Abstract
Mycophenolic acid (MPA) is an approved immunosuppressive agent widely prescribed to prevent rejection after kidney transplantation. Wide between-subject variability (BSV) in MPA exposure exists which in part may be due to variability in enterohepatic recirculation (EHC). Several modeling strategies were developed to evaluate EHC as part of MPA pharmacokinetics, however mechanistic representation of EHC is limited. These models have not provided a satisfactory representation of the physiology of EHC in their modeling assumptions. The aim of this study was i) to develop an integrated model of MPA (total and unbound) and its metabolites (MPAG and Acyl-MPAG) in kidney recipients, where this model provides a more physiological representation of EHC process, and ii) to evaluate the effect of donor and recipient clinical covariates and genotypes on MPA disposition. A five-compartment model with first-order input into an unbound MPA compartment connected to the MPAG, acyl-MPAG, and gallbladder compartment best fit the data. To represent the EHC process, the model was built based on the physiological concepts related to the hepatobiliary system and the gallbladder filling and emptying processes. The effect of cyclosporine versus tacrolimus on clearance of unbound MPA was included in the base model. Covariate analysis showed creatinine clearance to be significant on oral clearance of unbound MPA. The hepatic nuclear factor 1 alpha (HNF1A) genetic single nucleotide polymorphism (SNP) (rs2393791) in the recipient significantly affected the fraction of enterohepatically-circulated drug. Oral clearance of MPAG was affected by recipient IMPDH1 SNP (rs2288553), diabetes at the time of transplant, and donor sex.
Keywords: Mycophenolic acid, kidney transplant, pharmacokinetics, NONMEM, enterohepatic circulation
Introduction
Mycophenolate mofetil (MMF) is an ester pro-drug of mycophenolic acid (MPA) used as maintenance immunosuppressant to prevent allograft rejection after kidney transplantation 1. The pharmacokinetic process of MPA has been reported to be complex and to be associated with large inter- and intra-individual variability 2. On oral administration, MMF is rapidly and extensively metabolized to MPA via intestinal and liver esterases CES1 and CES2 3. Mycophenolic acid binding to blood cellular components is negligible (<0.01%); however, it is highly bound to serum albumin (∼ 98%) 4,5. Clearance of MPA occurs almost completely through metabolism with only ∼0.6% excreted unchanged in urine 4. The metabolism occurs primarily via the uridine 5′-diphosphate glucuronosyltransferase (UGTs) enzymes 1A9 producing the major inactive metabolite 7-0-MPA-β-glucuronide (MPAG) and to a lesser extent through UGT2B7 producing a minor metabolite, acyl-MPAG, which has comparable pharmacological potency to MPA 6-10. Both metabolites are readily excreted into the urine by glomerular filtration and active tubular secretion 11. MPA undergoes extensive enterohepatic recirculation (EHC) through biliary excretion of MPAG, followed by intestinal deglucuronidation by microflora and then reabsorption as MPA 4,12. The EHC process contributes to around 40% of the MPA area under the curve (AUC) and results in the appearance of one or more secondary peaks in the concentration-time profile of MPA 13,14.
In kidney transplant, associations of MPA pharmacokinetics with acute rejection have been conflicting and may be in part due to differing and inaccurate methods used to estimate MPA exposure 15,16.The presence of EHC likely results in a biased estimation of AUC, and therefore this may explain why some studies have been unable establish exposure response relationships 17. Therefore, an understanding of the EHC process is important when evaluating the pharmacokinetics and pharmacodynamics of MPA.
To address the complexity of EHC, modeling strategies have been developed to evaluate the EHC as part of MPA pharmacokinetics. However, these models do not provide a complete representation of the physiology of EHC in their modeling assumptions. The objectives of the study were to: 1) develop an integrated semi-physiologic model of MPA (total and unbound) and its metabolites in patients with kidney transplant; and 2) to evaluate transplant recipient and donor clinical characteristics and genetic SNPs as potential sources of variability in the MPA pharmacokinetics. Accurate models for MPA pharmacokinetics will improve our ability to predict systemic exposure and study the association between pharmacokinetics and clinical outcomes such as rejection and toxicity.
Methods
Subjects and Study Design
Ninety-two subjects were enrolled in the Deterioration of Kidney Allograft Function (DeKAF) Genomics study and intensive pharmacokinetic sampling with mycophenolate was conducted over one steady state dosing interval in these subjects. DeKAF Genomics is a multicenter observational trial aimed to evaluate the effect of individual genetic variability on clinical outcomes of kidney transplantation. Institutional Review Board approval was obtained at each participating center and all patients provided informed, written consents prior to enrollment. The study is registered at http://www.clinicaltrials.gov (NCT00270712).
Subjects were included if they met the following inclusion criteria: i) adults, ≥18 years, who had undergone kidney or kidney-pancreas transplantation and receiving MMF as a maintenance immunosuppression, ii) functioning kidney graft at the time of the pharmacokinetics visit with an estimated glomerular filtration rate (GFR) >50 ml/min/1.73m2 within two weeks prior to the pharmacokinetics visit. Patients were excluded if they simultaneously received another organ (other than the pancreas) with the qualifying kidney transplant, had post-transplant active gastroparesis or liver disease.
Subjects received oral MMF (doses of 500-1500 mg/day) divided into 2 equal doses administered every 12 hours. Supervised pharmacokinetic samples were obtained pre-dose and at 1, 2, 4, 6, 8, and 12 hours following the oral MMF dose. Sampling was obtained after maintaining the same MMF dosing regimen for at least 48 hours to assure steady state.
Bioanalysis of Mycophenolate
Blood samples were collected and processed within 30-60 minutes. Unbound and total MPA, MPAG and acyl-MPAG assay was based upon modifications to Streit et al. 18. Detection and quantification of unbound MPA, total MPA, MPAG and AcMPAG in plasma was performed using a high-performance liquid chromatography (Agilent 1200 Series, Santa Clara CA) coupled with a TSQ Quantum triple stage quadrupole mass spectrometer (Thermo-Electron, San Jose, CA). The chromatographic separation was performed with a Thermo BetaBasic C4 (2.1 × 100 mm), reversed phase column with a 3.0-micron particle size. The mobile phase used for gradient elution consisted of (A) 10 mM ammonium acetate in water, pH 3.0 (B) methanol. For unbound MPA, the concentrations were measured after membrane ultrafiltration. Plasma (1 mL) was centrifuged through a 30,000 MW microfiltration device (Centrifree; Amicon; Millipore, Milford, Mass) at 37°C for 1 hour at 2000 × g, with a fixed-angle centrifuge (Jouan, Winchester, Va). The mobile phase used was a mixture (by volume) of 10 mM ammonium acetate in water, pH 3.0 (40%) and methanol (60%) with a flow rate of 0.25 ml/min and total run time of 5 min. However, for total MPA, acyl-MPAG, and MPAG, the chromatographic conditions were isocratic from 0 to 1.76 min at 37% methanol followed by a linear gradient from 1.37 to 6.2 min of 37% - 61% methanol and returning to starting conditions (at 6.5 min) with a flow rate of 0.25 mL/min, for a total run time of 10 minutes. The column temperature was maintained at 35°C for the 4 analytes. For unbound MPA, the detector settings of the TSQ Quantum were: ESI with the stainless steel spray needle, positive polarity ionization, selective reaction monitoring mode (SRM); spray voltage, 4500 V; capillary temperature, 400 °C; argon collision gas pressure, 1.5 mTorr; unit resolution for Q1 and Q3, 0.7 u (FWHM); and ions detected (m/z), MPA precursor 338, product 207 and MPAC precursor 438, product 207. The collision energy for MPA and MPAG was 35eV. Following the addition of internal standard (10 ng of MPAG) to 0.25 ml of protein free plasma an aliquot (5 uL) was directly injected onto the LC-MS/MS for analysis. Whereas for total MPA, MPAG, and acyl-MPAG, the detector settings of the TSQ Quantum were: ESI with the stainless steel spray needle, positive polarity ionization, selective reaction monitoring mode (SRM); spray voltage, 4500 V; capillary temperature, 400 °C; argon collision gas pressure, 1.5 mTorr; unit resolution for Q1 and Q3, 0.7 u (FWHM); and ions detected (m/z), MPA precursor 338, product 207, MPAG and acyl-MPAG precursor 514, product 207, MPA-d3 precursor 341, product 210, MPAG-d3 and acyl-MPAG-d3 precursor 517, product 210. The collision energy for MPAG, acyl-MPAG was 34eV and MPA was set at 25 eV. Following the addition of internal standard (200 ng of MPAG-d3, 100 ng of MPA-d3 and 100 ng of acyl-MPAG-d3) 0.2 ml of plasma was extracted with protein precipitation using perchloric acid and sodium tungstate. MPA, MPAG and acyl-MPAG were obtained from Toronto Research Chemicals (Toronto, Ontario, CAN). The assay was linear in the range of 1-500, 25-20000, 1000-250000, and 25-20000 ng/mL for unbound MPA, total MPA, MPAG and acyl-MPAG, respectively. The assay accuracy for unbound MPA, total MPA, MPAG, and acyl-MPAG were 98, 103,102,106%. The assay total variability were 6.3, 6.1, 2.2, 5.8% for unbound MPA, total MPA, MPAG, and acyl-MPAG, respectively.
DNA Collection and Genotyping
DNA was isolated from peripheral blood collected from the donor and recipient prior to transplantation. Lymphocytes were isolated by centrifugation after red blood cell lysis. Isolated DNA was quantified at 260 nm absorbance. Single nucleotide polymorphisms were genotyped using a customized Affymetrix GeneChip and additional SNPs were measured using SNPlex (Applied Biosystems Inc, Foster City, California, USA) and Sequenom (Sequenom, Inc, San Diego, CA, USA) systems as previously described 19. Candidate polymorphisms (n=88) measured in both the donor and recipient potentially relevant to mycophenolate metabolism, transport and mechanism of action were selected for this analysis (Table S1).
Population Pharmacokinetic Modeling
The population pharmacokinetic parameters of MPA (unbound and total), MPAG and Acyl-MPAG were modeled simultaneously using nonlinear mixed effect modelling (NONMEM, version 7.2, ICON Dev. Soln., Ellicott City, MD) with Pirana® (http://www.pirana-software.com). First-order conditional estimation with η, ε-interaction (FOCE-I) was utilized throughout the modeling process. Population model development of unbound MPA, total MPA, MPAG, and acyl-MPAG was conducted as a series of steps that led eventually to a simultaneous population pharmacokinetic model of all four analytes.
Modeling Unbound MPA
Oral one-compartment and two-compartment models were tested. Several absorption models were tested including first order, zero order, mixed first order and zero order with and without lag time, and transit compartment model.
Inclusion of Metabolites
Unbound MPA was assumed to be eliminated only through metabolism. This metabolism results into two metabolites: MPAG and Acyl-MPAG. The fraction of unbound MPA metabolized to MPAG and Acyl-MPAG was fixed to literature values of 0.87, and 0.13, respectively 14. A one-compartment model with first order elimination was assumed for each metabolite.
Inclusion of the EHC Process
Evaluation of the EHC process of MPAG was modeled assuming a gallbladder-based EHC model. This model considers the physiological concepts related to the hepatobiliary system and the gallbladder filling and emptying processes. In the model, a gallbladder compartment was connected to MPAG and gut compartments. The extent of EHC (EHC%), which represents the percentage of the amount of MPAG that gets distributed via EHC, was described according to the following equation:
where kDG is the rate constant that describes the EHC distribution process out of the MPAG compartment, and k30 is MPAG elimination rate constant.
A continuous leak of a percentage of EHC% from the MPAG compartment to the gut was included where 75% of the EHC% was assumed to go to the gallbladder, while the remaining 25% drains directly from MPAG compartment to the gut compartment 20,21. The gallbladder emptying process was assumed to be triggered by meals, where various numbers of meals were evaluated, i.e., one meal, two meals, and three meals. Additionally, the timing of meals that stimulate the gallbladder contraction (mealtimes [mt]) was estimated. At each mealtime (mt), approximately 75% of the drug stored in the gallbladder is discharged to the gut within a duration (D) of 30 minutes 20,22-24. The transfer rate from the gallbladder to the gut was determined by the rate constant kGG according to the following equation:
where k31 is a rate constant and switch is a function that accounts for multiple cycles of gallbladder emptying. k31 was fixed to 2.77 hr-1 based on a simulation analysis which showed that this value results in eliminating 75% of the drug from the gallbladder, within the duration of 30 minutes. The switch function was defined according to the following equations:
where t is time after dose; mt1 represents the beginning of gallbladder emptying (occurs at the same time of mealtime); and Endmt1 represents the end of gallbladder emptying (and calculated as Endmt1 = mt1 + D, where D was fixed to 30 minutes.
The Switch function applies a double sigmoid function to regulate the gallbladder release rate constant (kGG). The equation Switch allows a rapid change in the kGG rate constant from zero to almost 2.77 around the time mt1. After approximately 30 minutes, Switch causes a rapid decline in kGG back to zero around time Endmt1. This rapid change of kGG was achieved by using a value of 20 in Hill1 and Hill2 in an attempt to mimic the toggle nature of gallbladder emptying. To represent the multiple cycles of gallbladder emptying, this set of equations was reapplied at each mealtime.
Inclusion of Total MPA
Total MPA concentrations were modeled assuming linear binding over the observed total MPA concentration according to the following:
where MPAtotal is the total MPA concentration, MPAunbound is the unbound MPA concentration, fu is the MPA fraction unbound that was estimated in the model. Nonlinear binding model was also evaluated.
Covariate Model Building
The concomitant calcineurin inhibitor (CNI; cyclosporine vs. tacrolimus) therapy (Cdrug) was included as part of the base model because previous reports demonstrated clinically significant differences on MPA pharmacokinetics 25. The effect of Cdrug was modeled on the unbound MPA clearance parameter as the following equation:
where TVCLu is the typical value of unbound MPA clearance; Cdrug equals 1 when the concomitant CNI therapy is cyclosporine, and 0 when it is tacrolimus; θ1 represents TVCLu when Cdrug is tacrolimus, and (1 + θ2) represents the fractional change in TVCLu when receiving concomitant cyclosporine rather than tacrolimus.
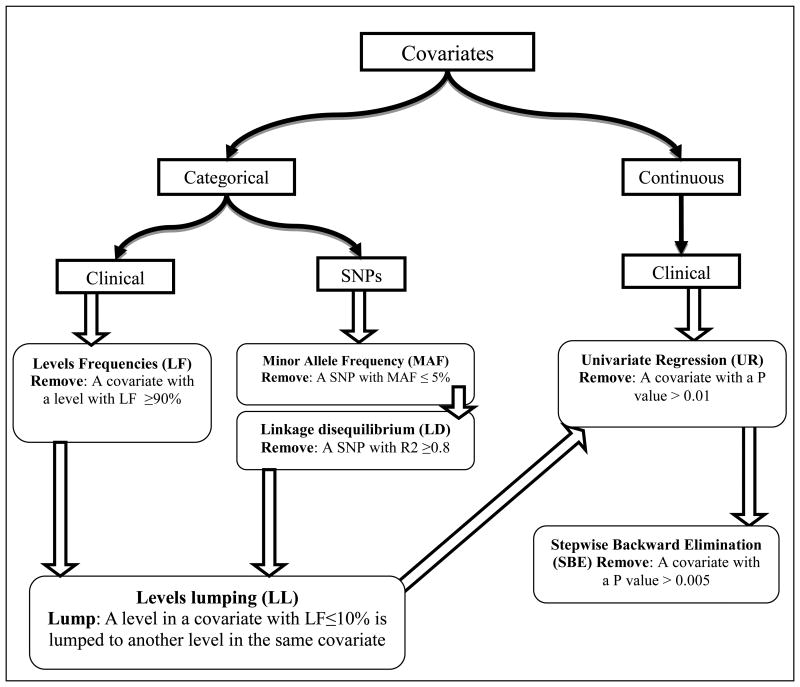
A total of 25 clinical characteristics (Table 1) and 88 SNPs (same SNPs measured in recipients and donors) (Table S1) were evaluated as covariates. As the covariate dataset was large, several data reduction steps were conducted according to Figure 1. A categorical clinical covariate was not considered for further analyses if one of its groups contained ≥ 90% of the sample patients. The minor allele frequency (MAF) was calculated for each SNP and those with a MAF ≤ 5% was excluded from any further analysis. This cutoff value was selected as it is generally used in the genetics studies literature 26. Remaining SNPs underwent linkage disequilibrium (LD) analysis where a pair of SNPs was considered highly linked when R2 ≥0.8. For high LD pairs, the decision of which SNP to exclude was based on the type and degree of importance of the SNP, i.e., a non-synonymous SNP is more important than a synonymous SNP, and a synonymous SNP is considered to be more important than an intronic SNP. If the 2 highly linked SNPs are of the same importance, one SNP was randomly selected for exclusion from further analyses. Then, to minimize type 1 error, SNPs and categorical clinical covariates underwent a “levels lumping” step, in which 2 covariate levels are lumped together (collapsed). In more details, a covariate level that contained ≤10% of the sample patients was considered infrequent and was lumped to another level. The choice of the level to be lumped to was based on the clinical and the physiological definitions of the levels. For example, if the infrequent level (≤10 of the patients) in a SNP was the “homozygous for the minor allele”, that level was lumped with the “heterozygous” level rather than the wild type.
Table 1. Patients' Characteristics and Clinical Covariates.
| Characteristic | Value |
|---|---|
|
| |
| Recipient Sex (Male/Female) | 62/27 |
| Recipient Age (Years)1 | 51 (12) |
| Recipient Race (White / Black /Asian) | 83/5/1 |
| Recipient Weight (Kg) 1 | 82.3 (17) |
| Recipient BMI* at Transplant1 | 28 (5.7) |
| Recipient Height (Cm) 1 | 173 (8.3) |
| Time to Pharmacokinetic Analysis# (Days) 1 | 36 (15) |
| Preemptive Transplantation + (Yes/No) | 42/47 |
| Previous Antibody Induction (IL2/ Monoclonal/Polyclonal) | 32/19/38 |
| Diabetes at Time of Transplant (Yes/No) | 26/63 |
| Number Of HLA† Mismatch (1/2/3/4/5/Other) | 9/12/27/9/18/13 |
| Prior Kidney Transplant ( No/ 1 Prior transplant) | 73/16 |
| Primary Cause of Kidney Disease (Glomerular disease/Diabetes/Hypertension/Polycystic kidney disease/Other/Unknown) | 21/19/6/21/17/5 |
| Serum Creatinine (mg/dL) 1 | 1.2 (0.3) |
| Serum Albumin (g/dL) 1,2 | 3.94 (0.5) |
| Serum Bilirubin (mg/dL) | 0.5 (0.3) |
| Serum Alanine Aminotransferase (IU/L) 1 | 27 (19.4) |
| Serum Alkaine Phosphatase (IU/L) 1 | 93.6 (95.8) |
| Creatinine Clearance (ml/min)1 | 82.8 (24.9) |
| Concomitant Calcineurin Inhibitor (Tacrolimus/ Cyclosporine) | 58/31 |
| Concomitant Steriod (Yes/No) | 57/32 |
| Concomitant Proton Pump Inhibitor (Yes/No) | 66/23 |
| Donor Status ( Living/ Deceased) | 88/1 |
| Donor Sex (Male/Female) | 52/37 |
| Donor Age1 | 42 (12) |
| Donor Race (White/ Black) | 28/2 |
Reported values for these characteristics are Mean (Standard deviation)
Reported value based on 91 subjects
Body Mass Index
Preemptive kidney transplantation is defined as transplantation before initiating any dialysis treatment
Refers to the duration of time between initiating MMF dosing and the pharmacokinetic visit
HLA: Human Leukocyte Antigen
Figure 1.
A schematic description of the covariate data reduction and analysis methods used in the study.
A univariate regression analysis was performed on the remaining covariates to identify a set of significant covariates. The univariate regression analysis was performed against the Empirical Bayes Estimates (EBEs) of the following important fixed effect parameters from the base model: unbound MPA oral clearance (CL/F), MPAG Clearance (CLm1), and EHC percent (EHC%). A linear regression model was used for each covariate parameter pair, where p-value ≤0.01 was considered significant. Covariates that had insignificant p-values were removed from further analysis.
The significant covariates from the previous step were further investigated using stepwise backward elimination method within the context of NONMEM using the SCM module in Perl Speaks NONMEM (PsN). Covariates were removed when its exclusion resulted in the smallest insignificant increase in the OFV [OFV change < 7.87 (chi- square test, p=0.005, df=1)]. This process is repeated until all remaining covariates are significant.
In the SCM analysis, continuous and categorical covariates were evaluated using different models. For categorical covariates, a linear proportional covariate model was used as the following:
where TVP is the typical value of the fixed effect parameter; θ1 is the population estimate of the structural parameter when the covariate is the reference level; (1 + θ2) represents the population fractional change in TVP when the covariate is a non-reference level; COV is the categorical covariate value.
Whereas, continuous covariates were evaluated in the SCM analysis using linear model as the following:
Where TVP is the typical value of the fixed effect parameter; θ1 is the population estimate of the structural parameter when the covariate has the median value; θ2 is the population proportional estimate of the covariate fixed effect; COV is the continuous covariate value; COVmedian is the median of the COV in the study sample.
Error Model
Modeling the between-subject variability (BSV) in the structural parameters was included using an exponential model as shown in the following equation:
where TVP is the typical value of the parameter in the population and ηj is the difference between the jth individual parameter value (Pj) and TVP on a lognormal scale. It was assumed that ηj values are independent and identically normally distributed with a mean of zero and a variance ω2, i.e. η ∼ N(0, ω2).
Proportional error models were used to characterize the residual unexplained variability (RUV) in the model, as the following:
where Cij is the observed jth plasma concentration in the ith individual and Cpred,ij is the predicted jth plasma concentration in the ith individual. Additionally, εij represents the proportional discrepancy between Cij and Cpred,ij. In these models, it is assumed that εij are normally distributed with a mean of zero and a variance σ2; i.e. ε ∼ N(0, σ2).
Model Qualification
Pharmacokinetic models were evaluated based on the objective function values, the stability of the model, and goodness of fit plots. For nested models, a decrease of more than 7.87 in the objective function value (chi-square; p <0.005; df=1) was considered significant. Additionally, biological plausibility and clinical importance of a statistically significant difference was considered. The final model was evaluated using visual predictive checks (VPC; 1000 simulations). Diagnostic plots were generated using Xpose4 package in R via Rstudio 27-29.
Results
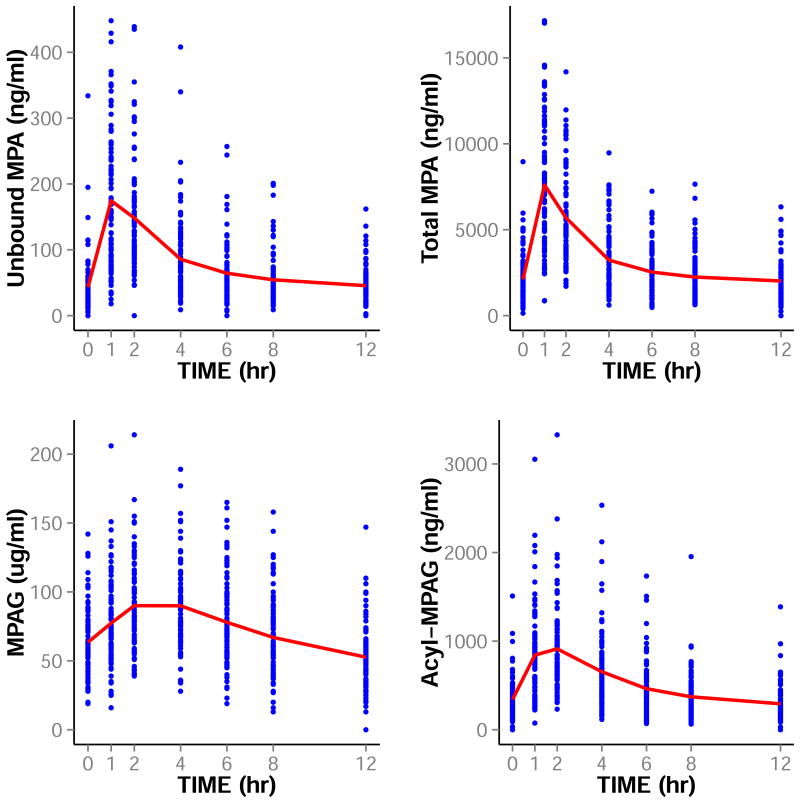
Six hundred and forty four concentration of each of unbound MPA, total MPA, MPAG, and acyl-MPAG concentrations from 92 kidney or kidney-pancreas transplant recipients were analyzed. Patient demographics are included in Table 1. The concentrations-time profiles of unbound MPA, total MPA, MPAG, and acyl-MPAG are shown in Figure 2.
Figure 2.
The Concentration-Time Profile of Unbound MPA, Total MPA, MPAG, and Acyl-MPAG. Points are the observed concentrations and lines are the means of these concentrations at each sampling time.
Base Model
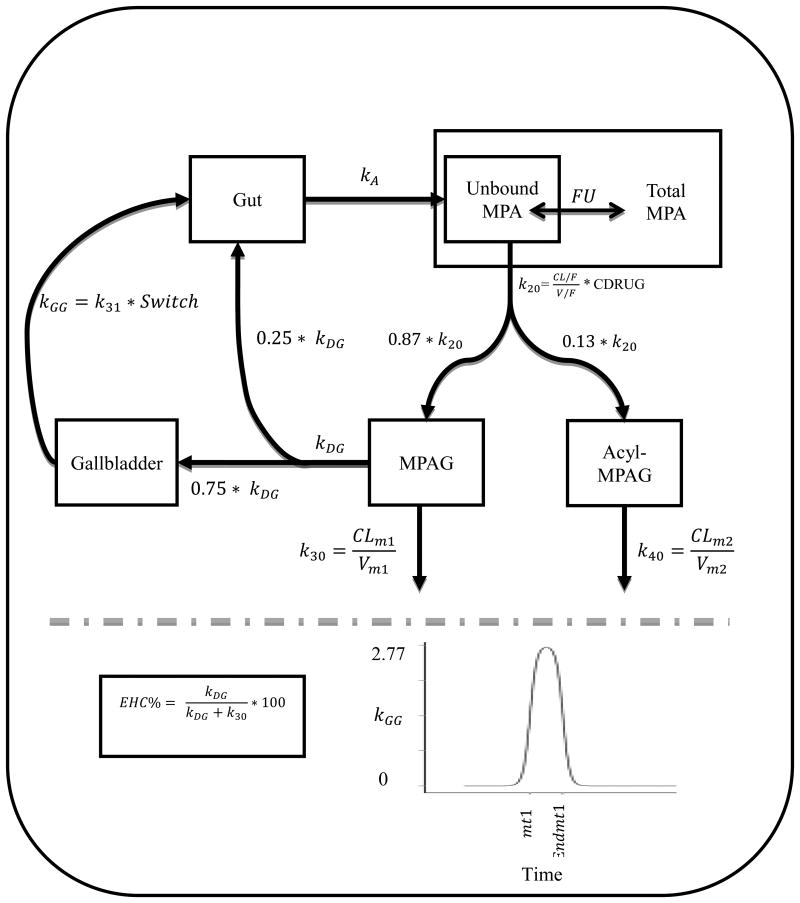
A schematic representation of the final pharmacokinetic model is presented in Figure 3. A five-compartment model with first-order absorption of unbound MPA, MPAG, acyl-MPAG, with an additional gallbladder compartments best fit the data. The EHC process was represented using the additional gallbladder compartment that forms a loop between MPAG and gut compartments. Modeling 2 or 3 mealtimes did not improve the fit over one mealtime; therefore, parameters assuming one mealtime were estimated. A linear binding model describing the relation between unbound MPA and total MPA best fit the data. A summary of the model parameter estimates are listed in Table 2.
Figure 3.
Schematic Representation of the Model for unbound MPA, total MPA, MPAG and acyl-MPAG. kA : absorption rate constant; FU: fraction unbound; CL/F: apparent oral clearance of unbound MPA; V/F: apparent volume of distribution of unbound MPA; K20: unbound MPA elimination rate constant; CDRUG: concomitant CNI treatment; kDG: EHC distribution rate constant; kGG: gallbladder emptying rate constant; CLm1:MPAG oral clearance; Vm1:MPAG volume of distribution; k30: MPAG elimination rate constant; CLm2: Acyl-MPAG oral clearance; Vm2: Acyl-MPAG volume of distribution; k40: Acyl-MPAG elimination rate constant; mt1: represents a mealtime (beginning of gallbladder emptying); Endmt1: represents the end of gallbladder emptying. K31: gallbladder emptying rate constant that result in emptying around 75% of gallbladder content and its fixed to 2.77 hr-1. The definition of switch is provided in the text.
Table 2.
Parameter estimates of the final model of umnbound MPA, total MPA, MPAG and acyl-MPAG in kidney transplant recipeints.
| Parameter | Estimate (%RSE) | % Between Subject Variability (%RSE) | % η-shrinkage | Significant covariates |
|---|---|---|---|---|
| kA (hr-1) | 2 (18.4) | 108.6 (29.3) | 16.4 | - |
| CL/F (L/hr) | 1450 (6.5) | 30.1 (25.4) | 8.8 | CrCl |
| V/F (L) | 5630 (7.9) | 35.5 (33.6) | 23.2 | - |
| EHC% (fraction) | 0.37 (5.9) | 27.8 (29.4) | 11.8 | rs2393791 |
| Mealtime (mt1) (hr) | 7.7 (0.1) | - | - | - |
| CLm1 (L/hr) | 0.96 (4.8) | 27.2 (20.6) | 1.1 | Diabetes at Time of Transplant, Donor Sex, rs2288553 |
| Vm1 (L) | 5.7 (4.9) | - | - | - |
| CLm2 (L/hr) | 32.3 (7.6) | 46.3 (19.8) | 2.2 | - |
| Vm2 (L) | 17.9 (9.4) | - | - | - |
| FU (fraction) | 0.024 (5.2) | 24 (24.3) | 17.5 | - |
|
| ||||
| Covariates Effects | ||||
| The marginal effect of CDRUG (cyclosporine instead of tacrolimus) on CL/F | 0.15 (61.1) | - | - | - |
| The marginal effect of CrCl on CL/F | 0.008 (30.5) | - | - | - |
| The marginal effect of having diabetes at time of transplant (versus not) on CLm1 | -0.19 (31.6) | - | - | - |
| The marginal effect of donor sex (female instead of male) on CLm1 | -0.18 (34.4) | - | - | - |
| The marginal effect of the IMPDH1 SNP rs2288553* (Heterozygous instead of Wild Type) on CLm1 | 0.33 (37.0) | - | - | - |
| The effect of the HNF1A SNP rs2393791 (Heterozygous or Homozygous Minor instead of wild type) on EHC% | 0.16 (55.0) | - | - | - |
|
| ||||
| Proportional Residual Variability | - | |||
| Unbound MPA | 40.5 (9) | - | - | - |
| Total MPA | 35.8 (10.9) | - | - | - |
| MPAG | 12.2 (6) | - | - | - |
| Acyl- MPAG | 24.8 (7.8) | - | - | - |
Estimates are the population estimates; %RSE is percent relative standard error calculated as %100*(SE/estimate); MPA: mycophenolic acid; MPAG: MPA glucuronide; kA : absorption rate constant; FU: fraction unbound; CL/F: apparent oral clearance of unbound MPA; V/F: apparent volume of distribution of unbound MPA; K20: unbound MPA elimination rate constant; CDRUG: concomitant CNI treatment; kDG: EHC distribution rate constant; kGG: gallbladder emptying rate constant; CLm1:MPAG oral clearance; Vm1:MPAG volume of distribution; CLm2: Acyl-MPAG oral clearance; Vm2: Acyl-MPAG volume of distribution; mt1: represents a mealtime (beginning of gallbladder emptying); EHC% is the percentage of MPAG that undergoes enterohepatic cycling.
Covariate Model
Recipient race, donor race and donor status (deceased vs. living donor) included one group that contained at least 90% of the population and therefore, were excluded from further covariate analysis. SNP data reduction based on MAF resulted in excluding 41 out of the 176 SNPs. Subsequent LD analyses on SNPs showed high LD (R2≥0.8) between rs7662029 and rs7438135. These are intronic SNPs in the UGT2B7 gene and only rs7438135 was used in the analysis. No clinical categorical covariates needed lumping, while level lumping was done for 82 of the SNPs. In all these SNPs, heterozygotes and homozygotes for the minor allele were lumped to form one level.
Univariate regression analysis was conducted on remaining 133 SNPs and 21 clinical covariates. Stepwise backward elimination identified 5 significant covariates (Table 2). For unbound MPA CL/F, after adjusting for type of CNI, only CrCl was identified as a significant covariate. The equation below shows the relation between unbound MPA oral clearance (CL/F), concomitant CNI treatment (Cdrug) and CrCl:
For MPAG oral clearance, diabetes at the time of transplant and female sex resulted in 19% and 18% marginal reduction in the oral clearance of MPAG, respectively. Additionally, a recipient who had 1 or 2 IMPDH1 (rs2288553) variant alleles, showed a 33% increase in oral clearance of MPAG.
Recipient hepatic nuclear factor 1 alpha (HNF1A) SNP (rs2393791) was found to be significant covariate on EHC%. The EHC% of MPAG was 37.4% in patients carrying two wild type alleles whereas patients carrying one or two variant alleles had an EHC% of 43.4%. Covariate's effect estimates are listed in Table 2.
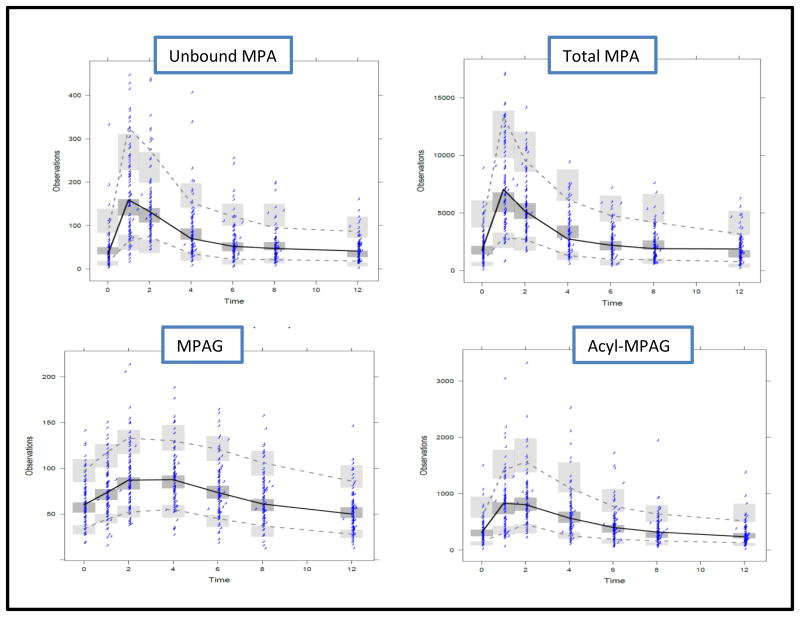
The final model diagnostic plots of unbound MPA, total MPA, MPAG, and acyl-MPAG are shown in Figures S1, S2, S3, S4, , respectively. Overall, the diagnostic plots suggest adequacy of the model. Visual predicative check (VPC) plots for unbound MPA, total MPA, MPAG, and acyl-MPAG are shown in Figure 4. The VPC plots show that 95% of the data largely falls within the 95% prediction intervals of the simulated data.
Figure 4.
Visual Predicative Check (VPC) Plots for Unbound MPA (upper left), Total MPA (upper right), MPAG (lower left), and Acyl-MPAG (lower right). Solid lines represent the medians of the observed data; dashed lines represent the 5th and 95th quantiles of the observed data; shaded areas represent the 95% prediction intervals for corresponding simulated data. Dots represent observed data.
Discussion
A population pharmacokinetic model that simultaneously describes the concentrations of unbound MPA, total MPA, MPAG, and acyl-MPAG following MMF administration in kidney transplant recipients was developed. The final population pharmacokinetic model describes the disposition of MPA and its metabolites, and characterizes the wide between subject variability by evaluating several recipients and donor related clinical characteristics and SNPs. Moreover, the model provides a physiological representation of the associated complex EHC. This is essential as EHC process accounts for around 40% of MPA AUC and, several EHC related factors add to the variability in the pharmacokinetic of MPA 4,11. To our knowledge this is the most comprehensive quantitative dose-exposure analysis of MMF that has evaluated a large number of clinical and genetic variables on it disposition in patients with kidney transplant.
Many population models have been developed to evaluate the pharmacokinetics of MPA 2,30-32; however, the majority of these studies utilize a conventional 2-compartment model without adequately representing the EHC process 33,34. This model most likely inadequately describes pharmacokinetics. Some studies have attempted to characterize the MPA EHC process using modeling strategies 35-37. However, these models do not provide a comprehensive physiological representation of the EHC parameterizations and assumptions which limit its applicability and ability to predict future data. Our model is advantageous as it incorporates the EHC process based on physiological aspects of the hepatobiliary system. Major parameterizations in the current EHC model are based on gallbladder cholescintigraphy studies, which showed that around 75% of bile enters the gallbladder after fasting 20,22-24. These studies also demonstrated that the gallbladder releases around 75% of its content after 30 minutes of cholecystokinin infusion. This physiological understanding was used in defining EHC processes including bile secretion, gallbladder filling and emptying, and duration of gallbladder emptying. The only study design-derived information used in our EHC model is the mealtimes. Therefore, the model is applicable to other MPA studies in kidney transplant recipients with similar EHC physiology. We speculate that a modification of the EHC model would also provide reasonable fit for other drugs that are impacted by EHC.
Our model incorporated both total and unbound MPA and estimated a 2.4% fraction unbound similar to what is reported in the literature 4. Although MPA has high protein binding and only the unbound MPA is thought to provide pharmacologic activity, studies suggest that both total MPA and unbound MPA are equally good predictors of the immunosuppressive response 5,38,39. In clinical practice though, total MPA is measured rather than unbound MPA concentrations. For these reasons, we attempted to characterize the relationship between unbound and total MPA using a linear binding model which produced a reasonable fit.
The inclusion of MPAG, although inactive, in the current model was done to allow the characterization of the EHC process of MPA as it primarily involves this metabolite 4. Acyl-MPAG is not substantially involved in the EHC process but has a pharmacological activity similar to its parent 6,7. Therefore, we also characterized the pharmacokinetics of this metabolite since it may have a role in immunosuppressive response.
Several clinical factors and genotypes influence the pharmacokinetics of MPA, and therefore inflate its between-subject variability 4,40. We attempted to reduce this variability by evaluating clinical covariates, concomitant drug therapy, and genotypes from donors and recipients. Given the large number of covariate and the long computational time for the model, it was extremely difficult to test all the covariates on all parameters. Parameters that were deemed clinically relevant for the maintenance dose adjustment were chosen. Although many studies have evaluated recipient genotypes, to our knowledge, this is the first population pharmacokinetic analysis of MPA that evaluates donor genotypes as potential covariates.
Mycophenolate in kidney transplant is used in conjunction with a calcineurin inhibitor (CNI) immunosuppressant (cyclosporine or tacrolimus). Cyclosporine has slowly been replaced by tacrolimus clinically and now accounts for over 90% of CNI use. It is well known that the type of CNI therapy differentially affects MPA disposition 11. For that reason, we incorporated the effect of concomitant CNI treatment in the base model as a covariate on the oral clearance (CL/F) of unbound MPA. Since all patients in our study received either tacrolimus or cyclosporine, we were only able to evaluate the effect of concomitant administration of cyclosporine as compared to concomitant administration of tacrolimus. Cyclosporine co-administration results in an estimated 15% increase in unbound MPA oral clearance compared to concomitant tacrolimus, which agrees with literature 41.
Creatinine clearance was found to be a significant covariate on the oral clearance (CL/F) of unbound MPA. Assuming mid-points of CrCL of 100, 85, 45, and 20 mL/min for normal, mild, moderate, and severe renal function, an estimated decrease in unbound MPA clearance by 10, 37, and 54% for mild, moderate, and severe renal impairment groups, respectively relative to the normal group. The same result was also observed in several other studies conducted on unbound MPA 41-43. This finding is interesting given the presumed limited effect of renal elimination on unbound MPA 4. However, there is a growing literature supporting the presence of an effect of renal impairment on the pharmacokinetics of some hepatically eliminated drugs 44.
Three covariates were found to be significant on MPAG clearance and they included diabetes at the time of transplant, donor sex, and recipient IMPDH1 SNP rs2288553. Controlling for the other 2 covariates, patients with diabetes at the time of transplantation demonstrated 19% lower MPAG clearance when compared to patients without diabetes. The estimated clearance of MPAG in our model is a function of both fraction formed from MPA probably through UGT1A9 and its systemic clearance. The systemic clearance occurs through excretion from kidney or through biliary excretion via ABCC2 transporter. We don't expect that the estimated reduction of clearance is occurring as a result of increasing formation of MPAG, as it has been shown that the expression and activity of UGT1A9 is not affected by diabetes 45. It has been shown that diabetic rats have decreased ABCC2 transporter 46. As such, we believe that the systemic clearance is truly reduced in diabetic patients as a result of reduced biliary excretion. Moreover, diabetes is known to reduce renal excretion as a result of nephropathy 47.
Our model estimated an 18% decrease in the oral clearance of MPAG when the kidney donor was female as compared to a male donor, while controlling for the other 2 covariates. This is physiologically possible since the male kidney is generally larger than females. Additionally, differences in the transporters activity between male and female kidneys have been demonstrated. For example, the expression of renal organic anion transporting polypeptide (OATP) had been demonstrated to be less in female rat kidney than in male rat kidney 48,49.
Our finding of the association between recipient IMPDH1 SNP rs2288553 and MPAG clearance is interesting but the biological mechanisms underlying the effect are unclear. Therefore this SNP will require validation in future studies.
Our model estimated 16.5% increase in EHC in recipients who are heterozygous or homozygous for the variant allele of rs2393791 on the HNF1A gene relative to wild type. This result is consistent with the proposed role of HNF1A being it is a transcription factor expressed in the liver, as well as the kidneys and is thought to regulate the expression of several genes including bile acid transporters 50,51.
Our study is complex and has some limitations. First, the use of a univariate regression analysis in the covariate modeling strategy could mask a hidden interaction between 2 covariates, and therefore does not allow capturing it. Second, in the univariate regression analysis, a linear relationship was imposed for evaluating the effect of covariates on the EBE. If the true association was nonlinear, the utilized univariate regression analysis may not be able to capture the covariate parameter association. Third, the use of model-based EBEs as the independent variable might introduce bias in identifying the significant covariates as it is known to be affected by model misspecification and shrinkage. Fourth, the authors acknowledge that Cmax is under-predicted by the proposed model. However, the clinical relevance of this under-prediction is believed to be minimal as several literature studies linked the AUC with efficacy and/or toxicity52-54. Fifth, the majority of subjects in this study were white, and several literature studies suggested differences between races that affects the enterohepatic circulation extent and therefore the PK of MPA55,56. Finally, the model results need to be externally validated in a second group of kidney transplant patients. Future studies should evaluate the effect of the gut microbiome on EHC of MPA.
Conclusion
The proposed model incorporates the physiological aspects associated with MPA disposition. The final population pharmacokinetic model included MPA (unbound and total) and its metabolites and adequately reflected the complex processes of EHC associated with MPA oral dosing. The model identified clinical and genetic covariates that explain part of MPA variability in this population. Incorporating individual specific covariates in this model, can improve prediction of the individual exposure for further development of individualized MPA dosing strategies in post-kidney transplant patients.
Supplementary Material
Acknowledgments
The authors wish to thank the research participants for their participation in this study. We acknowledge the dedication and hard work of our coordinators at each of the pharmacokinetic sites: University of Minnesota, Jill Nagorski and Mandi DeGrote; Hennepin County Medical Center, Lisa Berndt; Mayo Clinic, Tom DeLeeuw; University of Alabama, Jacquelin Vaughn and Tena Hilario. We also acknowledge the dedicated work of research scientist Marcia Brott. This study was supported in part by NIH/NIAID grants 5U19-AI070119 and 5U01-AI058013.
Funding: The study was supported by grants (U19-AI070119 and U01-AI058013) from the National Institute of Allergy and Infectious Disease (AM, PJ, AI, WO).
Footnotes
Indicate which authors, if any, are Fellows of the American College of Clinical Pharmacology (FCP): Richard C. Brundage, PharmD, PhD
Conflict of Interest: The authors have no conflicts of interest
References
- 1.Matas A, Smith J, Skeans M, et al. OPTN/SRTR 2013 annual data report: kidney. American Journal of Transplantation. 2015;15(S2):1–34. doi: 10.1111/ajt.13195. [DOI] [PubMed] [Google Scholar]
- 2.Sherwin CM, Fukuda T, Brunner HI, Goebel J, Vinks AA. The evolution of population pharmacokinetic models to describe the enterohepatic recycling of mycophenolic acid in solid organ transplantation and autoimmune disease. Clinical pharmacokinetics. 2011;50(1):1–24. doi: 10.2165/11536640-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee WA, Gu L, Miksztal AR, Chu N, Leung K, Nelson PH. Bioavailability improvement of mycophenolic acid through amino ester derivatization. Pharmaceutical research. 1990;7(2):161–166. doi: 10.1023/a:1015828802490. [DOI] [PubMed] [Google Scholar]
- 4.Bullingham RE, Nicholls AJ, Kamm BR. Clinical pharmacokinetics of mycophenolate mofetil. Clinical pharmacokinetics. 1998;34(6):429–455. doi: 10.2165/00003088-199834060-00002. [DOI] [PubMed] [Google Scholar]
- 5.Nowak I, Shaw LM. Mycophenolic acid binding to human serum albumin: characterization and relation to pharmacodynamics. Clinical chemistry. 1995;41(7):1011–1017. [PubMed] [Google Scholar]
- 6.Shipkova M, Armstrong VW, Wieland E, et al. Identification of glucoside and carboxyl-linked glucuronide conjugates of mycophenolic acid in plasma of transplant recipients treated with mycophenolate mofetil. British journal of pharmacology. 1999;126(5):1075–1082. doi: 10.1038/sj.bjp.0702399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schütz E, Shipkova M, Armstrong VW, Wieland E, Oellerich M. Identification of a pharmacologically active metabolite of mycophenolic acid in plasma of transplant recipients treated with mycophenolate mofetil. Clinical chemistry. 1999;45(3):419–422. [PubMed] [Google Scholar]
- 8.Picard N, Ratanasavanh D, Prémaud A, Le Meur Y, Marquet P. Identification of the UDP-glucuronosyltransferase isoforms involved in mycophenolic acid phase II metabolism. Drug metabolism and disposition. 2005;33(1):139–146. doi: 10.1124/dmd.104.001651. [DOI] [PubMed] [Google Scholar]
- 9.Dupuis R, Yuen A, Innocenti F. The influence of UGT polymorphisms as biomarkers in solid organ transplantation. Clinica Chimica Acta. 2012;413(17):1318–1325. doi: 10.1016/j.cca.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nowak I, Shaw LM. Effect of mycophenolic acid glucuronide on inosine monophosphate dehydrogenase activity. Therapeutic drug monitoring. 1997;19(3):358–360. doi: 10.1097/00007691-199706000-00018. [DOI] [PubMed] [Google Scholar]
- 11.Staatz CE, Tett SE. Clinical pharmacokinetics and pharmacodynamics of mycophenolate in solid organ transplant recipients. Clinical pharmacokinetics. 2007;46(1):13–58. doi: 10.2165/00003088-200746010-00002. [DOI] [PubMed] [Google Scholar]
- 12.Bullingham R, Monroe S, Nicholls A, Hale M. Pharmacokinetics and bioavailability of mycophenolate mofetil in healthy subjects after single-dose oral and intravenous administration. The Journal of Clinical Pharmacology. 1996;36(4):315–324. doi: 10.1002/j.1552-4604.1996.tb04207.x. [DOI] [PubMed] [Google Scholar]
- 13.Bullingham R, Nicholls A, Hale M. Pharmacokinetics of mycophenolate mofetil (RS61443): a short review. Paper presented at: Transplantation proceedings. 1996 [PubMed] [Google Scholar]
- 14.CellCept (Mycophenolate mofetil) [package insert] Hofmann-La Roche B S [Google Scholar]
- 15.Kuypers DR, Le Meur Y, Cantarovich M, et al. Consensus report on therapeutic drug monitoring of mycophenolic acid in solid organ transplantation. Clinical Journal of the American Society of Nephrology. 2010;5(2):341–358. doi: 10.2215/CJN.07111009. [DOI] [PubMed] [Google Scholar]
- 16.van Gelder T, Silva HT, de Fijter JW, et al. Comparing mycophenolate mofetil regimens for de novo renal transplant recipients: the fixed-dose concentration-controlled trial. Transplantation. 2008;86(8):1043–1051. doi: 10.1097/TP.0b013e318186f98a. [DOI] [PubMed] [Google Scholar]
- 17.Shepard TA, Reuning RH, Aarons LJ. Estimation of area under the curve for drugs subject to enterohepatic cycling. Journal of pharmacokinetics and biopharmaceutics. 1985;13(6):589–608. doi: 10.1007/BF01058903. [DOI] [PubMed] [Google Scholar]
- 18.Streit F, Shipkova M, Armstrong VW, Oellerich M. Validation of a rapid and sensitive liquid chromatography–tandem mass spectrometry method for free and total mycophenolic acid. Clinical chemistry. 2004;50(1):152–159. doi: 10.1373/clinchem.2003.024323. [DOI] [PubMed] [Google Scholar]
- 19.Jacobson PA, Schladt D, Oetting WS, et al. Genetic Determinants of Mycophenolate Related Anemia and Leukopenia Following Transplantation. Transplantation. 2011;91(3):309. doi: 10.1097/TP.0b013e318200e971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaffer E. Review article: control of gall-bladder motor function. Alimentary pharmacology & therapeutics. 2000;14(s2):2–8. doi: 10.1046/j.1365-2036.2000.014s2002.x. [DOI] [PubMed] [Google Scholar]
- 21.AA S. Overview of Biliary Function 2013. [updated November 2013]. Available from: https://www.merckmanuals.com/professional/hepatic-and-biliary-disorders/gallbladder-and-bile-duct-disorders/overview-of-biliary-function.
- 22.Shaffer E, McOrmond P, Duggan H. Quantitative cholescintigraphy: assessment of gallbladder filling and emptying and duodenogastric reflux. Gastroenterology. 1980;79(5 Pt 1):899–906. [PubMed] [Google Scholar]
- 23.Fisher RS, Stelzer F, Rock E, Malmud LS. Abnormal gallbladder emptying in patients with gallstones. Digestive diseases and sciences. 1982;27(11):1019–1024. doi: 10.1007/BF01391749. [DOI] [PubMed] [Google Scholar]
- 24.Clavien PA, Baillie J. Diseases of the gallbladder and bile ducts: diagnosis and treatment. John Wiley & Sons; 2008. [Google Scholar]
- 25.Shaw LM, Figurski M, Milone MC, Trofe J, Bloom RD. Therapeutic drug monitoring of mycophenolic acid. Clinical Journal of the American Society of Nephrology. 2007;2(5):1062–1072. doi: 10.2215/CJN.03861106. [DOI] [PubMed] [Google Scholar]
- 26.Consortium IH. A haplotype map of the human genome. Nature. 2005;437(7063):1299. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonsson EN, Karlsson MO. Xpose—an S-PLUS based population pharmacokinetic/pharmacodynamic model building aid for NONMEM. Computer methods and programs in biomedicine. 1998;58(1):51–64. doi: 10.1016/s0169-2607(98)00067-4. [DOI] [PubMed] [Google Scholar]
- 28.TEAM RC. R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria. 2010. URL: http://www.R-project.org.
- 29.RStudio: Integrated development environment for R [computer program] Version 0.97.551; Boston M: 2012. [Google Scholar]
- 30.Dong M, Fukuda T, Vinks AA. Optimization of mycophenolic acid therapy using clinical pharmacometrics. Drug metabolism and pharmacokinetics. 2014;29(1):4–11. doi: 10.2133/dmpk.dmpk-13-rv-112. [DOI] [PubMed] [Google Scholar]
- 31.Han N, Yun Hy, Kim IW, Oh YJ, Kim YS, Oh JM. Population pharmacogenetic pharmacokinetic modeling for flip-flop phenomenon of enteric-coated mycophenolate sodium in kidney transplant recipients. European journal of clinical pharmacology. 2014;70(10):1211–1219. doi: 10.1007/s00228-014-1728-4. [DOI] [PubMed] [Google Scholar]
- 32.Ling J, Shi J, Jiang Q, Jiao Z. Population pharmacokinetics of mycophenolic acid and its main glucuronide metabolite: a comparison between healthy Chinese and Caucasian subjects receiving mycophenolate mofetil. European journal of clinical pharmacology. 2015;71(1):95–106. doi: 10.1007/s00228-014-1771-1. [DOI] [PubMed] [Google Scholar]
- 33.Shum B, Duffull S, Taylor P, Tett S. Population pharmacokinetic analysis of mycophenolic acid in renal transplant recipients following oral administration of mycophenolate mofetil. British journal of clinical pharmacology. 2003;56(2):188–197. doi: 10.1046/j.1365-2125.2003.01863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Winter BC, Mathot RA, Sombogaard F, Vulto AG, van Gelder T. Nonlinear relationship between mycophenolate mofetil dose and mycophenolic acid exposure: implications for therapeutic drug monitoring. Clinical Journal of the American Society of Nephrology. 2011;6(3):656–663. doi: 10.2215/CJN.05440610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiao Z, Ding Jj, Shen J, et al. Population pharmacokinetic modelling for enterohepatic circulation of mycophenolic acid in healthy Chinese and the influence of polymorphisms in UGT1A9. British journal of clinical pharmacology. 2008;65(6):893–907. doi: 10.1111/j.1365-2125.2008.03109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sam WJ, Joy MS. Population pharmacokinetics of mycophenolic acid and metabolites in patients with glomerulonephritis. Therapeutic drug monitoring. 2010;32(5):594. doi: 10.1097/FTD.0b013e3181ee52e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Winter BC, van Gelder T, Sombogaard F, Shaw LM, van Hest RM, Mathot RA. Pharmacokinetic role of protein binding of mycophenolic acid and its glucuronide metabolite in renal transplant recipients. Journal of pharmacokinetics and pharmacodynamics. 2009;36(6):541–564. doi: 10.1007/s10928-009-9136-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smits TA, Cox S, Fukuda T, et al. Effects of unbound mycophenolic acid on inosine monophosphate dehydrogenase inhibition in pediatric kidney transplant patients. Ther Drug Monit. 2014;36(6):716–723. doi: 10.1097/FTD.0000000000000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reine PA, Vethe NT, Kongsgaard UE, et al. Mycophenolate pharmacokinetics and inosine monophosphate dehydrogenase activity in liver transplant recipients with an emphasis on therapeutic drug monitoring. Scandinavian journal of clinical and laboratory investigation. 2013;73(2):117–124. doi: 10.3109/00365513.2012.745947. [DOI] [PubMed] [Google Scholar]
- 40.Roberts MS, Magnusson BM, Burczynski FJ, Weiss M. Enterohepatic circulation. Clinical pharmacokinetics. 2002;41(10):751–790. doi: 10.2165/00003088-200241100-00005. [DOI] [PubMed] [Google Scholar]
- 41.Naito T, Mino Y, Otsuka A, et al. Impact of calcineurin inhibitors on urinary excretion of mycophenolic acid and its glucuronide in kidney transplant recipients. The Journal of Clinical Pharmacology. 2009;49(6):710–718. doi: 10.1177/0091270009335003. [DOI] [PubMed] [Google Scholar]
- 42.Frymoyer A, Verotta D, Jacobson P, Long-Boyle J. Population pharmacokinetics of unbound mycophenolic acid in adult allogeneic haematopoietic cell transplantation: effect of pharmacogenetic factors. British journal of clinical pharmacology. 2013;75(2):463–475. doi: 10.1111/j.1365-2125.2012.04372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim H, Long-Boyle J, Rydholm N, et al. Population pharmacokinetics of unbound mycophenolic acid in pediatric and young adult patients undergoing allogeneic hematopoietic cell transplantation. The Journal of Clinical Pharmacology. 2012;52(11):1665–1675. doi: 10.1177/0091270011422814. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Zhang L, Abraham S, et al. Assessment of the impact of renal impairment on systemic exposure of new molecular entities: evaluation of recent new drug applications. Clinical pharmacology and therapeutics. 2009;85(3):305–311. doi: 10.1038/clpt.2008.208. [DOI] [PubMed] [Google Scholar]
- 45.Dostalek M, Hazarika S, Akhlaghi F. Diabetes mellitus reduces activity of human UDP-glucuronosyltransferase 2B7 in liver and kidney leading to decreased formation of mycophenolic acid acyl-glucuronide metabolite. Drug Metabolism and Disposition. 2011;39(3):448–455. doi: 10.1124/dmd.110.036608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anger GJ, Magomedova L, Piquette-Miller M. Impact of Acute Streptozotocin-Induced Diabetes on ABC Transporter Expression in Rats. Chemistry & biodiversity. 2009;6(11):1943–1959. doi: 10.1002/cbdv.200900053. [DOI] [PubMed] [Google Scholar]
- 47.Gwilt PR, Nahhas RR, Tracewell WG. The effects of diabetes mellitus on pharmacokinetics and pharmacodynamics in humans. Clinical pharmacokinetics. 1991;20(6):477–490. doi: 10.2165/00003088-199120060-00004. [DOI] [PubMed] [Google Scholar]
- 48.Lu R, Kanai N, Bao Y, Wolkoff AW, Schuster VL. Regulation of renal oatp mRNA expression by testosterone. American Journal of Physiology-Renal Physiology. 1996;270(2):F332–F337. doi: 10.1152/ajprenal.1996.270.2.F332. [DOI] [PubMed] [Google Scholar]
- 49.Cerrutti JA, Brandoni A, Quaglia NB, Torres AM. Sex differences in p-aminohippuric acid transport in rat kidney: role of membrane fluidity and expression of OAT1. Molecular and cellular biochemistry. 2002;233(1-2):175–179. doi: 10.1023/a:1015563021602. [DOI] [PubMed] [Google Scholar]
- 50.Odom DT, Zizlsperger N, Gordon DB, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303(5662):1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shih DQ, Bussen M, Sehayek E, et al. Hepatocyte nuclear factor-1α is an essential regulator of bile acid and plasma cholesterol metabolism. Nature genetics. 2001;27(4):375–382. doi: 10.1038/86871. [DOI] [PubMed] [Google Scholar]
- 52.Shaw LM, Korecka M, Venkataramanan R, Goldberg L, Bloom R, Brayman KL. Mycophenolic acid pharmacodynamics and pharmacokinetics provide a basis for rational monitoring strategies. American Journal of Transplantation. 2003;3(5):534–542. doi: 10.1034/j.1600-6143.2003.00079.x. [DOI] [PubMed] [Google Scholar]
- 53.Weber LT, Shipkova M, Armstrong VW, et al. The pharmacokinetic-pharmacodynamic relationship for total and free mycophenolic acid in pediatric renal transplant recipients: a report of the German study group on mycophenolate mofetil therapy. Journal of the American Society of Nephrology. 2002;13(3):759–768. doi: 10.1681/ASN.V133759. [DOI] [PubMed] [Google Scholar]
- 54.Mourad M, Malaise J, Eddour DC, et al. Correlation of mycophenolic acid pharmacokinetic parameters with side effects in kidney transplant patients treated with mycophenolate mofetil. Clinical chemistry. 2001;47(1):88–94. [PubMed] [Google Scholar]
- 55.Fleming D, Mathew B, John G, Chandy S, Manivannan J, Jeyaseelan V. A six-hour extrapolated sampling strategy for monitoring mycophenolic acid in renal transplant patients in the Indian subcontinent. Journal of postgraduate medicine. 2006;52(4):248. [PubMed] [Google Scholar]
- 56.Young CJ, Gaston RS. Renal transplantation in black Americans. New England Journal of Medicine. 2000;343(21):1545–1552. doi: 10.1056/NEJM200011233432107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.