Significance
Approximately 20% of breast cancers have amplification of a cancer-causing signaling molecule known as human epidermal growth factor receptor 2 (HER2). Decreased mRNA expression of the autophagy gene, beclin 1/BECN1, increases the risk of HER2-positive breast cancer. However, the role of Beclin 1-dependent autophagy in regulating HER2-mediated tumorigenesis is unknown. Here, we show that a mutation in Becn1 that increases basal autophagy prevents HER2-mediated tumorigenesis in mice and prevents HER2-mediated inhibition of autophagy in cultured cells. Furthermore, treatment with a cell-penetrating, autophagy-inducing peptide derived from Beclin 1 inhibits growth of HER2-positive human breast tumor xenografts in mice as efficiently as a clinically used agent that inhibits HER2 receptor tyrosine kinase activity. These findings demonstrate that genetic and pharmacological activation of autophagy inhibits HER2-mediated breast tumorigenesis.
Keywords: autophagy, Beclin 1, HER2, breast cancer
Abstract
Allelic loss of the autophagy gene, beclin 1/BECN1, increases the risk of patients developing aggressive, including human epidermal growth factor receptor 2 (HER2)-positive, breast cancers; however, it is not known whether autophagy induction may be beneficial in preventing HER2-positive breast tumor growth. We explored the regulation of autophagy in breast cancer cells by HER2 in vitro and the effects of genetic and pharmacological strategies to increase autophagy on HER2-driven breast cancer growth in vivo. Our findings demonstrate that HER2 interacts with Beclin 1 in breast cancer cells and inhibits autophagy. Mice with increased basal autophagy due to a genetically engineered mutation in Becn1 are protected from HER2-driven mammary tumorigenesis, and HER2 fails to inhibit autophagy in primary cells derived from these mice. Moreover, treatment of mice with HER2-positive human breast cancer xenografts with the Tat-Beclin 1 autophagy-inducing peptide inhibits tumor growth as effectively as a clinically used HER2 tyrosine kinase inhibitor (TKI). This inhibition of tumor growth is associated with a robust induction of autophagy, a disruption of HER2/Beclin 1 binding, and a transcriptional signature in the tumors distinct from that observed with HER2 TKI treatment. Taken together, these findings indicate that the HER2-mediated inhibition of Beclin 1 and autophagy likely contributes to HER2-mediated tumorigenesis and that strategies to block HER2/Beclin 1 binding and/or increase autophagy may represent a new therapeutic approach for HER2-positive breast cancers.
The amplification of human epidermal growth factor receptor 2 (HER2), an oncogenic receptor tyrosine kinase (RTK), is a driver mutation in ∼20% of patients with breast cancer and is associated with a worse prognosis (1). In addition, activating mutations in the HER2 tyrosine kinase domain are found in breast and other cancers (2, 3). Targeted therapy with anti-HER2 agents, either monoclonal antibodies against HER2 (e.g., trastuzumab, pertuzumab) or a dual EGF receptor (EGFR)/HER2 tyrosine kinase inhibitor (TKI) (e.g., lapatinib), significantly improves outcomes in patients with HER2-positive breast cancer (reviewed in refs. 4, 5). However, resistance to targeted HER2 therapies or therapy-limiting side effects occur in many patients. Thus, there is a need to identify and target new cellular pathways that regulate HER2-mediated tumorigenesis.
Several clues suggest a possible link between decreased autophagy and the development of HER2-positive breast cancer, its clinical course, and/or its resistance to targeted HER2 therapies. The essential autophagy gene, beclin 1/BECN1, is a haploinsufficient tumor suppressor monoallelically deleted in ∼30% of human breast cancers (6, 7). In mice, its allelic loss results in increased mammary tumorigenesis (8, 9), and its enforced expression reduces the growth of human breast cancer xenografts (10, 11). There is an association between loss of beclin 1/BECN1 and HER2 amplification in breast cancer (12). In large breast cancer databases, decreased BECN1 mRNA expression is strongly associated with increased risk of HER2-positive breast cancer and worse disease-free survival (7). Moreover, clinical resistance to targeted HER2 therapies may occur as a result of mutations in signaling pathways downstream of the HER2 receptor that suppress autophagy, such as PI3K activating mutations or PTEN loss [which result in Akt and mechanistic target of rapamycin (mTOR) activation] (13, 14), and in preclinical models, tumor sensitivity to anti-HER2 agents can be restored by PI3K and/or mTOR inhibitors (15). In vitro, down-regulation of the essential autophagy protein ATG9 contributes to trastuzumab resistance in HER2-positive breast cancer cells (16).
Despite these lines of evidence, it remains unknown whether autophagy protects against HER2-mediated tumorigenesis. We previously reported a link between activating mutations in another oncogenic RTK HER family member, EGFR; Beclin 1 tyrosine phosphorylation and autophagy inhibition; the growth of non-small cell lung cancer (NSCLC) xenografts; and the chemoresistance of such tumors to targeted EGFR TKI therapy (erlotinib) (17). Moreover, we showed that Akt-mediated Beclin 1 phosphorylation and autophagy suppression contribute to the transforming and tumorigenic activity of Akt (18). In addition, HER2 has been reported to interact with Beclin 1 in cultured breast cancer cells, while lapatinib diminishes this interaction and induces autophagy (19). However, the in vivo significance of HER2/Beclin 1 interaction and of altered autophagy in HER2-driven tumorigenesis is unknown.
Here, we show that endogenous HER2 interacts with Beclin 1 in breast cancer cells and inhibits autophagy. A knock-in mutation in Beclin 1 (Becn1F121A) (20) that results in increased basal autophagy decreases mammary tumorigenesis in mice with mammary-specific expression of HER2 and blocks HER2-mediated suppression of autophagy. Moreover, Tat-Beclin 1, an autophagy-inducing peptide (21, 22), reduces Beclin 1/HER2 binding and induces autophagy in HER2-positive breast tumor xenografts, and is as effective as a clinically used HER2 TKI in preventing in vivo tumor growth. Together, these findings point to a fundamental role for Beclin 1 and/or the autophagy pathway in suppressing HER2-mediated tumorigenesis.
Results
Beclin 1/BECN1 Loss Is Associated with HER2 Amplification/Overexpression in Human Patients with Breast Cancer.
Although beclin 1/BECN1 and ERBB2/HER2 both map to chromosome 17q, they are located ∼3 million bp apart (Fig. S1A), allowing translocations within this region to result in a discordancy between BECN1 loss and HER2 amplification/overexpression. Utilizing the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) breast cancer database, we confirmed that ∼10% (217 of 2,173) of patients with breast cancer have net amplification of HER2 and net loss of BECN1 (Fig. S1B) and that ∼68% (146 of 215) of patients with HER2 overexpression have net loss of BECN1 (Fig. S1 C and D). The long-term survival among patients with HER2 overexpression is significantly worse in those with net BECN1 loss as compared to those with BECN1 diploid copy number (Fig. S1E). Thus, BECN1 allelic loss is common and associated with a worse prognosis in patients with HER2-overexpressing breast tumors.
HER2 Interacts with Beclin 1 and Inhibits Autophagy in a Manner That Requires Its Kinase Activity.
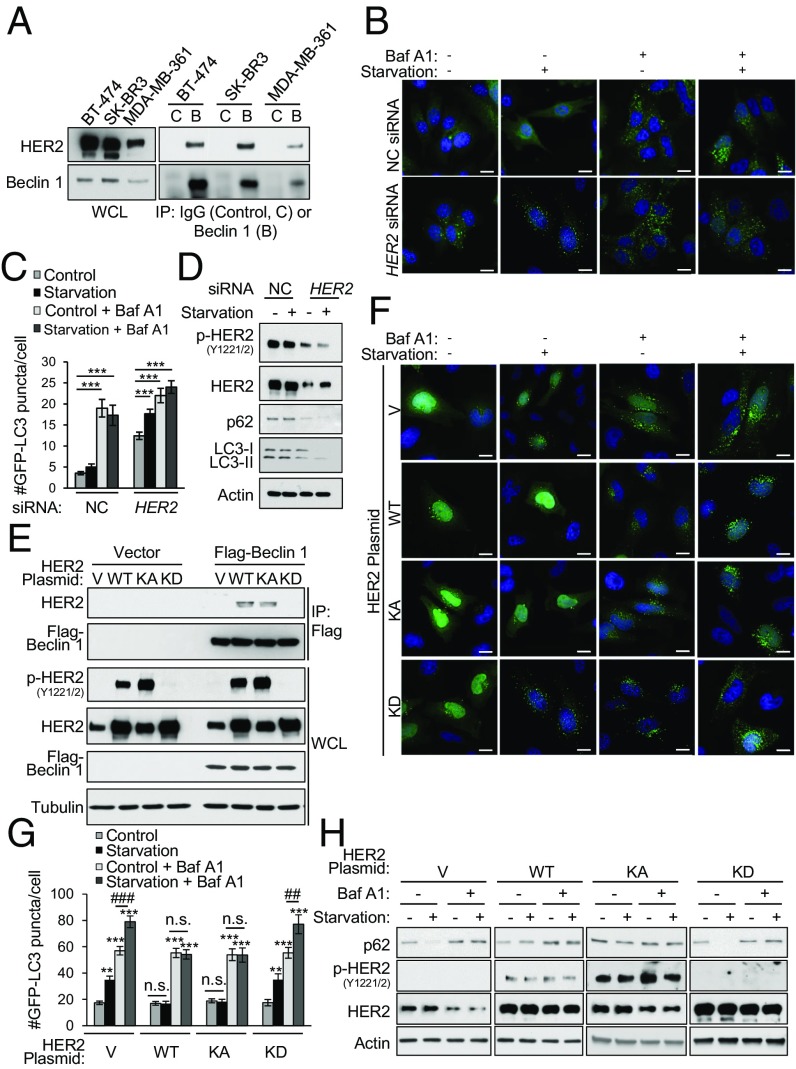
Given the evidence of an association between HER2 and Beclin 1 in breast cancer pathogenesis, we evaluated whether HER2 interacts with Beclin 1 and regulates autophagy in human HER2-positive breast cancer cell lines. In three HER2-positive breast cancer cell lines (BT-474, SK-BR3, and MDA-MB-361 cells), endogenous HER2 coimmunoprecipitated with endogenous Beclin 1 (Fig. 1A). To determine whether HER2 negatively regulates autophagy, we examined the effects of siRNA-mediated knockdown of HER2 on basal and starvation-induced autophagy in BT-474 cells, a cell line that is sensitive to lapatinib and able to form tumors in immunodeficient mice (23). Starvation, a potent physiological inducer of autophagy, failed to increase autophagy in cells treated with control siRNA, as measured by numbers of GFP-LC3 puncta (a marker of autophagosomes) (Fig. 1 B and C) and Western blot analysis of levels of p62 (a substrate of the autophagy pathway) and/or conversion of the LC3-I to the lipidated, autophagosome-associated form, LC3-II, or total LC3 degradation (Fig. 1D). However, knockdown of HER2 resulted in a significant increase in basal autophagy and rescue of starvation-induced autophagy measured by these same assays. The increased numbers of autophagosomes represented an increase in autophagic flux, rather than a block in autophagolysosomal maturation, as numbers of GFP-LC3 puncta increased further upon treatment with the lysosomal inhibitor bafilomycin A1 (Baf A1) (Fig. 1 B and C). Thus, endogenous HER2 interacts with Beclin 1 and negatively regulates autophagy in human breast cancer cells.
Fig. 1.
HER2 interacts with Beclin 1 and reduces starvation-induced autophagy. (A) Coimmunoprecipitation of endogenous HER2 with endogenous Beclin 1 in indicated HER2-positive breast cancer cell lines. B, Beclin 1 IgG; C, Control IgG; IP, immunoprecipitation; WCL, whole-cell lysates. (B and C) HER2 knockdown effects on autophagy in BT-474 cells cotransfected with GFP-LC3 and a nontargeting control (NC) or HER2 siRNA and grown in either normal media (starvation, −) or starvation conditions (starvation, +; HBSS, 3 h) in the presence or absence of 100 nM Baf A1. Representative images (B) and quantification (C) of GFP-LC3 puncta are shown. (D) Western blot analysis of autophagy (p62 and LC3) in BT-474 cells treated with NC or HER2 siRNA and grown in normal media or starvation conditions. (E) Coimmunoprecipitation of indicated HER2 constructs with Flag-Beclin 1 in transiently transfected HeLa cells. V, empty vector. (F and G) HER2 effects on autophagy in HeLa cells cotransfected with GFP-LC3 and the indicated HER2 expression plasmid and grown in normal media or starvation conditions (HBSS, 3 h) ± 50 nM Baf A1. Representative images (F) and quantification (G) of GFP-LC3 puncta. (H) Western blot analysis of autophagy (p62) in HeLa cells transfected with the indicated HER2 expression plasmid, and grown in normal media or starvation conditions for 3 h ± 100 nM Baf A1. Bars are mean ± SEM of triplicate samples (100–150 cells per condition). Similar results were observed in three independent experiments. n.s., not significant. **P < 0.01 and ***P < 0.001 vs. normal media control, one-way ANOVA; ##P < 0.01 and ###P < 0.001 for comparison of starvation vs. normal media in the presence of Baf A1, one-way ANOVA (see also Fig. S2). (Scale bars, 15 μm.)
To determine whether the kinase activity of HER2 is required for interaction with Beclin 1 and suppression of autophagy, we used HeLa cells, a cell line with an intact autophagy pathway (21) and low basal levels of HER2 expression (Fig. 1E). Exogenously expressed wild-type (WT) HER2 and a kinase-active (KA; A775_G776insYVMA) HER2 mutant, but not a kinase-dead (KD; D845A) HER2 mutant, were autophosphorylated at Y1221/2 and coimmunoprecipitated with Flag-Beclin 1 (Fig. 1E). The effects of WT HER2, KA HER2, and KD HER2 on starvation-induced autophagic flux correlated with their ability to interact with Beclin 1. WT HER2 and KA HER2, but not KD HER2, blocked starvation-induced increases in GFP-LC3 puncta (Fig. 1 F and G) and degradation of p62 (Fig. 1H). Thus, the kinase activity of HER2 is required for its interaction with Beclin 1 and inhibition of autophagy.
While both WT HER2 and KA HER2, but not KD HER2, interact with Beclin 1 and block starvation-induced autophagic flux, they do so by different mechanisms. Similar to EGFR (17), KA HER2 leads to Beclin 1 tyrosine phosphorylation (Fig. S2A); blocks autophagy in an mTOR-independent manner (Fig. S2 B and C); and fails to inhibit autophagy in cells that express a Beclin 1 mutant lacking the tyrosine phosphorylation sites Y229, Y233, and Y352 (Beclin 1 Y229F/Y233F/W353F) (17) (Fig. S2 D and E). In contrast, WT HER2 does not lead to Beclin 1 tyrosine phosphorylation (Fig. S2A), inhibits autophagy in an mTOR-dependent manner (Fig. S2 B and C), and retains the ability to inhibit autophagy in cells that express the Beclin 1 Y229F/Y233F/Y353F mutant (Fig. S2 D and E). Thus, active HER2 behaves similar to active EGFR and inhibits Beclin 1 autophagy by tyrosine phosphorylation in an mTOR-independent manner. The mechanism by which WT HER2 inhibits Beclin 1 autophagy function in an mTOR-dependent fashion remains unknown.
Mice with a Knock-In Mutation in Becn1 That Increases Mammary Epithelial Cell Autophagy Are Protected Against HER2-Mediated Tumorigenesis.
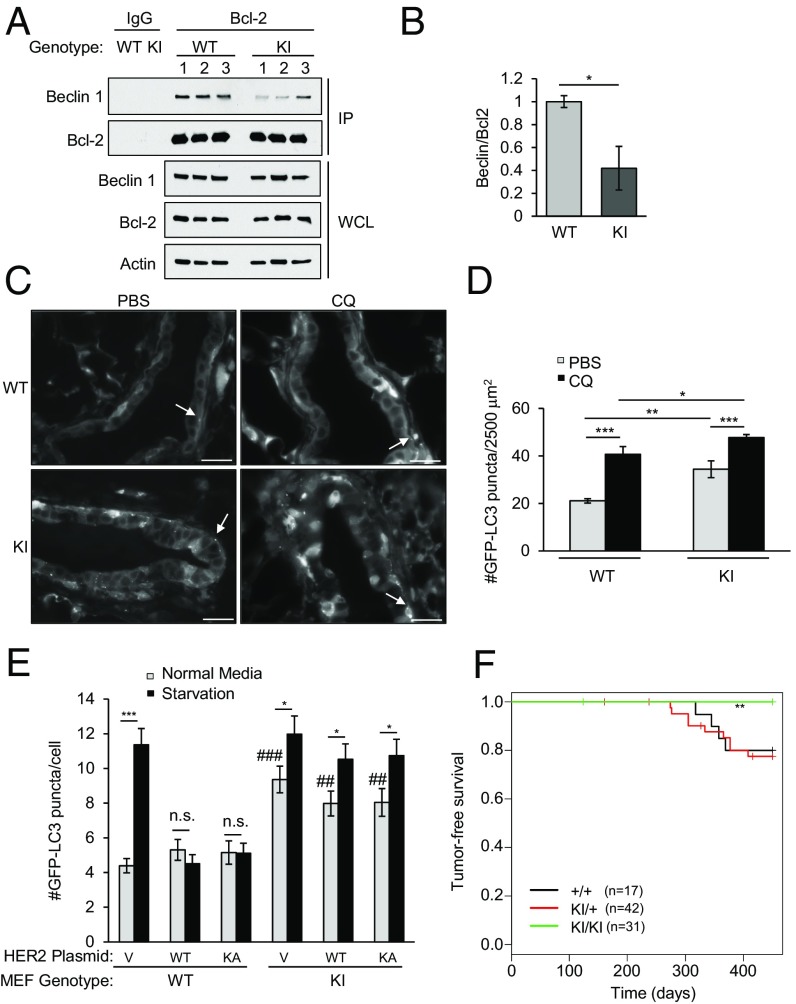
To study the significance of HER2-mediated suppression of autophagy in mammary tumorigenesis in vivo, we utilized a genetically engineered mouse model with increased systemic autophagy. We recently reported that mice (Becn1F121A) with a homozygous knock-in mutation that results in an F121A substitution in Beclin 1 have decreased interaction of Beclin 1 with Bcl-2 and increased autophagy in tissues such as skeletal muscle and brain (20). Likewise, in mammary glands, the coimmunoprecipitation of Beclin 1 with Bcl-2 is decreased in these mice despite similar levels of Beclin 1 expression (Fig. 2 A and B). Moreover, Becn1F121A mice crossed with transgenic mice that express GFP-LC3 (Becn1F121A:GFP-LC3) had increased numbers of GFP-LC3 puncta in their mammary epithelial cells compared with Becn1WT:GFP-LC3 mice in both control conditions and after treatment with chloroquine, a drug that blocks autophagolysosomal maturation (Fig. 2 C and D). Thus, Becn1F121A mice have increased basal levels of mammary epithelial cell autophagic flux and provide a good model for studying the effects of increased autophagy on HER2-mediated tumorigenesis.
Fig. 2.
Becn1F121A knock-in mice have decreased HER2-mediated tumorigenesis. (A) Coimmunoprecipitation of Beclin 1 with Bcl-2 in mammary glands of 2-mo-old female Becn1WT (WT) and homozygous Becn1F121A (KI) mice (three mice per genotype). IP, immunoprecipitation; WCL, whole-cell lysates. (B) Densimetric quantification of the ratio of Beclin 1 compared with Bcl-2 in each sample in A normalized to a 1.0 value in the mammary glands of WT mice. Bars are mean values ± SEM of mammary glands from three mice per group. *P < 0.05; ANOVA. Representative images (C) and quantification of GFP-LC3 puncta (D) are shown in mammary epithelial ducts of Becn1WT:GFP-LC3 (WT) and Becn1F121A:GFP-LC3 (KI) mice 6 h after treatment with PBS or 50 mg⋅kg−1 chloroquine (CQ). White arrows denote representative GFP-LC3 puncta in C. *P < 0.05; ***P < 0.001; ANOVA. (E) GFP-LC3 puncta (autophagosomes) in MEFs of the indicated genotype transfected with the indicated HER2 expression plasmids and GFP-LC3 and grown in normal media or subjected to starvation (HBSS, 3 h). Bars are mean ± SEM of triplicate samples (100–150 cells per condition). Similar results were observed in three independent experiments. n.s., not significant. *P < 0.01 and ***P < 0.001 vs. normal media control; ##P < 0.01 and ###P < 0.001 for comparison of value in normal media in the indicated condition in KI MEFs vs. WT MEFs. (F) Kaplan–Meier curves for tumor-free survival of Becn1WT (+/+) Becn1WT/F121A (KI/+), and Becn1F121A (KI/KI) mice crossed with MMTV-HER2 mice. **P < 0.01; log-rank test (see also Fig. S3). (Scale bars, 100 μm.)
To determine whether the Becn1F121A mutation can block HER2-mediated inhibition of autophagy, we cotransfected primary murine embryonic fibroblasts (MEFs) derived from Becn1WT or Becn1F121A homozygous mutant mice with GFP-LC3 and either WT HER2 or KA HER2. No differences in levels of Beclin 1 or HER2 expression were observed between MEFs of the two genotypes (Fig. S3). However, Becn1F121A MEFs transfected with empty vector and WT HER2 or KA HER2 had significantly more GFP-LC3 puncta in basal conditions than did Becn1WT MEFs (Fig. 2E). Moreover, unlike Becn1WT MEFs, Becn1F121A MEFs had a significant increase in numbers of GFP-LC3 puncta in response to starvation in the presence of WT HER2- or KA HER2-enforced expression (Fig. 2E). Thus, the Becn1F121A mutation bypasses the inhibitory effects of HER2 on autophagy.
We next examined the effects of bypassing HER2 suppression of autophagy on tumorigenesis in vivo by crossing mice that transgenically express HER2 under the control of a mouse mammary tumor virus (MMTV) mammary-specific promoter [FVB/N-Tg (MMTVneu)] with Becn1F121A mice. Approximately 25% of HER2-expressing Becn1WT mice (+/+) or mice with a single knock-in allele of Becn1F121A (KI/+) developed mammary tumors by 450 d of life. In contrast, no HER2-expressing mice with a homozygous knock-in mutation of Becn1F121A (KI/KI) developed tumors within this observation period (Fig. 2F). The lower incidence of mammary tumors in our study compared with some previous reports (24) is likely due to the mixed FVB:B6 background compared with the tumor incidence observed in a pure FVB background. However, the difference in mammary tumorigenesis in WT or KI/+ versus KI/KI mice cannot be attributed to the background strain, as all mice in this study were littermate offspring derived from MMTV-HER2 transgenic/WT (FVB); Becn1F121A/WT (C57/B6) × Becn1F121A/WT (C57/B6) intercrosses. Thus, a genetic mutation that increases basal autophagy in mammary epithelial cells blocks HER2-mediated tumorigenesis in vivo.
An HER2 TKI and an Autophagy-Inducing Peptide, Tat-Beclin 1, Disrupt HER2/Beclin 1 Binding and Induce Autophagy in HER2-Positive Breast Cancer Cells.
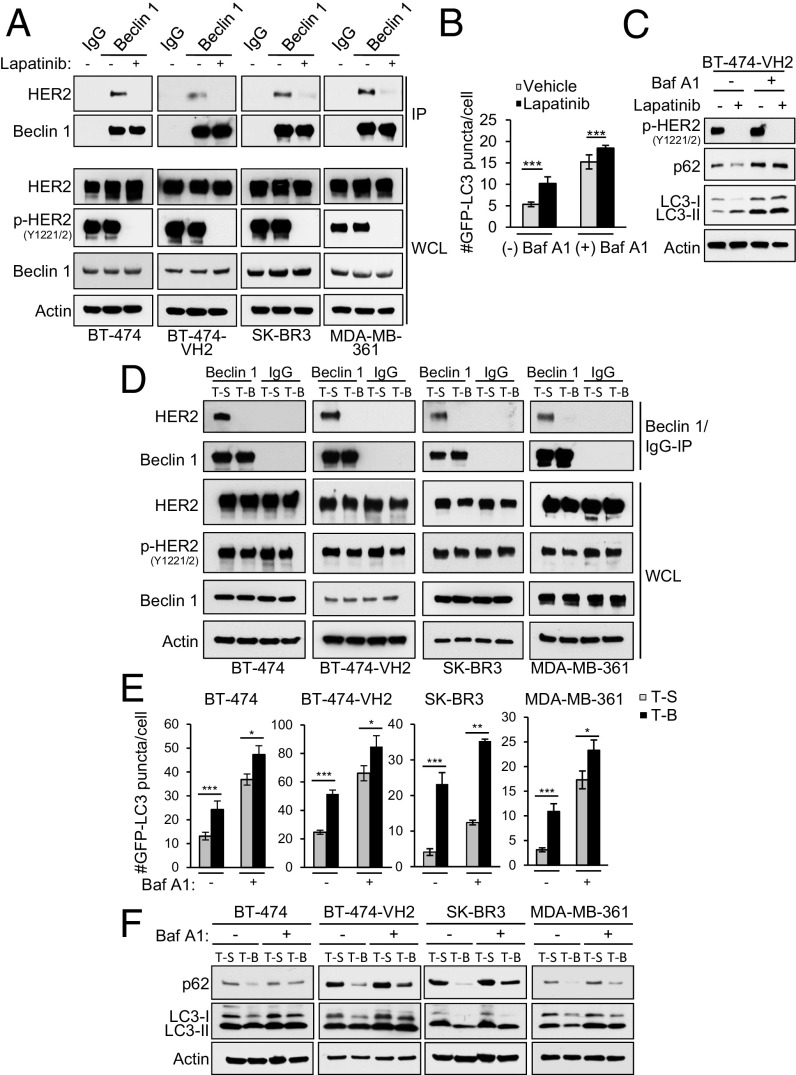
The dual EGFR/HER2 TKI, lapatinib, has been shown to disrupt HER2/Beclin 1 binding and increase autophagy in breast cancer cells (19). We confirmed that lapatinib decreased HER2 interaction with Beclin 1 in BT-474 cells and the more tumorigenic subclone, BT-474-VH2 (25) cells; in SK-BR3 cells; and in MDA-MB-361 cells in parallel with blocking HER2 phosphorylation (Fig. 3A). As predicted, lapatinib increased autophagic flux in BT-474-VH2 cells, as measured by numbers of GFP-LC3 puncta, p62 degradation, and LC3-I–to–LC3-II conversion in the absence or presence of Baf A1 (Fig. 3 B and C). Lapatinib also increased GFP-LC3 puncta (autophagosomal numbers) in BT-474, SK-BR3, and MDA-MB-361 cells (Fig. S4).
Fig. 3.
Lapatinib and Tat-Beclin 1 disrupt HER2/Beclin 1 binding and induce autophagy in breast cancer cells. (A) Effects of lapatinib on HER2/Beclin 1 coimmunoprecipitation in HER2-positive breast cancer cell lines treated with control (DMSO) or lapatinib (1 μM, 3 h). IP, immunoprecipitation; WCL, whole-cell lysates. (B) Quantification of GFP-LC3 puncta (autophagosomes) in BT-474-VH2 cells in the ±1 μM lapatinib and ±100 nM Baf A1. (C) Western blot analysis of p62 degradation in cells treated as in A. (D) Effects of Tat-Beclin 1 (T-B) or control Tat-Scrambled (T-S) peptides (5 μM, 2 h) on HER2/Beclin 1 coimmunoprecipitation in HER2-positive breast cancer cell lines. (E) Quantification of GFP-LC3 puncta (autophagosomes) in the indicated HER2-positive breast cancer cell line after treatment with T-B or T-S (10 μM, 1 h) ± 100 nM Baf A1. (F) Western blot analysis of p62 and LC3I/II levels in the indicated HER2-positive cancer cell line treated as in E. Bars are mean ± SEM of triplicate samples (100–150 cells per condition). Similar results were observed in three independent experiments. *P < 0.05, **< 0.01, ***P < 0.001 for indicated comparison; ANOVA.
To examine the effects of disruption of HER2/Beclin 1 binding on autophagy and HER2-mediated tumorigenesis independent of effects on HER2 dephosphoryation, which has multiple downstream effects on cellular oncogenic signaling pathways, we developed an alternative approach that involved the use of a cell-penetrating, autophagy-inducing peptide, Tat-Beclin 1. Tat-Beclin 1 contains an 11-amino acid cationic-rich, cell-penetrating sequence from HIV Tat, a diglycine linker, and 11 amino acids from the evolutionarily conserved region of Beclin 1 (22). This peptide (or a version containing 18 amino acids of Beclin 1) (21) induces autophagy in vitro and in vivo in a manner that requires Beclin 1 and downstream autophagy machinery (21); is well-tolerated in vivo (21); and has been shown to have beneficial effects in rodent models of infection (21, 26), cardiac disease (27, 28), bone disease (29), and axonal injury (30).
We found that Tat-Beclin 1, but not a control peptide, Tat-Scrambled (in which the 11 amino acids of the Beclin 1 sequence are scrambled), disrupted HER2/Beclin 1 interaction as effectively as lapatinib in BT-474 cells, BT-474-VH2 cells, SK-BR3 cells, and MDA-MB-361 cells (Fig. 3D). However, in contrast to lapatinib, this disruption of binding occurred in the presence of persistent HER2 tyrosine phosphorylation. Tat-Beclin 1 induced a strong autophagic flux response in all four HER-positive breast cancer cells, as measured by the same assays used to assess lapatinib-induced autophagy, including quantitation of numbers of GFP-LC3 puncta (Fig. 3E), degradation of p62, and LC3-II conversion and/or decreased total levels of LC3 (Fig. 3F) in the absence or presence of Baf A1.
An Autophagy-Inducing Peptide, Tat-Beclin 1, Prevents the Growth of HER2-Positive Xenografts in Mice.
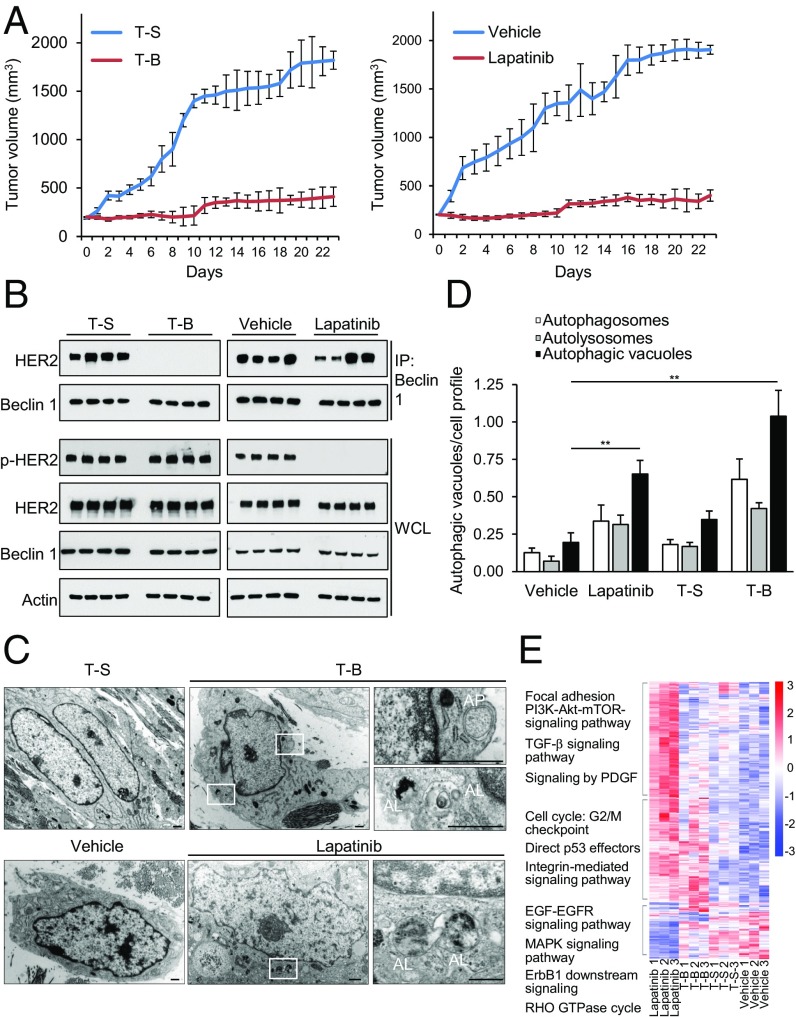
To evaluate the efficacy of Tat-Beclin 1 on HER2-positive breast cancer in a preclinical model, we studied the effects of Tat-Beclin 1 on the growth of HER2-positive BT-474-VH2 xenografts in nude mice (Fig. 4). Mice were randomized to receive daily i.p. administration of vehicle, lapatinib, Tat-Beclin 1, or Tat-Scrambled when their tumor volume reached 200 μm3. Strikingly, Tat-Beclin 1 decreased the rate of tumor progression as effectively as lapatinib, a drug that is clinically approved for the treatment of HER2-positive breast cancers (Fig. 4A).
Fig. 4.
Tat-Beclin 1 decreases HER2 breast cancer xenograft growth. (A) Effect of Tat-Beclin 1 (T-B) versus Tat-Scrambled (T-S) control peptide (Left) and vehicle versus lapatinib (Right) on growth of BT-474-VH2 xenografts in nude mice. P = 5.3E-94 (Left) and P = 1.0E-53 (Right), linear mixed-effect model. The difference between Tat-Beclin 1 and lapatinib was not significant (P = 0.236). Shown are results from one cohort of 40 mice randomized to four treatment groups. Similar results were observed in three independent cohorts. (B) Effect of lapatinib and T-B on Beclin 1/HER2 interaction and HER2 phosphorylation in BT-474-VH2 xenografts. Samples were harvested after 3 d of daily treatment with the indicated agent. IP, immunoprecipitation; WCL, whole-cell lysates. (C) Representative images of ultrastructural analyses of BT-474-VH2 xenografts with the indicated treatment. Representative autophagosomes (AP) and representative autolysosomes (AL) would be scored as positive in D. (D) Quantification of autophagic structures in xenografts with the indicated treatment. Bars are mean values ± SEM of three xenografts per treatment group. At least 50 cell profiles were counted per xenograft sample. **P = 0.01; t test. (E) Heat map profile of the expression data derived from RNAseq of BT-474-VH2 xenografts comparing lapatinib-, Tat-Beclin 1 (T-B)–, Tat-Scrambled (T-S)–, and vehicle-treated groups (n = 3 animals per group). Gene clusters were identified and annotated based on statistically (P < 0.01) significant pathways observed within each cluster (see also Fig. S5 and Datasets S1–S3).
To gain insight into mechanisms of the antitumorigenic effects of Tat-Beclin 1, we performed additional analyses of xenografts at 3 d after initiation of vehicle, lapatinib, Tat-Beclin 1, or Tat-Scrambled treatment. There were no increases in TUNEL-positive cells in either the lapatinib or Tat-Beclin 1 treatment groups (Fig. S5A). A mild decrease in Ki-67 staining observed in the lapatinib versus vehicle groups was not observed in the Tat-Beclin 1 versus Tat-Scrambled groups (Fig. S5B). Notably, at this time point, there was an almost complete block of HER2 coimmunoprecipitation with Beclin 1 in tumor xenografts from mice treated with Tat-Beclin 1 (Fig. 4B). Counter to expectations based on experiments in cultured cells (Fig. 3A), lapatinib did not detectably reduce HER2/Beclin 1 binding in BT-474-VH2 xenografts (Fig. 4B). The dephosphorylation of HER2 in xenografts from lapatinib-treated mice and the block in HER2/Beclin 1 interaction in xenografts from Tat-Beclin 1–treated mice were both associated with significant induction of autophagy, as measured by quantitative analyses of autophagosomes and autolysosomes at the ultrastructural level (Fig. 4 C and D). Finally, RNA sequencing (RNAseq) analysis of the transcriptome in xenografts revealed a unique, but overlapping, transcriptional signature of lapatinib treatment compared with Tat-Beclin 1 treatment (Fig. 4E). Specifically, pathways directly relevant to HER2 signaling, such as EGF-EGFR signaling, Erb1 downstream signaling, and PI3K-Akt-mTOR signaling, were differentially regulated in lapatinib- versus Tat-Beclin 1–treated xenografts, whereas pathways involving G2/M checkpoint, direct p53 effectors, and integrin-mediated signaling were enriched in both lapatinib- and Tat-Beclin 1–treated xenografts.
Taken together, our xenograft studies show that either blockade of HER2 phosphorylation or blockade of HER2/Beclin 1 interaction is sufficient to induce autophagy in HER2-positive human breast cancer xenografts and provide strong evidence that HER2-positive breast tumor growth suppression in vivo is associated with autophagy induction.
Discussion
The role of autophagy in progression of RTK-driven tumorigenesis and response to RTK inhibitor therapy has been debated (31, 32). In vitro studies have suggested that pharmacological inhibition or knockdown of autophagy genes results in reduced tumor cell viability, leading to the speculation that autophagy may be protumorigenic in the setting of RTK-driven tumorigenesis. Herein, we describe two approaches to assess the role of autophagy in HER2-positive breast tumorigenesis in vivo. First, we demonstrate that an activating mutation in Beclin 1 that increases basal autophagy (even in the setting of HER2 overexpression) blocks tumor formation in mice with mammary-specific expression of HER2. Second, we demonstrate that a specific autophagy inducer, Tat-Beclin 1, which induces autophagy independently of blocking HER2 phosphorylation, is as effective as a clinically used HER2 kinase inhibitor in impeding HER2-positive breast tumor growth in vivo.
Our studies suggest that enhanced autophagy not only prevents HER2-mediated tumorigenesis but also prevents the progression of established HER2-positive breast cancer xenografts. However, we cannot definitively exclude other antitumor effects of the Becn1 F121A mutation and the Tat-Beclin 1 peptide besides autophagy induction in preventing the initiation or progression, respectively, of HER2-positive breast tumors. Nonetheless, the decreased incidence of HER2-positive tumors in mice with increased autophagy due to an activating mutation in Beclin 1 is consistent with a tumor-suppressive role of Beclin 1 and autophagy in breast cancer (8–11, 33, 34). Importantly, our data with Tat-Beclin 1, which induces autophagy more specifically than RTK inhibitors (which block multiple downstream signaling events), strongly suggest that autophagy induction prevents, rather than promotes, the growth of established HER2-positive breast cancers, at least in the BT-474-VH2 xenograft model tested. Further preclinical studies are needed to determine whether Tat-Beclin 1 has similar effects in HER2-positive patient-derived xenografts.
A prior study concluded that lapatinib-induced autophagy contributed to therapeutic resistance in HER2-positive breast cancer cells (19). However, this conclusion was based on the findings that knockdown of Beclin 1 increased apoptosis in HER2-positive breast cancer cells preselected for lapatinib resistance. As it is well established that autophagy gene knockdown increases short-term susceptibility to apoptotic cell death (35), such findings do not directly address the role of autophagy in anti-HER2 treatment responses. Blockade of erlotinib-induced autophagy in NSCLCs, via expression of a Beclin 1 tyrosine phosphomimetic mutant, knockdown of the essential autophagy gene ATG7, or expression of a mutant viral Bcl-2 protein that blocks autophagy but not apoptosis, increases clonigenic survival of erlotinib-treated NSCLCs (despite an acute increase in apoptosis) and results in chemoresistance in NSCLC xenografts in vivo (17). Thus, it is not possible to predict the role of autophagy in chemotherapeutic responses to HER family inhibitors by relying on assessment of acute levels of apoptosis with autophagy gene knockdown. Moreover, it is not possible to extrapolate the role of autophagy in the pathogenesis of HER2-driven breast cancer in vivo from in vitro studies of cell death in HER2-positive breast cancer cells.
Our data suggest that novel strategies to activate autophagy warrant further preclinical investigation in the treatment of HER2-positive breast cancer. While the precise mechanism of antitumor action of the Tat-Beclin 1 peptide is unknown, it appears to function by a mechanism that is different from the clinically used HER2 TKI lapatinib. Tat-Beclin 1 does not result in HER2 dephosphorylation; it induces a transcriptional response in tumors that is distinct from that of lapatinib, and unlike lapatinib, it is able to dissociate HER2/Beclin 1 binding in tumor xenografts. The disruption of HER2/Beclin 1 binding and autophagy induction, independent of HER2 dephosphorylation, might be particularly useful for tumors that are resistant to conventional anti-HER2 agents and/or might provide additive benefit in combination with such agents for initial treatment.
Methods
Cell Culture.
HeLa, BT-474, SKBR3, and MDA-MB-361 cells were obtained from the American Type Culture Collection (ATCC) and cultured according to ATCC instructions. BT-474-VH2 cells were a generous gift from José Baselga, Memorial Sloan Kettering Cancer Center, New York.
Mouse Strains and Breeding.
Mice of the FVB/N-Tg (MMTVneu) 202 Mul/J strain were obtained from The Jackson Laboratory (002376). C57/B6 Becn1 knock-in Becn1F121A mice were generated as described (20). MMTVneu mice were crossed with Becn1F121A/F121A mice to generate MMTV/neu transgenic; Becn1F121A/WT mice. These mice were intercrossed, and offspring were monitored for tumor development weekly by mammary gland palpation. Becn1WT and Becn1F121A mice were crossed with GFP-LC3 transgenic mice (36), and mammary glands of the Becn1WT:GFP-LC3 and Becn1F121A:GFP-LC3 mice were analyzed for autophagy. Five- to 6-wk-old female nu/nu mice were obtained from Taconic Farms.
Peptides.
Tat-Beclin 1 (YGRKKRRQRRR-GG-VWNATFHIWHD) and Tat-Scrambled (YGRKKRRQRRR-GG-WNHADHTFVWI) were synthesized by the University of Texas Southwestern Protein Technology Center as reported by Peraro et al. (22).
Additional information on antibodies and chemical reagents, plasmids, plasmid and siRNA transfection, biochemical analyses, electron microscopy, RNAseq analyses, xenograft and other mouse experiments, autophagy analyses, histopathological analyses, and METABRIC breast cancer dataset analyses is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank J. Baselga for providing critical reagents, L. Nguyen for expert technical assistance, S. Peña-Llopis for statistical advice, all members of the B.L. laboratory for helpful discussions, and H. Smith for assistance with manuscript preparation. This work was supported by NIH Grants U19AI199725 (to B.L.), R01CA109618 (to B.L.), 5R01GM115473 (to Y.X.), and 5R01CA172211 (to G.X.) and by Cancer Prevention Research Institute of Texas Grants RP120718 (to B.L.) and RP150596 (to Y.X.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717800115/-/DCSupplemental.
References
- 1.Ross JS, et al. The HER-2 receptor and breast cancer: ten years of targeted anti-HER-2 therapy and personalized medicine. Oncologist. 2009;14:320–368. doi: 10.1634/theoncologist.2008-0230. [DOI] [PubMed] [Google Scholar]
- 2.Petrelli F, et al. Clinical and pathological characterization of HER2 mutations in human breast cancer: a systematic review of the literature. Breast Cancer Res Treat. 2017;166:339–349. doi: 10.1007/s10549-017-4419-x. [DOI] [PubMed] [Google Scholar]
- 3.Foster SA, et al. Activation mechanism of oncogenic deletion mutations in BRAF, EGFR, and HER2. Cancer Cell. 2016;29:477–493. doi: 10.1016/j.ccell.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Gingras I, Gebhart G, de Azambuja E, Piccart-Gebhart M. HER2-positive breast cancer is lost in translation: time for patient-centered research. Nat Rev Clin Oncol. 2017;14:669–681. doi: 10.1038/nrclinonc.2017.96. [DOI] [PubMed] [Google Scholar]
- 5.Higgins MJ, Baselga J. Targeted therapies for breast cancer. J Clin Invest. 2011;121:3797–3803. doi: 10.1172/JCI57152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aita VM, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 7.Tang H, et al. Decreased BECN1 mRNA expression in human breast cancer is associated with estrogen receptor-negative subtypes and poor prognosis. EBioMedicine. 2015;2:255–263. doi: 10.1016/j.ebiom.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu X, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cicchini M, et al. Autophagy regulator BECN1 suppresses mammary tumorigenesis driven by WNT1 activation and following parity. Autophagy. 2014;10:2036–2052. doi: 10.4161/auto.34398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang XH, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 11.Wei Y, et al. The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. eLife. 2015;4:e05289. doi: 10.7554/eLife.05289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Negri T, et al. Chromosome band 17q21 in breast cancer: significant association between beclin 1 loss and HER2/NEU amplification. Genes Chromosomes Cancer. 2010;49:901–909. doi: 10.1002/gcc.20798. [DOI] [PubMed] [Google Scholar]
- 13.Berns K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 14.Nagata Y, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 15.Serra V, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008;68:8022–8030. doi: 10.1158/0008-5472.CAN-08-1385. [DOI] [PubMed] [Google Scholar]
- 16.Nunes J, et al. ATG9A loss confers resistance to trastuzumab via c-Cbl mediated Her2 degradation. Oncotarget. 2016;7:27599–27612. doi: 10.18632/oncotarget.8504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei Y, et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154:1269–1284. doi: 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang RC, et al. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338:956–959. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, et al. Interaction between Her2 and Beclin-1 proteins underlies a new mechanism of reciprocal regulation. J Biol Chem. 2013;288:20315–20325. doi: 10.1074/jbc.M113.461350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rocchi A, et al. A Becn1 mutation mediates hyperactive autophagic sequestration of amyloid oligomers and improved cognition in Alzheimer’s disease. PLoS Genet. 2017;13:e1006962. doi: 10.1371/journal.pgen.1006962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shoji-Kawata S, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494:201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peraro L, et al. Diversity-oriented stapling yields intrinsically cell-penetrant inducers of autophagy. J Am Chem Soc. 2017;139:7792–7802. doi: 10.1021/jacs.7b01698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scaltriti M, et al. Lapatinib, a HER2 tyrosine kinase inhibitor, induces stabilization and accumulation of HER2 and potentiates trastuzumab-dependent cell cytotoxicity. Oncogene. 2009;28:803–814. doi: 10.1038/onc.2008.432. [DOI] [PubMed] [Google Scholar]
- 24.Guy CT, et al. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA. 1992;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baselga J, Norton L, Albanell J, Kim YM, Mendelsohn J. Recombinant humanized anti-HER2 antibody (Herceptin) enhances the antitumor activity of paclitaxel and doxorubicin against HER2/neu overexpressing human breast cancer xenografts. Cancer Res. 1998;58:2825–2831. [PubMed] [Google Scholar]
- 26.Miao Y, Li G, Zhang X, Xu H, Abraham SN. A TRP channel senses lysosome neutralization by pathogens to trigger their expulsion. Cell. 2015;161:1306–1319. doi: 10.1016/j.cell.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shirakabe A, et al. Drp1-dependent mitochondrial autophagy pays a protective role against pressure overload-induced mitochondrial dysfunction and heart failure. Circulation. 2016;133:1249–1263. doi: 10.1161/CIRCULATIONAHA.115.020502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.An M, et al. ULK1 prevents cardiac dysfunction in obesity through autophagy-meditated regulation of lipid metabolism. Cardiovasc Res. 2017;113:1137–1147. doi: 10.1093/cvr/cvx064. [DOI] [PubMed] [Google Scholar]
- 29.Cinque L, et al. FGF signalling regulates bone growth through autophagy. Nature. 2015;528:272–275. doi: 10.1038/nature16063. [DOI] [PubMed] [Google Scholar]
- 30.He M, et al. Autophagy induction stabilizes microtubules and promotes axon regeneration after spinal cord injury. Proc Natl Acad Sci USA. 2016;113:11324–11329. doi: 10.1073/pnas.1611282113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui J, et al. EGFR inhibitors and autophagy in cancer treatment. Tumour Biol. 2014;35:11701–11709. doi: 10.1007/s13277-014-2660-z. [DOI] [PubMed] [Google Scholar]
- 32.Shi H, Zhang W, Zhi Q, Jiang M. Lapatinib resistance in HER2+ cancers: Latest findings and new concepts on molecular mechanisms. Tumour Biol. 2016;37:15411–15431. doi: 10.1007/s13277-016-5467-2. [DOI] [PubMed] [Google Scholar]
- 33.Liang XH, Yu J, Brown K, Levine B. Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res. 2001;61:3443–3449. [PubMed] [Google Scholar]
- 34.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 35.Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.