Summary
Objective
This study aimed to identify non-invasive biomarkers of human epilepsy that can reliably detect and localize epileptic brain regions. Having non-invasive biomarkers would greatly enhance patient diagnosis, patient monitoring, and novel therapy development. At present time, only surgically invasive, direct brain recordings are capable of detecting these regions with precision, which severely limits the pace and scope of both clinical management and research progress in epilepsy.
Methods
We compared high versus low or non-spiking regions in 9 medically intractable epilepsy surgery patients by performing integrated metabolomic-genomic-histological analyses of electrically-mapped human cortical regions using high resolution magic angle spinning proton magnetic resonance spectroscopy, (HR-MAS 1H MRS), cDNA microarrays, and histological analysis.
Results
We found a highly consistent and predictive metabolite logistic regression model with reduced lactate and increased creatine plus phosphocreatine (Cr+PCr) and choline, suggestive of a chronically altered metabolic state in epileptic brain regions. Linking gene expression, cellular, and histological differences to these key metabolites using a hierarchical clustering approach predicted altered metabolic vascular coupling in the affected regions. Consistently, these predictions were validated histologically showing both neovascularization and newly discovered, millimeter-sized microlesions.
Significance
Using a systems biology approach on electrically-mapped human cortex provides new evidence for spatially-segregated, metabolic derangements in both neurovascular and synaptic architecture in human epileptic brain regions that could be a non-invasively detectable biomarker of epilepsy. These findings highlight both the immense power of a systems biology approach and identify a potentially important role that magnetic resonance spectroscopy can play in the research and clinical management of epilepsy.
Keywords: epilepsy, interictal spikes, human metabolite biomarkers, magnetic resonance spectroscopy
Introduction
Epilepsy is a disorder of recurrent seizures that affects 50 million people worldwide and medication management is ineffective in up to 30%1. In patients whose seizures start from focal brain regions, surgery that removes electrically active brain regions can be curative; however, identifying these regions can be challenging. Precise localization of these inciting regions is pivotal to treatment success, but frequently requires invasive implantation of recording electrodes2. While some patients develop epilepsy in response to a lesion (e.g. injury, tumor, or stroke), the regions of brain that produce seizures are often well outside the margins of the lesion and are also be resected for optimal outcome3,4. The identification of a reliable, non-invasive biomarker would greatly help in localizing epileptic brain regions for surgery in addition to being informative for clinical trials and diagnosis.
Epileptic regions of the brain produce abnormal synchronous discharges involving large populations of neurons. These discharges can be single, as in the case of interictal discharges, or they can become rhythmic and propagate over large regions of the brain, resulting in seizures. While the exact relationship between interictal spiking and seizures is not understood5, their localization is highly concordant6 and the removal of both regions is associated with improved surgical outcome7. Animal studies have shown that interictal activity may precede and hence be a biomarker of seizure development8.
We collected electrically-mapped regions of human neocortex from patients undergoing long-term invasive monitoring with electrocorticography (ECoG) using subdural grid electrodes to localize regions of the cortex that produce seizures and interictal spiking9 (Figure 1). Our systems biology program was designed to integrate quantitative clinical, neurochemical, electrical, and genomic signatures of different brain regions with and without significant epileptic activities10-13. By knowing the in vivo electrical behavior of each resected piece of tissue, we have the unique ability to compare highly epileptic brain regions to nearby electrically quiet regions serving as internal controls within each patient. Studies from this program have implicated layer-II/III-specific activation of MAPK and CREB signaling and the presence of deeper “microlesions” that show a dramatic reduction in axodendritic connectivity in brain regions with high levels of epileptic activity13. In the present study, we use the exact same brain regions that we have studied using transcriptional profiling and histology to assess metabolomic differences.
Figure 1. Experimental design.
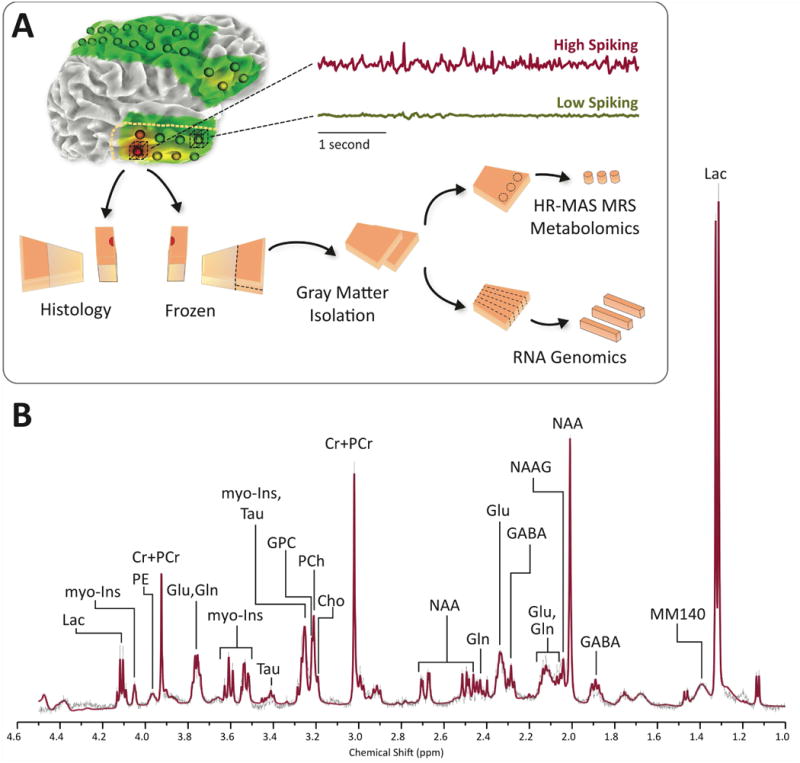
A) Human brain tissues were removed as part of planned surgery for refractory epilepsy. Regions of high and low spiking are identified based on long-term subdural electrocorticography. Each section of tissue is precisely mapped to the overlying subdural electrode and then split into two halves. One half is used for histology while cortical gray matter is isolated from the other half. The cortical gray matter is then further divided into two portions, half of which is used to generate RNA for microarray analysis while the other half is used for 1H HR-MAS MRS. B) Representative high resolution spectrum of 1H HR-MAS MRS of human brain performed on partial thickness neocortical sample (Layers I-III). LCModel fitted spectrum (red line) shows excellent correspondence with averaged raw signal (thin black line). Cho = free choline, Cr+PCr = creatine plus phosphocreatine, GABA = γ-aminobutyric acid, Gln = glutamine, Glu = glutamate, GPC = glycerophosphorylcholine, myo-Ins = myo-inositol, Lac = lactate, MM140 = macromolecules at 1.4 ppm, NAA = N-acetylaspartate, NAAG = N-acetylaspartylglutamic acid, PCh = phosphorylcholine, PE = phosphorylethanolamine, Tau = taurine.
Magnetic resonance spectroscopy (MRS) is a technique for characterizing compounds associated with tissue metabolism. This technique has high translational potential since it can be applied to both intact tissue samples ex vivo as well as in vivo. While there are numerous MRS visible nuclei, 1H spectroscopy is the most common by virtue of its high sensitivity and the numerous compounds it is capable of probing. Previous 1H MRS studies of epilepsy have demonstrated perturbations in the neurochemistry of epilepsy patients and have assisted in efforts to lateralize the epileptic focus14,15; however, a highly sensitive and specific set of non-invasive biomarkers have yet to emerge in part due to uncertainties in localization, poor spatial distinction of epileptic and non-epileptic brain regions, and use of relatively low field strength MR systems. Here, we addressed these challenges for the first time using resected human neocortical brain samples that were precisely localized and electrically characterized using long-term ECoG recordings. We then used the superior spectral resolution provided by an ultra-high field spectrometer at 11.7T to ask whether epileptic human neocortex has a unique metabolite signature. We found that epileptic human neocortex exhibits a unique metabolomic profile and that by using an integrative systems biology approach with genomic and histopathologic findings an altered metabolic state was found in epileptic neocortex that can be used as a non-invasive biomarker of epileptic activity.
Methods
Isolation of human tissue and electrophysiology
Human brain tissue samples from 9 human subjects with refractory epilepsy were obtained with informed consent as part of a research protocol approved by the Wayne State University Institutional Review Board and their participation in the study had no influence on their clinical care or treatment plan. All 9 subjects underwent a two-stage surgery, with long-term subdural ECoG, consisting of initial electrode placement (Stage 1) for long term recording and observation to optimize surgical resection (Stage 2) of seizure generating areas (Table 1). Detailed methods on how ECoG and surgical resection tissues were identified, processed, and analyzed have been described previously10,13. Briefly, interictal spiking activity was determined by averaging spike counts from 3 independent 10 min segments of ECoG recording, which were continuously recorded for at least 3 days prior to resection. To avoid contamination from ictal activity, we use the following criteria for selecting 10 minute segments of awake intracranial EEG data for analysis: 1) at least 3 hour interval between segments, 2) more than 2 hours since previous partial seizure, and 3) at least 8 hours since the previous generalized tonic-clonic seizure. Spike counts were carried out by an in-house Matlab program. Medications were tapered and discontinued for all patients to allow for adequate seizure capture, with the tapering schedule being determined by the clinical team. Tissue removed during surgery was immediately placed on ice. Electrode placements were mapped precisely to their corresponding location on the neocortex using a combination of intraoperative electrode placement photos, as well as pre- and post-placement head CT, MRI, and X-rays. Tissue samples localized to each electrode of interest was removed and divided into two halves. One half was fixed in 4% paraformaldehyde for use in histological studies while the other half was rapidly frozen on dry ice and stored frozen at -80°C until for future analysis (Figure 1A). From resection to -80°C storage, samples were processed in approximately 45-60 minutes, depending on the size and number of tissue samples. Because of the great variance between patients in absolute spike frequency between regions considered for high and low spiking, we dichotomized the spike frequencies into “high” and “low” spiking categories to reduce high-leverage effects. For each patient, the subdural electrode demonstrating the highest spiking frequency was classified as “high spiking” and the electrode with the lowest or no spiking was classified as “low spiking.” In this manner, high and low spiking electrodes for each patient were identified and analyzed using both microarray and HR-MAS 1H MRS at 11.7T. Critical for the present studies, nearby regions with differing levels of spiking were processed in parallel with the same processing times between resection and freezing so that metabolomics differences would not be attributed to processing times within a given patient.
Table 1. Patient clinical demographics and tissue information.
| Patient | ILAE | Age of Onset | Age at Surgery | Sex | Region | MRI | Outcome (Engel 6 months) | Other tissue diagnoses | Spike Frequency | Epilepto-genic | Microarray Characteristics (% genes) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | Low | High | Increase | Decrease | Total | |||||||||
| 1 | NA | NA | 10 | F | Temporal | PO | NA | DG, AI | 1 | 116 | N | N | 7.6 | 9.2 | 16.8 |
| 2 | FB | 9 | 11 | F | Frontal | H | NA | H | 0 | 5 | Y | Y | 2.6 | 5 | 7.5 |
| 3 | ES, F | 0.4 | 3 | F | Parietal | PE | I | MG, H | 66 | 141 | Y | Y | 6.2 | 4.2 | 10.3 |
| 4 | ES | 0.5 | 3 | F | Temporal | BGW | I | MG | 56 | 212 | Y | Y | 4.9 | 5.4 | 10.2 |
| 5 | FB | NA | 7 | F | Frontal | P, W | I | CD, MG | 25 | 215 | N | Y | 11.6 | 11.1 | 22.7 |
| 6 | ES | 2 | 6 | F | Frontal | TC, CD | I | MG | 26 | 124 | Y | Y | 16.5 | 15 | 31.5 |
| 7 | ES | 0.5 | 8 | M | Parietal | NA | III | MG | 3 | 172 | Y | Y | 10.2 | 9.8 | 19.9 |
| 8 | F, FB | 0 | 16 | M | Temporal | PO | I | DG | 44 | 176 | N | N | 6.9 | 10.6 | 17.5 |
| 9 | FB | 6 | 11 | F | Frontal | Normal | I | CD | 2 | 66 | N | N | 4.8 | 4.1 | 8.9 |
Table 1. Profile of patients with neocortical epilepsy and the corresponding tissue samples that were used for this study. Spike frequency, expressed as number spikes per 10 minute period, reflects epileptiform spike rates for both high and low spiking regions in the patient's brain, recorded in vivo using electrocorticography as part of their clinical treatment plan. Principal ILAE histopathology classifications of the brain lesions are described in the “ILAE” column. “Epileptogenic” column describes classification of tissue samples as either being part of the epileptogenic zone (Y = yes) or not part of the epileptogenic zone (N = no). Microarray characteristics show percent of genes that are at least 1.4 fold increased or decreased (after FDR correction to 1%) in expression in high spiking relative to low spiking samples. Documented tissue pathologies (“Other tissue diagnoses”) were made from separate clinical tissue samples and were not present in the tissues used for this study, which were normal appearing on histology. Abbreviations: AI = acute inflammation; BGW = blurring of grey-white junction; CD = cortical dysplasia; DG = diffuse gliosis; ES = epileptic spasms; H = heterotopia; MG = mild gliosis; NA = data not available; F = focal; PE = periventricular mild increase in FLAIR; PO = porencephalic cyst; FB = focal to bilateral tonic-clonic ; SupH = superficial heterotopia; TC = thickened cortex; W = increased white matter signal; WG = white matter gliosis.
Because the “low” spiking regions are used as internal controls for each patient, within-subject comparisons can reduce variance due to patient-specific differences in medications and brain regions.16 This internally-controlled study design is critical to rule out differences in tissue processing, inflammatory responses from recording electrodes, and time-dependent metabolomic changes related to the tissue resection. All tissue samples in this study were selected on the basis of their varying degrees of spiking activity and were free of apparent pathological changes, including inflammatory responses form the recording electrodes, and surgical hemorrhage based on gross examination and histology. Tissue histopathology identified in Table 1 reflects tissue diagnoses made from a separate region of the resected brain tissue. Tissue histology studies were done as described in our previous study13.
High Resolution Magic Angle Spinning 1H Magnetic Resonance Spectroscopy
For each patient, we compared multiple regions with high versus low (or no) epileptic activity using HR-MAS 1H MRS, which allows for metabolite profiling of intact brain tissue without the need for disruptive chemical extractions17,18. For metabolite acquisition using HR-MAS 1H-MRS, 2 mm punches were obtained from the apical neocortical gray matter (layers I-III), for each pair of high and low spiking samples within every patient in triplicate, at minimum, while frozen on solid CO2. Tissue samples that were larger in size may have more than 3 replicates. The median number replicates for low spiking tissue was 4 with a range from 3 to 8 and the median number of replicates for high spiking tissue was 6 with a range from 3 to 9. A total of 94 individual data samples were analyzed for this study, consisting of 52 high spiking samples and 42 low spiking samples. The frozen punches were analyzed using HR-MAS 1H-MRS on a 500 MHz (11.7T) Bruker Avance DRX-500 spectrometer equipped with a g-HR MAS 500 WB BL4 P-HD probe (Bruker BioSpin Corporation, Billerica, MA), as described by Ghoddoussi et al. (2010)19. The samples were placed directly into a Bruker zirconium rotor containing 5μL phosphate buffer (pH = 7.4), D2O, trimethylsilyl-proprionate (TSP) as the internal chemical shift reference (0.00 ppm), and formate for phase correction (8.44 ppm). The filled rotor was placed into a Bruker magic angle spinning probe maintained at 4°C and spun at 4.2 ± 0.002 kHz while positioned at 54.7° relative to the static magnetic field (B0). Semi-automated and manual first and second order shimming was used to reduce field inhomogeneities. A rotor-synchronized 1-D Carr-Purcell-Meiboom-Gill (CPMG) with [90°-(τ-180°-τ)n] pulse sequence was used to acquire tissue spectra20. Twelve echo pulses were applied (n = 12) with an inter-pulse delay (τ) of 150 μs for TE = 3.6 ms (echo time) and TR = 6.21 s (repetition time). All spectra were acquired at a spectral bandwidth of 7 kHz (14 ppm) with 128 averages for a total acquisition time of approximately 13 min per sample.
Metabolite fitting
The raw 1H MRS spectra were analyzed using LCModel21 with a custom experimentally derived basis-set containing 27 individual neurochemical metabolite model spectra combined with simulated lipid and macromolecule signals. The experimentally derived basis-set was obtained under identical experimental conditions as the tissue analysis. The concentration of the metabolites between 1.0 to 4.2 ppm were estimated using LCModel and ultimately expressed as normalized mole percent of total metabolites, allowing measurements to be relatively insensitive to changes in extracellular volume22. LCModel's estimated confidence of the fitted metabolite spectra (i.e. how well it matches the original data) was estimated using Cramér-Rao Lower Bounds (CRLB). For this study, only metabolites whose average CRLB were less than or equal to 10% were used for further analysis.
Microarray analysis and detection of differentially expressed genes
We further performed genome wide expression studies using Agilent microarrays and histology on the same tissue samples. Total RNA was isolated from pooled alternating strips of full-thickness (layers I-VI) neocortical gray matter, helpful in averaging out small local differences. A quadruplicate, flip-dye experimental design was used for each pair of high and low spiking samples within every patient and has been described previously10,11. Briefly, labeled antisense RNAs were spin column purified and hybridized to human, genome-wide 60-mer oligonucleotide arrays (Catalog #G411A, Agilent), in a two-color dye-swap fashion. Differentially expressed genes were identified with a two-step hierarchical linear mixed model, correcting for array, dye, patient, array-die interactions and within-patient effects. Genes that had more than a 1.4-fold change between high and low spiking samples with a false discovery rate of < 0.01 were considered to be differentially expressed. We identified 990 such differentially expressed genes from our 9-subject sample.
Tissue discrimination and clustering
In order to get a sense of whether or not there are detectable differences in metabolites between high and low spiking regions, two-sample t-tests were initially performed on all 14 metabolites of interest to determine if significant differences exist between the metabolite in high and low spiking tissues. Afterwards, more refined classification and discrimination of high and low spiking samples using the metabolite profiles of all 94 samples were performed using generalized estimating equation (GEE) logistic regression model with an exchangeable covariance matrix implemented in R with geepack23,24. This repeated measures approach help account for within-subject variability, since the samples were measured in multiplicates. Receiver Operating Characteristic (ROC) curves, a measure of model performance, were calculated using pROC package, together with a bootstrap estimated 95% confidence interval of the model sensitivity25. Because several metabolites were highly correlated (r > 0.8) with each other, principal components analysis was initially used to address the issue of multicollinearity. Since the goal of our PCA analysis was to minimize multicollinearity, we chose to include the maximum number of components possible (i.e. all 14 components) for our full model GEE logistic regression, in order to make predictions on whether a particular tissue sample is high or low spiking. The appropriateness of using the varimax orthogonal rotation method of PCA is validated by both an adequate Kaiser-Meyer-Olkin measure of sampling accuracy (MSA = 0.84) as well as significant Bartlett's test of sphericity, χ2(91) = 1109, p < .001, indicating that the data is appropriate for PCA analysis26. The resultant factor scores were used as predictors in our logistic regression model, which used a given sample's measured metabolite profile to determine a predicted probability (P̂) that the sample was a high spiking sample. The full 14 component model performed extremely well in discriminating high versus low spiking tissue with an ROC area under the curve (AUC) of 0.90, 95% CI [0.83, 0.96]. A maximum accuracy of 83% along with 85% sensitivity and 81% specificity was achieved using cutoff threshold of 0.57, where all P̂ ≥ 0.57 would be considered “high spiking” while those below the cutoff would be considered “low spiking.”
Gender was not included as a covariate in the model due to the highly uneven distribution between female (n = 7) and male (n = 2) participants. Effects of region (based on electrode placement), subject age, and whether brain regions were also epileptogenic zones were tested and determined to have insignificant effects on the model overall (χ2 (3) = 1.09), and hence were removed from all analyses to reduce over fitting. Components playing a significant role (Wald p-values < 0.10) in discriminating high from low spiking samples were identified and key metabolites were identified based on which metabolite had the highest loading factor on each component, where the loading factor was at least 0.80. A confirmatory 10-fold cross validation analysis with 10 repetitions with was also carried out to assess for over-fitting.
To better understand the mechanisms behind the differences in metabolite expression, these metabolites of interest are used for additional correlational clustering studies in order to observe how changes in key metabolites can correspond to changes in gene expression and cell type distribution. Pearson correlations between the 990 differentially expressed genes (described above) across the 18 high and low spiking samples obtained from our 9 subjects and their corresponding mean metabolite concentrations for each sample were calculated13. Linkages between two genes were created when Pearson correlations were greater than or equal to 0.70 (r ≥ 0.70), a cutoff that was also consistent with an FDR of less than 0.0727. Clusters were further analyzed using a combination of ConsensusPathDB28 and primary literature searches for functional and pathway enrichment. A cutoff threshold of FDR adjusted p < .05 was used to identify potentially significant pathways associated with each metabolite-gene cluster. Additional associations between metabolites and putative cell types are explored using Pearson correlation based hierarchical clustering using the average linkage method. Metabolite-cell cluster relationships were further characterized using Pearson correlation values, where Pearson correlations ≥ 0.70 (FDR adjusted p < 0.05) were considered statistically significant.
Results
Neocortical samples obtained from precisely localized brain regions demonstrating high and low spiking were identified and subdivided for metabolomic, genomic, and histological analyses (Figure 1A). These samples were obtained from 9 patients who underwent surgery for refractory epilepsy (Table 1). Transcriptional and histological data on these tissues had been gathered in our previous study13. For HR-MAS 1H MRS, the acquired spectra were of high quality (Figure 1B) with mean LCModel estimated signal to noise ratio (SNR) of 34.3 ± 8.41 (mean ± s.d). There were 14 metabolites with mean CRLB less than or equal to 10%, which were included for further analysis. These metabolites were: choline, glycerophosphorylcholine (GPC), γ-aminobutyric acid (GABA), glutamine, glutamate, myo-inositol, lactate, N-acetylaspartate (NAA), N-acetylaspartylglutamate (NAAG), phosphorylcholine (PCh), phosphorylethanolamine (PE), taurine, simulated macromolecule at resonance position 1.40 ppm (MM140) and creatine plus phosphocreatine (Cr+PCr).
Mean values for the 14 metabolites in high spiking and low spiking tissue samples are summarized in Table 2. Statistically significant (p < 0.05) differences in metabolite levels between high and low spiking tissues were found in 4 of 14 metabolites, namely choline, phosphorylcholine (PCh), lactate, and phosphorylethanolamine (PE) (Table 2). All were decreased in high spiking tissue. Further analysis of all 14 metabolites using a combination principal components analysis and generalized estimating equation (GEE)29 based logistic regression yielded a short list of 8 potentially important metabolites. Of these, 6 were identified to make significant contributions (Wald p < 0.05) in differentiating high versus low spiking samples (Figure 2B and Table 3). These metabolites were: choline, creatine plus phosphocreatine (Cr+PCr), glycerophosphorylcholine (GPC), myo-inositol, lactate, and N-acetylaspartylglutamate (NAAG). Our logistic regression model performed well in discriminating high and low spiking tissue samples (Figure 2A) with a Receiver Operating Characteristic (ROC) area under the curve (AUC) of 0.88, 95% CI [0.81, 0.95] (Figure 2C). Our optimal accuracy in classifying high versus low spiking using these metabolites was 82% with a sensitivity of 85% and specificity 79%. Our confirmatory 10-fold cross validation analysis of our model yielded a mean AUC of 0.83, consistent with a model that is not over-fitted. In summary, this metabolomic signature presents a highly sensitive and specific new way to differentiate epileptic brain regions from their non-epileptic counterparts and provides a potential approach to non-invasively “visualize” epileptic brain regions clinically using 1H MRS.
Table 2. Univariate analysis of metabolites.
| Metabolite | Low Spiking Mean (SE) |
High Spiking Mean (SE) |
Percent Change | t-Test (p, 2-tailed) |
|
|---|---|---|---|---|---|
| * | Cho | 0.61 (0.04) | 0.50 (0.02) | -19% | .01 |
| Cr+PCr | 8.55 (0.24) | 8.36 (0.21) | -2% | .56 | |
| GABA | 3.13 (0.11) | 3.10 (0.09) | -1% | .79 | |
| Gln | 3.75 (0.25) | 3.53 (0.15) | -6% | .44 | |
| Glu | 7.03 (0.19) | 6.81 (0.21) | -3% | .44 | |
| GPC | 0.58 (0.03) | 0.58 (0.03) | 0% | .98 | |
| m-Ins | 5.11 (0.22) | 4.63 (0.15) | -9% | .07 | |
| *** | Lac | 15.06 (0.38) | 12.98 (0.48) | -14% | < 0.001 |
| MM140 | 28.80 (1.62) | 32.17 (1.42) | 12% | .12 | |
| NAA | 5.40 (0.20) | 5.21 (0.18) | -3% | .49 | |
| NAAG | 0.99 (0.03) | 1.00 (0.03) | 1% | .78 | |
| *** | PCh | 0.93 (0.04) | 0.78 (0.03) | -16% | < 0.01 |
| * | PE | 2.36 (0.09) | 2.11 (0.08) | -11% | .03 |
| Tau | 2.02 (0.09) | 2.03 (0.08) | 1% | .90 | |
Table 2. Means and standard errors (SE) of key metabolites measured from tissue samples of high and low spiking regions. Statistically significant differences were found between the high and low spiking tissue samples in 4 out of 14 metabolites (in bold) based on two-sample t-tests. All metabolites are normalized to the total fitted signals and expressed as percent total signal. Percent signal changes in high spiking tissue samples relative to low spiking tissue are also indicated for each metabolite. Cho = choline, Cr+PCr = creatine plus phosphocreatine, GABA = γ-aminobutyric acid, Gln = glutamine, Glu = glutamate, GPC = glycerophosphorylcholine, myo-Ins = myo-inositol, Lac = lactate, MM140 = macromolecules at 1.4 ppm, NAA = N-acetylaspartate, NAAG = N-acetylaspartylglutamic acid, PCh = phosphorylcholine, PE = phosphorylethanolamine, Tau = taurine.
p < .05,
p < .01,
p < .001, uncorrected for multiple comparisons
Figure 2. Logistic regression demonstrates a consistent metabolomic signature.
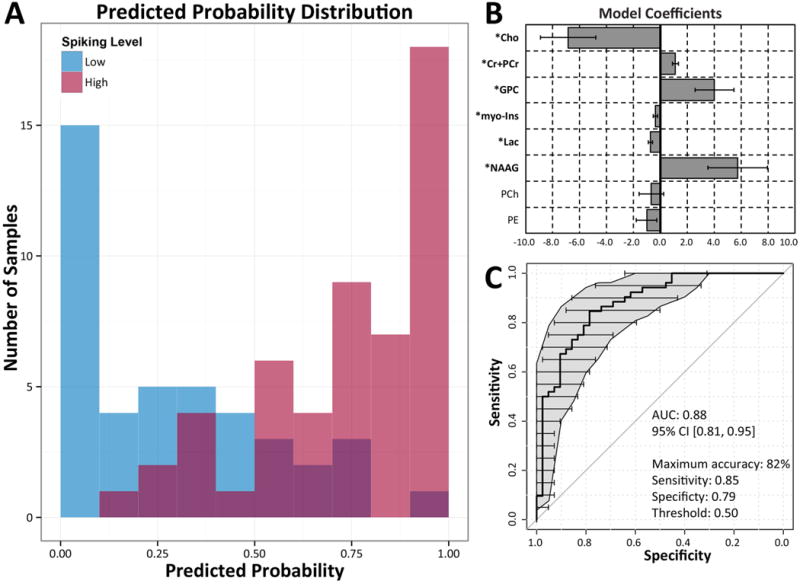
Logistic regression using 8 metabolites demonstrates high sensitivity and specificity in discriminating high spiking tissue samples from low spiking tissue samples. A) Distribution of high and low spiking tissue samples (red and blue shading, respectively) and their predicted probabilities of being high spiking are illustrated in the histogram. B) Model coefficients used for predicting high and low spiking tissue. Metabolites with significant (p < 0.05) contributions to the model are bolded. C) Overall model performance is characterized by a receiver operating characteristic (ROC) area under the curve (AUC) of 0.88, indicating excellent discrimination between high and low spiking tissues. Gray regions and horizontal error bars indicate the estimated 95% confidence intervals of sensitivity and specificity using non-parametric bootstrapping of 1000 samples.
Table 3. Metabolite based logistic regression.
| Metabolite | Estimate (SE) | SE | 95% CI for Odds Ratio | Adjusted OR | Wald p-value | |||
|---|---|---|---|---|---|---|---|---|
| OR | Lower | Upper | ||||||
| ** | (Intercept) | 0.41 (0.14) | 0.13 | 1.50 | 1.15 | 1.97 | 1.04 | < 0.01 |
| *** | Cho | -6.83 (2.06) | 1.83 | 1.08E-03 | 1.92E-05 | 6.06E-02 | 0.51 | < 0.001 |
| *** | Cr+PCr | 1.13 (0.22) | 0.34 | 3.09 | 2.02 | 4.72 | 1.12 | < 0.001 |
| ** | GPC | 4.01 (1.45) | 1.39 | 55.3 | 3.20 | 9.56E+02 | 1.49 | < 0.01 |
| * | myo-Ins | -0.37 (0.17) | 0.16 | 0.69 | 0.50 | 0.97 | 0.96 | .03 |
| *** | Lac | -0.74 (0.17) | 0.17 | 0.48 | 0.34 | 0.67 | 0.93 | < 0.001 |
| ** | NAAG | 5.75 (2.22) | 0.09 | 315 | 4.08 | 2.43E+04 | 1.78 | < 0.01 |
| PCh | -0.67 (0.91) | 2.09 | 0.51 | 0.09 | 3.06 | 0.94 | .46 | |
| PE | -1.02 (0.76) | 0.81 | 0.36 | 0.08 | 1.59 | 0.90 | .18 | |
Table 3. Logistic regression parameter estimates and odds ratios of metabolites included in the final predictive model. Significant metabolite predictors are indicated in bold and are in good agreement with results from the two-sample t-tests. Exponentiation of parameter estimates gives their corresponding odds ratios (OR). An OR greater than 1 indicates that for a given unit increase in the specified metabolite within a sample, there is also an increase in the probability of the tissue sample being high spiking. In contrast, OR less than 1 indicates decrease in the likelihood that a tissue sample is high spiking for a unit increase in the specified metabolite. Adjusted OR are odds ratios calculated for every 0.1 increment of a metabolite instead of the standard 1.0 increment used to calculated standard OR. It is important to note that the odds ratios are multiplied by for every unit increment in the predictor (i.e. metabolite levels) due to the exponential nature of logistic regression. For example, in the case of choline, a 0.20 unit increase in choline reduced the odds of the tissue being high spiking by a factor 0.26 (i.e. 0.51 × 0.51 = 0.26)
p < .05,
p < .01,
p < .001
In order to validate these findings and to better understand their significance, we performed an integrative analysis of metabolomic, transcriptional, and histological measures from each of the 18 brain samples. Two major clusters of gene-metabolite interactions emerged, centering on changes in energy state, with a down regulation of lactate and upregulation of Cr+PCr (Figure 3A). Down-regulated lactate clustered with a group of genes associated with G-protein coupled receptor signaling and angiogenesis pathways (FDR adjusted p < 0.01), specifically VEGFA, FLT1, RGS1, RGS2, RHOU, GNA13, and TFRC, all of which were up-regulated in high-spiking brain regions. Also notable from the cluster was the upregulation of multiple genes associated with ubiquitination, highly suggestive of the preferential involvement of the ubiquitin-proteasome pathway (UPP) in high spiking areas.
Figure 3. Correlational analysis between metabolites, genes, and histology suggest an alerted metabolic state in high spiking human brain.
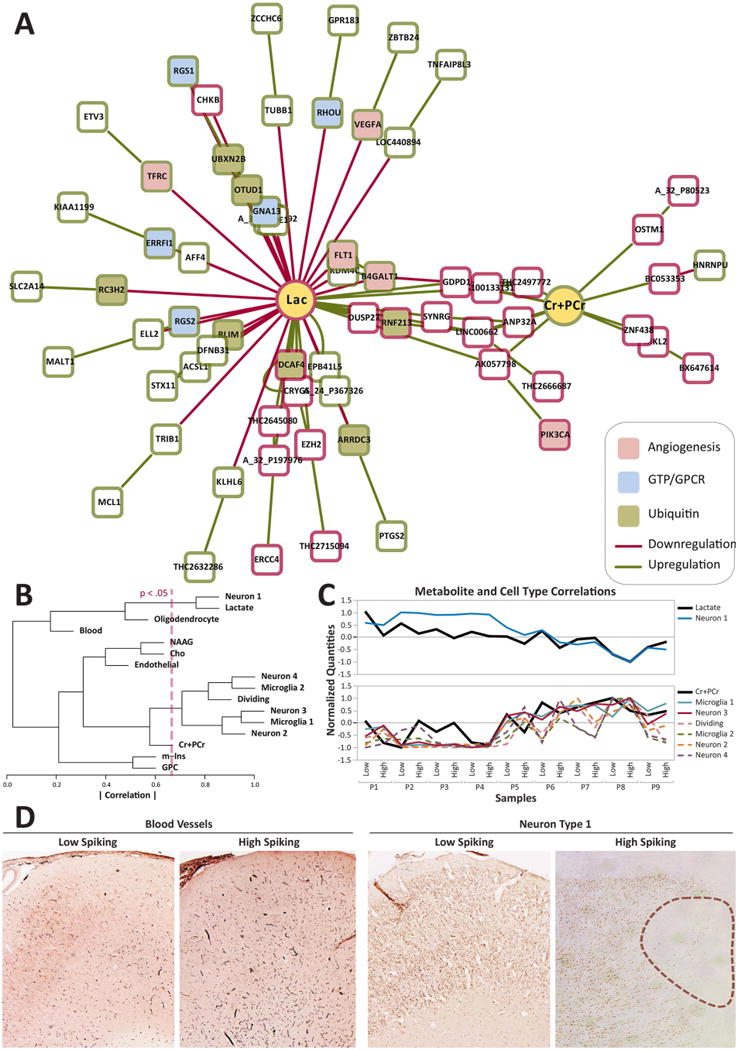
A) Correlational clustering between predictor metabolites and differentially expressed genes in high spiking tissue demonstrate a large cluster of up-regulated genes around lactate, a negative predictor of high spiking. Upregulated genes are indicated in green and downregulated genes are indicated in red. Pathway enrichment analyses indicate many of these differentially expressed genes to be involved in G-protein signaling, angiogenesis as well as ubiquitination. Relevant genes in the lactate cluster include: VEGFA, FLT1, RGS1, RGS2, RHOU, GNA13, PK3CA, and TFRC. B) Hierarchical clustering between metabolites and cell types show significant associations between lactate and Neuron Type 1 (Neuron 1) and also between Cr+PCr and Microglia Type 1 (Microglia 1). The red line indicates significance threshold of correlation, where paired relationships to the right of the line are significant. C) Normalized expression over all 18 samples between Lactate and Neuron Type 1 and between Cr+PCr and the cluster consisting of Microglia Type 1 and several other cell correlational clusters show stronger clustering between lactate, Cr+PCr and several cell types with the correlations between Microglia Type 1 and Neuron Type 3 being significant (solid lines). Other correlations in the same cluster, while not statistically significant, are also shown (dashed lines). D) Representative histology from Patient 1 demonstrating increased blood vessels and the presence of microlesions (outlined) showing reduced NeuN staining in high spiking tissue compared to its low spiking counterpart.
Using a larger set of brain samples, some of which were included in this study, we developed a novel clustering approach to predict cell-type specific changes in regions of high versus low epileptic spiking, creating what we called a “cellular interactome”13. In that study, we showed consistent differences in high spiking tissues that included increases in blood vessel density, inflammatory microglia, and millimeter-sized microlesions in deeper cortical layers. These microlesions contain a unique population of neurons with reduced NeuN staining (Type 1 Neurons) and increased microglia (Type 1 Microglia). While these cell populations do not correspond to any specific known cell type, they were previously identified in our microarray study by a statistical clustering technique, where these cell clusters exhibited a similar set of transcriptional features that corresponded to distinct histological differences13. Combining the cellular interactome with the present metabolomic dataset, we found significant correlations between reduced lactate and Neuron 1 (r = 0.76; p < 0.05) and increased Cr+PCr and Microglia 1 (r = 0.72; p < 0.05) (Figure 3B-C). Both cell populations correlated with the degree of epileptic activity and the number of microlesions13. Histological examination of the specific tissue samples used here consistently showed an increase of microlesions and increased vascular density in high versus low spiking regions (Figure 3D). Taken together, the combined genomic-metabolomic and cellular interactome suggest the existence of a unique metabolomic-genomic signature linked to altered tissue energy demand and consumption in high spiking brain tissue that could serve as a clinically translatable, non-invasive biomarker for the functional and structural abnormalities that underlie human neocortical epilepsy.
Discussion
The lack of non-invasive methods that can reliably detect and precisely localize epileptic brain regions has made epilepsy a challenging disease to diagnose and has hindered the development of novel therapeutics. Scalp EEG recordings are limited in their spatial resolution, and can only reliably detect epileptic discharges if they involve at least 10 cm2 of brain tissue30. Intracranial recordings are needed to increase spatial resolution. 1H MRS, with its ability to simultaneously measure multiple metabolites, offers a potentially powerful means to detect epileptic brain regions non-invasively. Previous 1H MRS epilepsy studies have focused on epilepsy associated differences in individual metabolites or the ratios of a few metabolites15,31, but have not precisely co-localized these changes with intracranial recordings. Here, by comparing human neocortical brain regions displaying high or low interictal activity from intracranial recordings, we found a highly consistent metabolomic-genomic profile with high predictive value. These differences suggest that high spiking regions of the neocortex have a unique metabolic and energetic signature that could be translated into a non-invasive 1H MRS approach to differentiate epileptic from normal brain regions clinically. Interestingly, whether a given brain region was also an epileptogenic zone did not affect the predictive ability of this model, suggesting that the metabolomic signature correlates mainly with interictal activity and is not altered by the presence or absence of an epileptogenic zone.
Here, we used a novel systems biology approach that linked human brain electrical activity to metabolomics, genomics, and cellular/histological changes. This offered a powerful, unbiased means to discover and simultaneously validate biomarkers of human epileptic brain activity while providing further insights into the underlying molecular and cellular abnormalities associated with epilepsy. Hierarchical clustering between metabolites and cellular transcriptional changes, showed a close parallel between the down-regulation of both lactate and Type 1 Neurons that we recently described and can be identified by reduced NeuN staining in high spiking regions of most patients with neocortical epilepsy13. Similarly, the upregulation of Cr+PCr correlated with an increase of other neurons and Microglia. Interestingly, microglia have also been shown to express high levels of PCr32, which may help explain Cr+PCr as a positive predictor of high spiking activity.
Exactly why lactate is consistently downregulated in high spiking brain regions is not clear. This may be due to unmet energy demands from the frequent interictal spiking leading to lactate consumption as an alternative energy source. Recent work has shown that inhibition of lactate dehydrogenase that coverts lactate to pyruvate can cause hyperpolarization in neurons and suppress epileptiform activity33, providing evidence for a possible role for lactate metabolism in epileptiform activity. The strong relationship between lactate and the cluster of genes enriched in angiogenesis, is further validated by histological evidence of blood vessel proliferation in high spiking samples that may be a compensatory response to higher energy demands13.
Our unique study design, using multiple paired samplings of both high and low spiking samples from within the same patient, attempts to reflect the differences in baseline metabolism in vivo. While post-resection tissue ischemia could contribute to our metabolite results, particularly by increasing lactate, our study design and analysis attempted to minimize these contributions by comparing samples that underwent the same post-resection processing. These results are not likely due to effects of tissue degradation since acetate levels, which can be expected to increase with tissue degradation34, remained minimal in our samples. Furthermore, re-analysis of our dataset excluding lactate as a predictor also yielded good discrimination between high and low spiking tissue samples. This allows us to be more confident that high and low spiking tissue exhibit different metabolic characteristics. Nevertheless, we cannot rule out the possibility that post-resection metabolism differences contributed to reduced lactate in the high spiking areas.
While numerous studies of human epilepsy using MRS have also implicated neuronal, mitochondrial, metabolic, and energetic derangements, which frequently extended beyond the regions of identified seizure onset 15,35, none of these have quantitatively correlated these findings with electrophysiology with high spatial resolution. Our study is the first of its kind to use a multi-modal approach to assess metabolic features in human epileptic neocortex using HR-MAS 1H MRS and incorporate complementary genetic and histological approaches to further inform our findings. Further validation of our studies using large and independent datasets would be insightful, particularly for those that also seek to identify the metabolic features of seizure onset regions as well as interictal spiking regions with an in vivo approach. Further advancements in equipment technology and spectral editing techniques may be needed to translate these ex vivo findings into in vivo epilepsy biomarkers.
A key feature of these epileptic brain regions, based on our recent findings, is that they contain microlesions that show dramatic, focal reductions in synaptic connectivity13. Consistently, the involvement of both choline and GPC in our predictive model suggests heightened cell membrane turnover in high spiking tissue. GPC and free fatty acids are key breakdown products of phosphatidylcholine36, a major membrane constituent. This breakdown process can be initiated under hypoxic conditions with the calcium-dependent activation of phospholipase A2 (PLA2), which is also responsible for the release of arachidonic acid, a potent inflammatory intermediate37. Furthermore, in recurrent seizures, cortical oxygenation level has been shown to be inversely related to free fatty acid release38. Choline, involved in membrane turnover, is active both as a membrane precursor and as a membrane breakdown product from GPC degradation. Given the comparative lack of free choline despite a relative increase in GPC in high spiking tissue, we suspect a re-distribution of choline into various anabolic pathways, such as regeneration of phosphatidylcholine for new membrane synthesis or acetylcholine synthesis. Elevated acetylcholine receptor activation is known to increase seizure potential39. In fact, pilocarpine, an acetylcholine receptor agonist, is a relatively common agent used to generate seizures in animal models40,41.
In summary, we have found a unique high resolution metabolite profile that can predict accurately whether or not a given tissue sample is high or low spiking and suggests a consistently altered metabolic state in epileptic human brain regions that show clear pathological and transcriptional differences. With further technical refinement using advanced MRS sequences that are capable of high resolution whole brain acquisition, and higher field strength to detect larger numbers of metabolites42, the measurement of these metabolites could be adapted for clinical use and become an invaluable tool in our efforts to better understand, localize, and treat epilepsy and the underlying processes leading to the disorder.
KEY POINTS.
Proton magnetic resonance spectroscopy (1H MRS) is able to identify a characteristic metabolite profile associated with the human epileptic neocortex
This characteristic epileptic metabolite profile can be useful as a non-invasive biomarker for the detection epileptogenic regions in the brain
Transcriptional and histological correlates of metabolite changes implicate a baseline neurovascular imbalance between energy demand and energy supply in epileptic neocortex
High resolution whole brain 1H MRS approaches in vivo could be a powerful way to localize epileptogenic regions in patients based on spatial metabolite profiles.
Acknowledgments
This work was funded by NIH/NINDS Grants R56 NS083527, R01NS045207, and R01NS058802 (JAL).
Footnotes
AUTHOR CONTRIBUTIONS: JAL is the corresponding author, conceived of and directed the project; HCW and FD performed data analysis with help from DF, JAS, and FG; MPG, SB, and FG performed the experiments; HCW and JAL wrote the manuscript with all other authors providing important comments.
CONFLICTS OF INTEREST: None of the authors has any conflict of interest to disclose.
ETHICAL PUBLICATION STATEMENT: We confirm that we have read the Journal's position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
References
- 1.Kwan P, Brodie MJ. Early identification of refractory epilepsy. New Engl J Med. 2000;342:314–319. doi: 10.1056/NEJM200002033420503. [DOI] [PubMed] [Google Scholar]
- 2.Engel J. Update on surgical treatment of the epilepsies. Summary of the Second International Palm Desert Conference on the Surgical Treatment of the Epilepsies (1992) Neurology. 1993;43:1612–1617. doi: 10.1212/wnl.43.8.1612. [DOI] [PubMed] [Google Scholar]
- 3.Mikuni N, Ikeda A, Takahashi JA, et al. A step-by-step resection guided by electrocorticography for nonmalignant brain tumors associated with long-term intractable epilepsy. Epilepsy Behav. 2006;8:560–564. doi: 10.1016/j.yebeh.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Mittal S, Barkmeier D, Hua J, et al. Intracranial EEG analysis in tumor-related epilepsy: Evidence of distant epileptic abnormalities. Clinical Neurophysiology. 2016;127:238–244. doi: 10.1016/j.clinph.2015.06.028. [DOI] [PubMed] [Google Scholar]
- 5.Gotman J. Relationships between interictal spiking and seizures: human and experimental evidence. Can J Neurol Sci. 1991;18:573–576. doi: 10.1017/s031716710003273x. [DOI] [PubMed] [Google Scholar]
- 6.Asano E, Muzik O, Shah A, et al. Quantitative interictal subdural EEG analyses in children with neocortical epilepsy. Epilepsia. 2003;44:425–434. doi: 10.1046/j.1528-1157.2003.38902.x. [DOI] [PubMed] [Google Scholar]
- 7.Bautista RE, Cobbs MA, Spencer DD, et al. Prediction of surgical outcome by interictal epileptiform abnormalities during intracranial EEG monitoring in patients with extrahippocampal seizures. Epilepsia. 1999;40:880–890. doi: 10.1111/j.1528-1157.1999.tb00794.x. [DOI] [PubMed] [Google Scholar]
- 8.White A, Williams PA, Hellier JL, et al. EEG spike activity precedes epilepsy after kainate-induced status epilepticus. Epilepsia. 2010;51:371–383. doi: 10.1111/j.1528-1167.2009.02339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loeb JA. Identifying targets for preventing epilepsy using systems biology. Neurosci Lett. 2011;497:205–212. doi: 10.1016/j.neulet.2011.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rakhade SN, Yao B, Ahmed S, et al. A common pattern of persistent gene activation in human neocortical epileptic foci. Ann Neurol. 2005;58:736–747. doi: 10.1002/ana.20633. [DOI] [PubMed] [Google Scholar]
- 11.Beaumont TL, Yao B, Shah A, et al. Layer-Specific CREB Target Gene Induction in Human Neocortical Epilepsy. J Neurosci. 2012;32:14389–14401. doi: 10.1523/JNEUROSCI.3408-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lipovich L, Dachet F, Cai J, et al. Activity-dependent human brain coding/noncoding gene regulatory networks. Genetics. 2012;192:1133–1148. doi: 10.1534/genetics.112.145128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dachet F, Bagla S, Keren-Aviram G, et al. Predicting novel histopathological microlesions in human epileptic brain through transcriptional clustering. Brain. 2015;138:356–370. doi: 10.1093/brain/awu350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peeling J, Sutherland G. 1H magnetic resonance spectroscopy of extracts of human epileptic neocortex and hippocampus. Neurology. 1993;43:589–594. doi: 10.1212/wnl.43.3_part_1.589. [DOI] [PubMed] [Google Scholar]
- 15.Pan JW, Kuzniecky RI. Utility of magnetic resonance spectroscopic imaging for human epilepsy. Quant Imaging Med Surg. 2015;5:313–322. doi: 10.3978/j.issn.2223-4292.2015.01.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loeb JA. A human systems biology approach to discover new drug targets in epilepsy. Epilepsia. 2010;51(Suppl 3):171–177. doi: 10.1111/j.1528-1167.2010.02635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng LL, Ma MJ, Becerra L, et al. Quantitative neuropathology by high resolution magic angle spinning proton magnetic resonance spectroscopy. Proc Natl Acad Sci USA. 1997;94:6408–6413. doi: 10.1073/pnas.94.12.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beckonert O, Coen M, Keun HC, et al. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat Protoc. 2010;5:1019–1032. doi: 10.1038/nprot.2010.45. [DOI] [PubMed] [Google Scholar]
- 19.Ghoddoussi F, Galloway MP, Jambekar A, et al. Methionine sulfoximine, an inhibitor of glutamine synthetase, lowers brain glutamine and glutamate in a mouse model of ALS. J Neurol Sci. 2010;290:41–47. doi: 10.1016/j.jns.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 20.Cheng LL, Lean CL, Bogdanova A, et al. Enhanced resolution of proton NMR spectra of malignant lymph nodes using magic-angle spinning. Magn Reson Med. 1996;36:653–658. doi: 10.1002/mrm.1910360502. [DOI] [PubMed] [Google Scholar]
- 21.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30:672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 22.Klunk WE, Xu CJ, Panchalingam K, et al. Analysis of magnetic resonance spectra by mole percent: comparison to absolute units. Neurobiol Aging. 1994;15:133–140. doi: 10.1016/0197-4580(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 23.Højsgaard S, Halekoh U, Yan J. The R Package geepack for Generalized Estimating Equations. J Stat Softw. 2005;15:1–11. [Google Scholar]
- 24.Team RC. Book R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2014. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 25.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dziuban CD, Shirkey EC. When is a correlation matrix appropriate for factor analysis? Some decision rules. Psychol Bull. 1974;81:358–361. [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995:289–300. [Google Scholar]
- 28.Kamburov A, Stelzl U, Lehrach H, et al. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013;41:D793–800. doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang K-Y, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 30.Tao JX, Ray A, Hawes-Ebersole S, et al. Intracranial EEG substrates of scalp EEG interictal spikes. Epilepsia. 2005;46:669–676. doi: 10.1111/j.1528-1167.2005.11404.x. [DOI] [PubMed] [Google Scholar]
- 31.Hammen T, Stefan H, Eberhardt KE, et al. Clinical applications of 1H-MR spectroscopy in the evaluation of epilepsies - What do pathological spectra stand for with regard to current results and what answers do they give to common clinical questions concerning the treatment of epilepsies? Acta Neurol Scand. 2003;108:223–238. doi: 10.1034/j.1600-0404.2003.00152.x. [DOI] [PubMed] [Google Scholar]
- 32.de Gannes FM, Merle M, Canioni P, et al. Metabolic and cellular characterization of immortalized human microglial cells under heat stress. Neurochem Int. 1998;33:61–73. [PubMed] [Google Scholar]
- 33.Sada N, Lee S, Katsu T, et al. Targeting LDH enzymes with a stiripentol analog to treat epilepsy. Science. 2015 doi: 10.1126/science.aaa1299. [DOI] [PubMed] [Google Scholar]
- 34.Opstad KS, Bell BA, Griffiths JR, et al. An assessment of the effects of sample ischaemia and spinning time on the metabolic profile of brain tumour biopsy specimens as determined by high-resolution magic angle spinning 1H NMR. NMR Biomed. 2008;21:1138–1147. doi: 10.1002/nbm.1296. [DOI] [PubMed] [Google Scholar]
- 35.Park EJ, Otaduy MCG, Lyra KPd, et al. Extratemporal abnormalities in phosphorus magnetic resonance spectroscopy of patients with mesial temporal sclerosis. Arq Neuropsiquiatr. 2016;74:93–98. doi: 10.1590/0004-282X20160005. [DOI] [PubMed] [Google Scholar]
- 36.Podo F. Tumour phospholipid metabolism. NMR Biomed. 1999;12:413–439. doi: 10.1002/(sici)1099-1492(199911)12:7<413::aid-nbm587>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 37.Farooqui AA, Yang HC, Rosenberger TA, et al. Phospholipase A2 and Its Role in Brain Tissue. J Neurochem. 2002;69:889–901. doi: 10.1046/j.1471-4159.1997.69030889.x. [DOI] [PubMed] [Google Scholar]
- 38.Visioli F, Rihn LL, Rodriguez de Turco EB, et al. Free fatty acid and diacylglycerol accumulation in the rat brain during recurrent seizures is related to cortical oxygenation. J Neurochem. 1993;61:1835–1842. doi: 10.1111/j.1471-4159.1993.tb09824.x. [DOI] [PubMed] [Google Scholar]
- 39.Priel MR, Albuquerque EX. Short-Term Effects of Pilocarpine on Rat Hippocampal Neurons in Culture. Epilepsia. 2002;43:40–46. doi: 10.1046/j.1528-1157.43.s.5.18.x. [DOI] [PubMed] [Google Scholar]
- 40.Cavalheiro EA, Leite JP, Bortolotto ZA, et al. Long-term effects of pilocarpine in rats: structural damage of the brain triggers kindling and spontaneous recurrent seizures. Epilepsia. 1991;32:778–782. doi: 10.1111/j.1528-1157.1991.tb05533.x. [DOI] [PubMed] [Google Scholar]
- 41.Pitkänen A, Schwartzkroin PA, Moshé SL. Models of seizures and epilepsy. Elsevier Academic; Oxford: 2005. [Google Scholar]
- 42.Chadzynski GL, Pohmann R, Shajan G, et al. In vivo proton magnetic resonance spectroscopic imaging of the healthy human brain at 9.4 T: initial experience. MAGMA. 2015;28:239–249. doi: 10.1007/s10334-014-0460-5. [DOI] [PubMed] [Google Scholar]


