Abstract
Apoptosis is a genetically directed process of programmed cell death. A variety of microRNAs (miRNAs), endogenous single-stranded non-coding RNAs of about 22 nucleotides in length have been shown to be involved in the regulation of the intrinsic or extrinsic apoptotic pathways. There is increasing evidence that the aberrant expression of miRNAs plays a causal role in the development of diseases such as cancer. This makes miRNAs promising candidate molecules as therapeutic targets or agents. MicroRNA (miR)-217-5p has been implicated in carcinogenesis of various cancer entities, including colorectal cancer. Here, we analyzed the pro-apoptotic potential of miR-217-5p in a variety of colorecatal cancer cell lines showing that miR-217-5p mimic transfection led to the induction of apoptosis causing the breakdown of mitochondrial membrane potential, externalization of phosphatidylserine, activation of caspases and fragmentation of DNA. Furthermore, elevated miR-217-5p levels downregulated mRNA and protein expression of atypical protein kinase c iota type I (PRKCI), BAG family molecular chaperone regulator 3 (BAG3), integrin subunit alpha v (ITGAV) and mitogen-activated protein kinase 1 (MAPK1). A direct miR-217-5p mediated regulation to those targets was shown by repressed luciferase activity of reporter constructs containing the miR-217-5p binding sites in the 3′ untranslated region. Taken together, our observations have uncovered the apoptosis-inducing potential of miR-217-5p through its regulation of multiple target genes involved in the ERK-MAPK signaling pathway by regulation of PRKCI, BAG3, ITGAV and MAPK1.
Keywords: Apoptosis, Cell death, Colorectal cancer, miR-217-5p, Target analysis
Introduction
MicroRNAs (miRNAs) are endogenous single-stranded, about 22 nucleotides long non-coding RNA molecules that regulate gene expression post-transcriptionally (Breving and Esquela-Kerscher 2010). MiRNAs are evolutionary conserved and generated in a multistep process starting with the RNA-polymerase II-mediated transcription and processing by Drosha/DGCR8 in the nucleus (Breving and Esquela-Kerscher 2010; Han et al. 2006). The precursor miRNA is translocated into the cytoplasm for further processing to generate the RNA duplex, comprising a 5p and 3p strand. Finally, the functional strand is loaded onto a miRNA-induced-silencing complex for its direction to the target mRNA (Kim et al. 2009). Mammalian miRNAs interact with the target mRNA by imperfect base pairing mostly leading to translational repression or degradation of the mRNA. MiRNAs are estimated to be involved in the expression control of 30–50% of human proteins (Breving and Esquela-Kerscher 2010; van Kouwenhove et al. 2011) and the regulation of fundamental processes including development, cell proliferation, differentiation, and apoptosis (Bushati and Cohen 2007).
Apoptosis is a genetically directed process of programmed cell death. Its execution is mainly regulated by two distinct but interrelated signaling cascades, the extrinsic and the intrinsic apoptosis death pathway. The extrinsic pathway involves binding of a pro-apoptotic inductor ligand like tumor necrosis factor related apoptosis inducing ligand (TRAIL) or tumor necrosis factor to a death receptor (MacFarlane 2003) whereas the intrinsic pathway is initiated by cytochrome c release from the mitochondria (Zimmermann et al. 2001). The signaling cascades culminate in activation of caspases and subsequent specific morphological and biochemical changes like nuclear condensation, cleavage of genomic DNA and cell shrinkage resulting in programmed cell death and elimination of degenerated cells by phagocytosis (Elmore 2007). A variety of miRNAs have been shown to be involved in the regulation of the intrinsic or extrinsic apoptotic pathways (Fischer et al. 2015; Jovanovic and Hengartner 2006; Lima et al. 2011; Lynam-Lennon et al. 2009) and there is increasing evidence that the aberrant expression of miRNAs plays a causal role in the development of diseases such as cancer. Due to these observations, miRNAs are promising candidate molecules as diagnostic or prognostic biomarkers and as therapeutic targets or agents (Lynam-Lennon et al. 2009).
Colorectal cancer (CRC) represents the third most common cancer in men and the second most common cancer in women globally (Cunningham et al. 2010; Debarros and Steele 2013; Qaseem et al. 2012). With the development of early diagnosis and treatment modalities, the 5-year survival rate of CRC has been improved over the past two decades (Cunningham et al. 2010; Ragnhammar et al. 2001). Research has been focused on tumor suppressor genes, oncogenes and cell signaling pathways, including their role in the proliferation, apoptosis and aggressiveness of these tumors (Moss 2014). Various miRNAs have been shown to be involved in CRC tumorigenesis, demonstrating that abnormal expression or mutations of miRNAs play a role in different stages of CRC development (Bader 2012; Cekaite et al. 2016; Slaby et al. 2009).
The involvement in the regulation of fundamental cellular processes such as apoptosis together with the increasing evidence for a potential function as tumor suppressor genes makes miRNAs highly interesting candidate molecules for the generation of novel anticancer therapeutics. Based on previous cellular high throughput screenings (Fischer et al. 2014; Kleemann et al. 2017) we identified miR-217-5p to strongly induce apoptosis in a CRC cell line. The aim of the current study was to further elucidate the role of miR-217-5p in programmed cell death. Since miRNAs are known to regulate the expression of a multiplicity of target genes and to influence cellular signaling pathways at various sites, we intended to identify target gene networks to elucidate the pro-apoptotic molecular mechanisms of miR-217-5p.
Materials and methods
Cell culture
HCT 116 and T98G cells were grown in RPMI-1640 medium, HT-29 cells in McCoy’s 5A and SW480 cells in Leibovitz’s L-15 medium. SKOV3 and HEK293T cells were grown in DMEM. All media were obtained from Thermo Fisher Scientific, Waltham, MA, USA and supplemented with 10% (v/v) heat inactivated fetal calf serum (Sigma-Aldrich, München, Germany). Cells were cultured at 37 °C and 5% CO2. The phenotype of all cell lines was proofed frequently by microscopy.
Cell confluence after miRNA mimic transfection was monitored by the fully automated cell imager NyOne (Synen-Tec Bio Services, Münster, Germany).
Transfection of miRNA mimics
The cells were seeded one day prior to transfection in 96-well cell culture plates (Greiner Bio-One). MiRNA mimics (Qiagen, Hilden, Germany) comprised the sequence of the human miR-217-5p (obtained from miRBase version 21 (Kozomara and Griffiths-Jones 2014)). For controls, a non-targeting small interfering RNA (siRNA) (NT) as well as a cell death inducing siRNA (DT) (Qiagen) were used. Cells were transfected with 62.5 nM miRNA mimic and ScreenFect®A (InCella, Eggenstein-Leopoldshafen, Germany).
Cell death assays
To quantify apoptosis induction by miRNA mimic transfection, cells were harvested, stained for different characteristics of cell death and analyzed via flow cytometry using the MACSQuant® Analyser (Miltenyi Biotec, Bergisch Gladbach, Germany). Assay validity of the cell death assays was tested by simultaneously accessing cells treated with the chemical apoptosis inducers Etoposide (25 μM) and TRAIL (80 ng/ml (HCT 116), 150 ng/ml (HT-29, SW480)) (Enzo Life Sciences, New York, USA).
To quantify the fragmentation of DNA and the reduction of detached cellular DNA content, cells were processed by Nicoletti staining comprising the resuspension in 80 μl hypotonic staining buffer (0.1% w/v sodium citrate (Biochemika, Fluka, Buchs, Switzerland), 0.05% v/v Triton X-100 (Carl Roth GmbH & Co. KG, Karlsruhe, Germany), 10 μg/ml Propidium Iodide and 3.3 μg/ml RNase A (Carl Roth GmbH & Co. KG) in phosphate-buffered saline (PBS, GE Healthcare, Buckinghamshire, UK). The stained cells were analyzed by flow cytometry.
For detection of phosphatidylserine (PS) externalization, cells were washed in PBS, resuspended in 100 μl binding buffer to which 2.5 μl Annexin V-FITC staining solution was added (Annexin V-FITC Apoptosis detection Kit, Affimetrix, Frankfurt am Main, Germany). After a washing step in 200 μl binding buffer and resuspension in 90 μl binding buffer, 5 μl propidium iodide (PI) solution was added. The cells were quantified by flow cytometry.
To access the breakdown of mitochondrial membrane potential, active mitochondria of healthy cells were stained with Tetramethylrhodamine ethyl ester perchlorate (TMRE). To this end, cells were resuspended in 80 μl fresh medium. As positive control for the loss of mitochondrial function, untreated cells were incubated with 5 μM carbonyl cyanide m-chlorophenyl hydrazine (CCCP). Then, TMRE (Santa Cruz Biotechnology, Dallas, Texas, USA) was added at a final concentration of 300 nM and cells were analyzed by flow cytometric analysis.
To measure caspase-3 and -7 activity, cells were incubated with CellEvent® Caspase-3/7 Green Detection Reagent at a final concentration of 500 nM (CellEvent® Caspase-3/7 Green Flow Cytometry Assay Kit, molecular probes™, Thermo Fisher Scientific) and analysed by flow cytometry.
Quantification of endogenous miR-217-5p expression
To determine the endogenous miR-217-5p expression in colorectal cancer cells, HCT 116, HT-29 and SW480 cells were stimulated with Etoposide (25 μM), TRAIL (80 ng/ml (HCT 116), 150 ng/ml (HT-29, SW480)). Then, cells were harvested and total RNA was isolated using the miRNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. CDNA was synthesized by reverse transcription of 1 μg RNA using the miScript II RT Kit and the included 5× miScript HiSpec Buffer to ensure reverse transcription from mature miRNA (Qiagen). CDNA was diluted 1:30 and used for miR-217-5p expression analysis by quantitative PCR. To this end, 2 x GreenMasterMix (Genaxxon Bioscience, Ulm, Germany) were mixed with miRNA-specific forward primer (5 μM stock) comprising the mature miR-217-5p (5′-TACTGCATCAGGAACTGATTGGA-3′) or miR-217-3p (5′-CATCAGTTCCTAAT GCATTGCCT -3′) sequence based on the information on miRBase, 10× miScript Universal Primer and 1:30 pre-diluted cDNA. For normalization of cDNA input, the U6 snRNA was co-amplified using U6 snRNA-specific forward primer (5′-AACGCTTCACGAATTTGCGT-3′) and U6 snRNA-specific forward primer (5′- CTCGCTTCGGCAGCACA-3′). Quantitative PCR was performed in the LightCycler® 480 (Roche Diagnostics GmbH). Each cDNA sample was measured in triplicates. At the end, a melting step was included to access amplification of one specific product and collected fluorescence data were analyzed by the ΔΔCT method to calculate the relative differences in miRNA expression.
Monitoring of mRNA expression of potential target genes
To determine mRNA expression of potential target genes 48 h upon miRNA mimic transfection, total RNA was isolated as described before and 1 μg RNA was used for cDNA synthesis via the Transcriptor High Fidelity cDNA Synthesis Kit (Roche Diagnostics GmbH) and the included anchored oligo(dT)18 Primer. The cDNA was diluted 1:30 and used for mRNA expression. To this end, 2 x GreenMasterMix (Genaxxon Bioscience) were mixed with target gene-specific forward primer and reverse primer (5 μM stock) and 1:30 pre-diluted cDNA. Peptidylprolyl isomerase A (PPIA) mRNA was amplified as housekeeping gene. Quantitative PCR was performed and analyzed as described in the preceding paragraph. The following primer sequences were used: MAPK1 forward (5′-TCTGCACCGTGACCTCAA-3′), MAPK1 reverse (5′-GCCAGGCCAAAGTCACAG-3′), MAPK3 forward (5′-CCCTAGCCCAGACAGACATC-3′), MAPK3 forward (5′-CCCTA GCCCAGACAGACATC-3′), MAPK3 reverse (5′-GCACAGTGTCCATTTTCTAACAGT-3′), ITGAV forward (5′-AAGCTGAGCTCATCGTTTCC-3′), ITGAV reverse (5′-GCACAGGAAAGTCTTGCTAAGG-3′), PRKCl forward (5′-TCCCTTGTGTACCAGAAC GTC-3′), PRKCl reverse (5′-GGCACAATAAAGCTTTCTCCA-3′), BAG3 forward (5’-CTCAGCCAGATAAACAGTGTGG-3′), BAG3 reverse (5′-GTCAGAGGCAGCTGGAGA CT-3′), PPIA forward (5′-ATGCTGGACCCAACACAAAT -3′) and PPIA reverse (5′-TCTTTCACTTTGCCAAACACC -3′).
Western blot analysis of protein expression of potential target genes and apoptosis induction
For protein analysis of potential target genes and assessment of apoptosis induction cells were harvested 60 h after miRNA mimic transfections and lysed with radioimmunoprecipitation assay (RIPA) buffer consisting of 1 mM ethylenediaminetetraacetic acid, 0.5 mM dithiothreitol, 0.5% sodium deoxycholate in PBS. The protein concentration was determined by bicinchoninic acid assay (BCA) and 20 μg protein were separated on a gradient SDS-PAGE (8–16%) and transferred to a polyvinylidene difluoride (PVD) membrane (Carl Roth GmbH & Co. KG). The membrane was blocked with 5% w/v bovine serum albumin in PBS-0.1%Tween (PBS-T) and probed with primary antibodies. These primary antibodies comprised rabbit monoclonal anti-integrin alpha V (#60896), anti-protein kinase C iota (PKCι/λ) (#2998), rabbit polyclonal anti-p44/42 MAPK (Erk1/2) (#9102) antibodies from Cell Signaling Technology (Cambridge, United Kingdom) and mouse anti-BAG3 (SAB1404732 from Sigma Aldrich). To access apoptosis induction by miR-217-5p mimic transfection, PVDF membranes were also probed with the rabbit polyclonal caspase-3 (#9662), anti-PARP (#9542), rabbit monoclonal anti-cleaved caspase-3 (#9664) and rabbit polyclonal anti-cleaved PARP (#9541) antibodies from Cell Signaling Technology. The mouse monoclonal anti-GAPDH antibody (MA5–15738, Thermo Fisher Scientific) was used as loading control. Bound antibody was revealed with the appropriate secondary HRP linked antibody (anti-rabbit IgG, (#7074, Cell Signaling) or anti-mouse IgG, (A4416, Sigma Aldrich, München, Germany)) and protein was visualized by enhanced chemiluminescence using Immobilon Western Chemiluminescent HRP Substrat from Merck Millipore and the Fusion FX image acquisition system (Vilber Lourmat, Eberhardzell, Germany) for detection.
In silico target prediction
Six different in silico target prediction tools were applied to identify potential miR-217-5p target genes, the prediction tools TargetScan Human (Agarwal et al. 2015), miRanda (Betel et al. 2010), Rna22 (Miranda et al. 2006), DIANA TOOLS (Vlachos et al. 2015), miRDB (Wong and Wang 2015) and miRWalk (Dweep et al. 2011) were used.
Employing the free-accessible online gene classification soft-ware PANTHER (Protein Analysis Through Evolutionary Relationships) (Thomas et al. 2003) and IPA (Ingenuity Pathway Analysis) (Qiagen Bioinformatics) suggested potential target genes were restricted to genes with anti-apoptotic or survival promoting functions. In addition, already experimentally validated miR-217-5p target genes listed in miRTarBase (Chou et al. 2016) and DIANA-TarBase (Vlachos et al. 2015) were excluded from the further investigations. Upon examination of tissue expression profiles of predicted potential target genes employing online databases as The Human Protein Atlas (Uhlen et al. 2015) or GeneCards® (Rebhan et al. 1997) a selection of potential target genes was chosen to access their potential post-transcriptional regulation by miR-217-5p.
Potential miR-217-5p binding sites were obtained from the database microRNA.org (Betel et al. 2010) by aligning miR-217-5p with the mRNA transcript of predicted potential target genes.
Luciferase reporter assay
Complementary oligonucleotide pairs comprising a portion of putative miRNA binding sites were synthesized, annealed and cloned into the pmirGlo® Dual Luciferase miRNA target expression vector (Promega Corporation, USA) between the NheI/NotI restriction sites of the multiple cloning site downstream of a luciferase gene.
For luciferase assays, HEK 293 T cells were co-transfected with 200 ng of the pmirGlo® Dual Luciferase miRNA target expression vector and miR-217-5p or microRNA inhibitor anti-miR-217-5p or non-targeting siRNA control (NT) at a final concentration of 50 nM using Lipofectamine® 3000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. Three days after transfection, cells were lysed with the Dual-Glo® Reagent (Dual-Glo® Luciferase Assay System; Promega Corporation) and luciferase activity was quantified on a SpectraMax M5e microplate reader (Molecular Devices, Sunnyvale, CA, USA). After calculating the ratio of firefly luminescence to the luminescence from Renilla, the experimental well ratio was normalized to the ratio of the control wells.
Statistical analysis
Data are presented as mean ± SD. Statistical analysis was carried out using GraphPad Prism (version 5.04). Apoptosis rates were statistical tested by a two-way ANOVA followed by Bonferroni post-test, whereas differences in miRNA expression after apoptosis induction were tested by one-way ANOVA followed by Bonferroni post-test. Differences between the mRNA expression of potential target genes in miR-217-5p mimic and NT transfected cells as well as luciferase reporter data were analyzed using the two-tailed unpaired t-test. A p-value <0.05 was considered to be statistically significant.
Results
Apoptosis screening identified miR-217-5p to act pro-apoptotic in colon carcinoma cells
MiRNAs are highly interesting candidate molecules for the generation of novel anticancer therapeutics due to their involvement in the regulation of fundamental cellular processes such as apoptosis together with the increasing evidence for a potential function as tumor suppressor-genes. Therefore, we investigated the potential effects of miRNAs identified in two previous apoptosis screenings (Fischer et al. 2014; Kleemann et al. 2017) in various human cancer cell lines including HCT 116, SKOV3 and the human glioblastoma cell line T98G. The different cell lines were used to test the apoptosis effect of these miRNAs to identify general and tissue specific acting miRNAs. Apoptosis induction by miRNAs was accessed by transient miRNA mimics transfection and subsequent apoptosis measurement by quantitative flow cytometry. In addition to described apoptosis inducing and tumor suppressor miRNAs such as miR-133a-3p (Yang et al. 2017; Zhang et al. 2015) and miR-185-5p (Liu et al. 2011) (Fig. 1a), microRNA 217-5p was identified to induce apoptosis in the CRC cell line HCT 116. Compared to a non-targeting siRNA control (NT), miR-217-5p significantly raised apoptosis (cells in subG0/G1) from 10.7% (± 2.4%) to 42.3% (± 1.3%). The previously reported pro-apoptotic miR-133a-3p and miR-185-5p were able to induce apoptosis up to 55.6% (± 0.9%) and 35.6% (± 3.6%) in these cells, respectively. Based on these initial data from the previous screening, we further focused on dissecting the pro-apoptotic effects of miR-217-5p in CRC cell lines. In this study, we aimed to reveal the mechanism of action leading to apoptosis by the sequence conserved miR-217.
Fig. 1.
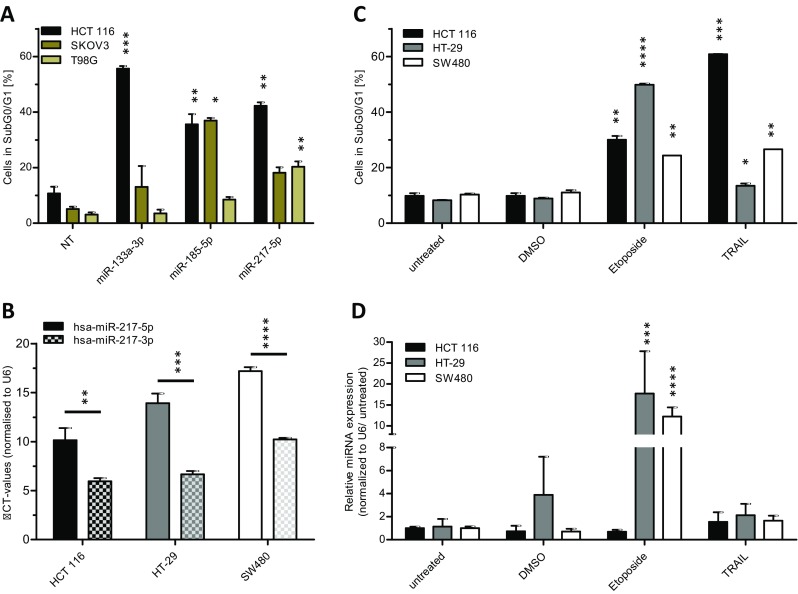
miR-217-5p expression is increased upon induction of the apoptosis. For validation screening HCT 116, SKOV3, and T98G cells were seeded 24 h before transfection with miRNA mimics (50 nM and 0.4 μl ScreenFect®A) or non-targeting siRNA (NT) control. Apoptosis rates 72 h after transfection were analyzed by Nicoletti staining followed by flow cytometric analysis (a). For miR-217-3p and -5p expression analysis total RNA was isolated from untreated HCT 116, HT-29 and SW480 cells and applied to cDNA synthesis followed by qRT-PCR. The expression analysis of both miR-217 strands was done by normalization to the CT value of U6 snRNA (b). For determination of miR-217-5p expression after induction of apoptosis, cells were seeded in 24 h prior treatment with Etoposide (25 μM), TRAIL (150 ng/ml, except HCT 116 with 80 ng/ml) or DMSO for additional 48 h. The apoptosis rates 48 h after treatment were analyzed by Nicoletti staining and flow cytometric analysis (c). The miRNA expression of miR-217-5p was normalized to the CT value of U6 snRNA and the untreated control (d). Statistical analyses for part (a, c and d) were performed by two-way ANOVA followed by Bonferroni post-test. Statistical differences for part B were tested using unpaired t-test. The treatments were compared to NT (a) or untreated cells (c and d) [n = 3 biological replicates; mean ± SD, *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001]
miR-217-5p expression was increased after apoptosis induction
To further access the functions of miR-217-5p in the regulation of the apoptotic process in CRC, the expression of miR-217-5p was determined by qRT-PCR in a set of CRC model cell lines comprising HTC 116, HT-29 and SW480. In all cell lines, the basal expression level of miR-217-5p was significant lower compared to the complementary miR-217-3p strand (Fig. 1b). To examine the expression pattern of miR-217-5p in all cell lines after induction of apoptosis, cells were incubated with Etoposide or TRAIL. Etoposide is an inductive stimulus for the intrinsic apoptotic pathway through inhibition of topoisomerase II (Montecucco et al. 2015), whereas TRAIL induces extrinsic apoptosis by binding to its receptor on the cell surface (Wang and El-Deiry 2003). Apoptosis rates were determined 48 h after treatment by flow cytometric analysis and were found to be increased in all cell lines tested. The induction of apoptosis in HT-29 cells was small due to its known resistance to TRAIL treatment (Lee et al. 2011) (Fig. 1c). Analysis of miR-217-5p expression by qRT-PCR revealed a highly significant increase in expression after treatment with Etoposide of about 20-fold in HT-29 and 14-fold in SW480 cell lines, whereas no effect was seen in HCT116 cells. In contrast, TRAIL did not elevate expression of miR-217-5p (Fig. 1d) in any of the cell lines examined. The observed upregulation of miR-217-5p expression after treatment with Etoposide suggested an involvement of the analyzed miRNA strand in the initiation or progression of the intrinsic apoptotic pathway.
Repression of cell growth and proliferation by miR-217-5p
In order to substantiate our observations on pro-apoptotic effects of miR-217-5p and to provide a better understanding of the underlying molecular mechanisms, we further dissected miRNA functions in the apoptotic pathways. Due to low basal miR-217-5p expression levels gain-of-function studies were conducted employing transient transfection of miR-217-5p mimic. In addition, the transfection with either miR-133a-3p mimic or treatment with Etoposide or TRAIL as well as transfection with a cell death inducing siRNA (DT) served as positive controls, while transfection with a non-targeting siRNA (NT) or treatment with DMSO were used as negative controls. Measurement of cell confluence 72 h after transient transfection and Etoposide or TRAIL treatment revealed a highly significant reduction in cell confluence especially for miR-217-5p mimic transfected HCT 116 cells (0.36 fold ±0.01 fold compared to NT control). Although the reduction of relative cell confluency was lower in miR-217-5p transfected HT-29 (0.84 fold ±0.01 fold) and SW480 cells (0.69 fold ±0.05 fold), it confirmed assay validity and in addition highlighted the strong miR-217-5p responsiveness of HCT 116 cells (Fig. 2a). The data point towards a potential role of miR-217-5p in the regulation of cellular growth and proliferation.
Fig. 2.
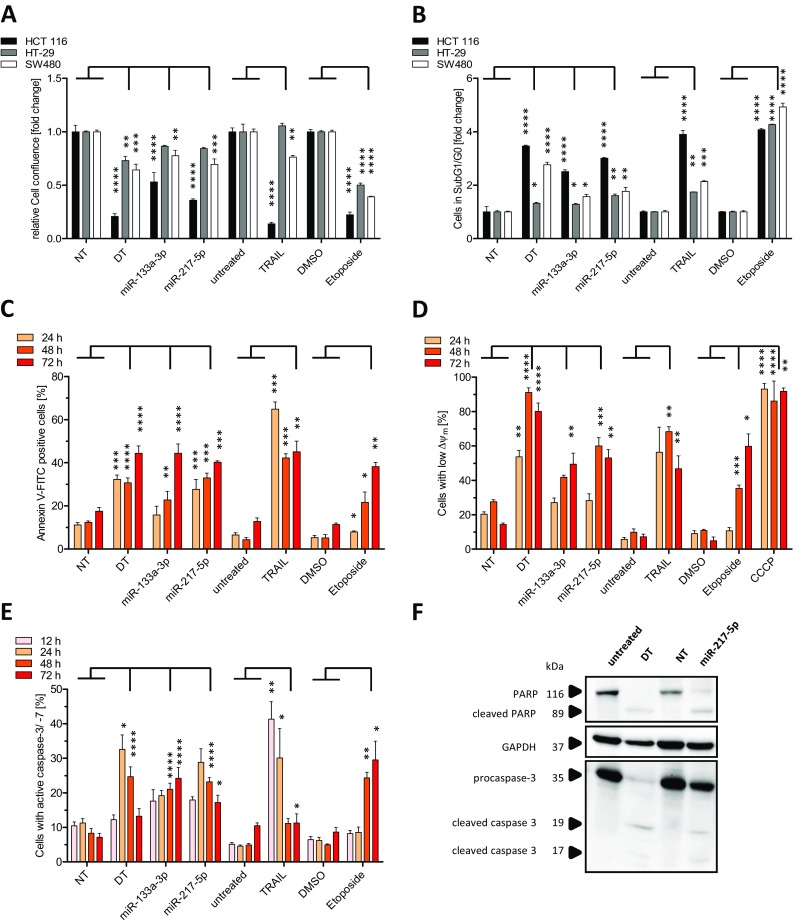
Apoptosis induction after miR-217-5p mimic transfection. HCT 116, HT-29 and SW480 cells were seeded one day prior transfection with miR-217-5p, miR-133a-3p mimics, non-targeting siRNA (NT) or treatment with Etoposide (25 μM) TRAIL (80 ng/ml) or DMSO in 96 well plates. 72 h after transfection of miR-217-5p and miR-133a-3p (62.5 nM) cell confluency was determined by automated microscopy using NyONE (a). The apoptosis rates 72 h after treatment were analyzed by Nicoletti staining and flow cytometric analysis (b). After 24 h, 48 h, and 72 h, molecular characteristics of apoptosis were detected in HCT 116 cells by measuring phosphatidylserine externalization (c) or the breakdown of mitochondrial membrane potential ΔΨm by TMRE staining during flow cytometric analysis (d). As a positive control for the breakdown of mitochondrial potential ΔΨm, 5 μM CCCP was used. 48 h after transfection HCT 116 cells were harvested and lysed followed by SDS-PAGE and immunoblotting with procaspase-3, cleaved caspase-3, PARP and cleaved PARP antibodies. GAPDH was used as loading control (f). Caspase-3 and -7 activities were detected by flow cytometry using a labeled fluorescent substrate (e). Statistical analysis was performed by two-way ANOVA followed by Bonferroni post-test [n = 3 biological replicates; mean ± SD, *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001]
Molecular changes during apoptosis induced by miR-217-5p
To investigate the pro-apoptotic potential of miR-217-5p, we applied several flow cytometric methods to access molecular changes in apoptotic pathways induced upon transient miR-217-5p mimic transfection into the three CRC cell lines. Nicoletti staining was performed to detect apoptotic cells in SubG1/G0. As in the case of reduction in cell confluency (Fig. 2a), HCT 116 cells showed the highest amount of cells in SubG1/G0 (3.01 fold ±0.03 vs 1.61 fold ±0.06 in HT-29 and 1.77 fold ±0.14 fold in SW480 cells) (Fig. 2b). Based on these data, only HCT116 cells were used for further detailed analysis on the time response of different apoptosis markers. A hallmark of apoptosis is the externalization of PS located on the cytoplasmic surface of the cell membrane. PS translocates to the outer leaflet of the membrane in the intermediate stages of apoptosis where it can be detected. Performing Annexin V-FITC staining 24 h, 48 h, and 72 h after miR-217-5p and miR-133a-3p mimic transfections revealed a highly significant time dependent increase in PS externalization of up to 40.24% (± 0.6%) and 44.36% (± 4.33%) after 72 h respectively, compared to 17.46% (± 1.81%) for the NT control (Fig. 2c). This observation provides evidence for the induction of apoptosis by elevated levels of miR-217-5p or miR-133a-3p.
As part of the intrinsic apoptotic pathway the mitochondrial membrane potential (MMP, ΔΨm) decreases and subsequently leads to cytochrome C release (Wang 2001). The MMP was traced by the lipophilic cationic fluorescent dye Tetramethylrhodamine ethyl ester perchlorate (TMRE), which is unable to accumulate in the mitochondrial matrix after MMP breakdown (Gottlieb and Granville 2002). To investigate the effects of miR-217-5p on MMP, HCT 116 cells were transfected with miRNA mimics and stained with TMRE 24 h, 48 h, and 72 h after transfection (Fig. 2d). In addition to the cell death siRNA control, cells were treated with Carbonyl cyanide m-chlorophenyl hydrazone (CCCP), which uncouples oxidative phosphorylation (Perry et al. 2011), to control assay validity. The increase in cells with reduced ΔΨ m upon miR-217-5p mimic transfection, shown by 60.03% (± 4.87%) low fluorescent cells compared to 27.56% (± 1.21%) for the NT control at 48 h post transfection, indicated a loss of mitochondrial integrity. These data give strong evidence for the induction of the intrinsic, mitochondrial apoptotic pathway upon miR-217-5p transfection.
Both intrinsic as well as extrinsic apoptotic pathways converge in the activation of the effector caspase-3 and -7 (Strasser et al. 2000). To access the effects of miR-217-5p on the cleavage and activation of effector caspases, we quantified the amount of cells with caspase-3 and -7 activity 12 h, 24 h, 48 h, and 72 h after transfection of miR-217-5p and miR-133a-3p mimics using fluorescent labeled caspase substrates and subsequent flow cytometric analysis. We observed highly significant caspase-3 and -7 activities at 48 h and 72 h after miR-217-5p (23.2% ± 1.23% and 17.21% ± 2.12%) and miR-133a-3p (21.04% ± 1.75% and 24.24% ± 3.09%) mimic transfections compared to NT control (8.3% ± 1.4% and 7.16% ± 1.14%). In addition, treatment with Etoposide led to highly significant caspase activities of 24.33% (± 1.64%) at 48 h and 29.56% (± 5.4%) at 72 h compared to cells treated with DMSO (4.97% ± 0.23% at 48 h and 8.68% ± 1.33% at 72 h) (Fig. 2e). At these time points, caspase activity was decreasing in TRAIL treated cells, which indicated that cells were already entering the state of late apoptosis or secondary necrosis.
To confirm the induction of apoptosis induced by miR-217-5p mimic and to validate the results received by flow cytometric analyses at the molecular level, Western blot analysis detecting cleaved caspase-3 as well as cleaved poly (ADP-ribose) polymerase (PARP), which is a substrate of activated caspase-3 (Cregan et al. 2004), were conducted in miR-217-5p-mimic, NT or cell death siRNA transfected HCT 116 cells. At 60 h after transfection, the protein level of PARP and procaspase-3 remained almost unchanged in control (NT) and untreated cells (Fig. 2f), whereas cell death siRNA led to an expected increase in the abundance of cleaved caspase-3 as well as cleaved PARP accompanied by reduced levels of procaspase-3 and undetectable levels of PARP. Comparable results were found for miR-217-5p, confirming the flow cytometry data (Fig. 2b).
In summary, the pro-apoptotic effects of miR-217-5p were confirmed by various methods detecting different characteristics of apoptosis after transient transfection in HCT 116 cells and point towards a possible role of miR-217 in the intrinsic apoptotic pathway.
miR-217-5p regulates genes relevant for survival and apoptosis
In order to elucidate critical downstream targets and signaling pathways controlled by miR-217-5p, accounting for the discovered pro-apoptotic effects in HCT 116 cells, in silico target gene prediction analysis for miR-217-5p was performed. Since the computational target prediction yielded a large number of putative target genes, all genes were subjected to a functional clustering analysis using PANTHER Analysis (Thomas et al. 2003) and Ingenuity® Pathway Analysis (Qiagen Bioinformatics) to assort them into functional groups. Focusing on survival promoting or anti-apoptotic functions, twenty-five target genes were selected for further analysis (Table 1). For functional validation, we transiently transfected HCT 116 cells with miR-217-5p mimic or NT and analyzed the mRNA expression after 48 h by qRT-PCR. Interestingly, twelve putative target genes were confirmed to be significantly downregulated after miR-217-5p transfection, including integrin subunit alpha V (ITGAV), mitogen-activated protein kinase 1 (MAPK1), protein kinase C iota 1 (PRKC1), baculoviral IAP repeat containing 2 (BIRC3), transforming growth factor beta receptor 2 (TGFBR2), BCL2 associated athanogene 3 (BAG3), catenin beta 1 (CTNNB1), MAPK3, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), epidermal growth factor receptor (EGFR), transcriptional activator Myb (MYB) and BRCA1 DNA repair associated (BRCA1), while four predicted target genes showed a significant upregulation after miR-217-5p transfection (Ras-related C3 botulinum toxin substrate 1 (RAC1), Raf-1 proto-oncogene, serine/threonine kinase (RAF1), RAD51 recombinase (RAD51), and BCL2 apoptosis regulator (BCL2)) (Fig. 3a). Focusing on the twelve downregulated genes, we next accessed whether this regulatory activity can be confirmed by reduced protein levels. For target genes showing the most dramatic downregulation (ITGAV, MAPK1, MAPK3, PRKC1 and BAG3), we quantified protein levels by Western blot analysis 60 h after miR-217-5p mimic transfection into HTC 116 cells. As expected, miR-217-5p led to a significant down-regulation of MAPK1/3, PRKC1 and BAG3 protein levels except for ITGAV which might be due to delayed protein degradation (Fig. 3b and c). Taken together, these findings strongly suggest a concerted regulatory function of miR-217-5p during apoptosis.
Table 1.
In silico target prediction of putative target genes of miR-217
| Putative target gene ID | Gene name | Cluster function |
|---|---|---|
| MAPK1/ ERK2 | mitogen-activated protein kinase 1/ extracellular signal-regulated kinase 2 | IPA: apoptosis of tumor cell lines |
| MAPK3/ ERK1 | mitogen-activated protein kinase 3/ extracellular signal-regulated kinase 2 | IPA: apoptosis of tumor cell lines |
| PIK3CA | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha | IPA: apoptosis of tumor cell lines |
| BRCA1 | BRCA1, DNA repair associated | IPA: apoptosis of tumor cell lines |
| RAD51 | RAD51 recombinase | IPA: apoptosis of tumor cell lines |
| ITGAV | integrin subunit alpha V | IPA: cell death of colorectal cancer cell lines |
| PRKCI | protein kinase C iota | IPA: cell death of colorectal cancer cell lines |
| TGFBR2 | transforming growth factor beta receptor 2 | IPA: cell death of colorectal cancer cell lines |
| BAG3 | BCL2 associated athanogene 3 | IPA: cell death of colorectal cancer cell lines |
| CTNNB1 | catenin beta 1 | IPA: cell death of colorectal cancer cell lines |
| EGFR | epidermal growth factor receptor | IPA: cell death of colorectal cancer cell lines |
| MYB | MYB proto-oncogene, transcription factor | IPA: cell death of colorectal cancer cell lines |
| CFLAR | CASP8 and FADD like apoptosis regulator | IPA: cell death of colorectal cancer cell lines |
| XIAP | X-linked inhibitor of apoptosis | IPA: cell death of colorectal cancer cell lines |
| MAPK14 | mitogen-activated protein kinase 14 | IPA: cell death of colorectal cancer cell lines |
| RUNX1 | runt related transcription factor 1 | IPA: cell death of colorectal cancer cell lines |
| MAP3K7 | mitogen-activated protein kinase kinase kinase 7 | IPA: cell death of colorectal cancer cell lines |
| ABL1 | ABL proto-oncogene 1, non-receptor tyrosine kinase | IPA: cell death of colorectal cancer cell lines |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 | IPA: cell death of colorectal cancer cell lines |
| RAF1 | Raf-1 proto-oncogene, serine/threonine kinase | IPA: cell death of colorectal cancer cell lines |
| BCL2 | BCL2, apoptosis regulator | IPA: cell death of colorectal cancer cell lines |
| BIRC2 | baculoviral IAP repeat containing 2 | PANTHER Analysis: Involved in apoptosis signaling |
| BAG5 | BCL2 associated athanogene 5 | PANTHER Analysis: negative regulation of apoptotic process |
| BDNF | brain derived neurotrophic factor | PANTHER Analysis: negative regulation of apoptotic process |
| AKT2 | AKT serine/threonine kinase 2 | PANTHER Analysis: negative regulation of apoptotic process |
Fig. 3.
Post-transcriptional regulations of potential miR-217-5p target genes. To experimentally validate miR-217-5p mediated regulation of potential target genes, HCT 116 cells were transfected with miR-217-5p mimic or non-targeting siRNA (NT) as described in Fig. 2. After 48 h, potential target gene expression was analyzed by qRT-PCR (a). The relative mRNA expression of potential target genes was normalized to PPIA and NT. In light blue highlighted candidate genes of (a) were further analyzed by Western blot (b – representative blots; n = 3). Densitometric analyses of the Western Blot were performed using the software FusionCapt Advance. GAPDH was used as loading control. The expression of the respective protein was normalized to NT. Differences in the expression of mRNA or target proteins between miRNA mimic transfected cells and NT transfected cells were accessed using unpaired t-test [n = 3 biological and technical replicates; mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001]
Direct regulation of the MAPK pathway by miR-217-5p in CRC cell lines
In order to test a direct miRNA/mRNA interaction of the downregulated genes described above, we searched for potential binding sites within the 3′ untranslated region (3′ UTR) of the mRNA using microRNA.org, RNA22 (Miranda et al. 2006) and TargetScanHuman (Agarwal et al. 2015). Two predicted miR-217-5p binding sites in the 3′ UTR of the ITGAV mRNA transcript, four predicted miR-217-5p binding sites in the 3′ UTR of the PRKCI mRNA transcript and six predicted binding sites in the 3′ UTR of the MAPK1 mRNA transcript were revealed (Fig. 4a and b). However, no binding sites in the 3′ UTR of MAPK3 mRNA were predicted. To demonstrate a direct miR-217-5p mediated regulatory effect on the potential target genes, each predicted miR-217-5p binding site located in the 3′ UTR of the potential target gene was cloned into the 3′ UTR of a luciferase reporter gene of the pmirGLO Dual-Luciferase miRNA Target Expression Vector. The three binding sides of MAPK1 which are already reported (Zhang et al. 2016) served as controls for the luciferase experiments.
Fig. 4.
Localization of putative miR-217-5p binding sites in selected predicted target genes. Schematic overview of the mRNA composition of PRKCI, BAG3, ITGAV and MAPK1 and with putative miR-217-5p binding sites (blue boxes) in the 3′ untranslated region (3′ UTR) of the respective target gene mRNA (a) and the miRNA:mRNA alignment based on the data in microRNA.org (b)
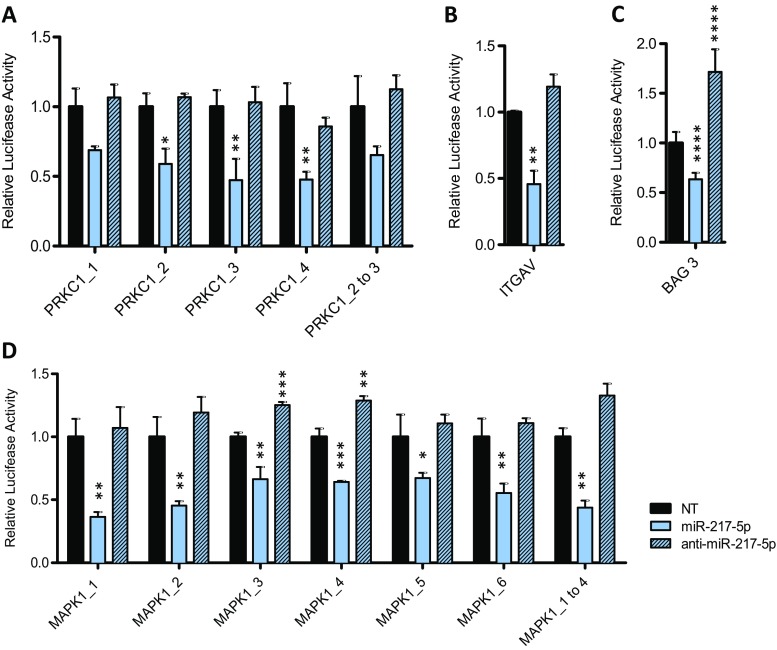
Since well transfectable HEK293T cells are mostly employed for miRNA luciferase reporter analyses (Chaudhuri et al. 2013; Fischer et al. 2014), we accessed the downregulation of relative luciferase activity in this well-established system. After co-transfection with pmirGlo® Dual Luciferase miRNA target expression vectors containing the predicted miRNA binding sites and miR-217-5p mimics, miRNA inhibitor anti-miR-217-5p or a non-targeting siRNA control, relative luciferase activity was determined. As shown in Fig. 5d, relative luciferase activity of the reporter containing the predicted miR-217-5p binding sites of the 3′UTR of MAPK1 mRNA transcript was significantly reduced when co-transfected with miR-217-5p compared to NT. MiR-217-5p binding to MAPK1 with its 6 binding sites, resulted in a comparable decreased luciferase activity of less than 0.36 ± 0.04 observed for binding site 1 of MAPK1. Additionally, binding sites 1 to 4, cloned into one pmirGlo® vector, revealed similar results for downregulated luciferase activity (0.44 ± 0.06). Similar results were obtained when the pmirGlo® Dual luciferase vector contained the predicted miR-217-5p binding sites in the 3′ UTR of PRKCI mRNA transcript (Fig. 5a). Interestingly, no reduction of the luciferase activity beyond that observed with the various single PRKCI binding sites could be achieved with PRKCI_2 to 3. Further, reduced luciferase activity of the reporter containing the predicted miR-217-5p binding site of the 3’UTR of ITGAV mRNA transcript in the presence of miR-217-5p was shown with a reduction to 0.46 ± 0.10 (Fig. 5b). Also for the binding site of BAG3 a direct interaction resulted in the reduction of luciferase activity up to 0.59 ± 0.05 (Fig. 5c). As a control, transfection of miRNA inhibitor anti-miR-217-5p, the complementary miRNA strand to miR-217-5p, revealed luciferase activity comparable to that observed with the non-targeting siRNA control. Occasionally, there was a slight increase in activity when compared to non-targeting siRNA pointing to some basal activity emanating from the endogenous miR-217-5p. Taken together, our data identify PRKCI, ITGAV, BAG3 and MAPK1 as direct targets of miR-217-5p.
Fig. 5.
PRKC1, ITGAV, BAG3 and MAPK1 as direct targets for miR-217-5p in HEK293T cells. Relative luciferase activity 3 days after co-transfection of the pMirGLO vector with the binding sites 1 to 4 and the fusion of binding sites 2 and 3 of PRKC1 (a), the binding sites of ITGAV (b) and BAG3 (c) as well with the binding sites MAPK1_1 to 6 and the fusion of binding sites 1–4 (d) with miR-217-5p mimic, miRNA inhibitor anti-miR-217-5p or non-targeting siRNA (NT). Statistical differences between means were tested using unpaired t-test [n = 3 biological and technical replicates; mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001]
Discussion
The involvement in the regulation of fundamental cellular processes such as apoptosis and the increasing evidence for a potential function as tumor suppressor genes makes miRNAs highly interesting candidate molecules for the generation of novel anticancer therapeutics. Based on previously performed high-throughput screenings for pro-apoptotic miRNAs (Fischer et al. 2014; Kleemann et al. 2017), we were able to identify miR-217-5p as a novel pro-apoptotic miRNA in CRC cell lines. In the current study, we validated the pro-apoptotic potential of miR-217-5p in HCT 116 CRC cells by detecting several molecular changes including the externalization of PS (Fig. 2c), the reduction of the ΔΨm (Fig. 2d), the activation of effector caspases and the cleavage of PARP (Fig. 2e and f), and the fragmentation of cellular DNA (Fig. 2b). Further, we identified a range of target genes whose miR-217-5p-mediated downregulation suggested to be involved in the induction of apoptosis in CRC cells.
Initial determination of the endogenous miR-217-5p and miR-217-3p strand expression level in different CRC cell lines showed a low basal miR-217-5p expression that was elevated by the stimulation with Etoposide, an inducer of the intrinsic apoptotic pathway (Fig. 1b and d). These data are in agreement with the findings of Wang et al. demonstrating a downregulation of miR-217-5p in CRC cell lines compared to the normal human colon mucosal epithelial cell line NCM460 and in CRC tissue samples compared to the respective adjacent noncancerous tissue. Further, low miR-217-5p expression correlates with poor CRC prognosis (Wang et al. 2015a). These findings imply a potential regulatory and inhibitory role for miR-217-5p in apoptosis and tumorigenesis.
Since miRNAs are known to have the potential to control a multiplicity of target genes, the aim of this study was to identify the target gene network regulated by miR-217-5p possibly underlying the observed pro-apoptotic mechanisms. By in silico target prediction and experimental validation we identified a complex network of genes to be regulated by miR-217-5p and further confirmed several direct target genes involved in apoptosis. MiR-217-5p was identified to directly regulate the ERK-MAPK signaling pathway by repressing MAPK1 directly via six binding sites in the 3′ UTR leading to direct or indirect MAPK1/3 mRNA downregulation. In addition to previously reported three binding sides (MAPK1_1 to _3; (Zhang et al. 2016)), we were able to identify three more direct binding sites of miR-217-5p, all of them leading to significantly reduced luciferase activity (Fig. 5). Since the stimulation of the ERK-MAPK pathway leads to the activation of survival and proliferation promoting transcription factors, including proto-oncogene c-myc (Zhang and Liu 2002), Ets like protein 1 (Elk1) or proto-oncogene c-Fos (Klein and Assoian 2008) and the stabilization of anti-apoptotic Bcl-2 family members such as Mcl-1 (Thomas et al. 2010) or the destabilization of pro-apoptotic molecules like BH3-only molecules (Akiyama et al. 2009), the miR-217-5p mediated downregulation of MAPK1/3 suggests a pathway mediating the observed pro-apoptotic effects (Fig. 6).
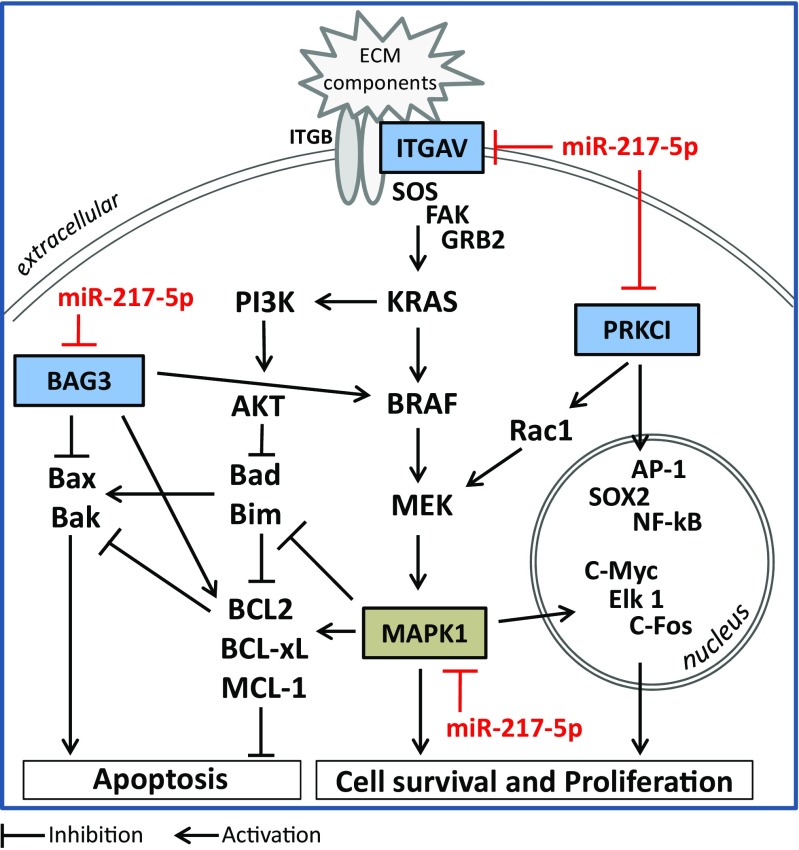
Fig. 6.
Potential pro-apoptotic mechanisms of miR-217-5p regulating the ERK-MAPK pathway at different sites. MiR-217-5p was shown to directly downregulate PRKCI, ITGAV, BAG3 and MAPK1, connected in a signaling network modulating the ERK-MAPK pathway. Binding of extracellular matrix components to integrins comprising a β (ITGB) and α subunit such as αv (ITGAV) may activate the ERK- MAPK signaling pathway via focal adhesion kinase (FAK) activation, growth factor receptor-bound protein 2 (GRB2), and guanine nucleotide exchange factor son of sevenless (SOS), leading to the activation of the kinase cascade including KRAS, BRAF, MEK, and MAPK1 and the activation of, Phosphoinositide 3-kinase (PI3K)/Akt. These pathways culminate in the activation of survival and proliferation promoting transcription factors including c- myc, c-fos or Ets like protein 1 (Elk1), in the induction and stabilization of anti-apoptotic members (Bcl-2, Bcl-xL, Mcl-1) of the Bcl-2 protein family and the inhibition of pro-apoptotic members including the BH3-only members Bad and Bim promoting the initiation of intrinsic apoptosis via Bax and Bak. PRKCI, ITGAV, BAG3 and MAPK1 promote cell survival and proliferation by induction of transcription factors including NK-κB, AP1 or SOX2, or by induction of the ERK-MAPK pathway via Rac1
Apart from directly influencing the ERK-MAPK signaling pathway by negatively regulating MAPK1/3) (Fig. 5; (Zhang et al. 2016)), we were able to show that miR-217-5p indeed regulates this pathway on multiple levels by additionally targeting the transmembrane protein ITGAV, the cytoplasmic localized BAG3 and the transcriptional activator PKCι/λ (Fig. 6). Integrins are a family of transmembrane receptors composed of two subunits, α and β. These receptors facilitate interactions between cells and the extracellular matrix, participate in cytoskeleton organization and play important roles in cell signaling (Stupack and Cheresh 2002). Upon ligand binding, the PI3K/Akt and ERK- MAPK signaling pathways are stimulated resulting in increased proliferation and survival of the cell (Fu et al. 2015; Stupack and Cheresh 2002). Thus, downregulation of integrin αV may subsequently lead to the induction of apoptosis, which has been shown in laryngeal cancer cells by the use of antisense oligonucleotide mediated repression of integrin αV leading to inhibition of proliferation and induction of apoptosis (Lu et al. 2009). In addition, dysregulated integrin expression has been associated with tumor cell growth as well as metastasis. In fact, integrin αV expression levels were demonstrated to be increased in non-small lung cancer (Fu et al. 2015), cervical squamous cell carcinoma (Hazelbag et al. 2007), laryngeal and hypopharyngeal squamous cell carcinoma (Lu et al. 2009) and to correlate with enhanced proliferation, invasion, metastasis and poor clinical outcome. These data again highlight the relevance of miRNAs as negative regulators and potential cancer therapeutics. The use of negative regulators such as miR-217-5p may display therapeutic potential especially in addition to the use of monoclonal antibodies as e.g. the anti-integrin αV antibody etaracizumab, which resulted in decreased ovarian cancer proliferation and invasion (Landen et al. 2008). Further, this combined approach might allow for lower therapeutic antibody concentration with reduced side effects.
Another direct target gene of miR-217-5p and promoter of the ERK-MAPK signaling pathway is BAG3. This co-chaperone was demonstrated to interact with BRAF to stabilize and protect it from proteasomal degradation (Chiappetta et al. 2007) and to be overexpressed and involved in the pathogenesis and progression of different types of cancer, including ovarian carcinoma (Suzuki et al. 2011), pancreatic adenocarcinoma (Rosati et al. 2012), hepatocellular carcinoma (Xiao et al. 2014), and CRC (Shi et al. 2016; Yang et al. 2013). Apart from stimulating the ERK-MAPK pathway via BRAF (Chiappetta et al. 2007) and facilitating the nuclear factor-kappa B (NF-κB) signaling by protecting IKKγ from proteasomal degradation (Ammirante et al. 2010), BAG3 was found to interact with members of the Bcl-2 protein family, including Bax to prevent its translocation to the mitochondrial outer membrane (Festa et al. 2011) and the anti-apoptotic family members Mcl-1 (Boiani et al. 2013), Bcl-xL and Bcl-2 (Jacobs and Marnett 2009; Zhang et al. 2012) resulting in their stabilization, the inhibition of apoptosis and resistance apoptosis-inducing therapeutic approaches (Chiappetta et al. 2007) implicating the benefit of miR-217-5p mediated BAG3 downregulation.
Finally, we identified the PRKCI encoding the protein PKCι/λ as a direct target gene of miR-217-5p. PKCι/λ was shown to play a pivotal role in cell proliferation and differentiation by activating transcription factors such as sex determining region Y-box 2 (SOX2) (Justilien et al. 2014), NF-κB, and activating protein-1 (AP-1) (Ishiguro et al. 2009) or by connecting to the MAPK-ERK pathway via Rac-1 (Regala et al. 2005; Scotti et al. 2010; Zhang et al. 2004). In addition, PKCι/λ was implicated in the carcinogenesis of several malignancies, including lung squamous cell carcinoma (Justilien et al. 2014), tongue squamous cell carcinoma (Song et al. 2014), and pancreatic adenocarcinoma (Scotti et al. 2010). Apart from miR-217-5p target genes in CRC cells identified within this study, only Wang et al. identified another direct target, astrocyte-elevated gene-1/ Metadherin to be directly downregulated by mir-217-5p in CRC (Wang et al. 2015a). Taken together, the collective post-transcriptional regulation of these target genes may mediate the pro-apoptotic effect of miR-217-5p.
Considering that the miRNA-mediated overall effect is dependent on the tissue specific expression of the different miRNA target genes, further target genes of miR-217-5p were identified by others in different tumor entities and may also be involved in the observed pro-apoptotic effect of miR-217-5p in CRC. In this regard, miR-217-5p was shown to directly target KRAS in lung cancer and pancreatic adenocarcinoma (Zhao et al. 2010), WAS Protein Family Member 3 in osteosarcoma (Shen et al. 2014), Insulin Like Growth Factor 1 Receptor in epithelial ovarian cancer (Li et al. 2016), Runt related transcription factor 2 in glioblastoma (Zhu et al. 2016), as well as enhancer of zeste homolog 2 (Chen et al. 2015) and glypican 5 (Wang et al. 2015b) in gastric cancer.
In summary, miR-217-5p was validated to induce apoptosis in CRC cells. A panel of multiple novel target genes was identified, contributing to the understanding of the molecular mechanisms of network regulation by miR-217-5p mediated apoptosis.
Acknowledgements
HCT 116 cells were kindly provided by Prof. Dr. Verena Jendrossek, IFZ, University of Duisburg-Essen and HT-29 and SW480 were kindly provided by Prof. Dr. Uwe Knippschild, University Hospital Ulm.
Abbreviations
- AEG-1
Astrocyte-elevated gene-1
- AP-1
Activating protein-1
- BAG3
BAG family molecular chaperone regulator 3
- BIRC3
Baculoviral IAP repeat containing 2
- BRCA1
BRCA1 DNA repair associated
- CCCP
Carbonyl cyanide m-chlorophenyl hydrazine
- CRC
Colorectal cancer
- CTNNB1
Catenin beta 1
- DMSO
Dimethyl sulfoxide
- DT
Death inducing
- EGFR
Epidermal growth factor receptor
- ElK1
Ets like protein 1
- ERK
Extracellular-signal regulated kinase
- FCS
Fetal calf serum
- FITC
Fluorescein isothiocyanate
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- HRP
Horseradish peroxidase
- ITGAV
Integrin subunit alpha v
- MAP K1
Mitogen-activated protein kinase 1
- miR
MicroRNA
- MMP
Mitochondrial membrane potential
- MYC
Transcriptional activator MYC
- NF-kB
Nuclear factor-kappa B
- NT
Non-targeting
- PARP
Poly (ADP-ribose) polymerase
- PBS
Phosphate-buffered saline
- PI
Propidium iodide
- PIK3CA
Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha
- PPIA
Peptidylprolyl isomerase A
- PRKCI
Protein kinase c iota type I
- PS
Phosphatidylserine
- RAC1
Ras-related C3 botulinum toxin substrate 1
- RAF1
Raf-1 proto-oncogene, serine/threonine kinase
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- siRNA
Small interfering RNA
- SOX2
Sex determining region Y-box 2
- TGFBR2
Transforming growth factor beta receptor 2
- TMRE
Tetramethylrhodamine ethyl ester perchlorate
- TRAIL
Tumor necrosis factor related apoptosis inducing ligand
- UTR
Untranslated region
Funding
This study was funded by the Postgraduate Scholarships Act of the Ministry for Science, Research and Arts of the federal state government of Baden-Wuerttemberg, Germany. Further acknowledgements address the International Graduate School in Molecular Medicine of Ulm University, Germany, for scientific encouragement and support to Michael Kleemann.
Compliance with ethical standards
Conflicts of interest
The authors declare no conflict of interest.
Footnotes
Marion Flum and Michael Kleemann contributed equally to this work.
References
- Agarwal V, Bell GW, Nam JW, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife 4. 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed]
- Akiyama T, Dass CR, Choong PF. Bim-targeted cancer therapy: a link between drug action and underlying molecular changes. Mol Cancer Ther. 2009;8:3173–3180. doi: 10.1158/1535-7163.MCT-09-0685. [DOI] [PubMed] [Google Scholar]
- Ammirante M, Rosati A, Arra C, Basile A, Falco A, Festa M, Pascale M, d'Avenia M, Marzullo L, Belisario MA, et al. IKK{gamma} protein is a target of BAG3 regulatory activity in human tumor growth. Proc Natl Acad Sci U S A. 2010;107:7497–7502. doi: 10.1073/pnas.0907696107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG (2012) miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet, 3:120 [DOI] [PMC free article] [PubMed]
- Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiani M, Daniel C, Liu X, Hogarty MD, Marnett LJ. The stress protein BAG3 stabilizes Mcl-1 protein and promotes survival of cancer cells and resistance to antagonist ABT-737. J Biol Chem. 2013;288:6980–6990. doi: 10.1074/jbc.M112.414177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breving K, Esquela-Kerscher A. The complexities of microRNA regulation: mirandering around the rules. Int J Biochem Cell Biol. 2010;42:1316–1329. doi: 10.1016/j.biocel.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Cekaite L, Eide PW, Lind GE, Skotheim RI, Lothe RA. MicroRNAs as growth regulators, their function and biomarker status in colorectal cancer. Oncotarget. 2016;7:6476–6505. doi: 10.18632/oncotarget.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri AD, Yelamanchili SV, Marcondes MC, Fox HS. Up-regulation of microRNA-142 in simian immunodeficiency virus encephalitis leads to repression of sirtuin1. FASEB J. 2013;27:3720–3729. doi: 10.1096/fj.13-232678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DL, Zhang DS, Lu YX, Chen LZ, Zeng ZL, He MM, Wang FH, Li YH, Zhang HZ, Pelicano H, et al. microRNA-217 inhibits tumor progression and metastasis by downregulating EZH2 and predicts favorable prognosis in gastric cancer. Oncotarget. 2015;6:10868–10879. doi: 10.18632/oncotarget.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappetta G, Ammirante M, Basile A, Rosati A, Festa M, Monaco M, Vuttariello E, Pasquinelli R, Arra C, Zerilli M, et al. The antiapoptotic protein BAG3 is expressed in thyroid carcinomas and modulates apoptosis mediated by tumor necrosis factor-related apoptosis-inducing ligand. J Clin Endocrinol Metab. 2007;92:1159–1163. doi: 10.1210/jc.2006-1712. [DOI] [PubMed] [Google Scholar]
- Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44:D239–D247. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregan SP, Dawson VL, Slack RS. Role of AIF in caspase-dependent and caspase-independent cell death. Oncogene. 2004;23:2785–2796. doi: 10.1038/sj.onc.1207517. [DOI] [PubMed] [Google Scholar]
- Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, Starling N. Colorectal cancer. Lancet. 2010;375:1030–1047. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- Debarros M, Steele SR. Colorectal cancer screening in an equal access healthcare system. J Cancer. 2013;4:270–280. doi: 10.7150/jca.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweep H, Sticht C, Pandey P, Gretz N. miRWalk--database: prediction of possible miRNA binding sites by "walking" the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festa M, Del Valle L, Khalili K, Franco R, Scognamiglio G, Graziano V, De Laurenzi V, Turco MC, Rosati A. BAG3 protein is overexpressed in human glioblastoma and is a potential target for therapy. Am J Pathol. 2011;178:2504–2512. doi: 10.1016/j.ajpath.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S, Buck T, Wagner A, Ehrhart C, Giancaterino J, Mang S, Schad M, Mathias S, Aschrafi A, Handrick R, Otte K. A functional high-content miRNA screen identifies miR-30 family to boost recombinant protein production in CHO cells. Biotechnol J. 2014;9:1279–1292. doi: 10.1002/biot.201400306. [DOI] [PubMed] [Google Scholar]
- Fischer S, Handrick R, Aschrafi A, Otte K. Unveiling the principle of microRNA-mediated redundancy in cellular pathway regulation. RNA Biol. 2015;12:238–247. doi: 10.1080/15476286.2015.1017238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S, Fan L, Pan X, Sun Y, Zhao H. Integrin alphav promotes proliferation by activating ERK 1/2 in the human lung cancer cell line A549. Mol Med Rep. 2015;11:1266–1271. doi: 10.3892/mmr.2014.2860. [DOI] [PubMed] [Google Scholar]
- Gottlieb RA, Granville DJ. Analyzing mitochondrial changes during apoptosis. Methods. 2002;26:341–347. doi: 10.1016/S1046-2023(02)00040-3. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Hazelbag S, Kenter GG, Gorter A, Dreef EJ, Koopman LA, Violette SM, Weinreb PH, Fleuren GJ. Overexpression of the alpha v beta 6 integrin in cervical squamous cell carcinoma is a prognostic factor for decreased survival. J Pathol. 2007;212:316–324. doi: 10.1002/path.2168. [DOI] [PubMed] [Google Scholar]
- Ishiguro H, Akimoto K, Nagashima Y, Kojima Y, Sasaki T, Ishiguro-Imagawa Y, Nakaigawa N, Ohno S, Kubota Y, Uemura H. aPKClambda/iota promotes growth of prostate cancer cells in an autocrine manner through transcriptional activation of interleukin-6. Proc Natl Acad Sci U S A. 2009;106:16369–16374. doi: 10.1073/pnas.0907044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs AT, Marnett LJ. HSF1-mediated BAG3 expression attenuates apoptosis in 4-hydroxynonenal-treated colon cancer cells via stabilization of anti-apoptotic Bcl-2 proteins. J Biol Chem. 2009;284:9176–9183. doi: 10.1074/jbc.M808656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic M, Hengartner MO. miRNAs and apoptosis: RNAs to die for. Oncogene. 2006;25:6176–6187. doi: 10.1038/sj.onc.1209912. [DOI] [PubMed] [Google Scholar]
- Justilien V, Walsh MP, Ali SA, Thompson EA, Murray NR, Fields AP. The PRKCI and SOX2 oncogenes are coamplified and cooperate to activate hedgehog signaling in lung squamous cell carcinoma. Cancer Cell. 2014;25:139–151. doi: 10.1016/j.ccr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kleemann M, Bereuther J, Fischer S, Marquart K, Hänle S, Unger K, Jendrossek V, Riedel CU, Handrick R, Otte K (2017) Investigation on tissue specific effects of pro-apoptotic micro RNAs revealed miR-147b as a potential biomarker in ovarian cancer prognosis. Oncotarget 8(12):18773–18791. 10.18632/oncotarget.13095 [DOI] [PMC free article] [PubMed]
- Klein EA, Assoian RK. Transcriptional regulation of the cyclin D1 gene at a glance. J Cell Sci. 2008;121:3853–3857. doi: 10.1242/jcs.039131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landen CN, Kim TJ, Lin YG, Merritt WM, Kamat AA, Han LY, Spannuth WA, Nick AM, Jennnings NB, Kinch MS, et al. Tumor-selective response to antibody-mediated targeting of alphavbeta3 integrin in ovarian cancer. Neoplasia. 2008;10:1259–1267. doi: 10.1593/neo.08740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Cheong HJ, Kim SJ, Yoon J, Kim HJ, Kim KH, Kim SH, Kim HJ, Bae SB, Kim CK, et al. Low-dose combinations of LBH589 and TRAIL can overcome TRAIL-resistance in colon cancer cell lines. Anticancer Res. 2011;31:3385–3394. [PubMed] [Google Scholar]
- Li J, Li D, Zhang W. Tumor suppressor role of miR-217 in human epithelial ovarian cancer by targeting IGF1R. Oncol Rep. 2016;35:1671–1679. doi: 10.3892/or.2015.4498. [DOI] [PubMed] [Google Scholar]
- Lima RT, Busacca S, Almeida GM, Gaudino G, Fennell DA, Vasconcelos MH. MicroRNA regulation of core apoptosis pathways in cancer. Eur J Cancer. 2011;47:163–174. doi: 10.1016/j.ejca.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Liu M, Lang N, Chen X, Tang Q, Liu S, Huang J, Zheng Y, Bi F. miR-185 targets RhoA and Cdc42 expression and inhibits the proliferation potential of human colorectal cells. Cancer Lett. 2011;301:151–160. doi: 10.1016/j.canlet.2010.11.009. [DOI] [PubMed] [Google Scholar]
- Lu JG, Sun YN, Wang C, Jin DJ, Liu M. Role of the alpha v-integrin subunit in cell proliferation, apoptosis and tumor metastasis of laryngeal and hypopharyngeal squamous cell carcinomas: a clinical and in vitro investigation. Eur Arch Otorhinolaryngol. 2009;266:89–96. doi: 10.1007/s00405-008-0675-z. [DOI] [PubMed] [Google Scholar]
- Lynam-Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos Soc. 2009;84:55–71. doi: 10.1111/j.1469-185X.2008.00061.x. [DOI] [PubMed] [Google Scholar]
- MacFarlane M. TRAIL-induced signalling and apoptosis. Toxicol Lett. 2003;139:89–97. doi: 10.1016/S0378-4274(02)00422-8. [DOI] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Montecucco A, Zanetta F, Biamonti G. Molecular mechanisms of etoposide. EXCLI J. 2015;14:95–108. doi: 10.17179/excli2015-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss JA. Gene therapy review. Radiol Technol. 2014;86:155–180. [PubMed] [Google Scholar]
- Perry SW, Norman JP, Barbieri J, Brown EB, Gelbard HA. Mitochondrial membrane potential probes and the proton gradient: a practical usage guide. BioTechniques. 2011;50:98–115. doi: 10.2144/000113610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qaseem A, Denberg TD, Hopkins RH, Jr, Humphrey LL, Levine J, Sweet DE, Shekelle P, Clinical Guidelines Committee of the American College of P Screening for colorectal cancer: a guidance statement from the American College of Physicians. Ann Intern Med. 2012;156:378–386. doi: 10.7326/0003-4819-156-5-201203060-00010. [DOI] [PubMed] [Google Scholar]
- Ragnhammar P, Hafstrom L, Nygren P, Glimelius B, Care SB-gSCoTAiH A systematic overview of chemotherapy effects in colorectal cancer. Acta Oncol. 2001;40:282–308. doi: 10.1080/02841860151116367. [DOI] [PubMed] [Google Scholar]
- Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: integrating information about genes, proteins and diseases. Trends Genet. 1997;13:163. doi: 10.1016/S0168-9525(97)01103-7. [DOI] [PubMed] [Google Scholar]
- Regala RP, Weems C, Jamieson L, Khoor A, Edell ES, Lohse CM, Fields AP. Atypical protein kinase C iota is an oncogene in human non-small cell lung cancer. Cancer Res. 2005;65:8905–8911. doi: 10.1158/0008-5472.CAN-05-2372. [DOI] [PubMed] [Google Scholar]
- Rosati A, Bersani S, Tavano F, Dalla Pozza E, De Marco M, Palmieri M, De Laurenzi V, Franco R, Scognamiglio G, Palaia R, et al. Expression of the antiapoptotic protein BAG3 is a feature of pancreatic adenocarcinoma and its overexpression is associated with poorer survival. Am J Pathol. 2012;181:1524–1529. doi: 10.1016/j.ajpath.2012.07.016. [DOI] [PubMed] [Google Scholar]
- Scotti ML, Bamlet WR, Smyrk TC, Fields AP, Murray NR. Protein kinase Ciota is required for pancreatic cancer cell transformed growth and tumorigenesis. Cancer Res. 2010;70:2064–2074. doi: 10.1158/0008-5472.CAN-09-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Wang P, Yang J, Li X. MicroRNA-217 regulates WASF3 expression and suppresses tumor growth and metastasis in osteosarcoma. PLoS One. 2014;9:e109138. doi: 10.1371/journal.pone.0109138. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Shi H, Xu H, Li Z, Zhen Y, Wang B, Huo S, Xiao R, Xu Z. BAG3 regulates cell proliferation, migration, and invasion in human colorectal cancer. Tumour Biol. 2016;37:5591–5597. doi: 10.1007/s13277-015-4403-1. [DOI] [PubMed] [Google Scholar]
- Slaby O, Svoboda M, Michalek J, Vyzula R. MicroRNAs in colorectal cancer: translation of molecular biology into clinical application. Mol Cancer. 2009;8:102. doi: 10.1186/1476-4598-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song KB, Liu WJ, Jia SS. miR-219 inhibits the growth and metastasis of TSCC cells by targeting PRKCI. Int J Clin Exp Med. 2014;7:2957–2965. [PMC free article] [PubMed] [Google Scholar]
- Strasser A, O'Connor L, Dixit VM. Apoptosis signaling. Annu Rev Biochem. 2000;69:217–245. doi: 10.1146/annurev.biochem.69.1.217. [DOI] [PubMed] [Google Scholar]
- Stupack DG, Cheresh DA. Get a ligand, get a life: integrins, signaling and cell survival. J Cell Sci. 2002;115:3729–3738. doi: 10.1242/jcs.00071. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Iwasaki M, Sugio A, Hishiya A, Tanaka R, Endo T, Takayama S, Saito T. BAG3 (BCL2-associated athanogene 3) interacts with MMP-2 to positively regulate invasion by ovarian carcinoma cells. Cancer Lett. 2011;303:65–71. doi: 10.1016/j.canlet.2011.01.019. [DOI] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LW, Lam C, Edwards SW. Mcl-1; the molecular regulation of protein function. FEBS Lett. 2010;584:2981–2989. doi: 10.1016/j.febslet.2010.05.061. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Vlachos IS, Paraskevopoulou MD, Karagkouni D, Georgakilas G, Vergoulis T, Kanellos I, Anastasopoulos IL, Maniou S, Karathanou K, Kalfakakou D, et al. DIANA-TarBase v7.0: Indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015;43:D153–D159. doi: 10.1093/nar/gku1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- Wang S, El-Deiry WS. TRAIL and apoptosis induction by TNF-family death receptors. Oncogene. 2003;22:8628–8633. doi: 10.1038/sj.onc.1207232. [DOI] [PubMed] [Google Scholar]
- Wang B, Shen ZL, Jiang KW, Zhao G, Wang CY, Yan YC, Yang Y, Zhang JZ, Shen C, Gao ZD, et al. MicroRNA-217 functions as a prognosis predictor and inhibits colorectal cancer cell proliferation and invasion via an AEG-1 dependent mechanism. BMC Cancer. 2015;15:437. doi: 10.1186/s12885-015-1438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Dong X, Gu X, Qin R, Jia H, Gao J. The MicroRNA-217 functions as a potential tumor suppressor in gastric cancer by targeting GPC5. PLoS One. 2015;10:e0125474. doi: 10.1371/journal.pone.0125474. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Cheng S, Tong R, Lv Z, Ding C, Du C, Xie H, Zhou L, Wu J, Zheng S. BAG3 Regulates epithelial-mesenchymal transition and angiogenesis in human hepatocellular carcinoma. Lab Investig. 2014;94:252–261. doi: 10.1038/labinvest.2013.151. [DOI] [PubMed] [Google Scholar]
- Yang X, Tian Z, Gou WF, Takahashi H, Yu M, Xing YN, Takano Y, Zheng HC. Bag-3 expression is involved in pathogenesis and progression of colorectal carcinomas. Histol Histopathol. 2013;28:1147–1156. doi: 10.14670/HH-28.1147. [DOI] [PubMed] [Google Scholar]
- Yang QS, Jiang LP, He CY, Tong YN, Liu YY (2017) Up-regulation of MicroRNA-133a inhibits the MEK/ERK signaling pathway to promote cell apoptosis and enhance radio-sensitivity by targeting EGFR in esophageal cancer in vivo and in vitro. J Cell Biochem 118(9):2625–2634. 10.1002/jcb.25829 [DOI] [PubMed]
- Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- Zhang J, Anastasiadis PZ, Liu Y, Thompson EA, Fields AP. Protein kinase C (PKC) betaII induces cell invasion through a Ras/Mek-, PKC iota/Rac 1-dependent signaling pathway. J Biol Chem. 2004;279:22118–22123. doi: 10.1074/jbc.M400774200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang JH, Lu Q, Wang YJ. Bag3 promotes resistance to apoptosis through Bcl-2 family members in non-small cell lung cancer. Oncol Rep. 2012;27:109–113. doi: 10.3892/or.2011.1486. [DOI] [PubMed] [Google Scholar]
- Zhang W, Liu K, Liu S, Ji B, Wang Y, Liu Y. MicroRNA-133a functions as a tumor suppressor by targeting IGF-1R in hepatocellular carcinoma. Tumour Biol. 2015;36:9779–9788. doi: 10.1007/s13277-015-3749-8. [DOI] [PubMed] [Google Scholar]
- Zhang N, Lu C, Chen L. miR-217 regulates tumor growth and apoptosis by targeting the MAPK signaling pathway in colorectal cancer. Oncol Lett. 2016;12:4589–4597. doi: 10.3892/ol.2016.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM, Chen J. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis. 2010;31:1726–1733. doi: 10.1093/carcin/bgq160. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Zhao H, Feng L, Xu S. MicroRNA-217 inhibits cell proliferation and invasion by targeting Runx2 in human glioma. Am J Transl Res. 2016;8:1482–1491. [PMC free article] [PubMed] [Google Scholar]
- Zimmermann KC, Bonzon C, Green DR. The machinery of programmed cell death. Pharmacol Ther. 2001;92:57–70. doi: 10.1016/S0163-7258(01)00159-0. [DOI] [PubMed] [Google Scholar]