Abstract
This review intends to uncover how information from large-scale genetic profiling (whole genome sequencing, and whole exome sequencing) of nonalcoholic fatty liver disease (NAFLD), as well as information from circulating transcriptomics (cell-free miRNAs) and metabolomics, contributes to the understanding of NAFLD pathogenesis. A further aim is to address the question of whether OMICs information is ready to be implemented in the clinics. The available evidence suggests that any new knowledge pertaining to molecular signatures associated with NAFLD and nonalcoholic steatohepatitis should be promptly translated into the clinical setting. Nevertheless, rigorous steps that must include validation and replication are mandatory before utilizing OMICs biomarkers in diagnostics to identify patients at risk of advanced disease, including liver cancer.
Keywords: Nonalcoholic steatohepatitis, Fibrosis, Liver biopsy, Genetics, PNPLA3, TM6SF2, Metabolomics, Proteomics, Transcriptomics, Nonalcoholic fatty liver disease, miR122
Core tip: It is expected that, in the near future, nonalcoholic fatty liver disease patients can be diagnosed and treated according to their own “molecular signature”. Specific focus should be placed on prevention and early diagnosis through the application of biomarkers of disease risk. Selection of “personalized drugs” as well as tailored therapy according to the specific molecular signature should be further guaranteed.
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) is a chronic liver disease that affects adult and children populations around the world, with prevalence reaching alarming levels[1,2].
NAFLD may progress from a benign histological disease stage characterized by plain fat accumulation, usually referred to as simple steatosis or nonalcoholic fatty liver (NAFL), to a more severe histological form characterized by liver cell injury, a mixed inflammatory lobular infiltrate, and variable fibrosis named nonalcoholic steatohepatitis (NASH)[3,4].
Precise histological diagnosis, including disease stages (NAFL and NASH), is commonly based on liver biopsy[2]. Nevertheless, because this method imposes certain limitations, including potential complications such as bleeding and patients’ abdominal discomfort, and needs to be performed in a special setting, noninvasive approaches are favored and have gained considerable attention. It is also noteworthy that the histological diagnosis of the severity of NAFLD might be potentially biased if a small portion of hepatic tissue is sampled.
Hence, significant clinical and research efforts are currently being directed toward the search for reliable biomarkers aimed at the prediction of the disease severity and prognosis.
Knowledge in the field of liver diseases, particularly NAFLD, has benefitted in the last ten years from the rapid development of high-throughput technologies, including genomics, transcriptomics, proteomics and metabolomics. This review intends to uncover how information from large-scale genetic profiling (whole genome sequencing and whole exome sequencing) of NAFLD, as well as information from transcriptomics and metabolomics, and the interplay of these personal characteristics with dietary factors may contribute to the diagnosis and risk prediction of NAFLD progression. In addition, the question of whether OMICs information is ready to be implemented in the clinics will be addressed.
A brief description of OMICs signatures, including their main applications as biomarkers in clinical practice, is provided in Figure 1. OMICs biomarkers may be considered either for screening purposes to assess the disease risk or exposure, or for the assessment of the disease severity and prognosis, and/or for monitoring treatment response (Figure 1).
Figure 1.
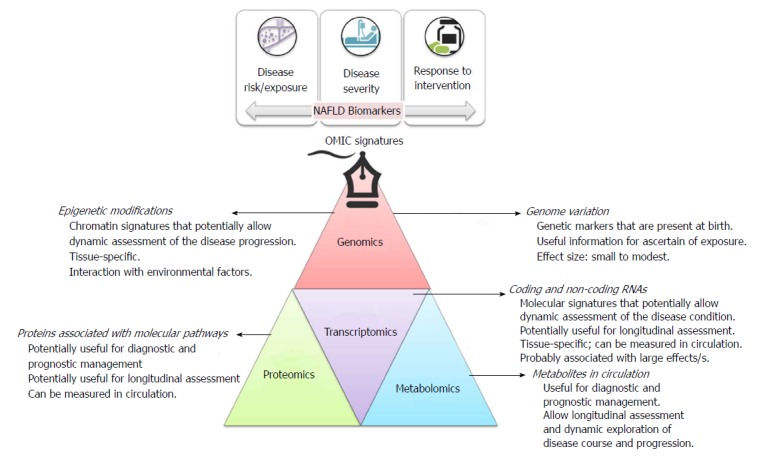
Brief description of OMICs signatures, including their main applications as biomarkers in clinical practice.
ROLE OF GENETIC MARKERS IN THE PREDICTION OF NAFLD RISK AND DISEASE SEVERITY
Although the pathogenesis of NAFLD is not understood fully, a growing body of evidence indicates that the disease develops from a complex process involving many factors, including genetic susceptibility and environmental insults[5,6].
In fact, the results yielded by the first genome-wide association study on NAFLD[7] on the role of rs738409 C/G -a variant nonsynonymous single nucleotide polymorphism (SNP) of PNPLA3 (patatin-like phospholipase domain containing 3, also known as adiponutrin or calcium-independent phospholipase A2-epsilon) have significantly contributed to the knowledge of the genetic component of NAFLD. This finding was subsequently widely replicated around the world, confirming that the G allele in the forward strand is significantly associated not only with an increased risk of fatty liver but the histological disease severity as well[8,9] (OR 1.88 per G allele). In fact, rs738409 explains about 5.3% of the total variance in NAFLD[9].
Furthermore, results of the first exome-wide association study of liver fat content indicate that rs58542926 (E167K), a nonsynonymous variant located in TM6SF2 (Transmembrane 6 Superfamily Member 2), is significantly associated with increased liver fat content[10]. Nevertheless, in contrast to the effect of the variant located in PNPLA3, the rs58542926 exerts a moderate effect on the risk of NAFLD (odds ratio: 2.13)[11]. Subsequent studies have also revealed an association of rs58542926 with the disease severity[12-14], as well as dual and opposite role in cardiovascular disease prevention[11,12,15].
Thus, it is reasonable to speculate that genetic markers, particularly the 738409-G risk allele, may be used for individual risk assessment either alone or as a part of multi-score biomarkers (Figure 2). For example, Kotronen and coworkers evaluated the performance of rs738409 in predicting the risk of NAFLD by combining routine clinical and laboratory data and the rs738409 genotypes[16]. The authors observed a sensitivity of 86% and a specificity of 71% in the estimation of increased liver fat content[16]. Surprisingly, addition of the genetic information to the score improved the accuracy of NAFLD prediction by less than 1%.
Figure 2.
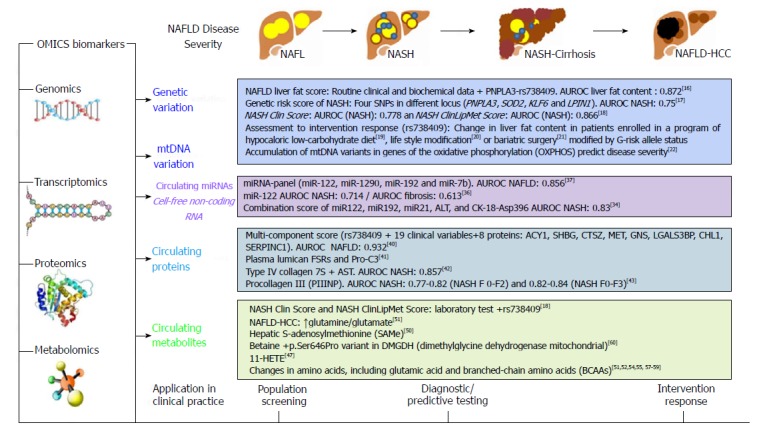
Summary of OMICs biomarkers in the prediction of nonalcoholic fatty liver disease severity.
The incorporation of genetic markers into noninvasive tests that discriminate between NAFL and NASH results in a more challenging strategy; despite these difficulties, there have been some interesting attempts. For instance, a risk score comprising of both clinical and genetic (PNPLA3 rs738409 C>G, SOD2 rs4880 C>T, KLF6 rs3750861 G>A, and LPIN1 rs13412852 C>T) risk factors resulted in an AUROC (Area Under the Receiver Operating Characteristic) of 0.80 to predict NASH in obese children with increased levels of liver enzymes[17], as shown in Figure 2.
Other examples include the NASH Clin Score that combines laboratory tests (AST, fasting insulin) and rs738409 genotypes, and the NASH ClinLipMet Score that combines laboratory test (AST, fasting insulin), circulating metabolites (glutamate, isoleucine, glycine, lyso PC 16:0; PE 40:6) and rs738409 genotypes[18], as depicted in Figure 2.
Furthermore, promising results have been reported on the use of genetic markers in predicting NAFLD-intervention response, as summarized in Figure 2. For example, it was observed that genetic variation in PNPLA3 might confer sensitivity to liver fat content decrease in obese patients undergoing weight loss[19]. The findings yielded by this study, though based on a small number of subjects, suggested that weight loss was more effective in decreasing liver fat in subjects who were homozygous for the rs738409-G allele[19]. Likewise, rs738409 correlated with changes in metabolic profile and intrahepatic triglyceride content (IHTG) as measured by proton magnetic resonance spectroscopy in patients enrolled in a lifestyle modification program[20]. Concordant results were reported regarding greater improvement in hepatic steatosis after bariatric surgery in the risk-G-rs738409 allele carriers[21] (Figure 2).
A different approach to the use of genetic testing based on single base variations in the DNA sequence requires search for variants in mitochondrial DNA (mtDNA). Mitochondria contain their own genetic information in the mtDNA (16.5 kb), which is maternally inherited; the 13 mtDNA-encoded proteins are all components of the oxidative phosphorylation (OXPHOS). A comprehensive exploration of the complete liver mtDNA-mutation spectrum in patients with NAFLD during different stages of the disease by next generation sequencing showed that the disease severity is associated with an increased liver mtDNA mutational burden, including point mutations in OXPHOS-genes that showed high degrees of heteroplasmy[22]. Given that the variability in the mt-genomes observed in NAFLD and NASH seems to originate from a common germline source, rather than from tissue-specific mutations, point mutations can also be assessed in samples of peripheral blood mononuclear cells[22].
ROLE OF EPIGENETIC MODIFICATIONS AS NONINVASIVE BIOMARKERS OF NAFLD AND NASH
The dynamic nature of epigenetic modifications is not only an ideal frame to explain the cross-talk between NAFLD and related phenotypes, including insulin resistance[23], but is also an attractive target for therapeutic intervention24. Treatment-induced epigenetic remodeling of liver tissue was observed in a cohort of obese patients with NAFLD who underwent bariatric surgery[24]. In addition, changes in DNA methylation could be used as a target of a biomarker that allows monitoring, for instance, effectiveness of pharmacotherapy. Interesting results have been reported in the context of other non-cancer complex diseases, including rheumatoid arthritis[25], pediatric asthma[26] or anxiety disorders[27].
It is worth noting that epigenetic modifications, i.e. DNA methylation, are not restricted to the nuclear genome, but can also be found in mt-genomes[28]. In fact, we found for the first time that hepatic methylation and transcriptional activity of the MT-ND6 (mt genome-encoded NADH deshydrogenase 6, a member of the OXPHOS complex 1) are associated with the histological severity of NAFLD[29]. This epigenetic change to mtDNA is potentially reversible by lifestyle interventional programs, as physical activity could modulate the methylation status of MT-ND6[29].
CELL-FREE DNA AND RNA AS NONINVASIVE BIOMARKERS OF NASH
Circulating molecular biomarkers, particularly cell-free DNA (cfDNA) and cell-free RNA (cfRNA) are focus of intensive research; however, the strategies employed in these studies are not necessarily novel. In fact, the first description of cell-free nucleic acids (cfNAs) was provided by Mandel and Métais in 1948[30]; indeed, these authors introduced the concept of liquid biopsy.
Basically, cfNAs refer to molecules of nucleic acids that circulate free of cells in the bloodstream and the source of which is primarily dying cells from distant tissues.
Considerable efforts have been dedicated to the use cfDNA for the prediction of liver fibrosis associated with NASH and alcoholic liver disease[31]; however, the preliminary results indicate substantial lack of specificity, as they can be completely unrelated to NASH-biology[32]. Furthermore, the fact that cfDNA circulates not only at very low concentrations but is also highly fragmented imposes analytical and technical challenges that are very difficult to overcome[33].
Conversely, detection of microRNAs (miRNAs), which are highly conserved noncoding small RNAs, has demonstrated quite robust performance, particularly in the circulating compartment. In addition, unlike cfDNA, cfmiRNAs are resistant to degradation as well as to several freeze–thaw cycles, making them ideal biomarkers for use in the clinical setting.
The circulating miRNA signature of NAFLD has been extensively explored in case-control studies, including patients with liver biopsy[34-37], Figure 2. Studies in which liver and circulating miRNA levels were compared demonstrated that cfmiRNAs are good predictors of NAFLD-disease stages[36]. Specifically, circulating miR122 and miR192 not only mirror histological and molecular events occurring in the liver, but have a reliable predictive power in differentiating simple steatosis from NASH[36]. Thus, it can be posited that cfmiRNAs are reliable candidates for incorporation into multi-panel scores for the prediction of NAFLD and NASH (Figure 2).
For example, a miRNA panel, composed by the detection of miR122-5p, miR1290, miR27b-3p, and miR192-5p) showed a high diagnostic accuracy for NAFLD[37] (Figure 2). A combination score that included miR122, miR192, miR21, ALT, and CK-18-Asp396 exhibited an AUROC of 0.83 for the prediction of NASH[34] (Figure 2).
ROLE OF CIRCULATING PROTEINS IN THE PREDICTION OF NASH SEVERITY
The use of proteins that circulate in serum or plasma for predicting liver-related histological outcomes, specifically liver fibrosis, has been largely relegated probably because such approaches are technically challenging, while offering low performance and poor accuracy. The most remarkable example of this strategy is based on the use of plasma caspase-generated cytokeratin-18 fragments (CK-18) as a noninvasive alternative biomarker of NASH. Results from a large multicenter study showed that plasma CK-18 has relatively good specificity for NAFLD (AUROC: 0.77), NASH (0.65) and fibrosis (0.68). Nevertheless, the overall sensitivity for NAFLD (63%), NASH (58%) and fibrosis (54%) is limited, making this test inadequate for use as a single noninvasive screening test[38].
Interesting attempts to develop multi-component tests that integrate clinical and laboratory data, including circulating proteins, have also been made. For example, we have tested a diagnostic model based on a composite index using clinical and laboratory data, including circulating biomarkers such as soluble intercellular adhesion molecule-1 (sICAM-1), which was able to differentiate between patients with simple steatosis and NASH with a post-test probability for NASH of 99.5% when all positive tests were present[39].
There are similar proposals - though restricted to the prediction of NAFLD but not NASH - based on OMICs-derived data, including genetic information (rs738409), clinical variables, and measurement of different proteins (ACY1, SHBG, CTSZ, MET, GNS, LGALS3BP, CHL1, SERPINC1), which - if combined - seem to be quite reliable in disease risk identification (AUROC for steatosis 0.935)[40]. Nevertheless, it seems that this approach has limited cost-effectiveness for NAFLD-screening programs.
Latest advancements in this field focus directly on disease phenotypes, for example liver fibrosis, which target the detection of excess collagen synthesis rate both directly in liver tissue and noninvasively in blood[41].
The combination of type IV collagen 7S and aspartate aminotransferase (AST) in a multi-test for the prediction of NASH-fibrosis showed promising results[42]. Likewise, measurement of circulating procollagen III (PIIINP) has been quite accurate in the prediction of NASH (AUROC 0.77-0.82) and NASH-fibrosis (0.82-0.84)[43].
Unfortunately, proteomic analysis using state of the art technology is currently poorly developed in the field of NAFLD. In fact, robust attempts to refine, replicate and follow-up on putative discovered proteins have not been done, even though some promising studies have been carried out. For example, using MALDI TOF/TOF and western blot analysis of coupled tissue and serum samples allowed the identification of two interesting protein candidates, including the mitochondrial enzyme CPS1 (Carbamoyl-Phosphate Synthase 1) and GRP78, also known as heat shock protein family A (Hsp70) member 5, which could stratify the different phenotypes associated with the disease severity[44]. Results obtained by using similar approaches, including SELDI-TOF mass spectrometry[45] and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI TOF-MS)[46] have been published. Still, the identified peaks require validation, replication and large-scale testing.
CIRCULATING METABOLITES IN NASH PREDICTION
Initial case-control studies on plasma metabolomics of NAFLD have been performed years ago by Puri et al[47], who conducted a comprehensive analysis of plasma lipids and eicosanoid metabolites quantified by mass spectrometry. The authors reported a stepwise increase in lipoxygenase (LOX) metabolites, 5(S)-hydroxyeicosatetraenoic acid (5-HETE), 8-HETE and 15-HETE that characterized the progression from normal liver to NAFL to NASH[47]. Puri and colleagues found that the level of 11-HETE, a nonenzymatic oxidation product of arachidonic (20:4) acid, was significantly and specifically increased in NASH but not in NAFL patients[47]. Subsequent studies that included untargeted global metabolomic analysis revealed marked changes in bile salts and glutathione-related metabolites, as well as higher levels of branched-chain amino acids, phosphocholine, carbohydrates (glucose, mannose), lactate and pyruvate, in subjects with severe NAFLD[48]. Regarding bile salts, a recent study indicated that total conjugated primary bile acids were significantly higher in NASH[49].
A novel study in which the authors combined metabolomic data from experimental animals and human samples introduced the interesting concept that NASH might be sub-classified into two major subtypes according to the circulating pattern of triglycerides, diglycerides, fatty acids, ceramides and oxidized fatty acids[50].
As mentioned earlier, interesting strategies that combine clinical, genetic and lipidomic-derived variables into a multi-score have shown good predictive values in differentiating NAFL from NASH. Specifically, Zhou and coworkers reported on the performance of the NASH Clin Score, obtained through backward stepwise logistic regression analyses of biochemical variables (glutamate, isoleucine, glycine, lysophosphatidylcholine 16:0, phosphoethanolamine 40:6, AST, and fasting insulin), along with rs738409 genotypes[18]; this score identified patients with NASH with an AUROC of 0.866 (Figure 2).
Recent explorations on changes in liver metabolism during NASH development[51,52], along with the findings from high-throughput circulating profiling of patients with metabolic syndrome[53] suggest that elevated levels of alanine (ALT) and aspartate (AST) aminotransaminases in patients with NAFLD are the consequence of impaired liver metabolism of amino acids, including glutamate and aromatic amino acids, rather than a mere biomarker of liver injury[14,52,54]. This observation is consistent with the fact that NASH is associated with changes in the level of circulating amino acids[55], including L-glutamic acid, 2-hydroxyglutarate and alanine / pyruvate ratio, which are significantly associated with NAFLD-disease severity[52,56]. Changes in the level of branched-chain amino acids were described in pediatric population[57], and these findings were replicated in studies on adults as well[58].
Interestingly, alterations in multiple aminoacids, gamma-glutamyl dipeptides and lipids may be related to common genetic variations associated with NAFLD, as observed in earlier in vitro studies based on knocking down or over-expression of the pIle148Met (rs738409) isoforms[59].
Finally, a two-stage multicenter case-control study that combined results of NAFLD-histological variables, levels of circulating metabolites and genetic markers indicated that NASH is associated with decreased levels of betaine in circulation. Furthermore, the disease severity is associated with genotypes of the missense variant p.Ser646Pro (rs1805074) in DMGDH gene, which encodes for the mitochondrial dimethylglycine dehydrogenase[60]. Betaine (N,N,N-trimethylglicine) performs a critical function in the pathway of methylogenesis by controlling the serum methionine levels; thus, the results of the aforementioned study[61] might be used to tailor therapeutic interventions based on metabolites that modulate the liver methylome.
INTEGRATION OF DATA DERIVED FROM GENOMICS/PROTEOMICS/TRANSCRIPTOMICS AND METABOLOMICS TO PREDICT BIOMARKERS ASSOCIATED WITH NAFLD AND NASH
Integration of analyses carried out across multiple biological measurements or OMIC-platforms represents an emerging approach aimed at addressing the challenges imposed by the complex biochemical regulation processes[62].
For example, application of Systems Biology approaches, i.e. Gene Set Enrichment Analysis (GSEA)[63], to the field of genomic data has rendered novel knowledge of shared disease-pathways between alcoholic and nonalcoholic liver disease[64]. Likewise, integration of genomic data has highlighted the shared genetic basis of metabolic syndrome and NAFLD[5].
A similar approach can be employed in the field of metabolomics to analyze the enrichment of metabolites that are overrepresented (ORA) in a query-sample against the whole set of metabolites in metabolic pathways. In this context, metabolite set enrichment analysis (MSEA) is the metabolomic counterpart of gene set enrichment. Such analysis, which can be performed by using either commercial or freely available software[65], has been applied to demonstrate alterations in metabolic pathways associated with NAFLD[66].
As a proof of principle, as a part of this work, we performed OMICs-integrative analysis using the IMPaLA (Integrated Molecular Pathway Level Analysis, http://impala.molgen.mpg.de)[67] platform. Briefly, the analysis was conducted by integrating the information on metabolites, genes and proteins, allowing the joint adjusted P-value (Q-value) to be calculated.
Specifically, we selected a list of genes previously associated with NAFLD[5,64], and metabolites that are known to be altered in NAFLD/NASH[68]. Names on metabolites were curated using the compound ID conversion of the web-based MetaboAnalyst tool (http://www.metaboanalyst.ca/)[69,70]. We found 2,827 pathways; however, only 219 of 347 input gene-identifiers were mapped to 219 distinct physical entities found in these pathways (with a gene background size of 12655). Similarly, only 32 of 51 input metabolite-identifiers were mapped to 32 distinct physical entities found in the pathways (with a metabolite background size of 5340). Relevant findings, excluding data that was exclusively and heavily dependent on genes or metabolites, are shown in Table 1; pathways and the Q-values for gene and/or metabolite enrichment were jointly calculated.
Table 1.
List of pathways involved in nonalcoholic fatty liver disease selected from significant Q-values that dependent on both genes and metabolites analyzed jointly
| Pathway name | Q-joint |
| Solute carriers -mediated transmembrane transport | 1.23E-12 |
| Transmembrane transport of small molecules | 9.66E-12 |
| Transport of glucose and other sugars bile salts and organic acids metal ions and amine compounds | 8.40E-10 |
| Leukotriene biosynthesis | 8.71E-10 |
| Transport of glucose and other sugars bile salts and organic acids metal ions and amine compounds | 1.91E-09 |
| Transport of inorganic cations-anions and amino acids-oligopeptides | 4.27E-09 |
| Amino acid and oligopeptide SLC transporters | 1.10E-08 |
| Transport of inorganic cations/anions and amino acids/oligopeptides | 2.40E-08 |
| tRNA Aminoacylation | 3.03E-08 |
| Gamma-glutamyl cycle | 3.61E-08 |
| tRNA charging | 5.96E-08 |
| mRNA protein and metabolite induction pathway by cyclosporine A | 8.47E-08 |
| Class I MHC mediated antigen processing & presentation | 1.73E-07 |
| Na+/Cl- dependent neurotransmitter transporters | 3.10E-07 |
| Amino acid transport across the plasma membrane | 3.72E-07 |
| S-methyl-5-thio-alpha;-D-ribose 1-phosphate degradation | 6.17E-07 |
| Amine compound solute carrier transporters | 6.17E-07 |
| Protein digestion and absorption - homo sapiens (human) | 2.13E-06 |
| Amino acid interconversion | 2.21E-06 |
| Biochemical pathways part I | 2.34E-06 |
| Amino acid metabolism | 3.96E-06 |
| Aminoacyl-tRNA biosynthesis - homo sapiens (human) | 6.88E-06 |
| Metabolism of amino acids and derivatives | 8.72E-06 |
| Mineral absorption - homo sapiens (human) | 1.47E-05 |
| Cytosolic tRNA aminoacylation | 2.86E-05 |
| Mitochondrial tRNA aminoacylation | 2.86E-05 |
| tRNA Aminoacylation | 2.86E-05 |
| Histidine, lysine, phenylalanine, tyrosine, proline and tryptophan catabolism | 0.000159 |
| Gene expression | 0.000181 |
| Tryptophan catabolism | 0.000275 |
| Phase II conjugation | 0.000426 |
| Phenylalanine and tyrosine catabolism | 0.003 |
| Glutamine and glutamate metabolism - homo sapiens (human) | 0.00376 |
| Glutaminolysis and cancer | 0.00493 |
| Glycine metabolism | 0.0052 |
| Glutamate glutamine metabolism | 0.00665 |
| Recycling of bile acids and salts | 0.00669 |
| Glycine serine alanine and threonine metabolism | 0.0101 |
| Branched-chain amino acid catabolism | 0.0103 |
OMICs-integrative analysis was performed using the IMPaLA (integrated molecular pathway level analysis, http://impala.molgen.mpg.de)[67] platform. A joined adjusted P-value (Q-value) was calculated to control for multiple testing by false discovery rate.
It is interesting to highlight and discuss a few examples in more detail. For instance, in the pathway “SLC-mediated transmembrane transport” (Reactome database), the overlapping genes and metabolites are CALM1 (Calmodulin 1), G6PC (Glucose-6-Phosphatase Catalytic Subunit), FGF21 (Fibroblast Growth Factor 21), GCK (Glucokinase) and GCKR (Glucokinase Regulator), and taurocholic acid, D-mannose, creatinine, L-lactic acid, L-valine, L-isoleucine, L-phenylalanine, L-aspartic acid, L-tyrosine, carnitine, betaine, L-glutamine, linoleic acid, oleic acid, L-leucine and glycocholic acid, respectively.
Another interesting example is the pathway “Transmembrane transport of small molecules” (Reactome database), in which the overlapping genes and metabolites are G6PC, CALM1, ATP1A1 (ATPase Na+/K+ Transporting Subunit Alpha 1), TF (Transferrin), ABCC1 (ATP Binding Cassette Subfamily C Member 1), FGF21, GCK, GCKR, HMOX1 (Heme Oxygenase 1), ABCB1 (ATP Binding Cassette Subfamily B Member 1), ABCC2 (ATP Binding Cassette Subfamily C Member 2), ABCC3 (ATP Binding Cassette Subfamily C Member 3) and ABCG2 ATP Binding Cassette Subfamily G Member 2), and L-glutamine, D-mannose, creatinine, L-lactic acid, L-valine, L-isoleucine, L-phenylalanine, taurocholic acid, L-aspartic acid, L-tyrosine, carnitine, betaine, linoleic acid, oleic acid, L-leucine and glycocholic acid, respectively.
Finally, in the pathway “Central carbon metabolism in cancer -Homo sapiens (human)” (KEGG database), the overlapping genes and metabolites are PTEN (Phosphatase and Tensin Homolog), EGFR (Epidermal Growth Factor Receptor), MET (MET Proto-Oncogene, Receptor Tyrosine Kinase), PIK3CA (Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha), MTOR (Mechanistic Target Of Rapamycin Kinase), AKT2 (AKT Serine/Threonine Kinase 2) and GCK, and L-glutamine, L-lactic acid, L-valine, L-isoleucine, L-phenylalanine, L-aspartic acid, L-tyrosine and L-leucine, respectively.
From these few examples, we may conclude that some pathways such as solute carrier (SLC) transporters should be further explored; in fact, available experimental data, while limited, support the participation of ABCC-family in NAFLD pathophysiology[71].
Nonetheless, the findings discussed above do not necessarily indicate that no other important pathways are potentially involved in the biology of NAFLD. In fact, Table 2 illustrates the myriad of processes involved in the pathogenesis of a complex disease such as NAFLD. In addition, Figure 3 depicts the complexity of the interactome among the whole set of genes, enzymes, chemical reactions and metabolites associated with NAFLD. Figure 4 shows a sub-network emphasizing the importance of the urea-cycle and metabolism of L-arginine, L-proline, L-glutamate, L-aspartate and L-asparagine. Specifically, features in Figure 4 highlight the central role played by aminotransferases and gamma-glutamyl transferases in the frame of altered L-glutamine/L-glutamate, glutathione and BCAA levels, as already mentioned.
Table 2.
List of pathways involved in nonalcoholic fatty liver disease selected from significant Q-values independently on whether they represent the effect of gene/s or metabolite/s only
| Pathway name | Pathway source | Q-joint |
| Adipogenesis | Wikipathways | 2.00E-17 |
| Non-alcoholic fatty liver disease (NAFLD) - homo sapiens (human) | KEGG | 2.33E-17 |
| Metabolism | Reactome | 3.72E-17 |
| AGE-RAGE pathway | Wikipathways | 4.22E-17 |
| Vitamin B12 Metabolism | Wikipathways | 5.24E-17 |
| Hepatitis B - homo sapiens (human) | KEGG | 1.79E-16 |
| Folate metabolism | Wikipathways | 1.29E-15 |
| Selenium micronutrient network | Wikipathways | 3.87E-15 |
| TNF signaling pathway - homo sapiens (human) | KEGG | 5.77E-15 |
| JAK-STAT-core | Signalink | 1.99E-14 |
| Adipocytokine signaling pathway - homo sapiens (human) | KEGG | 7.07E-14 |
| Nuclear receptors meta-pathway | Wikipathways | 1.26E-13 |
| IL1 and megakaryocytes in obesity | Wikipathways | 2.73E-13 |
| AGE-RAGE signaling pathway in diabetic complications - homo sapiens (human) | KEGG | 3.79E-13 |
| Spinal cord injury | Wikipathways | 5.44E-13 |
| Malaria - homo sapiens (human) | KEGG | 7.09E-13 |
| Metabolism of lipids and lipoproteins | Reactome | 7.09E-13 |
| SLC-mediated transmembrane transport | Reactome | 1.23E-12 |
| Pathways in cancer - homo sapiens (human) | KEGG | 1.41E-12 |
| Inflammatory bowel disease (IBD) - homo sapiens (human) | KEGG | 2.25E-12 |
| Lung fibrosis | Wikipathways | 2.63E-12 |
| Integrated pancreatic cancer pathway | Wikipathways | 3.10E-12 |
| PI3K-Akt signaling pathway - homo sapiens (human) | KEGG | 3.28E-12 |
| Chagas disease (American trypanosomiasis) - homo sapiens (human) | KEGG | 4.67E-12 |
| HIF-1 signaling pathway - homo sapiens (human) | KEGG | 4.67E-12 |
| AMPK signaling pathway - homo sapiens (human) | KEGG | 9.56E-12 |
| Transmembrane transport of small molecules | Reactome | 9.66E-12 |
| Central carbon metabolism in cancer - homo sapiens (human) | KEGG | 1.41E-11 |
| Jak-STAT signaling pathway - homo sapiens (human) | KEGG | 5.75E-11 |
| DNA damage response (only ATM dependent) | Wikipathways | 7.27E-11 |
| Cytokine-cytokine receptor interaction - homo sapiens (human) | KEGG | 1.01E-10 |
| Longevity regulating pathway - homo sapiens (human) | KEGG | 1.02E-10 |
| Toll-like receptor signaling pathway | Wikipathways | 2.12E-10 |
| Toll-like receptor signaling pathway - homo sapiens (human) | KEGG | 3.94E-10 |
| Toxoplasmosis - homo sapiens (human) | KEGG | 4.73E-10 |
| ABC transporters - homo sapiens (human) | KEGG | 5.94E-10 |
| Transport of glucose and other sugars bile salts and organic acids metal ions and amine compounds | Wikipathways | 8.40E-10 |
| Leukotriene biosynthesis | HumanCyc | 8.71E-10 |
| Insulin resistance - homo sapiens (human) | KEGG | 1.14E-09 |
| Transport of glucose and other sugars bile salts and organic acids metal ions and amine compounds | Reactome | 1.91E-09 |
| Sudden infant death syndrome (SIDS) susceptibility pathways | Wikipathways | 2.12E-09 |
| Cytokines and inflammatory response | Wikipathways | 2.17E-09 |
| AP-1 transcription factor network | PID | 2.22E-09 |
| FoxO signaling pathway - homo sapiens (human) | KEGG | 3.05E-09 |
| Leptin signaling pathway | Wikipathways | 3.57E-09 |
| Transport of inorganic cations-anions and amino acids-oligopeptides | Wikipathways | 4.27E-09 |
| Oncostatin M signaling pathway | Wikipathways | 5.72E-09 |
| Focal adhesion-PI3K-Akt-mTOR-signaling pathway | Wikipathways | 6.53E-09 |
| Amino acid and oligopeptide SLC transporters | Reactome | 1.10E-08 |
| Apoptosis | Wikipathways | 1.41E-08 |
| Apoptotic signaling pathway | Wikipathways | 1.41E-08 |
| Photodynamic therapy-induced NF-kB survival signaling | Wikipathways | 1.84E-08 |
| JAK STAT molecularvariation 1 | INOH | 2.04E-08 |
| MAPK signaling pathway | Wikipathways | 2.04E-08 |
| Aryl hydrocarbon receptor | Wikipathways | 2.35E-08 |
| Transport of inorganic cations/anions and amino acids/oligopeptides | Reactome | 2.40E-08 |
| tRNA aminoacylation | Wikipathways | 3.03E-08 |
| gamma-glutamyl cycle | HumanCyc | 3.61E-08 |
| Glucose homeostasis | Wikipathways | 4.08E-08 |
| Validated transcriptional targets of AP1 family members Fra1 and Fra2 | PID | 4.13E-08 |
| Hepatitis C and hepatocellular carcinoma | Wikipathways | 4.26E-08 |
| Calcineurin-regulated NFAT-dependent transcription in lymphocytes | PID | 4.29E-08 |
| Prostate cancer - homo sapiens (human) | KEGG | 4.29E-08 |
| Tuberculosis - homo sapiens (human) | KEGG | 4.45E-08 |
| Apoptosis - homo sapiens (human) | KEGG | 4.54E-08 |
| tRNA charging | HumanCyc | 5.96E-08 |
| Transcription factor regulation in adipogenesis | Wikipathways | 6.27E-08 |
| Sterol regulatory element-binding proteins (SREBP) signalling | Wikipathways | 6.27E-08 |
| Integrated lung cancer pathway | Wikipathways | 6.43E-08 |
| TNF related weak inducer of apoptosis (TWEAK) signaling pathway | Wikipathways | 8.14E-08 |
| mRNA protein and metabolite inducation pathway by cyclosporin A | Wikipathways | 8.47E-08 |
| PPAR signaling pathway | Wikipathways | 9.54E-08 |
| Immune system | Reactome | 9.57E-08 |
| Regulation of lipid metabolism by peroxisome proliferator-activated receptor alpha (PPARalpha) | Wikipathways | 1.13E-07 |
| AMP-activated protein kinase (AMPK) signaling | Wikipathways | 1.34E-07 |
| Photodynamic therapy-induced NFE2L2 (NRF2) survival signaling | Wikipathways | 1.52E-07 |
| Leptin insulin overlap | Wikipathways | 1.65E-07 |
| Class I MHC mediated antigen processing and presentation | Wikipathways | 1.73E-07 |
| Caspase cascade in apoptosis | PID | 1.99E-07 |
| Overview of nanoparticle effects | Wikipathways | 2.17E-07 |
| Alpha6Beta4Integrin | NetPath | 2.29E-07 |
| VEGFA-VEGFR2 signaling pathway | Wikipathways | 2.30E-07 |
| HIV-1 Nef: Negative effector of Fas and TNF-alpha | PID | 2.65E-07 |
| Innate immune system | Reactome | 2.69E-07 |
| Na+/Cl- dependent neurotransmitter transporters | Reactome | 3.10E-07 |
| Colorectal cancer - homo sapiens (human) | KEGG | 3.42E-07 |
| Regulation of toll-like receptor signaling pathway | Wikipathways | 3.64E-07 |
| stress induction of hsp regulation | BioCarta | 3.64E-07 |
| Amino acid transport across the plasma membrane | Reactome | 3.72E-07 |
| Programmed cell death | Reactome | 3.85E-07 |
| Apoptosis modulation and signaling | Wikipathways | 4.42E-07 |
| SREBF and miR33 in cholesterol and lipid homeostasis | Wikipathways | 4.84E-07 |
| JAK STAT pathway and regulation | INOH | 5.42E-07 |
OMICs-integrative analysis was performed using the IMPaLA (Integrated Molecular Pathway Level Analysis, http://impala.molgen.mpg.de)[67] platform. A joined adjusted P-value (Q-value) was calculated to control for multiple testing by false discovery rate.
Figure 3.
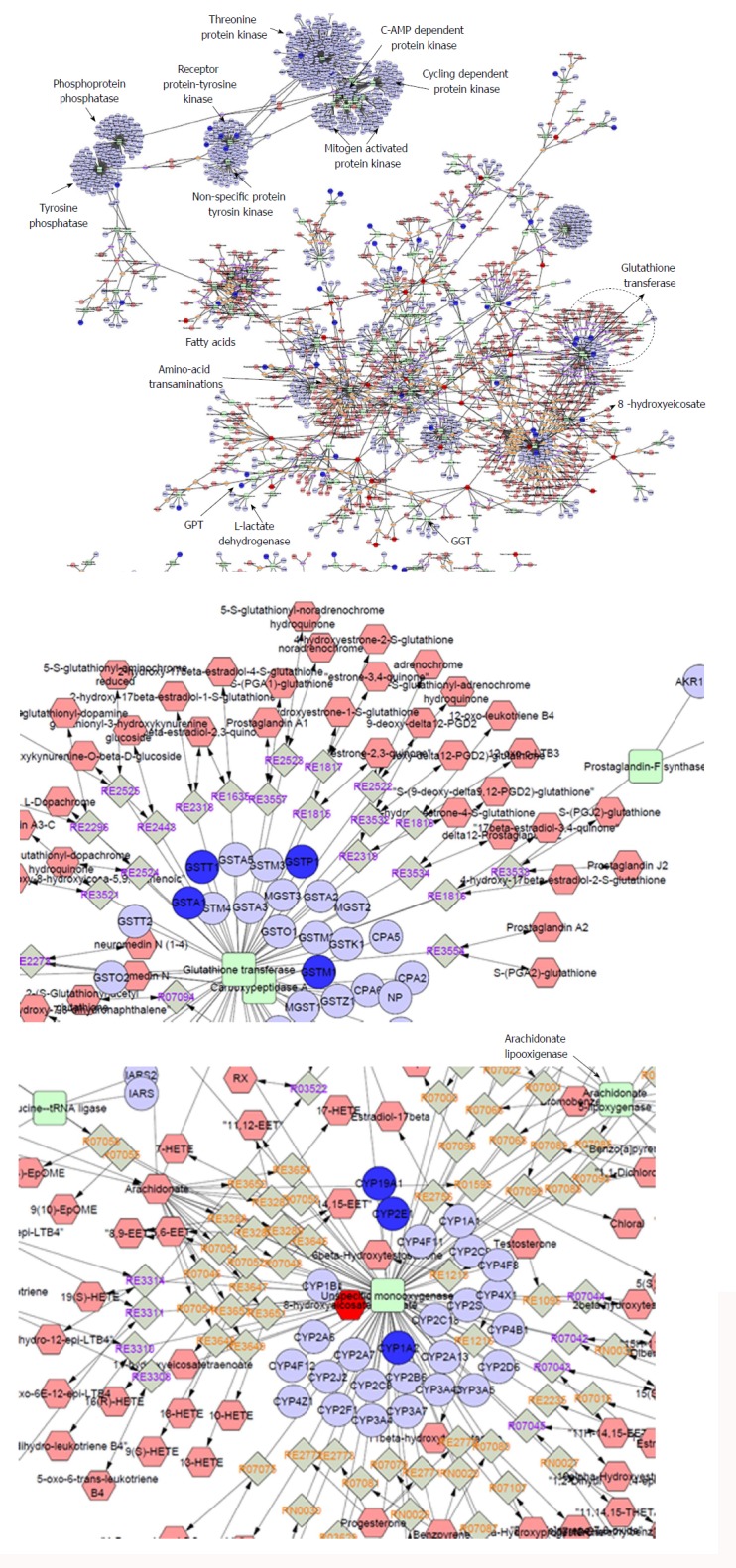
Whole interactome of compounds (hexagons), chemical reactions (diamonds), enzymes (squares) and genes (circles) associated with nonalcoholic fatty liver disease. Details on the set of genes and metabolites that were included in the analysis can be found in the main text; terms were filtered according to the ones already found in the databases. The interactome was built using Metscape[73], a plug-in for the widely used network analysis software Cytoscape[74] that supports calculation, analysis and visualization of gene-to-metabolite networks in the context of metabolism.
Figure 4.
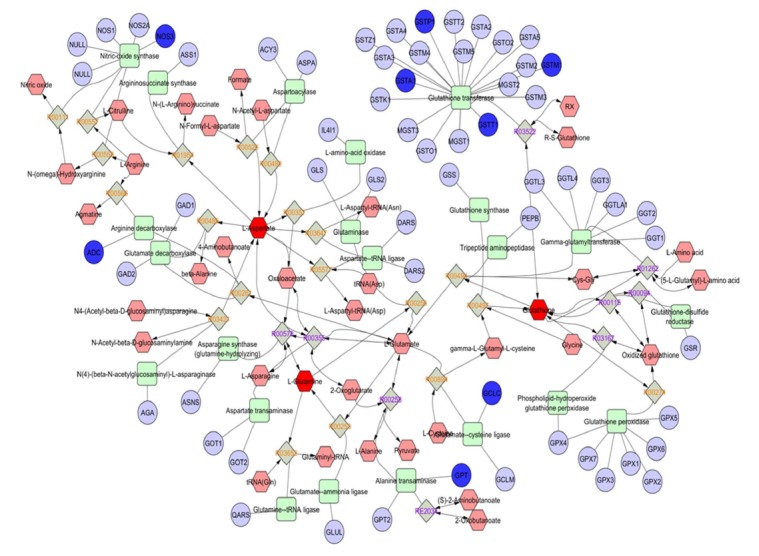
The urea-cycle, glutamate, and branched-chain amino acids in the biology of nonalcoholic fatty liver disease. Sub-network analysis showing the urea-cycle and metabolism of amino acids (L-arginine, L-proline, L-glutamate, L-aspartate and L-asparagine) that were extracted from the interactome shown in Figure 3. Compounds (common names in the Human Metabolome Database, http://www.hmdb.ca), chemical reactions, enzymes (KEGG database) and genes (HUGO symbols) are represented by hexagons, diamonds, squares and circles, respectively.
Finally, additional biomarkers that target immunity-related pathways, for example circulating levels of cytokines/chemokines, antibodies etc. might be useful in predicting NASH progression toward advanced phases[72].
CONCLUSION
Implementation of OMICs-derived biomarkers in the management and treatment of patients with NAFLD is still under extensive evaluation. Knowledge gained on genetic signatures associated with NAFLD and NASH, as well as the role of circulating cfmiRNAs and plasma metabolites, should be promptly translated into the clinical setting. Nevertheless, rigorous steps that must include validation and replication are mandatory before OMICs biomarkers are ready for use as diagnostic markers to identify patients at risk of advanced disease, including liver cancer.
What to expect for the near future: A personalized NAFLD approach by integration of OMICs - big data and clinical information (Figure 5): (1) It is expected that, in the near future, NAFLD patients can be diagnosed and treated according to their own “molecular signature”; (2) Specific focus should be placed on prevention and early diagnosis by the application of biomarkers of disease risk; (3) Selection of “personalized drugs” as well as tailored therapy should be made according to the specific molecular signature; and (4) Personalized lifestyle intervention is desirable but it is envisioned that the basic and general recommendations about alcohol restriction, healthy diet and exercise would remain the foundation of prevention and therapy.
Figure 5.
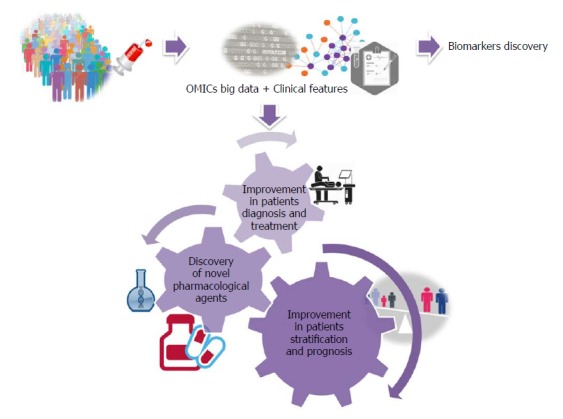
What to expect for the near future. A personalized nonalcoholic fatty liver disease approach by integrating OMICs big data with clinical information.
Footnotes
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Argentina
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B, B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Supported by Agencia Nacional de Promoción Científicay Tecnológica, No. PICT 2014-0432, No. PICT 2014-1816 and No. PICT 2015-0551.
Conflict-of-interest statement: No potential conflicts of interest.
Peer-review started: February 1, 2018
First decision: February 24, 2018
Article in press: March 31, 2018
P- Reviewer: Arslan N, Enomoto H, Jarcuska P, Sutti S, Toshikuni N S- Editor: Ma YJ L- Editor: A E- Editor: Huang Y
Contributor Information
Carlos J Pirola, Department of Genetics and Molecular Biology of Complex Diseases. University of Buenos Aires, Institute of Medical Research A Lanari, Buenos Aires, Argentina, National Scientific and Technical Research Council-University of Buenos Aires. Institute of Medical Research (IDIM), CABA 1427, Argentina.
Silvia Sookoian, Clinical and Molecular Hepatology, University of Buenos Aires, Institute of Medical Research A Lanari, Buenos Aires, Argentina, National Scientific and Technical Research Council-University of Buenos Aires. Institute of Medical Research (IDIM), CABA 1427, Argentina. sookoian.silvia@lanari.fmed.uba.ar.
References
- 1.Brunt EM, Wong VW, Nobili V, Day CP, Sookoian S, Maher JJ, Bugianesi E, Sirlin CB, Neuschwander-Tetri BA, Rinella ME. Nonalcoholic fatty liver disease. Nat Rev Dis Primers. 2015;1:15080. doi: 10.1038/nrdp.2015.80. [DOI] [PubMed] [Google Scholar]
- 2.Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, Harrison SA, Brunt EM, Sanyal AJ. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2018;67:328–357. doi: 10.1002/hep.29367. [DOI] [PubMed] [Google Scholar]
- 3.Brunt EM. Histopathology of non-alcoholic fatty liver disease. Clin Liver Dis. 2009;13:533–544. doi: 10.1016/j.cld.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Brunt EM. Pathology of nonalcoholic fatty liver disease. Nat Rev Gastroenterol Hepatol. 2010;7:195–203. doi: 10.1038/nrgastro.2010.21. [DOI] [PubMed] [Google Scholar]
- 5.Sookoian S, Pirola CJ. Nonalcoholic fatty liver disease and metabolic syndrome: Shared genetic basis of pathogenesis. Hepatology. 2016;64:1417–1420. doi: 10.1002/hep.28746. [DOI] [PubMed] [Google Scholar]
- 6.Sookoian S, Pirola CJ. Genetic predisposition in nonalcoholic fatty liver disease. Clin Mol Hepatol. 2017;23:1–12. doi: 10.3350/cmh.2016.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sookoian S, Castaño GO, Burgueño AL, Gianotti TF, Rosselli MS, Pirola CJ. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J Lipid Res. 2009;50:2111–2116. doi: 10.1194/jlr.P900013-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53:1883–1894. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 10.Kozlitina J, Smagris E, Stender S, Nordestgaard BG, Zhou HH, Tybjærg-Hansen A, Vogt TF, Hobbs HH, Cohen JC. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pirola CJ, Sookoian S. The dual and opposite role of the TM6SF2-rs58542926 variant in protecting against cardiovascular disease and conferring risk for nonalcoholic fatty liver: A meta-analysis. Hepatology. 2015;62:1742–1756. doi: 10.1002/hep.28142. [DOI] [PubMed] [Google Scholar]
- 12.Dongiovanni P, Petta S, Maglio C, Fracanzani AL, Pipitone R, Mozzi E, Motta BM, Kaminska D, Rametta R, Grimaudo S, et al. Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology. 2015;61:506–514. doi: 10.1002/hep.27490. [DOI] [PubMed] [Google Scholar]
- 13.Liu YL, Reeves HL, Burt AD, Tiniakos D, McPherson S, Leathart JB, Allison ME, Alexander GJ, Piguet AC, Anty R, et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat Commun. 2014;5:4309. doi: 10.1038/ncomms5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sookoian S, Pirola CJ. Liver enzymes, metabolomics and genome-wide association studies: from systems biology to the personalized medicine. World J Gastroenterol. 2015;21:711–725. doi: 10.3748/wjg.v21.i3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sookoian S, Castaño GO, Scian R, Mallardi P, Fernández Gianotti T, Burgueño AL, San Martino J, Pirola CJ. Genetic variation in transmembrane 6 superfamily member 2 and the risk of nonalcoholic fatty liver disease and histological disease severity. Hepatology. 2015;61:515–525. doi: 10.1002/hep.27556. [DOI] [PubMed] [Google Scholar]
- 16.Kotronen A, Peltonen M, Hakkarainen A, Sevastianova K, Bergholm R, Johansson LM, Lundbom N, Rissanen A, Ridderstråle M, Groop L, et al. Prediction of non-alcoholic fatty liver disease and liver fat using metabolic and genetic factors. Gastroenterology. 2009;137:865–872. doi: 10.1053/j.gastro.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Nobili V, Donati B, Panera N, Vongsakulyanon A, Alisi A, Dallapiccola B, Valenti L. A 4-polymorphism risk score predicts steatohepatitis in children with nonalcoholic fatty liver disease. J Pediatr Gastroenterol Nutr. 2014;58:632–636. doi: 10.1097/MPG.0000000000000279. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, Orešič M, Leivonen M, Gopalacharyulu P, Hyysalo J, Arola J, Verrijken A, Francque S, Van Gaal L, Hyötyläinen T, et al. Noninvasive Detection of Nonalcoholic Steatohepatitis Using Clinical Markers and Circulating Levels of Lipids and Metabolites. Clin Gastroenterol Hepatol. 2016;14:1463–1472.e6. doi: 10.1016/j.cgh.2016.05.046. [DOI] [PubMed] [Google Scholar]
- 19.Sevastianova K, Kotronen A, Gastaldelli A, Perttilä J, Hakkarainen A, Lundbom J, Suojanen L, Orho-Melander M, Lundbom N, Ferrannini E, et al. Genetic variation in PNPLA3 (adiponutrin) confers sensitivity to weight loss-induced decrease in liver fat in humans. Am J Clin Nutr. 2011;94:104–111. doi: 10.3945/ajcn.111.012369. [DOI] [PubMed] [Google Scholar]
- 20.Shen J, Wong GL, Chan HL, Chan RS, Chan HY, Chu WC, Cheung BH, Yeung DK, Li LS, Sea MM, et al. PNPLA3 gene polymorphism and response to lifestyle modification in patients with nonalcoholic fatty liver disease. J Gastroenterol Hepatol. 2015;30:139–146. doi: 10.1111/jgh.12656. [DOI] [PubMed] [Google Scholar]
- 21.Krawczyk M, Jiménez-Agüero R, Alustiza JM, Emparanza JI, Perugorria MJ, Bujanda L, Lammert F, Banales JM. PNPLA3 p.I148M variant is associated with greater reduction of liver fat content after bariatric surgery. Surg Obes Relat Dis. 2016;12:1838–1846. doi: 10.1016/j.soard.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Sookoian S, Flichman D, Scian R, Rohr C, Dopazo H, Gianotti TF, Martino JS, Castaño GO, Pirola CJ. Mitochondrial genome architecture in non-alcoholic fatty liver disease. J Pathol. 2016;240:437–449. doi: 10.1002/path.4803. [DOI] [PubMed] [Google Scholar]
- 23.Sookoian S, Rosselli MS, Gemma C, Burgueño AL, Fernández Gianotti T, Castaño GO, Pirola CJ. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: impact of liver methylation of the peroxisome proliferator-activated receptor γ coactivator 1α promoter. Hepatology. 2010;52:1992–2000. doi: 10.1002/hep.23927. [DOI] [PubMed] [Google Scholar]
- 24.Ahrens M, Ammerpohl O, von Schönfels W, Kolarova J, Bens S, Itzel T, Teufel A, Herrmann A, Brosch M, Hinrichsen H, et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18:296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Plant D, Webster A, Nair N, Oliver J, Smith SL, Eyre S, Hyrich KL, Wilson AG, Morgan AW, Isaacs JD, et al. Differential Methylation as a Biomarker of Response to Etanercept in Patients With Rheumatoid Arthritis. Arthritis Rheumatol. 2016;68:1353–1360. doi: 10.1002/art.39590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Biagini Myers JM, Yadagiri VK, Ulm A, Chen X, Weirauch MT, Khurana Hershey GK, Ji H. Nasal DNA methylation differentiates corticosteroid treatment response in pediatric asthma: A pilot study. PLoS One. 2017;12:e0186150. doi: 10.1371/journal.pone.0186150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts S, Lester KJ, Hudson JL, Rapee RM, Creswell C, Cooper PJ, Thirlwall KJ, Coleman JR, Breen G, Wong CC, et al. Serotonin transporter [corrected] methylation and response to cognitive behaviour therapy in children with anxiety disorders. Transl Psychiatry. 2014;4:e444. doi: 10.1038/tp.2014.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci U S A. 2011;108:3630–3635. doi: 10.1073/pnas.1012311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pirola CJ, Gianotti TF, Burgueño AL, Rey-Funes M, Loidl CF, Mallardi P, Martino JS, Castaño GO, Sookoian S. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut. 2013;62:1356–1363. doi: 10.1136/gutjnl-2012-302962. [DOI] [PubMed] [Google Scholar]
- 30.MANDEL P, METAIS P. [Not Available] C R Seances Soc Biol Fil. 1948;142:241–243. [PubMed] [Google Scholar]
- 31.Hardy T, Zeybel M, Day CP, Dipper C, Masson S, McPherson S, Henderson E, Tiniakos D, White S, French J, et al. Plasma DNA methylation: a potential biomarker for stratification of liver fibrosis in non-alcoholic fatty liver disease. Gut. 2017;66:1321–1328. doi: 10.1136/gutjnl-2016-311526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yiğit B, Boyle M, Özler O, Erden N, Tutucu F, Hardy T, Bergmann C, Distler JHW, Adalı G, Dayangaç M, et al. Plasma cell-free DNA methylation: a liquid biomarker of hepatic fibrosis. Gut. 2018 doi: 10.1136/gutjnl-2017-315668. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sookoian S, Pirola CJ. Cell-free DNA methylation as liquid biopsy for the assessment of fibrosis in patients with nonalcoholic steatohepatitis: a gap between innovation and implementation. Hepatobiliary Surg Nutr. 2017;6:117–121. doi: 10.21037/hbsn.2017.01.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becker PP, Rau M, Schmitt J, Malsch C, Hammer C, Bantel H, Müllhaupt B, Geier A. Performance of Serum microRNAs -122, -192 and -21 as Biomarkers in Patients with Non-Alcoholic Steatohepatitis. PLoS One. 2015;10:e0142661. doi: 10.1371/journal.pone.0142661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pirola CJ, Fernández Gianotti T, Castaño GO, Mallardi P, San Martino J, Mora Gonzalez Lopez Ledesma M, Flichman D, Mirshahi F, Sanyal AJ, Sookoian S. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut. 2015;64:800–812. doi: 10.1136/gutjnl-2014-306996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan Y, Ge G, Pan T, Wen D, Gan J. A pilot study of serum microRNAs panel as potential biomarkers for diagnosis of nonalcoholic fatty liver disease. PLoS One. 2014;9:e105192. doi: 10.1371/journal.pone.0105192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cusi K, Chang Z, Harrison S, Lomonaco R, Bril F, Orsak B, Ortiz-Lopez C, Hecht J, Feldstein AE, Webb A, et al. Limited value of plasma cytokeratin-18 as a biomarker for NASH and fibrosis in patients with non-alcoholic fatty liver disease. J Hepatol. 2014;60:167–174. doi: 10.1016/j.jhep.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 39.Sookoian S, Castaño G, Burgueño AL, Gianotti TF, Rosselli MS, Pirola CJ. A diagnostic model to differentiate simple steatosis from nonalcoholic steatohepatitis based on the likelihood ratio form of Bayes theorem. Clin Biochem. 2009;42:624–629. doi: 10.1016/j.clinbiochem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 40.Wood GC, Chu X, Argyropoulos G, Benotti P, Rolston D, Mirshahi T, Petrick A, Gabrielson J, Carey DJ, DiStefano JK, et al. A multi-component classifier for nonalcoholic fatty liver disease (NAFLD) based on genomic, proteomic, and phenomic data domains. Sci Rep. 2017;7:43238. doi: 10.1038/srep43238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Decaris ML, Li KW, Emson CL, Gatmaitan M, Liu S, Wang Y, Nyangau E, Colangelo M, Angel TE, Beysen C, et al. Identifying nonalcoholic fatty liver disease patients with active fibrosis by measuring extracellular matrix remodeling rates in tissue and blood. Hepatology. 2017;65:78–88. doi: 10.1002/hep.28860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okanoue T, Ebise H, Kai T, Mizuno M, Shima T, Ichihara J, Aoki M. A simple scoring system using type IV collagen 7S and aspartate aminotransferase for diagnosing nonalcoholic steatohepatitis and related fibrosis. J Gastroenterol. 2018;53:129–139. doi: 10.1007/s00535-017-1355-9. [DOI] [PubMed] [Google Scholar]
- 43.Tanwar S, Trembling PM, Guha IN, Parkes J, Kaye P, Burt AD, Ryder SD, Aithal GP, Day CP, Rosenberg WM. Validation of terminal peptide of procollagen III for the detection and assessment of nonalcoholic steatohepatitis in patients with nonalcoholic fatty liver disease. Hepatology. 2013;57:103–111. doi: 10.1002/hep.26030. [DOI] [PubMed] [Google Scholar]
- 44.Rodriguez-Suarez E, Mato JM, Elortza F. Proteomics analysis of human nonalcoholic fatty liver. Methods Mol Biol. 2012;909:241–258. doi: 10.1007/978-1-61779-959-4_16. [DOI] [PubMed] [Google Scholar]
- 45.Younossi ZM, Baranova A, Ziegler K, Del Giacco L, Schlauch K, Born TL, Elariny H, Gorreta F, VanMeter A, Younoszai A, et al. A genomic and proteomic study of the spectrum of nonalcoholic fatty liver disease. Hepatology. 2005;42:665–674. doi: 10.1002/hep.20838. [DOI] [PubMed] [Google Scholar]
- 46.Ulukaya E, Yilmaz Y, Moshkovskii S, Karpova M, Pyatnitskiy M, Atug O, Dolar E. Proteomic analysis of serum in patients with non-alcoholic steatohepatitis using matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Scand J Gastroenterol. 2009;44:1471–1476. doi: 10.3109/00365520903353379. [DOI] [PubMed] [Google Scholar]
- 47.Puri P, Wiest MM, Cheung O, Mirshahi F, Sargeant C, Min HK, Contos MJ, Sterling RK, Fuchs M, Zhou H, et al. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology. 2009;50:1827–1838. doi: 10.1002/hep.23229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kalhan SC, Guo L, Edmison J, Dasarathy S, McCullough AJ, Hanson RW, Milburn M. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metabolism. 2011;60:404–413. doi: 10.1016/j.metabol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puri P, Daita K, Joyce A, Mirshahi F, Santhekadur PK, Cazanave S, Luketic VA, Siddiqui MS, Boyett S, Min HK, et al. The presence and severity of nonalcoholic steatohepatitis is associated with specific changes in circulating bile acids. Hepatology. 2017 doi: 10.1002/hep.29359. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alonso C, Fernández-Ramos D, Varela-Rey M, Martínez-Arranz I, Navasa N, Van Liempd SM, Lavín Trueba JL, Mayo R, Ilisso CP, de Juan VG, et al. Metabolomic Identification of Subtypes of Nonalcoholic Steatohepatitis. Gastroenterology. 2017;152:1449–1461.e7. doi: 10.1053/j.gastro.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sunny NE, Parks EJ, Browning JD, Burgess SC. Excessive hepatic mitochondrial TCA cycle and gluconeogenesis in humans with nonalcoholic fatty liver disease. Cell Metab. 2011;14:804–810. doi: 10.1016/j.cmet.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sookoian S, Castaño GO, Scian R, Fernández Gianotti T, Dopazo H, Rohr C, Gaj G, San Martino J, Sevic I, Flichman D, et al. Serum aminotransferases in nonalcoholic fatty liver disease are a signature of liver metabolic perturbations at the amino acid and Krebs cycle level. Am J Clin Nutr. 2016;103:422–434. doi: 10.3945/ajcn.115.118695. [DOI] [PubMed] [Google Scholar]
- 53.Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang TP, Walter K, Menni C, Chen L, Vasquez L, Valdes AM, Hyde CL, Wang V, Ziemek D, Roberts P, Xi L, Grundberg E; Multiple Tissue Human Expression Resource (MuTHER) Consortium, Waldenberger M, Richards JB, Mohney RP, Milburn MV, John SL, Trimmer J, Theis FJ, Overington JP, Suhre K, Brosnan MJ, Gieger C, Kastenmüller G, Spector TD, Soranzo N. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yin X, Subramanian S, Willinger CM, Chen G, Juhasz P, Courchesne P, Chen BH, Li X, Hwang SJ, Fox CS, et al. Metabolite Signatures of Metabolic Risk Factors and their Longitudinal Changes. J Clin Endocrinol Metab. 2016;101:1779–1789. doi: 10.1210/jc.2015-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sookoian S, Pirola CJ. NAFLD. Metabolic make-up of NASH: from fat and sugar to amino acids. Nat Rev Gastroenterol Hepatol. 2014;11:205–207. doi: 10.1038/nrgastro.2014.25. [DOI] [PubMed] [Google Scholar]
- 56.Sookoian S, Pirola CJ. The nonalcoholic steatohepatitis metabotype: Imbalance of circulating amino acids and transamination reactions reflect impaired mitochondrial function. Hepatology. 2018;67:1177–1178. doi: 10.1002/hep.29705. [DOI] [PubMed] [Google Scholar]
- 57.Goffredo M, Santoro N, Tricò D, Giannini C, D’Adamo E, Zhao H, Peng G, Yu X, Lam TT, Pierpont B, et al. A Branched-Chain Amino Acid-Related Metabolic Signature Characterizes Obese Adolescents with Non-Alcoholic Fatty Liver Disease. Nutrients. 2017;9:pii: E642. doi: 10.3390/nu9070642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaggini M, Carli F, Rosso C, Buzzigoli E, Marietti M, Della Latta V, Ciociaro D, Abate ML, Gambino R, Cassader M, et al. Altered amino acid concentrations in NAFLD: Impact of obesity and insulin resistance. Hepatology. 2018;67:145–158. doi: 10.1002/hep.29465. [DOI] [PubMed] [Google Scholar]
- 59.Min HK, Sookoian S, Pirola CJ, Cheng J, Mirshahi F, Sanyal AJ. Metabolic profiling reveals that PNPLA3 induces widespread effects on metabolism beyond triacylglycerol remodeling in Huh-7 hepatoma cells. Am J Physiol Gastrointest Liver Physiol. 2014;307:G66–G76. doi: 10.1152/ajpgi.00335.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sookoian S, Puri P, Castaño GO, Scian R, Mirshahi F, Sanyal AJ, Pirola CJ. Nonalcoholic steatohepatitis is associated with a state of betaine-insufficiency. Liver Int. 2017;37:611–619. doi: 10.1111/liv.13249. [DOI] [PubMed] [Google Scholar]
- 61.Wanichthanarak K, Fan S, Grapov D, Barupal DK, Fiehn O. Metabox: A Toolbox for Metabolomic Data Analysis, Interpretation and Integrative Exploration. PLoS One. 2017;12:e0171046. doi: 10.1371/journal.pone.0171046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wanichthanarak K, Fahrmann JF, Grapov D. Genomic, Proteomic, and Metabolomic Data Integration Strategies. Biomark Insights. 2015;10:1–6. doi: 10.4137/BMI.S29511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sookoian S, Pirola CJ. Systems biology elucidates common pathogenic mechanisms between nonalcoholic and alcoholic-fatty liver disease. PLoS One. 2013;8:e58895. doi: 10.1371/journal.pone.0058895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cambiaghi A, Ferrario M, Masseroli M. Analysis of metabolomic data: tools, current strategies and future challenges for omics data integration. Brief Bioinform. 2017;18:498–510. doi: 10.1093/bib/bbw031. [DOI] [PubMed] [Google Scholar]
- 66.Dumas ME, Kinross J, Nicholson JK. Metabolic phenotyping and systems biology approaches to understanding metabolic syndrome and fatty liver disease. Gastroenterology. 2014;146:46–62. doi: 10.1053/j.gastro.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 67.Kamburov A, Cavill R, Ebbels TM, Herwig R, Keun HC. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics. 2011;27:2917–2918. doi: 10.1093/bioinformatics/btr499. [DOI] [PubMed] [Google Scholar]
- 68.Safaei A, Arefi Oskouie A, Mohebbi SR, Rezaei-Tavirani M, Mahboubi M, Peyvandi M, Okhovatian F, Zamanian-Azodi M. Metabolomic analysis of human cirrhosis, hepatocellular carcinoma, non-alcoholic fatty liver disease and non-alcoholic steatohepatitis diseases. Gastroenterol Hepatol Bed Bench. 2016;9:158–173. [PMC free article] [PubMed] [Google Scholar]
- 69.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xia J, Wishart DS. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr Protoc Bioinformatics. 2016;55:14.10.1–14.10.91. doi: 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
- 71.Tanaka N, Matsubara T, Krausz KW, Patterson AD, Gonzalez FJ. Disruption of phospholipid and bile acid homeostasis in mice with nonalcoholic steatohepatitis. Hepatology. 2012;56:118–129. doi: 10.1002/hep.25630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sutti S, Jindal A, Bruzzì S, Locatelli I, Bozzola C, Albano E. Is there a role for adaptive immunity in nonalcoholic steatohepatitis? World J Hepatol. 2015;7:1725–1729. doi: 10.4254/wjh.v7.i13.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karnovsky A, Weymouth T, Hull T, Tarcea VG, Scardoni G, Laudanna C, Sartor MA, Stringer KA, Jagadish HV, Burant C, et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012;28:373–380. doi: 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]


