Abstract
We have selected 143 independent Nicotiana plumbaginifolia cell lines that survive in the presence of 5-fluoroorotic acid. These lines show several diverse phenotypes. The majority of these cell lines showed reduced levels of UMP synthase. However, one particular phenotype, which represents 14% of the total independent lines (20 cell lines), showed an unexpected, high level of UMP synthase and was therefore analyzed in detail. The selected cell lines showed no differences with wild-type cells with respect to uptake of orotic acid, affinity of UMP synthase for its substrates, or UMP synthase gene-copy number. Alternative detoxification mechanisms were also excluded. The elevated enzyme activity was correlated with elevated UMP synthase protein levels as well as elevated UMP synthase mRNA levels. In contrast to wild-type cell lines, the fluoroorotic acid-selected cell lines did not respond to thymine or to other biochemicals that affect thymine levels. In addition, there was also a concomitant up-regulation of aspartate transcarbamoylase, however, dihydroorotase and dihydroorotate dehydrogenase are not up-regulated in these cell lines.
Pyrimidines play a central role in cellular regulation and metabolism. They are substrates for DNA/RNA biosynthesis, regulators of the biosynthesis of some amino acids, and cofactors in the biosynthesis of phospholipids, glycolipids, sugars, and polysaccharides. The classical de novo pyrimidine biosynthetic pathway ends with the synthesis of UMP. Other divergent pathways lead to the formation of CTP and TTP (Neuhard and Nygaard, 1987). Also, salvage pathways exist, which allow cells to utilize preformed nucleotides, thereby avoiding the metabolic cost of biosynthesis (Jones and Hahn, 1979; Neuhard and Nygaard, 1987).
The enzymatic activities of the de novo pyrimidine biosynthetic pathway are well known and invariant in all examined organisms; however, the gene organization of these several steps varies among organisms. Plants differ from other higher eukaryotes in that the first three steps of the de novo pathway are carried out by separate enzymes that are individually encoded (Doremus, 1986). In mammals, some fungi, and insects, however, the genes encoding these enzymes have been rearranged during evolution into a single transcriptional unit that encodes a single polyprotein termed CAD, having three enzymatic activities, carbamoylphosphate synthase, Aspartate transcarbamoylase, and dihydroorotase (Kim et al., 1992; van den Hoff et al., 1995).
An additional bifunctional protein, UMP synthase, is present in both plants and animals. This enzyme contains the last two steps of the de novo biosynthetic pathway (Jones, 1980; Nasr et al., 1994; Maier et al., 1995). These last two enzymatic steps are orotate phosphoribosyltransferase and orotidine decarboxylase. During evolution, the genes encoding these two enzymes have become fused into a single transcriptional unit encoding one protein having both enzymatic activities (Jones, 1980). UMP synthase is one of the key enzymes of the de novo biosynthesis of pyrimidines. It is the rate-limiting step of the pathway in both mammals (Traut and Jones, 1977) and in plants (Santoso and Thornburg, 1992).
Previously we have used a negative selection scheme with 5-fluoroorotic acid (FOA) to produce plant cell lines that have stable alterations in pyrimidine metabolism (Santoso and Thornburg, 1992, 1998). Because UMP synthase is the rate-limiting step in pyrimidine biosynthesis, alterations affecting its expression are frequently observed. By far the majority of the selected cell lines show reduced levels of UMP synthase as was expected from previous studies of selection in Saccharomyces cerevisiae (Boeke et al., 1984) and Dictyostelium discoideum (Kalpaxis et al., 1991). However, some of these cell lines showed elevated rather than reduced levels of UMP synthase activity. In an effort to understand the up-regulation of UMP synthase levels, we have investigated the expression of UMP synthase in these stable FOA-selected cell lines.
RESULTS
The lines described in this work were isolated from protoplast and cell suspension cultures derived from a haploid Nicotiana plumbaginifolia line, hNp28. After 5 to 7 weeks of selection, cell lines were obtained that continued to grow in the presence of FOA. These cell lines were routinely maintained in the presence of 0.1 mm FOA. Based upon earlier work in S. cerevisiae (Boeke et al., 1984), D. discoideum (Kalpaxis et al., 1991), and Nicotiana tabacum, (Santoso and Thornburg, 1992) we anticipated that selection of N. plumbaginifolia cell lines on FOA would produce cell lines with reduced levels of UMP synthase. Indeed, the majority of the cell lines isolated in these experiments show reduced levels of UMP synthase. However, we have also produced many cell lines that show elevated (>300% of wild type) levels of UMP synthase. Of the 143 cell lines isolated in eight separate replicates of selection in the presence of FOA, 14% of the resulting cell lines (20 cell lines) had elevated UMP synthase levels. To understand why some cell lines showed high, constitutive levels of UMP synthase in response to selection on the toxic fluoroorotic acid, we characterized the expression of UMP synthase in these cell lines.
Resistance to toxic compounds in cultured cell lines has been studied widely in animal cell systems (Schweitzer et al., 1990; Volkenandt et al., 1993). Such resistance can result from at least four distinct mechanisms: (a) altered transport of the toxic compound into cells; (b) altered affinity of the enzyme for the substrates; (c) biochemical modification leading to detoxification of the toxic compound; and (d) specific overproduction of the target enzyme. Each of these possibilities was examined to determine whether these mechanisms caused the ability of these cells to survive on fluoroorotic acid.
To evaluate whether the cell lines were affected in their ability to take up orotic acid, the wild-type and the FOA-selected cell lines were plated onto media containing 6-[14C]orotic acid. The level of radioactivity in both types of cells was monitored for 84 h. After an initial rapid uptake of approximately 7 pmol of orotic acid within the 1st h, thereafter approximately 1 pmol of orotic acid was taken up per hour for all cell lines. This rate of uptake did not differ significantly between the wild-type and FOA-resistant cell lines. From these studies we concluded that the FOA-resistant cell lines are unaffected in their ability to take up orotic acid.
To evaluate whether there is altered specificity of the UMP synthase enzyme for fluoroorotic acid we tested the UMP synthase from both the wild-type and from one of our fluoroorotic acid-selected cell lines, hNp28-umps820. The cell line, hNp28-umps820 was one of three cell lines with the highest levels of UMP synthase. The other two cell lines, hNp28-umps802 and hNp28-umps822, showed similar results throughout. UMP synthase was partially purified from cellular extracts of these cell lines by ammonium sulfate fraction [70% saturated (NH4)2SO4]. This enzyme was used to determine kinetic and inhibition constants of these enzymes for fluoroorotic acid. The results are shown in Table I. The Km for orotic acid was identical between the wild-type and hNp28-umps820 enzymes. The Vmax, in contrast, was found to differ, being higher for the hNp28-umps820 enzyme. As with the Km, the inhibition constants for fluoroorotic acid were not significantly different for the wild-type and hNp28-umps820 enzymes. Thus the mechanism of resistance to fluoroorotic acid cannot be attributed to altered specificity of the enzyme for this toxic compound. Further, because the Km for orotic acid does not differ from the Ki for fluoroorotic acid, it appears that both UMP synthases bind orotic acid and fluoroorotic acid equally well.
Table I.
Kinetic parameters of tobacco UMP synthases
| Wild Type | umps820 | |
|---|---|---|
| Vmax (units/mg) | 0.68 ± 0.14 | 1.18 ± 0.18 |
| Km (μm) | 1.22 ± 0.28 | 1.24 ± 0.30 |
| Ki (μm) | 1.15 ± 0.18 | 1.12 ± 0.13 |
The standard enzyme reaction contained 20 mm [Tris(hydroxy-methyl)-aminomethane]-HCl, pH 8.0, 2 mm MgCl2, 1 mm dithiothreitol, 2.0 μm 7-[14C]orotic acid, and extracted proteins from either wild-type or hNp28-umps820 cells. For determination of the Km and Vmax values for orotic acid, the concentration of orotic acid was varied over a 100-fold range from 0.1 × Km to 10 × Km. Phosphoribosyl pyrophosphate was held constant at 0.2 mm. The Km and Vmax values for orotic acid were determined by least-squares analysis of Eadie-Hofstee plots using the initial velocity rates (n = 4). The Ki for FOA was evaluated as a competitive inhibitor from Lineweaver-Burk plots displaying increasing concentration of the inhibitor using the formula Ki = [FOA]/α − 1, where the slope of each line = αKm/Vmax (n = 4). Data presented are average values ± sd.
Detoxification of fluoroorotic acid via any of several mechanisms could also result in escape from fluoroorotic acid toxicity. First, replacement of the fluorine atom with a hydrogen would yield orotic acid, but enzymes catalyzing dehalogenation are extremely rare (Mohn and Tiedje, 1992) and to our knowledge are unknown in plants. Second, destruction of the pyrimidine ring might yield non-toxic products. Degradation of the pyrimidine ring can occur by five different pathways (Wasternack, 1978). However, these pathways utilize uracil (or thymine) as the initial substrate rather than orotic acid. A few microorganisms such as Clostridium oroticum have an additional reductive pathway that starts from orotic acid. This alternate reductive pathway, which is found in organisms that overproduce orotic acid, is essentially the reverse of the biosynthetic pathway, but this pathway is unknown in plants. Finally, direct decarboxylation of orotic acid to uracil could occur. This pathway is well known in many bacteria, but again, unknown in plants.
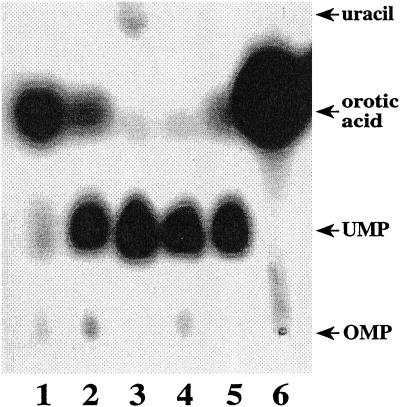
We utilized a thin-layer chromatography (TLC) assay to monitor whether orotic acid was converted directly into any compounds other than UMP. This assay is shown in Figure 1. The positions of migration of orotic acid, OMP, UMP, and uracil are indicated at the right of the figure. We have never observed any significant amount of 14C-labeled orotic acid converted directly into uracil, nor have we ever observed conversion of orotic acid into any other compound. In every case the major products are OMP and UMP. As controls, lanes 5 and 6 show extracts from wild-type and FOA-selected N. tabacum Tr25-umps121 cell lines, respectively. The Tr25-umps121 cell line (lane 6) was previously shown to have reduced levels of UMP synthase (Santoso and Thornburg, 1992). Lanes 2, 3, and 4 show extracts of the wild-type and two FOA-resistant N. plumbaginifolia cell lines. Under the conditions of this assay, the hNp28-umps820 and hNp28-umps822 cell lines converted >90% of the orotic acid into UMP. We further noticed that the FOA-selected cell lines showed an increased conversion of orotic acid into UMP, which is consistent with the selected cell lines having higher levels of the enzyme UMP synthase. It is also clear that the labeled orotic acid is not converted into other unidentified compounds. Based upon these results we have concluded that the mechanism permitting the cells to survive in the presence of fluoroorotic acid is not due to substrate detoxification or alterations in either substrate transport or enzyme structure. Instead it appears that escape from fluoroorotic acid toxicity may be due to elevated levels of UMP synthase.
Figure 1.
TLC analysis of UMP synthase from fluoroorotic acid-selected tobacco cell lines. Lane 1 shows purified orotic acid. Lane 2 and 5 contained radiolabeled orotic acid incubated with 100 μg of protein extracted from wild-type cell lines of N. plumbaginifolia and N. tabacum, respectively. Lanes 3 and 4 contain radiolabeled orotic acid incubated with 100 μg of protein extracted from the fluoroorotic acid-selected N. plumbaginifolia cell lines from hNp-umps820 and hNp-umps822, respectively. Lane 6 contains radiolabeled orotic acid incubated with 100 μg of protein extracted from the fluoroorotic acid-selected N. tabacum cell line Tr25-umps121.
Mechanisms that permit overproduction of specific enzymes are varied. In animal systems (Alt et al., 1978; Wahl et al., 1982; Andrulis et al., 1983) and a few plant systems (Shyr et al., 1992; Reinbothe et al., 1993), gene amplification has been shown to result in overexpression of target enzymes. Indeed in rat hepatoma cells, inhibition of UMP synthase leads to gene amplification leading to enzyme overproduction (Kanalas and Suttle, 1984; Suttle, 1989; Daniel et al., 1994). To examine whether these cell lines showed signs of gene amplification, we compared the wild-type and FOA-selected cell lines by Southern-blot analysis. The probe used for these studies was the N. tabacum UMP synthase cDNA (Maier et al., 1995). This cDNA recognizes a single 9.2-kb band in EcoRI digests of N. plumbaginifolia DNA and two bands of 23 and 11.2 kb in BamHI digests (Maier et al., 1995). Hybridization analysis revealed no differences between the wild-type and FOA-selected cell lines (data not shown), indicating that gene amplification was not responsible for the increase in UMP synthase levels in these UMP synthase overexpressing cell lines.
Although the UMP synthase gene is not amplified, it is nevertheless clear that UMP synthase activity is elevated in the fluoroorotic acid-selected cell lines. To understand the molecular mechanism responsible for the up-regulation of UMP synthase enzyme activity, we first examined whether UMP synthase protein levels were also elevated. Therefore we used western blots to examine the levels of UMP synthase protein present in the wild-type and fluoroorotic acid-selected cell lines.
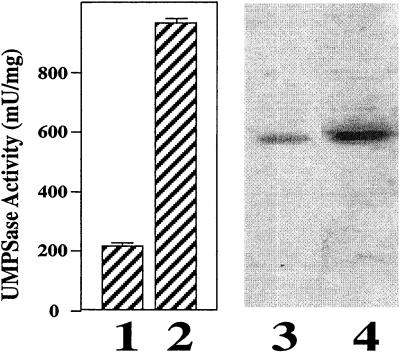
In Figure 2, lanes 1 and 2 show the levels of UMP synthase enzyme activity in wild-type and fluoroorotic acid-selected cell lines, respectively. As can be seen, the fluoroorotic acid-selected cell line shows a 3- to 4-fold increase in the level of enzyme activity. To determine whether this increase in enzyme activity was associated with a concomitant increase in protein levels, protein extracts of each cell line were processed for western blots. This analysis is shown in lanes 3 and 4 of Figure 2.
Figure 2.
The levels of UMPase of the N. plumbaginifolia wild-type and the hNp28-umps820 the cell lines. Lanes 1 and 2 show the UMP synthase activity of the cell lines assayed with the CO2-release method as described in “Materials and Methods.” Lane 1 contains proteins extracted from the wild-type cell line precultured in media without FOA. Lane 2 is from hNp28-umps820 cell lines without FOA. Data are averages and error bars represent the sd of the mean (n = 6). Lanes 3 and 4 are the western-blot analysis of the cellular extracts from the same pooled cell lines as lanes 1 and 2, respectively.
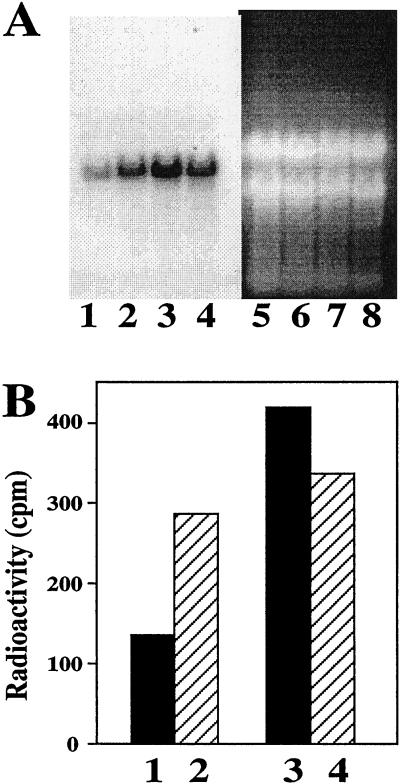
Our previous work showed that UMP synthase is transcriptionally up-regulated in plants by the addition of fluoroorotic acid or other compounds that result in a thymine starvation (Santoso and Thornburg, 1998). To evaluate whether transcript levels are expressed at higher levels in the fluoroorotic acid-selected cell lines than in wild-type cells, we performed northern blots. Figure 3A shows the level of UMP synthase transcript levels in wild-type and fluoroorotic acid-selected hNp28-umps820 cell line (compare lanes 1 and 3). Quantitation of these bands (B) was performed by excision of the labeled portions of the blot and counting in a liquid scintillation counter. This revealed that the fluoroorotic acid-selected cell line had approximately three times more UMP synthase transcripts than did the wild-type cells. As we had previously demonstrated, when wild-type cells are treated with fluoroorotic acid for 10 d, the UMP synthase transcript level increases 2- to 3-fold (compare lanes 1 and 2). In contrast, the hNp28-umps820 cell line fails to show a concomitant increase in UMP synthase transcript levels. Thus, the selection process has produced a stable alteration in these cell lines, which results in a high, constitutive level of UMP synthase transcript accumulation and a loss of sensitivity to compounds that result in a thymine starvation.
Figure 3.
The northern-blot analysis of transcripts isolated from wild-type and hNp28-umps820 cells. A, Lanes 1 to 4 are after hybridization. Lanes 5 to 8 are the same ethidium bromide-stained gel as lanes 1 to 4 before transfer and hybridization demonstrating equal loading of each lane. Lanes 1 and 5 are from the wild-type cells without FOA preculture. Lanes 2 and 6 are from the wild-type cells with FOA preculture. Lanes 3 and 7 are from the hNp28-umps820 cells without FOA preculture. Lanes 4 and 8 are from hNp28-umps820 cells with FOA preculture. B, Quantification of lanes 1 to 4 of A.
Do umps820 Cells Respond to Elevated Levels of Thymine?
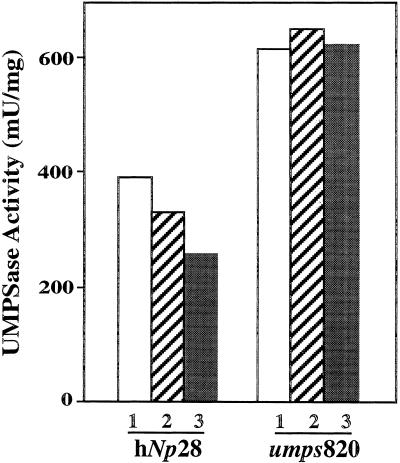
Because UMP synthase in fluoroorotic acid-selected hNp28-umps820 cells is not induced by exogenously added fluoroorotic acid, we decided to test the effect of thymine directly on these cells. For wild-type cells, high levels of UMP synthase was induced in the presence of fluoroorotic acid. These cells were then plated on media containing 200 μm thymine. Cells of the fluoroorotic acid-selected cell line, hNp28-umps820, were likewise plated onto media containing 200 μm thymine. Data from a representative experiment are shown in Figure 4. In this figure exogenously-added thymine reduces the level of UMP synthase activity in wild-type cells, but has no effect on the levels of UMP synthase activity in the hNp28-umps820 cells. Thus, the FOA-selected cells have apparently lost the ability to either up-regulate or to repress UMP synthase in response to thymine or to respond to compounds that alter intracellular thymine levels.
Figure 4.
The effect of thymine on the UMP synthase activity of the wild-type and FOA-selected hNp28-umps820 cells. In this representative experiment wild-type cells were precultured for 10 d in the presence of FOA. The cells were then replated onto media without FOA, but containing various concentrations of thymine. After 3 d, the levels of UMP synthase activity were assayed. The white bars, labeled 1 are from cells cultured without thymine. The hatched bars, labeled 2 are from cells cultured with a 0.2 mm thymine. The black bars, labeled 3 are from cells cultured with 2 mm thymine. Data are average values (n = 3).
Are Other Steps of Pyrimidine Biosynthesis Affected?
Because the umps820 cell line abnormally expresses high, constitutively levels of UMP synthase, we decided to examine whether other enzymes in the pyrimidine biosynthetic pathway were also similarly affected. Table II compares the enzyme activities for pyrimidine biosynthetic enzymes in wild-type and hNp28-umps820 cell lines. As can be seen, the activity of UMP synthase is up-regulated in the hNp28-umps820 cell line relative to the wild-type hNp28 cells. Similarly, the levels of Aspartate transcarbamoylase are also overexpressed to almost the same level as with UMP synthase. In contrast, both the dihydroorotase and the dihydroorotate dehydrogenase activities were unaffected in the hNp28-umps820 cell line and were indistinguishable from wild-type activities. Similar results were obtained for the hNp28-umps802 and hNp28-umps822 cell lines. We concluded that selection of these cell lines on fluoroorotic acid results in the constitutive up-regulation of two independent steps in the pyrimidine biosynthetic pathway.
Table II.
Enzyme activities of pyrimidine biosynthetic enzymes
| Wild Type | umps820 | Wild Type | |
|---|---|---|---|
| milliunits mg−1 protein | % | ||
| ATCaseb | 8.5 ± 1.8 | 16.4 ± 2.0 | 193 |
| DHOase | 11.8 ± 1.9 | 12.2 ± 2.0 | 103 |
| DHODH | 117 ± 17 | 111 ± 23 | 95 |
| UMPSase | 0.27 ± 0.06 | 0.67 ± 0.07 | 248 |
Enzyme assays were performed as described in “Materials and Methods.” All enzyme activities were normalized per milligram protein in extracts of wild-type or umps820 cell lines.
One unit is defined as the conversion of 1 μmol of product released per min. Data are average values ± sd (n = 6).
ATCase, Aspartate transcarbamoylase; DHOase, dihydroorotase; DHODH, dihydroorotate dehydrogenase; UMPSase, UMP synthase.
DISCUSSION
Plant cells normally respond to thymine starvation by up-regulating the rate-limiting step of pyrimidine biosynthesis, UMP synthase (Santoso and Thornburg, 1998). In this report we have characterized stabile fluoroorotic acid-selected cell lines. This analysis permits us to extend our observations in two ways. The first observation is that some stably selected fluoroorotic acid-resistant plant cell lines show very high, constitutive, and unregulatable levels of UMP synthase. Based upon this first observation it appears that wild-type tobacco cells normally express UMP synthase at relatively low levels, yet when necessary they can readily overexpress UMP synthase. This regulation is dependent upon the cellular levels of thymine and is transcriptionally controlled (Santoso and Thornburg, 1998). These fluoroorotic acid-selected cell lines that have constitutively elevated levels of UMP synthase have lost the ability to respond to thymine levels. That this loss of regulation by thymine in our fluoroorotic acid-selected cell lines is coupled with the complete, unbridled expression of UMP synthase implies that the normal low levels of UMP synthase found in wild-type cells are maintained by repression of the UMP synthase gene. Further, it appears that one method of escaping the toxic effects of FOA is the loss of this repression resulting in constitutive unregulated levels of some of the pyrimidine biosynthetic enzymes.
Our second observation is that the constitutive up-regulation of pyrimidine biosynthetic genes in the FOA-selected cell lines is not limited to UMP synthase, the rate-limiting step, but also includes the first committed step of de novo pyrimidine biosynthesis. Whereas other enzymes may also be up-regulated in these FOA-selected cell lines, we have demonstrated that dihydroorotase and dihydroorotate dehydrogenase are not. The normal cellular response to thymine starvation is to up-regulate de novo pyrimidine biosynthesis. This transcriptional up-regulation has been best studied for UMP synthase (Santoso and Thornburg, 1998).
When thymine is provided to normal cells following thymine-starvation, the level of the induced enzymes return to low levels. Thus, plant cells apparently have a mechanism to detect thymine levels within the cell and can communicate that information to the promoter of the UMP synthase gene. The finding in this work that stably selected cell lines showing a thymine starvation phenotype also have other pyrimidine biosynthetic enzymes that are up-regulated suggests that several genes are coordinately controlled by a similar mechanism. These include Aspartate transcarbamoylase (the first committed step of pyrimidine biosynthesis) and UMP synthase (the rate limiting step). Other pyrimidine metabolic enzymes may also be up-regulated.
That multiple enzymatic steps appear to be coordinately regulated suggests the existence of a regulatory factor. This factor can detect the level of thymine or perhaps a thymine metabolite and subsequently modify the expression of pyrimidine metabolic genes. In the case of UMP syntheses this regulation is transcriptional suggesting that this factor may be a DNA-binding protein or similar effecting trans-acting factor. The loss of this single factor from plant cells would result in the biochemical phenotype observed for the stable fluoroorotic acid-selected cell lines described in this work.
MATERIALS AND METHODS
Materials
Haploid Nicotiana plumbaginifolia plants (hNp28) in sterile culture were kindly provided by Dr. Laszlo Marton (Department of Biology, University of South Carolina, Columbia). Antiserum raised against squash-UMP synthase was previously described (Santoso and Thornburg, 1998).
Phosphoribosyl pyrophosphate, orotic acid, FOA, uracil, and nucleotide metabolites were purchased from Sigma Chemical Company (St. Louis). Media for plant tissue culture and plant hormones were purchased either from Sigma or from GIBCO Laboratories (Grand Island, NY). The radiochemicals, 7-[14C]orotic acid with a specific activity of 48.5 mCi/mMol, and [125I]-rProteinA (9.0 mCi/mg), were purchased from New England Nuclear (Boston). Other materials were of the highest purity available and were obtained either locally or from Fisher Chemical Company (Pittsburgh).
Tissue Culture, Induction, and Enzyme Assays
Methods for the selection of cell lines in the presence of fluoroorotic acid were performed as previously described (Santoso and Thornburg, 1992). Induction was accomplished by plating cells onto induction medium as described (Santoso and Thornburg, 1998). Aspartate transcarbamoylase activity was determined as described (Gerhart and Pardee, 1962). To increase the sensitivity of this assay, 2 μm orotate and 0.1 μm aza-UMP were added to the reaction mixture. Dihydroorotase and dihydroorotate dehydrogenase activities were assayed according to Beckwith et al. (1962) and Caroline (1969), respectively. To increase sensitivity, 2 μm orotate and 0.1 μm aza-UMP were added to the dihydroorotase reaction mixture and 0.1 μm aza-UMP was added to the dihydroorotate dehyrogenase reaction mixture. UMP synthase was determined by two different techniques: a CO2 release assay and a TLC assay. The CO2 release assay was previously described (Santoso and Thornburg, 1992). With the TLC assay, the reaction conditions were similar to those of the CO2-release assay except that a smaller volume of the reaction mixture (100 μL) containing ring-labeled orotic acid (6-[14C]orotic acid) was utilized. After stopping the reaction by boiling for 3 min, the reaction mixtures were spotted onto polyethylenei-mine-cellulose TLC plates (Sigma). The plates were developed with 0.3 m NaCl and autoradiography was done at −70°C for 3 d.
Blots
Western blots were performed as described (Timmons and Dunbar, 1990). Incubation of the transferred proteins in antiserum was performed overnight at 4°C. Total RNA was isolated from N. plumbaginifolia cell lines by the method of Wadsworth et al. (1988). The average yield was about 50 μg of RNA per gram of tissue. The RNA samples were denatured and electrophoresed as described (Tirimanne and Colbert, 1991). Northern blots were performed (Ausubel et al., 1987) using the previously described Nicotiana tabacum UMP synthase cDNA (Maier et al., 1995).
Footnotes
This work was sponsored by the U.S. Department of Agriculture (grant no. 91–37301–6208). This is journal paper no. J–16512 of the Iowa Agriculture and Home Economics Experiment Station, Ames, Iowa and project no. 3202.
LITERATURE CITED
- Alt FW, Kellems RE, Bertino JR, Schimke RT. Selective multiplication of dihydrofolate reductase genes in methotrexate-resistant variants of cultured murine cells. J Biol Chem. 1978;253:1357–1370. [PubMed] [Google Scholar]
- Andrulis I, Duff C, Evans-Blackler S, Worton R, Siminovitch L. Chromosomal alterations associated with overproduction of asparagine synthetase in albizziin-resistant Chinese hamster ovary cells. Mol Cell Biol. 1983;3:391–398. doi: 10.1128/mcb.3.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Smith JA, Seidman JG, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1987. [Google Scholar]
- Beckwith JR, Pardee AB, Austrian R, Jacob F. Coordination of the synthesis of the enzymes in the pyrimidine pathway of E. coli. J Mol Biol. 1962;5:618–634. doi: 10.1016/s0022-2836(62)80090-4. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Lacroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoroorotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Caroline DF. Pyrimidine biosynthesis in Neurospora crassa: gene enzyme relationships. J Bacteriol. 1969;100:1371–1377. doi: 10.1128/jb.100.3.1371-1377.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel DJ, Tonzetich J, Chernin M, McMaster J, Novak J. Acquisition of resistance to 6-azauridine through DNA amplification in neoplastic but not normal osteoblasts. Anticancer Res. 1994;14:937–942. [PubMed] [Google Scholar]
- Doremus HD. Organization of the pathway of de novo pyrimidine nucleotide biosynthesis in pea (Pisum sativum L. cv Progress No. 9) leaves. Arch Biochem Biophys. 1986;250:112–119. doi: 10.1016/0003-9861(86)90707-1. [DOI] [PubMed] [Google Scholar]
- Gerhart JC, Pardee AB. The enzymology of control by feedback inhibition. J Biol Chem. 1962;237:891–896. [PubMed] [Google Scholar]
- Jones GE, Hahn J. Haplopappus gracilis cell strains resistant to pyrimidine analogues. Theor Appl Genet. 1979;54:81–87. doi: 10.1007/BF00265474. [DOI] [PubMed] [Google Scholar]
- Jones ME. Pyrimidine nucleotide biosynthesis in animals: genes, enzymes and regulation of UMP biosynthesis. Annu Rev Biochem. 1980;49:253–279. doi: 10.1146/annurev.bi.49.070180.001345. [DOI] [PubMed] [Google Scholar]
- Kalpaxis D, Zundorf I, Werner H, Reindl N, Boy-Marcotte E, Jacquet M, Dingermann T. Positive selection for Dictyostelium discoideum mutants lacking UMP synthase activity based on resistance to 5-fluoroorotic acid. Mol Gen Genet. 1991;225:492–500. doi: 10.1007/BF00261692. [DOI] [PubMed] [Google Scholar]
- Kanalas J, Suttle D. Amplification of the UMP synthase gene and enzyme overproduction in pyrazofurin-resistant rat hepatoma cells: molecular cloning of a cDNA for UMP synthase. J Biol Chem. 1984;259:1848–1853. [PubMed] [Google Scholar]
- Kim H, Kelly RE, Evans DR. The structural organization of the hamster multifunctional protein CAD J. Biol Chem. 1992;267:7717–7784. [PubMed] [Google Scholar]
- Maier T, Zhou L, Thornburg R. Nucleotide sequence of a cDNA encoding UMP synthase from Nicotiana tabacum (GenBank U22260) Plant Physiol. 1995;108:1747. [Google Scholar]
- Mohn WW, Tiedje JM. Microbial reductive dehalogenation. Microbiol Rev. 1992;56:482–507. doi: 10.1128/mr.56.3.482-507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasr F, Berthauche N, Dufour M-E, Minet M, Lacroute F. Heterospecific cloning of Arabidopsis thaliana cDNAs by direct complementation of pyrimidine auxotrophic mutants of Saccharomyces cerevisiae: I. Cloning and sequence analysis of two cDNAs catalyzing the second, fifth and sixth steps of the de novo pyrimidine biosynthesis pathway. Mol Gen Genet. 1994;244:23–32. doi: 10.1007/BF00280183. [DOI] [PubMed] [Google Scholar]
- Neuhard J, Nygaard P. Purines and pyrimidines. In: Neidhardt FC, Ingraham JL, Low BK, Magasanik B, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Washington, DC: American Society for Microbiology; 1987. pp. 445–473. [Google Scholar]
- Reinbothe S, Ortel B, Parthier B. Overproduction by gene amplification of the multifunctional arom protein confers glyphosate tolerance to a plastid-free mutant of Euglena gracilis. Mol Gen Genet. 1993;239:416–424. doi: 10.1007/BF00276940. [DOI] [PubMed] [Google Scholar]
- Santoso D, Thornburg R. UMP synthase is transcriptionally regulated by pyrimidine levels in Nicotiana plumbaginifolia. Plant Physiol. 1998;116:815–821. doi: 10.1104/pp.116.2.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoso D, Thornburg RW. Isolation and characterization of UMP synthase mutants from haploid cell suspensions of Nicotiana tabacum. Plant Physiol. 1992;99:1216–1225. doi: 10.1104/pp.99.3.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer B, Dicker A, Bertino J. Dihydrofolate reductase as a therapeutic target. FASEB J. 1990;4:2441–2452. doi: 10.1096/fasebj.4.8.2185970. [DOI] [PubMed] [Google Scholar]
- Shyr Y, Hepburn A, Widholm J. Glyphosate selected amplification of the 5-enolpyruvylshikimate-3-phosphate synthase gene in cultured carrot cells. Mol Gen Genet. 1992;232:377–382. doi: 10.1007/BF00266240. [DOI] [PubMed] [Google Scholar]
- Suttle D. A reversible selection system for UMPsynthase gene amplification and deamplification. Somat Cell Mol Genet. 1989;15:435–443. doi: 10.1007/BF01534894. [DOI] [PubMed] [Google Scholar]
- Timmons ED, Dunbar BS. Protein blotting and immunodetection. Method Enzymol. 1990;182:679–687. doi: 10.1016/0076-6879(90)82053-5. [DOI] [PubMed] [Google Scholar]
- Tirimanne TS, Colbert JT. Transient down-regulation of phytochrome mRNA abundance in etiolated cucumber cotyledons in response to continuous white light. Plant Physiol. 1991;97:1581–1584. doi: 10.1104/pp.97.4.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut TW, Jones ME. Inhibitors of orotate phosphoribosyl-transferase and orotidine-5′-phosphate decarboxylase from mouse ehrlich ascites cells: a procedure for analyzing the inhibition of a multi-enzyme complex. Biochem Pharmacol. 1977;26:2281–2291. doi: 10.1016/0006-2952(77)90293-3. [DOI] [PubMed] [Google Scholar]
- van den Hoff MJ, Jonker A, Beintema JJ, Lamers WH. Evolutionary relationships of the carbamoylphosphate synthase genes. J Mol Evol. 1995;41:813–832. doi: 10.1007/BF00173161. [DOI] [PubMed] [Google Scholar]
- Volkenandt M, Schweitzer B, Otter G, Schmid F, Sirontnak F, Bertino J. Methotrexate resistance in vivo mouse tumor due to a non-active-site dihydrofolate reductase mutation. Proc Natl Acad Sci USA. 1993;90:11797–11801. doi: 10.1073/pnas.90.24.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth GJ, Redinbaugh MG, Scandalios JG. A procedure for the small-scale isolation of plant RNA suitable for RNA blot analysis. Anal Biochem. 1988;172:279–283. doi: 10.1016/0003-2697(88)90443-5. [DOI] [PubMed] [Google Scholar]
- Wahl G, Vitto L, Padgett R, Stark G. Single-copy and amplified CAD genes in Syrian hamster chromosomes localized by a highly sensitive method for in situ hybridization. Mol Cell Biol. 1982;2:308–319. doi: 10.1128/mcb.2.3.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C. Degradation of pyrimidines: enzymes, localization and role in metabolism. Biochem Physiol Pflanzen. 1978;173:467–499. [Google Scholar]