Abstract
Lung cancer is a leading cause of cancer mortality worldwide. Promoter methylation of transcription factor 21 (TCF21) was frequently observed in the early stage of non-small cell lung cancer (NSCLC). However, clinical relevance and molecular functions of TCF21 in NSCLC progression remain unclear. In this study, we analyzed the associations between TCF21 expression and clinicopathological features in 100 patients with NSCLC and revealed the underlying molecular mechanisms of TCF21 methylation on cell viability, apoptosis and invasion of H1299 cells. We found that the expression of TCF21 was significantly regulated by its methylation level in patients with NSCLC and was associated with tumor stage, metastasis and invasion. Demethylation of H1299 cells by 5-aza-2′-deoxycytine (5-Aza) demonstrated that a higher level of TCF21 expression led to remarkable decreases of cell viability and invasion ability but an increase of cell apoptosis. Accordingly, TCF21 knockdown showed converse results to high expression of TCF21. TCF21 knockdown cells exhibited significantly upregulated ATG-9, BECLIN-1, and LC3-I/II expressions but decreased p62 expression compared to wildtype cells. Inhibition of autophagy by 3-methyladenine (3-MA) elevated TCF21 expression and increased cell apoptosis. TCF21 expression is clinically related to the progress of lung cancer and may inhibit autophagy by suppressing ATG-9 and BECLIN-1. In turn, autophagy may also play an important role in regulation TCF21 expression.
Electronic supplementary material
The online version of this article (10.1007/s12079-017-0418-2) contains supplementary material, which is available to authorized users.
Keywords: Transcription factor 21 (TCF21), Methylation, Non-small cell lung cancer (NSCLC), Autophagy
Introduction
Lung cancer is a leading cause of cancer mortality worldwide. During last two decades, a steady increase in the lung cancer survival rate was observed in United States (Siegel et al. 2016) as a result of less tobacco use (Jemal et al. 2008) and development of earlier detection using low-dose computed tomographic (Aberle et al. 2011). However, lung cancer remains the most common diagnosed cancer with high mortality rate in China where accounts for up to 50% of global burden (Ferlay et al. 2004; Howlader et al. 2015; Siegel et al. 2016). It is mainly due to the continuing use of x-rays screening detection and long term of tobacco exposure (Hong et al. 2015). Lung cancer is divided into two subtypes, small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). More than 85% are NSCLC which is less aggressive than SCLC. Efficient early detection methods may remarkably improve the survival rate of patients with NSCLC.
DNA methylation has emerged as a potential source of biomarkers for different cancers. Numerous genes become hypermethylated in patients with cancers, leading to the silence of the genes, among which some are tumor suppressor genes (Ahmad et al. 2017). This hypermethylation often occurs in a region containing repeats of phosphate-linked cytosine and guanine nucleotides, the ‘CpG’ islands. Among all of the about 45,000 CpG islands per haploid genome, up to 90% are methylated. Methylation status is associated with repression or promotion of gene expressions. Thus, by comparing gene methylation profiles between tumor and non-tumor tissues, several cancers such as breast, lung, and prostate cancers show significant differences in gene methylation (Conway et al. 2015; Tsou et al. 2007; Wang et al. 2012). In 2006, Smith et al. evidenced that the transcription factor 21 (TCF21) is inactivated by aberrant promoter hypermethylation in head and neck, and lung cancers (Smith et al. 2006). Restoration of TCF21 expression led to a significant decrease of proliferation of lung cancer cells and tumor growth in a mouse xenograft model.
TCF21 is a specific class II basic helix-loop-helix transcription factor that is encoded TCF21 gene located on chromosome 6q23–q24. TCF21 was firstly identified to enhance the differentiation of mesenchymal cells into epithelial cells and plays important roles in embryonic development. TCF21 expression level is high during embryonic development and loss of TCF21 gene results in perinatal death due to its role in the development of kidney and lung (Guarino et al. 2007). After birth, its expression rapidly decreased in most tissues but maintained high in interstitial cells in several organs such as lung, intestine and kidney (Quaggin et al. 1999). Promoter methylation of TCF21 was frequently observed in the early stage of NSCLC (Richards et al. 2011), leading TCF21 to a potential candidate biomarker of lung cancer. However, clinical relevance and molecular functions of TCF21 in NSCLC progression remain unclear. In this study, we assessed methylation status of patients with lung cancer and found that the methylation level of TCF21 was associated with tumor progression. Deeper insights into the molecular mechanism were investigated.
Materials and methods
Patients and sample collections
A total of 100 patients who were diagnosed with NSCLC between June 2010 and October 2014 at the Peking University Shenzhen Hospital were enrolled in this study. Informed consent was signed by each patient. Medical records including age, gender and pathology outcomes such as tumor size, stage, metastasis, invasion were used for analysis in this study. Diagnosis was confirmed by at least two pathologists and staging was achieved by the results of hematoxylin and eosin stain according to the World Health Organization Classification of Tumors. All specimens were collected under the protocol approved by Peking University Shenzhen Hospital and fixed with formalin or embedded with paraffin. For each patient, we obtained 4 puncture points from each case of lung cancer tissue.
Cell culture and treatments
A549 and H1299 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA) and 100 U/mL streptomycin and 100 mg/mL penicillin at 37 °C in a humidified atmosphere with 5% CO2. A549 and H1299 cells were treated with or without 5 μM of 5-aza-2′-Deoxycytidine (5-Aza, Sigma–Aldrich, St Louis, MO, USA) for 3 days and 5-Aza was replenished every 24 h. For 3-methyladenine (3-MA, Sigma-Aldrich, USA) treatment, cells were incubated with or without 3 mmol/L 3-MA for 2 days.
Immunohistochemistry
Immunohistochemical analysis employed 4 μm sections sliced from the specimens. The sections were deparaffinized by Xylene and subjected to antigen retrieval in 10 mmol/L of sodium citrate for 30 min in a boiling water bath. Then, the sections were incubated with a polyclonal rabbit antihuman TCF21 antibody (#ab32981, Abcam, Cambridge, UK), overnight at 4 °C, and then incubated with a goat antirabbit Envision System Plus-HRP (Dako Cytomation, Carpinteria, CA, USA) for 30 min at room temperature. After washed with PBS for 3 times, the sections were stained by DAB for 1 min and then counterstained with Mayer hematoxylin (Sigma-Aldrich, USA). Serial sections were selected for hematoxylin and eosin staining as reference. According to the staining results, the intensity was estimated by compared to the control and percentage of positive-stained cells was calculated. The scores were allocated as follow: 0: negative (0%), 1: weak (1–25%), 2: moderate (25–50%), 3: strong (51–100%). For statistical analysis, we divided the TCF21 expression into low expression with scores of 0–2 and high expression with a score of 3.
RNA extraction and quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted using a Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The purity and quality of RNA were assessed by NanoDrop system (Thermo Scienti c, Wilmington, DE, USA). Reverse transcription to cDNA was performed using a PrimeScript™ 1st Strand cDNA Synthesis Kit (Takara Bio, Otsu, Japan). SYBR green qPCR assay (BioRad, Hercules, CA, USA) was used to detect the mRNA level of TCF21 on ABI 7500 system (Applied Biosystems, Foster City, CA, USA). β-actin was used as control to normalize the starting quantity of RNA. Primers used for TCF21 and β-actin are as follows: TCF21 Forward: 5′-GCCTTCTCCAGACTCAAGACCAC-3′, Reverse: 5′-CATAAAGGGCCACGTCAGGTTG-3′; β-actin Forward: 5′-GTCATTCCAAATATGAGATGCGT-3′, Reverse: 5′-GCTATCACCTCCCCTGTGTG-3′.
DNA extract, methylation-specific PCR and global DNA methylation
DNA from tissues or cell line was extracted Wizard Genomic DNA purification kit (Promega, Madison, WI, USA). Bisulfite converted with the kit EZ DNA Methylation-Gold Kit (Zymo Research, CA, USA) according to the manufacturer’s protocol. Methylation-specific PCR was employed for detection of TCF21 promoter methylation. The primers for methylated DNA are as follow: forward, 5′-TTTGGTTAACGATAAATACGAGAACG-3′, reverse, 5′-CCTAAAAACTCTAAACCCGCGAT-3′, which produced a 198-bp band. The primers for unmethylated DNA are as follow: forward, 5′-TTTGGTTAATGATAAAT ATGAGAATGG-3′, reverse, 5′-TCCCTAAAAACTCTAAACCCACAAT-3′ (antisense), which produced a 200-bp band. The reaction mix comprised 0.5 μl bisulfited DNA under a condition of 94 °C for 5 min followed by 35 cycles of 95 °C for 30 s, 52 °C for 30 s, 72 °C for 40 s, and 72 °C for 10 min. PCR products were subjected to 2% agarose gel electrophoresis at 120 V for 40 min. For global methylation detection, liquid chromatography-electrospray ionization/multi-stage mass spectrometry (LC-ESI/MS/MS) technique was employed as described previously stated using an Agilent 1200 series HPLC system (Dwi Putra et al. 2014).
Cell viability and apoptosis assays
Cell viability was measured using the Cell Counting Kit-8 (Dojindo, Kamimashiki-gun Kumamoto, Japan). In brief, after treatment, cells were incubated in fresh medium containing 10% CCK8 at 37 °C for 2 h. The results were detected at the absorbance of 450 nm using the Microplate Reader (Thermo Fisher, Waltham, Massachusetts, USA). For cell apoptosis assay, Annexin V-FITC Apoptosis Detection kit (BD Biosciences, San Diego, CA, USA) was used to measure the cell apoptosis ratio according to the manufacturer’s instructions. Protein levels of cleaved PARP and Caspase-3 were detected by western blotting.
Activity of autophagy by testing p62 was verified by inhibiting the activity of apoptosis using inhibitor Z-VAD-FMK (20 mM for 20 min).
Cell invasion assay
Invasion assay was assesed using the Transwell Chamber (Transwell Permeable Supports, Corning, Inc., Corning, NY, USA). Briefly, 200 μl of cells suspended in cell medium containing 0.1% FBS, were added into the upper chamber while 500 μl of medium with 10% FBS was filled in lower chamber. After 24 h, cells migrated through the membrane were fixed with 70% alcohol, stained with crystal violet, and imaged using an inverted microscope (Nikon, Japan).
TCF21 knockdown
To establish a stable TCF21 knockdown cell line, oligonucleotides encoding shTCF21 as previous stated (Nurnberg et al. 2015) were inserted into pWPI lentiviral backbone (#12254, Addgene, Inc. MA, USA). ShTCF21 sequence is shown as follow: GaattcgaacgctgacgtcatcaacccgctccaaggaatcgcgggcccagtgtcactaggcgggaacacccagcgcgcgtgcgccctggcaggaagatggctgtgagggacaggggagtggcgccctgcaatatttgcatgtcgctatgtgttctgggaaatcaccataaacgtgaaatgtctttggatttgggaatcttataagttctgtatgagaccacagatctCCCTGGAGATGTTGGAATGTGACGGGTTGATTCAAGAGATCAACCCGTCACATTCCAACATCTCCATTTTTGGAAaagcttATCGAT. The shTCF21 constructs and pWPI vector were separately transfected with packaging vectors and Mission Lentiviral packaging mix (Sigma-Aldrich, USA) into HEK293T cells. After 48 h, cell media was harvested and centrifuged 3000×g for 15 min to remove dead cells. The virus was filtered through a 0.45 μm Steri-flip filter and then aliquoted and stored at −80 °C until use. The collected viruses were used to transfect H1299 and A549 cells and 2 μg/mL puromycin was used after 48 h of transfection. After selection, these cells were plated into 96-well plate using limiting dilution. Clones were formed after about one week of cell culture. All formed clones were separately cultured in new plates and each of them was tested for TCF21 expression using western blotting. Clone with TCF21 knockdown was selected for further experiments.
Gene array
The extracted RNA was labelled with Cy3 dye using Agilent Low-Input QuickAmp Labeling kit (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s protocol. The human genome expression assay was performed using the Whole Human Genome 4x44K Oligomicroarray kit containing 41,193 probes (60-mer oligo DNA, including control probes). Labeled RNAs were hybridized using a Gene Expression Hybridization kit (Agilent Technologies, USA). The microarray slides were scanned with the Agilent Microarray scanner and images were processed using the Agilent Feature Extractions version 9.5.
Western blotting
Protein was extracted by RIPA Lysis Buffer (Beyotime, Jiangsu, China) containing 1% phenylmethylsulfonyl fluoride (PMSF) (Sigma, USA). Equal amounts of samples separated by 10% SDS-PAGE and transferred onto a NC membrane (Sigma, USA). Membranes were blocked with 8% skim milk. Primary antibodies include anti-β-actin rabbit pAb (#A2668, Sigma-Aldrich, USA), anti-human ATG9A rabbit pAb (#SAB2100173, Sigma-Aldrich, USA), anti-beclin-1 rabbit monoclonal antibody (#3495, Cell Signaling Technology, Beverly, MA, USA), anti-TCF21 antibody (#ab32981, Abcam), anti-LC3-I/II rabbit polyclonal antibody (ABC929, Sigma), anti-p62 (#5114, Cell Signaling, MA, USA). In addition, anti-caspase-3, anti-cleaved caspase-3, anti-PARP, anti-cleaved PARP were purchased from Santa Cruz Biotechnology (USA). All antibodies were diluted in tris-buffered saline containing 0.1% (v/v) Tween-20 (PBST) and incubated with the membrane overnight at 4 °C. After washing with PBST, membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse IgG (Amersham Pharmacia Biotech, Piscataway, NJ, USA). Enhanced Chemiluminescence kit (GE Healthcare, UK) was used to detect the signals and exposed to X-ray film.
Statistical analysis
All data were shown as the mean ± standard deviation (SD). SPSS 15.0 (IBM, Armonk, NY, USA) was used for statistical analysis. The χ2 test was used to analyze the correlation between TCF21 expression and clinicopathologic characteristics. Student t-test was employed to compare the differences between treatment and control groups. A p < 0.05 was considered statistically significant.
Results
Association between TCF21 expression and clinicopathological characteristics
To investigate the role of TCF21 in the progress of NSCLC, we firstly analyzed the association between TCF21 transcriptional expression and clinicopathological features in 100 patients with NSCLC whose tumor biopsies were collected over the last several years. Table 1 shows that TCF21 expression was significantly associated with tumor stage (p = 0.001), metastasis (p = 0.001) and invasion (p < 0.001). Lower expression of TCF21 significantly exhibited higher level of TCF21 promoter methylation (p < 0.001).
Table 1.
TCF21 expression and clinicopathologic characteristics of lung cancer samples
| Characteristics | Total | TCF21 expression | P value | |
|---|---|---|---|---|
| Low | High | |||
| Age (years) | 0.567 | |||
| < 60 | 46 | 22 | 21 | |
| > 60 | 54 | 26 | 28 | |
| Gender | 0.541 | |||
| Female | 58 | 28 | 30 | |
| Male | 52 | 26 | 26 | |
| Tumor stages | 0.001* | |||
| I | 21 | 15 | 6 | |
| II | 26 | 19 | 7 | |
| III | 29 | 28 | 1 | |
| IV | 24 | 24 | 0 | |
| Tumor size (cm) | 0.341 | |||
| < 5 | 35 | 30 | 7 | |
| > 5 | 65 | 60 | 5 | |
| Tumor metastasis | 0.001* | |||
| Negative | 51 | 45 | 6 | |
| Positive | 49 | 48 | 1 | |
| Tumor invasion | 0.000* | |||
| Negative | 27 | 20 | 7 | |
| Positive | 73 | 70 | 3 | |
| Methylation level | 0.000* | |||
| Low | 56 | 30 | 26 | |
| High | 44 | 44 | 0 | |
Statistical analyses were performed by a χ2 test
*P < 0.05 was considered significant
The effects of demethylation induced by 5-Aza treatment on H1299 cells
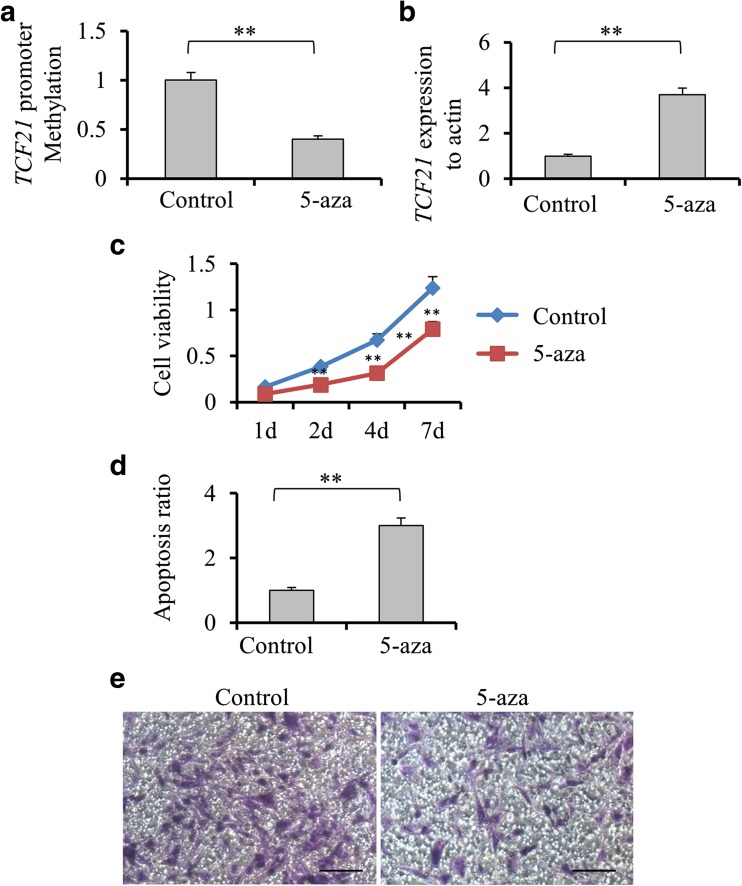
To reveal that DNA methylation is responsible for regulation of TCF21 transcriptional expression, H1299 cells were treated with 5-Aza, a DNA methyltransferase inhibitor. We found that cells treated with 5-Aza showed significantly lower level of TCF21 methylation and higher level of TCF21 expression compared with untreated cells (p < 0.01, Fig. 1a, b). Additionally, 5-Aza treatment led to a remarkable decrease of cell viability (p < 0.01, Fig. 1c) and increase of cell apoptosis (p < 0.01, Fig. 1d). Reduced cell invasion ability was observed in cells treated by 5-Aza compared with untreated cells (Fig. 1e).
Fig. 1.
The effects of demethylation induced by 5-Aza treatment on TCF21 expression and tumor development in H1299 cells. a Comparison of the levels of transcription factor 21 (TCF21) promoter methylation in the H1299 cells treated with or without 5-aza-2′-deoxycytine (5-Aza) by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR). b The expression level of TCF21 in the H1299 cells with or without 5-Aza treatment by qRT-PCR. c The H1299 cells with or without 5-Aza treatment were placed on 96-well plates (2.5 × 104 cells/well). Cell viability was detected as 1, 2, 4 and 7 days by CCK-8 assay. d Annexin V/propidium iodide double-staining assay was performed to detect the apoptosis levels of H1299 cell line with or without treatment of 5-aza. E. The relation of demethylation and invasion capacity was tested at 72 h after culturing cells by transwell assays. All values were represented as mean + SD from three independent experiments with triple replicates per experiment. Scale bar, 20 μm. ** p < 0.01
TCF21 knockdown enhances the growth and invasion of H1299 and A549 cells
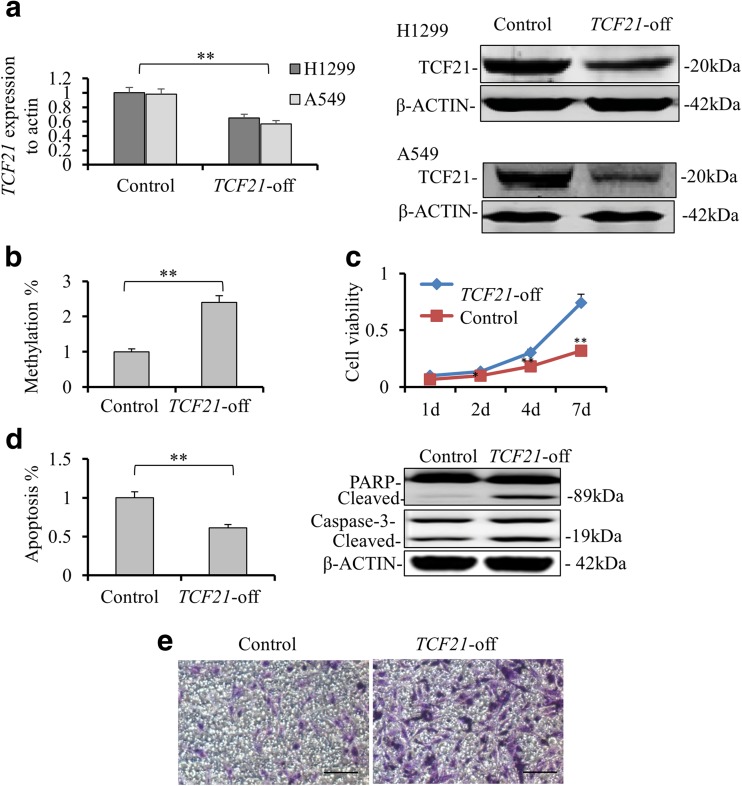
To further confirm the role of TCF21 in cell viability, we next investigated the influence of TCF21 knockdown on the growth of H1299 cells. ShTCF21-expressing lentivirus was used to establish stable H1299 and A549 cell lines with TCF21 knockdown. The efficiency of TCF21 knockdown was assessed by qRT-PCR and western blotting. Significantly lower mRNA level and protein expression of TCF21 were detected in TCF21 knockdown cells compared with control (p < 0.01; Fig. 2a). The methylation level of TCF21 promoter was downregulated in A549 cells with TCF21 knockdown (Fig. S1A). Significantly elevated global DNA methylation level was observed in TCF21 knockdown cells compared with it in control (p < 0.01, Fig. 2b and Fig. S1B). Conversely, downregulation of TCF21 increased cell viability (p < 0.01, Fig. 2c and Fig. S1C), reduced cell apoptosis (p < 0.01, Fig. 2d and Fig. S1D) and promoted cell invasion (Fig. 2e and Fig. S1E). Several apoptotic relative proteins were measured by western blot. The protein levels of cleaved PARP and cleaved caspase-3 were lower in TCF knockdown cells compared to them of its control cell line (Fig. 2d).
Fig. 2.
TCF21 knockdown affects the global methylation level and enhances the growth and invasion in H1299 cells. a The expression level of transcription factor 21 (TCF21) in H1299 and A549 cells with or without TCF21 knockdown by qRT-PCR (left, **p < 0.01). The protein expressions of TCF21 in both H1299 and A549 cell lines with or without TCF21 knockdown were tested by Western blotting. b The level of global methylation in H1299 cells with or without TCF21 knockdown were detected by HPLC. c The H1299 cells with or without TCF21 knockdown were placed on 96-well plates (2.5 × 104 cells/well). Cell viability was detected as 1, 2, 4 and 7 days by CCK-8 assay. d Annexin V/propidium iodide double-staining assay was performed to detect the apoptosis levels of H1299 cell line with or without TCF21 knockdown. Cleaved PARP and Caspase-3 after TCF21 knockdown was detected by western blotting (on the right). E. The relation of demethylation and invasion capacity was tested at 72 h after culturing cells by transwell assays. All values were represented as mean + SD from three independent experiments with triple replicates per experiment. Scale bar, 20 μm. (**p < 0.01)
TCF21 knockdown increased autophagy in lung cancer cells
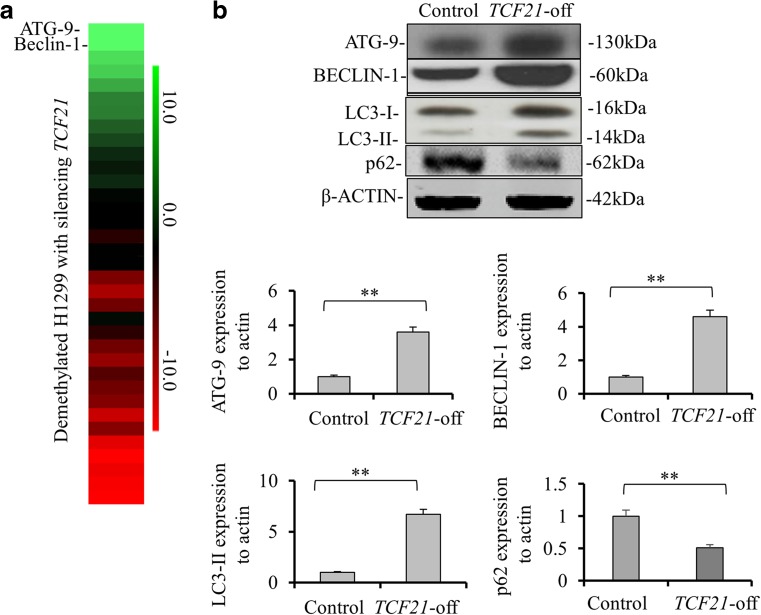
By comparing the gene expressions between TCF21 knockdown and wildtype cells, we found that ATG9 and BECLIN-1 genes were significantly upregulated in TCF21 knockdown cells (Fig. 3a). We then tested their protein expressions and the results showed that TCF21 knockdown in H1299 cells markedly enhanced the protein expressions of ATG9 (p < 0.01) and BECLIN-1 (p < 0.01) compared to control cells (Fig. 3)b. These results were also observed in A549 cells (Fig. S1F). To further confirm the increase of autophagy induced by TCF21 knockdown, we measured the protein levels of LC3-I, LC-3II and p62 in H1299 and A549 cells. It was observed that the expression levels of LC3-I and -II were significantly elevated in TCF21 knockdown cells compared to it in control cells (p < 0.01, Fig. 3b). However, p62 expression was significantly down-regulated in TCF21 knockdown cells compared with its control cells (p < 0.01).
Fig. 3.
Autophagy is increased in lung cancer cells after silencing TCF21. a The heatmap of differentially expressed genes identified in H1299 cells with TCF21 knockdown. The decreased and increased genes are indicated by red and green intensities, respectively. b ATG9, BECLIN-1, p62, LC3-I and LC3-II protein levels in H1299 with TCF21 knockdown were detected by Western blotting. Data were presented as the means ± SD from three independent experiments with triple replicates per experiment. **p < 0.01, indicate significant difference compared to normal group
Inhibition of autophagy by 3-methyladenine promoted TCF21 expression and cell apoptosis
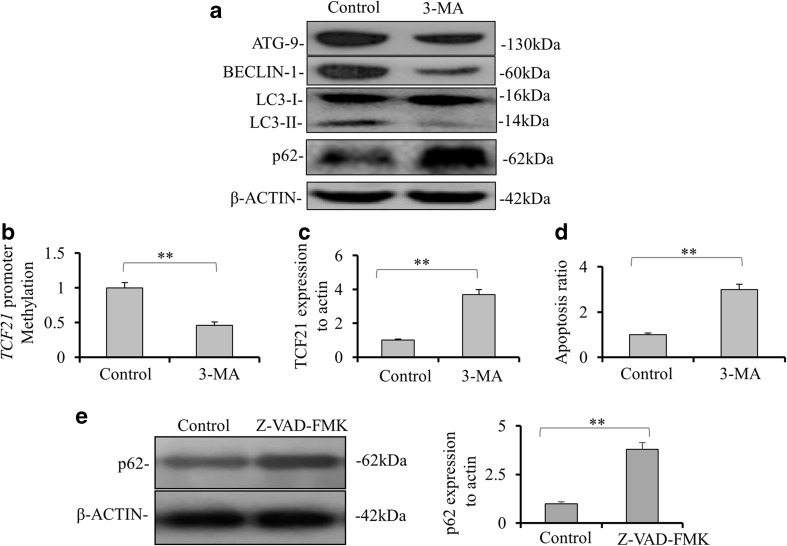
To confirm that TCF21 transcriptional expression was regulated by ATG9 and BECLIN-1 proteins, we treated H1299 cells with 3-MA autophagy inhibitor. Both ATG9 and BECLIN-1 protein levels were decreased in H1299 cells treated with 3-MA compared to them in control cells (Fig. 4a). Although no significant change in protein level of LC3-I was observed after 3-MA treatment, the expression of LC3-II was significantly repressed. Interestingly, 3-MA treatment also significantly decreased the level of TCF21 promoter methylation (p < 0.01, Fig. 4b). Accordingly, 3-MA treatment led to a significant increase of TCF21 expression in H1299 cells (p < 0.01, Fig. 4c). The apoptosis activity was remarkably elevated in cells treated by 3-MA compared with it in control cells (p < 0.01, Fig. 4d). These results suggest that autophagy may play a role in regulation of TCF21. We further investigated the correlation between autophagy and apoptosis using a caspase inhibitor, Z-VAD-FMK. We found that p62 expression level was significantly increased in Z-VAD-FMK-treated cells compared to untreated cells (p < 0.01, Fig 4e).
Fig. 4.
Inhibition of autophagy by 3-methyladenine promoted TCF21 expression and cell apoptosis. a The expressions of ATG9, BECLIN-1, LC3-I and LC3-II and p62 were tested in H1299 with or without 3-methyladenine (3-MA) treatment by Western blotting. b The TCF21 promoter methylation levels in the H1299 cells with or without 3-MA treatment were detected by methylation-specific PCR. c The mRNA level of TCF21 in the H1299 cells with or without 3-MA treatment by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR). d Annexin V/propidium iodide double-staining assay was performed to detect the apoptosis levels of H1299 cells with or without 3-MA treatment. e Activity of autophagy by testing p62 was verified by inhibiting the activity of apoptosis using inhibitor Z-VAD-FMK (20 mM for 20 min). Data were presented as the means ± SD from three independent experiments with triple replicates per experiment. ** p < 0.01, indicate significant difference compared to untreated cells
Discussion
Clinical analysis revealed that more than 80% of patients with NSCLC showed hypermethylated TCF21 with reduced TCF21 mRNA level (Richards et al. 2011) in lung cancer patients (Anglim et al. 2008). Evidence has proven the level of TCF21 methylation was correlated with tumor stage, metastasis and invasion in gastric cancer (Yang et al. 2015). In the present study, we found that TCF21 expression was significantly correlated with its promoter methylation level. The expression level of TCF21 is related to the tumor stages of patients with NSCLC. Our results also revealed that patients with lower expression of TCF21 were more prone to develop tumor metastasis and invasion. Although overexpression of TCF21 in mouse xenografts led to a decrease of tumor size was observed in a previous study (Smith et al. 2006), another clinical data demonstrated there was no correlation between TCF21 methylation and tumor size (Dai et al. 2016), which is consistent with our results. These results imply that TCF21 contributes to suppression of the progression of lung cancer.
Downregulation of TCF21 was observed in almost all cancer cell lines (Richards et al. 2011). A previous study reported that restoration of TCF21 expression inhibited the proliferation and promoted apoptosis of colorectal cancer cells (Dai et al. 2016). Little is known about the effects of TCF21 on NSCLC cells. We treated H1299 cells with 5-Aza, a DNA methyltransferase inhibitor, and found that 5-Aza upregulated the expression level of TCF21 in H1299 cells. The expression level of TCF21 is negatively related with its promoter methylation level. Reduced cell viability and increased apoptosis were found in H1299 cells treated with 5-Aza compared with them in non-treated cells. It is well known that epithelial-to-mesenchymal transition (EMT) plays a critical role in cancer progression (Thiery 2002). Given the function of TCF21 in promoting the transition of mesenchymal into epithelial (MET) cells, silencing TCF21 by DNA hypermethylation may lead to a reduced cell ability of MET and therefore give rise to EMT and promote cancer cell invasion. Several studies have demonstrated that downregulation of TCF21 by methylation resulted in cell invasion and migration of melanoma, colorectal cancer and renal cancer. (Arab et al. 2011; Dai et al. 2016). We found that cells treated with 5-Aza were more aggressive than non-treated cells. Accordingly, knockdown TCF21 reversed these observations. Silencing TCF21 resulted in increased expression levels of ATG9, BECLIN-1, and LC3-I/II. It is possible that TCF21 exerts an inhibitory function on ATG9 and BECLIN-1, and therefore may repress autophagy. ATG9 is a multimembrane-spanning protein and services as an important component in the formation of membranes for autophagosome (Noda et al. 2000; Yamamoto et al. 2012). BECLIN-1 was first identified by Liang XH et al. in Liang et al. 1999 as an autophagic function protein in mammalian (Liang et al. 1999). It was sequentially revealed that BECLIN-1 plays a critical role in the initial stage of autophagosome formation through the interaction with VPS15, the Class III PI3K VPS34, and ATG14 (Abrahamsen et al. 2012; Fu et al. 2013; Funderburk et al. 2010; He and Levine 2010).
Autophagy could play either positive or negative roles in the progression of lung cancer. The regulation between cell apoptosis and autophagy is strikingly controlled. Normally autophagy could elicit cell death via activating apoptosis whereas increasing evidence has demonstrated that it also promotes cancer cell survival during therapy (Kondo et al. 2005). In this study, it is difficult to conclude whether the increased autophagy is related with less TCF21 expression. It is possible that knockdown TCF21 promotes global DNA methylation, which further leads to increased autophagy that in turn enhances cancer progression. On the other hand, when we repressed autophagy by 3-MA treatment, it was found that global methylation level and TCF21 promoter methylation lever were both decreased. However, TCF21 promoter methylation was also suppressed by 3-MA treatment. Thus, autophagy may be positively related to DNA methylation and TCF21 promoter methylation downregulates the expression of TCF21, leading to the alterations in apoptosis. However, the methylation decrease may be caused by the 3-MA treatment or the change of ATG9 and BECLIN-1. No previous studies have demonstrated whether 3-MA, ATG9 and BECLIN-1 play roles in demethylation process. However, BECLIN-1 gene was found to be silenced in 40–75% of sporadic breast tumors (Li et al. 2010) and low expression of BECLIN-1 was associated with lymph node metastasis, advanced TNM stage and poor prognosis of gastric cancer (Cao et al. 2016). Spontaneous lung cancer was observed in BECLIN-1+/− mice (Qu et al. 2003; Yue et al. 2003). Similar as TCF21, BECLIN-1 is also a tumor suppressor and low expression of BECLIN-1 may lead to autophagy deficiency and tumorigenesis. Recent clinical studies demonstrated that BECLIN-1 expression was predominant in heavy-smokers and significantly related to lymph node metastasis of NSCLC (Lv et al. 2015; Wang et al. 2015). The controversial results mainly are due to the dual roles of autophagy in oncogenesis, which correlated with the stage of cancers (Zhou et al. 2016). Few research has concentrated on the relation of ATG9 with lung cancer but high expression of ATG9 was observed in gastric cancer (Cao et al. 2016). A previous study reported that 5-Aza treatment significantly downregulated BECLIN-1 in prostate cancer cell lines (Liao et al. 2016). According to our results on H1299 cells treated by 5-Aza, it is possible that increased TCF21 expression by demethylation reduces BECLIN-1 expression. Further investigation is needed to confirm whether TCF21 directly suppresses BECLIN-1 and ATG9 expressions.
In summary, the expression of TCF21 was significantly regulated by the methylation level of TCF21 in patients with NSCLC and was associated with tumor stage, metastasis and invasion. TCF21 knockdown showed higher expressions of ATG9 and BECLIN-1. Inhibition of autophagy by 3-MA treatment significantly suppressed ATG and BECLIN-1 expressions, but elevated TCF21 expression and increased cell apoptosis. These results suggest that there is a correlation between autophagy and TCF21 promoter methylation. TCF21 expression is related to cell viability and invasion ability. However, the detailed relations between TCF21, autophagy and apoptosis need further investigations.
Electronic supplementary material
(DOCX 328 kb)
Abbreviations
- TCF21
transcription factor 21
- NSCLC
non-small cell lung cancer
- 5-Aza
5-aza-2′-deoxycytine
- 3-MA
3-methyladenine
Funding
This study was funded by the Science and Technology Development Fund Project of Shenzhen (No. JCYJ 20150403091443278 and JCYJ 20150403091443310).
Compliance with ethical standards
Conflict of interest
The authors have no conflict of interest to disclose.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s12079-017-0418-2) contains supplementary material, which is available to authorized users.
References
- Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM, Sicks JD. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahamsen H, Stenmark H, Platta HW. Ubiquitination and phosphorylation of Beclin 1 and its binding partners: tuning class III phosphatidylinositol 3-kinase activity and tumor suppression. FEBS Lett. 2012;586:1584–1591. doi: 10.1016/j.febslet.2012.04.046. [DOI] [PubMed] [Google Scholar]
- Ahmad A, Azim S, Zubair H, Khan MA, Singh S, Carter JE, Rocconi RP, Singh AP. Epigenetic basis of cancer health disparities: looking beyond genetic differences. Biochim Biophys Acta. 2017;1868:16–28. doi: 10.1016/j.bbcan.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglim PP, Galler JS, Koss MN, Hagen JA, Turla S, Campan M, Weisenberger DJ, Laird PW, Siegmund KD, Laird-Offringa IA. Identification of a panel of sensitive and specific DNA methylation markers for squamous cell lung cancer. Mol Cancer. 2008;7:62. doi: 10.1186/1476-4598-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arab K, Smith LT, Gast A, Weichenhan D, Huang JP, Claus R, Hielscher T, Espinosa AV, Ringel MD, Morrison CD, Schadendorf D, Kumar R, Plass C. Epigenetic deregulation of TCF21 inhibits metastasis suppressor KISS1 in metastatic melanoma. Carcinogenesis. 2011;32:1467–1473. doi: 10.1093/carcin/bgr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao QH, Liu F, Yang ZL, XH F, Yang ZH, Liu Q, Wang L, Wan XB, Fan XJ. Prognostic value of autophagy related proteins ULK1, Beclin 1, ATG3, ATG5, ATG7, ATG9, ATG10, ATG12, LC3B and p62/SQSTM1 in gastric cancer. Am J Transl Res. 2016;8:3831–3847. [PMC free article] [PubMed] [Google Scholar]
- Conway K, Edmiston SN, Tse C-K, Bryant C, Kuan PF, Hair BY, Parrish EA, May R, Swift-Scanlan T. Racial variation in breast tumor promoter methylation in the Carolina breast cancer study. Cancer Epidemiology and Prevention Biomarkers:cebp. 2015;24:921–930. doi: 10.1158/1055-9965.EPI-14-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Duan H, Duan C, Zhou R, He Y, Tu Q, Shen L. Down-regulation of TCF21 by hypermethylation induces cell proliferation, migration and invasion in colorectal cancer. Biochem Biophys Res Commun. 2016;469:430–436. doi: 10.1016/j.bbrc.2015.09.109. [DOI] [PubMed] [Google Scholar]
- Dwi Putra SE, Neuber C, Reichetzeder C, Hocher B, Kleuser B. Analysis of genomic DNA methylation levels in human placenta using liquid chromatography-electrospray ionization tandem mass spectrometry. Cell Physiol Biochem. 2014;33:945–952. doi: 10.1159/000358666. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Bray F, Pisani P, Parkin D (2004) Cancer incidence, mortality and prevalence worldwide. IARC Cancer Base No. 5, version 2.0. IARCPress, Lyon
- Fu LL, Cheng Y, Liu B. Beclin-1: autophagic regulator and therapeutic target in cancer. Int J Biochem Cell Biol. 2013;45:921–924. doi: 10.1016/j.biocel.2013.02.007. [DOI] [PubMed] [Google Scholar]
- Funderburk SF, Wang QJ, Yue Z. The Beclin 1–VPS34 complex–at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39:305–318. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- He C, Levine B. The beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong QY, GM W, Qian GS, CP H, Zhou JY, Chen LA, Li WM, Li SY, Wang K, Wang Q, Zhang XJ, Li J, Gong X, Bai CX. Prevention and management of lung cancer in China. Cancer. 2015;121(Suppl 17):3080–3088. doi: 10.1002/cncr.29584. [DOI] [PubMed] [Google Scholar]
- Howlader N, Noone A, Krapcho M, Garshell J, Miller D, Altekruse S, Kosary C, Yu M, Ruhl J, Tatalovich Z. SEER cancer statistics review, 1975–2012. Bethesda: National Cancer Institute; 2015. [Google Scholar]
- Jemal A, Thun MJ, Ries LA, Howe HL, Weir HK, Center MM, Ward E, Wu X-C, Eheman C, Anderson R. Annual report to the nation on the status of cancer, 1975–2005, featuring trends in lung cancer, tobacco use, and tobacco control. J Natl Cancer Inst. 2008;100:1672–1694. doi: 10.1093/jnci/djn389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- Li Z, Chen B, Wu Y, Jin F, Xia Y, Liu X. Genetic and epigenetic silencing of the beclin 1 gene in sporadic breast tumors. BMC Cancer. 2010;10:98. doi: 10.1186/1471-2407-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Liao H, Xiao Y, Hu Y, Xiao Y, Yin Z, Liu L, Kang X, Chen Y. Methylation-induced silencing of miR-34a enhances chemoresistance by directly upregulating ATG4B-induced autophagy through AMPK/mTOR pathway in prostate cancer. Oncol Rep. 2016;35:64–72. doi: 10.3892/or.2015.4331. [DOI] [PubMed] [Google Scholar]
- Lv ZQ, Han JJ, Liu YQ, Wang LL, Tang QL, Sun Q, Li HG. Expression of beclin 1 in non-small cell lung cancer: an immunohistochemical study. Clin Respir J. 2015;9:359–365. doi: 10.1111/crj.12148. [DOI] [PubMed] [Google Scholar]
- Noda T, Kim J, Huang W-P, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberg ST, Cheng K, Raiesdana A, Kundu R, Miller CL, Kim JB, Arora K, Carcamo-Oribe I, Xiong Y, Tellakula N. Coronary artery disease associated transcription factor TCF21 regulates smooth muscle precursor cells that contribute to the fibrous cap. PLoS Genet. 2015;11:e1005155. doi: 10.1371/journal.pgen.1005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaggin SE, Schwartz L, Cui S, Igarashi P, Deimling J, Post M, Rossant J. The basic-helix-loop-helix protein pod1 is critically important for kidney and lung organogenesis. Development. 1999;126:5771–5783. doi: 10.1242/dev.126.24.5771. [DOI] [PubMed] [Google Scholar]
- Richards KL, Zhang B, Sun M, Dong W, Churchill J, Bachinski LL, Wilson CD, Baggerly KA, Yin G, Hayes DN. Methylation of the candidate biomarker TCF21 is very frequent across a spectrum of early-stage nonsmall cell lung cancers. Cancer. 2011;117:606–617. doi: 10.1002/cncr.25472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- Smith LT, Lin M, Brena RM, Lang JC, Schuller DE, Otterson GA, Morrison CD, Smiraglia DJ, Plass C. Epigenetic regulation of the tumor suppressor gene TCF21 on 6q23-q24 in lung and head and neck cancer. Proc Natl Acad Sci U S A. 2006;103:982–987. doi: 10.1073/pnas.0510171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Tsou JA, Galler JS, Siegmund KD, Laird PW, Turla S, Cozen W, Hagen JA, Koss MN, Laird-Offringa IA. Identification of a panel of sensitive and specific DNA methylation markers for lung adenocarcinoma. Mol Cancer. 2007;6:70. doi: 10.1186/1476-4598-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Dorsey TH, Terunuma A, Kittles RA, Ambs S, Kwabi-Addo B. Relationship between tumor DNA methylation status and patient characteristics in African-American and European-American women with breast cancer. PLoS One. 2012;7:e37928. doi: 10.1371/journal.pone.0037928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Du Z, Li L, Shi M, Yu Y. Beclin 1 and p62 expression in non-small cell lung cancer: relation with malignant behaviors and clinical outcome. Int J Clin Exp Pathol. 2015;8:10644–10652. [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Li DM, Xie Q, Dai DQ. Protein expression and promoter methylation of the candidate biomarker TCF21 in gastric cancer. J Cancer Res Clin Oncol. 2015;141:211–220. doi: 10.1007/s00432-014-1809-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Yuan M, Yu Q, Zhou X, Min W, Gao D. Autophagy regulation and its role in gastric cancer and colorectal cancer. Cancer Biomark. 2016;17:1–10. doi: 10.3233/CBM-160613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 328 kb)