Abstract
Background
Subclinical necrotic enteritis (SNE) widely outbreaks in chickens which inflicted growth-slowing, causing enormous social and economic burdens. To better understand the molecular underpinnings of SNE on lipid metabolism and explore novel preventative strategies against SNE, we studied the regulatory mechanism of a potential probiotic, Lactobacillus johnsonii BS15 on the lipid metabolism pathways involved in chickens with SNE.
Methods
One hundred eighty one-day-old chickens were randomly divided into three groups and arranged with basal diet (control and SNE group). Added with BS15 (1 × 106 cfu/g) or Man Rogosa Sharpe (MRS) liquid medium for 28 days. The hepatic gene expression of each group was then measured using high-throughput analysis methods (RNA-Seq). Quantitative real-time PCR (qRT-PCR) was used to detect the expression changes of the related genes.
Results
The results showed that there are eleven lipid metabolic pathways were found during the prevention of BS15 treatment in SNE chickens by RNA-Seq, including the peroxisome proliferator-activated receptor (PPAR) signaling pathway and arachidonic acid metabolism. BS15 notably facilitated the expressions of fatty acid binding protein 2 (FABP2), acyl-CoA synthetase bubblegum family member 1 (ACSBG1), perilipin 1 (PLIN1) and perilipin 2 (PLIN2), which were involved in PPAR signaling pathway of SNE chickens. Besides, suppression of phospholipase A2 group IVA (PLA2G4A) in arachidonic acid metabolism was observed in SNE chickens after BS15 prevention. The expression patterns of FABP2, ACSBG1, PLIN1, PLIN2 and PLA24G in qRT-PCR validation were consistent with RNA-Seq results.
Conclusions
These findings indicate that SNE may affect the hepatic lipid metabolism of chickens. Meanwhile, BS15 pretreatment may provide a prospective natural prophylaxis strategy against SNE through improving the PPAR signaling pathway and arachidonic acid metabolism.
Electronic supplementary material
The online version of this article (10.1186/s12944-018-0741-5) contains supplementary material, which is available to authorized users.
Keywords: Lactobacillus johnsonii, Subclinical necrotic enteritis, Hepatic transcriptome, Lipid metabolism pathways
Background
Subclinical necrotic enteritis (SNE) of broiler chickens is characterized by the intestinal damage and growth-slowing without mortality [1]. The residues of enterotoxigenic Clostridium perfringens that is the most predominant causes of SNE menaces the public health via the food chain [2, 3]. As a result of the ban of subtherapeutic antibiotics usage in the European Union, the incidence of SNE has further increased in recent years [4], which made numerous social economic losses via chronic impairing the lipid metabolism of chicken [5] and decreasing the quality of chicken meat [6]. Accordingly, finding valid alternatives to antibiotics has gained in importance [7, 8]. To date, as one of the best ideal alternatives, probiotics exhibit positive influences on the growth development and necrotic enteritis diseases in broiler chickens [9–12], which are proposed as an attractive option for treating SNE. Based on this, we firstly discovered that Lactobacillus johnsonii BS15 (CCTCC M2013663) strain exhibited the beneficial effects on lipid metabolism in the previous studies, resulting in preventing non-alcoholic fatty liver disease in obese mice [13]. Then we applied this strain to chickens and found similar conditions, including it improved the meat nutritional value through altering the fatty acid composition [14], and promoted growth performance and lowered fat deposition in broilers [15]. Subsequently, we demonstrated that the aberrations of lipid metabolism during subclinical Clostridium perfringens infection was obviously ameliorated after BS15 prevention through controlling the lipid deposits and fatty acid composition [6].
Based on the rationale mentioned above, we speculated that L. johnsonii BS15 may prevent SNE associated with regulating the hepatic lipid metabolism. However, information is still limited on the molecular underpinnings of SNE pathogenesis in hepatic transcriptome, and how BS15 exerts its beneficial effects on hepatic lipid metabolism of SNE chickens has not been reported. To verify this speculation, we have indicated that BS15 prevention really improved the hepatic lipid metabolism of chickens with SNE [5], nevertheless, its molecular mechanism remains unrevealed. Therefore, RNA sequencing was undertaken to characterize the hepatic transcriptome in the present study for better understanding the molecular mechanism about lipid metabolism in SNE chickens and providing a novel preventative strategy against SNE.
Results
Preventive effect of BS15 on the SNE infection chicken model
To explore the molecular preventive mechanism of BS15, we carried out a hepatic gene expression study with a SNE infection chicken model. There is no death records during the whole experiment, and the some rudimentary parameters results of growth performance and serum were described in the previous report [5]. Besides, the screened genes among control, SNE and BS15 was showed in Fig. 1.
Fig. 1.
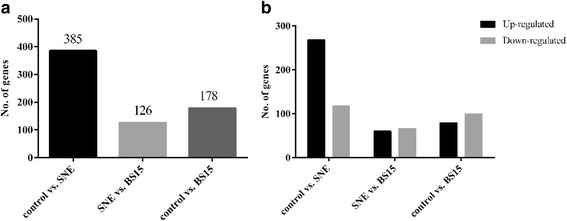
RNA-seq results among control, subclinical necrotic enteritis (SNE) and Lactobacillus johnsonii BS15 (BS15) groups. a, screened genes among control, SNE and BS15. 385 differentially expressed genes were screened in control vs. SNE groups, 126 differentially expressed genes were screened in SNE vs. BS15 groups, 178 differentially expressed genes were screened in control vs. BS15. b, up-regulated and down-regulated genes in hepatic transcriptome. 268 genes were up-regulated from control to SNE, 60 genes were up-regulated from SNE to BS15; while 117 genes were down-regulated from control to SNE, 66 genes were down-regulated from SNE to BS15
Genes and pathways related to lipid metabolism associated with SNE disease
We analyzed alterations in lipid metabolism gene expression in three SNE and three normal chicken liver tissue specimens using the powerful RNA sequencing technology. 385 genes as being differentially expressed in the SNE liver samples as compared with control animals’ (p < 0.05) were observed (Fig. 2, Additional file 1: Table S1). Gene ontology (GO) functional analysis was used to decipher the major biological processes affected among the differentially expressed genes. From this analysis, we found that SNE infection altered a multitude of biological processes relating to the metabolic regulation of lipid, including fatty acid, cholesterol, lipid storage, phosphatidylinositol, steroid, phospholipid and triglyceride (p < 0.05) (Additional file 2: Table S2). In addition, some biological processes were also found to be dysregulated and included regulation of cellular response to oxidative stress, calcium ion transmembrane transport, inflammatory response, and so on, manifesting that there exists inflammation of the liver, which was responsible for successful SNE infection model. More importantly, the kyoto encyclopedia of genes and genomes (KEGG) database revealed ten genes that were enriched in the hepatic transcriptome that participated in 12 pathways related to lipid metabolism, including steroid biosynthesis, insulin signaling pathway, adipocytokine signaling pathway, PPAR signaling pathway, and so on (Table 1).
Fig. 2.
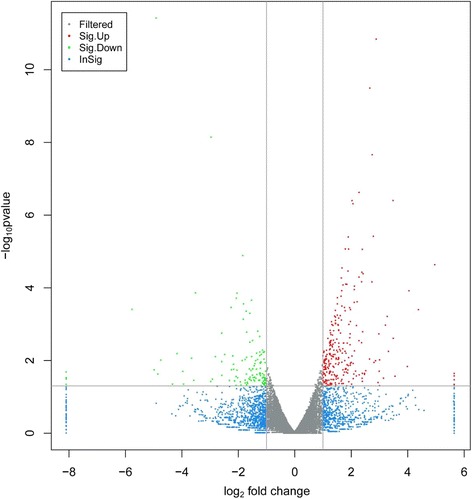
Volcano plot of control group vs. subclinical necrotic enteritis group. The 385 differentially expressed genes that fall above our threshold value are pictured in red (Up-regulated) and green (Down-regulated). The blue dots are the insignificantly different expressed genes
Table 1.
Important regulatory genes and pathways related to lipid metabolism in comparison of control group and SNE group
| Pathway | P-value | Pathway ID | Genes |
|---|---|---|---|
| Steroid hormone biosynthesis | 0.05007 | gga00140 | CYP17A1; CYP1B1 |
| Steroid biosynthesis | 0.07212 | gga00100 | LIPA |
| Ether lipid metabolism | 0.08339 | gga00565 | LPCAT1; PLA2G4A |
| alpha-Linolenic acid metabolism | 0.10632 | gga00592 | PLA2G4A |
| Linoleic acid metabolism | 0.13425 | gga00591 | PLA2G4A |
| beta-Alanine metabolism | 0.16359 | gga00410 | ALDH1A3 |
| Arachidonic acid metabolism | 0.34997 | gga00590 | PLA2G4A |
| Glycerophospholipid metabolism | 0.41918 | gga00564 | LPCAT1; PLA2G4A |
| PPAR signaling pathway | 0.49544 | gga03320 | PLIN2 |
| Insulin resistance | 0.50696 | gga04931 | PIK3R5; PRKCB |
| Adipocytokine signaling pathway | 0.52206 | gga04920 | TNFRSF1B |
| Insulin signaling pathway | 0.84921 | gga04910 | PIK3R5 |
Genes and pathways mediating the preventive effects of BS15 treatment
In order to identify the certain genes and pathways associated with BS15 prevention, KEGG analysis of pathway enrichment was conducted with the differentially expressed genes between the SNE infection and BS15 treatment groups. BS15 prevention created far-reaching influence on genes expression in the livers of SNE chickens. Compared with the SNE group, significantly elevated expression of 60 genes and obviously decreased expression of 66 genes were found in the livers of BS15-disposed chickens (Fig. 3. Additional file 3: Table S3). What’s more, the results of enriched pathways analysis highlighted the changes related to lipid metabolism, including glycerophospholipid metabolism, adipocytokine signaling pathway, glycerolipid metabolism, fatty acid metabolism, PPAR signaling pathway, and so on (Table 2).
Fig. 3.
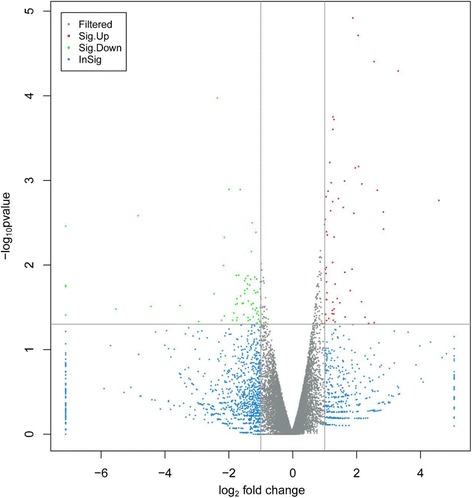
Volcano plot of subclinical necrotic enteritis group vs. Lactobacillus johnsonii BS15 group. The 126 differentially expressed genes that fall above our threshold value are pictured in red (Upregulated) and green (Down-regulated). The blue dots are the insignificantly different expressed genes
Table 2.
Genes and pathways relevant to lipid metabolism mediating the preventive effects of BS15 treatment bases on SNE chickens
| Pathway | P-value | Pathway ID | Genes |
|---|---|---|---|
| PPAR signaling pathway | 0.00107 | gga03320 | ACSBG1; PLIN1; PLIN2 |
| Fatty acid biosynthesis | 0.00443 | gga00061 | ACSBG1 |
| Arachidonic acid metabolism | 0.00472 | gga00590 | PLA2G4A; PTGES2 |
| alpha-Linolenic acid metabolism | 0.01043 | gga00592 | PLA2G4A |
| Linoleic acid metabolism | 0.01368 | gga00591 | PLA2G4A |
| Fatty acid degradation | 0.02567 | gga00071 | ACSBG1 |
| Ether lipid metabolism | 0.03364 | gga00565 | PLA2G4A |
| Fatty acid metabolism | 0.04244 | gga01212 | ACSBG1 |
| Glycerolipid metabolism | 0.06227 | gga00561 | LIPC |
| Adipocytokine signaling pathway | 0.08463 | gga04920 | ACSBG1 |
| Glycerophospholipid metabolism | 0.14045 | gga00564 | PLA2G4A |
We also compared the control group with the BS15 group, and found that there were 15 lipid metabolic pathways enriched (Table 3). According to the literature and the above data analysis, We found phospholipase A2 group IVA (PLA2G4A) was persistently significant differentially expressed in two comparisons. Besides, we also found some other genes differential expression relevant to lipid metabolism in comparison of control versus SNE groups or SNE versus BS15 groups, such as fatty acid binding protein 2 (FABP2), acyl-CoA synthetase bubblegum family member 1 (ACSBG1), perilipin 1 (PLIN1), perilipin 2 (PLIN2), and so on.
Table 3.
Important regulatory genes and pathways related to lipid metabolism in comparison of control group and BS15 group
| Pathway | P-value | Pathway ID | Genes |
|---|---|---|---|
| Steroid biosynthesis | 7.64E-09 | gga00100 | CYP51A1; DHCR7; LIPA; MSMO1; SQLE |
| Fatty acid metabolism | 0.00093 | gga01212 | ACSBG1; HADHA; SCD5 |
| Fatty acid elongation | 0.00113 | gga00062 | ELOVL1; HADHA |
| Biosynthesis of unsaturated fatty acids | 0.00131 | gga01040 | HADHA; SCD5 |
| PPAR signaling pathway | 0.00332 | gga03320 | ACSBG1; FABP2; SCD5 |
| Steroid hormone biosynthesis | 0.00374 | gga00140 | CYP17A1; CYP1B1 |
| Fatty acid degradation | 0.00450 | gga00071 | ACSBG1; HADHA |
| Fatty acid biosynthesis | 0.00801 | gga00061 | ACSBG1 |
| Adipocytokine signaling pathway | 0.02749 | gga04920 | ACSBG1; SOCS3 |
| Butanoate metabolism | 0.02841 | gga00650 | HADHA |
| beta-Alanine metabolism | 0.03058 | gga00410 | HADHA |
| Ether lipid metabolism | 0.05823 | gga00565 | LPCAT1 |
| Glycerophospholipid metabolism | 0.22438 | gga00564 | LPCAT1 |
| Insulin resistance | 0.27196 | gga04931 | SOCS3 |
| Insulin signaling pathway | 0.35875 | gga04910 | SOCS3 |
Validation of next-generation sequencing data using qRT-PCR
To validate the RNA sequencing results, qRT-PCR analysis was used to determine the expression patterns of some target genes that were normalized by the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). These genes consisted of overexpressed and underexpressed genes (FABP2, ACSBG1, PLIN1, PLIN2, PLA2G4A) in SNE or BS15 groups. The results showed in Fig. 4, confirming these genes are crucial in regulation of lipid metabolism in SNE infection chickens.
Fig. 4.
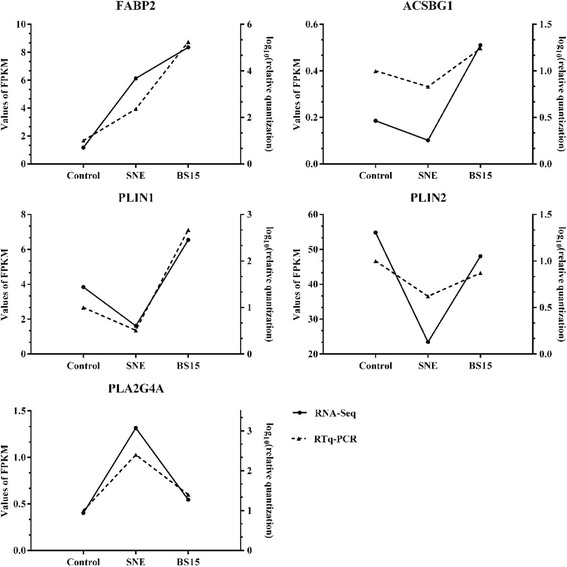
Expression changes of FABP2, ACSBG1, PLIN1, PLIN2 and PLA2G4A. The expression levels of FABP2, ACSBG1, PLIN1, PLIN2 and PLA2G4A during chicken liver from RNA-Seq results are same with the expression trends of these genes from quantitative real-time (qRT)-PCR results. FPKM, fragments per kb per million reads
Discussion
Subclinical necrotic enteritis, a enteric bacterial disease, is known to widely hamper the growth of chicken, contaminate chicken meat, and cause huge economic losses, suggesting that not only the poultry but also the public health is exposed to this harmful subclinical disease. In previous study, we have demonstrated that SNE infection could cause the deregulation of lipid metabolism in chicken liver, and L. johnsonii BS15 pretreatment can alleviate this damage [5]. Then, we hypothesized that BS15 may prevent SNE by ameliorating hepatic lipid metabolism. To test this hypothesis, we used chickens as models to excavate the key functional genes and pathways related to hepatic lipid metabolism of SNE chickens following BS15 supplementation in daily diet. Our results were consistent with this assumption, and more importantly, we found that adding BS15 in the whole experiment could improve the lipid metabolism, mainly by regulation of the peroxisome proliferator-activated receptor (PPAR) signaling pathway and arachidonic acid metabolism.
The rudimentary parameters values of growth status, such as feed conversion rate and the abdominal rate, as well as some concentrations of serum biochemical indicators (alanine aminotransferase, aspartate transaminase, total cholesterol, high-density lipoprotein cholesterol), were significantly increased in the SNE chickens [5], manifesting that the success of SNE experimental model which is indispensable for the follow-up trial. Meanwhile, chicken characteristics have showed that SNE infection could indeed bring adverse effects on the lipid metabolism of chickens. Similar phenomenon was also observed by Wang et al. (2017) [6] that demonstrated subclinical C. perfringens infection elevated fatty acid and lipid production, resulting in lipid accumulation in the muscle. However, there was a detente after BS15 preconditioning, which caught our attention. The liver of chickens is a vital organ where the majority of lipid metabolism process occurs, and considerable evidence support that the interrelationships between liver and some bowel diseases. For example, ascites can lead broiler liver oxidation damage and energy generation obstruction [16]. Also, Coble et al. (2013) [17] found that Salmonella enteritidis infection spawned many pronounced response on inflammation, metabolic pathways, and mitochondria-mediated apoptosis in broiler liver. Based on the above multi-factors, we focused on the lipid metabolism in hepatic transcriptome of SNE chickens prevented by BS15.
Numerous researches showed that the activation of PPAR signaling pathway at the transcriptomic level is closely associated with fatty acids biosynthesis and metabolism [18], and PPAR signaling pathway also touches on ameliorating insulin sensitivity [19], maintaining energy balance [20], regulating the fatty acid oxidation and ketogenesis [21]. In addition, adipocyte differentiation is adjusted by miscellaneous transcription factors, notably PPARs [22], whose expressions are susceptible due to altering different diets in broiler livers [10, 23]. In present study, our data showed that BS15 treatment boosted the gene expression levels of enzymes responsible for oxidation and transportation of fatty acid in the PPAR signaling pathway, including FABP2, ACSBG1, PLIN1 and PLIN2.
FABP2 predominantly expressed in enterocytes belongs to FABPs, which is cytoplasmic proteins touched upon intracellular fatty acid transport and metabolism [24, 25]. Previous studies have identified that FABP2 plays a key role in the absorption and intracellular transport of dietary long-chain fatty acids [26, 27], and the explanation of observed fatty acids alterations in Major Depressive Disorder [28]. Further, FABP2 Ala54Thr polymorphism, a mutant phenotype, is closely related to insulin resistance and abnormal lipid metabolism, which are the main risk factors for cardiovascular disease [29]. These observations were consistent with our findings that the expression of FABP2 was statistically significantly up-regulated (Fold Chang, FC: 6.372) by BS15 compared with the control group, although there was no apparent response in the SNE chicken group. This phenomenon presumably results from the indubitable facilitation of BS15 on hepatic fatty acid transport and metabolism in SNE chickens.
Before participating in most catabolic and anabolic reactions, free fatty acids must be activated to their CoA thioesters [30, 31]. Processes such as incorporation of fatty acids into fatty acid unsaturation or elongation, degradation of fatty acids by oxidation, which all need to activate the fatty acid substrates. Most activations are catalyzed by acyl-CoA synthetases that play a virtual role in fatty acid and lipid metabolism in animals [32, 33]. Further analysis in this study, we found ACSBG1 is another key gene which persistently differentially expressed in three groups. ACSBG1 was proved to be the one has robust acyl-CoA synthetase activity and capable of activating both long- and very long-chain fatty acid substrates [34]. Fruit flies lacking this gene have elevated tissue levels of saturated very long-chain fatty acids [35], more importantly, ACSBG1 characterized by regulation of nerve impulses [36]. In current study, ACSBG1 overexpressed (FC: 2.506) in the BS15 chickens compared the SNE group, involving in PPAR signaling pathway enrichment relevant to lipid metabolism, suggesting the acceleration of fatty acid transport in SNE chickens liver controlled by BS15.
However, as the analysis of transcriptome goes on, we found two other key genes, PLIN1 and PLIN2, which may have an important regulatory effect on PPAR signaling pathway. PLIN1 and PLIN2 are both the members of PAT protein family which is a key protein that regulates the formation of lipid droplet [37, 38]. Previous researches demonstrated that phosphorylated PLIN1 activated lipolytic activity and hydrolysis of triacylglycerides [39, 40]. Relatively lower expression of PLIN2 can promote the reduction of hepatic fibrosis and enhance insulin sensitivity, while simultaneously suppressing PLIN2 and PLIN3 can lead to insulin resistance [41]. Whence the role of the PAT family protein in the disease associated with lipid droplet and dyslipidemia is still not really clearly in the current researches. Our sequencing data showed that the up-regulated of PLIN1 (FC: 0.267) and PLIN2 (FC: 0.493) were observed in BS15 chickens group compared to the SNE group, and there were no significant differences between control and BS15 groups in terms of these two genes comparison. To some extent, these results suggest that L. johnsonii BS15 addition may slow down the fat cracking to recuperate the metabolic rate and body resistance, inasmuch as the growth of SNE chickens was generally poor [5, 6]. However, this feature of BS15 is far from sufficient to cover the characteristic of BS15 facilitating the transfer of fatty acids in statistics.
On the other hand, the membranes of cells and suborganelles are rich in easily oxidized phospholipids that is the initiation site of fatty acid oxidation. Phospholipase A2 (PLA2) with special selectivity of acyl are the key rate-limiting enzymes of phospholipids oxidation [42]. Besides, PLA2 can catalyze the hydrolysis of membrane phospholipid glycerin to produce free fatty acids and lysophospholipid, such as arachidonic acid and linoleic acid [43]. As a n-6 highly unsaturated fatty acid, arachidonic acid is an important inflammatory lipid media and can participate in the systemic stress responses and inflammation [44, 45], activate PPARγ with its metabolites [46], affect the transcription of lipid metabolism related genes and regulate the synthesis and storage of fatty acids [47]. PLA2G4A is the main isotype of the release of arachidonic acid. Our results manifested that the significantly overexpression of PLA2G4A (FC: 2.989) in SNE chickens liver compared to the control group, suggesting that SNE infection accelerated the accumulation of arachidonic acid, which may cause large amount of fatty acid oxidation and the formation of lipid peroxides, aggravating liver cell injury. This situation was consistent with the previous researches that indicated that SNE may induce the inflammation in liver [3]. However, an obviously suppression of PLA2G4A (FC: 2.462) in SNE chickens under the BS15 pretreatment was observed, and there was no significantly differential between BS15 and control groups, implying BS15 supplementation can certainly alleviate inflammation through regulating the arachidonic acid metabolism of SNE chickens liver.
Methods
Insolation and cultivation of strains
The insolation of Lactobacillus johnsonii BS15 (CCTCC M2013663) strain was carried out from homemade yogurt collected from the Hongyuan Prairie, Aba Autonomous Prefecture, China. And the quantities of BS15 were assessed by heterotrophic plate counts after cultivating in MRS liquid medium at 37 °C for 36 h in the anaerobic cabinet. Afterwards, the probiotic cells were collected, washed with saline, and suspended in phosphate buffered saline (PBS, pH 7.0) for further trial use. The premium additive concentration of BS15 in diet was maintained at a level of 1 × 106 colony-forming unit (cfu) per gram to ensure the availability of cells throughout the experimental period.
A Clostridium perfringens (CVCC2030) strain, originated from the intestine of a chicken with severe necrotic enteritis, was obtained from China Veterinary Culture Collection Center and characterized as a NetB toxin positive type A strain. The bacteria were cultivated in cooked meat medium at 37 °C for 24 h under anaerobic environment, and then the strain was aseptically stored in fluid thioglycollate broth overnight at the same environment before undergoing the inoculation of chicken.
Animal maintenance and subclinical necrotic enteritis trials
Cobb 500 male chickens were purchased from the Chia Tai broiler hatchery (Chengdu, China). The protocols for animals studies were reviewed and approved by the Institutional Animal Care and Use Committee of Sichuan Agricultural University. Chickens were maintained, with 24 h a day light, under 24 °C after gradually decreased by 3 °C per a week from 33 °C. Chickens (one-day-old) were randomized into three groups (six replicates per group, ten individuals per replicate) which are as follows: normal control group (NC), SNE group (SNE, SNE experiment model), BS15 preventative group (BS15, L. johnsonii BS15 at a dose of 106 cfu per gram). All groups of chickens were allowed drinking water and feed ad libitum unless otherwise stated. The feed was based on the NRC (1994) and shown in Table 4.
Table 4.
Composition of the basal diets for broilers
| Ingredienta | diet (%) |
|---|---|
| Ground yellow corn | 56.0 |
| Soybean meal | 37.0 |
| Soybean oil | 3.66 |
| Ground limestone | 0.57 |
| Dicalcium phosphate | 1.80 |
| Salt | 0.30 |
| Choline chloride | 0.10 |
| DL-Met | 0.24 |
| Micronutrientsb | 0.33 |
| Calculated nutrients level (%) | |
| ME (MJ kg-1) | 12.39 |
| CP | 21.17 |
| Lys | 1.19 |
| Met | 0.50 |
| Met + Cys | 0.86 |
| Ca | 0.85 |
| Nonphytate P | 0.44 |
aIngredient and nutrient composition are reported on as-fed basis
bMicronutrients are provided per kilogram of diet: vitamin A (all-trans retinol acetate), 12,500 IU; cholecalciferol, 2500 IU; vitamin E (all-rac-a-tocopherol acetate), 18.75 IU; vitamin K (menadione Na bisulfate), 5.0 mg; thiamin (thiamin mononitrate), 2.5 mg; riboflavin, 7.5 mg; vitamin B6, 5.0 mg; vitamin B12, 0.0025 mg; pantothenate, 15 mg; niacin, 50 mg; folic acid, 1.25 mg; biotin, 0.12 mg; Cu (CuSO4·5H2O), 10 mg; Mn (MnSO4·H2O), 100 mg; Zn (ZnSO4·7H2O), 100 mg; Fe (FeSO4·7H2O), 100 mg; I (KI), 0.4 mg; Se (Na2SeO3), 0.2 mg
All chickens were fed on normal diet for first seven days to stabilize their metabolic condition. From day 8 onwards, L. johnsonii BS15 bacteria were mixed in the feed of the BS15 preventative group at a concentration of 106 cfu per gram. The dose of 20,000 Eimeria acervulina oocysts and 5000 Eimeria maxima oocysts (Guangdong Academy of Agricultural Sciences, Guangzhou, China) was given in chickens by gastric infusion as the preliminary infection of SNE on day 15, while the NC group chickens received the same amount of sterile PBS instead. From day 18 to 22, all Eimeria oocysts infected chickens were orally challenged (two times a day) with 1 ml of a fresh medium culture containing 2.2 × 108 cfu C. perfringens per milliliter, meanwhile, the unchallenged chickens received sterile fluid thioglycollate medium instead. On day 28 all chickens were euthanized for tissue collection under the the institutional animal care guidelines.
RNA library construction and illumina sequencing.
Total RNA was extracted from chicken livers using RNAiso Plus reagent (TaKaRa, Dalian, China). Fifty nanogram of RNA samples with an RNA integrity number (RIN) greater than seven were used for library construction (Agilent, CA, USA). Nine libraries (three each for the control, SNE treatment and BS15 preventative groups) were established using the TruSeq Stranded mRNA LT Sample Prep Kit in the light of the specifications (Illumina, San Diego, USA). And then paired-end reads sequencing was performed on the HiSeq X Ten system (Illumina, Inc., USA) by Shanghai OE Biotech. Co., Ltd. (Shanghai, China). The obtained results were compared with the database and annotations of every gene for subsequent experimental analysis.
Quantitative real time polymerase chain reaction (qRT-PCR) verification.
The total extracted RNA of chicken livers was then reverse transcribed into cDNA immediately using a Prime ScriptTM RT reagent kit (TaKaRa, Dalian, China) to serve as a template for qRT-PCR verification. qRT-PCR was implemented to determine the expression of differentially target genes according to the the fluorescence quantitative PCR kit instructions, using a CFX96 Real-Time system (Bio-Rad, Hercules, CA, USA) with SYBR® Premix Ex Taq™ II (TaKaRa, Dalian, China). The primer sequences for qRT-PCR verification are listed in Table 5. Fold change relative to normal control group was evaluated by using the 2-ΔΔCt method in Microsoft Excel software.
Table 5.
Primer sequences used in quantitative qRT-PCR analysis
| Gene name | Primer sequence (5 → 3) | Size (bp) |
|---|---|---|
| FABP2 | F: ATACAGGTGAGTTGAACAGTCGCTT R: TGAAGATAAGTGAGGCTGATTGGT |
127 |
| ACSBG1 | F: CGAATCAGTGCTGTGTGCTT R: GGCTGAGCGGAAGATAACTG |
171 |
| PLIN1 | F: TGCTGCTTGTTGAAGAACCACT R:AGGCATTCTGTGATGATTATGTGGT |
109 |
| PLIN2 | F: GTTGCCAATGCTAAGGGTGT R: ACCACACGACTTCCCAAGAC |
194 |
| PLA2G4A | F: ACTTGACCACTTCCCGTGAC R: GGGTTGTGACTGACCGAGTT |
250 |
| GADPH | F:GGTGAAAGTCGGAGTCAACGG R:CGATGAAGGGATCATTGATGGC |
108 |
Statistical analysis
The differentially expressed genes between different samples were compared by the method of reads per kb fragments per million reads (FRKM). False Discovery Rate (FDR) was set at 0.0001 to determine the threshold of the p-value in multiple tests. An absolute log2 fold-change between conditions ≥0.5 was used as the threshold for significance of the gene expression difference. qRT-PCR data were analyzed by one-way analysis of variance (ANOVA), and multiple comparisons were tested using Duncan’s multiple-range test. A p-value of < 0.05 was considered statistically significant differences, and all statistical analyses were performed using SigmaPlot for Social Sciences version 13.
Conclusion
In conclusion, the data presented here highlights the preventative mechanism of Lactobacillus johnsonii BS15 that may contribute to lipid metabolism, mainly by regulation of the PPAR signaling pathway and the arachidonic acid metabolism. And this lipid-targeting BS15 precaution may be a good match for the multi-hit driven SNE pathogenesis. We hope that this study lays the foundation for the innovation of natural products for treating SNE.
Data availability
The raw counts data obtained in this study were deposited in the NCBI Sequence Read Archive (SRA) under the accession number SRP118769.
Additional files
Table S1. Details for regulatory genes and pathways related to lipid metabolism in comparison of control group and SNE group. (XLSX 120 kb)
Table S2. Details for genes and pathways relevant to lipid metabolism mediating the preventive effects of BS15 treatment bases on SNE chickens. (XLSX 13 kb)
Table S3. Details for regulatory genes and pathways related to lipid metabolism in comparison of control group and BS15 group. (XLSX 42 kb)
Acknowledgments
Funding
This study was supported by Chengdu Agricultural Technology Research and Development Project (2015-NY02–00295-NC) and International Cooperation Project of Sichuan Science and Technology Department (2018HH0103).
Availability of data and materials
All data generated or analysed during this study are included in the Additional files of this article.
Abbreviations
- ACSBG1
acyl-CoA synthetase bubblegum family member 1
- FABP2
fatty acid binding protein 2
- FC
Fold Chang
- FDR
False Discovery Rate
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- KEGG
kyoto encyclopedia of genes and genomes
- PLA2G4A
phospholipase A2 group IVA
- PLIN1
perilipin 1
- PLIN2
perilipin 2
- PPAR
proliferator-activated receptor
- qRT-PCR
Quantitative real-time polymerase chain reaction
- RIN
RNA integrity number
- SNE
Subclinical necrotic enteritis
Authors’ contributions
XQ, XN and DZ designed the experiments. HW, LL, JL, KP and BJ performed the experiments. HW, LL, and AK analyzed the experiment data. XQ and HW wrote this paper. All authors read and approved the final manuscript.
Ethics approval
All animal experiments were performed according to the guidelines for the care and use of laboratory animals approved by Institutional Animal Care and Use Committee of the Sichuan Agricultural University.
Competing interests
The authors declare that they have no competing interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12944-018-0741-5) contains supplementary material, which is available to authorized users.
Contributor Information
Xiaodan Qing, Email: XiaoDQing@hotmail.com.
Dong Zeng, Email: zend@sicau.edu.cn.
Hesong Wang, Email: 236432291@qq.com.
Xueqin Ni, Email: xueqinni@foxmail.com.
Jing Lai, Email: 2358864988@qq.com.
Lei Liu, Email: 1558034850@qq.com.
Abdul Khalique, Email: abdulkhalique36@gmail.com.
Kangcheng Pan, Email: pankangcheng71@126.com.
Bo Jing, Email: 2909374240@qq.com.
References
- 1.Skinner J, Bauer S, Young V, Pauling G, Wilson J. An economic analysis of the impact of subclinical (mild) necrotic enteritis in broiler chickens. Avian Dis. 2010;54:1237–1240. doi: 10.1637/9399-052110-Reg.1. [DOI] [PubMed] [Google Scholar]
- 2.Nowell V, Poppe C, Parreira V, Jiang Y, Reid-Smith R, Prescott J. Clostridium perfringens in retail chicken. Anaerobe. 2010;16:314–315. doi: 10.1016/j.anaerobe.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Cooper K, Bueschel D, Songer J. Presence of Clostridium perfringens in retail chicken livers. Anaerobe. 2013;21:67–68. doi: 10.1016/j.anaerobe.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 4.Tsiouris V. Poultry management: a useful tool for the control of necrotic enteritis in poultry. Avian Pathol. 2016;45:323–325. doi: 10.1080/03079457.2016.1154502. [DOI] [PubMed] [Google Scholar]
- 5.Qing X, Zeng D, Wang H, Ni X, Liu L, Lai J, Khalique A, Pan K, Jing B. Preventing subclinical necrotic enteritis through lactobacillus johnsonii BS15 by ameliorating lipid metabolism and intestinal microflora in broiler chickens. AMB Express. 2017;7:139. doi: 10.1186/s13568-017-0439-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, Ni X, Liu L, Zeng D, Lai J, Qing X, Li G, Pan K, Jing B. Controlling of growth performance, lipid deposits and fatty acid composition of chicken meat through a probiotic, Lactobacillus johnsonii during subclinical Clostridium perfringens infection. Lipids Health Dis. 2017;16:38. doi: 10.1186/s12944-017-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fallah R, Kiani A, Azarfar A. A review of the role of five kinds of alternatives to infeed antibiotics in broiler production. J Vet Med Anim Health. 2013;5:317–321. [Google Scholar]
- 8.Immerseel F, Rood J, Moore R, Titball R. Rethinking our understanding of the pathogenesis of necrotic enteritis in chickens. Trends Microbiol. 2009;17:32–36. doi: 10.1016/j.tim.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Peng Q, Zeng X, Zhu J, Wang S, Liu X, Hou C, Thacker P, Qiao S. Effects of dietary lactobacillus plantarum B1 on growth performance, intestinal microbiota, and short chain fatty acid profiles in broiler chickens. Poult Sci. 2016;95:893–900. doi: 10.3382/ps/pev435. [DOI] [PubMed] [Google Scholar]
- 10.Zhou M, Zeng D, Ni X, Tu T, Yin Z, Pan K, Jing B. Effects of Bacillus licheniformis on the growth performance and expression of lipid metabolism-related genes in broiler chickens challenged with Clostridium perfringens -induced necrotic enteritis. Lipids Health Dis. 2016;15:38. doi: 10.1186/s12944-016-0204-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venessa E, Wang J, Alexander V, Freddy H, Marie J, Gwen F, Jeroen R, Richard D, Filip V. The ProbioticButyricicoccus pullicaecorumReduces feed conversion and protects from potentially harmful intestinal microorganisms and necrotic enteritis in broilers. Front Microbiol. 2016;7:1416. doi: 10.3389/fmicb.2016.01416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin Y, Xu S, Zeng D, Ni X, Zhou M, Zeng Y, Wang H, Zhou Y, Zhu H, Pan K, Li G. Disruption in the cecal microbiota of chickens challenged with Clostridium perfringens and other factors was alleviated by Bacillus licheniformis supplementation. PLoS One. 2017;12:e0182426. doi: 10.1371/journal.pone.0182426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xin J, Zeng D, Wang H, Ni X, Yi D, Pan K, Jing B. Preventing non-alcoholic fatty liver disease through lactobacillus johnsonii BS15 by attenuating inflammation and mitochondrial injury and improving gut environment in obese mice. Appl Microbiol Biotechnol. 2014;98:6817–6829. doi: 10.1007/s00253-014-5752-1. [DOI] [PubMed] [Google Scholar]
- 14.Liu L, Ni X, Zeng D, Wang H, Jing B, Yin Z, Pan K. Effect of a dietary probiotic, lactobacillus johnsonii BS15, on growth performance, quality traits, antioxidant ability, and nutritional and flavour substances of chicken meat. Anim Prod Sci. 2016;57:920–926. doi: 10.1071/AN15344. [DOI] [Google Scholar]
- 15.Wang H, Ni X, Qing X, Zeng D, Luo M, Liu L, Li G, Pan K, Jing B. Live probiotic lactobacillus johnsonii BS15 promotes growth performance and lowers fat deposition by improving lipid metabolism, intestinal development, and gut microflora in broilers. Front Microbiol. 2017;8:1073. doi: 10.3389/fmicb.2017.01073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Guo Y, Ning D, Peng Y, Yang Y, Liu D. Analysis of liver transcriptome in broilers with ascites and regulation by L-carnitine. J Poult Sci. 2013;50:126–137. doi: 10.2141/jpsa.0120124. [DOI] [Google Scholar]
- 17.Coble D, Sandford E, Ji T, Abernathy J, Fleming D, Zhao H, Lamont S. Impacts of Salmonella enteritidis infection on liver transcriptome in broilers. Genesis. 2013;51:357–364. doi: 10.1002/dvg.22351. [DOI] [PubMed] [Google Scholar]
- 18.He K, Wang Q, Wang Z, Pan Y. Association study between gene polymorphisms in PPAR signaling pathway and porcine meat quality traits. Mamm Genome. 2013;24:322–331. doi: 10.1007/s00335-013-9460-4. [DOI] [PubMed] [Google Scholar]
- 19.Zhou M, Yu P, Sun Q, Li Y. Expression profiling analysis: uncoupling protein 2 deficiency improves hepatic glucose, lipid profiles and insulin sensitivity in high-fat diet-fed mice by modulating expression of genes in peroxisome proliferator-activated receptor signaling pathway. J Diabetes Invest. 2016;7:179–189. doi: 10.1111/jdi.12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupont J, Reverchon M, Cloix L, Froment P, Ramé C. Involvement of adipokines, AMPK, PI3K and the PPAR signaling pathways in ovarian follicle development and cancer. Int J Dev Biol. 2012;56:959–967. doi: 10.1387/ijdb.120134jd. [DOI] [PubMed] [Google Scholar]
- 21.Nakagawa Y, Satoh A, Tezuka H, Han S, Takei K, Iwasaki H, Yatoh S, Yahagi N, Suzuki H, Iwasaki Y, Sone H, Matsuzaka T, Yamada N, Shimano H. CREB3L3 controls fatty acid oxidation and ketogenesis in synergy with PPARα. Sci Rep. 2016;6:39182. doi: 10.1038/srep39182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lefterova M, Haakonsson A, Lazar M, Mandrup S. PPARγ and the global map of adipogenesis and beyond. Trends Endocrinol Metab. 2014;25:293–302. doi: 10.1016/j.tem.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goh Y, Sazili A, Othman F, Royan M, Navishad B. Effects conjugated linoleic acid, fish oil and soybean oil on PPARs (0709 and 0701) mRNA expression in broiler chickens and its relation to the body fat deposits. Int J Mol Sci. 2011;12:8581–8595. doi: 10.3390/ijms12128581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stamey J, Robert M, Benjamin A, Matthew W, Christina S, Mark D. Systemic and Intracellular Trafficking of Long-chain Fatty Acids in Lactating Dairy Cattle. Virginia Tech; 2012. http://hdl.handle.net/10919/38689.
- 25.Alharbi K, Bazzi M, Al-daghri N, Khan I, Al-nbaheen M, Alharbi F, Al-sheik Y, Syed R, Atieh M, Ghouse M, Ansari A. Codon 54 in the fatty acid–binding Protein-2 (A54T) polymorphism studies in type 2 diabetes mellitus in Saudi population. Jokull. 2013;63(5). ISSN:0449-0576. https://s3.amazonaws.com/academia.edu.documents/43767542/Codon_54_in_the_Fatty_AcidBinding_Protei20160315-20927-z2b7a3.pdf?AWSAccessKeyId=AKIAIWOWYYGZ2Y53UL3A&Expires=1524104080&Signature=UO%2By2eUw0qw1WBktxUu5Y2gp9L4%3D&response-content-disposition=inline%3B%20filename%3DCodon_54_in_the_Fatty_Acid_binding_Prote.pdf.
- 26.Alharbi K, Khan I, Bazzi M, Al-daghri N, Hasan T, Alnbaheen M, Alharbi Y, Syed R, Aboul-soud M. A54T polymorphism in the fatty acid binding protein 2 studies in a Saudi population with type 2 diabetes mellitus. Lipids Health Dis. 2014;13:61. doi: 10.1186/1476-511X-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klapper M, Böhme M, Nitz I, Döring F. Transcriptional regulation of the fatty acid binding protein 2 (FABP2) gene by the hepatic nuclear factor 1 alpha (HNF-1α) Gene. 2008;416:48–52. doi: 10.1016/j.gene.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 28.Mocking R, Lok A, Assies J, Koeter M, Visser I, Ruhé H, Bockting C, Schene A. Ala54Thr fatty acid-binding protein 2 (FABP2) polymorphism in recurrent depression: associations with fatty acid concentrations and waist circumference. PLoS One. 2013;8:e82980. doi: 10.1371/journal.pone.0082980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Luis D, Sagrado M, Aller R, Izaola O, Conde R, Romero E. Ala54Thr polymorphism of fatty acid binding protein 2, role on insulin resistance and cardiovascular risk factors in presurgical morbid obesity patients. Obes Surg. 2009;19:1691–1696. doi: 10.1007/s11695-009-9859-x. [DOI] [PubMed] [Google Scholar]
- 30.Sun P, Xia S, Lal B, Shi X, Yang K, Watkins P, Laterra J. Lipid metabolism enzyme ACSVL3 supports glioblastoma stem cell maintenance and tumorigenicity. BMC Cancer. 2014;14:1–11. doi: 10.1186/1471-2407-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bu S, Mashek D. Hepatic long-chain acyl-CoA synthetase 5 mediates fatty acid channeling between anabolic and catabolic pathways. J Lipid Res. 2010;51:3270–3280. doi: 10.1194/jlr.M009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyares R. Fatty acid metabolism in the zebrafish: the role of acyl-CoA synthetases during embryonic development. Dissertations and Theses - Gradworks. 2012. [Google Scholar]
- 33.Dong B, Kan C, Singh A, Liu J. High-fructose diet downregulates long-chain acyl-CoA synthetase 3 expression in liver of hamsters via impairing LXR/RXR signaling pathway. J Lipid Res. 2013;54:1241–1254. doi: 10.1194/jlr.M032599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pei Z, Oey N, Zuidervaart M, Jia Z, Li Y, Steinberg S, Smith K, Watkins P. The acyl-CoA synthetase "bubblegum" (lipidosin): further characterization and role in neuronal fatty acid beta-oxidation. J Biol Chem. 2003;278:47070. doi: 10.1074/jbc.M310075200. [DOI] [PubMed] [Google Scholar]
- 35.Pei Z, Jia Z, Watkins P. The second member of the human and murine bubblegum family is a testis- and brainstem-specific acyl-CoA synthetase. J Biol Chem. 2006;281:6632–6641. doi: 10.1074/jbc.M511558200. [DOI] [PubMed] [Google Scholar]
- 36.Hess J, Kawaguchi D, Wagner K, Faraone S, Glatt S. The influence of genes on “positive valence systems” constructs: a systematic review. Am J Med Genet B. 2016;171:92–110. doi: 10.1002/ajmg.b.32382. [DOI] [PubMed] [Google Scholar]
- 37.Lecchi C, Invernizzi G, Agazzi A, Modina S, Sartorelli P, Savoini G, Ceciliani F. Effects of EPA and DHA on lipid droplet accumulation and mRNA abundance of PAT proteins in caprine monocytes. Res Vet Sci. 2013;94:246–251. doi: 10.1016/j.rvsc.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 38.Tsai T, Chen E, Li L, Saha P, Lee H, Huang L, Shelness G, Chan L, Chang B. The constitutive lipid droplet protein PLIN2 regulates autophagy in liver. Autophagy. 2017;13:1130–1144. doi: 10.1080/15548627.2017.1319544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhai W, Xu C, Ling Y, Liu S, Deng J, Qi Y, Londos C, Xu G. Increased lipolysis in adipose tissues is associated with elevation of systemic free fatty acids and insulin resistance in perilipin null mice. Horm Metab Res. 2010;42:247–253. doi: 10.1055/s-0029-1243599. [DOI] [PubMed] [Google Scholar]
- 40.Dalen K, Londos C, Kimmel A. Adipocyte fat mobilization: the regulatory roles of Perilipin 1, adipose triglyceride lipase, and hormone-sensitive lipase. Encyclopedia Biol Chem. 2013:47–51.
- 41.Greenberg A, Coleman R, Kraemer F, McManaman J, Obin M, Puri V, Yan Q, Miyoshi H, Mashek D. The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest. 2011;121:2102–2110. doi: 10.1172/JCI46069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schröter J, Süß R, Schiller J. MALDI-TOF MS to monitor the kinetics of phospholipase a 2 -digestion of oxidized phospholipids. Methods. 2015;104:41–47. doi: 10.1016/j.ymeth.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 43.Ee S, Lo Y, Shui G, Wenk M, Shin E, Kim H, Ong W. Distribution of secretory phospholipase A2 XIIA in the brain and its role in lipid metabolism and cognition. Mol Neurobiol. 2014;50:60–75. doi: 10.1007/s12035-014-8635-7. [DOI] [PubMed] [Google Scholar]
- 44.Carrier J, Watanabe W, Harel M, Rezek T, Seaton P, Shafer T. Effects of dietary arachidonic acid on larval performance, fatty acid profiles, stress resistance, and expression of Na +/K + ATPase mRNA in black sea bass Centropristis striata. Aquaculture. 2011;319:111–121. doi: 10.1016/j.aquaculture.2011.06.027. [DOI] [Google Scholar]
- 45.Furne M, Holen E, Araujo P, Lie K, Moren M. Cytokine gene expression and prostaglandin production in head kidney leukocytes isolated from Atlantic cod (Gadus morhua) added different levels of arachidonic acid and eicosapentaenoic acid. Fish Shellfish Immunol. 2013;34:770–777. doi: 10.1016/j.fsi.2012.11.044. [DOI] [PubMed] [Google Scholar]
- 46.Sun L, Xu Y, Han J, Liang H, Wang N, Cheng Y. 12/15-lipoxygenase metabolites of arachidonic acid activate PPARγ: a possible neuroprotective effect in ischemic brain. J Lipid Res. 2015;56:502–514. doi: 10.1194/jlr.M053058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian J, Ji H, Oku H, Zhou J. Effects of dietary arachidonic acid (ARA) on lipid metabolism and health status of juvenile grass carp. Ctenopharyngodon idellus Aquaculture. 2014;430:57–65. doi: 10.1016/j.aquaculture.2014.03.020. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Details for regulatory genes and pathways related to lipid metabolism in comparison of control group and SNE group. (XLSX 120 kb)
Table S2. Details for genes and pathways relevant to lipid metabolism mediating the preventive effects of BS15 treatment bases on SNE chickens. (XLSX 13 kb)
Table S3. Details for regulatory genes and pathways related to lipid metabolism in comparison of control group and BS15 group. (XLSX 42 kb)
Data Availability Statement
The raw counts data obtained in this study were deposited in the NCBI Sequence Read Archive (SRA) under the accession number SRP118769.
All data generated or analysed during this study are included in the Additional files of this article.


