Abstract
An extracellular lipase of a newly isolated S. aureus strain ALA1 (SAL4) was purified from the optimized culture medium. The SAL4 specific activity determined at 60 °C and pH 12 by using olive oil emulsion or TC4, reached 7215 U/mg and 2484 U/mg, respectively. The 38 NH2-terminal amino acid sequence of the purified enzyme starting with two extra amino acid residues (LK) was similar to known staphylococcal lipase sequences. This novel lipase maintained almost 100% and 75% of its full activity in a pH range of 4.0–12 after a 24 h incubation or after 0.5 h treatment at 70 °C, respectively. Interestingly, SAL4 displayed appreciable stability toward oxidizing agents, anionic and non-ionic surfactants in addition to its compatibility with several commercial detergents. Overall, these interesting characteristics make this new lipase promising for its application in detergent industry.
Abbreviations: HPLC, high-performance liquid chromatography; NaDC, sodium deoxycholic acid; NaTDC, sodium taurodeoxy cholic acid; OD, optical density; PCR, polymerase chain reaction; rDNA, ribosomal deoxy ribo nucleic acid; rpm, revolutions per minute; SHyL, Staphylococcus hyicus lipase; SAL, Staphylococcus aureus lipase; SEL, Staphylococcus epidermidis lipase; SL1, Staphylococcus sp. lipase; SSL, Staphylococcus simulans lipase; SXL, Staphylococcus xylosus lipase; S. aureus, Staphylococcus aureus; SDS, sodium dodecyl sulfate; TFA, tri fluoroacetic acid; TC3, tripropionin; TC4, tributryin; TC8, trioctanoin; TC18, triolein
Keywords: Staphylococcus aureus lipase, Purification, Characterization, Thermo-alkaline, Detergent-stable
1. Introduction
Lipases are water-soluble enzymes regulating the hydrolysis of ester bonds in insoluble acylglycerols at lipid-water interface (Jaeger et al., 1999). They are serine hydrolases composed of Ser-Asp/Glu-His. Besides, a consensus sequence (Gly-x-Ser-x-Gly) that is usually found encircling serine, the active site (Pleiss et al., 2000). Recently, new lipolytic enzymes have shown to have a different pattern (Gly-Asp-Ser-Leu) (Akoh et al., 2004). Lipases three-dimensional (3-D) structures reveal the α/β-hydrolase fold (Nardini and Dijkstra, 1999). Indeed, the active site in most lipases cannot be reached due to the coverage by surface loops or helical structures. The structures of lipases binding substrate and those inhibited by transition-state analogs (Egloff et al., 1995, Satsuki and Watanabe, 1990) revealed that when the active site interacts with micelles or substrate molecules, it becomes exposed to substrate. These findings have given a structural basis of the interfacial activation compared to nearly forty years ago (Rosenstein and Gotz, 2000, Brzozowsky et al., 1991). In addition to their carboxylic ester bond hydrolysis, lipases also catalyze the reverse esterification, amidation or transesterification processes in aqueous and anhydrous organic solvents (Bornscheuer and Kazlauskas, 1999).
Microbial Lipases are lately of great commercial importance thanks to their high versatility and high stability as well as the advantage of being readily produced in high yields (Hasan et al., 2009). In fact, there has been now more interest in studying enzymes from extremophiles, not only due to their thermo stability but also to their high resistance to chemical agents and extreme pH values besides their mesophilic homologs (Sharma et al., 2001, Illanes, 1999). Some lipases from fungi and bacteria were exploited as cheap and versatile catalysts in various industries, such as food, dairy, detergent and pharmaceutics, along with the degradation of fatty wastes and biodiesel production (Gupta et al., 2002, Laemmli, 1970).
In 1994, the recombinant lipase “Lipolase”, isolated from the fungus Thermomyces lanuginosus and expressed in Aspergillus oryzae was first introduced by NovoNordisk. Then In 1995, two bacterial lipases ‘Lumafast’ from Pseudomonas mendocina and ‘Lipomax’ from Pseudomonas alcaligenes correspondingly were introduced by Genencor International (Jaeger and Reetz, 1998). Detergent lipases also were originated from Candida (Novak et al., 1990) and Chromobacterium (Nawani et al., 1998). As for Laundering, that is generally carried out in alkaline media, microbial lipases were found to be active too (Satsuki and Watanabe, 1990; Handelsman and Shoham, 1994).
An increased interest of recently isolated staphylococcal lipases results from their potential in modern biotechnology. The immobilized lipases are used in non aqueous media as biocatalyst to catalyze the alcoholysis, the transesterification, and the esterification of the alcohols with organic acids in various industries (Horchani et al., 2012).
However, thermostable and alkaline tolerant lipases extracted from Staphylococcus sp. have been rarely described in the literature (Cherif et al., 2011, Chauhan et al., 2013). Furthermore, their practical application is somehow limited due to their relatively lower stabilities and catalytic activities under high temperature and extreme pH values particularly. Since each application has specific biocatalytic properties related to specificity, stability, optimal temperature, and pH-dependence, there is still an interest in researching new lipases with novel applications.
In our previous study, we have screened, isolated and optimized the production of extracellular lipase by a new Staphylococcus aureus (S. aureus) strain ALA1 (GenBank no. KF 678862) from dromedary milk (Ben Bacha et al., 2015). Here, we reported the purification and the characterization of a thermoactive, alkaline and detergent-stable lipase (SAL4), as well as its stability in the presence of several commercial solid and liquid detergents, oxidizing agents and surfactants.
2. Materials and methods
2.1. Production and purification of SAL4
S. aureus strain ALA1 lipase was produced as previously reported by Ben Bacha et al. (2015). Cells were discarded after 30 h of culture by centrifugation (30 min, 12,000 rpm) and the resulting crude enzyme solution (250 mL) was precipitated with solid ammonium sulfate (65% saturation) at 4 °C. The precipitate obtained after centrifugation was then resuspended in 25 mM Tris–HCl, pH 8 containing 50 mM NaCl and 2 mM benzamidine (buffer A) and treated for 15 min at 70 °C. After centrifugation for 30 min at 12,000 rpm, the treated supernatant was (7.5 mL, 22040U) dialyzed overnight at 4 °C against buffer A.
The obtained sample was then loaded on a C18 HPLC column pre-equilibrated with 0.1% TFA. It was then eluted with a pure acetonitrile linear gradient 0–80% at a flow rate of 1 mL/min. The lipase activity was checked as previously described and the elution profile of proteins was monitored at 280 nm. After 15%-SDS–PAGE analysis, pure and active fractions were stored at −20 °C until used for more biochemical characterization.
2.2. Determination of lipase activity
Lipolytic activity was tested titrimetrically with a pH-stat using tributyrin (TC4) or olive oil emulsion at 60 °C and pH 12 (Rathelot et al., 1981). Enzyme activity was also measured using tripropionin (TC3) as substrate to investigate the interfacial activation of the pure SAL4 while trioctanoin (TC8) and triolein (C18) were used for its kinetic studies (Abdelkafi et al., 2009). Lipase activity was expressed in international units (U) where 1 U is 1 μmol of fatty acid produced/minute.
2.3. Protein analysis
Protein content was determined following the Bradford protocol (Bradford, 1976). The purified lipase was analyzed electrophoretically by SDS–PAGE (15%) according to the of Laemmli method (Laemmli, 1970). The N-terminal amino acid sequence was identified by automated Edman’s degradation (Hewick et al., 1981).
2.4. pH and temperature effects on SAL4 stability and activity
Lipase activity was measured at 60 °C and at different pH (8–13) using appropriate buffers. The pH stability of the enzyme was studied by pre-incubating the SAL4 at various pH values ranging from 2 to 13 at room temperature for one day using various buffer solutions [200 mM glycine-NaOH buffer (pH 10–13); 200 mM tris–HCl buffer (pH 8–9); 200 mM potassium phosphate buffer (pH 6–7); 200 mM sodium acetate buffer (pH 2–5)]. After centrifugation, the residual lipase activity was measured in triplicate at different time intervals.
Furthermore, the enzyme assay was carried out at different temperatures (25–65 °C) and at pH 12 in order to determine the optimal temperature of the SAL4 activity. The thermal stability of the pure lipase was also investigated by pre-incubating the enzyme for 30 min at various temperatures ranging from 30 to 75 °C and measuring the residual activity with time.
2.5. Effect of surfactants, oxidizing agents and commercial detergents on lipase stability
The SAL4 sample (120 U from the heat treated fraction) was incubated in the presence of surfactants (Triton X-100, Tween-80, Tween-20, SDS, NaDC or sodium taurocholate (NaTDC)), and oxidants (hydrogen peroxide, sodium perborate or sodium hypochlorite) at 1% for 60 min at 40 °C.
The compatibility of the SAL4 preparation with different kinds of commercial solid (Dixan (Henkel, Spain), Nadhif (Henkel-Alki, Tunisia), Ariel (Procter and Gamble, Switzerland) and Tide (Procter & Gamble, Saudi Arabia)) and liquid laundry detergents (Dixan (Henkel, Spain), Nadhif (Henkel-Alki, Tunisia), DAC (Henkel, Saudi Arabia), and Fairy (Modern industries, Saudi Arabia)) was also studied as described by Cherif et al. (2011) and compared to the commercial lipolase (Novo Nordisk, Denmark). All the experiments of characterization study were performed three times.
3. Results
3.1. Production and purification of lipase
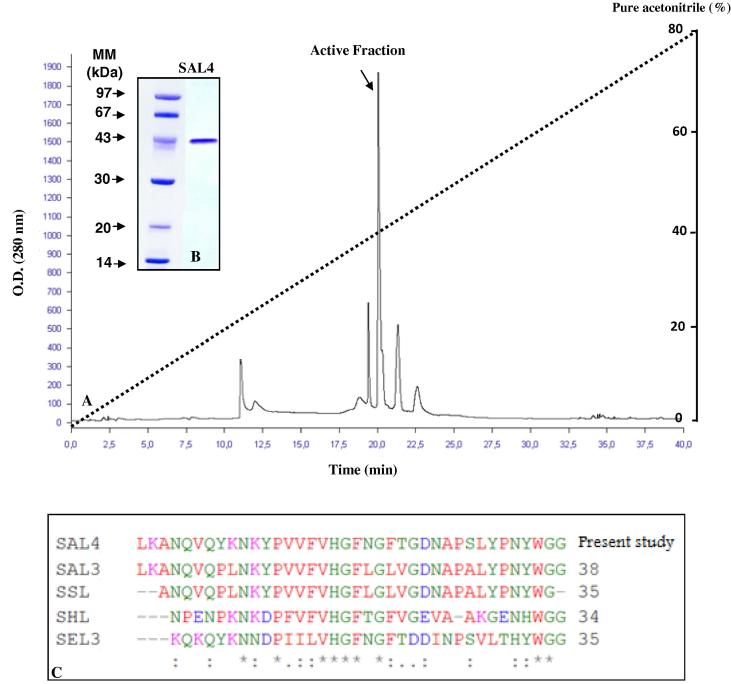
Production and purification protocol of SAL4 was detailed in Section 2. Fig. 1A showed the protein elution profile recorded at the HPLC step of the enzyme purification. The active fractions of the peak that emerged at 40% acetonitrile were pooled and analyzed on SDS–PAGE (Fig. 1). Thus, the obtained data clearly showed that lipase was purified to homogeneity with an apparent molecular mass (MM) of about 43 kDa.
Figure 1.
Chromatography on RP-HPLC column and SDS–PAGE (15%) of SAL4. (A) RP-HPLC on a C-18 column, elution was performed at room temperature within 40 min using a gradient from 0 to 80% acetonitrile at a flow rate of 1 mL/min. The gradient is indicated by the dotted line. The absorbance was measured at 280 nm. AU: Arbitrary Units. (B) 15%-SDS–PAGE of pure SAL4. Lane 1, molecular mass markers (Pharmacia); lane 2, 7 μg of purified SAL4 eluted from RP-HPLC. (C) Alignment of the amino acid sequences of the mature forms of SAL4 (Present study, KF67886), SAL3 (Horchani et al., 2009), SSL (Sayari et al., 2001), SEL3 (Rosenstein et al., 2000) and SHL (Tiesinga et al., 2007).
As shown in Table 1, a purification factor of 81.45-fold was reached at the last purification step with an appreciable recovery yield of 51.75% of the starting lipase activity and a specific activity of the purified enzyme reaching 7215 U/mg using olive oil as substrate. Moreover, the N-terminal amino acid sequencing of SAL4 definitely allowed the identification of 38 amino acid residues of the purified lipase (Fig. 1C).
Table 1.
Flow sheet of the SAL4 purification.
| Purification step | Totala activity (units) | Proteinb (mg) | Specific activity (U/mg) | Activity recovery (%) | Purification factor |
|---|---|---|---|---|---|
| Extraction | 32,000 | 362 | 88.4 | 100 | 1 |
| (NH4)2SO4 Precipitation (25–65%) | 25,600 | 115 | 222.6 | 80 | 2.5 |
| Heat treatment (70 °C) | 22,040 | 18.5 | 1191.3 | 68.9 | 13.5 |
| RP-HPLC (C-18) | 16,594.5 | 2.3 | 7215 | 51.7 | 81.4 |
1 Unit: μmole of fatty acid released per min using olive oil emulsion as a substrate in the presence of 1 mM NaDC and 2 mM CaCl2.
Proteins were estimated using the Bradford method (Bradford, 1976). The experiments were conducted three times.
3.2. Specificity and kinetic studies of SAL4
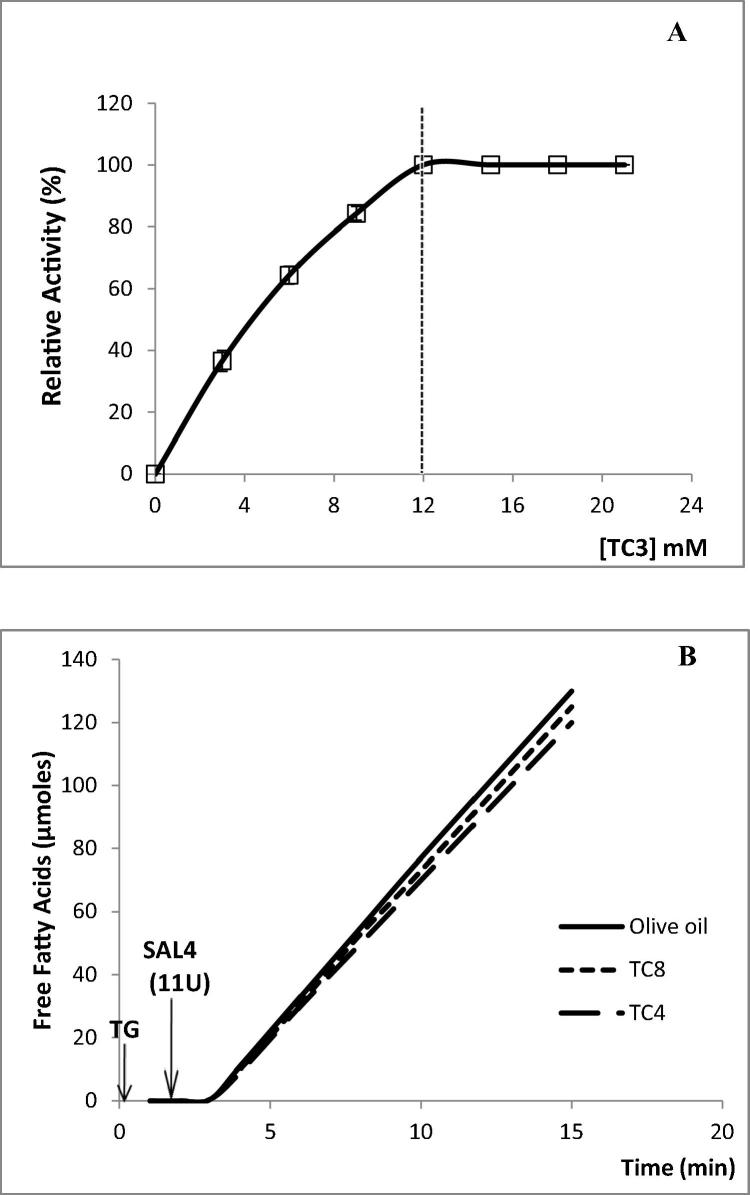
The interfacial activation phenomenon of SAL4 was investigated by studying the hydrolysis rate of TC3 emulsion by SAL4 as a function of substrate concentration. A normal Michaelis–Menten dependence of the activity on the substrate concentration was recorded (Fig. 2A).
Figure 2.
(A) Hydrolysis rate of TC3 by SAL4 as function of substrate concentration. The release of propionic acid was recorded continuously at pH 7 and 37 °C using a pH-stat. The CMC of TC3 (12 mM) is indicated by vertical dotted lines. (B) Kinetics of hydrolysis of TC4, TC8, or TC18 emulsions by SAL4 (11U). Lipase activity was followed at pH 12 and 60 °C. Each data point represents an average of at least two independent experiments, each in triplicate.
The lipase activities of SAL4 were also tested on TC4, TC8 and olive oil emulsion. (Fig. 2B and Table 2). The representative curve of the hydrolysis rate of all tested triacylglycerols substrates remained linear more than 15 min (Fig. 2B). A specific activity of 7215, 4480.3 and 2484.5 U/mg was measured on TC18, TC8 and TC4 as substrate, respectively, under optimal assay conditions (In the presence of 1 mM NaDC and 2 mM CaCl2, pH 12, 60 °C) (Table 2).
Table 2.
Chain length selectivity and kinetic parameters of SAL4. Enzyme activity was determined at pH 12 and 60 °C.
| Substrate | Km app. (mM) | Vmax (μmol/min/mg) | kcat (s−1) | Kcat/Km app. (s−1 mM−1) |
|---|---|---|---|---|
| TC4 | 18.9 | 2484.5 | 1781 | 94.3 |
| TC8 | 10.3 | 4480.3 | 5011.5 | 486.3 |
| TC18 | 3.9 | 7215 | 8070.5 | 2069.5 |
The kinetic constants: apparent Michaelis constant (Km app.), the catalytic constant (kcat) and the deduced catalytic efficiency (Kcat/km app.) of the SAL4 were determined and compared by means of Lineweaver–Burk plots using pure TC4, TC8 orTC18 as substrate in the presence of 1 mM NaDC and 2 mM CaCl2 (pH 12, 60 °C) (Table 2).
3.3. Temperature and pH effects on SAL4 activity and stability
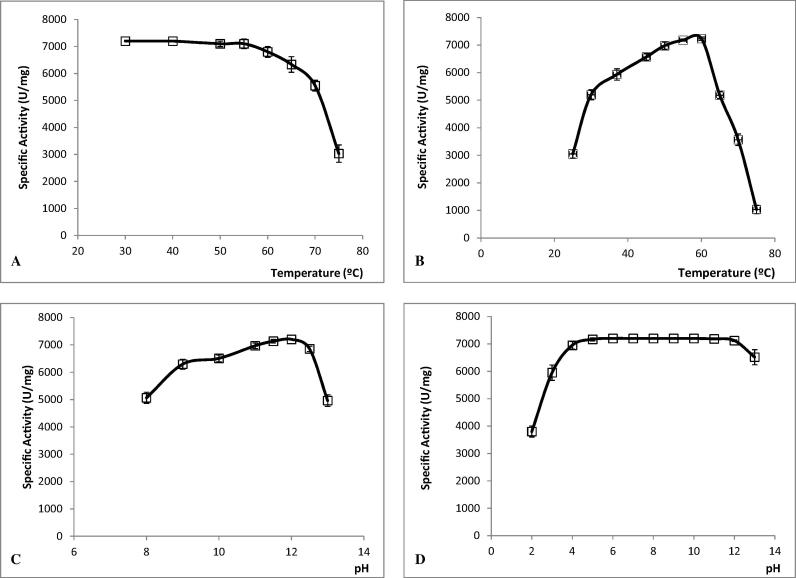
Results presented in Fig. 3A indicate that SAL4 is still active with around 75% of its maximum activity after a 30-min incubation at 70 °C. On the other hand, the maximum activity of SAL4 was observed in the range of 55 to 60 °C whereas the enzyme lost almost its full activity at 75 °C (Fig. 3B).
Figure 3.
Temperature effect of on SAL4 stability (A) and activity (B). pH effect on enzyme activity (C) and stability (D) of SAL4. All experiments were repeated at least three times.
As shown in Fig. 3C, pure SAL4 was highly active with a pH ranging from 8 to 13, with a maximal activity observed at pH 12. Furthermore, the purified lipase maintained its full activity when incubated between pH 4.0 and 12 for 24 h and it lost 50% of its activity at pH 2.0 (Fig. 3D).
3.4. Metal ions influence on the activity of SAL4
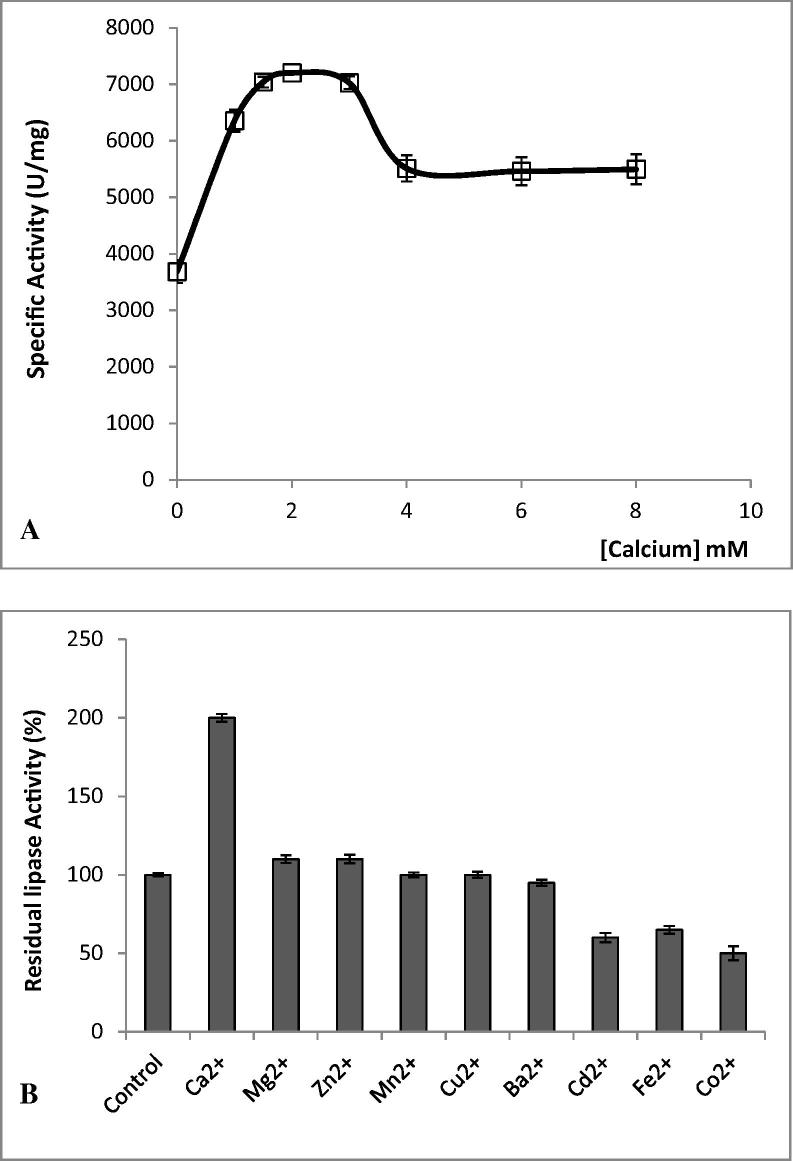
The variation in the hydrolysis rates of olive oil emulsion by the SAL4 was investigated with the addition of different concentrations of Ca2+ (Fig. 4A). Data depicted in Fig. 4A have proven that SAL4 activity was significantly detectable in the absence of Ca2+ with a specific activity of 3500 U/mg reached in the presence of calcium chelators (10 mM EDTA or EGTA). However, the specific activity of the SAL4 was found to increase, reaching a maximum (7215 U/mg) at 2 mM CaCl2 (Fig. 4A) in the absence of EDTA or EGTA.
Figure 4.
(A) Influence of Ca2+ concentration on SAL4 activity. The star symbol indicates the absence of lipase activity checked in the absence of CaCl2 and in the presence of 10 mM EDTA. (B) Influence of metal ions concentrations (2 mM each) on SAL4 activity was studied using olive oil emulsion as substrate under optimal conditions of pH and temperature. The control represents 100% of lipase activity in the absence of metal ions under the same condition. Values represent the mean of three replicates.
The influence of other metal ions such as Cd2+, Cu2+, Fe2+, Mn2+ and Zn2+ (at a final concentration of 2 mM) on the activity of pure SAL4 was also determined under the same conditions (Fig. 4B). The obtained data showed that whereas the addition of Cd2+, Co2+ and Fe2+ caused a significant inhibition of the SAL4 residual activity (>35% inhibition) with Co2+ the strongest inhibitor (50% of inhibition), Cu2+, Zn2+ and Mn2+ had no notable effect on the ALA1 lipase activity (Fig. 4B).
3.5. Effects of various detergent ingredients, oxidizing agents and surfactants on SAL4 activity
As shown in Table 3, SAL4 retained almost its maximal activity after 1 h incubation at 40 °C in the presence of the tested non-ionic surfactants (1% Triton X-100, Tween-80 or Tween-20). Likewise, the addition of 1% NaDC or NaTDC was found to enhance the enzyme activity significantly. Interestingly, the SAL4 retained around 86% of its original lipolytic activity when incubated with the strong anionic surfactant, SDS (1%). Moreover, under the same conditions the SAL4 was significantly more stable than the Lipolase®.
Table 3.
Lipase stability of SAL4 and the commercial detergent Lipolase® in the presence of surfactants, detergents and oxidizing agent.
| Detergent components | Residual activity (%) |
|
|---|---|---|
| SAL4 | Lipolase® | |
| None | 100 | 100 |
| Surfactants (1%) | ||
| Tween-20 | 102.3 ± 2.5 | 82.4 ± 1.6 |
| Tween-80 | 100.7 ± 2 | 72.71 ± 2.3 |
| Triton-X100 | 100.7 ± 1.2 | 49.67 ± 4.2 |
| SDS | 86.7 ± 1.7 | 69 ± 1.2 |
| NaDC | 110 ± 2.5 | 87.9 ± 3.1 |
| NaTDC | 108 ± 1.7 | 80.2 ± 2 |
| Oxidizing agents (1%) | ||
| Hydrogen peroxide | 89.3 ± 4 | 55.3 ± 2.5 |
| Sodium perborate | 91 ± 3.6 | 61.3 ± 4.2 |
| Sodium hypochlorite | 79 ± 2.5 | 52 ± 2 |
| Commercial detergents | ||
| Solid detergent (5mg/mL) | ||
| Ariel | 100 ± 0 | 63 ± 3.6 |
| Dixan | 97.7 ± 2.5 | 59.7 ± 5.5 |
| Nadhif | 92.3 ± 2.1 | 49.3 ± 4 |
| Tide | 87 ± 3 | 53 ± 4.3 |
| Liquid detergent (1/100) | ||
| Dixan | 85. 7 ± 4 | 54 ± 1.5 |
| Nadhif | 85. 7 ± 1.2 | 46 ± 3.5 |
| Dac | 88.3 ± 2.5 | 52 ± 1.7 |
| Fairy | 83.3 ± 3.5 | 80.3 ± 1.5 |
SAL4 preparation was incubated with different detergents components for 60 min at 40 °C and the remaining activity was measured under standard assay conditions. Residual SAL4 activity was determined at pH 12 and 60 °C. Results are the relative lipase activity expressed as the percentage of the maximum activity recorded without the addition of compound; data are means of triplicate determinations ± standard deviation.
On the other hand, as shown in Table 3, ALA1 lipase retained between 79% and 91% of the initial lipolytic activity when pre-incubated with 1% H2O2 or sodium perborate or sodium hypochlorite. It is worthy to note that SAL4 exhibited higher stability compared to the commercial detergent Lipolase®, in the presence of strong oxidizing agent sodium hypochlorite (Residual enzyme activity: 79% vs. 52%) (Table 3).
Moreover, as can be seen from Table 3, SAL4 was found to be highly stable in the presence of several commercial liquid and solid laundry detergents (60 min at 40 °C). To allow further comparison the results obtained by the Lipolase® under the same conditions are reported in the same table (Table 3). Data presented in Table 3 demonstrated that SAL4 had negligible loss in the activity (⩽15% inhibition) after exposure to the different commercial detergents. Remarkably, this novel lipase showed higher stability compared to the commercially used one which lost more than 40% of its original activity after pre-incubation with Dixan, Ariel, Tide or Nadhif. However, both lipases showed a similar stability toward Fairy liquid detergents (Table 3).
4. Discussion
In a previous study, we have isolated an alkaline lipase-producing strain ALA1 from dromedary milk which has been identified as S. aureus from the microscopic appearance, biochemical tests and 16S rDNA sequence analysis (Ben Bacha et al., 2015). After the optimization of various factors like incubation time, culture conditions (temperature and pH) and medium composition (nitrogen, carbon, surfactants and lipid compounds) the lipase production by S. aureus strain ALA1 (128 U/mL) was around 4 and 40-times higher than the previously isolated ones produced by the S. aureus strains (Ben Bacha et al., 2015, Daoud et al., 2013, Horchani et al., 2009).
SAL4 was then purified 81-fold with 52% yield by a single step of RP-HPLC on C-18 column (Fig. 1A, Table 1). The recorded yield is approximately 2 to 5-fold higher than that obtained with lipases previously isolated from S. aureus (SAL3) (Horchani et al., 2009) and S. xylosus (SXL 1 and SXL2), respectively (Mosbah et al., 2005, Bouaziz et al., 2011). Furthermore, lipase from the ALA1 bacterium was purified after only one chromatography step while the previously described staphylococcal lipases needed 3 chromatography steps (Mosbah et al., 2005, Horchani et al., 2009).
SDS–PAGE analysis of purified lipase from S. aureus strain ALA1 yielding a single 43 kDa band (Fig. 1B) was confirmed by native conditions gel filtration suggesting that SAL4 is a monomeric protein with a molecular mass of about 43 kDa. However, all previously-reported staphylococcal lipases, except SXL1 (Mosbah et al., 2005), aggregate under native conditions (Rosenstein and Gotz, 2000, Sayari et al., 2001).
As shown in Fig. 1C, the first 38 N-terminal residues of the pure enzyme exhibited a significant similarity degree with previously characterized staphylococcal lipases (Nikoleit et al., 1995, Sayari et al., 2001, Horchani et al., 2009, Bouaziz et al., 2011). Interestingly, an extra amino dipeptide leu-Lys was observed at the N-terminus of ALA1 lipase as compared to S. simulans lipase (SSL) (Sayari et al., 2001). Similar results were obtained with extracellular SAL3 (Horchani et al., 2009) and SXL2 (Bouaziz et al., 2011). Authors suggested that the different biochemical characteristics of both lipases might due to these two residues probably originating from a different proteolytic cleavage along the mature lipase processing. In fact, Mosbah et al. (2010) have shown an amelioration of the temperature stability and activity of SXL1 after the insertion of a dipepetide (Leu-Lys) at the N-terminus of the enzyme (Mosbah et al., 2010).
It is well known that lipases differ from classical esterases by their ability to be activated by interfaces as first described by Sarda and Desnuelle (1958). TC3 was used to study the interfacial activation phenomenon of SAL4 as reported by Ferrato et al. (1997). As shown in Fig. 2A, SAL4 does not present this phenomenon as reported for SAL3, Staphylococcus sp. lipase (SL1), SSL and SXL1 (Sayari et al., 2001, Mosbah et al., 2005, Horchani et al., 2009, Cherif et al., 2011). However, independently of the absence, or the presence, of the interfacial activation phenomenon and according to Ferrato et al. (1997) and Chahinian and Sarda (2009), SAL4 which hydrolyzes olive oil efficiently, is a true lipase.
Pure SAL4 was found to be capable to efficiently hydrolyze the pure TC4, TC8 and the olive oil emulsion alone (Fig. 2B). Accordingly, the studied lipase may exhibit a tri-dimensional structure allowing it not only to efficiently hydrolyze TC4 at high interfacial energy without any denaturation, but also to tolerate the presence of long-chain free fatty acids in the absence of any amphipathic compound at the olive oil/water interface.
Furthermore, it is well recognized that the physical state of the substrate significantly affects the enzymatic activity of lipases. In fact, staphylococcal lipases showed different chain length selectivity. S. hyicus lipase, SXL2 and S. aureus lipase exhibited a potent preference for short-chain triglycerides (Simons et al., 1998, Rosenstein and Gotz, 2000, Bouaziz et al., 2011, Daoud et al., 2013), whereas SXL1 (Mosbah et al., 2005) and SSL (Sayari et al., 2001) hydrolyze triacyglycerols regardless of their chain length.
The results presented in Table 2 show that in contrast to the previously described staphylococcal lipases, pure SAL4 hydrolyzes long-chain triacylglycerols more efficiently than short-chain ones. Moreover, the SAL4 is among the most active lipases in the staphylococcal family with a specific activity 3.5-fold and 7-fold higher than SXL1 (Mosbah et al., 2005) and the SSL (Sayari et al., 2001), respectively. Moreover, the catalytic efficiency (kcat/Km app.) of SAL4 was 2069.5 when using TC18 as substrate while it decreased by 4.3 or 21.9-fold when using TC8 or TC4 as substrate, respectively, which confirms that lipase hydrolyzes more efficiently long- than short-chain triacylglycerols and acts as a true lipase.
Thermostable and alkaline lipases exhibit higher activity and stability at high temperatures and under alkaline conditions. Besides, they are frequently resistant to chemical denaturation which makes them attractive tools for chemical and industrial processes generally carried out at relatively extreme conditions of pH and temperatures and/or in the presence of organic solvents.
Our results revealed that, as opposed to the majority of staphylococcal lipases reported so far (Rosenstein and Gotz, 2000, Sayari et al., 2001, Guncheva et al., 2010), SAL4 lipase is highly stable over a broad pH range and against heat. The highest catalytic activity was indicated at the range of 55–60 °C and pH 12 (Fig. 3). Horcahni et al. (2009) and Bouaziz et al. (2011) showed that the optimum pH of the SAL3 and SXL2 activities were found to be ranging between pH 8.0 and 9.5. The same lipases (SAL3 and SXL2) were highly stable between pH 5.0 and 12.0 (Horchani et al., 2009, Bouaziz et al., 2011) while the SXL1 and SSL were found to be stable only between pH 5–8.5and pH 4–9, respectively (Mosbah et al., 2005, Sayari et al., 2001).
Therefore, alkaline and thermostable lipases are extremely attractive to the synthesis of biodiesel and biopolymers in addition to their potential use in the production of agrochemicals, cosmetics, detergent, flavor and pharmaceuticals (Chen et al., 1998, Illanes, 1999). Hence, one can conclude that the SAL4 could be an ideal candidate for various industrial and biotechnological applications.
These differences between SAL4 and other staphylococcal lipases may be explained by structural variation of the determined amino acid residues among these lipases in particular the two extra Leu-Lys residues at the N terminus of SAL4 as previously demonstrated by Mosbah et al. (2010). Moreover, single amino acid mutation in the SAL4 sequence may significantly modify its activity and stability as reported by Parameswaran et al. (2010). The authors suggest that mutation can be carried out at some positions in the protein sequence of SAL3 in the direction of emulating protein stability (Parameswaran et al., 2010).
It is well established that Ca2+ ions are essential for both the catalysis and binding of staphylococcal lipases to their substrates (Rosenstein and Gotz, 2000). Contrarily to previous research studies reporting that the presence of Ca2+ is absolutely required to trigger the triacylglycerols hydrolysis by several staphylococcal lipases such as SAL, SEL and SHL (Rosenstein and Gotz, 2000), Ca2+ seemed to be not necessary for SAL4 activity when using olive oil emulsion as substrate. Interestingly, its enzymatic activity was enhanced up to 52% by 2 mM Ca2+ (Fig. 4A). Previous studies reported that the activities of lipases from S. epidermidis and Pseudomonas glumae are dramatically promoted by the Ca2+ ion binding to a site near the active-site and formed by two conserved Asp residues (Simons et al., 1999, Noble et al., 1993).
Nevertheless, Fig. 4B shows that the substitution of Ca2+ by other divalent ions (Zn2+, Mn2+ Fe2+ Cu2+, Co2+, Cd2+ or Ba2+) decreased the catalytic activity.
During washing an effective detergent lipase should tolerate the presence of different surfactants besides its temperature and pH stability (Tiesinga et al., 2007). As can be seen from Table 3, SAL4 was found to be able to hydrolyze its substrate in the presence of various oxidizing agents (sodium hypochlorite, hydrogen peroxide and sodium perborate) as well as some non-ionic (Tween-80, Tween-20 and Triton X-100) and ionic (sodium cholate, sodium taurocholate and SDS) surfactants and some solid (Ariel, Dixan, Nadhif and Tide) and liquid (Dixan, Nadhif, DAC and Fairy) commercial detergents. As a matter of fact, SAL4 displayed better stability in the presence of the tested agents as compared to Lipolase®, (Table 3). Horchani et al. (2009) demonstrated that SAL3 is highly stable and fully active in the presence some known surfactants such Triton X-100, Tween 80, NaTDC and NaDC at a final concentration of 20 mM. However, a total loss of lipase activity was reported by Dannert et al. (2010) after incubation with Tween-80 or Tween-20 whereas no effect was recorded in the presence of Triton X-100. As regards Prazeres et al. (2006), they showed that in contrast to Triton X-114 and Triton X-100 which displayed an activating effect, lipase activity decreased by approximately 30% after incubation with surfactin, Tween-80 or Tween-40.
In addition, because SDS stable lipases are rarely described, the stability of SAL4 solution toward SDS is very important (Nawani et al., 1998, Cherif et al., 2011). In the same direction, as in washing conditions some oxidizing reactions exerted by sodium perborate, sodium hypochlorite and hydrogen peroxide may take place, the oxidizing stability is thus highly recommended for an enzyme to be used in detergent formulations. This important characteristic had been achieved by protein engineering and site-directed mutagenesis for Lipolase® and many proteases (Outtrup et al., 1995, Wolff et al., 1996, Gupta et al., 2002).
In the present study and as shown in Table 3, the SAL4 was found to be resistant against all the tested oxidizing agents (sodium hypochlorite, sodium perborate and hydrogen peroxide) which slightly affected its activity.
The compatibility of SAL4 with several commercially important detergents was investigated as well. Remarkably, the novel isolated enzyme was more stable than the commercially used lipase (Lipolase®) toward all detergents tested except Fairy liquid detergent (Table 3). These findings strongly demonstrate that S. aureus strain ALA1 lipase can be considered as a prospective candidate to be essentially used for the application in laundry detergents.
5. Conclusion
In the present study, the lipase from S. aureus is found to display encouraging properties for biotechnological applications. It hydrolyzes three times more effectively long-chain triglycerides than short-chain ones. In addition, the high stability and activity of SAL4 at extreme temperatures and pHs and toward SDS, Triton X-100 and Tween 20 and oxidizing agents together with its excellent compatibility with some commercial laundry detergents are especially encouraged. Overall, the obtained data suggested that SAL4 may be used in the detergent industry.
Acknowledgements
This project was funded by the National Plan for Science, Technology and Innovation (MAARIFAH), King Abdulaziz City for Science and Technology, Kingdom of Saudi Arabia, Award number (12-ENV2528-02).
Footnotes
Peer review under responsibility of King Saud University.
References
- Abdelkafi S., Fouquet B., Barouh N., Durner S., Pina M., Scheirlinckx F., Villeneuve P., Carrière F. In vitro comparisons between Carica papaya and pancreatic lipases during test meal lipolysis: potential use of CPL in enzyme replacement therapy. Food Chem. 2009;115:488–494. [Google Scholar]
- Akoh C.C., Lee G.C., Liaw Y.C., Huang Shaw T.H.J.F. GDSL family of serine esterases/lipases. Prog. Lipid Res. 2004;43:534–552. doi: 10.1016/j.plipres.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Ben Bacha A., Moubayed N.M.S., Al Assaf A. An organic solvent-stable lipase from a newly isolated Staphylococcus aureus ALA1 strain with potential for use as an industrial biocatalyst. Biotechnol. Appl. Biochem. 2015 doi: 10.1002/bab.1381. [DOI] [PubMed] [Google Scholar]
- Bornscheuer U.T., Kazlauskas R.J. Wiley-VCH; Weinheim: 1999. Hydrolases in Organic Synthesis – Regio- and Stereoselective Biotransformations. 336. [Google Scholar]
- Bouaziz A., Horchani H., Ben Salem N., Gargouri Y., Sayari A. Expression, purification of a novel alkaline Staphylococcus xylosus lipase acting at high temperature. Biochem. Eng. J. 2011;54:93–102. [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantification of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brzozowsky A.M., Derewenda U., Derewenda Z.S., Dodson G.G., Lawson D.M., Turkenburg J.P., Bjorkling F., Huge-Jensen B., Patkar S.A., Thim L. A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex. Nature. 1991;351:491–494. doi: 10.1038/351491a0. [DOI] [PubMed] [Google Scholar]
- Chahinian H., Sarda L. Distinction between esterases and lipases: comparative biochemical properties of sequence-related carboxylesterases. Protein Pept. Lett. 2009;16:116–1149. doi: 10.2174/092986609789071333. [DOI] [PubMed] [Google Scholar]
- Chauhan M., Chauhan R.S., Garlapati V.K. Evaluation of a new lipase from Staphylococcus sp. for detergent additive capability. Biomed. Res. Int. 2013 doi: 10.1155/2013/374967. 374967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.J., Cheng C.Y., Chen T.L. Production of an alkaline lipase by Acinetobacter radioresistens. J. Ferment. Bioeng. 1998;86:308–312. [Google Scholar]
- Cherif S., Mnif S., Hadrich F., Abdelkafi S., Sayadi S. A: newly high alkaline lipase: an ideal choice for application in detergent formulations. Lipids Health Dis. 2011;10:221. doi: 10.1186/1476-511X-10-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daoud L., Kamoun J., Ali M.B., Jallouli R., Bradai R., Mechichi T., Gargouri Y., Ali Y.B., Aloulou A. Purification and biochemical characterization of a halotolerant Staphylococcus sp. extracellular lipase. Int. J. Biol. Macromol. 2013;57:232–237. doi: 10.1016/j.ijbiomac.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Egloff M.P., Ransac S., Marguet F., Rogalska E., van Thilburg H., Buono G., Cambillau C., Verger R. The 2,46 A resolution structure of the pancreatic lipase-colipase complex inhibited by a C11 alkyl phosphonate. Biochemistry. 1995;34:2751–2762. doi: 10.1021/bi00009a003. [DOI] [PubMed] [Google Scholar]
- Ferrato F., Carrière F., Sarda L., Verger R. A critical reevaluation of the phenomenon of interfacial activation. Methods Enzymol. 1997;286:327–346. doi: 10.1016/s0076-6879(97)86018-1. [DOI] [PubMed] [Google Scholar]
- Guncheva M., Zhiryakova D., Radchenkova N., Kambourova M. Properties of immobilized lipase from Bacillus stearothermophilus MC7. Acidolysis of triolein with caprylic acid. World J. Microbiol. Biotechnol. 2010;25:727–731. [Google Scholar]
- Gupta R., Beg O., Lorenz P. Bacterial alkaline proteases: molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 2002;59:15–32. doi: 10.1007/s00253-002-0975-y. [DOI] [PubMed] [Google Scholar]
- Handelsman T., Shoham Y. Production and characterization of an extracellular thermostable lipase from a thermophilic Bacillus sp. J. Gen. Appl. Microbiol. 1994;40:435–443. [Google Scholar]
- Hasan F., Shah A.A., Hameed A. Methods for detection and characterization of lipases: a comprehensive review. Biotechnol. Adv. 2009;27:782–798. doi: 10.1016/j.biotechadv.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Hermoso J., Pignol D., Kerfelec B., Crenon I., Chapus C., Fontecilla-Camps J.C. Lipase activation by non ionic detergents: the crystal structure of the porcine lipase-colipase tetrahethylene glycol monooctyl ether complex. J. Biol. Chem. 1996;271:18007–18016. doi: 10.1074/jbc.271.30.18007. [DOI] [PubMed] [Google Scholar]
- Horchani H., Mosbah H., Ben Salem N., Gargouri Y., Sayari A. Biochemical and molecular characterisation of a thermoactive, alkaline and detergent-stable lipase from a newly isolated Staphylococcus aureus strain. J. Mol. Catal. B: Enzym. 2009;56:237–245. [Google Scholar]
- Horchani H., Aissa I., Ouertani S., Zarai Z., Gargouri Y., Sayari A. Staphylococcal lipases: biotechnological applications. J. Mol. Catal. B Enzym. 2012;76:125–132. [Google Scholar]
- Illanes A. Stability of biocatalysts. J. Biotechnol. 1999;2:7–15. [Google Scholar]
- Jaeger K.E., Reetz T.M. Microbial lipases from versatile tools for biotechnology. Trends Biotechnol. 1998;16:396–403. doi: 10.1016/s0167-7799(98)01195-0. [DOI] [PubMed] [Google Scholar]
- Jaeger K., Dijkstra B.W., Reetz M.T. Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Ann. Rev. Microbiol. 1999;53:315–351. doi: 10.1146/annurev.micro.53.1.315. [DOI] [PubMed] [Google Scholar]
- Jaeger K.E., Dijkstra B.W., Reetz M.T. Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu. Rev. Microbiol. 1999;53:315–351. doi: 10.1146/annurev.micro.53.1.315. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mosbah H., Sayari A., Mejdoub H., Dhouib H., Gargouri Y. Biochemical and molecular characterization of Staphylococcus xylosus lipase. Biochim. Biophys. Acta. 2005;1723:282–291. doi: 10.1016/j.bbagen.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Mosbah H., Horchani H., Sayari A., Gargouri Y. The insertion of (LK) residues at the N-terminus of Staphylococcus xylosus lipase affects its catalytic properties and its enantioselectivity. Process Biochem. 2010;45:777–785. [Google Scholar]
- Nardini M., Dijkstra B.W. α/β Hydrolase fold enzymes: the family keeps growing. Curr. Opin. Struct. Biol. 1999;9:732–737. doi: 10.1016/s0959-440x(99)00037-8. [DOI] [PubMed] [Google Scholar]
- Nawani N., Dosanjh N.S., Kaur J. A novel thermostable lipase from a thermophilic Bacillus sp.: characterization and esterification studies. Biotechnol. Lett. 1998;20:997–1000. [Google Scholar]
- Nikoleit K., Rosenstein R., Verheij H.M., Götz F. Comparative biochemical and molecular analysis of the Staphylococcus hyicus, Staphylococcus aureus and a hybrid lipase. Eur. J. Biochem. 1995;228:732–738. [PubMed] [Google Scholar]
- Noble M.E.M., Cleasby A., Johnson L.N., Egmond M.R., Frenken L.G.J. The crystal structure of triacylglycerol lipase from Pseudomonas glumae reveals a partially redundant catalytic aspartate. FEBS Lett. 1993;331:123–128. doi: 10.1016/0014-5793(93)80310-q. [DOI] [PubMed] [Google Scholar]
- Outtrup, H., Dambmann, C., Christiansen, M., Aaslyng, D.L., 1995. Bacillus sp. JP395, method of making and detergent composition, US Patent number. 55, 466–594.
- Parameswaran S.1., Throat A.A., Patra S. Deciphering role of amino acids for the stability of Staphylococcus aureus lipase (SAL3) Interdiscip. Sci. 2010;2:271–279. doi: 10.1007/s12539-010-0029-6. [DOI] [PubMed] [Google Scholar]
- Prazeres J.N., Cruz J.A.B., Pastore G.M. Characterization of alkaline lipase from Fusarium oxysporum and the effect of different surfactants and detergents on the enzyme activity. Braz. J. Microbiol. 2006;37:505–509. [Google Scholar]
- Rosenstein R., Gotz F. Staphylococcal lipases: biochemical and molecular characterization. Biochimie. 2000;82:1005–1014. doi: 10.1016/s0300-9084(00)01180-9. [DOI] [PubMed] [Google Scholar]
- Sarda L., Desnuelle P. Action de la Iipase pancréatique sur les esters en émulsion. Biochim. Biophys. Acta. 1958;30:513–521. doi: 10.1016/0006-3002(58)90097-0. [DOI] [PubMed] [Google Scholar]
- Satsuki T., Watanabe T. Application of lipase to laundry detergents. Bio. Ind. 1990;7:501–507. [Google Scholar]
- Sayari A., Agrebi N., Jaoua S., Gargouri Y. Biochemical and molecular characterization of Staphylococcus simulans lipase. Biochimie. 2001;83:863–871. doi: 10.1016/s0300-9084(01)01327-x. [DOI] [PubMed] [Google Scholar]
- Sharma R., Chisti Y., Banerjee U.C. Production, purification, characterization, and applications of lipases. Biotechnol. Adv. 2001;19:627–662. doi: 10.1016/s0734-9750(01)00086-6. [DOI] [PubMed] [Google Scholar]
- Simons J.W., Götz F., Egmond M.R., Verheij H.M. Biochemical properties of staphylococcal (phospho)lipases. Chem. Phys. Lipids. 1998;93:27–37. doi: 10.1016/s0009-3084(98)00026-7. [DOI] [PubMed] [Google Scholar]
- Simons J.W., van Kampen M.D., Ubarretxena-Belandia I., Cox R.C., Alves dos Santos C.M., Egmond M.R., Verheij H.M. Cloning, purification and characterization of the lipase from Staphylococcus epidermidis. Biochemistry. 1999;38:2–10. doi: 10.1021/bi981869l. [DOI] [PubMed] [Google Scholar]
- Tiesinga J.W., van Pouderoyen G., Nardini M., Ransac S., Dijkstra B.W. Structural basis of phospholipase activity of Staphylococcus hyicus lipase. J. Mol. Biol. 2007;371:447–456. doi: 10.1016/j.jmb.2007.05.041. [DOI] [PubMed] [Google Scholar]
- Wolff A.M., Showell M.S., Venegas M.G., Barnett B.L., Wertz W.C. Laundry performance of subtilisin protease. In: Bott R.C., editor. Subtilisin Enzymes: Practical Protein Engineering. Plenum Press; New York: 1996. pp. 113–120. [Google Scholar]