Abstract
Legionella pneumophila invades protozoa with an “accidental” ability to cause pneumonia upon transmission to humans. To support its nutrition during intracellular residence, L. pneumophila relies on host amino acids as the main source of carbon and energy to feed the TCA cycle. Despite the apparent lack of a requirement for glucose for L. pneumophila growth in vitro and intracellularly, the organism contains multiple amylases, which hydrolyze polysaccharides into glucose monomers. Here we describe one predicted putative amylase, LamB, which is uniquely present only in L. pneumophila and L. steigerwaltii among the ~60 species of Legionella. Our data show that LamB has a strong amylase activity, which is abolished upon substitutions of amino acids that are conserved in the catalytic pocket of amylases. Loss of LamB or expression of catalytically-inactive variants of LamB results in a severe growth defect of L. pneumophila in Acanthamoeba polyphaga and human monocytes-derived macrophages. Importantly, the lamB null mutant is severely attenuated in intra-pulmonary proliferation in the mouse model and is defective in dissemination to the liver and spleen. Our data show an essential role for LamB in intracellular replication of L. pneumophila in amoeba and human macrophages and in virulence in vivo.
Introduction
The accidental human pathogen, Legionella pneumophila, causes an atypical pneumonia when water droplets, stemming from a contaminated water source such a cooling tower or humidifier, are inhaled by humans, which are considered as accidental host1–3. Over 20 protozoa species are known to harbor Legionella species, likely with more yet to be identified4. Growth within the natural protozoan host serves as a “training grounds”, priming for infection of human alveolar macrophages, as these bacteria are more infectious than their free-living counterparts5–7. Success in replicating in macrophages may have been facilitated by the exploitation of evolutionarily conserved host processes, which allow L. pneumophila to modulate conserved pathways in both macrophages and protozoa4,8–10. Inhaled bacteria enter into the lungs where they primarily reside and proliferate within alveolar macrophages11–13. The intracellular lifecycle in the evolutionarily distant host cells is nearly identical4. Once the bacterium enters the host cell, it actively evades lysosomal fusion and intercepts ER-derived secretory vesicles to generate and ER-derived vacuole, known as the Legionella-containing vacuole (LCV)14–17, and modulate a plethora of cellular and innate immune processes18–21.
Essential to intracellular replication is the Dot/Icm Type 4b secretion system (T4SS), which inject proteins, known as “effectors”, from the bacterium to the host cytoplasm to modulate host processes22–24. Because of the broad host range, L. pneumophila has evolved over 320 effectors that are translocated by the Dot/Icm system and utilized as a “toolbox” to modulate cellular processes of various environmental hosts25–29. Many unique mechanisms of interfering with host processes, such as lysosomal-evasion and trafficking, have been identified and attributed to specific Dot/Icm effectors23,25,30–33. The effector’s ability to interfere with the function of eukaryotic host target proteins, comes from their evolutionary history; many effectors are derived from eukaryotic proteins acquired by inter-kingdom horizontal gene transfer (HGT)26,34–36.
The primary food source for L. pneumophila is amino acids which are used for carbon and energy through feeding the TCA cycle37–40.The generation of host amino acids by L. pneumophila is an effector-driven process41. Substantial generation of host amino acids is required in human macrophages and amoebae where the effector AnkB hijacks the host ubiquitin-proteasome protein degradation machinery, which is required for successful pathogen replication in the host41–44. In contrast to human macrophages and amoebae, during infection of mouse macrophages with the L. pneumophila LP02 strain, the mammalian target of rapamycin complex 1 (mTORC1), a nutrient/energy sensor, is inhibited by multiple effectors to prevent protein synthesis, thus liberating amino acids for bacterial consumption45. Distinct pathogen mechanisms of generating host cell amino acids may be employed within diverse host cells and the pathogen mechanism may differ by various strains of L. pneumophila to acquire the high levels of host amino acids needed for replication.
Glucose is minimally metabolized by L. pneumophila through the Entner-Doudoroff (ED) pathway39,46–48. Traditional glycolysis through the Embden-Meyeroff-Parnas (EMP) pathway is also minimal, despite all the necessary genes being present in the L. pneumophila genome39,46. Glucose does not support growth of L. pneumophila, but it is predominantly imported by L. pneumophila upon termination of growth39,46, as the bacterium is preparing for cellular egress, and used mainly for the generation of the storage molecule, poly-3-hydroxybutyrate (PHB) through the ED pathway46,49. During nutrition deprivation, PHB is converted to acetyl-CoA that feeds the TCA cycle50–52. Genes involved in glucose metabolism and glucose uptake are up-regulated during growth in amoebae and may play a role in infection52,53.
Amylases are a conserved group of enzymes that catalyze hydrolysis of starch and glycogen into glucose54. They are members of a larger family, called glucosidases, and include other enzymes such as cellulase and lactase55. Interestingly, despite the minimal need of glucose by L. pneumophila, four putative amylases have been identified in the L. pneumophila genome, Lpg0422, Lpg1669, Lpg1671, and Lpg2528. The Lpg0422 (GamA) enzyme is the only characterized amylase56. It is secreted by the Type II secretion system (T2SS) and expressed during exponential growth but not required for intracellular growth53,56. Lpg1669 is a putative amylase that lacks putative secretion signals for the T4SS or T2SS, based on bioinformatical analysis. Lpg1671 is predicted to be a T4SS substrate but its role in intracellular infection is not known57,58. The gene for gamA is found in most Legionella species and lpg1669 and lpg1671 are found in three species of Legionella.
The predicted putative amylase, Lpg2528, has been designated as LamB. Among the 60 Legionella species, L. pneumophila and L. steigerwaltii are the only two Legionella species to harbor lamB. Since L. pneumophila is responsible for 85% of Legionnaire’s disease cases, we decided to determine the role of LamB in the intracellular infection of amoebae and human macrophages2,59. Here we show that despite the minimal role of glucose in L. pneumophila metabolism, the LamB amylase is surprisingly necessary for intracellular replication in amoebae and human macrophages, and is required for virulence in vivo, in the A/J mouse model.
Results
Identification of amylases in L. pneumophila
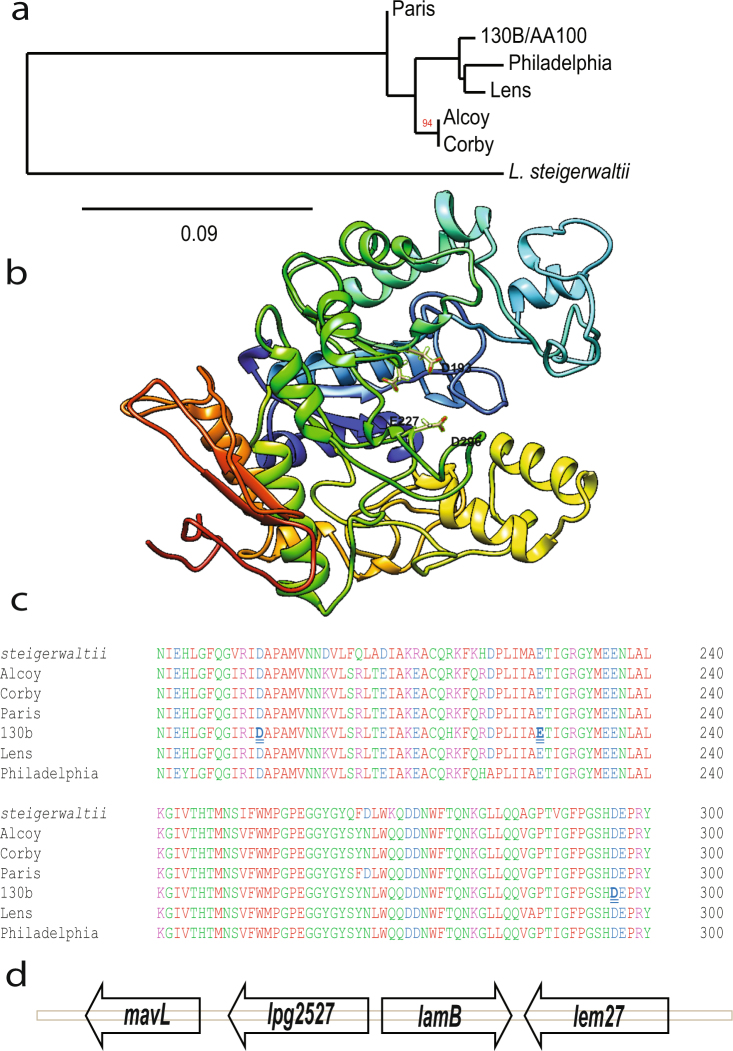
Based on domain sequence homology, three new putative amylases were identified in the L. pneumophila genome, in addition to the one described (GamA) amylase (Fig. S1a–c)56,60. The Lpg2528 putative amylase is designated as LamB, which is encoded by a monocistronic gene (Fig. 1d). Considering LamB is only present in L. pneumophila and L. steigerwaltii, of the 60 Legionella species, it is more likely that LamB has been acquired after the speciation event of L. pneumophila and suggests that L. steigerwaltii may have arisen recently from L. pneumophila (Fig. 1a). The evolution of this gene in L. pneumophila mirrors that of the strain evolution (Fig. 1a). L. pneumophila strain Lens is most related in genome sequence homology to strain 130b/AA100, which is seen with lamB (Branch length, 99)61. Similarly, L. pneumophila strain Alcoy is most homologous to strain Corby, as also seen with lamB (Branch length, 94)61. LamB shares amino acids sequence homology only with other soil and freshwater organisms such as, Methylobacterium and Insolitispirillum (see Supplementary Fig. S2). Thus, it is likely that lamB may have been acquired by HGT from other intra-amoebal or planktonic, environmental organisms. Because L. pneumophila is responsible for 85% of Legionnaire’s disease cases, we characterized the role of this enzyme in the intracellular infections of human monocyte-derived macrophages (hMDMs) and A. polyphaga2,59.
Figure 1.
LamB is a putative amylase unique to L. pneumophila. (a) Phylogram representation of LamB divergence in L. pneumophila strains and L. steigerwaltii. Measure of node support was determined by aLRT using Phylogeny.fr (b) LamB is conserved among L. pneumophila strains and is only found in one other Legionella species, L. steigerwaltii. (c) The structure of LamB, generated from I-TASSER server, which suggests it is an amylase96. Highlighted within the catalytic binding pocket of amylases are resides critical for catalytic activity, D193, E227, and D296. The lamB gene is found on a monocistronic operon within the L. pneumophila genome, this area of the chromosome is shown in (d).
Structure of LamB and its potential secretion
LamB has an α-amylase domain (residues 18–376) that is structurally similar to the crystalized glucosidase of Streptococcus mutants, SmDG (Fig. S1d)62,63. The putative catalytic site, which is located within the predicted catalytic pocket of the enzyme, is conversed amongst amylases and in LamB of L. pneumophila and L. steigerwaltii (Figs 1b,c and S1d). Iterative Threading Assembly Refinement (I-TASSER) is a bioinformatics method of predicting the three-dimensional structure of proteins based on fold recognition64,65. The server also predicts ligand binding sites, and gene ontology. Structural modeling of LamB using the I-TASSER database shows structural similarity with other crystalized glucosidases (Fig. 1b).
L. pneumophila proteins can access the host cytosol by two major routes, translocation via the T4SS or through secretion into the LCV lumen by the type II secretion system (T2SS) and into the cytosol through the semipermeable LCV membrane22,66. L. pneumophila secretes many proteins via the T2SS, which is critical for intracellular growth and pulmonary disease67–69. Twenty proteins were identified to be secreted by the T2SS of L. pneumophila while an additional 250+ proteins have been suggested to contain a putative T2SS signal70. LamB was not identified by any of these methods as potential type-II substrate70. In addition, LamB lacks the N-terminal secretion signal characteristic of T2SS substrates69.
In order to be translocated by the Dot/Icm translocation system, effectors are recognized by a translocation signal on the C-terminus; alterations at the C-terminus of effectors causes a failure in translocation57,71,72. However, no translocation consensus sequence exists for all effectors of the Dot/Icm translocation system. Machine learning techniques have identified conserved bi- and tri-residues at the C-terminal end of L. pneumophila effector proteins, 10% of known effectors do not contain at any of these motifs and some proteins harboring these motifs are not translocated effectors57,73. LamB contains seventeen bi-residues identified to be heavily enriched in the last 100 amino acids of the C-terminus of T4SS effector proteins (see Supplementary Fig. S3)73. To determine whether LamB was Dot/Icm-translocated, the adenylate cyclase (CyaA) reporter function was used74,75. Transformation of plasmids expressing reporters CyaA-LamB or the positive control, CyaA-RalF, as fusion proteins was performed in WT L. pneumophila and the translocation-deficient mutant, dotA. The data showed that the CyaA-LamB reporter was not translocated by the Dot/Icm translocation system, as there was no significant difference between the secretion of CyaA-LamB by WT L. pneumophila or the dotA mutant (Student t-test, p > 0.2); whereas, the control, CyaA-RalF, was readily translocated into the host cells during infection with WT L. pneumophila but not the dotA mutant (Student t-test, p < 0.01) (Fig. 2).
Figure 2.
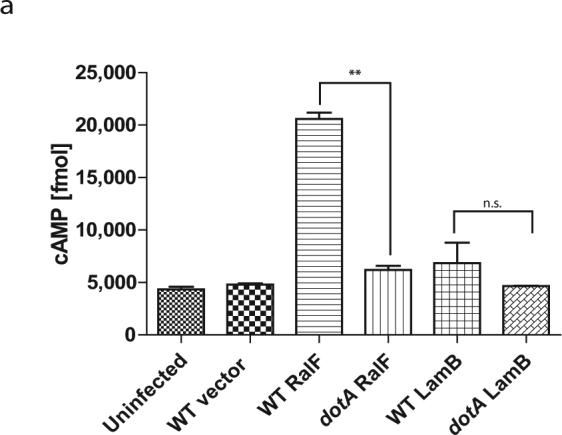
CyaA-LamB reporter is not translocated by the Dot/Icm T4SS. (a) Adenylate cyclase fusions of LamB expressed in L. pneumophila and infected into hMDMs for 1 hr, in triplicate, using known T4SS effector, RalF, as a positive control. Production of cAMP was assessed by ELISA. Data is shown as mean cAMP concentration ± SD, n = 3 independent infections.
The amylase activity of LamB
To confirm the putative enzymatic activity of LamB as an amylase, in vitro biochemical activity was determined by using the standard amylase activity colorimetric assay, quantifying the cleavage of ethylidene-pNP-G7 to p-nitrophenol, which can be measured at 405 nm76,77. Three highly conserved residues, D193, E227, and D296 were identified in the catalytic pocket of LamB and confirmed with domain alignment to other amylases (Fig. 1b,c). Constructs harboring native LamB or three LamB variants of single amino acid substitutions in the catalytic pocket were expressed in E. coli as GST fusion proteins, controlled by an IPTG inducible promotor. Expression of these proteins was confirmed by western blot (Fig. S4). With IPTG induction, amylase activity was highest for the wild type protein compared to uninduced (Student t-test, p < 0.001) (Fig. 3). The three catalytic mutants showed no amylase activity after inducing with IPTG. These data confirmed that LamB is indeed an amylase and that the identified catalytic pocket is essential for enzymatic activity.
Figure 3.
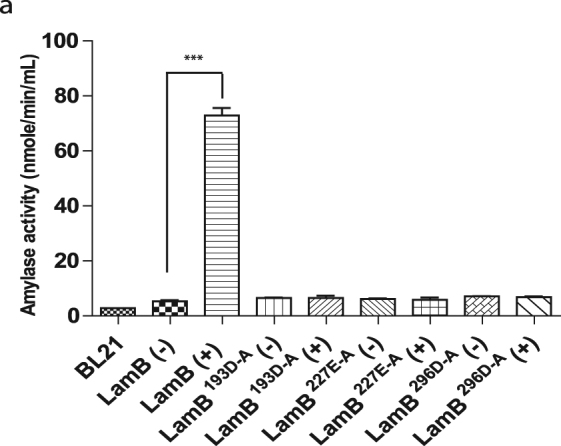
LamB is a functional amylase. (a) Amylase activity of ITPG-inducible (+), GST-LamB fusions and catalytic site mutants, expressed in E. coli, was assessed by colormetric assay of cleavage of an artificial compound. Data are representative of three independent experiments represented by mean amylase activity, of triplicate sample, as measured by cleavage of ethylidene-pNP-G7 into p-nitrophenol, shown as mean amylase activity ± SD, n = 3 independent cultures.
Requirement of LamB for growth in amoebae and hMDMs
In order to test the role of LamB in intracellular growth, a lamB null mutant was generated. Complementation was achieved by expression of lamB on a plasmid, lamB/C. The LamB variants with amino acid substitutions in the catalytic domain of LamB, D193A, E227A, and D296A were also introduced into the lamB null mutant (Fig. 1c). Infections of Acanthamoeba polyphaga or hMDMs were performed, as we previously described78.
The lamB mutant was severely defective for intracellular growth in hMDMs and A. polyphaga (Figs 4 and 5). At 24 h post-infection, there was a significant difference in the replication of the null mutant and the three catalytically inactive mutants compared to WT L. pneumophila (Two-way ANOVA, p < 0.001). The growth defect was partially restored to the mutant by in trans-complementation of the gene, which is likely due to loss of the plasmid. However, complementation of the null mutant with any of the three catalytic variants did not restore any growth to the lamB mutant in A. polyphaga or hMDMs (Figs 4 and 5). This defect is not attributed to a growth defect in vitro, as the lamB mutant grows just as well as the WT strain in broth (see Supplementary Fig. S5). These data show that LamB is necessary for intracellular replication of L. pneumophila in both hMDMs and A. polyphaga. Indeed, it is the amylase activity of LamB that contributes to its essential role in intracellular growth, indicating the requirement for degradation of polysaccharides by L. pneumophila. This is surprising, considering the minimal role of glucose in metabolism of L. pneumophila, and that it is mainly utilized during late stages of growth to synthesize the PHB storage compound.
Figure 4.
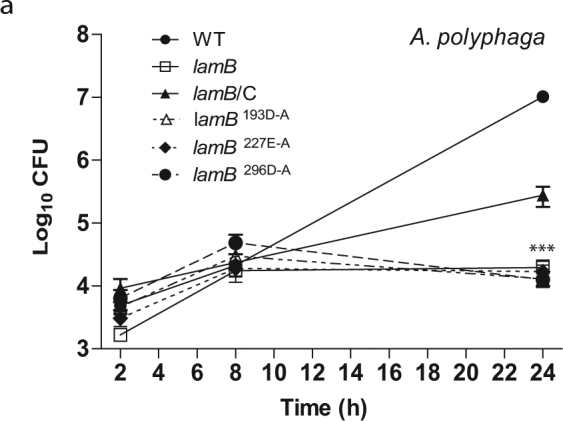
LamB is required for growth in amoebae. To determine intra-vacuolar replication of the WT strain, the dotA mutant, the lamB mutant, catalytic mutants (D193A, E227A, and D296A), and complemented lamB mutant (lamB/C), A. polyphaga were infected and number of CFUs were determined at 2, 8, and 24 h post-infection. Data points represent (mean CFUs ± SD, n = 3) and are representative of three independent experiments.
Figure 5.
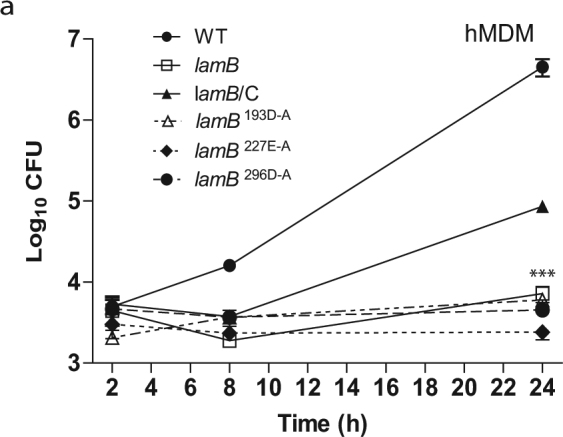
LamB is required for growth in hMDMs. To determine intra-vacuolar replication of the WT strain, the dotA mutant, the lamB mutant, catalytic mutants (D193A, E227A, and D296A), and complemented lamB mutant (lamB/C), hMDMs were infected and number of CFUs were determined at 2, 8, and 24 h post-infection. Data points represent (mean CFUs ± SD, n = 3) and are representative of three independent experiments.
To determine if generation of host glucose by LamB was necessary for intracellular replication, A. polyphaga was supplemented with exogenous glucose during infection (Fig. S6). Glucose supplementation did not rescue the lamB mutant for its defect in intracellularly replication nor did it alter the growth of the WT strain or the complemented mutant (Fig. S6).
Role of LamB in vivo
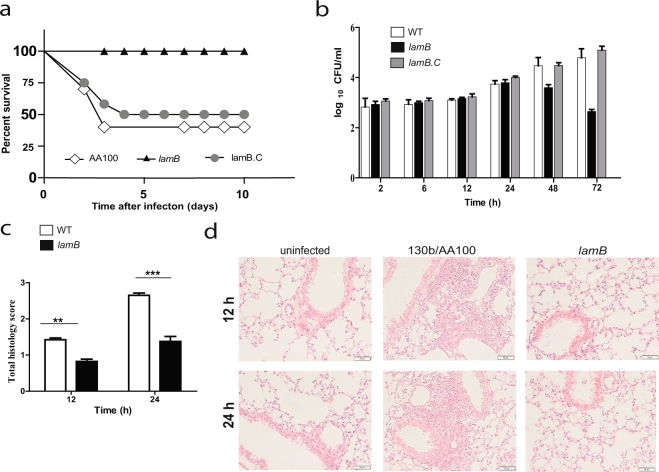
Given that A. polyphaga and hMDMs restrict the lamB mutant, we sought to determine the role of LamB in intrapulmonary growth in the mouse model, in vivo. Intra-trachael infection of A/J mice with WT L. pneumophila, the lamB mutant, or the complemented mutant (lamB/C) was performed, as we described previously43. Within 10 days, 50% of the mice infected with WT or the complemented mutant had died. However, 100% of mice infected with the lamB mutant survived for the 10 days of the study (Fig. 6a). Analysis of bacterial burden in the lungs of surviving mice, at 24, 48, and 72 hrs showed that the lamB mutant had significantly decreased numbers of bacteria within the lungs, compared to the WT strain (Student t-test, p < 0.05) (Fig. 6b). The defective phenotype was completely recovered by complementation, indicating minimal loss of plasmid in vivo compared to ex vivo infection (Figs 4, 5). Histopathology on pulmonary biopsies taken at 12 and 24 hrs post-infection, with wild type L. pneumophila showed severe inflammatory infiltrates of mononuclear cells (Fig. 6c,d). In contrast, following challenge with the lamB mutant, minimal inflammatory infiltration into the alveolar, bronchial, or peribronical spaces was observed (Fig. 6c,d).
Figure 6.
Role of LamB in virulence in A/J mice. Mice were infected intra-trachaelly with 106 CFUs of WT (white), the lamB mutant (black), or the completed, lamB/C (grey). (a) Survival of the mice over days and (b) CFU organ burden in the lungs was assessed at the various time points. Pulmonary histopathology scores at 12 h (Student t-test, p < 0.01) and 24 h (Student t-test, p < 0.005) are shown in (c) and representative images of uninfected, WT, and lamB are shown in (d).
The lamB mutant was less efficient in disseminating to the liver and spleen compared to the WT strain (Fig. 7a,b). At 48 hrs post-infection, there was a significant decrease in the amount of bacteria in the liver of mice infected with the lamB mutant compared to the WT strain (Student t-test, p < 0.01). Compared to the WT strain, fewer lamB mutants disseminated into the spleen at 48 hrs (Student t-test, p < 0.05) and 72 hrs (Student t-test, p < 0.01) post-infection compared to the WT strain. The reduced dissemination of the lamB mutant was completely restored upon complementation by lamB.
Figure 7.
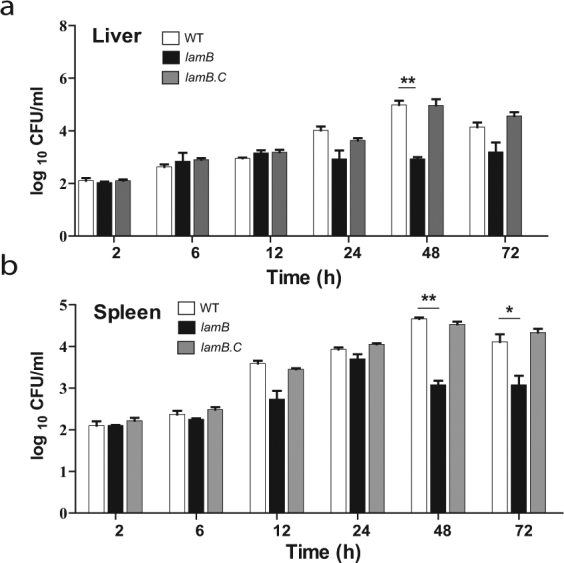
The role of LamB in dissemination of L. pneumophila in A/J mice. Mice were infected intra-trachaelly with 106 CFUs of WT (white bars), the lamB mutant (black bars), or the completed, lamB/C (grey bars). Dissemination of the bacteria to the (a) liver and (b) spleen, as measured by CFU organ burden was assessed at the various time points.
Discussion
A “bipartite” metabolism has been used to describe the nutritional needs and metabolic regulation of L. pneumophila4,47,50,79. During early intracellular replication within human macrophages or amoebae, L. pneumophila relies on amino acids to generate carbon and energy from the TCA cycle37,80. Once amino acid levels become low, the bacteria undergo growth phase transition, switching from the replicative phase to the transmissive phase81–84. At this point, L. pneumophila increases uptake and utilization of glucose and converts it into the storage compound PHB46,47. Experiments with 13C-glucose demonstrated that glucose is used for de novo synthesis of amino acids and PHB during late stages of infection39. Additionally, labeling of glucose demonstrated a carbon flux from glucose to pyruvate via the Enter Doudoroff (ED) pathway but not the Pentose Phosphate Pathway (PPP)39. However, addition of excess exogenous glucose does not result in increased growth of the organism during any stage46. Therefore, generation of excess glucose in the host, as a source of carbon and energy, through degradation of polysaccharides by LamB is unlikely to support growth. Thus, it is surprising to identify a major role for LamB in intracellular growth, since L. pneumophila mainly utilizes amino acids for growth85. We speculate LamB is involved in processes other than nutritional virulence85,86.
Uptake of glucose is increased by L. pneumophila during post-exponential growth, most notably for the generation of the storage molecule, PHB39,46,49, and nutrient importers are important for intracellular growth of L. pneumophila6,87. Having large stores of PHB allows the organism to persist outside of the host for extended periods of time51.Transcriptomic studies in the human macrophage cell line THP-1 have demonstrated that expression of lamB is highest early in infection (8 hrs) rather than later (14 hrs), opposite of when the organism starts increasing consumption of glucose46,88. Faucher et al. also classified this gene as “highly induced in cells”88. LamB may be involved in increasing availability of glucose in the host ahead of when the organism prepares to utilize it during the late stages of infection. However, our data excludes that possibility. Increasing the availability of glucose with an amylase could allow L. pneumophila to generate more PHB, promoting long-term survival. It is possible that failure to store sufficient amounts of PHB by the lamB mutant results in an early defect in intracellular growth, due to the lack of a rapid influx of acetyl-CoA from reduced levels of PHB. Alternatively, since amylases are known to act on the glycosylation of proteins, LamB may be acting on the post-translational modification of host proteins to control processes important for replication, independent of nutrition or PHB storage89. This could allow the bacterium to evade some aspect of the innate host immune response necessary for successful intracellular replication. Future studies are aimed at identification of target(s) of LamB and how they contribute to infection. Considering LamB is unique to L. pneumophila and its loss causes complete defect in intracellular growth in macrophages and amoebae, and attenuation in vivo, it may contribute to the enhanced virulence of L. pneumophila and its prevalence as a disease-causing species compared to other Legionella species.
Bioinformatical analysis indicates that LamB does not contain a T2SS secretion signal, but it does however contain putative T4SS translocation signals57,58,70. However, through the CyaA reporter assay our data show that LamB is not translocated. Previous reports have shown effectors that not translocated as CyaA reporter assay, were actually translocated T4SS effectors25,58. This reporter could interfere with the translocation of LamB, like seen with other effectors25. Predicted strength of the translocation signal is not a definitive answer to whether a protein is translocated, low-scoring predicted effectors have been shown to be translocated by the Dot/Icm System and high-scoring proteins have been shown to not be translocated58. Lifshitz et al. identified LamB to be a high-scoring putative effector and in the same study tested 10 new high-scoring putative effectors, of which three were confirmed to not be translocated by the Dot/Icm system using the CyaA reporter assay, but LamB was not tested58. Despite being a high-scoring putative effector, LamB may not be translocated, as observed in our CyaA reporter assay. Loss of lamB does not affect L. pneumophila’s ability to grow in vitro, supporting the idea that LamB is likely secreted into the host cytosol or into the lumen of the LCV, but the mechanism remains to be determined.
In summary, we report an amylase essential for intracellular proliferation of L. pneumophila within the two evolutionarily distant hosts, human macrophages and amoebae. Given its uniqueness to L. pneumophila, LamB serves as an interesting enzyme that may contribute to the prevalence and virulence of L. pneumophila compared to other Legionella species.
Materials and Methods
Strains and cell lines
L. pneumophila strain AA100/130b (ATCC BAA-74) and the dotA T4SS-deficient mutant, were grown on Buffered Charcoal Yeast Extract (BCYE) agar, as we previously described78. To generate the isogenic mutant lamB (lpg2528), 2 kb flanking DNA on either side of lamB, was amplified using PCR with primers listed in Table S1. The resulting amplicon was cloned into the shuttle vector, pBCSk+, to generate pBCSK + lamBKO. To delete the entire gene of lamB, inverse PCR was employed using the primers listed in Table S1, resulting in pBCSK + lamBKO2. The kanamycin resistance cassette from the Ez-Tn5 transposon was amplified using primers listed in Table S1. The resulting PCR product was subcloned in pBSCK + lamBKO2 between the lamB flanking regions using standard molecular procedures, resulting in pBCSK + lamBKO3. This plasmid was introduced into L. pneumophila AA100/130b via natural transformation, as we previously described90. Natural transformants were recovered by plating on BCYE agar supplemented with 50 μg/ml kanamycin. To complement the lamB mutant, PCR was used to amplify the lamB gene and its upstream promotor region, using primers listed in Table S1, and subcloned into pBCSK + , generating pBCSK + lamB/C. Complement mutants of lamB with mutations in the catalytic pocket were made by substituting the amino acid for alanine, to generate pBCSK + lamBD193A, pBCSK + lamBD296A, and pBCSK + lamBE227A, using primers listed in Table S1. These plasmid was introduced into the lamA mutant, via electroporation, as previously described91. Complemented lamB mutants were selected on BCYE plates supplemented with 5 μg/ml chloramphenicol, resulting in the complemented strains, lamB/C, lamB/D193A, lamB/D296A, and lamB/E227A.
Intracellular replication
For infection of cell monolayers, L. pneumophila strains were grown in BYE broth with appropriate antibiotic selection, at 37 °C with shaking, to post-exponential phase (OD550nm 2.1–2.2). A. polyphaga was cultured in PYG media at 22 °C, experiments were performed in PY media at 35 °C, as previously described78. Glucose supplementation experiments were done in presence of 100 mM glucose in the media. Human monocyte-derived macrophages (hMDMs) were isolated from healthy donors and cultured in RPMI 1640, supplemented with 10% fetal bovine serum, as previously described78,92. All methods were approved and carried out in accordance to the University of Louisville Institutional Review Board guidelines and blood donors gave informed consent as required by the University of Louisville Institutional Review Board (IRB # 04.0358).
The wild type strain; the isogenic mutants, dotA and lamB; and complements lamB/C, lamB/D193A, lamB/E227A, and lamB/D296A were grown to post-exponential phase in BYE broth at 37 °C with shaking, prior to infection and used to infect hMDMs and A. polyphaga, as previously described78,92. A total of 1 × 105 host cells were plated in 96 well plates and infected with L. pneumophila at an MOI of 10. Plates were centrifuged at 200 × g (5 mins), to synchronize infection. After 1 h, cells were treated for 1 h with gentamicin to kill extracellular bacteria, as previously described78,92. Over a 24 h time course, host cell were lysed with sterile water (hMDMs) or 0.02% v/v Triton X-100 (A. polyphaga). L. pneumophila CFUs were determined by plating serial dilutions onto BCYE agar.
Bioinformatics analysis of LamB
Protein domain analysis was performed using NCBI’s Search for Conserved Domains (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Phylogenetic analysis was determined using amino acid sequences of LamB, with the Phyologeny.fr platform. Branch length was determined by aLRT93,94. Predicted structures were generated via I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/)60. Structures generated from I-TASSER were aligned using TM-align to generate a TM-score of structural similarity (https://zhanglab.ccmb.med.umich.edu/TM-align/).
Translocation Assay
To assess translocation of LamB by L. pneumophila T4SS, during infection of host cells, an adenylate cyclase fusion74 was generated using standard biology techniques with primers listed in Table S1. A total of 1 × 106 hMDMs were infected with wild type or dotA mutant L. pneumophila harboring plasmids expressing various adenylate cyclase fusions at an MOI of 20 for 1 h, as previously described74,92. Following infection, the cell monolayers were lysed and processed to assess cAMP concentration by ELISA using the Direct cAMP ELISA kit (Enzo) according to the manufacturer’s protocol and measure with a Synergy H1 microplate reader (BioTek).
Amylase activity
To determine if LamB is a functional amylase, the lamB gene was cloned into the IPTG-inducible GST-fusion expression vector pGEX-6p-1 (Amersham) and expressed in E. coli BL21 using primers listed in Table S1. Additionally, residues within the predicted catalytic pocket were substituted to alanine using inverse PCR using primers listed in Table S1. E. coli cultures (5 ml) harboring either the empty vector, lamB, or the various catalytic inactive mutants were grown in LB broth at 37 °C with shaking until the OD600nm reached 0.8. The cultures were spilt and one half was induced with 0.1 mM IPTG for 2.5 h at room temperature. One ml of each culture was pelleted by centrifugation and subjected to lysis with 0.5 ml buffer (0.1% v/v Triton X-100, 150 nM NaCl, 10 mM Tris pH7.5), containing protease inhibitors. Insoluble material was pelleted by centrifugation (16000 × g, 10 min, 4 °C) and the resulting supernatant was retained. Expression of fusion proteins was similar in all cultures (see Supplementary Fig. S3). To measure amylase activity, 25 μl of supernatant was analysed using an Amylase Assay Kit (Sigma), following the manufacturer’s instructions. This kit utilizes an artificial substrate, ethylidene-pNP-G7, which when cleaved by an amylase generates a colorimetric product detectable at 405 nm.
Mouse model
For testing the virulence of the lamB mutant, specific pathogen-free, 6–8 weeks old A/J mice were used, as previously described43,92. Groups of 3 A/J mice, for each time point, were infected intratracheally with 1 × 106 CFUs. At 2, 12, 24, 48, and 72 h after infection mice were humanely sacrificed and lungs, liver, and spleen were harvested and homogenized in sterile saline (5 ml) followed by cell lysis in distilled water. To determine CFUs, serial 10-fold dilutions were plated on BCYE agar and incubated at 37 °C. For histopathology, lungs of infected mice were fixed in 10% neutral formalin and embedded in paraffin. Serial 5 μm sections were cut, stained with haematoxylin and eosin (H&E), for light microscopy analysis. Twenty random high-powered fields (HPFs) were assessed to grade inflammation severity including alveolar and bronchial damage, as well as percentage of parenchyma involved. The histology assessment included the number of the mononuclear cells and percent of parenchyma involved by using modification of double-blind scoring method at a magnification of 40x, as we described previously95. The inflammation process was graded normal (score of 0), when there were 0–19 monocular cells infiltrates per HPF with no alveolar and bronchial involvement; mild (score of 1), for 20 to 49 cells per HPF, including mild damage of alveolar and bronchial regions; moderate (score of 2), for 50 to 99 cells per HPF with moderate alveolar and bronchial inflammation; or severe (score of 3), for 100 to 200 mononuclear cells per HPF with severe effacement of alveolar and bronchial regions. The murine lung section was examined in sagittal direction and percent of parenchyma involved was scored as 0 when no area was compromised. The involvement of the parenchyma was scored as 1 when up to 25% of the total area was occupied by inflammatory exudate, or scored as 2 when 26 to 50% of parenchyma area was occupied with inflammatory cells, and 3 if comprised of more than 51% of the total area. The total histology score was calculated as an average of individual criteria scores. Uninfected tissue was used as a baseline score. All the experimental procedures were in accordance with National guidelines and were approved by the Institutional Animal Care and Use committee (IACUC) at Faculty of Medicine, University of Rijeka.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
Electronic supplementary material
Acknowledgements
The YAK lab is supported by Public Health Service Awards R01AI120244 from the NIAID and by the Commonwealth of Kentucky Research Challenge Trust Fund. AB was supported by a National Science Foundation Graduate Research Fellowship (NSF GRFP) under Grant No. DGE- 1144204.
Author Contributions
C.T., Y.A.K., A.B., M.S., and S.J. conceived the ideas and designed the experiments. A.B., C.P., M.O., and S.J. performed the experiments. A.B. analysed all the data. A.B. and Y.A.K. wrote the main manuscript text and A.B. prepared all figures. All authors have read and approved of the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Ashley Best and Christopher Price contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-24724-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.McDade JE, et al. Legionnaires’ disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N.Engl.J.Med. 1977;297:1197–1203. doi: 10.1056/NEJM197712012972202. [DOI] [PubMed] [Google Scholar]
- 2.Fields BS, Benson RF, Besser RE. Legionella and Legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev. 2002;15:506–526. doi: 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boamah, D. K., Zhou, G., Ensminger, A. W. & O’Connor, T. J. From Many Hosts, One Accidental Pathogen: The Diverse Protozoan Hosts of Legionella. Frontiers in Cellular and Infection Microbiology7, 10.3389/fcimb.2017.00477 (2017). [DOI] [PMC free article] [PubMed]
- 4.Richards AM, Von Dwingelo JE, Price CT, Abu Kwaik Y. Cellular microbiology and molecular ecology of Legionella-amoeba interaction. Virulence. 2013;4:307–314. doi: 10.4161/viru.24290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y. Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 2005;71:20–28. doi: 10.1128/AEM.71.1.20-28.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lama A, Drennan SL, Johnson RC, Rubenstein GL, Cambronne ED. Identification of Conserved ABC Importers Necessary for Intracellular Survival of Legionella pneumophila in Multiple Hosts. Front Cell Infect Microbiol. 2017;7:485. doi: 10.3389/fcimb.2017.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swart, A. L., Harrison, C. F., Eichinger, L., Steinert, M. & Hilbi, H. Acanthamoeba and Dictyostelium as Cellular Models for Legionella Infection. Frontiers in Cellular and Infection Microbiology8, 10.3389/fcimb.2018.00061 (2018). [DOI] [PMC free article] [PubMed]
- 8.Segal G, Shuman HA. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infection and immunity. 1999;67:2117–2124. doi: 10.1128/iai.67.5.2117-2124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cianciotto NP, Fields BS. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. USA. 1992;89:5188–5191. doi: 10.1073/pnas.89.11.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia MT, Jones S, Pelaz C, Millar RD, Abu Kwaik Y. Acanthamoeba polyphaga resuscitates viable non-culturable Legionella pneumophila after disinfection. Environmental microbiology. 2007;9:1267–1277. doi: 10.1111/j.1462-2920.2007.01245.x. [DOI] [PubMed] [Google Scholar]
- 11.Baskerville A, Dowsett AB, Fitzgeorge RB, Hambleton P, Broster M. Ultrastructure of pulmonary alveoli and macrophages in experimental Legionnaires’ disease. J.Pathol. 1983;140:77–90. doi: 10.1002/path.1711400202. [DOI] [PubMed] [Google Scholar]
- 12.Nash TW, Libby DM, Horwitz MA. Interaction between the legionnaires’ disease bacterium (Legionella pneumophila) and human alveolar macrophages. Influence of antibody, lymphokines, and hydrocortisone. J.Clin.Invest. 1984;74:771–782. doi: 10.1172/JCI111493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horwitz MA, Silverstein SC. Legionnaires’ disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. J.Clin.Invest. 1980;66:441–450. doi: 10.1172/JCI109874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horwitz MA. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J.Exp.Med. 1983;158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swanson MS, Isberg RR. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect.Immun. 1995;63:3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abu Kwaik Y. The phagosome containing Legionella pneumophila within the protozoan Hartmanella vermiformis is surrounded by the rough endoplasmic reticulum. Appl. Environ. Microbiol. 1996;62:2022–2028. doi: 10.1128/aem.62.6.2022-2028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson CG, Roy CR. Attachment and fusion of endoplasmic reticulum with vacuoles containing Legionella pneumophila. Cell Microbiol. 2006;8:793–805. doi: 10.1111/j.1462-5822.2005.00666.x. [DOI] [PubMed] [Google Scholar]
- 18.Bärlocher, K., Welin, A. & Hilbi, H. Formation of the Legionella Replicative Compartment at the Crossroads of Retrograde Trafficking. Frontiers in Cellular and Infection Microbiology7, 10.3389/fcimb.2017.00482 (2017). [DOI] [PMC free article] [PubMed]
- 19.Speir, M. et al. Legionella pneumophila Strain 130b Evades Macrophage Cell Death Independent of the Effector SidF in the Absence of Flagellin. Frontiers in Cellular and Infection Microbiology7, 10.3389/fcimb.2017.00035 (2017). [DOI] [PMC free article] [PubMed]
- 20.Casson, C. & Shin, S. Inflammasome-mediated cell death in response to bacterial pathogens that access the host cell cytosol: lessons from legionella pneumophila. Frontiers in Cellular and Infection Microbiology3, 10.3389/fcimb.2013.00111 (2013). [DOI] [PMC free article] [PubMed]
- 21.Abu Khweek, A. et al. Biofilm-derived Legionella pneumophila evades the innate immune response in macrophages. Frontiers in Cellular and Infection Microbiology3, 10.3389/fcimb.2013.00018 (2013). [DOI] [PMC free article] [PubMed]
- 22.Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coers J, et al. Identification of icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for legionella pneumophila intracellular growth. Molecular microbiology. 2000;38:719–736. doi: 10.1046/j.1365-2958.2000.02176.x. [DOI] [PubMed] [Google Scholar]
- 24.Dumenil G, Montminy TP, Tang M, Isberg RR. IcmR-regulated membrane insertion and efflux by the Legionella pneumophila IcmQ protein. The Journal of biological chemistry. 2004;279:4686–4695. doi: 10.1074/jbc.M309908200. [DOI] [PubMed] [Google Scholar]
- 25.Zhu W, et al. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS One. 2011;6:e17638. doi: 10.1371/journal.pone.0017638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burstein D, et al. Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat Genet. 2016;48:167–175. doi: 10.1038/ng.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schroeder, G. N. The Toolbox for Uncovering the Functions of Legionella Dot/Icm Type IVb Secretion System Effectors: Current State and Future Directions. Frontiers in Cellular and Infection Microbiology7, 10.3389/fcimb.2017.00528 (2018). [DOI] [PMC free article] [PubMed]
- 28.Qiu, J. & Luo, Z.-Q. Hijacking of the Host Ubiquitin Network by Legionella pneumophila. Frontiers in Cellular and Infection Microbiology7, 10.3389/fcimb.2017.00487 (2017). [DOI] [PMC free article] [PubMed]
- 29.Ghosh S, O’Connor TJ. Beyond Paralogs: The Multiple Layers of Redundancy in Bacterial Pathogenesis. Front Cell Infect Microbiol. 2017;7:467. doi: 10.3389/fcimb.2017.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asare R, Abu Kwaik Y. Early trafficking and intracellular replication of Legionella longbeachaea within an ER-derived late endosome-like phagosome. Cell Microbiol. 2007;9:1571–1587. doi: 10.1111/j.1462-5822.2007.00894.x. [DOI] [PubMed] [Google Scholar]
- 31.de Felipe KS, et al. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS pathogens. 2008;4:e1000117. doi: 10.1371/journal.ppat.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horwitz MA. The Legionnaires’ disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J.Exp.Med. 1983;158:2108–2126. doi: 10.1084/jem.158.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allgood SC, et al. Legionella Effector AnkX Disrupts Host Cell Endocytic Recycling in a Phosphocholination-Dependent Manner. Front Cell Infect Microbiol. 2017;7:397. doi: 10.3389/fcimb.2017.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Felipe KS, et al. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. Journal of bacteriology. 2005;187:7716–7726. doi: 10.1128/JB.187.22.7716-7726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomez-Valero L, et al. Extensive recombination events and horizontal gene transfer shaped the Legionella pneumophila genomes. BMC Genomics. 2011;12:536. doi: 10.1186/1471-2164-12-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price C, et al. Host FIH-Mediated Asparaginyl Hydroxylation of Translocated Legionella pneumophila Effectors. Front Cell Infect Microbiol. 2017;7:54. doi: 10.3389/fcimb.2017.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tesh MJ, Morse SA, Miller RD. Intermediary metabolism in Legionella pneumophila: utilization of amino acids and other compounds as energy sources. J.Bacteriol. 1983;154:1104–1109. doi: 10.1128/jb.154.3.1104-1109.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schunder E, et al. Amino Acid Uptake and Metabolism of Legionella pneumophila Hosted by Acanthamoeba castellanii. The Journal of biological chemistry. 2014;289:21040–21054. doi: 10.1074/jbc.M114.570085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eylert E, et al. Isotopologue profiling of Legionella pneumophila: role of serine and glucose as carbon substrates. The Journal of biological chemistry. 2010;285:22232–22243. doi: 10.1074/jbc.M110.128678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fonseca MV, Swanson MS. Nutrient salvaging and metabolism by the intracellular pathogen Legionella pneumophila. Front Cell Infect Microbiol. 2014;4:12. doi: 10.3389/fcimb.2014.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price CT, Al-Quadan T, Santic M, Rosenshine I, Abu Kwaik Y. Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science. 2011;334:1553–1557. doi: 10.1126/science.1212868. [DOI] [PubMed] [Google Scholar]
- 42.Lomma M, et al. The Legionella pneumophila F-box protein Lpp2082 (AnkB) modulates ubiquitination of the host protein parvin B and promotes intracellular replication. Cell Microbiol. 2010;12:1272–1291. doi: 10.1111/j.1462-5822.2010.01467.x. [DOI] [PubMed] [Google Scholar]
- 43.Price CT, Al-Quadan T, Santic M, Jones SC, Abu Kwaik Y. Exploitation of conserved eukaryotic host cell farnesylation machinery by an F-box effector of Legionella pneumophila. J Exp Med. 2010;207:1713–1726. doi: 10.1084/jem.20100771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kubori, T., Bui, X. T., Hubber, A. & Nagai, H. Legionella RavZ Plays a Role in Preventing Ubiquitin Recruitment to Bacteria-Containing Vacuoles. Frontiers in Cellular and Infection Microbiology7, 10.3389/fcimb.2017.00384 (2017). [DOI] [PMC free article] [PubMed]
- 45.De Leon JA, et al. Positive and Negative Regulation of the Master Metabolic Regulator mTORC1 by Two Families of Legionella pneumophila Effectors. Cell reports. 2017;21:2031–2038. doi: 10.1016/j.celrep.2017.10.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harada E, Iida K, Shiota S, Nakayama H, Yoshida S. Glucose metabolism in Legionella pneumophila: dependence on the Entner-Doudoroff pathway and connection with intracellular bacterial growth. Journal of bacteriology. 2010;192:2892–2899. doi: 10.1128/JB.01535-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hauslein I, Manske C, Goebel W, Eisenreich W, Hilbi H. Pathway analysis using (13) C-glycerol and other carbon tracers reveals a bipartite metabolism of Legionella pneumophila. Molecular microbiology. 2016;100:229–246. doi: 10.1111/mmi.13313. [DOI] [PubMed] [Google Scholar]
- 48.Eisenreich W, Heesemann J, Rudel T, Goebel W. Metabolic host responses to infection by intracellular bacterial pathogens. Front Cell Infect Microbiol. 2013;3:24. doi: 10.3389/fcimb.2013.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gillmaier, N. et al. Growth-Related Metabolism of the Carbon Storage Poly-3-Hydroxybutyrate in Legionella pneumophila. The Journal of biological chemistry, 10.1074/jbc.M115.693481 (2016). [DOI] [PMC free article] [PubMed]
- 50.Gillmaier N, et al. Growth-related Metabolism of the Carbon Storage Poly-3-hydroxybutyrate in Legionella pneumophila. The Journal of biological chemistry. 2016;291:6471–6482. doi: 10.1074/jbc.M115.693481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.James BW, Mauchline WS, Dennis PJ, Keevil CW, Wait R. Poly-3-hydorxyburyrate in Legionella pneumophila, an energy source for survival in low-nutrient environments. Appl.Environ.Microbiol. 1999;65:822–827. doi: 10.1128/aem.65.2.822-827.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oliva, G., Sahr, T. & Buchrieser, C. The Life Cycle of L. pneumophila: Cellular Differentiation Is Linked to Virulence and Metabolism. Frontiers in Cellular and Infection Microbiology8, 10.3389/fcimb.2018.00003 (2018). [DOI] [PMC free article] [PubMed]
- 53.Bruggemann H, et al. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell Microbiol. 2006;8:1228–1240. doi: 10.1111/j.1462-5822.2006.00703.x. [DOI] [PubMed] [Google Scholar]
- 54.Kuriki T, Imanaka T. The concept of the α-amylase family: structural similarity and common catalytic mechanism. Journal of bioscience and bioengineering. 1999;87:557–565. doi: 10.1016/S1389-1723(99)80114-5. [DOI] [PubMed] [Google Scholar]
- 55.Gurung N, Ray S, Bose S, Rai V. A Broader View: Microbial Enzymes and Their Relevance in Industries, Medicine, and Beyond. BioMed Research International. 2013;2013:329121. doi: 10.1155/2013/329121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herrmann V, et al. GamA is a eukaryotic-like glucoamylase responsible for glycogen- and starch-degrading activity of Legionella pneumophila. International journal of medical microbiology: IJMM. 2011;301:133–139. doi: 10.1016/j.ijmm.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 57.Burstein D, et al. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS pathogens. 2009;5:e1000508. doi: 10.1371/journal.ppat.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lifshitz Z, et al. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E707–715. doi: 10.1073/pnas.1215278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muder RR, Yu VL. Infection due to Legionella species other than L. pneumophila. Clin Infect Dis. 2002;35:990–998. doi: 10.1086/342884. [DOI] [PubMed] [Google Scholar]
- 60.Marchler-Bauer A, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic acids research. 2017;45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reuter, S. et al. A pilot study of rapid whole-genome sequencing for the investigation of a Legionella outbreak. BMJ Open3, 10.1136/bmjopen-2012-002175 (2013). [DOI] [PMC free article] [PubMed]
- 62.de Souza PM, de Oliveira Magalhães P. Application of microbial α-amylase in industry – A review. Brazilian Journal of Microbiology. 2010;41:850–861. doi: 10.1590/S1517-83822010000400004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kobayashi M, et al. Structural insights into the catalytic reaction that is involved in the reorientation of Trp238 at the substrate-binding site in GH13 dextran glucosidase. FEBS letters. 2015;589:484–489. doi: 10.1016/j.febslet.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 64.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang J, Zhang Y. Protein Structure and Function Prediction Using I-TASSER. Curr Protoc Bioinformatics. 2015;52:5 8 1–5 8 15. doi: 10.1002/0471250953.bi0508s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Truchan, H. K., Christman, H. D., White, R. C., Rutledge, N. S. & Cianciotto, N. P. Type II Secretion Substrates of Legionella pneumophila Translocate Out of the Pathogen-Occupied Vacuole via a Semipermeable Membrane. mBio8, 10.1128/mBio.00870-17 (2017). [DOI] [PMC free article] [PubMed]
- 67.Rossier O, Starkenburg SR, Cianciotto NP. Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires’ disease pneumonia. Infection and immunity. 2004;72:310–321. doi: 10.1128/IAI.72.1.310-321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rossier O, Dao J, Cianciotto NP. The type II secretion system of Legionella pneumophila elaborates two aminopeptidases, as well as a metalloprotease that contributes to differential infection among protozoan hosts. Applied and environmental microbiology. 2008;74:753–761. doi: 10.1128/AEM.01944-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hales LM, Shuman HA. Legionella pneumophila contains a type II general secretion pathway required for growth in amoebae as well as for secretion of the Msp protease. Infect.Immun. 1999;67:3662–3666. doi: 10.1128/iai.67.7.3662-3666.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DebRoy S, Dao J, Soderberg M, Rossier O, Cianciotto NP. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19146–19151. doi: 10.1073/pnas.0608279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nagai H, et al. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:826–831. doi: 10.1073/pnas.0406239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang L, et al. The E Block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol. 2011;13:227–245. doi: 10.1111/j.1462-5822.2010.01531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y, Wei X, Bao H, Liu SL. Prediction of bacterial type IV secreted effectors by C-terminal features. BMC Genomics. 2014;15:50. doi: 10.1186/1471-2164-15-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sory MP, Cornelis GR. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Molecular microbiology. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 75.Bardill JP, Miller JL, Vogel JP. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Molecular microbiology. 2005;56:90–103. doi: 10.1111/j.1365-2958.2005.04539.x. [DOI] [PubMed] [Google Scholar]
- 76.Kruse-Jarres JD, et al. Evaluation of a new alpha-amylase assay using 4.6-ethylidene-(G7)-1-4-nitrophenyl-(G1)-alpha-D-maltoheptaoside as substrate. J Clin Chem Clin Biochem. 1989;27:103–113. [PubMed] [Google Scholar]
- 77.Hagele EO, Kratzer M, Schaich E, Rauscher E. Mechanism of action of human pancreatic and salivary alpha-amylase on 4,6-ethylidene-alpha-4-nitrophenyl-maltoheptaoside substrate. Clin Chem. 1989;35:188–189. [PubMed] [Google Scholar]
- 78.Al-Khodor S, Price CT, Habyarimana F, Kalia A, Abu Kwaik Y. A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Molecular microbiology. 2008;70:908–923. doi: 10.1111/j.1365-2958.2008.06453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eisenreich W, Heuner K. The life stage-specific pathometabolism of Legionella pneumophila. FEBS letters. 2016;590:3868–3886. doi: 10.1002/1873-3468.12326. [DOI] [PubMed] [Google Scholar]
- 80.Tesh MJ, Miller RD. Amino acid requirements for Legionella pneumophila growth. J.Clin.Microbiol. 1981;13:865–869. doi: 10.1128/jcm.13.5.865-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bachman MA, Swanson MS. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Molecular microbiology. 2001;40:1201–1214. doi: 10.1046/j.1365-2958.2001.02465.x. [DOI] [PubMed] [Google Scholar]
- 82.Hammer BK, Swanson MS. Co-ordination of legionella pneumophila virulence with entry into stationary phase by ppGpp. Molecular microbiology. 1999;33:721–731. doi: 10.1046/j.1365-2958.1999.01519.x. [DOI] [PubMed] [Google Scholar]
- 83.Dalebroux ZD, Yagi BF, Sahr T, Buchrieser C, Swanson MS. Distinct roles of ppGpp and DksA in Legionella pneumophila differentiation. Molecular microbiology. 2010;76:200–219. doi: 10.1111/j.1365-2958.2010.07094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hales LM, Shuman HA. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. Journal of bacteriology. 1999;181:4879–4889. doi: 10.1128/jb.181.16.4879-4889.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Abu Kwaik Y, Bumann D. Host Delivery of Favorite Meals for Intracellular Pathogens. PLoS pathogens. 2015;11:e1004866. doi: 10.1371/journal.ppat.1004866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Abu Kwaik Y, Bumann D. Microbial quest for food in vivo: ‘Nutritional virulence’ as an emerging paradigm. Cell Microbiol. 2013;15:882–890. doi: 10.1111/cmi.12138. [DOI] [PubMed] [Google Scholar]
- 87.Li, L. & Faucher, S. P. The Membrane Protein LasM Promotes the Culturability of Legionella pneumophila in Water. Frontiers in Cellular and Infection Microbiology, 6, 10.3389/fcimb.2016.00113 (2016). [DOI] [PMC free article] [PubMed]
- 88.Faucher SP, Mueller CA, Shuman HA. Legionella Pneumophila Transcriptome during Intracellular Multiplication in Human Macrophages. Frontiers in microbiology. 2011;2:60. doi: 10.3389/fmicb.2011.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Quintarelli G, Dellovo MC, Balduini C, Castellani AA. The effects of alpha amylase on collagen-proteoglycans and collagen-glycoprotein complexes in connective tissue matrices. Histochemie. 1969;18:373–375. doi: 10.1007/BF00279887. [DOI] [PubMed] [Google Scholar]
- 90.Stone BJ, Abu Kwaik Y. Natural competency for DNA uptake by Legionella pneumophila and its association with expression of type IV pili. J.Bacteriol. 1999;181:1395–1402. doi: 10.1128/jb.181.5.1395-1402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen DQ, Huang SS, Lu YJ. Efficient transformation of Legionella pneumophila by high-voltage electroporation. Microbiol Res. 2006;161:246–251. doi: 10.1016/j.micres.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 92.Price CT, et al. Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS pathogens. 2009;5:e1000704. doi: 10.1371/journal.ppat.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Systematic biology. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 94.Dereeper A, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic acids research. 2008;36:W465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Santic M, et al. A Francisella tularensis pathogenicity island protein essential for bacterial proliferation within the host cell cytosol. Cell Microbiol. 2007;9:2391–2403. doi: 10.1111/j.1462-5822.2007.00968.x. [DOI] [PubMed] [Google Scholar]
- 96.Yang J, et al. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).