Abstract
Drugs with ideal pharmacokinetic profile require long half-life but little organ accumulation. Generally, PK and organ accumulation are contradictory factors: smaller size leads to faster excretion and shorter half-lives and thus a lower tendency to reach targets; larger size leads to longer circulation but stronger organ accumulation that leads to toxicity. Organ accumulation has been reported to be size dependent due in large part to engulfing by macrophages. However, publications on the size effect are inconsistent because of complication by the effect of shape that varies from nanoparticle to nanoparticle. Unique to RNA nanotechnology, size could be tuned without a change in shape, resulting in a true size comparison. Here we investigated size effects using RNA squares of identical shape but varying size and shape effects using RNA triangles, squares, and pentagons of identical size but varying shape. We found that circulation time increased with increasing RNA nanoparticle size from 5–25 nm, which is the common size range of therapeutic RNA nanoparticles. Most particles were cleared from the body within 2 hr after systemic injection. Undetectable organ accumulation was found at any time for 5 nm particles. For 20 nm particles, weak signal was found after 24 hr, while accumulation in tumor was strongest during the entire study.
Keywords: RNA nanoparticles, pRNA 3WJ motif, nanobiotechnology, phi29 DNA packaging motor, phi29 pRNA, RNA Nanotechnology, RNA nanostructure, viral DNA packaging, motor pRNA, bacteriophage phi29
Jasinski et al. explore the effects of RNA nanoparticle size and shape on biodistribution using RNA polygons. Animal imaging demonstrated a strong correlation between increased size and increased circulation time. RNA nanoparticle size and shape are precisely controlled, so different-size nanoparticles with identical shape could be studied.
Introduction
The past decades have seen the emergence of nanotechnology as the next potential solution for advancing the field of medicine.1, 2 Diverse types of nanoparticle platforms have been developed, including but not limited to polymer-based nanoparticles,3, 4, 5 dendrimer-based nanoparticles,6, 7, 8 lipid-based nanoparticles,9, 10, 11 metal-based nanoparticles (such as gold,12, 13, 14 silver,15, 16, 17 and iron18, 19, 20), and nucleic acid-based nanoparticles.21, 22, 23, 24 Nanotechnology displays distinct advantages over traditional treatments because of the increased circulation of nanoparticles over their small-molecule counterparts.25, 26 When combined with targeting ligands, peptides, or aptamers, nanoparticles possess the potential for cell-specific drug delivery.27, 28 The enhanced permeability and retention (EPR) effect allows even non-targeted nanoparticles to accumulate in tumor tissue because of the large amount of leaky vascularization at the tumor site.29 It has been reported that accumulation of nanoparticles into tumors is based on the EPR. This is true in preclinical animal models, but data on the EPR effect in human clinical trials are limited, and the observation is controversial, pending further investigation. However, many nanoparticles, although effectively targeting tumor and cancerous cells, accumulate non-specifically in healthy tissues, causing toxicity and side effects.30 Non-biocompatible materials cause toxicity because they are never fully excreted from the body.
Size and shape have been shown to greatly affect the pharmacokinetics and biodistribution profiles of nanoparticles.31, 32, 33 Small particles (<5 nm) are filtered by the kidneys and excreted in the urine.34 Larger particles (20–100 nm) are engulfed by macrophages or sequestered in healthy tissue, reducing the rate of successful navigation to the desired target.35 Large, rigid particles that get stuck in healthy organs have even more difficulty escaping, remaining trapped.
RNA nanotechnology, in which the scaffold and ligands can be composed solely of RNA, is an emerging technology that has shown promise to overcome several current challenges in nanotechnology.36, 37, 38, 39, 40, 41, 42 The concept of RNA nanotechnology was first shown in 199821 and since then has been expanding rapidly.36, 43, 44, 45 RNA nanoparticles can be engineered to diverse size and shape while retaining the high thermostability necessary for in vivo applications.46, 47, 48, 49, 50 Simple sequence engineering can be applied to use an identical RNA motif for the construction of multiple RNA nanoparticles, reducing the number of RNA oligomers needed for the construction of myriad RNA nanoparticles.51, 52, 53 Furthermore, modifications to the ribose sugar (such as 2′-fluorine, 2′-O-methyl, 2′-amine, or locked nucleic acid [LNA] modification54) of RNA bases confers stability in serum for more than 24 hr.55, 56 The unique base-pairing mechanism that occurs during the assembly of RNA nanoparticles leads to consistent assembly properties, resulting in defined-size nanoparticles. Thus, a particular RNA nanoparticle will display homogeneity from batch to batch of assembly. In order to optimize drug delivery, it is important to have nanoparticles that are consistent in assembly and will display a narrow size distribution, as this will lead to consistent pharmacokinetics and biodistribution in vivo.57 Furthermore, RNA is a flexible biomaterial by nature, therefore allowing some structural deformity, unlike many rigid nanoparticles under development.58, 59
The pRNA three-way junction (3WJ) derived from the packaging RNA (pRNA) of bacteriophage phi29 has recently been shown to display high thermostability37, 60 and fast kinetic assembly61 and has been successfully used in vivo for microRNA (miRNA) and small interfering RNA (siRNA) delivery and tumor growth suppression.62, 63, 64 The 3WJ is an RNA motif that has been extensively used for drug delivery and cancer treatment.65, 66, 67 It is an RNA nanoparticle composed of three component strands: 3WJ-a, 3WJ-b, and 3WJ-c.37 These three strands interact to form a unique 3WJ motif. This 3WJ motif was used as a module for construction of the RNA polygons used in this study.46, 68, 69 The addition of aptamers and targeting ligands allow the 3WJ to successfully enter tumor cells specifically. Interestingly, 3WJ particles have repeatedly displayed little to no accumulation in healthy tissue, including the liver, kidneys, lung, and spleen.37, 62, 63 Despite the small size of the 3WJ (<5 nm), total clearance by the kidneys does not occur, while accumulation in tumor tissue is still observed.62, 63, 64
In this study, we investigated the effects of RNA nanoparticle size and shape on biodistribution using previously constructed and characterized RNA triangles, squares, and pentagons.46, 68, 69 Analysis by fluorescence imaging of whole mice and their organs demonstrated a strong correlation between increased size and increased circulation time using RNA nanosquares. Unlike many nanoparticle systems, with which it is hard to study size and shape independently, RNA nanoparticle size and shape are precisely controlled from nanoparticle conception. Therefore, different-size nanoparticles that have identical shape could be studied. This study demonstrates the effect RNA nanoparticle size and shape have on their biodistribution and elimination pathways.
Results
Design and Assembly of RNA Polygons with Variable Size and Shape
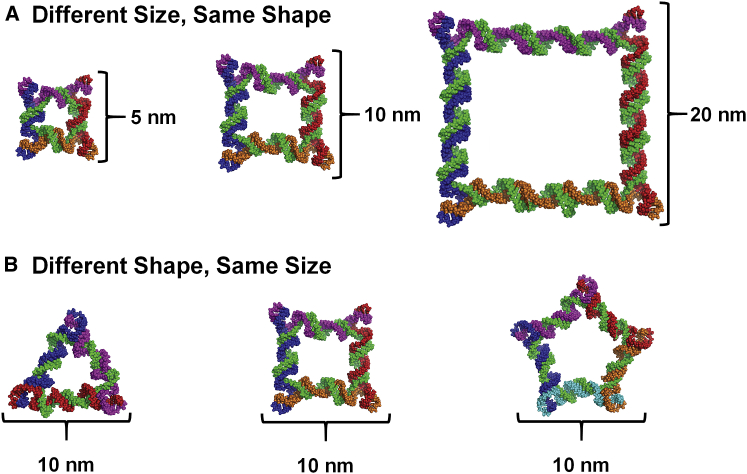
In vivo biodistribution studies of RNA nanoparticles were carried out using previously designed, constructed, and characterized RNA polygons.46, 69 The nanosquares tested were designed to be 5, 10, and 20 nm along each edge (Figure 1A), while RNA triangle, square, and pentagons were each designed to be 10 nm along each edge (Figure 1B). Throughout this report, nanosquares are referred to by their designed edge length (5, 10, or 20 nm). As previously described, RNA polygon size is determined by selecting the number of base pairs between each 3WJ at each vertex, and the shape is determined by selecting the number of total 3WJs in the particle. Each polygon is constructed from multiple single-stranded (ss) RNA oligonucleotides: short external strands (one along each edge) and one long core strand connecting each external strand (Figure 1). RNA strands were chemically synthesized with fluorine modifications at the 2′ sugar position (2′F) of cytosine and uracil, which confers enzymatic and thermodynamic stability.55 Polygons were assembled by mixing equimolar amounts of each strand in buffer, followed by thermal denaturation at 85°C and slow cooling to 4°C over 1 hr.
Figure 1.
RNA Nanoparticle Design
(A) Structures of the 5, 10, and 20 nm 2′F nanosquares. (B) Structures of 10 nm RNA triangle, square, and pentagon.
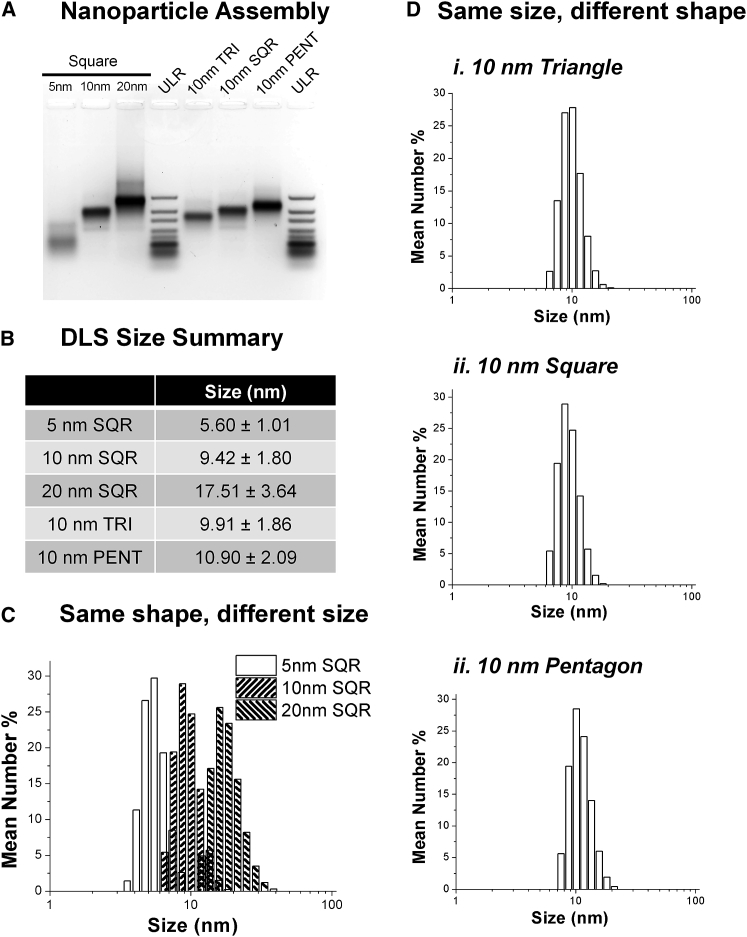
Assembly of the particles was analyzed by visualization of total RNA using agarose gel electrophoresis (Figure 2A). Gel analysis indicates successful formation of RNA nanoparticles, as distinct RNA bands are seen for each nanoparticle. Further size characterization was performed using dynamic light scattering (DLS) (Figures 2B–2D). Sizes for the small, medium, and large nanosquares were found to be 5.60 ± 1.01, 9.42 ± 1.80, and 17.51 ± 3.64 nm, respectively, and 9.91 ± 1.86 and 10.90 ± 2.09 nm for the triangle and pentagon, respectively.
Figure 2.
Physical Properties of RNA Nanoparticles
(A) Assembly of 2′F RNA nanoparticles. (B) DLS size summary of RNA nanoparticles. (C) DLS graphs of same-shape, different-size RNA nanosquares. (D) DLS graphs of same-size, different-shape RNA polygons. PENT, pentagon; SQR, square; TRI, triangle; ULR.
Labeling only one of five strands with Alexa Fluor 647 was used to confirm that the particles were not dissociating in vivo (Figures 3, 4, and 5). It has been shown previously that ssRNA is eliminated rapidly through the kidneys after intravenous (i.v.) injection.70, 71 Therefore, if the particles were indeed dissociating, signal would have been lost within 5 min of polygon injection.
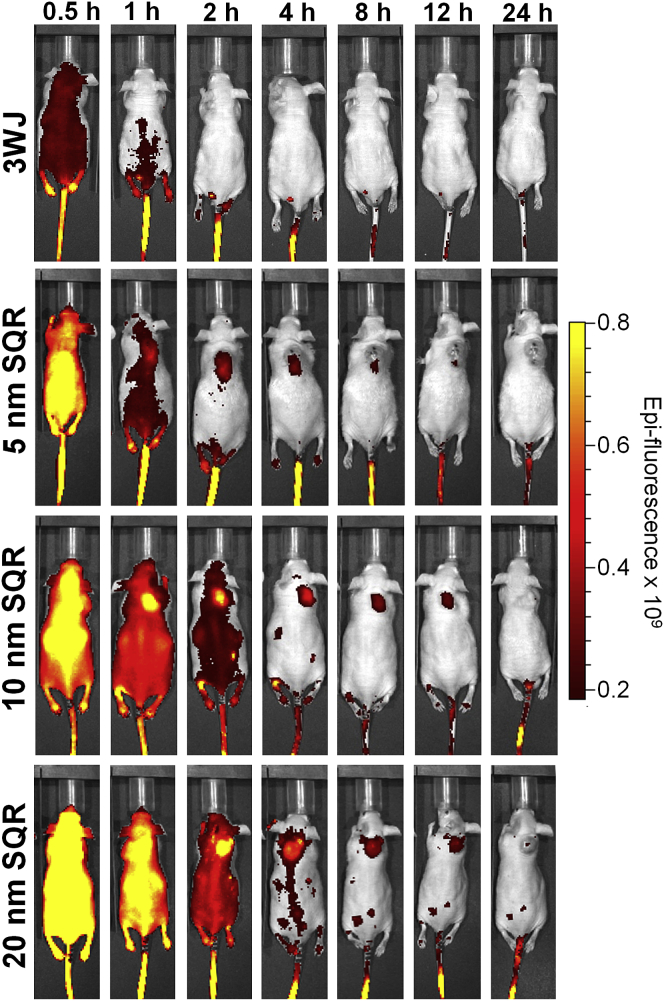
Figure 3.
In Vivo Biodistribution of Different-Size RNA Nanoparticles
Time-course fluorescence images of 5, 10, and 20 nm 2′F nanosquares. SQR, square.
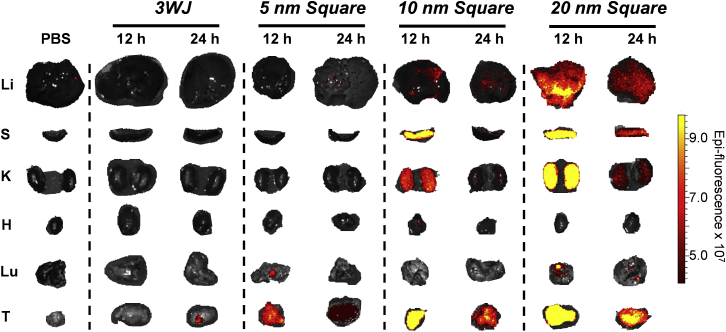
Figure 4.
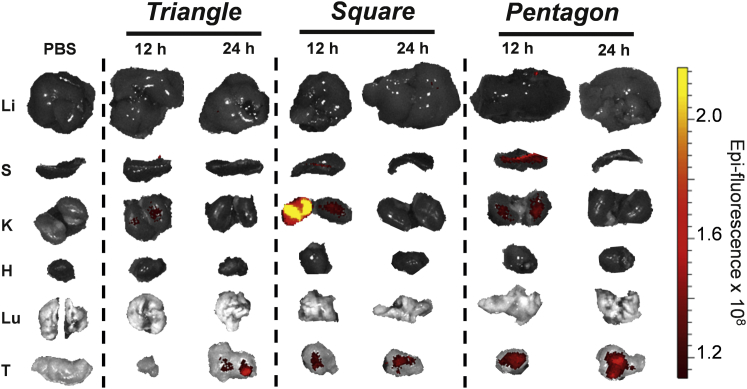
Organ Images of 5, 10, and 20 nm 2′F Nanosquares
Fluorescent organ images of diverse-size RNA nanosquares after 12 and 24 hr of circulation. H, heart; K, kidneys; Li, liver; Lu, lung; S, spleen; T, tumors.
Figure 5.
In Vivo Biodistribution of Different-Shape RNA Nanoparticles
Time-course fluorescence images of RNA triangle, square, and pentagon. H, heart; K, kidneys; Li, liver; Lu, lung; S, spleen; T, tumors.
In Vivo Biodistribution of RNA Polygons with Variable Sizes
Subcutaneous xenograft tumors were developed in male nude mice with KB cells. After tumors were fully developed (∼2 weeks), 100 μL of 15 μM nanosquares harboring chemically conjugated Alexa Fluor 647 were injected i.v. via the tail vein. It is important to note that the same volume of the same concentration nanoparticles of variable sizes was injected, thus resulting in the same number of particles injected per sample. Additionally, single-fluorophore labeling of each particle among all size particles further demonstrates comparable in vivo results among all nanoparticles. Whole-body fluorescent images were taken at 0.5, 1, 2, 4, 8, 12, and 24 hr (Figure 3). A diverse set of biodistribution profiles is seen when comparing different size 2′F nanosquares. 20 nm nanosquare fluorescence is observed throughout the mice at 4 hr, while both the 5 and 10 nm nanosquares are sequestered mainly in the tumor. As seen previously, the 3WJ nanoparticle shows no accumulation in vital organs in mice.37, 62, 63, 64
The same trend is seen from the organ images as from whole-body scans of the mice. At 12 and 24 hr, mice were sacrificed, and their liver, spleen, kidneys, heart, lung, and tumors were collected (Figure 4). The 5 nm nanosquare is eliminated quickly from vital organs, localizing in only the tumor after 12 hr. For the 10 and 20 nm nanosquares, fluorescence is much more intense in organs, especially after 12 hr. However, the 10 nm nanosquares localize only to the tumor after 24 hr. Some fluorescence is still seen in the liver and kidneys of mice injected with 20 nm nanosquares after 24 hr, but we expect that at longer time points, the nanosquares would circulate out of all healthy organs.
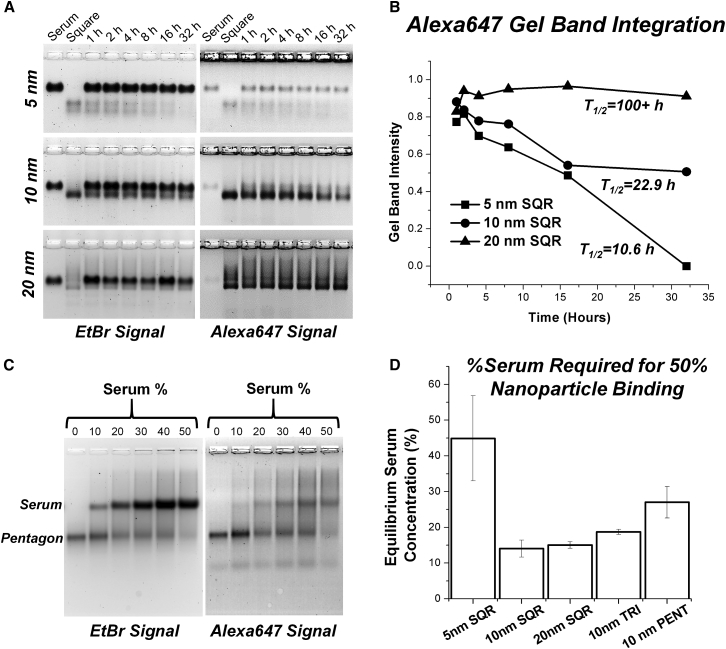
Serum testing was performed on the 2′F RNA nanoparticles (Figure 6). Nanosquares labeled with Alexa Fluor 647 were incubated at 37°C in 10% fetal bovine serum (FBS) solution, and time points were taken by immediately freezing the samples on dry ice to stop enzymatic degradation. After incubation, nanosquares were analyzed using 2% agarose gels scanned for Alexa Fluor 647 fluorescent signal and ethidium bromide signal (Figure 6A). ImageJ was used to quantify gel band intensity, and values were plotted (Figure 6B). Gel band intensity was normalized to the initial band intensity at time zero. The plots were fit to exponential decay curves, and half-lives (T1/2) were calculated to be 10.6, 22.9, and >100 hr for the 5, 10, and 20 nm squares, respectively. It is important to note that the half-lives of each nanoparticle are far greater than their apparent fluorescent half-lives seen during in vivo biodistribution experiments.
Figure 6.
Serum Assays
(A) Gel images of serum degradation of 5, 10, and 20 nm 2′F nanosquares. Different sizes of squares result in different intensities seen by gel band visualization. (B) Plots of gel band intensity from serum degradation gels of 5, 10, and 20 nm 2′F nanosquares. (C) Representative gel of serum binding experiments. (D) Equilibrium serum binding concentrations for 2′F RNA nanoparticles. Error bars indicate SD from three independent experiments. PENT, pentagon; SQR, square; TRI, triangle.
Serum binding to the nanoparticles was tested by titrating the percentage of serum incubation to determine an equilibrium serum concentration (i.e., the percentage of serum solution at which 50% of the particle is bound by serum). A gel shift assay was used to determine serum binding to the particles, and a representative gel image is shown in Figure 6C. Alexa 647-labeled 2′F RNA nanoparticles were incubated in 0%, 10%, 20%, 30%, 40%, and 50% serum for 5 min and then directly loaded on agarose gels for analysis. Both a decrease in EtBr signal of the lower nanoparticle-only band and an increase in the Alexa 647 signal of the upper nanoparticle-serum band indicate nanoparticle binding with serum. Interestingly, different equilibrium serum concentrations are seen among the nanoparticles (Figure 6D). These observed differences in serum binding also contribute to differences seen in the biodistribution of the different size and shape nanoparticles.
In Vivo Biodistribution of RNA Polygons with Variable Shapes
Next, we investigated the role shape plays in biodistribution profiles of RNA nanoparticles using previously constructed RNA polygons: triangle, square, and pentagon. The shape of RNA nanoparticles is easily controlled by modulating the inner angle of the 3WJ motif, which was used as the module for polygon construction. The inner angle is increased by extending the core strand by 22 nt, corresponding to two helix turns of A-type RNA duplex, and adding one external strand.46 RNA polygons were assembled using the same method as the nanosquares mentioned above. 2′F-modified polygons were used for in vivo biodistribution analysis.
Subcutaneous xenograft tumors were developed in male nude mice with KB cells. After tumors were fully developed (∼2 weeks), 100 μL of 15 μM 2′F polygons harboring chemically conjugated Alexa Fluor 647 were injected i.v. via the tail vein. After 12 and 24 hr, mice were sacrificed, and their organs were collected and imaged for Alexa Fluor 647 signal (Figure 5).
It is evident that shape plays some part in the excretion routes and times of 2′F nanoparticles. Triangular nanoparticles appear to clear the fastest, showing only low fluorescent signal in the kidneys after 12 hr and only fluorescence in the tumor after 24 hr. Square-shaped nanoparticles show high fluorescent signal in the kidneys and low fluorescent signal in the spleen after 12 hr. However, after 24 hr, fluorescent signal remains only in the tumor. Pentagon-shaped nanoparticles show low signal in the kidneys after 12 hr, with higher fluorescence seen in the spleen. Importantly, pentagonal along with triangular and square nanoparticles only show fluorescence in the tumors after 24 hr of circulation.
Discussion
Effect of Size on RNA Nanoparticle Biodistribution
In cancer treatment, nanoparticles accumulate at solid tumors by taking advantage of the EPR effect. However, EPR is principally affected by the circulation time of nanoparticles.29 Larger nanoparticles generally show much higher circulation times, allowing them to capitalize on the EPR effect. Therefore, nanoparticle size is an important factor for nanoparticles that rely on the EPR effect to localize at tumors.
RNA nanoparticle size is easily controlled, and the assembly process is highly reproducible and homogeneous, resulting in narrow size distributions and batch-to-batch reproducibility. This is an advantage of RNA nanoparticles over other systems, whose assembly processes are less controlled. Demonstrated by in vivo biodistribution results, the size of RNA nanoparticles greatly affects circulation time and accumulation in healthy organs and tumors. A strong correlation between increased nanosquare size and increased circulation time is seen, indicated by higher fluorescent signal in whole-body and organ images of mice (Figures 3 and 4). There are numerous reasons for these observations. Particles less than 15 nm, such as the 5 nm nanosquare, are excreted primarily through the kidneys, a quicker route of elimination compared with liver excretion.34 Larger particles up to 100 nm, such as the 10 and 20 nm nanosquares, have more interaction with macrophages and are excreted more slowly through the liver.35 Another reason for different observed rates of RNA nanoparticle clearance is protein binding to the nanosquares, referred to as the protein corona.72 Differences in nanoparticle size have been shown to dictate the amount and identity of proteins bound to nanoparticles, affecting their circulation time and excretion pathways.57 It is possible that differences in protein binding to the nanosquares greatly affect their elimination routes and therefore how long they stay in circulation. Additionally, increasing nanoparticle size increases its hydrodynamic viscosity. Thus, increased interactions of the larger squares with its surrounding environment (i.e., water) will slow down its diffusion, contributing to the increase in circulation time of larger particles.
Because of the high-fidelity assembly of RNA nanoparticles, they have a high likelihood to display consistent and controllable in vivo properties. Therefore, the same nanoparticle system can be used, easing nanoparticle preparation strategies while displaying diverse but controllable in vivo properties. On the basis of the trend found in this study, it is possible that the construction of larger RNA nanoparticles would extend their circulation times even further.
The biodistribution is the consequence resulting from two factors. Small RNA nanoparticles, from one side, are less prone than the larger particles to be captured by macrophages of the reticulo-endothelial system. On the other hand, small nanoparticles are more rapidly eliminated by the kidneys, as is the opposite with the larger RNA nanoparticles. Thus, the final blood circulation properties of RNA nanoparticles will result from the balance between these two elimination pathways (i.e., macrophages and urinary excretion).
Effect of Shape on RNA Nanoparticle Biodistribution
Although size is a determining factor of in vivo circulation properties of nanoparticles, shape is also very important. Like size, the shape of RNA nanoparticles can be easily tuned. In this study, RNA polygons of different shape but the same size along each edge were analyzed in mouse models. The organ biodistribution was similar among the particles after 24 hr, but after 12 hr, there appear to be differences in biodistribution, per the intensity of fluorescent signal in different organs (Figure 5). For example, nanosquares show high fluorescence in kidneys after 12 hr, whereas the triangle shows none and the pentagon very little. And in the spleen, the highest fluorescence is seen in mice injected with pentagonal nanoparticles.
Different shape nanoparticles interact very differently with the in vivo environment.33 The protein corona is drastically changed by the shape of nanoparticles, which will affect the elimination pathways of nanoparticles. Additionally, previous studies have shown that symmetric nanoparticles can increase interaction with the immune system and trigger toxicity because of similarity in size and shape to pathogenic microorganisms.73 Although RNA nanoparticles have been proved to trigger very low immune responses,46 shape is still something to consider, and each nanoparticle should be tested to determine immune responses.
Additionally, when considering entry into cells, both size and shape will be very important. Cell receptors vary in size and shape, and it will be beneficial to design nanoparticles that can fit precisely into the receptor binding pockets, enhancing recognition and uptake.74 This is an area in which RNA nanoparticles could potentially excel, as size, shape, and properties of RNA nanoparticles can be controlled and finely tuned to fit specific protein receptors.
Conclusions
The size of RNA nanoparticles has a large effect on circulation time and biodistribution in vivo. A strong correlation between increased size and increased circulation time was observed, evidenced by the increased duration of fluorescent signal in whole-body images of tumor-bearing nude mice over 24 hr. Despite increased size from 5–25 nm, there was still minimal fluorescent signal in healthy organs such as the liver and kidneys after 24 hr compared with tumors. Here, shape was seen to have less effect on the biodistribution of RNA nanoparticles. However, the observance of fluorescent signal after 12 hr suggests that elimination pathways could be different on the basis of the shape of RNA nanoparticles. This study demonstrates a unique property of RNA nanoparticles that allows them to evade accumulation in healthy organs while still accumulating in tumor tissue by the EPR effect. Importantly, we have shown that the circulation time of RNA nanoparticles can be tuned simply by modulating their size. Circulation time is an important factor in drug delivery, as sometimes a short-lived particle is preferred to reduce toxicity, and other times a long-circulating particle could be beneficial for slow-acting therapies. RNA is an advantageous biomaterial for nanoparticle construction, as the base-pairing self-assembly mechanism results in nanoparticles with defined size and shape. Thus, in vivo drug delivery attributes, such as circulation time, can be controlled and tuned via construction of the nanoparticle. It will be possible to use this unique property of RNA nanoparticles to enhance the current state of nucleic acid nanotechnology and drug delivery.
Materials and Methods
RNA Nanoparticle Synthesis and Assembly
RNA strands were prepared as previously detailed75 by either T7 in vitro transcription (strands longer than 70 nt) or typical phosphoramidite oligonucleotide chemical synthesis (strands shorter than 70 nt) using an automated oligo-synthesizer. DNA strands used as primers for double-stranded DNA (dsDNA) transcription templates were ordered from Integrated DNA Technologies (IDT). Assembly was carried out by mixing equimolar amounts of RNA strands in PBS buffer containing 1 mM MgCl2 followed by thermal denaturation at 85°C for 5 min and then slow cooling to 4°C over 1 hr. For each nanoparticle, one strand was fluorescent-labeled with Alexa Fluor 647 for fluorescent tracking in vivo. To confirm nanoparticle assembly, each nanoparticle was run on 2% agarose gel electrophoresis in TAE buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA) for 1 hr at 90 V and room temperature. Sequences for all nanoparticles used in this study can be found in the Supplemental Information.
DLS Measurements
The hydrodynamic diameter of the RNA nanoparticles was determined at a concentration of 5 μM in 150 μL PBS buffer using a Zetasizer nano-ZS (Malvern Instruments) at 25°C. Average size was determined by fitting data points to a Gaussian distribution.
In Vivo Biodistribution Experiments
KB cells were cultured in vitro and subcutaneously injected under the skin of 4-week-old male nude mice (NU/NU; Taconic). A total of 2 × 106 cells were injected into each mouse. Tumors were grown for 2 weeks before mice were injected i.v. through the tail vein. Mice were administered PBS as a blank control. Mice were imaged for whole-body fluorescence at time points of 0.5, 1, 2, 4, 8, 12, and 24 hr with an In Vivo Imaging System (IVIS) imager (Caliper Life Sciences). For this study, two mice were used for the 0.5 through 12 hr time points. At the 12 hr time point, one mouse was sacrificed, and its organs were removed. At 24 hr, the second mouse was sacrificed, and its organs were removed and imaged. Tumors, hearts, kidneys, livers, spleen, and lungs were collected and imaged on the IVIS system. Two mice were used per particle time point. All animal experiments were housed and performed in accordance with the Subcommittee on Research Animal Care of The Ohio State University guidelines approved by the institutional review board.
Serum Stability Assays
2′F nanosquares were assembled and purified using PAGE prior to serum degradation testing. After purification, 0.5 μM fluorescently labeled squares were incubated at 37°C in 10% FBS. Enzymatic degradation was stopped by freezing on dry ice at 0, 1, 2, 4, 8, 16, and 32 hr. Samples were then analyzed using 2% agarose gel electrophoresis. Gels were then scanned for Alexa Fluor 647 fluorescent signal. Gel band intensity was integrated using ImageJ software.76 Values were normalized to initial band intensity and then plotted using OriginPro.
Serum Binding Assays
RNA nanoparticles at a concentration of 500 nM were incubated at 0%, 10%, 20%, 30%, 40%, and 50% concentrations of FBS solution in 1× PBS buffer for 5 min at 37°C followed by analysis on 2% agarose gel electrophoresis. Serum binding experiments for each particle were repeated twice, and error is indicated by error bars. To measure serum binding, EtBr signal from the RNA nanoparticle bands (lower bands) was integrated using ImageJ software. EtBr signal for each lane was normalized to the 0% serum band, and values were then plotted (the y axis is the percentage of nanoparticles bound by serum, and the x axis is the percentage of serum used in incubation) and fit to an exponential decay model with the y intercept set to 1, representing 0% serum binding at 0% serum incubation. Equilibrium serum concentration, at which 50% of the particles are bound, was calculated using the plots by setting the y value to 50%.
Author Contributions
P.G. and D.L.J. conceived and designed the experiments. D.L.J. and H.L. performed the experiments. P.G. provided the funding and resources. D.L.J., H.L., and P.G. co-wrote the manuscript, and all authors refined the manuscript.
Conflicts of Interest
P.G.’s Sylvan G. Frank Endowed Chair in Pharmaceutics and Drug Delivery is funded by the CM Chen Foundation. P.G. is a consultant to Oxford Nanopore Technologies and Nanobio Delivery Pharmaceutical Co. Ltd, as well as a cofounder of Shenzhen P&Z Bio-medical Co. Ltd. and its subsidiary US P&Z Biological Technology LLC.
Acknowledgments
The research in P.G.’s lab was supported by NIH grants R01EB019036 and U01CA207946. The authors would like to thank Mario Vieweger and Daniel W. Binzel for their insight during manuscript preparation.
Footnotes
Supplemental Information includes one table and can be found with this article online at https://doi.org/10.1016/j.ymthe.2017.12.018.
Supplemental Information
References
- 1.Duncan R. The dawning era of polymer therapeutics. Nat. Rev. Drug Discov. 2003;2:347–360. doi: 10.1038/nrd1088. [DOI] [PubMed] [Google Scholar]
- 2.Ferrari M. Cancer nanotechnology: opportunities and challenges. Nat. Rev. Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 3.Quintanar-Guerrero D., Allémann E., Fessi H., Doelker E. Preparation techniques and mechanisms of formation of biodegradable nanoparticles from preformed polymers. Drug Dev. Ind. Pharm. 1998;24:1113–1128. doi: 10.3109/03639049809108571. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee R., Parida S., Maiti C., Mandal M., Dhara D. pH-degradable and thermoresponsive water-soluble core cross-linked polymeric nanoparticles as potential drug delivery vehicle for doxorubicin. RSC Advances. 2015;5:83565–83575. [Google Scholar]
- 5.Lv S., Tang Z., Zhang D., Song W., Li M., Lin J., Liu H., Chen X. Well-defined polymer-drug conjugate engineered with redox and pH-sensitive release mechanism for efficient delivery of paclitaxel. J. Control. Release. 2014;194:220–227. doi: 10.1016/j.jconrel.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Kang H., DeLong R., Fisher M.H., Juliano R.L. Tat-conjugated PAMAM dendrimers as delivery agents for antisense and siRNA oligonucleotides. Pharm. Res. 2005;22:2099–2106. doi: 10.1007/s11095-005-8330-5. [DOI] [PubMed] [Google Scholar]
- 7.Sharma A., Haque F., Pi F., Shlyakhtenko L.S., Evers B.M., Guo P. Controllable self-assembly of RNA dendrimers. Nanomedicine (Lond.) 2016;12:835–844. doi: 10.1016/j.nano.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang M.E., Keswani R.K., Pack D.W. Dependence of PEI and PAMAM gene delivery on clathrin- and caveolin-dependent trafficking pathways. Pharm. Res. 2015;32:2051–2059. doi: 10.1007/s11095-014-1598-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westesen K., Siekmann B., Koch M.H.J. Investigations on the physical state of lipid nanoparticles by synchrotron-radiation x-ray-diffraction. Int. J. Pharm. 1993;93:189–199. [Google Scholar]
- 10.Yingchoncharoen P., Kalinowski D.S., Richardson D.R. Lipid-based drug delivery systems in cancer therapy: what is available and what is yet to come. Pharmacol. Rev. 2016;68:701–787. doi: 10.1124/pr.115.012070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tam Y.Y., Chen S., Cullis P.R. Advances in lipid nanoparticles for siRNA delivery. Pharmaceutics. 2013;5:498–507. doi: 10.3390/pharmaceutics5030498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eigler D.M., Schweizer E.K. Positioning single atoms with a scanning tunneling microscope. Nature. 1990;344:524–526. [Google Scholar]
- 13.Jensen S.A., Day E.S., Ko C.H., Hurley L.A., Luciano J.P., Kouri F.M., Merkel T.J., Luthi A.J., Patel P.C., Cutler J.I. Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci. Transl. Med. 2013;5:209ra152. doi: 10.1126/scitranslmed.3006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Unak G., Ozkaya F., Medine E.I., Kozgus O., Sakarya S., Bekis R., Unak P., Timur S. Gold nanoparticle probes: design and in vitro applications in cancer cell culture. Colloids Surf. B Biointerfaces. 2012;90:217–226. doi: 10.1016/j.colsurfb.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 15.Munro C.H., Smith W.E., Garner M., Clarkson J., White P.C. Characterization of the surface of a citrate-reduced colloid optimized for use as a substrate for surface-enhanced resonance raman-scattering. Langmuir. 1995;11:3712–3720. [Google Scholar]
- 16.Liu J., Zhao Y., Guo Q., Wang Z., Wang H., Yang Y., Huang Y. TAT-modified nanosilver for combating multidrug-resistant cancer. Biomaterials. 2012;33:6155–6161. doi: 10.1016/j.biomaterials.2012.05.035. [DOI] [PubMed] [Google Scholar]
- 17.Pastoriza-Santos I., Liz-Marzan L.M. Colloidal silver nanoplates. State of the art and future challenges. J. Mater. Chem. 2008;18:1724–1737. [Google Scholar]
- 18.Babes L., Denizot B., Tanguy G., Le Jeune J.J., Jallet P. Synthesis of iron oxide nanoparticles used as MRI contrast agents: a parametric study. J. Colloid Interface Sci. 1999;212:474–482. doi: 10.1006/jcis.1998.6053. [DOI] [PubMed] [Google Scholar]
- 19.Sun W., Mignani S., Shen M., Shi X. Dendrimer-based magnetic iron oxide nanoparticles: their synthesis and biomedical applications. Drug Discov. Today. 2016;21:1873–1885. doi: 10.1016/j.drudis.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 20.Huang H.C., Barua S., Sharma G., Dey S.K., Rege K. Inorganic nanoparticles for cancer imaging and therapy. J. Control. Release. 2011;155:344–357. doi: 10.1016/j.jconrel.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Guo P., Zhang C., Chen C., Garver K., Trottier M. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol. Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 22.Lee J.B., Hong J., Bonner D.K., Poon Z., Hammond P.T. Self-assembled RNA interference microsponges for efficient siRNA delivery. Nat. Mater. 2012;11:316–322. doi: 10.1038/nmat3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Afonin K.A., Viard M., Koyfman A.Y., Martins A.N., Kasprzak W.K., Panigaj M., Desai R., Santhanam A., Grabow W.W., Jaeger L. Multifunctional RNA nanoparticles. Nano Lett. 2014;14:5662–5671. doi: 10.1021/nl502385k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osada E., Suzuki Y., Hidaka K., Ohno H., Sugiyama H., Endo M., Saito H. Engineering RNA-protein complexes with different shapes for imaging and therapeutic applications. ACS Nano. 2014;8:8130–8140. doi: 10.1021/nn502253c. [DOI] [PubMed] [Google Scholar]
- 25.Morgen M., Bloom C., Beyerinck R., Bello A., Song W., Wilkinson K., Steenwyk R., Shamblin S. Polymeric nanoparticles for increased oral bioavailability and rapid absorption using celecoxib as a model of a low-solubility, high-permeability drug. Pharm. Res. 2012;29:427–440. doi: 10.1007/s11095-011-0558-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mudshinge S.R., Deore A.B., Patil S., Bhalgat C.M. Nanoparticles: emerging carriers for drug delivery. Saudi Pharm. J. 2011;19:129–141. doi: 10.1016/j.jsps.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehto T., Simonson O.E., Mäger I., Ezzat K., Sork H., Copolovici D.M., Viola J.R., Zaghloul E.M., Lundin P., Moreno P.M. A peptide-based vector for efficient gene transfer in vitro and in vivo. Mol. Ther. 2011;19:1457–1467. doi: 10.1038/mt.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellington A.D., Szostak J.W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature. 1992;355:850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 29.Maeda H., Nakamura H., Fang J. The EPR effect for macromolecular drug delivery to solid tumors: improvement of tumor uptake, lowering of systemic toxicity, and distinct tumor imaging in vivo. Adv. Drug Deliv. Rev. 2013;65:71–79. doi: 10.1016/j.addr.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Wilhelm S., Tavares A.J., Dai Q., Ohta S., Audet J., Dvorak H.F., Chan W.C.W. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 2016;1:1–12. [Google Scholar]
- 31.Gustafson H.H., Holt-Casper D., Grainger D.W., Ghandehari H. Nanoparticle uptake: the phagocyte problem. Nano Today. 2015;10:487–510. doi: 10.1016/j.nantod.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobrovolskaia M.A., Shurin M., Shvedova A.A. Current understanding of interactions between nanoparticles and the immune system. Toxicol. Appl. Pharmacol. 2016;299:78–89. doi: 10.1016/j.taap.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Canton I., Battaglia G. Endocytosis at the nanoscale. Chem. Soc. Rev. 2012;41:2718–2739. doi: 10.1039/c2cs15309b. [DOI] [PubMed] [Google Scholar]
- 34.Longmire M., Choyke P.L., Kobayashi H. Clearance properties of nano-sized particles and molecules as imaging agents: considerations and caveats. Nanomedicine (Lond.) 2008;3:703–717. doi: 10.2217/17435889.3.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hardonk M.J., Harms G., Koudstaal J. Zonal heterogeneity of rat hepatocytes in the in vivo uptake of 17 nm colloidal gold granules. Histochemistry. 1985;83:473–477. doi: 10.1007/BF00509211. [DOI] [PubMed] [Google Scholar]
- 36.Guo P. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 2010;5:833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shu D., Shu Y., Haque F., Abdelmawla S., Guo P. Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat. Nanotechnol. 2011;6:658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Afonin K.A., Viard M., Tedbury P., Bindewald E., Parlea L., Howington M., Valdman M., Johns-Boehme A., Brainerd C., Freed E.O., Shapiro B.A. The use of minimal RNA toeholds to trigger the activation of multiple functionalities. Nano Lett. 2016;16:1746–1753. doi: 10.1021/acs.nanolett.5b04676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bindewald E., Afonin K.A., Viard M., Zakrevsky P., Kim T., Shapiro B.A. Multistrand structure prediction of nucleic acid assemblies and design of RNA switches. Nano Lett. 2016;16:1726–1735. doi: 10.1021/acs.nanolett.5b04651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Severcan I., Geary C., Chworos A., Voss N., Jacovetty E., Jaeger L. A polyhedron made of tRNAs. Nat. Chem. 2010;2:772–779. doi: 10.1038/nchem.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Afonin K.A., Bindewald E., Yaghoubian A.J., Voss N., Jacovetty E., Shapiro B.A., Jaeger L. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol. 2010;5:676–682. doi: 10.1038/nnano.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Severcan I., Geary C., Verzemnieks E., Chworos A., Jaeger L. Square-shaped RNA particles from different RNA folds. Nano Lett. 2009;9:1270–1277. doi: 10.1021/nl900261h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jasinski D., Haque F., Binzel D.W., Guo P. Advancement of the emerging field of RNA nanotechnology. ACS Nano. 2017;11:1142–1164. doi: 10.1021/acsnano.6b05737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H., Lee T., Dziubla T., Pi F., Guo S., Xu J., Li C., Haque F., Liang X.J., Guo P. RNA as a stable polymer to build controllable and defined nanostructures for material and biomedical applications. Nano Today. 2015;10:631–655. doi: 10.1016/j.nantod.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo P., Coban O., Snead N.M., Trebley J., Hoeprich S., Guo S., Shu Y. Engineering RNA for targeted siRNA delivery and medical application. Adv. Drug Deliv. Rev. 2010;62:650–666. doi: 10.1016/j.addr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khisamutdinov E.F., Li H., Jasinski D.L., Chen J., Fu J., Guo P. Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square and pentagon nanovehicles. Nucleic Acids Res. 2014;42:9996–10004. doi: 10.1093/nar/gku516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shu Y., Haque F., Shu D., Li W., Zhu Z., Kotb M., Lyubchenko Y., Guo P. Fabrication of 14 different RNA nanoparticles for specific tumor targeting without accumulation in normal organs. RNA. 2013;19:767–777. doi: 10.1261/rna.037002.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Afonin K.A., Lindsay B., Shapiro B.A. Engineered RNA nanodesigns for applications in RNA nanotechnology. DNA RNA Nanotechnol. 2013;1:1–15. doi: 10.2478/rnan-2013-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boerneke M.A., Dibrov S.M., Hermann T. Crystal-structure-guided design of self-assembling RNA nanotriangles. Angew. Chem. Int. Ed. Engl. 2016;55:4097–4100. doi: 10.1002/anie.201600233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dibrov S.M., McLean J., Parsons J., Hermann T. Self-assembling RNA square. Proc. Natl. Acad. Sci. U S A. 2011;108:6405–6408. doi: 10.1073/pnas.1017999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petrov A.I., Zirbel C.L., Leontis N.B. Automated classification of RNA 3D motifs and the RNA 3D Motif Atlas. RNA. 2013;19:1327–1340. doi: 10.1261/rna.039438.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leontis N.B., Lescoute A., Westhof E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leontis N.B., Westhof E. Analysis of RNA motifs. Curr. Opin. Struct. Biol. 2003;13:300–308. doi: 10.1016/s0959-440x(03)00076-9. [DOI] [PubMed] [Google Scholar]
- 54.Watts J.K., Deleavey G.F., Damha M.J. Chemically modified siRNA: tools and applications. Drug Discov. Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 55.Liu J., Guo S., Cinier M., Shlyakhtenko L.S., Shu Y., Chen C., Shen G., Guo P. Fabrication of stable and RNase-resistant RNA nanoparticles active in gearing the nanomotors for viral DNA packaging. ACS Nano. 2011;5:237–246. doi: 10.1021/nn1024658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawasaki A.M., Casper M.D., Freier S.M., Lesnik E.A., Zounes M.C., Cummins L.L., Gonzalez C., Cook P.D. Uniformly modified 2′-deoxy-2′-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J. Med. Chem. 1993;36:831–841. doi: 10.1021/jm00059a007. [DOI] [PubMed] [Google Scholar]
- 57.Bae Y.H., Park K. Targeted drug delivery to tumors: myths, reality and possibility. J. Control. Release. 2011;153:198–205. doi: 10.1016/j.jconrel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lipfert J., Skinner G.M., Keegstra J.M., Hensgens T., Jager T., Dulin D., Köber M., Yu Z., Donkers S.P., Chou F.C. Double-stranded RNA under force and torque: similarities to and striking differences from double-stranded DNA. Proc. Natl. Acad. Sci. U S A. 2014;111:15408–15413. doi: 10.1073/pnas.1407197111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herrero-Galán E., Fuentes-Perez M.E., Carrasco C., Valpuesta J.M., Carrascosa J.L., Moreno-Herrero F., Arias-Gonzalez J.R. Mechanical identities of RNA and DNA double helices unveiled at the single-molecule level. J. Am. Chem. Soc. 2013;135:122–131. doi: 10.1021/ja3054755. [DOI] [PubMed] [Google Scholar]
- 60.Binzel D.W., Khisamutdinov E.F., Guo P. Entropy-driven one-step formation of Phi29 pRNA 3WJ from three RNA fragments. Biochemistry. 2014;53:2221–2231. doi: 10.1021/bi4017022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Binzel D.W., Khisamutdinov E., Vieweger M., Ortega J., Li J., Guo P. Mechanism of three-component collision to produce ultrastable pRNA three-way junction of Phi29 DNA-packaging motor by kinetic assessment. RNA. 2016;22:1710–1718. doi: 10.1261/rna.057646.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Binzel D.W., Shu Y., Li H., Sun M., Zhang Q., Shu D., Guo B., Guo P. Specific delivery of MiRNA for high efficient inhibition of prostate cancer by RNA nanotechnology. Mol. Ther. 2016;24:1267–1277. doi: 10.1038/mt.2016.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shu D., Li H., Shu Y., Xiong G., Carson W.E., 3rd, Haque F., Xu R., Guo P. Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology. ACS Nano. 2015;9:9731–9740. doi: 10.1021/acsnano.5b02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cui D., Zhang C., Liu B., Shu Y., Du T., Shu D., Wang K., Dai F., Liu Y., Li C. Regression of gastric cancer by systemic injection of RNA nanoparticles carrying both ligand and siRNA. Sci. Rep. 2015;5:10726. doi: 10.1038/srep10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee T.J., Haque F., Shu D., Yoo J.Y., Li H., Yokel R.A., Horbinski C., Kim T.H., Kim S.H., Kwon C.H. RNA nanoparticle as a vector for targeted siRNA delivery into glioblastoma mouse model. Oncotarget. 2015;6:14766–14776. doi: 10.18632/oncotarget.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rychahou P., Haque F., Shu Y., Zaytseva Y., Weiss H.L., Lee E.Y., Mustain W., Valentino J., Guo P., Evers B.M. Delivery of RNA nanoparticles into colorectal cancer metastases following systemic administration. ACS Nano. 2015;9:1108–1116. doi: 10.1021/acsnano.5b00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haque F., Shu D., Shu Y., Shlyakhtenko L.S., Rychahou P.G., Evers B.M., Guo P. Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today. 2012;7:245–257. doi: 10.1016/j.nantod.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khisamutdinov E.F., Jasinski D.L., Guo P. RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry. ACS Nano. 2014;8:4771–4781. doi: 10.1021/nn5006254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jasinski D.L., Khisamutdinov E.F., Lyubchenko Y.L., Guo P. Physicochemically tunable polyfunctionalized RNA square architecture with fluorogenic and ribozymatic properties. ACS Nano. 2014;8:7620–7629. doi: 10.1021/nn502160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Behlke M.A. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18:305–319. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- 71.Behlke M.A. Progress towards in vivo use of siRNAs. Mol. Ther. 2006;13:644–670. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gunawan C., Lim M., Marquis C.P., Amal R. Nanoparticle-protein corona complexes govern the biological fates and functions of nanoparticles. J. Mat. Chem. B. 2014;2:2060–2083. doi: 10.1039/c3tb21526a. [DOI] [PubMed] [Google Scholar]
- 73.Fadeel B. Clear and present danger? Engineered nanoparticles and the immune system. Swiss Med. Wkly. 2012;142:w13609. doi: 10.4414/smw.2012.13609. [DOI] [PubMed] [Google Scholar]
- 74.Vindigni G., Raniolo S., Ottaviani A., Falconi M., Franch O., Knudsen B.R., Desideri A., Biocca S. Receptor-mediated entry of pristine octahedral DNA nanocages in mammalian cells. ACS Nano. 2016;10:5971–5979. doi: 10.1021/acsnano.6b01402. [DOI] [PubMed] [Google Scholar]
- 75.Shu Y., Shu D., Haque F., Guo P. Fabrication of pRNA nanoparticles to deliver therapeutic RNAs and bioactive compounds into tumor cells. Nat. Protoc. 2013;8:1635–1659. doi: 10.1038/nprot.2013.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Collins T.J. ImageJ for microscopy. Biotechniques. 2007;43(1, Suppl):25–30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.