Significance
The KRAS gene is frequently mutated in human cancer, especially in pancreatic, lung, and colorectal tumors. We developed an intact protein assay for the detection and quantitation of KRAS protein forms (proteoforms), enabling the measurement of how genetically encoded mutations affect posttranslational modifications on the same protein molecule. The modifications found are known to activate KRAS or interfere with membrane attachment, suggesting cellular mechanisms that control downstream KRAS activity. Analysis of wild-type and mutant-specific forms of the KRAS protein with complete molecular specificity is shown to be possible by top-down proteomics and enables future tests of how an individual’s KRAS proteoforms are linked to disease stage and chance of survival.
Keywords: RAS, KRAS, posttranslational modification, top-down proteomics, immunoprecipitation
Abstract
Mutations of the KRAS gene are found in human cancers with high frequency and result in the constitutive activation of its protein products. This leads to aberrant regulation of downstream pathways, promoting cell survival, proliferation, and tumorigenesis that drive cancer progression and negatively affect treatment outcomes. Here, we describe a workflow that can detect and quantify mutation-specific consequences of KRAS biochemistry, namely linked changes in posttranslational modifications (PTMs). We combined immunoaffinity enrichment with detection by top-down mass spectrometry to discover and quantify proteoforms with or without the Gly13Asp mutation (G13D) specifically in the KRAS4b isoform. The workflow was applied first to isogenic KRAS colorectal cancer (CRC) cell lines and then to patient CRC tumors with matching KRAS genotypes. In two cellular models, a direct link between the knockout of the mutant G13D allele and the complete nitrosylation of cysteine 118 of the remaining WT KRAS4b was observed. Analysis of tumor samples quantified the percentage of mutant KRAS4b actually present in cancer tissue and identified major differences in the levels of C-terminal carboxymethylation, a modification critical for membrane association. These data from CRC cells and human tumors suggest mechanisms of posttranslational regulation that are highly context-dependent and which lead to preferential production of specific KRAS4b proteoforms.
The RAS oncogenes are frequently mutated in human cancer and are therefore often the focus of diagnostic and therapeutic strategies (1–3). The three RAS genes in humans give rise to four base protein sequences: HRAS, NRAS, and two KRAS isoforms, KRAS4a and KRAS4b, resulting from alternative splicing (4). The RAS proteins belong in the GTPase family and function as GTP-regulated molecular switches controlling pathways involved in critical functions such as signaling, cell growth, and proliferation (5). Guanine nucleotide exchange factors (GEFs) promote activation of the RAS GTPases while GTPase-activating proteins (GAPs) inactivate RAS by enhancing GTP hydrolysis (6–8). Once activated, RAS-GTP can bind and activate several downstream effectors. The 21 kDa RAS proteins are highly similar in primary sequence (>80% sequence identity) and share 100% sequence identity in their N-terminal amino acids from 1 to 86 (SI Appendix, Fig. S1 A and B). The first 165 amino acids form the G domain, which contains the catalytic and switching portion of the protein responsible for GDP/GTP binding and association with regulators and downstream effectors (9, 10). The RAS proteins differ in their C-terminal hypervariable regions (illustrated in SI Appendix, Fig. S1C), which contain a C-terminal CaaX motif, where “a” is an aliphatic amino acid and X is any amino acid. The C terminus of KRAS is posttranslationally processed in three steps: farnesylation of the Cys in the CaaX motif, proteolysis of the aaX sequence, and carboxymethylation (COOMe) of the resulting C terminus. Farnesylation and COOMe of the C-terminal Cys are essential for association with the plasma membrane (4, 11–13), where KRAS4b can interact with upstream regulators and activate downstream effectors.
Of the two KRAS isoforms, KRAS4a contains a palmitoylation site at Cys180 that has been shown to assist membrane localization (14). In contrast, the KRAS4b isoform is present at ∼10× higher levels than KRAS4a (14) and lacks the palmitoylation site, containing in its stead a polybasic sequence that facilitates membrane association by electrostatic interaction with negatively charged lipid heads in the membrane (15). In cancers, KRAS mutations usually result in decreased rates of intrinsic GTP hydrolysis or impaired binding to GAPs (16). Mutagenesis studies have demonstrated that different amino acid substitutions have distinct functional consequences (17). Moreover, comparison of cell lines harboring different, allele-specific KRAS mutations has provided an additional level of complexity regarding KRAS-dependent effector signaling (18, 19). Recent studies with isogenic cell lines revealed that mitochondrial translation is essential for KRAS Gly13Asp (G13D) mutant cell proliferation (20, 21). Despite intense research on KRAS, a complete mechanistic basis for this remains unclear. We posited that the presence of different mutations might result in distinct proteoforms with mutation-specific posttranslational modification (PTM) profiles, each with the potential to generate distinct downstream signaling or phenotypic outputs.
Top-down (TD) proteomics is emerging as a high-value method of protein analysis because it directly measures the composition of whole proteoforms (i.e., the various protein forms arising from coding mutations, differential splicing, and PTMs; ref. 22), and associates them with normal and diseased phenotypes (23). Intact mass measurements (referred to as MS1), along with fragmentation in the second step of tandem mass spectrometry (called MS/MS or MS2), provide precise information about the gene product, sequence, length, and composition of individual proteoforms, including the relative abundance of degradation products, sequence variants, and PTMs co-occurring on the same molecule. A typical TD workflow consists of single or multistep protein separations, with the resulting protein fractions separated by liquid chromatography in-line with a mass spectrometer (LC-MS) (24). Previous analyses of KRAS in complex proteomes used the complementary bottom-up (BU) approach after immunoprecipitation (IP) (25). However, the BU approach entails proteolytic digestion, thus severing the linkage between mutations and PTMs occurring in disparate regions of the same protein molecule (SI Appendix, Fig. S1). The high sequence identity among the protein products of the three RAS genes (SI Appendix, Fig. S1A) translates to many identical proteolytic peptides (SI Appendix, Fig. S1B) that complicate precise isoform and proteoform measurement due to the protein inference problem (26–28). Conversely, the TD approach does not require a protease, and allows for the analysis of co-occurring mutations and PTMs on each individual proteoform, but is less sensitive and underdeveloped relative to BU (29, 30).
Here, we report a targeted TD workflow (Fig. 1) for the allele- and isoform-specific interrogation of proteoforms expressed from KRAS genes within human colorectal cancer (CRC) cell lines and tumor samples. We combined IP for protein enrichment with the TD approach for intact protein analysis to directly measure KRAS proteoforms with high molecular specificity. As proof of principle, isogenic CRC cell lines were used to investigate the effect of the KRAS G13D mutation on PTMs, resulting in the detection of KRAS4b nitrosylation exclusively within two cell lines expressing a single wild-type (WT) KRAS allele. Application of the TD workflow to other cancer cell lines with three mutations at Gly12 along with primary colorectal tumor samples uncovered 11 proteoforms of KRAS4b and quantified mutant versus WT KRAS4b expression ratios to reveal a highly variable degree of KRAS4b C-terminal COOMe, a PTM critical for membrane association and productive RAS-mediated signaling (27). This study highlights the importance of precise proteoform mapping in the discovery of the dominant and dynamic PTMs underlying key regulatory mechanisms operating in both human cells and patients.
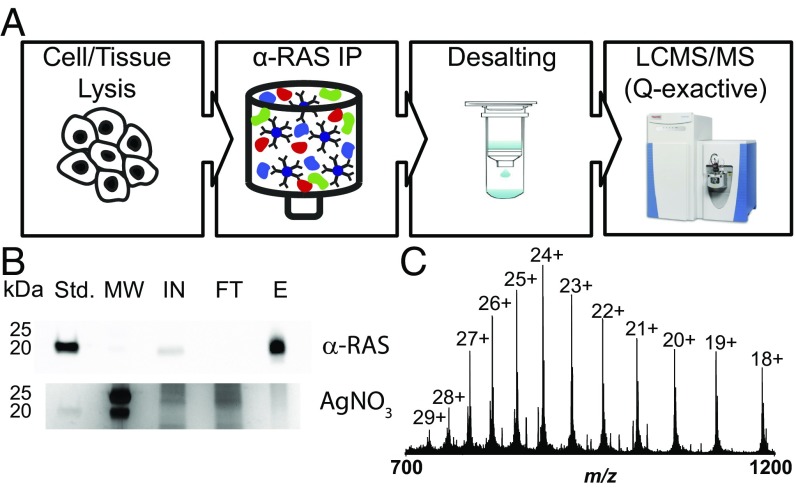
Fig. 1.
Workflow for the enrichment and detection of KRAS proteoforms. (A) Schematic of the approach. (B) Silver-stained gel and corresponding Western blot illustrating enrichment of RAS proteins. E, eluate; FT, flow through; IN, input; MW, molecular weight markers; Std, KRAS4b standard. (C) Isotopically resolved mass spectrum of intact KRAS4b.
Results
Enrichment of KRAS Proteins for Proteoform Analysis.
The RAS family of proteins present a range of analytical challenges, including copy number and biochemical properties, which lead to expectations of poor solubility and recovery before whole protein mass spectrometry using electrospray (25). Indeed, a previous study that identified >1,800 proteoforms from the DLD-1 cell line used here did not detect evidence of any RAS family proteoforms (31). Therefore, an enrichment strategy for KRAS proteoforms before TD MS was required. We used a pan-RAS IP approach to purify and increase the concentration of RAS proteins before chromatographic separation directly coupled to TD proteomics (Fig. 1A). Based on densitometric quantitation of the RAS-specific band within Western blots performed to assess enrichment level, the eluted RAS proteins were enriched by more than 10-fold in the elution step compared with the lysate used as input (Fig. 1B). While KRAS, NRAS, and HRAS could all be detected with the optimized IP protocol (SI Appendix), LC-MS measurements were also tailored to enable detection of KRAS proteoforms with good signals in intact (MS1) and fragmentation (MS2) spectra. Consistent with transcriptomic data on cancer cell lines (14), KRAS4a is only a small fraction of total KRAS (<10%) relative to the 4b isoform and was not detected by our assay.
Optimization and Validation of TD Assays Using Recombinant KRAS4b Proteoforms.
Recombinant but fully processed KRAS4b proteoforms (32) harboring either WT or mutant G13D sequences were used to optimize LC and MS parameters before analysis of endogenous KRAS4b proteoforms (see extended details in SI Appendix) (33, 34). By this analysis, we were able to visualize the complex charge state distribution of KRAS4b (Fig. 1C), typical for analysis of denatured intact proteins at pH ∼ 3 due to protonation of multiple basic residues within the protein sequence. Farnesylation has been demonstrated to be a labile modification under certain MS conditions (35). Therefore, we first investigated whether selecting different precursor charge states could preserve and localize this modification on an intact, farnesylated proteoform (SI Appendix, Fig. S2). It was noted that farnesylation was fully ejected during fragmentation of higher charge states (e.g., the 24+), resulting in the lack of y-type ions bearing this labile modification (SI Appendix, Fig. S2A). However, when lower charge states (e.g., the 20+) were fragmented, several y-type ions partially retained the farnesyl group, allowing for confirmation of its C-terminal localization (SI Appendix, Fig. S2 B and C). Similarly, we also investigated the effect of collision energy. Lower collision energy spectra contained farnesylated y-ions, while increasing the collision energy resulted in complete defarnesylation of KRAS4b in the gas phase.
The same recombinant proteoforms were used to clearly distinguish between different sequence variants of KRAS4b by measuring abundances of several b-type ions, which contain the N-terminal region and, therefore, report on the site of mutation. Representative mass spectra of the three KRAS standards are shown in SI Appendix, Fig. S3A. The standards differ from endogenous KRAS by the addition of an extra Gly-Gly (GG) sequence at the N terminus. As expected, the fully processed WT KRAS4b standard (GG-KRAS4b-FMe WT) has a monoisotopic mass of 21,268.92 Da, which is 58 Da lighter than the fully processed G13D KRAS4b standard (GG-KRAS4b-FMe G13D, 21,326.92 Da) and 14 Da heavier than the farnesylated (but not carboxymethylated) WT KRAS4b standard (GG-KRAS4b-Farn WT, 21,254.90 Da). The mass shift due to the presence of the G13D mutation can also be observed in b-ions generated during fragmentation, such as the abundant fragment ion b55 (SI Appendix, Fig. S3B). Note that the b55 from GG-KRAS4b corresponds to the b53 from endogenous KRAS4b. Similarly, C-terminal COOMe can be monitored by measuring y-ions, such as the fragment ion y12 (SI Appendix, Fig. S3C). Relative abundance information for each proteoform can be obtained by monitoring either the precursor ions (selected ion monitoring scan) or the fragment ions [targeted MS2 (tMS2) scan]. This process functions well due to the linear relationship between peak intensities of proteoform specific precursor ions (SI Appendix, Fig. S3D) or b- and y-ions (SI Appendix, Fig. S3 E and F) and the amount of KRAS4b protein injected in the mass spectrometer.
Discovery and Mapping of KRAS4b Proteoforms from Isogenic Cell Lines.
Having optimized an enrichment strategy and targeted assay for KRAS4b proteoforms, we sought to assess the effects of mutational differences upon the composition of endogenous KRAS proteoforms. Therefore, we selected DLD-1 CRC cell lines, as a wealth of prior data has been generated within this system (36–38). The parental cell line (DLD-1 PAR) is heterozygous at the KRAS locus, containing both a WT and a mutant G13D allele (WT/G13D). Two cell lines derived from the DLD-1 parental cells (DLD-1 WT and DLD-1 MUT) express either the single WT KRAS allele (WT/-) or the single KRAS G13D allele (G13D/-) and were found to express only WT or mutant KRAS4b proteoforms, respectively (Table 1).
Table 1.
Detection of KRAS4b proteoforms in cell lines
| Cell lines | KRAS genotype | % of G13D KRAS4b detected* | % of Cys118 nitrosylated† |
| DLD-1 PAR | WT/G13D | 45 (±3)‡ | 0 |
| DLD-1 MUT | -/G13D | 100 | 0 |
| DLD-1 WT | WT/- | 0 | >91† |
| HCT116 PAR | WT/G13D | 47 (±4)‡ | 0 |
| HCT116 WT | WT/- | 0 | >90† |
| SW48 PAR | WT/WT | 0 | 0 |
| SW48 MUT | WT/G13D | 49% (±3)‡ | 0 |
Percentage of G13D KRAS4b listed is the average of nine experimental replicates.
Percentage of nitrosylation listed is the average of nine experimental replicates.
Each average is listed as the mean ± SEM.
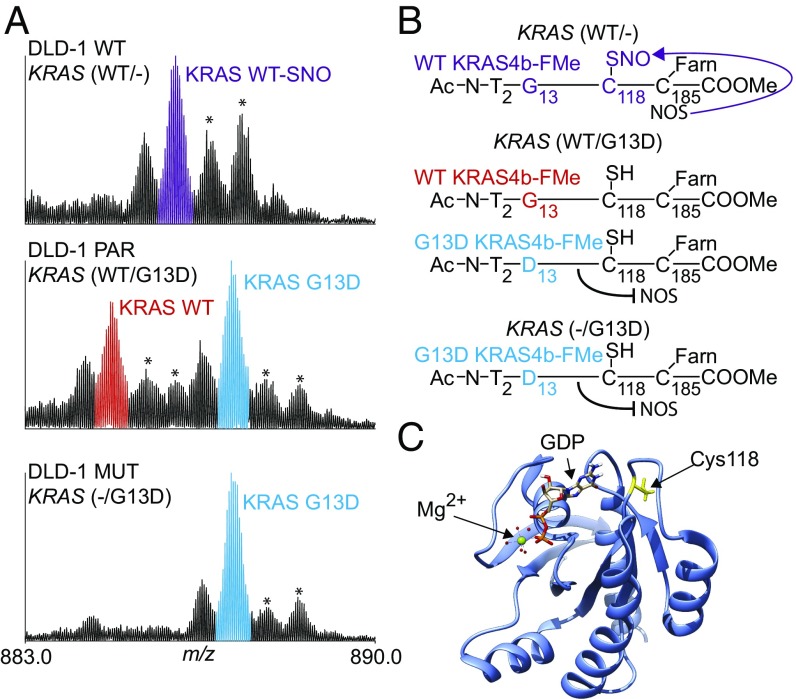
Four milligrams of total protein from DLD-1 PAR cell lysate (from ∼2e6 cells) were subjected to the IP-TDMS assay described above. Two major proteoforms differing in mass by 58 Da were detected, corresponding to WT and G13D mutant versions of KRAS4b (Fig. 2A, Middle). Both proteoforms were modified at the N terminus with removal of the initiating Met and acetylation of the following Thr residue. The C-terminal Cys was both farnesylated and carboxymethylated due to complete CaaX processing. Use of a 5- to 8-m/z isolation window enabled the coisolation of KRAS4b proteoforms and the quantitative measurement of fragment ions from related proteoforms in the same spectrum (procedure described in SI Appendix, SI Materials and Methods). In brief, the ratio of WT and G13D alleles of KRAS4b was determined from the relative abundances of both intact proteoforms and fragment ions (tMS2 scans). Each method found the expression ratio to be near 1:1 (Table 1, first row), indicating equal expression of both alleles. Analysis of the DLD-1 MUT cell line (G13D/-) identified a single major proteoform matching the fully processed G13D KRAS4b proteoform present in DLD-1 PAR cells (Fig. 2A, Bottom). No WT KRAS4b proteoforms were detected (>1% limit of detection), in agreement with the genotype.
Fig. 2.
KRAS4b proteoform detection in cell lines. (A) Isotopically resolved mass spectrum of KRAS4b, charge state 24+, isolated from three isogenic cell lines (DLD-1 PAR, DLD-1 WT, DLD-1 MUT) and illustrating the effect of KRAS mutational status on posttranslational modifications. Asterisks denote known electrospray oxidation products. (B) Schematic of KRAS4b and its PTM profile. (C) Depiction of the GDP/GTP binding pocket within the tertiary structure of KRAS4b (yellow, Cys118; magenta, Asp13; cyan, GDP; green, Mg2+). This figure was based on PDB ID code 4OBE.
When analyzing DLD-1 WT cells with the WT/- genotype, no G13D KRAS4b proteoforms were detected (Table 1, row 3). However, the major proteoform detected was not shared with the parental cell line. Rather, this species was 29 Da heavier than the WT proteoform observed in the DLD-1 PAR cells (Fig. 2A, Top). Targeted fragmentation of this proteoform localized the mass shift to Cys118 (SI Appendix, Fig. S4A), a residue known to be posttranslationally modified in HRAS by a nitrosyl group (R-NO) (39). The experimentally determined mass of this proteoform was 21,225.92 Da and matched within tolerance the theoretical monoisotopic mass of S-nitrosylated KRAS4b (21,225.88 Da, Δm = 1.9 ppm). To corroborate this finding, recombinant and fully processed KRAS4b was incubated with S-nitrosoglutathione (GSNO, a nitric oxide-donating reagent), and the product was analyzed by TD MS. Incubation with GSNO resulted in the observation of a peak from a proteoform 29 Da heavier than the recombinant standard (SI Appendix, Fig. S4C). Targeted fragmentation of this species localized the mass shift at the same Cys residue (Cys118), providing a reference spectrum and further evidence for the endogenous nitrosylation of KRAS4b at this site (SI Appendix, Fig. S4D).
Nitrosylation of Cysteine 118 Is Constitutively Present in Cell Lines with KRAS4b WT/- Genotypes.
All nine technical replicates of DLD-1 WT cells contained >90% nitrosylated KRAS4b (SI Appendix, Fig. S5A). In addition, varying the cell lysate input for IP did not affect the levels of nitrosylation (SI Appendix, Fig. S5B). To eliminate the possibility of a DLD-1 cell line-specific result, we obtained two more sets of isogenic CRC cell lines. The HCT116 parental cell line (HCT116 PAR) carries the heterozygous KRAS G13D mutation (as in DLD-1 PAR) and the derived WT HCT116 (HCT116 WT) results from a targeted knockout of the G13D mutant allele. Here, the TD assay identified the same major proteoforms as in the DLD-1 cells (SI Appendix, Fig. S4B). Nitrosylation levels of KRAS4b in HCT116 cells with WT/- KRAS were calculated to be greater than >90%, providing additional evidence in a different genetic background for the up-regulation of Cys118 nitrosylation in the absence of mutant KRAS4b. Interestingly, no nitrosylation of WT KRAS4b was observed in either DLD-1 PAR or HCT116 PAR cells expressing both WT and G13D KRAS4b. SW48 CRC cells, which are WT/WT at the KRAS gene, were also investigated using this assay. KRAS4b proteoforms detected in SW48 cells showed no detectable levels of nitrosylation. These results are summarized in Table 1.
KRAS4b Proteoforms from Cell Lines Harboring Gly12 Mutations.
For two lung cancer cell lines with G12C/WT and G12V/G12V genotypes (H1792 and COR-L23, respectively), we applied a different version of the IP-TDMS assay using a narrower 3-m/z window for precursor ion selection to independently isolate WT or mutant proteoforms (SI Appendix, Fig. S6). This improved assay revealed no evidence for nitrosylation of Cys118 or differential levels of C-terminal COOMe on WT vs. mutant KRAS4b (SI Appendix, Table S1). Together with a SW48 colorectal cell line with a G12D/WT genotype, a total of seven additional KRAS4b proteoforms were characterized. These proteoforms were given individual proteoform record numbers and are reported in SI Appendix, Table S2 along with their PTMs.
KRAS4b Proteoforms in Colorectal Tumors.
Four deidentified samples of colorectal tumors with defined KRAS genotypes were obtained from the Biospecimen Core Resource of the NCI Clinical Proteomic Tumor Analysis Consortium (CPTAC). Two of the tumor samples were WT/WT (subjects 1 and 2), and two were KRAS G13D/WT (subjects 3 and 4; Table 2). Tumor material contained >80% cancerous cells as determined by H&E staining (40), and the weights of the tumor samples ranged from 0.15 to 0.25 g. Frozen tumor sections were first cryo-pulverized and then subjected to the IP-TDMS assay illustrated in Fig. 1A. Representative mass spectra obtained from tumors are shown in Fig. 3A and SI Appendix, Fig. S7. While low protein signal and high relative oxidation prevented allele-specific PTM determination in these four tumors, we used ratios of b-type product ions to determine that the relative amounts of mutant versus WT KRAS4b present at the protein level for subjects 3 and 4 were 11% and 29% of mutant KRAS4b, respectively (Fig. 3B and Table 2).
Table 2.
Relative levels of WT and mutant KRAS4b proteoforms in human colorectal tumor samples
| CRC tumor sample | KRAS genotype | % MUT KRAS detected* | % C-terminal COOMe† | Cancer stage |
| Subject 1 | WT/WT | 3 (±2) | 27 (±4) | IIIB |
| Subject 2 | WT/WT | 0 | 69 (±6) | IIA |
| Subject 3 | WT/G13D | 11 (±1) | 18 (±1) | IVA |
| Subject 4 | WT/G13D | 29 (±1) | 79 (±7) | IIIB |
Calculated using the relative abundances of sequence variant specific b-ions; errors are SEM from three replicate injections of each nonrenewable clinical specimen.
Calculated using the relative abundances of y-ions carrying this PTM; errors are SEM from three replicate injections of each nonrenewable clinical specimen.
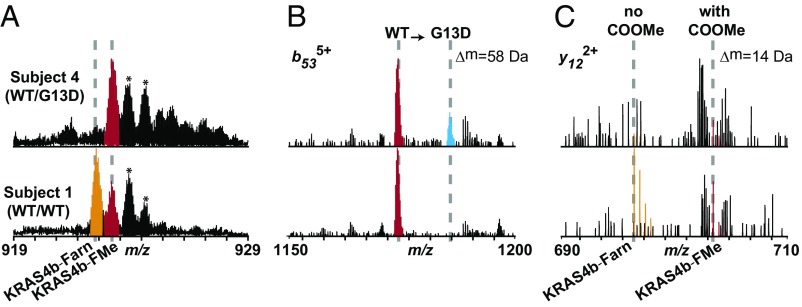
Fig. 3.
KRAS4b proteoform detection in tumor samples. (A) MS1 data for the 23+ charge state of KRAS4b detected from a tumor carrying the G13D mutation (Upper) and a tumor containing no KRAS mutations (Lower). Fully processed KRAS4b is highlighted in red, and KRAS4b lacking the COOMe is highlighted in orange. Asterisks denote known ESI oxidation products. (B) Zoomed-in fragmentation spectrum illustrating detection of the b53 ion from two sequence variants, WT (red) and G13D (cyan). Intensities of these ions were used to calculate relative abundances of mutant and WT KRAS in Table 2. (C) Zoomed-in fragmentation spectrum illustrating detection of the y12 ion from KRAS4b proteoforms with and without the C-terminal methyl ester, KRAS4b-FMe (red) and KRAS4b-Farn (orange), respectively. Intensities of these ions were used to calculate percentage of C-terminal COOMe in Table 2.
Beyond the assessment of allele-specific KRAS4b expression in these tissues, we also observed major differences in C-terminal processing of KRAS4b proteoforms. For subjects 1, 2, and 3, their tumor samples contained new proteoforms 14 Da lighter than expected (SI Appendix, Fig. S7A). Targeted fragmentation allowed localization of this −14 Da mass shift to the last 12 C-terminal residues (Fig. 3C). Specifically, this fragmentation pattern is precisely that observed for recombinant proteoforms lacking the C-terminal COOMe. Thus, this −14 Da mass shift indicates the marked absence of the C-terminal COOMe, a modification that has been shown recently to be essential for proper membrane association of KRAS (27, 28). By measuring fragment ions containing the C terminus (Fig. 3C), we could quantify the relative levels of C-terminal COOMe in individual tumor samples, the results of which are summarized in Table 2 (fourth column). Two subjects with WT/WT genotypes for their KRAS genes had very different tissue profiles of KRAS proteoforms. Subject 2 had 69% C-terminal COOMe while subject 1 retained only 27% of the C-terminal COOMe (SI Appendix, Fig. S7A, top two images). Of these two patients, the tumor sample with 69% C-terminal COOMe had a lower disease stage (IIA) compared with the tumor sample with 27% C-terminal COOMe (IIIB, Table 2). Interestingly, the two patients with WT/G13D genotypes (subjects 3 and 4) had disparate levels of C-terminal COOMe: One had 79% retention and the other had only 18% (Table 2 and SI Appendix, Fig. S7). Furthermore, subject 3 with the WT/G13D genotype and 18% C-terminal COOMe had a higher stage of disease (stage IVA) compared with subject 4 with the WT/G13D genotype with 79% C-terminal COOMe (stage IIIB).
To determine the relative amount of C-terminal COOMe present on mutant and WT proteoforms of KRAS4b, the modified IP-TDMS assay was applied to two additional tumor samples with G12V/WT and WT/WT genotypes (subjects 5 and 6, respectively). The results from this allele-specific analysis of tumor proteoforms are summarized in SI Appendix, SI Results, SI Discussion, and Table S3. With this more precise IP-TDMS workflow, we determined that only 9% of the mutant G12V KRAS4b was C-terminally carboxymethylated while 49% of the WT KRAS4b contained the C-terminal COOMe (SI Appendix, Fig. S7 and Table S3). This shows that the measurement can be made to probe differential PTM occupancy on mutant and WT KRAS4b.
Discussion
Discovery and Measurement of Protein Cross-Talk at the Proteoform and Peptide Levels.
The results described above illustrate certain advantages of mapping WT and mutant proteoforms with high molecular specificity using the TD approach (SI Appendix, Fig. S8). This process discovers what modifications are present, their relative abundance, and how they relate to each other. Without the need for antibodies specific to RAS isoforms (SI Appendix, Fig. S8A), we were able to (i) precisely elucidate gene-, isoform- and allele-specific PTM patterns; (ii) discover unexpected PTM changes; and (iii) determine the relative amount of specific allele-dependent isoforms expressed at the protein level in cancer cell lines and tumors. These advantages flow directly from use of the TD approach, which can be difficult to export to the wider community. Once mapped, proteoform-level details can inform the development of targeted assays using the BU approach, with all its associated advances in interlab validation and assay portability made over the past decade (SI Appendix, Fig. S8B) (29, 30). This complementarity between the BU and TD approaches should prove applicable to the discovery of mutation/modification cross-talk in other oncogene products (e.g., TP53, IDH1). The use of TD and BU can thus be combined (SI Appendix, Fig. S8D) to advance our understanding of protein expression from key oncogenes endogenously present within human cells or primary tissue.
Contextualization of Results from Cell Lines.
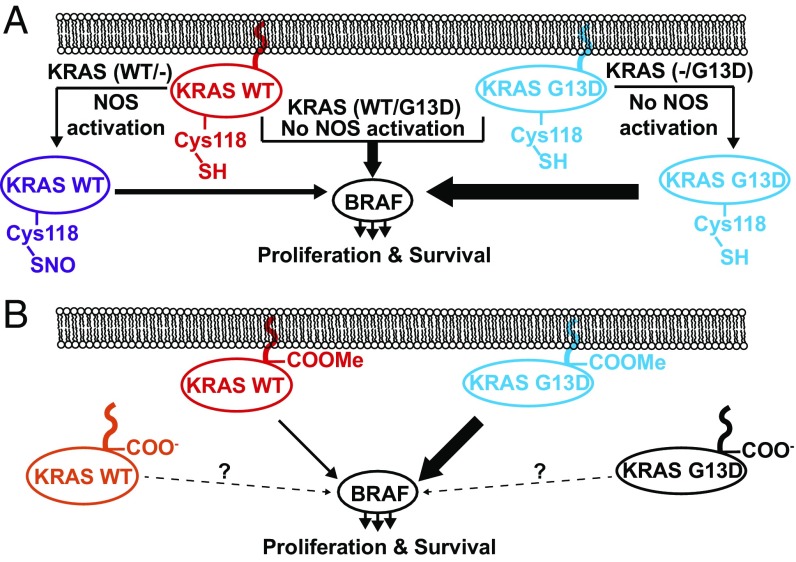
Analysis of isogenic cell lines revealed a direct linkage between the knockout of mutant alleles and the nitrosylation of WT KRAS4b proteoforms in KRAS-driven cancer cell lines (Fig. 4A). This supports the idea that WT KRAS may control cell growth and proliferation by activating downstream pathways through a mechanism different from that of mutant KRAS, but having lower proliferative potential. Interestingly, nitrosylation of HRAS at Cys118 in vitro increases both the intrinsic rate of GTP hydrolysis and GDP exchange for HRAS, and moreover, glutathione addition to Cys118 via a disulfide or Cys118 mutation to Ser eliminates these effects (39, 41). Furthermore, in rabbit aortic endothelial cells, Cys118 nitrosylation stimulates downstream MAPK signaling through ERK1/2 phosphorylation, while mutation of Cys118 to Ser abrogates this effect (42). Indeed, while nitrosylated KRAS is a more efficient enzyme relative to its unmodified form, KRAS4b WT/- cells divide at approximately half the rate of DLD-1 cells with a WT/G13D genotype (43). The stoichiometric nitrosylation of Cys118 may therefore be a mechanism for immortalized cell lines with a single WT KRAS allele to stimulate downstream signaling pathways and, thereby, promote proliferation. We postulate that the Cys118Ser mutation in hemizygous cell types that are WT/- for KRAS would lead to growth rate defects due to a lack of KRAS-mediated signaling.
Fig. 4.
Basic models for the role of Cys118 nitrosylation and C-terminal methylation in modulating the signaling level of KRAS proteoforms in cell lines and primary tumors. (A) In cell lines, generation of nitrosylated KRAS4b proteoforms is completely linked to the KRAS genotype. In heterozygous cells (e.g., DLD-1 PAR), mutant G13D KRAS4b (blue) signals robustly through BRAF. In hemizygous cells where G13D KRAS has been knocked out, NOS is posited to activate WT KRAS through Cys118 nitrosylation (purple), up-regulating BRAF-mediated signaling. In cells that have WT KRAS knocked out, G13D KRAS4b is not observed to be nitrosylated. (B) In primary tumors and cell lines, mutant KRAS4b proteoforms are believed to robustly signal through BRAF, while WT KRAS4b is believed to weakly activate downstream BRAF and other effector pathways. While there is a relatively high abundance of KRAS4b proteoforms (WT and mutant) lacking the C-terminal COOMe in tumors, the effects on downstream signaling have yet to be determined.
Findings from Primary Tumors.
Analysis of tumor samples from subjects 3, 4, and 5 revealed that the proportion of mutant KRAS4b expressed was a small fraction of the total (11, 29, and 18%, respectively). Furthermore, we identified differences in CaaX processing of KRAS4b. C-terminal COOMe of KRAS enhances hydrophobicity and promotes membrane association (27). Selective removal of the COOMe could affect KRAS subcellular localization, which could in turn impair downstream signaling and even tumor growth rate. For subject 5, the C-terminal COOMe levels differed widely for each allele (SI Appendix, Fig. S7B), suggesting a posttranslational regulatory mechanism to differentially modulate the tumorigenic activity of WT vs. mutant KRAS4b (Fig. 4B). We note that these tumor samples could have exhibited cells harboring a majority of KRAS4b proteoforms without C-terminal COOMe (e.g., subject 3 in Table 2 with only 18% of KRAS4b fully processed) or included a mixed KRAS4b allele (and therefore proteoform) population due to intratumoral heterogeneity. Future iterations of the IP-TDMS assay on a larger cohort of genotyped subjects will include the more precise proteoform isolation and enable the study of tissue from pancreatic cancers where >90% are known to harbor one of three mutations in the KRAS gene (G12, G13, and Q61) (44).
Despite the lack of concordance between relative PTM abundance observed in isogenic cell lines compared with those identified within primary tumors, it is notable that in both contexts, a high degree of control over KRAS proteoform production was observed. Near-complete stoichiometric control over Cys118 nitrosylation in KRAS4b was detected in two different isogenic cell line models with different genetic backgrounds. This suggests that a tight signaling logic is at play, perhaps involving a membrane-associated source of NO, such as eNOS (Fig. 4A) (45). Given that we did not observe Cys118 nitrosylation in WT/WT SW48 cells or in WT/WT tumors, it is likely that Cys118 nitrosylation is exclusive to KRAS WT/- cells. Combined with the fact that that WT/- cells proliferate at a slower rate than mutant counterparts and are therefore at a selective disadvantage for tumor growth, KRAS nitrosylation is unlikely to be found in CRC patients that are homozygous for WT KRAS.
This study highlights the feasibility and importance of measuring PTMs on mutant-specific proteoforms. We also highlight the complementary roles that TD and BU proteomics can play in interrogating protein composition. Proteoform mapping by TD followed by targeted measurement of peptides by multiple reaction monitoring/parallel reaction monitoring type assays provide a guide for complementary use of MS technologies in protein biochemistry (SI Appendix, Fig. S8D). In the case of KRAS, these mutation-specific PTMs may potentiate downstream effects, including the penetrance of driving mutations into a given tissue sample or the extent of disease progression. With the adoption of personalized medicine, the ability to measure mutation- and proteoform-specific differences might prove essential in gaining a precise understanding of effects of oncogenic mutations and the development of mutation-selective anti-RAS strategies (46).
Materials and Methods
The DLD-1, HCT116, and SW48 cell lines were obtained from Horizon Discovery. Primary colorectal tumor samples were obtained from the Biospecimen Core Resource of the NCI CPTAC. KRAS protein standards were provided by the Fredrick National Lab RAS program. Following lysis, protein extracts were enriched for RAS proteins using an anti-RAS antibody (Millipore) as previously described (47), with a few modifications. Immunoenriched samples were further separated on a PLRP-S (Agilent) column and introduced with an UltiMate 3000 RSLCnano System (Thermo Scientific) into a modified Thermo Fisher Q-Exactive HF mass spectrometer for intact protein measurements. Data were processed and analyzed using ProSightPC 4.0, ProSight Lite, and QualBrowser, part of the Xcalibur software packaged with the Q-Exactive HF. Detailed methods of the procedures described in the text are provide in SI Appendix, SI Materials and Methods. Raw data files produced in this work have been made publicly available in the MassIVE online repository (ftp://massive.ucsd.edu/MSV000082030).
Supplementary Material
Acknowledgments
We thank Kevin Haigis for providing an IP protocol and the following members of the Kelleher Research Group and Proteomics Center of Excellence for helpful discussions and experimental assistance: Nicole Haverland, Jeff Anderson, and Joseph Greer. This work was supported by Federal Funds from the National Cancer Institute (Office of Cancer Clinical Proteomics Research), National Institutes of Health, under Contract HHSN261200800001E, and by Paul G. Allen Family Foundation Grant Award 11715 (to N.L.K.), and was carried out in collaboration with the National Resource for Translational and Developmental Proteomics under Grant P41 GM108569 from the National Institute of General Medical Sciences, National Institutes of Health. We also thank Wenan Qiang at the Center for Developmental Therapeutics at Northwestern University (funding provided by Grants CCSG P30 CA060553 and U54 CA193419) for technical assistance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Raw data files produced in this work have been made publicly available in the MassIVE online repository (ftp://massive.ucsd.edu/MSV000082030).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716122115/-/DCSupplemental.
References
- 1.Ellis CA, Clark G. The importance of being K-Ras. Cell Signal. 2000;12:425–434. doi: 10.1016/s0898-6568(00)00084-x. [DOI] [PubMed] [Google Scholar]
- 2.Hobbs GA, Der CJ, Rossman KL. RAS isoforms and mutations in cancer at a glance. J Cell Sci. 2016;129:1287–1292. doi: 10.1242/jcs.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 5.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 6.Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- 7.Hennig A, Markwart R, Esparza-Franco MA, Ladds G, Rubio I. Ras activation revisited: role of GEF and GAP systems. Biol Chem. 2015;396:831–848. doi: 10.1515/hsz-2014-0257. [DOI] [PubMed] [Google Scholar]
- 8.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nussinov R, Tsai CJ, Chakrabarti M, Jang H. A new view of Ras isoforms in cancers. Cancer Res. 2016;76:18–23. doi: 10.1158/0008-5472.CAN-15-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 11.Willumsen BM, Christensen A, Hubbert NL, Papageorge AG, Lowy DR. The p21 ras C-terminus is required for transformation and membrane association. Nature. 1984;310:583–586. doi: 10.1038/310583a0. [DOI] [PubMed] [Google Scholar]
- 12.Wright LP, Philips MR. Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J Lipid Res. 2006;47:883–891. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Cox AD, Der CJ, Philips MR. Targeting RAS membrane association: Back to the future for anti-RAS drug discovery? Clin Cancer Res. 2015;21:1819–1827. doi: 10.1158/1078-0432.CCR-14-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai FD, et al. K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc Natl Acad Sci USA. 2015;112:779–784. doi: 10.1073/pnas.1412811112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 16.Hunter JC, et al. Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol Cancer Res. 2015;13:1325–1335. doi: 10.1158/1541-7786.MCR-15-0203. [DOI] [PubMed] [Google Scholar]
- 17.Bandaru P, et al. Deconstruction of the Ras switching cycle through saturation mutagenesis. eLife. 2017;6:e27810. doi: 10.7554/eLife.27810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammond DE, et al. Differential reprogramming of isogenic colorectal cancer cells by distinct activating KRAS mutations. J Proteome Res. 2015;14:1535–1546. doi: 10.1021/pr501191a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young A, Lou D, McCormick F. Oncogenic and wild-type Ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer Discov. 2013;3:112–123. doi: 10.1158/2159-8290.CD-12-0231. [DOI] [PubMed] [Google Scholar]
- 20.Martin TD, et al. A role for mitochondrial translation in promotion of viability in K-Ras mutant cells. Cell Reports. 2017;20:427–438. doi: 10.1016/j.celrep.2017.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandra A, et al. The GDI-like solubilizing factor PDEδ sustains the spatial organization and signalling of Ras family proteins. Nat Cell Biol. 2011;14:148–158. doi: 10.1038/ncb2394. [DOI] [PubMed] [Google Scholar]
- 22.Smith LM, Kelleher NL. Consortium for Top Down Proteomics Proteoform: a single term describing protein complexity. Nat Methods. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savaryn JP, Catherman AD, Thomas PM, Abecassis MM, Kelleher NL. The emergence of top-down proteomics in clinical research. Genome Med. 2013;5:53. doi: 10.1186/gm457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toby TK, Fornelli L, Kelleher NL. Progress in top-down proteomics and the analysis of proteoforms. Annu Rev Anal Chem (Palo Alto, Calif) 2016;9:499–519. doi: 10.1146/annurev-anchem-071015-041550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, et al. Mutant proteins as cancer-specific biomarkers. Proc Natl Acad Sci USA. 2011;108:2444–2449. doi: 10.1073/pnas.1019203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Mol Cell Proteomics. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Dharmaiah S, et al. Structural basis of recognition of farnesylated and methylated KRAS4b by PDEδ. Proc Natl Acad Sci USA. 2016;113:E6766–E6775. doi: 10.1073/pnas.1615316113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergo MO, et al. Inactivation of Icmt inhibits transformation by oncogenic K-Ras and B-Raf. J Clin Invest. 2004;113:539–550. doi: 10.1172/JCI18829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whiteaker JR, et al. Clinical Proteomic Tumor Analysis Consortium (CPTAC) CPTAC Assay Portal: a repository of targeted proteomic assays. Nat Methods. 2014;11:703–704. doi: 10.1038/nmeth.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whiteaker JR, et al. Using the CPTAC assay portal to identify and implement highly characterized targeted proteomics assays. Methods Mol Biol. 2016;1410:223–236. doi: 10.1007/978-1-4939-3524-6_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson LC, et al. Identification and characterization of human proteoforms by top-down LC-21 tesla FT-ICR mass spectrometry. J Proteome Res. 2017;16:1087–1096. doi: 10.1021/acs.jproteome.6b00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillette WK, et al. Farnesylated and methylated KRAS4b: high yield production of protein suitable for biophysical studies of prenylated protein-lipid interactions. Sci Rep. 2015;5:15916. doi: 10.1038/srep15916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fornelli L, et al. Advancing top-down analysis of the human proteome using a benchtop quadrupole-orbitrap mass spectrometer. J Proteome Res. 2017;16:609–618. doi: 10.1021/acs.jproteome.6b00698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheltema RA, et al. The Q Exactive HF, a Benchtop mass spectrometer with a pre-filter, high-performance quadrupole and an ultra-high-field Orbitrap analyzer. Mol Cell Proteomics. 2014;13:3698–3708. doi: 10.1074/mcp.M114.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wotske M, Wu Y, Wolters DA. Liquid chromatographic analysis and mass spectrometric identification of farnesylated peptides. Anal Chem. 2012;84:6848–6855. doi: 10.1021/ac301437m. [DOI] [PubMed] [Google Scholar]
- 36.Hutton JE, et al. Oncogenic KRAS and BRAF drive metabolic reprogramming in colorectal cancer. Mol Cell Proteomics. 2016;15:2924–2938. doi: 10.1074/mcp.M116.058925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demory Beckler M, et al. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol Cell Proteomics. 2013;12:343–355. doi: 10.1074/mcp.M112.022806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halvey PJ, et al. Proteogenomic analysis reveals unanticipated adaptations of colorectal tumor cells to deficiencies in DNA mismatch repair. Cancer Res. 2014;74:387–397. doi: 10.1158/0008-5472.CAN-13-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lander HM, et al. A molecular redox switch on p21(ras). Structural basis for the nitric oxide-p21(ras) interaction. J Biol Chem. 1997;272:4323–4326. doi: 10.1074/jbc.272.7.4323. [DOI] [PubMed] [Google Scholar]
- 40.Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part I. The utility of immunohistochemistry. J Mol Diagn. 2008;10:293–300. doi: 10.2353/jmoldx.2008.080031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hobbs GA, Bonini MG, Gunawardena HP, Chen X, Campbell SL. Glutathiolated Ras: characterization and implications for Ras activation. Free Radic Biol Med. 2013;57:221–229. doi: 10.1016/j.freeradbiomed.2012.10.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliveira CJ, et al. Nitric oxide and cGMP activate the Ras-MAP kinase pathway-stimulating protein tyrosine phosphorylation in rabbit aortic endothelial cells. Free Radic Biol Med. 2003;35:381–396. doi: 10.1016/s0891-5849(03)00311-3. [DOI] [PubMed] [Google Scholar]
- 43.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260:85–88. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 44.Eser S, Schnieke A, Schneider G, Saur D. Oncogenic KRAS signalling in pancreatic cancer. Br J Cancer. 2014;111:817–822. doi: 10.1038/bjc.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta A, et al. PARK2 depletion connects energy and oxidative stress to PI3K/Akt activation via PTEN S-Nitrosylation. Mol Cell. 2017;65:999–1013.e7. doi: 10.1016/j.molcel.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Papke B, Der CJ. Drugging RAS: Know the enemy. Science. 2017;355:1158–1163. doi: 10.1126/science.aam7622. [DOI] [PubMed] [Google Scholar]
- 47.Yang MH, et al. Regulation of RAS oncogenicity by acetylation. Proc Natl Acad Sci USA. 2012;109:10843–10848. doi: 10.1073/pnas.1201487109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.