Significance
After myocardial infarction in the mammalian heart, millions of cardiomyocytes are lost and replaced by fibrotic scar tissue. While fibrosis is persistent in adult mammals, there are some vertebrates, including zebrafish, with the capacity for regeneration. This process does not occur in the absence of fibrosis. Here we studied subpopulations of collagen-producing cells and analyzed their fate after complete regeneration of the zebrafish myocardium. Our data show that fibroblasts persisted in the regenerated heart but shut down the profibrotic program. While fibrosis could be considered as detrimental to the regeneration process, our study reveals a positive effect on cardiomyocyte proliferation. Accordingly, a fibrotic response can be beneficial for heart regeneration.
Keywords: heart regeneration, fibroblast inactivation, zebrafish, fibrosis, cardiomyocyte proliferation
Abstract
In the zebrafish (Danio rerio), regeneration and fibrosis after cardiac injury are not mutually exclusive responses. Upon cardiac cryoinjury, collagen and other extracellular matrix (ECM) proteins accumulate at the injury site. However, in contrast to the situation in mammals, fibrosis is transient in zebrafish and its regression is concomitant with regrowth of the myocardial wall. Little is known about the cells producing this fibrotic tissue or how it resolves. Using novel genetic tools to mark periostin b- and collagen 1alpha2 (col1a2)-expressing cells in combination with transcriptome analysis, we explored the sources of activated fibroblasts and traced their fate. We describe that during fibrosis regression, fibroblasts are not fully eliminated but become inactivated. Unexpectedly, limiting the fibrotic response by genetic ablation of col1a2-expressing cells impaired cardiomyocyte proliferation. We conclude that ECM-producing cells are key players in the regenerative process and suggest that antifibrotic therapies might be less efficient than strategies targeting fibroblast inactivation.
Cardiac fibrosis is the main cause of heart failure and other cardiovascular pathologies, including those associated with myocardial infarction (MI). Fibroblasts accumulate at the injury site, where they form an extracellular matrix (ECM)-rich scar that protects the heart from ventricular wall rupture. Fibroblasts can have pleiotropic effects after MI, producing ECM proteins and influencing the electrical coupling and contractility of cardiomyocytes.
Fibroblasts are defined as mesenchymal cells that express ECM proteins such as collagens (1). Expression of collagens, however, cannot be used as the sole criterion to define a fibroblast, since these proteins are expressed by several other cell types (1). Indeed, the search for the origin of cardiac fibroblasts during pathological fibrosis has been hindered by the lack of specific fibroblast markers (2). So far, marker genes that have been shown to be more specific for profibrotic fibroblasts include Collagen 1 alpha1 (Col1a1) (3) and Periostin (Postn) (4, 5). A lineage tracing study using the Cre/lox system showed that epicardial Tcf21-derived cells give rise to Postn-expressing activated fibroblasts in response to MI in the adult mouse (6). Moreover, ablation of Postn+ cells decreased post-MI survival, confirming that fibrosis is an efficient mechanism for repairing the myocardium after acute cardiomyocyte loss (6).
Unlike mammals, zebrafish hearts can fully regenerate cardiac tissue upon injury (7, 8). After controlled cryoinjury of a quarter of the zebrafish cardiac ventricle, myocardial regeneration is preceded by the accumulation of ECM at the injury site (9), indicating that a fibrotic response is not incompatible per se with regeneration. The origin and fate of fibroblasts in the context of heart regeneration are not well-known. Possible sources of cardiac fibroblasts in the injured zebrafish heart include the epicardium and epicardial-derived cells (EPDCs) (10, 11). However, they might not be the only contributors to a fibrotic response, since collagen 1 has also been detected at the endocardial border of the injury area (IA) (12).
Mechanistically, adult cardiomyocytes reenter the cell cycle to proliferate and give rise to de novo cardiomyocytes, which then repopulate the injured myocardium (8). In vitro coculture experiments using murine cells indicate that fibroblasts can have a beneficial impact on cardiomyocytes by promoting their proliferation (13), but their effect on regeneration has not been reported so far. Furthermore, inhibition of Tgf-beta signaling during zebrafish heart regeneration impairs cardiomyocyte proliferation, suggesting that fibrosis could play a role in heart regeneration (14). Since Tgf-beta also directly activates Smad3 phosphorylation in cardiomyocytes (14), the effect of fibrosis on cardiomyocyte proliferation and heart regeneration remains an open question.
Here, we examined the origin of cardiac fibroblasts in the zebrafish and characterized activated fibroblast populations in the injured heart. We also analyzed the mechanisms of fibroblast clearance during zebrafish heart regeneration. Using genetic ablation of collagen-producing cells, we show that rather than being detrimental for heart regeneration, a fibrotic response is in fact necessary for cardiomyocyte proliferation.
Results
Endogenous Fibroblasts Contribute to ECM Deposition in the Cryoinjured Zebrafish Heart.
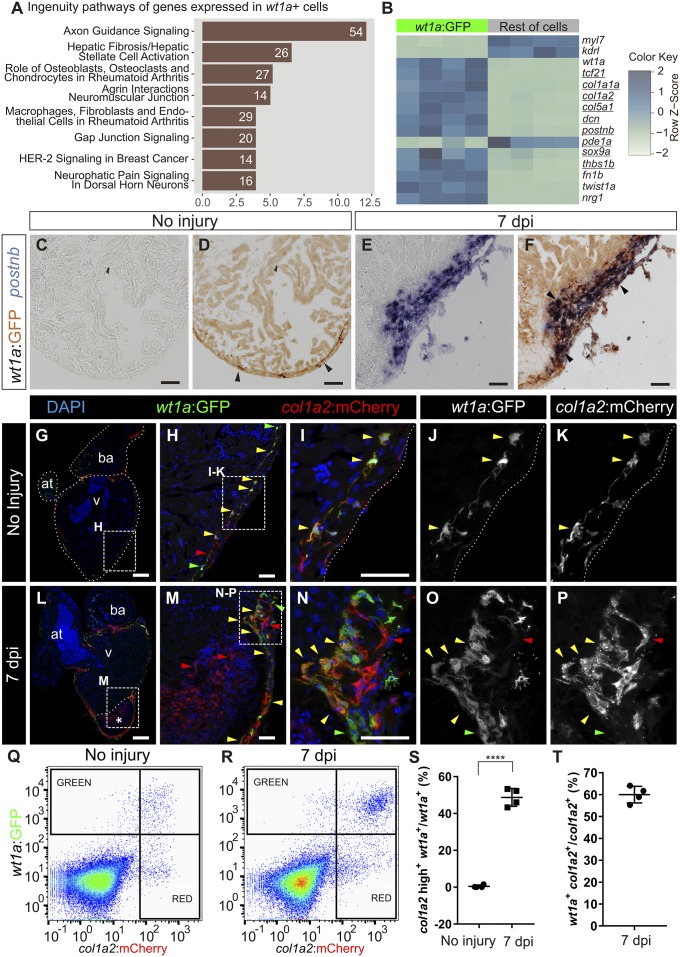
A population of collagen-producing cells located between the cortical and trabecular myocardium in the zebrafish heart was identified by transmission electron microscopy (15). This region was further shown to harbor a cell population labeled in the -6.8kbwt1a:GFP line (hereafter termed wt1a:GFP) (16) and to be positive for collagen XII expression (17). To unequivocally identify them as cardiac fibroblasts, we compared the gene expression profile of wt1a:GFP+ cells with the remainder of cardiac cells using whole-transcriptome analysis (Fig. 1 A and B and Dataset S1). Pathway analysis revealed an association of wt1a:GFP+ cells with fibrosis (Fig. 1A). Moreover, wt1a:GFP+ cells expressed eight out of nine genes described as the most specific mammalian cardiac fibroblast marker genes (18) (Fig. 1B). We next studied whether wt1a:GFP+ cells up-regulate a fibrotic gene program in response to injury. Expression of postnb was undetectable by mRNA in situ hybridization (ISH) on sections of uninjured wt1a:GFP hearts (Fig. 1 C and D; n = 3/3), but upon cryoinjury wt1a:GFP+ cells expressing postnb were detected surrounding the IA (Fig. 1 E and F; n = 4/4; 66 ± 7% of wt1a:GFP+ cells expressed high levels of postnb mRNA of a total of 660 cells counted in four hearts). We next crossed wt1a:GFP with col1a2:mCherry-NTR, a line allowing marking of collagen-producing cells (Fig. S1 A–E; n = 4/4). In uninjured hearts, 57 ± 8% of wt1a:GFP+ cells expressed col1a2:mCherry-NTR (360 cells analyzed from five hearts; Fig. 1 G–K and Q; colocalization was seen in n = 5/5). At 7 days postinjury (dpi), a more intense mCherry expression was observed in 48 ± 5% of wt1a:GFP+ cells (6,675 wt1a:GFP+ cells analyzed from four hearts; Fig. 1 L–P, R, and S; n = 4/4). The mean fluorescence level in wt1a:GFP+ cells changed from 190 ± 50 to 1,210 ± 180 a.u. (n = 4). In addition, 60 ± 4% of all col1a2:mCherry+ cells were wt1a:GFP+ (Fig. 1T; n = 4/4). In sum, a large proportion of wt1a:GFP+ cells express high levels of postnb and col1a2 upon injury. However, not all col1a2+ cells in the cryoinjured heart are wt1a:GFP+, suggesting that other cell types also contribute to fibrosis.
Fig. 1.
Intracardiac fibroblasts contribute to transient fibrosis during zebrafish heart regeneration. (A and B) Transcriptome analysis of wt1a:GFP+ cells isolated from adult zebrafish hearts. (A) Ingenuity pathways enriched in wt1a:GFP+ ventricular cells compared with all GFP− cells. A number of differentially expressed genes are shown; the x axis shows −log10[Benjamini–Hochberg adjusted (B.H.) P values]. (B) Heatmap indicating up-regulation of fibrotic marker genes (underlined) in the wt1a:GFP+ cell fraction. (C–F) postnb mRNA in situ hybridization followed by anti-GFP immunohistochemistry on sections of wt1a:GFP ventricles without injury (C and D) and at 7 d postinjury (E and F). Arrowheads indicate wt1a:GFP+ cells. (G–P) Immunofluorescence staining of sections of wt1a:GFP;col1a2:mCherry-NTR double-transgenic hearts without injury (G–K) or at 7 dpi (L–P). Red arrowheads mark col1a2:mCherry+ cells; green arrowheads mark wt1a:GFP+ cells; and yellow arrowheads mark double-positive cells. Asterisk in L indicates injured area. (Q and R) FACS-sorted cells from the wt1a:GFP;col1a2:mCherry ventricular apex without injury (Q) or at 7 dpi (R). Representative examples from a total of four hearts per condition were analyzed. (S) The percentage of wt1a:GFP+ cells that express more than 1,000 a.u. of mCherry, with the threshold corresponding to the maximum value of mCherry detected in nonactivated fibroblasts. (T) The percentage of col1a2:mCherry+;wt1a:GFP+ cells according to the thresholds shown in R. Graphs show individual measurements and means ± SD. ****P < 0.0001 by two-tailed t test. at, atrium; ba, bulbus arteriosus; v, ventricle. [Scale bars, 25 μm (H, I, M, and N), 50 μm (C–F), and 100 μm (G and L).]
To unambiguously demonstrate that intracardiac wt1a+ cells contribute to fibrosis, a wt1a:CreERT2 line was generated and crossed with ubb:loxP-GFP-loxP-STOP-mCherry (ubb:Switch). Recombination was induced by 4-hydroxytamoxifen (4-OHT) administration at 4 and 3 d before injury (Fig. S1F). In uninjured hearts, wt1a-derived cells were observed between the trabecular and cortical myocardium, colocalizing with low levels of col1a1 immunofluorescence staining (Fig. S1 G–J; n = 4/4). By contrast, in cryoinjured hearts, wt1a-derived cells were surrounded by a strong signal of col1a1 immunostaining (Fig. S1 K–N; n = 4/4). Furthermore, recombination in the double-transgenic line wt1a:CreERT2; col1a2:loxP-tagBFP-loxP-mCherry-NTR allowed mCherry labeling of cells close to the IA (Fig. S1 O–S; n = 3/3). In the mouse and chicken, intracardiac fibroblasts have been described to derive mainly from the embryonic epicardium (19).
We therefore lineage-traced embryonic wt1a-derived cells in the zebrafish (Fig. S2A). In the embryo, wt1a:GFP marks a population of proepicardial cells (16). Recombination in 2-d-old embryos yielded labeled epicardial cells on the surface of the myocardium at 5 d postfertilization (dpf) (Fig. S2 B–E; n = 8/8). In the adult, while some embryonic wt1a-derived cells remained within the epicardial layer, they could also be observed in deeper cell layers within the myocardium (Fig. S2 F–I; n = 3/3). In line with single-cell analysis of tcf21-derived cells in zebrafish, this suggests that intracardiac fibroblasts in the zebrafish also derived from the embryonic epicardium (20). To confirm that EPDCs contribute to cardiac fibrosis, we crossed tcf21:CreERT2 with col1a2:loxP-tagBFP-loxP-mCherry-NTR to label the col1a2+ cells that expressed tcf21 at the time of 4-OHT administration (Fig. S2J). The presence of mCherry in injured hearts confirmed that tcf21-derived cells contribute to collagen deposition (Fig. S2 K–N; n = 5/5). Overall, these results show that EPDC-derived resident cardiac fibroblasts secrete collagen in response to injury in the zebrafish heart.
Kidney Marrow-Derived Cells Do Not Contribute to Cardiac Fibrosis in the Zebrafish.
We next assessed whether cells derived from the main hematopoietic organ in the zebrafish, the kidney marrow, contribute to cardiac fibrosis in the zebrafish. Kidney marrow-derived cells (KMDCs) from ubb:GFP;col1a2:mCherry-NTR fish were transplanted into irradiated wild types to reconstitute the hematopoietic system with fluorescently labeled cells (Fig. S3A). After heart cryoinjury and fixing at 7 dpi, GFP+ KMDCs colonized the injured heart (n = 5/5). No mCherry expression could be detected in these hearts at 7 dpi (Fig. S3 B–E; n = 5/5), a stage at which col1a2:mCherry expression was readily observed in a control group of donors (Fig. S3 F–I; n = 4/4). This finding suggests that KMDCs do not contribute to ECM production during cardiac fibrosis in the zebrafish.
Endocardial Cells Have a Limited Contribution to ECM Deposition in the Cryoinjured Zebrafish Heart.
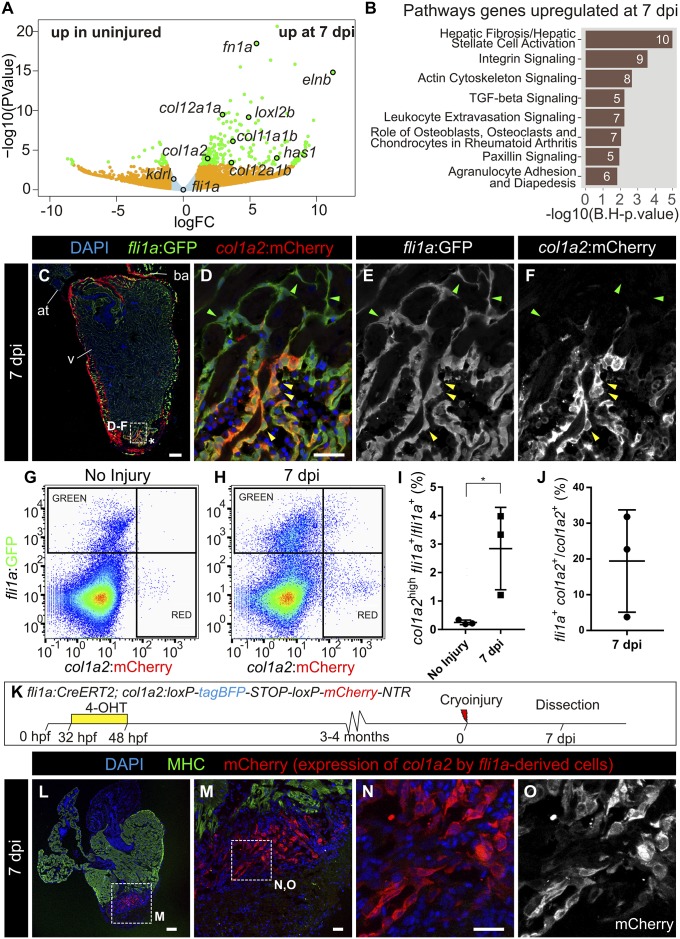
To further characterize the contribution of the endocardium to fibrosis, we analyzed the gene expression signature of endocardial cells in response to injury. We utilized the kdrl:mCherry transgenic line, which predominantly labels endocardial cells and relatively few coronary arteries (21), and performed RNA-seq (sequencing) analysis of fluorescence-activated cell sorter (FACS)-sorted mCherry+ cells from the apical region of control hearts and the injured ventricular apex of hearts at 7 dpi (Fig. 2 A and B and Dataset S2). The most up-regulated genes in kdrl+ cells were related to fibrosis. To validate the RNA-seq results, we crossed the endothelial and endocardial reporter line fli1a:GFP with col1a2:mCherry-NTR fish. At 7 dpi, expression of GFP and mCherry colocalized at the IA but not in the border zone (Fig. 2 C–F; n = 5/7). Cell sorting revealed that upon injury, a small proportion of fli1a:GFP+ cells expressed high levels of mCherry (Fig. 2 G–I): 19 ± 14% of fli1a:GFP+ cells were col1a2:mCherry-NTR+ (Fig. 2J). Next, fli1a+ cells were traced with a newly generated fli1a:CreERT2 line crossed with ubb:Switch (Fig. S4A). In uninjured hearts, mCherry+ cells lined the endocardial border, revealing efficient recombination (Fig. S4 B–E; n = 5/5). At 7 dpi, fli1a-derived cells were found in close proximity to col1a1 staining at the IA (Fig. S4 F–I). We also crossed fli1a:CreERT2 with the col1a2:loxP-tagBFP-loxP-mCherry-NTR line (Fig. 2K). At 7 dpi, mCherry+ cells were present at the IA but not the periphery, revealing that fli1a-derived cells produce collagen locally at the IA (Fig. 2 L–O; n = 7/12). Although endocardial cells close to the IA became more rounded, they did not appear to adopt a mesenchymal phenotype and remained connected, forming a layer around the luminal border of the site of injury. In agreement, while epithelial–mesenchymal transition (EMT) markers were up-regulated in wt1a:GFP+ cells upon injury, kdrl:mCherry+ cells revealed low levels of twist1a, prrx1a, and snail2 expression (Fig. S4 J–L).
Fig. 2.
Endocardium-derived cells contribute to transient fibrosis during zebrafish heart regeneration. (A) Analysis of the transcriptome of kdrl:mCherry+ cells FACS-sorted from the ventricular apex of hearts with no injury or at 7 dpi was performed. Volcano plot. Light blue, false discovery rate (FDR) >0.05, abs[log fold change (LFC)] <1; orange, FDR >0.05, abs(LFC) >1; green, FDR <0.05, abs(LFC) >1. (B) Ingenuity pathway analysis including the number of differentially expressed genes. (C–F) Immunofluorescence with anti-GFP and anti-mCherry of a heart section from an adult fli1a:GFP;col1a2:mCherry-NTR zebrafish. Yellow arrowheads mark double-positive cells at the injury area (IA; asterisk). Green arrowheads mark fli1a:GFP+ cells negative for mCherry. (G and H) FACS-sorted cells from fli1a:GFP;col1a2:mCherry hearts without injury or at 7 dpi. Shown are two representative examples from a total of four hearts per condition analyzed. (I) The percentage of fli1a:GFP+ cells that express more than 100 a.u. of mCherry, with the threshold corresponding to the maximum value of mCherry detected in endocardial cells from uninjured hearts. (J) The percentage of col1a2:mCherry+;fli1a:GFP+ cells according to the thresholds in H. Graphs show individual measurements from hearts as well as mean ± SD values. *P = 0.0363 by two-tailed t test. (K) Experimental scheme for visualizing collagen-producing endocardial cells. (L–N) Immunostaining of a heart section close to the IA. M is a zoomed-in view of L. (N and O) Zoomed-in views of M. mCherry marks fli1a-derived cells expressing col1a2, myosin heavy chain (MHC) marks the myocardium, and nuclei are DAPI-counterstained. [Scale bars, 25 µm (D, M, and N) and 100 µm (C and L).]
postnb Expression Reports Activated Fibroblasts in the Injured Zebrafish Heart.
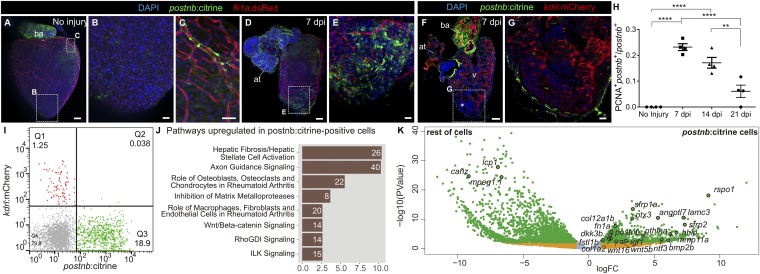
To characterize postnb-expressing cells that appear in response to injury, we generated a postnb:citrine reporter line. In adult uninjured hearts, postnb:citrine expression was detected in the bulbus arteriosus, valves, some perivascular cells associated with large coronary arteries in the basal ventricle, and isolated epicardial cells (Fig. 3 A–C; n = 3/3) but not in the ventricular apex (Fig. 3B). At 7 dpi, postnb:citrine+ cells were surrounding the IA in the cryoinjured apex and exhibited a mesenchymal morphology (Fig. 3 D and E, n = 3/3 and Fig. 3 F and G, n = 7/7). col1a2+ and postnb+ cells did not colocalize completely (Fig. S5 A and B, n = 3/3 hearts and Fig. S5 C–G, n = 6/6 hearts). Moreover, we did not observe cells coexpressing postnb:citrine and fli1a:DsRed or kdrl:mCherry (Fig. 3 F and G, n = 4/4 and Fig. 3I, n = 5/5), further confirming that at least some of the collagen-producing cells have an endocardial origin. The postnb+ population expanded during the first week postinjury, as revealed by proliferator cell nuclear antigen (PCNA) staining in postnb+ cells sampled at 7 and 14 dpi (Fig. 3H). After 3 wk, the number of postnb/PCNA double-positive cells significantly decreased. To further characterize this population, we analyzed the transcriptome of postnb:citrine+ cells sorted from the injured ventricle, identifying pathways related to fibrosis and axon guidance (Fig. 3J and Dataset S3). Compared with the remainder of cells at the IA, postnb:citrine+ cells were highly enriched for genes encoding secreted proteins. In total, 128 out of 917 differentially expressed genes up-regulated in the postnb:citrine+ population encoded secreted molecules, while only 12 out of 2,206 enriched in the negative population belonged to this category (P < 0.00001 by χ2 test). postnb:citrine+ cells expressed several ECM proteins in addition to 12 matrix metalloproteases (Fig. 3K and Fig. S5 H and I). Interestingly, postnb:citrine+ cells also expressed several signaling molecules (Fig. 3K and Fig. S5J). While some of these molecules have been described to influence heart development or regeneration (22–26), several others have not been studied in the context of heart regeneration to date, including wnt5a, wnt16, rspo1, and hhip (Fig. 3K).
Fig. 3.
periostin b expression marks an activated cardiac fibroblast population upon ventricular cryoinjury. (A–E) Whole-heart immunofluorescence in the postnb:citrine; fli1a:dsRedEx double-transgenic line. Whole-heart (A) and zoomed-in views (B and C) of the ventricular apex of an uninjured heart and a heart at 7 dpi (D and E). Perivascular cells can be observed in C. postnb:citrine, green; fli1a:dsRedEx, red. (F and G) Immunofluorescence staining of a sagittal heart section of a postnb:citrine;kdrl:mCherry zebrafish (F). Asterisk in F marks the injury area. (G) Zoomed-in view of the injured ventricular apex. postnb:citrine, green; kdrl:mCherry, red. (H) Quantification of proliferating postnb+ cells postinjury (mean ± SD; ****P < 0.0001, **P < 0.01 by one-way ANOVA followed by Tukey’s multiple comparisons test). (I) FACS-sorted cells from kdrl:mCherry;postnb:citrine hearts. No double-positive cells were detected. (J and K) Transcriptome analysis of postnb:citrine+ cells isolated from the ventricular apex. (J) Ingenuity pathway analysis. The x axis shows −log10(B.H. P values). (K) Volcano plot. Light blue, FDR >0.05, abs(LFC) <1; orange, FDR <0.05, abs(LFC) <1; green, FDR <0.05, abs(LFC) >1. [Scale bars, 25 µm (B, C, E, and G) and 100 µm (A, D, and F).]
Overall, these results reveal that the postnb:citrine zebrafish line marks activated fibroblasts, which express gene-encoding ECM proteins as well as proteins responsible for ECM degradation. Furthermore, they express secreted signaling genes that could influence heart regeneration.
Fibroblast Inactivation Leads to Fibrosis Regression During Cardiac Regeneration.
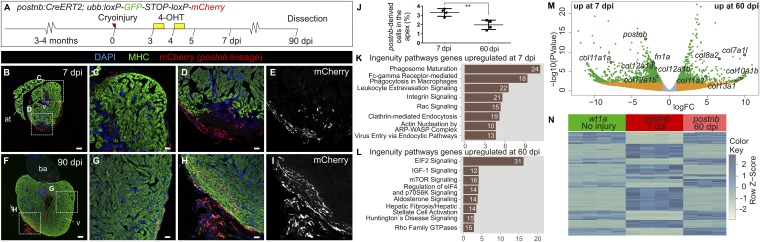
Whereas cardiac fibrosis is irreversible in adult mammals, degradation of fibrotic tissue occurs in the zebrafish, and the expression of fibrosis-related genes drops to near-baseline levels at 90 dpi (27) (Fig. S6 A and B). Elimination of activated fibroblasts has been suggested to progress via a mechanism of programmed cell death (1). However, terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining of postnb:citrine heart sections at different postinjury stages revealed a low number of apoptotic postnb+ cells at 7 and 14 dpi and none at 21 dpi (Fig. S6 C–G). Given the low number of apoptotic fibroblasts identified at the three stages analyzed, fibroblasts might remain in the heart even after complete regeneration. To test this, we generated a postnb:CreERT2 line and crossed it with the ubb:Switch line (Fig. 4A). Two pulses of 4-OHT at 3 and 4 dpi allowed permanent mCherry labeling of postnb+ cells. postnb-derived cells were detected at the IA at 7 dpi (Fig. 4 B–E; n = 4/4). Surprisingly, at the ventricle apex, postnb-derived cells could still be detected within the regenerated myocardium at 90 dpi, albeit in lower numbers (Fig. 4 F–J; n = 5/5). To assess whether fibroblasts revert to a quiescent phenotype during regeneration, we performed RNA-seq analysis of postnb-derived cells at 7 and 60 dpi (Fig. 4 K–N and Dataset S4). Pathway analysis revealed differences in the expression profiles of postnb-derived cells between these stages (Fig. 4 K and L). Although overall the level of ECM gene expression decreased from 7 to 60 dpi, there were also collagen-encoding genes that were up-regulated at 60 dpi, such as col7a1l and col8a2 (Fig. 4 M and N and Fig. S7 A–I). These genes were not expressed by wt1a+ resident fibroblasts in the uninjured heart, indicating that the inactivation of fibroblasts does not fully revert the expression profile to a homeostatic baseline. Overall, the gene signature of postnb-derived cells at 60 dpi resembled more the signature of wt1a:GFP cells than that of postnb-derived cells at 7 dpi (Fig. 5O and Fig. S7 J–L). Indeed, postnb− wt1a:GFP+ cells were found at 130 dpi in the regenerated myocardium (Fig. S8).
Fig. 4.
Persistence of postnb-derived cells in the regenerated myocardium. (A) 4-Hydroxytamoxifen was added to postnb:CreERT2;ubb:Switch fish at 3 and 4 dpi, and hearts were dissected at different dpi. (B–I) Immunofluorescence staining of heart sections at 7 (B–E) or 90 dpi (F–I). (J) Percentage of postnb-derived cells at the injury area. Symbols show individual measurements, and boxes and whiskers show mean ± SD; **P = 0.0064 by two-tailed unpaired t test. (K–M) postnb-derived mCherry+ cells were sorted from the ventricular apex at 7 and 60 dpi, and transcriptome analysis was performed on isolated mCherry+ cells. (K and L) Ingenuity pathway analysis. Bars represent pathways enriched in postnb-derived cells compared with the remainder of cells in the injury area at 7 and 60 dpi. Numbers of differentially expressed genes are indicated. The x axis shows −log10(B.H. P values). (M) Volcano plot. Light blue, FDR >0.05, abs(LFC) <1; orange, FDR >0.05, abs(LFC) >1; green, FDR <0.05, abs(LFC) >1. (N) Heatmap of the top 200 differentially expressed genes between 7 and 60 dpi in postnb-traced cells and their expression in wt1a:GFP+ cells from uninjured hearts. [Scale bars, 25 µm (C–E and G–I) and 100 µm (B and F).]
Fig. 5.
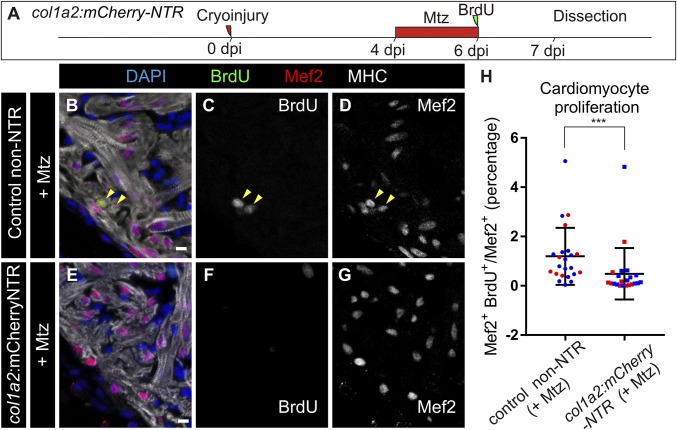
Genetic ablation of collagen 1a2-expressing cells impairs cardiomyocyte proliferation in the cryoinjured heart. (A) col1a2:mCherry-NTR adult animals were cryoinjured and treated with metronidazole (Mtz) from 4 to 6 dpi. BrdU injection was performed 1 d before fixation. (B–G) Immunofluorescence using anti-mef2 and anti-MHC to mark cardiomyocytes, and anti-BrdU in col1a2:loxP-tagBFP-loxP-mCherry-NTR (control) and col1a2:mCherry-NTR fish treated with Mtz and BrdU as described in A. Arrowheads mark BrdU+ cardiomyocytes. (H) Quantification of BrdU+ cardiomyocytes in col1a2:mCherry-NTR and control hearts. Shown are individual measurements and median ± interquartile range; ***P = 0.0004 by Mann–Whitney test, n = 23 fish per condition, from two different experiments (blue and red dots). Three whole-heart ventricle sections were quantified per point. [Scale bars, 10 µm (C and E–G) and 100 µm (B and D).]
Altogether, the data indicate that during fibrosis regression, activated fibroblasts partially return to a quiescent stage without fully resembling endogenous cardiac fibroblasts from the uninjured heart.
Fibroblasts Influence Cardiomyocyte Proliferation During Heart Regeneration.
To test whether cardiac fibrosis influences heart regeneration, we ablated col1a2-expressing cells using a col1a2:mCherry-NTR line, in which col1a2+ cells express nitroreductase (NTR) and are ablated upon metronidazole (Mtz) treatment (Fig. S9A). Fragmented col1a2+ cell bodies and nuclei were found in Mtz-treated animals at 7 dpi, indicating efficient ablation (Fig. S9 B and C; n = 8/8). This was not detected in the untreated control group (Fig. S9 D and E; n = 3/3). No significant differences in heart regeneration were observed between the experimental and control groups at 35 dpi, suggesting that ablation of ECM-producing cells does not enhance regenerative capacity (Fig. S9 F–M). Nonetheless, collagen 1 could still be detected upon genetic ablation (Fig. S10). However, we found a fourfold reduction in cardiomyocyte proliferation in col1a2:mCherry-NTR fish compared with control siblings (Fig. 5).
In sum, our results not only suggest that fibrosis is compatible with regeneration but also indicate that transient fibrosis upon cryoinjury is necessary for cardiomyocyte proliferation.
Discussion
In the mouse, cardiac repair upon MI or ventricular overload has been shown to occur mainly by ECM deposition from intracardiac fibroblasts (3, 6, 28, 29) and the epicardium (30, 31), whereas a contribution from the bone marrow is controversial (3, 28, 32–37). Endothelial cells have been suggested to contribute to cardiac fibrosis in a pressure overload model (3, 28, 36). Our data reveal that in the context of heart regeneration, mostly preexisting fibroblasts but also endocardial cells contribute to collagen production. Coexpression of fli1a+ or fli1a-derived cells and col1a2:mCherry was not observed in all analyzed hearts, suggesting that the endocardium is not the principal contributor to fibrosis. Of note, endocardial cells at the injury border failed to undergo full EMT, and thus do not adopt a complete fibroblast-like phenotype. Cells from the epicardial border were found to produce both periostin and collagen, whereas the endocardial cells only produced collagen. These two distinct ECM environments surrounding the IA may play an important role in guiding heart regrowth.
In contrast to mammals, where ECM persists after MI, it is degraded in the zebrafish heart. We found that down-regulation of fibroblast ECM production is a key step for fibrosis regression. Interestingly, clearance does not involve the complete elimination of ECM-producing cells. Indeed, our data suggest that postnb+ cells might not only be actively involved in ECM production but may also participate in its degradation during regeneration.
tcf21-derived cells detected in the regenerated myocardium are epicardial cells or EPDCs that were already present in the uninjured myocardium, and whether they represent inactivated fibroblasts has not been previously explored (38). Our results suggest that this population represents tcf21+ cells that activate postnb in response to injury. The herein reported long-term permanence of postnb-derived cells in the regenerated heart might also provide an explanation for why there is only a partial recovery of ventricular wall contraction after cryoinjury (39).
Our data also demonstrate that ablation of ECM-producing cells at an early stage of the injury response impairs cardiomyocyte proliferation in the injured ventricle. Thus, fibrosis has a beneficial effect on heart regeneration. Interestingly, col1a2+ cells affect cardiomyocyte proliferation through mechanisms not directly linked to collagen deposition. postnb-derived cells express several secreted molecules that promote cardiomyocyte proliferation in the zebrafish. Furthermore, heart regeneration requires heart reinnervation (40, 41), and our gene expression analysis suggests that fibroblasts might also contribute to axon pathfinding. While cardiomyocyte proliferation was impaired, cardiac regeneration recovered after the ablation of collagen-producing cells. This may suggest compensatory mechanisms, or indicate that a recovery occurring after ablation can overcome the delayed regeneration.
In the adult mouse, ablation of Postn-derived cells impairs the recovery of cardiac pumping efficiency upon MI, revealing an important role for fibroblasts during cardiac repair (6). The lack of fibroblasts was proposed as an explanation for the regenerative capacity of the zebrafish heart (42). However, here we confirm the presence of cardiac fibroblasts in the zebrafish, and reveal that they not only contribute to the fibrotic response after injury but are also necessary for cardiomyocyte proliferation during regeneration. This new information on how fibrosis influences myocardial regeneration and how it is eliminated in a species with endogenous regenerative potential could have important implications for regenerative medicine strategies.
Materials and Methods
Experiments were approved by the Community of Madrid “Dirección General de Medio Ambiente” in Spain and the “Amt für Landwirtschaft und Natur” from the Canton of Bern, Switzerland. Animals were housed and experiments were performed in accordance with Spanish and Swiss bioethical regulations for the use of laboratory animals. Animals used for experiments were randomly selected from siblings of the same genotype. Detailed information on the lines used and generated can be found in SI Materials and Methods.
ISH on paraffin sections and on whole-mount larvae was performed as described (10, 43). A detailed protocol and list of riboprobes and antibodies used for immunofluorescence staining can be found in Supporting Information.
Apoptosis was detected by TUNEL staining using the In Situ Cell Death Detection Kit from Roche.
A Leica TCS SP5 confocal microscope was used for immunofluorescence imaging on sections and whole-mount hearts, whereas a Nikon 90i was used to image nonfluorescent sections. Images were quantified manually in ImageJ (NIH) using the Cell Counter plugin.
RNA-seq data have been deposited in the Gene Expression Omnibus database with accession nos. GSE101204, GSE101200, and GSE101199. Raw data have been deposited at https://data.mendeley.com/datasets/z7ypxkgv2m/1. A detailed protocol can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to the Animal Facility, Histology, Microscopy, Cellomics, Bioinformatics, and Genomics units of the CNIC, and the Microscopy Imaging Center of the University of Bern. We thank C. Helker for sharing reagents, and O. Kanisicak for discussions. H.S.-I. and N.M. were funded by the Spanish Ministry of Economy and Competitiveness (MINECO) by Grants FPU12/03007 and BFU2014–56970–P, Plan Estatal 2013–2016, Programa Estatal de I+D+i: Proyectos I+D+i 2016, and Fondo Europeo de Desarrollo Regional. N.M. is supported by Swiss National Science Foundation Grant 31003A_159721 and ERC Starting Grant 337703–Zebra–Heart. A.E. is funded through ANR-SNF Project 320030E-164245 (to N.M.). The CNIC is supported by MINECO and the ProCNIC Foundation, and is a Severo Ochoa Center of Excellence (MINECO Award SEV-2015-0505).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The RNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE101204, GSE101200, and GSE101199). Raw data from this manuscript have been deposited at https://data.mendeley.com/datasets/z7ypxkgv2m/1. Fish line information has been deposited at www.zfin.org.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716713115/-/DCSupplemental.
References
- 1.Travers JG, Kamal FA, Robbins J, Yutzey KE, Blaxall BC. Cardiac fibrosis: The fibroblast awakens. Circ Res. 2016;118:1021–1040. doi: 10.1161/CIRCRESAHA.115.306565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tallquist MD, Molkentin JD. Redefining the identity of cardiac fibroblasts. Nat Rev Cardiol. 2017;14:484–491. doi: 10.1038/nrcardio.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore-Morris T, et al. Resident fibroblast lineages mediate pressure overload-induced cardiac fibrosis. J Clin Invest. 2014;124:2921–2934. doi: 10.1172/JCI74783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Norris RA, et al. Neonatal and adult cardiovascular pathophysiological remodeling and repair: Developmental role of periostin. Ann N Y Acad Sci. 2008;1123:30–40. doi: 10.1196/annals.1420.005. [DOI] [PubMed] [Google Scholar]
- 5.Snider P, et al. Origin of cardiac fibroblasts and the role of periostin. Circ Res. 2009;105:934–947. doi: 10.1161/CIRCRESAHA.109.201400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanisicak O, et al. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat Commun. 2016;7:12260. doi: 10.1038/ncomms12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kikuchi K. Advances in understanding the mechanism of zebrafish heart regeneration. Stem Cell Res (Amst) 2014;13:542–555. doi: 10.1016/j.scr.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 8.González-Rosa JM, Burns CE, Burns CG. Zebrafish heart regeneration: 15 years of discoveries. Regeneration (Oxf) 2017;4:105–123. doi: 10.1002/reg2.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.González-Rosa JM, Martín V, Peralta M, Torres M, Mercader N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development. 2011;138:1663–1674. doi: 10.1242/dev.060897. [DOI] [PubMed] [Google Scholar]
- 10.González-Rosa JM, Peralta M, Mercader N. Pan-epicardial lineage tracing reveals that epicardium derived cells give rise to myofibroblasts and perivascular cells during zebrafish heart regeneration. Dev Biol. 2012;370:173–186. doi: 10.1016/j.ydbio.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Karra R, Dickson AL, Poss KD. Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration. Dev Biol. 2013;382:427–435. doi: 10.1016/j.ydbio.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Münch J, Grivas D, González-Rajal Á, Torregrosa-Carrión R, de la Pompa JL. Notch signalling restricts inflammation and serpine1 expression in the dynamic endocardium of the regenerating zebrafish heart. Development. 2017;144:1425–1440. doi: 10.1242/dev.143362. [DOI] [PubMed] [Google Scholar]
- 13.Ieda M, et al. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev Cell. 2009;16:233–244. doi: 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chablais F, Jazwinska A. The regenerative capacity of the zebrafish heart is dependent on TGFβ signaling. Development. 2012;139:1921–1930. doi: 10.1242/dev.078543. [DOI] [PubMed] [Google Scholar]
- 15.Lafontant PJ, et al. Cardiac myocyte diversity and a fibroblast network in the junctional region of the zebrafish heart revealed by transmission and serial block-face scanning electron microscopy. PLoS One. 2013;8:e72388. doi: 10.1371/journal.pone.0072388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peralta M, González-Rosa JM, Marques IJ, Mercader N. The epicardium in the embryonic and adult zebrafish. J Dev Biol. 2014;2:101–116. doi: 10.3390/jdb2020101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marro J, Pfefferli C, de Preux Charles AS, Bise T, Jaźwińska A. Collagen XII contributes to epicardial and connective tissues in the zebrafish heart during ontogenesis and regeneration. PLoS One. 2016;11:e0165497. doi: 10.1371/journal.pone.0165497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLaughter DM, et al. Single-cell resolution of temporal gene expression during heart development. Dev Cell. 2016;39:480–490. doi: 10.1016/j.devcel.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furtado MB, Nim HT, Boyd SE, Rosenthal NA. View from the heart: Cardiac fibroblasts in development, scarring and regeneration. Development. 2016;143:387–397. doi: 10.1242/dev.120576. [DOI] [PubMed] [Google Scholar]
- 20.Cao J, et al. Single epicardial cell transcriptome sequencing identifies caveolin 1 as an essential factor in zebrafish heart regeneration. Development. 2016;143:232–243. doi: 10.1242/dev.130534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison MR, et al. Chemokine-guided angiogenesis directs coronary vasculature formation in zebrafish. Dev Cell. 2015;33:442–454. doi: 10.1016/j.devcel.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barandon L, et al. Reduction of infarct size and prevention of cardiac rupture in transgenic mice overexpressing FrzA. Circulation. 2003;108:2282–2289. doi: 10.1161/01.CIR.0000093186.22847.4C. [DOI] [PubMed] [Google Scholar]
- 23.Gibb N, Lavery DL, Hoppler S. sfrp1 promotes cardiomyocyte differentiation in Xenopus via negative-feedback regulation of Wnt signalling. Development. 2013;140:1537–1549. doi: 10.1242/dev.088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y, et al. Igf signaling is required for cardiomyocyte proliferation during zebrafish heart development and regeneration. PLoS One. 2013;8:e67266. doi: 10.1371/journal.pone.0067266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei K, et al. Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature. 2015;525:479–485. doi: 10.1038/nature15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu CC, et al. Spatially resolved genome-wide transcriptional profiling identifies BMP signaling as essential regulator of zebrafish cardiomyocyte regeneration. Dev Cell. 2016;36:36–49. doi: 10.1016/j.devcel.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 27.Rodius S, et al. Analysis of the dynamic co-expression network of heart regeneration in the zebrafish. Sci Rep. 2016;6:26822. doi: 10.1038/srep26822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali SR, et al. Developmental heterogeneity of cardiac fibroblasts does not predict pathological proliferation and activation. Circ Res. 2014;115:625–635. doi: 10.1161/CIRCRESAHA.115.303794. [DOI] [PubMed] [Google Scholar]
- 29.Teekakirikul P, et al. Cardiac fibrosis in mice with hypertrophic cardiomyopathy is mediated by non-myocyte proliferation and requires Tgf-β. J Clin Invest. 2010;120:3520–3529. doi: 10.1172/JCI42028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duan J, et al. Wnt1/βcatenin injury response activates the epicardium and cardiac fibroblasts to promote cardiac repair. EMBO J. 2012;31:429–442. doi: 10.1038/emboj.2011.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou B, et al. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J Clin Invest. 2011;121:1894–1904. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haudek SB, et al. Bone marrow-derived fibroblast precursors mediate ischemic cardiomyopathy in mice. Proc Natl Acad Sci USA. 2006;103:18284–18289. doi: 10.1073/pnas.0608799103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Möllmann H, et al. Bone marrow-derived cells contribute to infarct remodelling. Cardiovasc Res. 2006;71:661–671. doi: 10.1016/j.cardiores.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 34.Ruiz-Villalba A, et al. Interacting resident epicardium-derived fibroblasts and recruited bone marrow cells form myocardial infarction scar. J Am Coll Cardiol. 2015;65:2057–2066. doi: 10.1016/j.jacc.2015.03.520. [DOI] [PubMed] [Google Scholar]
- 35.van Amerongen MJ, et al. Bone marrow-derived myofibroblasts contribute functionally to scar formation after myocardial infarction. J Pathol. 2008;214:377–386. doi: 10.1002/path.2281. [DOI] [PubMed] [Google Scholar]
- 36.Zeisberg EM, et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat Med. 2007;13:952–961. doi: 10.1038/nm1613. [DOI] [PubMed] [Google Scholar]
- 37.Moore-Morris T, et al. Infarct fibroblasts do not derive from bone marrow lineages. Circ Res. 2018;122:583–590. doi: 10.1161/CIRCRESAHA.117.311490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kikuchi K, et al. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development. 2011;138:2895–2902. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.González-Rosa JM, et al. Use of echocardiography reveals reestablishment of ventricular pumping efficiency and partial ventricular wall motion recovery upon ventricular cryoinjury in the zebrafish. PLoS One. 2014;9:e115604. doi: 10.1371/journal.pone.0115604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahmoud AI, et al. Nerves regulate cardiomyocyte proliferation and heart regeneration. Dev Cell. 2015;34:387–399. doi: 10.1016/j.devcel.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White IA, Gordon J, Balkan W, Hare JM. Sympathetic reinnervation is required for mammalian cardiac regeneration. Circ Res. 2015;117:990–994. doi: 10.1161/CIRCRESAHA.115.307465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ausoni S, Sartore S. From fish to amphibians to mammals: In search of novel strategies to optimize cardiac regeneration. J Cell Biol. 2009;184:357–364. doi: 10.1083/jcb.200810094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mercader N, Fischer S, Neumann CJ. Prdm1 acts downstream of a sequential RA, Wnt and Fgf signaling cascade during zebrafish forelimb induction. Development. 2006;133:2805–2815. doi: 10.1242/dev.02455. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.