Significance
Discrimination of clinically relevant mutations from neutral mutations is of paramount importance in precision medicine and pharmacogenomics. Our study shows that current computational predictions of pathogenicity, mostly based on analysis of sequence conservation, may be improved by considering the changes in the structural dynamics of the protein due to point mutations. We introduce and demonstrate the utility of a classifier that takes advantage of efficient evaluation of structural dynamics by elastic network models.
Keywords: structural dynamics, missense variants, elastic network models, machine learning
Abstract
Accurate evaluation of the effect of point mutations on protein function is essential to assessing the genesis and prognosis of many inherited diseases and cancer types. Currently, a wealth of computational tools has been developed for pathogenicity prediction. Two major types of data are used to this aim: sequence conservation/evolution and structural properties. Here, we demonstrate in a systematic way that another determinant of the functional impact of missense variants is the protein’s structural dynamics. Measurable improvement is shown in pathogenicity prediction by taking into consideration the dynamical context and implications of the mutation. Our study suggests that the class of dynamics descriptors introduced here may be used in conjunction with existing features to not only increase the prediction accuracy of the impact of variants on biological function, but also gain insight into the physical basis of the effect of missense variants.
Significant advances have been made in recent years in collecting data on single-nucleotide polymorphisms (SNPs) and developing computational strategies for identifying disease-causing variants among sequence alterations induced by nonsynonymous SNPs (1–11). Such investigations greatly benefited from the creation of publicly available databases of mutations found in humans and computational tools developed for pathogenicity prediction (2, 11–14). Sequence conservation/evolution analyses using machine learning methods is a common approach in those tools. Mainly, the relative frequencies of wild-type (WT) and mutated amino acids at the given mutation site are evaluated from multiple sequence alignments (MSAs) of homologous proteins. While this approach has proved effective, it suffers from the usual shortcomings associated with the creation of sufficiently populated and variegated MSAs. Besides, the intuitive correlation between sequence conservation and low tolerance to mutations does not depict the whole scenario: In many cases, variants at nonconserved positions exhibit a complex pattern of functional modifications, stemming from the very fact that the lack of conservation may be a source of variability allowing for the functional specialization of proteins belonging to the same family. Functional/dysfunctional effects induced by mutations at nonconserved positions often do not correlate with the evolutionary frequencies of the amino acids (15).
In view of these limitations, the need for devising prediction tools that take account of the physical properties of the protein, complementing those implied by evolutionary statistics, has become clear. Currently (February 2018), there are more than 6,300 UniProt Knowledgbase entries out of 71,772 proteins reported in the Homo sapiens proteome that have structural data in the Protein Data Bank (PDB). Several studies demonstrated the benefits of considering structural features in the evaluation of the impact of single amino acid variants (SAVs) on functionality and in the identification of disease-causing mutations (16–19). Among them, PolyPhen-2 (2) is a broadly used tool that showed significant success by including various structural properties as parameters in the evaluation of a variant’s pathogenicity. However, there is a need to further improve our ability to discriminate between disease-causing and neutral mutations, given the discrepancy that still exists between predicted and actual consequences of many missense mutations (20).
The present study shows the utility of considering the equilibrium dynamics of the protein as a means of improving the predictive ability of current pathogenicity predictors. The underlying assumption is that the mutations that interfere with structural flexibility or conformational mechanics are likely to cause dysfunction. Efficient screening of protein dynamics at the structural proteome scale requires the adoption of simple but robust methods. To this aim, we evaluated a set of features uniquely defined by 3D topology, generated by elastic network models (ENMs) for proteins (21), using the application programming interface ProDy (22). Earlier work demonstrated the relevance of ENM-predicted motions to biomolecular mechanisms of function and interactions (23, 24).
ProDy outputs include the identity of sites implicated in mediating collective motions, conveying allosteric signals, or ensuring adaptability to promiscuous functionality (25). To evaluate the contributions of such dynamic features, we used as a benchmarking set an updated collection of labeled variants previously employed for comparing other prediction methods (26). Our study shows that a measurable improvement can be achieved in the classification of variants upon considering their impact on the structural dynamics. Furthermore, the approach provides a framework for gaining insight into the possible origins of the observed effects of mutations, beyond those inferred by sequence or structure analyses alone.
Results and Discussion
Datasets of Variants and Pathogenicity Prediction Tools.
We used five datasets, including HumVar (2), ExoVar (11), VariBenchSelected, predictSNPSelected, and SwissVarSelected, which have been manually curated to minimize possible overlaps and proposed to serve as a benchmarking set (26). The latter three, indicated by the suffix “Selected”, are subsets of VariBench (12), predictSNP (13), and SwissVar (14), respectively, obtained upon clearing entries already represented in the former two most populated datasets (i.e., HumVar and ExoVar). Such preliminary filtering has been performed to allow for a fair comparison of the performances of pathogenicity predictors and to remove “training bias”—that is, any bias that might originate from partial overlap between the corresponding training and testing datasets. The five datasets differ considerably in their size and proportion of deleterious vs. neutral variants (SI Appendix, Table S1). They also use different criteria for assigning the deleterious/neutral label to a given variant (26). The construction of the datasets, their main differences, the peculiarities of the prediction tools, and methods for accurate comparison of their performance are described in previous work (26).
In addition, we generated an Integrated Dataset using the content of these five datasets, composed of all 20,413 nonoverlapping SAVs that have been structurally characterized. This dataset is proposed to serve as a benchmark dataset for structure-based evaluations of the functional consequences of mutations.
The binary classification of variants into deleterious or neutral might be an oversimplification, being inadequate for capturing the variegate spectrum of effects induced by a mutation [see, for instance, the distinction between “rheostat” and “toggles” in an earlier study (15)]. However, our aim is to demonstrate in a quantitative way the utility of adopting new features based on structure-encoded dynamics and providing a classifier that permits the assessment of the pathogenicity of SAVs in light of protein dynamics. This type of binary measure allows us to have access to a sufficiently large source of data as input. The outputs, on the other hand, help shed light on possible structural and dynamic origins of the impact of mutations at the molecular level.
Dynamics-Derived Features and DYN/SEQ-Based Predictors.
To explore whether features derived from structural dynamics, referred to as dynamical (DYN) features, can help improve the overall accuracy of variant classification, we considered the following features: (i) gaussian network model (GNM)-based mean-square fluctuations (MSF) of residues, where minima indicate the sites that potentially act as hinges for supporting the protein’s mechanics; (ii) the propensity of residues to act as sensors or as effectors of allosteric signals (27) based on perturbation-response scanning (PRS) analysis (28) of their ability to sense or transmit local perturbations; (iii) the mechanical bridging score (MBS) (29), which quantifies the role of each residue in maintaining the stability of the protein modeled as an anisotropic network model (ANM); and (iv) the mechanical stiffness associated with each residue, as computed by MechStiff (30). In addition to these features, we included a structural property that is a major determinant of conformational flexibility, the solvent-accessible surface area (SASA), computed using the DSSP program (31). See SI Appendix, Supplementary Methods for more details on the physical meaning and origin of these features.
DYN features capture the global properties of the protein—that is, those originating from the overall 3D topology of interresidue contacts. As such, they provide a metric for assessing the effect of mutation on the overall (global) dynamics, as opposed to physicochemical features such as SASA, which depend on the local geometry.
DYN features are purely position dependent: They do not depend on the identity of the amino acid at that position. To distinguish between variants that occur at the same site but involve different amino acid mutations, we used two sequence-dependent (SEQ) features extracted from PolyPhen-2 (2, 32): (i) the conservation score of the WT amino acid represented by the position-specific independent counts (PSIC) score (WT PSIC); and (ii) the difference between the PSIC scores of the WT and mutant amino acids (ΔPSIC). The classification of SAVs into deleterious or neutral was performed using a Random Forest (RF) metaestimator, implemented in the open-source machine learning Python library Scikit-learn (33). The method is robust to overfitting and requires minimal parameter optimization (see SI Appendix, Supplementary Methods and Fig. S1 for details).
Comparative Analysis Highlights the Utility of DYN Features for Accurate Assessment of the Effect of Mutations.
For assessing the importance of including protein dynamics in pathogenicity prediction, we compared the output from the DYN/SEQ-based predictors to those obtained with 11 pathogenicity prediction tools: (i) Mutation Taster-2; (ii) PolyPhen-2; (iii) Mutation Assessor; (iv) Combined Annotation Dependent Depletion; (v) SIFT; (vi) likelihood ratio test; (vii) FatHMM-U; (viii) GERP++ and (ix) phyloP, which have been developed independently; and (x) Condel and (xi) Logit, which are metapredictors that combine the predictions from PolyPhen-2, SIFT, and Mutation Assessor. Three of these tools (Mutation Taster-2, PolyPhen-2, and Mutation Assessor) have been partially trained on some of the benchmark datasets. Thus, their predictions, and those of the two metapredictors that utilize them, are affected by training bias (26). Each tool returns a pathogenicity score (accessible in the supplementary data of ref. 26) for each variant, which represents the expected probability of having a deleterious effect on function. In addition to these 11 predictors, we also considered three predictors—FatHMM-W, Condel+, and Logit+—that have been reported to suffer from the so-called type 2 bias (26): in these three cases, the classifier is biased toward assigning one dominant class of SAVs, deleterious or neutral, to all mutations in a given protein. These predictors benefit from the fact that many proteins in the available datasets contain almost exclusively one class of variants (either neutral or deleterious). The set of 14 tools, including these additional three, is called the extended set of predictors.
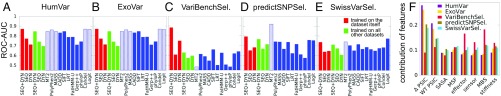
Results are presented in Fig. 1 A–E. For each dataset, shown on a separate panel, we report the results from testing our predictor based on SEQ or DYN features exclusively and on their combination (SEQ+DYN). Two sets of results are presented: one for the RF classifiers trained/tested through cross-validation on the same dataset (red bars in Fig. 1 A–E) and the other for a classifier trained on the four other datasets (green bars). The results from the 11 predictors listed above are shown in solid blue bars in Fig. 1 A–E, and those benefiting from training bias in dashed blue bars. SI Appendix, Fig. S2 displays the counterpart of Fig. 1 A–E for the extended set of predictors, with the results from the three additional predictors shown in gray bars. The prediction accuracy is measured by the area under the curve (AUC) evaluated for the receiver operating characteristic (ROC) curve. The AUC is 0.5 for random classification (main diagonal), and 1.0 for perfect classification. SI Appendix, Fig. S3 illustrates the ROC curves [i.e., true-positive (TP) rate (sensitivity) against false-positive (FP) rate (specificity)] obtained using our classifiers (SI Appendix, Fig. S3A) and the extended set of classifiers (SI Appendix, Fig. S3B) on the Integrated Dataset.
Fig. 1.
Comparison of the performance of the introduced Random Forest (RF) classifiers (SEQ+DYN, SEQ, and DYN) and existing tools for pathogenicity prediction. (A–E) AUC values derived from ROC plots (SI Appendix, Fig. S3) are presented for five datasets as indicated. The red bars refer to a 10-fold cross-validated classification on the dataset used for learning the RF classifiers; green bars refer to the RF classifiers trained on the other four datasets combined and tested on the given dataset. Solid blue bars show the AUC values from existing tools, obtained from ref. 26; dashed blue bars refer to those predictors potentially trained on the testing dataset (training bias). See SI Appendix, Fig. S2 for the results from an extended set of tools. (F) Relative contribution of eight features used in RF classifiers to pathogenicity assessment. Results for each dataset are shown in a different color. The first two features (SEQ) are residue specific, based on conservation (WT PSIC) score and its change upon mutation (ΔPSIC); the last six (DYN) are nonspecific. They account for flexibility and accessibility (SASA and MSF), allosteric properties (effector and sensor), and mechanical properties (MBS and stiffness) of sites on the 3D structure, regardless of amino acid identity. CADD, Combined Annotation Dependent Depletion; LRT, likelihood ratio test; MASS, Mutation Assessor; MT2, Mutation Taster-2.
Several observations are made in Fig. 1 A–E and SI Appendix, Fig. S2. First, despite its simplicity [e.g., being trained on a reduced set of structurally known proteins (about 25% of the complete set); SI Appendix, Table S1] and the use of a small number of easily computed features, SEQ+DYN predictions exhibit accuracy levels comparable to, and in some cases better than, those obtained by the other advanced methods. The AUCs for SEQ+DYN rank always among the top when excluding the cases affected by training bias. Second, SEQ+DYN performance shows little dependency on the training procedure (red vs. green bars), whereas other methods generally show a pronounced decrease in AUC when tested against datasets other than their training datasets (compare the dashed and solid blue bars for the same method across different panels in Fig. 1 A–E). An outlier is the VariBenchSelected dataset in Fig. 1C, which will be discussed later.
Closer examination shows that the SEQ-only classifier outperforms DYN-only in the cases of the HumVar dataset (red and green bars in Fig. 1 A–E), the ExoVar dataset (red bars), and the specialized SwissVarSelected dataset (green bars), distinguished by a low population of deleterious SAVs. This dominant role of SEQ features is also supported by the analysis of the relative contributions (weights) of features, presented in Fig. 1F. A plausible explanation is the consideration of the specific type of amino acid substitution by SEQ features, whereas DYN features are solely based on the position of the mutated residue. On the other hand, the usefulness of SEQ features depends crucially on the quality of the MSA used for computing them, which explains why their contribution is particularly strong in the two datasets (HumVar and ExoVar) specifically designed for training PolyPhen-2. In contrast, the DYN classifier outperforms the SEQ classifier when tested against VariBenchSelected and predictSNPSelected.
As previously mentioned, VariBenchSelected exhibits a unique behavior: the AUC plot in Fig. 1C shows an unusually high accuracy in the SEQ+DYN cross-validation analysis. The disparity between the red and green bars in Fig. 1C suggests that this behavior originates from the nature of the dataset itself. A closer investigation shows that a considerable fraction of SAVs (∼40%; see SI Appendix, Table S1) in this dataset are in the form of multiple mutations at the same site in a given protein—that is, there is a preponderance of variants with different types of amino acid substitutions at the same position. In addition, nearly all such same-site variants are assigned the same pathogenicity class (see SI Appendix, Table S1, column 6). This leads to a situation where the classifier is trained to assign less weight to amino acid identity and more to its position; hence, the success of DYN features (red bars in Fig. 1F), which are agnostic to amino acid identity but take account of the position in the 3D structure. This also explains the high AUC values in SEQ+DYN cross-validation.
RF Classifier Trained on the Integrated Dataset Outperforms Existing Unbiased Predictors.
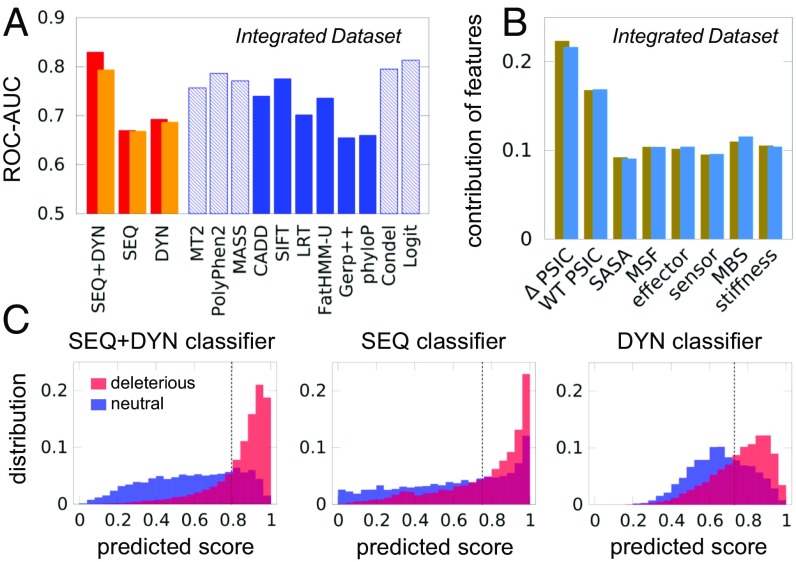
We also evaluated the level of accuracy obtained by the RF classifiers applied to the Integrated Dataset, using a 10-fold cross-validation procedure. Fig. 2A presents the results in comparison with other prediction tools. The SEQ+DYN classifier outperforms all others in this case. The accuracy (AUC) obtained by SEQ+DYN (first red bar in Fig. 2A) is 0.83, the highest among all the considered tools, except for those benefiting from type 2 bias (SI Appendix, Fig. S4A). Since same-site SAVs amount to a significant fraction in some datasets (SI Appendix, Table S1), we repeated the analysis by making sure that same-site variants were not simultaneously present in both the training and test sets. The results (orange bars in Fig. 2A), show a slight decrease in the AUC (0.79) for the SEQ+DYN classification. The method’s accuracy remains higher, however, than all other unbiased methods.
Fig. 2.
Performance of pathogenicity prediction tools tested against the Integrated Dataset. (A) The 10-fold cross-validated accuracy measure for the three RF classifiers (red bars) is compared with those of other predictors (blue bars). Dashed blue bars refer to cases possibly affected by training bias; orange bars describe analogous results after excluding same-site variants from training and test sets. See SI Appendix, Fig. S4 for additional comparisons, and SI Appendix, Table S3 for the results from additional performance metrics. (B) Feature importance plot for the SEQ+DYN classification (tan bars). Blue bars correspond to orange bars in A. (C) Distribution of predicted scores for neutral (shown in blue) and deleterious (shown in red) variants in the Integrated Dataset, for the three classifiers. The dashed vertical line represents the cutoff based on Youden’s index. CADD, Combined Annotation Dependent Depletion; LRT, likelihood ratio test; MASS, Mutation Assessor; MT2, Mutation Taster-2.
It is interesting to note in Fig. 2A that the DYN-based classifier slightly outperformed the SEQ-based classifier; this may be attributed to limitations in MSA quality and the inclusion of only two SEQ features, as opposed to six DYN features. Indeed, the individual SEQ features make larger contributions to decision making (Fig. 2B, tan bars) than individual DYN features. Exclusion of same-site SAVs had a minimal effect on the contribution of features (Fig. 2B, blue bars). Fig. 2C depicts in a more comprehensible manner the discriminatory power of the three classifiers, displaying the histograms of predictions (scores) collected during cross-validation. The SEQ+DYN histogram is used to evaluate pathogenicity probabilities (SI Appendix, Fig. S5).
We further compared the performance of the different tools using an expanded list of metrics, listed in SI Appendix, Table S2. Since the class imbalance might skew a few specific metrics, like the AUC of the precision-recall curve (SI Appendix, Fig. S6), we also provide as a reference the comparison with random classifications, artificially biased toward either deleterious or neutral classes. Results in SI Appendix, Table S3 show that the SEQ+DYN classifier ranks among the top performers across all metrics, even when considering those quantities centered on predictions of neutrals [e.g., specificity = TN/(FP+TN) and negative predictive value = TN/(TN+FN), where TN is true negative and FN is false negative].
Additional comparison reveals the decrease in the performance of the three tools benefiting from type 2 bias when datasets of proteins with more balanced distributions of deleterious and neutral mutations are used as benchmark. SI Appendix, Fig. S4B displays the results for the complete set of “mixed” proteins, which have both neutral and deleterious mutations. The performances of our RF classifiers and the 11 prediction tools are only moderately lowered, if any, compared with those observed for the original (Integrated) dataset, whereas there is a drop in the AUC values of the three tools (gray bars in SI Appendix, Fig. S4B) that no longer benefit from type 2 bias. This effect is further pronounced when considering increasingly smaller subsets of proteins, with the deleterious-to-neutral ratio progressively approaching 1 (SI Appendix, Fig. S7). These results demonstrate that our classifiers are robust against the changes in the dataset composition.
Examination of Significance of DYN Features Reveals the Competing Roles of Allosteric Signaling Sites.
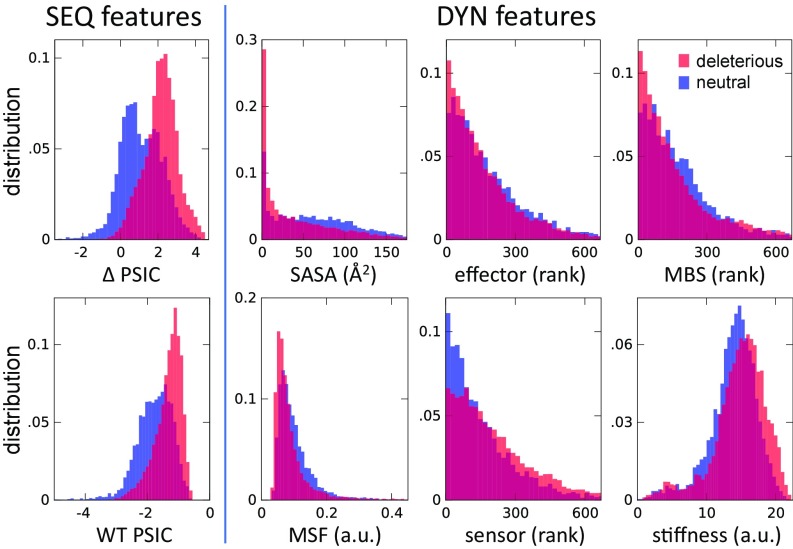
Our analysis provides insight into the role of structural dynamics in general, and individual dynamic features in particular, in shaping the effect of missense variants. Fig. 3 provides a visual assessment of the ability of each of the SEQ and DYN features to discriminate between deleterious and neutral SAVs. In each case, we display two histograms, representing the values (or rank orders) observed for deleterious (shown in red) or neutral (shown in blue) variants. These data reveal several features. First, the strong discriminatory power of SEQ features (WT PSIC and ΔPSIC) is confirmed. Second, neutral SAVs exhibit relatively higher SASA and MSF values, consistent with the adaptability of the structure to accommodate spatial changes with increasing conformational flexibility. Conversely, the distribution of deleterious SAVs is skewed toward lower MSFs. Similarly, the sites distinguished by a high mechanical stiffness are likely to give rise to deleterious SAVs, and top-ranking residues involved in mechanical bridging tend to induce deleterious effects if mutated.
Fig. 3.
Histograms of SEQ and DYN features for neutral and deleterious variants. Each panel refers to a distinctive feature (see abscissa). The two histograms refer to the subgroups of deleterious (shown in red) and neutral (shown in blue) mutations. Sharper differences indicate higher discrimination power of the feature.
The effect of sensors and effectors is more complex. As described earlier, these play a role in substrate recognition and/or allosteric regulation. One might expect them to impair functionality if mutated. Top-ranking effectors indeed show such an effect. On the other hand, sensors exhibit the opposite behavior (i.e., mutations at those sites are instead correlated with nonpathogenic effects), and this effect is quite pronounced at the top-ranking sensors. A careful consideration of the role of sensors can explain this somewhat surprising result. A previous application to molecular chaperones (27) showed that sensors are frequently found close to substrate or cochaperone recognition sites and are characterized by strong coevolutionary propensities, which has been attributed to the necessity of adapting to binding different substrates. Based on current data, we can also deduce that such variability is a symptom of an intrinsic ability to accommodate diverse interactions, especially in promiscuous proteins. Therefore, mutations at those sites do not necessarily cause a loss of function; on the contrary, they may be essential to gain of function. This is in stark contrast to the behavior of effectors (28), which are presumed to play a central role in mediating allosteric signals (27) and are sensitive to mutations.
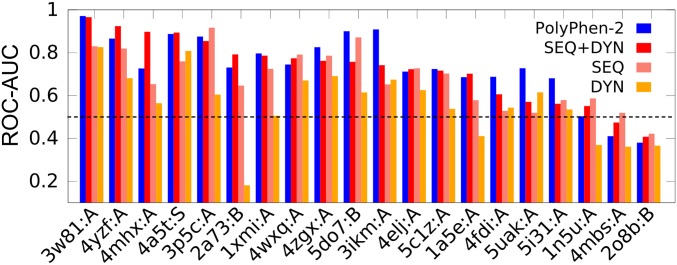
The DYN-based classifier can detect a new class of deleterious sites while assisting in improving pathogenicity predictions. To illustrate the type of information one can obtain from DYN-based predictors, we present a few applications. For a critical assessment, we focus on mixed proteins that contain at least six neutral and six deleterious mutations. The Integrated Dataset contains 20 such cases (SI Appendix, Table S4). We classified the variants in each case using a RF classifier trained on the SAVs belonging to all other proteins in the Integrated Dataset. The results are presented in Fig. 4. The figure also shows the AUC values obtained with PolyPhen-2, selected as a reference. Note that the PolyPhen-2 dataset benefits from training bias, as its training datasets (HumVar and ExoVar) contain data for most of the variants of these 20 proteins. Despite the large variability in accuracy levels between these 20 proteins, SEQ+DYN (red bars in Fig. 4) and PolyPhen-2 (blue bars) share comparable AUC values. Specifically, the pathogenicity of SAVs in certain proteins cannot be accurately predicted using either approach (the rightmost three cases in Fig. 4, where results are poorer than random); whereas others (on the left) lend themselves to accurate prediction. Of the 17 cases where the results are better than random, 11 yielded more accurate results when the SEQ+DYN classifier was adopted as opposed to SEQ-only, consistent with the overall improvement in pathogenicity prediction upon incorporation of DYN features.
Fig. 4.
Comparative analysis of prediction accuracy of DYN- and/or SEQ-based RF classifiers and PolyPhen-2 applied to 20 mixed proteins. The proteins (listed in SI Appendix, Table S4) are organized along the abscissa in the order of decreasing AUC obtained by the SEQ+DYN classifier.
A closer examination of the lower performance observed in the other six cases suggests that the degree of collectivity of global modes, as well as environmental effects, affects the accuracy of DYN predictions. SI Appendix, Fig. S8 presents a careful analysis of the performance of the DYN classifier as a function of the collectivity of the most probable (soft) motions accessible to the 20 proteins. A preference for higher AUC values is discernable with increasing collectivity, suggesting that proteins whose soft modes are dominated by localized fluctuations (e.g., isolated loop motions rather than cooperative domain rearrangements) may lead to misinterpretations of DYN features. We furthermore analyzed two striking cases: (i) the human complement component C3 (PDB ID code 2a73, chain B), where SEQ+DYN outperforms PolyPhen-2, although the DYN classifier alone shows a very poor performance (Fig. 4); and (ii) the human sterol transporter (PDB ID code 5do7, chain B), which yields a lower AUC using the SEQ+DYN classifier compared with that obtained by SEQ-only. In both cases, the biological assembly comprises multiple chains. Reevaluation of DYN features by considering the intact structures of the assemblies instead of the single chains leads to improved AUC values (SI Appendix, Fig. S8). This analysis highlights the importance of considering the biological assembly for improved evaluation of DYN features.
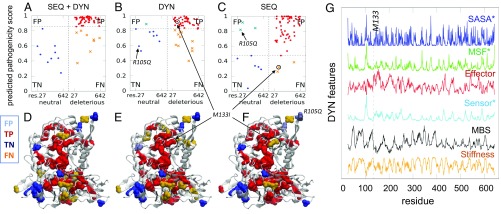
In Fig. 5 and SI Appendix, Fig. S9, we examine more closely two other cases. The confusion matrices in Fig. 5 A–C and SI Appendix, Fig. S9 A–C display the predicted pathogenicity score as a function of residue index, organized in two classes: neutral and deleterious residues. Each class is further divided into two subgroups, depending on predicted pathogenicity scores: TPs and FNs (for deleterious sites) and FPs and TNs (for neutral sites). The threshold scores that separate these subgroups are optimized to maximize the differentiation between the subgroups. A “perfect” classifier would populate the TP block (Fig. 5 A–C and SI Appendix, Fig. S9 A–C, Upper Right, red dots) and TN block (Lower Left, blue dots) of the confusion matrix, and exclude the FP block (Upper Left, cyan x’s) and FN block (Lower Right, orange x’s). It is interesting to note that among SEQ+DYN misclassifications, FNs are much more common than FPs—that is, the classifier misses a few deleterious SAVs, while it correctly predicts almost all neutral SAVs.
Fig. 5.
Illustration of detailed analysis for human α-l-iduronidase (PDB ID code 3w81, chain A). (A–C) Confusion matrices for the three RF classifiers—that is, the predicted pathogenicity score for the two subgroups of variants, ordered by residue index along the abscissa. Lower Left and Upper Right submatrices represent the accurately predicted neutral variants (TN, blue dots) and deleterious variants (TP, red dots). The off-diagonal entries are the FPs (cyan x’s) and FNs (orange x’s). The threshold value (horizontal dashed line) for pathogenicity is chosen according to the Youden’s index (39) (SI Appendix, Supplementary Methods). (D–F) Corresponding ribbon diagrams, where mutation sites (spheres) are color-coded by the dots in the confusion plots (i.e., blue or red refers to correctly predicted neutral or deleterious residues, respectively). A number of FNs (shown in orange) in E occur on the surface of the protein, where amino acid-specific features presumably weigh more than structural/dynamical ones in defining functionality (e.g., recognition). The prediction for variant R105Q, on the other hand, estimated to be an FP (shown in cyan) based on sequence only, is now corrected upon inclusion of DYN features. (G) DYN feature profiles as a function of residue index. The dashed vertical line indicates the position of M133, recognized as a TP by the DYN classifier and misclassified by the SEQ classifier. The asterisks indicate those features for which low values give rise to deleterious mutations.
For a closer examination of the outcomes, we generated color-coded diagrams (Fig. 5 D–F and SI Appendix, Fig. S9 D–F) that enable the comparative visualization of the accurate predictions (TP shown in red and TN shown in blue) and inaccurate predictions (FP shown in cyan and FN shown in orange). Deleterious SAVs (red and orange) are usually located in the protein’s interior, and those incorrectly predicted to be neutral (orange) are usually on the surface, signaling that more discriminative classifiers are needed to detect those deleterious sites. We note in this respect that the DYN classifier assists in such cases. An example is M133I in the hydrolase illustrated in Fig. 5. The latter is misclassified as neutral by SEQ-based predictor because M133 is not evolutionarily conserved. On the other hand, it is correctly recognized to be potentially deleterious, if mutated, by the DYN classifier, as it satisfies many DYN criteria: high propensity to act as effector, low propensity to act as a sensor, low conformational flexibility (probed by SASA and MSF), and high stiffness and mechanical bridging ability (Fig. 5G); and the DYN features dominate the outcome in the DYN+SEQ classifier. SI Appendix, Fig. S9 illustrates a case where a mutation (R589H in an anion transport protein) inaccurately assessed to be neutral by either the SEQ or DYN classifier is correctly predicted to be deleterious using SEQ+DYN. These examples suggest that the SEQ+DYN classifier can synergistically predict the actual effects of the missense variants when SEQ and/or DYN classifiers fail to do so.
A Test Case: CFTR Variants.
A recent study of cystic fibrosis transmembrane conductance regulator (CFTR) variants (34) presents a list of variants organized into three categories: (i) those commonly associated with cystic fibrosis (CF); (ii) those associated with a bicarbonate defect in channel function, leading to disorders like pancreatitis but not cystic fibrosis (BD); and (iii) those reported in previous chronic pancreatitis genetic studies, but without strong evidence of pathogenicity (“others”); see SI Appendix, Fig. S10A. Results from our evaluation of these variants are presented in SI Appendix, Table S5 and Fig. S10B. The most striking observation is that 9 of 13 “other” variants are classified as neutral, in contrast to most of the assignments listed in the Integrated Dataset (SI Appendix, Table S5, column 11) and most of the predictions from PolyPhen-2. Moreover, 6 of 10 CF/BD variants are classified as deleterious, in line with the results of the pancreatitis study (34). It is remarkable that most of deleterious variants predicted by our classifier fall in the CF/BD categories. The remaining variants, whose functional impact is still debated, mostly predicted to be neutral, will need future studies for possible verification.
Conclusion
With the steady increase in genome-scale data made available in recent years, it has become essential to develop tools that can extract useful information in a systematic, efficient, and robust way. In this study, we built on past research in the field of pathogenicity prediction of SAVs, as well as recent advances in genome-scale characterization of protein dynamics (35), to test and demonstrate the validity of our hypothesis: that structural dynamics, not only sequence or structure, might be considered a determinant of the effect of missense variants on biological function. Our analysis showed that a measurable improvement is achieved when DYN and SEQ features are combined.
Our study also provided insight into the interpretation of the functional impact of variants in the light of the intrinsic dynamics of the mutated site. We could confirm the current understanding of such quantities with regard to the localization of dynamically important residue positions that are more likely to incur detrimental mutations. In the specific case of the sensitivity measurements obtained from PRS analysis, however, we produced evidence in support of the concept that residues identified as sensors are usually associated with neutral variants (i.e., they are able to accommodate amino acid substitutions despite their role in allosteric signaling). This behavior contrasts that of effectors of signaling, whose mutations were predominantly deleterious. While the current DYN features have been evaluated for individual proteins/subunits, detailed examination suggests that the DYN predictions may be further refined upon consideration of environmental effects (SI Appendix, Fig. S8).
Lastly, we focused on a few case studies that highlighted the tendency of deleterious mutations to localize in the core of a protein. A corollary would be that variants at exposed regions would be neutral, but this is not the case. The frequent occurrence of FNs at those regions indicates that accurate prediction of (dys)functional regions remains a challenge. This is mainly due to a competition between adaptability to promiscuous interactions (which are functional in a given organism/pathway and need to be retained) and the inherent conformational malleability (which can tolerate substitutions without affecting other regions). The combination of DYN- and SEQ-based features emerges as a useful tool for improving the accuracy of predictions at such challenging sites.
Materials and Methods
SAV datasets for training and testing of the RF classifiers and evaluating pathogenicity scores and labels for the 14 prediction tools were extracted from previous work (26) and summarized in SI Appendix, Table S1. The variants used in this work are those SAVs for which an associated PDB structure exists. We mapped between SAV sequences and PDB structures using the UniProt database (36). GNM- and ANM-predicted DYN properties were calculated using the ProDy application programming interface (22). MBS (29) was computed with code adapted from ref. 37, and SASAs were computed using the DSSP program (31). The SEQ features based on the PSIC score (38) were extracted from PolyPhen-2 (2, 32). The details on the DYN/SEQ features and the RF algorithm are presented in SI Appendix, Supplementary Methods. The method presented in this paper has been implemented on the web server RAPSODY (Re-Assessment of Pathogenicity of SAVs based On Dynamics; rapsody.csb.pitt.edu/). The integrated dataset used for training and the source code are available at rapsody.csb.pitt.edu/download.html.
Supplementary Material
Acknowledgments
We acknowledge useful discussions with Dr. David Whitcomb on CFTR variants and with Dr. Sean Mooney on PolyPhen-2. Support from NIH Grants P41 GM103712 and U54 HG008540 (to I.B.) is gratefully acknowledged.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.L.J. is a guest editor invited by the Editorial Board.
Data deposition: The method presented in this paper has been implemented on the web server RAPSODY (Re-Assessment of Pathogenicity of SAVs based On Dynamics; rapsody.csb.pitt.edu/). The integrated dataset used for training and the source code are available at rapsody.csb.pitt.edu/download.html.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715896115/-/DCSupplemental.
References
- 1.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 2.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shihab HA, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davydov EV, et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper GM, Shendure J. Needles in stacks of needles: Finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12:628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 10.González-Pérez A, López-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88:440–449. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li MX, et al. Predicting mendelian disease-causing non-synonymous single nucleotide variants in exome sequencing studies. PLoS Genet. 2013;9:e1003143. doi: 10.1371/journal.pgen.1003143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasidharan Nair P, Vihinen M. VariBench: A benchmark database for variations. Hum Mutat. 2013;34:42–49. doi: 10.1002/humu.22204. [DOI] [PubMed] [Google Scholar]
- 13.Bendl J, et al. PredictSNP: Robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput Biol. 2014;10:e1003440. doi: 10.1371/journal.pcbi.1003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mottaz A, David FPA, Veuthey AL, Yip YL. Easy retrieval of single amino-acid polymorphisms and phenotype information using SwissVar. Bioinformatics. 2010;26:851–852. doi: 10.1093/bioinformatics/btq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller M, Bromberg Y, Swint-Kruse L. Computational predictors fail to identify amino acid substitution effects at rheostat positions. Sci Rep. 2017;7:41329. doi: 10.1038/srep41329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlsson J, Soussi T, Persson B. Investigation and prediction of the severity of p53 mutants using parameters from structural calculations. FEBS J. 2009;276:4142–4155. doi: 10.1111/j.1742-4658.2009.07124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujimoto A, et al. Systematic analysis of mutation distribution in three dimensional protein structures identifies cancer driver genes. Sci Rep. 2016;6 doi: 10.1038/srep26483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamburov A, et al. Comprehensive assessment of cancer missense mutation clustering in protein structures. Proc Natl Acad Sci USA. 2015;112:E5486–E5495. doi: 10.1073/pnas.1516373112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solomon O, et al. G23D: Online tool for mapping and visualization of genomic variants on 3D protein structures. BMC Genomics. 2016;17:681. doi: 10.1186/s12864-016-3028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miosge LA, et al. Comparison of predicted and actual consequences of missense mutations. Proc Natl Acad Sci USA. 2015;112:E5189–E5198. doi: 10.1073/pnas.1511585112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahar I, Lezon TR, Yang L-W, Eyal E. Global dynamics of proteins: Bridging between structure and function. Annu Rev Biophys. 2010;39:23–42. doi: 10.1146/annurev.biophys.093008.131258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakan A, Meireles LM, Bahar I. ProDy: Protein dynamics inferred from theory and experiments. Bioinformatics. 2011;27:1575–1577. doi: 10.1093/bioinformatics/btr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakan A, Bahar I. The intrinsic dynamics of enzymes plays a dominant role in determining the structural changes induced upon inhibitor binding. Proc Natl Acad Sci USA. 2009;106:14349–14354. doi: 10.1073/pnas.0904214106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tobi D, Bahar I. Structural changes involved in protein binding correlate with intrinsic motions of proteins in the unbound state. Proc Natl Acad Sci USA. 2005;102:18908–18913. doi: 10.1073/pnas.0507603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haliloglu T, Bahar I. Adaptability of protein structures to enable functional interactions and evolutionary implications. Curr Opin Struct Biol. 2015;35:17–23. doi: 10.1016/j.sbi.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grimm DG, et al. The evaluation of tools used to predict the impact of missense variants is hindered by two types of circularity. Hum Mutat. 2015;36:513–523. doi: 10.1002/humu.22768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.General IJ, et al. ATPase subdomain IA is a mediator of interdomain allostery in Hsp70 molecular chaperones. PLoS Comput Biol. 2014;10:e1003624. doi: 10.1371/journal.pcbi.1003624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atilgan C, Atilgan AR. Perturbation-response scanning reveals ligand entry-exit mechanisms of ferric binding protein. PLoS Comput Biol. 2009;5:e1000544. doi: 10.1371/journal.pcbi.1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ponzoni L, et al. Unifying view of mechanical and functional hotspots across class A GPCRs. PLoS Comput Biol. 2017;13:e1005381. doi: 10.1371/journal.pcbi.1005381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eyal E, Bahar I. Toward a molecular understanding of the anisotropic response of proteins to external forces: Insights from elastic network models. Biophys J. 2008;94:3424–3435. doi: 10.1529/biophysj.107.120733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Touw WG, et al. A series of PDB-related databanks for everyday needs. Nucleic Acids Res. 2015;43:D364–D368. doi: 10.1093/nar/gku1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;Chapter 7:Unit7.20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pedregosa F, et al. Scikit-learn: Machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 34.LaRusch J, et al. North American Pancreatitis Study Group Mechanisms of CFTR functional variants that impair regulated bicarbonate permeation and increase risk for pancreatitis but not for cystic fibrosis. PLoS Genet. 2014;10:e1004376. doi: 10.1371/journal.pgen.1004376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Chang Y-Y, Lee JY, Bahar I, Yang L-W. DynOmics: Dynamics of structural proteome and beyond. Nucleic Acids Res. 2017;45:W374–W380. doi: 10.1093/nar/gkx385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The UniProt Consortium UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ponzoni L, Polles G, Carnevale V, Micheletti C. SPECTRUS: A dimensionality reduction approach for identifying dynamical domains in protein complexes from limited structural datasets. Structure. 2015;23:1516–1525. doi: 10.1016/j.str.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 38.Sunyaev SR, et al. PSIC: Profile extraction from sequence alignments with position-specific counts of independent observations. Protein Eng. 1999;12:387–394. doi: 10.1093/protein/12.5.387. [DOI] [PubMed] [Google Scholar]
- 39.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.