Significance
In mammals, the suprachiasmatic nucleus (SCN) synchronizes circadian rhythms in cells throughout the body to the environmental light/dark cycle. We have developed a fluorescence-based technology that enables long-term monitoring of clock gene expression in particular brain nuclei and specific types of neurons at high temporal resolution, in freely moving mice, in a light/dark cycle. Using this system, we demonstrate the dynamics of Per2 and Cry1 transcriptional rhythms in SCN vasoactive intestinal peptide (VIP) neurons following an 8-h phase advance in the light/dark cycle.
Keywords: circadian clock, in vivo fluorescence recording, suprachiasmatic nucleus, light/dark cycle, phase advance
Abstract
Endogenous circadian clocks control 24-h physiological and behavioral rhythms in mammals. Here, we report a real-time in vivo fluorescence recording system that enables long-term monitoring of circadian rhythms in the brains of freely moving mice. With a designed reporter of circadian clock gene expression, we tracked robust Cry1 transcription reporter rhythms in the suprachiasmatic nucleus (SCN) of WT, Cry1−/−, and Cry2−/− mice in LD (12 h light, 12 h dark) and DD (constant darkness) conditions and verified that signals remained stable for over 6 mo. Further, we recorded Cry1 transcriptional rhythms in the subparaventricular zone (SPZ) and hippocampal CA1/2 regions of WT mice housed under LD and DD conditions. By using a Cre-loxP system, we recorded Per2 and Cry1 transcription rhythms specifically in vasoactive intestinal peptide (VIP) neurons of the SCN. Finally, we demonstrated the dynamics of Per2 and Cry1 transcriptional rhythms in SCN VIP neurons following an 8-h phase advance in the light/dark cycle.
In mammals, the suprachiasmatic nucleus (SCN), a small bilateral nucleus located in the hypothalamus that functions as the “master clock,” governs the whole body’s circadian rhythm and coordinates appropriate responses to environmental changes (1, 2). Core molecular components of the circadian clock include the genes Per1-3, Cry1-2, Clock, and Bmal1. Heterodimers of CLOCK/BMAL1 activate the transcription of Per and Cry, and the PER and CRY proteins then suppress the function of CLOCK/BMAL1, forming a transcription–translation negative feedback loop that takes about 24 h to complete a single cycle (3).
Previous studies have mainly used ex vivo slice cultures (4), in situ hybridization (5, 6), or quantitative real time polymerase chain reaction (qPCR) (7) to reveal the dynamics of circadian clock gene expression during circadian misalignment or in response to environmental time cues. But in vivo longitudinal studies have been infrequent. SCN multiunit neural activity (MUA) rhythms have been monitored in freely moving rats or mice, either in constant conditions (8) or during an 8-h phase advance of the light cycle (9). Bioluminescent reporters have also been used to monitor circadian clock gene expression in freely moving mice, in constant darkness (10–13) or after a light pulse (14). However, bioluminescent reporters are too dim to permit continuous monitoring in a light/dark cycle. Because light is the predominant input signal for SCN-mediated entrainment of circadian clocks (15), there is a pressing need for in vivo monitoring of circadian clocks in a light/dark cycle.
In this study, we developed a fluorescence-based technology to monitor, in the brains of freely moving mice, circadian rhythms of transcription of the clock genes Per2 and Cry1 in a light/dark cycle and constant darkness. Using this technology, we monitored circadian clock gene transcription specifically in vasoactive intestinal peptide (VIP) neurons of the SCN for a period of several weeks. Additionally, as an example of the power of this technology, we demonstrated the dynamics of Per2 and Cry1 transcriptional rhythms in VIP neurons of the SCN following an 8-h phase advance in the light/dark cycle.
Results
Methodology for Long-Term in Vivo Recording of Circadian Rhythms in the SCN of Freely Moving Mice Using Fluorescent Probes.
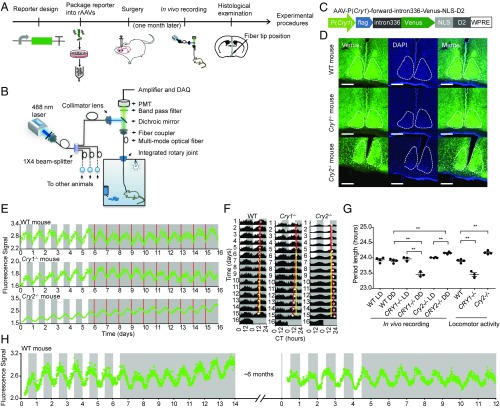
Our goal was to develop a fluorescence-based method for long-term monitoring of circadian clock gene transcription in neurons of freely moving mice. An overview of the experimental procedures is schematized in Fig. 1A. In this system, we designed fluorescent reporters and used recombinant adeno-associated viruses (rAAVs) to deliver genes encoding them into particular regions of the mouse brain (Fig. S1A, Left). The precise schematics of the in vivo monitoring instrument are depicted in Fig. 1B. We used a 1 × 4 beam splitter to divide a single laser beam into four beams, increasing the number of recording channels from one to four, which enabled us to record four mice simultaneously. After the beam splitter, the four recording channels were all separated, to prevent cross-talk among recording channels.
Fig. 1.
In vivo monitoring setup and in vivo recording of Cry1 transcription in the SCN of freely moving mice. (A) Overview of the experimental workflow. (B) Schematic diagram of the in vivo monitoring setup. (C) Cry1 transcription reporter design. (D) Fluorescence microscopy analysis of Cry1 transcription reporter expression (green) in the SCN of WT (Top), Cry1−/− (Middle), and Cry2−/− (Bottom) mice. (Scale bar: 200 µm.) (E) In vivo recording results of Cry1 transcription reporter rhythms in the SCN of WT (Top), Cry1−/− (Middle), and Cry2−/− (Bottom) mice. Recording started 1 mo after the surgery; the day of the recording start was defined as 0. The white and gray areas indicate the light-on and light-off periods, respectively. The red lines indicate the subjective lights-on time. Raw data are plotted at 10-min intervals. (F) Double-plotted data of the in vivo recording in the SCN of WT (Left), Cry1−/− (Middle), and Cry2−/− (Right) mice. The solid red lines indicate the peak times in the LD condition, the dashed red lines indicate the peak times in the DD condition, and the solid yellow lines are the extension of the solid red lines. (G) In vivo recording and locomotor activity period length of WT, Cry1−/−, and Cry2−/− mice; n = 4 for WT and Cry2−/− mice, n = 3 for Cry1−/− mice, mean ± SEM, one-way ANOVA, **P < 0.01. (H) Long-term recording of Cry1 transcription in the SCN of WT mouse. CT, circadian time; DAQ, data acquisition; PMT, photomultiplier tube.
In Vivo Recording of Cry1 Transcription Rhythm in the SCN.
Considering the central role of Cryptochrome (Cry) genes in the circadian clock (16) and guided by detailed studies of Cry1 transcription (17), we first designed a fluorescent reporter of Cry1 transcription (Fig. 1C). The Cry1 promoter was used to drive expression of the fluorescent protein Venus. To maintain the signal in the cell body and to increase turnover kinetics (18, 19), a nuclear localization signal (NLS) and residues 422 to 461 of mouse ornithine decarboxylase (D2), respectively, were fused to the C-terminal end of Venus (designated “dVenus”). The functional relevance of the intronic enhancer (intron336) for Cry1 transcription was demonstrated in cultured cells (17) and in peripheral tissues (20), and its possible functional importance in the SCN is not clear. Nevertheless, given its potential importance for the circadian clock, the reporter we designed contains both the Cry1 promoter and the intron. The complete reporter, AAV-P(Cry1)-forward-intron336-Venus-NLS-D2, was then introduced by stereotactic injection into the SCN of WT, Cry1−/−, and Cry2−/− mice. The reporter led to strong expression of Venus in the SCN (Fig. 1D).
With the use of fluorescence recording in a long-term experiment, phototoxicity is always a concern. A terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) assay was conducted 1 wk after the fiber recording to test if there was any phototoxicity-induced cell apoptosis. We found that, with 15-µW laser power turned on for 15 s every 10 min, there was no detectable cell apoptosis (Fig. S1B). Thus, to minimize potential phototoxicity, the total laser power used at the following in vivo recording experiments was reduced to a low level (10 to 20 µW), and the laser was only turned on briefly for each time point (15 s at 10-min intervals).
To determine at which level ambient light may interfere with the recording, optical fibers were inserted into mouse brains (Fig. S1A, Right) at seven different depths. After 10 postsurgery recovery days, light noise was measured during the dark–light transition under three different light intensities [150 lux (lx), 500 lx, 1,000 lx] either with or without the application of black nail polish (Fig. S1C). The result shows that, when the depth of probe in the brain was shallower than 2 cm and the ambient light intensity was stronger than 150 lx, the ambient light introduced some noise to the recording data, which can be eliminated by the application of black nail polish. Thus, in the remainder of the recording experiments, to avoid the influence of ambient light, all of the in vivo recordings were performed under an ambient light intensity of 100 lx and with the application of black nail polish.
To test whether our system was sensitive and stable enough to detect rhythmic fluorescence, the Cry1 transcription reporter was delivered into the SCN, and an optical fiber was targeted into the SCN at the same time; mice without AAV injection were used as a negative control. One month after the surgery, mice were recorded for 1 d in the LD condition (12 h light, 12 h dark) and 1 d in the DD condition (constant darkness). In both lighting conditions, we observed a very low, nonrhythmic background level in the control mouse while mice with the Cry1 transcription fluorescence reporter showed robust circadian rhythm in both LD and DD conditions (Fig. S1D). These results indicate that our recording system is sensitive enough to monitor approximately 24-h rhythms of fluorescence in the SCN of freely moving mice in either a light/dark cycle or constant darkness.
To determine when the Cry1 transcription reporter gives an obvious circadian rhythm after injection of the reporter and optical fiber implantation surgery, a mouse with Cry1 transcription reporter virus injection in the SCN and optical fiber implantation targeted into the SCN was recorded 8 d after the surgery (Fig. S1E). The result showed that, with increased extension of the AAV infection and increased expression of the reporters, the intensity of the fluorescence signal increased each day, the rapid increase in the signal intensity itself masked our ability to discern the circadian rhythm, and we could not clearly isolate the signal for the rhythm from day 8 to day 17 postsurgery. Later, as the reporter expression reached a relatively stable (albeit still slightly increasing) state, we were able to clearly discern the circadian rhythm. Thus, in the remainder of the recording experiments, in vivo recordings were performed 1 mo after the surgery.
We next recorded the rhythm of the Cry1 transcription reporter in the SCN of WT, Cry1−/−, and Cry2−/− mice for 6 d in the LD condition and then 10 d in the DD condition (Fig. 1E). Double-plotted approximately 24-h rhythms of Cry1 transcription reporter rhythm are shown in Fig. 1F. The reporter shows robust rhythms in the SCN in both LD and DD conditions. After optical recording, wheel-running activity was measured in LD and DD conditions (Fig. S2A), showing patterns typical for the respective genotypes. To examine whether AAV infection or implantation of the optical probe affected animal circadian rhythm, wheel-running activity was measured between uninfected mice and virus infected mice with or without fiber insertion (Fig. S2B). These results showed that neither virus infection nor fiber insertion affected the circadian phenotype; the gene expression and locomotor periods correlate (Fig. S2 A and B and Table S1). After optical recording and behavior experiments, histology examination was conducted to confirm the optical fiber tip position in the SCN (Fig. S2 A and B).
The in vivo recorded Cry1 transcription reporter rhythms of Cry1−/− mice in the DD condition were significantly shorter than its period in the LD condition and also significantly shorter than the period of WT mice in the DD condition (Fig. 1G). The in vivo recorded Cry1 transcription reporter rhythms of Cry2−/− mice in the DD condition was significantly longer than its period in the LD condition and also significantly longer than the period of WT mice in the DD condition (Fig. 1G). The in vivo recordings were consistent with the genotype-specific periods of locomotor activity rhythms (Fig. 1G and Fig. S2A). Recordings in Cry1−/− and Cry2−/− mice showed that the in vivo recorded rhythms were not somehow induced by environmental factors. Additionally, recording in Cry1−/− and Cry2−/− mice showed that the in vivo recording system can distinguish the period difference between different genotypes of mice.
Our system also enabled us to monitor Cry1 transcription rhythms in the SCN of freely moving mice for more than 6 mo (Fig. 1H and Fig. S2C, n = 3 mice). Cry1 transcription was first monitored in LD and DD conditions; about 6 mo later, the same mouse was then monitored in LD and DD conditions.
All of these results indicate that our recording system can be used for accurate long-term monitoring of circadian clock reporter rhythm in the SCN of freely moving mice at a high temporal resolution, either in constant darkness or in a light/dark cycle.
In Vivo Recording of Cry1 Transcription Rhythm in the Subparaventricular Zone and Hippocampus.
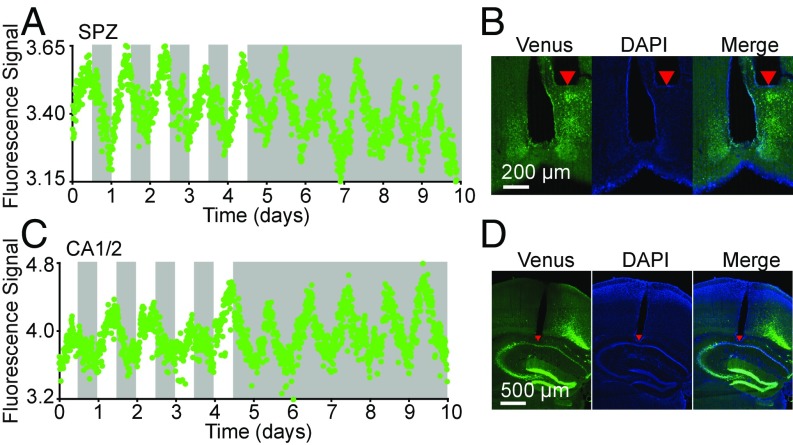
To monitor circadian dynamics in different brain nuclei, the Cry1 transcription reporter virus was injected into the hypothalamic subparaventricular zone (SPZ) or the hippocampal CA1/2 region. At the same time of the virus injection, optical fibers were implanted into the corresponding brain nuclei. We then recorded Cry1 transcription reporter rhythms in these brain nuclei for 5 d in the LD condition and then 5 d in the DD condition (Fig. 2 A and C and Fig. S3A).
Fig. 2.
In vivo recording of Cry1 transcription rhythms in the SPZ and hippocampus CA1/2 regions. (A and C) Cry1 transcription rhythms in the SPZ (A) and CA1/2 (C). Recording started 1 mo after the surgery; the day of the recording start was defined as 0. The white and gray areas indicate the light-on and light-off periods, respectively. Raw data are plotted at 10-min intervals. (B and D) The photomicrographs show expression of the Cry1 transcription reporter in the SPZ (B) and CA1/2 (D). The red triangles in the brain slices show the tip position of the optical fiber.
Unlike ex vivo slice culture (4), in both LD and DD conditions the transcription of Cry1 in SPZ and CA1/2 showed a robust and sustained circadian rhythm without damping of the circadian amplitude. Histological examination after these recordings confirmed that the optical fiber tip had been correctly implanted at the targeted sites (Fig. 2 B and D and Fig. S3B). These results indicated the ability of our recording system for in vivo recording of the circadian clocks in various brain nuclei of freely moving mice under normal physiological conditions.
In Vivo and ex Vivo Recordings of Per2 and Cry1 Transcription Rhythms Specifically in SCN VIP Neurons.
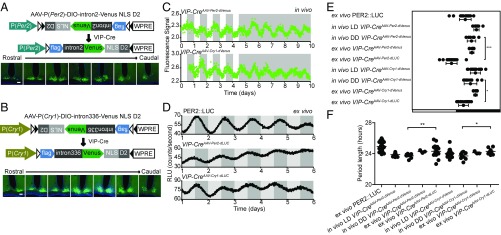
Brain nuclei typically comprise multiple populations of functionally distinct neurons. We therefore used the Cre-loxP system to further improve the targeting specificity of our rAAV-based circadian reporter. Considering the importance of VIP neurons in the SCN (21), we designed two reporters (Fig. 3 A and B, Top), which, combined with VIP-Cre mice (22), can be used to monitor transcription of Per2 and Cry1 specifically in VIP neurons of the SCN. As in earlier experiments, we used the Cry1 promoter to drive expression of dVenus. For Per2, we found that the Per2 promoter alone could drive expression in the mouse cerebral cortex but not in the SCN (Fig. S4A). Considering the constraints of AAV vector-packaging capacity and Per2 gene information, we added intron2 of Per2 as an enhancer to improve rhythmic expression in the SCN. Infusion of reporter rAAVs into the SCN of VIP-Cre mice led to expression of Venus in SCN VIP neurons (Fig. 3 A and B, Bottom) (these mice are henceforth referred to as VIP-CreAAV-Per2-dVenus, VIP-CreAAV-Cry1-dVenus). The Venus-NLS-D2 protein turnover kinetics was verified by Western blotting (Fig. S4B): The two reporters each had a half-life of about 2 h. Immunolabeling with VIP antibody 2 wk after unilaterally injecting the reporter into the SCN of VIP-Cre mice showed that about half of VIP immunopositive neurons in the SCN were labeled by the reporter and that, of neurons expressing the reporter, about 95% were VIP-immunopositive (Fig. S4 C and D). The VIP-staining positive neurons in the cortex were mainly restricted in layer V (Fig. S4E).
Fig. 3.
In vivo and ex vivo recordings of circadian clock gene expression in the SCN. (A and B) The design of the Per2 (A) and Cry1 (B) in vivo fluorescent transcription reporter (Top), and fluorescence expression of Per2 (A) and Cry1 (B) reporter in the SCN VIP neurons in a series of rostral-to-caudal brain slices (Bottom). (Scale bars: 100 µm.) (C) In vivo recording Per2 (Top) and Cry1 (Bottom) transcription in SCN VIP neurons of VIP-Cre mice. The white and gray areas indicate the lights-on and lights-off periods, respectively. Raw data are plotted at 10-min intervals. (D) Luminescence recording of ex vivo SCN slices. The light gray and dark gray areas indicate the relative lights-on and lights-off periods, respectively, of the light cycle before dissection. Raw data are plotted at 10-min intervals. (E) Circadian peak phase of Per2 and Cry1. (F) Circadian periods of Per2 and Cry1. *P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant; mean ± SD, one-way ANOVA.
To compare our fluorescence reporters with the most commonly used bioluminescence reporters, dVenus of the fluorescence reporters were changed by the standard destabilized versions of firefly luciferase (Fig. S4 F and G, Left). One month after infusing the bioluminescence reporters rAAVs into the SCN of VIP-Cre mice (henceforth referred to as VIP-CreAAV-Per2-dLUC, VIP-CreAAV-Cry1-dLUC), ex vivo SCN slice cultures were prepared and imaged. The results showed that the bioluminescence reporters give an expression of luciferase in the SCN ventral part (Fig. S4 F and G, Right). SCN slice culture imaging showed that both the fluorescence and bioluminescence reporters exhibited approximately 24-h rhythms (Fig. S4H and Movies S1 and S2) (PER2::LUC mice SCN slice as a control).
We next in vivo recorded the rhythm of VIP-CreAAV-Per2-dVenus and VIP-CreAAV-Cry1-dVenus mice for 5 d in the LD condition and 5 d in the DD condition. The reporters showed robust rhythms in the SCN VIP neurons of VIP-CreAAV-Per2-dVenus and VIP-CreAAV-Cry1-dVenus mice in both LD and DD conditions (Fig. 3C). Ex vivo luminescence analysis of VIP-CreAAV-Per2-dLUC and VIP-CreAAV-Cry1-dLUC showed that all these cultures exhibited robust circadian rhythms in SCN VIP neurons (Fig. 3D) (PER2::LUC mice SCN slice as a control). Single cell analysis of VIP-CreAAV-Per2-dVenus and VIP-CreAAV-Cry1-dVenus ex vivo SCN slice cultures showed that individual labeled VIP neurons exhibited a circadian rhythm (Fig. S4I).
The peak phases of Per2 and Cry1 transcription measured in ex vivo cultures using bioluminescence were advanced compared with the peaks observed in our fluorescence ex vivo and in vivo recording results (Fig. 3E and Table S2). This difference of apparent phase with bioluminescent and fluorescent reporters could be attributed to the following: First, the chromophore maturation time of Venus (∼1 h) is longer than the luciferase maturation time (23); second, the effective half-life of dVenus (∼2 h) is longer than dLUC (∼15 m). The circadian periods of Per2 and Cry1 transcription, as measured in the ex vivo cultures, were slightly longer than those measured for these genes in the in vivo recordings (Fig. 3F and Table S3).
Collectively, these results indicate that, although our reporters for in vivo recording introduce some delay in apparent circadian phase, they still accurately reflect the period of transcription rhythms of these genes in SCN VIP neurons. Combined with the VIP-Cre mouse line, the in vivo recording system can record Per2 and Cry1 transcription rhythm specifically in the VIP neurons of the SCN in both LD and DD conditions in freely moving mice.
In Vivo Monitoring of Per2 and Cry1 Transcription Rhythms in SCN VIP Neurons During Experimental Jet Lag.
Both jet lag and rotating shift work are known to misalign internal circadian clocks and disrupt their responses to environmental time cues, altering the transcription of circadian clock genes (2, 5). Repeated misalignment of the circadian clock greatly increases the risk of various human diseases, including sleep disorders and depression (24, 25).
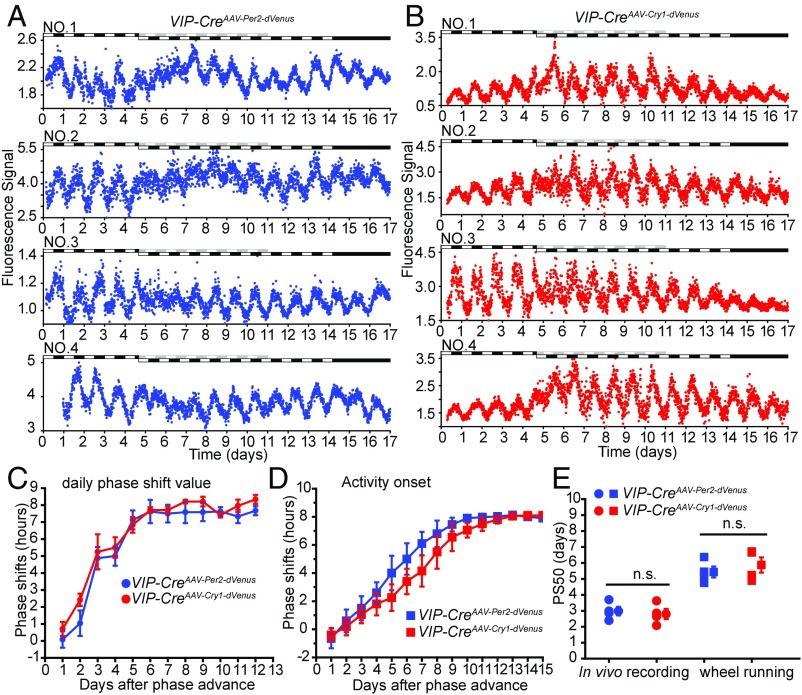
To study how circadian clock genes reentrained to the new environmental condition, we recorded Per2 and Cry1 transcription rhythm in SCN VIP neurons after a “phase-advance” treatment. VIP-CreAAV-Per2-dVenus and VIP-CreAAV-Cry1-dVenus mice were initially monitored for 4 d in the LD condition, and for 10 d after an 8-h phase advance of the light/dark cycle. Finally, they were monitored for two more days in the DD condition (Fig. 4 A and B). We found that the 8-h phase advance of the light/dark cycle temporarily disrupted the robustness of the Per2 and Cry1 rhythms in the SCN for 1 to 3 d.
Fig. 4.
In vivo monitoring of circadian clock genes reentrainment in the SCN. (A) Per2 transcription profiles in SCN VIP neurons. The light condition is indicated at the top of the figure: open bars indicate lights on, and closed bars indicate lights off. Raw data are plotted at 10-min intervals. (B) Cry1 transcription profiles in SCN VIP neurons. The light condition is indicated at the top of the figure: open bars indicate lights on, and closed bars indicate lights off. Raw data are plotted at 10-min intervals. (C) Daily phase shift value analysis of the reentrainment of Per2 and Cry1 transcription in SCN VIP neurons. (D) Daily phase shift of the wheel running activity onset time-based analysis of reentrainment of VIP-CreAAV-Per2-dVenus and VIP-CreAAV-Cry1-dVenus mice. (E) Half-time of the daily phase shift value transitions [50% phase shift value (PS50 values)] of Per2 and Cry1 transcription in SCN VIP neurons and the wheel running activity onset time. n = 4 for each gene, mean ± SEM; n.s., not significant, Student’s t test.
To get each day’s phase shift value after the 8-h phase advance of the light/dark cycle, the data of each mouse were first normalized (Fig. S5 A and B, Left). Then, 3 d (days 1 to 4) of normalized data on the initial LD condition were averaged and used as a template. For each day after the 8-h phase advance, the normalized data were cross-correlated with the template (Fig. S5 C and E), and the phase shift value was obtained from the x coordinate of the peak of the cross-correlation curve (Fig. S5 D and F).
Phase transition kinetics analysis was then performed to evaluate the phase change of Per2 and Cry1 during the 8-h phase advance of the light/dark cycle (Fig. 4C). We found that both Per2 and Cry1 transcription rhythm phases advanced quickly at the first 3 d after the 8-h phase advance, Per2 and Cry1 rhythm gave about 5 h phase advance at the first 3 d after the 8-h phase advance, and then Per2 and Cry1 rhythm gave another 3 h phase advance at the following 3 d. Fifty percent phase shift value (PS50) analysis showed that two genes take about 6 d to fully adjust to the new circadian phase (Fig. 4E). After optical recording, mice were tested on running wheels (Fig. S5 A and B, Right). Activity onset analysis (Fig. 4D) confirmed that the fiber insertion did not disrupt mice locomotor rhythms, and animals showed a normal phase shift of locomotor activity rhythms after the 8-h phase advance of the light/dark cycle (Fig. 4E).
Collectively, these results indicate that this in vivo recording system can be used to monitor how the circadian clock gene changes in SCN VIP neurons during the phase shift. Combined the use of other Cre mouse lines and other light conditions, this in vivo recording system in theory can also be used to monitor how the circadian clock gene changes in other conditions, such as under long photoperiod-caused depression and during sleep deprivation.
Discussion
In circadian biology, light is the primary environmental stimulus for synchronizing rhythms in the SCN (2); this system using fluorescent reporters for in vivo recording allowed us to record the circadian clock rhythm in a light/dark cycle. Combining the system with other Cre mouse lines (e.g., AVP-Cre mouse or Nms-iCre), long-term monitoring of these specific neurons under different physiological and environmental conditions will give us more information about how the clock gene changes in these neurons.
We tested the in vivo recording system by monitoring real-time transcription of Cry1 in the SCN, SPZ, and CA1/2 in WT mice. Our in vivo recording in VIP-CreAAV-Per2-dVenus mice showed that—limited by the penetration depth of the excitation laser (∼15-µW laser powers) and by the intensity of emission fluorescence in brain tissue—the optical fiber we used can detect the fluorescence signals within a target zone of about 150 µm away from the end of the fiber (Fig. S6A). The fiber end for recording in the SPZ was at least 200 µm away from the dorsal part of the SCN (Fig. 2B) so it is unlikely that the SPZ signal the optic fiber recorded could have been from the dorsal part of the SCN. On the other hand, because we did not target the reporter to specific neurons in the SPZ or CA1/2, we cannot exclude a contribution to the fluorescence signal from neurons just outside those regions. In the current study, we demonstrate successful in vivo recording of circadian rhythms in various brain nuclei; studies using this technology will be able to use Cre mouse lines to target specific neurons of interest, as we did with the VIP-Cre mouse line.
Previous studies have used GCaMP, a genetically encoded fluorescent calcium indicator, to record circadian Ca2+ rhythms in mouse SCN slices ex vivo (26, 27). Using our in vivo recording system, we demonstrated use of GCaMP6m for long-term monitoring of Ca2+ levels in SCN VIP neurons (Fig. S6B).
There are several previous cross-sectional studies of circadian clock gene expression during phase advances. A time course of mRNA in situ hybridization (6) showed that circadian patterns of Per expression in mouse SCN react rapidly to a 6-h phase advance of the light cycle whereas rhythmic Cry1 expression advances more slowly, more similar to the gradual resetting of the activity/rest cycle. A time course of SCN laser microdissection and qPCR (7) showed that circadian patterns of Per1, Per2, Bmal1, and Dbp expression in mouse SCN shift similarly after an 8-h phase advance of the lighting schedule. In vivo bioluminescence recording in the SCN (14) showed that a phase-delaying light pulse could dissociate circadian rhythms of Per1 and Bmal1. In the present study, we found that both Per2 and Cry1 take about 6 d to fully adjust to the new circadian phase after an 8-h phase advance of the lighting schedule. Differences among these studies could be due to differences in which genes were studied, and particularly in which cells, or due to cross-sectional vs. longitudinal recording methods. For example, we specifically recorded VIP neurons, mostly in the ventral part of the SCN, which is known to shift more quickly than the dorsal part (5). Interestingly, in longitudinal bioluminescence recording of circadian clock gene expression after a phase-shifting light stimulus in Drosophila, a transient weakening of rhythms was observed in neurons of the fly circadian pacemaker network (28), similar to our present results in mice.
Several other fluorescent circadian reporter knock-in (29) or transgenic mice (30, 31) have been reported previously, and these can be used directly with our recording methodology to monitor circadian clock gene expression in the brain. Use of the in vivo system we developed to monitor various genes in various types of neurons longitudinally under physiological conditions in mice will definitely lead to important insights and deepen our understanding of entrainment (and other) mechanisms of the circadian clock.
There are also some limitations of this in vivo recording method. For example, like other invasive methods, our method requires implantation of a fiber optic probe in brain tissue, which can induce neuroinflammation (Fig. S6C). Notably, we demonstrated that laser illumination had minimal phototoxicity in our recordings; but continuous exposure to a high-power laser can induce an appreciable level of cell apoptosis (Fig. S1B).
It is important to reiterate that this technique is by no means limited to circadian rhythm-related research. By designing reporters to assess the expression of other genes, or by using transgenic mouse strains that carry fluorescence reporters of gene expression in specific brain regions or specific neurons, this in vivo recording system can be extended to monitor expression of many other genes in many other brain regions.
Materials and Methods
Animals.
Mice used in the present study were all of the C57BL/6 background. All procedures were conducted with the approval of the Institutional Animal Care and Use Committee (IACUC) of the National Institute of Biological Sciences, Beijing.
Stereotactic Surgery.
Details for virus injection and fiber implantation surgeries can be found in SI Materials and Methods.
In Vivo Recording System.
A homemade four-channel recording instrument was used to acquire fluorescence signals from the brains of freely moving mice. To record signals from four animals simultaneously, a 488-nm laser (OBIS 488LS; Coherent) was coupled to a 1 × 4 beam splitter that divided the light into four laser excitation outputs of equal power. The excitation laser light was reflected by a dichroic mirror (MD498; Thorlabs) and was then delivered the brain via an optical fiber with a multimode fiber cable (200-µm-diameter core, N.A. = 0.39, RJPSF2; Thorlabs). The emission fluorescence signals were collected via the same optical fiber and filtered with a band pass filter (MF525-39; Thorlabs) and eventually detected by a photomultiplier tube (R3896; Hamamatsu). The voltage signals were then low pass filtered (30 Hz) and recorded with customized software.
Data Processing and Statistical Analysis.
For in vivo recording, 15 s of the fluorescence signal was collected once every 10 min, with a sample read frequency of 100 Hz (1,500 sample points per reading); the average fluorescence for this 15-s period was used as the signal value for a given time point. Mice without signal (6 mice of the 79 surgery mice in this study) or that did not pass the histological examination (5 mice of the 79 surgery mice in this study) were excluded from further analysis. The circadian period and the phase of gene transcription intensity were determined based on the converged sine curve [] using the sine curve fit function of OriginPro 8 (analysis > fitting > nonlinear curve fitting > origin basic functions > sine >fit till converged; OriginLab). For the data in the 8-h phase advance experiment, each day’s data were normalized using the following equation: [], where x is the original value and x′ is the normalized value. Phase shift value was calculated using cross-correlation analysis in Matlab. PS50 values were calculated using a sigmoidal dose–response curve with variable slope: , as reported before (7). Student’s t test was used when two groups were compared. One-Way ANOVA was used when three or more groups were being compared.
Supplementary Material
Acknowledgments
We thank Drs. Ying Xu (Soochow University) for providing Cry1−/− and Cry2−/− mice; Z. Josh Huang (Cold Spring Harbor Laboratory) for VIP-Cre mice; and Hiroki Ueda (RIKEN/University of Tokyo) for the pcDNA3.1-P(Cry1)-intron336-CRY1-6HIS construct. We are also grateful to members in the E.E.Z. and C.Z. labs for stimulating discussions and technical assistance. This research was supported by the National Natural Science Foundation of China Grants (31500860 to C.Z. and 61475059 to X.L.), the Ministry of Science and Technology of China (973 Program Grant 2012CB837700 to E.E.Z. and C.Z.), and funding from the Beijing Municipal Government (to E.E.Z.). D.K.W. is supported by a US Veterans Affairs Merit Award BX001146, and E.E.Z. was supported by the Chinese “Recruitment Program of Global Youth Experts.”
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The generated AAV constructs reported in this paper have been deposited to Addgene (www.addgene.org).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717735115/-/DCSupplemental.
References
- 1.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: Cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18:164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoo SH, et al. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagano M, et al. An abrupt shift in the day/night cycle causes desynchrony in the mammalian circadian center. J Neurosci. 2003;23:6141–6151. doi: 10.1523/JNEUROSCI.23-14-06141.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy AB, Field MD, Maywood ES, Hastings MH. Differential resynchronisation of circadian clock gene expression within the suprachiasmatic nuclei of mice subjected to experimental jet lag. J Neurosci. 2002;22:7326–7330. doi: 10.1523/JNEUROSCI.22-17-07326.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi Y, et al. Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science. 2013;342:85–90. doi: 10.1126/science.1238599. [DOI] [PubMed] [Google Scholar]
- 8.Inouye ST, Kawamura H. Persistence of circadian rhythmicity in a mammalian hypothalamic “island” containing the suprachiasmatic nucleus. Proc Natl Acad Sci USA. 1979;76:5962–5966. doi: 10.1073/pnas.76.11.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamura W, et al. In vivo monitoring of circadian timing in freely moving mice. Curr Biol. 2008;18:381–385. doi: 10.1016/j.cub.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 10.Yamaguchi S, et al. View of a mouse clock gene ticking. Nature. 2001;409:684. doi: 10.1038/35055628. [DOI] [PubMed] [Google Scholar]
- 11.Ono D, Honma K, Honma S. Circadian and ultradian rhythms of clock gene expression in the suprachiasmatic nucleus of freely moving mice. Sci Rep. 2015;5:12310. doi: 10.1038/srep12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saini C, et al. Real-time recording of circadian liver gene expression in freely moving mice reveals the phase-setting behavior of hepatocyte clocks. Genes Dev. 2013;27:1526–1536. doi: 10.1101/gad.221374.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamada T, et al. In vivo imaging of clock gene expression in multiple tissues of freely moving mice. Nat Commun. 2016;7:11705. doi: 10.1038/ncomms11705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ono D, et al. Dissociation ofPer1andBmal1circadian rhythms in the suprachiasmatic nucleus in parallel with behavioral outputs. Proc Natl Acad Sci USA. 2017;114:E3699–E3708. doi: 10.1073/pnas.1613374114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 16.van der Horst GT, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 17.Ukai-Tadenuma M, et al. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell. 2011;144:268–281. doi: 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 18.Li X, et al. Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- 19.Nagoshi E, et al. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 20.Fustin JM, O’Neill JS, Hastings MH, Hazlerigg DG, Dardente H. Cry1 circadian phase in vitro: Wrapped up with an E-box. J Biol Rhythms. 2009;24:16–24. doi: 10.1177/0748730408329267. [DOI] [PubMed] [Google Scholar]
- 21.Harmar AJ, et al. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell. 2002;109:497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- 22.Taniguchi H, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iizuka R, Yamagishi-Shirasaki M, Funatsu T. Kinetic study of de novo chromophore maturation of fluorescent proteins. Anal Biochem. 2011;414:173–178. doi: 10.1016/j.ab.2011.03.036. [DOI] [PubMed] [Google Scholar]
- 24.Drake CL, Roehrs T, Richardson G, Walsh JK, Roth T. Shift work sleep disorder: Prevalence and consequences beyond that of symptomatic day workers. Sleep. 2004;27:1453–1462. doi: 10.1093/sleep/27.8.1453. [DOI] [PubMed] [Google Scholar]
- 25.Soria V, et al. Differential association of circadian genes with mood disorders: CRY1 and NPAS2 are associated with unipolar major depression and CLOCK and VIP with bipolar disorder. Neuropsychopharmacology. 2010;35:1279–1289. doi: 10.1038/npp.2009.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enoki R, et al. Synchronous circadian voltage rhythms with asynchronous calcium rhythms in the suprachiasmatic nucleus. Proc Natl Acad Sci USA. 2017;114:E2476–E2485. doi: 10.1073/pnas.1616815114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noguchi T, et al. Calcium circadian rhythmicity in the suprachiasmatic nucleus: Cell autonomy and network modulation. eNeuro. 2017;4:ENEURO.0160-17.2017. doi: 10.1523/ENEURO.0160-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts L, et al. Light evokes rapid circadian network oscillator desynchrony followed by gradual phase retuning of synchrony. Curr Biol. 2015;25:858–867. doi: 10.1016/j.cub.2015.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smyllie NJ, et al. Visualizing and quantifying intracellular behavior and abundance of the core circadian clock protein PERIOD2. Curr Biol. 2016;26:1880–1886. doi: 10.1016/j.cub.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng HY, et al. Segregation of expression of mPeriod gene homologs in neurons and glia: Possible divergent roles of mPeriod1 and mPeriod2 in the brain. Hum Mol Genet. 2009;18:3110–3124. doi: 10.1093/hmg/ddp252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LeSauter J, et al. A short half-life GFP mouse model for analysis of suprachiasmatic nucleus organization. Brain Res. 2003;964:279–287. doi: 10.1016/s0006-8993(02)04084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.