Abstract
Microbes have their own communication systems. Secretion and reception of chemical signaling molecules and ion-channels mediated electrical signaling mechanism are yet observed two special ways of information transmission in microbial community. In this article, we address the aspects of various crucial machineries which set the backbone of microbial cell-to-cell communication process such as quorum sensing mechanism (bacterial and fungal), quorum sensing regulated biofilm formation, gene expression, virulence, swarming, quorum quenching, role of noise in quorum sensing, mathematical models (therapy model, evolutionary model, molecular mechanism model and many more), synthetic bacterial communication, bacterial ion-channels, bacterial nanowires and electrical communication. In particular, we highlight bacterial collective behavior with classical and quantum mechanical approaches (including quantum information). Moreover, we shed a new light to introduce the concept of quantum synthetic biology and possible cellular quantum Turing test.
Keywords: Quorum sensing, Biofilm, Ion-channels, Synthetic biology, Quantum biology
Introduction
The three-domain system of cellular life corresponds to archaea, bacteria and eukaryota. In a recent report, it has been confirmed that unicellular microorganisms archaea existed in 3456 million year ago (Schopf et al. 2018) and bacteria are the first evidence of life in earth (Schopf 1994). We can find different microbes in each and every corner of the entire world. Antonie Van Leeuwenhoek (father of microbiology) studied microbes for the first time with his self designed microscope and put forwarded the concepts of bacteria and their survival in a diverse range of environment in his famous paper “letter on the protozoa” (Leewenhoeck 1677). Leeuwenhoek’s pioneering discovery and remarkable observations (nicely captured in the article by Lane 2015) gave birth to a new branch in science and a new era of microbiology began. Since then microbiologists have extensively studied single bacterial cell structures (Gahlmann and Moerner 2014) and their molecular mechanisms (Liu et al. 2015; Dangkulwanich et al. 2014) using powerful microscopes to understand the flow and processing of information required by the cell to maintain stability and to perform in a collective fashion. In the last four decades, the collective behaviors of the bacteria have caught the attention of microbiologists and the intriguing collective phenomenon of regulation of gene expression as a called quorum sensing has been discovered. Since then, it has been the central point of attraction and several researches have been focused on it (Fuqua et al. 1994; Gray et al. 1994; Miller and Bassler 2001).
Due to high adapting capability with the diverse environments, bacteria are considered very intelligent among the family of microorganisms. The way bacteria communicates among themselves to regulate their survival strategies is the manifestation of their intelligence. Even, bacteria can talk!
They talk to each other with chemical signaling molecules or autoinducers and coordinate the complex biochemical processes, which is known as quorum sensing mechanism (see Fig. 1). In human world, inter-human conversation is considered the most advantageous form of information transmission. Similarly, in bacterial community, the transmission of information is regulated via quorum sensing mechanism, which is not only limited in intraspecies communication but is extended to interspecies communication purpose also. Bacterium (e.g. E. coli, B. subtilis, V. fischeri, P. aeruginosa) release autoinducers from the cells and other bacterium in their vicinity are received this molecules. When autoinducers reach a threshold concentration, a coordinated change in bacterial behavior is initiated which stimulates cascade of signaling events resulting in the activation of quorum sensing genes. Bacteria make their own optimal survival strategy by this cell-to-cell communication mechanism (Shapiro 1998; Williams et al. 2007; Majumdar and Mondal 2016).
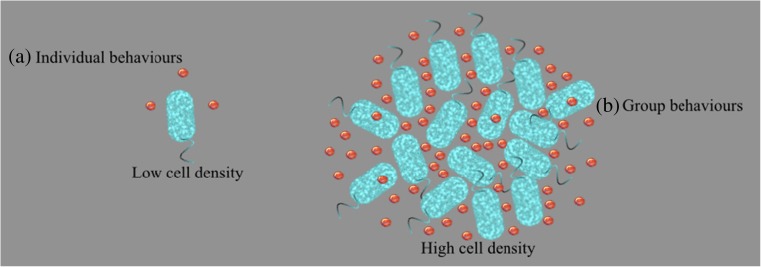
Fig. 1.
Schematic diagram of bacterial quorum sensing mechanism. (a) Single bacterial cell emits quorum sensing molecules but no quorum sensing occurs in low cell density, (b) Bacteria are secreted out quorum sensing molecules, which are sensed by the other bacterial cells in their vicinity. After reaching a threshold concentration of molecules, a coordinated change in bacterial behavior is initiated
To understand the information flow in the bacterial quorum sensing mechanism, researchers have engineered artificial bacterial cells. This fascinating branch of investigation, which is formally known as synthetic biology, is interdisciplinary in nature and is of current interest. In synthetic biology, the basic science is combined with engineering which enables us to design artificial biological systems for deeper understanding of different types of complex biological phenomena and to apply it in medical sciences and industries (Nandagopal and Elowitz 2011; Khalil and Collins 2010; Bashor et al. 2010) to discover the underlying mechanisms of different microscopic and macroscopic biological systems. But, the important challenge behind it is to recognize general, scalable strategies that accredit fabrication of increasingly complex gene circuits with reliable performance, as well as to construct original and significant technological platforms for the quantitative circuit characterization (Simpson 2004; Endy 2005; Marguet et al. 2007). In the early twenty first century, Prof. J. J. Collins and his co-workers constructed a synthetic genetic toggle switch, a bistable gene regulatory network in E. coli (quorum sensing bacteria) and proposed a noble scheme, which predicts necessary condition for bistability. This is a remarkable achievement because this toggle switch concepts can build a practical device (addressable cellular memory unit) which is useful for medical science and technology like biocomputing, biotechnology and many more (Gardner et al. 2000). Another contemporary letter showed the design principle and construction of synthetic network in E. coli, where three transcriptional repressor systems were used, which were not a part of any natural biological clock. This interdisciplinary approach leads us to understanding of cellular behaviours as well as naturally occurring networks (Elowitz and Leibler 2000). Following this pathway, researchers have been able to better understand the cellular interactions, which shed a new light in cell-to-cell communication (You et al. 2004; Basu et al. 2005; Brenner et al. 2007; Shou et al. 2007; Stricker et al. 2008; Danino et al. 2010; Chen et al. 2015; Scott and Hasty 2016).
On the other hand, synthetic microecology allow us to study some important fact as biodiversity and the coexistence of populations using various mathematical models. Most of the previous researches in the context of microbial association did not consider spatial relationship, which has fundamental importance to study pattern formation. A predator-prey synthetic system was studied in 2008, which consisted of oscillatory population dynamics due to bacterial chemical communication mechanism (quorum sensing) (Balagaddé et al. 2008). Then this approach was extended to study the spatiotemporal dynamics of biodiversity between two-engineered E. coli populations (Song et al. 2009). Moreover, spatiotemporal dynamics (evolution of microbial) of single motile E. coli was studied in 2016 with antibiotic background (Baym et al. 2016). In recent investigation, a synthetic Eschericha coli killer-prey (killer strain NB003 and prey strain DZ10) microecological system is developed where the killer strain emits autoinduces (AHL), which can bind with LuxR, after autoinducers enter into prey. AHL-LuxR complex then produces cell lysis protein and kills the prey (see Fig. 2). This is a very robust synthetic design to study the spatiotemporal dynamics of the microecological system in a nutrient rich environment (Datla et al. 2017).
Fig. 2.
Schematic diagram of E. coli killer-prey system: Killer strain kills the prey strain and the prey can evolve to be resistant to the killer
In general, biofilms are defined as “aggregates of microbes in which cells are often embedded in a self-produced matrix of extracellular polymeric substances that are adherent to each other and/or a surface” (Flemming et al. 2016) (see Fig. 3). Bacterial cell-to-cell communication process is responsible for conciliating a wide variety of social activities in biofilms, which includes biofilm growth (Chopp et al. 2003; García-Aljaro et al. 2012), biofilm dispersion (Solano et al. 2014; Cárcamo-Oyarce et al. 2015), antimicrobial resistance (Thompson et al. 2015), swarming motility and gene expression (Quiñones et al. 2005). Moreover this chemical signaling mechanism regulates the extracellular polymeric substances production during biofilm formation (Waters and Bassler 2005; Vuong et al. 2003; Marketon et al. 2003). P. aeruginosa has two different types of cell- to- cell communication process (lasR-lasI and rhlR-rhlI), which are also involved in bacterial biofilm development. It has been observed that for certain population densities, these biochemical signals reach the required level of concentration for the gene activation which are involved in biofilm differentiation (Davies et al. 1998).
Fig. 3.
Schematic visualization of multistage process of bacteria biofilm formation
Bacterial biofilm, the emergent form of bacterial lifestyle on surfaces, are very organized communities where millions of densely packed cells are accommodated and have coordinated motion inside it. This co-ordinated motion of bacteria inside the biofilm is a consequence of chemical signaling mechanism. Apart from chemical signaling, recently, another type of signaling mechanism has been realized which is mediated by bacterial ion-channels (electrical signaling) (see Fig. 4).
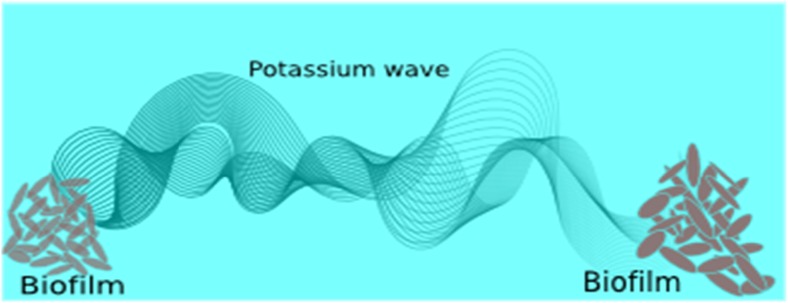
Fig. 4.
Illustration of potassium ion-channels mediated wave (electrical communication) in bacterial communities
Prof. G. M. Süel and his collaborators observed that biofilm communities exhibit fascinating microscopic potassium wave mechanism (Prindle et al. 2015; Beagle and Lockles 2015). Bacterial cells residing within a community can cooperate and compete with each other for nutrients. It also has been observed that the metabolic oscillation of bacterial membrane is triggered by resource limitation and the oscillatory dynamics resulted from long-range metabolic co-dependence between cells in the interior and periphery of the Bacillus subtilis biofilm. It has been reported that the growth of the biofilm halts periodically and the collective oscillation in the biofilm growth is an advantage to bacterial community to secure bacterial life from the chemical attack (Liu et al. 2017). Moreover, this finding suggests that bacteria use K+ ion-channel mediated electrical signaling to coordinate metabolism within the biofilm and hence conduct a long range electrical signaling within bacterial biofilm communities through the propagating wave of potassium (Prindle et al. 2015).
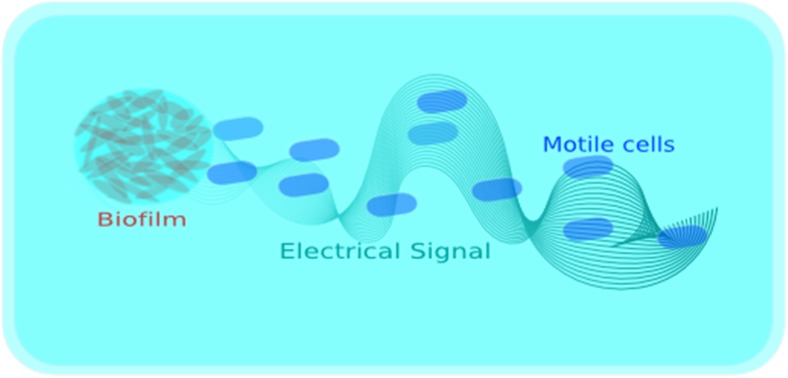
In a very recent studies, an attraction of motile cells (P. aeruginosa) towards biofilm is observed (see Fig. 5), where bacterial ion-channels play an important role. Researchers demonstrated that changes in extracellular potassium gradients are sufficient to direct motile cell behaviour and the attraction depends on membrane potential and mediated sensitivity of the motile cells to the potassium signals generated by the B. subtilis biofilm (Humphries et al. 2017; Majumdar and Pal 2017). Liu et al. (2017) extended the idea and showed a time sharing behaviour, which maintains biofilms growth under limited nutrient supply.
Fig. 5.
Illustration of species independent attraction of motile bacterial cells toward biofilm through electrical signaling
Talking about talking microbes
Bacterial quorum sensing mechanism not only studied by microbiologist but also investigated by the mathematician and physicist. Last 18 years, several mathematical models have been proposed to quantify this biological phenomena, which can be divided into five different class of models: (i) Models of quorum sensing molecular mechanisms, (ii) Therapy related models, (iii) Evolutionary models, (iv) Mathematical models of specific quorum sensing regulated process and (v) Other approach models (Pérez-Velázquez et al. 2016).
Mathematical models of bacterial cell-to-cell communication
Models of quorum sensing molecular mechanisms
The first model of quorum sensing system of V. fischeri was developed by James et al., where the model had two stable metabolic states corresponding to phenotypes and it had three steady states (one is stable, another two is stable under certain conditions). This conditions leads to a switch-like behaviour of the regulatory quorum sensing system (James et al. 2000). Dockery and Keener (2001) developed a quorum sensing model of P. aeruginosa, where the kinetics of the Las system is emphasized by an eight dimensional system with Michaelis-Menten type kinetics. Moreover, this model was extended into a more realistic type model by adding a spatial variable. Ward et al. (2001) approached talking bacterial mechanism as a population dynamics and examined the bacterial population growth (up-regulation and down-regulation system) and autoinducer production. This is the first attempt, where experiments were designed to estimate model parameters.
(See more details in Ward et al. 2001 and Majumdar et al. 2012)
(Fagerlind et al. 2003) proposed a model of two quorum sensing systems of P. aeruginosa (las/rhl) and showed that mono stability of the system can occur due to high concentration of autoinducers. (Gustafsson et al. 2004) investigated the quorum sensing system of S. aureus to determine the specific role of SarA in the agr system. (Goyachev et al. 2006) studied a cell communication model, which consists of two positive feedback loops to revealing relevant facts of the relationship between observable phenotype changes and the structural organization of intra cellular networks. They also studied the system including molecular noise. (Li et al. 2006) formulated luxS system of E. coli using stochastic approach to unveil an alternative pathway for AI-2 synthesis. Latter, another group of researchers discovered that the rate of synthesis is dependent on glucose (Barrios et al. 2009; Barrios and Achenie 2010). (Fekete et al. 2010) studied quantitative information, which can be utilized to estimate parameters such as production rate of the autoinducers, threshold concentration to achieve activation and the fundamental role of an autoinducer regulated enzyme, which degrades autoinducers. Further information about this types of models, are described in Refs (Quan et al. 2016; Hunter et al. 2013; Hense and Schuster 2015; Ward et al. 2004).
Therapy related models
Alternative of antibiotics is one of the major areas of research now a days. Since, pathogenic quorum sensing bacteria are intimately connected with the human health, several mathematical models have been recently proposed whose major concern is to use quorum sensing mechanism as a target for therapy --- scientifically known as quorum quenching. As an example, (Anguige et al. 2004) considered P. aeruginosa, one of the important human pathogenic bacteria, and developed a new model to explore an alternative way of antibiotic treatment. They discovered that autoinducer concentration can be reduced by the application of anti-quorum sensing agent in specific dose while the rest of the parameters were also important for successful treatment. Later, in the next year, they extended their work to incorporate well mixed bacterial populations (Anguige et al. 2005). On the other hand, (Viretta and Fussenegger 2004) proposed a deterministic model (three quorum sensing systems las, rhl and mvfR-PQS) to understand virulence and the quorum sensing response to pharmacological interference. They confirmed that the interference of the PQS signaling system can act as an efficient anti-virulence agent. (Fagerlind et al. 2005) also developed a model (quorum quenching) for the autoinducer antagonist (quorum sensing blockers) which predicted that quorum sensing blockers induced degradation of LasR is crucial for developing successful quorum quenching. In addition, (Ward 2008) studied anti-quorum sensing model in batch culture and biofilms and demonstrated that the effeciency of the treatment is probably dependent on the time of application of QSI and putative anti-LasI treatment is vey potent. Apart from these specific examples, there are various therapy related models (Beckmann et al. 2012; Anand et al. 2013) (see more details in Pérez-Velázquez et al. 2016), which can provide an alternative direction of antibiotics in near future.
Evolutionary models
In the first evolutionary model, quorum sensing mechanism was presented as a particular case and the evolutionary stability of host was studied in the context of cooperation (Brown 1999). (Brown and Johnstone 2001) presented a partial differential equation based model to describe the optimal level of cooperation. An agent-based model of biofilm development was investigated by (Nadell et al. 2008) which showed the production of extracellular polymeric substances regulated through quorum sensing mechanism. In particular, it was observed that four strains interacted and differed in their ability to produce extracellular polymeric substances: (1) no polymer secretion and no quorum sensing; (2) polymer secretion but no quorum sensing; (3) polymer secretion under negative quorum sensing control; and (4) polymer secretion under positive quorum sensing control. These observations immediately led to the conclusion that quorum sensing mechanism controls biofilm formation. Besides, Stochastic cellular automata model of quorum sensing showed that bacterial communication mechanism evolves from non-cooperating strains when the population is low and surprisingly, cheating and exploitation were found in communication process (Czárán and Hoekstra 2009). In short, the evolutionary model of bacterial communication system (regulated by different signaling mechanism) has potential to provide some fundamental insights of the collective mechanism such as colony formation, regrowth, mixing, colony dispersal, stabilize cooperation and many more. There are other findings also like cheater punishment (Friman et al. 2013) and stress produced by the quorum sensing proficient individuals (Wang et al. 2015) are some of the interesting new outcomes.
Mathematical models of specific quorum sensing regulated process
Biofilms
Since last 32 years, biofilms have been the focus of major studies and several mathematical models of biofilms are investigated. Some of the models incorporate one dimensional partial differential equation approaches while the others are multi-dimensional (spatial coordinates) to explore the possibility of life on a surface. (Eberl et al. 2001) presented a nonlinear density dependent reaction-diffusion system to exploit quorum sensing regulated biofilm formation and spatiotemporal quorum sensing induction patterns. The autoinducer concentration profile can also change and effect biofilm growth (Nilsson et al. 2001). (Chopp et al. 2002) studied in detail about growing biofilms together with cell communication. In related other works, biofilm was treated as two parts (1) active biomass and (2) inactive biomass and the concept of critical biofilm depth together with the corresponding approximation time was developed (Chopp et al. 2003). (Ward et al. 2003) found the travelling wave mechanism of the quorum sensing mechanism. There are several other mathematical frameworks related to quorum sensing regulated biofilms formation, which can be found in detail in (Pérez-Velázquez et al. 2016).
Swarming
The two dimensional swarming behaviour was emphasized by Netotea et al., which showed the control mechanism of secreted factors and chemical signaling schemes. The model included chemotactic agents, cell displacement towards nutrients, well-defined quorum sensing genes and their properties. It also explored dendritic growth patterns (Netotea et al. 2009) (for detail discussions follow Pérez-Velázquez et al. 2016).
Virulence
It has been observed that the virulence is regulated by the quorum sensing mechanism of various bacterium such as P. aeruginosa, V. cholerae and S. aureus. The process of endosome escape of S. aureus was modeled by (Koerber et al. 2005), which presented a brief asymptotic analysis using Monte Carlo simulations. (Karlsson et al. 2007) showed that competence appears as waves. (Haseltine and Arnold 2008) explained how inducting density tune the virulence of V. fischeri. (Anand et al. 2013) studied LuxI/LuxR quorum sensing system using a computational modeling approach and predicted that the virulence may be inhibited by the combination of LuxI and LuxR (non competitive inhibitor).
Other approach models
Researchers make some other different mathematical frame works to understand this cell-to -cell communication mechanism. However, in this article, our motivation is to accumulate few significant and fundamental recently developed biophysical models to explore the specific features of chemical and electrical bacterial communication. Prof. Bonnie L. Bassler and her collaborator recently explored material properties of extracellular matrix in presence of environmental perturbation (osmotic pressure difference) (Yan et al. 2017). They also showed that RhlR controls the biofilm formation and genes encoding virulence factors in absence of Rhll. (Mukherjee et al. 2017). Quorum sensing induced biofilm detachment was studied by (Emerenini et al. 2015; Emerenini et al. 2017). (Dilanji et al. 2012) presented autoinducers spatiotemporal patterns and propagating wave phenomena. (Zhao and Wang, 2017) numerically investigated quorum sensing regulated biofilm morphology. They suggested that the structure formation of biofilms is affected by cell communication. Recently, the experimentally observed ion-channel mediated electrical signaling was also verified by Hodgkin-Huxley model (Hodgkin and Huxley 1952). This electro-physiological model concerns about the changes of membrane in response to extracellular potassium ion. Biofilm was treated as oscillation source of extracellular potassium ion, which can periodically attract motile cells P. aeruginosa) by changing their membrane potential (Prindle et al. 2015; Beagle and Lockles 2015; Liu et al. 2015a, b; Humphries et al. 2017; Majumdar and Pal 2017a, b).
The role of noise in quorum sensing mechanism
Noise plays a significant role in gene circuit. Cox et al. studied the role of noise is quorum sensing circuit operation with a set of tool like frequency domain analysis, stochastic simulation, power spectral density and whitening effect. They demonstrated noise in quorum sensing circuit as a positive feedback function in entire population. Autoregulatory gene circuit could optimize the performance by shifting noise into frequency domain and made significant and dynamic modification to the noise spectra (Cox et al. 2003). (Weber and Buceta, 2011) explored the transcriptional noise in cell-to-cell communication system near the threshold. The dynamics of the autoinducer is dependent on diffusion coefficient and the total noise has a non monotonic behavior. Moreover, they showed that bacteria had adapted their communication processes in order to improve the signal-to-noise ratio. (Bressloff 2016) analyzed ultrasensitivity and noise amplification of V. harveyi quorum sensing system and showed that quorum sensing can protect against the noise amplification of fast environmental fluctuations. Other research works also showed that quorum sensing networks can mitigate the effects of noise (Russo and Slotine 2010; Tabareau et al. 2010). One of the present author (SM) together with Sisir Roy proposed a theoretical framework of this complex biochemical phenomenon by using noisy Burgers equation (Majumdar and Roy 2017a, b). They assumed that a collection of bacteria (densely packet bacteria inside biofilm) behaves as dense granular system. The finite size of the bacteria indicates the existence of an intermediate length scale, which leads to the introduction of a source of fluctuation (non-local noise) in the system. The swimming induced stress on the bacteria can also change the local arrangement of bacteria induced stress fluctuations. Based on this assumption they have formulated cell communication process, which can be described as
where, ∇η = − ν∇2(∆uk) and ∆uk = u − uk .They also emphasized that the non-local hydrodynamical model (based on Ginzburg-Landau framework) can explain the quorum sensing mechanism quite consistently. The noise induced kinematic viscosity ν plays the crucial role to understand the quorum sensing phenomena. Spatiotemporal behaviour of the system and pattern formation (Majumdar et al. 2017) is also studied in detail.
Fungal quorum sensing
Quorum sensing mechanism are not only limited to bacteria. It has been observed in pathogenic fungus Candida albicans also. The communication process of C. albicans is mediated by farnesol (quorum sensing molecule) (Hornby et al. 2001). Other than farnesol, there are other quorum sensing molecules such as tyrosol (second quorum sensing molecule in C. albicans) (Chen et al. 2004), phenylethanol and tryptophol (quorum sensing molecules of Saccharomyces cerevisiae) (Chen and Fink 2006). Fungal communication process are also explored in different fungi such as Histoplasma capsulatum (thermo-dimorphic pathogenic fungus) (Kügler et al. 2000), Ceratocystis ulmi (phytopathogenic fungus) (Hornby et al. 2004), Neurospora crassa (Roca et al. 2005), Saccharomyces cerevisiae (Severin et al. 2008) and Cryptococcus neoformans (Lee et al. 2007). Fernesol affects the regulation of C. albicans filamentation, biofilm formation, oxidative stress, modulation of drug efflux and other microbes (Aspergillus nidulans, Saccharomyces cerevisiae, Aspergillus niger, Aspergillus fumigatus, Fusarium graminearum, Candida dubliniensis, Candida parapsilosis, Paracoccidioides brasiliensis, Mycobacterium smegmatis, Pseudomonas aeruginosa) (Albuquerque & Casadevall, 2012). For recent updated research, one can consider the review article by (Wongsuk et al. 2016).
Quantum information and cell communication
Quantum biology is a concept where quantum physics and biology intersects each other. In the early twentieth century researchers began to understand various complex biological phenomena using powerful microscopes and biophysical theories. Erwin Schrödinger in his famous book “What is Life?” mentioned that quantum mechanics accounts for the stability of living things and their cellular processes through our understanding of quantum mechanics of the stability of molecules and the fact that quantum effects create sometimes large energy gaps between different states of chemical systems (Schrödinger 1944; Majumdar and Pal 2016). Recent experimental evidences have proved the significant and fundamental role of quantum physics in vision, electron and proton tunneling, photosynthesis, magneto-reception and olfactory sensing (Lambert et al. 2013) etc. These ingenuous findings are of paramount importance and have certainly opened up some basic questions: Is there any phenomenon that is triggered by the non-trivial quantum effect in microbiology? Does the probabilistic world of quantum mechanics have any role in microbial communication? If yes then how quantum information theory decode biological information? Can quantum mechanical aspects shed some lights on the survival strategies and communication process of some microorganism? All of these issues are yet not resolved and still are the open challenges to meet. However, in room temperature, to understand the role of quantum effects, we discuss bacterial ion-channels, bacterial nanowire and the perspective of quantum synthetic biology and quantum Turing test in bacterial cell-to-cell communication system.
Bacterial ion-channels and quantum interpretations
Neurophysiology, biophysics, electrophysiology and cognitive science are very much interlinked to each other and a very active field of research to emphasize the activity of human brain, where ion-channels mediated neural signaling play the dominant role. The structural configurations of different ion-channels in human brain are similar to bacterial ion-channels, which provide a pivotal role of the structural basic of signaling mechanism (Doyle et al. 1998). Bacterial ion-channels are protein complexes, which regulate the ions’ flow across the cell membrane and have been indispensable medium in long-range communication. Bacteria have many types of ion-channel such as potassium ion-channel KcsA (Prindle et al. 2015; Doyle et al. 1998) (See Fig. 6), sodium channels (Ren et al. 2001), chloride channels (Iyer et al. 2002), calcium-gated potassium ion channels (Jiang et al. 2002), ionotropic glutamate receptors (Chen et al. 1999).
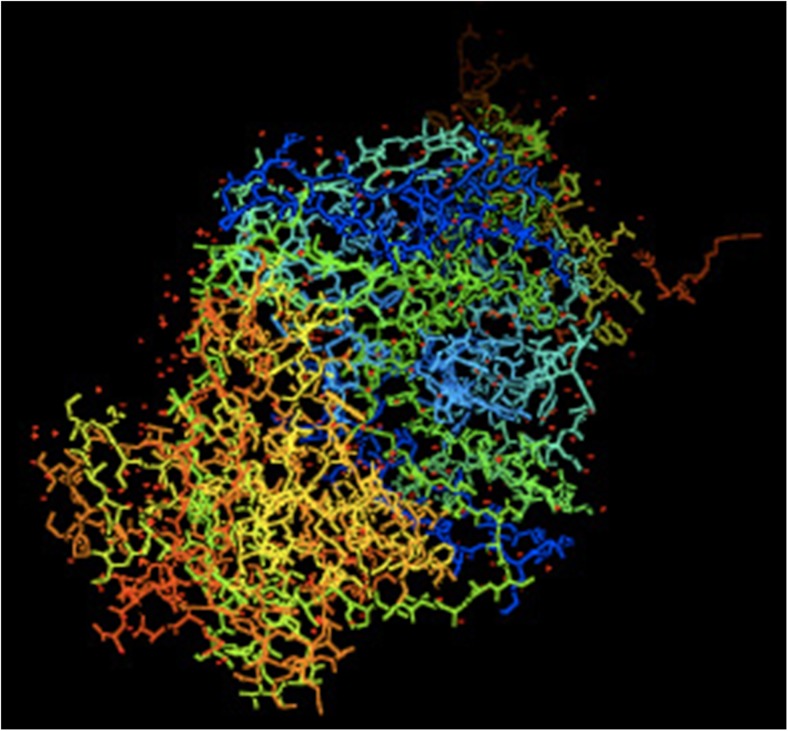
Fig. 6.
Illustration of potassium ion-channel from PDB 1K4C
Ion-channels have been crystallized using x-ray crystallography for better understanding of complex structure protein on atomic level and mechanisms of ion selectivity and conductance are explored as well (Dutzler et al. 2002; Berneche and Roux 2001; Noskov et al. 2004). (Vaziri and Plenio 2010) studied the ion-channels and suggested that the selectivity filter ion-channels may exhibit quantum coherence. (Salari et al. 2017) recently simulated neighboring two ion-channels with a double slit experiment and got decoherence timescales (ion survives approximately 100 ps in channel and approximately 17-53 ps out the channels), which indicates that due to environmental decoherence quantum interference of similar ions is not observed. (Bernroider and Roy 2005) explained that the physical action behind ion permeation though pores requires action orders at a quantum scale and represented a superposition of ion states in the permeation filter of the channels which is close to the filter from its environment as
(where, | ψ⟩ is the ions state vector for the low potassium ion structure and | ai, j⟩ represents the ket vector of either (i = 1, j = 3) or (i = 2, j = 4) ion location). Moreover, they presented an entangled state between the two subsystems of the channel filter as
(The probability of finding K+ in the state a1, 3 and a2, 4 can be calculated by |β|2 = 1 − |α|2 and O is the subsystem) (See details in (Bernroider and Roy 2005)). So, one can find the state transition from above equation, which is equivalent to a fundamental mapping of quantum computation (c-not two-qubit quantum gate).
Majumdar and Roy investigated K+ channel mediated bacterial communication process using nonlinear Schrödinger of the from (Roy & Llinás, 2009)
(where ψ is the wave function of Potassium ion) and arrived Complex Ginzburg-Landau equation by adding perturbation to the above nonlinear Schrödinger equation. They explained potassium wave mechanism as
where ψ(x, t) is a complex field and ϵ > 0 while h = h(Ξ) and g = g(Ξ) are real analytic functions over [0, inf) (Majumdar and Roy 2017). Latter they analyzed this communication system and numerically showed spatiotemporal disordered regimes (defect-mediated turbulence, phase turbulence and spatiotemporal intermittency) and chaos in non-equilibrium bacterial conversation system (Majumdar and Roy 2017a, b).
Bacterial nanowires
Electron transfer is crucial in biology. Microorganisms capture electrons from different sources and transfer them to electron acceptors. Extracellular appendages of bacteria is known as bacterial nanowires, which are the recommended pathways for electron transport in different microbes including metal reducing bacteria and cyanobacteria (photosynthetic bacteria) (El-Naggar et al. 2010). El-Naggar et al. 2010 studied bacterial nanowires and electrical transport along it from Shewanella oneidensis and reported a microbial strategy for extracellular electron transport while (Reguera et al. 2006) presented long range electron transfer across multilayer biofilm.
Quantum synthetic biology: a perspective
Quantum gate circuit can be constructed using quantum information theory, which includes classical information as well. In many cases bacteria communicate to each others using more than one autoinducers and they integrate the information and convey it. Bacteria can learn about the environment and can evolve different signals and finally are able to merge the signals using signal integration circuits. (Karafyllidis 2012) proposed a quantum circuit model of bacterial quorum sensing mechanism, then simulated quorum sensing circuit (based on quantum information theory) and analyzed autoinducers variations, which contain a crucial information about environment. Following this research trajectory, a design principle of the communication process can be devised, which may open up a new paradigm for synthetic biology. We can think it as a quantum synthetic biology, where quantum gate circuit concept allows us to make synthetic bacterial cell to study information processing inside single bacterial cell and its communication system to reveal unknown information processing mechanism in microbial communications (inter and intra species). On other hand, one can also make a synthetic quantum quorum quenching scheme to develop therapy model.
Quantum Turing test in biology
The assertion of Church-Turing hypothesis dictates a close connection between physical systems and model computing machines. Following the hypothesis, a strong possibility of two- way cellular communication between artificial and natural cells opens up through the chemical signaling mechanism in biological systems. The recent experimental observation by (Lentini et al. 2017) has already estimated the efficiency of artificial cells in efficiently imitating the natural cells through cellular Turing test.
Recent experiments also have progressed in this direction to reconstruct the well characterized pathways based on quorum sensing mechanism to build an artificial cells that can imitate the ability of natural cells like V. fisheri, E. coli, P. aeruginusa. The artificial cells of these bacteria can send and sense the chemical information and hence can interact with natural bacterial cells. Luminescence, fluorescence, RT-qPCR and RNA-seq have been observed in artificial cells (Lentini et al. 2017; Majumdar and Pal 2017a, b).
The artificial cells containing DNA can support transcription and translation process (Noireaux and Libchaber 2004), which develops the concept of one way synthetic communication. The quest for quantum basis of information gave birth to the digital data and computers in past. But one can’t deny the scopes of quantum computers. Particularly, to unveil unidentified underlying signaling pathways of bacterial decision making and/or optimal survival strategy we propose cellular quantum Turing test (Deutsch 1985) for digital signal processing as the possible route towards this two-way synthetic communication.
Conclusions
Bacteria understands that being united they stand, divided they misunderstand. We accumulate major portion of bacterial quorum sensing mechanism (experimentally, theoretically) and quorum sensing regulated behaviors such as extracellular polymeric substances and biofilm development, swarming, virulence together with recently discovered potassium ion-channel mediated communication system. In case of bacterial electrical communication, we still don’t know what are the exact informations bacteria can process through this mechanism. Stochastic fluctuation (noise) can also play a very significant role in biochemical conversation. This ongoing research on bacterial collective behavior is highly significant for our every day life as we are surrounded by bacterial world. Even, the numbers of bacterial cells are ten times more than human cells in our body. Thus different kind of bacterial infections are quite common and can be life threatening. (National Institutes of Health 2007) stated
"Biofilms are clinically important, accounting for over 80 percent of microbial infections in the body. Examples include: infections of oral soft tissues, teeth and dental implants; middle ear; gastrointestinal tract; urogenital tract; airway/lung tissue; eye; urinary tract prostheses; peritoneal membrane and peritoneal dialysis catheters, in-dwelling catheters for hemodialysis and for chronic administration of chemotherapeutic agents (Hickman catheters); cardiac implants such as pacemakers, prosthetic heart valves, ventricular assist devices, and synthetic vascular grafts and stents; prostheses, internal fixation devices, percutaneous sutures; and tracheal and ventilator tubing.”
We also demonstrate some key facts of fungal quorum sensing system, which has a major role in agricultural research and food science and technology. Synthetic bacterial communication can reveal different unseen phenomenon in biochemical communication, which is applicable in biotechnology, biocomputing and medical sciences. Bacterial nanowires, ion-channels and quantum approach (including quantum information) of this conversation game are till now has been explored. Researchers can design their experiments based on the proposed quantum phenomena. We extend the subject matter with a new perspective of quantum synthetic system and aim towards the cellular quantum Turing test, which can be one of the major challenges for future research in this context and hopefully will be able to disclose unfinished symphony of microbial communication.
Contributor Information
Sarangam Majumdar, Email: majumdarsarangam@yahoo.in.
Sukla Pal, Email: sukla.ph10@gmail.com.
References
- Albuquerque P, Casadevall A. Quorum sensing in fungi – a review. Med Mycol. 2012;50(4):337–345. doi: 10.3109/13693786.2011.652201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand R, Rai N, Thattai M. Interactions among quorum sensing inhibitors. PLoS One. 2013;8(4):e62254. doi: 10.1371/journal.pone.0062254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguige K, King JR, Ward JP (2005) Modelling antibiotic- and anti-quorum sensing treatment of a spatially-structured Pseudomonas aeruginosa population. J Math Biol 51(5):557–594. 10.1007/s00285-005-0316-8 [DOI] [PubMed]
- Anguige K, King JR, Ward JP, Williams P. Mathematical modelling of therapies targeted at bacterial quorum sensing. Math Biosci. 2004;192(1):39–83. doi: 10.1016/j.mbs.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Balagaddé FK, Song H, Ozaki J, Collins CH, Barnet M, Arnold FH, Quake SR, You L. A synthetic Escherichia coli predator–prey ecosystem. Mol Syst Biol. 2008;4(1):187. doi: 10.1038/msb.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrios AFG, Achenie LE. Escherichia coli autoinducer-2 uptake network does not display hysteretic behavior but AI-2 synthesis rate controls transient bifurcation. Biosystems. 2010;99(1):17–26. doi: 10.1016/j.biosystems.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Barrios AFG, Covo V, Medina LM, Vives-Florez M, Achenie L. Quorum quenching analysis in Pseudomonas aeruginosa and Escherichia coli: network topology and inhibition mechanism effect on the optimized inhibitor dose. Bioprocess Biosyst Eng. 2009;32(4):545–556. doi: 10.1007/s00449-008-0276-7. [DOI] [PubMed] [Google Scholar]
- Bashor CJ, Horwitz AA, Peisajovich SG, Lim WA. Rewiring cells: synthetic biology as a tool to interrogate the organizational principles of living systems. Annu Rev Biophys. 2010;39:515–537. doi: 10.1146/annurev.biophys.050708.133652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434(7037):1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- Baym M, Lieberman TD, Kelsic ED, Chait R, Gross R, Yelin I, Kishony R. Spatiotemporal microbial evolution on antibiotic landscapes. Science. 2016;353(6304):1147–1151. doi: 10.1126/science.aag0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagle SD, Lockles SW (2015) Electical signaling goes bacterial. Nature 527 [DOI] [PubMed]
- Beckmann BE, Knoester DB, Connelly BD, Waters CM, McKinley PK. Evolution of resistance to quorum quenching in digital organisms. Art&Life. 2012;18(3):291–310. doi: 10.1162/artl_a_00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berneche S, Roux B. Energetics of ion conduction through the K+ channel. Nature. 2001;414(6859):73. doi: 10.1038/35102067. [DOI] [PubMed] [Google Scholar]
- Bernroider G, Roy S. Quantum entanglement of K+ ions, multiple channel states, and the role of noise in the brain. SPIE. 2005;5841:205–213. [Google Scholar]
- Brenner K, Karig DK, Weiss R, Arnold FH. Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium. Proc Natl Acad Sci. 2007;104(44):17300–17304. doi: 10.1073/pnas.0704256104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressloff PC (2016) Ultrasensitivity and noise amplification in a model of V. harveyi quorum sensing, Physical Review E. 93(6):062418 [DOI] [PubMed]
- Brown SP. Cooperation and conflict in host-manipulating parasites. Proc R Soc B. 1999;266:1899–1899. [Google Scholar]
- Brown SP, Johnstone RA. Cooperation in the dark: signalling and collective action in quorum-sensing bacteria. Proc R Soc B. 2001;268:961–965. doi: 10.1098/rspb.2001.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárcamo-Oyarce G, Lumjiaktase P, Kümmerli R, Eberl L. Quorum sensing triggers the stochastic escape of individual cells from Pseudomonas putida biofilms. Nat Commun. 2015;6:5945. doi: 10.1038/ncomms6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GQ, Cui C, Mayer ML, Gouaux E. Functional characterization of a potassium-selective prokaryotic glutamate receptor. Nature. 1999;402(6763):817–821. doi: 10.1038/45568. [DOI] [PubMed] [Google Scholar]
- Chen H, Fink GR. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 2006;20(9):1150–1161. doi: 10.1101/gad.1411806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Fujita M, Feng Q, Clardy J, Fink GR. Tyrosol is a quorum-sensing molecule in Candida albicans.Proceedings of the. National academy of Sciences of the United States of America. 2004;101(14):5048–5052. doi: 10.1073/pnas.0401416101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Kim JK, Hirning AJ, Josić K, Bennett MR. Emergent genetic oscillations in a synthetic microbial consortium. Science. 2015;349(6251):986–989. doi: 10.1126/science.aaa3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopp DL, Kirisits MJ, Moran B, Parsek MR. A mathematical model of quorum sensing in a growing bacterial biofilm. J Ind Microbiol Biotechnol. 2002;29(6):339–346. doi: 10.1038/sj.jim.7000316. [DOI] [PubMed] [Google Scholar]
- Chopp DL, Kirisits MJ, Moran B, Parsek MR. The dependence of quorum sensing on the depth of a growing biofilm. Bull Math Biol. 2003;65(6):1053–1079. doi: 10.1016/S0092-8240(03)00057-0. [DOI] [PubMed] [Google Scholar]
- Cox CD, Peterson GD, Allen MS, Lancaster JM, McCollum JM, Austin D, Yan L, Sayler GS, Simpson ML. Analysis of noise in quorum sensing. OMICS A Journal of Integrative Biology. 2003;7(3):317–334. doi: 10.1089/153623103322452422. [DOI] [PubMed] [Google Scholar]
- Czárán T, Hoekstra RF. Microbial communication, cooperation and cheating: quorum sensing drives the evolution of cooperation in bacteria. PLoS One. 2009;4(8):e6655. doi: 10.1371/journal.pone.0006655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danino T, Mondragón-Palomino O, Tsimring L, Hasty J. A synchronized quorum of genetic clocks. Nature. 2010;463(7279):326–330. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datla US, Mather WH, Chen S, Shoultz IW, Täuber UC, Jones CN, Butzin NC. The spatiotemporal system dynamics of acquired resistance in an engineered microecology. Sci Rep. 2017;7(1):16071. doi: 10.1038/s41598-017-16176-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280(5361):295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- Deutsch D (1985) Quantum theory, the Church–Turing principle and the universal quantum computer. Proceedings of the Royal Society A 400(1818):97–117. 10.1098/rspa.1985.0070
- Dilanji GE, Langebrake JB, De Leenheer P, Hagen SJ. Quorum activation at a distance: spatiotemporal patterns of gene regulation from diffusion of an autoinducer signal. J Am Chem Soc. 2012;134(12):5618–5626. doi: 10.1021/ja211593q. [DOI] [PubMed] [Google Scholar]
- Dockery JD, Keener JP. A mathematical model for quorum sensing in Pseudomonas aeruginosa. Bull Math Biol. 2001;63(1):95–116. doi: 10.1006/bulm.2000.0205. [DOI] [PubMed] [Google Scholar]
- Doyle DA, Cabral JM, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280(5360):69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- Dutzler R, Campbell EB, Cadene M, Chait BT, MacKinnon R. X-ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature. 2002;415(6869):287–294. doi: 10.1038/415287a. [DOI] [PubMed] [Google Scholar]
- Eberl HJ, Parker DF, Van Loosdrecht M. A new deterministic spatio-temporal continuum model for biofilm development. Computational and Mathematical Methods in Medicine. 2001;3(3):161–175. [Google Scholar]
- El-Naggar MY, Wanger G, Leung KM, Yuzvinsky TD, Southam G, Yang J, Lau WM, Nealson KH, Gorby YA. Electrical transport along bacterial nanowires from Shewanella oneidensis MR-1. Proc Natl Acad Sci. 2010;107(42):18127–18131. doi: 10.1073/pnas.1004880107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403(6767):335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Emerenini BO, Hense BA, Kuttler C, Eberl HJ. A mathematical model of quorum sensing induced biofilm detachment. PLoS One. 2015;10(7):e0132385. doi: 10.1371/journal.pone.0132385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerenini BO, Sonner S, Eberl HJ. Mathematical analysis of a quorum sensing induced biofilm dispersal model and numerical simulation of hollowing effects.Mathematical. Biosciences & Engineering. 2017;14(3):625–653. doi: 10.3934/mbe.2017036. [DOI] [PubMed] [Google Scholar]
- Endy D. Foundations for engineering biology. Nature. 2005;438(7067):449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- Fagerlind MG, Nilsson P, Harlén M, Karlsson S, Rice SA, Kjelleberg S. Modeling the effect of acylated homoserine lactone antagonists in Pseudomonas aeruginosa. Biosystems. 2005;80(2):201–213. doi: 10.1016/j.biosystems.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Fagerlind MG, Rice SA, Nilsson P, Harlén M, James S, Charlton T, Kjelleberg S. The role of regulators in the expression of quorum-sensing signals in Pseudomonas aeruginosa. J Mol Microbiol Biotechnol. 2003;6(2):88–100. doi: 10.1159/000076739. [DOI] [PubMed] [Google Scholar]
- Fekete A, Kuttler C, Rothballer M, Hense BA, Fischer D, Buddrus-Schiemann K, Lucio M, Müller J, Schmitt-Kopplin P, Hartmann A. Dynamic regulation of n-acyl-homoserine lactone production and degradation in Pseudomonas putida ISOF. FEMS Microbiol Ecol. 2010;72:22–34. doi: 10.1111/j.1574-6941.2009.00828.x. [DOI] [PubMed] [Google Scholar]
- Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14(9):563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- Friman VP, Diggle SP, Buckling A. Protist predation can favour cooperation within bacterial species. Biol Lett. 2013;9(5):20130548. doi: 10.1098/rsbl.2013.0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacte- ria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahlmann A, Moerner WE. Exploring bacterial cell biology with single-molecule tracking and super-resolution imaging. Nat Rev Microbiol. 2014;12(1):9–22. doi: 10.1038/nrmicro3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Aljaro C, Melado-Rovira S, Milton DL, Blanch AR. Quorum-sensing regulates biofilm formation in Vibrio scophthalmi. BMC Microbiol. 2012;12(1):287. doi: 10.1186/1471-2180-12-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403(6767):339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- Goryachev AB, Toh DJ, Lee T. Systems analysis of a quorum sensing network: design constraints imposed by the functional requirements, network topology and kinetic constants. Biosystems. 2006;83(2-3):178–187. doi: 10.1016/j.biosystems.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Gray KM, Passador L, Iglewski BH, Greenberg EP. Interchangeability and specificity of components from the quorum-sensing regulatory systems of Vibrio fischeri and Pseudomonas aeruginosa. J Bacteriol. 1994;176:3076–3080. doi: 10.1128/jb.176.10.3076-3080.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson E, Nilsson P, Karlsson S, Arvidson S. Characterizing the dynamics of the quorum-sensing system in Staphylococcus aureus. J Mol Microbiol Biotechnol. 2004;8(4):232–242. doi: 10.1159/000086704. [DOI] [PubMed] [Google Scholar]
- Haseltine EL, Arnold FH. Implications of rewiring bacterial quorum sensing. Appl Environ Microbiol. 2008;74(2):437–445. doi: 10.1128/AEM.01688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hense BA, Schuster M. Core principles of bacterial autoinducer systems. Microbiol Mol Biol Rev. 2015;79(1):153–169. doi: 10.1128/MMBR.00024-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117(4):500. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornby JM, Jacobitz-Kizzier SM, McNeel DJ, Jensen EC, Treves DS, Nickerson KW. Inoculum size effect in dimorphic fungi: extracellular control of yeast-mycelium dimorphism in Ceratocystis ulmi. Appl Environ Microbiol. 2004;70(3):1356–1359. doi: 10.1128/AEM.70.3.1356-1359.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornby JM, Jensen EC, Lisec AD, Tasto JJ, Jahnke B, Shoemaker R, Dussault P, Nickerson KW. Quorum sensing in the dimorphic fungusCandida albicans is mediated by farnesol. Appl Environ Microbiol. 2001;67(7):2982–2992. doi: 10.1128/AEM.67.7.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries J, Xiong L, Liu J, Prindle A, Yuan F, Arjes HA, Tsimring L, Süel GM. Species-independent attraction to biofilms through electrical signaling. Cell. 2017;168(1):200–209. doi: 10.1016/j.cell.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter GAM, Vasquez FG, Keener JP (2013) A mathematical model and quantitative comparison of the small RNA circuit in the Vibrio harveyi and Vibrio cholerae quorum sensing systems. Phys Biol 10: 046007. http://stacks.iop.org/1478-3975/10/i=4/a=046007 [DOI] [PubMed]
- Iyer R, Iverson TM, Accardi A, Miller C. A biological role for prokaryotic ClC chloride channels. Nature. 2002;419(6908):715–718. doi: 10.1038/nature01000. [DOI] [PubMed] [Google Scholar]
- James S, Nilsson P, James G, Kjelleberg S, Fagerström T. Luminescence control in the marine bacterium Vibrio fischeri: an analysis of the dynamics of lux regulation1. J Mol Biol. 2000;296(4):1127–1137. doi: 10.1006/jmbi.1999.3484. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002;417(6888):515–522. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- Karafyllidis IG. Quantum gate circuit model of signal integration in bacterial quorum sensing. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2012;9(2):571–579. doi: 10.1109/TCBB.2011.104. [DOI] [PubMed] [Google Scholar]
- Karlsson D, Karlsson S, Gustafsson E, Normark BH, Nilsson P. Modeling the regulation of the competence-evoking quorum sensing network in Streptococcus pneumoniae. Biosystems. 2007;90(1):211–223. doi: 10.1016/j.biosystems.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat Rev Genet. 2010;11(5):367. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerber AJ, King JR, Williams P. Deterministic and stochastic modelling of endosome escape by Staphylococcus aureus:“quorum” sensing by a single bacterium. J Math Biol. 2005;50(4):440–488. doi: 10.1007/s00285-004-0296-0. [DOI] [PubMed] [Google Scholar]
- Kügler S, Sebghati TS, Eissenberg LG, Goldman WE. Phenotypic variation and intracellular parasitism by Histoplasma capsulatum. Proc Natl Acad Sci. 2000;97(16):8794–8798. doi: 10.1073/pnas.97.16.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert N, Chen YN, Cheng YC, Li CM, Chen GY, Nori F. Quantum biology. Nat Phys. 2013;9(1):10–18. [Google Scholar]
- Lane N (2015) The unseen world: reflections on Leeuwenhoek (1677) Concerning little animals. Philosophical Transactions of the Royal Society B: Biological Sciences 370(1666): 20140344. 10.1098/rstb.2014.0344 [DOI] [PMC free article] [PubMed]
- Lee H, Chang YC, Nardone G, Kwon-Chung KJ. TUP1 disruption in Cryptococcus neoformans uncovers a peptide-mediated density-dependent growth phenomenon that mimics quorum sensing. Mol Microbiol. 2007;64:591–601. doi: 10.1111/j.1365-2958.2007.05666.x. [DOI] [PubMed] [Google Scholar]
- Leewenhoeck A. Observation, communicated to the publisher by Mr. Antony van Leewenhoeck, in a Dutch letter of the 9 Octob. 1676 here English'd: concerning little animals by him observed in rain-well-sea and snow water; as also in water wherein pepper had lain infused. Philos Trans. 1677;12:821–831. [Google Scholar]
- Lentini R, Yeh Martín N, Forlin M, Belmonte L, Fontana J, Cornella M, Martini L, Tamburini S, Bentley EW, Jousson O, Mansy SS. Two-way chemical communication be- tween artificial and natural cells. ACS Central Science. 2017;3(2):117. doi: 10.1021/acscentsci.6b00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang L, Hashimoto Y, Tsao CY, Wood TK, Valdes JJ, Zafiriou E, Bentley WE. A stochastic model of Escherichia coli AI-2 quorum signal circuit reveals alternative synthesis pathways. Mol Syst Biol. 2006;2(1):67. doi: 10.1038/msb4100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Arthur P, Jacqueline H, Marçal G-S, Munehiro A, Lee Dong-yeon D, San L, Jordi G-O, Süel GM. Metabolic co-dependence gives rise to collective oscillations within biofilms. Nature. 2015;523(7562):550–554. doi: 10.1038/nature14660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Corral MR, Prindle A, Dong-yeon LD, Larkin J, Sagarra GM, Ojalvo GJ, Süel MG. Coupling between distant biofilms and emergence of nutrient time-sharing. Science. 2017;356(6338):638–642. doi: 10.1126/science.aah4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Lavis LD, Betzig E. Imaging live-cell dynamics and structure at the single-molecule level. Mol Cell. 2015;58(4):644–659. doi: 10.1016/j.molcel.2015.02.033. [DOI] [PubMed] [Google Scholar]
- Majumdar S, Datta S, Roy S. Mathematical modelling of quorum sensing and bioluminescence in bacteria. Int J Adv Appl Sci. 2012;1(3):139–146. [Google Scholar]
- Majumdar S, Mondal S. Conversation game: talking bacteria. J Cell Commun Signal. 2016;10(4):331–335. doi: 10.1007/s12079-016-0333-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar S, Pal S. Quorum sensing: a quantum perspective. Journal of cell communication and signaling. 2016;10(3):173–175. doi: 10.1007/s12079-016-0348-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar S, Pal S. Cross-species communication in bacterial world. Journal of cell communication and signaling. 2017;11(2):187–190. doi: 10.1007/s12079-017-0383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar S, Pal S. Bacterial intelligence: imitation games, time-sharing, and long-range quantum coherence. Journal of cell communication and signaling. 2017;11(3):281–284. doi: 10.1007/s12079-017-0394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar S, Roy S (2017a) Relevance of quantum mechanics in bacterial communication. NeuroQuantology. Accepted
- Majumdar S, Roy S (2017b) Spatiotemporal patterns and chaos in non-equilibrium bacterial communication. 17th BIOMAT International Symposium on Mathematical and Computational Biology, Moscow
- Majumdar S, Roy S, Llinas R (2017) Bacterial conversations and pattern formation. bioRxiv. 10.1101/098053
- Marguet P, Balagadde F, Tan C, You L. Biology by design: reduction and synthesis of cellular components and behaviour. J R Soc Interface. 2007;4(15):607–623. doi: 10.1098/rsif.2006.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marketon MM, Glenn SA, Eberhard A, González JE. Quorum sensing controls exopolysaccharide production in Sinorhizobium meliloti. J Bacteriol. 2003;185(1):325–331. doi: 10.1128/JB.185.1.325-331.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Moustafa D, Smith CD, Goldberg JB, Bassler BL. The RhlR quorum-sensing receptor controls Pseudomonas aeruginosa pathogenesis and biofilm development independently of its canonical homoserine lactone autoinducer. PLoS Pathog. 2017;13(7):e1006504. doi: 10.1371/journal.ppat.1006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadell CD, Xavier JB, Levin SA, Foster KR. The evolution of quorum sensing in bacterial biofilms. PLoS Biol. 2008;6(1):e14. doi: 10.1371/journal.pbio.0060014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandagopal N, Elowitz MB. Synthetic biology: integrated gene circuits. Science. 2011;333(6047):1244–1248. doi: 10.1126/science.1207084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institutes of Health (2007) Immunology of Biofilms (R01), grants.nih.gov/grants/guide/pa-files/PA-07-288.html
- Netotea S, Bertani I, Steindler L, Kerényi Á, Venturi V, Pongor S. A simple model for the early events of quorum sensing in Pseudomonas aeruginosa: modeling bacterial swarming as the movement of an" activation zone". Biol Direct. 2009;4(1):6. doi: 10.1186/1745-6150-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson P, Olofsson A, Fagerlind M, Fagerström T, Rice S, Kjelleberg S, Steinberg P. Kinetics of the AHL regulatory system in a model biofilm system: how many bacteria constitute a “quorum”? J Mol Biol. 2001;309(3):631–640. doi: 10.1006/jmbi.2001.4697. [DOI] [PubMed] [Google Scholar]
- Noireaux V, Libchaber A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc Natl Acad Sci U S A. 2004;101(51):17669–17674. doi: 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov SY, Berneche S, Roux B. Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature. 2004;431(7010):830–834. doi: 10.1038/nature02943. [DOI] [PubMed] [Google Scholar]
- Pérez-Velázquez J, Gölgeli M, García-Contreras R. Mathematical modelling of bacterial quorum sensing: a review. Bull Math Biol. 2016;78(8):1585–1639. doi: 10.1007/s11538-016-0160-6. [DOI] [PubMed] [Google Scholar]
- Prindle A, Jintao L, Munehiro A, San L, Jordi G-O, Süel GM. Ion channels enable electrical communication in bacterial communities. Nature. 2015;527(7576):59–63. doi: 10.1038/nature15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan DN, Tsao CY, Wu HC, Bentley WE. Quorum sensing desynchronization leads to bimodality and patterned behaviors. PLoS Comput Biol. 2016;12(4):e1004781. doi: 10.1371/journal.pcbi.1004781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiñones B, Dulla G, Lindow SE. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol Plant-Microbe Interact. 2005;18(7):682–693. doi: 10.1094/MPMI-18-0682. [DOI] [PubMed] [Google Scholar]
- Reguera G, Nevin KP, Nicoll JS, Covalla SF, Woodard TL, Lovley DR. Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Appl Environ Microbiol. 2006;72(11):7345–7348. doi: 10.1128/AEM.01444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Navarro B, Xu H, Yue L, Shi Q, Clapham DE. A prokaryotic voltage-gated sodium channel. Science. 2001;294(5550):2372–2375. doi: 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- Roca MG, Arlt J, Jeffree CE, Read ND. Cell biology of conidial anastomosis tubes in Neurospora crassa. Eukaryot Cell. 2005;4(5):911–919. doi: 10.1128/EC.4.5.911-919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Llinás R. Relevance of quantum mechanics on some aspects of ion channel function. Comptes rendus biologies. 2009;332(6):517–522. doi: 10.1016/j.crvi.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo G, Slotine JJE. Global convergence of quorum-sensing networks. Phys Rev E. 2010;82(4):041919. doi: 10.1103/PhysRevE.82.041919. [DOI] [PubMed] [Google Scholar]
- Salari V, Naeij H, Shafiee A. Quantum Interference and Selectivity through Biological Ion Channels. Sci Rep. 2017;7:41625. doi: 10.1038/srep41625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopf JW. Disparate rates, differing fates: tempo and mode of evolution changed from the Precambrian to the Phanerozoic. Proc Natl Acad Sci U S A. 1994;91(15):6735–6742. doi: 10.1073/pnas.91.15.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopf JW, Kitajima K, Spicuzza MJ, Kudryavtsev AB, Valley JW. SIMS analyses of the oldest known assemblage of microfossils document their taxon-correlated carbon isotope compositions. Proc Natl Acad Sci U S A. 2018;115(1):53–58. doi: 10.1073/pnas.1718063115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger E. What is life? Cambridge: Cambridge University Press; 1944. [Google Scholar]
- Scott SR, Hasty J. Quorum sensing communication modules for microbial consortia. ACS Synth Biol. 2016;5(9):969–977. doi: 10.1021/acssynbio.5b00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin FF, Meer MV, Smirnova EA, Knorre DA, Skulachev VP. Natural causes of programmed death of yeast Saccharomyces cerevisiae. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 2008;1783(7):1350–1353. doi: 10.1016/j.bbamcr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Shapiro JA. Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol. 1998;52:81–104. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- Shou W, Ram S, Vilar JM. Synthetic cooperation in engineered yeast populations. Proc Natl Acad Sci. 2007;104(6):1877–1882. doi: 10.1073/pnas.0610575104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson ML. Rewiring the cell: synthetic biology moves towards higher functional complexity. Trends Biotechnol. 2004;22(11):555–557. doi: 10.1016/j.tibtech.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Solano C, Echeverz M, Lasa I. Biofilm dispersion and quorum sensing. Curr Opin Microbiol. 2014;18:96–104. doi: 10.1016/j.mib.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Song H, Payne S, Gray M, You L. Spatiotemporal modulation of biodiversity in a synthetic chemical-mediated ecosystem. Nat Chem Biol. 2009;5(12):929–935. doi: 10.1038/nchembio.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456(7221):516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabareau N, Slotine JJ, Pham QC, Breakspear M (2010) How synchronization protects from noise. PLoS Comput Biol 6(1):e1000637. 10.1371/journal.pcbi.1000637 [DOI] [PMC free article] [PubMed]
- Thompson JA, Oliveira RA, Djukovic A, Ubeda C, Xavier KB. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 2015;10(11):1861–1871. doi: 10.1016/j.celrep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- Vaziri A, Plenio MB. Quantum coherence in ion channels: resonances, transport and verification. New J Phys. 2010;12(8):085001. [Google Scholar]
- Viretta AU, Fussenegger M. Modeling the quorum sensing regulatory network of human-pathogenic Pseudomonas aeruginosa. Biotechnol Prog. 2004;20:670–678. doi: 10.1021/bp034323l. [DOI] [PubMed] [Google Scholar]
- Vuong C, Gerke C, Somerville GA, Fischer ER, Otto M. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J Infect Dis. 2003;188(5):706–718. doi: 10.1086/377239. [DOI] [PubMed] [Google Scholar]
- Wang M, Schaefer AL, Dandekar AA, Greenberg EP. Quorum sensing and policing of Pseudomonas aeruginosa social cheaters. Proc Natl Acad Sci. 2015;112(7):2187–2191. doi: 10.1073/pnas.1500704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. Mathematical modeling of quorum-sensing control in biofilms. In: Balaban N, editor. Control of biofilm infections by signal manipulation, vol. 2 of Springer Series on Biofilms. Berlin: Springer; 2008. pp. 79–108. [Google Scholar]
- Ward JP, King JR, Koerber AJ, Croft JM, Sockett RE, Williams P. Early development and quorum sensing in bacterial biofilms. J Math Biol. 2003;47(1):23–55. doi: 10.1007/s00285-002-0190-6. [DOI] [PubMed] [Google Scholar]
- Ward JP, King JR, Koerber AJ, Croft JM, Sockett RE, Williams P. Cell-signalling repression in bacterial quorum sensing. Mathematical Medicine and Biology. 2004;21(3):169–204. doi: 10.1093/imammb/21.3.169. [DOI] [PubMed] [Google Scholar]
- Ward JP, King JR, Koerber AJ, Williams P, Croft JM, Sockett RE. Mathematical modelling of quorum sensing in bacteria. Mathematical Medicine and Biology. 2001;18(3):263–292. [PubMed] [Google Scholar]
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- Weber M, Buceta J. Noise regulation by quorum sensing in low mRNA copy number systems. BMC Syst Biol. 2011;5(1):11. doi: 10.1186/1752-0509-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P, Winzer K, Chan WC, Camara M. Look who’s talking: communication and quorum sensing in the bacterial world. Philos Trans R Soc Lond Ser B Biol Sci. 2007;362(1483):1119–1134. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongsuk T, Pumeesat P, Luplertlop N. Fungal quorum sensing molecules: role in fungal morphogenesis and pathogenicity. J Basic Microbiol. 2016;56(5):440–447. doi: 10.1002/jobm.201500759. [DOI] [PubMed] [Google Scholar]
- Yan J, Nadell CD, Stone HA, Wingreen NS, Bassler BL. Extracellular-matrix-mediated osmotic pressure drives Vibrio cholerae biofilm expansion and cheater exclusion. Nat Commun. 2017;8(1):327. doi: 10.1038/s41467-017-00401-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L, Cox RS, III, Weiss R, Arnold FH. Programmed population control by cell–cell communication and regulated killing. Nature. 2004;428(6985):868–871. doi: 10.1038/nature02491. [DOI] [PubMed] [Google Scholar]
- Zhao J, Wang Q. Three-dimensional numerical simulations of biofilm dynamics with quorum sensing in a flow cell. Bull Math Biol. 2017;79(4):884–919. doi: 10.1007/s11538-017-0259-4. [DOI] [PubMed] [Google Scholar]