10.1242/dev.121533. [DOI] [PubMed] [Google Scholar]
- 62.Karali M, et al. High-resolution analysis of the human retina miRNome reveals isomiR variations and novel microRNAs. Nucleic Acids Res. 2016;44(4):1525–40. doi: 10.1093/nar/gkw039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hughes AE, et al. Mutation altering the miR-184 seed region causes familial keratoconus with cataract. Am J Hum Genet. 2011;89(5):628–33. doi: 10.1016/j.ajhg.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conte I, et al. MiR-204 is responsible for inherited retinal dystrophy associated with ocular coloboma. Proc Natl Acad Sci U S A. 2015;112(25):E3236–45. doi: 10.1073/pnas.1401464112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan L, et al. Single and Compound Knock-outs of MicroRNA (miRNA)-155 and Its Angiogenic Gene Target CCN1 in Mice Alter Vascular and Neovascular Growth in the Retina via Resident Microglia. J Biol Chem. 2015;290(38):23264–81. doi: 10.1074/jbc.M115.646950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lumayag S, et al. Inactivation of the microRNA-183/96/182 cluster results in syndromic retinal degeneration. Proc Natl Acad Sci U S A. 2013;110(6):E507–16. doi: 10.1073/pnas.1212655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krol J, et al. A network comprising short and long noncoding RNAs and RNA helicase controls mouse retina architecture. Nat Commun. 2015;6:7305. doi: 10.1038/ncomms8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fan J, et al. Maturation arrest in early postnatal sensory receptors by deletion of the miR-183/96/182 cluster in mouse. Proc Natl Acad Sci U S A. 2017;114(21):E4271–E4280. doi: 10.1073/pnas.1619442114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krol J, et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141(4):618–31. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 70.Busskamp V, et al. miRNAs 182 and 183 are necessary to maintain adult cone photoreceptor outer segments and visual function. Neuron. 2014;83(3):586–600. doi: 10.1016/j.neuron.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 71.Briggs JA, et al. Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution. Neuron. 2015;88(5):861–77. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- 72.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rapicavoli NA, et al. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC developmental biology. 2010;10:49. doi: 10.1186/1471-213X-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Young TL, et al. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol. 2005;15(6):501–12. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 75.Yan B, et al. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res. 2015;116(7):1143–56. doi: 10.1161/CIRCRESAHA.116.305510. [DOI] [PubMed] [Google Scholar]
- 76.Liu C, et al. RNCR3 knockdown inhibits diabetes mellitus-induced retinal reactive gliosis. Biochem Biophys Res Commun. 2016;479(2):198–203. doi: 10.1016/j.bbrc.2016.09.032. [DOI] [PubMed] [Google Scholar]
- 77.Mustafi D, et al. Photoreceptor phagocytosis is mediated by phosphoinositide signaling. FASEB J. 2013;27(11):4585–95. doi: 10.1096/fj.13-237537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alfano G, et al. Natural antisense transcripts associated with genes involved in eye development. Hum Mol Genet. 2005;14(7):913–23. doi: 10.1093/hmg/ddi084. [DOI] [PubMed] [Google Scholar]
- 79.Rapicavoli NA, et al. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural development. 2011;6:32. doi: 10.1186/1749-8104-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meola N, et al. The long noncoding RNA Vax2os1 controls the cell cycle progression of photoreceptor progenitors in the mouse retina. RNA. 2012;18(1):111–23. doi: 10.1261/rna.029454.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Michaelides M, et al. Macular dystrophy associated with the A3243G mitochondrial DNA mutation. Distinct retinal and associated features, disease variability, and characterization of asymptomatic family members. Arch Ophthalmol. 2008;126(3):320–8. doi: 10.1001/archopht.126.3.320. [DOI] [PubMed] [Google Scholar]
- 82.Charif M, et al. Optic neuropathy, cardiomyopathy, cognitive disability in patients with a homozygous mutation in the nuclear MTO1 and a mitochondrial MT-TF variant. Am J Med Genet A. 2015;167A(10):2366–74. doi: 10.1002/ajmg.a.37188. [DOI] [PubMed] [Google Scholar]
- 83.Mansergh FC, et al. Retinitis pigmentosa and progressive sensorineural hearing loss caused by a C12258A mutation in the mitochondrial MTTS2 gene. Am J Hum Genet. 1999;64(4):971–85. doi: 10.1086/302344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Crimi M, et al. A mitochondrial tRNA(His) gene mutation causing pigmentary retinopathy and neurosensorial deafness. Neurology. 2003;60(7):1200–3. doi: 10.1212/01.wnl.0000055865.30580.39. [DOI] [PubMed] [Google Scholar]
- 85.Puffenberger EG, et al. Genetic mapping and exome sequencing identify variants associated with five novel diseases. PLoS One. 2012;7(1):e28936. doi: 10.1371/journal.pone.0028936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.DeLuca AP, et al. Hypomorphic mutations in TRNT1 cause retinitis pigmentosa with erythrocytic microcytosis. Hum Mol Genet. 2016;25(1):44–56. doi: 10.1093/hmg/ddv446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stephenson ML, Zamecnik PC. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc Natl Acad Sci U S A. 1978;75(1):285–8. doi: 10.1073/pnas.75.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Joglekar MV, et al. Circulating microRNA Biomarkers of Diabetic Retinopathy. Diabetes. 2016;65(1):22–4. doi: 10.2337/dbi15-0028. [DOI] [PubMed] [Google Scholar]
- 89.Berber P, et al. An Eye on Age-Related Macular Degeneration: The Role of MicroRNAs in Disease Pathology. Mol Diagn Ther. 2017;21(1):31–43. doi: 10.1007/s40291-016-0234-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Villegas-Ruiz V, et al. Genome-wide mRNA analysis reveals a TUBD1 isoform profile as a potential biomarker for diabetic retinopathy development. Exp Eye Res. 2017;155:99–106. doi: 10.1016/j.exer.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 91.Pelletier R, et al. RNA based gene therapy for dominantly inherited diseases. Curr Gene Ther. 2006;6(1):131–46. doi: 10.2174/156652306775515592. [DOI] [PubMed] [Google Scholar]
- 92.Petersen CP, et al. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21(4):533–42. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 93.Amadio M, et al. Targeting VEGF in eye neovascularization: What’s new?: A comprehensive review on current therapies and oligonucleotide-based interventions under development. Pharmacol Res. 2016;103:253–69. doi: 10.1016/j.phrs.2015.11.027. [DOI] [PubMed] [Google Scholar]
- 94.Millington-Ward S, et al. Suppression and replacement gene therapy for autosomal dominant disease in a murine model of dominant retinitis pigmentosa. Mol Ther. 2011;19(4):642–9. doi: 10.1038/mt.2010.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.O’Reilly M, et al. RNA interference-mediated suppression and replacement of human rhodopsin in vivo. American journal of human genetics. 2007;81(1):127–35. doi: 10.1086/519025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Phylactou LA, et al. Ribozymes as therapeutic tools for genetic disease. Hum Mol Genet. 1998;7(10):1649–53. doi: 10.1093/hmg/7.10.1649. [DOI] [PubMed] [Google Scholar]
- 97.Lewin AS, et al. Ribozyme rescue of photoreceptor cells in a transgenic rat model of autosomal dominant retinitis pigmentosa. Nat Med. 1998;4(8):967–71. doi: 10.1038/nm0898-967. [DOI] [PubMed] [Google Scholar]
- 98.Slijkerman RW, et al. Antisense Oligonucleotide-based Splice Correction for USH2A-associated Retinal Degeneration Caused by a Frequent Deep-intronic Mutation. Mol Ther Nucleic Acids. 2016;5(10):e381. doi: 10.1038/mtna.2016.89. [DOI] [PubMed] [Google Scholar]
- 99.Garanto A, et al. In vitro and in vivo rescue of aberrant splicing in CEP290-associated LCA by antisense oligonucleotide delivery. Hum Mol Genet. 2016;25(12):2552–2563. doi: 10.1093/hmg/ddw118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bonifert T, et al. Antisense Oligonucleotide Mediated Splice Correction of a Deep Intronic Mutation in OPA1. Mol Ther Nucleic Acids. 2016;5(11):e390. doi: 10.1038/mtna.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Roundtree IA, et al. Dynamic RNA Modifications in Gene Expression Regulation. Cell. 2017;169(7):1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Helm M, Motorin Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat Rev Genet. 2017;18(5):275–291. doi: 10.1038/nrg.2016.169. [DOI] [PubMed] [Google Scholar]
- 103.Roignant JY, Soller M. m6A in mRNA: An Ancient Mechanism for Fine-Tuning Gene Expression. Trends Genet. 2017;33(6):380–390. doi: 10.1016/j.tig.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 104.Zhao BS, et al. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18(1):31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu N, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518(7540):560–4. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fustin JM, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155(4):793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 108.Hu W, et al. Dynamic landscape of alternative polyadenylation during retinal development. Cell Mol Life Sci. 2017;74(9):1721–1739. doi: 10.1007/s00018-016-2429-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nelson JW, Breaker RR. The lost language of the RNA World. Sci Signal. 2017;10(483) doi: 10.1126/scisignal.aam8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Smalheiser NR. Exosomal transfer of proteins and RNAs at synapses in the nervous system. Biol Direct. 2007;2:35. doi: 10.1186/1745-6150-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bellingham SA, et al. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012;40(21):10937–49. doi: 10.1093/nar/gks832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guo L, Shorter J. It’s Raining Liquids: RNA Tunes Viscoelasticity and Dynamics of Membraneless Organelles. Mol Cell. 2015;60(2):189–92. doi: 10.1016/j.molcel.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fathinajafabadi A, et al. CERKL, a retinal disease gene, encodes an mRNA-binding protein that localizes in compact and untranslated mRNPs associated with microtubules. PLoS One. 2014;9(2):e87898. doi: 10.1371/journal.pone.0087898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kwong JM, et al. RNA binding protein with multiple splicing: a new marker for retinal ganglion cells. Invest Ophthalmol Vis Sci. 2010;51(2):1052–8. doi: 10.1167/iovs.09-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Castello A, et al. Metabolic Enzymes Enjoying New Partnerships as RNA-Binding Proteins. Trends Endocrinol Metab. 2015;26(12):746–57. doi: 10.1016/j.tem.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jin M, et al. Glycolytic Enzymes Coalesce in G Bodies under Hypoxic Stress. Cell Rep. 2017;20(4):895–908. doi: 10.1016/j.celrep.2017.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hurley JB, et al. Glucose, lactate, and shuttling of metabolites in vertebrate retinas. J Neurosci Res. 2015;93(7):1079–92. doi: 10.1002/jnr.23583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chinchore Y, et al. Glycolytic reliance promotes anabolism in photoreceptors. Elife. 2017:6. doi: 10.7554/eLife.25946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kooragayala K, et al. Quantification of Oxygen Consumption in Retina Ex Vivo Demonstrates Limited Reserve Capacity of Photoreceptor Mitochondria. Invest Ophthalmol Vis Sci. 2015;56(13):8428–36. doi: 10.1167/iovs.15-17901. [DOI] [PMC free article] [PubMed] [Google Scholar]